Abstract
Recently the gene encoding a member of the RecQ helicase family, RecQ5, was cloned from the fruit fly, Drosophila melanogaster [J.J.Sekelsky, M.H.Brodsky, G.M.Rubin and R.S.Hawley (1999) Nucleic Acids Res., 27, 3762–3769]. The Drosophila RecQ5 transcript is alternatively spliced, like its human counterpart, to yield three protein isoforms. Two of these isoforms are almost identical and have a predicted molecular weight of 54 kDa. The third isoform is larger and contains, in addition to the helicase domain shared by all three isoforms, a long highly charged C-terminal region. A small isoform of the Drosophila RecQ5 protein (RECQ5) has been expressed in Escherichia coli and purified. The purified protein is a single-stranded DNA-stimulated ATPase (dATPase) and a 3′→5′ DNA helicase. Hydrolysis of the nucleotide cofactor is required for unwinding activity and dATP supported the unwinding reaction better than other NTPs. The turnover number for the single-stranded DNA-stimulated dATPase activity was 1380 min–1, ~1.5-fold higher than that observed for the ATPase activity (900 min–1). The purified protein catalyzed unwinding of partial duplex substrates up to at least 93 bp, however, unwinding of an 89 bp blunt duplex substrate was not detected.
INTRODUCTION
Three autosomal recessive human genetic disorders, Bloom Syndrome, Werner Syndrome and a subset of the Rothmund–Thomson Syndrome, have been linked to mutations in human homologs of the Escherichia coli recQ gene, the founding member of the RecQ family of DNA helicases (1). The proteins belonging to the RecQ family all contain an ∼450 amino acid core sequence containing the seven so-called ‘helicase’ motifs, including the DExH box in motif II. In addition, many of the RecQ family members contain N- and/or C-terminal sequences that flank the core RecQ helicase domain. So far no function(s) have been assigned to these regions except in the case of the human Wrn protein and the yeast Sgs1 protein. Both Wrn protein and Sgs1 contain long N- and C-terminal regions on either side of the helicase domain. Wrn protein has been shown to contain sequences homologous to RNase D in the N-terminal region (2,3) and the protein has been shown to possess an intrinsic exonuclease activity (4–9). The N-terminal region of the Sgs1 protein, on the other hand, has been shown to be critical for interacting with topoisomerases II and III (10–13). The function of the C-terminal region in Sgs1 protein remains unknown.
To date, recQ homologs have been identified in a wide range of organisms. Like bacteria, budding yeast and fission yeast have a single recQ homolog each, SGS1 and rqh1+, respectively (11,14). SGS1 was isolated originally as a suppressor of slow growth in top3 mutants (11). The completion of the Caenorhabditis elegans genome revealed the presence of four recQ homologs in this organism (15) and five recQ homologs have been identified in humans thus far (16). As indicated above, mutations in three of the human recQ homologs, BLM, WRN and RecQ4, have been linked to rare human genetic disorders. Although these syndromes are characterized by chromosomal instability and predisposition to malignancies in general, they differ from each other phenotypically at both the cellular and organismal level (17,18). The other two human recQ homologs, RecQ1 and RecQ5, have not been linked to any genetic disorders but roles in maintaining genomic stability seem likely based on studies with other recQ homologs.
Recently, the RecQ5 homolog in Drosophila melanogaster was identified and the gene has been cloned (19). Studies have shown that the transcript is alternatively spliced resulting in three different isoforms of the RECQ5 protein. Two isoforms are almost identical 54 kDa proteins and the other is a 121 kDa protein with a long C-terminal region of unknown function. Although the human RecQ5 protein was originally thought to consist of 410 amino acids that make up the core helicase domain (16), recent data suggests that it too has isoforms similar to those found in Drosophila (19,20). The sequence alignment of the human and fly proteins reveals significant similarity in the C-terminal region of the large isoforms in addition to strong homology within the helicase domain.
Transient expression of the human proteins showed nuclear localization of the large isoform (RecQ5β) but not the smaller isoforms in tissue culture cells (20). The large isoform of the Drosophila RECQ5 (RECQ5a) was also localized in the nucleus of Schneider2 cells when transiently expressed (19). In addition, human RecQ5β was shown to colocalize and coimmunoprecipitate with topoisomerases 3α and β, but not with topoisomerase I. However, the cellular role of RecQ5 remains unknown, as does the reason for multiple isoforms of this protein.
To begin an investigation into the role of RECQ5 in the cell, and to discover the significance of the multiple isoforms of this protein, we have purified and characterized RECQ5 from D.melanogaster. Here we present the results of expression, purification and initial biochemical characterization of the small isoform of RECQ5. The purified protein is a 3′→5′ DNA helicase that demonstrates a modest preference for dATP as a nucleotide cofactor over ATP. The dATPase (ATPase) activity is maximally stimulated by a single-stranded DNA (ssDNA) effector; double-stranded DNA (dsDNA) or RNA do not stimulate the dATPase activity. The purified enzyme unwinds partial duplex DNA substrates containing a 3′ ssDNA tail but does not unwind blunt duplex substrates. This protein has many of the biochemical characteristics of the Wrn helicase and the Blm helicase.
MATERIALS AND METHODS
Bacterial strains, plasmids and DNAs
Escherichia coli ER2566 and pTYB4 were from New England Biolabs. pTYB4-dRecQ5 was constructed by amplifying the Drosophila RecQ5 gene in a PCR with primers that contained NcoI and BsrBI sites and inserting the PCR product, after digestion with NcoI and BsrBI, between the NcoI and SmaI sites in pTYB4. This created a translational fusion of RECQ5 with the intein and chitin binding protein present in the pTYB4 vector. The RecQ5 gene in pTYB4-dRecQ5 was sequenced to eliminate the possibility of any mutations that may have been introduced by PCR amplification. The fusion protein was designed such that cleavage between RECQ5 and the intein will release RECQ5b protein (19) with a glycine in place of the last glutamic acid. The K49M mutant form of RECQ5 was constructed by PCR using primers that changed codon 49 from AAA to ATG. The resulting clone was sequenced to confirm the presence of the K49M mutation and the absence of other mutations. pFastBac was from Life Technologies. Poly(dT) (average range 1500–2500 bases) was from US Biochemicals. M13mp18 ssDNA was prepared as previously described (21). rRNA was from Boehringer Mannheim. All nucleotides were from Amersham Pharmacia Biotech.
Antibody
The RECQ5 antibody was raised in rabbits against the peptide CLGNPKEFKDTPKPQ, which corresponds to RECQ5 residues 241–255, a region of the protein between helicase-associated motifs III and IV.
Purification of RECQ5
The recombinant RECQ5-intein-chitin binding domain fusion protein was expressed in E.coli ER2566. Two liters of cells were grown at 42°C in LB media containing 100 µg/ml ampicillin to an OD600 of ∼1.5. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mM and induction was at 27°C for 4 h. The cells were harvested by centrifugation, suspended in 50 ml of lysis buffer (20 mM Tris–HCl pH 8, 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 20 µM phenylmethylsulfonyl fluoride) and lysed by sonication (20 bursts at 10 s intervals). The lysate was clarified by centrifugation at 27 000 g for 45 min at 4°C. The soluble cell lysate was loaded on a chitin column (New England Biolabs, packed bed volume = 5 ml) and washed extensively (∼25 column volumes) with column buffer (20 mM Tris–HCl pH 8, 500 mM NaCl, 1 mM EDTA). Cleavage of the intein from RECQ5 was induced by incubating the column in cleavage buffer (column buffer + 50 mM DTT) at 16°C for 24 h. The protein was eluted with column buffer and 1 ml fractions were collected. The fractions were analyzed by SDS–PAGE and fractions containing RECQ5 were pooled (∼5 ml). The pool was dialyzed against heparin agarose column buffer (20 mM Tris–HCl pH 8, 100 mM NaCl, 0.1 mM EDTA, 0.01% NP-40, 10% glycerol) to reduce the NaCl concentration to 100 mM and to include 10% glycerol and 0.01% NP-40. The dialyzed pool was loaded onto a heparin agarose column (packed bed volume = 1 ml) equilibrated with heparin agarose column buffer containing 100 mM NaCl. The protein was eluted with a gradient from 100 to 600 mM NaCl in column buffer. RECQ5 eluted at ~350 mM NaCl and was >90% homogeneous as determined by SDS–polyacrylamide gel analysis. Individual fractions were also analyzed using a helicase activity assay (described below) to identify the fractions containing active helicase and lacking contaminating exonuclease. Fractions with no apparent exonuclease activity were pooled and dialyzed against storage buffer (50% glycerol, 20 mM Tris–HCl pH 8, 200 mM NaCl, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM DTT). The concentration of purified RECQ5 was determined using the Bradford protein assay (Bio-Rad) with bovine serum albumin as the standard.
DNA helicase substrates
The partial duplex DNA helicase substrates used in this study were prepared as previously described (22). The 20 bp partial duplex substrate was generated by radioactively labeling a 20 base oligonucleotide using T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP followed by annealing to M13mp18 ssDNA. The 42 and 93 bp partial duplex substrates were generated using 40 and 91 base oligonucleotides, respectively. The appropriate oligonucleotide was annealed to circular M13mp18 ssDNA and radioactively labeled using a DNA polymerase I (large fragment)-catalyzed extension reaction in the presence of [α-32P]dCTP. The blunt duplex DNA substrate was prepared by digesting pLitmus28 (New England Biolabs) with RsaI to completion and isolating the resulting 89 bp fragment using Geneclean II (Bio101). The phosphate on each 5′-end of the blunt ended fragment was removed using Calf Intestinal Phosphatase (Boehringer Mannheim) and radioactive labeling was accomplished using T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP.
The helicase directionality substrate (see Fig. 5) was constructed using the 91 base oligonucleotide which is complementary to M13mp18 ssDNA between bases 2490 and 2581. After annealing the oligonucleotide to M13 ssDNA, the partial duplex DNA was digested with ClaI. A DNA polymerase I (large fragment)-catalyzed extension reaction was carried out in the presence of [α-32P]dCTP and dGTP to fill in the staggered ends resulting from the ClaI digest and the 3′-end of the annealed oligonucleotide. The resulting DNA molecule has a long internal ssDNA region and partial duplex regions (43 bp on the 3′-end and 52 bp on the 5′-end) on both ends.
Figure 5.
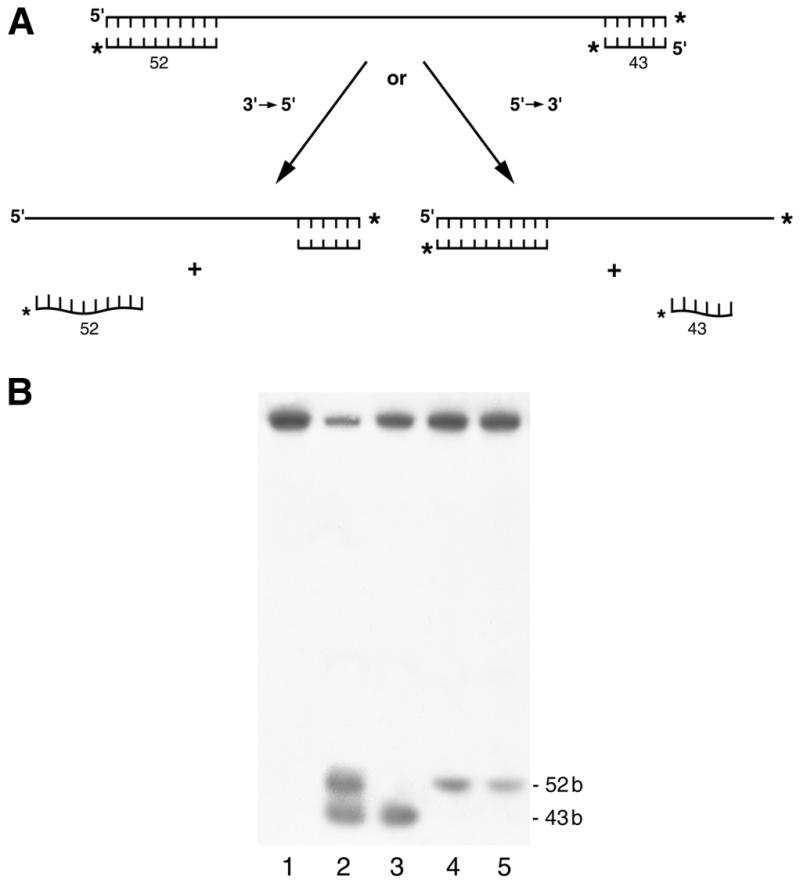
RECQ5 unwinds DNA in the 3′→5′ direction. Helicase assays were performed as described in Materials and Methods using the substrate for determining the polarity of unwinding. (A) Schematic depiction of the helicase polarity substrate and the anticipated results. (B) Lanes 1 and 2 contained no helicase, lane 2 was heat denatured by boiling for 3 min; lane 3, helicase I at 14 nM; lane 4, helicase II at 5 nM; lane 5, RECQ5 at 850 nM.
Helicase assays
Helicase reaction mixtures (20 µl) contained 25 mM Tris–HCl pH 8, 20 mM NaCl, 1 mM DTT, 50 µg/ml bovine serum albumin, 5 mM MgCl2, 3 mM ATP (or dATP) and the indicated DNA substrate (~1.7 µM nucleotide phosphate for partial duplex substrates and 0.023 µM nucleotide phosphate for the blunt duplex substrate). The reaction was initiated with enzyme and incubation was at 30°C for 30 min unless otherwise indicated. The reactions were quenched with 10 µl of stop solution (37.5% glycerol, 50 mM EDTA, 0.05% each of xylene cyanol and bromophenol blue, 0.3% SDS). Reaction products were resolved on a non-denaturing polyacrylamide gel (8% for partial duplex substrates and 15% for blunt duplex substrate, 20:1 cross-linking ratio). The results were visualized using a phosphorimager and analyzed with ImageQuant software (Molecular Dynamics).
ATPase/dATPase assays
The ssDNA-stimulated hydrolysis of ATP and dATP was measured as previously described (23). The reaction mixtures were the same as helicase reaction mixtures except M13mp18 ssDNA (at 30 µM nucleotide phosphate) was substituted for the [32P]DNA substrate and [α- 32P]ATP or [α-32P]dATP was substituted for unlabeled ATP or dATP. For Km measurements, the reaction volume was 20 µl and two 5 µl aliquots were removed after a 10 min incubation at 30°C. The ATP (or dATP) concentration was varied between 50 µM and 2.5 mM. For kcat measurements, the reaction volume was 30 µl and 5 µl aliquots were removed at 2, 4, 6, 8 and 10 min when ssDNA was present, and 10, 20, 30, 40 and 50 min when ssDNA was absent from the reaction mixtures. Reactions were initiated by the addition of enzyme (5 nM for Km experiments, 14.5 and 50 nM for kcat experiments in the presence of ssDNA, 50 nM for kcat experiments in the absence of ssDNA). The aliquots were quenched with 5 µl ATPase stop solution (33 mM EDTA, 7 mM ATP, 7 mM ADP). The resulting 10 µl aliquots were spotted on polyethyleneimine cellulose thin layer chromatography plates and developed in 1.0 M HCOOH, 0.8 M LiCl. Visualization was accomplished using a phosphorimager and analyzed with ImageQuant software (Molecular Dynamics).
For experiments involving various DNA effector molecules, solutions of M13mp18 ssDNA, poly(dT), supercoiled plasmid (pFastBac), linear dsDNA (pFastBac digested with EcoRV) and rRNA were prepared at 300, 150, 75, 37.5, 18.75, 9.38, 4.69 and 2.34 µM (DNA phosphate) and used at 1:10 dilution in reactions to achieve the final concentrations indicated. The reactions were initiated by the addition of enzyme (5 nM) and were incubated at 30°C for 35 min. The results are plotted as a function of increasing DNA (RNA) concentration (nucleotide phosphate).
RESULTS
A new member of the RecQ family of DNA helicases, RecQ5, has been identified in both Drosophila and humans (16,19). The role of RecQ5 protein is currently unknown but it seems likely that this protein will be involved in maintaining genomic stability based on the proposed roles for other RecQ helicases (18,24). Of special interest is the fact that the RecQ5 mRNA is alternatively spliced in both humans and flies to yield three different isoforms of the RecQ5 protein. This is the second example of a RecQ DNA helicase that exists in multiple isoforms in the cell (25) and we have initiated a biochemical characterization of the isoforms of the Drosophila homologs of RecQ5 by purifying and characterizing a small isoform.
Purification of RECQ5
The gene encoding the small isoform of RECQ5 was cloned in pTYB4 to construct a translational fusion of RECQ5 with a C-terminal intein and chitin binding domain. The fusion protein was expressed and purified from E.coli ER2566. The purification (Materials and Methods) consisted of two columns, the first a chitin column to separate the expressed fusion protein from the bulk of the cellular proteins and then a heparin agarose column to eliminate the contaminants present in the chitin column elution. The purified protein was >90% homogeneous (Fig. 1A) and had a Mr of ~54 kDa consistent with its identity as a small isoform of RECQ5. In addition, the purified protein reacted with a polyclonal antibody directed against the RECQ5 protein (Fig. 1B).
Figure 1.
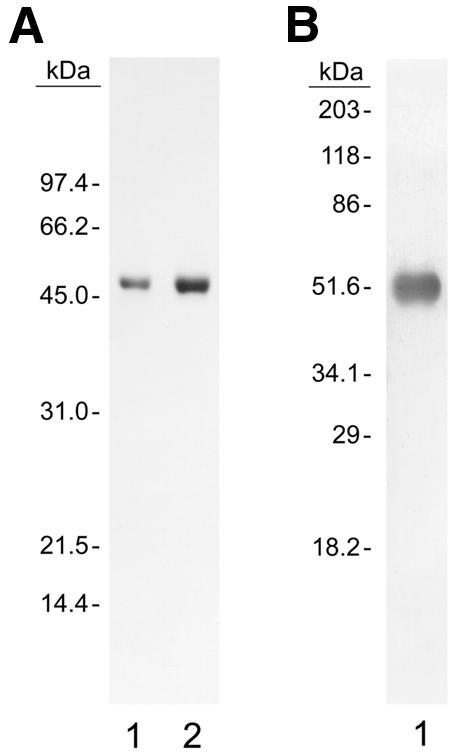
Analysis of purified RECQ5 protein. (A) Purified RECQ5 was resolved on a 9.6% SDS–polyacrylamide gel and stained with Coomassie blue. Lane 1, 2 µg purified RECQ5; lane 2, 4 µg of purified RECQ5. (B) An identical gel was run using 2 µg of purified RECQ5, transferred to nitrocellulose and probed with antibody directed against RECQ5.
Requirements for the helicase activity of RECQ5 protein
Previous reports suggested that RECQ5 exhibited helicase activity (26). We sought to extend this observation using purified protein by characterizing the unwinding reaction catalyzed by RECQ5. Our initial characterization used a 42 bp partial duplex substrate. The unwinding reaction catalyzed by RECQ5 required a nucleoside 5′-triphosphate (in this case ATP) as expected (Fig. 2). Omitting ATP or Mg++ from the reaction mixture eliminated helicase activity (Fig. 2, lanes 4 and 6). Moreover, the hydrolysis of the nucleotide cofactor was essential for unwinding since none of the poorly hydrolyzable or non-hydrolyzable ATP analogs (ATPγS, AMP-PNP and AMP-PCP) could support the helicase activity (Fig. 2, lanes 8, 10 and 12). The addition of ATPγS to the complete reaction inhibited the helicase, as expected (Fig. 2, lane 7), since ATPγS is presumably bound by the protein but not hydrolyzed. Interestingly, the other two ATP analogs (AMP-PNP and AMP-PCP) did not have the same effect. Their addition to a complete unwinding reaction did not abolish unwinding of the 42 bp partial duplex substrate (Fig. 2, lanes 9 and 11). We presume that AMP-PNP and AMP-PCP fail to bind RECQ5 and thus do not act as competitive inhibitors of the ATPase/helicase activity.
Figure 2.
ATP hydrolysis is required for helicase activity. Helicase reaction mixtures (20 µl) contained the 42 bp partial duplex substrate, 1.5 µM RECQ5, 20 mM NaCl, 5 mM MgCl2 and 3 mM ATP unless otherwise specified. The reactions were initiated by the addition of enzyme and were incubated at 30°C for 30 min. Lanes 1 and 2 were controls and contained no enzyme, the reaction shown in lane 2 was boiled for 3 min; lane 3, complete reaction; lane 4, MgCl2 was omitted; lane 5, MgCl2 was omitted and 5 mM EDTA was included; lane 6, ATP was omitted; lane 7, 3 mM ATPγS was included; lane 8, 3 mM ATPγS was substituted for ATP; lane 9, 3 mM AMP-PCP was included; lane 10, AMP-PCP was substituted for ATP; lane 11, 3 mM AMP-PNP was included; lane 12, 3 mM AMP-PNP was substituted for ATP. The substrate and product are shown schematically on the left. An asterisk denotes a radioactive label.
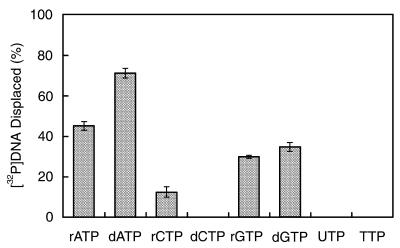
To determine the nucleotide preference of RECQ5, all eight nucleotides were tested for their ability to support unwinding of the 42 bp partial duplex substrate (Fig. 3). The results indicate that, under the reaction conditions used, >70% of the substrate was unwound in the presence of dATP. ATP, dGTP, GTP and CTP also supported the helicase reaction, although to a lesser extent than dATP, with ATP being second best (∼45%) followed by dGTP (∼35%), GTP (∼30%) and finally CTP (∼10%). None of the other nucleotides could support the helicase reaction. Since dATP was preferred over ATP as a nucleotide cofactor in the unwinding reaction we have used dATP in many of the experiments reported below.
Figure 3.
Nucleotide cofactors. Helicase assays were as described in Materials and Methods using the 42 bp partial duplex substrate. Each reaction contained 680 nM RECQ5 and one of the eight nucleotides at a final concentration of 3 mM. The data presented is the average of three independent experiments. Error bars represent the standard deviation about the mean.
The NTPase activity of RECQ5
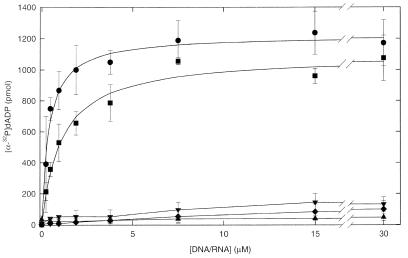
The NTPase activity of helicases is generally strongly stimulated by the presence of a nucleic acid cofactor (27). To determine the nucleic acid cofactor preference of RECQ5, dATPase reactions were evaluated using a variety of nucleic acid effectors (Fig. 4). Circular M13mp18 ssDNA, poly(dT), linear dsDNA, supercoiled plasmid DNA and rRNA were tested as effectors of the dATPase reaction at concentrations ranging from 0 to 30 µM nucleotide phosphate. In the absence of a nucleic acid effector dATPase activity could be detected. However, the rate of hydrolysis was very low (~1100-fold lower than hydrolysis in the presence of ssDNA). In the presence of rRNA or dsDNA (both supercoiled plasmid and linear dsDNA) the dATPase activity of the protein was slightly higher than that observed in the absence of effector (Fig. 4). Both ssDNA cofactors, M13mp18 ssDNA and poly(dT), strongly stimulated the dATPase activity of the protein. The stimulation was not significantly different when using circular M13mp18 ssDNA or poly(dT). Essentially identical results were obtained when ATP was substituted for dATP in these reactions.
Figure 4.
The dATPase activity is stimulated by ssDNA. dATPase reactions were as described in Materials and Methods. In each reaction RECQ5 was present at 5 nM, and dATP was included at a final concentration of 2 mM. Circles, M13 ssDNA; squares, poly(dT); inverse triangles, linear dsDNA; triangles, rRNA; diamonds, supercoiled dsDNA. The data represent the average of at least three independent experiments. The equation for a rectangular hyperbola was fitted to the data obtained for M13 ssDNA and poly(dT) (solid lines). Error bars represent the standard deviation about the mean.
The steady-state kinetic parameters, Km and kcat, of the ATPase and dATPase reactions catalyzed by RECQ5, were measured in the presence and absence of M13mp18 ssDNA (Table 1). For determination of Km values, the ATP (dATP) concentration was varied from 50 µM to 2.5 mM. The Km values were 250 and 150 µM for dATP and ATP, respectively, in the presence of ssDNA. The low rate of NTP hydrolysis in the absence of ssDNA made it technically difficult to determine Km values under these conditions. kcat values were determined in the absence and in the presence of the ssDNA cofactor in reactions that contained 2 mM ATP or dATP (∼10× Km). The kcat values in the absence of ssDNA were 15 ± 1 and 2.6 ± 0.1 min–1 for dATPase and ATPase activities, respectively. In the presence of ssDNA the hydrolysis rates for both nucleotides increased dramatically to 1380 ± 120 and 900 ± 120 min–1. Thus, RECQ5 hydrolyzed dATP more rapidly than ATP regardless of the presence of ssDNA.
Table 1. Steady-state kinetic constants for ssDNA-stimulated ATP (dATP) hydrolysis catalyzed by RECQ5.
| ssDNA | ATP | dATP | ||
| |
– |
+ |
– |
+ |
|
kcat (min–1) |
2.6 ± 0.1 |
900 ± 120 |
15 ± 1 |
1380 ± 120 |
| Km (µM) | nd | 150 ± 14 | nd | 250 ± 50 |
ATPase (dATPase) assays were as described in Materials and Methods. The results represent the average of at least three independent determinations ± the standard deviation. nd, not determined.
The helicase activity of RECQ5
All RecQ homologs characterized so far have been shown to unwind duplex DNA with a 3′→5′ directionality. The polarity of RECQ5 was determined using the substrate shown in Figure 5A. A helicase with 5′→3′ polarity, like E.coli DNA helicase I (28), will catalyze displacement of the 43 nt DNA fragment (Fig. 5B, lane 3). A helicase with 3′→5′ polarity, like E.coli helicase II (22), will catalyze displacement of the 52 nt DNA fragment (Fig. 5B, lane 4). The data shown in Figure 5B indicate that RECQ5 unwinds duplex DNA with a 3′→5′ polarity (lane 5).
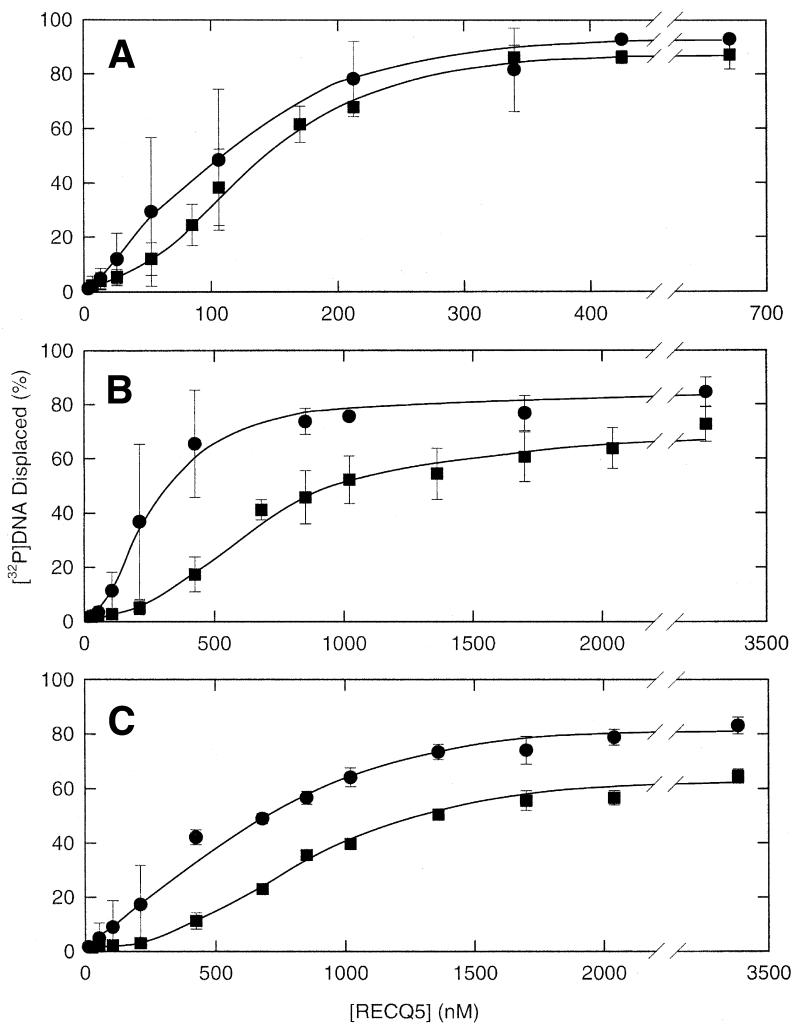
The helicase reaction catalyzed by RECQ5 was further investigated using three partial duplex substrates with increasing duplex regions (20, 42 and 93 bp) (Fig. 6A, B and C) and a blunt duplex substrate (89 bp, data not shown). Both ATP and dATP were used as nucleotide cofactors in these reactions since dATP was hydrolyzed faster than ATP. The 20 bp partial duplex substrate was unwound with essentially the same efficiency in the presence of either ATP or dATP. However, both the 42 and 93 bp partial duplex substrates were unwound more efficiently in the presence of dATP than in the presence of ATP. In addition, unwinding of the longer partial duplex DNA substrates required a higher protein concentration than did unwinding of the 20 bp partial duplex substrate.
Figure 6.
Unwinding of partial duplex substrates. Partial duplex substrates containing three different lengths of the duplex region were used in helicase assays to characterize the unwinding reaction catalyzed by RECQ5. Helicase reaction mixtures were as described in Materials and Methods and unwinding reactions were initiated by the addition of enzyme. Each data point is the average of at least three independent experiments. Circles, unwinding measured in the presence of dATP; squares, unwinding measured in the presence of ATP. Error bars represent the standard deviation about the mean.
The 89 bp blunt duplex substrate was not unwound at any concentration of RECQ5 tested (up to 3.5 µM). Since the protein was clearly able to unwind duplex regions of this length (i.e. the 93 bp partial duplex substrate), this suggests that the protein requires a 3′ ssDNA tail to efficiently load onto and unwind a DNA substrate. This is in contrast to the RecQ helicase from E.coli which has been shown to unwind partial duplex and blunt duplex substrates with essentially equal efficiency (29).
To ensure that the helicase reaction described above was due to the activity of RECQ5 and not a minor contaminant present in the preparations of RECQ5, a mutant form of RECQ5 was purified and characterized. RECQ5 contains the seven helicase-associated motifs commonly found in Superfamily II DNA helicases. Motif I contains the highly conserved GXGKT/S sequence found in all DNA helicases (30). The invariant lysine residue in motif I (K49) was changed to methionine by site-directed mutagenesis to construct the K49M mutant form of RECQ5. This protein was expressed and purified using exactly the same procedure used to express and purify the wild-type protein. The purified RECQ5-K49M protein lacked dATPase (ATPase) activity and failed to catalyze an unwinding reaction (data not shown). Thus, the helicase activity measured using purified RECQ5 must be an intrinsic activity of the protein and not due to a minor contaminant present in the preparations of purified protein.
DISCUSSION
The mRNA product of the Drosophila RecQ5 gene is alternatively spliced to produce three isoforms of the RECQ5 protein; two small (54 kDa) isoforms differing only in the last few amino acids and a large isoform (121 kDa) with a C-terminal extension containing a large percentage of charged residues. The mRNA transcripts of all three isoforms are present in all developmental stages examined, from embryos to adults (J.J.Sekelsky, unpublished results), and preliminary data from immunoblotting experiments suggests that adult flies express the small isoform(s) as well as the large isoform of the RECQ5 protein (M.Adams, personal communication). The human RECQ5 large isoform contains several potential nuclear localization signals in the C-terminus whereas the smaller isoforms do not. This is in agreement with the results reporting nuclear localization of the large isoform but not the small isoforms (20). The fly large isoform also localizes to the nucleus when transiently expressed (19). However, the cellular localization of the small isoforms has not been determined. In contrast to the human RECQ5, both of the small isoforms in the fly contain a putative C-terminal nuclear localization signal (KRPK or RPKK at amino acids 432 or 433) which may indicate nuclear localization for these proteins. Further studies are necessary to investigate the expression levels of the three isoforms in the tissues of the whole organism as well as subcellular localization of Drosophila RECQ5.
Here, we report the purification of a small isoform of the protein that has been shown to catalyze a 3′→5′ DNA helicase reaction. The unwinding polarity is consistent with the fact that RECQ5 has been classified as a member of the RecQ helicase family based on sequence similarity with other RecQ helicases and it is likely that all three isoforms of this protein will catalyze a 3′→5′ helicase reaction. The impact of the C-terminal extension present in the large isoform, if any, on the helicase reaction catalyzed by RECQ5 is currently under investigation.
As expected, purified RECQ5 protein also catalyzed a ssDNA-stimulated ATPase (dATPase) reaction. Somewhat surprising was the fact that the kcat for dATP hydrolysis was ∼1.5-fold higher than for ATP hydrolysis suggesting that dATP may be the preferred NTP for this protein. However, since the Km for dATP was slightly higher than the Km for ATP (250 versus 150 µM) in the hydrolysis reaction, and ATP is more abundant in the cell than dATP, it is possible that ATP is used in vivo. Nonetheless, both dATP and ATP were used in the characterization of the unwinding reaction presented here. It should be noted that other helicases have also been shown to utilize an NTP other than ATP as the source of energy for the helicase reaction. For example, the bacteriophage T7 gene 4 protein utilizes TTP (23). The Wrn protein, another member of the RecQ family of helicases, has also been shown to utilize dATP preferentially in vitro as the nucleotide substrate for its helicase reaction (5). Finally, it has been shown that E.coli RecQ hydrolyzes dATP as well as ATP, although recently Harmon and Kowalczykowski (31) have shown that ATP supports the initial rate and extent of RecQ-catalyzed unwinding ~2-fold better than dATP. Our data with RECQ5 suggest that dATP supports the unwinding of duplex DNA better than ATP. The biological significance of this observation, if any, remains unknown.
In the absence of an appropriate DNA cofactor (effector), the rate of RECQ5-catalyzed ATP (dATP) hydrolysis was quite low as expected. Both circular M13 ssDNA and linear poly(dT) proved to be good effectors of the dATPase reaction with no significant difference in the Vmax reached at high concentrations of the effector. The fact that the kDNA (effector concentration required for one-half maximal velocity) for poly(dT) was ~3-fold higher than that for M13 ssDNA (1.06 versus 0.38 µM) may suggest RECQ5 has a higher affinity for M13 DNA due to the presence of secondary structure in M13 ssDNA. An alternative explanation would suggest that RECQ5 translocates processively along ssDNA using NTP hydrolysis to fuel translocation. In this case, a circular molecule lacking ends might support a higher rate of NTP hydrolysis than a linear molecule since the protein would have to dissociate and seek a new effector molecule less frequently than would be the case on a linear DNA effector. In either case it should be possible to ‘drive’ the reaction in the presence of poly(dT) to the same Vmax as the reaction in the presence of M13 ssDNA, which was observed at high concentrations of poly(dT). Supercoiled DNA, linear duplex DNA and RNA all proved to be extremely poor effectors of the ATPase reaction suggesting that RECQ5 did not interact productively with any of these RNA/DNA molecules. The E.coli RecQ protein has been shown to unwind duplex DNAs with blunt ends. Apparently, the RECQ5 protein does not share this property with E.coli RecQ. This observation is consistent with the fact that the RECQ5 protein did not unwind blunt duplex substrates.
The RECQ5 protein catalyzed the unwinding of partial duplex substrates containing duplex regions as long as 93 bp; longer partial duplex DNAs have not been tested as substrates for this protein at this time. The enzyme failed to unwind an 89 bp blunt duplex DNA suggesting that it does not bind and initiate unwinding at a blunt end. This is consistent with the results obtained in the dATPase assay and, thus, a 3′ ssDNA tail appears to be essential for unwinding. This region of ssDNA may provide a binding/loading site for RECQ5 as is the case for many other DNA helicases. As indicated above, dATP hydrolysis supported duplex DNA unwinding better than ATP hydrolysis, at least with the 42 and 93 bp partial duplex DNA substrates (see Fig. 6). We observed no significant difference between ATP and dATP when unwinding of a 20 bp partial duplex substrate was measured. This may be due to RECQ5 not catalyzing a bona fide unwinding reaction using this particular substrate (see below).
The unwinding reaction catalyzed by RECQ5 required unexpectedly high protein concentrations, particularly when the 42 and 93 bp partial duplex substrates were used. There are several possible explanations for this result. First, since this is a recombinant protein isolated from E.coli it is possible that our preparations contain significant amounts of inactive protein. We do not favor this explanation since the kcat for ATP (dATP) hydrolysis is similar to that measured for other members of the RecQ family. This would not be the case if a significant fraction of the protein were inactive. Moreover, multiple preparations of RECQ5 exhibited nearly identical kcat values in the ATP (dATP) hydrolysis assay and comparable unwinding activities. However, we cannot completely rule out the possibility that a large fraction of the protein is inactive as a helicase but active as an ATPase. Secondly, it is possible that RECQ5 requires another protein to serve as an accessory protein for optimal helicase activity. For example, the Wrn protein displays nearly an absolute requirement for hRPA when the unwinding of long partial duplex DNA substrates (≥69 bp partial duplex) is measured (32). Preliminary experiments with yeast RPA did not alter the requirement for high concentrations of RECQ5 (A.Z.Ozsoy and S.W.Matson, unpublished observations). However, we have not tested RPA from Drosophila and, therefore, an RPA requirement cannot be ruled out. It is also possible that some other stimulatory protein that remains to be identified is required. Thirdly, it is possible that RECQ5 prefers a substrate other than the partial duplex substrates tested here. Many helicases are known to have a preference for a DNA substrate containing a fork. A forked DNA has not yet been fully tested as a substrate for RECQ5. Finally, and perhaps the most likely alternative, RECQ5 may require multimerization to catalyze unwinding of duplex DNA. This is an intriguing possibility for which there is some experimental support. Recent studies have indicated that the Blm helicase, another member of the RecQ family, is capable of forming hexamers which may be the active agent of unwinding (33). This may also be true for E.coli RecQ helicase (31). If RECQ5 also formed hexamers and the Kd for multimerization were high, at least in vitro, then a high concentration of RECQ5 would be required to unwind duplex DNA. It should be noted that in the presence of ATP the titration of RECQ5 displays a significant sigmoidal character consistent with the possibility of multimerization. Interestingly, this sigmoidicity was reduced when dATP was used as the NTP cofactor. If an NTP was required for multimerization, and dATP was more effective than ATP, then just such a result would be expected. Additional biophysical studies will be required to determine whether or not RECQ5 is active as a multimer.
It is important to note that unwinding of the 20 bp partial duplex substrate is indifferent to the NTP present in the reaction and fails to display a sigmoidal response to increasing protein concentration. Moreover, the reaction requires significantly less protein than does unwinding of the longer partial duplex DNA substrates. The reaction observed in this case may not represent a bona fide unwinding reaction. The RECQ5 protein may be simply ‘pushing’ the short oligonucleotide off the DNA as a consequence of active translocation along the DNA molecule. This has been observed for the PriA helicase from E.coli when short partial duplex substrates were used (34). If this is the case, then the protein may be capable of translocation along the DNA as a monomer. These issues remain to be resolved.
This study is the first extensive biochemical characterization of a RecQ homolog from Drosophila, as well as the first characterization of a RecQ5 protein. Biochemical characterization of the large isoform of this protein and comparison of the two isoforms together with genetic data will shed light on the role of RECQ5 in the cell.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Susan Whitfield for help with preparation of the artwork and members of the laboratory for helpful discussions. This investigation was supported by National Institutes of Health Grant GM33476 to S.W.M.
References
- 1.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 2.Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mushegian A.R., Douglas,E., Bassett,J., Boguski,M.S., Bork,P. and Koonin,E.V. (1997) Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc. Natl Acad. Sci. USA, 94, 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen J.-C., Gray,M.D., Oshima,J., Kamath-Loeb,A.S., Fry,M. and Loeb,L.A. (1998) Werner syndrome protein I. DNA helicase and DNA exonuclease reside on the same polypeptide. J. Biol. Chem., 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- 5.Shen J.-C., Gray,M.D., Oshima,J. and Loeb,L.A. (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res., 26, 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S., Li,B., Gray,M.D., Oshima,J., Mian,I.S. and Campisi,J. (1998) The premature ageing syndrome protein, WRN, is a 3′ to 5′ exonuclease. Nature Genet., 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamath-Loeb A.S., Shen,J.-C., Loeb,L.A. and Fry,M. (1998) Werner syndrome protein III. Characterization of the integral 3′ to 5′ exonuclease. J. Biol. Chem., 273, 34145–34150. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N., Shiratori,M., Goto,M. and Furuichi,Y. (1999) Werner syndrome helicase contains a 5′ to 3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res., 27, 2361–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J.-C. and Loeb,L.A. (2000) Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res., 28, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E.coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- 11.Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricke W.M., Kaliraman,V. and Brill,S.J. (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem., 276, 8848–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett R.J., Noirot-Gros,M.-F. and Wang,J.C. (2000) Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem., 275, 26898–26905. [DOI] [PubMed] [Google Scholar]
- 14.Stewart E., Chapman,C.R., Al-Khodairy,F., Carr,A.M. and Enoch,T. (1997) rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J., 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusano K., Berres,M.E. and Engels,W.R. (1999) Evolution of the RECQ family of helicases: a drosophila homolog, Dmblm, is similar to the human Bloom syndrome gene. Genetics, 151, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitao S., Ohsugi,I., Ichikawa,K., Goto,M., Furuichi,Y. and Shimamoto,A. (1998) Cloning of the two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics, 54, 443–452. [DOI] [PubMed] [Google Scholar]
- 17.Lindor N.M., Furuichi,Y., Kitao,S., Shimamoto,A., Arndt,C. and Jalal,S. (2000) Rothmund–Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet., 90, 223–228. [DOI] [PubMed] [Google Scholar]
- 18.van Brabant A.J., Stan,R. and Ellis,N.A. (2000) DNA helicases, genomic instability, and human genetic disease. Annu. Rev. Genomics Hum. Genet., 1, 409–459. [DOI] [PubMed] [Google Scholar]
- 19.Sekelsky J.J., Brodsky,M.H., Rubin,G.M. and Hawley,R.S. (1999) Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res., 27, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimamoto A., Nishikawa,K., Kitao,S. and Furuichi,Y. (2000) Human RecQ5β, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res., 28, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechner R.L. and Richardson,C.C. (1983) A preformed, topologically stable replication fork. J. Biol. Chem., 258, 11185–11196. [PubMed] [Google Scholar]
- 22.Matson S.W. (1986) Escherichia coli helicase II (uvrD gene product) translocates unidirectionally in a 3′ to 5′ direction. J. Biol. Chem., 261, 10169–10175. [PubMed] [Google Scholar]
- 23.Matson S.W. and Richardson,C.C. (1983) DNA-dependent nucleoside 5′-triphosphatase activity of the Gene4 protein of bacteriophage T7. J. Biol. Chem., 258, 14009–14016. [PubMed] [Google Scholar]
- 24.Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- 25.Wang W.-S., Seki,M., Yamaoka,T., Seki,T., Tada,S., Katada,T., Fujimoto,H. and Enomoto,T. (1998) Cloning of two isoforms of mouse DNA helicase Q1/RecQL cDNA; α form is expressed ubiquitously and β form specifically in the testis. Biochim. Biophys. Acta, 1443, 198–202. [DOI] [PubMed] [Google Scholar]
- 26.Jeong S.M., Kawasaki,K., Juni,N. and Shibata,T. (2000) Identification of Drosophila melanogaster RECQE as a member of a new familiy of RecQ homologues that is preferentially expressed in early embryos. Mol. Gen. Genet., 263, 183–193. [DOI] [PubMed] [Google Scholar]
- 27.Matson S.W., Bean,D.W. and George,J.W. (1994) DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays, 16, 13–22. [DOI] [PubMed] [Google Scholar]
- 28.Lahue E.E. and Matson,S.W. (1988) Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem., 263, 3208–3215. [PubMed] [Google Scholar]
- 29.Umezu K. and Nakayama,H. (1993) RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J. Mol. Biol., 230, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 30.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 31.Harmon F.G. and Kowalczykowski,S.C. (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem., 276, 232–243. [DOI] [PubMed] [Google Scholar]
- 32.Brosh R.M., Orren,D.K., Nehlin,J.O., Ravn,P.H., Kenny,M.K., Machwe,A. and Bohr,V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- 33.Karow J.K., Newman,R.H., Freemont,P.S. and Hickson,I.D. (1999) Oligomeric ring structure of the Bloom’s syndrome helicase. Curr. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- 34.Lee M.S. and Marians,K.J. (1990) Differential ATP requirements distinguish the DNA translocation and DNA unwinding activities of the Escherichia coli PRI A protein. J. Biol. Chem., 265, 17078–17083. [PubMed] [Google Scholar]