Abstract
Defective autophagy has been linked to lipotoxicity in several cellular models. We aimed to investigate autophagy in lipid-stimulated hepatoma (Huh7) cells and tested whether 4-phenyl butyric acid (4-PBA), a chemical chaperone, has a beneficial role in hepatic fat accumulation and lipotoxicity. We report that long-term (24 h) exposure of hepatocytes to palmitate block autophagic flux that leads to lipid accumulation and cell death. Western blotting analysis showed increased accumulation of SQSTM1/p62, and decreased expression of Beclin1 and Atg7 in palmitate-treated cells. Autophagy inhibition by 3-methyladenine (3-MA) in palmitate-treated cells neither increased SQSTMI/p62 accumulation nor cell death, thus suggesting complete blockade of autophagy by palmitate. 4-PBA reduced lipid accumulation and cell death that were associated with restoration of autophagy. siRNA-mediated knockdown of Atg7 and presence of autophagy inhibitors, 3-MA and chloroquine, resulted in the decrease in lipid-lowering effect of 4-PBA, suggesting that 4-PBA mediates its lipid-lowering effect via autophagy. Apoptotic parameters, including altered Bcl2:Bax ratio and PARP1 cleavage induced by palmitate, were improved by 4-PBA. Our results indicate that palmitate impairs autophagy and increases lipid accumulation in Huh7 cells, whereas 4-PBA plays a protective role in lipid accumulation and lipotoxicity through activation of autophagy.
Keywords: lipid droplet, Atg7, cell death
Macroautophagy (hereinafter referred to as autophagy) is a lysosomal degradation pathway that cells use to degrade long-lived proteins and organelles. Autophagy plays a vital role in normal development, in adaptation to stress, and in a wide range of disease states (1–3). Autophagy occurs at basal levels in almost all cells to perform homeostatic functions. It is upregulated when cells require intracellular nutrients and energy, for example, during starvation, growth factor withdrawal, high bioenergetic demands, oxidative stress, infection, or protein aggregate accumulation (4). One of the main regulators of autophagy in mammals is mTOR (mammalian target of rapamycin, TOR kinase), which is an inhibitory signal and shuts off autophagy in the presence of sufficient nutrients and growth factors (2, 5, 6). Other regulatory players that control autophagy include adenosine 5′-monophosphate-activated protein kinase, which responds to the low energy status of a cell; eukaryotic initiation factor 2α (eIF2α), which responds to nutrient deprivation; endoplasmic reticulum (ER) stress; and Bcl-2 homology-3 (BH3)-only protein containing BH3 domain and disrupting Bcl-2/Bcl-XL inhibition of Beclin1/class III PI3K complex (3, 6, 7). Downstream of the main regulator mTOR, there are more than 35 genes known as ATG genes (many of which are evolutionarily conserved) that encode proteins that are essential for the execution of autophagy (2, 8). The molecular cascade that regulates the initiation and execution of autophagy has been the subject of immense interest and is now emerging as a central biological pathway that functions to promote health and longevity.
Nonalcoholic fatty liver disease (NAFLD) is a pathological condition that is associated with the accumulation of lipid droplets (LDs) and lipotoxicity in liver cells (9, 10). Recent studies have demonstrated that LDs are not only simple cytosolic structures passively storing triglycerides (TGs) and cholesterol but rather mobile and dynamic organelles that perform a variety of biological functions (11–13). Although the primary cause of the accumulation of lipids and subsequent formation of LDs in liver cells is not yet clear, it may arise from increased supply of lipids, de novo lipogenesis, impaired lipoprotein synthesis, or secretion or reduced fatty acid oxidation (14). Interestingly, these LDs have been identified as a substrate for autophagy, which mobilizes the lipids from LDs for metabolism through a process called lipophagy (15, 16). Pharmacological inhibition of autophagy by 3-methyladenine (3-MA) or genetic knockdown of Atg5 markedly increased TG and cholesterol content in lipid-stimulated cells (16). Recently chaperone-mediated autophagy has been shown to degrade LDs associated with proteins and facilitate lipolysis (17). Further evidence for the role of autophagy in hepatocyte lipid accumulation is that mice with a specific knockout of Atg7 developed massively enlarged livers mainly due to increased TG and cholesterol content (15). These findings suggest that impaired autophagy or defective lipophagy may underlie the development of lipid-associated disease such as NAFLD. Another line of evidence that suggests autophagy is impaired by exogenous lipid stimulus comes from β cells treated with saturated fatty acids (18, 19). However, there are controversial reports regarding the induction of autophagy in lipid-stimulated hepatocytes. Furthermore, it has been suggested that pharmacological agents that can improve ER folding capacity and stabilize misfolded proteins, as well as target the autophagy machinery, could provide a promising strategy to treat human diseases (19); one such agent is 4-phenyl butyric acid (4-PBA), which is a short-chain fatty acid chemical chaperone and known to improve insulin sensitivity in in vivo settings (20). 4-PBA has been shown to stabilize protein conformation, improve the capacity of ER folding, and facilitate proper trafficking of mutant proteins (21). More recently, it has been shown that inhibition of autophagy-mediated lipotoxic state causes increased cytosolic calcium levels (22). Previously, we have also reported increased levels of cytosolic calcium in palmitate-treated immortalized hepatoma cells (9).
On the basis of these reports, we hypothesized that LD formation and subsequent lipotoxicity could be the result of impaired autophagy in hepatocytes and could be reduced not only through inhibition of lipogenesis but also through the degradation of lipids in hepatocytes by restoring the impaired autophagy. Therefore, we investigated autophagic flux in lipid-stimulated hepatocytes and tested whether 4-PBA has a beneficial effect on LD formation and subsequent lipophagy in lipid-stimulated hepatocytes.
MATERIALS AND METHODS
Cell culture and treatments
Huh7 cell line was obtained as a kind gift from Michael Charlton (Department of Gastroenterology and Hepatology, Mayo Clinic, Rochester, NY). AML-12 cells (normal mouse liver cells) were purchased from ATCC. Cells were grown and maintained in DMEM [containing L-glutamine, glucose (3.5 g/l), 15 mM HEPES, 200 U/ml penicillin, 270 µg/ml streptomycin, obtained from Sigma-Aldrich (St. Louis, MO), and 10% FBS, obtained from Gibco], at 37°C in a humidified atmosphere of 5% CO2. However, for AML-12 cells, an additional 1% insulin-selenium-transferrin supplementation (catalog no. I1884, Sigma-Aldrich) was used as recommended. For all experiments, cells were used at a density of 5 × 104 cells per 24-well plate, 7 × 105 cells per 6-well plate, or 1.2 × 106 cells per 60 mm dish, unless otherwise mentioned. All the studies were conducted using 70%–80% confluent cells, which were treated with indicated concentrations of palmitate (Sigma-Aldrich) for 24 h. For 4-PBA (Sigma-Aldrich), cells were pretreated with 4-PBA 4 h prior to palmitate exposure. In the case of everolimus (a kind gift from Michael Charlton), 3-MA, bafilomycin, and chloroquine (CQ; Sigma-Aldrich), the pretreatment time was 1 h.
FFA-BSA complex preparation
Palmitate was complexed to fatty acid-free BSA (Sigma-Aldrich), as was mentioned previously (9). Briefly, a 20% stock solution of fatty acid-free BSA was prepared by dissolving 750 mg of BSA in 3.75 ml of PBS (pH 7.4). To begin with, BSA was layered on top of the 2–2.5 ml PBS. BSA was allowed to dissolve on its own (without stirring) at 4°C. The final volume was adjusted to 3.75 ml. For 20 mM palmitic acid solution, 5.6 mg of sodium palmitate was added to 1 ml water (preheated at 70°C). The mixture was incubated at 70°C for 20–30 min with constant vertexing. FFA-BSA complex was prepared by mixing 1 ml of the 20 mM palmitic acid solution to 3.3 ml of 20% BSA (prewarmed at 37°C). The complex formed was immediately added to 15.7 ml DMEM prewarmed to 37°C. A final solution prepared had 1 mM palmitic acid. The solution was sterile filtered and is stable for about 1 week at 4°C.
Nile red staining and TG content measurement
Nile red (also known as Nile blue oxazone), a lipophilic stain, was used to assess the lipid accumulation, as was mentioned (9). Briefly, cells were grown on sterilized coverslips and incubated with indicated concentrations of palmitate alone or pretreated with 4-PBA, 3-MA, and CQ, as mentioned. Cells were washed with PBS and fixed in methanol:acetic acid (3:1) solution overnight at 4°C. After fixation, cells were again washed and incubated in PBS containing 500 ng/ml Nile red (Sigma-Aldrich) for 30 min at 37°C. Slides were prepared, and imaging was done using Olympus FluoView FV-1000 fluorescence microscope, ×20 dry lens. For TG content measurement, cells were grown in 60-mm dishes and incubated with indicated concentrations of palmitate alone or pretreated with 4-PBA, 3-MA, and CQ, as mentioned. Cells were collected, and TG content was measured by using an LPL buffer assay, as described previously (9).
MDC and lysotracker red staining
Monodansylcadaverine (MDC; Sigma-Aldrich), a fluorescent dye, was used to detect the formation of autophagosomes by fluorescence microscopy. MDC has been proposed as a selective fluorescent tracer for autophagosomes (23). The autophagosomes were labeled with MDC by incubating the cells with 50 µM MDC in PBS at 37°C for 1 h. After incubation, cells were washed three times with PBS and immediately analyzed by fluorescence microscope using a ×40 lens. Lysotracker red, an acidotrophic probe for labeling and tracking acidic organelles (usually lysosomes) in living cells, was used for the detection of lysosomes. After 24 h of corresponding treatments as mentioned, cells were stained with 0.3 µM Lysotracker red (catalog no. L-7525; Invitrogen, Life Technologies) for 45 min at 37°C. The cells were washed with PBS twice and immediately analyzed by fluorescence microscope using a ×60 oil immersion objective lens. The imaging in all the cases was done using an Olympus FluoView FV-1000 fluorescence microscope.
Protein isolation and Western blotting
Cells were trypsinized, harvested in PBS (pH 7.4), centrifuged, and resuspended in RIPA buffer (Sigma-Aldrich). After incubation for 45 min at 4°C, lysate was centrifuged at 17,530 g for 30 min at 4°C to remove cellular debris. Protein concentrations were determined by Bradford reagent (catalog no. B6916; Sigma-Aldrich). For Western blotting, 30–50 µg protein was denatured at 100°C for 3 min in Laemmli buffer. Protein samples were resolved on 7%–15% SDS gels at 70 V. Proteins were electrotransferred to PVDF membrane (Bio-Rad, Hercules, CA) using Bio-Rad Mini Transblot Electrophoretic Transfer Cell. Membranes were blocked in 5% fat-free dry milk in 50 mM Tris, pH 8.0, with 150 mM sodium chloride, 2.6 mM KCl, and 0.05% Tween20 for 2 h. The following antibodies were used as primary antibodies in fat-free milk incubated overnight at 4°C: Rabbit, anti-GRP78, anti-SQSTM1/p62, anti-Nrf2, anti Bcl2, anti-Bax, anti-PARP1, anti-mTOR, anti-P110α, anti-CHOP, anti-elf2α, anti-p-elf2α (Santa Cruz Biotechnologies, CA), anti-LC3I/II, anti-Beclin1, anti-Atg7, anti-Atg3, anti-Atg5, anti-Atg12, anti-Atg16 (Cell Signaling Technology, Danvers, MA); and mouse, anti-β actin (Sigma-Aldrich). Goat anti-rabbit and goat anti-mouse immunoglobulin G antibodies conjugated with HRP (Santa Cruz Biotechnologies) were used as secondary antibodies. Chemiluminescence was detected by Immobilon chemiluminescent HRP substrate (EMD-Millipore, Billerica, MA) and visualized by Molecular Image ChemiDocTM XRS+ (Bio-Rad). Densitometric measurement of the bands was performed using Image LabTM software (version 3.0; Bio-Rad).
siRNA-mediated knockdown of Atg7 and Atg5 expression
Validated Atg7 siRNA and Atg 5 siRNA were purchased from Santa Cruz Biotechnology. siRNAs and lipofectamine (Invitrogen) were diluted into Opti-MEM I reduced serum medium (Invitrogen) per the manufacturer’s instructions. Huh7 cells were incubated for 16 h with a transfection mixture at a final siRNA concentration of 50 pmol and then supplemented with fresh medium and subsequent palmitate and 4-PBA treatments as described.
P62 and Beclin1 immunostaining
Cultured cells were seeded on coverslips in six-well plates and incubated in the presence or absence of indicated concentrations of palmitate and 4-PBA for 24 h. Cells were washed twice with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. Cells were permeabilized in PBS with 0.1% TritonX-100 at room temperature for 10 min. Nonspecific binding sites were blocked by incubating the cells with 10% normal goat serum (Santa Cruz Biotechnologies). Cells were incubated with p62 and Beclin1 antibody (Santa Cruz Biotechnologies) at a dilution of 1:100 in 0.1% Triton X-100 in PBS for 1 h at room temperature, then washed and incubated with Alexa Fluor 488 conjugated anti-rabbit secondary antibody (Invitrogen) at a dilution of 1:500 in PBS for 1 h at room temperature. Cells were then washed three times with PBS and stained with DAPI 1 µg/ml in PBS. The coverslips were mounted on glass slides, and cells were imaged by a laser-scanning confocal microscope (Olympus Fluoview FV1000) by using an ×60 oil immersion objective lens.
MTT assay
Cytotoxicity test was done to identify the cytotoxic potential of palmitate and effect of 4-PBA on lipotoxicity (24). Huh7 and AML12 cells were seeded in 24-well plates at a density of 5 × 104 cells per well and incubated overnight. The cells were pretreated with 0.5 mM 4-PBA for 4 h and exposed to palmitate (0.5 mM in the case of Huh7 cells and 0.25 mM in the case of AML12 cells) for 24 h. After 24 h, the medium was removed, and the cells were incubated in fresh medium without serum, containing 250 μg/ml 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) for 3 h at 37°C. Cytotoxicity/cell viability was evaluated by assaying the ability of mitochondria to catalyze the reduction of thiazolyl blue tetrazolium bromide (MTT) to a formazan salt by mitochondrial dehydrogenases.
Determination of ROS
2′,7′-Dichlorodihydrofluorescein diacetate (H2DCF-DA) staining was used to assess reactive oxygen species (ROS), as was mentioned previously (25). Briefly, Huh7 cells were seeded in six-well plates and allowed to attach overnight. The cells were pretreated with 0.5 mM 4-PBA for 4 h and then incubated with indicated concentrations of palmitate for 24 h. After corresponding treatments for 24 h, cells were washed three times with PBS and further incubated with 5 µM H2DCF-DA for 30 min at 37°C. Cells were again washed three times with PBS and imaged under a thin film of PBS. Imaging was done by fluorescence microscopy (Nikon TE 2000U, Tokyo, Japan) using green filters. Image quantification was done by Image J analysis, as was mentioned previously (26).
Statistical analysis
Data are presented as mean ± SD from three independent experiments. Statistical comparisons between the groups were determined by using a one-way ANOVA, followed by a Bonferroni multiple-comparisons test using INSTAT statistical software. P < 0.05 was considered statistically significant.
RESULTS
Palmitate blocked autophagy in a hepatocellular model of lipotoxicity
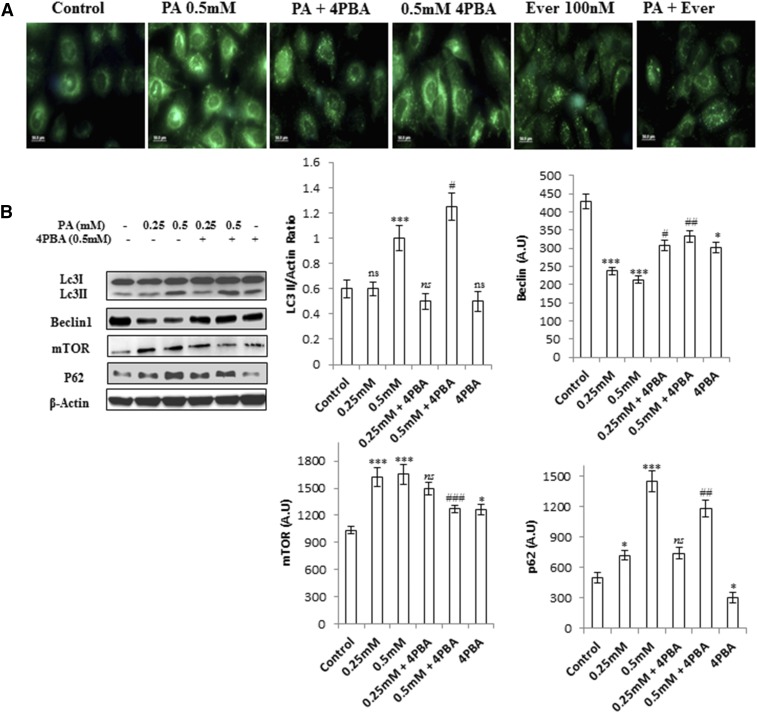
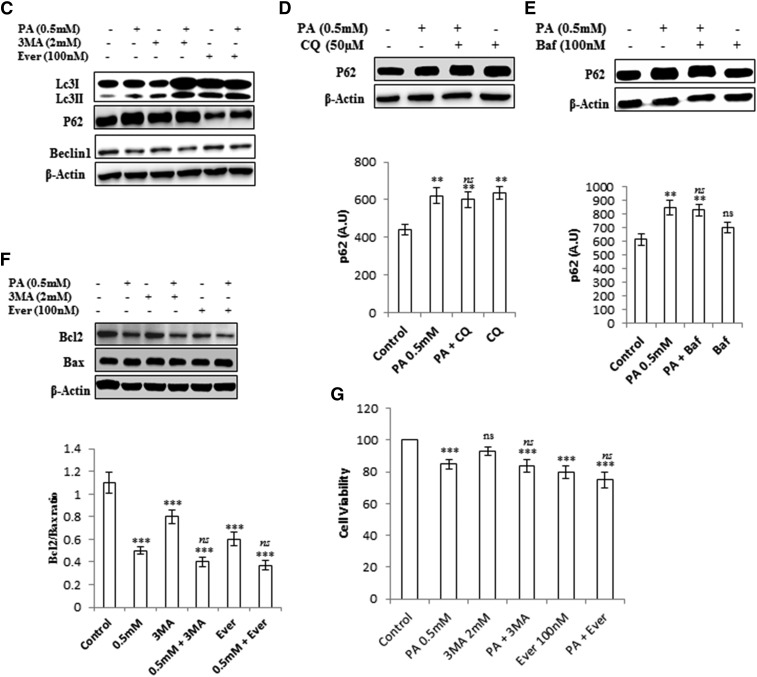
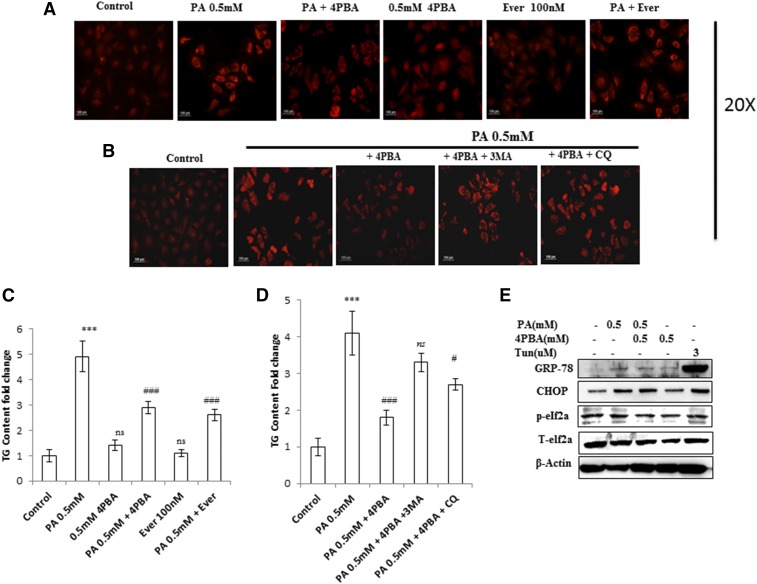
It has been previously reported that long-term palmitate treatment impairs autophagy in a cellular model of β-cell lipotoxicity (18, 19). We tested whether a similar phenomenon could be reproduced in lipid-stimulated hepatocytes (human hepatoma Huh7 and AML12 hepatocytes). To test this hypothesis, we first exposed Huh7 cells to palmitate bound to fatty acid-free BSA for 24 h at concentrations of 0.25 and 0.5 mM. Autophagosomes were analyzed by fluorescence microscopy using MDC staining. Results showed increased accumulation of autophagosomes in 0.5 mM palmitate-treated cells, whereas fewer autophagosomes were observed in control cells (Fig. 1A). Next, the cells were treated with palmitate, and levels of LC3B-II, which is a characterized marker to visualize autophagosomes and represents the membrane-bound form of LC3B on autophagosomes, were measured. Results indicate increased LC3B-II:actin ratio in 0.5 mM palmitate-treated cells (Fig. 1B, supplemental Fig. S2), thus showing an increase in autophagosomes. Pretreatment of everolimus (100 nM), an mTOR inhibitor and inducer for autophagy, showed decreased autophagosomes, as measured by MDC staining in palmitate-treated cells (Fig. 1A), thus suggesting impairment in autophagic degradation. To confirm this, we treated the cells with palmitate and measured the levels of SQSTM1/p62, a protein known to be degraded exclusively by autophagy. Consistently, p62 was accumulated by palmitate treatment, as observed by Western blotting and immunostaining (Figs. 1B, 2D, supplemental Fig. S2). Blockade of autophagy by pretreatment of bafilomycin (100 nM), which inhibits the fusion of lysosomes, and CQ (50 µM), which inhibits lysosomal degradation, further increased LC3B-II in palmitate-treated cells (supplemental Fig. S3C). Pretreatment of another autophagy inhibitor, 3-MA (2 mM) also increased LC3B-II in palmitate-treated cells and had no effect on the levels of SQSTM1/p62 (Fig. 1C–E). Beclin1, the mammalian ortholog of Atg6, which plays a central role in autophagy, was also decreased in palmitate-treated cells. In addition, most Atgs involved in initiation and execution of autophagy were decreased in palmitate-treated cells (supplemental Fig. S2B). Because mTORC1 is an important player in the regulation of autophagy and SQSTM1/p62-mTORC1-autophagy connections have been well established (27), we measured the levels of mTORC1 in palmitate-treated cells, and results show increased expression of mTORC1 in palmitate-treated cells in comparison with untreated cells (supplemental Fig. S2C), thus correlating with the blockade of autophagy. Pretreatment of everolimus, an inhibitor of mTORC1 in palmitate-treated cells, decreased the accumulation of SQSTM1/p62 and autophagosomes, thereby restoring the autophagy (Fig. 1A, C). Imaging of cells stained with Lysotracker Red showed decreased intensity in palmitate-treated cells in comparison with control cells (Fig. 2A). So taken collectively, accumulation of autophagosomes and p62 in palmitate-treated cells reflected the defect in the autophagic degradation machinery. Further cellular viability results showed that inhibition of autophagy by 3-MA in palmitate-treated cells does not aggravate the lipotoxicity (Fig. 1F, G), suggesting complete blockade of autophagy by palmitate in Huh7 cells. Bafilomycin and CQ pretreatment in palmitate-treated cells did not significantly increase SQSTM1/p62 in comparison to palmitate-treated cells alone, thereby again suggesting blockade of autophagic degradation by palmitate (Fig. 1D, E).
Fig. 1.
Palmitate-impaired autophagic process in human hepatocellular carcinoma cells (Huh7 cells). Inhibition of autophagy in palmitate-treated cells does not aggravate the autophagic block and had no significant effect on cell death induced by palmitate. A: Huh7 cells were treated with palmitate and palmitate + 4-PBA for 24 h. MDC staining showed palmitate-treated cells with increased accumulation of autophagosomes in comparison with control. Pretreatment of 4-PBA in palmitate-treated cells reduced the accumulation of autophagosomes. Palmitate + everolimus-treated cells showed decreased autophagosomes in comparison with the only palmitate-treated cells. B: Huh7 cells were treated with palmitate and palmitate + 4-PBA for 24 h. Western blotting analysis of indicated proteins was performed and shown as an average of three independent experiments. Palmitate significantly increased the expression of mTORC1 (P < 0.001 at 0.25 mM, and 0.5 mM vs. control) and SQSTM1/p62 (P < 0.05 at 0.25 mM, and P < 0.001 at 0.5 mM vs. control) and decreased Beclin1 (P < 0.001 at 0.25 mM and 0.5 mM vs. control). Palmitate increased the LC3II/actin ratio (not significant at 0.25 mM and P < 0.001 at 0.5 mM vs. control). Pretreatment of 4-PBA in palmitate-treated cells further increased the LC3II:actin ratio (not significant at 4-PBA + 0.25 mM, and P < 0.05 at 4-PBA + 0.5 mM). The expression of Beclin1 also increased in 4-PBA + palmitate-treated cells (P < 0.05 at 4-PBA + 0.25 mM, and P < 0.01 at 4-PBA + 0.5 mM). The mTORC1 level was decreased by pretreatment of 4-PBA in palmitate-treated cells (not significant at 4-PBA + 0.25 mM and P < 0.001 at 4-PBA + 0.5 mM). SQSTM1/p62 expression was also brought down by 4-PBA pretreatment in palmitate-treated cells (not significant at 4-PBA + 0.25 mM and P < 0.01 at 4-PBA + 0.5 mM). C: Huh7 cells were treated with palmitate (0.5 mM), palmitate + 3-MA (2 mM), and palmitate + everolimus (100 nM), as was mentioned in the experimental procedures. Western blot analysis revealed that pretreatment of 3-MA in palmitate-treated cells increased the LC3II/actin in comparison with the only palmitate-treated cells. No significant effect was observed on the levels of SQSTM1/p62 and Beclin1 in palmitate + 3-MA-treated cells in comparison with the only palmitate-treated cells. Pretreatment of the autophagy inducer everolimus (100 nM) in palmitate-treated cells decreased LC3II/actin, decreased SQSTM1/p62, and increased Beclin1 in comparison with the only palmitate-treated cells. D–E: SQSTM1/p62 showed increased accumulation in palmitate-treated cells (P < 0.01, control vs. palmitate) in the presence of the autophagy inhibitors bafilomycin and CQ. Pretreatment of Baf/CQ failed to increase p62 levels in palmitate-treated cells (not significant palmitate vs. palmitate + Baf/CQ). F: Palmitate reduced the Bcl2:Bax ratio in palmitate-treated cells (0.5 mM) after 24 h (P < 0.001 control vs. palmitate). Pretreatment with 3-MA and everolimus had no further effect on the ratio (not significant palmitate vs. palmitate + 3-MA/everolimus ). G: Cellular viability was assessed by using MTT assay as mentioned in the experimental procedures. Huh7 cells were treated with palmitate (0.5 mM), palmitate + 3-MA (2 mM), and palmitate + everolimus (100 nM), as has been mentioned. Palmitate reduced the cellular viability (P < 0.001 control vs. palmitate). Pretreatment with 3-MA and everolimus had no further effect on cell viability in palmitate + 3-MA/everolimus-treated cells in comparison with the only palmitate-treated cells (P < 0.001 control vs. palmitate + 3-MA/everolimus and not significant palmitate vs. palmitate + 3-MA/everolimus). Baf, bafilomycin; Ever, everolimus; PA, palmitate; ns, not significant. *P < 0.05. **P < 0.01. ***P < 0.001: control vs. palmitate. #P < 0.05. ##P < 0.01. ###P < 0.001: palmitate vs. palmitate + 4-PBA.
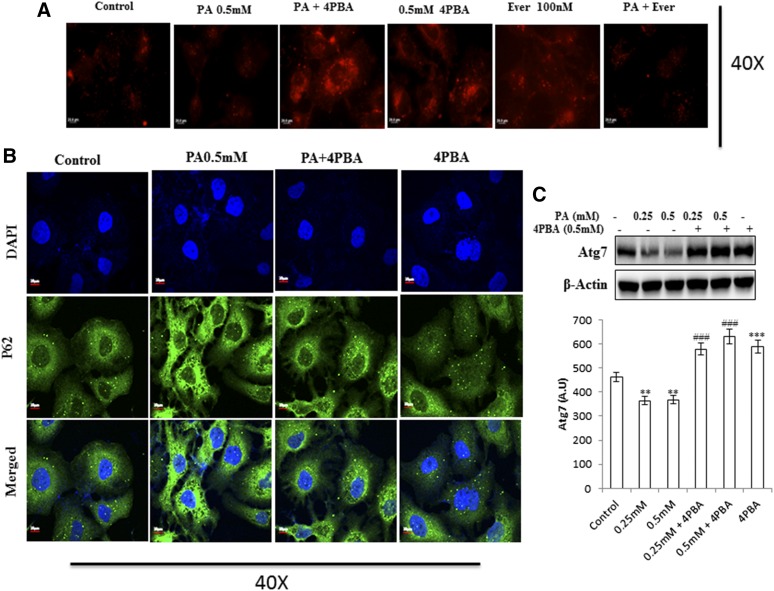
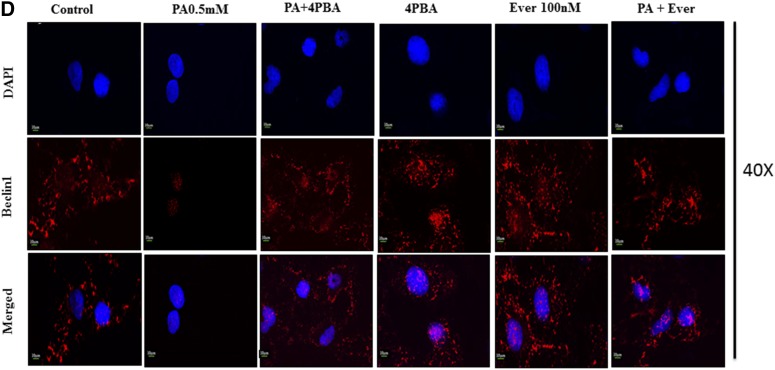
Fig. 2.
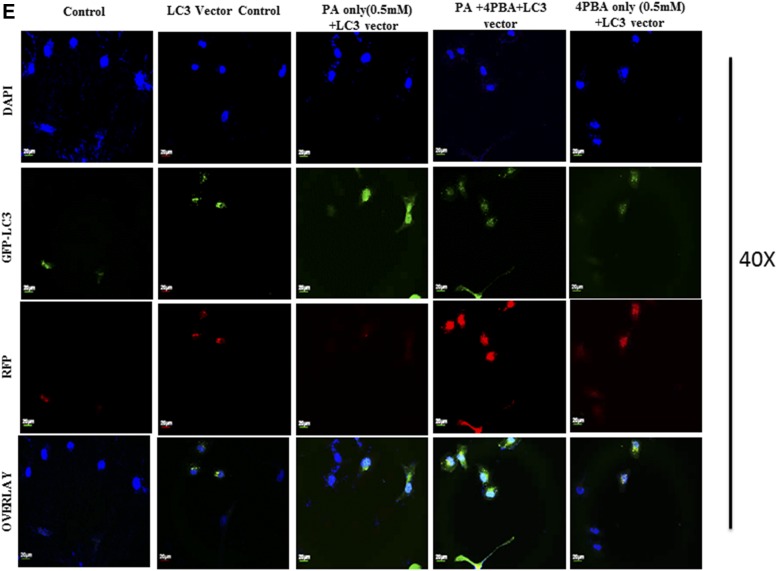
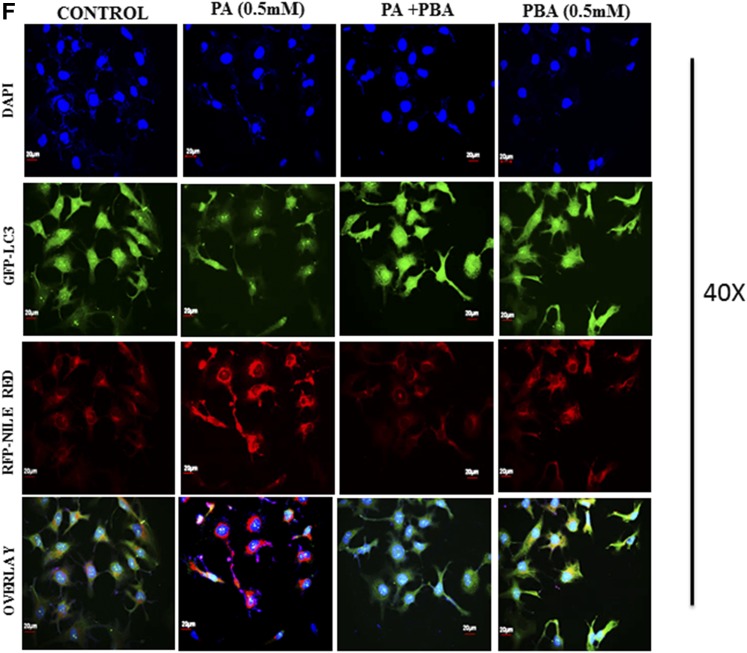
Pretreatment of 4-PBA and everolimus suppressed the autophagic block and restored autophagic flux in palmitate (PA)-treated Huh7 cells. A: Huh7 cells were stained with lysotracker red, as has been described. Pretreatment of 4-PBA and everolimus in palmitate-treated cells increased the fluorescence in comparison with the only palmitate-treated cells. B: Immunostaining of SQSTM1/p62 in Huh7 cells. Palmitate-treated cells showed increased fluorescence of p62 in comparison with untreated control cells. Pretreatment of 4-PBA in palmitate cells showed decreased fluorescence of p62 in comparison with the only palmitate-treated cells. C: Expression of Atg7 protein in palmitate and palmitate + 4-PBA-treated cells. Palmitate treatment significantly decreased Atg7 expression in Huh7cells after 24 h (P < 0.01 at 0.25 and 0.5 mM). Pretreatment of 4-PBA in palmitate-treated cells increased its expression (P < 0.001 at 4-PBA + 0.25 mM and 4-PBA + 0.5 mM). D: Immunostaining of Beclin1 in Huh7 cells. Palmitate-treated cells showed reduced Beclin-1 fluorescence in comparison with control cells. 4-PBA/ever + PA-treated cells showed increased fluorescence in comparison with the only palmitate-treated cells. E: Immunostaining of autophagosomes via GFP-LC3 puncta vector assay. PA-exposed cells showed increased green fluorescence (impaired autophagy, autophagosomes accumulation), whereas 4-PBA-pretreated cells, with palmitate, showed increased red fluorescence (shift to autolysosomes, autophagy normalcy). F: Immunostaining of cells for lipid droplets (Nile red) and LC-3 protein (green) in palmitate-exposed cells, 4-PBA-pretreated cells, or both. Palmitate-treated cells showed reduced green fluorescence and enhance red fluorescence (LD accumulation), whereas 4-PBA-pretreated cells showed reduced LD formation and enhanced LC-3 green fluorescence (autophagy restoration). RFP, red fluorescent protein. **P < 0.01. ***P < 0.001. ###P < 0.001.
Impaired autophagy in hepatocytes is associated with increased LD formation
Pharmacological inhibition of autophagy is known to increase triglycerides and LD formation (16). We therefore tested whether inhibition of autophagy by lipid stimulus in hepatocytes leads to lipid accumulation. Intracellular lipid accumulation was analyzed by microscopy with Nile red staining. Because TGs are the major component of the lipid droplets, we also measured the TG content of cells in this experimental setup. In comparison with untreated control cells, Huh7 cells treated with palmitate for 24 h showed increased lipid accumulation and TG content (Fig. 3A, C, supplemental Fig. S1), thus correlating with the blockade of autophagy. Pretreatment with everolimus (100 nM) in palmitate-treated cells significantly decreased the lipid accumulation and TG content (Fig. 3A, C, supplemental Fig. S1). In addition, a key autophagic gene, Atg7, has been shown to play an important role in lipophagy. Our results show decreased expression of Atg7 in palmitate-treated cells in comparison with untreated control cells (Fig. 2C). These results clearly suggest that palmitate-induced impairment of autophagic degradation is associated with impaired lipophagy, resulting in increased lipid accumulation and TG content in Huh7 cells. Furthermore, to study the effect of oleic acid (unsaturated fatty acid) on lipophagy, we treated the Huh7 cells with 0.5 mM oleic acid (oleate) for 24 h and studied the autophagic protein markers and lipid accumulation. Unlike palmitate, oleate did not impair autophagic flux, because protein expression of p62, Beclin1, and Atg7 were not significantly affected by oleate exposure in comparison with unexposed cells (supplemental Fig. S3B). Further, Nile red analysis suggested a nonsignificant LD accumulation after 24 h exposure of oleate (0.5 mM) to Huh7 cells, in comparison with nontreated cells (supplemental Fig. S3A).
Fig. 3.
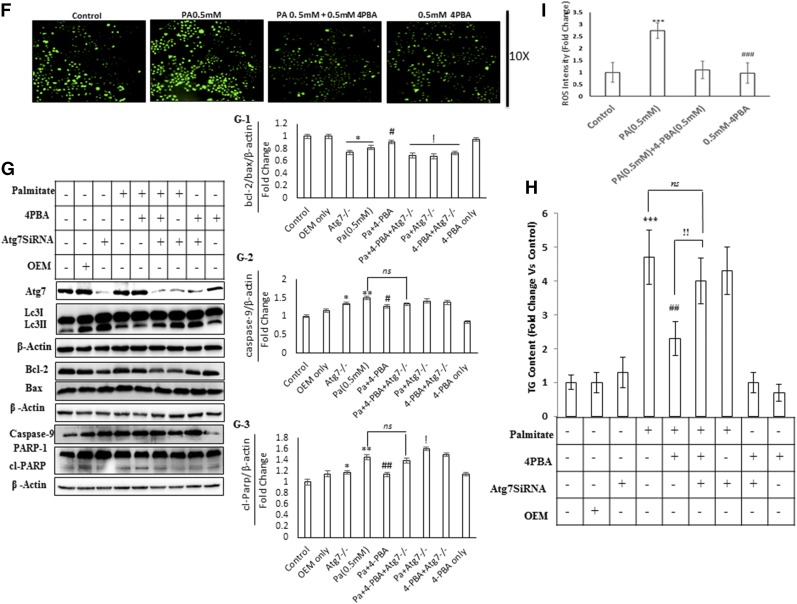
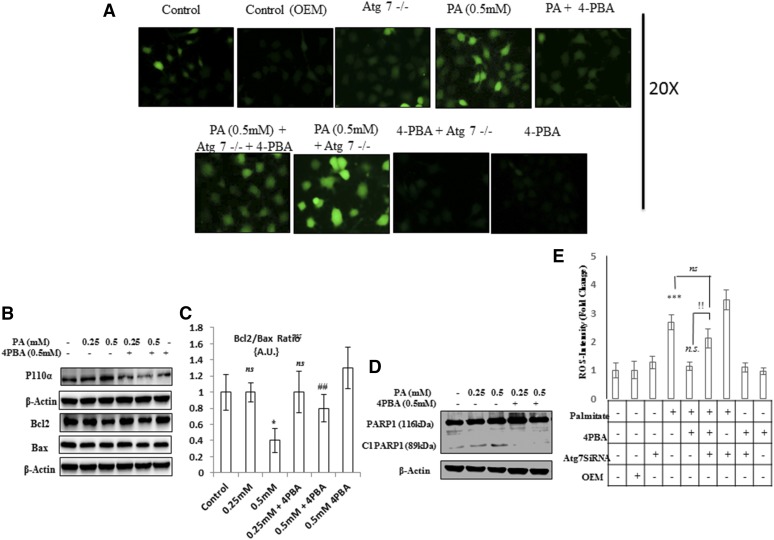
Impaired autophagy in palmitate-treated cells is associated with increased LD and TG accumulation. 4-PBA reduces the LD and TG content in palmitate-treated cells via autophagy. 4-PBA also prevents palmitate-induced ER stress and oxidative stress in Huh7 cells. A, C: Huh7 cells were treated with palmitate + 4-PBA and palmitate + everolimus for 24 h. Nile red staining to assess the lipid accumulation was performed, as mentioned in the experimental procedures. Palmitate-treated cells showed increased fluorescence in comparison with untreated control cells. Pretreatment of 4-PBA in palmitate-treated (0.5 mM) cells reduced the lipid accumulation as shown by decreased staining in 4-PBA + palmitate-treated cells, in comparison with the only palmitate-treated cells. Pretreatment of everolimus in palmitate-treated cells reduced the lipid accumulation. The bar graph represents TG content as measured by LPL assay buffer, as described in the experimental procedures. Palmitate-treated cells showed significantly increased TG content in comparison with untreated control cells (P < 0.001, control vs. palmitate). Pretreatment of 4-PBA in palmitate-treated cells showed significant reduction in TG content in comparison with the only palmitate-treated cells (P < 0.001, palmitate vs. 4-PBA + palmitate). Everolimus also reduced the TG content significantly in palmitate-treated cells (P < 0.001, palmitate vs. everolimus + palmitate). B, D: Huh7 cells were treated with palmitate, palmitate + 4-PBA, and palmitate + 4-PBA + 3-MA/CQ (autophagy inhibitors). Palmitate + 4-PBA + 3-MA/CQ showed more lipid accumulation than did the palmitate + 4-PBA-treated cells. TG content was also measured. Palmitate + 4-PBA + 3-MA showed more TG content than did the palmitate + 4-PBA-treated cells (!!P < 0.01, palmitate + 4-PBA + 3-MA vs. palmitate + 4-PBA, and ns (not significant), palmitate + 4-PBA + 3-MA vs. palmitate alone). Palmitate + 4-PBA + CQ also showed increased TG content as compared with palmitate + 4-PBA treated cells (!P < 0.05 palmitate + 4-PBA + CQ vs. palmitate + 4-PBA, and P < 0.05, palmitate + 4-PBA + CQ + palmitate alone). E: Huh7 cells were treated with palmitate (0.5 mM) for 12 h in the presence and absence of 4-PBA. Western blot was done for indicated proteins. Palmitate induced the upregulation of GRP-78, CHOP, and p-elf2α, and 4-PBA pretreatment prevented their upregulation. F: Huh7 cells were treated with palmitate and palmitate + 4-PBA for 24 h. ROS levels were observed by H2DCF-DA staining, as mentioned in the experimental procedures. Palmitate (0.25 and 0.5 mM) increased ROS levels in Huh7 cells as shown by increased staining in these cells. G: Huh7 cells were treated with Atg7 siRNA, palmitate, and 4-PBA in different combinations, as indicated. Western blotting analysis of indicated proteins was performed. Silencing of Atg7 increased the LC3II-B levels in both palmitate and palmitate + 4-PBA-treated cells. G-1: Densitometric analysis of bcl-2/bax (P < 0.05, control vs. Atg7−/− and control vs. Pa [0.5 mM]; P < 0.05, Pa [0.5 mM] vs. Pa + 4-PBA; P < 0.05, Pa [0.5 mM] vs. Pa + 4-PBA + Atg7−/−, Pa [0.5 mM] vs. Pa + Atg7−/−, and Pa [0.5 mM] vs. 4-PBA + Atg7−/−). G-2: Densitometric analysis of caspase-9 (P < 0.05, control vs. Atg7−/−; P < 0.01 control vs. Pa [0.5 mM]; P < 0.05, Pa [0.5 mM] vs. Pa + 4-PBA). G-3: Densitometric analysis of cl-PARP (P < 0.05, control vs. Atg7−/−; P < 0.01, control vs. Pa [0.5 mM]; P < 0.01, Pa [0.5 mM] vs. Pa + 4-PBA; P < 0.05, Pa [0.5 mM] vs. Pa + Atg7−/−). H: Huh7 cells were treated with Atg7 siRNA, palmitate, and 4-PBA in different combinations, as is indicated. Inhibition of autophagy by knockdown of Atg7 significantly reduced the lipid-lowering capacity of 4-PBA (P < 0.01, palmitate + 4-PBA vs. palmitate + 4-PBA + Atg7 siRNA). I: Intensity fold change for the ROS study by H2DCF-DA staining; images were analyzed by Image J software (National Institutes of Health [NIH], USA) (P < 0.001, control vs. palmitate; P < 0.001, palmitate vs. 4-PBA). OEM, Opti-MEM I. *P < 0.05. **P < 0.01. ***P < 0.001. #P < 0.05. ##P < 0.01. ###P < 0.001. !P < 0.05. !!P < 0.01.
4-PBA, a chemical chaperone, decreased LD formation by restoring autophagy
Previously, we have shown that lipid stimulus to hepatocytes led to the imbalance in Ca2+ homeostasis and development of unfolded protein response (9). In the present study, we hypothesized whether the use of chemical chaperones such as 4-PBA, which assist in protein folding, could prove beneficial for the restoration of autophagy and degradation of lipids. To test this hypothesis, we performed different experiments. Pretreatment with 0.5 mM 4-PBA, for 4 h before exposure to palmitate, significantly reduced FFA-induced lipid accumulation and TG content (Fig. 3A, supplemental Fig. S1), thereby suggesting a modulating role of lipogenic and lipid degradation pathways. To elucidate the possible mechanism of clearance of LDs by 4-PBA and analyze whether it is autophagy mediated, we monitored steps of autophagy in cells treated with both 4-PBA and palmitate. Western blotting results in palmitate plus 4-PBA-treated cells show increased LC3B-II and Beclin1 and decreased SQSTMI/p62 accumulation in comparison with palmitate-treated cells alone (Fig. 1B, supplemental Fig. S2). Immunostaining also confirmed these events, in palmitate plus 4-PBA-treated cells, by revealing reduced intensity of SQSTM1/p62 and increased intensity of Beclin-1, in comparison with palmitate-treated cells only (Fig. 2B, D; see palmitate vs. palmitate + 4-PBA). MDC staining showed a decrease in autophagosome formation in palmitate plus 4-PBA-treated cells in comparison with palmitate-treated cells alone (Fig. 1A). Pretreatment of everolimus (100 nM) also decreased autophagosome formation in palmitate-treated cells. Furthermore, to confirm the autophagosome-related events, we performed the green fluorescent protein (GFP)-LC3 puncta assay, expressing the GFP-LC3 autophagosomes marker. Only palmitate-treated cells showed accumulation of autophagosomes (indicated by enhanced green fluorescence), hence reflecting disrupted autophagy in which autophagosomes are not being converted into autolysosomes. However, in cells pretreated with 4-PBA, there was an enhanced red fluorescence (possibly in acidic pH, showing autolysosomes), suggesting normal autophagic functioning (Fig. 2E). Furthermore, the level of mTORC1 decreased significantly because of pretreatment of 4-PBA in palmitate-treated cells (Fig. 1B). These results clearly suggest that autophagy is restored by pretreatment of 4-PBA in palmitate-treated cells. As previously mentioned, the level of key autophagic gene, Atg7 (which also regulates LD formation), is decreased by palmitate. Pretreatment with 4-PBA in palmitate-treated cells significantly restored the levels of Atg7 (Fig. 1E). We further studied the LC3 and LDs correlation via fluorescence microscopy by coincubating the treated cells with the LC-3 antibody and Nile red stain (Fig. 2F). The palmitate-treated cells showed increased LD formation and reduced fluorescence of LC3; an overlay image showed enhanced red color, along with reduced green LC3, indicating the disrupted autophagy associated with LD formation, and 4-PBA pretreated cells showed less LD formation and enhanced LC3 fluorescence, indicating normal autophagy, with reduced LD formation in cells. These results gave us the insight that 4-PBA mediated its LD-reducing capacity by inducing autophagy in palmitate-treated cells. To confirm the statement that 4-PBA could attenuate FFA-induced lipid accumulation via autophagy, we investigated the lipid accumulation and TG content in the presence and absence of autophagy inhibitor 3-MA and lysosomal inhibitor CQ in palmitate-stimulated cells pretreated with 4-PBA. Our results indicate that restoration of autophagy by 4-PBA caused a reduction in lipid accumulation and formation of LDs. There was a significant difference in lipid content between cells treated with palmitate and 4-PBA and those treated with palmitate and 4-PBA in the presence of 3-MA and CQ (Fig. 3B, D), clearly indicating that 4-PBA failed to reduce lipid content in the presence of autophagy inhibitors.
4-PBA prevented ER stress and ROS generation in palmitate-exposed Huh7 cells
As described above, 4-PBA exerted its lipid-lowering effect, via restoring autophagy. 4-PBA being a chemical chaperone is known to exert its effect through the prevention of ER stress mediated unfolded protein response. We, therefore, studied the effect of 4-PBA on prevention of ER stress and ROS generation. As was expected, palmitate-exposed Huh7 cells (0.5 mM for 12 h) induced upregulation of ER stress markers (GRP-78, CHOP, and p-elf2α) in comparison with control cells (Fig. 3E), whereas 4-PBA pretreatment prevented upregulation of these markers. We also investigated the effect of 4-PBA on the ROS induced by palmitate exposure. Pretreatment of 4-PBA in palmitate-stimulated Huh7 cells significantly reduced the ROS generation, as measured by H2DCFDA staining (Fig. 3F, I). These results indicate that 4-PBA-mediated reduced LD formation and reduced lipotoxicity were also linked to the prevention of ROS generation and ER stress in palmitate-exposed cells.
Lipid lowering and ROS prevention by 4-PBA was reduced in Atg7 knockdown cells
Because the expressions of Atg7 (Fig. 2C) and Atg5 (supplemental Fig. S2B) were reduced by palmitate exposure in Huh7 cells, therefore, we wanted to explore the role of Atg7 and Atg5 in the lipid-lowering effect of 4-PBA. To answer this question, we investigated the lipid-lowering capacity of 4-PBA under Atg7 and Atg5 knockdown conditions. We transfected Huh7 cells with siRNA against Atg7 and Atg5 and then treated the cells with palmitate in the presence and absence of 4-PBA. The treatment with Atg7 siRNA decreased the expression of Atg7 and increased the LC3B-II levels. The increased LC3B-II levels in Atg7-deficient cells represent the blockade of autophagy, and this is in accordance with the guidelines of autophagy (28). Furthermore, the 4-PBA failed to prevent cell death induced by palmitate in Atg7−/− cells, as shown by increased PARP1 cleavage, increased caspase-9, and reduced bcl2-bax ratio ( ). Next, we analyzed the TG content in Atg7 knockdown conditions. As is shown in Fig. 3H, 4-PBA reduced the lipid accumulation in palmitate-treated cells, but its lipid-lowering capacity was significantly reduced by the silencing of the Atg7 gene. Similar results were obtained when we examined the TG content in Atg5−/− cells, induced by palmitate (supplemental Fig. S3E). 4-PBA failed to reduce the lipid content in Atg5−/− cells exposed to palmitate, and furthermore, the TG content rose significantly in Atg5−/− cells exposed to palmitate in comparison with wild-type cells exposed to palmitate (supplemental Fig. S3E). We also evaluated the effect of 4-PBA in preventing ROS generation, in Atg7−/− cells (Fig. 4A). ROS generation due to palmitate was significantly higher in Atg7−/− cells than in normal wild cells exposed to the similar concentration of palmitate. This rise in ROS levels in Atg7−/− cells could not be prevented by 4-PBA treatment in ATG7−/− cells, exposed to palmitate (Fig. 4A, E). Taken collectively, inhibition of autophagy either by Atg7 knockdown or Atg5 knockdown or 3-MA or CQ significantly reduced the 4-PBA- mediated lipid-lowering effect in Huh7 cells, thus again suggesting that 4-PBA reduced lipid accumulation via autophagy.
Fig. 4.
4-PBA protects against lipoapoptosis in palmitate-treated cells. ATG7 silencing decreases 4-PBA effects in palmitate-induced oxidative stress. A: Huh7 cells were treated with Atg7 siRNA, palmitate, and 4-PBA in different combinations, as indicated. ROSs were observed by H2DCF-DA staining, as mentioned in the experimental procedures. B-C: Huh7 cells were treated with palmitate and palmitate + 4-PBA for 24 h. Western blotting showed that palmitate increased the expression of p110α and decreased Bcl2/Bax ratio (not significant at 0.25 mM and P < 0.05 at 0.5 mM). Pretreatment with 4-PBA decreased the expression of p110α and improved Bcl2:Bax ratio in palmitate-treated cells (P < 0.01, palmitate 0.5 mM vs. palmitate 0.5 mM + 4-PBA). D: Palmitate treatment increased the levels of cleaved PARP1, whereas pretreatment of 4-PBA reduced its levels in palmitate-treated cells. E: Intensity fold change for the ROS study by H2DCF-DA staining; images were analyzed by Image J software (NIH) (P < 0.01, palmitate + 4-PBA vs. palmitate + 4-PBA + Atg7 SiRNA). *P < 0.05. ***P < 0.001. ##P < 0.01. !!P < 0.01.
4-PBA protected against lipotoxicity and improved Bcl2/Bax ratio in palmitate-treated Huh7 and AML-12 cells
Previously, we have shown that palmitic acid alters lipid metabolism, causes oxidative stress, and causes lipoapoptosis in Huh7 cells (9). In the present study, we aimed to investigate the effect of 4-PBA on these toxic manifestations induced by palmitate. Pretreatment of 4-PBA in palmitate-treated Huh7 and AML-12 cells restored the cell viability by about 40%–50% as measured by MTT assay (supplemental Figs. S1C, S2D). Palmitate treatment increased the expression of p110α, decreased Bcl2 /Bax ratio, and increased PARP1 cleavage. Pretreatment of 4-PBA decreased the expression of P110α, improved Bcl2/Bax ratio, and decreased PARP1 cleavage (Fig. 4B–D). Furthermore, we measured one of the transcription factors, Nrf2, an essential protein constituting the antioxidant defense system/antioxidant response element. Palmitate reduced the levels of Nrf2 after 24 h, whereas pretreatment of 4-PBA significantly restored its levels (supplemental Fig. S3D). So taken collectively, 4-PBA protected the cells from palmitate-induced lipoapoptosis.
DISCUSSION
Impaired autophagy in the liver results in protein inclusions in cytosol, such as misfolded proteins and excess accumulation of damaged organelles, which lead to various pathological conditions, including liver disorders (29). Recent investigations have shown autophagy as a new pathway for LD degradation, and its inhibition has been implicated in the development of steatosis in mice (15, 16). Another report concluded that impaired autophagic flux is associated with increased ER stress during the development of NAFLD (30). Therefore, therapeutic strategies to increase autophagic function may provide a new approach to prevent the lipotoxicity and development of NAFLD. In view of these facts, we aimed to assess the autophagy in FFA-treated (palmitate treated) human liver (hepatoma) cells and mouse hepatocytes (AML-12 cells), and attention was drawn to the antilipophagic effects of a chemical chaperone, 4-PBA, on restoration of autophagy and clearance of LD accumulation in lipid-stimulated hepatocytes (Huh7 and AML-12 cells).
Lipotoxicity is the main contributor to the progression of disease such as NAFLD (9). Previously, it was demonstrated that autophagic flux is impaired in livers from both nonalcoholic steatohepatitis patients and murine models of NAFLD, suggesting that disruption of autophagy could lead to cellular dysfunction (30). We tested the status of autophagy in palmitate-treated human liver (Huh7) cells and questioned whether LD accumulation is autophagy dependent. Our results showed that exposure of palmitate caused a significant accumulation of lipids and impaired autophagy in Huh7 cells. We found that defective autophagy due to nutrient overload (palmitate exposure) leads to accumulation of LC3B-II and autophagosomes (early autophagic markers), which were previously attributed to increase in autophagic flux (Fig. 1A, B, supplemental Fig. S2). However, recent evidence has questioned this interpretation and suggested that LC3B-II and autophagosome accumulation is in fact a consequence of the blockade of autophagic flux (28). This was also accompanied by significant accumulation of SQSTMI/p62 (Figs. 1B, 2B, supplemental Fig. S2), a key autophagic substrate, whose accumulation reflects the decrease in autophagic flux. Our observation of the decrease of autophagic turnover in palmitate-treated cells, leading to the accumulation of SQSTM1/p62, was also supported by downregulation of a key autophagic gene Atg7 in palmitate-treated cells (Fig. 2C). Deficiency of Atg7 led to a decrease in autophagic flux and accumulation of SQSTM1/p62 (31). Beclin1, a mammalian ortholog of yeast Atg6, is another key autophagic gene playing a central role in autophagy and mediates the cross-talk between apoptosis and autophagy (32). Beclin1 plays an essential role in the nucleation step of autophagy. The embryonic phenotype of Beclin1 null mice (beclin1−/−) has been shown to be more severe than other autophagy gene-deficient mice, as they die in early embryonic development (33). We tested the expression of Beclin1 in palmitate-treated cells and found that exposure (24 h) of palmitate significantly decreased its expression (Figs. 1B, ), thus providing further evidence that autophagy is impaired in palmitate-treated cells.
As is evident from the present results, defective autophagy was associated with LD accumulation and increased TG content (Fig. 3A, supplemental Fig. S1); we hypothesized that restoration of autophagy could lead to decreased LD accumulation and reduced lipotoxicity. Attention was drawn toward 4-PBA, a chemical chaperone, which in one of our previous studies was shown to reduce lipotoxicity in palmitate-treated cells and restored the cell viability by about 40%–50% in palmitate-treated cells (9). In the present study, we investigated its role with regard to autophagy and LD accumulation in palmitate-treated cells. Our results indicated that pretreatment of 4-PBA, 4 h prior to palmitate treatment, significantly restored the autophagic flux and reduced LD accumulation and TG content in Huh7 cells. MDC staining showed a significant decrease in autophagosomes in the cells treated with 4-PBA plus palmitate in comparison with palmitate-treated cells alone (Fig. 1A). Furthermore, increased LC3B-II, increased Beclin1, and decreased SQSTM1/p62 were observed in lipid-stimulated but 4-PBA-pretreated Huh7 cells (Figs. 1B, 2B, D, supplemental Fig. S2). Moreover, LC3-Nile red costaining confirmed that the 4-PBA clears LDs via inducing autophagy. In LC3-Nile red costaining, 4-PBA-treated cells showed a significant decrease in LDs and increased staining of LC3, reflecting activated autophagy, unlikely of the only palmitate-treated cells, which showed enhanced LDs and reduced fluorescence LC3 (Fig. 2F) (34). 4-PBA-mediated restoration of autophagy was further confirmed by GFP-LC3 puncta assay, revealing autophagosome accumulation in palmitate-treated cells (impaired autophagic flux, enhanced GFP) and enhanced red fluorescent protein in 4-PBA-pretreated cells, indicating the formation of autolysosomes. Lysotracker red showed increased intensity in palmitate-exposed cells pretreated with 4-PBA in comparison with cells exposed to palmitate only (Fig. 2A). These results clearly provide enough evidence that autophagic blockade induced by palmitate treatment is restored by 4-PBA pretreatment in Huh7 cells. One of the strong regulators of autophagy in a mammalian system is mammalian target of rapamycin (mTOR), whose activation by growth factors and nutrients leads to inhibition of autophagy (35). We tested whether nutrient overload (palmitate exposure) in our case had any significant effect on the levels of mTOR in Huh7 cells. Our results showed increased levels of mTORC1 in palmitate-treated cells, thus inhibiting autophagy (Fig. 1A). Inhibition of mTORC1 by its potent inhibitor, everolimus, restored the autophagy in palmitate-exposed cells, suggesting the fact that autophagic block is mTOR dependent. Pretreatment of 4-PBA in palmitate-exposed cells decreased the mTORC1 levels (Fig. 1A). As is mentioned above, 4-PBA significantly reduced LD accumulation and TG content; we next asked whether the LD-reducing capacity of 4-PBA is autophagy mediated. We used the well-known inhibitors of autophagy, 3-MA, CQ, and Baf, and assessed the lipid accumulation in cells treated with palmitate alone, palmitate + 4-PBA and palmitate + 4-PBA + 3-MA/CQ/Baf. Our results showed that 3-MA and CQ significantly reduced the LD- reducing capacity of 4-PBA (Fig. 3B, D). These results indicated that increased LD accumulation was a direct consequence of impaired autophagy in palmitate-treated cells. Atg7, a key autophagic gene that plays a role in lipophagy and has a prosurvival role in lipotoxic conditions (16, 36), was downregulated in palmitate-exposed Huh7 cells. Pretreatment of 4-PBA again increased its expression (Fig. 1E), suggesting that LD accumulation is due to the autophagic blockade and that 4-PBA mediated its LD-lowering effect via autophagy.
The role of 4-PBA has been established as an alleviator of ER stress. Furthermore, ER stress is known for inducing impaired autophagy and LD accumulation (37–39). We also studied this phenomenon in our cellular model (Huh7 cells), in which ER stress was induced by palmitate, after 12 h of treatment, as confirmed by Western blot analysis of ER stress- related proteins (Fig. 3E). As was expected, 4-PBA prevented the palmitate-induced ER stress and generation of ROS, which is in correlation with an earlier study (20). Contrary to our findings, the studies by Kim et al and Qin et al (38, 39) have shown that ER stress induces autophagy, leading to cell death, and that 4-PBA treatment prevented it. However, these studies have used potent ER stress-inducing agents such as Thapsigargin, Tunicamycin, DTT, and MG-132, whereas in our work, we used palmitate, which is a highly relevant fatty acid to human physiology and has a crucial role in developing steatosis and, subsequently, NAFLD. Furthermore, in Qin et al’s study (39), only LC3IIB accumulation has been studied as a marker of autophagic flux. However, as per the revised guidelines for monitoring autophagy, LC-3II is not the universal marker for autophagy process (40), but it can be used as supportive evidence only, whereas in this study, we have based our findings on multiple and more reliable markers of autophagy (Beclin-1, p62, Atg7, Atg5). Our hypothesis that 4-PBA reduces lipid accumulation via autophagy is again supported by knockdown of Atg7 and Atg5 in Huh7 cells, in which we observed that 4-PBA failed to reduce lipid accumulation in the presence of Atg7 siRNA and Atg5 siRNA (Fig. 3G, H, supplemental Fig. S3E). Everolimus, a positive control for autophagy induction, also decreased the LD accumulation and TG content in palmitate-treated cells (Fig. 3A, C). Taken collectively, our findings suggest that accumulation of autophagy cargo SQSTM1/p62 and autophagosomes and Atg7 downregulation correlate directly with impaired autophagy and increased fat content in liver cells and the use of the chemical chaperone 4-PBA restored the autophagy. This restoration of autophagy correlates inversely with total lipid content in liver cells. With regard to 4-PBA lipid-lowering effect, it has been shown that 4-PBA inhibited hepatic denovolipogenesis induced by ER stress, as a result of fructose consumption in rats (37). However, the current study provides an alternative insight of 4-PBA action in alleviating lipid accumulation, by restoring the palmitate-impaired autophagic flux, related to elevated ER stress in hepatocytes.
Finally, the effect of 4-PBA on lipotoxic parameters, including cleaved PARP1 and Bcl2/Bax ratio, were studied. 4-PBA efficiently reduced the levels of ROS in palmitate-treated cells (Fig. 3F). Disruption of Bcl2/Bax and PARP1 cleavage induced by palmitate was improved by 4-PBA (Fig. 4B–D), suggesting antilipotoxic properties of 4-PBA.
In conclusion, nutrient overload results in the impairment of autophagic flux, leading to accumulation of autophagic substrates such as SQSTM1/p62, accumulation of autophagosomes, Atg7 downregulation, and lysosomal dysfunction, thus contributing to cell death. This autophagic blockade is associated with LD accumulation and increased TG content. Stimulation of autophagy by a chemical chaperone 4-PBA alleviated the suppression of autophagic turnover, decreased LD accumulation and TG content, and reduced cell death (Fig. 5). Thus, therapeutic intervention using a chemical chaperone such as 4-PBA, which stabilizes misfolded proteins, could provide a promising strategy for treating lipotoxic conditions and diseases such as NAFLD.
Fig. 5.
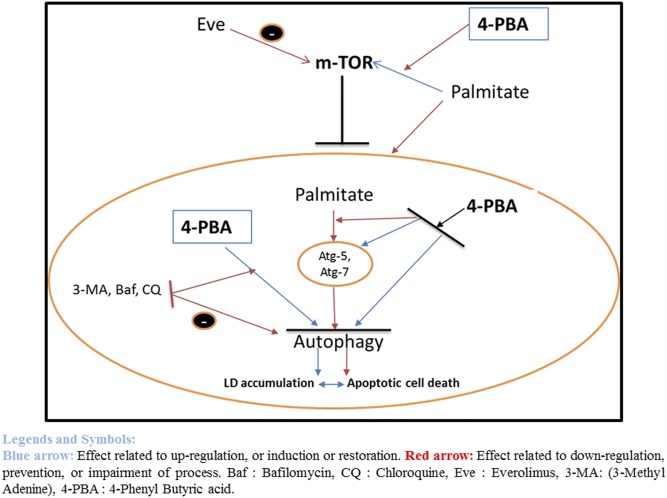
Palmitate impaired autophagy, resulting in increased lipid accumulation and apoptotic cell death. 4-PBA restored the autophagic blockade and reduced lipid accumulation and cell death. Palmitate upregulated the mTOR, and the upregulated mTOR resulted in the blockade of autophagy. Palmitate also impaired autophagic flux, shown by the reduced expression of Atg-7, Atg-5, and Beclin-1, leading to LD accumulation and cell death (red arrows). Use of autophagy inhibitor viz 3-MA, bafilomycin, and CQ, further blocked the autophagy, and synergized the palmitate-related autophagic block (red arrows). Everolimus, being an mTOR inhibitor and autophagy inducer, prevented the palmitate-induced impaired autophagy. On similar lines, the chemical chaperone 4-PBA prevented the palmitate-induced impaired autophagy by preventing upregulation of mTOR (red arrows) and restoring the functions of Atg-7 and Atg-5 (blue arrows), as is shown in this study, hence preventing palmitate-induced LD formation and lipotoxicity. However, use of autophagy inhibitors, along with 4-PBA, diminished the effect of 4-PBA as an autophagy restorer. Eve, everolimus. Blue arrows, effects related to upregulation, or induction or restoration; red arrows, effects related to downregulation, prevention, or impairment of process.
Supplementary Material
Acknowledgments
Michael R. Charlton is acknowledged for his mentorship and input and for providing Huh7 cells and everolimus.
Footnotes
Abbreviations:
- CQ
- chloroquine
- ER
- endoplasmic reticulum
- LD
- lipid droplet
- 3-MA
- 3-methyladenine
- MDC
- monodansylcadaverine
- MTT
- 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- NAFLD
- nonalcoholic fatty liver disease
- 4-PBA
- 4-phenyl butyric acid
- ROS
- reactive oxygen species
- TG
- triglyceride
Financial assistance was provided by the Council of Scientific and Industrial Research (CSIR), New Delhi, India; by University Grants Commission, New Delhi, India (A.U.N. and L.A.N.); and by the Department of Science and Technology, New Delhi, India (L.S.). The authors declare no conflict of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Choi A. M., Ryter S. W., and Levine B.. 2013. Autophagy in human health and disease. N. Engl. J. Med. 368: 651–662. [DOI] [PubMed] [Google Scholar]
- 2.Levine B., and Kroemer G.. 2008. Autophagy in the pathogenesis of disease. Cell. 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein D. C., Gestwicki J. E., Murphy L. O., and Klionsky D. J.. 2007. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6: 304–312. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky D. J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8: 931–937. [DOI] [PubMed] [Google Scholar]
- 5.Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., and Thompson C. B.. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 120: 237–248. [DOI] [PubMed] [Google Scholar]
- 6.Meijer A. J., and Codogno P.. 2006. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 27: 411–425. [DOI] [PubMed] [Google Scholar]
- 7.Maiuri M. C., Zalckvar E., Kimchi A., and Kroemer G.. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8: 741–752. [DOI] [PubMed] [Google Scholar]
- 8.Klionsky D. J. 2012. Look people, “Atg” is an abbreviation for “autophagy-related.” That’s it. Autophagy. 8: 1281–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissar A. U., Sharma L., and Tasduq S. A.. 2015. Palmitic acid induced lipotoxicity is associated with altered lipid metabolism, enhanced CYP450 2E1 and intracellular calcium mediated ER stress in human hepatoma cells. Toxicol. Res. (Camb.). 4: 1344–1358. [Google Scholar]
- 10.Nissar A. U., and Tasduq S. A.. 2015. Endoplasmic reticulum stress and oxidative stress in the pathogenesis of non-alcoholic fatty liver disease. Free Radic. Res. 49: 1405–1418. [DOI] [PubMed] [Google Scholar]
- 11.Martin S., and Parton R. G.. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7: 373–378. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., and Shinohara Y.. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiele C., and Spandl J.. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20: 378–385. [DOI] [PubMed] [Google Scholar]
- 14.Anderson N., and Borlak J.. 2008. Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharmacol. Rev. 60: 311–357. [DOI] [PubMed] [Google Scholar]
- 15.Dong H., and Czaja M. J.. 2011. Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. 22: 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J.. 2009. Autophagy regulates lipid metabolism. Nature. 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaushik S., and Cuervo A. M.. 2015. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Las G., Serada S. B., Wikstrom J. D., Twig G., and Shirihai O. S.. 2011. Fatty acids suppress autophagic turnover in beta-cells. J. Biol. Chem. 286: 42534–42544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mir S. U., George N. M., Zahoor L., Harms R., Guinn Z., and Sarvetnick N. E.. 2015. Inhibition of autophagic turnover in beta-cells by fatty acids and glucose leads to apoptotic cell death. J. Biol. Chem. 290: 6071–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Q., Xu L., Li H., Sun H., Wu S., and Zhou B.. 2017. 4-PBA reverses autophagic dysfunction and improves insulin sensitivity in adipose tissue of obese mice via Akt/mTOR signaling. Biochem. Biophys. Res. Commun. 484: 529–535. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim S. H., Kohli R., and Gores G. J.. 2011. Mechanisms of lipotoxicity in NAFLD and clinical implications. J. Pediatr. Gastroenterol. Nutr. 53: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H. W., Park H., Semple I. A., Jang I., Ro S. H., Kim M., Cazares V. A., Stuenkel E. L., Kim J. J., Kim J. S., et al. 2014. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat. Commun. 5: 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederbick A., Kern H. F., and Elsasser H. P.. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 66: 3–14. [PubMed] [Google Scholar]
- 24.Caro A. A., and Cederbaum A. I.. 2001. Synergistic toxicity of iron and arachidonic acid in HepG2 cells overexpressing CYP2E1. Mol. Pharmacol. 60: 742–752. [PubMed] [Google Scholar]
- 25.Eruslanov E., and Kusmartsev S.. 2010. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594: 57–72. [DOI] [PubMed] [Google Scholar]
- 26.Love S., Mudasir M. A., Bhardwaj S. C., Singh G., and Tasduq S. A.. 2017. Long-term administration of tacrolimus and everolimus prevents high cholesterol-high fructose-induced steatosis in C57BL/6J mice by inhibiting de-novo lipogenesis. Oncotarget. Epub ahead of print. February 8, 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscat J., and Diaz-Meco M. T.. 2011. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 147: 724–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J. A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N., Levine B., Cuervo A. M., and Klionsky D. J.. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González-Rodriguez A., Mayoral R., Agra N., Valdecantos M. P., Pardo V., Miquilena-Colina M. E., Vargas-Castrillón J., Lo Iacono O., Corazzari M., Fimia G. M., et al. 2014. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 5: e1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., and Mizushima N.. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang R., Zeh H. J., Lotze M. T., and Tang D.. 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Z., Jin S., Yang C., Levine A. J., and Heintz N.. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 100: 15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N., Yoshimori T., and Levine B.. 2010. Methods in mammalian autophagy research. Cell. 140: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y. C., and Guan K. L.. 2015. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 125: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan S. H., Shui G., Zhou J., Li J. J., Bay B. H., Wenk M. R., and Shen H. M.. 2012. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin). J. Biol. Chem. 287: 14364–14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren L-P., Song G-Y., Hu Z-J., Zhang M., Peng L., Chen S-C., Wei L., Li F., and Sun W.. 2013. The chemical chaperon 4-phenylbutyric acid ameliorates hepatic steatosis through inhibition of de novo lipogenesis in high-fructose-fed rats. Int. J. Mol. Med. 32: 1029–1036. [DOI] [PubMed] [Google Scholar]
- 38.Kim D-S., Li B., Rhew K. Y., Oh H-W., Lim H-D., Lee W., Chae H-J., and Kim H-R.. 2012. The regulatory mechanism of 4-phenylbutyric acid against ER stress-induced autophagy in human gingival fibroblasts. Arch. Pharm. Res. 35: 1269–1278. [DOI] [PubMed] [Google Scholar]
- 39.Qin L., Wang Z., Tao L., and Wang Y.. 2010. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 6: 239–247. [DOI] [PubMed] [Google Scholar]
- 40.Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C. M., Adams P. D., Adeli K., et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.