Abstract
Objectives
This study was intended to analyze effects of miR-199a-3p and Smad1 on proliferation, migration and invasion of prostate cancer (PCa) cells.
Results
MiR-199a-3p was significantly decreased in PCa tissues in comparison to that in adjacent normal tissues (P < 0.05). Over-expressed miR-199a-3p markedly suppressed proliferation and invasion of PCa cells (P < 0.05). MiR-199a-3p was negatively correlated with Smad1 expression, and overexpression of Smad1 could antagonize the effects of miR-199a-3p on PCa cells.
Materials and methods
The PCa tissues and their adjacent normal tissues were collected from 54 PCa patients. Expressions of miR-199a-3p and Smad1 mRNA in tissues and cells were evaluated with real-time quantitative polymerase chain reaction (RT-qPCR), and immunohistochemistry assay was used to detect Smad1 protein expressions. The target relationship between miR-199a-3p and Smad1 was assessed by luciferase reporter assay. The PCa cell lines (i.e. PC-3 cells) were transfected with miR-199a-3p mimics and Smad1-cDNA. MTT and Transwell assays were applied to detect proliferative, migratory and invasive abilities of PCa cells.
Conclusions
MiR-199a-3p suppressed proliferation and invasion of PCa cells by targeting Smad1.
Keywords: MiR-199a-3p, Smad1, prostate cancer, proliferation, invasion
INTRODUCTION
Prostate cancer (PCa) is the second major cause of cancer-related deaths among American males, and the proportion of it among all cancers achieves up to 43% [1, 2]. Although the survival rate of the clinically localized PCa patients has been increasing owing to treatments of surgery and radiotherapy, the recurrence rate of PCa within 5 years remains high at 25% [3, 4]. As a result, the molecular mechanisms underlying PCa development need to be further explored, which allows us to find novel therapeutic targets for PCa.
MicroRNAs (miRNAs), a kind of short non-coding RNAs that are approx 22–25nt long, can modify mRNA translations within carcinogenic pathways through binding to 3′-UTR region of the target mRNA [5–7]. Accumulating studies have also demonstrated remarkable associations of miRNAs expressions with occurrence and progression of PCa [8–11]. Specifically, it was indicated that miR-345 was differentially expressed between PCa and benign prostatic hypertrophy based on miRNA microarray analysis [12]. Wang et al. reported that serum miR-19 and miR-519c-5p levels can predict negative pathology of PCa patients [13]. In addition, miR-199b might impose inhibitive effects on PCa development since lower miR-199a expressions were observed with aggravation of PCa [14, 15].
Interestingly, miR-199a directly targeting Smad1 (Gene accession number: X69881) was demonstrated to modulate the activation of pancreatic stellate cells (PSCs) and PSC-induced pro-tumoral effects in pancreatic cancer [16]. Besides, Lin et al. also indicated that miR-199a is a BMP2 reactive miRNA which negatively regulated the differentiation of early chondrocyte by directly targeting Smad1 transcription factors [17]. In fact, Smad1 not only served as a substrate of mitogen-activated protein kinases (MAPKs), but also exerted a major role in transferring signals from bone morphogenetic proteins (BMPs), thereby inducing development of various disorders, including PCa, fibrosis, cardiovascular diseases and so on [18–22]. Furthermore, the ALK2/Smad1 pathway activated by endoglin was found to inhibit motility of PCa cells, and down-regulated Smad1 expressions due to target of miR-345 could inhibit growth, invasion and migration of human PCa cells [12, 23]. Thus, the synthetic effects of miR-199a and Smad1 were hypothesized to be correlated with PCa development.
However, this research pointed that whether miR-199a-3p might regulate PCa development by targeting Smad1 was still lacking up to now. In the present study, we conducted comprehensive research on the molecular effect of miR-199a-3p targeting Smad1 on the proliferation and invasion of PCa cells.
RESULTS
Expressions of miR-199a-3p and Smad1 in PCa tissues and PCa cell lines
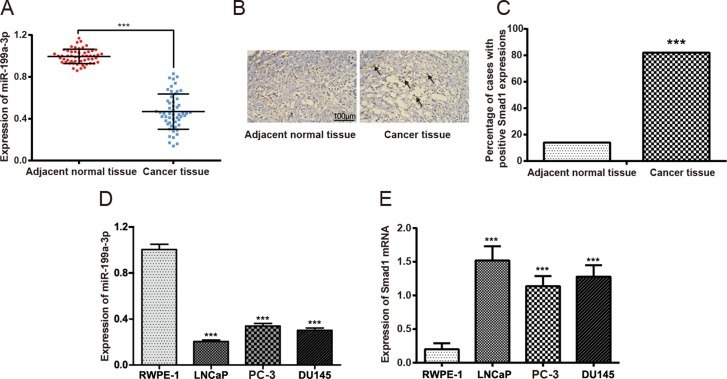
The expression of miR-199a-3p was significantly lower in PCa tissues than in adjacent normal tissues (P < 0.05) (Figure 1A). As displayed in Figure 1B–1C, the proportion of positively expressed Smad1 cells in PCa tissues (75.9%) significantly exceeded that in adjacent normal prostate tissues (12.9%). In addition, significantly down-regulated miR-199a-3p expressions and elevated Smad1 expressions were observed in PCa cell lines (i.e. LNCaP, PC-3 and DU145) in comparison to the normal prostate epithelial cell line (i.e. RWPE-1) (P < 0.05) (Figure 1D–1E). As PC-3 cells had higher expression of miR-199a-3p, it was used to perform subsequent cell experiments.
Figure 1. The expressions of miR-199a-3p and Smad1 in prostate tissue samples.
(A) Difference in the expression of miR-199a-3p between prostate cancer tissues and corresponding adjacent normal prostate tissues. Data were presented as mean ± SD. (B) Immunohistochemistry results showed the protein expression of Smad1 in prostate cancer tissues and cancer adjacent normal tissues. The red rectangles and black arrows have been pictured to direct Smad 1 positive staining (i.e. the brown colors). (C) Number of cases with positive Smad1 expressions. ***P < 0.001 compared with adjacent normal tissues. (D) The expression of miR-199a-3p in prostate cancer cell lines (i.e. LNCaP, PC-3 and DU145) and normal prostate epithelial cells (i.e. RWPE-1). ***P < 0.001 compared with RWPE-1. (E) The expression of Smad1 mRNA in prostate cancer cell lines (LNCaP, PC-3 and DU145) and normal prostate epithelial cells (i.e. RWPE-1). ***P < 0.001 compared with RWPE-1.
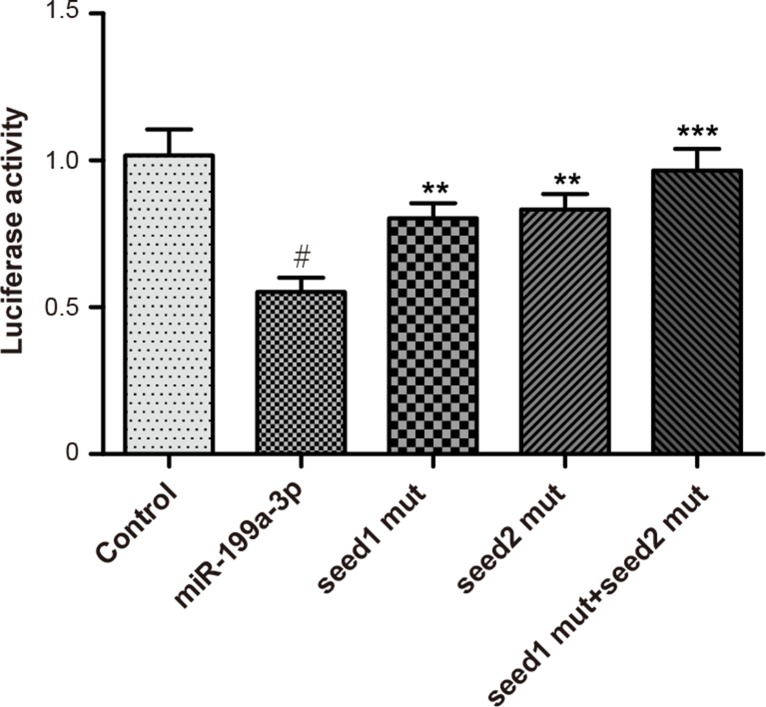
MiR-199a-3p targeted to Smad1
Perfect base pairing was observed between the sequence of mature miR-199a-3p and the 3′-UTR of Smad1 mRNA [17]. As suggested by luciferase reporter gene assay, miR-199a-3p group had a significantly lower relative light unit (RLU) value than the control group, whereas the RLU values of Smad1 seed1 mut group, Smad1 seed2 mut group, and Smad1 seed1 + seed2 mut group were significantly elevated in comparison to the miR-199a-3p group (P < 0.05) (Figure 2).
Figure 2. Luciferase activities were detected among groups of control, miR-199a-3p, Smad1 seed1 mut, Smad1 seed2 mut and Smad1 seed1+seed2 mut.
Data were presented as mean ± SD. #P < 0.01 compared with control group; **P < 0.01 compared with miR-199a-3p group; ***P < 0.001 compared with miR-199a-3p group.
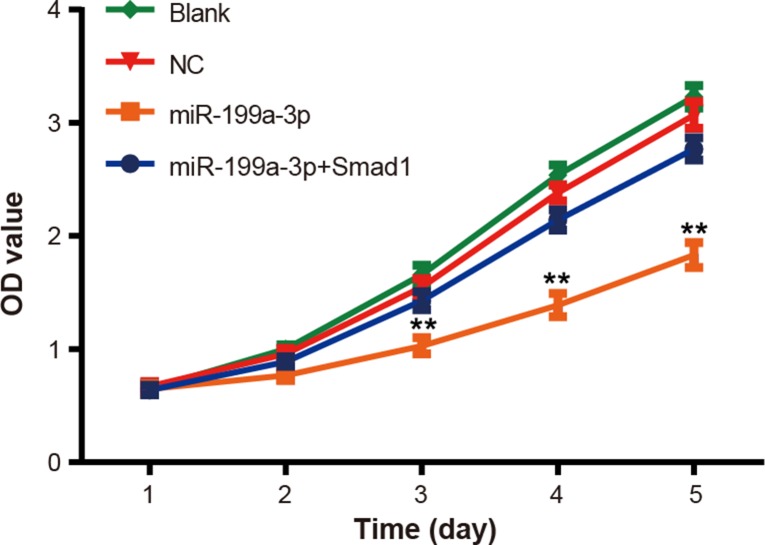
MiR-199a-3p suppressed proliferation of PCa cells
Significantly constrained proliferation of PCa cells transfected with miR-199a-3p mimics was observed when compared with the normal control group (P < 0.05) (Figure 3). Nonetheless, addition of Smad1 can reverse the inhibitory effects of miR-199a-3p on proliferation of PCa cells (P < 0.05). Moreover, no notable difference regarding motility of PCa cells was present among the blank group and normal control group at each time point (all P > 0.05).
Figure 3. The effect of miR-199a-3p overexpression on PC-3 cell proliferation was evaluated by MTT assay.
Data were presented as mean ± SD. **P < 0.01 compared with NC group.
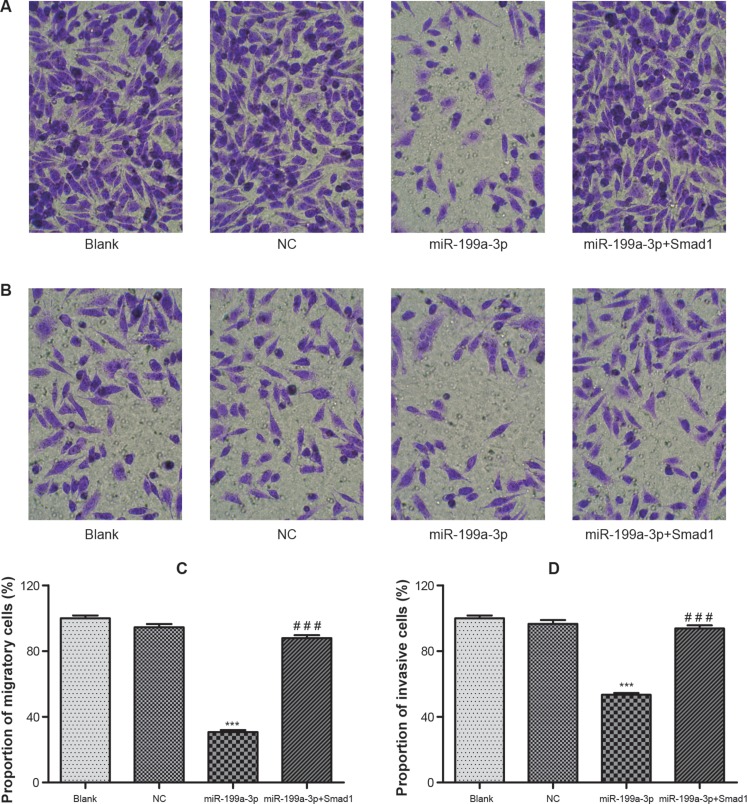
MiR-199a-3p inhibited migration and invasion of PCa cells
The results of Transwell experiments indicated that up-regulation of Smad1 or down-regulation of miR-199a-3p could restrict the migratory and invasive ability of PC-3 cells (Figure 4A–4B). The average proportion of migratory cells in the miR-199a-3p mimics [(30.7 ± 2.15)%] group was significantly smaller than that in the normal control group (P < 0.05), while the normal control group [(94.5 ± 3.61)%] was nearly no different from blank control group (P > 0.05) (Figure 4C). As for invasion of PCa cells, the difference between the normal control group [(96.7 ± 3.92)%] and blank control group was insignificant (P > 0.05). However, the percentage of invasive cells in the miR-199a-3p mimics group [(53.4 ± 2.02)%] was significantly below that in the normal control group (P < 0.05) (Figure 4D).
Figure 4. The migration and invasion of transfected PC-3 cells were detected by Transwell assay (× 200).
(A) The migration of cells in groups of blank, NC, miR-199a-3p and miR-199a-3p+Smad1 was visualized under the microscope. (B) The invasion of cells in groups of blank, NC, miR-199a-3p and miR-199a-3p+Smad1 was visualized under the microscope. (C) Histogram of the migration rate of PC-3 cells. (D) Histogram of the invasion rate of PC-3 cells. Data were presented as mean ± SD. ***P < 0.001 compared with NC group; ###P < 0.001 compared with miR-199a-3p group.
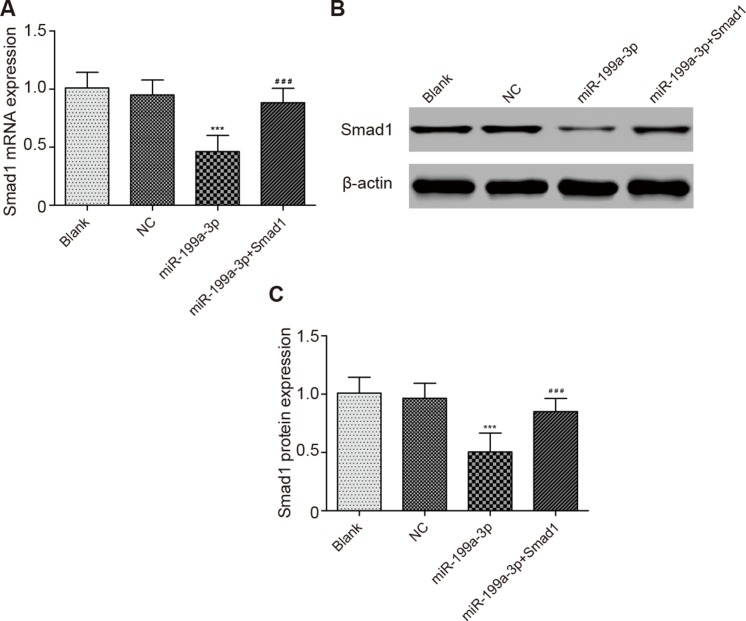
MiR-199a-3p suppressed Smad1 expression
Scarcely any notable difference in Smad1 expressions was observed among the blank group, normal control group and miR-199a-3p + Smad1 group (P > 0.05). Furthermore, miR-199a-3p significantly down-regulated Smad1 expressions in comparison with the normal control group (P < 0.05), whereas the Smad1 expressions in the miR-199a-3p + Smad1 group was notably higher than those in the miR-199a-3p group (P < 0.05) (Figure 5).
Figure 5. Comparison of Smad1 expressions in different groups.
(A) Comparison of Smad1 mRNA expression in different groups. (B) The effect of miR-199a-3p transfection on the PC-3 cells was detected by Western blotting assay. (C) Comparison of Smad1 protein expression in different groups. Data were presented as mean ± SD. ***P < 0.001 compared with NC group; ###P < 0.001 compared with miR-199a-3p group.
DISCUSSION
PCa now became a major public health problem within developed countries [24]. Its established risk factors mainly included aging, family history, hormones and genetic susceptibility [25, 26]. Accumulating evidences have indicated that miRNAs (e.g. miR-182, miR-130b and miR-345) may act as novel modulators for PCa development through regulation of mRNA of specific genes [e.g. metalloproteinase-2 (MMP-2) and Smad1] [9, 11, 12].
Notably, miRNA-199a could trigger tumor progression through inhibiting functions of MMP9, a protein that could degrade the extracellular matrix [27]. Additionally, the reduction in miR-199a-3p expressions and thereby non-suppression of mammalian target of rapamycin (mTOR) could result in advanced forms of cancers [28]. A documentary also showed that miR-199a could serve to regulate metastasis of PCa cells through transforming growth factor-beta1 (TGF-β1)-dependant way [29]. Similarly, our study found that the expression level of miR-199a-3p was significantly lower in PCa tissues than that in adjacent normal tissues. We also discovered that expressions of miR-199a-3p in LNCaP, DU145and PC-3 cell lines were significantly decreased compared with human normal prostate epithelial cells. Besides, over-expressed miR-199a-3p suppressed the proliferation and invasion of PCa cells, implying that miR-199a-3p potentially played a restrained role in PCa development.
Smad proteins were recognized as central modulators of TGF-β and BMP signaling pathways, which modulated cell growth and differentiation [30, 31]. It was found that miR-155 decreased Smad1 and Smad5 expressions in the lung epithelial cell line A549, modulating the cyclin-dependent kinase suppressor p21 [32]. In addition, Liu et al. provided evidence that the Smad1/Akt/GSK3β pathway was related with Nanog expression induced by Snail and cancer stem cell-like transformation in non-small-cell lung cancer cells [33]. In our study, we found that the expression of Smad1 was significantly higher in PCa tissues than in adjacent normal tissues. Moreover, the expressions of Smad1 and Smad1 mRNA were both reduced after transfection of PC-3 cells with miR-199a-3p.
The targeting relationship between miR-199a-3p and Smad1 has also been emphasized to date, for instance, miR-199a-3p was found to negatively modulate differentiation of early chondrocyte by directly targeting Smad1 transcription factors [17]. Besides, it was documented that miR-199a-3p might modulate activation of pancreatic stellate cells (PSCs) and PSC-induced pro-tumoral effects by directly targeting Smad1 transcription factor [16]. In the present study, we used the luciferase reporter assay to confirm that Smad1 is a target gene of miR-199a-3p, and over-expression of Smad1 would antagonize the effects of miR-199a-3p on PCa cells. Therefore, our study revealed the potentially molecular mechanism of miR-199a-3p targeting Smad1 with respect to proliferation and invasion of PCa cells.
However, there still existed some limitations in our experiments. For example, only 54 pairs of clinical samples and one human PC cell line PC 3 were used to detect the influence of miR-199a-3p targeting Smad1 on tumor proliferation, migration and invasion. And it was necessary to study the relative molecular pathway, as miR-199a-3p had other targeting gene. Hence, the mechanism of miR-199a and Smad1 on development and progression of PCa needs to be studied further. In conclusion, miR-199a-3p exerts anti-growth, anti-invasion and anti-migration effects on human PCa by partially down-regulating Smad1, suggesting that miR-199a-3p might be a potential diagnostic and therapeutic target for patients with advanced PCa.
MATERIALS AND METHODS
Subjects
This study included 54 PCa patients which were treated with radical prostatectomy at the Affiliated Nanjing Drum Tower Hospital, Medical School of Nanjing University from March 2015 to September 2015. All patients were not treated with preoperative chemotherapy or radiotherapy treatment. All subjects underwent pathological diagnoses, and were confirmed to be PCa. The PCa tissues and corresponding adjacent normal tissues were frozen in liquid nitrogen and were preserved in −70°C refrigerator. Of note, the adjacent normal tissues were representative of tissues that were located about > 2 cm distant from tumors or in another lobe that was confirmed with no cancer cells. Biological biopsy has been performed to check that the adjacent normal tissues were not contaminated by PCa cells. All procedures were in accordance with the Declaration of Helsinki [34] and were approved by the ethics committee of Human Experimentation of the Affiliated Nanjing Drum Tower Hospital, Medical School of Nanjing University. All patients included in this study have signed consent forms.
Immunohistochemistry for detecting expression of Smad1 protein
Smad1 protein expression was detected with PV-9000 two-step immunohistochemical method according to manufacturers’ instructions (ZSGB-BIO Cor., Beijing; catalog number: PV-9000). Specifically, conventional dewaxing and antigen microwave thermal retrieval were performed on four-micron-thick serial sections of formalin-fixed and paraffin-embedded tissues. The sections were incubated in 3% hydrogen peroxide to block endogenous peroxidase activity. Primary antibodies (Rabbit anti-human Smad1 polyclonal antibody, 1:400) (Bioss Cor., Beijing; catalog number: bs-1619R) were applied to sections at 4°C and refrigerated overnight. The sections were incubated for 20 minutes at room temperature after addition of polymer helper and incubated for another 30 minutes at room temperature after applying secondary antibodies (Bioss Cor., Beijing; catalog number: bs-0295M-HRP). The reaction product was visualized by incubation with the substrate/chromogen 3,3′-diaminobenzidine (DAB). Finally, the sections were counterstained with hematoxylin (0.02%). Phosphate buffer solution (PBS) was served as the negative control (NC). Each couple of sections, respectively, included one prostate cancer tissue and one adjacent tissue of the same patient. The five couples were randomly selected from five patients. Five random fields were selected from each slice (×400) to record positive cell number under an optical microscope (magnification, ×400; BX51; Olympus Corporation, Japan). The counting field would be selected if more than 100 cells were present therein. Smad1 expressions would be regarded as 1) positive (+) when more than 10% cells were immuno-reactive, and 2) negative (−) when the proportion of immuno-reactive cells was less than 10%. The results of immunohistochemistry were recorded and calculated blindly by two investigators.
Cell culture
Three kinds of human PCa cell lines [i.e. LNCaP, PC-3 and DU145 (ATCC-HTB-81)] and the normal prostate epithelial cell line (i.e. RWPE-1) were purchased from Insitute of Biochemistry and Cell Biology (Shanghai). Cells were cultivated in the mixed solution of RPMI-1640 medium (Gibco, USA), 10% newborn bovine serum, 100 IU/mL of penicillin and 100 μg/mL streptomycin within a 5% CO2 humidified chamber at 37°C.
Luciferase reporter assay
Smad1 3′ UTR had two binding sites of miR-199a-3p (Seed1: 5′-ACCTCTGTGACCAAC TATTG-3′; Seed2: 5′-AACTGTATGCTGGCTGTATTACTGT-3′). We constructed mutant Smad1 3′ UTR by site-directed mutagenesis kit (Stratagene, USA) (Seed1 mut: 5′-ACCTCTGTGACCATAATTTG-3′; Seed2 mut: 5′-AACTGTATGCTGGCTGTATATAAGT-3′). Smad1 3′ UTR, Smad1 seed1 mut and Smad1 seed 2 mut were inserted into pRL-CMV vector. After being seeded in a 48-well plate (concentration: 1.0 × 104 per well), PCa cells were co-transfected 24 h later with miR-199a-3p and 1) Smad1 3′ UTR, 2) Smad1 seed1 mut (100 ng), or 3) Smad1 seed2 mut (100 ng), or 4) Smad1 seed1 mut + seed2 mut (100 ng). Lipofectamine 2000 was simultaneously utilized with 1 μL luciferase reporter plasmid and 1 ng pRL-CMV per well (Invitrogen; catalog number: GM021043). On the other hand, the NC group was transfected with empty control (Invitrogen, USA; catalog number: GM-0210NC). Cells were lysed after 48 h of transfection, and luciferase activity was measured using the dual-luciferase reporter assay system according to the manufacturers’ instructions (Promega, USA). Data were normalized by dividing firefly luciferase activity by that of Renilla luciferase for the purpose of transfection efficiency.
Cell transfection
Cells were divided into four groups of blank, negative control (NC), miR-199a-3p transfection group, and miR-199a-3p + Smad1 transfection group. Transfection was performed by using Lipofectamine 2000 (Invitrogen, USA). Cells in the blank group were not given any treatment. Cells in other groups were transfected with scrambled miRNA mimics (GenePharma Cor., Shanghai; catalog number: E-S-4/5), miR-199a-3p mimics (GenePharma Cor., Shanghai; catalog number: E-S-4/5) and Smad1-complementary DNA (cDNA, Molbase, USA). Complete medium was replaced after 6 h and then cell culture was continued for further experiments.
MTT cell proliferation assay
When the density of transfected cells reached about 80%, cells were washed for 2 times using PBS. After being digested with trypsin into cell suspension, cells were inoculated into 96-well plates. Each well (volume: 200 μL) included 3 × 103−6 × 103 cells, and a total of 6 wells were replicated. Subsequently, MTT (20 μL, 5 mg/mL, Sigma, USA) was placed into each well and the cells were then cultured in an incubator with 5% CO2 at 37°C for 4 h. Dimethyl sulfoxide (DMSO, 150 μL) was added to each well and cells were gently shaken for about 10 minutes in order to dissolve the crystal. A microplate reader was used to detect the absorbance value of each well at a wavelength of 490 nm at the same time of five days. The experiment was repeated for three times.
Transwell migration and invasion assays
After 72 h of transfection, cells were placed in the upper chamber, and FBS-containing medium was put in the lower chamber as a chemoattractant. Cells were incubated in the 5% CO2 chamber at 37°C for 48 h. Non-migratory cells on the upper surface of the membrane were scraped off with cotton swabs. Cells that migrated to the bottom of the membrane were stained for 30 min (Costar Corporation, USA), and the stained cells were viewed under the optical microscope (Nikon, Japan). Five randomly selected fields were quantified using × 200 magnification, and the average number of cells was calculated.
Transwell invasion was performed utilizing 24-well Transwell plates with 8 μm pores, and coating Matrigel (BD Bioscience, US) was introduced to measure the invasive conditions of cells. Briefly, cells were trypsinized and resuspended in a serum-free medium after 48-hour transfection. A total of 2 × 104 cells were added to the upper transwell chamber, and then complete RPMI-1640 medium (volume: 0.5mL) was added into the lower chamber as the chemoattractant. After cells were incubated at 37°C for 24 h, non-invasive cells in the top chamber were removed by cotton swabs. Invasive cells at the bottom of the membrane were fixed with methanol and then stained with crystal violet. The number of invasive cells was finally counted.
Real time-quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction was conducted with aid of Trizol reagent kit (Ambion, USA) in line with the manufacturers’ descriptions. After added poly(A) tail on miRNA, the cDNAs were acquired based on oligo(dT)18 primers and M-MLV reverse transcriptase utilizing the reverse transcription Kit (Fermentas, Canada). RT-qPCR assay was conducted with qPCR instrument (Fermentas, Canada) to detect relative expression levels of miR-199a-3p and Smad1 mRNA. The primers synthesized by Invitrogen were displayed in Table 1. The reaction conditions were set as follows: 1) pre-degeneration at 95°C for 10 minutes, 2) degeneration at 95°C for 10 seconds, and 3) annealing at 60°C for 20 seconds and extension at 72°C for 35 seconds. The relative expression quantity was calculated using the 2−△△Ct method. The experiment was replicated for three times.
Table 1. Primers sequences used for quantitative RT-PCR.
| Genes | Primer pair sequences |
|---|---|
| MiR-199a | 5′-CAATCGCTTTCAAATAG-3′ |
| 5′-CAGGAGATGCTGTCATC-3′ | |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ |
| 5′-AACGCTTCACGAATTTGCGT-3′ | |
| Smad1 | 5′-AACCGTTTGAAGTGTAA-3′ |
| 5′-AAACAAACTGGGAAAGA-3′ | |
| GAPDH | 5′-GTGGACCTGACCTGCGTCT-3′ |
| 5′-GGAGGAGTGGGTGTCGCTGT-3′ |
Western blotting assay
Cells were washed with PBS for 3 times and were lysed with M-PER protein extraction reagent with protease inhibitors included (Pierce, USA). Lysates were then collected after centrifugation at 12,000 rpm for 10 min. Protein concentration was determined by a bicinchoninic acid (BCA) protein assay kit. Equal proteins were firstly separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to equilibrated polyvinylidene difluoride membranes, and finally blocked by 5% non-fat milk Tris-buffered saline (pH 7.4) containing Tween 20 for 2 h. Subsequently, the primary antibody anti-Smad1 (Sino Biological Inc., USA; catalog number: 101176-T08) were incubated over night at 4°C, while secondary antibodies (Abcam, USA; catalog number: ab98624) were incubated for 1 h at room temperature. Ultimately, proteins were visualized using ECL plus Western blotting detection reagents (Amersham Biosciences, USA) and detected with enhanced chemiluminescence (Amersham Pharmacia Biotechnology, USA).
Statistical analysis
Statistical analyses were analyzed using the GraphPad Prism 5.0 software (GraphPad Software Inc., USA). Counting data was analyzed by chi-square test. Measurement data were showed as mean ± standard deviation (SD). Difference in measurement data between groups was analyzed using the t-test or the rank sum test. It would be considered statistically significant when P < 0.05.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81572512) and Medical Science and Technology Development Foundation, Nanjing Department of Health (YKK15081).
Abbreviations
- PCa
prostate cancer
- BMPs
bone morphogenetic proteins
- DAB
diaminobenzidine
- PBS
phosphate buffer solution
- miRNAs
micro RNAs
- MAPKs
Mitogen-activated protein kinases
- PBS
phosphate buffer solution
- NC
negative control
- cDNA
complementary DNA
- RT-qPCR
real-time quantitative polymerase chain reaction
- TBST
Tris buffered saline with Tween
- GRP78
glucose-modulated protein 78
- mTOR
mammalian target of rapamycin
- TGFβ
transforming growth factor beta
- BMP
bone morphogenetic protein
Footnotes
Authors’ contributions
FQ, HL and HH designed and performed the research; XY, TY, JZ and YY conducted the experiments; XL, CJ and LF analyzed and interpreted the data; FQ, HG and WG wrote the manuscript and approved the final version.
CONFLICTS OF INTEREST
None.
ETHICAL APPROVAL
54 PCa patients included were treated with radical prostatectomy without any history of preoperative chemotherapy or radiotherapy which was approved by the Affiliated Nanjing Drum Tower Hospital, Medical School of Nanjing University.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Moore TH, King AJ, Evans M, Sharp D, Persad R, Huntley AL. Supportive care for men with prostate cancer: why are the trials not working? A systematic review and recommendations for future trials. Cancer Med. 2015;4:1240–1251. doi: 10.1002/cam4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson SV, O’Malley DM, Miller SM. Achieving optimal delivery of follow-up care for prostate cancer survivors: improving patient outcomes. Patient Relat Outcome Meas. 2015;6:75–90. doi: 10.2147/PROM.S49588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki HI, Miyazono K. Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway. J Mol Med (Berl) 2010;88:1085–1094. doi: 10.1007/s00109-010-0650-1. [DOI] [PubMed] [Google Scholar]
- 8.Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new biomarkers for prostate cancer. Br J Cancer. 2013;108:1925–1930. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Zhao X, Zhang H, Yuan H, Zhu M, Sun Q, Lai X, Wang Y, Huang J, Yan J, Yu J. MiR-130b suppresses prostate cancer metastasis through down-regulation of MMP2. Mol Carcinog. 2015;54:1292–1300. doi: 10.1002/mc.22204. [DOI] [PubMed] [Google Scholar]
- 10.Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS One. 2013;8:e83991. doi: 10.1371/journal.pone.0083991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R, Li J, Teng Z, Zhang Z, Xu Y. Overexpressed microRNA-182 promotes proliferation and invasion in prostate cancer PC-3 cells by down-regulating N-myc downstream regulated gene 1 (NDRG1) PLoS One. 2013;8:e68982. doi: 10.1371/journal.pone.0068982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen QG, Zhou W, Han T, Du SQ, Li ZH, Zhang Z, Shan GY, Kong CZ. MiR-345 suppresses proliferation, migration and invasion by targeting Smad1 in human prostate cancer. J Cancer Res Clin Oncol. 2016;142:213–224. doi: 10.1007/s00432-015-2016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SY, Shiboski S, Belair CD, Cooperberg MR, Simko JP, Stoppler H, Cowan J, Carroll PR, Blelloch R. miR-19, miR-345, miR-519c-5p serum levels predict adverse pathology in prostate cancer patients eligible for active surveillance. PLoS One. 2014;9:e98597. doi: 10.1371/journal.pone.0098597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang W, Chen X, Nie L, Xu M, Chen N, Zeng H, Zhou Q. MiR199b suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in prostate cancer cells. Int J Mol Sci. 2013;14:8422–8436. doi: 10.3390/ijms14048422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu Y, Huang X, Li Z, Liu J, Wu J, Chen D, Zhao F, Mu D. miR-199a-3p inhibits aurora kinase A and attenuates prostate cancer growth: new avenue for prostate cancer treatment. Am J Pathol. 2014;184:1541–1549. doi: 10.1016/j.ajpath.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Kuninty PR, Bojmar L, Tjomsland V, Larsson M, Storm G, Ostman A, Sandstrom P, Prakash J. MicroRNA-199a and -214 as potential therapeutic targets in pancreatic stellate cells in pancreatic tumor. Oncotarget. 2016;7:16396–16408. doi: 10.18632/oncotarget.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long X, Miano JM. Transforming growth factor-beta1 (TGF-beta1) utilizes distinct pathways for the transcriptional activation of microRNA 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286:30119–30129. doi: 10.1074/jbc.M111.258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald RA, Hata A, MacLean MR, Morrell NW, Baker AH. MicroRNA and vascular remodelling in acute vascular injury and pulmonary vascular remodelling. Cardiovasc Res. 2012;93:594–604. doi: 10.1093/cvr/cvr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol. 2013;229:274–285. doi: 10.1002/path.4119. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda H, Fuentealba L, Ikeda A, Reversade B, De Robertis EM. Default neural induction: neuralization of dissociated Xenopus cells is mediated by Ras/MAPK activation. Genes Dev. 2005;19:1022–1027. doi: 10.1101/gad.1306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu T, Grizzle WE, Oelschlager DK, Shen X, Cao X. Control of prostate cell growth: BMP antagonizes androgen mitogenic activity with incorporation of MAPK signals in Smad1. EMBO J. 2007;26:346–357. doi: 10.1038/sj.emboj.7601499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craft CS, Romero D, Vary CP, Bergan RC. Endoglin inhibits prostate cancer motility via activation of the ALK2-Smad1 pathway. Oncogene. 2007;26:7240–7250. doi: 10.1038/sj.onc.1210533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Semenas J, Allegrucci C, Boorjian SA, Mongan NP, Persson JL. Overcoming drug resistance and treating advanced prostate cancer. Curr Drug Targets. 2012;13:1308–1323. doi: 10.2174/138945012802429615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YC, Page JH, Chen R, Giovannucci E. Family history of prostate and breast cancer and the risk of prostate cancer in the PSA era. Prostate. 2008;68:1582–1591. doi: 10.1002/pros.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Zhang D, Wu GQ, Feng ZY, Zhu SM. Propofol inhibits the adhesion of hepatocellular carcinoma cells by upregulating microRNA-199a and downregulating MMP-9 expression. Hepatobiliary Pancreat Dis Int. 2013;12:305–309. doi: 10.1016/s1499-3872(13)60048-x. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Huang HJ, He CN, Wang KY. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR) Int J Gynecol Cancer. 2013;23:1191–1197. doi: 10.1097/IGC.0b013e31829ea779. [DOI] [PubMed] [Google Scholar]
- 29.Santos JI, Teixeira AL, Dias F, Gomes M, Nogueira A, Assis J, Medeiros R. Restoring TGFbeta1 pathway-related microRNAs: possible impact in metastatic prostate cancer development. Tumour Biol. 2014;35:6245–6253. doi: 10.1007/s13277-014-1887-z. [DOI] [PubMed] [Google Scholar]
- 30.Wakefield LM, Hill CS. Beyond TGFbeta: roles of other TGFbeta superfamily members in cancer. Nat Rev Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, Cheng HW, Raghuram S, Arany Z, et al. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res. 2013;113:1231–1241. doi: 10.1161/CIRCRESAHA.113.301780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin Q, Wang X, Fewell C, Cameron J, Zhu H, Baddoo M, Lin Z, Flemington EK. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J Virol. 2010;84:6318–6327. doi: 10.1128/JVI.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CW, Li CH, Peng YJ, Cheng YW, Chen HW, Liao PL, Kang JJ, Yeng MH. Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3beta signaling pathway in non-small-cell lung cancer. Oncotarget. 2014;5:3880–3894. doi: 10.18632/oncotarget.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Medical Association I Declaration of Helsinki Ethical principles for medical research involving human subjects. J Indian Med Assoc. 2009;107:403–405. [PubMed] [Google Scholar]