Abstract
Advanced backcrosses (ABs) and introgression lines (ILs) of eggplant (Solanum melongena) can speed up genetics and genomics studies and breeding in this crop. We have developed the first full set of ABs and ILs in eggplant using Solanum incanum, a wild eggplant that has a relatively high tolerance to drought, as a donor parent. The development of these ABs and IL eggplant populations had a low efficiency in the early stages, because of the lack of molecular markers and genomic tools. However, this dramatically improved after performing genotyping-by-sequencing in the first round of selfing, followed by high-resolution-melting single nucleotide polymorphism genotyping in subsequent selection steps. A set of 73 selected ABs covered 99% of the S. incanum genome, while 25 fixed immortal ILs, each carrying a single introgressed fragment in homozygosis, altogether spanned 61.7% of the S. incanum genome. The introgressed size fragment in the ILs contained between 0.1 and 10.9% of the S. incanum genome, with a mean value of 4.3%. Sixty-eight candidate genes involved in drought tolerance were identified in the set of ILs. This first set of ABs and ILs of eggplant will be extremely useful for the genetic dissection of traits of interest for eggplant, and represents an elite material for introduction into the breeding pipelines for developing new eggplant cultivars adapted to the challenges posed by the climate-change scenario.
Keywords: Solanum melongena, Solanum incanum, advanced backcrosses, introgression lines, drought tolerance, high-throughput genotyping
Introduction
Eggplant (Solanum melongena L., 2n = 2x = 24) is a major vegetable crop of increasing importance during the last few years; while other major crops in the genus Solanum and in the family Solanaceae, like tomato, potato, pepper, tobacco, and petunia, have been widely studied from the genetic and genomic points of view, developments in eggplant genomics have lagged behind. In fact, the first draft of the eggplant genome was published only in 2014 (Hirakawa et al., 2014), while in the other major solanaceous crops high-quality genome sequences were available much earlier (Potato Genome Sequencing Consortium, 2011; Tomato Genome Consortium, 2012). While other eggplant genomic tools are slowly being published, like the first transcriptome (Yang et al., 2014), genetic and association maps (Barchi et al., 2012; Fukuoka et al., 2012; Gramazio et al., 2014; Portis et al., 2015) and quantitative trait locus (QTL) studies (Miyatake et al., 2016; Rinaldi et al., 2016; Toppino et al., 2016), there is a general lack of eggplant experimental populations, such as recombinant inbred lines (RILs), advanced backcrosses (ABs), introgression lines (ILs) or multiparent advanced generation intercross (MAGIC) populations.
However, although a great phenotypic diversity can be found in the common eggplant (Portis et al., 2015; Kaushik et al., 2016), its genetic base is quite narrow due to the genetic bottlenecks undergone during the domestication and subsequent crop evolution processes (Muñoz-Falcón et al., 2009; Meyer et al., 2012). This low genetic diversity could be a serious constraint for the development of new highly performing cultivars, which have to be able to contribute to increasing yield under the environmental changes resulting from a climate-change scenario (Tilman et al., 2011; Ray et al., 2013).
Although, many resistances and tolerances to biotic and abiotic stresses have been described in eggplant wild relatives (Rotino et al., 2014), just a few attempts have been made to transfer them to the cultivated eggplant (Toppino et al., 2008; Liu et al., 2015). Recent efforts in enhancing wild relatives for the adaptation of eggplant to climate change have resulted in the development of over 50 combinations of hybrids and 30 first backcross (BC1) generations (Kouassi et al., 2016; Plazas et al., 2016). These materials were obtained from six S. melongena parents and 14 wild species from different genepools, including the primary genepool (GP1, which includes relatives that can be easily crossed with the cultivated species to produce a highly fertile progeny), secondary genepool (GP2, which includes species that can be crossed with the cultivated species, although with some difficulties due to reproductive barriers, and that may give a low fertility progeny) and the tertiary genepool (GP3, which is made of phylogenetically distant species where pre- and post-zygotic barriers require specific breeding techniques, like embryo rescue, for hybridization and the progeny is often sterile).
The development of ABs and ILs in the eggplant genepool would make a major contribution to enhancing the use of crop wild relatives in eggplant breeding (Prohens et al., 2017). ABs consist of materials obtained through a backcross breeding scheme containing fixed or non-fixed single or multiple introgressions from a donor in the genetic background of a recurrent parent (Fulton et al., 1997). ABs represent powerful tools that allow more precise mapping of QTLs compared to balance populations (e.g., F2, BC1, RIL) and an elite material for breeding (Tanksley and Nelson, 1996). IL collections are a special type of AB population, consisting of fixed lines with single introgressed fragments from a donor parent (Eshed and Zamir, 1995; Tian et al., 2006). ILs are immortal populations making them more suitable for direct incorporation into breeding pipelines. The general aim for developing sets of ILs is to represent the whole genome of a donor parent, almost always an exotic or wild genotype, in the genetic background of a recipient cultivated parent through a set of lines carrying overlapping homozygous chromosome fragments of the former in the latter (Eshed and Zamir, 1994; Zamir, 2001; Eduardo et al., 2005). Several studies have demonstrated the higher efficiency of ILs and near isogenic lines (NILs) in QTL estimation than other segregating populations, such as F2, double haploid lines and RILs (Wehrhahn and Allard, 1965; Eshed and Zamir, 1995; Kearsey and Farquhar, 1998; Lebreton et al., 1998; Alonso-Blanco et al., 2006; Yin et al., 2016).
Although, the availability of IL populations in crop species is still limited, in the last two decades many have been developed, mainly in major cultivated crops such as maize, wheat, rice, barley, soybean, and melon (Koester et al., 1993; Concibido et al., 2003; Von Korff et al., 2004; Eduardo et al., 2005; Liu et al., 2006; Pestsova et al., 2006; Tian et al., 2006). The availability of a large number of IL populations in tomato has promoted and facilitated studies of the genetics of morphology (Chitwood et al., 2013), physiology (Ranjan et al., 2016), biotic resistance (Verlaan et al., 2013), abiotic tolerance (Albacete et al., 2015), chemical composition (Ikeda et al., 2017), enzyme activity (Caruso et al., 2016), metabolic profiling (Schauer et al., 2006), transcriptional profiles (Baxter et al., 2005), and many more traits.
In the case of eggplant, up until now no sets of ABs or ILs covering the whole genome of a relative have been obtained. In order to obtain the first set of introgression materials of this type in eggplant, we chose Solanum incanum L. as a donor parent. This wild species belongs to the secondary genepool of common eggplant (Syfert et al., 2016), and is naturally distributed in the desertic and dryland areas from western Pakistan, Afghanistan, and Iran across the Middle East and northern Africa (Ranil et al., 2015; Vorontsova and Knapp, 2016). S. incanum is reported as having considerable drought tolerance (Lester and Hasan, 1991; Daunay, 2008; Knapp et al., 2013), which has been confirmed under experimental conditions (Savarino, 2017). Drought tolerance is a trait of major interest for adaptation to climate change, where new drought-tolerant varieties are required (Dempewolf et al., 2014). In addition, S. incanum presents resistance to some fungal diseases, such as Fusarium oxysporum and Phomopsis vexans (Yamakawa and Mochizuki, 1979; Collonnier et al., 2001). Also, S. incanum exhibits a high content of bioactive phenolic compounds, mostly of chlorogenic acid type (Stommel and Whitaker, 2003; Ma et al., 2011; Prohens et al., 2013; Meyer et al., 2015). These favorable features led us to use S. incanum for the construction of an interspecific genetic map with S. melongena, which is linked to four eggplant and one tomato genetic maps (Gramazio et al., 2014). Furthermore, the transcriptome of S. incanum was sequenced to create a large set of molecular markers directly applicable in breeding programs and for a better understanding of the target genes involved in metabolic pathways relevant for eggplant breeding (Gramazio et al., 2016). Also, recently, the entire genome of S. incanum has been sequenced (Gramazio, unpublished data).
Here, we present the development of the first full set of ABs and ILs in the eggplant genetic background, from the first generations, where little genomic information was available and low–medium throughput genotyping techniques were used, to the last ones, where more markers were available and affordable high-throughput genotyping-by-sequencing (GBS) helped in the characterization of the ABs and in obtaining a set of ILs with introgressions containing a single fragment from the S. incanum donor.
Materials and methods
Plant material
The donor parent selected for the development of the set of ABs and ILs was S. incanum accession number MM577, collected in the wild in Israel and provided by the INRA Avignon (France) germplasm bank. MM577 plants are prickly, with no anthocyanin pigmentation and produce small green rounded fruits (Figure 1). This MM577 plant has been used in several genetic and genomic studies, as it has been characterized for morphological traits and phenolic acids content (Stommel and Whitaker, 2003), for androgenic competence through anther culture (Salas et al., 2011), as a rootstock for eggplant yield and its effects on fruit quality (Gisbert et al., 2011), for phylogeographic studies (Meyer et al., 2015), for breeding for adaptation to climate change (http://eggplantprebreeding.upv.es/), as a parental of an interspecific genetic map (Gramazio et al., 2014), for transcriptome sequencing (Gramazio et al., 2016) and for whole genome resequencing (Gramazio, unpublished data).
Figure 1.

Parents (Solanum incanum MM577 and S. melongena AN-S-26) of the AB and IL sets, their hybrid (F1) and the segregating first backcross (BC1) generation to the recipient parent S. melongena AN-S-26.
The recipient parent was S. melongena accession number AN-S-26, a Spanish local variety from Andalusia obtained from the Universitat Politècnica de València (UPV), Valencia, Spain, germplasm bank. The AN-S-26 plant is non-prickly, with anthocyanin pigmentation and large obovoid dark purple fruits (Figure 1). The morphological characterization and phenolics content of S. incanum MM577 and S. melongena AN-S-26 revealed important differences in plant and fruit morphology and phenolic concentration between them (Prohens et al., 2013). The genetic characterization of both parents with simple sequence repeat (SSR) and conserved ortholog set of genes II (COSII) markers also revealed a high level of genetic polymorphism betwee them (Gramazio et al., 2014). Both accessions numbersare maintained at the UPV germplasm genebank.
Seeds were germinated in Petri dishes and were subsequently transferred to seedling trays. These were kept in a climatic chamber under a photoperiod and temperature regime of 16 h light (25°C): 8 h dark (18°C). The plantlets were then transplanted to a pollinator-free glasshouse situated in the campus of the UPV, Valencia, Spain (GPS coordinates: latitude, 39° 28′ 55″ N; longitude, 0° 20′ 11″ W; 7 m above sea level). Plants were grown in 15 L pots filled with coconut fiber, irrigated and fertilized using a drip irrigation system, and pruned and trained with vertical strings. Phytosanitary treatments against spider mites and whiteflies were performed when necessary. Plants were hand pollinated and crosses were performed between the recurrent parent and the selected descendants of a family (hereafter referred to as progeny). S. melongena AN-S-26 was used mainly as the male parent, except for when the F1 and BC1 generations were obtained, where AN-S-26 was the female parent.
The hand pollinations were carried out as follows: the flowers of the female parent were emasculated before anthesis (i.e., before flower opening), then pollen was deposited on the stigma by gently rubbing it with a glass slide covered with the pollen of the male parent. Pollinated flowers were covered with a paper or mesh bag in order to avoid undesired cross-pollination, and tagged with information on the cross or selfing performed and the date.
Breeding scheme
The breeding scheme used to develop the ABs and ILs is depicted in Figure 2. The F1 hybrid was obtained in 2008 using the recipient parent as a female. The hybrid was completely fertile and in the following year was backcrossed to the AN-S-26 parent to generate the BC1 population.
Figure 2.
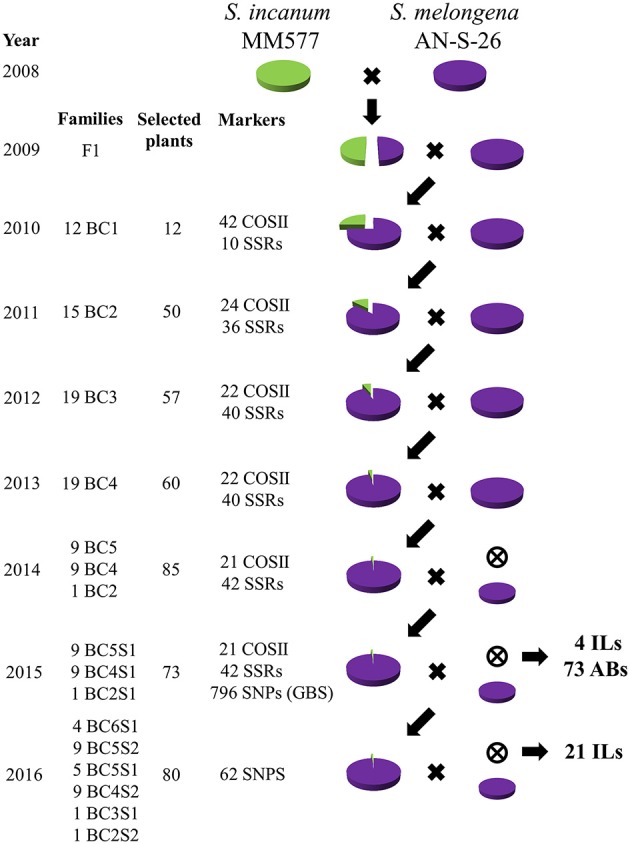
Breeding scheme used to develop the first set of eggplant ABs and ILs in the genetic background of Solanum melongena AN-S-26 (purple circle) by using S. incanum MM577 (green circle) as a donor. For each year (one generation per year), the number of families, selected plants, molecular markers assessed and type of cross are shown, the backcrosses with the recurrent parents are represented by an × and a purple circle, and the selfing rounds by an encircled ×.
The BC1 population, which consisted of 91 plants, was used to build the interspecific genetic map (Gramazio et al., 2014). A total of 12 selected BC1 plants (hereafter referred to as families) were backcrossed to generate the BC2 population. A subset of 50 selected BC2 plants, four to five per BC1 family, were grown in the greenhouse and backcrossed to obtain the BC3 population.
Twenty plants of each BC3 family were genotyped and three of each family were selected for the next round of backcross. Fifteen to twenty BC4 plants were genotyped at the seedling stage and 60 of them were selected for producing the next backcross generation. Some BC4 families did not set fruit; thus in 2014, in addition to the BC5 plants, some BC4 and BC2 plants, which had not given any progeny, were genotyped and selfed for a total of 85 plants.
After a preliminary manual genotyping of the progenies obtained, and driven by the recent publication of the first draft of the eggplant genome (Hirakawa et al., 2014), a high-throughput GBS was performed for the selected selfed genotypes. A total of 73 genotyped plants, including BC5S1, BC4S1, and BC2S1, were grown in the greenhouse and selfed to produce seeds in order to obtain the set of ABs and a first subset of ILs with single homozygous introgressions. In addition, the plants that presented two or more homozygous introgressions were backcrossed in order to obtain single homozygous introgressions.
A last round of selection for obtaining fixed ILs, including BC6S1, BC5S2, BC5S1, BC4S2, BC3S1, and BC2S2 families, was made with a new set of single nucleotide polymorphisms (SNPs) obtained by the GBS method based on the physical position. An additional 21 ILs with a single homozygous introgression, were identified. A total of 80 selected plants, around three to four per family, were grown in the greenhouse. Depending on the introgressions carried, they were selfed to produce seeds for the new ILs identified and to reduce the residual heterozygosity, or backcrossed to reduce the number of homozygous and off-target introgressions.
DNA extraction
Total genomic DNA was isolated from young leaves of plantlets at the 3–4 true leaves stage according to the CTAB method (Doyle and Doyle, 1987), with slight modifications. The extracted DNA was dissolved in Milli-Q water and its quality was checked in a 0.8% agarose gel. DNA concentration was measured with a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA) and adjusted to 30 ng/μL for PCR and high resolution melting (HRM) amplification (Wittwer et al., 2003) and to 100 ng/μL for GBS.
Genotyping methods
At the same time as the ABs and ILs were developed, a genetic linkage map was built from a cross between the parents, and was anchored to four eggplant and one tomato genetic maps (Gramazio et al., 2014). This map comprised 42 COSII, 99 SSRs, 88 amplified fragment length polymorphisms (AFLPs), 9 cleaved amplified polymorphic sequences (CAPS), 4 SNPs, and one morphological polymorphic markers, and encompassed 1,085 cM distributed in 12 linkage groups. A subset of these markers was used to assist in the selection of the plants during the development of the ABs and ILs.
Forty-two universal primers of COSII markers (Wu et al., 2006), which were previously mapped in an interspecific genetic map between S. linnaeanum and S. melongena (Wu et al., 2009), and 10 SSRs, developed by Nunome et al. (2009), which corresponded to the first set of markers mapped, were used to genotype the 91 BC1 plants and to select the 12 BC1 used to generate the BC2 population. The 12 plants selected presented all the markers from the donor for the target chromosome, each one different from the other 11 BC1 plants. The 240 BC2 progeny were genotyped at the seedling stage with 60 markers, 24 COSII and 36 SSRs, some of which were developed by our group (Vilanova et al., 2012). The details of the marker types, polymorphism and segregation distortion were reported in Gramazio et al. (2016). The new SSRs substituted for some of the COSII markers for reaching new genetic positions uncovered by the latter. For the BC2 population, not the whole background of each plant was screened, but just the target chromosome of each family. So, for example, the BC2 plants derived from the BC1 plant selected for chromosome 1 were genotyped only for chromosome 1 and not for chromosomes 2–12.
This strategy was employed to reduce the time and resources required to assess all the markers in all the chromosomes, and to perform genotyping at the seedling stage in order to grow only the selected plants to the reproductive stage. In fact, the approach was to develop, as a first step and as long as possible, a set of chromosome substitution lines in order to reduce the number of plants to be genotyped, grown, and backcrossed. The set of markers used for genotyping the BC3, BC4, BC5, and BC5S1 generations was practically the same, except for a few changes where SSRs replaced some COSII markers.
The selected plants from the first round of selfing were also genotyped with a GBS platform. A total of 796 SNPs was selected to improve the estimation of recombination breakpoints and to detect non-target introgressions that were dragged during the ABs and ILs development. From the whole collection of SNPs detected by GBS, 62 of them were selected, from four to eight markers per chromosome, and adapted for HRM analysis to genotype the plants involved in the second round of selfing. The number of SNPs assessed for each progeny was variable, depending on the size and the type (target or not-target) of the introgressions. The size and the position of the introgressions in megabases (Mb) and the percentage of coverage were calculated according to the physical position of the new eggplant genome assembly developed by the Italian Eggplant Genome Sequencing Consortium (Barchi et al., 2016). Primers pairs were designed using the Primer3 tool (v. 0.4.0, http://bioinfo.ut.ee/primer3-0.4.0/primer3/) with a mean amplicon range of 80–120 bp.
COSII marker analysis
COSII primer pairs were amplified by PCR in a 12 μL reaction volume comprising 7.21 μL water, 1.2 μL 1 × PCR buffer, 0.6 μL MgCl2 (50 mM), 0.24 μL dNTPs (10 mM), 0.3 μL of each primer (10 μM), 0.15 μL Taq DNA Polymerase (5 U/μL), and 2 μL DNA template (20 ng/μL), under the following cycling conditions: denaturation at 95°C for 3 min; followed by 30 cycles of 30 s at 95°C, 30 s at 65°C, and of 30 s at 72°C; with a final extension at 72°C for 5 min. Then, restriction enzymes were used to cut in the polymorphic regions and the different size bands were visualized in 2–3% agarose gel.
SSR marker analysis
The amplification of SSRs was performed by PCR in a final volume of 12 μL: 7.21 μL water, 1.2 μL 1 × PCR buffer, 0.6 μL MgCl2 (50 mM), 0.24 μL dNTPs (10 mM), 0.3 μL reverse primer (10 μM), 0.06 μL forward primer with M13 tail (10 μM), 0.24 μL fluorochrome (FAM, VIC, NED, and PET) (10 μM), 0.15 μL Taq DNA Polymerase (5 U/μL), and 2 μL DNA template (20 ng/μL). The PCR amplification program consisted of: a first step of denaturation at 95°C for 3 min; followed by 30 cycles of 30 s at 95°C, 30 s at 65°C, and of 30 s at 72°C; and a last step of extension at 72°C for 5 min. Subsequently, the PCR products were diluted in formamide and analyzed with an automated ABI PRISM 3100-Avant DNA sequencer, with a GeneScan 600LIZ (Applied Biosystems, California, USA) size standard. The fragments were analyzed using GeneScan software (Applied Biosystems) to obtain the electropherograms and polymorphisms were analyzed with Genotyper DNA Fragment Analysis software (Applied Biosystems).
SNP marker analysis
SNPs were validated in a LightCycler 480 Real-Time PCR (Roche, Basel, Switzerland). The reactions were performed in a 10 μL volume comprising 5 μL Master Mix (2×), 0.8 μL MgCl2 (25 mM), 0.25 μL each primer, 1.7 μL water, and 2 μL DNA (30 ng/μL) with the following touchdown PCR program: denaturation at 95°C for 10 min; followed by 55 cycles of 10 s at 95°C, 15 s at 65°C (decreasing 1°C each cycle until 55°C) and 15 s at 72°C; and a final melting step of 1 min at 95°C, 1 min at 40°C, and 1 s at 60°C with the temperature being raised by 0.02°C/s until 95°C was reached.
GBS analysis
GBS was performed in 83 plants comprising 73 pre-selected plants from different generations (BC5S1, BC4S1, and BC2S1), 2 samples of each parent, 2 additional samples of doubled haploids of the recurrent parent and 2 samples of the hybrid. After DNA extraction and dilution, the samples were sent to the Cornell University genomic facilities in August 2015 for library preparation (a PstI enzyme was used) and sequencing. The raw data were demultiplexed and the Illumina barcode removed using the software GBS Barcode Splitter (https://sourceforge.net/projects/gbsbarcode/). Subsequently, the sequences were filtered at a 25 Phred quality score and the Illumina adapters removed with the Fastq-mcf program (Aronesty, 2013). A quality control before and after the trimming was performed with the FastQC tool (Andrews, 2010) and general statistics were obtained using Fastq-stats (Aronesty, 2013). Then, the clean reads were mapped against the reference published genome (Hirakawa et al., 2014) and against another eggplant genome with improved quality, kindly provided by the Italian Eggplant Genome Sequencing Consortium (Barchi et al., 2016). Due to the better assembly of the latter (12 pseudomolecules vs. 33,873 scaffolds of the former), we decided to use it for the further analysis. The mapping was performed using BWA-MEM software (Li, 2013) with default settings and the SNP calling using the FreeBayes program (Garrison and Marth, 2012), ignoring those variants that presented less coverage than 15, a mapping quality of 20 or a base quality of 20. In order to select the most reliable SNPs for genotyping the plants with HRM, different filters from the Seq Crumbs tools (https://bioinf.comav.upv.es/seq_crumbs/) were applied to the VCF file for maximizing the polymorphism validation. The filters CS60 and CL60 identified polymorphisms that were at a distance of < 60 nucleotides to another polymorphism or to the alignment edge, respectively. The filter HV0.05 determined whether the region had more than 5 polymorphisms per 100 bases, while the filter VKS discriminated INDELs from SNPs. GBS raw data were deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) repository under the accession number SAMN07249022.
Positioning candidate genes for drought tolerance
Candidate genes involved in pathways strictly related to drought tolerance were gathered from the current scientific literature (Bolger et al., 2014; Zhang N. et al., 2014; Krannich et al., 2015; Landi et al., 2016). Orthologous genes of these candidates were searched for in tomato (Solanum lycopersicum) and potato (Solanum tuberosum) through EnsemblPlants (http://plants.ensembl.org/index.html/) and NCBI (National Center for Biotechnology Information; https://www.ncbi.nlm.nih.gov/) databases, and the corresponding Fasta sequences were retrieved. Subsequently, a Blast search (with a cut-off value of 1e-60) was performed with the Fasta sequences to position the candidate genes on the eggplant genome (Barchi et al., 2016) and to assign them to the corresponding ILs.
Results
Development of the ABs and ILs
A total of 91 BC1 plants were obtained by crossing the F1 hybrid S. melongena AN-S-26 × S. incanum MM577 to the recurrent parent (AN-S-26), generating a highly diverse population in plant morphology and in fruit color, shape, size, and prickles (Figure 1). Based on the information of the 52 molecular markers assessed, 12 BC1 plants were selected. Each of them was heterozygous for all the scored markers of the target chromosome and had a reduced percentage of donor parent genetic background in the rest of their chromosomes. The BC2 progeny, obtained from the 12 BC1 plants, were genotyped with a mean of four to five molecular markers (24 COSII and 36 SSRs). This allowed selection of 15 BC2 families, which were taken to the greenhouse in order to obtain the BC3 population. For chromosomes 5, 7, and 8, no individual plants displayed a chromosome with all scored markers completely heterozygous; thus, two complementary individuals for each of them were selected to cover all the markers for the target chromosomes. As in the previous year (2011), the selected plants were transplanted to the greenhouse at the end of October with the aim of obtaining two cycles of backcross per year (winter–spring and summer–fall). For several reasons, including seed dormancy, slow growth, poor flowering with low-quantity pollen during winter and parthenocarpy, just one cycle per year was achievable.
The BC3 progeny were genotyped with 22 COSII and 38 SSRs. Four chromosomes (1, 2, 4, and 10) were not completely heterozygous for the markers used in any of the selected plants; hence, the number of progeny rose to nineteen. The genotyping of the BC4 plants (22 COSII and 42 SSRs) allowed selection of the progeny for the last round of backcross. Unfortunately, nine BC4 families (two from chromosome 1, two from chromosome 2, one from chromosome 6, two from chromosome 7, one from chromosome 10, and one from chromosome 11) did not produce fruit due to poor fruit flowering and setting. Thus, in order to avoid loss of these introgressions, new BC4 families for those from which we could not obtain seeds were genotyped again, together with the progenies of the BC5 families. Moreover, for chromosome 6, the few remaining seeds of the BC4 and BC3 generations did not germinate and the BC2 progeny were used to continue the process.
The first round of selfing was performed for nine BC5, nine BC4, and one BC2 families. In addition, the BC4 and BC2 families were backcrossed for a backup, due to the fact that the BC2 and most of the BC4 generations that were re-genotyped had few remaining seeds. The progenies obtained from selfing were manually genotyped with 24 COSII and 44 SSRs. Subsequently, 73 plants that were pre-selected from the BC5S1, BC4S1, and BC2S1 families were additionally sequenced through GBS. The selfed offspring of these 73 plants constituted the ABs set.
The genotyping by GBS allowed a very accurate detection of introgression length, precisely defining the recombination breakpoints and the double recombination events undetected due to the low–medium density molecular marker assisted selection used in the previous generations. The sequencing of the 83 genotypes (73 pre-selected plants from BC5S1, BC4S1, and BC2S1, 2 samples of each parent, 2 additional samples of doubled haploids of the recurrent parent and 2 samples of the F1 hybrid) generated a total of 201,181,531 reads, with a mean of 2,423,873 reads per genotype. After the filtering steps, more than 85% clean reads were aligned to the 12 pseudomolecules of the new eggplant genome developed by the Italian Eggplant Genome Sequencing Consortium (Barchi et al., 2016). The SNP calling allowed identification of a total of 58,401 polymorphisms. Several filters were applied to the VCF file, using NGS_CRUMBS software (https://bioinf.comav.upv.es/ngs_crumbs/), in order to select the most suitable variants for assessment using the HRM technique. Although, all polymorphisms matched the quality criteria, a subset of 796 was selected for the genotyping.
The mean number of SNPs per chromosome was 66, with a range of 38 (chromosomes 9 and 11) to 92 (chromosome 3). Using the subset of 796 filtered SNPs, increasing the number of markers for genotyping over elevenfold, more than 90% of introgressions were confirmed by GBS, as well as the segregation of the heterozygous regions previously analyzed by manual genotyping. A total of 36% of the introgressions was totally homozygous for the donor parent, 21.3% were still totally heterozygous, and 33.3% of them displayed a combination of homozygous and heterozygous regions in the first round of selfing (Figure 3).
Figure 3.
Graphical genotypes of the selected plants genotyped by genotyping-by-sequencing (GBS) after four to five (two for P-36 to P-43) rounds of backcross (typically five, but two or four for some genotypes) and one round of selfing, using Solanum incanum MM577 as the donor parent in the genetic background of S. melongena AN-S-26. The first row indicates chromosome number and the first column the plant code. Homozygous introgressions of the donor parent are depicted in red, while the genetic background of the recipient parent is depicted in blue and the heterozygous regions in yellow.
Double recombinations, which were practically undetected in the previous manual genotyping, were identified in the regions with the highest density of molecular markers. In fact, the distribution of SNPs was very uneven along the chromosomes, being generally greater in the euchromatin and less in the centromeric regions. The development of the backcross generations without screening the genetic background of the selected progenies until the first round of selfing led to the dragging of non-target introgressions. In fact, 84% of the genotyped plants presented at least one non-target introgression, with the most common case being two non-targeted introgressions per plant (37.3%), followed by one (26.6%) and three (8%) non-target introgressions per plant. Obviously, the plants selected for chromosome 6 were those that presented the highest number of non-targeted introgressions (from seven to eleven) due to the fact that they were backcrossed just twice instead of being subjected to four or five rounds of backcrosses of the others.
However, in most cases, the non-target introgressions of the 73 ABs progenitors (i.e., selected BC5S1, BC4S1, and BC2S1 plants) were smaller than 5 Mb and positioned on the edges of the chromosomes, while few others were larger, especially those of chromosome 6, and located in the centromeric regions where generally the recombination is low. Most of the 159 non-target introgressions in the 73 ABs progenitors were heterozygous (60.3%), which meant an easier elimination by segregation through selfing, while 52 non-target introgressions (32.7%) were homozygous for the donor parent, and 11 (7%) were both heterozygous and homozygous in different percentages. The negative selection against the homozygous non-target introgressions is more complicated compared to the heterozygous ones, requiring at least an additional round of backcross before the elimination by segregation in a subsequent selfing generation.
However, genetic characterization by GBS revealed the high coverage of the donor parent across the ABs progenitors (Table 1). In fact, thanks also to the non-target introgressions, the representation of S. incanum was around 99% in the AB materials, while the other 1% belonged to non-polymorphic regions between the two parentals. This means that no region of the donor parental had been lost during the backcross process and all the S. incanum genome had been introgressed into the ABs, although some materials will need more rounds of backcrossng and selfing to produce ILs with just one introgression in homozygosis.
Table 1.
Statistics of Solanum incanum MM577 genome coverage in the 73 progenitors (corresponding to selected plants of the BC5S1, BC4S1, and BC2S1 generations) of the advanced backcrosses (ABs) set, after the genotyping by genotyping-by-sequencing (GBS).
| Chrom. | Chrom. size (Mb) | S. incanum size (Mb) | S. incanum (%) | Heter. regions size (Mb) | Heterozygous (%) | S. melongena size (Mb) | S. melongena (%) |
|---|---|---|---|---|---|---|---|
| 1 | 136 | 20.8 | 15.3 | 109.7 | 80.7 | 5.5 | 4.0 |
| 2 | 83 | 25.1 | 30.2 | 57.9 | 69.8 | – | – |
| 3 | 97 | 92.4 | 95.2 | 4.6 | 4.8 | – | – |
| 4 | 105 | 105 | 100.0 | – | – | – | – |
| 5 | 43 | 43 | 100.0 | – | – | – | – |
| 6 | 108 | 108 | 100.0 | – | – | – | – |
| 7 | 142 | 142 | 100.0 | – | – | – | – |
| 8 | 109 | 106.8 | 97.9 | 2.2 | 2.1 | – | – |
| 9 | 43 | 43 | 100.0 | – | – | – | – |
| 10 | 106 | 95.4 | 90.1 | 5.6 | 5.2 | 5.0 | 4.7 |
| 11 | 72 | 72 | 100.0 | – | – | – | – |
| 12 | 100 | 100 | 100.0 | – | – | – | – |
| Total | 1144 | 953.5 | 83.4 | 180.0 | 15.7 | 10.5 | 0.9 |
Solanum melongena AN-S-26 regions were assigned when no heterozygous or S. incanum introgressions were present, while heterozygous regions were assigned when no donor parent introgressions were present at the same time. All other regions were considered as S. incanum regions.
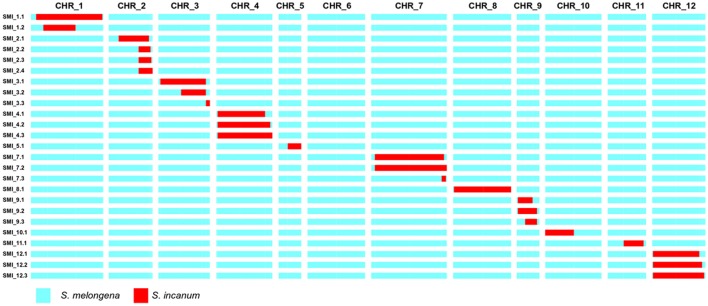
The first round of selfing and the high-throughput genotyping by GBS allowed identification of the first four fixed ILs (SMI_2-1, SMI_7-1, SMI_7-3, SMI_8-1), with two of them (SMI_7-1 and SMI_8-1) covering most of the entire chromosome physical size (Table 2 and Figure 4). Based on the genotype of the target introgressions (homozygous, heterozygous, or both), and the number and the genotype of the non-target introgressions, selected plants of the ABs were selfed or backcrossed in order to obtain new ILs. The progenies obtained were screened with a set of 62 SNPs derived from the GBS and converted for assessment using the HRM technique, the mean marker density being higher in the euchromatin regions and lower in the centromeres. The design of the new markers was based on fixing the single-introgression ILs and reducing the number of non-target introgressions. Hence, the number of SNPs was different for each chromosome and depended on the size of the latter, resulting in four to eight SNPs per chromosome.
Table 2.
Statistics of the S. incanum MM577 introgression lines (ILs) in the genetic background of S. melongena AN-S-26 using a 12 pseudomolecule eggplant genome (Italian Eggplant Genome Sequencing Consortium; Barchi et al., 2016), and a number of candidate genes for drought tolerance.
| ILs | Chrom. | Donor parent (%) | IL size (Mb) | IL position (Mb) | Chrom. IL size (%) | Chrom. total ILs size (Mb) | Chrom. total ILs size (%) | Drought-tolerance related genesa |
|---|---|---|---|---|---|---|---|---|
| SMI_1.1 | 1 | 9.9 | 114 | 19–133 | 83.8 | 114 | 83.8 | – |
| SMI_1.2 | 0.7 | 9 | 27–36 | 6.5 | – | |||
| SMI_2.1 | 2 | 3.2 | 37 | 38–75 | 44.5 | 43 | 51.8 | 1 |
| SMI_2.2 | 0.1 | 2 | 75–77 | 2.4 | – | |||
| SMI_2.3 | 0.4 | 5 | 75–80 | 6.0 | 2 | |||
| SMI_2.4 | 0.5 | 6 | 75–81 | 7.2 | 3 | |||
| SMI_3.1 | 3 | 6.9 | 79 | 7–86 | 81.4 | 82 | 84.5 | 7 |
| SMI_3.2 | 0.6 | 8 | 78–86 | 8.2 | 2 | |||
| SMI_3.3 | 0.2 | 3 | 93–96 | 3.0 | 1 | |||
| SMI_4.1 | 4 | 7.0 | 81 | 4–85 | 75.2 | 101 | 96.1 | 3 |
| SMI_4.2 | 8.2 | 94 | 4–98 | 89.5 | 4 | |||
| SMI_4.3 | 8.8 | 101 | 4–105 | 96.1 | 6 | |||
| SMI_5.1 | 5 | 0.6 | 8 | 35–43 | 18.6 | 8 | 18.6 | 11 |
| SMI_7.1 | 7 | 10.5 | 121 | 14–135 | 85.2 | 125 | 88.0 | 4 |
| SMI_7.2 | 10.9 | 125 | 14–139 | 88.0 | 7 | |||
| SMI_7.3 | 0.8 | 10 | 129–139 | 7.0 | 4 | |||
| SMI_8.1 | 8 | 9.2 | 106 | 3–109 | 97.2 | 106 | 97.2 | 11 |
| SMI_9.1 | 9 | 1.8 | 21 | 5–26 | 48.3 | 29 | 64.4 | 3 |
| SMI_9.2 | 2.5 | 29 | 5–34 | 64.4 | 3 | |||
| SMI_9.3 | 0.7 | 9 | 26–34 | 20.9 | 1 | |||
| SMI_10.1 | 10 | 0.1 | 2 | 0–2 | 1.8 | 2 | 1.8 | – |
| SMI_11.1 | 11 | 0.2 | 3 | 60–63 | 4.1 | 3 | 4.1 | – |
| SMI_12.1 | 12 | 7.2 | 83 | 3–86 | 83.0 | 93 | 93.0 | 11 |
| SMI_12.2 | 7.7 | 89 | 3–92 | 89.0 | 14 | |||
| SMI_12.3 | 8.1 | 93 | 3–96 | 93.0 | 18 | |||
| Mean | 4.3 | 49.5 | 48.2 | 64 | 62.1 | 5.8 | ||
| Total | 706 | 68 |
The positions and size of the ILs are represented in megabases (Mb) according to the physical position in each chromosome (Chrom.).
Orthologous loci related to drought tolerance are reported in the Supplementary Table.
Figure 4.
Graphical genotypes of ILs developed using Solanum incanum MM577 as the donor parent in the genetic background of S. melongena AN-S-26. The first row indicates the chromosome number and the first column the IL code. Homozygous introgressions of the donor parent are depicted in red, while the genetic background of the recipient parent is depicted in blue.
The genotyping of an additional generation of selfing from the ABs allowed identification of 21 more fixed single-introgression homozygous ILs, raising the total set of ILs to 25 different lines (Table 2). On average, the ILs contained 4.3% of the donor parent genome, ranging from 0.1 to 10.9%, altogether covering 61.7% of the S. incanum genome. The mean size of the introgressions was 49.5 Mb (ranging from 2 to 125 Mb), encompassing between 1.8 and 97.2% of the target chromosome length. Chromosome 2 presented the largest number of ILs (4), while chromosome 6 had none. The most covered chromosome was 8, with 97.2% of the total length, and the least covered among those having at least one IL was chromosome 10, with 1.8%.
Candidate genes for drought tolerance
Due to the lack of genomic and transcriptomic studies in the eggplant genepool, candidate genes from different crops related to drought tolerance were retrieved from the current scientific literature in order to detect the potential genomic region in eggplant and in the recently developed ILs. A total of 142 candidate genes gave a positive Blast hit (cut-off value of 1e-60) in the eggplant genome, most of them involved in the abscisic acid biosynthesis, catabolism and signaling network, in ethylene synthesis and signaling, in osmolyte biosynthesis and cell protection, in aquaporins and detoxification regulation (Supplementary Table). The chromosomes containing most candidate genes for drought tolerance were chromosome 12 with 28 genes, chromosome 5 with 20 genes, and chromosome 10 with 18 genes. The S. incanum alleles of these candidate genes were present in the AB materials, while 68 of them were present in the IL set. For the IL set, SMI_12.3 was the line carrying the most candidate genes (18), followed by SMI_12.2 (14 genes), and SMI_5.1, SMI_8.1, and SMI_12.1 (11 genes each) (Table 2).
Discussion
S. incanum is a close relative of eggplant and its hybrids and backcrosses with S. melongena are fully fertile (Knapp et al., 2013; Kouassi et al., 2016; Plazas et al., 2016).
The development of the ABs and ILs of S. incanum in the genetic background of S. melongena, in its early stages, was characterized by a lack of information available for genotyping the progenies efficiently. This led, at the beginning of the process, to the use of markers for low–medium throughput genotyping from information obtained in other eggplant studies and to the generation of new molecular markers more suitable for our materials. Although, in general some molecular markers can be useful and polymorphic across species (Saha et al., 2004; Barbará et al., 2007), their identification and amplification can be very challenging. In fact, the markers used for genotyping the BC1 generation were developed in other eggplant segregating populations (Nunome et al., 2009; Wu et al., 2009), although they were progressively substituted by markers identified in our population. From the initial set of markers, just one fifth were polymorphic in our BC1 population, which led to a low saturation of some chromosomal regions, which reduced the efficiency for selecting the most appropriate genotypes.
Our strategy for this first step was to select 12 BC1 plants, each one with one chromosome completely heterozygous for the markers from the donor, and at the same time with a higher percentage of the recurrent parent background in the other 11 chromosomes. This approach, consisting of the development of a set of chromosome substitution lines, accelerated the process of recovering the recipient parent background using a small set of selected plants, avoiding the tracking many families until the first round of selfing.
Unfortunately, this strategy did not last long, because, during the selection of the BC2 generation, no plants with complete selected chromosomes heterozygous for the markers screened were found in the progeny of 3 out of the 12 BC1 plants. The same drawback occurred also in further selection steps in the BC3 and BC5 generations. In addition, to reduce the time and resources needed during selection, we genotyped the plants only for the target chromosome instead of for the whole set of chromosomes. The strategy was to develop ILs without screening the genetic background, for the sake of time and resource saving, until AB generations or even selfing rounds were performed also by other authors (Eichten et al., 2011; Pea et al., 2013). In fact, no affordable high-throughput genotyping platforms or SNP arrays were available in eggplant to genotype a large number of plants with a wide set of SNPs in a very short time, as in the major crops like maize (Ganal et al., 2011), wheat (Wang et al., 2014), tomato (Sim et al., 2012), and potato (Vos et al., 2015); so, in our case, all the markers were assessed manually using low–medium throughput genotyping. Moreover, more than half of the markers assessed were SSRs (24 COSII and 36 SSRs), which are very robust and reliable (Varshney et al., 2005), but compared to SNPs are generally more difficult to automate and more time-consuming and expensive, due to the need of detection through an agarose or polyacrylamide gel or capillary sequencing (Jones et al., 2007). Some authors, Yang et al. (2011), calculated that assessing SNPs through a high-throughput genotyping platform can be 100-fold faster and 75% less expensive than SSR gel-based or capillary methods.
All these combined circumstances made the genotyping and selection steps the bottlenecks in the AB and IL development process, more than the interspecific crossing of two genetically distant species, which probably discouraged attempts to start sets of ILs with eggplant genepool materials. In addition, in part due to the excessive amount of time needed for the genotyping and selection steps, and also due to difficulties in obtaining seeded fruit set during the winter–spring season (e.g., slow plant growth, poor flowering with reduced production of pollen, low pollen viability, and parthenocarpy), just one cycle of backcrossing per year was feasible. This meant a very long process of 8 years has been required to obtain a first set of ILs.
For other crops, where two or even three growing cycles per year are possible, the entire set of ILs could be obtained in 3 or 4 years (Eduardo et al., 2005; Barrantes et al., 2014). The strategy to bypassing the screening of the non-targeted chormosomes of the selected plants, even if it has the advantage of reducing the time and the cost for genotyping, increases the number of rounds of backcrosses required to recover the almost totality of the recipient parent background (Frisch et al., 1999). In fact, although generally five rounds of backcross are recommended to recover more than 98% of the recurrent parent, due to the fact that the donor genome proportion is halved with each backcross generation, an adequately high percentage of the recipient parent can be recovered after three generations of marker-assisted backcrossing (Tanksley et al., 1989; Hospital et al., 1992; Hospital and Charcosset, 1997), although this depends on the number of plants screened and the number of markers adequately distributed along the genome (Frisch and Melchinger, 2005).
High-throughput genotyping in the first generations of backcrossing not only accelerates the process of obtaining ILs, but also provides early detection of non-target introgressions and easy elimination of them during the backcross process. When the genetic background is not screened until advanced generations of backcrossing or even after the final collection of ILs, the presence of non-target introgressions is almost certain, as happened in several studies (Shen et al., 2001; Eichten et al., 2011; Pea et al., 2013; Arbelaez et al., 2015). However, non-target introgressions can go unnoticed, until a massive genotyping is performed, even when a low-density genotyping is performed during the IL development process, due to double recombination events undetected by two adjacent markers (Xu et al., 2010; Schmalenbach et al., 2011; Zhang et al., 2011).
Our case was similar to the first scenario. In fact, the genetic background of the progenies was not screened until the first round of selfing when a GBS was performed. The majority of the plants genotyped contained at least one non-target introgression (84%), although most of them were small and regularly placed in certain regions, which mostly corresponded with low-recombination regions. This finding allowed the characterization of the ABs, but caused indirectly a further delay in the development of a set of nearly isogenic lines due to the necessity to clean the genetic background of the non-target introgressions.
Given the high representation of the donor genome in the AB materials, the removal of non-target introgressions will allow obtainment of a complete set of single-introgression fixed ILs where more than 95% of the donor genome is represented. Also, double recombinants were identified by GBS both in target and non-target introgressions. The medium-density genotyping applied during the population development did not allow detect of all the double-crossing-over events, mainly in the euchromatin regions where the recombination rate is higher (Wang et al., 2006; Gaut et al., 2007). In fact, although the selection of the markers before GBS was based on the genetic map, which tends to represent more the euchromatic regions and less the centromeric regions (Zhang W. et al., 2014), some high-recombination regions were not properly saturated due to the fact that the map was already under development. Then, after the GBS, for a more efficient genotyping design, the selection of the new SNPs was based on a combination of the genetic and physical position, plus the necessary markers to screen the non-target introgressions.
Selection based just on genetic maps could under-represent telomeric regions and drag large centromeric regions (Wang et al., 2000), as happened in our case, while a selection based exclusively on physical maps tends to reduce the adequate marker density of high-recombination regions, running the risk of not detecting double recombinants and of over-representing centromeric regions (Barrantes et al., 2014), which is not cost-effective due to the low-recombination ratio. The most efficient approach for reducing the time for the IL population development, avoiding most of the non-target introgressions and double recombinants, and detecting regions undetected by low–medium genotyping, could be to perform high-throughput genotyping in the first cycles of backcrossing. In this way, most of the ILs can be developed just in four generations, as was the case for Barrantes et al. (2014), who developed a set of tomato ILs with Solanum pimpinellifolium introgressions, where most of the lines derived from a BC3S1 generation. Even though a few years ago the cost of massive genotyping techniques was high and prohibitive for many labs, currently, the decreasing cost of sequencing and the emergence of new genotyping technologies, like GBS and similar (Campbell et al., 2015), have made the task of genotyping more affordable and precise. This makes possible the development of experimental populations for non-model crops for which genomic resources are limited (Sallam et al., 2015).
The ABs and ILs developed may be of great interest not only for dissecting the genetic basis of domestication traits, but also for developing varieties of eggplant with new traits, including complex ones like drought resistance, introgressed from S. incanum. Drought is considered by many experts the stress that presents the strongest negative impact on yield and crop productivity (Krannich et al., 2015). Plants have developed different strategies, involving several different genes and pathways, to mitigate the negative effects of drought (Reguera et al., 2012). In this respect, S. incanum, being distributed from northern Africa to the Middle East, is able to survive in desert and dryland areas (Ranil et al., 2015; Vorontsova and Knapp, 2016) and, therefore, is a potential source of variation for tolerance to drought, as demonstrated by Savarino (2017). However, almost nothing is known about the mechanisms and strategies used by S. incanum to tolerate this abiotic stress. For the major crops, like rice and wheat (Tang et al., 2016; Valluru et al., 2016), several mechanisms have been described involving many genes and pathways required for tolerance to drought. In this study, we have identified 142 eggplant genome regions that are orthologs of candidate genes related to drought tolerance in other crops; while all these genome regions are represented in the AB materials, 68 of these regions are present in the ILs newly developed using a donor parent (S. incanum MM577) that is much more resistant to drought than the recipient parent (S. melongena AN-S-26) (Savarino, 2017). Some of these ILs are very promising, carrying several orthologs related to drought tolerance from different crops. Monitoring the expression of these regions during a screening test for drought tolerance using the AB materials and the subset of ILs could pave the way for understanding the regulatory mechanisms for drought tolerance in eggplant. At the same time, it could facilitate the development of new eggplant varieties in a scenario when drought and desertification are increasingly a serious constrain for crop production.
A preliminary drought screening test realized by Savarino (2017), using intermediate backcross materials during the IL development, showed significant drought-tolerance levels among the parents, hybrid and the backcross materials. Although, the results were promising, the screening was exploratory and a further analysis is required. In addition, Sotomayor (2016) realized a phenotypic characterization of 62 AB materials for morphological traits related to fruit, flower, prickles, and chlorogenic acid content. A great variation among the materials was found for almost all the traits (Supplementary Image). Once again, these results are preliminary and more extensive analyses are being undertaken to discover QTLs and dissect the genetic basis of important agronomic traits. In the meantime, and without any further delay, the collection of ABs and ILs is available for the scientific community.
In conclusion, in this study, using the available genomic information (reduced at the beginning and more comprehensive at the later stages), we have developed the first set of ABs and ILs in eggplant that cover the whole genome of a wild donor species (S. incanum). These materials will have a tremendous impact in a wide range of studies in this genepool, which was lagging behind compared to other important crops of the same genus, like tomato and potato. In fact, precision QTL mapping in the ABs and ILs will allow confirmation of existing QTLs (Buerstmayr et al., 2011; Sayed et al., 2012) and detection of new ones, which has been complicated until now by the lack of an experimental population for this crop. In addition, new genetic diversity has been introduced in a crop that had suffered a reduction in the genetic base due to repeated genetic bottlenecks during the domestication process (Muñoz-Falcón et al., 2009; Meyer et al., 2012). The ABs and ILs developed could be used directly by breeders in their breeding pipelines, as most of them do not display undesirable wild traits because they just have a small percentage of the donor parent genome. We have provide further evidence that the implementation of high-throughput genotyping technologies can have an extraordinary impact on the development and precision of IL development, reducing the time and, probably, the cost of obtaining this segregating population (Schmalenbach et al., 2011; Arbelaez et al., 2015). Also, the ABs and ILs developed up to now will allow isolation of improved versions of the IL population, which will further contribute to the enhancement of eggplant breeding.
Author contributions
JP and SV planned the study. PG, JP, and SV supervised the research. PG, GM, MP, and FH generated the ABs and ILs collection. PG managed the seed collections. PG, GM, and MP performed the genotyping. FH contributed to the GBS analysis. PG, JP, and SV drafted the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- AB
advanced backcross
- COSII
conserved ortholog set of genes II
- HRM
high resolution melting
- IL
introgression line
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- SNP
single nucleotide polymorphism
- SSR
simple sequence repeat
- UPV
Universitat Politècnica de València.
Footnotes
Funding. This work has been funded in part by the initiative “Adapting Agriculture to Climate Change: Collecting, Protecting and Preparing Crop Wild Relatives,” which is supported by the Government of Norway. This project is managed by the Global Crop Diversity Trust with the Millennium Seed Bank of the Royal Botanic Gardens, Kew and implemented in partnership with national and international gene banks and plant breeding institutes around the world. For further information see the project website: http://www.cwrdiversity.org/. Funding has also been received from the European Union's Horizon 2020 Research and Innovation Programme under grant agreement No 677379 (G2P-SOL project: Linking genetic resources, genomes and phenotypes of Solanaceous crops) and from Spanish Ministerio de Economía, Industria y Competitividad and Fondo Europeo de Desarrollo Regional (grant AGL2015-64755-R from MINECO/FEDER). PG is grateful to Universitat Politècnica de València for a pre-doctoral (Programa FPI de la UPV-Subprograma 1/2013 call) contract. MP is grateful to Ministerio de Economía, Industria y Competitividad for a post-doctoral grant within the Juan de la Cierva programme (FCJI-2015-24835). GM is grateful to Conselleria d'Educació, Investigació, Cultura i Esport de la Generalitat Valenciana for a pre-doctoral grant within the Santiago Grisolía programme (GRISOLIAP/2016/012). Authors also thank the Italian Eggplant Genome Sequencing Consortium for providing access to an improved version of the eggplant genome.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01477/full#supplementary-material
References
- Albacete A., Andújar C., Dodd I., Giuffrida F., Hichri I., Lutts S., et al. (2015). Rootstock-mediated variation in tomato vegetative growth under drought, salinity and soil impedance stresses. Acta Hortic. 1086, 141–146. 10.17660/ActaHortic.2015.1086.17 [DOI] [Google Scholar]
- Alonso-Blanco C., Koornneef M., van Ooijen J. W. (2006). QTL analysis. Methods Mol. Biol. 323, 79–99. 10.1385/1-59745-003-0:79 [DOI] [PubMed] [Google Scholar]
- Andrews (2010). Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Arbelaez J. D., Moreno L. T., Singh N., Tung C. W., Maron L. G., Ospina Y., et al. (2015). Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O.sativa cv. Curinga. Mol. Breed. 35:81. 10.1007/s11032-015-0276-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinform. J. 7, 1–8. 10.2174/1875036201307010001 [DOI] [Google Scholar]
- Barbará T., Palma-Silva C., Paggi G. M., Bered F., Fay M. F., Lexer C. (2007). Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol. Ecol. 16, 3759–3767. 10.1111/j.1365-294X.2007.03439.x [DOI] [PubMed] [Google Scholar]
- Barchi L., Delledonne M., Lanteri S., Dal Molin A., Minio A., Ferrarini A., et al. (2016). A high quality eggplant genome sequence: a new tool for the analysis of the Solanaceae family evolution and for the molecular deciphering of complex traits, in Plant Breeding: The Art of Bringing Science to Life - 20th EUCARPIA General Congress, eds Kölliker R., Boller B. (Zurich: Agroscope; ), 122. [Google Scholar]
- Barchi L., Lanteri S., Portis E., Valè G., Volante A., Pulcini L., et al. (2012). A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS ONE 7:e43740. 10.1371/journal.pone.0043740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes W., Fernández-del-Carmen A., López-Casado G., González-Sánchez M. Á., Fernández-Muñoz R., Granell A., et al. (2014). Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Mol. Breed. 34, 1817–1831. 10.1007/s11032-014-0141-0 [DOI] [Google Scholar]
- Baxter C. J., Sabar M., Quick W. P., Sweetlove L. J. (2005). Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J. Exp. Bot. 56, 1591–1604. 10.1093/jxb/eri154 [DOI] [PubMed] [Google Scholar]
- Bolger A., Scossa F., Bolger M. E., Lanz C., Maumus F., Tohge T., et al. (2014). The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46, 1034–1038. 10.1038/ng.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstmayr M., Lemmens M., Steiner B., Buerstmayr H. (2011). Advanced backcross QTL mapping of resistance to fusarium head blight and plant morphological traits in a Triticum macha x T. aestivum population. Theor. Appl. Genet. 123, 293–306. 10.1007/s00122-011-1584-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell N. R., Harmon S. A., Narum S. R. (2015). Genotyping-in-Thousands by sequencing (GT-seq): a cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Resour. 15, 855–867. 10.1111/1755-0998.12357 [DOI] [PubMed] [Google Scholar]
- Caruso G., Gomez L. D., Ferriello F., Andolfi A., Borgonuovo C., Evidente A., et al. (2016). Exploring tomato Solanum pennellii introgression lines for residual biomass and enzymatic digestibility traits. BMC Genet. 17:56. 10.1186/s12863-016-0362-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D. H., Kumar R., Headland L. R., Ranjan A., Covington M. F., Ichihashi Y., et al. (2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25, 2465–2481. 10.1105/tpc.113.112391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collonnier C., Fock I., Kashyap V., Rotino G. L., Daunay M. C., Lian Y., et al. (2001). Applications of biotechnology in eggplant. Plant Cell. Tissue Organ Cult. 65, 91–107. 10.1023/A:1010674425536 [DOI] [Google Scholar]
- Concibido V., La Vallee B., McLaird P., Pineda N. (2003). Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. 106, 575–582. 10.1007/s00122-002-1071-5 [DOI] [PubMed] [Google Scholar]
- Daunay M. C. (2008). Eggplant, in Vegetables II: Handbook of Plant Breeding, eds Prohens J., Nuez F. (New York, NY: Springer; ), 163–220. [Google Scholar]
- Dempewolf H., Eastwood R. J., Guarino L., Khoury C. K., Muller J. V., Toll J. (2014). Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agroecol. Sustain. Food Syst. 38, 369–377. 10.1080/21683565.2013.870629 [DOI] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Eduardo I., Arús P., Monforte A. J. (2005). Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor. Appl. Genet. 112, 139–148. 10.1007/s00122-005-0116-y [DOI] [PubMed] [Google Scholar]
- Eichten S. R., Foerster J. M., de Leon N., Kai Y., Yeh C.-T., Liu S., et al. (2011). B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiol. 156, 1679–1690. 10.1104/pp.111.174748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1994). A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79, 175–179. 10.1007/BF00022516 [DOI] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M., Melchinger A. (2005). Selection theory for marker-assisted backcrossing. Genetics 170, 909–917. 10.1534/genetics.104.035451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M., Bohn M., Melchinger A. (1999). Comparison of seletion strategies for marker-assisted backcrossing of a gene. Crop Sci. 39, 1295–1301. 10.2135/cropsci1999.3951295x [DOI] [Google Scholar]
- Fukuoka H., Miyatake K., Nunome T., Negoro S., Shirasawa K., Isobe S., et al. (2012). Development of gene-based markers and construction of an integrated linkage map in eggplant by using Solanum orthologous (SOL) gene sets. Theor. Appl. Genet. 125, 47–56. 10.1007/s00122-012-1815-9 [DOI] [PubMed] [Google Scholar]
- Fulton T. M., Beck-Bunn T., Emmatty D., Eshed Y., Lopez J., Petiard V., et al. (1997). QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor. Appl. Genet. 95, 881–894. 10.1007/s001220050639 [DOI] [Google Scholar]
- Ganal M. W., Durstewitz G., Polley A., Bérard A., Buckler E. S., Charcosset A., et al. (2011). A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6:e28334. 10.1371/journal.pone.0028334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E., Marth G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. [Google Scholar]
- Gaut B. S., Wright S. I., Rizzon C., Dvorak J., Anderson L. K., Rizzon C., et al. (2007). Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8, 77–84. 10.1038/nrg1970 [DOI] [PubMed] [Google Scholar]
- Gisbert C., Prohens J., Raigón M. D., Stommel J. R., Nuez F. (2011). Eggplant relatives as sources of variation for developing new rootstocks: effects of grafting on eggplant yield and fruit apparent quality and composition. Sci. Hortic. 128, 14–22. 10.1016/j.scienta.2010.12.007 [DOI] [Google Scholar]
- Gramazio P., Blanca J., Ziarsolo P., Herraiz F. J., Plazas M., Prohens J., et al. (2016). Transcriptome analysis and molecular marker discovery in Solanum incanum and S. aethiopicum, two close relatives of the common eggplant (Solanum melongena) with interest for breeding. BMC Genomics 17:300. 10.1186/s12864-016-2631-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramazio P., Prohens J., Plazas M., Andujar I., Herraiz F. J., Castillo E., et al. (2014). Location of chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map of eggplant. BMC Plant Biol. 14:350. 10.1186/s12870-014-0350-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Shirasawa K., Miyatake K., Nunome T., Negoro S., Ohyama A., et al. (2014). Draft genome sequence of eggplant (Solanum melongena L.): the representative Solanum species indigenous to the old world. DNA Res. 21, 649–660. 10.1093/dnares/dsu027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F., Charcosset A. (1997). Marker-assisted introgression of quantitative trait loci. Genetics 147, 1469–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F., Chevalet C., Mulsant P. (1992). Using markers in gene introgression breeding programs. Genetics 132, 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Shibuya T., Nishiyama M., Nakata Y., Kanayama Y. (2017). Physiological mechanisms accounting for the lower incidence of blossom-end rot in tomato introgression line IL8-3 fruit. Hortic. J. [Epub ahead of Print]. 10.2503/hortj.OKD-015 [DOI] [Google Scholar]
- Jones E., Sullivan H., Bhattramakki D., Smith J. (2007). A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for the genotypic analysis of maize (Zea mays L.). Theor. Appl. Genet. 115, 361–371. 10.1007/s00122-007-0570-9 [DOI] [PubMed] [Google Scholar]
- Kaushik P., Prohens J., Vilanova S., Gramazio P., Plazas M. (2016). Phenotyping of eggplant wild relatives and interspecific hybrids with conventional and phenomics descriptors provides insight for their potential utilization in breeding. Front. Plant Sci. 7:677. 10.3389/fpls.2016.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey M. J., Farquhar G. L. (1998). Short Review QTL analysis in plants ; where are we now? Heredity 80, 137–142. 10.1046/j.1365-2540.1998.00500.x [DOI] [PubMed] [Google Scholar]
- Knapp S., Vorontsova M. S., Prohens J. (2013). Wild relatives of the eggplant (Solanum melongena L.: Solanaceae): new understanding of species names in a complex group. PLoS ONE 8:e57039. 10.1371/journal.pone.0057039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester R. P., Sisco P. H., Stuber C. W. (1993). Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 33, 1209–1216. 10.2135/cropsci1993.0011183X003300060020x [DOI] [Google Scholar]
- Kouassi B., Prohens J., Gramazio P., Kouassi A. B., Vilanova S., Galán-Ávila A., et al. (2016). Development of backcross generations and new interspecific hybrid combinations for introgression breeding in eggplant (Solanum melongena). Sci. Hortic. 213, 199–207. 10.1016/j.scienta.2016.10.039 [DOI] [Google Scholar]
- Krannich C. T., Maletzki L., Kurowsky C., Horn R. (2015). Network candidate genes in breeding for drought tolerant crops. Int. J. Mol. Sci. 16, 16378–16400. 10.3390/ijms160716378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi S., Nurcato R., De Lillo A., Lentini M., Grillo S., Esposito S. (2016). Glucose-6-phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant Physiol. Biochem. 105, 79–89. 10.1016/j.plaphy.2016.04.013 [DOI] [PubMed] [Google Scholar]
- Lebreton C. H., Visscher P. M., Haley C. S., Semikhodskii A., Quarrie S. A. (1998). A nonparametric bootstrap method for testing close linkage vs. pleiotrophy of coincident quantitative trait loci. Genetics 150, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R., Hasan S. (1991). Origin and domestication of the brinjal eggplant, Solanum melongena, from Solanum incanum, in Africa and Asia, in Solanaceae III: Taxonomy, Chemistry and Evolution, eds Hawkes J. G., Lester R. N., Nee M., Estrada N. (London: Royal Botanic Gardens; ), 369–387. [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr. arXiv 0 3. [Google Scholar]
- Liu J., Zheng Z., Zhou X., Feng C., Zhuang Y. (2015). Improving the resistance of eggplant (Solanum melongena) to Verticillium wilt using wild species Solanum linnaeanum. Euphytica 201, 463–469. 10.1007/s10681-014-1234-x [DOI] [Google Scholar]
- Liu S., Zhou R., Dong Y., Li P., Jia J. (2006). Development, utilization of introgression lines using a synthetic wheat as donor. Theor. Appl. Genet. 112, 1360–1373. 10.1007/s00122-006-0238-x [DOI] [PubMed] [Google Scholar]
- Ma C., Dastmalchi K., Whitaker B. D., Kennelly E. J. (2011). Two new antioxidant malonated caffeoylquinic acid isomers in fruits of wild eggplant relatives. J. Agric. Food Chem. 59, 9645–9651. 10.1021/jf202028y [DOI] [PubMed] [Google Scholar]
- Meyer R. S., Karol K. G., Little D. P., Nee M. H., Litt A. (2012). Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Mol. Phylogenet. Evol. 63, 685–701. 10.1016/j.ympev.2012.02.006 [DOI] [PubMed] [Google Scholar]
- Meyer R. S., Whitaker B. D., Little D. P., Wu S. B., Kennelly E. J., Long C. L., et al. (2015). Parallel reductions in phenolic constituents resulting from the domestication of eggplant. Phytochemistry 115, 194–206. 10.1016/j.phytochem.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Miyatake K., Saito T., Negoro S., Yamaguchi H., Nunome T., Ohyama A., et al. (2016). Detailed mapping of a resistance locus against Fusarium wilt in cultivated eggplant (Solanum melongena). Theor. Appl. Genet. 129, 357–367. 10.1007/s00122-015-2632-8 [DOI] [PubMed] [Google Scholar]
- Muñoz-Falcón J. E., Prohens J., Vilanova S., Nuez F. (2009). Diversity in commercial varieties and landraces of black eggplants and implications for broadening the breeders' gene pool. Ann. Appl. Biol. 154, 453–465. 10.1111/j.1744-7348.2009.00314.x [DOI] [Google Scholar]
- Nunome T., Negoro S., Kono I., Kanamori H., Miyatake K., Yamaguchi H., et al. (2009). Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theor. Appl. Genet. 119, 1143–1153. 10.1007/s00122-009-1116-0 [DOI] [PubMed] [Google Scholar]
- Pea G., Aung H. H., Frascaroli E., Landi P., and Pè, M. E. (2013). Extensive genomic characterization of a set of near-isogenic lines for heterotic QTL in maize (Zea mays L.). BMC Genomics 14:61. 10.1186/1471-2164-14-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestsova E., Börner A., Röder M. (2006). Development and QTL assessment of Triticum aestivum–Aegilops tauschii introgression lines. Theor. Appl. Genet. 112, 634–647. 10.1007/s00122-005-0166-1 [DOI] [PubMed] [Google Scholar]
- Plazas M., Vilanova S., Gramazio P., Rodríguez-Burruezo A., Fita A., Herraiz F. J., Ranil R., et al. (2016). Interspecific hybridization between eggplant and wild relatives from different genepools. J. Am. Soc. Hortic. Sci. 141, 3–44. [Google Scholar]
- Portis E., Cericola F., Barchi L., Toppino L., Acciarri N., Pulcini L., et al. (2015). Association mapping for fruit, plant and leaf morphology traits in eggplant. PLoS ONE 10:e0135200. 10.1371/journal.pone.0135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. 10.1038/nature10158 [DOI] [PubMed] [Google Scholar]
- Prohens J., Gramazio P., Plazas M., Dempewolf H., Kilian B., Díez M. J., et al. (2017). Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213:158 10.1007/s10681-017-1938-9 [DOI] [Google Scholar]
- Prohens J., Whitaker B. D., Plazas M., Vilanova S., Hurtado M., Blasco M., et al. (2013). Genetic diversity in morphological characters and phenolic acids content resulting from an interspecific cross between eggplant, Solanum melongena, and its wild ancestor (S. incanum). Ann. Appl. Biol. 162, 242–257. 10.1111/aab.12017 [DOI] [Google Scholar]
- Ranil R., Niran H., Plazas M., Fonseka R. M., Fonseka H. H., Vilanova S., et al. (2015). Improving seed germination of the eggplant rootstock Solanum torvum by testing multiple factors using an orthogonal array design. Sci. Hortic. 193, 174–181. 10.1016/j.scienta.2015.07.030 [DOI] [Google Scholar]
- Ranjan A., Budke J., Rowland S., Chitwood D. (2016). eQTL in a precisely defined tomato introgression population reveal genetic regulation of gene expression patterns related to physiological and developmental. bioRxiv. 10.1101/040592 [DOI] [Google Scholar]
- Ray D., Mueller N., West P., Foley J. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. 10.1371/journal.pone.0066428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera M., Peleg Z., Blumwald E. (2012). Targeting metabolic pathways for genetic engineering abiotic stress-tolerance.pdf. Biochim. Biophys. Acta 1819, 186–194. 10.1016/j.bbagrm.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Rinaldi R., Van Deynze A., Portis E., Rotino G. L., Toppino L., Hill T., et al. (2016). New insights on eggplant/tomato/pepper synteny and identification of eggplant and pepper orthologous QTL. Front. Plant Sci. 7:1031. 10.3389/fpls.2016.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotino G. L., Sala T., Toppino L. (2014). Eggplant, in Alien Gene Transfer in Crop Plants, Vol. 2, eds Pratap A., Kumar J. (New York, NY: Springer; ), 381–409. 10.1007/978-1-4614-9572-7_16 [DOI] [Google Scholar]
- Saha M. C., Mian M. A. R., Eujayl I., Zwonitzer J. C., Wang L., May G. D. (2004). Tall fescue EST-SSR markers with transferability across several grass species. Theor. Appl. Genet. 109, 783–791. 10.1007/s00122-004-1681-1 [DOI] [PubMed] [Google Scholar]
- Salas P., Prohens J., Seguí-Simarro J. M. (2011). Evaluation of androgenic competence through anther culture in common eggplant and related species. Euphytica 182, 261–274. 10.1007/s10681-011-0490-2 [DOI] [Google Scholar]
- Sallam A. H., Endelman J. B., Jannink J., Smith K. P. (2015). Assessing genomic selection prediction accuracy in a dynamic barley breeding population. Plant Genome 8. 10.3835/plantgenome2014.05.0020 [DOI] [PubMed] [Google Scholar]
- Savarino F. (2017). Sviluppo di Popolazioni Biparentali e Multiparentali in Melanzana e Prove di Resistenza Alla Siccitá. Master thesis, Tuscia University. [Google Scholar]
- Sayed M. A., Schumann H., Pillen K., Naz A., Leon J. (2012). AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet. 13:61. 10.1186/1471-2156-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454. 10.1038/nbt1192 [DOI] [PubMed] [Google Scholar]
- Schmalenbach I., March T. J., Bringezu T., Waugh R., Pillen K. (2011). High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the illumina goldengate assay. G3 1, 187–196. 10.1534/g3.111.000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Courtois B., McNally K. L., Robin S., Li Z. (2001). Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor. Appl. Genet. 103, 75–83. 10.1007/s001220100538 [DOI] [Google Scholar]
- Sim S. C., Durstewitz G., Plieske J., Wieseke R., Ganal M. W., van Deynze A., et al. (2012). Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 7:e40563. 10.1371/journal.pone.0040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor I. (2016). Desarrollo y Fenotipado de Dos Generaciones Para la Construcción de Líneas de Introgresión de S. incanum en el Fondo Genético de la Berenjena (S. melongena). Available online at: https://riunet.upv.es/handle/10251/68716 (Accessed June 22, 2017). [Google Scholar]
- Stommel J. R., Whitaker B. D. (2003). Phenolic Acid Content and composition of eggplant fruit in a germplasm core subset. J. Am. Soc. Hort. Sci. 128, 704–710. [Google Scholar]
- Syfert M. M., Castañeda-Álvarez N. P., Khoury C. K., Sarkinen T., Sosa C. C., Achicanoy H. A., et al. (2016). Crop wild relatives of the brinjal eggplant (Solanum melongena): poorly represented in genebanks and many species at risk of extinction. Am. J. Bot. 103, 635–651. 10.3732/ajb.1500539 [DOI] [PubMed] [Google Scholar]
- Tang N., Ma S., Zong W., Yang N., Lv Y., Yan C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28, 2161–2177. 10.1105/tpc.16.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S. D., Nelson J. C. (1996). Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92, 191–203. 10.1007/BF00223376 [DOI] [PubMed] [Google Scholar]
- Tanksley S. D., Young N. D., Paterson A. H., Bonierbale M. W. (1989). RFLP mapping in plant breeding: new tools for an old science. Bio/Technology 7, 257–264. 10.1038/nbt0389-257 [DOI] [Google Scholar]
- Tian F., De J. L., Fu Q., Zuo F. Z., Yong C. F., Xiang K. W., et al. (2006). Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112, 570–580. 10.1007/s00122-005-0165-2 [DOI] [PubMed] [Google Scholar]
- Tilman D., Balzer C., Hill J., Befort B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. 10.1073/pnas.1116437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppino L., Barchi L., Lo Scalzo R., Palazzolo E., Francese G., Fibiani M., et al. (2016). Mapping quantitative trait loci affecting biochemical and morphological fruit properties in Eggplant (Solanum melongena L.). Front. Plant Sci. 7:256. 10.3389/fpls.2016.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppino L., Valè G., Rotino G. L. (2008). Inheritance of Fusarium wilt resistance introgressed from Solanum aethiopicum Gilo and Aculeatum groups into cultivated eggplant (S. melongena) and development of associated PCR-based markers. Mol. Breed. 22, 237–250. 10.1007/s11032-008-9170-x [DOI] [Google Scholar]
- Valluru R., Davies W. J., Reynolds M. P., Dodd I. C. (2016). Foliar abscisic acid-to-ethylene accumulation and response regulate shoot growth sensitivity to mild drought in wheat. Front. Plant Sci. 7:461. 10.3389/fpls.2016.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Graner A., Sorrells M. E. (2005). Genic microsatellite markers in plants:features and applications. Trends Biotechnol. 23, 48–55. 10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Verlaan M. G., Hutton S. F., Ibrahem R. M., Kormelink R., Visser R. G. F., Scott J. W., et al. (2013). The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-Class RNA-Dependent RNA Polymerases. PLoS Genet. 9:e1003399. 10.1371/journal.pgen.1003399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova S., Manzur J. P., Prohens J. (2012). Development and characterization of genomic simple sequence repeat markers in eggplant and their application to the study of diversity and relationships in a collection of different cultivar types and origins. Mol. Breed. 30, 647–660. 10.1007/s11032-011-9650-2 [DOI] [Google Scholar]
- Von Korff M., Wang H., Léon J., Pillen K. (2004). Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor. Appl. Genet. 109, 1736–1745. 10.1007/s00122-004-1818-2 [DOI] [PubMed] [Google Scholar]
- Vorontsova M. S., Knapp S. (2016). A revision of the spiny solanums, Solanum subgenus Leptostemonum (Solanaceae) in Africa and Madagascar. Syst. Bot. Monogr. 99, 1–436. [Google Scholar]
- Vos P. G., Uitdewilligen J. G., Voorrips R. E., Visser R. G. F., van Eck H. J. (2015). Development and analysis of a 20K SNP array for potato (Solanum tuberosum): an insight into the breeding history. Theor. Appl. Genet. 128, 2387–2401. 10.1007/s00122-015-2593-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang J., Jiang J., Zhang Q. (2000). Mapping of centromeric regions on the molecular linkage map of rice (Oryza sativa L.) using centromere-associated sequences. Mol. Gen. Genet. 263, 165–172. 10.1007/s004380050044 [DOI] [PubMed] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., Huang B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang X., Cheng Z., Mueller L., Giovannoni J., Tanksley S. D. (2006). Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics 172, 2529–2540. 10.1534/genetics.106.055772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrhahn C., Allard R. (1965). The detection and measurement of the effects of individual genes involved in the inheritance of a quantitative character in wheat. Genetics 51, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer C. T., Reed G. H., Gundry C. N., Vandersteen J. G., Pryor R. J. (2003). High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49, 853–860. 10.1373/49.6.853 [DOI] [PubMed] [Google Scholar]
- Wu F., Eannetta N. T., Xu Y., Tanksley S. D. (2009). A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor. Appl. Genet. 118, 927–935. 10.1007/s00122-008-0950-9 [DOI] [PubMed] [Google Scholar]
- Wu F., Mueller L. A., Crouzillat D., Patiard V., Tanksley S. D. (2006). Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174, 1407–1420. 10.1534/genetics.106.062455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhao Q., Du P., Xu C., Wang B., Feng Q., et al. (2010). Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 11:656. 10.1186/1471-2164-11-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K., Mochizuki H. (1979). Nature and inheritance of Fusarium-wilt resistance in eggplant cultivars and related wild Solanum species. Bull. Veg. Ornam. Crop. Res. Stn. 6, 19–27. [Google Scholar]
- Yang X., Cheng Y. F., Deng C., Ma Y., Wang Z. W., Chen X. H., et al. (2014). Comparative transcriptome analysis of eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.): phylogenomics and disease resistance analysis. BMC Genomics 15:412. 10.1186/1471-2164-15-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Xu Y., Shah T., Li H., Han Z., Li J., et al. (2011). Comparison of SSRs and SNPs in assessment of genetic relatedness in maize. Genetica 139, 1045–1054. 10.1007/s10709-011-9606-9 [DOI] [PubMed] [Google Scholar]
- Yin X., Struik P., Gu J., Wang H. (2016). Modelling QTL-trait-crop relationships: past experiences and future prospects, in Crop Systems Biology, eds Yin X., Stuik C. (Berlin: Springer; ), 193193–218.218. [Google Scholar]
- Zamir D. (2001). Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989. 10.1038/35103590 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao Q., Sun Z. Z., Zhang C. Q., Feng Q., Tang S. Z., et al. (2011). Development and high-throughput genotyping of substitution lines carring the chromosome segments of indica 9311 in the background of japonica Nipponbare. J. Genet. Genomics 38, 603–611. 10.1016/j.jgg.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Zhang N., Liu B., Ma C., Zhang G., Chang J., Si H., et al. (2014). Transcriptome characterization and sequencing-based identification of drought-responsive genes in potato. Mol. Biol. Rep. 41, 505–517. 10.1007/s11033-013-2886-7 [DOI] [PubMed] [Google Scholar]
- Zhang W., Cao Y., Wang K., Zhao T., Chen J., Pan M., et al. (2014). Identification of centromeric regions on the linkage map of cotton using centromere-related repeats. Genomics 104, 587–593. 10.1016/j.ygeno.2014.09.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.