Abstract
Most Foxp3+ regulatory T (Treg) cells develop in the thymus as a functionally mature T cell subpopulation specialized for immune suppression. Their cell fate appears to be determined before Foxp3 expression; yet molecular events that prime Foxp3− Treg precursor cells are largely obscure. We found that Treg cell–specific super-enhancers (Treg-SEs), which were associated with Foxp3 and other Treg cell signature genes, began to be activated in Treg precursor cells. T cell–specific deficiency of the genome organizer Satb1 impaired Treg-SE activation and the subsequent expression of Treg signature genes, causing severe autoimmunity due to Treg cell deficiency. These results suggest that Satb1-dependent Treg-SE activation is crucial for Treg cell lineage specification in the thymus and that its perturbation is causative of autoimmune and other immunological diseases.
The majority of Treg cells are produced in the thymus as a functionally distinct and mature T cell subpopulation that is actively involved in the maintenance of immunological self-tolerance and homeostasis1. They specifically express the transcription factor Foxp3, which has crucial roles in Treg cell development and function2–4. In addition, Treg cells acquire specific DNA hypomethylation patterns that are enriched at Treg cell signature genes including Foxp3, Il2ra, Ctla4 and Ikzf2. Acquisition of this epigenetic feature is independent of Foxp3 and associated with stable Treg-specific gene expression required for Treg cell lineage commitment and maintenance5,6. It is not clear, however, how Treg-specific gene transcription and epigenetic changes are coordinately controlled in developing Treg cells in the thymus.
Thymus-derived Treg (tTreg) cells develop mainly from immature CD24hiCD4+CD8− (CD4 single-positive (CD4SP)) thymocytes, with a minor fraction arising from CD4+CD8+ double-positive (DP) thymocytes7. Relatively strong agonistic T cell antigen receptor (TCR) stimulation and CD28 costimulation appear to generate CD25+GITR+Foxp3−CD4SP tTreg precursor cells8. TCR and IL-2 stimulation drive CD25+GITR+Foxp3−CD4SP tTreg precursor cells to differentiate into Foxp3+ tTreg cells that show the Treg cell–type DNA hypomethylation pattern8,9, but such stimulation does not generate Treg cells from CD25−Foxp3−CD4SP thymocytes. These observations indicate that whether thymocytes will differentiate into Treg cells is already determined at the tTreg precursor stage, before Foxp3 expression, posing the question of how tTreg precursor cells are primed to differentiate into tTreg cells at the transcriptional and epigenetic levels.
Cell differentiation is generally determined by the formation of the cell type–specific epigenetic landscape and the network of transcription factors10,11. Given that binding of most transcription factors depends on chromatin status, it is thought that the cell type–specific epigenetic landscape needs to be established before or concurrently with the expression of lineage-specifying transcription factors. In forming an epigenetic landscape, enhancer activation precedes promoter activation and associated gene expression12. Moreover, a number of studies have identified cell type–specific super-enhancers (SEs), which are defined as genomic regions with dense clustering of highly active enhancers, and demonstrated their association with the genes that define cell identity and determine cell lineage specification13–16. These findings suggest that the formation of a Treg cell–specific enhancer landscape may be a key determinant of priming tTreg precursor cells for tTreg cell differentiation.
Here we address how Treg cell lineage specification occurs before the expression of Foxp3 and other Treg signature genes. We show that Treg-SEs, which are associated with Treg signature genes, are gradually established and activated in early stages of tTreg cell development. Furthermore, we found that developmental stage–specific deletion of the genome organizer Satb1 impairs Treg-SE activation and fails to induce Treg cell signature genes, thereby causing autoimmune diseases and IgE hyperproduction via tTreg cell deficiency. Our results suggest that Satb1-dependent Treg-SE activation is a crucial epigenetic event guiding tTreg cell development.
RESULTS
Association of Treg-SEs with Treg signature genes
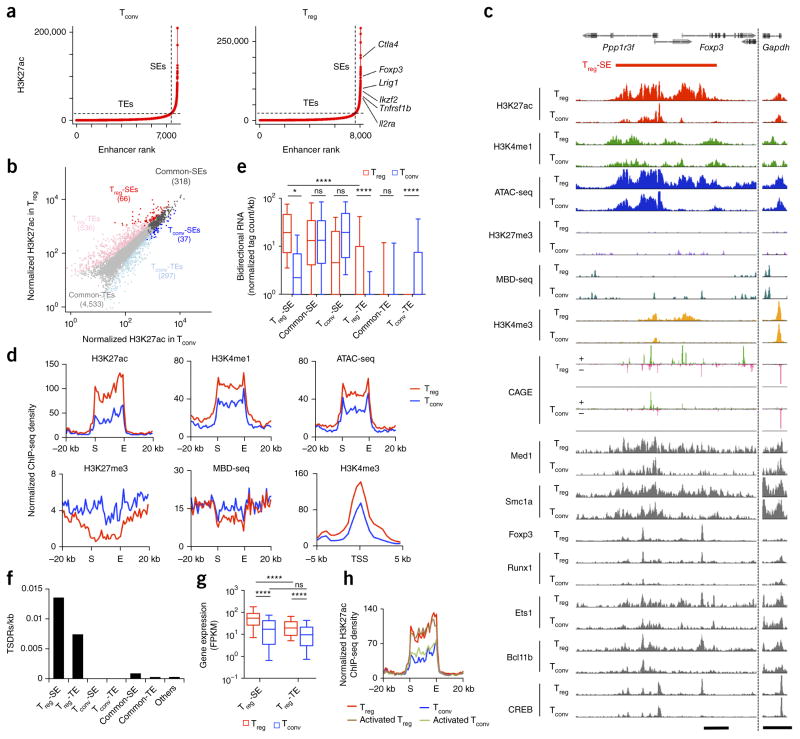
We first searched for SEs in the genomes of mouse CD4+CD25+Foxp3+ Treg and CD4+CD25−Foxp3− ‘conventional’ T (Tconv) cells by chromatin immunoprecipitation sequencing (ChIP-seq) of histone H3 acetylated at Lys27 (H3K27ac), an indicator of active enhancers. We used the algorithm ROSE13,14 to stitch peaks within 12.5 kb and rank them by signal intensity. We plotted the H3K27ac signal against stitched enhancer rank and used the tangent of the resulting curve to distinguish between SEs and typical enhancers (TEs) (Fig. 1a) (details of SE definitions are given in Online Methods). Among 384 SEs thus defined in Treg cells, 66 showed significantly (false discovery rate (FDR) < 0.05) higher H3K27ac signal intensity in Treg cells (Treg-SEs) and 318 were common to Treg and Tconv cells (common-SEs); 37 among 355 SEs found in Tconv cells were specific to these cells (Tconv-SEs) (Fig. 1b). TEs were similarly grouped as Treg-specific (Treg-TEs), common (common-TEs) or Tconv-specific (Tconv-TEs).
Figure 1.
Identification of Treg-specific SEs. (a) Distribution of H3K27ac signals across the genome in peripheral Tconv and Treg cells. Cumulative H3K27ac signals at stitched enhancers are plotted against enhancer rank. Dotted lines show the boundary between TEs and SEs. (b) Scatter plot showing normalized H3K27ac ChIP-seq tag counts at stitched enhancer regions in Tconv and Treg cells. Numbers of regions in each category are shown in parentheses. (c) H3K27ac, H3K4me1, H3K27me3 and H3K4me3; ATAC-seq and MBD-seq signals; RNA transcription start sites assessed by cap analysis gene expression (CAGE) method; and binding of various transcription factors at the Foxp3 Treg-SE. Positive (+) and negative (−) strands are indicated for CAGE analysis. Peak heights are normalized at the Gapdh locus (right). Scale bars, 5 kb. (d) H3K27ac, H3K4me1 and H3K27me3; ATAC-seq; and MBD-seq signal at global Treg-SE regions and H3K4me3 signal around transcription start sites (TSS) of Treg-SE-associated genes in Treg and Tconv cells. Average normalized ChIP-seq density of 66 Treg-SEs is plotted for merged Treg-SE regions ± 20 kb or TSS ± 5 kb. Merged ends of Treg-SEs are marked as S (start) and E (end). (e) Relative expression of bidirectional RNA produced from indicated regions in Treg and Tconv cells. Box plots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers). ns, P > 0.05; *P ≤ 0.05; ****P ≤ 0.0001 (Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (f) Frequency of Treg-specific DNA hypomethylated regions (TSDRs). (g) Expression of genes associated with Treg-SEs (61 genes) and Treg-TEs (287 genes) in Tconv and Treg cells. Average fragments per kilobase of transcript per million reads mapped (FPKM) of 2 independent RNA-seq experiments. Box plots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers). ns, P > 0.05 and ****P ≤ 0.0001 (Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (h) H3K27ac signals at merged Treg-SE ± 20 kb (as in d) in Tconv and Treg cells, before and after in vitro TCR stimulation with IL-2. Data are from 1 experiment (transcription factor ChIP-seq, ATAC-seq, H3K4me1 and H3K27me3 ChIP-seq), are representative of 2 independent experiments (H3K27ac ChIP-seq, H3K4me3 ChIP-seq and MBD-seq, a,c,d–f,h) or are the average of 2 independent experiments (RNA-seq, b,g).
When we compared the Treg-SE region at the Foxp3 locus in Treg and Tconv cells, the former showed stronger H3K27ac and monomethylation of H3K4 (H3K4me1, an active enhancer mark when combined with H3K27ac)17, greater chromatin accessibility (as determined by assay for transposase-accessible chromatin using sequencing (ATAC-seq)) and weaker H3K27me3 (an inactive enhancer mark) and DNA methylation (as indicated by methyl-CpG binding domain protein-enriched genome sequencing (MBD-seq)) (Fig. 1c). Average intensities of these signals at the 66 Treg-SEs showed the same trends (Fig. 1d). Bidirectional enhancer RNAs, which are produced by active enhancers18, showed significantly higher transcription at Treg-SEs than at Treg-TEs or the corresponding regions in Tconv cells (Fig. 1c,e). Multiple transcription factors, including Foxp3, Runx1, Bcl11b, Ets1 and CREB, which contribute to Treg cell function in various ways19, bound densely to Treg-SEs (Fig. 1c and Supplementary Fig. 1a). Med1 and Smc1a, components of mediator and cohesin complexes, respectively, frequently co-occupied Treg-SEs, indicating possible occurrences of promoter–enhancer looping within Treg-SEs20 (Fig. 1c and Supplementary Fig. 1a,b). Treg-SEs were also enriched for Treg-specific DNA demethylated regions, including hallmarks of Treg cell identity at the Foxp3, Ctla4 and Ikzf2 loci6 (Fig. 1f). Similarly to these findings with Treg-SEs, H3K27ac density correlated with that of other ‘permissive’ epigenetic modifications at common-SEs, Tconv-SEs and TEs (Fig. 1e,f and Supplementary Fig. 1a–c).
Many Treg-SEs were found in close proximity to Treg signature genes, such as Foxp3, Ctla4, Il2ra and Ikzf2 (Fig. 1a and Supplementary Data). Such Treg-SE-associated genes showed higher expression in Treg cells than in Tconv cells and higher expression than Treg-TE-associated genes (Fig. 1g). Most of them also showed higher specificity for thymic and peripheral Treg cells than for various immune cell types (Supplementary Fig. 1d). In addition, promoters of Treg-SE-associated genes showed higher intensity of H3K4me3 (a promoter activation mark) in Treg cells than in Tconv cells (Fig. 1c,d). In contrast, common-SEs were associated with genes expressed similarly in Treg and Tconv cells; further, the genes associated with Tconv-SEs included those downregulated in Treg cells (Supplementary Fig. 1e).
Notably, the corresponding Treg-SE regions in Tconv cells possessed H3K4me1 and H3K27me3 marks, with chromatin accessibility and binding of transcription factors, indicating their poised states17 (Fig. 1c,d). However, TCR and IL-2 stimulation did not activate Treg-SEs in Tconv cells or alter them in Treg cells (Fig. 1h). These results, taken together, demonstrate highly specific and robust establishment of Treg-SEs in Treg cells and a strong correlation between Treg-SE activity and associated gene expression, which suggests important roles of Treg-SEs in Treg-specific gene transcription.
Treg-SE activation before Foxp3 expression
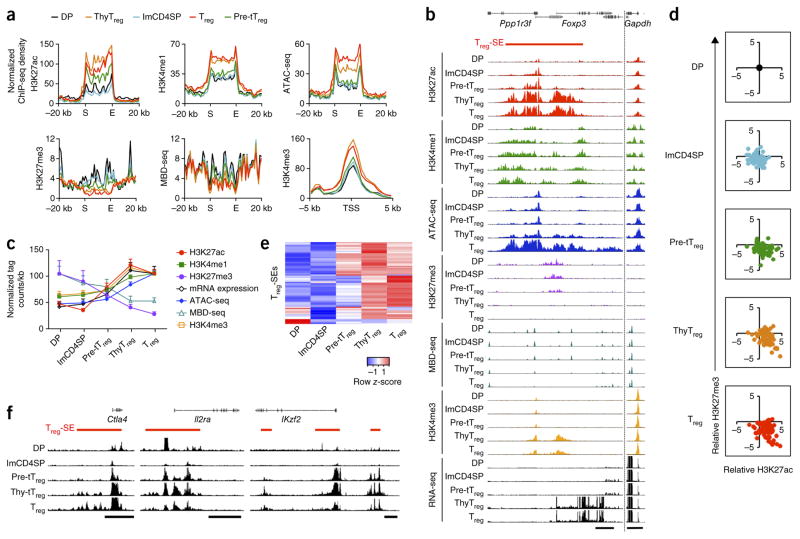
We next examined the timing of Treg-SE establishment in the course of in vivo tTreg cell development. In DP and immature CD4SP thymocytes, Treg-SE regions were marked with permissive H3K4me1 and repressive H3K27me3 modifications, which together indicated their poised state17, as shown at the merged Treg-SEs (Fig. 2a) and at the representative Foxp3 gene locus (Fig. 2b). In tTreg precursor cells, Treg-SEs showed increased H3K27ac, decreased H3K27me3 and no changes in H3K4me1, indicating that they had begun to be activated (Fig. 2a–d and Supplementary Figs. 2a and 3a). These changes were intensified at the thymic Treg cell stage. More detailed analyses revealed that approximately half of the Treg-SEs showed significant (FDR < 0.05) increases in H3K27ac signal in tTreg precursor cells before Foxp3 expression, whereas others were activated at the thymic Treg cell stage (Fig. 2d,e). The former included Treg-SEs at Foxp3 and other Treg cell signature gene loci (Fig. 2b,f). These changes at Treg-SE regions preceded chromatin loosening, DNA demethylation, activation of associated promoters and transcription of associated genes (Fig. 2a–c and Supplementary Figs. 2a and 3a). Furthermore, from early stages of tTreg cell development, enhancer activity was stronger at Treg-SEs than at Treg-TEs, which probably facilitates robust induction of Treg-SE-associated genes (Supplementary Fig. 2b). In contrast with Treg-SEs, common-SEs were active in all tTreg differentiation stages, whereas most Tconv-SEs were activated only in peripheral Tconv cells and maintained at a poised state in developing tTreg cells, as shown for merged SEs (Supplementary Fig. 2c) and for each representative SE (Supplementary Fig. 3b,c). Thus, a series of chromatin configuration changes occurred specifically at Treg-SEs in the course of tTreg cell development, before the expression of Foxp3.
Figure 2.
Establishment of Treg-specific SEs in developing Treg cells. (a) H3K27ac, H3K4me1 and H3K27me3 and ATAC-seq and MBD-seq signals at global Treg-SE regions and H3K4me3 signal around transcription start sites (TSS) of Treg-SE-associated genes in DP thymocytes, immature CD4SP (imCD4SP) thymocytes and tTreg precursor (pre-tTreg), thymic Treg (thyTreg) and peripheral Treg cells. Average normalized ChIP-seq density of 66 Treg-SEs is plotted for merged Treg-SE regions ± 20 kb or TSS ± 5 kb. Merged ends of Treg-SEs are marked as S (start) and E (end). (b) Chromatin configuration changes of the Treg-SE as in a and mRNA expression at the Foxp3 locus during Treg cell development. Peak heights are normalized at the Gapdh locus (right). Scale bars, 5 kb. (c) Changes in H3K27ac, H3K27me3, H3K4me1, ATAC-seq and MBD-seq signals at Treg-SE regions, in H3K4me3 signal at TSS ± 5 kb of Treg-SE-associated genes and in mRNA transcription of these genes during Treg cell development. Mean normalized tag counts per kb ± s.e.m. of 66 Treg-SE regions are plotted. (d) Scatter plots showing changes in H3K27ac (x axis) and H3K27me3 (y axis) signals at each Treg-SE during Treg cell development relative to those at the DP stage. Each dot indicates a Treg-SE region. (e) Heat map of H3K27ac intensity of each Treg-SE (rows) in indicated cell types (columns). Row z-score is shown in color gradient. (f) Changes in H3K27ac modifications in Treg-SEs at Treg signature gene loci during tTreg cell development. Scale bars, 25 kb. Data are from 1 experiment (transcription factor ChIP-seq, ATAC-seq, H3K4me1 and H3K27me3 ChIP-seq), are representative of 2 independent experiments (H3K27ac ChIP-seq, H3K4me3 ChIP-seq and MBD-seq), or are the average of 2 independent experiments (RNA-seq) (a–f).
Potential of Satb1 as a pioneer factor
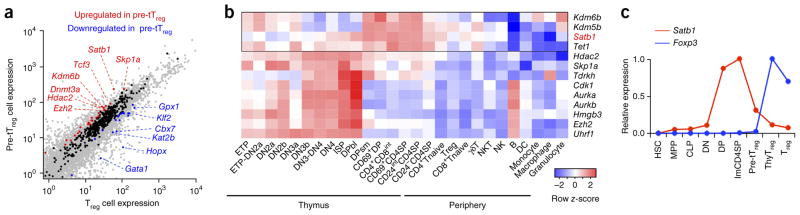
Next, to investigate the roles of Treg-SEs during tTreg cell differentiation in vivo, we searched for potential regulators of Treg-SEs by expression analysis of epigenetic modifiers (GO terms chromatin organization (QuickGO GO:0006325) and epigenetic regulation of gene expression (QuickGO GO:0040029)). On the basis of the assumption that pioneer factors able to open up condensed chromatin would be required for an initial phase of cell differentiation21,22, we selected epigenetic modifiers that showed significantly higher mRNA expression in tTreg precursor cells than in peripheral Treg cells (Fig. 3a). The most differentially expressed epigenetic modifier was Satb1, which encodes a genome organizer that regulates both transcriptional and epigenetic changes in T cells by forming long-range chromatin loops, bringing distal genes together and recruiting epigenetic modifying enzymes and transcriptional machineries to target gene loci23,24. Satb1 was also one of the few candidate genes that were upregulated toward the CD4SP stage, where most Treg cells developed (Fig. 3b). It was expressed from the DP stage, further upregulated in immature CD4SP thymocytes and downregulated as Treg cells differentiated (Fig. 3c). This pattern of Satb1 expression, which precedes Foxp3 expression, suggests a potential role of Satb1 in priming Treg-SEs for Treg cell differentiation.
Figure 3.
Satb1 expression in Treg precursor cells and binding to Treg-SEs. (a) Differential expression of genes encoding epigenetic modifiers in tTreg precursor (pre-tTreg) and peripheral Treg cells. Epigenetic modification–associated genes that are upregulated and downregulated in pre-tTreg cells are highlighted in red and blue, respectively, and those that show similar expression are shown in black. (b) Relative expression of epigenetic modifiers upregulated in pre-tTreg cells throughout thymocyte development and in various immune cells. Those with uniform gene expression patterns (variance < 0.5) were excluded, and row z-score is shown in heat map. (c) Relative mRNA expression of Satb1 and Foxp3 during Treg cell development. ETP, early T cell lineage progenitor; ISP, immature SP; DPbl, DP blasts; DPsm, small DP; HSC, hematopoietic stem cell; MPP, multipotent progenitor; CLP, common lymphoid progenitor. Average normalized values from 2 independent RNA-seq experiments (a,c) and averages of 3 independent microarray experiments for each cell type (b) are shown.
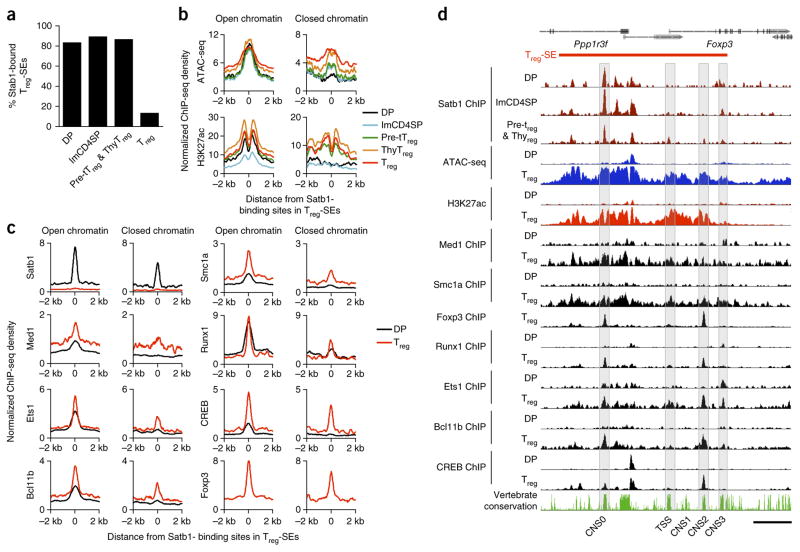
We then examined the potential of Satb1 (or a Satb1-containing complex) as a pioneer factor for establishing Treg-SEs. Satb1 ChIP-seq revealed that Satb1 bound to most Treg-SEs, common-SEs and Tconv-SEs in DP, CD4SP and developing tTreg cells (tTreg precursor and thymic Treg cells combined) but not in mature Treg cells (Fig. 4a and Supplementary Fig. 4a). Moreover, 27% of Satb1-binding sites within Treg-SEs at the DP stage were present in closed chromatin regions where ATAC-seq signal was low, indicating that Satb1, unlike typical transcription factors, can bind to closed chromatin. The Satb1-binding sites in open chromatin regions at the DP stage accompanied H3K27ac modification at flanking nucleosomes (Fig. 4b–d). In contrast, those in closed chromatin regions, notably present in Treg-SEs at most Treg cell signature gene loci, were not yet H3K27ac-modified and scarcely bound by transcription factors at the DP stage, showing gradual chromatin opening and H3K27ac modification from the tTreg cell precursor stage. The former Satb1-bindng sites, which were enriched for Ets family binding motifs, were indeed bound by Ets1 and other transcription factors from the DP stage, whereas the latter was not enriched for any of ~2,000 motifs examined (Supplementary Fig. 4b). At the Foxp3 locus, for example, Satb1 binding to closed chromatin first occurred at a newly identified conserved noncoding sequence (CNS), denoted as CNS0, in DP thymocytes. Satb1 subsequently bound to CNS3, an enhancer required for efficient Foxp3 transcription25, in CD4SP thymocytes, and to the TSS and CNS2, an enhancer specifically demethylated in Treg cells and stabilizing Foxp3 expression5, in tTreg precursor and thymic Treg cells. This sequential binding of Satb1 along tTreg cell development suggested that Satb1 might be involved in chromatin looping among regulatory regions initiated at CNS0.
Figure 4.
Potential roles of Satb1 in activating Treg-SEs. (a) Percentage of Treg-SEs bound by Satb1 in DP, imCD4SP, a mixture of tTreg precursor (pre-tTreg) and thymic Treg (thyTreg) cells, and peripheral Treg cells. (b) Chromatin accessibility and H3K27ac changes during Treg cell development around sites where Satb1 bound at the DP stage in open or closed chromatin within Treg-SEs. Average signal intensity of Satb1-binding sites (172 and 65 regions at open and closed chromatin, respectively) is shown for indicated cell types. (c) Density of indicated transcription factor binding in DP and peripheral Treg cells around Satb1-binding sites at the DP stage in open or closed chromatin within Treg-SEs. Normalized ChIP-seq signal density is plotted for Satb1-binding sites ± 2 kb. (d) Binding of Satb1 during tTreg cell development and chromatin accessibility and transcription factor binding in DP and Treg cells at the Treg-SE of the Foxp3 locus. Sequence conservation among vertebrates is also shown. The transcription start site (TSS) of the Foxp3 gene and CNS are highlighted. Scale bar, 5 kb. Histone ChIP-seq data are representative of 2 independent experiments (b,d) and transcription factor ChIP-seq and ATAC-seq data are from 1 experiment (a–d).
These results demonstrate that Satb1 binds to Treg-SEs and other SEs from the DP stage and suggest that a Satb1-containing complex could serve as a pioneer factor that initiates chromatin configuration changes of key Treg-SE regions and regulates subsequent transcriptional and epigenetic changes of Treg signature genes.
Defective tTreg cell development by Satb1 deletion
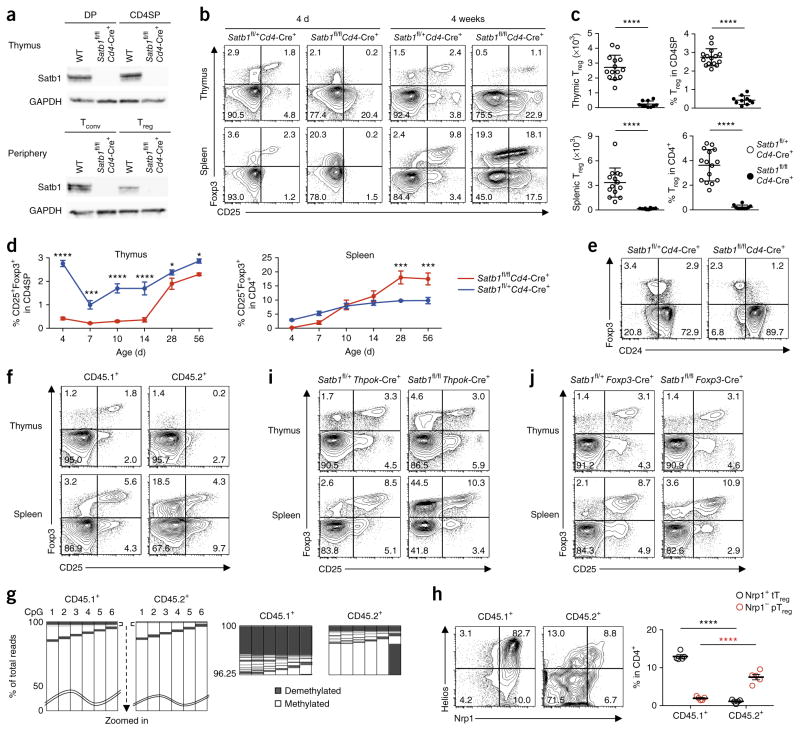
To further elucidate the role of Satb1 in Treg-SE establishment and tTreg cell development, we ablated Satb1 protein expression from the DP stage onwards by crossing mice bearing a loxP-flanked Satb1 (Satb1fl/fl) with mice expressing Cre recombinase from the Cd4 promoter (Cd4-Cre+)26 (Fig. 5a). Notably, CD25+Foxp3+ Treg cells were almost completely absent in both the thymus and the spleen of neonatal Satb1fl/flCd4-Cre+ mice (Fig. 5b). Satb1 deletion reduced the efficiency of thymocyte differentiation into CD4SP thymocytes by approximately 50% (data not shown), but it particularly affected tTreg cell development, as indicated by a significant reduction in the number and percentage of Foxp3+ cells among CD4SP thymocytes (Fig. 5c). The deficiencies of CD25+Foxp3+CD4SP thymocytes and splenic CD25+Foxp3+CD4+ T cells persisted for 14 d and 7 d, respectively, then recovered (Fig. 5d). Despite the age-dependent recovery of Treg cell percentages, CD25+Foxp3+ T cells in the thymus of adult Satb1fl/flCd4-Cre+ mice were confined to the CD24− mature fraction (i.e., lacking CD24+ immature Treg cells found in wild-type thymus) and highly activated (Fig. 5e and Supplementary Fig. 5a,b). Mixed bone marrow chimeras prepared by reconstituting irradiated Rag2−/− mice with T cell–depleted bone marrow cells from CD45.1+ wild-type mice and CD45.2+ Satb1fl/flCd4-Cre+ mice also showed that Satb1 deletion severely reduced thymic production of Foxp3+ T cells (Fig. 5f). The production remained impaired 12 weeks after bone marrow reconstitution, indicating that both early and late waves of thymic Treg cell development27 were defective in Satb1fl/flCd4-Cre+ mice (data not shown). In addition, amplicon sequencing of bisulfite-treated DNA from wild-type or Satb1-deficient thymocytes in the chimeras revealed that Satb1 deletion not only impaired Foxp3 induction but also Treg cell–specific DNA demethylation (Fig. 5g).
Figure 5.
Indispensable roles of Satb1 in tTreg cell development. (a) Satb1 and GAPDH protein expression assessed by immunoblotting in DP thymocytes, CD4SP thymocytes and peripheral Tconv and Treg cells from 4-week-old wild-type (WT) and Satb1fl/flCd4-Cre+ mice. (b) Flow cytometry of CD4SP thymocytes and CD4+ splenocytes from 4-d-old and 4-week-old Satb1fl/flCd4-Cre+ mice and littermate controls for identification of Treg cells by the expression of Foxp3 and CD25. (c) Numbers and percentages of CD25+Foxp3+ Treg cells in the thymus and spleen of 4-d-old Satb1fl/flCd4-Cre+ and littermate controls (mean ± s.e.m., n = 14, 10, 15 and 10). ****P ≤ 0.0001 (two-tailed unpaired t-test). (d) Kinetics of Treg cell accumulation in the thymus and spleen of Satb1fl/flCd4-Cre+ mice and littermate controls. Percentage of CD25+Foxp3+ Treg cells among CD4SP thymocytes (left) and CD4+ splenocytes (right) are plotted (mean ± s.e.m., n = 6 per group). *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001 (two-way ANOVA followed by Holm–Šídák multiple comparison test). (e) Flow cytometry of CD4SP thymocytes from 4-week-old Satb1fl/flCd4-Cre+ mice and littermate controls for identification of CD24+Foxp3+ immature Treg cells. (f) Flow cytometry of CD45.1+ or CD45.2+ CD4SP thymocytes and CD4+ splenocytes from bone marrow chimeras generated with T cell–depleted bone marrow cells from WT (CD45.1+) and Satb1fl/flCd4-Cre+ (CD45.2+) mice for identification of Treg cells by Foxp3 and CD25 expression. (g) DNA methylation status of six CpG residues within Foxp3 CNS2 in WT and Satb1-deficient CD4SP thymocytes from mixed bone marrow chimeras as in f, assessed by amplicon sequencing. CpGs are numbered 1–6 from the 5′ end (columns) and amplicons are ordered according to degree of demethylation (rows). Demethylated or methylated status is indicated by color code; waved lines (top) indicate the 0–50% of fully methylated amplicons omitted for presentation. The top 3.75% of total reads, when ordered from most to least demethylated clones, are magnified (bottom). (h) Flow cytometry of CD25+Foxp3+ Treg cells from bone marrow chimeras as in f for the expression of Nrp1 and Helios (top) and percentages of Nrp1+CD25+Foxp3+ tTreg and Nrp1−CD25+Foxp3+ pTreg cells among CD45.1+ and CD45.2+ CD4+ T cells (mean ± s.e.m., n = 4) (bottom). ****P ≤ 0.0001 (two-way ANOVA followed by Holm–Šídák multiple comparison test). (i) Flow cytometry of CD4SP thymocytes and CD4+ splenocytes from 4-week-old Satb1fl/+Thpok-Cre+ and Satb1fl/flThpok-Cre+ mice for identification of Treg cells by the expression of Foxp3 and CD25. (j) Flow cytometry of CD4SP thymocytes and CD4+ splenocytes from 4-week-old Satb1fl/+Foxp3-Cre+ and Satb1fl/flFoxp3-Cre+ mice for identification of Treg cells by the expression of Foxp3 and CD25. Data are representative or the summary of 3 independent experiments with 3 or more mice (a–f,h–j) or are representative of 2 independent experiments with 2 mice (g). Quadrant numbers indicate the percentages of gated cells (b,e,f,h–j).
The presence of mature peripheral Foxp3+ cells despite impaired thymic Treg cell production prompted us to examine their functional and phenotypic properties. Peripheral Foxp3+ cells in Satb1fl/flCd4-Cre+ mice consisted of CD25+ and CD25− populations (Fig. 5b). Satb1-deficient CD25+Foxp3+ cells, when compared with wild-type Treg cells, showed similar in vitro suppressive activity, expression of Treg cell signature genes, Treg cell–specific DNA hypomethylation patterns and stability of Foxp3 expression after in vitro TCR stimulation (Supplementary Fig. 5b–e). In contrast, Satb1-deficient CD25−Foxp3+ cells did not show these properties, which suggests that they might be Tconv cells with de-repressed Foxp3 expression. In addition, CD25+Foxp3+ cells from Satb1fl/flCd4-Cre+ mice contained Nrp1−Helios− cells at an increased frequency (Supplementary Fig. 5f). As the low expression of Nrp1 and Helios marks peripherally induced Treg (pTreg) cells under a noninflammatory condition28–30, we examined the mixed bone marrow chimeras and found that Satb1-deficient CD25+Foxp3+ cells were mostly Nrp1−Helios− (Fig. 5h). These results suggest that, although Satb1 deletion severely impaired tTreg cell development, it allowed and even enhanced the development of pTreg cells, and that some of them had recirculated to the thymus as CD24−Foxp3+ T cells31 (Fig. 5e).
To further elucidate the differential effects of Satb1 on tTreg and pTreg cell development and the possible role of Satb1 for Foxp3 repression in Tconv cells, we ablated Satb1 specifically in mature CD4+ T cells after tTreg cell development but before pTreg cell differentiation by preparing Satb1fl/flThpok-Cre+ mice32 (Supplementary Fig. 6a). This Satb1 deletion did not affect tTreg cell development (Fig. 5i) but de-repressed Foxp3 expression in Tconv cells without evoking expression of other Treg cell signature molecules or inducing Treg cell–specific DNA demethylation (Supplementary Fig. 6b–d). Notably, despite no Satb1 binding at the Foxp3 locus in wild-type Tconv cells, H3K27ac in the Treg-SE of Foxp3 was enhanced in both CD25−Foxp3− and CD25−Foxp3+ cells from Satb1fl/flThpok-Cre+ mice, which suggests that Satb1 deficiency indirectly activated Treg-SE to de-repress Foxp3 expression (Supplementary Fig. 6e). Satb1fl/flThpok-Cre+ mice also showed an increased proportion of Nrp1−Helios−CD25+Foxp3+ pTreg cells, which possessed Treg cell–specific DNA hypomethylation at the Foxp3 CNS2 region to a similar extent as in wild-type pTreg cells (Supplementary Fig. 6d,f). Moreover, when colitis was induced in Rag2−/− mice by cell transfer of wild-type and Satb1-deficient naive T cells, a significantly higher percentage (P < 0.0001) of Treg cells with both Foxp3 expression and Treg cell–specific DNA hypomethylation was generated from the Satb1-deficient fraction (Supplementary Fig. 6g,h). These results confirm that Satb1 deletion de-represses Foxp3 expression in mature Tconv cells and facilitates pTreg cell differentiation.
Next, to address whether Satb1 is also required for Treg cell maintenance, we deleted Satb1 in differentiated Treg cells from the Foxp3+ thymic Treg cell stage onwards by generating Satb1fl/flFoxp3-Cre+ mice. Those mice did not show any significant changes in Treg cell ratio or phenotype, which indicates that Satb1 is not required for the maintenance of Treg cells (Fig. 5j and Supplementary Fig. 6i–k).
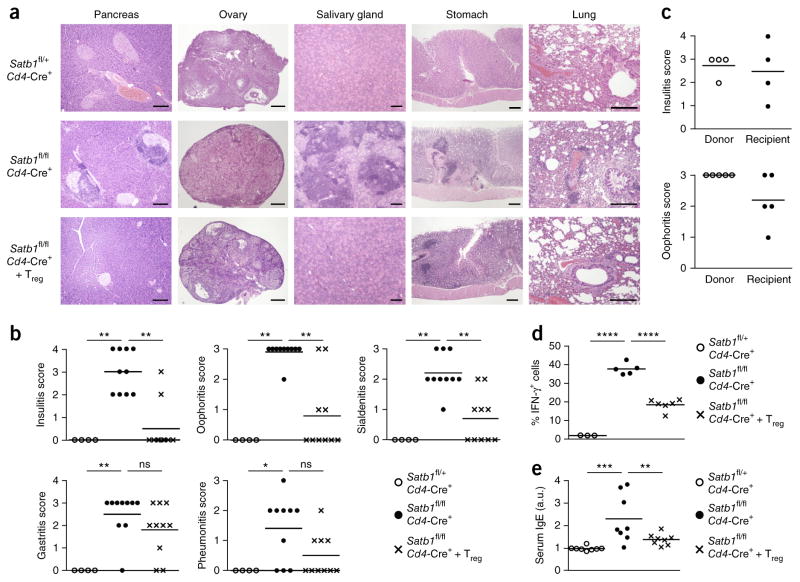
Notably, Satb1fl/flCd4-Cre+ but not Satb1fl/flThpok-Cre+ or Satb1fl/fl Foxp3-Cre+ mice spontaneously developed histologically and serologically evident autoimmune or inflammatory disease in various organs including the ovaries, Langerhans islets, the salivary glands, the lung and the stomach (Fig. 6a,b). The tissue lesions could be adoptively transferred by splenocytes into syngenic Rag2−/− mice (Fig. 6c). They also showed significant elevation of serum IgE and increase of interferon-γ (IFN-γ)-producing CD4+ T cells (Fig. 6d,e). We observed severe reductions in total Treg cells, especially during the neonatal period as shown above, whereas wild-type and Satb1fl/flCd4-Cre+ mice showed no substantial difference in the efficiency of negative selection of self-reactive Tconv cells (Supplementary Fig. 5g). Moreover, transfer of wild-type Foxp3+ Treg cells into 4-d-old Satb1fl/flCd4-Cre+ mice at a marginal dose (1 × 106 cells) significantly prevented histological development of autoimmune disease and IgE hyper-production (Fig. 6a,b,d,e).
Figure 6.
Induction of autoimmunity by T cell–specific deletion of Satb1. (a) Hematoxylin and eosin staining of tissue sections from 16-week-old Satb1fl/+Cd4-Cre+ mice (n = 4), Satb1fl/flCd4-Cre+ mice (n = 10) and Satb1fl/flCd4-Cre+ mice after 1 × 106 wild-type (WT) Treg cell transfer on day 4 after birth (n = 10). Scale bars, 200 μm. (b) Severity of disease in mice shown in a. Horizontal lines indicate mean. ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01 (Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (c) Oophoritis and insulitis score of donor Satb1fl/flCd4-Cre+ mice and Rag2−/− mice transferred with splenocytes from donor. Each recipient received cells from each donor. Horizontal lines indicate means. (d) Percentages of IFN-γ-producing cells among peripheral CD4+ non-Treg cells in mice described in a. Horizontal lines indicate means (n = 3, 5 and 6 for Satb1fl/+Cd4-Cre+, Satb1fl/flCd4-Cre+ and Treg-transferred Satb1fl/flCd4-Cre+ mice, respectively). ****P ≤ 0.0001 (ordinary one-way ANOVA, followed by Holm–Sidak multiple comparisons test). (e) Relative levels (arbitrary units (a.u.)) of serum IgE in mice shown in a (n = 8 per group). Horizontal lines indicate means. **P ≤ 0.01 and ***P ≤ 0.001 (ordinary one-way ANOVA, followed by Holm–Šídák multiple comparisons test). Data are representative and summary of 4, 10 and 10 experiments for Satb1fl/+Cd4-Cre+, Satb1fl/flCd4-Cre+ and Satb1fl/flCd4-Cre+ with WT Treg cell transfer independent experiments (a,b), summary of 4 or 5 independent experiments (c) or summary of 3 independent experiments (d,e).
Taken together, these results indicate that the timing of Satb1 deletion determines its effect on Treg cell development. Its ablation in thymocytes before Foxp3 expression predominantly impairs tTreg cell development, whereas its deficiency in mature Tconv cells seems to promote pTreg cell differentiation. In addition, impairment of tTreg cell differentiation via Satb1 deletion results in the development of autoimmune and other immunological diseases, despite enhanced generation of pTreg cells.
Treg-SE activation for Treg cell development
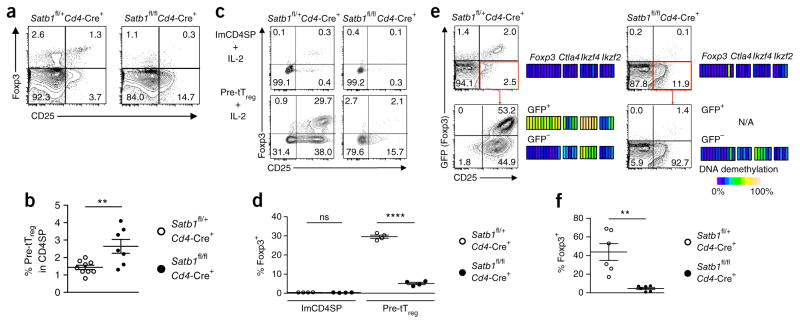
Next, to determine how Satb1 deletion would impair tTreg cell development, we assessed the potential of Satb1-deficient tTreg precursor cells to differentiate into thymic Treg cells. Despite tTreg cell deficiency, the tTreg precursor cell fraction was higher in percentage in Satb1fl/flCd4-Cre+ mice than in Satb1fl/+Cd4-Cre+ littermates and had a similar cell surface phenotype (Fig. 7a,b and Supplementary Fig. 7a). Upon in vitro stimulation with IL-2 for 24 h (ref. 8), ~30% of tTreg precursor cells from Satb1fl/+Cd4-Cre+ mice were Foxp3+; however, a significantly lower percentage of the Satb1-deficient counterparts became Foxp3+ (Fig. 7c,d). After IL-2 and TCR stimulation for 5 d (refs. 8,9), tTreg precursor cells from Satb1fl/+Cd4-Cre+ mice differentiated into Foxp3+ T cells with Treg cell–specific DNA hypomethylation, whereas those from Satb1fl/flCd4-Cre+ mice did not (Fig. 7e,f). In addition, CD25−Foxp3+CD4SP thymocytes, another Treg precursor population33, were substantially reduced in Satb1fl/flCd4-Cre+ mice (Fig. 7a and Supplementary Fig. 7b). Moreover, Satb1-deficient CD25−Foxp3+CD4SP cells showed unstable Foxp3 expression after in vitro TCR stimulation, unlike their wild-type counterparts (Supplementary Fig. 7c).
Figure 7.
Loss of Treg differentiation potential in Satb1-deficient tTreg precursor cells. (a) Flow cytometry of CD24+CD4SP thymocytes from 4-week-old Satb1fl/flCd4-Cre+ mice and littermate controls for identificationof tTreg precursor (pre-tTreg) cells by expression of CD25 and Foxp3. (b) Percentages of CD24+CD25+GITR+Foxp3−CD4SP tTreg precursor cells in mice shown in a (mean ± s.e.m.; n = 9 and 7 Satb1fl/+Cd4-Cre+ and Satb1fl/flCd4-Cre+ mice, respectively). **P ≤ 0.01 (two-tailed unpaired t-test). (c,d) Flow cytometry of IL-2-stimulated immature CD4SP (imCD4SP) thymocytes and pre-tTreg cells from Satb1fl/+Cd4-Cre+Foxp3GFP or Satb1fl/flCd4-Cre+Foxp3GFP mice for the expression of CD25 and Foxp3. Representative dot plots (c) and summary graph for the percentage of Foxp3+ cells (d) are shown (mean ± s.e.m., n = 4). ns, P > 0.05; ****P ≤ 0.0001 (ordinary one-way ANOVA, followed by Holm–Šídák multiple comparisons test). (e,f) Flow cytometry of IL-2- and TCR-stimulated pre-tTreg cells from Satb1fl/+Cd4-Cre+Foxp3GFP or Satb1fl/flCd4-Cre+Foxp3GFP mice for the expression of CD25 and Foxp3. Representative dot plots (e) and summary graph (f) for the percentage of Foxp3+ cells are shown (mean ± s.e.m., n = 6). Heat maps in e show DNA methylation status of Treg signature genes in GFP+ and GFP− cells before and after stimulation (N/A, data not available). Each block indicates an individual CpG in PCR-amplified regions. **P ≤ 0.01 (two-tailed unpaired t-test). Data are representative or summary of 3 (a,b and DNA methylation results in e), 2 (c,d) or 6 (e,f) independent experiments. Quadrant numbers indicate the percentages of gated cells (a,c,e).
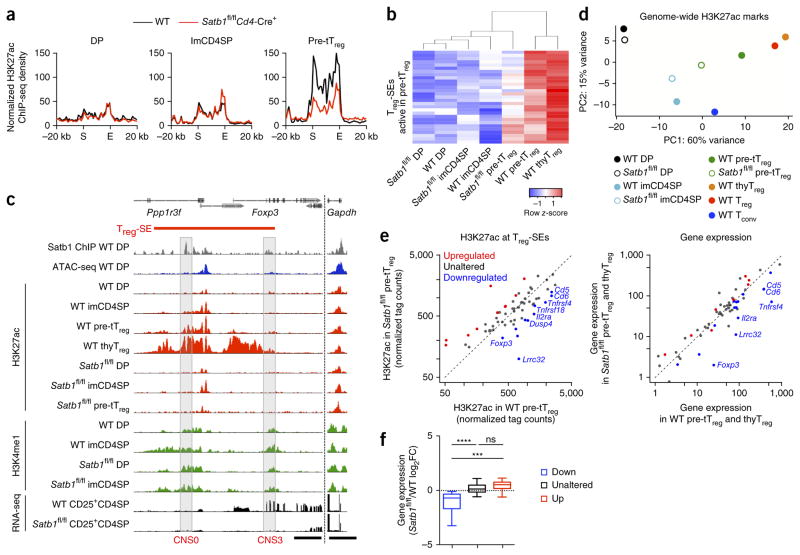
To determine then whether the loss of Treg cell differentiation potential in Satb1-deficient tTreg precursor cells could be attributed to impaired Treg-SE activation, we assessed the activation status of Treg-SEs, which are normally active at the tTreg precursor cell stage. Wild-type and Satb1-deficient DP cells and immature CD4SP cells showed similar intensities of H3K4me1, whereas Satb1-deficient tTreg precursor cells showed much lower H3K27ac intensity than their wild-type counterparts (Fig. 8a,b and Supplementary Fig. 7d). The reduction occurred ‘preferentially’ at Treg-SE regions to which Satb1 bound before chromatin opening (Fig. 8c and Supplementary Fig. 7e,f). For example, activation of Treg-SE at the Foxp3 locus did not increase beyond the level observed at the DP stage in Satb1-deficient thymocytes, most notably around CNS0, where Satb1 initially bound when chromatin was closed. In contrast, the effects of Satb1 deletion on common-SEs and Tconv-SEs were less pronounced, making Satb1-deficient tTreg precursor cells closer to immature CD4SP thymocytes in the principal component analysis of global H3K27ac pattern (Fig. 8d and Supplementary Fig. 7g,h). These results collectively indicate that stage-specific Satb1 deletion ‘preferentially’ impairs activation of Treg-SEs at the tTreg precursor stage.
Figure 8.
Satb1-dependent Treg-SE establishment and control of transcriptional changes in developing tTreg cells. (a) H3K27ac signals of Treg-SE regions in DP, immature CD4SP (imCD4SP) and tTreg precursor (pre-tTreg) cells from wild-type (WT) and Satb1fl/flCd4-Cre+ mice. Average normalized ChIP-seq density of 29 Treg-SE regions that are normally active in pre-tTreg cells were calculated and plotted for merged Treg-SE regions ± 20 kb. Merged ends of Treg-SEs are marked as S (start) and E (end). (b) Heat map showing H3K27ac signal at each Treg-SE (rows) in indicated cell types (columns), which are clustered by similarity. Row z-score is shown by color gradient. (c) H3K4me1 and H3K27ac modifications and mRNA transcription of the Foxp3 locus in indicated cell types from WT and Satb1fl/flCd4-Cre+ mice. Satb1-binding and chromatin accessibility in WT DP thymocytes are also shown. CNS0 and CNS3 regions are highlighted. Peak heights are normalized at the Gapdh locus. Scale bars, 5 kb. (d) Principal component analysis of global H3K27ac peaks in indicated cell populations. (e) Scatter plots showing changes in H3K27ac modifications of Treg-SEs at the pre-tTreg cell stage by Satb1 deficiency (left) and changes in Treg-SE-associated gene expression in CD24+CD25+GITR+CD4SP thymocytes (a mixture of pre-tTreg and immature thymic Treg (thyTreg) cells) (right). Treg-SEs are grouped according to the effects of Satb1 deficiency on their H3K27ac signal at the pre-tTreg cell stage, and associated genes are highlighted in the gene expression plot. (f) Summary of effects of Satb1 deletion on the expression of genes associated with Treg-SEs whose H3K27ac signal is downregulated, unaltered or upregulated by Satb1 deletion as in e (15, 34 and 7 genes, respectively). Box plots show median (center line), interquartile range (box) and tenth and ninetieth percentiles (whiskers) of log2 fold-change between Satb1-deficient and WT mice. ns, P > 0.05; ***P ≤ 0.001; ****P ≤ 0.0001 (Kruskal–Wallis test, followed by Dunn’s multiple comparisons test). H3K27ac ChIP-seq and RNA-seq data are representative of 2 independent experiments (a–d) or average of 2 independent experiments (e,f), and H3K4me1 ChIP-seq data are from 1 experiment (c).
To examine the effects of this defective Treg-SE activation on gene transcription, we compared the Treg-SE activity in tTreg precursor cells and CD24+CD25+CD4SP thymocyte gene expression (including tTreg precursor and immature tTreg cells) between wild-type and Satb1-deficient mice. Notably, among Treg-SEs to which associated genes were assigned (59 out of 66 regions), there was a significant correlation between reduced Treg-SE activity and impaired induction or upregulation of associated genes (P < 0.00005 in Fisher’s exact test). Moreover, the genes associated with Treg-SEs whose activity was downregulated by Satb1 deletion showed significantly reduced expression when compared to the genes associated with other Treg-SEs (Fig. 8e,f and Supplementary Fig. 8a,b). These genes included Foxp3, Tnfrsf4 (encoding OX40), Il2ra (encoding CD25) and Lrrc32 (encoding glycoprotein A repetitions predominant (GARP)).
Collectively, these data demonstrate a strong association between Treg-SE establishment and subsequent Treg cell–specific transcriptional changes during tTreg cell development, which suggests a role for Treg-SE activation in guiding Treg cell lineage specification (Supplementary Fig. 8c).
DISCUSSION
The main findings of this report are that Treg-SEs, associated with key Treg cell signature genes, begin to be established in parallel before the expression of Foxp3 in the course of tTreg cell development and that Satb1 deficiency before, but not after, the Foxp3+ thymic Treg stage impairs Treg-SE activation and, consequently, the expression of Treg cell signature genes, causing severe autoimmunity.
SEs have been suggested to control the expression of the associated lineage-specifying genes in a number of cell types13–16. We found that Treg-SEs were associated with the genes defining Treg cell identity, such as Foxp3, Ctla4 and Il2ra. In addition to the enriched transcription factor binding, we observed high transcription of enhancer RNAs from Treg-SEs. Their contribution to increasing chromatin accessibility and promoting associated gene transcription34 is one explanation for the enhanced transcription of associated genes. Moreover, frequent co-binding of Med1 and Smc1a, which indicates enhancer–promoter chromatin looping, suggests that multiple enhancers located within SEs regulate associated gene transcription via chromatin loop formation. These findings support the role of Treg-SEs in inducing high expression of Treg cell signature genes.
In the course of thymic Treg cell development, Treg-SE activation was initiated in parallel at most Treg signature gene loci before their expression or Treg-specific DNA demethylation9. This finding supports the contribution of Treg-SEs in the induction of Treg signature genes, and is in accord with findings that Foxp3 is not required for the induction of most Treg signature genes but augments a pre-established pattern of gene expression in addition to its role as a repressor6,35,36. Moreover, Treg-SEs were poised for activation in DP, immature CD4SP and peripheral Tconv cells, reflecting the potential of these cells to differentiate into Treg cells with additional signals. These findings also suggest that certain molecules involved in Treg-SE activation may be the true lineage specifying factors.
The pioneering effect of a Satb1-containing complex in Treg-SE activation was corroborated by several observations. First, Satb1 was required for de novo activation of Treg-SEs but not for the maintenance of established common-SEs. Second, the molecule could bind to closed chromatin at the DP stage, and the bound sites later became accessible and occupied by transcription factors in Treg cells. Third, it was required for early stages of Treg cell development but dispensable in differentiated Treg cells. Given that Satb1 is highly expressed and bound to the Treg-SEs from the DP stage23,24, it is likely that Satb1 binding is a prerequisite for subsequent Treg-SE activation upon appropriate signal transduction in developing thymic Treg cells. In contrast, in differentiated Treg cells, the repression of Satb1 appears to be necessary to prevent Treg cells from acquiring helper T cell characteristics, presumably because Satb1 might cause unwanted chromatin reorganization when unduly expressed37.
Our studies in T cell–specific Satb1-deficient mice revealed a correlation between impaired Treg-SE activation and defective induction of Treg signature genes, including Foxp3. Within the Treg-SE at the Foxp3 locus, Satb1 strongly bound to CNS0 before chromatin opening, and various transcription factors occupied this site along with Satb1-dependent enhancer activation. These properties of CNS0 suggest a role in initiating Treg-SE activation to induce Foxp3 expression. Indeed, deletion of this genomic region substantially impaired Foxp3 induction ((Y. Kitagawa, K.H., H. Watanabe, G. Kondoh and S.S., unpublished data). Other Treg signature gene loci at which Treg-SE activation and gene transcription were impaired by Satb1 deletion included Tnfrsf4, which was shown to promote Foxp3 induction38. Although CD25 expression was similar between wild-type and Satb1-deficient tTreg precursor cells, Treg-SE activity and gene transcription at the Il2ra locus were substantially reduced by Satb1 deletion, potentially destabilizing CD25 expression in vivo and interfering with tTreg cell differentiation, survival and proliferation39. These findings altogether suggest that Satb1-dependent Treg-SE activation is required for priming tTreg precursor cells to respond to cytokine and other signals and guiding relevant downstream transcription factors to the Treg signature gene loci.
Satb1 deficiency in mature CD4SP thymocytes and Tconv cells derepressed Foxp3 expression. Stage-specific Satb1 deletion elicited Treg-SE activation at the Foxp3 locus, particularly around CNS0, before Foxp3 de-repression. In addition, Satb1-deficient Tconv cells with derepressed Foxp3 expression were converted more efficiently in vivo into stable pTreg cells than were wild-type Tconv cells. Thus, despite the distinct functions of Satb1 in tTreg and pTreg cell differentiation, Treg-SE activation is a common mechanism triggering Foxp3 expression. This finding can be exploited in a clinical setting to generate Treg cells from Tconv cells via targeting Satb1.
Last, our findings indicate that defects in Treg-SE formation could be a potential cause of autoimmune and other immunological diseases. Satb1fl/flCd4-Cre+ mice developed multi-organ autoimmunity similar to that observed with neonatal tTreg cell deficiencies induced by other means27,40. The reduction of disease severity by neonatal transfer of Treg cells suggests that tTreg cell deficiency and consequent reduction in total Treg cell percentage in neonatal period was a major cause of the disease. Furthermore, the human SATB1 locus contains single nucleotide polymorphisms (SNPs) associated with autoimmune diseases such as inflammatory bowel disease, psoriasis and multiple sclerosis41 (GWAS Central, http://www.gwascentral.org, accessed 23 September 2015). Given that disease-associated SNPs are also enriched at cell type–specific SEs in various human cell types14,16, these findings suggest that altered establishment of Treg-SEs might contribute to genetic susceptibility to various immunological diseases via affecting Treg cell differentiation.
In conclusion, the present study strongly suggests that Satb1-dependent establishment of Treg-SEs controls the expression of Treg cell signature molecules, including Foxp3, during tTreg cell development. Impairment of this epigenetic event in developing Treg cells is associated with autoimmunity similar to that induced by tTreg cell depletion. Further study of how Treg-SEs are primed and activated would aid understanding of the molecular basis of Treg cell differentiation and of autoimmune and other immunological diseases.
ONLINE METHODS
Mice
Male, 4-week-old C57BL/6J mice (CLEA Japan) were used for preparing thymocyte subpopulations, and peripheral Tconv and Treg cells for transcription factor chromatin immunoprecipitation sequencing (ChIP-seq). Male, 4-week-old Foxp3-IRES-DTR/GFP knock-in (Foxp3GFP) mice (C57BL/6J), which express GFP under the endogenous Foxp3 promoter without disrupting Foxp3 expression42, were used to purify subpopulations of thymocytes and peripheral T cells for histone ChIP-seq, MBD-seq, ATAC-seq and RNA-seq. Satb1 conditional knockout mice were prepared by crossing previously described Satb1fl/fl mice26 with Cd4-Cre+ mice43, Foxp3-Cre+ mice44 or Thpok-Cre+ mice32 (C57BL/6J). Satb1fl/flCd4-Cre+ and Satb1fl/flThpok-Cre+ mice were crossed with Foxp3GFP mice to isolate thymocyte or peripheral T cell subpopulations. For the Foxp3 induction assay using tTreg precursor cells, Satb1fl/flCd4-Cre+ mice were backcrossed to BALB/c background 10 times and further crossed with previously described Foxp3-eGFP fusion knock-in mice45 to create a system where Foxp3 induction is perfectly mirrored by GFP expression. OT-II and RIP-OVA mice were previously described46,47. All mice used were maintained under specific pathogen-free conditions and all experiments were performed in accordance with guidelines for animal welfare set by Osaka University.
Cell preparation
For the preparation of double-positive (DP) thymocytes, CD4+CD8+ cells were sorted from total thymocytes using FACSAria II (BD Biosciences). For sorting immature CD4 single-positive (imCD4SP; CD3ε+CD4+CD8−CD24+CD25−GFP−), tTreg precursor (pre-tTreg; CD3ε+CD4+CD8−CD24+CD25+GITR+GFP−), thymic Treg (thyTreg; CD3ε+CD4+CD8−CD25+GFP+) cells from the thymus of Foxp3-DTR/GFP knock-in mice, CD8− thymocytes were first enriched by ‘panning’—i.e., thymocytes stained with rat anti-CD8α antibody (BD Biosciences, clone 53-6.7) were incubated on a dish coated with goat anti-rat IgG antibody (MP Biomedicals). Nonadherent cells were stained with relevant antibodies and sorted by FACSAria II. Peripheral conventional T (Tconv; CD4+CD25−GFP−) and Treg (CD4+CD25+GFP+) cells were sorted similarly, with prior removal of CD8+ and B220+ cells from lymphocytes and splenocytes. For the preparation of thymocyte subpopulations for Satb1 ChIP-seq, thymocytes were enriched and FACS-sorted from wild-type mice, and CD25−CD4SP thymocytes and CD25+CD4SP thymocytes (mixture of pre-tTreg and thymic Treg cells) were treated as immature CD4SP thymocytes and developing tTreg cells, respectively. The following antibodies were used for FACS sorting: anti-CD4-APC (1:200, BD Biosciences), anti-CD8α-PerCP (1:200, BD Biosciences), anti-CD3ε-V500 (1:200, BD Biosciences), anti-CD24-BV421 (1:200, BioLegend), anti-CD25-PE (1:100, BD Biosciences) and anti-GITR-PE-Cy7 (1:200, BD Biosciences). Clone numbers were as follows: anti-CD4-APC (clone RM4-5), anti-CD8-PerCP (clone 53-6.7), anti-CD3e-V500 (clone 500A2), anti-CD24-BV421 (clone M1/69), anti-CD25-PE (clone PC61), anti-GITR-PE-Cy7 (clone DTA-1).
For the separation of peripheral Tconv and Treg cells from pooled lymphocytes and splenocytes for transcription factor ChIP-seq, CD4+CD25+ Regulatory T Cell Isolation Kit (Miltenyi Biotec) and CD4 (L3T4) Microbeads (Miltenyi Biotec) were used according to the manufacturer’s instructions.
Activated Tconv and Treg cells (Fig. 1h) were prepared by stimulating FACS-sorted Tconv and Treg cells with Dynabeads Mouse T-Activator CD3/CD28 (Thermo Fisher) in RPMI 1640 supplemented with 10% FBS, penicillin–streptomycin, 2-mercaptoethanol and 30 or 100 U/ml IL-2, respectively, for 72 h.
ChIP-seq
For histone ChIP-seq and transcription factor ChIP-seq, 0.5–3 × 105 and 2–10 × 106 cells were used, respectively. Antibodies used were anti-H3K27ac (GeneTex, GEX60815), anti-H3K4me1 (ActiveMotif, 39297), anti-H3K27me3 (Millipore, 07-449), anti-H3K4me3 (Abcam, ab1012), anti-Satb1 (Abcam, ab70004), anti-Foxp3 (Abcam, 150743), anti-Runx1 (Abcam, ab23980), anti-CREB (Abcam, ab31387), anti-Ets1 (Santa Cruz, sc-350X), anti-Bcl11b (Bethyl Laboratories, A300-383A), anti-MED1 (Bethyl Laboratories, A300-793A), and anti-Smc1a (Bethyl Laboratories, A300-055A). Sorted cells were cross-linked in 1% (wt/vol) formaldehyde solution for 5 min (histone ChIP-seq) or 30 min (transcription factor ChIP-seq) and lysed. Cross-linked DNA was then fragmented by sonication using Digital Sonifier (Branson). The lysate was incubated overnight at 4 °C with 50–100 μl DynaBeads IgG magnetic beads (Thermo Fisher) that had been preincubated with 2.5–5 μg appropriate antibodies. Samples were washed, eluted, reverse cross-linked at 65 °C overnight, and purified using MinElute PCR Purification Kit (Qiagen). For transcription factor ChIP-seq, purified ChIP DNA was fragmented using Covaris Focused-ultrasonicator S220 (Covaris) before library preparation. Library was prepared using KAPA Library Preparation Kit Ion Torrent (KAPA Biosystems) according to the manufacturer’s instructions and sequenced using Ion Proton (Thermo Fisher).
ChIP-seq reads were mapped to the mouse genome mm9 illumina iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.html) using Bowtie2 (version 2.2.1). For visualization of ChIP peaks, MACS2.0 (version 10)48 was used for peak calling, with input reads as control. ChIP-seq tracks were presented in GenomeJACK Browser (version 3.1, Mitsubishi Space Software). Owing to the variable signal-to-noise ratio among cell types (for example, tTreg precursor cells are more apoptotic, and this tends to reduce signal-to-noise ratio), histone ChIP-seq peaks were normalized on the basis of the peak heights at the Gapdh locus when applicable, whereas other ChIP-seq peaks were normalized by total mapped read counts. For global analyses of histone ChIP-seq, raw tag counts were normalized using DESeq2 package (version 1.6.3) in R (version 3.1.2) (details are given below).
Identification of SEs and associated genes
H3K27ac ChIP-seq peaks were identified using FindPeaks in Homer package (version 4.7.2) with -region option, FDR set at 0.0001 and 40-fold enrichment over input. After removing peaks at promoter regions (within 2 kb of transcription start sites), H3K27ac peak density and clustering was assessed using ROSE13,14. Briefly, H3K27ac peaks within 12.5 kb of each other were stitched. Cumulative H3K27ac signal was determined for stitched enhancers and ranked by H3K27ac signal strength in ascending order. When stitched enhancer rank and H3K27ac signal strength were plotted in a lined scatter plot and both axes were scaled to 0 to 1, the point where the tangent of the curve = 1 was used to distinguish between SEs and TEs. Regions with enhancer rank higher and lower than this point were categorized as SEs and TEs, respectively. This process was carried out in duplicate, and regions intersecting in duplicates were defined as SEs for each cell type.
To identify SEs with statistically significant differences in H3K27ac intensity between Tconv and Treg cells, we first normalized ChIP-seq tag counts in these enhancers as follows. On the basis of the assumption that the global H3K27ac signal is similar among cell types (as similarly assumed in quantitative ChIP-seq data analysis programs such as MAnorm)49, mapped tags with MAPQ quality > 10 were counted for global H3K27ac peak regions and normalized using DESeq2 package in R. Tag counts at combined stitched enhancers in the two cell types were normalized using the same normalization ratio as estimated by DESeq2 on the global peak regions. Using the resulting normalized tag counts, the data set was then analyzed for differential intensities of H3K27ac between the two populations, prepared in duplicate, using DESeq2. Differentially regulated SEs or TEs with FDR < 0.05 were defined as cell type–specific regions, and the rest were categorized as common regions. Treg-SEs active in tTreg precursor cells (Fig. 8a,b) were defined as those that showed a statistically significant increase in H3K27ac level in tTreg precursor cells compared to DP thymocytes.
Enhancer-associated genes were defined as those located within 5 kb from both ends of the enhancer, on the basis of the calculation that median distance between genes in the mouse genome (mm9) is 13,726 bp and setting the limit at value smaller than half of median should theoretically determine which of the two neighboring genes an enhancer is assigned to. FPKM values <5 in all examined T cell populations (DP, imCD4SP, pre-tTreg, thyTreg, Treg and Tconv cells) were filtered out, and this cutoff value was determined by distribution of FPKM values of global genes and the relative size of s.d. in duplicated samples.
Metagene representation
Similarly to the described method14, average ChIP-seq density at merged SEs was presented by dividing SEs into 20 bins regardless of size, obtaining normalized tag counts for each bin and calculating the average for a group of SEs. Flanking 20-kb regions were also divided into 20 bins and average of normalized tag counts were calculated.
RNA-seq
For RNA-seq of DP, immature CD4SP, tTreg precursor, thymic Treg, Tconv and peripheral Treg cells, 1 × 105 cells were sorted from Foxp3GFP mice by FACSAria II and prepared for RNA-seq in duplicate. CD4+CD25− cells from Foxp3GFP and Satb1fl/flThpok-Cre+ Foxp3GFP mice were also sorted by FACSAria II, and 1 × 105 cells were used for RNA-seq. RNA was extracted using TRIzol Reagent (Thermo Fisher) and column purified using miRNeasy Micro Kit (Qiagen). RNA was subjected to library preparation with Ion Total RNA-Seq Kit v2 (Thermo Fisher) and sequenced by Ion Proton. Sequences were mapped to mm9 with TopHat2 (version 2.0.11). Normalized FPKM was generated with Cuffnorm (version 2.2.0) and differentially expressed genes (FDR < 0.05) were selected using Cuffdiff (version 2.2.0). Treg up and down signature genes were defined as those that (i) are differentially expressed in peripheral Tconv and Treg cells with FDR less than 0.05, (ii) have FPKM greater than 5 in either population, and (iii) have greater than threefold change in average FPKM of duplicates between Tconv and Treg cells. For the selection of differentially expressed genes between tTreg precursor and peripheral Treg cells (Fig. 3a), FDR <0.05 was used. A list of genes associated with epigenetic modification was obtained from QuickGO GO:0006325 and GO:0040029.
For the comparison of Treg-SE-associated gene expression (Supplementary Fig. 1g), RNA-seq data of various immune cell types were obtained from the NCBI database (GEO GSE60103)50 and analyzed using the procedure described above.
RNA-seq of CD24+CD25+GITR+CD4SP thymocytes (developing Treg cells, which include tTreg precursor and immature thymic Treg cells) and peripheral CD4+CD25+GFP+ cells from Satb1fl/+Cd4-Cre+Foxp3GFP and Satb1fl/flCd4-Cre+Foxp3GFP mice was performed in duplicate with 103 cells. Cells were lysed in RLT buffer (Qiagen) and reverse transcribed using SMART-seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech). cDNA was then fragmented by Covaris Focused-ultrasonicator S220, subjected to library preparation with KAPA Library Preparation Kit and sequenced by Ion Proton. Sequences were mapped to mm9 using Tophat2. Normalized FPKM was generated with Cuffnorm. For differential gene expression analyses, tag counts obtained by HT-seq (version 0.6.1) were analyzed using DESeq2 package in R (version 3.1.2).
Assay for transposase-accessible chromatin (ATAC)-seq
ATAC-seq was performed as described51, with modifications to sequence using Ion Proton sequencing platform. Native chromatin transposed with sequencing adaptors using Nextera DNA Library Prep Kit (Illumina) was purified using a MinElute PCR Purification Kit (Qiagen). Purified DNA was amplified using NEBNext High-Fidelity PCR Master Mix (New England BioLabs) with the following primers: forward 5′-TCGTCGGCAGCGTCAGATGTG-3′ and reverse 5′-GTCTCGTGGGCTCGGAGATGT-3′. PCR fragments were size-selected (150–500 bp) with Agencourt AMPure XP (Beckman Coulter), sonicated by Covaris Focused-ultrasonicator S220, subjected to library preparation with KAPA Library Preparation Kit, and sequenced by Ion Proton. Sequence reads were mapped to mm9 using Bowtie2 after removal of PCR primer sequences by Cutadapt. ATAC-seq peaks were detected by MACS2.0 and visualized using GenomeJACK Browser, with peak height normalized at the Gapdh locus. Similarly to histone ChIP-seq, raw tag counts were normalized using DESeq2 package in R for global analyses.
Methyl-CpG binding domain protein (MBD)-seq
Genomic DNA, extracted from 106 FACS-sorted cells, was fragmented by Covaris focused-ultrasonicator S220, and fragments with methylated CpG were immunoprecipitated using MethylMiner Methylated DNA Enrichment Kit (Thermo Fisher). After purification with MinElute PCR Purification Kit, fragments were subjected to library preparation with KAPA Library Preparation Kit and sequenced by Ion Proton. Sequences were mapped to mm9, and peaks detected by MACS2.0 were visualized using GenomeJack Brower, with peak height normalized at the Gapdh locus. Global analyses were conducted based on normalized tag counts obtained using DESeq2 package in R. Treg-specific DNA demethylated regions in Figure 1f were defined by comparing duplicated MBD-seq results in Tconv and Treg cells using DiffBind package (version 1.12.3) in R. Regions with FDR < 0.005 and fold change > 3 were selected as differentially methylated regions.
Comparison between SEs and TEs
For the comparison of enhancer activity between Treg-SEs and Treg-TEs (Supplementary Fig. 2b), normalized tag counts at their constituents (unstitched H3K27ac peak regions) were used so that the sizes of included enhancer regions are normalized (median of Treg-SE constituent size = 230 bp; median of Treg-TE constituent size = 229 bp).
Detection of transcription factor-binding sites
Transcription factor ChIP-seq peaks were identified using FindPeaks, with their sizes fixed at 500 bp, minimum distance between peaks being 500 bp, and FDR set at 0.001. The overlap of SEs with ChIP-seq peaks or among transcription factor ChIP-seq peaks was defined by ≥ 1 bp overlap.
Categorization of Satb1-binding sites by the status of chromatin accessibility was based on the intersection between ATAC-seq peak sites (identified using FindPeaks, with -region option, minimum distance between peaks 100 bp, and FDR set at 0.00001) and Satb1-binding sites (Fig. 4b).
Binding of transcription factors at Satb1-binding sites was examined by calculating average tag counts per 10 million reads at ± 2 kb from the center of Satb1-binding sites, using annotatePeaks in Homer package (Fig. 4e).
Effects of Satb1 deletion on Treg-SE activation
H3K27ac ChIP-seq data of wild-type and Satb1fl/flCd4-Cre+ tTreg precursor cells, prepared in duplicates, were analyzed for differential enhancer activity. Normalized tag counts at stitched enhancers, calculated following the procedure described above, were subjected to differential regulation test in DESeq2 package. Regions differentially regulated with FDR < 0.1 and fold change > 1.5 were defined as regions upregulated or downregulated by Satb1 deletion.
Bidirectional enhancer RNA identification
Processed cap analysis gene expression (CAGE) data were obtained from FANTOM5 database (http://fantom.gsc.riken.jp/data/). Expression of bidirectional enhancer RNA was determined using the algorithm previously described18, with tags mapped to exons and transcription start sites (TSS) excluded. Expression of bidirectional enhancer RNA was normalized by total read counts (Fig. 1c,e).
Microarray analysis
For analysis of gene expression in thymocyte fractions and other immune cells shown in Figure 3b, raw data from the NCBI database (GEO GSE15907) were analyzed52.
Motif analysis
Satb1-binding sites (±100 bp) were subjected to known motif analysis, using findMotifsGenome in Homer package. Those with P > 10−5 were considered as unreliable results (Supplementary Fig. 4b).
Flow cytometric analysis
Lymphocytes, splenocytes or thymocytes were subjected to Fc receptor blocking using purified anti-CD16/32 antibody (BioLegend), surface molecule staining with appropriate antibodies, and fixation and permeabilization using Foxp3/Transcription Factor Staining Buffer Set (Affymetrix eBioscience), followed by intracellular staining. Cytokine staining was carried out after stimulation with 20 ng/ml phorbol myristate acetate (PMA), 709 ng/ml ionomycin and 0.67 μl/ml GolgiStop (BD Bioscience) for 4 h. Stained cells were analyzed using LSRFortessa (BD Bioscience). The following antibodies were used in addition to those mentioned above: anti-CD25-APC (1:100, eBioscience), anti-CD45.1-PE-Cy7 (1:200, BD Bioscience), anti-Foxp3-eFluor450 (1:100, eBioscience), anti-CTLA4-PE (1:100, BD Bioscience), anti-Helios-PE (1:100, BioLegend), anti-Nrp1-biotin (1:100, R&D Systems), and anti-IFN-γ-APC (1:200, Affymetrix eBioscience). Clone and catalog numbers were as follows: anti-CD25-APC (clone PC61.5) anti-CD45.1-PE-Cy7 (clone A20), anti-Foxp3-eFluor 450 (clone FJK-16s), anti-CTLA4-PE (clone UC10-4F10-11), anti-Helios-PE (clone 22F6), anti-Nrp1-biotin (catalog BAF566), anti-IFN-γ (clone XMG1.2). Dead cells were excluded from analyses by using Live/Dead Cell Viability Kit (Thermo Fisher).
Cell culture
The method for in vitro suppression assay (Supplementary Fig. 5a) was described6. Briefly, 5 × 104 peripheral Tconv cells, labeled with CellTrace Violet Proliferation Kit (Thermo Fisher) were incubated with 1 μg/ml anti-CD3ε antibody (BD Biosciences, 553057), 1 × 105 irradiated splenocytes and various ratios of CD4+CD25+GFP+ Treg cells from Foxp3GFP or Satb1fl/flCd4-Cre+Foxp3GFP mice in RPMI 1640 (Gibco) supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol (Sigma-Aldrich), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco). Percentages of proliferated Tconv (responder) cells were analyzed using LSRFortessa after 4 d.
For the Foxp3 induction assay from tTreg precursor cells, two protocols were used— one with IL-2 stimulation (Fig. 7c,d) and the other with IL-2 and TCR stimulation (Fig. 7e,f). For the former, similarly to the method described8, CD3ε+CD24+CD25+GITR+GFP−CD4SP thymocytes from Satb1fl/+Cd4-Cre+Foxp3-eGFP and Satb1fl/flCd4-Cre+Foxp3-eGFP mice were incubated with 100 U/ml IL-2 for 24 h. For the latter, these cells were incubated with Dynabeads Mouse T-Activator CD3/CD28 (Thermo Fisher) in RPMI 1640 supplemented with 10% FBS, penicillin–streptomycin, 2-mercaptoethanol and 100 U/ml IL-2 for 5 d as described9. Percentages of Foxp3+ cells were examined, and GFP+ and GFP− cells were sorted by FACSAria II for DNA demethylation assay.
The Foxp3 stability assay (Supplementary Figs. 5c and 7c) was performed by incubating target cells with Dynabeads Mouse T-Activator CD3/CD28 (Thermo Fisher) in RPMI 1640 supplemented with 10% FBS, penicillin–streptomycin and 2-mercaptoethanol, with or without 100 U/ml IL-2. Percentages of Foxp3+ cells were analyzed after 6 d.
Immunoblotting
FACS-sorted cells were lysed and sonicated using Bioruptor UCD-200 (CosmoBio). Extracted protein was reduced, subjected to SDS-PAGE and transferred onto a PVDF membrane using iBlot2 (Thermo Fisher). Satb1 and GAPDH proteins were detected using anti-SATB1 antibody (BD Biosciences, 611182), HRP-conjugated anti-GAPDH antibody (Cell Signaling Technologies, 3683), HRP-conjugated anti-mouse IgG (GE Healthcare, NA931VS), ECL Prime Western Blotting Detection Reagent (GE Healthcare) and LAS-4000 mini (GE Healthcare).
ELISA
Serum samples from 16-week-old mice were collected by cardiac puncture and assessed for the level of serum immunoglobulin subtypes. Serum immunoglobulin was captured by anti-mouse Ig (Southern Biotech) and measured using HRP-conjugated anti-IgM, anti-IgG1, anti-IgG2c or anti-IgE (Southern Biotech) and TMB Substrate Reagent Set (BD Bioscience).
Generation of mixed bone marrow chimeric mice
T cell–depleted bone marrow cells from CD45.1+ and CD45.2+ Satb1fl/flCd4-Cre+ mice were mixed at a ratio of 2:1 (owing to the ‘preferential’ expansion of Satb1-deficient cells when mixed at 1:1 ratio) and injected intravenously in Rag2−/− mice, which had been irradiated at 3.5 Gy less than 24 h before transplantation.
Adoptive transfer of Treg cells
FACS-sorted 1 × 106 peripheral CD4+CD25+ Treg cells from 4- to 6-week-old wild-type mice (C57BL/6J) were adoptively transferred into 4-d-old Satb1fl/flCd4-Cre+ mice by intraperitoneal injection.
Histological analysis
Organs were harvested from 16-week-old mice and fixed in 10% formaldehyde for 1 week. They were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E). Stained sections were subjected to scoring of disease severity, in a double-blinded manner, based on the following criteria.
Insulitis: 0, no insulitis; 1, peri-islet inflammation; 2, mild intra-islet insulitis; 3, severe intra-islet insulitis; 4, complete destruction of islets.
Oophoritis: 0, no oophoritis; 1, inflammatory cell infiltration; 2, tissue destruction with reduced oocytes; 3, tissue destruction with no oocytes or corpus luteum.
Sialdenitis: 0, no sialdenitis; 1, mild inflammation; 2, intermediate inflammation with up to 50% of area infiltrated; 3, severe inflammation with more than 50% of area infiltrated.
Gastritis: 0, no gastritis; 1, submucosal inflammation; 2, mild mucosal inflammation; 3, intermediate mucosal inflammation with destruction of gastric glands; 4, severe mucosal inflammation with loss of parietal cells.
Pneumonitis: 0, no pneumonitis; 1, mild inflammation; 2, intermediate inflammation; 3, severe inflammation and tissue destruction.
DNA methylation analysis by Sanger sequencing
Methods and primers for DNA methylation analysis for hypomethylated regions (Fig. 7e and Supplementary Figs. 5d and 6f) were described6. Briefly, genomic DNA extracted from 104–105 FACS-sorted cells was subjected to bisulfite treatment using MethylEasy Xceed (Human Genetic Signatures), followed by PCR amplification of target regions and subcloning into pTAC-1 plasmid in DynaExpress TA PCR Cloning Kit (BioDynamics Laboratory Inc). Amplicon sequence in plasmids were directly amplified from colonies using Illustra TempliPhi Amplification Kit (GE Healthcare) and sequenced. 16 colonies per region were examined and average percentage of demethylated clones for each CpG residues in the amplicon sequence are shown in color code. Representative data of at least two independent experiments are shown.
DNA methylation analysis by deep sequencing
Genomic DNA was extracted from 1–5 × 105 FACS-sorted cells. Following bisulfite treatment with MethylEasy Xceed, Foxp3 CNS2 region was amplified by PCR using KAPA HiFi HotStart Uracil+ ReadyMix (KAPA Biosystems) and the following PCR primers: forward 5′-TTTTGGGTTTTTTTGGTATTTAAGA-3′ and reverse 5′-ACAAAT AATCTACCCCACAAATTTC-3′. PCR products were gel purified using MinElute Gel Extraction Kit (Qiagen). Approximately 500 ng purified amplicons was used for library construction with KAPA Library Preparation Kit according to the manufacturer’s instructions (KAPA Biosystems) and sequenced using MiSeq (Illumina). To maximize the accuracy of sequencing, amplicons were mixed with random sequences so that the final amplicon percentage within a run was <5%. After quality trimming, sequence reads were mapped to mm9 using Bismark (version 0.13.1). Methylation status of 6 CpG residues contained in the amplicon sequence was visualized in color code using Methplot package (version 1.0) in R. Representative data of two independent experiments are shown (Fig. 5g and Supplementary Fig. 6h).
Colitis induction
CD4+CD25−CD45RBhi T cells were FACS-sorted from CD45.1+ wild-type or CD45.2+ Satb1fl/flThpok-Cre+ lymphocytes. 2.5 × 105 cells of each population were mixed and intravenously transferred into Rag2−/− mice. After 17 d, cells from mesenteric lymph nodes were isolated for FACS analysis, and CD45.1+ or CD45.2+ CD4+ T cells were purified for DNA methylation analysis by amplicon sequencing.
Statistical analysis
Investigators estimated sample sizes on the basis of prior experience. Animals were selected randomly except for genotype and sex and were analyzed in a blinded manner. Experiments were independently replicated at least twice, and representative and/or summary data are shown. Flow cytometric analyses and DNA methylation analyses were independently repeated three or more times with three or more mice. All H3K27ac ChIP-seq and RNA-seq were prepared independently in two replicates, whereas other ChIP-seq experiments were performed once owing to the cost of sequencing. The number of experiments for representative images is stated in each figure legend. Variation in sample distribution was examined by Kolmogorov–Smirnov test. Statistical differences were determined by statistical tests stated in each Figure legend. P < 0.05 level of confidence was accepted for statistical significance.
Data availability
ChIP-seq, RNA-seq, MBD-seq and ATAC-seq data sets were deposited in DNA Data Bank of Japan under accession numbers DRA003955, DRA004738 and DRA005202.
Supplementary Material
Acknowledgments
We thank Y. Nakamura for DNA sequencing support and assistance with RNA-seq experiments, S. Kojo for providing technical advice regarding ChIP-seq experiments, and K. Chen for reading the manuscript. Bioinformatics analyses were conducted using the computer system at the Genome Information Research Center of the Research Institute for Microbial Diseases at Osaka University. This work was supported by Grants-in-Aid for Japanese Society for the Promotion of Science (JSPS) Fellows 261560 from the JSPS to Y.K. and Core Research for Evolutional Science and Technology from the Japan Science and Technology Agency to S.S. and JSPS Grants-in-Aid for Scientific Research B 15H04744 to N.O.
Footnotes
AUTHOR CONTRIBUTIONS
Y. Kitagawa designed, performed and analyzed most experiments, including flow cytometric analyses, in vivo and in vitro experiments, ChIP-seq, library preparation for sequencing and bioinformatics analyses. N.O. performed ATAC-seq and MBD-seq, Y. Kidani assisted with bioinformatical analyses and performed immunoblotting. A.V. and K.H. provided crucial advice. R.K. performed H3K4me3 ChIP-seq. K.Y. assisted with histological analysis. D.M. and S.N. performed amplicon sequencing. I.T. and T.K.-S. provided helpful suggestions. T.K.-S. and M.K. provided Satb1 conditional knockout mouse. I.T. provided Thpok-Cre mouse. Y. Kitagawa and S.S. wrote the manuscript, and all authors reviewed it. T.K.-S. and N.O. critically read the manuscript and provided advice. S.S. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 3.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 4.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 5.Floess S, et al. Epigenetic control of the Foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Lee HM, Hsieh CS. Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. J Immunol. 2009;183:2261–2266. doi: 10.4049/jimmunol.0901304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toker A, et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180–3188. doi: 10.4049/jimmunol.1203473. [DOI] [PubMed] [Google Scholar]
- 10.Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. Allen and Unwin; 1957. [Google Scholar]
- 11.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arner E, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vahedi G, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huehn J, Beyer M. Epigenetic and transcriptional control of Foxp3+ regulatory T cells. Semin Immunol. 2015;27:10–18. doi: 10.1016/j.smim.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 24.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature. 2015;528:132–136. doi: 10.1038/nature16141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao B, et al. An anti-silencer- and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. J Exp Med. 2015;212:809–824. doi: 10.1084/jem.20142207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh K, Hjort M, Thorvaldson L, Sandler S. Concomitant analysis of Helios and neuropilin-1 as a marker to detect thymic derived regulatory T cells in naïve mice. Sci Rep. 2015;5:7767. doi: 10.1038/srep07767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiault N, et al. Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat Immunol. 2015;16:628–634. doi: 10.1038/ni.3150. [DOI] [PubMed] [Google Scholar]
- 32.Mucida D, et al. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II–restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai X, et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity. 2013;38:1116–1128. doi: 10.1016/j.immuni.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 36.Morikawa H, et al. Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell–specific transcriptional regulation. Proc Natl Acad Sci USA. 2014;111:5289–5294. doi: 10.1073/pnas.1312717110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyer M, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmud SA, et al. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol. 2014;15:473–481. doi: 10.1038/ni.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng G, Yu A, Dee MJ, Malek TR. IL-2R signaling is essential for functional maturation of regulatory T cells during thymic development. J Immunol. 2013;190:1567–1575. doi: 10.4049/jimmunol.1201218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 43.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 44.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 45.Ito Y, et al. Detection of T cell responses to a ubiquitous cellular protein in autoimmune disease. Science. 2014;346:363–368. doi: 10.1126/science.1259077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurts C, et al. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J Exp Med. 1996;184:923–930. doi: 10.1084/jem.184.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 48.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mingueneau M, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq, RNA-seq, MBD-seq and ATAC-seq data sets were deposited in DNA Data Bank of Japan under accession numbers DRA003955, DRA004738 and DRA005202.