Abstract
Clustered protocadherins mediate neuronal self-recognition and non-self discrimination—neuronal “barcoding”—which underpin neuronal self-avoidance in vertebrate neurons. Recent structural, biophysical, computational, and cell-based studies on protocadherin structure and function have led to a compelling molecular model for the barcoding mechanism. Protocadherin isoforms assemble into promiscuous cis-dimeric recognition units and mediate cell-cell recognition through homophilic trans-interactions. Each recognition unit is composed of two arms extending from the membrane proximal EC6 domains. A cis-dimeric recognition unit with each arm coding adhesive trans homophilic specificity can generate a zipper-like assembly that in turn suggests a chain termination mechanism for self-vs-non-self-discrimination among vertebrate neurons.
Keywords: Clustered protocadherins, Neuronal self-avoidance, Cell-cell recognition, Protein interaction specificity, Crystal structure
1. Introduction
The establishment of functional neural circuits in the human brain involves highly specific connections among billions of neurons through trillions of synapses [1]. The formation of such complex neural circuits depends on a limited repertoire of guidance cues and cell surface receptors. Clustered protocadherins (Pcdhs) are a family of highly diverse cell-surface receptors that are thought to provide individual neurons with single-cell-specific molecular “barcodes”, which provide unique cell surface identities required for neurite self-avoidance [2–4]. Although recent studies have demonstrated that Pcdhs have additional roles in regulating neuronal survival, synaptogenesis, dendritic arborization, and neuronal tiling [2–17] —this review focuses primarily on the role of Pcdhs in neuronal self-avoidance that in turn requires that neurons be able to distinguish “self” from “non-self”.
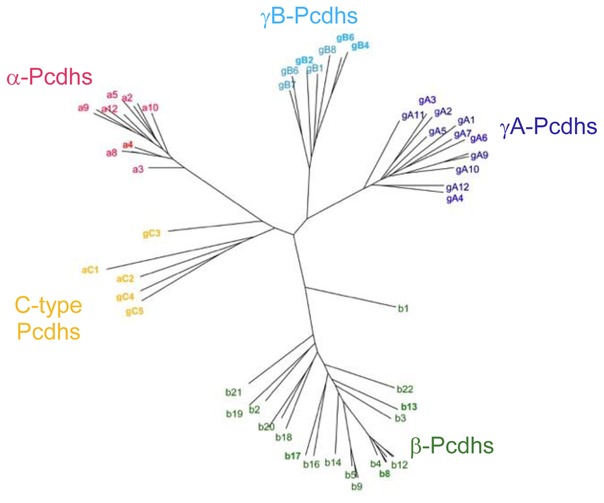
Mammalian genomes contain 50–60 Pcdh genes that are arranged in three contiguous gene clusters designated α, β, and γ [18, 19]. Other vertebrates, such as the fugu and elephant shark, also have Pcdh genes but with varying numbers of isoforms and distinct cluster organizations [20, 21]. Each Pcdh isoform has a distinct extracellular region, single pass transmembrane helix, and short cytoplasmic region encoded by a single “variable” exon. Additionally, the Pcdh α- and γ-gene clusters each contain three constant exons that encode a cluster-specific constant cytoplasmic region. Phylogenetic analysis of the 58 clustered Pcdh mouse isoforms revealed that they fall into five distinct subfamilies (Figure 1): alternate α-Pcdhs (1–12), alternate β-Pcdhs (1–22), alternate γA-Pcdhs (1–12), alternate γB-Pcdhs (1–2 & 4–8), and C-type Pcdhs (αC1, αC2, γC3, γC4, and γC5) (Figure 1). Alternate (non-C-type) Pcdh isoforms are chosen for expression in each neuron by a stochastic promoter choice mechanism [19, 22–26]. Individual neurons appear to express a small subset of the ~50 alternate isoforms [19, 22–26]. The C-type Pcdhs are expressed ‘deterministically’ rather than stochastically [22, 24].
Figure 1.
Phylogenetic tree of mouse Pcdh isoforms based on the sequences of their EC1–EC4 domains which mediate the homophilic trans-interactions. The a, b, gA, and gB isoforms are grouped into four separate clusters. Although part of the b-cluster, the b1 isoform is significantly divergent from other members of the b-cluster. The two aC and the three gC isoforms are divergent from all other isoforms, including the other members of the a and g clusters
In neurite self-avoidance, an essential feature of neural circuit assembly, branching neurites (axons and dendrites) from the same neuron avoid one another, while neurites from different neurons do not. This assures that neurites from the same neuron can arborize extensively without crossing or clumping, while neurites from different neurons can interdigitate and occupy the same field. This phenomenon requires a mechanism that allows individual neurons to distinguish self from non-self interactions [27, 28]. It appears that, for both vertebrates and insects, neuronal self-avoidance relies on generating unique individual cell surface identities through the stochastic expression of diverse repertoires of cell surface protein isoforms [27–30]. In the fly neuronal identity is defined by the expression of single-cell-specific subsets of Dscam1 protein isoforms, generated by stochastic alternative RNA splicing [31–34]. In vertebrates, neuronal identity is provided by stochastic expression of single-cell-specific subsets of Pcdh isoforms [4, 22–24].
Counter-intuitively, in both insects and vertebrates the process of self-avoidance begins with adhesive homophilic interactions required for recognition [27, 35–38]. In the fly, there are 19,008 possible Dscam isoforms with distinct extracellular domains, of which ~10–50 are expressed in each neuron [31, 33–35, 39]. The majority of these isoforms bind in trans in a strictly homophilic manner [35, 36]. In mammals, the 50–60 Pcdh isoforms have been shown to bind with homophilic specificity, as will be discussed below. Current thinking posits that identical Dscam/Pcdh isoforms located on the surface of neurites emanating from the same cell bind to each other homophilically in trans (different neurites) and this interaction triggers a signaling process that requires the intracellular domains [40], which leads to repulsion. In contrast, when two neurons expressing a sufficiently diverse set of Dscam/Pcdh isoforms come into contact, their different isoform composition will not lend itself to homophilic binding and hence an avoidance mechanism will not be triggered [27].
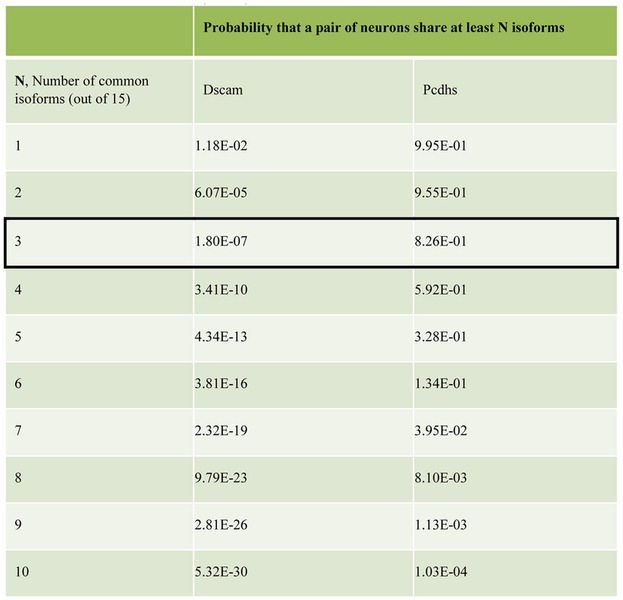
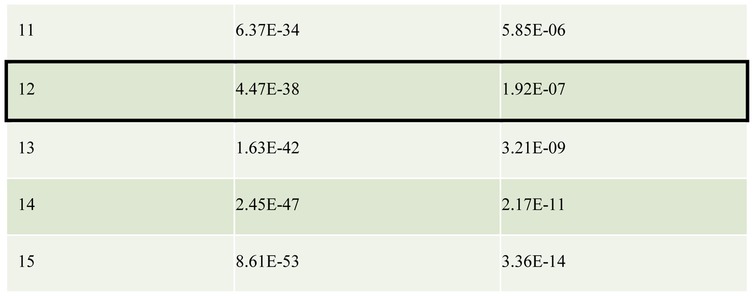
The large number of distinct Dscam1 isoforms generated in individual neurons by alternative RNA splicing decreases the probability that any two interacting fly neurons have an identical or even a similar isoform repertoire [29]. Assuming, for example, that 15 distinct isoforms are produced per cell, the probabilities that two cells will express three or more isoforms in common (thereby presumably leading to inappropriate initial adhesion and then repulsion) is ~10−7 (Table 1). These numbers are small enough to ensure that inappropriate repulsion will be a rare event [29]. How do Pcdhs, with far fewer isoforms, provide sufficient diversity for a single-cell identity within mammalian nervous systems, which are far more complex than that of the fly? Recent structural and biophysical studies combined with cell-based aggregation assays have provided a surprising mechanism that appears to solve this problem. Here, we review recent studies that have transformed our understanding of Pcdh structure and function, and have led to the proposal of a structure-based mechanism for neuronal barcoding, which provides greater neuronal diversity than that of Drosophila Dscam1.
Table 1.
The table shows the probability that a pair of neurons share N or more common Dscam/Pcdh isoforms. The total number of distinct isoforms expressed by each neuron is assumed to be 15 in this example. Isoforms are chosen from a pool of 19 008 or 58 for Dscam and Pcdhs respectively. For Dscam the ‘tolerance’ for common isoforms expressed by a pair of interacting neurons was assumed to be 20%, or 3 out of 15 isoforms expressed in this example (boxed). Any more common isoforms and it was assumed that a pair of neurons would incorrectly recognize one another as self and result in repulsion. There is only a ~10−7 probability of a pair of neurons sharing three or more isoforms, and therefore the chances of inappropriate repulsion between pairs of Dscam-expressing neurons is very low. To reach a similar probability of ~10−7 pairs of neurons expressing Pcdhs need to be able to tolerate at least 12 common isoforms out of 15 (boxed).
2. Homophilic cell-cell recognition specificity
In common with many other cadherin superfamily members [41], Pcdhs function in cell-cell recognition through calcium-dependent binding between their extracellular regions [37]. The Pcdh extracellular region contains six extracellular cadherin (EC) domains, each of which is composed of approximately 100 residues that form a two-layered anti-parallel β-sheet structure. Binding three Ca2+ ions to cadherin-conserved calcium-binding motifs stabilizes Pcdh EC interdomain junctions.
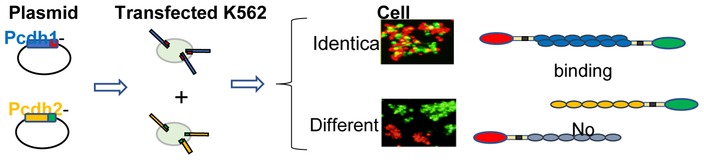
In an important study, using quantitative cell aggregation assay with K562 cells, Schreiner & Weiner [38] tested seven γ-Pcdhs and showed that they exhibit isoform-specific homophilic binding. Schreiner and Weiner [38] showed that K562 cells were a suitable cell line for protocadherin expression, as they are non-adherent in culture and do not endogenously express protocadherins or other cell adhesion molecules. In these assays plasmids expressing individual Pcdh isoforms are transfected into the K562 cells and trans-binding is assayed by cell aggregation (Figure 2). We note that using cell aggregation assays to report on Pcdh recognition may appear counterintuitive due to their in-vivo function of self-avoidance. However, repulsion depends on both the Pcdh ectodomains’ adhesive properties and intracellular signaling cascade. The non-neuronal kidney K562 cells that were used in these assays appear to lack the repulsive signaling function, as only adhesion is observed. Moreover, several studies have found that in some neuron-neuron and neuron-astrocyte interactions Pcdhs may play an adhesive role [42–44].
Figure 2.
Schematic diagram of the cell aggregation assays used to assess the trans-binding properties of Pcdh isoforms. The binding properties of two exemplary isoforms are summarized. Cells transfected with Pcdh isoforms tagged with either red or green fluorescent proteins are mixed. Aggregates containing both red and green cells form when both cell populations are expressing identical isoforms. Separate red and green aggregates form when the two cell populations are expressing different isoforms, demonstrating Pcdh trans interactions are preferentially homophilic.
Also using K562 cells Thu et al. [37] showed that Pcdh isoforms from all three gene clusters mediate specific homophilic interactions. In these studies, mixing two cell populations transfected with identical isoforms results in mixed aggregates. In contrast, mixing two cell populations transfected with different isoforms resulted in separate homophilic aggregates (Figure 2). Remarkably, even when the transfected isoforms had greater than 90% sequence identity there was no observed cross binding [37], demonstrating highly specific homophilic interactions.
Notably, Pcdh γC4 and all alternate α-Pcdhs fail to reach the cell surface when expressed alone [37, 45, 46] and therefore cannot mediate cell aggregation [37]. However, when these isoforms are co-transfected with any β, γ or some C-type Pcdh isoforms (carrier Pcdhs) they are able to reach the cell surface, and are thus able to mediate cell aggregation [37]. In fact, co-transfection of γC4 or any α-Pcdh with fragments that include the EC5–EC6 domains of carrier Pcdhs is sufficient for cell-surface delivery [37]. Importantly, the observation that γC4 and α-Pcdhs are carried to the cell surface by other Pcdhs reveals a cis (same cell) interaction between γC4 or α-Pcdhs and carrier Pcdhs, and this interaction is dependent on EC5–EC6 domains (this phenomenon will be further elaborated below) [37]. Overall, with the exception of Pcdh-αC1, all mouse Pcdh isoforms are able to engage in highly specific homophilic interactions on the apposing cell membranes [37, 38].
3. Crystal structures of Pcdh trans dimers
The most thoroughly characterized cadherins are the classical cadherins, which mediate calcium dependent cell-cell adhesion through trans (cell-cell) homodimerization of their membrane-distal EC1 domains. In contrast to classical cadherins the first Pcdh structures obtained, which included the membrane-distal EC1 domains [47] or EC1–EC3 domain fragments [48, 49], were found to be monomeric in solution. Consistently, constructs of corresponding size did not mediate cell-cell binding in cell aggregation assays [49]. These early Pcdh structures revealed that despite containing cadherin domains, Pcdhs are structurally distinct from classical cadherins. Most notably, the first β-strand (A-strand) of EC1 lacks the critical Trp-2 residue, which is conserved among classical and desmosomal cadherins and anchors the strand-swap trans-binding interface of these cadherins [47–51]. In addition, the inter-domain orientation of the three EC domains within each structure results in an overall straight architecture. This is in contrast to the curved architecture of classical cadherins which facilitates the formation of a parallel EC1/EC1 interaction forming between molecules from opposed cell surfaces [48, 49, 51].
Rubinstein et al. [49] demonstrated through solution biophysical measurements and cell aggregation assays with a Pcdh-ectodomain truncation series that EC1–EC4 was required for Pcdh trans-binding. They then used docking calculations of the EC1–EC3 structures, constrained by sequence and mutagenesis experiments to determine that the EC1–EC4 domains form an extended trans-binding interface that is topologically similar among all clustered Pcdhs. Moreover, the analysis also strongly suggested that the trans-interaction occurs in a head-to-tail (anti-parallel) arrangement, with EC1 interacting with EC4 and EC2 interacting with EC3. Independently, Nicoludis et al. [48] used Pcdh EC1–EC3 fragment structures which they had determined along with correlated mutation analysis to arrive at a similar conclusion.
More recent papers have presented crystal structures of Pcdh EC1–EC4-mediated trans-dimers describing the trans adhesive interface in atomic detail. Crystal structures of ectodomain fragments corresponding to EC domains 1–4 or 1–5 for nine different isoforms, including at least two representative isoforms from each of the four Pcdh subfamilies (α, β, γA, and γB), have been solved [52–54] (Figure 3A). Despite significant diversity in their sequences, isoforms from all subfamilies formed structurally similar dimers (Figure 3A). For almost all structures, membrane-distal EC domains 1–4 dimerize in a head-to-tail orientation in which residues from EC1 domains contact residues from EC4 domains and residues from EC2 domains contact residues from EC3 domains (Figure 3A). Individual molecules from each complex were found to be highly structurally similar to the monomeric EC1–EC3 structures, indicating that complex formation did not involve significant structural rearrangements [52]. The structures revealed that the interface is not continuous but is instead divided between an EC1/EC4 interface and an EC2/EC3 interface (Figure 3B and 3C). The average buried surface area upon trans dimer formation was found to be 4666 Å2 with the interface between EC1/EC4 and EC2/EC3 burying on average 2062 and 2604 Å2 respectively [52, 53].
Figure 3.
A) Trans-dimer structures of representative isoforms from the a, b, gA and gB clusters. The structures are shown in ribbon depiction with transparent molecular surfaces. Bound calcium ions are shown as green spheres. Glycans are shown as red, white, and blue spheres. B) Surface view of the gB7 trans-dimer. C) Open book depiction of the gB7 dimer revealing the interacting faces. Interfacial residues are colored grey if they are constant among all gB isoforms or colored green if they vary among gB isoforms. The EC1/EC4 interaction was absent in the gA8 crystal structure and one of two dimers observed in the gA1 crystal structure. While this suggests some flexibility in Pcdh structure, the lack of the EC1/EC4 interactions is likely due to crystallization artifacts since mutagenesis studies show that all four domains are required for dimerization and cell-cell recognition for these isoforms
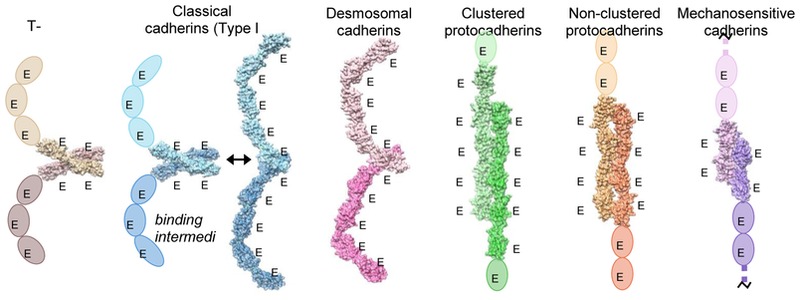
Despite having a similar domain structure, the homodimeric antiparallel EC1–EC4 interface formed by the clustered Pcdhs is fundamentally different from the homodimeric interfaces of classical cadherins and the heterodimeric interfaces of cadherin-23/Pcdh-15 and desmosomal cadherins (Figure 4) [52–54]. The classical and desmosomal cadherins bind by swapping their N-terminal β-strand (A-strand) between two interacting EC1 domains (Figure 4) [50, 51, 55]. In addition, an “X-dimer” interface, located in the linker region between EC1 and EC2, functions as a binding intermediate in classical cadherin and is the actual adhesive interface in the classical cadherin related protein T-cadherin (Figure 4) [56, 57]. Similar to the Pcdhs, the hetero-dimeric complex between cadherin-23 and Pcdh-15 exhibits an antiparallel interface; however, this interface comprises the EC1 and EC2 domains and hence is distinct from the clustered Pcdh interfaces (Figure 4) [58]. Recently, the crystal structure of the homodimeric complex of Pcdh-19, a non-clustered δ2-Pcdh, revealed an anti-parallel EC1–EC4 interface that is highly similar to that of the clustered Pcdh trans-dimer, thereby indicating that in addition to clustered Pcdhs and Pcdh-19 this interface may be used by other non-clustered δ-Pcdhs [59]. Overall, the cadherin domain has demonstrated a remarkable diversity in binding mechanisms.
Figure 4.
Trans-binding interactions of cadherin superfamily proteins. Representative crystal structures of the trans-binding mode of each cadherin family for which there is structural information are shown. Crystal structures are shown as molecular surfaces, which each protomer engaged in the dimer interaction colored in a light or dark shade. Extracellular domains that are not present in the crystal structure are shown as ovals. T-cadherin PDB:3K5S [51]; Type I classical cadherin X-dimer encounter complex, E-cadherin W2A mutant PDB:3LNH [52]; Type I classical cadherin strand-swap interface, C-cadherin PDB:1L3W; Desmosomal cadherin desmocolin1 PDB:5IRY [46]; Clustered protocadherin a7 PDB:5DZV [47]; Non-clustered d-protocadherin, protocadherin 19 PDB:5IU9 [54]; Tip-link proteins, cadherin-23 (dark purple) and protocadherin-15 (light purple), PDB:4APX [53].
4. Structural basis of Pcdh homophilic specificity
In order to identify the Pcdh trans-homophilic specificity-determining domains Pcdh chimeras with shuffled EC domains between different isoforms were used [38, 49]. Studies using Pcdh chimeras with multiple domains shuffled simultaneously demonstrated that chimeras with non-matching EC1 and EC4 domains do not bind to each other even when their EC2 and EC3 domains are identical [49]. Similarly, chimeras with non-matching EC2 and EC3 domains do not bind to each other even when their EC1 and EC4 domains are identical [49]. By contrast, when chimeras in which all four EC1 through EC4 domains are identical they do, in fact, bind to each other [49]. Together these and other data from mutagenesis experiments showed that all four membrane distal domains EC1–EC4 contribute to binding specificity [49, 52, 53].
The atomic-resolution structures of Pcdh trans-dimers of representative isoforms from α, β, and γ clusters accompanied by bioinformatics analyses have yielded significant insights into how Pcdhs achieve their remarkable trans-homophilic specificity. The structures of Pcdh trans-dimer complexes are similar overall among all isoforms, most notably in the antiparallel interaction between EC1–EC4 domains (Figure 3A). However, isoforms from different clusters generally exhibit prominent local structural differences in their dimerization interfaces. These are likely the primary reason why α/β, α/γA, β/γA, and β/γB heterodimers do not form [52, 53].
By contrast, the homodimeric structures of isoforms from the same cluster are structurally similar both globally and locally [52, 53]. The basis of specific homophilic binding preferences within subfamilies is therefore not predominantly architectural, but rather due to differences in the interfacial residues. In the mouse, over 90% of the interface residues, across all four interfacial domains, exhibit sequence variation among isoforms of the β and γ cluster (Figure 3C) [52, 53]. Importantly, many of these residues that vary among the mouse isoforms are conserved among different species suggesting that these residues play an important role in recognition specificity [52, 53]. Interfacial residues within the EC2–EC3 domains of α isoforms also exhibit high sequence variability among mouse isoforms with a similar isoform specific conservation pattern among different species. However, the EC1 and EC4 domains of α isoforms are exceptionally conserved with over 90% of interfacial residues conserved among all 12 mouse isoforms [52]. Specifically, for the EC1 domain, only Pcdh-α8 exhibits variability in its interfacial residues compared to the other isoforms, with two interfacial residues showing Pcdh-α8-specific conservation [49, 52]. The conservation of the EC1/EC4 interface in mouse α-Pcdh isoforms is suggestive of a functional role unique to α-isoforms.
Residue-swap experiments, in which interfacial residues that exhibit isoform-specific conservation were shuffled between isoforms, confirmed that such residues underpin Pcdh trans recognition specificity. These experiments also demonstrated that generation of new homophilic specificities often requires swapping pairs or small groups of residues that interact with one another in the trans-interface [49, 52, 53]. Mutated isoforms bind homophilically, but no longer bind to their wild-type parental isoforms [52]. The overall logic of generating strict homophilic specificity between closely related isoforms involves a relatively small number of interactions that are favorable in homodimers, but unfavorable in heterodimers. In some cases, these correspond to stabilizing salt bridges in the homodimer that would be disrupted in the putative heterodimer resulting in electrostatic repulsion. In other isoforms, shape-complementarity in the homodimer is replaced by steric hindrance in the heterodimer. Overall, these results are consistent with the free energy of binding being distributed over four interfaces, where the presence of all four is necessary to generate sufficient affinity to produce a stable homodimer [49, 52, 53].
5. Interference and Tolerance
It is critical that two interacting neurons do not erroneously recognize each other as “self” and avoid each other. However, since both Dscams and Pcdhs are stochastically expressed, there is a finite probability that any pair of neurons will express one or more common isoform, which will then bind to each other and potentially signal both cells to move apart. How can this inappropriate repulsion be avoided? Table 1 reports probabilities that two cells will randomly express one or more of the same isoforms for both Dscams and Pcdhs (assuming 15 different isoforms are expressed per cell). Even for Drosophila Dscam1, with thousands of isoforms to select from, the probability that two neurons will select at least one identical isoform is relatively high [29] while for Pcdhs, with only 58 isoforms to choose from it is essentially a certainty (Table 1). Therefore, it is critical for interacting neurons to be able to tolerate the presence of some common isoforms without triggering repulsion. What is the maximum proportion of common expressed isoforms between two interacting neurons that can be tolerated (the “tolerance”) before the two cells recognize each other erroneously as self? Figure 5A illustrates two extreme cases: the first is where a single isoform that is shared between two interacting neurons is sufficient to trigger repulsion, even if all other isoforms are different (no tolerance to common isoforms, panel i); the second is when two interacting neurons repel each other only if all their expressed isoforms are identical and where a single isoform mismatch is sufficient to prevent erroneous repulsion (high tolerance to common isoforms, panel ii).
Figure 5.
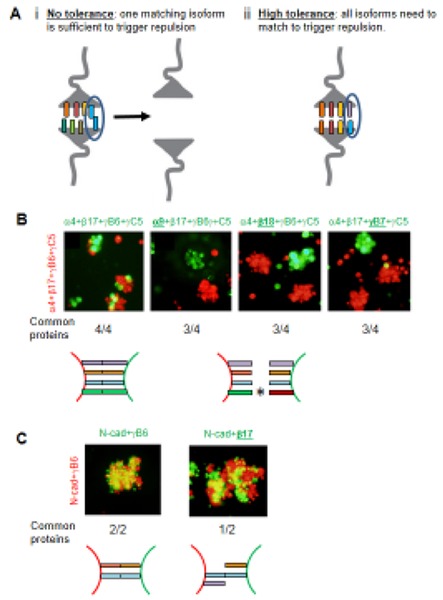
Tolerance and Interference. A) Schematic diagrams illustrating two extreme ‘tolerance to common isoforms’ levels. i) With no tolerance the recognition of one isoform in common between two interacting neurons is sufficient to mediate Pcdh recognition and to trigger repulsion even when all other isoforms are different. ii) With high tolerance one Pcdh mismatch between two interacting neurons is sufficient to interfere with Pcdhs recognition and ultimately repulsion even if all other Pcdhs isoforms are the same. B) Aggregation assays with K562 cells transfected with four Pcdh isoforms labeled in green or red. The underlined isoform in green indicates a mismatched isoform between the red and the green cells, which, as can be seen, results in the red and green cells forming separate aggregates. Experiments with two to five transfected isoforms gave similar results [36]. C) Control assays showing that N-cadherin does not “interfere” with Pcdh adhesion.
It is important to note that for Drosophila Dscam1, with thousands of isoforms, a tolerance of 20% was assumed [29]. As seen in Table 1, two interacting neurons will have a probability of only about 10−7 to share more than 20% (3 out of 15) of isoforms in common, which appears to be sufficiently rare to prevent inappropriate repulsion between interaction neurons in Drosophila [29]. In contrast, Pcdhs would have to have a tolerance of about 80% (12 of 15 isoforms) to achieve a similar probability (~10−7) of inappropriate pairwise repulsion.
A key question is therefore: What is the mechanism that underlies tolerance? For Dscams it is not hard to imagine that a small fraction of common isoforms (e.g. 3 out of 15) is too small for two cells to adhere with sufficient strength or time to lead to repulsion. For example, there may simply not be enough Dscams expressed on the cell surface to achieve a functional binding/repulsion complex. But this logic clearly fails for Pcdhs. Thus, another mechanism must be involved. An important clue to understanding this mechanism is provided by cell aggregation assays, which revealed that cells expressing multiple Pcdh isoforms co-aggregate only with cells expressing the identical set of isoforms [37] (Figure 5B). Remarkably, when 4 isoforms are expressed per cell, only a single mismatch is sufficient to “interfere” with cell-cell aggregation (Figure 5B). In striking contrast, coexpression of N-cadherin did not interfere with Pcdh-mediated aggregation (Figure 5C) [37]. This striking observation of Pcdh interference is fundamental to understanding how only 60 clustered Pcdh isoforms are sufficient to achieve functional diversity for neurite self-avoidance [36].
The interference observed in cell aggregation assays (Figure 5B) suggests that Pcdhs have a tolerance of at least 75%, which generates a pairwise probability of inappropriate recognition similar to that of Dscams with a tolerance of 20% (Table 1, [37]). It is possible that the tolerance is even higher, e.g. if a single mismatch would be enough to interfere with recognition when 10 isoforms are expressed per cell (tolerance of 90%). In this way, two different cells would incorrectly recognize each other as “self” only if their full complement of isoforms was identical (rather than merely “similar” as is the case for Dscam1). We now turn to a discussion of structural studies that have revealed how this level of tolerance can be achieved.
6. Cis-dimeric recognition units
In addition to their homophilic trans interactions, Pcdhs also interact in cis [37, 38, 49, 53]. Solution biophysical measurements of purified recombinant Pcdh ectodomains and cell aggregation studies showed that Pcdhs form cis dimers mediated by EC5 and EC6. Specifically, ectodomain fragments containing the EC5–EC6 domains of β and γB isoforms dimerize in solution independent of the EC1–EC4 trans dimer interactions, but do not aggregate cells indicating that this interaction occurs in cis [49, 53] (Figure 6A). Larger cis-multimers (e.g. tetramers) had been inferred based on gel filtration measurement in earlier studies [38, 60] however only cis-dimers have been conclusively observed.
Figure 6.
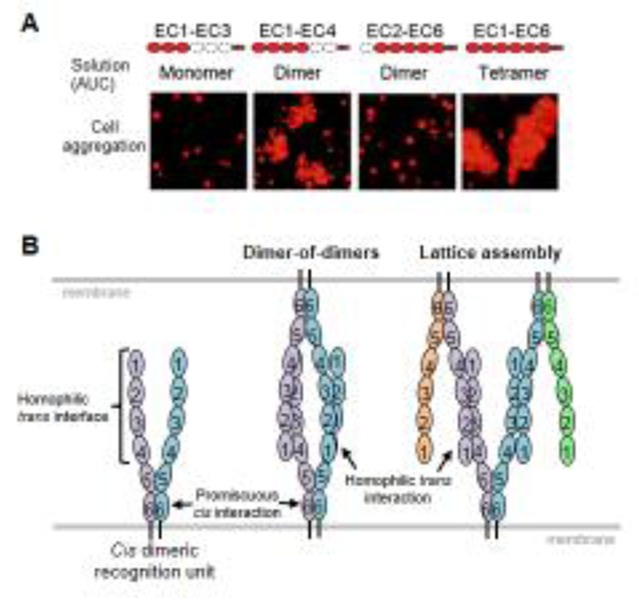
A) Analytical ultracentrifugation (AUC) and cell aggregation results for Pcdh gB6 domain deletion constructs. Domains that are present are shown in red. B) Alternative models for Pcdh trans interactions based on a cis dimeric recognition units (left). In the discrete tetramer model (center), a “two-armed” trans dimer forms between identical cis dimers. In the lattice model, cis-dimeric recognition units interact to form a zipper-like structure.
The important role of EC6 for cis-dimer formation can be seen from the fact that while wild-type Pcdh γB6 behaves as a tetramer in solution (a trans dimer of cis dimers), a single point mutation in the EC6 domain of this Pcdh isoform, which breaks the cis interaction, behaves as a dimer in solution [53]. In cell aggregation assays this point-mutation prevents γB6 Pcdh from both self-delivering and delivering α-Pcdhs to the cell surface, thereby suggesting that cis dimerization is required for cell surface delivery [53].
A large body of evidence demonstrates that Pcdh cis-interactions are promiscuous, with evidence for the formation of both homo- and hetero-cis dimers: First, co-immunoprecipitation experiments involving isoforms from different clusters showed promiscuous interactions between isoforms of the β and γB cluster, isoforms of γA and γB clusters, and isoforms of the γA cluster and the γC3 isoform [37, 38]. Second, in isolation, neither Pcdh-γC4 nor any α-Pcdh isoforms can reach the cell surface; however, both isoforms are delivered to the cell surface by co-expression with αC2, γC3, or any β or γ isoform (‘carrier Pcdhs’) [37]. Cell aggregation assays with either truncated isoforms or isoforms with shuffled domains have demonstrated that cell surface delivery of the clustered Pcdhs depends on the EC5–EC6 domains of both the alpha and the carrier Pcdhs. Importantly, the identity of the specific α-isoform or the specific carrier isoform used in the cell assays does not appear to impact the outcome, thereby revealing promiscuous interactions between α (or γC4) Pcdhs and carrier Pcdhs [37]. Third, the amino acid sequences of the EC6 domains, which control cis dimerization, are highly similar among β-isoforms and among γ-isoforms [37], a finding that is consistent with the idea that promiscuous interactions occur within each family.
Notably, in spite of their general promiscuity, not all possible cis-dimers form, and the homophilic binding affinities of cis-dimers from different subfamilies are highly variable. As previously mentioned, α-Pcdhs and Pcdh-γC4 do not reach the cell surface when singly expressed. This is likely due to a failure of these isoforms to form cis-homodimers. In addition, while β-Pcdhs, γB-Pcdhs and Pcdh-αC2 form strong cis-homodimers in solution with comparable affinities to that of the trans interaction affinity of Pcdhs and other cell adhesion molecules (with dimerization dissociation constants of 8.92 – 80.1 μM);γA-isoforms and Pcdh-γC3 do not form cis-homodimers with measureable affinities in solution [49, 53]. Cis interactions of classical cadherins are similarly weak in solution, however in the two-dimensional environment of the plasma membrane the Pcdh cis interactions will likely be further enhanced [61, 62]. Indeed, unlike α-isoforms, γA-isoforms and Pcdh-γC3 can reach the cell surface on their own, presumably because they are able to dimerize in the context of a cellular membrane. Together these data suggest that cis associations between Pcdh isoforms from different gene clusters will manifest distinct preferences [37, 49, 53]. Identifying preferences of cis interactions and their molecular origins is of great importance since, alongside Pcdh expression data, it will determine the repertoire of Pcdh cis-dimer recognition units presented on the cell surface [37, 49, 53].
7. Two models of the Pcdh recognition complex and associated functional implications
Two molecular models have been proposed to account for the role of Pcdhs in neuronal recognition. The first was based on the assumption (which we now know to be incorrect) that Pcdhs form cis-tetrameric recognition units that interact in trans to form discrete octamers between apposed cells. In this model, interference is caused by the dilution of matched isoform pairs on different cells through their incorporation into a large number of cis tetramers with isoforms that are not matched [60]. However, although the tetramer/octamer dilution model provides a sufficient level of diversity to account for non-self discrimination [60], it fails in that there is also dilution within a single cell such that the probability of two sister neurites containing the same tetramer, and hence repelling, would be far too small [49]. Of course, this model is in any case invalid based on the fact that clustered Pcdhs form cis-dimers, not tetramers [49, 53].
The dilution model can also be applied to dimeric recognition units which form a trans tetramer between apposed cells. However, cis-dimers do not appear to provide sufficient diversity to account for non-self recognition, and indeed all models based on the existence of discrete oligomeric recognition units encounter difficulties in explaining both self recognition and non-self discrimination [49]. Thus, it appears that the notion of achieving diversity through the formation of discrete Pcdh multimeric recognition units (where each unit plays the same role as a Dscam monomer, (Figure 6B, middle) cannot account for the role of Pcdhs in neuronal barcoding.
In the second model, the “isoform-mismatch chain termination” model (Figure 6B, right), each “arm” of a Pcdh cis-dimeric recognition unit interacts with one “arm” from two different recognition units on the apposed cell surface to form a one-dimensional zipper or lattice-like structure [49]. When identical isoforms are present in both cells, the length of the zipper-like chain is limited only by the copy number of the expressed isoforms. In contrast, cells with even a single mismatch will have the growing chain terminated by the incorporation of an isoform with no match on the apposing cell so that only small Pcdh assemblies will be formed (Figure 7) [49]. Statistical modeling illustrates that this chain-termination model exhibits step-function-like behavior which could yield a binary on/off signal (Figure 7) [49]. Underlying this model is the hypothesis that assembly size plays a crucial role in intracellular signaling, such that large assemblies would transduce an intracellular signal initiating repulsion, while the signal from small assemblies formed in the presence of a mismatch would remain below a critical threshold (Figure 7) [49]. The ‘isoform-mismatch chain termination’ model, in principle, can provide a mechanism for self-avoidance with self/non-self discrimination power higher than that achieved by the 19,008 distinct isoforms of Drosophila Dscam1 [49].
Figure 7.
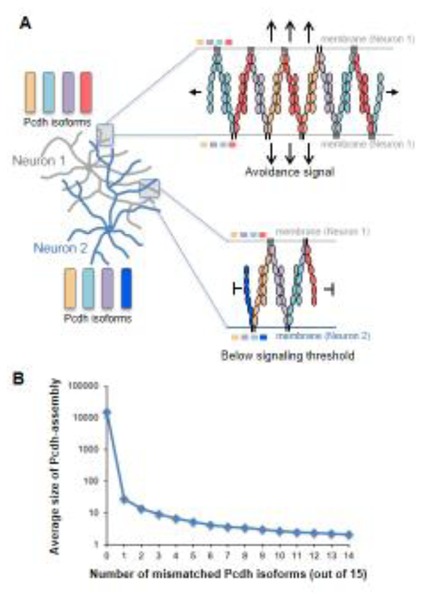
Chain termination model for neuronal recognition based on a Pcdh zipper-assembly. A) Top – For apposed neurites originating from the same neuron shown with four identical isoforms, Pcdh zipper assembly is limited only by Pcdh copy number. Bottom – For apposed neurites from different neurons with single isoform mismatch chain termination occurs quickly, through incorporation into the assembly of the mismatched isoforms. B) Monte-Carlo calculations demonstrate a striking dependence of Pcdh assembly size on the number of mismatched isoforms between interacting cells. Calculations assume 1000 copies per isoform. Note the step-function shape of the curve.
As noted above, studies have found that Pcdh interactions function in neuronal patterning through potentially distinct mechanisms. The neuronal self-avoidance and tiling functions are consistent with Pcdhs initiating repulsion [2–4], while Pcdhs also play other roles such as promoting the complexity of a neuron’s dendritic arbor [44]. We note that although this review focuses on the role of Pcdhs in neuronal self-avoidance in promoting repulsion, the cis and trans interactions formed between Pcdhs that we have described here are also relevant to the adhesive functions of Pcdhs.
8. Conclusion
Comparison of the molecular logic of Dscams and Pcdhs reveals a number of remarkable insights as to how vertebrates and many invertebrates have evolved to solve the problem of neuronal barcoding. Drosophila use alternative splicing to generate diversity that is coded on three independent domains, each of which exhibits homophilic binding specificity. Since each domain presents a separate interface, the 19,008 distinct isoforms simply correspond to the product of the number of alternative exons that can be expressed for each domain. Dscams appear to interact in trans as monomeric recognition units; there is no evidence they form ordered assemblies at interfacial regions. By contrast, vertebrate diversity is based on stochastic promoter choice which does not lend itself to the level of combinatorial diversity that can be generated via alternative splicing. Consequently, vertebrates have had to evolve a very different mechanism for neuronal barcoding.
A combination of structure determination, biophysical measurements, cell aggregation studies and computational analysis has revealed a likely molecular mechanism for Pcdh barcoding. Pcdhs appear on the cell surface as cis dimers that contain two distinct arms, each with its own trans homophilic binding specificity. Two sister neurites from the same neuron will contain the same complement of Pcdh isoforms and hence an identical, or near identical, population of cis dimers. When two sister neurites come into contact the two sets of cis dimers will form anti-parallel trans interactions that produce a linear assembly whose dimensions depend on the total number of expressed Pcdhs. The model assumes that this large assembly then produces a signal for the two cells to move apart, with the assembly itself likely destroyed by proteolysis. In fact, the clustered Pcdhs have been shown to be cleaved by a metalloprotease and γ-secretase during development [45, 63–65]. In contrast, when neurites from different neurons come into contact, there will be a high probability of at least one mismatched isoform which is enough to limit assembly size, and no repulsion signal will be produced. Remarkably, this mechanism produces an essentially binary signal that could be used to distinguish self from non-self (Figure 7).
Of course, much remains to be done to validate and refine the zipper/chain-termination model, and ultimately to test it in neurons. Key steps would determine the structure of the Pcdh cis dimer and visualize the assembly formed by Pcdhs between contacting membranes. Furthermore, Pcdhs have been shown to form cis interactions with other cell surface molecules such as RET, Neuroligin, and ROR2, [66–69] which may add an additional layer of complexity to the Pcdh cell surface assembly and function. However, this model is consistent with all available data and explains how 58 mouse Pcdhs can code for greater diversity than 19,008 fly Dscams. This model explains the function of the stochastically expressed alternate Pcdhs. The C-type Pcdhs, which engage in similar protein interactions to the alternate Pcdhs, appear to be expressed deterministically rather than stochastically, consistent with the possibility that they play distinct functional roles which may not involve the chain termination mechanism. For example, recent studies have shown that unique interaction of PcdhγC3 with Axin1 inhibits Wnt signaling [70]. In general, a more complex picture of Pcdh gene expression is emerging, in which distinct Pcdh gene clusters are expressed in different types of neurons, and the alternate and C-type isoforms are differentially expressed during the development of neural circuits [4, 16, 60].
A better understanding of the logic of Pcdh barcoding will require further study. However, the mechanistic insights obtained in past few years illustrate the importance of a greater integration of structural biology, molecular biophysics and neurobiology, as molecular structure can suggest novel mechanisms that would be hard to otherwise imagine.
Acknowledgments
This work was supported by a National Science Foundation grant to B.H. (MCB-1412472), an NIH grant to L.S. (R01GM062270), and a joint NIH grant to T.M. and L.S. (R01GM107571). The computing in this project was supported by two NIH instrumentation grants (S10OD012351 and S10OD021764) received by the Department of Systems Biology at Columbia University.
Abbreviations
- Pcdhs
Clustered Protocadherins
- EC
Extracellular Cadherin Domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–41. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–21. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostadinov D, Sanes JR. Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. Elife. 2015:4. doi: 10.7554/eLife.08964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mountoufaris G, Chen WV, Hirabayashi Y, O’Keeffe S, Chevee M, Nwakeze CL, et al. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science. 2017;356:411–4. doi: 10.1126/science.aai8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, et al. Functional Significance of Isoform Diversification in the Protocadherin Gamma Gene Cluster. Neuron. 2012;75:402–9. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–51. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad T, Weiner JA. Direct and indirect regulation of spinal cord la afferent terminal formation by the gamma-Protocadherins. Frontiers in Molecular Neuroscience. 2011:4. doi: 10.3389/fnmol.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasad T, Wang X, Gray PA, Weiner JA. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development. 2008;135:4153–64. doi: 10.1242/dev.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin C, Meng S, Zhu T, Wang X. PDCD10/CCM3 acts downstream of {gamma}-protocadherins to regulate neuronal survival. J Biol Chem. 2010;285:41675–85. doi: 10.1074/jbc.M110.179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett AM, Weiner JA. Control of CNS synapse development by {gamma}-protocadherin-mediated astrocyte-neuron contact. J Neurosci. 2009;29:11723–31. doi: 10.1523/JNEUROSCI.2818-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner JA, Wang XZ, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa S, Hamada S, Kumode Y, Esumi S, Katori S, Fukuda E, et al. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci. 2008;38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa S, Hirabayashi T, Kondo T, Inoue K, Esumi S, Okayama A, et al. Constitutively expressed Protocadherin-alpha regulates the coalescence and elimination of homotypic olfactory axons through its cytoplasmic region. Front Mol Neurosci. 2012;5:97. doi: 10.3389/fnmol.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, et al. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. 2008;28:1362–76. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36:843–54. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 16.Chen WV, Nwakeze CL, Denny CA, O’Keeffe S, Rieger MA, Mountoufaris G, et al. Pcdhalphac2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science. 2017;356:406–11. doi: 10.1126/science.aal3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa S, Kumagai M, Hagihara M, Nishimaru H, Hirano K, Kaneko R, et al. Distinct and Cooperative Functions for the Protocadherin-α,-β and-γ Clusters in Neuronal Survival and Axon Targeting. Frontiers in Molecular Neuroscience. 2016;9:155. doi: 10.3389/fnmol.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–51. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–90. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 20.Yu WP, Rajasegaran V, Yew K, Loh WL, Tay BH, Amemiya CT, et al. Elephant shark sequence reveals unique insights into the evolutionary history of vertebrate genes: A comparative analysis of the protocadherin cluster. Proc Natl Acad Sci U S A. 2008;105:3819–24. doi: 10.1073/pnas.0800398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu WP, Yew K, Rajasegaran V, Venkatesh B. Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts. BMC Evol Biol. 2007;7:49. doi: 10.1186/1471-2148-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nature Genetics. 2005;37:171–6. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 23.Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, Yagi T. Single-neuron diversity generated by Protocadherin-beta cluster in mouse central and peripheral nervous systems. Frontiers in Molecular Neuroscience. 2012:5. doi: 10.3389/fnmol.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, et al. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. Journal of Biological Chemistry. 2006;281:30551–60. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 25.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, et al. Promoter choice determines splice site selection in protocadherin alpha and -gamma pre-mRNA splicing. Molecular Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 26.Wang XZ, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: Evidence for a multiple promoter and cis-alternative splicing model. Genes & Development. 2002;16:1890–905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zipursky SL, Grueber WB. The Molecular Basis of Self-Avoidance. In: Hyman SE, editor. Annual Review of Neuroscience. Vol. 36. 2013. pp. 547–68. [DOI] [PubMed] [Google Scholar]
- 28.Zipursky SL, Sanes JR. Chemoaffinity Revisited: Dscams, Protocadherins, and Neural Circuit Assembly. Cell. 2010;143:343–53. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Hattori D, Chen Y, Matthews BJ, Salwinski L, Sabatti C, Grueber WB, et al. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461:644–U87. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-Mediated Cell Recognition Regulates Neural Circuit Formation. Annual Review of Cell and Developmental Biology. 2008:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genetics. 2004;36:240–6. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 32.Sun W, You X, Gogol-Doring A, He H, Kise Y, Sohn M, et al. Ultra-deep profiling of alternatively spliced Drosophila Dscam isoforms by circularization-assisted multi-segment sequencing. EMBO J. 2013;32:2029–38. doi: 10.1038/emboj.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–86. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic Splicing of Dscam1 Establishes Identity at the Level of Single Neurons. Cell. 2013;155:1166–77. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–33. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding Specificities arises from modular interactions of variable ig domains. Cell. 2007;130:1134–45. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, et al. Single-Cell Identity Generated by Combinatorial Homophilic Interactions between alpha, beta, and gamma Protocadherins. Cell. 2014;158:1045–59. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14893–8. doi: 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–84. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 40.Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–69. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Monreal M, Kang S, Phillips GR. Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mol Cell Neurosci. 2009;40:344–53. doi: 10.1016/j.mcn.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, et al. Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci. 2005;29:603–16. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Molumby MJ, Keeler AB, Weiner JA. Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity. Cell Reports. 2016;15:1037–50. doi: 10.1016/j.celrep.2016.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonn S, Seeburg PH, Schwarz MK. Combinatorial expression of alpha- and gamma-protocadherins alters their presenilin-dependent processing. Mol Cell Biol. 2007;27:4121–32. doi: 10.1128/MCB.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murata Y, Hamada S, Morishita H, Mutoh T, Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J Biol Chem. 2004;279:49508–16. doi: 10.1074/jbc.M408771200. [DOI] [PubMed] [Google Scholar]
- 47.Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, et al. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. Journal of Biological Chemistry. 2006;281:33650–63. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- 48.Nicoludis JM, Lau S-Y, Schaerfe CPI, Marks DS, Weihofen WA, Gaudet R. Structure and Sequence Analyses of Clustered Protocadherins Reveal Antiparallel Interactions that Mediate Homophilic Specificity. Structure. 2015;23:2087–98. doi: 10.1016/j.str.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, et al. Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions. Cell. 2015;163:629–42. doi: 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7160–5. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–13. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 52.Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsen G, et al. Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins. Neuron. 2016;90:709–23. doi: 10.1016/j.neuron.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodman KM, Rubinstein R, Thu CA, Mannepalli S, Bahna F, Ahlsen G, et al. γ-Protocadherin structural diversity and functional implications. eLife. 2016;5:e20930. doi: 10.7554/eLife.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicoludis JM, Vogt BE, Green AG, Scharfe CPI, Marks DS, Gaudet R. Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1–4. eLife. 2016:5. doi: 10.7554/eLife.18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciatto C, Bahna F, Zampieri N, VanSteenhouse HC, Katsamba PS, Ahlsen G, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nature Structural & Molecular Biology. 2010;17:339–U110. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison OJ, Bahna F, Katsamba PS, Jin X, Brasch J, Vendome J, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–57. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature. 2012;492:128. doi: 10.1038/nature11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper SR, Jontes JD, Sotomayor M. Structural determinants of adhesion by Protocadherin-19 and implications for its role in epilepsy. Elife. 2016:5. doi: 10.7554/eLife.18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Front Mol Neurosci. 2012;5:45. doi: 10.3389/fnmol.2012.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–U107. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, Honig B, Ben-Shaul A. Theory and Simulations of Adhesion Receptor Dimerization on Membrane Surfaces. Biophysical Journal. 2013;104:1221–9. doi: 10.1016/j.bpj.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchanan SM, Schalm SS, Maniatis T. Proteolytic processing of protocadherin proteins requires endocytosis. Proc Natl Acad Sci U S A. 2010;107:17774–9. doi: 10.1073/pnas.1013105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haas IG, Frank M, Veron N, Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. J Biol Chem. 2005;280:9313–9. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- 65.Hambsch B, Grinevich V, Seeburg PH, Schwarz MK. {gamma}-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J Biol Chem. 2005;280:15888–97. doi: 10.1074/jbc.M414359200. [DOI] [PubMed] [Google Scholar]
- 66.Han M-H, Lin C, Meng S, Wang X. Proteomics Analysis Reveals Overlapping Functions of Clustered Protocadherins. Molecular & Cellular Proteomics. 2010;9:71–83. doi: 10.1074/mcp.M900343-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molumby MJ, Anderson RM, Newbold DJ, Koblesky NK, Garrett AM, Schreiner D, et al. gamma-Protocadherins Interact with Neuroligin-1 and Negatively Regulate Dendritic Spine Morphogenesis. Cell Rep. 2017;18:2702–14. doi: 10.1016/j.celrep.2017.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schalm SS, Ballif BA, Buchanan SM, Phillips GR, Maniatis T. Phosphorylation of protocadherin proteins by the receptor tyrosine kinase Ret. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13894–9. doi: 10.1073/pnas.1007182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–60. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mah KM, Houston DW, Weiner JA. The gamma-Protocadherin-C3 isoform inhibits canonical Wnt signalling by binding to and stabilizing Axin1 at the membrane. Scientific Reports. 2016:6. doi: 10.1038/srep31665. [DOI] [PMC free article] [PubMed] [Google Scholar]