Abstract
DNA sequence analysis by oligonucleotide binding is often affected by interference with the secondary structure of the target DNA. Here we describe an approach that improves DNA secondary structure prediction by combining enzymatic probing of DNA by structure-specific 5′-nucleases with an energy minimization algorithm that utilizes the 5′-nuclease cleavage sites as constraints. The method can identify structural differences between two DNA molecules caused by minor sequence variations such as a single nucleotide mutation. It also demonstrates the existence of long-range interactions between DNA regions separated by >300 nt and the formation of multiple alternative structures by a 244 nt DNA molecule. The differences in the secondary structure of DNA molecules revealed by 5′-nuclease probing were used to design structure-specific probes for mutation discrimination that target the regions of structural, rather than sequence, differences. We also demonstrate the performance of structure-specific ‘bridge’ probes complementary to non-contiguous regions of the target molecule. The structure-specific probes do not require the high stringency binding conditions necessary for methods based on mismatch formation and permit mutation detection at temperatures from 4 to 37°C. Structure-specific sequence analysis is applied for mutation detection in the Mycobacterium tuberculosis katG gene and for genotyping of the hepatitis C virus.
INTRODUCTION
Sequence analysis of nucleic acids by oligonucleotide binding has traveled a long road from the pioneer works by Southern (1) and Wallace et al. (2) to an idea of large-scale sequence analysis with arrays of short oligonucleotides (3–5) and the development of arrays containing up to hundreds of thousands of different oligonucleotides (6). The basic idea of these methods is that the efficiency of probe binding depends on similarity between the target and probe, with the most stable duplexes being formed by fully complementary sequences. Any variations in the target sequence, such as substitutions, deletions or insertions, produce imperfect, less stable duplexes. This differential stability between the perfect and imperfect duplexes can be used as a means for DNA sequencing and mutation detection.
To achieve maximal mutation discrimination, the oligonucleotide binding should be performed under high stringency conditions, so that the perfect duplex remains stable but the mismatched duplex is unstable. Depending on the type of mismatch, its position in the duplex and probe length, a single mismatch will decrease the stability of the 16–22 nt probes typically used for mutation analysis by 3–10°C (7,8). Therefore, for successful mutation detection, any parameters affecting duplex stability, such as temperature, pH or concentrations of salt and destabilizing agents, must be carefully selected and tightly controlled during the analysis.
The secondary structure of nucleic acids is another well-documented factor that affects probe binding for both DNA and RNA molecules (9–16). Formation of secondary structure can reduce the binding constant of a specific probe by as much as 105–106 (11). This effect can obscure the relation between probe affinity and its similarity to the target. Although high stringency conditions reduce the effect of secondary structures on probe binding (9), decreasing the length of target molecules by additional chemical, enzymatic or thermal fragmentation is usually required to minimize the probability of secondary structure formation (6). Despite these measures, secondary structure still remains an unknown parameter that complicates sequence analysis (17). Evaluating this parameter is possible only if detailed knowledge of secondary structures is available.
Most experimental methods for secondary structure analysis rely on the differential reactivity of single- and double-stranded regions of nucleic acids with chemical or enzymatic agents (18,19). However, deducing structural information from these data is complicated by the fact that such reactivity is usually sequence dependent and is averaged over an ensemble of possible structures adopted by the molecule. Among computational methods, the comparative or phylogenetic methods for secondary structure prediction (20) provide the most reliable information, although they apply only to families of sequences with the same function. Secondary structure prediction for any given sequence relies on energy minimization algorithms, such as the well-known mfold program (21). The probability of correctly predicting a secondary structure by these algorithms is quite poor because of the limitations of the mathematical model and the uncertainties in the thermodynamic parameters used in these methods. To partially overcome this problem, mfold predicts multiple suboptimal structures with close free energy values. To select likely actual candidate structures thus requires further analysis.
Incorporating constraint parameters obtained from experimental data into computational methods used to predict secondary structures can greatly improve the results (22), but this approach is limited by the sensitivity of current structure probing techniques. In this work, we have used enzymatic cleavage of single-stranded DNA with the 5′-nuclease TaqExo derived from Taq DNA polymerase I (23,24) to obtain information about hairpin structures formed by DNA molecules. TaqExo specifically recognizes hairpin structures with stem duplexes longer than 6 bp and cleaves them between the first two base pairs at the 5′-end of the hairpin, thus creating a pattern of fragments unique for each sequence. It has been previously demonstrated (23) that TaqExo fragment patterns are extremely sensitive to small changes in the secondary structure of DNA, thus providing a useful tool to detect the point mutations responsible for such changes.
We used TaqExo cleavage sites as mfold constraint parameters to determine the secondary structures of Mycobacterium tuberculosis wild-type and mutant katG genes and hepatitis C virus (HCV) cDNAs. We demonstrate that a single mutation can significantly alter the folding of DNA molecules, as has been previously observed for RNA molecules (25), and that even relatively short molecules can adopt multiple mutually exclusive conformations. The revealed structural differences between similar DNA molecules were used to design structure-specific probes for mutation discrimination that target the conformationally different regions rather than the mismatch sites. We also designed structure-specific ‘bridge’ probes that bind to two non-contiguous regions in the target molecule whose relative positions are strongly affected by mutations. The bridge probes are similar to tethered (26,27) and stem-bridging oligonucleotides (28), which have been previously described for recognition of structured RNA and DNA molecules. The bridge probes we designed, based upon the TaqExo/mfold secondary structure prediction, showed a level of mutation discrimination similar to or exceeding that of linear mismatch discriminating probes. Finally, structure-specific probe binding requires no target fragmentation and can be performed under low stringency conditions that favor secondary structure formation, e.g. room temperature, reducing the adverse effects of temperature and binding buffer variations on mutation discrimination.
MATERIALS AND METHODS
Materials
Chemicals and buffers were from Fisher Scientific unless otherwise noted. Restriction enzymes were purchased from New England Biolabs. PCR amplification was done using a GeneAmp kit with AmpliTaq DNA polymerase (Perkin Elmer). TaqExo and MjaFEN 5′-nucleases were prepared as described (23,29).
Oligonucleotide synthesis and purification
All oligonucleotides were synthesized on an Expedite 8909 synthesizer (PerSeptive Biosystems) using standard phosphoramidite chemistry including biotin, fluorescein (Fl), and tetrachlorofluorescein (TET) modifications (Glen Research) and purified as described previously (30). Oligonucleotide concentrations were determined by measuring absorption at 260 nm and using specific extinction coefficients for A, T, G and C (31).
Mycobacterium tuberculosis katG gene DNA fragments
Genomic DNAs isolated from wild-type and mutant isoniazide-resistant strains of M.tuberculosis were the gift of Dr Cockerill (Mayo Clinic). A fragment of the catalase–peroxidase (katG) gene (GenBank accession no. U06263) corresponding to codons 302–507 was PCR amplified for each genomic DNA with sense and antisense strand primers 5′-AGCTCGTATGGCACCGGAAC and 5′-TTGACCTCCCACCCGACTTG, respectively, and cloned into a TA vector (Invitrogen). Presence of the G→C mutation at position 41 (G41C) of the mutant fragment was confirmed by sequencing. The 379, 391, 423 and 504 bp katG DNA fragments were generated by PCR amplification of the cloned fragments with the same sense strand primer labeled with Fl or TET at the 5′-end and one of the antisense strand primers 5′-CAAGGTATCTGGCAAGGGGA, 5′-GGACCAGCGGCCCAAGGTAT, 5′-GACCGGATCCTGCCACAGCA or 5′-GACAGTCAATCCCGATGCCC, respectively. For the wild-type and mutant 391 bp katG DNA fragment carrying the C385G substitution the antisense strand primer 5′-GGACCACCGGCCCAAGGTAT was used. The 423 bp katG DNA fragment, internally labeled with dUTP-Fl (Roche Molecular Biochemicals), was PCR amplified as described above, except that a mixture of 150 µM dTTP and 50 µM dUTP-Fl was used instead of 200 µM dTTP. The PCR products were purified by denaturing gel electrophoresis as described previously (23).
HCV 5′-untranslated region (5′-UTR) DNA fragments
The 244 bp DNA fragments of the 5′-UTR of HCV genotypes 1a, 1b, 2a/c and 3a were PCR amplified with negative strand primer 5′-CTCGCAAGCACCCTATCAGGCAGT labeled with biotin or TET at the 5′-end, and positive strand primer 5′-GCAGAAAGCGTCTAGCCATGGCGT using in-house HCV cDNA clones. The PCR products were purified as described above.
Sequencing reactions
Sequencing reactions were performed with a Thermosequenase kit (Amersham Pharmacia Biotech) using 250–500 ng PCR product as template and 0.25 µM sequencing primers labeled with TET at the 5′-end. The sequencing products were resolved on an 8% denaturing polyacrylamide gel and scanned on an FMBIO-100 fluorescence scanner (Hitachi) using a 585 nm emission filter.
TaqExo probing of DNA secondary structure
This was performed as described previously (23). Briefly, 100–500 fmol PCR product, with the analyzed strand labeled with TET at the 5′-end, was heat denatured in 13 µl of 5 mM MOPS, pH 7.5, for 15 s at 95°C and then cooled to 55°C. The reaction samples were mixed with 7 µl of a solution containing 1 µl of 25 ng/µl TaqExo, 2 µl of 100 mM MOPS, pH 7.5, 0.5% Tween-20, 0.5% Nonidet-P40, 2 µl of 2 mM MnCl2, 2 µl of water and the mixtures were then incubated for 90 s at 55°C. The reactions were terminated by addition of 16 µl of 95% formamide, 10 mM EDTA, pH 8.0, 0.05% crystal violet (Sigma). Aliquots of TaqExo cleavage products (5 µl) were resolved on a 10% denaturing polyacrylamide gel and scanned on an FMBIO-100 fluorescence scanner (Hitachi) using a 585 nm emission filter. A 25 bp DNA ladder (Promega) labeled at the 5′-end with a FluoroReporter Bodipy TMR-C5 oligonucleotide labeling kit (Molecular Probes) was used as molecular weight markers.
Secondary structure prediction using mfold and TaqExo constraints
DNA folding was performed at the DNA mfold server (bioinfo.math.rpi.edu/~mfold/dna/) using DNA free energy parameters (7). Each TaqExo cleavage site, selected as a constraint in DNA folding, was encoded according to the constraint options provided by the program. For example, a cleavage site at position n was represented by parameters F n 0 2 to force the nucleotides located directly 3′ and 5′ of the cleavage site to be base paired and P n – n + 1 1 – n + 16 to prohibit base pairing of nucleotides n and n + 1 with a region 1 – n + 16, and therefore to ensure that in the predicted structures the cleavage site n is located at the 5′-end of a hairpin with an at least 7 bp duplex region. Compatibility between constraints was investigated by a trial-and-error method and the sets of compatible constraints were determined manually.
Oligonucleotide probe binding under low stringency conditions
For each binding reaction, the 423 bp katG PCR product (10–30 nM), labeled with Fl either at the 5′-end of the target strand or internally, was denatured in 0.2 M NaOH, 5 mM EDTA for 10 min. The denatured target (1 µl) was mixed with 1.5 pmol of one of the probes labeled with biotin in 150 µl of a buffer containing 0.8 M NaCl, 45 mM NaH2PO4, pH 7.4, 4.5 mM EDTA, 0.2% Ultrapure BSA (Panvera) and 10 ng/µl tRNA (Sigma) and incubated at room temperature for 30 min before 100 µl of the reaction mixture was transferred to a well of a streptavidin-coated microtiter plate (Boehringer Mannheim, catalog no. 1,734,784). After incubation for 20 min at room temperature, the plate was washed three times with TBS buffer containing 25 mM Tris–HCl, pH 7.2, 0.15 M NaCl, 0.05 mg/ml NaN3, 0.1% Tween-20, using an Autostrip ELX50 (Bio-Tek Instruments). Each well of the washed plate was incubated for 20 min at room temperature with 100 µl of SuperBlock solution (Pierce) in TBS buffer containing 0.015 U Anti-Fluorescein-AP (Boehringer Mannheim). The unbound conjugate was washed out as described above and 100 µl of 0.6 mg/ml AttoPhos in AttoPhos buffer (JBL Scientific) was added to each well. After incubation for 30 min at 37°C, the chemiluminescence signal in each well was measured using a CytoFluor 4000 (PerSeptive Biosystems) equipped with a 450/50 excitation filter and a 580/50 emission filter. For signal normalization, the control katG probe, 5′-CGT CCT TGG CGG TGT ATT labeled at the 5′-end with biotin, was used.
The probe binding reactions at room temperature for the 244 nt HCV 5′-UTR DNA fragments were performed as described above, except that the DNA targets were 5′-labeled with biotin and probes were 5′-labeled with Fl. For the probe binding experiments at 4 and 37°C, both the probe binding and microtiter plate binding steps were performed at the selected temperature and the subsequent steps were carried out at room temperature. The sequences of the HCV structure-specific probes were 5′-TTG GGC GTT GCT TGT GGT (probe 3-2), 5′-AGT GTC GTT TGG AAC CGG (probe 5-4), 5′-AGT GTC GTT TCT TGT GGT (probe 5-2) and 5′-GCA GAA AGT TCT TGC GAG (probe 6-1). The spacer dinucleotides of the structure-specific probes are shown in italic. For signal normalization, the control probe, 5′-GCG AAA GGC CTT GTG G labeled at the 5′-end with Fl, was used.
RESULTS
Changes in secondary structure of single-stranded DNA induced by a single mutation can be detected by TaqExo probing
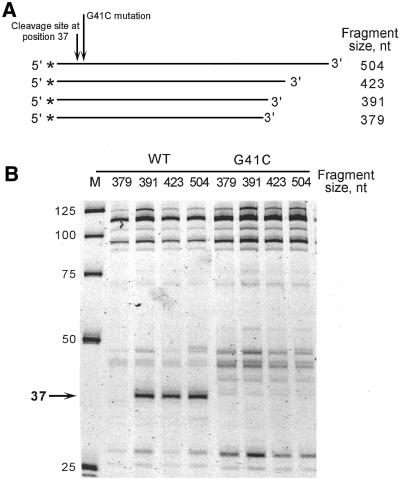
The ability of structure-specific 5′-nucleases to create a unique pattern of cleavage products has been applied to mutation analysis of numerous DNA molecules (23,32–36). Because 5′-nuclease cleavage patterns are sensitive to single nucleotide substitutions, they can be used to identify structural differences between closely related DNA sequences. An example of such an analysis is shown in Figure 1 for wild-type and mutant DNA fragments of the M.tuberculosis katG gene that differ by only a single G→C mutation at position 41 (G41C) (Fig. 1A). The TaqExo cleavage patterns of the wild-type and mutant 504 nt fragments (Fig. 1B) show that the G41C mutation eliminates the 37 nt cleavage product present in the wild-type molecule with little or no effect on the rest of the pattern (for brevity, only the portion of the gel corresponding to cleavage products that are 25–125 nt long is shown). Thus, TaqExo probing suggests that the G41C mutation destroys the hairpin structure responsible for the 37 nt cleavage product of the wild-type katG DNA fragment.
Figure 1.
Analysis of structural changes in katG DNA by TaqExo probing and the ‘PCR walk’ method. (A) Schematic presentation of the 504, 423, 391 and 379 nt fragments of katG gene DNA. The fragments have identical sequences at the 5′-ends labeled with TET (*). The TaqExo cleavage site at position 37 and the G37C mutation are shown by short and long arrows, respectively. (B) The TaqExo cleavage products of the wild-type (WT) and mutant (G41C) katG DNA fragments shown in (A). The 37 nt product is shown by an arrow. M, size markers.
Long-range interactions in the secondary structure of the wild-type katG DNA
According to the substrate specificity of TaqExo (29,37), the cleavage site at position 37 defines the 5′-end of a hairpin structure formed in the wild-type DNA molecule. Mfold predicts that nucleotide A37 of the 504 nt wild-type fragment can pair with either T254 or T389, if it is forced to be at the 5′-end of a hairpin structure (data not shown). To determine experimentally which of these two nucleotides defines the 3′-end of the hairpin, we developed a technique called the ‘PCR walk’. Using nested PCR primers, two sets of DNA fragments, which have identical 5′-ends but progressively truncated 3′-ends, were prepared for the wild-type and mutant katG genes by PCR amplification, as shown in Figure 1A. Analysis of the TaqExo cleavage products demonstrates that truncation of the wild-type DNA from 504 to 391 nucleotides does not affect the cleavage site at position 37, however, a further reduction from 391 to 379 nt causes the 37 nt product to disappear (Fig. 1B).
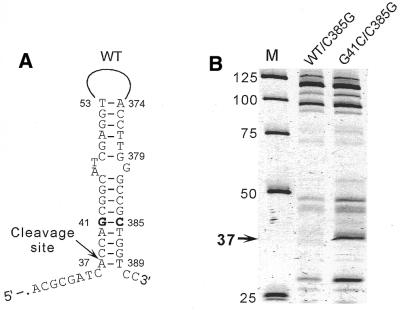
Disappearance of the 37 nt product was the only major change in the cleavage pattern of the 379 nt wild-type fragment, indicating that the length reduction did not affect other elements of the secondary structure. Thus, the PCR walk analysis suggests that the 3′-end of the hairpin that gives rise to the 37 nucleotides product is located between nucleotides 379 and 391. This result is consistent with the mfold structure prediction that indicates formation of an A37·T389 base pair in a potential hairpin duplex consisting of regions 37–53 and 374–390 of the wild-type sequence, as shown in Figure 2A. The G41C mutation would substitute the G41·C385 base pair in the wild-type structure with a C·C mismatch in the mutant structure, which increases the calculated free energy of hairpin duplex formation from –17.3 to –11.7 kcal/mol.
Figure 2.
Confirmation of the long-range interactions in wild-type katG DNA by compensatory substitutions. (A) Proposed hairpin structure of the wild-type (WT) 391 nt katG DNA fragment. The G41·C385 base pair is shown in bold. (B) The TaqExo cleavage products of the WT/C385G and G41C/C385G 391 nt katG DNA fragments carrying the C385G substitution. The 37 nt cleavage product and its position in the structures are shown by arrows. M, size markers. The nucleotide positions discussed in the text are indicated.
To experimentally confirm the existence of long-range base pairing between nucleotides G41 and C385, a C→G substitution was introduced at position 385 of the 391 nt wild-type and mutant katG DNA fragments. If the proposed hairpin structure was correct, the C385G substitution would favor base pairing between nucleotides 41 and 385 in the mutant G41C molecule, but would also substitute a G·G mismatch for the G41·C385 base pair in the wild-type DNA. In agreement with this prediction, TaqExo analysis showed that the C385G substitution restores the cleavage site at position 37 in the mutant fragment, but eliminates this site in the wild-type fragment (compare Figs 2B and 1B).
Mutation detection in the katG gene by structure-specific probes
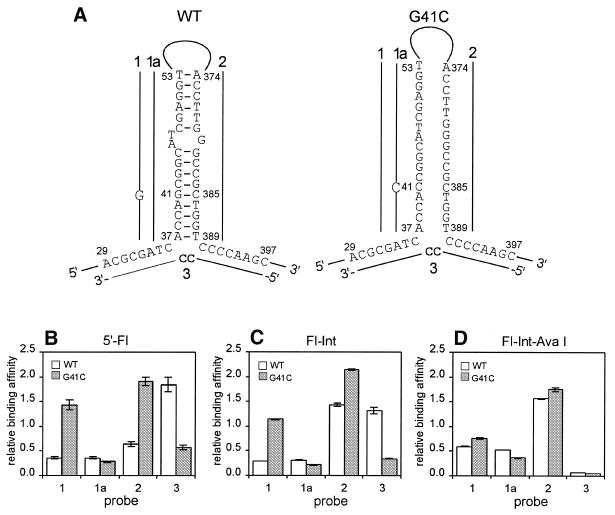
Using the proposed hairpin structure of the wild-type katG DNA, we designed four structure-specific probes for G41C mutation discrimination (Fig. 3A). Two probes, 1 and 1a, are complementary to region 37–53 of the mutant and wild-type DNAs, respectively, and their binding should be affected by both mismatch formation and secondary structure. Probe 2 is complementary to region 374–389, which comprises the 3′-part of the hairpin duplex formed in the wild-type structure. Bridge probe 3 is complementary to two non-contiguous regions, 29–36 and 390–397, which are presumably brought into close proximity in the secondary structure of the wild-type DNA (Fig. 3A). The target-specific regions of probe 3 are linked by a 5′-CC dinucleotide to stabilize the potential three-way helical DNA junction formed by probe 3 and the wild-type katG target (38). Because probes 2 and 3 are complementary to regions that do not include the G41C mutation, their binding should depend only on structural differences between the wild-type and mutant katG DNAs.
Figure 3.
Detection of the G41C mutation in katG DNA by structure-specific probe binding under low stringency conditions. (A) Proposed structures of the wild-type (WT) and mutant (G41C) 423 nt fragments of katG DNA. Probes 1, 1a, 2 and 3 are shown by solid lines at the regions complementary to the targets. Non-complementary nucleotides in the probes are indicated. (B) Relative binding affinities of probes 1, 1a, 2 and 3 with WT and G41C targets labeled with fluorescein at the 5′-end (5′-Fl). (C) Identical to (B) but with the targets internally labeled with fluorescein (Fl-Int). (D) Identical to (C) but with the internally labeled targets treated with AvaI (Fl-Int-Ava I). The binding affinities for WT and G41C targets are shown by white and gray rectangles, respectively. Error bars indicate the standard deviations obtained from triplicate measurements.
Binding of the designed structure-specific probes was studied by allowing complex formation between a FI-labeled single-stranded DNA target and a 5′-biotin-labeled probe under low stringency conditions at room temperature and then capturing the complex in streptavidin-coated microtiter plates. The relative binding of each probe was determined from the absolute fluorescence signal of the captured DNA target, normalized to the signal of a control probe (see Materials and Methods). Figure 3B shows the binding affinities of probes 1, 1a, 2 and 3 for the 423 nt fragments of the wild-type and mutant katG DNAs. Probe 1 shows a high level of discrimination between the two targets, possibly due to the combined negative effect of the mismatch and secondary structure on its binding with the wild-type fragment. Probe 1a binds weakly to both targets, presumably due to C·C mismatch formation with the mutant target and a competing secondary structure in the probe-binding region of the wild-type target. Probe 2 binds more efficiently with the mutant target, in agreement with the assumption that the probe-binding region in the mutant target is less structured compared to the wild-type target. Conversely, the presence of a hairpin in the wild-type target explains its high binding affinity for probe 3, probably due to formation of a stable three-way junction. The high binding affinity of probe 3 for the wild-type target and the high level of discrimination between the two targets demonstrate that bridge probes can be at least as efficient as linear probes.
Target fragmentation affects binding of structure-specific probes
Fragmentation of target DNA is often used to reduce the interference of secondary structures with probe binding (6). Consequently, target fragmentation should decrease the ability of structure-specific probes to discriminate mutations. To investigate this hypothesis, we studied the effect of AvaI treatment of wild-type and mutant katG DNA targets on the binding affinities of probes 1, 1a, 2 and 3 (Fig. 3A). The 423 nt katG targets, both internally labeled with FI, were cleaved with AvaI at positions 95 and 299, neither of which overlap with the binding sites of either the structure-specific or control probes. The internal labeling, required for detection of the fragmented DNA, did not significantly affect discrimination between the intact wild-type and mutant katG targets (compare Fig. 3B and C). However, the structure-specific probes were no longer able to discriminate the G41C mutation when AvaI-treated targets were used instead of intact ones (Fig. 3D). The binding affinities of bridge probe 3 were affected the most and decreased essentially to a background level.
Structural analysis of the 5′-UTR of hepatitis C virus
Analysis of the katG DNA described above was limited to a single hairpin structure. As a second example, we predicted the complete secondary structures of four DNA molecules corresponding to the 5′-UTR of HCV RNA. The 5′-UTR is highly conserved compared to the overall sequence of the viral RNA and is commonly used for HCV genotyping (32,39).
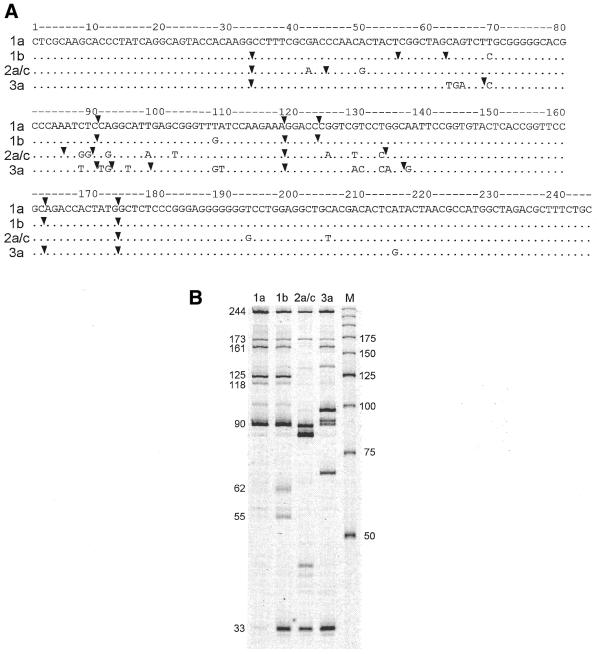
Figure 4A shows the sequences of the 244 nt DNA fragments corresponding to the region from –31 to –274 of the type 1a, 1b, 2a/c and 3a negative strand HCV RNAs chosen for this study. TaqExo cleavage products obtained by partial digestion of the four DNA molecules labeled at their 5′-ends are shown in Figure 4B. The positions of the TaqExo major cleavage sites were determined by sequencing gel analysis with single nucleotide resolution (data not shown) and indicated by the arrowheads in Figure 4A. These sites were then used as constraint parameters in mfold (21) to select only those structures that are consistent with the TaqExo data, as described in Materials and Methods. Briefly, each cleavage site was forced to be located between the first two nucleotides at the 5′-end of a hairpin structure with a duplex region of at least 7 bp. For convenience, we refer to the constraint imposed by a cleavage site at position n as constraint n.
Figure 4.
TaqExo analysis of types 1a, 1b, 2 and 3 5′-UTR HCV DNA fragments. (A) Alignment of DNA sequences corresponding to nucleotides –31 to –274 of the 5′-UTR of type 1a, 1b, 2 and 3 negative strand HCV RNAs (40). Major cleavage sites are indicated by arrows. (B) TaqExo cleavage products of type 1a, 1b, 2 and 3 DNAs shown in (A). M, size markers. The sizes of major cleavage products of type 1b DNA are indicated on the left.
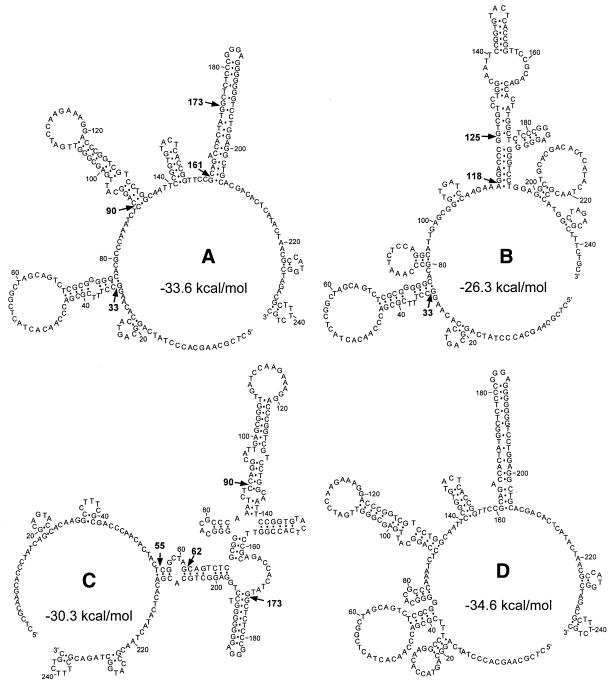
Figure 5A–C shows some of the structures predicted for the type 1b DNA molecule using mfold with constraints 33, 55, 62, 90, 118, 125, 161 and 173. Interestingly, the analysis could not predict any structure consistent with all constraints. For example, the structure shown in Figure 5A conforms to constraints 33, 90, 161 and 173, but is not compatible with constraints 55, 62, 118 and 125, because the positions of cleavage sites 55, 62, 118 and 125 in this structure contradict the substrate specificity of TaqExo (29). To satisfy constraints 118 and 125, the hairpin structures defined by constraints 90, 161 and 173 should be completely rearranged, resulting in the structure shown in Figure 5B. The structure shown in Figure 5C explains cleavage sites 55 and 62 and is compatible with constraints 90 and 173, but not with constraints 33 and 161. The calculated free energies of the structures proposed in Figure 5A–C at 1 M NaCl and 37°C are –33.6, –26.3 and –30.3 kcal/mol, respectively. In comparison, the optimal secondary structure predicted by mfold without any imposed constraints (Fig. 5D) has a free energy of –34.6 kcal/mol and is very similar to the structure shown in Figure 5A.
Figure 5.
Predicted secondary structures of the type 1b DNA fragment. Alternative secondary structures predicted for type 1b DNA by mfold using constraints (A) 33, 90, 161, 173, (B) 33, 118, 125 and (C) 55, 62, 90, 173. The positions of major cleavage sites are indicated by arrows. (D) Optimal structure of the type 1b DNA fragment predicted by mfold. The calculated free energies for each structure at 1 M NaCl and 37°C are shown.
Some cleavage sites, e.g. sites 173 and 125 in Figure 5A and B, respectively, are located at internal positions of the proposed hairpin structures. Since TaqExo cannot cleave duplexes internally, we assume that long hairpin structures can be partially melted under the reaction conditions (55°C, 10 mM MOPS, pH 7.5, 0.2 mM MnCl2) to generate truncated substrates for TaqExo (24,37).
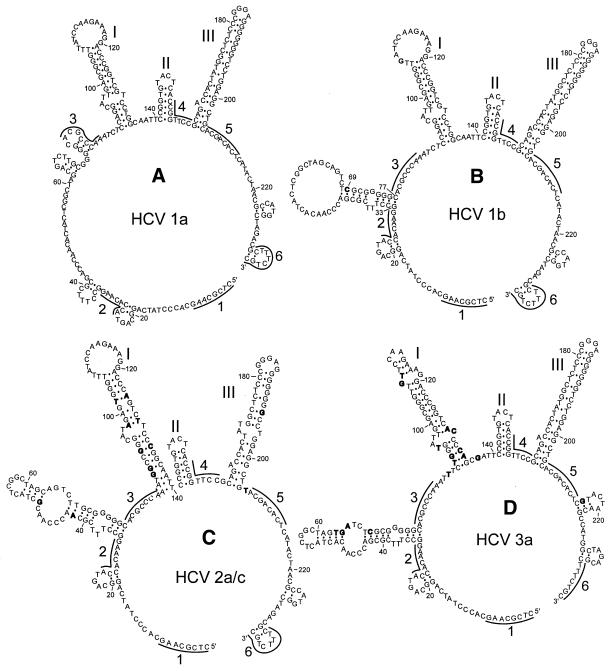
A similar analysis performed on types 1a, 2a/c and 3a DNA molecules using TaqExo cleavage sites (Fig. 4A) also revealed multiple alternative structures (data not shown), of which the most stable for each type are depicted in Figure 6, including the type 1b structure shown in Figure 5A. All four structures exhibit structurally conserved hairpins I, II and III. TaqExo does not detect hairpin II, probably because this enzyme cannot recognize hairpin duplexes shorter than 7–8 bp (29). However, the calculated free energy –6.1 kcal/mol of hairpin II and the observed cleavage at site 141 of all four types of DNA molecules with MjaFEN 5′-nuclease, which recognizes shorter hairpin duplexes (29), confirms the existence of hairpin II in the proposed structures (data not shown). Besides these conserved elements, the structures also exhibit structurally variable regions. For example, the T→C substitution at position 69 that discriminates HCV types 1a and 1b results in formation of a stable hairpin structure in region 33–77 of type 1b DNA (Fig. 6B). This difference in otherwise identical structures is supported by the strong TaqExo cleavage at position 33 of type 1b DNA, which is greatly reduced in type 1a DNA (Fig. 4B) and has not been used as a constraint for type 1a DNA structure prediction.
Figure 6.
Optimal secondary structures of type 1a, 1b, 2 and 3 5′-UTR DNA fragments predicted by mfold using TaqExo constraints. The structures were predicted using constraints (A) 90, 161, 173 for type 1a, (B) 33, 90, 161, 173 for type 1b, (C) 33, 85, 89, 173 for type 2a/c and (D) 33, 90, 92, 98, 161, 173 for type 3a (see Fig. 4A) and conditions of 1 M NaCl and 37°C. Polymorphic substitution identifying each genotype are shown in bold. Conserved hairpin structures I–III are indicated. Regions used for designing bridge probes are shown by solid lines and denoted by Arabic numerals.
Structure-specific bridge probes for HCV genotyping
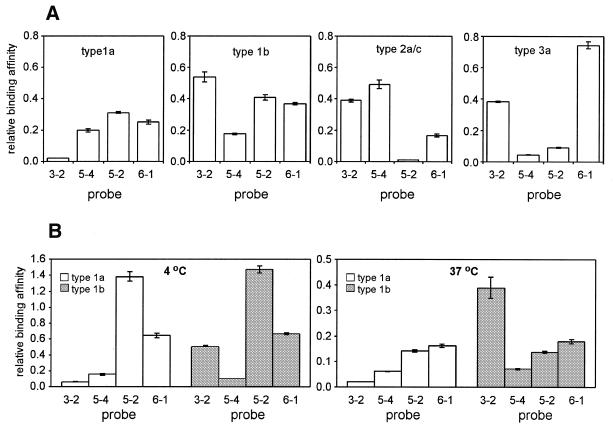
The proposed structures of the HCV cDNA molecules (Fig. 6) were used to design structure-specific bridge probes for HCV genotyping. We selected six 8 nt regions, shown in Figure 6, which are likely to be accessible according to the proposed structures. To exclude the effect of mismatch formation on probe binding, the regions were selected from sequences conserved among all four types. Each bridge probe consists of 5′ and 3′ 8 nt regions, complementary to two of the six selected regions, linked together by a spacer dinucleotide. The bridge probes were classified according to the target regions used for their design. For example, a probe with 5′ and 3′ regions complementary to target regions 3 and 2, respectively, is referred to as probe 3-2. Among 30 possible combinations, we tested 10 probes (data not shown) and finally selected a minimal set of four probes, 3-2, 5-4, 5-2 and 7-1, sufficient to identify the 1a, 1b, 2a/c and 3a genotypes. Figure 7A demonstrates that these probes exhibit unique and highly reproducible binding profiles with the four DNA targets under low stringency conditions, with a coefficient of variation of <6%, thus ensuring reliable discrimination of all four targets.
Figure 7.
HCV genotype identification using structure-specific probe binding under low stringency conditions. (A) Relative binding affinities of bridge probes 3-2, 5-4, 5-2 and 6-1 for type 1a, 1b, 2a/c and 3a 5′-UTR DNA targets at room temperature. (B) Identical to (A) but with the type 1a and 1b targets at 4 and 37°C. The binding affinities for the type 1a and 1b targets are shown by white and gray rectangles, respectively. Error bars indicate the standard deviations obtained from triplicate measurements.
Sequence analysis by structure-specific probe binding under low stringency conditions is temperature independent
Because secondary structure is the major factor affecting binding between target and structure-specific probes, the ability of probes to discriminate targets should be temperature independent under conditions favoring the formation of secondary structure. We tested this hypothesis by comparing the binding of probes 3-2, 5-4, 5-2 and 6-1 to type 1a and 1b DNA targets at room temperature, 4 and 37°C. Figure 7B shows that probe 3-2, which binds with type 1b DNA more strongly than with type 1a DNA at room temperature (Fig. 7A), exhibits the same level of discrimination at both 4 and 37°C, whereas the binding affinities of probes 5-4, 5-2 and 6-1 are practically independent of type at all temperatures. Thus, structure-specific probes can be used for reliable sequence analysis over a temperature range of >30°C, including room temperature.
DISCUSSION
Combined use of TaqExo enzymatic probing with the energy minimization mfold algorithm (21) was used in this work to improve the prediction of DNA secondary structures. The unique specificity of TaqExo, which cleaves hairpin structures precisely at their 5′-ends, permits straightforward conversion of TaqExo cleavage data into formal mfold constraint parameters. It can also be used with the PCR walk method described in this work. The power of TaqExo probing is demonstrated by detection of structural changes caused by single nucleotide variations in the katG gene DNA (Fig. 3A) and the HCV 5′-UTR cDNA (Fig. 6A and B) and by prediction of multiple alternative structures for the 244 nt type 1b HCV DNA fragment (Fig. 5). The latter example underscores the advantage of TaqExo probing when compared with other enzymatic and chemical methods by providing a superposition of discrete cleavage patterns rather than an average reactivity of each nucleotide over an ensemble of all alternative structures.
The prediction of alternative structures can be helpful in optimizing the thermodynamic parameters used in energy minimization programs. For example, the calculated free energies of two alternative structures adopted by type 1b DNA (Fig. 5A and B) differ by >7 kcal/mol. Such a difference would make formation of the first structure >105-fold more probable than formation of the second. Because this conclusion is not consistent with the fact that cleavage sites 118 and 125 are clearly detectable (Fig. 4B), additional interactions that are not accounted for in mfold likely stabilize the structure shown in Figure 5B.
The ability to identify structural changes caused by a single mutation opens new opportunities in designing oligonucleotide probes for sequence analysis based on structural differences rather than mismatch formation. Because structure-specific probes are not restricted to the site of mutation, a large number of these probes, especially bridge probes, can be designed, as opposed to mismatch discriminating probes, thus increasing the probability of achieving the desired level of mutation discrimination. The structure-specific approach has the further advantage of reducing the number of probes required for sequence analysis of multiple targets. For example, we used only four probes to reliably identify four HCV genotypes (Fig. 7A). Standard mismatch detecting methods would require at least twice as many probes for such an analysis. Potentially, structure-specific probes can substantially improve mutation discrimination compared with probes designed solely on the mismatch formation principle. For example, binding of probe 3-2 with types 1a and 1b HCV targets exhibits a very high discrimination factor of 25:1 (Fig. 7A).
The predicted secondary structures of target molecules can provide significant insight in designing highly selective, structure-specific probes. For example, probe 3-2 binds to type 1b, 2a/c and 3a HCV DNA molecules significantly more strongly than to the type 1a target (Fig. 7A). This finding agrees with the proposed hairpin structure in region 33–77 of type 1b, 2a/c and 3a molecules that brings regions 2 and 3 into close proximity to each other (Fig. 6). On the other hand, proposed structural differences among the HCV targets cannot explain in such simple terms the differential binding of the other three probes, suggesting that more elaborate models are required for more accurate prediction of the correlation between nucleic acid structure and the binding affinity of structure-specific probes.
An important feature of the structure-specific probes is their ability to detect mutations under low stringency conditions. We observed nearly identical levels of discrimination between the type 1a and 1b 5′-UTR DNAs with probe 3-2 at 4°C, room temperature and 37°C (Fig. 7). This result supports the hypothesis that structural differences between the targets remain practically unchanged under these conditions. Absolute binding efficiencies of the bridge probes, measured as the intensities of the fluorescence signals generated by the same amount of 1a and 1b 5′-UTR DNAs, were several-fold higher at room temperature than at 4 or 37°C (data not shown). Thus, temperature-induced variations in secondary structures can influence binding of bridge probes, but have no apparent effect on their ability to discriminate mutations. The negligible effect of temperature on the discrimination factor contributes to the high reproducibility of the results, permitting reliable sequence analysis even when the discrimination factor of structure-specific probes is only 2:1, e.g. the katG-specific probe 2 (Fig. 3B). In contrast, the strong temperature effect on relative binding of complementary and mismatched probes often adversely affects the reproducibility of mismatch discriminating methods operating under high stringency conditions.
We have demonstrated in this work that available secondary structure prediction programs, such as mfold, can benefit from the structural information provided by TaqExo 5′-nuclease, thus improving the probability of correct structure prediction. In addition, TaqExo data can be used to refine energy parameters for certain structural elements, e.g. three-way junctions, hairpins and internal loops, eventually simplifying the design of highly efficient structure-specific probes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Olke Uhlenbeck, James Dahlberg, Ted Ullman, Michael Zuker and John SantaLucia for discussions, Peggy Eis, Kafryn Lieder and Andrew Lukowiak for critically reading the manuscript, Frank Cockerill and Manual Altamirano for DNA samples and Natasha Lyamicheva for katG DNA clones. The work was supported by Cooperative Agreement 70NANB7H3015 from the Department of Commerce Advance Technology Program to Dr Lance Fors (Third Wave Technologies).
References
- 1.Southern E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol., 98, 503–517. [DOI] [PubMed] [Google Scholar]
- 2.Wallace R.B., Shaffer,J., Murphy,R.F., Bonner,J., Hirose,T. and Itakura,K. (1979) Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res., 6, 3543–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bains W. and Smith,G.C. (1988) A novel method for nucleic acid sequence determination. J. Theor. Biol., 135, 303–307. [DOI] [PubMed] [Google Scholar]
- 4.Drmanac R., Labat,I., Brukner,I. and Crkvenjakov,R. (1989) Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics, 4, 114–128. [DOI] [PubMed] [Google Scholar]
- 5.Southern E.M. (1996) DNA chips: analysing sequence by hybridization to oligonucleotides on a large scale. Trends Genet., 12, 110–115. [DOI] [PubMed] [Google Scholar]
- 6.Fodor S.P., Rava,R.P., Huang,X.C., Pease,A.C., Holmes,C.P. and Adams,C.L. (1993) Multiplexed biochemical assays with biological chips. Nature, 364, 555–556. [DOI] [PubMed] [Google Scholar]
- 7.SantaLucia J.,Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 9.Gamper H.B., Cimino,G.D. and Hearst,J.E. (1987) Solution hybridization of crosslinkable DNA oligonucleotides to bacteriophage M13 DNA. Effect of secondary structure on hybridization kinetics and equilibria. J. Mol. Biol., 197, 349–362. [DOI] [PubMed] [Google Scholar]
- 10.Fedorova O.S., Podust,L.M., Maksakova,G.A., Gorn,V.V. and Knorre,D.G. (1992) The influence of the target structure on the efficiency of alkylation of single-stranded DNA with the reactive derivatives of antisense oligonucleotides. FEBS Lett., 302, 47–50. [DOI] [PubMed] [Google Scholar]
- 11.Lima W.F., Monia,B.P., Ecker,D.J. and Freier,S.M. (1992) Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry, 31, 12055–12061. [DOI] [PubMed] [Google Scholar]
- 12.Godard G., Francois,J.C., Duroux,I., Asseline,U., Chassignol,M., Nguyen,T., Helene,C. and Saison-Behmoaras,T. (1994) Photochemically and chemically activatable antisense oligonucleotides: comparison of their reactivities towards DNA and RNA targets. Nucleic Acids Res., 22, 4789–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarrinkar P.P. and Williamson,J.R. (1994) Kinetic intermediates in RNA folding. Science, 265, 918–924. [DOI] [PubMed] [Google Scholar]
- 14.Parkhurst K.M. and Parkhurst,L.J. (1995) Kinetic studies by fluorescence resonance energy transfer employing a double-labeled oligonucleotide: hybridization to the oligonucleotide complement and to single-stranded DNA. Biochemistry, 34, 285–292. [DOI] [PubMed] [Google Scholar]
- 15.Schwille P., Oehlenschlager,F. and Walter,N.G. (1996) Quantitative hybridization kinetics of DNA probes to RNA in solution followed by diffusional fluorescence correlation analysis. Biochemistry, 35, 10182–10193. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nature Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 17.Williams J.C., Case-Green,S.C., Mir,K.U. and Southern,E.M. (1994) Studies of oligonucleotide interactions by hybridisation to arrays: the influence of dangling ends on duplex yield. Nucleic Acids Res., 22, 1365–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehresmann C., Bauldin,F., Mougel,M., Romby,P., Ebel,J.P. and Ehresmann,B. (1987) Probing the structure of RNAs in solution. Nucleic Acids Res., 15, 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knapp G. (1989) Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol., 180, 192–212. [DOI] [PubMed] [Google Scholar]
- 20.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 22.Gaspin C. and Westhof,E. (1995) An interactive framework for RNA secondary structure prediction with a dynamical treatment of constraints. J. Mol. Biol., 254, 163–174. [DOI] [PubMed] [Google Scholar]
- 23.Brow M.A., Oldenburg,M.C., Lyamichev,V., Heisler,L.M., Lyamicheva,N., Hall,J.G., Eagan,N.J., Olive,D.M., Smith,L.M., Fors,L. and Dahlberg,J.E. (1996) Differentiation of bacterial 16s rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage. J. Clin. Microbiol., 34, 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyamichev V., Brow,M.A., Varvel,V.E. and Dahlberg,J.E. (1999) Comparison of the 5′ nuclease activities of Taq DNA polymerase and its isolated nuclease domain. Proc. Natl Acad. Sci. USA, 96, 6143–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L.X., Basilion,J.P. and Stanton,V.P. (1999) Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl Acad. Sci. USA, 96, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson P.L. and Schepartz,A. (1991) Tethered oligonucleotide probes. A strategy for the recognition of structured RNA. J. Am. Chem. Soc., 113, 5109–5111. [Google Scholar]
- 27.Cload S.T., Richardson,P.L., Huang,Y.H. and Schepartz,A. (1993) Kinetic and thermodynamic analysis of RNA binding by tethered oligonucleotide probes: alternative structures and conformational changes. J. Am. Chem. Soc., 115, 5005–5014. [Google Scholar]
- 28.Francois J.C., Thuong,N.T. and Helene,C. (1994) Recognition and cleavage of hairpin structures in nucleic acids by oligodeoxynucleotides. Nucleic Acids Res., 22, 3943–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser M.W., Lyamicheva,N., Ma,W., Miller,C., Neri,B., Fors,L. and Lyamichev,V.I. (1999) A comparison of Eubacterial and Archaeal structure-specific 5′-exonucleases. J. Biol. Chem., 274, 21387–21394. [DOI] [PubMed] [Google Scholar]
- 30.Reynaldo L.P., Vologodskii,A.V., Neri,B.P. and Lyamichev,V.I. (2000) The kinetics of oligonucleotide replacements. J. Mol. Biol., 297, 511–520. [DOI] [PubMed] [Google Scholar]
- 31.Richards E.G. (1975) In Fasman,G.P. (ed.) Handbook of Biochemistry and Molecular Biology, 3rd Edn. CRC Press, Cleveland, OH, Vol. 1, p. 597. [Google Scholar]
- 32.Marshall D.J., Heisler,L.M., Lyamichev,V., Murvine,C., Olive,D.M., Ehrlich,G.D., Neri,B.P. and de Arruda,M. (1997) Determination of hepatitis C virus genotypes in the United States by cleavase fragment length polymorphism analysis. J. Clin. Microbiol., 35, 3156–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreevatsan S., Bookout,J.B., Ringpis,F.M., Mogazeh,S.L., Kreiswirth,B.N., Pottathil,R.R. and Barathur,R.R. (1998) Comparative evaluation of cleavase fragment length polymorphism with PCR-SSCP and PCR-RFLP to detect antimicrobial agent resistance in Mycobacterium tuberculosis. Mol. Diagn., 3, 81–91. [DOI] [PubMed] [Google Scholar]
- 34.Eisinger F., Jacquemier,J., Charpin,C., Stoppa-Lyonnet,D., Bressac-de Paillerets,B., Peyrat,J.P., Longy,M., Guinebretiere,J.M., Sauvan,R., Noguchi,T., Birnbaum,D. and Sobol,H. (1998) Mutations at BRCA1: the medullary breast carcinoma revisited. Cancer Res., 58, 1588–1592. [PubMed] [Google Scholar]
- 35.Killeen A.A., Jiddou,R.R. and Sane,K.S. (1998) Characterization of frequent polymorphisms in intron 2 of CYP21: application to analysis of segregation of CYP21 alleles. Clin. Chem., 44, 2410–2415. [PubMed] [Google Scholar]
- 36.Oldenburg M.C. and Siebert,M. (2000) New cleavase fragment length polymorphism method improves the mutation detection assay. Biotechniques, 28, 351–357. [DOI] [PubMed] [Google Scholar]
- 37.Lyamichev V., Brow,M.A. and Dahlberg,J.E. (1993) Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science, 260, 778–783. [DOI] [PubMed] [Google Scholar]
- 38.Leontis N.B., Kwok,W. and Newman,J.S. (1991) Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res., 19, 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young K.K., Resnick,R.M. and Myers,T.W. (1993) Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol., 31, 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds P., McOmish,F., Yap,P.L., Chan,S.W., Lin,C.K., Dusheiko,G., Saeed,A.A. and Holmes,E.C. (1993) Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J. Gen. Virol., 74, 661–668. [DOI] [PubMed] [Google Scholar]