Abstract
Liquid chromatography coupled to tandem mass spectrometry is the main method for high-throughput identification and quantification of peptides and inferred proteins. Within this field, data-independent acquisition (DIA) combined with peptide-centric scoring, exemplified by SWATH-MS, emerged as a scalable method to achieve deep and consistent proteome coverage across large-scale datasets. Here we discuss the adaptation of statistical concepts developed for discovery proteomics based on spectrum-centric scoring to large-scale DIA experiments analyzed with peptide-centric scoring strategies and provide guidance on their application. We show that optimal tradeoffs between sensitivity and specificity require careful considerations of the relationship between proteins in the samples and proteins represented in the spectral library. We propose the application of a global analyte constraint to prevent accumulation of false positives across large-scale datasets. Furthermore, to increase the quality and reproducibility of published proteomic results, well-established confidence criteria should be reported for detected peptide queries, peptides and inferred proteins.
Introduction
Technological advances in liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) have greatly advanced our capabilities to explore proteomes. In bottom-up proteomics, the most widely used approach, proteins are proteolytically digested into peptides to increase their accessibility by LC-MS/MS. These peptides are then ionized and processed to generate fragment ion spectra (i.e. MS/MS spectra) which can be used to derive the amino acid sequences. Several classes of bottom-up proteomic methods have been developed that differ in the way the peptide ions are selected for fragmentation and how the resulting spectra are processed computationally. Currently, three main data acquisition strategies are applied: data-dependent acquisition (DDA), targeted acquisition by selected or parallel reaction monitoring (SRM or PRM) and data-independent acquisition (DIA). Each class of methods has specific strengths and weaknesses that have been extensively discussed1–3. The acquired data can be analyzed by different strategies, where the two main approaches differ in their query unit and are referred to as spectrum-centric and peptide-centric scoring methods, respectively4. In spectrum-centric scoring approaches, implemented for DDA and DIA5–8 data analysis, a spectrum or pseudo spectrum (when generated from DIA data), is queried against a peptide sequence database to associate the most likely peptide sequence. In peptide-centric scoring methods, mainly applied to SRM, PRM or DIA9–13 data, a peptide of interest is queried with specific peptide query parameters against the data to find the best candidate peptide signal(s)4. Peptide query parameters are also referred to as transition lists or “Tier 3” assays14 that include sets of precursor and product ion m/z pairs that, in combination, enable selective and sensitive detection of a peptide by a “peak group” of co-eluting fragment ion chromatograms (Supplementary Table 1).
While these signal processing and scoring systems are applicable to datasets of varying size and complexity, special attention needs to be paid to appropriate methods of error rate control to prevent accumulation of false positive identifications, particularly in cases in which large sample cohorts are analyzed. The false discovery rate (FDR)15 is a metric used for controlling the error rate of identified or detected analytes in experiments affected by the multiple testing problem. It is currently the most commonly employed metric within the field of mass-spectrometry-based proteomics and can be estimated by different methods, including derivation from posterior error probabilities estimated without16 or with the help of decoys17 or by using non-parametric q-value estimation by the target-decoy approach18. Conversely, the false non-discovery rate (FNR)19–21 representing the rate of “missed” discoveries at selected thresholds, provides a controllable metric for sensitivity under the same assumptions as the FDR (Supplementary Note 1).
Error rate control originally emerged as a critical issue in DDA-based discovery proteomics as a result of advances in experimental design and instrumentation that generate datasets of increasing size22. Peptide identification is generally established by spectrum-centric database searches and statistical modeling provides error rate control at different levels, including peptide-spectrum matches (PSMs)16,23 and inferred proteins24,25, and in different experimental contexts26–29. While the underlying algorithms for error rate control are different, their results have been shown to converge within the boundaries of their assumptions18,30.
In contrast, targeted proteomic methods are commonly used in cases where specific peptides need to be quantified across large sample cohorts with a high degree of reproducibility and quantitative accuracy31. In this type of measurement, it is expected that the majority of targeted peptides are detected in most samples, thus reducing the detection challenge mainly to selecting and quantifying the correct peptide fragment signals, also referred to as “peak groups”32. Data generated by SRM or PRM targeted proteomic measurements are therefore not affected by the same statistical challenges as typical spectrum-centric discovery proteomics experiments.
Recent developments in MS technology, specifically the development of DIA methods2,3 and their application to cohorts consisting of hundreds of samples, have led to the generation of complex datasets, consisting of large numbers of measured peptides (typically thousands to tens of thousands per sample), the presence and quantity of which need to be established and compared over many samples. This presents challenges for peptide and protein-level error rate control in peptide-centric analysis of DIA data, particularly in cases in which comprehensive spectral libraries, i.e. covering a substantial fraction of the proteome33–36, are being used36. Such analyses conduct 10,000s of peptide queries per sample across tens to hundreds of runs, leading to substantial error accumulation when the resulting multiple testing challenges are not addressed appropriately.
Here we propose that the criteria established for confidence assessment of identified peptides and inferred proteins in spectrum-centric analysis should also be applied to peptide-centric scoring methods on both peptide- and protein-levels for such studies. We show that data interpretation is dependent on the experimental context and offer considerations for designing an optimal analysis strategy. The applicability of the described concepts is demonstrated on the examples of the SWATH-MS inter-laboratory reproducibility study37 and a human blood plasma dataset comprising hundreds of samples38. In this context, the tradeoffs between spectral library comprehensiveness and sample-specificity are discussed in light of their respective requirements for appropriate error rate control.
Results
Peptide queries based on sample-specific versus combined spectral libraries
Most published studies employing DIA with targeted data extraction have used sample-specific spectral libraries generated either from corresponding DDA runs9,10,13,38–41 or from the DIA6 data itself. When sample-specific spectral libraries are used, it is usually sufficient to perform error rate control on the peptide query-level only. Because the content of spectral libraries is restrictively filtered during the process of generating the library42, putative false positive proteins are unlikely to be included in the targeted data extraction step (Supplementary Note 2). This is not the case when spectral libraries are generated from multiple heterogeneous samples, e.g. different cell or tissue types. In such cases, the spectral library contains a large fraction of ‘false targets’ that are not detectable in a specific sample. This value is also referred to as π043. The π0 value is directly coupled to the error estimation within a dataset, where larger π0 require stricter multiple hypothesis testing as well as strategies to control for error accumulation from the PSM- or peptide query-level to the protein-level, as has been demonstrated for discovery proteomics27,44. This effect is further accentuated when repository-scale spectral libraries such as our combined human assay library (CAL) 36 are used to analyze large sample cohorts.
In light of these considerations, the ideal case would be peptide queries that exactly match the set of detectable targets in the DIA dataset. However, comprehensive libraries can substantially increase the sensitivity of peptide-centric scoring approaches36,37 and are required to quantitatively compare heterogeneous samples in larger sized cohorts such as clinical studies (Supplementary Note 3). Thus, it is crucial to apply robust error rate control methods in peptide-centric scoring workflows, similar to the situation in discovery proteomics, particularly in cases of high π0.
Protein FDR assessment
As stated above, error rate control on the peptide query-level only is insufficient to infer sets of proteins in workflows employing comprehensive spectral libraries leading to high π0 values. For these cases, we previously suggested that the error rate should be controlled not only on the peptide query-level, but also on the peptide- and protein-levels36. This can be achieved, for example, by adapting a target-decoy approach as initially implemented for protein-level spectrum-centric analyses in MAYU27 or recently in SWATH2stats45. Another option is the application of non-parametric modeling strategies for computing posterior probabilities at the peptide- and protein-level17, as have been adapted for DIA analyses in DIA-Umpire6,46 and SWATHProphet47.
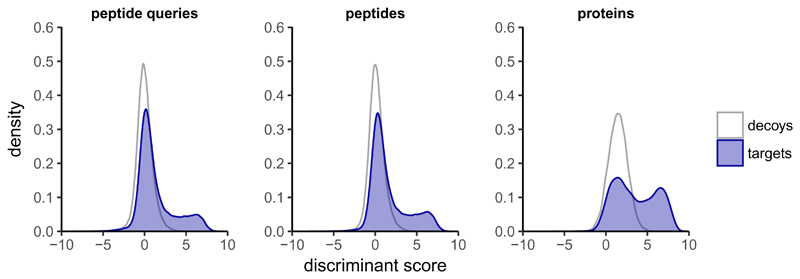
Alternatively, the q-value43 has been proposed for error estimation at the PSM-level as well as on the protein-level18. The q-value is a significance measure for analyte detection comparable to the p-value, but accounting for multiple testing analogously to the FDR. We have investigated whether peptide- and protein-level q-values could be estimated similar to the peptide query-level in our workflow consisting of OpenSWATH10 and PyProphet11, a reimplementation of the mProphet32 algorithm for DIA data. While OpenSWATH and related tools compute a set of scores for each peptide query, PyProphet combines these scores to a single discriminant score by applying semi-supervised learning to best separate decoys from high-scoring targets. The following peptide query-level q-value estimation step further uses the decoys to model a null distribution32. This concept can be extended to the protein-level by applying a similar strategy as has been suggested for discovery proteomics, only considering the best scoring PSM (or peptide query) for each peptide or protein for q-value estimation22,28,29,48. The applicability of this extended q-value estimation approach is illustrated on an exemplary sample (one run) of the SWATH-MS inter-laboratory reproducibility study37 that was analyzed with the CAL, containing 194,052 proteotypic peptide queries (Figure 1, Supplementary Figures 1-2). Here, only the best scoring peak group per protein (N=10,316) is considered for protein-level q-value estimation. The discriminant score distributions and p-value histograms43 indicate that, in analogy to the peptide query-level, peptide- and protein-level q-values can be applied as confidence metrics to avoid error-accumulation from the peptide query- to the protein-level.
Figure 1. Q-value estimation on peptide query-, peptide- and protein-level.
The peptide query-, peptide- and protein-level discriminant score density plots for one DIA run of the SWATH-MS inter-laboratory study analyzed with the combined human assay library (CAL) are depicted. The distributions indicate a large false target to total target ratio (π0 ≈ 0.6) on peptide query-level. The false target to total target ratio decreases slightly on peptide-level and more on protein-level (π0 ≈ 0.5), compared to the peptide query-level.
Context-dependent estimation of error rates
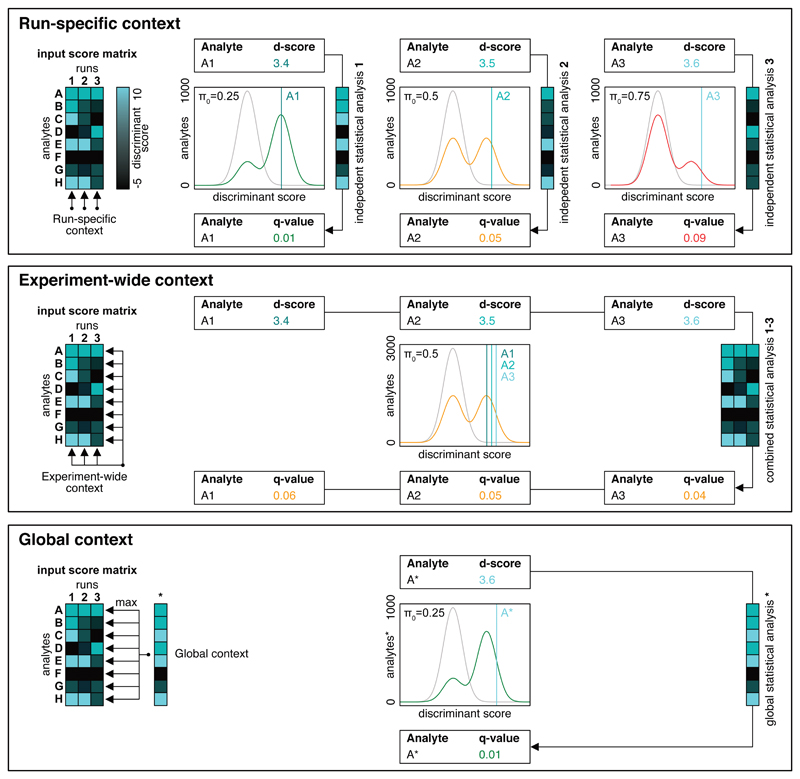
The q-value estimation for individual peptide queries (or proteins) is dependent on the context of the query, i.e. on other queries to the data18. This encompasses all peptide queries in the same LC-MS/MS run, but, in the context of a multi-sample study, also peptide queries in different LC-MS/MS runs. In an individual run, the question is asked: ”Is the query peptide detected in this sample?”4. If several runs are compared, this question might be extended to: “In which subset of samples is the query peptide detected?”. Alternatively, it might be of interest whether the query peptide was detected in any one of the samples. Depending on which question should be answered, the context of the hypothesis and the method for estimating an appropriate q-value needs to be adjusted. In analogy to the situation in spectrum-centric approaches (Supplementary Note 4), we suggest considering three scenarios for DIA peptide-centric scoring and error rate control: run-specific, experiment-wide and global context.
Run-specific context
For the research question “Which peptides can be detected within one LC-MS/MS run (i.e. one sample injection)?”, the run-specific context applies. Q-values or the FDR are therefore estimated from the single best scoring peak group per peptide query within one specific run, independently from the other runs that may have been acquired in the course of an experiment. Given a specified confidence threshold, the number of detectable peak groups, peptides or inferred proteins per run can be compared to the numbers achieved in other runs. This mode offers granularity for different levels of target peptide prevalence, since π0 values are estimated for each run separately. Samples with a low π0 thus benefit in sensitivity, because only limited multiple testing correction is required. In contrast, samples with a high π0 are more strictly corrected for multiple testing43. This has various implications for the analysis of comparative studies containing heterogeneous samples with truly different π0 between runs, e.g. AP-MS experiments or fractionated samples. This means that if peptides are queried using parameters based on the same spectral library against two runs that result in the same π0, peak groups with identical discriminant scores will also have the same estimated q-values. However, if the same peptides are queried against two runs with truly different π0, peak groups with identical discriminant scores will have substantially different q-values (Fig. 2).
Figure 2. Schematic illustration of the different context-dependent error-estimation strategies.
The run-specific context conducts separate q-value estimation for each sample. This method results in run-specific q-values which can represent different peak group qualities between runs with varying π0. This means that if the same peptide is queried in two samples using the same parameters, run 1 with a low π0 and run 2 with a high π0, and the scored peak groups have a similar discriminant score (d-score), they might get a low q-value in run 1 and a high q-value in run 2. The experiment-wide context considers all runs of an experiment for error rate control. The resulting q-values can be compared in terms of peak group quality between runs but should not be considered outside the context of the whole experiment. The global context only considers the best scoring peak group per analyte across the entire experiment. This approach enables the total set of detectable peptides or inferred proteins to be determined within the experiment. The global set of proteins can optionally be used as a constraint for the experiment-wide context to obtain the number of detected analytes in single runs.
Experiment-wide context
The experiment-wide context asks the question: “In which subset of samples is the query peptide detected?” In contrast to the run-specific, the experiment-wide context assesses detected peptides and inferred proteins within an experiment consisting of multiple runs, estimating π0 from the best scoring peak group matrix over all peptide queries and runs. A main assumption of this type of analysis is that the runs and π0 are different, because the samples represent different proteome subsets (e.g. comparison of whole cell lysate and fractionated samples) but not the quality of the samples or runs. These conditions are more frequently met in peptide-centric than in spectrum-centric scoring methods, because in comparative studies individual runs are queried for many peptides that might not be detectable in the sample. In this case, peptides with identical q-values will have an identical discriminant score (Fig. 2).
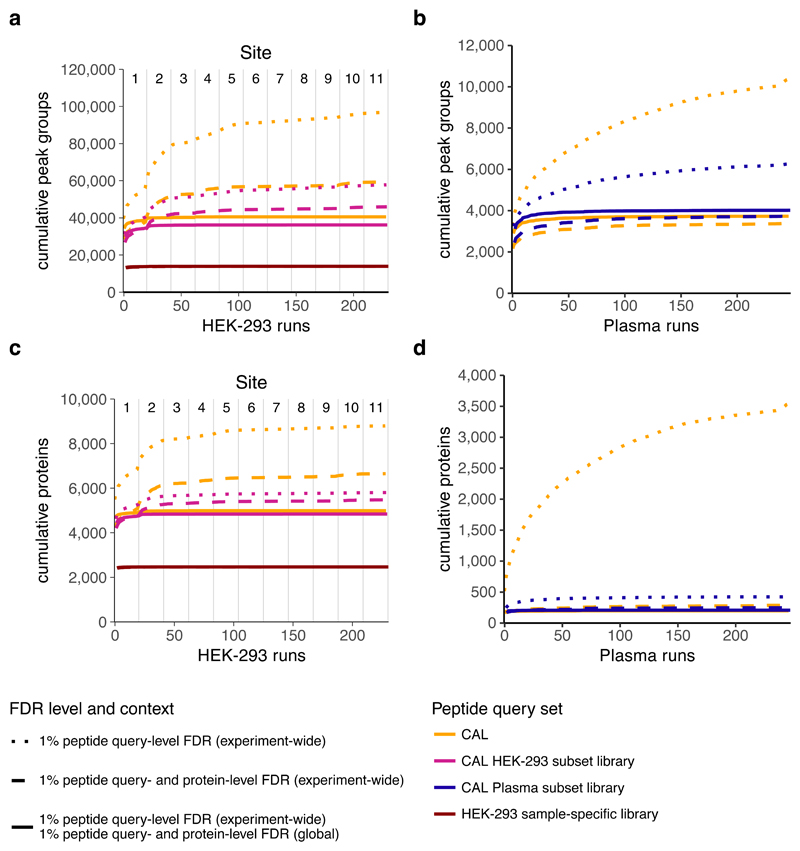
Both the run-specific and experiment-wide contexts can be used to generate matrices of detectable peak groups, peptides, or inferred proteins while controlling the error rate. However, when the analytes are summarized across a large study, false positive detections are accumulated. This effect is illustrated in Figure 3, which shows the cumulatively detected peak groups, peptides and inferred proteins across the 229 runs that constitute the inter-laboratory SWATH-MS study37 and independently, across the 246 runs of a previously published study measuring undepleted human blood plasma samples of 116 individuals38. The corresponding decoy accumulation rate is shown in Supplementary Figures 3-6. When using the CAL and applying a q-value cutoff of 1% on the peptide query-level, as estimated within the experiment-wide context, the cumulative number of target proteins inferred reaches almost the respective number of proteins covered by the spectral library. Applying an experiment-wide context with a q-value cutoff of 1% on peptide query- and 1% on protein-level decreases the number of inferred proteins, but still results in an accumulation of detected peptides and inferred proteins in the HEK-293 samples. This is not the case for the samples of the plasma dataset, which contain in average more peptides per inferred protein, but a much lower total number of proteins. To prevent such accumulation of potentially false positives in studies where this accumulation is problematic, the global context can be applied.
Figure 3. Analyte accumulation across multiple runs.
The number of cumulatively detected peak groups (a: inter-laboratory SWATH-MS study, b: plasma dataset), and inferred proteins (c: inter-laboratory SWATH-MS study, d: plasma dataset) is shown. While the different approaches for error rate control show the same result for the sample-specific spectral library (SSL) in the inter-laboratory SWATH-MS study, the accumulation of putative false positive analytes using peptide queries with higher fractions of non-detectable targets is largely influenced by the applied filtering strategies (combined human assay library (CAL), HEK-293 subset of the CAL (HEK), plasma subset of the CAL). Error rate control on peptide query-level in the experiment-wide context (dotted lines) shows accumulation of targets on all levels, almost until library saturation. Further, additional strict filtering on protein-level reduces the number of detected peak groups and inferred proteins, highlighting the importance of considering accumulation of putative false positives (dashed lines). The third strategy shows how applying a global analyte constraint on peptide query- and protein-level in addition to experiment-wide peptide query-level error rate control lowers the accumulation of proteins with low confidence in the global context, reaching an early saturation of inferred proteins across all runs (solid lines).
Global context
The global context asks the question: “Which peptides can be detected in at least one LC-MS/MS run of the experiment?” For this purpose, it considers only the best scoring detected peak groups, peptides or inferred proteins over all runs for the error-rate control. The resulting global protein master list can then be used to define a set of overall inferred proteins in the entire study which can be used to filter the matrix obtained by using either the run-specific or the experiment-wide context. The effect of applying constraints based on the global context is shown in Figure 3. Applying a peptide query- and protein-level global FDR cutoff of 1% (in addition to the 1% peptide query-level, experiment-wide FDR cutoff) results in a consistent number of cumulatively detected analytes across all 229 runs of the inter-laboratory SWATH-MS study, even when using the large CAL. In the plasma dataset, accumulation at the inferred protein level is already reasonably well controlled by the experiment-wide FDR on protein level and the application of the global context constraint further reduces the observed accumulation.
Tradeoff between spectral library specificity and comprehensiveness
As discussed above, sample-specific spectral libraries have the benefit of less error rate control being required (low π0), but the achievable proteome coverage depends on the completeness of the library. In contrast, repository-scale spectral libraries covering additional peptides that are detectable in the sample but not found in the sample-specific spectral library can reach higher coverage of the studied proteome when additional detections are not lost to the stricter multiple testing adjustments (high π0) required. Adding new undetectable targets only will reduce sensitivity when multiple testing correction is correctly applied, as was demonstrated in a recent study49 (Supplementary Note 3).
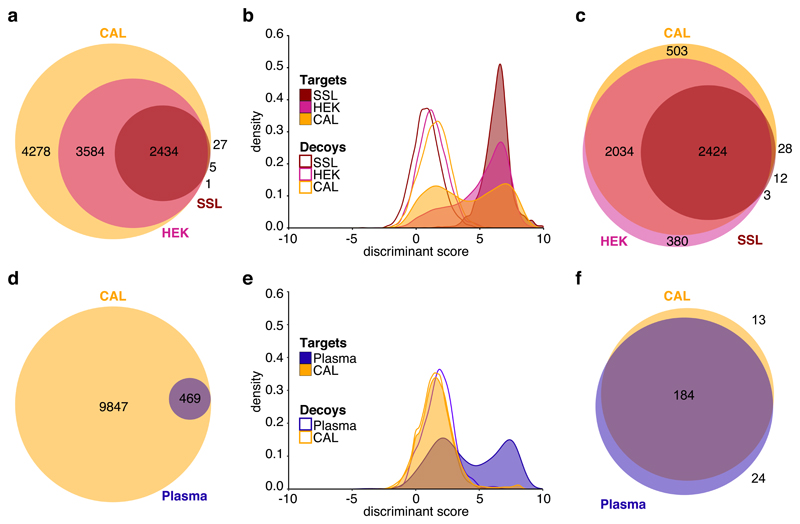
To further illustrate these effects we have applied three spectral libraries of different levels of sample-specificity and comprehensiveness to query the inter-laboratory SWATH-MS study37. A sample-specific library (SSL) was generated from the spectra obtained in six DDA runs of the SWATH-MS study sample. The CAL was used as a second, repository-scale library, which consists of 331 runs, of which 134 were acquired from fractionated and unfractionated HEK-293 samples. The third library applied was a HEK-293 subset of the CAL (HEK), only containing spectra observed in unfractionated and fractionated HEK-293 samples that were included in the original library. To assess the effects of library size and specificity on a real-world dataset, we applied the CAL to the plasma dataset38 and additionally generated a plasma-specific subset of the results. This is an extreme scenario, because the CAL itself contains only 8 runs acquired from plasma samples and the vast majority of peptides in the CAL is not expected to be detectable from unfractionated plasma samples. Figure 4a illustrates the size and protein overlap between the different libraries used for the analysis of the inter-laboratory SWATH-MS study. In Figures 4b, the global protein-level discriminant score distributions of targets and decoys are shown, illustrating the different π0 between the libraries. The reported proteins were compared after independent q-value estimation at a global protein-level cutoff of 1% (Figure 4c). When applying peptide queries based on the HEK-293 sample-specific spectral library, all proteins could be recovered from the DIA data of the inter-laboratory SWATH-MS study. For the queries based on the CAL, a global set of 4989 proteins was inferred at 1% protein FDR. This corresponds to a protein-level recovery of roughly 50% compared to the CAL and is almost twice the number of proteins that could be inferred by using the sample-specific spectral library, indicating that the additional proteins were not identified in the sample-specific DDA runs or did not fulfill the requirements for peptide query parameter generation. For the HEK-293 subset of the CAL, 4841 proteins out of the 6019 proteins queried were confidently inferred. The relatively small discrepancy between the proteins inferred using the CAL and its HEK-293 subset illustrates the tradeoff of a larger but more comprehensive query space requiring strict multiple testing correction. The 380 (7.8% of total) proteins exclusively found with the HEK-293 subset illustrates a loss of sensitivity, while the additional 503 (10% of total) proteins illustrates the opportunity gained. Figures 4d-f illustrate the size and protein overlap between the different libraries, the global protein-level discriminant score distributions, and the reported protein overlap at 1% global protein-level FDR for the plasma dataset. Even though the subset of proteins that can be inferred in the plasma dataset is smaller compared to the ones from the inter-laboratory SWATH-MS study, the relative results are qualitatively similar. This analysis shows that large comprehensive spectral libraries can achieve sensitive results at appropriate error rate control. On the other hand, decreasing the number of peptide queries can lower the requirement for multiple testing adjustments at the potential cost of proteome coverage. The optimal tradeoff for a study depends on how well the spectral library represents the actual sample content.
Figure 4. Comparison between peptide queries with varying target prevalence.
a) Number and overlap between proteotypic proteins in the combined human assay library (CAL), a HEK-293 subset of the CAL (HEK), and a sample-specific spectral library (SSL). b) The discriminant score distributions illustrate the different π0 values between the CAL, HEK and SSL. c) Comparison of the sets of proteins inferred at 1% protein FDR in the global context of all 229 DIA runs of the SWATH-MS inter-laboratory comparison study using the CAL, HEK and SSL spectral libraries. Using the CAL and HEK spectral libraries, a substantially higher number and overlapping set of proteins can be inferred compared to using the SSL. The CAL enables the detection of 503 proteins that are not detectable when using the HEK subset library. In contrast, using the HEK subset library allows the detection of 380 proteins that are not detected using the CAL, despite all of the peptides for these proteins being part of the CAL. Panels d-f) show the corresponding results of the analysis on the plasma dataset.
Discussion
With the increasing numbers of peptides queried in samples acquired in data-independent acquisition mode by peptide-centric targeted data extraction, it is imperative to adopt strict quality assessment metrics such as the established criteria from spectrum-centric discovery proteomics to ensure reproducible reporting of results. Here, we have discussed the challenges associated with error rate control in the analysis of DIA data. We have demonstrated that the FDR should be controlled not only on peptide query-, but also on peptide- and protein-level in peptide-centric scoring workflows applying comprehensive spectral libraries. Furthermore, we propose the application of different context-dependent error rate estimation strategies. While the run-specific context offers per-run granularity, the experiment-wide context provides comparable result matrices across large heterogeneous datasets. The global context can be used to generate a list of detected peak groups, peptides, and inferred proteins that can be confidently detected in a study. We suggest that a practical method to control the error rate is to filter the result matrices generated from either the run-specific or experiment-wide contexts using the set of analytes confidently detected in the global context. We have shown that this results in a uniform set of inferred proteins with negligible accumulation of false positives over a large number of samples. The error rate control strategies we have described are implemented and available in an updated PyProphet version (Online Methods) and are available in Spectronaut 1113. Future developments might extend the statistical models to adjust probabilities for the detection of peptides and inference of proteins across multiple runs to improve detection sensitivity26,47. Other extensions and adaptations may be necessary if heterogeneous datasets, e.g. acquired on different instrument types, are analyzed together or if the parameters and assumptions of the algorithms are changed (Supplementary Note 5, Supplementary Figure 7). Despite the herein proposed strategies to control error rates in large-scale targeted proteomics experiments, the increased query space in repository-scale spectral libraries compromises the detection sensitivity. We have illustrated by means of the inter-laboratory SWATH-MS study37 and the plasma dataset38 that different spectral library specificity and comprehensiveness have profound effects on the importance of multiple-testing corrections and the respective analysis results. Therefore, it might be interesting for future applications to consider strategies for reducing the query space to provide an optimal tradeoff between proteome coverage and the fraction of undetectable targets. For this purpose, several different strategies have been suggested previously (Supplementary Note 6); however, further investigations are required to evaluate the optimal tradeoffs for different studies and future algorithmic development will continue to abolish the borders between spectrum-centric and peptide-centric scoring approaches to provide fully integrated workflows.
The development and application of DIA as an enabling tool in quantitative proteomics has undergone rapid expansion in recent years and this is set to continue for the foreseeable future. We hope that this article will serve to stimulate community discussion on these topics, and to aid researchers in choosing appropriate strategies for error rate control broadly improving the quality of data emerging from DIA-based quantitative proteomics studies.
Online Methods
PyProphet
We have implemented the described error rate control strategies in an updated and extended version of PyProphet11. PyProphet is a Python-based reimplementation of the mProphet32 algorithm originally developed for semi-supervised learning and statistical validation of targeted proteomics data. The PyProphet implementation reported here extends the original approach by the following options:
Semi-supervised learning
Instead of conducting independent iterations of learning and statistical validation separately per run, PyProphet conducts subsampling of paired target and decoy peak groups29 over all runs to learn a single, experiment-wide linear discriminant analysis (LDA) scoring model. From the LDA function, a discriminant score is derived by computation of the z-score using the decoy peak group mean and standard deviation as described previously32. The purpose of this integrated step is to ensure that the peak groups can be sorted according to their quality in a unified way across heterogeneous samples or samples of variable quality.
Statistical validation
In addition to the original parametric assumptions32, PyProphet now also supports non-parametric, empirical estimation of p-values43. To estimate q-values on different levels, PyProphet enables aggregation over peptide- or protein-level groups by selection of the best scoring peak group. For each level, q-values, FDR15/FNR19,20 or pFDR/pFNR21 are computed independently using the corresponding decoys as null model. For the different contexts, PyProphet supports different modes to either conduct q-value estimation per run (run-specific context), across all runs (experiment-wide context) or in a global fashion (global context).
Multi-run and high-throughput processing
To process large datasets, for example the inter-laboratory SWATH-MS study37, we improved the scalability of PyProphet under conditions where hundreds of runs each with a file size of 5 – 10 Gb need to be analyzed concurrently. The new PyProphet version is optimized for parallel processing in a cluster environment (IBM® Platform LSF™ or OpenLava) but can be readily adopted to other environments by Python extensions. Using subsampling and integrated scoring, q-value estimation can be conducted using very large numbers of peptide queries for hundreds of runs within hours using a common cluster or cloud environment: A full analysis of the 229 reports of OpenSWATH (9 GB per run) using 1-32 CPUs (depending on the individual step), 4-48 GB RAM (depending on the individual step) required a processing time of 1.5h, using several sequential and parallel jobs. Because the OpenSWATH results are stored as text files, the main requirement for the processing is throughput of filesystem input / output operations and temporary storage capacity.
Code availability
Our software is implemented in Python, available for all major platforms and released under the 3-clause BSD license. PyProphet is available along with detailed instructions from https://github.com/PyProphet. Further documentation of our workflow is available on http://openswath.org.
Analysis of the SWATH-MS inter-laboratory reproducibility dataset
Spectral library and peptide query parameter generation
Combined human assay library: The combined human assay library (CAL) for the 64 variable windows setting36 was filtered for proteotypic peptides and complemented by 30 additional SIS peptides as described previously37.
HEK-293 subset library: The HEK-293 subset library (HEK) was generated by filtering the CAL to only contain spectra from HEK-293 samples. The peptide query parameters were derived from the HEK-293 filtered spectral library as described previously36.
Sample-specific library: The sample-specific library (SSL) was generated from the spectra collected by six LC-MS/MS runs in DDA mode of the identical unfractionated HEK-293 tryptic digest, as described previously36.
Combined human + M. tuberculosis library: Based on the SpectraST consensus library of the CAL36 and the M. tuberculosis34,41 libraries, we generated a merged library by appending the M. tuberculosis to the CAL library using SpectraST (TPP 5.0). The protein identifiers were updated using the combined original FASTA files of the two libraries, to later exclude any shared peptides between the two organisms. Peptides and fragment ions were selected identically as described previously36,42 (msproteomicstools: master@c10a2b8) and OpenMS (version 2.1) was used with OpenSwathDecoyGenerator to generate combined target-decoy libraries (method: shuffle, similarity_threshold: 0.05, identity_threshold 0.7, exclude_similar: true, append: true).
DIA data analysis
The analysis of the inter-laboratory SWATH-MS dataset was conducted identically as described previously37. The SWATH-MS data analysis was performed using OpenSWATH (OpenMS v2.0) essentially as described10 with the following modified parameters: m/z extraction window = 75 ppm, RT extraction window = 900 seconds. The analysis was performed separately for the four different spectral libraries described above: combined human assay library, HEK-293 subset library, sample-specific library, and combined human + M. tuberculosis library.
Semi-supervised learning and statistical validation were performed using the above described extended version of PyProphet (PyProphet-cli v0.19 - https://github.com/PyProphet). PyProphet was run for all three available contexts to conduct q-value estimation per run (run-specific context), across all runs (experiment-wide context) or in a global fashion (global context), with a fixed λ of 0.4. The set of peptide peak groups used for learning the score weights of OpenSWATH sub-scores to produce a single discriminant score were sampled with a ratio ≈ 1/(no. of samples), for aggregated analysis of all sites a ratio of 0.005 was used.
A global “master list” of detected peak groups and proteins across the entire dataset was generated by filtering the results from the global context at 1% peptide query FDR and 1% protein FDR.
The results from the experiment-wide context were filtered on three different stringency levels: 1% peptide query FDR, 1% peptide query FDR and 1% protein FDR, and 1% peptide query FDR and additional filtering based on the global “master list” of peptide queries and proteins.
For the analysis of the three different libraries (Figure 4), separate scoring models were trained.
Analysis of the plasma dataset
The combined human assay library (CAL) for the 32 fixed windows setting36 was used to analyze the plasma dataset38 identically as described above with the following differences for OpenSWATH: m/z extraction window = 0.05 Da, RT extraction window = 600 seconds. The following set of scores was used: xx_lda_prelim_score, intensity_score, isotope_correlation_score, isotope_overlap_score library_corr, library_rmsd, log_sn_score, massdev_score, massdev_score_weighted, norm_rt_score, xcorr_coelution, xcorr_coelution_weighted, xcorr_shape, xcorr_shape_weighted.
The OpenSWATH results were filtered to only contain proteotypic peptides. To generate the results for the plasma subset analysis, the OpenSWATH results were filtered to only contain peptides mapping to proteins that were confidently detected (confidence threshold for inclusion in original library36) in at least one of the eight DDA plasma runs part of the CAL. This approach is equivalent to using a subset library for data extraction by OpenSWATH.
PyProphet was executed as described above, however the scoring model (LDA weights) of the plasma subset analysis was applied to the whole CAL analysis to ensure that the differences of the comparison originated only from the different library sizes. The analysis was conducted independently for both the parametric and the non-parametric methods.
Data availability
The raw data and processed results of the analysis of the SWATH-MS inter-laboratory reproducibility study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository50 (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD004884.
The processed results of the analysis of the twin plasma study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository50 (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD006625.
Supplementary Material
Acknowledgements
M.H. was supported by a grant from the Institut Mérieux. A.I.N. was founded by the National Institutes of Health (NIH grant R01GM094231). H.L.R was founded by the Swiss National Science Foundation (SNSF grant P2EZP3 162268). B.C.C. was supported by a Swiss National Science Foundation Ambizione grant (PZ00P3_161435). R.A. was supported by European Research Council grant ERC-2008-AdG 233226 and ERC-2014-AdG 70821 and, the Swiss National Science Foundation (grant #31003A-130530).
We would like to thank the authors of the SWATH-MS inter-laboratory study and of the human blood plasma dataset for providing the data to conduct this study. We would like to further thank the Scientific IT Support (ID SIS) of ETH Zurich for support and maintenance of the lab-internal computing infrastructure and the HPC team (Brutus).
Footnotes
Author contributions
G.R., I.B. and R.A. wrote the paper with feedback from all authors. G.R and B.C.C. developed the methods. I.B. analyzed the dataset. U.S. and G.R. implemented the PyProphet extension. M.H., C.H., Y.L., M.J.M., B.X.M, A.I.N., P.G.A.P., L.R., H.L.R., S.T. and Y.S.T. provided critical input on the project. B.C.C. and R.A. designed and supervised the study.
Competing financial interests
C.H. and S.T. are employees of SCIEX, which operates in the field covered by the article. L.R. is employee of Biognosys AG, which operates in the field covered by the article. R.A. holds shares of Biognosys AG. The remaining authors declare no competing financial interest.
References
- 1.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 2.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 3.Gillet LC, Leitner A, Aebersold R. Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annu Rev Anal Chem. 2016;9:449–472. doi: 10.1146/annurev-anchem-071015-041535. [DOI] [PubMed] [Google Scholar]
- 4.Ting YS, et al. Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Mol Cell Proteomics. 2015;14:2301–2307. doi: 10.1074/mcp.O114.047035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 6.Tsou C-C, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat Methods. 2015;12:1106–1108. doi: 10.1038/nmeth.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files. Nat Methods. 2015;12:1105–1106. doi: 10.1038/nmeth.3593. [DOI] [PubMed] [Google Scholar]
- 9.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717–O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 11.Teleman J, et al. DIANA-algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2014;31:555–562. doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- 12.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruderer R, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr SA, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 16.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 17.Choi H, Nesvizhskii AI. Semisupervised Model-Based Validation of Peptide Identifications in Mass Spectrometry-Based Proteomics. J Proteome Res. 2008;7:254–265. doi: 10.1021/pr070542g. [DOI] [PubMed] [Google Scholar]
- 18.Käll L, Storey JD, MacCoss MJ, Noble WS. Posterior Error Probabilities and False Discovery Rates: Two Sides of the Same Coin. J Proteome Res. 2007;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 19.Genovese C, Wasserman L. Operating characteristics and extensions of the false discovery rate procedure. J R Statist Soc B. 2002;64:499–517. [Google Scholar]
- 20.Iyer V, Sarkar S. An adaptive single-step FDR procedure with applications to DNA microarray analysis. Biom J. 2007;49:127–135. doi: 10.1002/bimj.200610316. [DOI] [PubMed] [Google Scholar]
- 21.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat. 2003;31:2013–2035. [Google Scholar]
- 22.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 24.Serang O, Noble W. A review of statistical methods for protein identification using tandem mass spectrometry. Stat Interface. 2012;5:3–20. doi: 10.4310/sii.2012.v5.n1.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The M, Tasnim A, Käll L. How to talk about protein-level false discovery rates in shotgun proteomics. Proteomics. 2016;16:2461–2469. doi: 10.1002/pmic.201500431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shteynberg D, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiter L, et al. Protein Identification False Discovery Rates for Very Large Proteomics Data Sets Generated by Tandem Mass Spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savitski MM, Wilhelm M, Hahne H, Kuster B, Bantscheff M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M114.046995. mcp.M114.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The M, MacCoss MJ, Noble WS, Käll L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J Am Soc Mass Spectrom. 2016;27:1–9. doi: 10.1007/s13361-016-1460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi H, Ghosh D, Nesvizhskii AI. Statistical Validation of Peptide Identifications in Large-Scale Proteomics Using the Target-Decoy Database Search Strategy and Flexible Mixture Modeling. J Proteome Res. 2007;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 31.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 32.Reiter L, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 33.Karlsson C, Malmström L, Aebersold R, Malmstrom J. Proteome-wide selected reaction monitoring assays for the human pathogen Streptococcus pyogenes. Nat Commun. 2012;3:1301. doi: 10.1038/ncomms2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert OT, et al. The Mtb Proteome Library: A Resource of Assays to Quantify the Complete Proteome of Mycobacterium tuberculosis. Cell Host Microbe. 2013;13:602–612. doi: 10.1016/j.chom.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picotti P, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberger G, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins BC, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nature communications. 2017 doi: 10.1038/s41467-017-00249-5. doi:(in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786–786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selevsek N, et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-MS. Mol Cell Proteomics. 2015;14 doi: 10.1074/mcp.M113.035550. mcp.M113.035550–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo T, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert OT, et al. Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium tuberculosis. Cell Host Microbe. 2015;18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Schubert OT, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 43.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serang O, Käll L. Solution to Statistical Challenges in Proteomics Is More Statistics, Not Less. J Proteome Res. 2015;14:4099–4103. doi: 10.1021/acs.jproteome.5b00568. [DOI] [PubMed] [Google Scholar]
- 45.Blattmann P, Heusel M, Aebersold R. SWATH2stats: An R/Bioconductor Package to Process and Convert Quantitative SWATH-MS Proteomics Data for Downstream Analysis Tools. PLOS ONE. 2016;11:e0153160. doi: 10.1371/journal.pone.0153160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou C-C, Tsai CF, Teo G, Chen YJ, Nesvizhskii AI. Untargeted, spectral library-free analysis of data independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics. 2016 doi: 10.1002/pmic.201500526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller A, Bader SL, Shteynberg D, Hood L, Moritz RL. Automated Validation of Results and Removal of Fragment Ion Interferences in Targeted Analysis of Data-independent Acquisition Mass Spectrometry (MS) using SWATHProphet. Mol Cell Proteomics. 2015;14:1411–1418. doi: 10.1074/mcp.O114.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta N, Pevzner PA. False Discovery Rates of Protein Identifications: A Strike against the Two-Peptide Rule. J Proteome Res. 2009;8:4173–4181. doi: 10.1021/pr9004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muntel J, et al. Advancing Urinary Protein Biomarker Discovery by Data-Independent Acquisition on a Quadrupole-Orbitrap Mass Spectrometer. J Proteome Res. 2015;14:4752–4762. doi: 10.1021/acs.jproteome.5b00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vizcaíno JA, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and processed results of the analysis of the SWATH-MS inter-laboratory reproducibility study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository50 (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD004884.
The processed results of the analysis of the twin plasma study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository50 (http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD006625.