Abstract
Genetic discovery from the multitude of phenotypes extractable from routine healthcare data can transform our understanding of the human phenome and accelerate progress towards precision medicine. However, a critical question when analysing high-dimensional and heterogeneous data is how to best interrogate increasingly specific subphenotypes whilst retaining statistical power to detect genetic associations. Here we develop and employ a novel Bayesian analysis framework that exploits the hierarchical structure of diagnosis classifications to analyse genetic variants against UK Biobank disease phenotypes derived from self-reporting and hospital episode statistics. Our method displays a more than 20% increase in power to detect genetic effects over other approaches and identifies novel associations between classical human leukocyte antigen (HLA) alleles and common immune-mediated diseases (IMDs). By applying the approach to genetic risk scores (GRSs) we reveal the extent of genetic sharing between IMDs and expose differences in disease perception or diagnosis with potential clinical implications.
Large-scale, hypothesis-free approaches for identifying genetic risk variants, including genome-wide association studies (GWAS) and next generation sequencing analyses, have greatly advanced our understanding of complex traits, with implications for drug development and clinical practice1–5. These approaches typically involve genetic discovery from case-control cohorts where clinically derived phenotypes are considered one at a time. By contrast, resources such as the UK Biobank6,7, which has prospectively collected extensive health-relevant phenotypic and genotypic information from 500,000 participants, allow for simultaneous investigation of multiple traits and are set to lead to a step-change in the rate of genetic discovery8,9.
However, capitalizing on availability of population-based cohorts for biomedical research is complicated by the scale and nature of the data: the phenotypic space is multi-dimensional and heterogeneous as data can be subject to observational predilections, non-uniform recording practices, and longitudinal biases, and phenotype prevalence is variable10–16. This creates new challenges that are not addressed by existing analytical methods for GWAS and phenome-wide association studies (PheWAS). An open question is how to interrogate the many precise phenotypes obtainable from routine healthcare data at a resolution that reveals associations above and beyond those identified through GWAS, but without sacrificing statistical power. Making use of disease classification hierarchies, such as the tree of International Classification of Diseases, Tenth Revision (ICD-10) codes, provides a tractable solution. Here we have developed a novel Bayesian analysis framework for identifying genetic associations across the entire health phenotype space by taking advantage of the relative topology of nodes within two tree-structured phenotypic datasets from the UK Biobank - the self-reported (SR) diagnoses that are organised using the UK Biobank classification tree which includes 531 diagnostic terms, and the hospitalisation episode statistics (HES) data that utilise ICD-10 codes and contain 16,310 diagnostic terms.
Results
Tree analysis approach
To test the association of genetic variation with any given UK Biobank clinical phenotype, we want to construct a statistical framework that meets a set of fundamental requirements. Firstly, the method must accommodate different types of genetic variation, such as (i) single nucleotide polymorphisms (SNPs), (ii) haplotypes in a highly polymorphic region like the HLA gene region, or (iii) GRSs constructed using multiple SNPs or haplotypes known to be associated with a quantitative trait or complex disease. Secondly, for single locus variation, any genetic model (e.g. additive, dominant or full) must be accommodated. Thirdly, the method must allow for joint analysis and quantification of evidence of association at each clinical phenotype, and must estimate the genetic coefficients of effects. Next, the method must allow identification of independent genetic effects through conditional analysis. Lastly, the method must model correlation structure of genetic effects across observed clinical phenotypes using a priori knowledge of phenotype relationships obtained from a diagnosis classification tree.
To meet these requirements, we have developed a novel Bayesian analysis framework, termed TreeWAS, which models genetic coefficients across all phenotypes as a set of random variables. To model the correlation structure we allow coefficients to evolve down a tree in a Markov process (Fig. 1). A known classification hierarchy determines the tree structure, where each node is a clinical term in the classification, and observations can be made at terminal and internal nodes. The prior θ determines the expected correlation between genetic coefficients across phenotypes. The coefficient at a parent node can be inherited by a child node with probability e−θ, or can transition to a new uncorrelated value, with probability 1 − e−θ. This new value will be zero with a probability 1 − π1, or non-zero with a probability π1. Thus, parameters θ and π1 define transition probabilities controlling the Markov process. Given the model structure and the Markov process assumption, we can calculate the likelihood over genetic coefficients across all clinical phenotypes using dynamic programming (details are provided in the Supplementary Note), and we estimate a Bayes Factor statistic (BFtree) for the evidence that genetic coefficients are non-zero for at least one node. Similarly, because of the model’s properties, using dynamic programming and the forward and backward algorithms, we can determine the marginal posterior probability (PP) at each node that the genetic coefficient is non-zero, and the magnitude of this effect using the maximum a posteriori (MAP) estimator (see Supplementary Note).
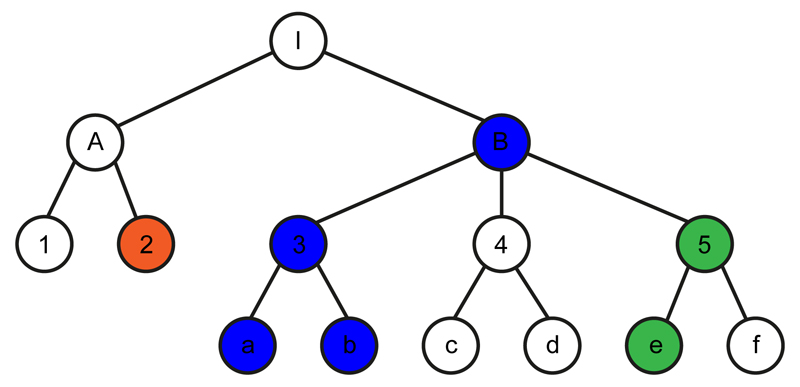
Figure 1. Schematic of diagnosis classification tree and genetic coefficient transition scenarios tested.
Each node in the tree represents a clinical diagnosis and nodes are ordered in a hierarchical structure based on a classification criterion (such as similarities in clinical manifestations). White nodes represent the null state whereby there is no genetic association with the clinical phenotype. Green, red and blue nodes represent the alternative state whereby there is a genetic association with the clinical phenotype, with the different colours corresponding to different, uncorrelated genetic coefficients of association. A genetic coefficient can transition from the null state to a non-zero coefficient as in the I→B and A→2 pairs. From the non-zero state a genetic coefficient can remain in a correlated non-zero state (as in the B→3, 3→a, 3→b and 5→e pairs); it can transition back to the null state (as in the B→ 4 and 5→f pairs); or it can transition to a new, uncorrelated non-zero state (as in the B→5 pair). An in-depth description of the method is provided in the Supplementary Note.
HLA-B*27:05 TreeWAS and PheWAS comparison
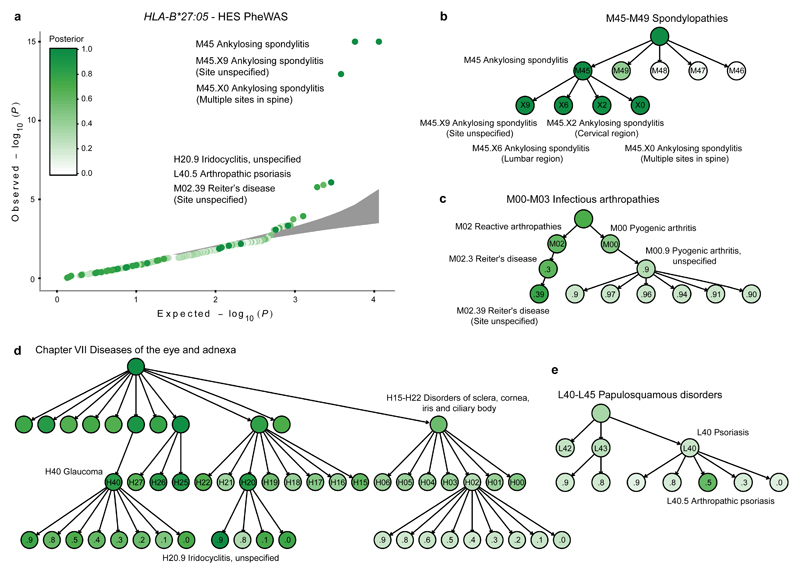
We illustrate the advantages of the TreeWAS approach compared to existing PheWAS tests by analysing the association of the HLA-B*27:05 allele against the UK Biobank HES dataset. The HLA-B*27:05 association with ankylosing spondylitis (AS) is one of the strongest genetic effects observed in human complex diseases, with an odds ratio of 46 (ref. 17), and this allele also confers risk for reactive arthritis18, psoriatic arthritis19 and anterior uveitis (iridocyclitis/iritis)20. Using PheWAS, where evidence of genetic association for each clinical term is estimated independently, HLA-B*27:05 is significantly associated with six ICD-10 terms after correcting for multiple testing (P-adj < 0.05; using the Benjamini & Hochberg procedure21), including M45 AS and M45.X9 AS (Site unspecified) (Fig. 2a). However, this approach fails to identify associations with terms with a greater granularity of clinical description and a relatively low prevalence, such as M45.X6 AS with lumbar spine involvement (P = 0.01, P-adj = 1.0), which is 17 times less prevalent than M45.X9 (0.08%). By contrast, when employing TreeWAS with priors θ = 1/3 and π1 = 0.001 we observed HLA-B*27:05 associations with 145 ICD-10 terms (PP ≥ 0.75; the level of significance used throughout the analysis), clustered in different branches of the classification tree (Fig. 2b-e and Supplementary Table 1). These prior values were chosen to maximise power and sensitivity after exploring the variability of the BFtree statistic and the number of non-zero nodes at a threshold of PP = 0.75 over the parameter space of θ and π1 (Supplementary Fig. 1). As for PheWAS, there was a significant association with M45 AS (PP = 1), but TreeWAS additionally revealed associations with four M45 subcategories (M45.X0, M45.X2, M45.X6 and M45.X9) rather than two (M45.X0 and M45.X9) (Fig. 2a,b). Moreover, there was an association with the broader Spondylopathies category (M45-M49) (PP = 1.0), which was likely driven by associations with M45 (PP = 1.0) and M49 (PP = 0.43), but not M47 Spondylosis (PP = 0.07), despite the latter being ten times more prevalent than M45 (Fig. 2b). As spondylosis occurs due to age-related disk degeneration22, lack of an HLA-B*27:05 association with M47 is consistent with its non-immunological aetiology.
Figure 2. Evidence of HLA-B*27:05 allele association with risk for clinical diagnoses in the HES dataset.
a, Quantile-quantile plot of association test P-values of the HLA-B*27:05 allele with each diagnosis term in the ICD-10 classification tree performed with maximum likelihood estimation using a logistic regression model. Grey area depicts the 95% confidence interval of sampling variance. Results are coloured-coded based on the posterior probability (PP) that HLA-B*27:05 is associated with each diagnosis term as estimated with the TreeWAS model. b-e, Branches of the ICD-10 classification tree where significant associations between HLA-B*27:05 and clinical diagnoses were identified (PP>0.75). Results are tabulated in Supplementary Table 1. AS, ankylosing spondylitis; PS, psoriasis.
Associations with reactive arthritis (e.g. M02.39 Reiter’s disease; PP = 0.78) and anterior uveitis (H20.9 Iridocyclitis, unspecified; PP = 0.98) were also observed (Fig. 2c,d), and we detected a previously unreported HLA-B*27:05 association with H40 Glaucoma (PP = 0.84) (Fig. 2d). As glaucoma is a common complication of chronic uveitis23, comorbidity may explain this association. Lastly, we observed a weak effect on L40.5 Arthropathic psoriasis (PS) susceptibility (PP = 0.60), but not non-arthropathic PS (PP ≤ 0.25 for L40 child nodes except L40.5), consistent with prior studies24 (Fig. 2e). Therefore, our TreeWAS analysis of HLA-B*27:05 in the HES dataset recapitulates known associations, and demonstrates that our method can identify additional genuine associations compared to PheWAS.
Sensitivity and specificity analysis of TreeWAS approach using simulated data
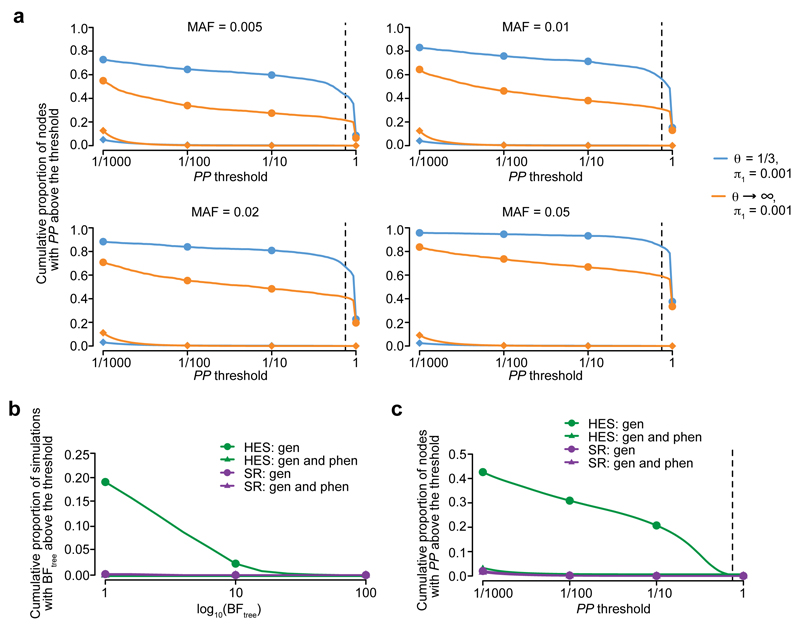
Given the capacity of TreeWAS to identify multiple associations with HLA-B*27:05 we wanted to further investigate the method’s sensitivity and specificity. To assess the relative power of TreeWAS, and to explore its robustness and accuracy, we performed two sets of simulations. In the first set, we assessed power by simulating data from a simple scenario where genetic coefficients are non-zero for a set of five clinical annotations in the tree. These were chosen to occur within a single branch of the tree (clustered nodes), or across distant branches (distributed nodes). We compared the power obtained under these two scenarios when considering a range of allele frequencies. We fitted the TreeWAS model under a two-parameter setting with default parameters θ = 1/3 and π1 = 0.001. For the alternative PheWAS model we assumed complete independence across annotations, equivalent to setting θ → ∞. Under the clustered nodes simulations, the relative gain in power for identifying active nodes, where the genetic coefficients are non-zero, of TreeWAS compared to PheWAS was 20-25% across the allele frequencies tested (Fig. 3a). This gain in power was not associated with an increased false positive rate (< 0.001), as observed in nodes simulated with zero genetic coefficients (Fig. 3a). When we simulated non-zero genetic coefficients in distributed nodes there was a 1-3% reduction in power to identify active nodes for TreeWAS compared to PheWAS (Supplementary Fig. 2). We also observed an increase in power in quantifying the overall evidence for association with clustered nodes (3.4-5.4%), but a small decrease with distributed nodes (0.2-1.0%) (Supplementary Fig. 3 and 4). Therefore, when genetic coefficient correlation is captured by the classification tree the gain in power with TreeWAS relative to PheWAS is substantial, and if the correlation is not well-represented by the tree then the cost incurred with the former method is minimal.
Figure 3. Sensitivity and specificity analysis of TreeWAS on simulated data.
a, Rate of active node identification at increasing posterior probability (PP) thresholds and different simulated minor allele frequencies (MAF) of the causal genetic variant, for the TreeWAS method (θ = 1/3 and π1 = 0.001;orange), and for the PheWAS method (a model assuming complete independence among phenotypes with θ → ∞ and π1 = 0.001; blue). For each simulation replicate (N=500) we simulated five clustered nodes with non-zero genetic coefficients (•) and for the remaining nodes, phenotype counts were simulated to match observed disease prevalence and zero genetic coefficients (♦). Vertical dashed line denotes the PP = 0.75 threshold used in the analysis. Rate of false positives in the BFtree statistic (b) and active node identification (c) when genotypes for the HLA-B*27:05 allele are permuted in both phenotypic datasets. Gen, genotype; phen, phenotype.
In the second simulation set we assessed the impact of non-independence between annotations arising from the clinical data collection approach. For example, recording of a specific disease subtype for an individual may mean that other subtypes are less likely to be recorded for the same patient. We performed simulations under the null using the individual-level phenotype data from both UK Biobank phenotype datasets. For each simulation we permuted the observed genotypes of HLA-B*27:05, representative of a common genetic variant (given its 4.05% allele frequency in the UK Biobank), whilst maintaining non-independence between annotations in the tree. For comparison, we also performed permutations of individual-level phenotype data in addition to the genetic data, where all correlation is removed. With these permutations we quantified the rate of false positives in our approach. When we permuted genotypes only, we observed an inflation of the BFtree statistic and the node-level PP with the HES dataset, consistent with the more prominent correlation structure in the ICD-10 compared to the SR diagnosis trees (Fig. 3b,c). Through these simulations we estimated a false positive rate of 0.05 and 0.01 with a log10 BFtree threshold of 10 and 20, respectively, in the HES dataset, when substantial non-independence exists between nodes. For the SR dataset, the false positive rate at these thresholds was below 0.01. Thus, although non-independence between nodes can artificially increase test statistics, this can be countered by using conservative significance thresholds to maintain the false positive rate at an appropriate level.
The effects on HLA allelic variation in the phenome
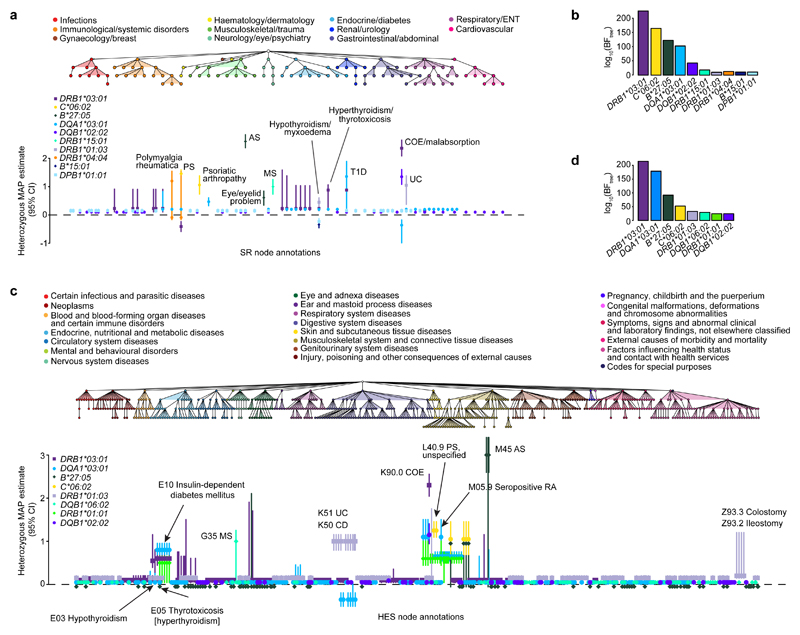
HLA region genetic variation is associated with numerous human disorders, in particular autoimmune and autoinflammatory diseases. Hence, we sought to interrogate HLA effects on the full range of SR and HES phenotypes using TreeWAS. Through conditional analysis (Online Methods and Supplementary Note), we identified independent associations for ten HLA alleles in the SR data (log10 BFtree ≥ 10) and eight in the HES data (log10 BFtree ≥ 20) (Fig. 4 and Supplementary Tables 2 and 3). Seven of these alleles or alleles in high linkage disequilibrium (LD; r > 0.98) were associated in both datasets (Supplementary Fig. 5).
Figure 4. Genetic analysis of HLA allelic variation in the risk of clinical phenotypes from the UK Biobank SR diagnosis and HES datasets.
a, The tree depicts the hierarchical structure of self-reported clinical phenotypes as determined by the UK Biobank classification. Only nodes with a significant association (PP > 0.75) with at least one HLA allele are shown, along with their parent nodes. The graph shows estimated effect sizes for the heterozygous genotype of the different HLA alleles on susceptibility to each clinical phenotype. Bars show the 95% credible interval. b, Evidence of association for each HLA allele with at least one node in the tree (BFtree) in the conditional TreeWAS analysis for the SR dataset (Supplementary Table 9). c, The tree depicts the hierarchical structure of HES-derived clinical phenotypes as determined by the ICD-10 classification (showing nodes with PP > 0.75 and their parent nodes). The graph shows estimated effect sizes for the heterozygous genotype of the different HLA alleles on susceptibility to each clinical phenotype. d, Evidence of association for each HLA allele with at least one node in the tree in the conditional TreeWAS analysis using the HES data (Supplementary Table 10). Estimates for heterozygous and homozygous genotype effect sizes and descriptions of all phenotypes shown are available in Supplementary Tables 2 and 3. AS, ankylosing spondylitis; CI, confidence interval; COE, coeliac disease; ENT, ear, nose, throat; MAP, maximum a posteriori; MS, multiple sclerosis; PS, psoriasis; RA, rheumatoid arthritis; T1D, type 1 diabetes; UC, ulcerative colitis.
These associations were fine-mapped, and the majority of the strongest effects were with IMDs, as reported previously through GWAS17,25–30 (Fig. 4). For class I alleles, we observed associations with PS (HLA-C*06:02) and AS (HLA-B*27:05), with the genetic coefficients of the latter being the largest observed in the SR and HES datasets (Fig. 4a,c). For class II alleles, HLA-DRB1*03:01 and HLA-DQB1*02:02 were observed to be independently associated with coeliac disease (COE) in both datasets; these alleles tag two of the strongest known COE HLA risk haplotypes, DR3-DQ2 and DR7-DQ2 (ref. 26). In both datasets, HLA-DQA1*03:01 was identified and fine-mapped to rheumatoid arthritis (RA); this allele is in moderate LD with HLA-DRB1*04:01 (r = 0.71), which is the likely causal allele driving this association27. Similarly, HLA-DQA1*03:01 was associated with type 1 diabetes (T1D), noting that this allele is in LD with HLA-DQB1*03:02 (r = 0.67), which has been indicated as the most significantly associated T1D class II allele26. In the SR dataset we identified an HLA-DRB1*15:01 association and fine-mapped it to multiple sclerosis (MS) (Fig. 4a). In the HES dataset HLA-DQB1*06:02 was identified instead and also fine-mapped to MS (PP = 1; Fig. 4c), but this allele is in strong LD with HLA-DRB1*15:01 (r = 0.97) (Supplementary Fig. 5). Lastly, HLA-DRB1*01:03 was fine-mapped to ulcerative colitis (UC) and Crohn’s disease (CD) in both datasets, and it is the likely causal allele for these two types of inflammatory bowel disease (IBD)30.
Apart from established HLA associations with common IMDs, we also confirmed HLA effects for conditions where GWAS have not been performed, detected associations with clinical annotations linked to disease complications, and identified novel HLA associations with other IMDs. For example, in the SR dataset, we confirmed the association of HLA-DRB1*04:04 with polymyalgia rheumatic and giant cell arteritis, which has been previously identified only through small candidate gene studies31,32 (Fig. 4a). The UC- and CD-associated HLA-DRB1*01:03 allele was found to also be associated with surgical procedures linked to complications of IBD, such as Z93.3 Colostomy status (PP = 1) and Z93.2 Ileostomy status (PP = 1), consistent with findings by the International IBD Genetics Consortium33 (Fig. 4c). Of the ten HLA alleles independently associated with clinical phenotypes in the SR dataset, five were associated with hypothyroidism/myxoedema, and three of the eight alleles from the HES data were associated with the E03 hypothyroidism code. This disease is thus the phenotype with the largest number of independent HLA associations across both UK Biobank datasets. Associations have been reported with hypothyroidism for both HLA class I and II loci, but the specific alleles driving these are not well resolved34,35, apart from a recently reported HLA-DQA1*05:01-HLA-DQB1*02:01-HLA-DRB1*03:01 (HLA-DR3-DQ2 haplotype) association36. Further to HLA-DRB1*03:01, we refined the HLA associations with hypothyroidism to two additional independent risk alleles, HLA-DQA1*03:01 and HLA-DRB1*01:03, and two independent protective alleles, HLA-B*15:01 and HLA-DPB1*01:01 (Fig. 4 and Supplementary Table 4). Our HLA analysis therefore demonstrates the validity of our method as it can identify known genetic associations, and can facilitate discovery of new associations for relatively understudied diagnoses.
Genetic risk score associations with IMDs
Outside of the HLA, over the last decade our understanding of genetic susceptibility to the common IMDs has increased tremendously, with tens to hundreds of risk loci being identified per disease37. However, given the prevalence of IMDs in the UK Biobank and the typically small effect sizes estimated, we expect low power at individual loci. For example, when considering nine of the most common autoimmune and auto-inflammatory diseases (see Online Methods) we observed evidence of association (log10 BFtree > 0) for 64 individual SNPs (12.96% of GWAS SNPs tested) in the SR and 125 SNPs (25.30%) in the HES datasets. Nevertheless, we can gain power by combining the effects of multiple typed and imputed susceptibility variants as a GRS (see Online Methods), and using the TreeWAS approach to assess their relationship with the UK Biobank phenome (Fig. 5).
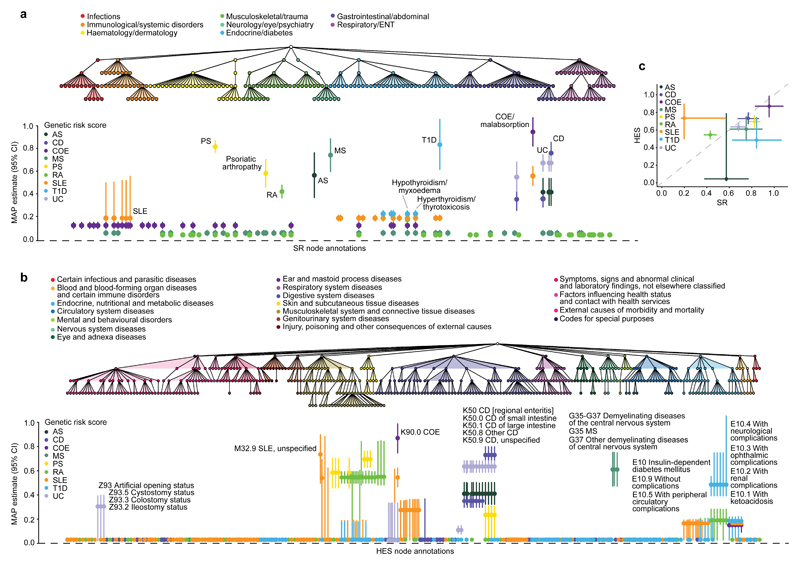
Figure 5. Association analysis of genetic risk for multiple IMDs derived from clinical phenotypes in the UK Biobank SR diagnosis and HES datasets.
a, The tree depicts the hierarchical structure of SR clinical phenotypes as determined by the UK Biobank classification. Only nodes with a significant association (posterior probability > 0.75) with at least one IMD genetic risk score (GRS) are shown, along with their parent nodes. The graph shows estimated effect size of GRS on susceptibility to each clinical phenotype with posterior probability > 0.75. Bars show the 95% credible interval. b, The tree depicts the hierarchical structure of HES-derived clinical phenotypes as determined by the ICD-10 classification (showing nodes with posterior probability > 0.75 and their parent nodes). The graph shows estimated effect sizes of GRS on susceptibility to each clinical phenotype. c, Comparison of estimated genetic coefficients for each GRS and the respective clinical annotation in both phenotypic datasets. Estimates of effect sizes and description of all phenotypes shown are available in Supplementary Tables 6 and 7 and evidence of association for each GRS with at least one node in the tree are available in Supplementary Tables 11 and 12. AS, ankylosing spondylitis; CD, Crohn’s disease; CI, confidence interval; COE, coeliac disease; ENT, ear, nose, throat; MAP, maximum a posteriori; MS, multiple sclerosis; PS, psoriasis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type 1 diabetes; UC, ulcerative colitis; MAP.
Typically the GRSs best identified those clinical annotations from which they were constructed, with secondary associations being detected for conditions with shared genetic risk. For example, CD and UC have a high genetic correlation38, although disease-specific susceptibility loci have been identified for each and heterogeneity in effect sizes has been observed39. The GRS for CD was thus associated with both CD itself as well as UC, but the magnitude of genetic coefficients was greater for CD as expected (β = 0.86 vs. β = 0.44 in SR and β = 0.73 vs. β = 0.35 in HES for CD and UC, respectively). However, the GRS for UC could not differentiate these two clinical annotations, with estimated genetic coefficients of the same magnitude for both CD and UC (β = 0.68 in SR and β = 0.64 in HES; Fig. 5a,b). This indicates some level of variation in the precision of different GRSs to identify specific phenotypes, such that the discriminatory capacity of GRSs will depend on the degree of genetic sharing between conditions and may require the consideration of additional clinical features33.
For all associations, genetic coefficients were less than 1, demonstrating a degree of dilution in phenotype detection across both the SR and HES datasets, and noting that simulation analyses estimated an expected dilution of ~15% due to the winner’s curse (Supplementary Note and Supplementary Table 5). The least dilution was observed for the association of the COE GRS and this disease (β = 0.96 and β = 0.87 in the SR and HES datasets, respectively). The COE phenotypes derived from the UK Biobank healthcare data are thus highly comparable to the clinically ascertained disease phenotype used in the GWAS40 from which the variants for the COE GRS were obtained. Across both datasets the greatest dilution of a GRS and its respective disease was observed for RA (β = 0.43 and β = 0.55 in the SR and HES data, respectively), whilst in the HES data specifically the AS GRS was not associated with the disease (PP = 0.01), potentially due to the small number of AS patients in this dataset (n = 146), and in the SR data the SLE GRS association with SLE had a genetic coefficient of only 0.20 (Fig. 5a,b).
Overall the GRS associations were largely consistent between the SR and HES datasets, and for the GRSs and their respective diseases the estimated genetic coefficients were weakly positively correlated (rcorrected = 0.23, correcting for measurement error) (Fig. 5c). Strikingly, although the SLE GRS capacity to identify SLE itself in the SR data was so diluted that the SLE GRS was in fact a better predictor of COE (β = 0.57) (Fig. 5a), in the HES dataset this was not the case. The SLE GRS was most predictive of M32.9 SLE (β = 0.50; PP = 1.00), and to a lesser extent of K90.0 COE (β = 0.47; PP = 1.00) (Fig. 5b). This discrepancy between the SR and HES datasets suggests differences in the diseases annotated as SLE in the two datasets, which may in turn reflect differences in disease perception or diagnosis that could have clinical implications. Notably, in the SR data SLE was also associated with the COE GRS (β = 0.13), but this was not the case in the HES data, further supporting a distinction between SLE phenotypes in the two datasets.
Secondary associations of the GRSs were identified either with known complications of the disease with which the primary association was observed, or with other IMDs. For example, as for the HLA-DRB1*01:03 associations, the UC GRS was associated with colostomy and ileostomy events (β = 0.31 and PP = 0.98, and β = 0.31 and PP = 1, respectively), as was the CD GRS, although the effect size magnitude was lower (β = 0.03 and PP = 0.91, and β = 0.03 and PP = 0.87, respectively). Also paralleling the HLA analysis, hypothyroidism was associated with several GRSs: five and four of the nine GRSs tested were associated with the disease in the SR and HES datasets, respectively, with those for COE, RA, SLE and T1D being found in both datasets. Hence, hypothyroidism is the single phenotype with the largest number of different GRS associations (Fig. 5a,b and Supplementary Table 6 and 7).
Discussion
By exploiting the inherent hierarchical structure of diagnostic classifications, our Bayesian analysis framework addresses a fundamental challenge for the analysis of high-dimensional, heterogeneous routine healthcare data - how to identify statistically significant genetic associations when interrogating thousands of diagnoses without employing methods11,13 that sacrifice phenotypic resolution. When applying TreeWAS to interrogate the effect of HLA on the UK Biobank phenome, associations were identified with 143 and 966 nodes in the SR and HES datasets, respectively. Assessing the impact of IMD GRSs also revealed associations with 151 and 810 nodes in the two respective datasets. The total number of nodes identified demonstrates the power of TreeWAS for detecting associations in datasets where numerous weak but correlated effects are present across the classification tree.
Amongst the many active nodes for which genetic associations were observed, previously established effects of HLA alleles on specific IMDs were detectable, as were effects for relatively understudied conditions. Notably, multiple novel associations with HLA alleles were discovered for hypothyroidism. Although not all previously reported HLA associations could be detected for any single IMD - such as AS41 or MS29 - due to limited power with the current UK Biobank datasets, the capacity for genetic discovery will improve with increasing cohort size, and associations with nodes displaying a substantial granularity of clinical description were already identifiable.
In the GRS analysis, associations between GWAS-derived GRSs and their respective diseases were typically the strongest effects observed, even without HLA allele inclusion, demonstrating that non-HLA variants can provide precision for detecting specific IMDs. Cross-disease associations of GRSs were also identified, particularly for hypothyroidism, and this previously unappreciated extent of genetic sharing indicates a common, genetically determined pathogenesis. For all GRS associations, dilution of the capacity for phenotype detection was observed but was largely comparable between the SR and HES datasets. An intriguing exception was the differential association of the SLE GRS with the respective SLE terms in the two datasets: this GRS could not precisely predict the self-reported disease, but could accurately detect the hospitalisation record-derived phenotype. Compared to other the IMDs investigated, SLE is a more heterogeneous, systemic condition which consequently presents a substantial diagnostic challenge42. Therefore, this discrepancy in the magnitude of SLE GRS associations could reflect incorrect reporting of the disease, disease over-diagnosis not discernible in the HES data if hospitalisation is associated with more clear-cut diagnosis, or greater disease heterogeneity whereby SLE as defined in GWAS and in the HES data represents only a subset of a more genetically variable syndrome.
Identifying misclassification, misdiagnosis and miscoding in routine healthcare data is an on-going challenge, although there are recognised instances, such as inaccuracy in T1D and type 2 diabetes (T2D) differentiation43. In the UK Biobank, the T1D GRS is not associated with T2D terms in the SR data (PP = 0.0002), and shows weak evidence of association with the HES data (PP = 0.52). However, the T2D GRS, which can accurately detect T2D terms (β = 0.80 and PP = 1.00 and β = 0.71 and PP = 1.00 in the SR and HES datasets, respectively), is also associated with T1D in the HES (β = 0.71 and PP = 1.00) but not SR data (PP = 0.30; and see Supplementary Note and Supplementary Table 8). These cross-disease associations may be attributable to T1D/T2D misclassification, misdiagnosis and miscoding43 (Supplementary Note and Supplementary Figures 6 and 7), but also to genetic sharing44, and poor distinction of latent autoimmune diabetes of adulthood patients45, whose genetic profiles comprise a mixture of T1D and T2D risk loci46. Thus, the SLE and diabetes examples demonstrate how exploring the genetic basis of the healthcare phenome can expose disease areas where improvements are required to ameliorate disease perception or strengthen diagnostic practices. Digital phenotyping using genetic data, combined with longitudinal clinical information, physical measures and biomarkers43,47, could help to rectify misclassification, misdiagnosis and miscoding present in healthcare data and to infer missing phenotypes. This could in turn facilitate patient management, particularly if it enables correction of treatment strategies within an actionable time frame.
Integration of genomic data with routine healthcare information offers much potential to learn about differences in disease risk, diagnosis, and reporting within and between healthcare systems, including between countries. Moreover, increased incorporation of correlated, high-dimensional phenotypes (e.g. from molecular, cytometry and imaging readouts), including measures of temporal disease progression48, may come to lead to a genetically driven understanding of the architecture of the human phenome and of causal relationships. The value of TreeWAS lies in enhancing power to identify groups of endpoints affected by specific genetic risk factors, by exploiting the encoding of medical ontologies. A corollary is that structures that better capture the underlying biological process affecting the origin and progression of disease should be better correlated with genetic risk factors. Although generalising the TreeWAS method to structures reflecting temporal progression and associated quantitative data modalities requires future development, we believe that it is an important step towards the goal of learning a genetically motivated classification of disease and associated phenotypes.
Online Methods
UK Biobank data
The UK Biobank is a prospective cohort of over 500,000 men and women aged 40 to 69 years when recruited in 2006-2010. Participants have provided: data on lifestyle, environment, and medical history through an interview and completion of a questionnaire; physical measures; biological samples for genotyping and biochemical assays; and informed consent to long-term medical follow-up through linkage of national health registries. UK Biobank data is available under open access to conduct health-related research after approval of a project proposal6. The UK Biobank has obtained ethical approval covering this study from the National Research Ethics Committee (REC reference 11/NW/0382).
Phenotypic data
We analysed two phenotypic datasets available through the UK Biobank. The first included the SR diagnosis data, ascertained through the completion of questionnaires and interviews with study participants (data field 20002 Non-cancer illness code, self-reported); the second dataset included the HES registry dataset ascertained through linkage of health registries (data fields 41142 and 41078; accessed on September 2016). Clinical diagnoses in these datasets are described with different classification schemes, both of which follow a hierarchical structure. The diagnosis terms used to store the medical history of UK Biobank participants were proposed by the UK Biobank team (data-coding 6), and this classification tree is organised into 11 sub-classes with a total of 561 clinical terms, 531 of which are selectable. Diagnosis terms used to store hospitalisation events follow the ICD-10 list compiled by the World Health Organisation. The ICD-10 classification tree is organised into 22 Chapters and containing a total of 19,855 clinical terms, 16,310 of which are selectable. Each hospitalisation episode in the dataset has a primary diagnosis associated with the event and an event may be annotated with one or more secondary diagnoses. Disease outcomes for each individual, as a binary trait, were generated for the combined primary and secondary diagnoses annotations. Individuals were considered unaffected for any given diagnostic term unless the diagnosis was reported in the questionnaires and interviews, or a hospitalisation event with that diagnostic term was observed.
Genetic dataset
The interim release of the UK Biobank genetic data used for this study includes 152,732 individuals, 120,286 of which were determined to be of British Isles ancestry (Supplementary Fig. 8) and included in the analysis. The initial 50,000 individuals were genotyped on the Affymetrix UK BiLEVE Axiom array as part of a pilot study described elsewhere49 and the remaining 102,732 individuals were genotyped on the Affymetrix UK Biobank Axiom array. The quality control of SNP data and whole-genome SNP imputation was performed by the UK Biobank analysis team and described in the UK Biobank website (http://www.ukbiobank.ac.uk/scientists-3/genetic-data). We imputed 356 classical HLA alleles for the HLA-A, -B, -C, -DRB5, -DRB4, -DRB3, -DRB1, -DQB1, -DQA1, -DPB1 and -DPA1 loci at four digit resolution with the HLA*IMP:02 algorithm50,51 using data from a multi-population reference panel. The imputation panel contained 2,263 SNPs in the MHC region (GRCh37 coordinates chr6:29500000-33500000) which overlapped UK Biobank genotyped SNPs. This SNP set was selected to optimize MHC coverage and imputation performance and the HLA*IMP:02 algorithm was trained on this SNP set. Genetic risk scores, weighted by effect sizes, were generated for nine IMDs using genome-wide associated variants compiled from previous studies: AS17, CD39, COE40, MS52, PS25, rheumatoid arthritis53, SLE54, T1D55, and UC39. SNP genotypes for the UK Biobank individuals were extracted from the imputed genotype data and maintained if the imputation information score was above 0.85; if a SNP was not typed or imputed successfully it was not included in the GRS calculation.
Simulated data
To assess the accuracy of the method, we simulated case-control status for 120,000 individuals and the 531 selectable phenotypes in the diagnosis tree used for the self-reported dataset and with disease prevalence as observed in the UK Biobank cohort. Simulations were generated under two scenarios. For the first, we assumed a causal relationship between a genetic variant and five clinical terms under the same parent node in the tree (disease prevalence in these nodes ranged between 0.01 and 0.4%). These simulations are referred to as clustered clinical phenotypes. The second set of simulations, termed distributed phenotypes, consisted of five clinical terms with a causal relationship distributed under different branches of the classification tree; these clinical terms were selected with matching disease prevalence, as for the clustered simulations. For each scenario we simulated genotypes sampled from a multinomial distribution with a fixed allele frequency and genetic coefficients sampled from the prior (Supplementary Figure 9). Case-control status was determined by using logistic risk with a y-intercept matching the observed disease prevalence. Sets of simulations were performed for the allele frequencies 0.005, 0.01, 0.02 and 0.05. For each simulation we computed the evidence of association in the tree (BFtree), and the evidence of association at each individual node with the parameters θ = 1/3 and π1 = 0.001. We compared the power to detect association with at least one node in the tree with an analysis where we assume no correlation in the genetic coefficients between nodes in the tree, equivalent to setting θ → ∞ in the TreeWAS method (see Supplementary Note). 500 simulation replicates were performed for each combination of parameters and settings. To assess the robustness of the algorithm to the non-independence between annotations unaccounted by the tree structure, we performed simulations where we permuted the genotypes whilst leaving the observed phenotypes in the UK Biobank cohort intact. Simulations were performed with the observed self-reported and HES datasets, and we permuted the observed genotype.
HLA analysis
For each HLA locus we derived highest confidence genotypes by taking the allele at each chromosome with the highest imputation posterior probability. Genotypes were used to generate count distributions in affected and unaffected individuals at each terminal node in the tree. To identify independent HLA associations we performed sequential conditional analysis using an approximation to the likelihood function as described in the Supplementary Note. At each step, BFtree statistics were generated for each allele and the allele with the largest was selected for conditioning in the next iteration. Conditional analysis was repeated until all observed BFtree statistics were below 1010 in the self-reported diagnosis dataset and 1020 in the HES dataset, ensuring a false discovery rate below 0.01, as determined through the simulation analysis. For each significant allele association we computed the marginal PP for the genetic coefficient being not equal to 0 and the MAP estimate using posterior decoding as described in the Supplementary Note. Association with a clinical annotation was deemed significant if the PP was above 0.75.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource (application number 10625). The research has been supported by the Wellcome Trust (100956/Z/13/Z and 090532/Z/09/Z to G.M. and 100308/Z/12/Z to L.F.), the Danish National Research Foundation (L.F.), the Wellcome Trust/Royal Society (204290/Z/16/Z to C.A.D.), and Takeda Ltd (L.F. and C.A.D.). This work was supported by the Australian National Health and Medical Research Council (NHMRC), Career Development Fellowship ID 1053756 (S.L.); and by the Victorian Life Sciences Computation Initiative (VLSCI) grant number VR0240 on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government, Australia (S.L.). Research at the Murdoch Childrens Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program.
Footnotes
Author Contributions
A.C. and G.M. performed the analyses with contributions from C.A.D. A.C., C.A.D., L.J., P.D., L.F. and G.M. conceived the study. HLA imputation was performed by A.M., D.V., A.D. and S.L. A.C., C.A.D. and G.M. wrote the manuscript and all other authors reviewed the manuscript.
Competing Financial Interests
G.M. and P.D. are cofounders of, holder of shares in, and consultants to Genomics PLC. G.M., P.D. and S.L. are partners in Peptide Groove LLP. Peptide Groove has licensed HLA typing technology to Affymetrix Ltd. The other authors declare no competing financial interests.
Code availability. Code to perform TreeWAS analysis is available from the authors upon request or through the code repository at https://github.com/mcveanlab.
URLs. UK Biobank, http://www.ukbiobank.ac.uk/; UK Biobank genotyping procedure and genotype calling protocols, http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580; UK Biobank internal quality control procedures, http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580; HLA*IMP, https://oxfordhla.well.ox.ac.uk/hla/; World Health Organisation ICD-10 disease classification codes, http://www.who.int/classifications/icd/en/.
References
- 1.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 2.Mallal S, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 3.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 4.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 5.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–20. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 6.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson SG, Willeit P. UK Biobank comes of age. Lancet. 2015;386:509–10. doi: 10.1016/S0140-6736(15)60578-5. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson T, et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–9. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 9.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnes JH, et al. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17:129–45. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 12.Chan KS, Fowles JB, Weiner JP. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev. 2010;67:503–27. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 13.Denny JC, Bastarache L, Roden DM. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annual Review of Genomics and Human Genetics. 2016;17:353–373. doi: 10.1146/annurev-genom-090314-024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hersh WR, et al. Caveats for the Use of Operational Electronic Health Record Data in Comparative Effectiveness Research. Medical Care. 2013;51:S30–S37. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20:117–21. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, et al. Regional variations in diagnostic practices. N Engl J Med. 2010;363:45–53. doi: 10.1056/NEJMsa0910881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Genetics of Ankylosing Spondylitis C et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–8. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colmegna I, Cuchacovich R, Espinoza LR. HLA-B27-associated reactive arthritis: pathogenetic and clinical considerations. Clin Microbiol Rev. 2004;17:348–69. doi: 10.1128/CMR.17.2.348-369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eastmond CJ, Woodrow JC. The HLA system and the arthropathies associated with psoriasis. Ann Rheum Dis. 1977;36:112–20. doi: 10.1136/ard.36.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin TM, Rosenbaum JT. An update on the genetics of HLA B27-associated acute anterior uveitis. Ocul Immunol Inflamm. 2011;19:108–14. doi: 10.3109/09273948.2011.559302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300. [Google Scholar]
- 22.Takagi I, Eliyas JK, Stadlan N. Cervical spondylosis: an update on pathophysiology, clinical manifestation, and management strategies. Dis Mon. 2011;57:583–91. doi: 10.1016/j.disamonth.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Gritz DC, Wong IG. Incidence and prevalence of uveitis in Northern California; the Northern California Epidemiology of Uveitis Study. Ophthalmology. 2004;111:491–500. doi: 10.1016/j.ophtha.2003.06.014. discussion 500. [DOI] [PubMed] [Google Scholar]
- 24.Okada Y, et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am J Hum Genet. 2014;95:162–72. doi: 10.1016/j.ajhg.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez-Achury J, et al. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat Genet. 2015;47:577–8. doi: 10.1038/ng.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X, et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet. 2015;47:898–905. doi: 10.1038/ng.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moutsianas L, et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet. 2015;47:1107–13. doi: 10.1038/ng.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyette P, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–9. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Taboda VM, et al. HLA-DRB1 allele distribution in polymyalgia rheumatica and giant cell arteritis: influence on clinical subgroups and prognosis. Semin Arthritis Rheum. 2004;34:454–64. doi: 10.1016/j.semarthrit.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Haworth S, et al. Polymyalgia rheumatica is associated with both HLA-DRB1*0401 and DRB1*0404. Br J Rheumatol. 1996;35:632–5. doi: 10.1093/rheumatology/35.7.632. [DOI] [PubMed] [Google Scholar]
- 33.Cleynen I, et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–67. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson N, et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosley JD, et al. Identifying genetically driven clinical phenotypes using linear mixed models. Nat Commun. 2016;7:11433. doi: 10.1038/ncomms11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 38.Chen GB, et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet. 2014;23:4710–20. doi: 10.1093/hmg/ddu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortes A, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–21. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 43.de Lusignan S, et al. A method of identifying and correcting miscoding, misclassification and misdiagnosis in diabetes: a pilot and validation study of routinely collected data. Diabet Med. 2010;27:203–9. doi: 10.1111/j.1464-5491.2009.02917.x. [DOI] [PubMed] [Google Scholar]
- 44.Nogueira TC, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostergaard JA, Laugesen E, Leslie RD. Should There be Concern About Autoimmune Diabetes in Adults? Current Evidence and Controversies. Curr Diab Rep. 2016;16:82. doi: 10.1007/s11892-016-0780-0. [DOI] [PubMed] [Google Scholar]
- 46.Cervin C, et al. Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes. 2008;57:1433–7. doi: 10.2337/db07-0299. [DOI] [PubMed] [Google Scholar]
- 47.Shields BM, et al. Can clinical features be used to differentiate type 1 from type 2 diabetes? A systematic review of the literature. BMJ Open. 2015;5:e009088. doi: 10.1136/bmjopen-2015-009088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen AB, et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014;5:4022. doi: 10.1038/ncomms5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–81. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dilthey A, et al. Multi-population classical HLA type imputation. PLoS Comput Biol. 2013;9:e1002877. doi: 10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motyer A, et al. Practical Use of Methods for Imputation of HLA Alleles from SNP Genotype Data. bioRxiv. 2016 [Google Scholar]
- 52.International Multiple Sclerosis Genetics C et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–6. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.