Abstract
Prioritizing molecular alterations that act as drivers of cancer remains a crucial bottleneck in therapeutic development. Here we introduce HIT'nDRIVE, a computational method that integrates genomic and transcriptomic data to identify a set of patient-specific, sequence-altered genes, with sufficient collective influence over dysregulated transcripts. HIT'nDRIVE aims to solve the “random walk facility location” (RWFL) problem in a gene (or protein) interaction network, which differs from the standard facility location problem by its use of an alternative distance measure: “multihitting time,” the expected length of the shortest random walk from any one of the set of sequence-altered genes to an expression-altered target gene. When applied to 2200 tumors from four major cancer types, HIT'nDRIVE revealed many potentially clinically actionable driver genes. We also demonstrated that it is possible to perform accurate phenotype prediction for tumor samples by only using HIT'nDRIVE-seeded driver gene modules from gene interaction networks. In addition, we identified a number of breast cancer subtype-specific driver modules that are associated with patients’ survival outcome. Furthermore, HIT'nDRIVE, when applied to a large panel of pan-cancer cell lines, accurately predicted drug efficacy using the driver genes and their seeded gene modules. Overall, HIT'nDRIVE may help clinicians contextualize massive multiomics data in therapeutic decision making, enabling widespread implementation of precision oncology.
Genomic and transcriptomic alterations are the major contributors of tumorigenesis and progression of cancer. Over the past decade, high-throughput sequencing efforts have provided an unprecedented opportunity to identify such genomic alterations that can lead to changes in gene regulation, protein structure, and function (Stratton et al. 2009). Genomic and transcriptomic data provide unique and complementary information about a particular tumor, but the translation of “big” molecular data into insightful and impactful patient outcomes is extraordinarily challenging (Vogelstein et al. 2013). During tumor progression, cancer cells accumulate a multitude of genomic alterations; however, most are inconsequential “passenger” alterations that are effectively neutral. Nevertheless, a small fraction provide mission-critical “hallmark” functions and are known as “driver” alterations that modify transcriptional programs and therefore drive and sustain tumor progression (Greenman et al. 2007; Stratton et al. 2009; Vogelstein et al. 2013). Improving our knowledge on driver alterations, possibly through an integrative analysis of various omics data, is critical to better understand cancer mechanisms and select appropriate therapies for specific cancer patients.
There are several computational methods for identifying cancer drivers. However, many of them rely on the recurrence frequency of single-nucleotide variants (SNVs) with respect to the background mutation rate (Greenman et al. 2006; Youn and Simon 2011; Lawrence et al. 2013; Korthauer and Kendziorski 2015). As a result, these methods are restricted to identifying only highly recurrent mutations as driver events. Recent studies have implicated novel drivers that affect only a small subset of cancer patients. Notable examples include SPOP mutations and CHD1 deletions that are present in <20% of prostate cancer patients (Barbieri et al. 2012; Grasso et al. 2012). Whereas recurrent drivers are hypothesized to initiate carcinogenesis and are therefore present in the majority of tumor cells, rare drivers can arise during tumor evolution and be isolated to a smaller fraction of cells due to clonal expansion (Ding et al. 2012; Greaves and Maley 2012). These rare driver genes may be functionally important but are likely to be missed by a frequency-based approach.
Perhaps the first computational method to consider large-scale genomic alterations as driver events is CONEXIC (Akavia et al. 2010), which correlates genes with highly recurrent copy number alterations (CNAs) with variation in gene-expression profiles within a Bayesian network. Similarly, with no prior knowledge of pathways or protein interactions, MOCA correlates gene mutation information with expression profile changes in other genes (Masica and Karchin 2011). Suo et al. (2015) also prioritize highly mutated genes that interact with a large number of differentially expressed genes in a gene network. Another approach, (Multi) Dendrix (Leiserson et al. 2013) aims to simultaneously identify multiple driver pathways, assuming mutual exclusivity of mutated genes among patients, using either a Markov chain Monte Carlo algorithm or integer linear programming (ILP). XSEQ (Ding et al. 2015) uses probabilistic model to compute influence of mutated genes over expression profile changes in other genes by considering direct gene interactions. Finally, MEMo (Ciriello et al. 2012) identifies sets of proximally located genes from interaction networks, which are also recurrently altered and exhibit patterns of mutual exclusivity across the patient population.
Simultaneously with the above methods, several approaches were developed outside of cancer research to correlate the presence of casual genes with gene expression. For example, Tu et al. (2006) used a random walk approach on a molecular interaction network to associate causal genes and pathways. Similarly, ResponseNet (Yeger-Lotem et al. 2009; Lan et al. 2011) relates genetic perturbations to transcriptomic response in the yeast model, thereby identifying a subnetwork of regulators mediating the interactions. ResponseNet formulates a minimum-cost flow optimization problem that aims to maximize the flow between the source and target while minimizing the cost of the connecting paths. Similarly, expression quantitative trait loci (eQTL) electrical diagrams (eQEDs) (Suthram et al. 2008) integrate eQTL analysis with molecular interaction network using the circuit network model. To the best of our knowledge, NetQTL (Kim et al. 2011) is the first method to link CNAs to expression profile changes within an interaction network and connect specific “causal” aberrant genes with potential targets in the interaction network. EPoC (Jornsten et al. 2011) links CNAs to expression changes in an interaction network assuming steady-state perturbation effects. Similarly, PARADIGM (Vaske et al. 2010) computes gene-specific inferences using factor graphs to integrate different genomic changes and infer pathways altered in a patient. MAXDRIVER (Chen et al. 2013) uses maximum information flow to identify potential causal genes (CNAs) in an interaction network.
More recently, HotNet (Vandin et al. 2011) was the first tool to use a network diffusion approach to compute a pairwise influence measure between the genes in the (protein interaction) network and identify subnetworks enriched for mutations in cancer. TieDIE (Paull et al. 2013) also uses the diffusion model to identify a collection of pathways and subnetworks that associate a fixed set of driver genes to expression profile changes in other genes. Briefly, the network diffusion approach aims to measure the influence of one node over another by calculating the stationary proportion of a “flow” originating from the starting node that ends up in the destination node. Since this is based on the stationary distribution, the inferences that can be made by the diffusion model are time independent. In that sense, the diffusion approach is very similar to Rooted PageRank, the stationary probability of a random walk originating at a source node, being at a given destination node. Shi et al. (2016) also prioritizes genes based on diffusion score matrix (derived from a tripartite graph of mutations, outliers, and patients) rank aggregation. A final method, DriverNet (Bashashati et al. 2012), also aims to correlate genomic alterations with target genes’ expression profile changes, but only among direct interaction partners; the novel feature of DriverNet is that it aims to find the minimum number of potential drivers that can “cover” targets.
Among the above strategies, the ones based on mutual exclusivity still focus on frequent events. The others, based on “information flow” in gene/protein interaction networks, do not aim to discover cancer drivers but rather are designed to identify dysregulated subnetworks or modules. In addition, the notion of influence they employ is based on stationary distribution of “information” originating at a particular gene/protein. As a result, none of the available methods aim to identify rare, patient-specific driver events based on a time-dependent notion of influence. Finally, none of the available techniques aim to simultaneously consider different types of genomic alterations as potential drivers.
To address the above challenges, we have developed a novel combinatorial method, HIT'nDRIVE (a preliminary version was presented at the Research in Computational Molecular Biology [RECOMB] conference) (Shrestha et al. 2014). HIT'nDRIVE jointly analyzes genome and transcriptome data for identifying and prioritizing sequence-altered genes as potential cancer drivers. Because HIT'nDRIVE integrates patient-specific genomic alterations with the associated transcriptome profile, identifying driver genes that dysregulate large portion of each patient's transcriptome. Drawing upon the domain knowledge of molecular interactions presented as a gene/protein interaction network, HIT'nDRIVE uses network topology to derive the influence of one (sequence-altered) gene over another (expression-outlier) gene and aims to identify the most parsimonious set of patient-specific driver genes that have sufficient “influence” over a large proportion of the expression outliers.
Results
The primary goal of HIT'nDRIVE is to link alterations at the genomic level to changes at transcriptome level through a gene/protein interaction network. Intuitively, it aims to find the smallest set of altered genes that can explain most of the observed transcriptional changes in the cohort (for details, see Methods). In other words, HIT'nDRIVE aims to identify the minimum number of potential driver genes that can cause a user-defined proportion of the downstream expression effects observed. HIT'nDRIVE formulates this as a “random walk facility location” (RWFL) problem, a combinatorial optimization problem that we introduce in this article. RWFL generalizes the classical “facility location” (FL) problem by changing the notion of distance it uses. Given a network, FL problem defines the distance between a potential driver gene and an outlier gene as the length of the shortest path between them. The RWFL problem, in contrast, uses “hitting time” (or “first passage time”) (Condamin et al. 2007; Liben-Nowell and Kleinberg 2007), the expected length of a random walk between the two nodes, as their distance. Under the use of hitting time, the FL problem completely changes nature: In the classical FL formulation, the goal is to associate each outlier gene in the network with exactly one (the closest) driver gene. In the RWFL formulation, each outlier gene is associated with multiple driver genes (whose collective distance to the outlier will no longer be the shortest pairwise distance), forming a many-to-many relation.
As per the standard FL problem, RWFL is NP hard, even to approximate. As a result, we reduce it to the weighted multiset cover (WMSC) problem, for which we give an ILP formulation (for details, see Methods). Intuitively, in this new formulation, HIT'nDRIVE associates the genomic alterations with transcriptomic changes in the form of a bipartite graph with nodes on one partition representing the set of aberrant genes and nodes on the other partition representing the set of expression-altered genes, and each edge has an influence value equal to the inverse pairwise hitting time between the two nodes it connects (Fig. 1A). The WMSC problem on this representation of data asks to find the smallest subset of aberrant genes (as potential drivers) whose total influence (sum of pairwise influence values) over a user-defined fraction of expression-altered genes (for each patient) is sufficiently high.
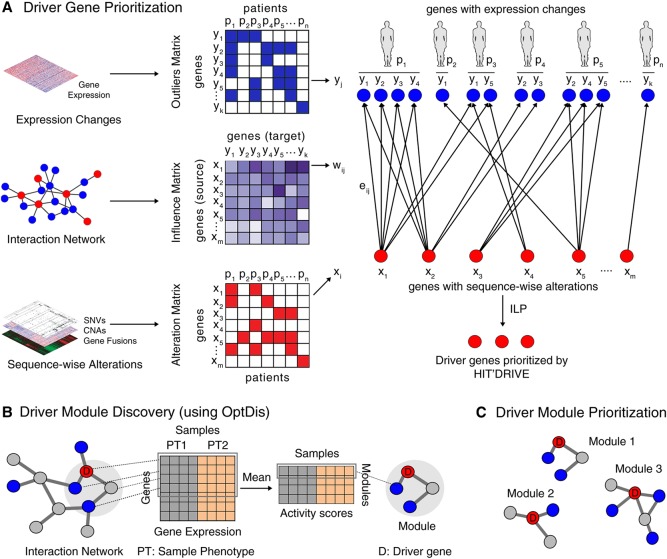
Figure 1.
Overview of HIT'nDRIVE algorithmic framework. (A) HIT'nDRIVE integrates sequence-wise changes in genome with expression changes in transcriptome obtained from patients’ tumor samples. The influence values derived from the protein interaction network indicate how likely a driver gene influences its downstream target genes in the network. (B) The predicted driver genes are used as seeds to discover modules of genes that discriminate between the sample phenotypes using OptDis. (C) Based on this, the driver modules are ranked and thus prioritized.
In order to quantitatively assess the genes identified by HIT'nDRIVE, we extended our previously developed algorithm, OptDis (Dao et al. 2011), for de novo identification of modules of small size inside the interaction network that are seeded by at least one predicted driver. The modules are chosen so that their discriminative power (for phenotype classification) is the greatest among connected subnetworks of similar size that contain the individual predicted driver genes (Fig. 1B,C). We report the classification accuracy based on the identified driver-seeded modules as means of quantitative validation of our results (in the absence of ground truth). We also look at the genes that build the chosen modules (of high classification accuracy) in an attempt to identify cancer-related pathways.
We have implemented HIT'nDRIVE in C++ and solved the ILP using IBM CPLEX version 12.5.1. HIT'nDRIVE uses three different user-defined input parameters (for details, see Methods ): (1) α determines the fraction of outliers to be covered overall (across all patients); (2) β determines the fraction of outliers to be covered in each patient; and (3) γ controls the fractional lower bound on the sum of the incoming edge weights (influence values). HIT'nDRIVE is robust with respect to the changes in α and β but is somewhat sensitive to the value of γ, as expected. However, as γ grows, the driver genes identified by HIT'nDRIVE do not change but simply grow in number by the addition of new driver genes, which indicates robustness of our method with respect to γ, too (Supplemental Fig. S1).
We used STRING v10 (Szklarczyk et al. 2015) functional protein-interaction network for our analysis. HIT'nDRIVE is not sensitive to topological biases in the network (Supplemental Fig. S3). Although, the number of driver genes predicted by HIT'nDRIVE differed slightly when different networks were used (and was proportional to the number of nodes in the network), the proportion of overlap between the driver genes predicted on different networks was quite robust (Supplemental Fig. S4; for details, see Supplemental Results).
HIT'nDRIVE predicts frequent as well as infrequent driver genes in multiomics cancer data sets
We applied HIT'nDRIVE to prioritize driver genes in four major cancer types: glioblastoma multiforme (GBM) (The Cancer Genome Atlas Research Network 2008), ovarian serous cystadenocarcinoma (OV) (The Cancer Genome Atlas Research Network 2011), breast adenocarcinoma (BRCA) (The Cancer Genome Atlas Research Network 2012), and prostate adenocarcinoma (PRAD) (The Cancer Genome Atlas Research Network 2015) obtained from The Cancer Genome Atlas (TCGA) Data Portal. Only samples with matched genomic alterations (SNVs and/or CNAs and/or gene fusions) and transcriptomic changes (outlier genes from gene-expression profile) were used in our study. We used the fusion prediction calls as reported in TCGA Fusion Gene Data Portal (Yoshihara et al. 2014).
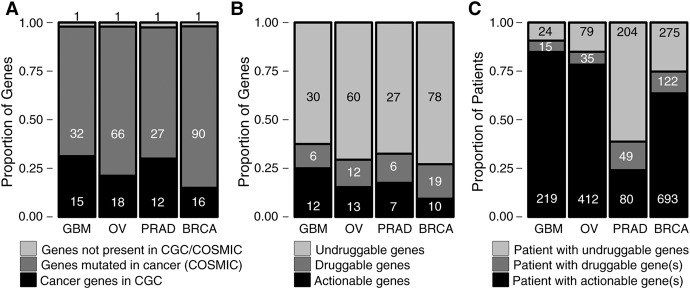
In GBM, we obtained 48 unique candidate driver genes altered at varying frequencies across 258 GBM patients (Supplemental Figs. S8, S9; Supplemental Tables S1, S5). EGFR (36%), TP53 (29.5%), PTEN (28%), and CHEK2 (26%) were the most frequently altered driver genes in GBM followed by CDKN2A (16%), RB1 (13%), and SEC61G (12%). Previous efforts in GBM genome characterization identified amplification in EGFR and PDGFRA; mutations in CHEK2, TP53, PTEN, RB1, and NF1; and deletions in CDKN2A to be associated with GBM (The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008; Verhaak et al. 2010). HIT'nDRIVE prioritized all of the above alterations. Alterations in EGFR is characteristic of a classical subtype; NF1, mesenchymal subtype; and PDGFRA, IDH1 with proneural subtype of GBM (Verhaak et al. 2010). Fifteen out of 48 driver genes predicted by HIT'nDRIVE (P-value = 8 × 10−4) were present in the Cancer Gene Census (CGC) database (Futreal et al. 2004), which contains genes for which mutations have been causally implicated in cancer (Fig. 2A). GSTT1 (deleted in 21 patients), a key player in drug metabolism, was found neither in the CGC nor in the Catalog of Somatic Mutations in Cancer (COSMIC) (Forbes et al. 2017) databases. Twelve GBM driver genes were found to be actionable targets. Actionable genes were extracted from TARGET database (Van Allen et al. 2014), which contains genes directly linked to a clinical action. In addition to the above list, six other driver genes were druggable (Fig. 2B). We extracted the list of druggable genes from the Drug–Gene Interaction Database (DGIdb) (Griffith et al. 2013). Interestingly, ∼85% of the patients in GBM cohort harbor at least one actionable driver gene, and an additional 5% of patients have druggable targets (Fig. 2C). HIT'nDRIVE also identified 12 infrequent driver genes, which we define as genes altered in, at most, 2% of the cases. Among the infrequent genes, SACS is known to be associated with neurological functions, NLRP3 is involved in apoptosis, and TIAM2 is involved in invasion and metastasis.
Figure 2.
Summary of driver genes prioritized by HIT'nDRIVE. (A) Distribution of predicted driver genes in cancer genes databases. The CGC database contains genes for which mutations have been causally implicated in cancer. Genes curated in the CGC database represent likely drivers of cancer. COSMIC is a comprehensive database of somatic mutations that have been reported in different cancers. However, every gene present in COSMIC database may not represent drivers of cancer. (B) Distribution of driver genes in druggable genes databases. Actionable genes in cancer therapy were derived from the TARGET database. List of druggable genes were extracted from DGI database. (A,B) The numbers in the panel represent the number of genes in respective categories. (C) Distribution of patient druggability. Patient druggability was accessed using information in the TARGET and DGI databases. The numbers in the panel represent the number of patients in respective categories.
The 526 OV patients harbored a total of 85 unique driver alterations (Supplemental Figs. S10, S11; Supplemental Tables S2, S6). TP53 mutations were prevalent in more than half (58%) of the patients in the cohort. Consistent with the previous findings, we found OV patients to be driven by genomic copy-number changes rather than recurrent point mutations (Ciriello et al. 2013; Patch et al. 2015). Recurrent somatic CNAs were observed in GSTT1 (32.3%), WWOX (28.1%), FAM49B (15.0%), UGT2B17 (14.6%), CCNE1 (13.1%), SLC39A4 (13.1%), and MYC (12.5%). Mutations in TP53 and BRCA1/2, and loss of RB1, NF1, and CCNE1 were previously associated with OV (The Cancer Genome Atlas Research Network 2011; Patch et al. 2015). HIT'nDRIVE revealed 18 CGC driver genes (P-value = 2 × 10−5) (Fig. 2A), among which 13 genes were actionable targets and other 12 genes were at least druggable (Fig. 2B). More than 75% of OV patients harbored at least one actionable target, and an additional 6% of patients have a druggable target (Fig. 2C). GSTT1 (altered in 170 patients), in OV, is involved in estrogen and drug metabolism. It was not found in the CGC or COSMIC databases. We identified 13 infrequent genes, among which MAPK1 is known to play an important role in oncogenic pathways in cancer.
HIT'nDRIVE identified 40 driver genes across 333 PRAD patients (Supplemental Figs. S12, S13; Supplemental Tables S3, S7). Copy number loss of SPECC1L (23.7%), STEAP1B (13%), and WWOX (10%) and amplification of NSD1 (16.2%) and SIRPB1 (16.2%) were the most recurrent events in PRAD patients. We also found recurrent somatic mutation in MUC4 (11%), SPOP (10.5%), and TP53 (10%). The most common alterations in PRAD genomes are fusion of androgen-regulated promoters with ERG and other members of ETS family of transcription factors: mainly TMPRSS2–ERG fusions (Tomlins et al. 2005). Since we relied on the gene-fusion predictions obtained from TCGA Fusion Gene Data Portal (Yoshihara et al. 2014), which analyzed only 178 (out of 333) patients, we observed ERG gene fusion in only 5.7% cases. The more recent TCGA publication (The Cancer Genome Atlas Research Network 2015) reported ERG fusions in almost half of the patients in the cohort. Moreover, the tools used for gene-fusion detection, in the two studies, were different; as a result, we observed a much smaller number of ERG fusions than reported previously. SPOP, TP53, FOXA1, and PTEN are the most frequently mutated genes that have been previously associated with prostate cancer (Barbieri et al. 2012). PRAD patients harbored 12 driver genes present in the CGC database (P-value = 9 × 10−4) (Fig. 2A), out of which eight driver genes were actionable (Fig. 2B). Approximately a quarter of PRAD patients could benefit with actionable targeted therapy (Fig. 2C). Moreover, an additional 14% of patients harbored druggable genes, which warrants deeper investigation of drug repurposing opportunities. NBPF1 (mutated in 17 patients), a known tumor suppressor gene known to have neural function and also to be involved in cell-cycle arrest, was not found in the CGC or COSMIC databases. We identified 11 infrequent genes in PRAD, among which IDH1-mutant patients were recently identified as a distinct molecular-subtype of PRAD (The Cancer Genome Atlas Research Network 2015), NKX3-1 is required for normal prostate tissue development, and CDKN1B was previously associated with PRAD.
In BRCA, HIT'nDRIVE identified 107 driver genes across 1090 patients (Supplemental Figs. S14, S15; Supplemental Tables S4, S8). Somatic mutation of PIK3CA (30.5%) and TP53 (30.2%) were the most recurrent events in BRCA. This was followed by somatic mutation of CHD1 (11.2%), GATA3 (10.5%), MUC16 (6.9%), and MAP3K1 (6.9%) and CNA amplification of NSD1 (8.7%) and MED1 (6.9%). BRCA patients harbored 16 genes present in the CGC database (P-value = 9.3 × 10−3) (Fig. 2A) among which 10 genes were actionable targets (Fig. 2B). More than 60% of BRCA patients could benefit with the actionable targeted therapy. Furthermore, additional 11% of BRCA patients harbored at least one of the 19 potentially druggable genes (Fig. 2C). ACACA (altered in 36 patients mostly from HER2 subtype), involved in fatty-acid metabolism, was not found in the CGC or COSMIC databases. We identified 46 infrequent driver genes, among which BRCA2 and GNAS have been previously linked to BRCA.
Although the driver events per tumor sample greatly varied, the median number of driver genes among the 2207 tumor samples in all four cancer types was three (Supplemental Fig. S16). Twenty-three percent of 2207 tumor samples harbored just a single driver gene. The remaining 77% of tumor samples harbored two or more driver events, which may indicate either the existence of multiple subclonal populations within the tumor or the presence of collaboration among multiple sequence altered genes in an oncogenic pathway.
To evaluate the sensitivity of HIT'nDRIVE to infrequent driver genes, we performed an in silico experiment using 1000 TCGA-BRCA tumors (referred here as the “original set”). Different subsets of tumor samples with varying sample size were chosen such that the subsample tumor population has similar alteration frequency distribution to that of the original 1000 tumor samples (Supplemental Fig. S21A; see Supplemental Methods). HIT'nDRIVE analysis was performed on the chosen subsets of tumor samples independently to identify driver genes and then compared the frequency of driver genes detected. HIT'nDRIVE detected driver genes that were prevalent in just a single patient tumor with a sample size of 15, 25, or 50 tumors representing 6.5%, 4%, and 2% of the tumor population, respectively (Supplemental Fig. S21B,C). Even when the sample size was increased to 700–900 samples, HIT'nDRIVE was able to detect driver genes prevalent in just four patients, representing <0.5% of the tumor population. This demonstrates that HIT'nDRIVE prioritizes driver genes independent of the selected sample size and that HIT'nDRIVE is very sensitive in detecting infrequent driver genes.
Network properties of cancer driver genes
Centrality of driver genes in the interactome
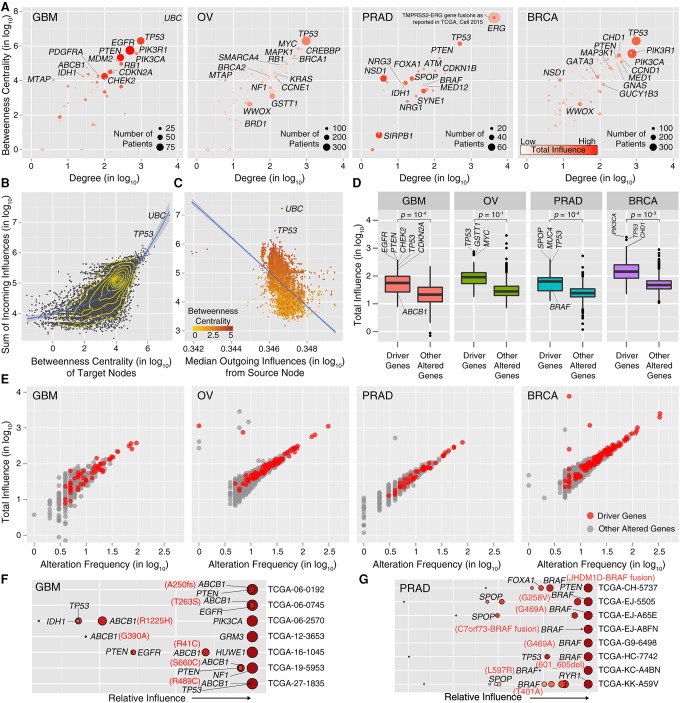
Cancer driver genes are known to occupy critical positions in the interactome. To check whether HIT'nDRIVE-predicted driver genes also occupy similar positions in the interaction network, we used the node degree as a “local measure” and used node betweenness (the number of shortest paths between node pairs that pass through the node) as a “global measure of centrality.” The driver genes predicted by HIT'nDRIVE include a number of well-known high-degree hubs—TP53, EGFR, RB1, MYC, PIK3CA, ERG, and CHD1—that are “central” in the interactome with high degree and high betweenness (Fig. 3A). Although there was very weak correlation between the number of edges (i.e., degree centrality) of a node and the number of samples/patients in which it is identified as a driver, remarkably each hub gene was typically altered in a large fraction of patients. Because of their centrality perturbations, hub genes are likely to dysregulate several other genes and the associated signaling pathways. Interestingly, HIT'nDRIVE also identified low-degree genes (IDH1, MTAP, NF1, NRG1, NSD1) that reside in the periphery of the interaction network. In particular, in prostate cancer, there seems to be an inverse correlation between the degree and how often the gene is picked as a driver. Most of these low-degree genes are altered in a small fraction of patients, indicating that HIT'nDRIVE, unlike many other methods, does not primarily return hubs that are altered in a large number of patients but is capable of identifying rare driver genes without trivial topological biases.
Figure 3.
Network properties of driver genes. (A) The centrality of the predicted drivers in STRING v10 network. The size of the circles is proportional to the alteration frequency of the driver gene. The color scale represents the total influence of the driver gene on the expression outliers. (B) Correlation between influence and centrality. Each dot represents a target node receiving a certain amount of influence from all source nodes in the network. A lowess regression line is represented in blue. (C) Correlation between incoming and outgoing influence of a node. Each dot represents a node in the network, and the color scale represents its betweenness centrality. A linear regression line is represented in blue. (D) Boxplot of the total influence of driver genes predicted by HIT'nDRIVE on the expression outliers compared with that of other altered genes (genes not predicted as drivers). (E) Correlation between gene influence and its alteration frequency in the respective patient cohort. (F) Relative influence of driver genes in each patient in GBM cohort with mutation in ABCB1. (G) Relative influence of driver genes in each patient in PRAD cohort with mutation in BRAF. All gene influence values have been multiplied by 105 before log transformation.
As discussed in previous section, we used hitting time to compute the influence of a node in an interaction network. The influence from a source to a target node depends on the topological position of the target node in the network. We observed that the nodes occupying central positions in the network, i.e., with high betweenness centrality, tend to receive more influence than the nodes in the periphery of the network (Pearson correlation coefficient [R] = 0.61) (Fig. 3B). This is because the distance between any source node and a central target node (i.e., a hub) is usually very short, implying a low hitting time and, thus, high influence of the source node on the target. We also observed negative correlation between a node's total incoming influence and the median outgoing influence (R = −0.54) (Fig. 3C). Although the central hub nodes (e.g., UBC, TP53) are good receivers of influence, when individual influences are considered, they do not contribute a lot.
Influential nodes prioritized as cancer driver genes
Next, we examined the influential driver genes that are responsible for driving cancer. For this, we computed the total outgoing influence from each altered gene (which has been chosen as a driver), defined as the weighted sum of all influence values from the source to all outlier genes it is connected to (targets), weighted by the corresponding outlier weights. First, we investigated driver genes with high influence values within each cancer type. We observed that on average the total influence of driver genes was higher than that of other altered genes in all cancer types (Fig. 3D). EGFR, PTEN, CHEK2, TP53, and CDKN2A were the most influential driver genes in GBM, which together exerted 38.5% of the total influence on the GBM patient cohort. In OV, TP53, GSTT1, and MYC together exerted 20% of the total influence. Similarly, in PRAD cohort, SPOP, MUC4, and TP53 were the most influential genes, exerting 23.7% of the total influence. PIK3CA, TP53, and CHD1 were the most influential genes, exerting 23% of the total influence on the BRCA patient cohort. Moreover, the gene influence was positively correlated to its alteration frequency (Fig. 3E).
We investigated influence of the predicted driver genes within individual patients. Many recurrently altered driver genes had a higher influence compared with other driver genes: e.g., EGFR in GBM; TP53 in OV; ERG in PRAD; TP53, PIK3CA, and PTEN in BRCA.
Interestingly, among the highly influential genes, there were also less-recurrent but functionally important and actionable driver genes. For example, somatic mutations in ATP binding cassette subfamily B member 1 (ABCB1) were influential driver genes in seven GBM patients (Fig. 3F). ABCB1 is a membrane-bound protein present in the endothelial cells of the blood–brain barrier. It harnesses the energy of ATP hydrolysis to drive the unidirectional transport of exogenous and xenobiotic substances (drug compounds) from the cytoplasm to the extracellular space. It is known to transport many anticancer compounds, including temozolomide (TMZ), which is used as a first-line treatment for GBM patients. Mutations and overexpression of ABCB1 in GBM have been associated with resistance to TMZ (Lin et al. 2014). It was intriguing that some of these GBM patients had undergone treatment prior to tissue collection and were initially mislabeled as untreated patients. Treatment-induced selection pressure in the drug transporter might be a plausible reason for high influence exerted by ABCB1.
Similarly, HIT'nDRIVE predicted BRAF as driver genes in eight PRAD patients (six somatic mutations and two gene fusions) (Fig. 3G). These patients harbored BRAF as a highly influential driver gene. None of these patients harbored BRAFV600E mutation that is prevalent in cutaneous melanomas, thyroid cancer, and many other cancer types. However, BRAFL597R can be targeted using MEK inhibitors (Dahlman et al. 2012; Bowyer et al. 2014). BRAF plays important roles in growth factor signaling pathways, which affect cell division and differentiation. These results serve as proof of concept that HIT'nDRIVE can prioritize functionally relevant cancer driver genes.
Phenotype classification using dysregulated modules seeded with the predicted driver genes
Evaluating computational methods for predicting cancer driver genes is challenging in the absence of the ground truth (i.e., follow-up biological experiments). Therefore, we mainly focused on testing whether our predictions provide insight into the cancer phenotype and improve classification accuracy on an independent cancer data set. To test association of the driver genes identified by HIT'nDRIVE with the cancer phenotype, as explained in the earlier section, we used the driver gene–seeded gene modules, a set of functionally related genes (e.g., in a signaling pathway), from the protein interaction network, as features for classifying the cancer phenotype. By using OptDis (here referred to as HIT'nDRIVE-OptDis), we identified small connected subnetworks that include (i.e., are seeded by) predicted driver genes in a greedy fashion. More specifically, we prioritized subnetworks (of at most seven genes) iteratively so that in each iteration, we identified the subnetworks that maximally discriminate sample phenotypes in a gene-expression matrix, among the subnetworks that share very few genes (at most 20%) with the subnetworks already prioritized.
Furthermore, we have also developed an unsupervised method for module identification (here referred to as HIT'nDRIVE-unsupervised), i.e., one that does not depend on any phenotype information. This unsupervised method seeds each module with one HIT'nDRIVE-identified driver gene and includes outlier genes that it has influence over and co-occurs with significantly across patients (Supplemental Fig. S22). For this, we perform a hypergeometric test to identify significant driver–outlier interaction (i.e., mutual presence) pairs across the patient cohort (P-value <10−3).
Here we compare HIT'nDRIVE-OptDis and HIT'nDRIVE-unsupervised to another network-based driver genes prioritization method: DriverNet (Bashashati et al. 2012). DriverNet itself does not aim to identify modules that we can use to compare against HIT'nDRIVE-OptDis or HIT'nDRIVE-unsupervised modules. Rather, DriverNet identifies driver genes in an iterative fashion, where in each iteration, DriverNet picks the driver genes that “cover” the maximum number of uncovered outliers. We use this driver and the outlier genes it covers as the “next” DriverNet module.
We used the set of prioritized subnetworks, i.e., the driver modules, first, to perform binary sample classification: tumor versus normal. For this, we used gene-expression data for each of the four cancer types (GBM, OV, PRAD, and BRCA) from TCGA as discovery data sets to calculate the mean gene-expression value for each subnetwork/driver module for each patient (Supplemental Table S10–S13). On these subnetworks, we used the k-nearest neighbor (KNN) classifier (with k = 1) to perform classification on both the expression values from TCGA and additional validation gene-expression data sets (Supplemental Fig. S4A–C; Supplemental Table S9). The additional validation data sets were used in order to assess the capability of the modules identified on TCGA cohort in classifying other cohorts.
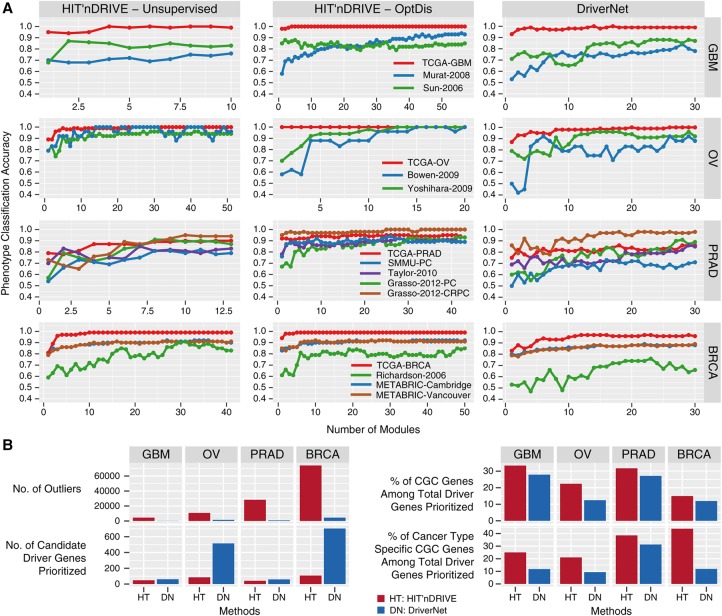
For every data set analyzed, the maximum classification accuracy achieved by HIT'nDRIVE modules (either HIT'nDRIVE-unsupervised or HIT'nDRIVE-OptDis), for any number of modules considered, was higher than that achieved by the DriverNet modules (Fig. 4A). Moreover, in most data sets, HIT'nDRIVE methods achieve maximum or near-maximum accuracy using a smaller fraction of modules (Supplemental Table S14). All three methods achieved perfect or near-perfect classification accuracy in the TCGA-GBM, TCGA-OV, and TCGA-BRCA data sets except for the TCGA-PRAD data set (where the maximum classification accuracy achieved was 90% by HIT'nDRIVE-unsupervised, 95% by HIT'nDRIVE-OptDis, and 86% by DriverNet). Overall, the driver modules (identified in one cohort) were able to distinguish the tumor phenotype from normal very well in validation data sets (on other cohorts), supporting the relevance of the identified driver genes to the cancer phenotype.
Figure 4.
Phenotype classification using driver-seeded modules. (A) Phenotype (tumor vs. normal) classification accuracy in gene-expression data sets of different cancer types using three different methods: HIT'nDRIVE-unsupervised (left), HITn'DRIVE-OptDis (middle), and DriverNet (right). (B) Comparison of HIT'nDRIVE with DriverNet.
Finally, we compared the classification accuracy of randomly formed modules in the TCGA-PRAD data set (which is the single most challenging data set), compared against modules identified using HIT'nDRIVE-OptDis. To generate the random modules, 60 random sets of genes, of random size (up to seven genes), were selected 20,000 times. Phenotype classification was then performed using a KNN (k = 1) classifier. HIT'nDRIVE-OptDis modules demonstrated superior classification accuracy compared with that of randomly selected modules (Supplemental Fig. S25).
CGC cancer type–specific gene enrichment
Next, we looked into the list of prioritized driver genes by both HIT'nDRIVE and DriverNet and their overlap with the known CGC genes (Fig. 4B). DriverNet selects a much larger number of driver genes, compared with HIT'nDRIVE, to cover most outlier genes (across all four cancer types) due to its model considering only direct interactions in the network. In particular, in OV and BRCA, the number of HIT'nDRIVE-identified driver genes are an order of magnitude smaller than that of DriverNet. Although in GBM and PRAD data sets, the number of driver genes identified by DriverNet is somewhat lower and comparable to that identified by HIT'nDRIVE (primarily because most outliers were filtered out due to sharing no interaction edge with candidate altered genes), HIT'nDRIVE-identified driver genes cover a significantly larger number of outliers. More importantly, even though HIT'nDRIVE identifies a smaller number of driver genes, a larger fraction of these driver genes can be found in the CGC database in comparison to the DriverNet-identified driver genes. In fact, even a larger fraction of CGC genes specific to the relevant cancer type can be found among HIT'nDRIVE-identified driver genes. Specifically, HIT'nDRIVE predicted four glioblastoma-specific CGC genes (IDH1, PDGFRA, PIK3CA, and PIK3R1) in the TCGA-GBM data set. Among them, IDH1, PDGFRA, and PIK3CA were not identified by DriverNet. Similarly, four ovarian cancer–specific CGC genes (BRCA1, BRCA2, CCNE1, and MAPK1) were predicted in the TCGA-OV data set. CCNE1 was not identified by DriverNet. Five prostate cancer–specific CGC genes (BRAF, ERG, FOXA1, PTEN, and SPOP) were predicted in the TCGA-PRAD data set. BRAF and SPOP were not identified by DriverNet. And seven breast cancer–specific CGC genes (BRCA2, CCND1, CDH1, GATA3, MAP3K1, PIK3CA, and TP53) were predicted in the TCGA-BRCA data set. Among them, CDH1 and MAP3K1 were not identified by DriverNet.
Breast cancer subtype classification using driver modules
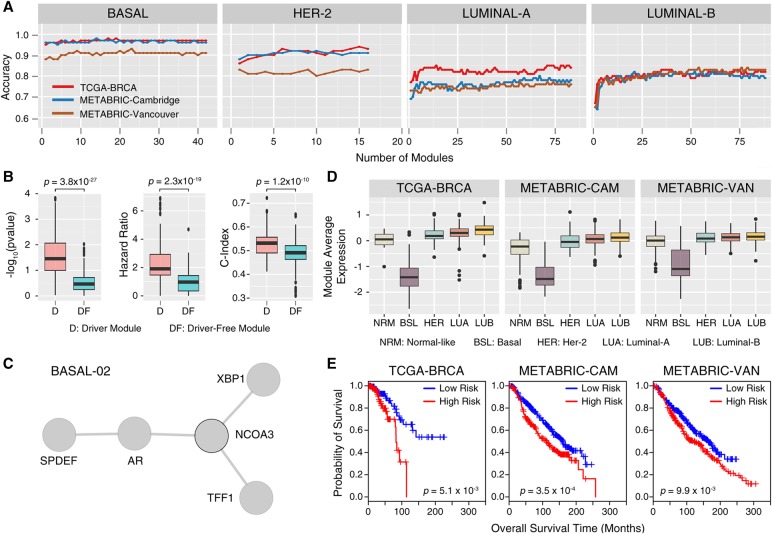
Our next goal was to classify four major subtypes of breast cancer: Basal, HER2, Luminal-A, and Luminal-B. For that purpose, we performed binary classification for each subtype: e.g., Basal versus non-Basal (including the normal samples). This was achieved through the use of HIT'nDRIVE-identified driver genes from TCGA-BRCA as seed genes, with which we identified subtype-specific driver modules from TCGA-BRCA gene-expression data (as described for tumor classification). We respectively obtained 37, 16, 43, and 39 subtype-specific driver modules for the Basal, HER2, Luminal-A, and Luminal-B subtypes (Supplemental Figs. S27–S29; Supplemental Table S15). As described above, by using these subtype-specific driver modules as features, we performed independent classification of BRCA subtypes in the TCGA-BRCA, METABRIC-Cambridge, and METABRIC-Vancouver data sets (Curtis et al. 2012).
The majority of Basal-like tumors constitute triple-negative breast cancers (TNBCs), which are highly aggressive tumors characterized by lack of expression of estrogen receptor 1 (ESR1), progesterone receptor (PGR), and erb-b2 receptor tyrosine kinase 2 (ERBB2). Molecular mechanisms driving TNBC are the least understood, and hence, no targeted therapies for TNBC yet exist (Bianchini et al. 2016). Interestingly, HIT'nDRIVE-seeded driver modules were able to classify Basal-like tumors with much higher accuracy (98%) compared with other BRCA-subtypes: HER2 (94%), Luminal-A (85%), and Luminal-B (83%) (Fig. 5A; Supplemental Table S16). As expected, ESR1 and PGR were highly expressed in Luminal-A/B but not in the Basal and HER2 subtypes. Modules containing ESR1 were consistently down-regulated in the Basal subtype and up-regulated in the Luminal-A/B subtype, whereas module LUMB-03 was up-regulated in the Luminal-B subtype (Supplemental Fig. S31). The ESR1 network neighborhood included 11 known transcriptional targets of ESR1 (TFF1, PGR, SLC9A3R1, GNAS, RARA, WWP1, WNT5A, TCF7L2, FKBP4, SPRY2, and RAD54B). These results were consistent with previous findings (Dutta et al. 2012). ERBB2 was expressed only in nine (of 16) HER2 modules and was the most prominent hub in the large interactome of HER2 modules. All modules containing ERBB2 were up-regulated in the HER2 subtype, and the module expression patterns were consistent in different BRCA data sets (Supplemental Fig. S32). PGR was present in two modules (BASAL-26 and HER2-12), both of which were down-regulated in the Basal subtype but up-regulated in Luminal-A/B. These results strongly suggest that HIT'nDRIVE can capture subtype-specific driver genes and that the driver-seeded modules we identified can indeed differentiate BRCA subtypes.
Figure 5.
BRCA subtype classification using driver modules. (A) Performance accuracy of classifying different subtypes for breast cancer using the activity score of subtype-specific driver modules as features in three distinct data sets. (B) Box plot comparing subtype-specific driver-seeded modules and driver-free modules with respect to three distinct measures: log-rank test P-value, hazard ratio (HR), and concordance index (c-index). (C) A BRCA subtype-specific driver module (BASAL-02) seeded by NCOA3 that distinguished the Basal subtype from rest of the BRCA subtypes. (D) Activity score of the BASAL-02 module across different BRCA subtypes. (E) Kaplan-Meier plot showing the significant association of the BASAL-02 module with patients’ clinical outcome in the three data sets considered.
Subtype-specific breast cancer driver modules are associated with survival outcome
To test for association of subtype-specific driver modules with patient survival outcome, we developed a risk score defined as a linear combination of the normalized gene-expression values of the component genes in the module weighted by their estimated univariate Cox proportional-hazard regression coefficients (see Methods). Based on the risk-score values, patients were stratified into low-risk (risk-score <33 percentile) and high-risk (risk-score >66 percentile) groups. Both Cox regression coefficients of each gene and risk-score cutoff values for each module were estimated from the TCGA-BRCA cohort (training data set); later these values were applied to the METABRIC cohorts (test data set). To assess whether the risk-score assignment to high/low categories was valid, a log-rank test was performed for each module in both training and test data sets.
We first compared driver-seeded modules against driver gene–free modules that, according to OptDis, have the best discriminative score for the TCGA-BRCA data set. For each module, we calculated three distinct indices: log-rank test P-value, hazard ratio (HR), and concordance-index (c-index). We found driver-seeded modules to outperform driver-free modules on all three indices, demonstrating that the driver-seeded modules were better correlated with survival (Fig. 5B). Motivated by this, we identified the top modules for each of the BRCA subtypes that do well based on all three indices, and checked whether they can return meaningful results with respect to survival. We found nine driver modules significantly associated with patients’ survival outcome (P-value < 0.01, HR > 1.5 and c-index > 0.5) in the TCGA-BRCA cohort (Supplemental Table S17). These nine modules were also significantly associated with patient survival outcome (P-value < 0.01) in two additional cohorts (METABRIC cohorts) (Supplemental Figs. S34–S41). It is interesting to note that two of these modules (BASAL-02 and HER2-01) were seeded by an oncogene-nuclear receptor coactivator 3 (NCOA3) driver gene. NCOA3 driver module was the second-topmost module (Fig. 5C) to separate Basal from other subtypes and was the top-most module (Supplemental Fig. S34) to separate HER2 subtype. The NCOA3 driver module was down-regulated in Basal subtype and associated with patients’ overall survival (Fig. 5D,E). A fraction of breast (and ovarian) cancer patients are known to harbor NCOA3 mutation, amplification, or deletion (Gupta et al. 2016). NCOA3 alone cannot distinguish the basal subtype. NCOA3 requires other component genes in the module (AR, XBP1, TFF1, and SPDEF) to collectively distinguish the basal subtype (Supplemental Fig. S33), which, per our knowledge, is a novel finding. However, the interactions within the module are well known. NCOA3 is a coactivator of steroid hormone receptors AR and ESR1 and is a transcriptional target of XBP1 (Gupta et al. 2016). NCOA3 is known to stimulate many intracellular signaling pathways that are critical for cancer proliferation and metastasis. The activity of NCOA3 is known to be associated with reduced responsiveness to tamoxifen in patients (Osborne et al. 2003). SPDEF is associated with regulation of AR activity (Lehmann et al. 2011).
HIT'nDRIVE-seeded driver genes accurately predict drug efficacy
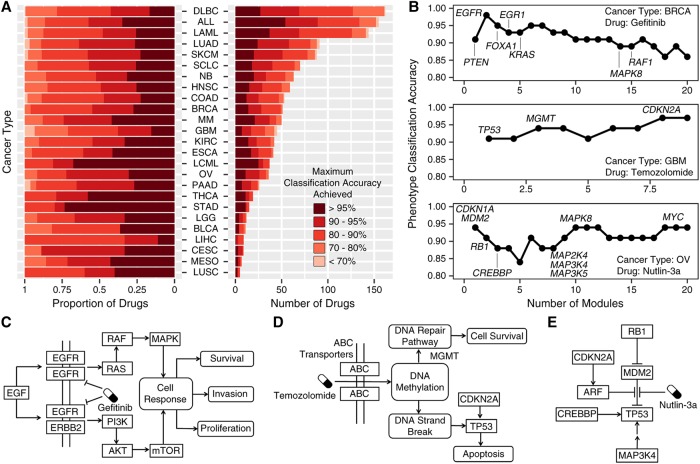
Next, we obtained somatic mutation, copy number aberration (CNA), and gene-expression data of pan-cancer cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) project (Iorio et al. 2016). We used HIT'nDRIVE (Supplemental Fig. S46) to identify driver genes of individual cancer cell lines (Supplemental Table S18). Following up on the premise by Iorio et al. (2016) that potential driver genes (i.e., cancer genes, which include the CGC genes) alone could predict drug efficacy fairly well, the predicted driver genes were used as seeds in the network (STRING v10) to identify subnetworks that discriminate between the drug-response phenotypes (i.e., sensitive vs. resistant cell lines). As available in GDSC, 265 different drug treatments were tested on each cell line provided. We present results for 25 cancer types (the remaining five cancer types for which only a very limited number of cell lines are available are statistically insignificant and thus have not been used).
Perhaps our most interesting result is that, for many drugs, the top HIT'nDRIVE-predicted driver module for a specific cancer type (more specifically, OptDis modules seeded by HIT'nDRIVE-identified driver genes, prioritized with respect to drug efficacy) includes not only the drug target but also the associated (downstream) signaling pathway. As importantly, we measured the accuracy of drug-response phenotype classification using HIT'nDRIVE-OptDis for each drug treatment in different cancer types (Fig. 6A; Supplemental Table S19). In most cancer types, HIT'nDRIVE-OptDis correctly predicted the response to >25% of the drugs in ≥95% of the patients. Specifically, stomach adenocarcinoma (STAD) and chronic myelogenous leukemia (LCML) are the cancer types with the highest fraction of drugs predicted with an accuracy of ≥95%, whereas liver hepatocellular carcinoma (LIHC) and GBM are the cancer types with the lowest fraction of drugs predicted with the same accuracy. Below we provide some of our observations on three well-known/promising cancer drugs for which we obtained high accuracy on specific cancer types.
Figure 6.
Drug efficacy predicted by HIT'nDRIVE-seeded driver genes. (A) Accuracy of drug-response phenotype classification for all 265 drugs used in the GDSC study across 25 cancer types (the remaining five cancer types for which only a very limited number of cell lines have been made available are statistically insignificant and thus have not been used). The classification accuracy for each drug on each cancer type is measured based on the collective use of at most 10 best-discriminating modules; i.e., the accuracy is maximized across the range of one to 10 (best-discriminating) modules. Note that many of the drugs were not tested on all cancer types; in fact, for the vast majority of cancer types, only a handful of drugs were tested. (B) Classification accuracy of modules that distinguish the drug-response phenotypes after treatment with Gefitinib in BRCA cell lines (top), temozolomide in GBM cell lines (middle), and Nutlin-3a in OV cell lines (bottom). Important genes identified in the modules and involved in the dysregulated signaling pathways have been highlighted. (C–E) The figures represent the dysregulated signaling pathways in the respective drug perturbation.
Gefitinib is a clinically approved (for patients with non-small-cell lung cancer) protein kinase inhibitor that selectively inhibits EGFR. Interestingly, in BRCA, EGFR copy-number amplification or overexpression primarily activates the RAS-RAF-MAPK pathway and PI3K-AKT-mTOR pathway triggering response for cell proliferation, invasion, and survival. By use of HIT'nDRIVE, EGFR was found as a driver gene of BRCA cell lines. Furthermore, the EGFR-seeded driver module was the second highest-scoring module to distinguish the drug-response phenotype, increasing the classification accuracy to 98% (Fig. 6B,C).
Another example, Nutlin-3a, is a promising preclinical stage compound that inhibits the interaction between MDM2 and TP53, inducing apoptosis. MDM2 was predicted as a driver gene in OV cell lines by HIT'nDRIVE. The MDM2-seeded module was the top predictor (maximum accuracy, 94%) of the drug-response phenotype when treated with Nutlin-3a (Fig. 6B,E). Our method predicted many other interacting partners (both as seed or component genes in the module) of MDM2 and TP53, which are known to play a critical role in TP53 pathway.
Finally, TMZ is a clinically approved first-line therapy for GBM. ABC transporters (including ABCB1) help to transport TMZ from the extracellular space to the cytoplasm of a cell. TMZ methylates selective nucleotides of the DNA triggering DNA repair pathway. MGMT specifically removes the methyl groups from the methylated nucleotides escaping from DNA strand breaks. MGMT was predicted as a component gene in the third top-scoring module. Failure to repair DNA strand breaks triggers the DNA damage response pathway, further activating TP53 and apoptosis. Interestingly, TP53 was predicted as the seed of the top-scoring module by HIT'nDRIVE-OptDis. Furthermore, another gene in the DNA damage response pathway, CDKN2A, seeds another top-ranking module, which improves the overall classification accuracy to 97% (Fig. 6B,D). Note that both CDKN2A and TP53 are the most frequently altered genes in GBM.
Discussion
In recent years, there has been an unprecedented increase in the multidimensional high-throughput data profiling (especially genome and transcriptome) of cancer patients. This has revealed extensive mutational heterogeneity observed in the cancer (sub)types, yielding a long-tailed distribution of mutated genes across the patients, implying the existence of many rare/private driver genes. Thus, there is a great need for computational methods to mine these massive data sets and prioritize clinically actionable driver events to aid treatment modalities using precision oncology.
Here, we have presented a network-based combinatorial method, HIT'nDRIVE, which models the collective effects of sequence altered genes on expression altered genes. HIT'nDRIVE aims to solve the RWFL problem on a gene/protein interaction network—which differs from the standard FL problem by its use of “hitting time,” the expected minimum number of hops in a random walk originating from any sequence altered gene (i.e., a potential driver) to reach an expression altered gene, as a distance measure. We introduced the notion of “multihitting time” and presented efficient and accurate methods to estimate it based on single-source hitting time in large-scale networks. HIT'nDRIVE reduces RWFL (with multihitting time as the distance) to a weighted multiset cover problem, which it formulates and solves as an ILP.
As a measure of influence, hitting time—the expected length of a random walk between two nodes—or its general version, the multihitting time, is quite different from the diffusion-based measures or Rooted PageRank, which are based on asymptotic distributions. We argue that hitting time is a better measure for our purposes as it is (1) parameter free (diffusion model introduces at least one additional parameter: the proportion of incoming flow “consumed” at a node in each time step), (2) it is time dependent (while the diffusion model and PageRank measure the stationary behavior), and (3) it is more robust with respect to small perturbations in the network (Hopcroft and Sheldon 2007).
In this article, we have demonstrated that, first, HIT'nDRIVE increases our ability to identify potential genomic driver alterations and, second, HIT'nDRIVE prioritizes clinically actionable driver genes, many of which happen to be private drivers. This implies that it is possible to replicate the lengthy and costly experimental approaches for detecting driver genes in common tumor types by HIT'nDRIVE in silico, strongly supporting the biological relevance of HIT'nDRIVE's algorithmic framework. The fact that a high portion of HIT'nDRIVE prioritized drivers in well-studied cancer types overlap with known driver genes increases our confidence in the calls made by HIT'nDRIVE in rarer tumor types for which driver genes are mostly unknown. In fact, the initial results of the Pan Cancer Analysis of Whole Genomes (PCAWG) project reveal that >20% of tumors do not have a single (genomically altered) driver gene from CGC (data not shown). HIT'nDRIVE is thus being used for the analysis of PCAWG data to reduce this gap. Results on new driver genes identified by HIT'nDRIVE, especially in rare tumor types will be made available by PCAWG. Third, HIT'nDRIVE prioritizes driver genes present in both the center and periphery of an interaction network. Fourth, our analysis revealed that driver genes have higher collective influence on the transcriptome than other altered genes. Some of these driver genes are central and naturally have high influence; however, there are also many noncentral driver genes with high influence over other genes in the network. Fifth, HIT'nDRIVE is especially suitable for identifying such noncentral driver genes or infrequent/private drivers. Sixth, we demonstrated that it is also possible to perform accurate phenotype prediction for tumor samples by only using HIT'nDRIVE-implied driver genes and their “network modules of influence” (small subnetworks involving each driver gene where the aggregate expression profile correlates well with the cancer phenotype) as features, providing additional evidence that these genes may be driving the cancer phenotype. The network modules we identified may provide new insights into the biological mechanisms underlying tumor progression. Seventh, HIT'nDRIVE can capture subtype-specific driver genes and such driver-seeded modules can indeed differentiate between different subtypes of a cancer. Eighth, we have demonstrated that subtype-specific driver modules are also associated with patients’ survival outcome, providing additional evidence that these driver genes have clinical significance. Ninth, we also demonstrated that HIT'nDRIVE-seeded driver genes (more specifically, OptDis modules seeded by HIT'nDRIVE-identified driver genes, prioritized with respect to drug efficacy) include not only the drug target but also the associated (downstream) signaling pathway. This provides us with the possibility of identifying and clinically targeting multiple genes (not necessarily sequence-wise altered but nevertheless in the module identified by HIT'nDRIVE) dysregulating critical oncogenic or metabolic pathways.
We also note that targeted therapeutics are being extensively used in clinical trials, but the drug response rate is very poor (only ∼5% of patients in clinical trials have good response to targeted therapeutics) (Prasad 2016). This is most likely because even if a cancer patient harbors an alteration for which targeted therapeutics are available, we do not know if that alteration is responsible for driving the tumor (Beltran et al. 2015). HIT'nDRIVE could potentially play a key role by prioritizing potential driver alterations from a vast pool of passenger alterations. In our study, we have used drug efficacy data from pan-cancer cell lines in order to demonstrate that the potential genomic drivers (more precisely driver gene–seeded modules) of the cell lines can be used as features to predict drug efficacy. Following a similar procedure in clinical trials, we believe that the application of HIT'nDRIVE to predict drug efficacy would likely improve the drug response rate.
HIT'nDRIVE predicted ABCB1 as the most influential driver gene in seven TCGA-GBM cases that were treated with TMZ prior to tissue collection. By using the GDSC data set, we demonstrated that HIT'nDRIVE-OptDis can predict mechanisms of drug sensitivity for TMZ and other drugs (Fig. 6B–E) Since ABCB1 was not mutated in any of the GBM cell lines in the analysis, it was not identified as a driver gene of GBM cell lines. However, the top seed driver gene, TP53, is an interaction partner of ABCB1 (in STRING v10 network). Other seed driver genes and their component genes in the module that are direct interaction partners of ABCB1 are UBC, CAV1, WDTC1, and DNAH8. ABC transporters (including ABCB1) help to transport TMZ from the extracellular space to the cytoplasm of a cell. On the other hand, DNA damage caused by TMZ activates TP53, thereby dysregulating apoptotic pathways. Thus, the presented analysis demonstrates that the downstream expression changes are, most likely, the manifestation of the selection pressure in ABCB1 induced by TMZ treatment.
Protein–protein interaction (PPI) networks representing physical interactions now include thousands of proteins and over 1 million (undirected) interactions between them. Regulatory networks, on the other hand, represent gene/protein regulation occurring at multiple levels of biological systems through directed links. Since available regulatory networks are very limited in size and scope, our study focuses on PPI networks. However, HIT'nDRIVE can easily be applied to regulatory networks as they grow in size and scope. In addition, the use of multihitting time as a distance measure between two or more driver genes and a target gene enables HIT'nDRIVE to capture synthetic rescue like scenarios; this is ideally suited for undirected PPI networks, but in principle can be extended to regulatory networks in the future.
HIT'nDRIVE is a driver gene prioritization tool that is flexible enough to incorporate different types of omics data. Both principles under RWFL and HIT'nDRIVE can be utilized to identify the causal genes in different complex disease facing analogous problems to cancer. Finally, we believe that applications of RWFL problem may extend beyond its application to driver gene identification to influence analysis in social networks, disease networks and others.
Methods
An overview of the HIT'nDRIVE algorithmic framework
HIT'nDRIVE links alterations at the genomic level to changes at transcriptome level through a gene/protein interaction network. More specifically, HIT'nDRIVE identifies the minimum number of potential driver genes that can cause a user-defined proportion of the downstream expression effects observed. We formulate this as a RWFL problem, a new combinatorial optimization problem that generalizes the classical FL problem by the use of a novel distance measure. Given a network, FL problem defines the distance between a potential driver gene and an outlier gene as the length of the shortest path between them. The RWFL problem, in contrast, uses “hitting time” (or “first passage time”) (Condamin et al. 2007; Liben-Nowell and Kleinberg 2007), the expected length of a random walk between the two nodes, as their distance. Under the use of hitting time, the FL problem completely changes nature: In the classical FL formulation, the goal is to associate each outlier gene in the network with exactly one (the closest) driver gene. In the RWFL formulation, each outlier gene is associated with multiple driver genes (whose collective distance to the outlier will no longer be the shortest pairwise distance), forming a many-to-many relation. Intuitively, hitting time measures how accessible a particular outlier gene is from potential driver genes. Thus, RWFL problem asks to find the smallest set of sequence-altered genes from which one can reach (a good proportion of) outliers within a user-defined “multihitting time” (the expected length of the shortest random walk originating from any of the sequence altered genes) and ending at an outlier.
As per the standard FL problem, RWFL is NP hard. In fact, even the problem of computing the multihitting time between a set of nodes in a network and a particular target node is difficult. In what follows, we summarize how we approach this problem within the HIT'nDRIVE framework (for details, see Supplemental Methods).
For simplicity, we first describe how HIT'nDRIVE works on single patient data. Given an interaction network with X denoting the set of sequence-altered genes (through SNVs or CNAs) and Y denoting the set of expression-altered genes, HIT'nDRIVE computes the smallest subset of X whose joint “influence” over (a user-defined fraction of) expression-altered genes is sufficiently high (i.e., above a user-defined threshold). The influence of a set of (sequence-altered) genes X over an expression-altered gene g is defined as 1/MHT(X, g), where MHT(X, g) denotes the multihitting time, the expected length of the shortest random walk originating at each one of the genes in X that ends at g. Therefore, HIT'nDRIVE aims to solve the RWFL problem in a network where X are the “potential facilities” and Y are the “requests.”
Since RWFL is a computationally hard problem and cannot be solved in a reasonable amount of time in its original formulation, we reduce the RWFL problem to the WMSC problem, for which we give an ILP formulation. Intuitively, in this new formulation, HIT'nDRIVE associates the genomic alterations with transcriptomic changes in the form of a bipartite graph Gbip(X, Y, E), where X is the set of aberrant genes, Y is the set of patient-specific expression-altered genes, and E is the set of edges. If gene xi is mutated in a patient p, we set edges between xi and all of the expression altered genes in the same patient (yj, p), where the edges are weighted by the inverse pairwise hitting times (Fig. 1A). The WMSC problem on this representation of data asks to find the smallest subset of X (as potential drivers) whose total influence (sum of pairwise influence values) over a user-defined fraction of expression-altered genes (for each patient) is sufficiently high.
The reduction from RWFL problem to the WMSC problem is achieved by estimating the multihitting time as a function of independent hitting times of the drivers to an outlier, which provides an upper bound on the multihitting time. The exact individual hitting times are calculated by a matrix inversion method (for details, see Supplemental Methods) (Tetali 1999). The resulting WMSC problem can then be formulated as the ILP below, which is efficiently solvable by IBM CPLEX (within minutes) for all data sets we considered:
The above ILP formulation for the WMSC problem introduces binary variables xi, yj, eij, respectively, for each potential driver, expression alteration event, and edge in the bipartite graph. The objective of the ILP is to minimize the number of drivers (i.e., the sum of xi values) subject to four constraints. The first constraint ensures that a selected driver contributes to the coverage of each of the expression alteration events it is connected to (in each patient, if multiple patients are available). The second constraint ensures that selected (patient-specific) driver genes contribute enough to cover at least a (γ) fraction of the sum of all incoming edge weights to each expression alteration event. This constraint corresponds to setting an upper bound on our estimate on the inverse of multihitting time of the selected (patient-specific) drivers on an expression alteration event. The third constraint ensures that the selected driver genes collectively cover at least an α fraction of the set of expression alteration events. And the fourth constraint ensures that for each patient, the top β fraction of the expression altered genes with highest weights (λj) are always covered.
As indicated above, our ILP formulation for the WMSC problem can be generalized to multiple patients with the objective of minimizing the total number of driver genes across all patients, subject to the constraint that a user-defined proportion of outlier genes in each of the patients are covered by the subset of driver genes present in that patient (for details, see Supplemental Methods).
In order to quantitatively assess the genes identified by HIT'nDRIVE, we extended our previously developed algorithm, OptDis (Dao et al. 2011), for de novo identification of modules of small size inside the interaction network that contain (i.e., are seeded by) at least one predicted driver gene. The modules are chosen so that their discriminative power (for phenotype classification) is the greatest among connected subnetworks of similar size that contain the individual predicted driver genes. In general, OptDis performs supervised dimensionality reduction on the set of connected subnetworks. It projects the high-dimensional space of all connected subnetworks to a user-specified lower-dimensional space of subnetworks such that, in the new space, the samples belonging to the same class are closer and the samples from different classes are more distant to each other (i.e., minimize in-class distance and maximize out-class distance) with respect to a normalized distance measure (typically L1). Then we use module features (average expression of genes in the module) for phenotype classification (Fig. 1B,C). By using such module features, we hope that the classifier in use does not overfit on rare driver genes and is able to generalize the signal coming from rare drivers to new patients. We report the classification accuracy based on the identified driver-seeded modules as means of quantitative validation of our results (in the absence of ground truth). We also look at the genes that build the chosen modules (of high classification accuracy) in an attempt to identify cancer-related pathways.
Data sets and analysis
We used publicly available data sets of four major cancer types: GBM, OV, BRCA, and PRAD from TCGA project. All data were obtained from TCGA Data Portal in May 2014, which were mapped to GRCh37 genome build. Although TCGA has recently made available all data realigned to the newer GRCh38 genome build, to ensure compatibility, all TCGA data we have used in this study has been mapped to GRCh37.
Somatic mutation
Calls (level 2 data) from all available platforms/centers were merged. Only missense, nonsense, and splice-site mutations were marked as somatic-mutation alteration events.
CNAs
For GBM and OV, Agilent human genome CGH microarray 244A (level 1) data files were used, and for PRAD and BRCA, Affymetrix genome-wide human SNP array 6.0 (level 3) data files were used to generate the copy number profiles.
These Agilent FE format sample files were loaded into BioDiscovery Nexus Copy Number software v7.0, where quality was assessed and data were visualized and analyzed. All samples were mapped to the most recent genome build (hg 19, GRCh37) via Agilent probe identifiers and annotation (downloaded from Agilent's website) based on the 1M SurePrint G3 human CGH microarray 1×1M design platform. BioDiscovery's FASST2 segmentation algorithm, a hidden Markov model–based approach, was used to make copy number calls. The FASST2 algorithm, unlike other common HMM methods for copy number estimation, does not aim to estimate the copy number state at each probe but uses many states to cover more possibilities, such as mosaic events. These state values are then used to make calls based on a log-ratio threshold. The significance threshold for segmentation was set at =5 × 10−6, also requiring a minimum of three probes per segment and a maximum probe spacing of 1000 between adjacent probes before breaking a segment. The log ratio thresholds for single copy gain and single copy loss were set at 0.2 and −0.23, respectively. The log ratio thresholds for two or more copy gains and homozygous losses were set at 1.14 and −1.1, respectively. Upon loading of raw data files, signal intensities are normalized via division by mean. All samples are corrected for GC wave content using a systematic correction algorithm. Only the high-confidence CNAs, i.e., high copy number gain or homozygous deletions, were marked as copy number aberrant events. Finally, genes that harbor either a somatic-mutation aberrant event or a copy number aberrant event were taken to be the final list of aberrant genes at the genomic level.
Gene expression
We used microarray-based gene expression (Affymetrix HT human genome U133 array plate set, level 1) for GBM and OV data sets, whereas for BRCA and PRAD data sets, RNA-seq–derived gene expression was used (level 3). Gene-expression profiles of normal and tumor phenotypes were used as sample groups.
Gene fusions
Transcript fusions prediction calls for GBM, OV, BRCA, and PRAD were obtained from TCGA Fusion Gene Data Portal (http://www.tumorfusions.org) (Yoshihara et al. 2014). The fusion partner genes were tagged for gene-fusion alteration.
Genomics of drug sensitivity in cancer
Somatic mutation, copy number alterations and gene expression, and drug screening data of cancer cell lines were downloaded from the GDSC (Iorio et al. 2016) website (http://www.cancerrxgene.org/downloads). Data were downloaded on August 2016.
Interaction networks
We used STRING v10 (Szklarczyk et al. 2015) protein-interaction network that contains high-confidence functional PPIs (for details, see Supplemental Methods).
Pathway enrichment analysis
The selected set of genes were tested for enrichment against gene sets of pathways present in Molecular Signature Database (MSigDB) v5.0 (Subramanian et al. 2005). A Fisher's exact test–based gene set enrichment analysis was used for this purpose (for details, see Software and Code Availability). A cut-off threshold of false-discovery rate (FDR) ≤ 0.01 was used to obtain the significantly enriched pathways. The same procedure, as above, is used to assign biological functional to the gene modules.
Derivation of outlier-genes
We used a generalized extreme Studentized deviate (GESD) test (Rosner 1983) to obtain the outlier genes (for details, see Supplemental Methods and Software and Code Availability).
Association of driver modules with patients’ survival outcome
To test for association of driver modules with patients’ survival outcome, we developed a risk-score based on multigene (component genes of the module) expression. The risk-score (S) defined as a weighted sum of the normalized gene-expression values of the component genes in the module weighted by their estimated univariate Cox proportional-hazard regression coefficients (Beer et al. 2002) as given in the equation below:
Here i and j represent a gene and a patient, respectively; βi is the coefficient of cox regression for gene i; xij is the normalized gene expression of gene i in patient j; and k is the number of component genes in a gene module. The normalized gene-expression values were fitted against overall survival time with living status as the censored event using univariate Cox proportional-hazard regression (Exact method).
Based on the risk-score values, patients were stratified into two groups: low-risk group (patients with S < 33 percentile of S) and high-risk group (patients with S > 66 percentile of S). Patients that fall in between (i.e., patients with S ≥ 33 percentile of S and ≤66 percentile of S) were discarded from the further analysis as these patients fall into the intermediate-risk group and are bound to introduce noise while performing log-rank test.
Both Cox regression coefficients of each gene and risk-score cutoff values for each module were estimated from the TCGA-BRCA cohort (training data set); later these values were applied to METABRIC cohorts (test data set). To assess whether the risk-score assignment to high/low categories was valid, a log-rank test was performed for each module in both training and test data sets.
Finally, to identify the significant list of driver-modules that were robust enough to predict patients’ survival, we calculated log-rank test P-value, HR (Wald test), and c-index (Wald test).
Software and code availability
The hitting time–based influence-matrix generation and HIT'nDRIVE algorithm are implemented in C++. They can be accessed from GitHub (https://github.com/sfu-compbio/hitndrive) and in Supplemental Archive 1. The GESD test–based outlier detection method is available at GitHub (https://github.com/raunakms/GESD) and as Supplemental Archive 2. A Fisher's exact test–based gene-set enrichment analysis is available at GitHub (https://github.com/raunakms/GSEA-Fisher) and as Supplemental Archive 3.
Supplementary Material
Acknowledgments
We thank Dr. Shancheng Ren (Second Military Medical University, Shanghai, China) for providing us with RNA-seq data for prostate cancer patients. We thank Dr. Oktay Gunluk (IBM T.J. Watson Research Center) and the IBM Academic Initiative for providing us with free license to IBM ILOG CPLEX Optimization Studio (CPLEX). This project was funded by the following: Canadian Cancer Society Research Institute (S.C.S., C.C.C.), Natural Sciences and Engineering Research Council of Canada (NSERC) Discover Grants (S.C.S.), and Terry Fox Research Institute (C.C.C.). R.S. is supported by Mitacs Accelerate Awards (IT07445), Prostate Cancer Foundation–BC Research Award and Canadian Institutes of Health Research (CIHR) Bioinformatics Training Program. E.H. is supported by NSERC-CREATE Computational Methods for the Analysis of the Diversity and Dynamics of Genomes (MADD-Gen) program. We thank all members of the Sahinalp and Collins laboratories for helpful suggestions. The results published here are in part based upon data generated by TCGA Research Network: http://cancergenome.nih.gov/. The funding agencies had no role in study design, data collection, and analysis; decision to publish; or preparation of the manuscript.
Author contributions: R.S., E.H., G.H., and S.C.S. conceived the study. R.S. and E.H. developed the HIT'nDRIVE algorithm to prioritize driver genes, performed entire analysis, and wrote the paper. T.S. provided mathematical proofs for estimating multisource hitting time. P.D. and K.W. developed OptDis algorithm to identify driver modules. F.V., J.Y., and S.A. helped in data analysis. C.C.C. helped in biological interpretation of the results. All authors contributed to preparation of the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.221218.117.
References
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe'er D. 2010. An integrated approach to uncover drivers of cancer. Cell 143: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. 2012. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashashati A, Haffari G, Ding J, Ha G, Lui K, Rosner J, Huntsman DG, Caldas C, Aparicio SA, Shah SP. 2012. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol 13: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SLR, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. 2002. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8: 816–824. [DOI] [PubMed] [Google Scholar]
- Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, Kossai M, Pauli C, Faltas B, Fontugne J, et al. 2015. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 1: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. 2016. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 13: 674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer SE, Rao AD, Lyle M, Sandhu S, Long GV, McArthur GA, Raleigh JM, Hicks RJ, Millward M. 2014. Activity of trametinib in K601E and L597Q BRAF mutation-positive metastatic melanoma. Melanoma Res 24: 504–508. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2015. The molecular taxonomy of primary prostate cancer. Cell 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hao J, Jiang W, He T, Zhang X, Jiang T, Jiang R. 2013. Identifying potential cancer driver genes by genomic data integration. Sci Rep 3: 3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. 2012. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 22: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45: 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamin S, Bénichou O, Tejedor V, Voituriez R, Klafter J. 2007. First-passage times in complex scale-invariant media. Nature 450: 77–80. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, Atefi M, Su Z, Branch S, Lyle PL, et al. 2012. BRAFL597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao P, Wang K, Collins C, Ester M, Lapuk A, Sahinalp SC. 2011. Optimally discriminative subnetwork markers predict response to chemotherapy. Bioinformatics 27: i205–i213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, McConechy MK, Horlings HM, Ha G, Chun Chan F, Funnell T, Mullaly SC, Reimand J, Bashashati A, Bader GD, et al. 2015. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat Commun 6: 8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta B, Pusztai L, Qi Y, André F, Lazar V, Bianchini G, Ueno N, Agarwal R, Wang B, Shiang CY, et al. 2012. A network-based, integrative study to identify core biological pathways that drive breast cancer clinical subtypes. Br J Cancer 106: 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. 2006. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173: 2187–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB, Eldred JM, et al. 2013. DGIdb: mining the druggable genome. Nat Methods 10: 1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Hossain MM, Miller N, Kerin M, Callagy G, Gupta S. 2016. NCOA3 coactivator is a transcriptional target of XBP1 and regulates PERK-eIF2α-ATF4 signalling in breast cancer. Oncogene 35: 5860–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcroft J, Sheldon D. 2007. Manipulation-resistant reputations using hitting time. In Algorithms and models for the web-graph: proceedings of the fifth international workshop, WAW 2007, San Diego, CA, USA, December 11–12, 2007 (ed. Bonato A, Chung FRK), pp. 68–81. Springer, Berlin, Heidelberg. [Google Scholar]
- Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H, et al. 2016. A landscape of pharmacogenomic interactions in cancer. Cell 166: 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jornsten R, Abenius T, Kling T, Schmidt L, Johansson E, Nordling TE, Nordlander B, Sander C, Gennemark P, Funa K, et al. 2011. Network modeling of the transcriptional effects of copy number aberrations in glioblastoma. Mol Syst Biol 7: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YA, Wuchty S, Przytycka TM. 2011. Identifying causal genes and dysregulated pathways in complex diseases. PLoS Comput Biol 7: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthauer KD, Kendziorski C. 2015. MADGiC: a model-based approach for identifying driver genes in cancer. Bioinformatics 31: 1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A, Smoly IY, Rapaport G, Lindquist S, Fraenkel E, Yeger-Lotem E. 2011. ResponseNet: revealing signaling and regulatory networks linking genetic and transcriptomic screening data. Nucleic Acids Res 39: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. 2011. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121: 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MDM, Blokh D, Sharan R, Raphael BJ. 2013. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput Biol 9: e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liben-Nowell D, Kleinberg J. 2007. The link-prediction problem for social networks. J Am Soc Inform Sci Technol 58: 1019–1031. [Google Scholar]
- Lin F, De Gooijer MC, Roig EM, Buil LCM, Christner SM, Beumer JH, Würdinger T, Beijnen JH, Van Tellingen O. 2014. ABCB1, ABCG2, and PTEN determine the response of glioblastoma to temozolomide and ABT-888 therapy. Clin Cancer Res 20: 2703–2713. [DOI] [PubMed] [Google Scholar]
- Masica DL, Karchin R. 2011. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res 71: 4550–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SAW, Wong J, Allred DC, Clark GM, Schiff R, et al. 2003. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst 95: 353–361. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science (New York, N.Y.) 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey PJ, et al. 2015. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521: 489–494. [DOI] [PubMed] [Google Scholar]
- Paull EO, Carlin DE, Niepel M, Sorger PK, Haussler D, Stuart JM. 2013. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics 29: 2757–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V. 2016. Perspective: the precision-oncology illusion. Nature 537: S63 Outlook. [DOI] [PubMed] [Google Scholar]
- Rosner B. 1983. Percentage points for a generalized ESD many-outlier procedure. Technometrics 25: 165–172. [Google Scholar]
- Shi K, Gao L, Wang B. 2016. Discovering potential cancer driver genes by an integrated network-based approach. Mol BioSyst 12: 2921–2931. [DOI] [PubMed] [Google Scholar]
- Shrestha R, Hodzic E, Yeung J, Wang K, Sauerwald T, Dao P, Anderson S, Beltran H, Rubin MA, Collins CC, et al. 2014. HIT'nDRIVE: Multi-driver gene prioritization based on hitting time. In Research in computational molecular biology: proceedings of the 18th Annual International Conference, RECOMB 2014, Pittsburgh, PA, USA, April 2–5, 2014 (ed. Sharan R), pp. 293–306. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo C, Hrydziuszko O, Lee D, Pramana S, Saputra D, Joshi H, Calza S, Pawitan Y. 2015. Integration of somatic mutation, expression and functional data reveals potential driver genes predictive of breast cancer survival. Bioinformatics 31: 2607. [DOI] [PubMed] [Google Scholar]
- Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. 2008. eQED: an efficient method for interpreting eQTL associations using protein networks. Mol Syst Biol 4: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. String v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetali P. 1999. Design of on-line algorithms using hitting times. SIAM J Comput 28: 1232–1246. [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, N.Y.) 310: 644–648. [DOI] [PubMed] [Google Scholar]
- Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. 2006. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics 22: 489–496. [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, et al. 2014. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 20: 682–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. 2011. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol 18: 507–522. [DOI] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. 2010. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26: i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science (New York, N.Y.) 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat Genet 41: 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RGW. 2014. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34: 4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Simon R. 2011. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics 27: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.