ABSTRACT
As a species, Streptococcus pneumoniae (the pneumococcus) utilizes a diverse array of capsular polysaccharides to evade the host. In contrast to large variations in sugar composition and linkage formation, O-acetylation is a subtle capsular modification that nonetheless has a large impact on capsular shielding and recognition of the capsule by vaccine-elicited antibodies. Serotype 15B, which is included in the 23-valent pneumococcal polysaccharide vaccine (PPV23), carries the putative O-acetyltransferase gene wciZ. The coding sequence of wciZ contains eight consecutive TA repeats [(TA)8]. Replication slippage is thought to result in the addition or loss of TA repeats, subsequently causing frameshift and truncation of WciZ to yield a nonacetylated serotype, 15C. Using sensitive serological tools, we show that serotype 15C isolates whose wciZ contains seven or nine TA repeats retain partial O-acetylation, while serotype 15C isolates whose wciZ contains six TA repeats have barely detectable O-acetylation. We confirmed by inhibition enzyme-linked immunosorbent assay that (TA)7 serotype 15C is ∼0.1% as acetylated as serotype 15B, while serotype 15X is nonacetylated. To eliminate the impact of genetic background, we created isogenic serotype 15B, (TA)7 serotype 15C, and 15BΔwciZ (15X) strains and found that reduction or absence of WciZ-mediated O-acetylation did not affect capsular shielding from phagocytes, biofilm formation, adhesion to nasopharyngeal cells, desiccation tolerance, or murine colonization. Sera from PPV23-immunized persons opsonized serotype 15B significantly but only slightly better than serotypes 15C and 15X; thus, PPV23 may not result in expansion of serotype 15C.
KEYWORDS: O-acetyltransferase, O-acetylation, capsule diversity, pneumococcal vaccine, capsular polysaccharide, serotyping, replication slippage
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus), a Gram-positive human pathogen, mediates pneumonia and invasive diseases such as septicemia and meningitis and colonizes 6 to 76% of the world's pediatric population (1–3). Pneumococcal diseases are the leading cause of death in children under 5 years old (4), and pneumococcal capsular polysaccharide is the organism's most significant virulence factor as it shields the pneumococcus from various chemical and immune assaults (5). As a result, nonencapsulated pneumococci rarely cause pneumonia or invasive pneumococcal disease (6–8). Pneumococcal capsular types differ in their shielding abilities; therefore, some capsule types are more virulent than others (9, 10). Our previous studies have indicated that small chemical modifications of the capsular repeat unit result in differential shielding of closely related serotypes (11, 12).
An important capsular modification is O-acetylation: an uncharged, but polar, acetyl functional group that exerts a broad effect on polysaccharides. In many bacteria, O-acetylation alters the physicochemical properties of capsule by changing the conformation of capsular repeat units (13) or increasing viscosity (14). O-acetylation also alters the host-pathogen interaction by creating immunogenic epitopes (11, 13, 15), by neutralizing reactive chlorine species (16, 17), and/or by mediating resistance to lysozyme and complement deposition (18–21). Many bacteria have membrane-bound O-acetyltransferases (MOATs) that are thought to attach O-acetyl groups to the capsular repeat unit at the end of the capsular biosynthetic process (22–24). Accordingly, the loss of MOAT-mediated O-acetylation may not disrupt global capsule biosynthesis (12, 25–27).
Serotypes 15B and 15C contain the MOAT-encoding gene wciZ within their capsular polysaccharide synthesis loci (cps loci). WciZ is functional in serotype 15B but is nonfunctional in serotype 15C, and this functionality is based on a tract of TA repeats within wciZ which permits replication slippage at a low frequency (28). The serotype 15B wciZ contains eight TA repeats, resulting in an enzymatically active WciZ with 326 amino acids and 10 membrane domains. In contrast, the serotype 15C wciZ contains more or fewer TA repeats [(TA)6, (TA)7, or (TA)9], resulting in frameshifts that lead to a nonfunctional WciZ with ∼150 amino acids and only four transmembrane domains (29). Consistent with these genetic findings, chemical studies concluded that serotype 15C polysaccharide is nonacetylated (29, 30). In addition, it was reported using a few immune sera that vaccination with serotype 15B polysaccharide does not elicit antibodies opsonizing serotype 15C (31). Despite this, our in-house serotype 15B-specific monoclonal antibody (Hyp15BG5) was partially reactive with serotype 15C polysaccharide (32), suggesting that serotype 15C capsule is partially O-acetylated and, therefore, that the truncated serotype 15C WciZ retains some activity. As serotypes 15B and 15C have increased in prevalence following the clinical use of pneumococcal conjugate vaccines (33) and since serotype 15B polysaccharide may be included in many future vaccines (34), cross-protection against serotype 15C should be clearly determined. Therefore, we have investigated the basis for the partial functionality of the serotype 15C WciZ and cross-opsonization of 23-valent pneumococcal polysaccharide vaccine (PPV23)-elicited antibodies against serotype 15C.
RESULTS
Hyp15BG5 and factor serum 15b (fs15b) recognize WciZ-mediated acetyl groups.
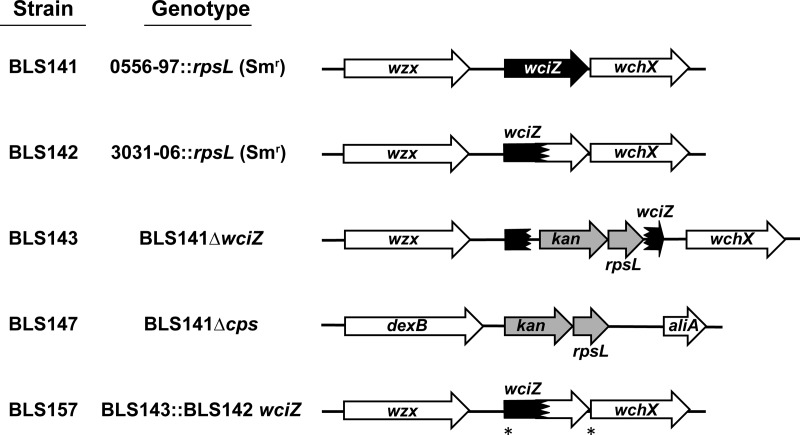
Parental strains were created using clinical isolates representing serotypes 15B (isolate 0556-97) and 15C (isolate 3031-06) (Table 1 and Fig. 1). Both clinical isolates belong to sequence type 199 (a genotype commonly associated with serotypes 15B and 15C [35–39]) and contain mixed populations of transparent and opaque colonies (see Table S1 in the supplemental material). BLS141 and BLS142 were produced by inserting rpsL (Smr), a gene conferring streptomycin (Sm) resistance, into the background of strains 0556-97 and 3031-06, respectively. Additional clinical isolates were used in initial experiments to confirm serological differences between serotypes 15B and 15C. To study the impact of WciZ-mediated O-acetylation on the biological properties of serotype 15B and 15C capsule, isogenic variants were created in the BLS141 genomic background. BLS143 (designated serotype 15X) was created by irreversibly disrupting wciZ of BLS141 with a Janus cassette (JS), and BLS147 (isogenic nonencapsulated strain) was created by deleting the BLS141 cps locus with JS. BLS157, an isogenic serotype 15C strain, was created by transformation of the wciZ from serotype 15C strain BLS142 into the wciZ-null strain BLS143.
TABLE 1.
Strains used in this study
| Strain | wciZ (TA)n | Serotype | Descriptionb | WciZ O-acetylationc | GenBank accession no. | Source |
|---|---|---|---|---|---|---|
| BLS141 | 8 | 15B | 0556-97::rpsL (Smr) | ++ | KY750633 | This study |
| BLS142 | 7 | 15C | 3031-06::rpsL (Smr) | + | KY750634 | This study |
| BLS143 | NAa | 15X | BLS141ΔwciZ | − | KY750635 | This study |
| BLS147 | NA | Nonencapsulated | BLS141Δcps | − | KY750636 | This study |
| BLS157 | 7 | 15C | BLS143::BLS142 wciZ | + | KY750637 | This study |
| BLS171 | 6 | 15C | BLS143::(TA)6 wciZ | +/− | KY750638 | This study |
| 0556-97 | 8 | 15B | Clinical isolate | ++ | KY750640 | Atlanta, GA |
| MHI-224 | 8 | 15B | Clinical isolate | ++ | KY750641 | Wakayama, Japan |
| MNZ161 | 8 | 15B | Clinical isolate | ++ | KY750642 | Pittsburgh, PA |
| SSISP15B/3 | 8 | 15B | Clinical isolate | ++ | KY750643 | Copenhagen, Denmark |
| 3031-06 | 7 | 15C | Clinical isolate | + | KY750644 | Atlanta, GA |
| 3933-06 | 7 | 15C | Clinical isolate | + | KY750645 | Atlanta, GA |
| MNK1031 | 9 | 15C | Clinical isolate | + | KY750646 | Seoul, South Korea |
| SSISP15C/2 | 9 | 15C | Clinical isolate | + | KY750647 | Copenhagen, Denmark |
| ST1058/03 | 6 | 15C | Clinical isolate | +/− | KY750648 | São Paulo, Brazil |
NA, not applicable.
Smr, streptomycin resistance.
Based on mean fluorescence intensity (MFI) of Hyp15BG5 binding in flow cytometry. ++, binding of >1,200 MFI; +, binding of 200 to 1,200 MFI; +/−, binding of 45 to 200 MFI; −, binding of <45 MFI.
FIG 1.
Genetic summary of serotype 15B and serotype 15C variants. BLS141 and BLS142 were created by inserting rpsL (Smr) into the background of clinical isolate 0556-97 (serotype 15B) and 3031-06 (serotype 15C), respectively. The white region of wciZ arrowheads in BLS142 and BLS157 indicate the region of wciZ after the premature stop. The asterisks in BLS157 denote the wciZ region amplified from BLS142 which was transformed into BLS143 (as described in Materials and Methods). Gene fragments or prematurely truncated genes are denoted by arrows with jagged edges.
To investigate the epitopes targeted by the serotype 15B-specific monoclonal antibody, Hyp15BG5, we utilized an inhibition enzyme-linked immunosorbent assay (ELISA) with serotype 15B polysaccharide-coated ELISA plates. As depicted in Fig. 2A, two serotype 15B lysates (BLS141 and SSISP15B) inhibited Hyp15BG5 binding at approximately 1,000-fold greater dilution than serotype 15C lysates (BLS142 and SSISP15C). This finding strongly suggested that the antibodies are targeting the O-acetyl group but that serotype 15C capsular polysaccharide has a smaller amount of O-acetylation. To test this possibility, we removed the O-acetyl groups from lysates of serotype 15B and 15C strains by mild alkaline hydrolysis and retested them for inhibition. Neither the hydrolyzed lysates (dotted lines) nor the lysate of a serotype 15X strain (solid inverted triangle), which has an irreversibly disrupted wciZ gene, inhibited Hyp15BG5 (Fig. 2A). The same observations were made with factor serum 15b (fs15b) (Fig. 2B). These experiments suggest that Hyp15BG5 and fs15b are specific for WciZ-mediated O-acetylation and that serotype 15C strains produce partially O-acetylated capsular polysaccharide. Using additional clinical isolates (Table 1), we confirmed the partial O-acetylation of serotype 15C isolates compared to serotype 15B isolates (Fig. 2C). However, one serotype 15C isolate, ST1058/03, showed approximately 200-fold less inhibition than the other serotype 15C strains (shown with an arrow in Fig. 2C). Therefore, the degree of partial O-acetylation varies among serotype 15C strains.
FIG 2.
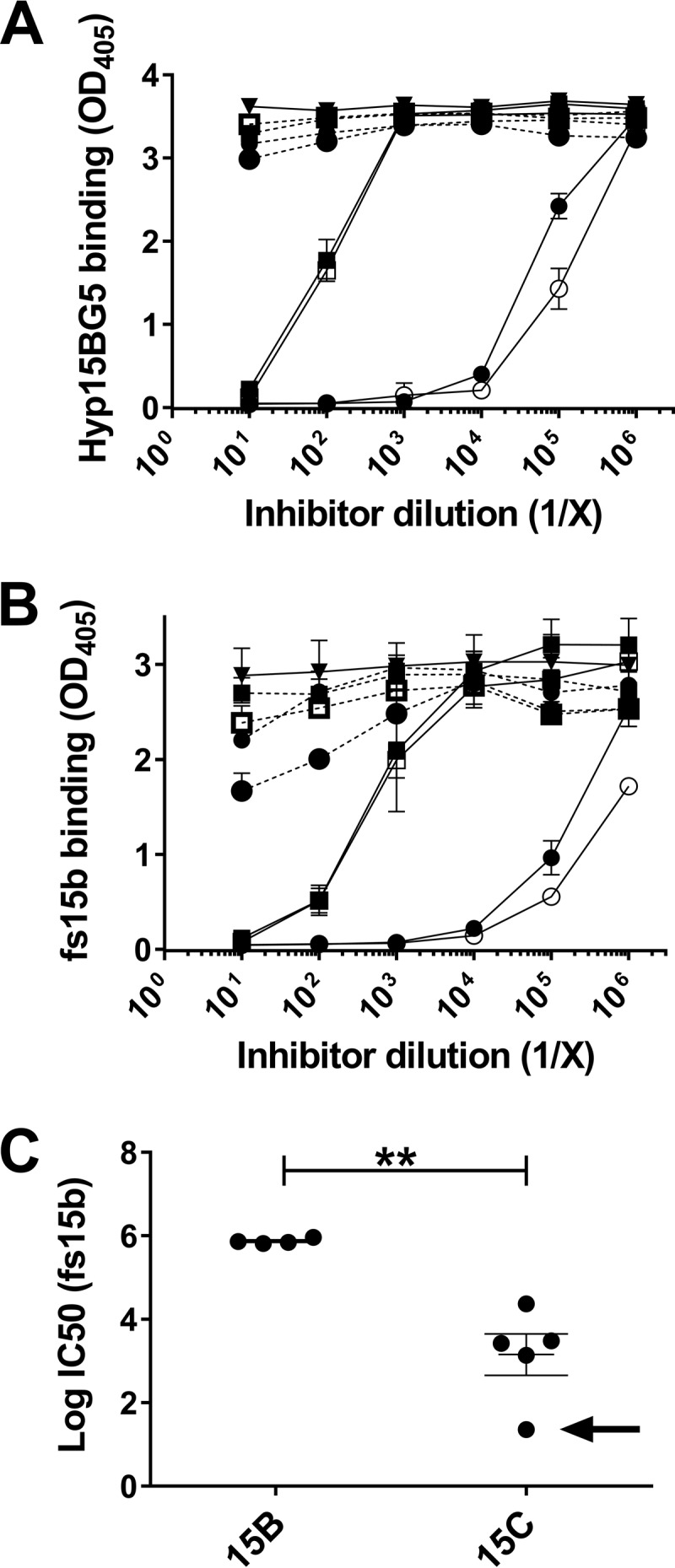
Inhibition ELISA showing the specificity of Hyp15BG5 and fs15b for WciZ-mediated O-acetylation. (A and B) Inhibition of Hyp15BG5 (A) or fs15b (B) binding to serotype 15B polysaccharide-coated plates by reference serotype 15B and serotype 15C strain lysates and control lysates. The solid circle indicates inhibition by SSISP15B/3 lysate, and the open circle indicates inhibition by BLS141 lysate (serotype 15B). The solid square indicates inhibition by SSISP15C/2 lysate, and the open square indicates inhibition by BLS142 lysate (serotype 15C). The solid inverted triangle indicates inhibition by serotype 15X lysate. Data depict one experiment performed in triplicate, and results are representative of three independent experiments. Error bars indicate standard deviations from the three intra-assay replicates. Dashed lines indicate alkaline-hydrolyzed lysates. (C) The IC50 (the concentration at which lysates inhibited fs15b binding by 50%) was calculated for the reference serotype 15B and 15C strains used in panels A and B, for two additional serotype 15B clinical isolates, and three additional serotype 15C clinical isolates. Data points represent the averages from two independent experiments, each performed in triplicate, and data are represented as log(IC50). The average log(IC50) of serotype 15B strains was significantly higher than that of serotype 15C strains (**, P < 0.01 by unpaired t test). The black arrow indicates ST1058/03 lysate.
Phenotypes of O-acetylation vary among serotype 15C isolates using flow cytometry.
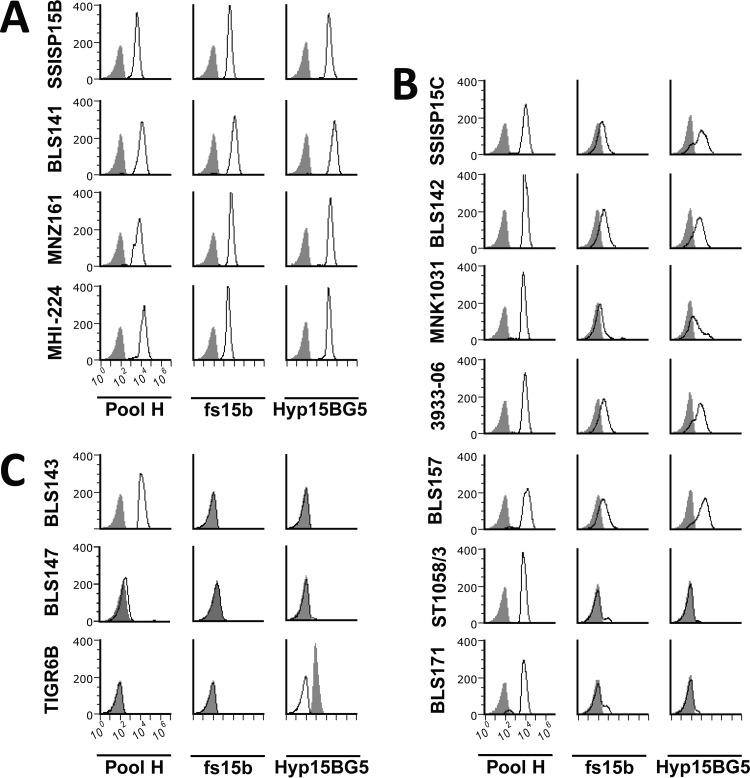
Having shown that both fs15b and Hyp15BG5 target WciZ-mediated O-acetylation, we investigated serotype 15C strain heterogeneity at the single-cell level using flow cytometry. All serotype 15B clinical isolates examined reacted with pool H serum (containing antibodies recognizing all serogroup 15 capsule), fs15b, and Hyp15BG5 (Fig. 3A). Pool H did not bind to unrelated serotype 6B (TIGR6B) or to nonencapsulated BLS147 but did bind to serotype 15B, 15C, and 15X (BLS143) strains equally well. Therefore, based on mean fluorescent intensity, these strains appear to express equivalent amounts of capsule. However, serotype 15X did not react with fs15b or Hyp15BG5 at all, similar to the nonencapsulated strain or TIGR6B (Fig. 3C). Thus, our flow cytometry assay can clearly distinguish serotype 15B strains expressing heavily acetylated capsule from a serotype 15X strain expressing capsule with no O-acetylation.
FIG 3.
Serotype 15B and 15C strains have serologically distinguishable levels of WciZ-mediated O-acetylation by flow cytometry. Histograms depict binding of antibodies (listed in the bottom row) to pneumococcal strains (listed at the left of each row). Binding to serotype 15B clinical isolates (A), binding to serotype 15C clinical isolates (B), and binding to negative-control strains (C) is shown. The gray shaded peaks represent negative controls obtained with normal rabbit serum or monoclonal antibody, Hyp6BG9, which binds to strain TIGR6B. All rabbit sera were adsorbed against nonencapsulated TIGRJS prior to serological characterization of these isolates. Pool H serum contains antibodies which bind to serogroup 15 capsules. fs15b and Hyp15BG5 specifically bind WciZ-mediated O-acetylation, as shown in Fig. 2.
When serotype 15C isolates were examined, four isolates (SSISP15C, BLS142, MNK1031, and 3393-06) were bound weakly by fs15b and Hyp15BG5 (Fig. 2C). However, ST1058/3, the strain with the lowest level of O-acetylation in the inhibition ELISA (Fig. 2C), again showed almost no binding by either probe. Thus, the heterogeneity of O-acetylation among serotype 15C isolates observed by ELISA was confirmed by flow cytometry.
Genetic basis for partial serotype 15C WciZ activity.
To investigate the genetic basis for the observed variability in O-acetylation among serotype 15C strains, we sequenced the wciZ genes of the serotype 15B and 15C isolates (Table 1). All serotype 15B strains had eight TA repeats [(TA)8], as expected. The serotype 15C isolates demonstrating low levels of O-acetylation had seven or nine repeat units [(TA)7 or (TA)9]. Upon replacement of the serotype 15B wciZ (in BLS141) with the serotype 15C (TA)7 wciZ (from strain BLS142) to create strain BLS157, the partial O-acetylation phenotype was observed (Fig. 3B). Thus, the partial O-acetylation is conferred by the (TA)7 repeat within wciZ.
ST1058/03, the serotype 15C isolate with minimal reactivity to fs15b and Hyp15BG5, had a wciZ gene with only six TA repeat units [(TA)6] but also had two missense mutations early in the gene (resulting in changes of amino acids 7 and 87). To demonstrate that the two missense mutations were not responsible for the minimal O-acetylation phenotype, a (TA)6 strain was created in the BLS141 background. The new strain (named BLS171) also minimally reacted with fs15b or Hyp15BG5 (Fig. 3B), suggesting that the decrease in O-acetylation was conferred by the (TA)6 repeat, not by the missense mutations. Taken together, serotype 15C strains with (TA)7 or (TA)9 wciZ can O-acetylate the capsule partially, but serotype 15C strains with (TA)6 wciZ can O-acetylate the capsule at barely detectable levels.
Biological consequences of WciZ-mediated O-acetylation.
Having identified the genetic basis of heterogeneity among serotype 15C isolates, we investigated the impact of O-acetylation on carriage and spreading by studying the ability of serotype 15B, 15C [(TA)7], and 15X capsule to shield their cell walls from antibody, form biofilms, adhere to nasopharyngeal cells, survive short-term drying, and colonize mice. These parameters are indicators of the serotypes' fitness to colonize the nasopharynx and to spread among different individuals (40–44). Only isogenic strains were used in these biological assays, and they are referred to as 15B (BLS141), 15C [BLS157, (TA)7], 15X (BLS143), and the nonencapsulated strain (BLS147).
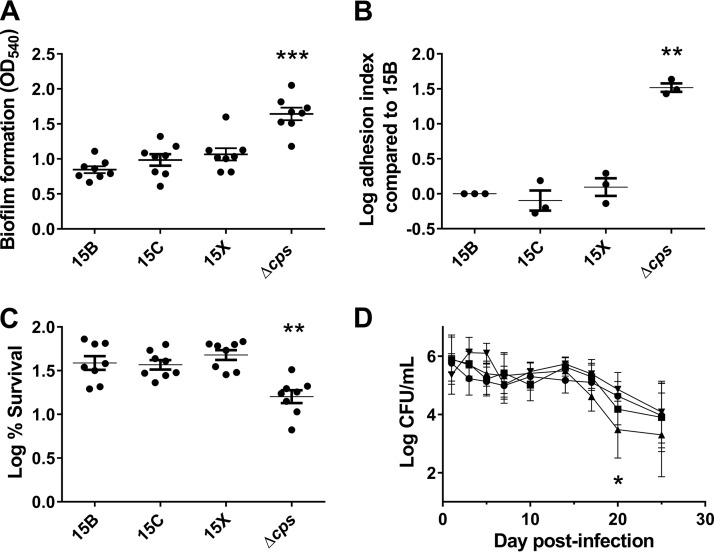
The capsular shielding abilities of 15B, 15C, and 15X were equivalent, as indicated by the ability of anti-phosphocholine antibodies to access the cell wall (data not shown). 15B, 15C, and 15X formed equivalent amounts of biofilm on polystyrene surfaces and adhered equally to nasopharyngeal cells, but all formed less biofilm and adhered to nasopharyngeal cells less effectively than the nonencapsulated strain (Fig. 4A and B). When survival after a short period of drying was assessed, 15B, 15C, and 15X survived better than the nonencapsulated strain (Fig. 4C). Finally, 15B, 15C, 15X, and the nonencapsulated strain colonized the murine nasopharynx equally well (Fig. 4D). No mice became ill, became septic, or died during the 25-day experiment. Taking these findings together, WciZ-mediated O-acetylation does not seem to influence the biological properties of serotype 15B or 15C capsule.
FIG 4.
Biological properties of pneumococcal serotypes 15B, 15C, and 15X. (A) Biofilm formation (OD540) by various pneumococcal serotypes on polystyrene surfaces. Each dot represents one experiment, and eight independent experiments were performed. Repeated-measures one-way ANOVA statistical analysis found a significant difference between strains (P < 0.001), and Dunnett's multiple-comparison test indicated a difference in the Δcps strain compared to 15B (P < 0.001). (B) Log adhesion index [log(% adhesion of a given serotype/% adhesion of 15B)] of serotype 15B variant strains. Three independent experiments were performed, each in triplicate. Repeated-measures one-way ANOVA statistical analysis of log adhesion index found a significant difference between strains (P < 0.01), and Dunnett's multiple-comparison test indicated a difference in the Δcps strain compared to 15B (P < 0.01). (C) Log percent bacterial survival after drying [log(% survival of 15B variants)] for indicated strains. One hundred percent survival equals a value of 2 on the y axis. Eight independent experiments were performed. Repeated-measures one-way ANOVA statistical analysis of log-transformed percentages found a significant difference between strains (P < 0.0001), and Dunnett's multiple-comparison test indicated a difference in the Δcps strain compared to 15B (P < 0.01). D) Nasopharyngeal carriage of 15B variants. Female 6-week-old BALB/cJ mice were intranasally inoculated with 107 CFU of 15B (•; 10 mice per group), 15C (■; nine mice per group), 15X (▲; nine mice per group), or 15BΔcps (▼; nine mice per group) bacteria. NALF was collected, and CFU were quantified at 1, 3, 5, 7, 10, 14, 17, 20, and 25 days postinoculation. Two-way ANOVA found significant differences between days (P < 0.0001) and between bacterial strains (P < 0.05). Dunnett's multiple-comparison test indicated a difference in CFU counts in NALF of mice infected with 15X compared to those infected with 15B on day 20 postinfection (P < 0.05). All data points shown in panels A to D represent means with standard errors. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Impact of reduced O-acetylation on opsonophagocytic killing.
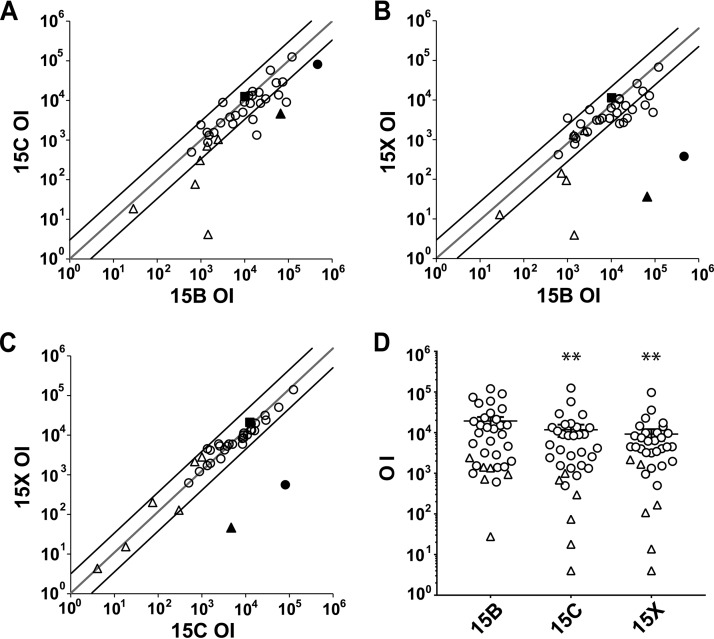
Previous studies have shown that serotype 15B-specific antibodies induced by PPV23 predominantly target WciZ-mediated O-acetylation (31); therefore, we tested opsonophagocytic killing of serotype 15B (BLS141), 15C [BLS157, (TA)7], and 15X (BLS143) target strains (Fig. 5). Opsonization indices (OIs) were calculated for each serum sample or antibody against each target strain and are defined as the serum dilution at which 50% of the target bacteria are killed. When opsonization assays were performed with antibodies that specifically target the WciZ-mediated O-acetyl group (Hyp15BG5 and fs15b), serotype 15X was not opsonized (OIs below the limit of detection) and serotype 15C was weakly opsonized (OIs for serotype 15C were 5- to 15-fold less than OIs for serotype 15B). This again suggests that the WciZ-mediated acetyl group, expressed at a lower density on serotype 15C isolates, is the target of the opsonic antibodies. Using a human serum pool (007sp) from PPV23-vaccinated adults, the OI for serotype 15B was not significantly different from the OIs for serotypes 15C and 15X (P = 0.45 by repeated-measures one-way analysis of variance [ANOVA] with Dunnett's multiple comparisons) (Fig. 5A to C, square symbol). Further, when OIs were obtained with 28 individual sera from adults immunized with PPV23 (open circles) and six individual sera from unvaccinated adults (open triangles), titers against serotype 15B and 15C strains were correlated (r = 0.77) (Fig. 5A), as were titers against serotype 15B and 15X strains (r = 0.75) (Fig. 5B). OIs were significantly higher for serotype 15B than for serotypes 15C and 15X (1.6-fold [P < 0.01] and 2.1-fold [P < 0.01], respectively) (Fig. 5D). OIs for serotypes 15C and 15X were highly correlated (r = 0.99) (Fig. 5C), and OIs for serotype 15C were slightly higher than those for serotype 15X (1.3-fold; not significant) (Fig. 5D). Thus, the degree of WciZ-mediated O-acetylation has a small but measurable effect on the opsonic titers induced with PPV23.
FIG 5.
Pairwise correlations (A to C) and comparisons (D) of opsonic indices (OIs) against serotypes 15B (BLS141), 15C [BLS157, (TA)7], and 15X (BLS143). (A to C) Target serotypes are indicated on each axis. Each open circle indicates a human serum sample from PPV23-vaccinated individuals (n = 28), and each open triangle indicates an unvaccinated human serum sample. The solid triangle indicates Hyp15BG5, the solid square indicates 007sp, and the solid circle indicates fs15b. Hyp15BG5 was 20-fold prediluted and fs15b was 225-fold prediluted; thus, they have different minimum detection thresholds. No opsonic activity was detected against serotype 15X using Hyp15BG5 or fs15b, and those data points are assigned as half the minimum detectable titer. Therefore, the minimum detectable titer for fs15b is high because the predilution was high. The 45° gray line indicates the line of identity, and the 45° black lines indicate 3-fold deviations from the identity. Open circles and open triangles are mostly below the line of identity shown in panels A and B, suggesting that both PPV23-vaccinated and unvaccinated individuals have generally higher OIs for serotype 15B than serotype 15C or 15X. (D) Comparison of OIs between serotype 15B, 15C, and 15X target strains. Each open circle represents one of the 28 sera from PPV23-vaccinated individuals, and each open triangle represents one of the six sera from unvaccinated individuals. Repeated-measures one-way ANOVA of log-transformed OIs found significant differences between strains (P = 0.0011), and Tukey's multiple-comparison test indicated a difference between serotypes 15B and 15C (P < 0.01) and between serotypes 15B and 15X (P < 0.01) but not between serotypes 15C and 15X (P = 0.094). Error bars represent standard errors. (A to D) Each point indicates the average from two independent experiments with one serum or antibody, performed in duplicate. **, P < 0.01.
DISCUSSION
In the past, serotype 15C capsule was thought to be nonacetylated based on chemical and serological studies that are insensitive to small degrees of O-acetylation (29, 30). Here, using antibodies targeting the O-acetyl group, we show that serotype 15C isolates express small and variable amounts of O-acetylation. We also demonstrate that the number of TA repeats within wciZ affects the amount of capsular O-acetylation. wciZ genes containing ±1 TA repeat relative to the serotype 15B sequence [(TA)7 or (TA)9] result in low (~0.1%) but detectable capsular O-acetylation by inhibition ELISA, while wciZ genes containing two fewer TA repeats [(TA)6] result in almost undetectable O-acetylation.
Based on the geometric mean of fluorescence intensities of Hyp15BG5 and fs15b binding (normalized to pool H binding) by flow cytometry, O-acetylation of the serotype 15C capsule was approximately 2% relative to serotype 15B, 20-fold higher than the value calculated by inhibition ELISA. The difference may reflect confounding factors in these assays. While flow cytometry is more biologically relevant because binding is detected on intact cells and may predict opsonophagocytosis, the mean fluorescence intensity (MFI) may be affected by confounding factors, such as pneumococcal chain length. Since O-acetylation is relatively unstable, it is also possible that the O-acetyl groups were reduced during preparation of lysates for the inhibition ELISA. However, the inhibition ELISA produces reliable results; therefore, we have reported the percent O-acetylation of serotype 15C capsule based on the ELISA data. Despite the apparent discrepancy in percentages of serotype 15C O-acetylation amount by inhibition ELISA and flow cytometry, the trend of decreased but detectable O-acetylation in serotype 15C capsule is confirmed in both immunoassays.
Although O-acetylation has been shown to alter physical and biological properties of polysaccharide, O-acetylation of serotype 15B and 15C capsule had little effect on biofilm formation, adhesion to tumor necrosis factor (TNF)-treated nasopharyngeal cells, survival after drying, and nasopharyngeal carriage in mice. Some have reported that cell activation by TNF can mask the nasopharyngeal cell adhesion differences observed between bacterial strains under resting conditions (45); therefore, slight differences may be observed between serotypes 15B, 15C, and 15X if adhesion were tested under resting conditions. Taken together, however, these data suggest that O-acetylation has little effect on the biological properties of serotype 15B and 15C capsule. These data reinforce our recent observation that the effects of O-acetylation are polysaccharide context dependent (12), and these data are consistent with epidemiological studies, which found that both serotypes 15B and 15C are commonly found in the nasopharynx (46).
To our surprise, we observed that the nonencapsulated and encapsulated strains colonized mice equally well (Fig. 4). While this is discrepant from published literature that suggests that capsule is important for colonization (47, 48), most colonization studies have used laboratory strains, such as D39 or TIGR4, and their derivatives (47, 48), and very few studies have examined colonization in mice with recent clinical isolates. Since proteins such as PspK or AliD are known to enable colonization of nonencapsulated pneumococci (49), the specific clinical isolate background used here (BLS141) may express molecules that aid colonization. We failed to detect the presence of pspK or aliD in the BLS141 genomic background (data not shown). Further genomic studies may identify new molecules providing an apparent colonization advantage.
Rajam et al. reported, based on a small number of vaccinated individuals (n = 7), that PPV23 elicits high titers of opsonic antibodies against serotype 15B with almost undetectable titers of opsonic antibodies against serotype 15C (31). This finding strongly implied that serotype 15C replaces serotype 15B following vaccination with serotype 15B capsular polysaccharide. However, when we examined a larger number of sera from immunized individuals (n = 28), we found that PPV23-elicited antibodies opsonized serotype 15B only slightly better than serotype 15C or 15X (1.6- and 2.1-fold, respectively). These data suggest that a conjugate vaccine containing serotype 15B polysaccharide would elicit antibodies targeting the O-acetyl group as well as cross-reactive antibodies targeting the core structure of the capsular polysaccharide; therefore, it is unlikely that serotype 15C will expand. If the serotype 15B-containing conjugate vaccine does cause expansion of serotype 15C, however, then serotype 15C polysaccharide could be used instead to more equally target both serotypes, since there was no compelling reason for choosing serotype 15B polysaccharide over serotype 15C polysaccharide when PPV23 was designed (50).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The pneumococcal strains used in this study are listed in Table 1. Clinical isolates were obtained from the Centers for Disease Control and Prevention (CDC), Atlanta, GA; University of Alabama at Birmingham (UAB), Birmingham, AL; Korea University Guro Hospital, Seoul, South Korea; Adolfo Lutz Institute, São Paulo, Brazil; Children's Hospital of Pittsburgh, University of Pennsylvania Medical Center, Pittsburgh, PA; and Wakayama Medical University, Wakayama, Japan. Two reference isolates were obtained from Statens Serum Institut (SSI), Copenhagen, Denmark.
Pneumococci were routinely grown in Todd Hewitt broth with 0.5% yeast extract (THY) for liquid culture and on blood agar plates (Remel) or THY agar plates with THY agar overlay for cultivation on solid medium as previously described (51). Kanamycin (Km; 100 μg/ml) and streptomycin (Sm; 300 μg/ml) were added to THY agar overlay where indicated. Liquid aliquots of mid-log culture supplemented to 16% glycerol were stored at −80°C as working stocks.
Genetic manipulation of pneumococci.
Transformation of DNA constructs into pneumococcal strains was performed as previously described (52, 53). Competence was induced in a parental pneumococcal strain using 50 μg/ml CSP-1 and/or CSP-2 for 9 min at 37°C, and a 1:1 mixture of construct DNA and lysate of the parental strain (prepared as described in reference 54) was added to the competent pneumococcal cells and incubated for 2 h. Transformations were plated as described above with antibiotic (Sm or Km) and 2,3,5-triphenyl tetrazolium chloride (TTC).
We created parental serotype 15B and 15C strains with an Sm resistance gene, rpsL (Smr), to facilitate subsequent genetic modifications (55). rpsL (Smr) was amplified from strain BLS101 (12, 56) and transformed as described above with selection for Sm resistance into serotype 15B clinical isolate 0556-97 to create strain BLS141 or into serotype 15C clinical isolate 3031-06 to create strain BLS142. We detected no differences in the O-acetylation (fs15b) or capsular amount (pool H) between the parental clinical isolates (0556-97 and 3031-06) and the transformed reference strains (BLS141 and BLS142, respectively) by inhibition ELISA (see Fig. S1 in the supplemental material). Isogenic wciZ variants were created in the BLS141 background and are summarized in Fig. 1 and Table 1.
A wciZ deletion strain was made by amplifying JS from TIGRJS lysate and homologous flanking sequence from BLS141 lysate. The construct was assembled by overlap extension PCR and transformed into BLS141 as described above to create the wciZ-null strain BLS143. Similarly, a cps deletion construct (dexB-JS-aliA region amplified from TIGRJS lysate) was transformed into BLS141 to create BLS147. Km-resistant BLS143 and BLS147 transformants were analyzed by PCR and flow cytometry for their genetic and serological properties. To create a (TA)7 serotype 15C isolate in the BLS141 background, 3031-06 wciZ [(TA)7] was amplified from BLS142 lysate and transformed into BLS143 with selection for Sm resistance, resulting in strain BLS157. A (TA)6 serotype 15C isolate was created in the BLS141 background by amplifying the (TA)6 region from isolate ST1058/03 using primers 5256 and 3164. The remaining portion of the gene was amplified from BLS141 to avoid two missense mutations found early in wciZ of ST1058/03 using primers 5168 and 3229. These fragments were joined by overlap extension PCR, the resulting construct was transformed into BLS143 with selection for Sm resistance, and the resulting strain was named BLS171. Primers used to create these strains are detailed in Table S2. The Heflin Center Genomics Core Laboratory at the University of Alabama at Birmingham performed DNA sequencing.
Detection of WciZ-mediated O-acetylation by inhibition ELISA.
Inhibition ELISAs were performed as previously described, with some modifications (57). Plates were coated with 0.25 μg/ml serotype 15B polysaccharide (SSI; Copenhagen, Denmark) in phosphate-buffered saline (PBS) for 3 h at room temperature, washed, and blocked with 1% bovine serum albumin in phosphate-buffered saline with Tween (PBST) for 30 min. Lysates used for inhibition were made by growing strains to an optical density at 600 nm (OD600) of 1.0 and lysing as described in reference 54. Half of each lysate was treated with 0.2 M NaOH for 2 h at room temperature to hydrolyze the acetyl groups. Hydrolyzed and nonhydrolyzed lysates were dialyzed against water overnight. Lysates were 10-fold serially diluted, added to the ELISA plates, and allowed to incubate for 5 min before addition of either Hyp15GB5 or adsorbed fs15b. Factor serum was adsorbed against heat-killed TIGRJS bacteria as described previously to remove antibodies targeting other surface molecules, such as teichoic acids or surface proteins (54). Plates were shaken for a few seconds to mix and incubated at room temperature for 2 h. After washing, either goat anti-rabbit Ig or goat anti-mouse IgG secondary antibody conjugated to alkaline phosphatase (1/3,000 dilution for each; Southern Biotech, Birmingham, AL) was added to plates and incubated for 30 min at room temperature. After extensive washing, 1 mg/ml 4-nitrophenyl phosphate disodium salt hexahydrate (Sigma; St. Louis, MO) was added, and color was allowed to develop for several hours before reading absorbance at 405 nm.
Flow cytometry analyses of bacterial strains.
Serological properties of serotype 15B, 15C, and 15X strains were determined by flow cytometry as previously described (11, 27, 58). Pool H and fs15b were obtained from SSI (Copenhagen, Denmark), and normal rabbit serum was included as a negative control. All sera were adsorbed against TIGRJS bacteria as described in reference 54. Hyp15BG5 and Hyp6BG9 are monoclonal antibodies specific for serotypes 15B and 6B, respectively (32). Anti-mouse IgG and anti-rabbit Ig secondary antibodies conjugated to phycoerythrin were used to detect binding by the above-mentioned probes (1/5,000 and 1/1,000 dilutions, respectively; Southern Biotech; Birmingham, AL). Data were collected on a BD Accuri C6 plus (San Jose, CA), and analysis was performed using FCS Express software, version 6.
In vitro biological assays.
Biofilm formation, adhesion to nasopharyngeal cells, and survival after short-term drying were performed as previously described (44, 59–61) using BLS141 (15B), BLS157 (15C), BLS143 (15X), and BLS147 (Δcps) isogenic strains. Biofilm formation was measured on polystyrene surfaces and was quantified using crystal violet staining and spectrophotometric reading at OD540. Biofilm experiments were performed eight independent times. Percent adhesion to Detroit 562 cells (ATCC [Manassas, VA]) was quantified as the quotient of CFU after 1 h of incubation with TNF-treated nasopharyngeal cells divided by CFU before 1 h of incubation with TNF-treated nasopharyngeal cells. Data are represented as log10 fold change in adhesion from that of parental strain 15B. Each experiment was performed three independent times in triplicate. Survival after drying was quantified by plating bacteria in PBS on polystyrene surfaces, drying for 1 h, allowing further desiccation for 1 h, scraping the bacteria from the surface, and quantifying the surviving CFU. Percent survival was calculated as the quotient of the CFU surviving drying divided by the CFU added to the polystyrene plate initially. Data are represented as log10 percent survival. Drying experiments were performed eight independent times.
Intranasal murine infections.
Female 6-week-old BALB/cJ mice (The Jackson Laboratory, Bar Harbor, ME) were infected intranasally in the left nare with 107 CFU of BLS141 (15B), BLS157 (15C), BLS143 (15X), and BLS147 (Δcps) isogenic strains in 25 μl sterile PBS at day 0. Serotype 15B was given to 10 mice, but all other strains were given to 9 mice each. At days 1, 3, 5, 7, 10, 14, 17, 20, and 25, nasopharyngeal lavage fluid (NALF) was collected and the CFU were determined by serial dilution. NALF was collected by quickly pipetting 10 μl PBS in and out of the left nare of the mouse. Mice were euthanized at the end of the experiment on day 25.
In vitro opsonophagocytic killing assay.
The in vitro opsonophagocytosis assay was performed as described in reference 62 with sera from six unvaccinated humans and 28 humans immunized with PPV23. All unvaccinated human sera were 2-fold prediluted, all postvaccination human sera were 20-fold prediluted, fs15b was 225-fold prediluted, and Hyp15GB5 was 20-fold prediluted. Colonies were counted using NICE software (63), and OIs were calculated using Opsotiter (A1.0 NICE) (62).
Statistical analyses.
Statistical analysis was performed using Prism7 for Windows (version 7.02; GraphPad Software, La Jolla, CA) as described in the figure legends.
Accession numbers.
The multilocus sequence type (MLST) loci of 0556-97 and 3031-06 were amplified according to reference 64, and strains and their sequence types were deposited in the PubMLST database. The PubMLST numbers are presented in Table S1 in the supplemental material. The full cps loci of 0556-97, 3031-06, BLS141, BLS142, BLS143, BLS147, BLS171, and ST1058/03 and the wciZ region of clinical isolates MHI-224, MNZ161, 3933-06, and MNK1031 were sequenced and deposited in GenBank. The accession numbers are presented in Table 1. The cps locus sequence of 0556-97 contains differences from the published serotype 15B sequence in GenBank (accession no. CR931664.1) (65), and the differences are summarized in Table S3.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institutes of Health grant R01AG050607-01 (M.H.N.) and by the National Institutes of Health grant R01AI114800 (C.J.O.).
We thank B. Beall at the CDC, J. Y. Song at Korea University, A. Hoberman at Children's Hospital of Pittsburgh, A. Brandao at Instituto Adolfo Lutz, Brazil, M. Hotomi at Wakayama Medical University, and S. Hollingshead at UAB for providing us with bacterial strains. We also thank C. L. Turnbough, Jr., for advice relating to reiterative transcription in this study, K. A. Geno for critical review of the manuscript, and the UAB Heflin Center Genomics Core Laboratory for performing the sequencing (Comprehensive Cancer Center Core grant P30 CA013148, Center for AIDS Research grant P30 AI027767).
The University of Alabama at Birmingham has intellectual property rights to some reagents developed in the laboratory of M. H. Nahm, and all study authors are UAB employees. We have no additional conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00099-17.
REFERENCES
- 1.Navne JE, Borresen ML, Slotved HC, Andersson M, Melbye M, Ladefoged K, Koch A. 2016. Nasopharyngeal bacterial carriage in young children in Greenland: a population at high risk of respiratory infections. Epidemiol Infect 144:3226–3236. doi: 10.1017/S0950268816001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soysal A, Karabag-Yilmaz E, Kepenekli E, Karaaslan A, Cagan E, Atici S, Atinkanat-Gelmez G, Boran P, Merdan S, Hasdemir U, Soyletir G, Bakir M. 2016. The impact of a pneumococcal conjugate vaccination program on the nasopharyngeal carriage, serotype distribution and antimicrobial resistance of Streptococcus pneumoniae among healthy children in Turkey. Vaccine 34:3894–3900. doi: 10.1016/j.vaccine.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Hjalmarsdottir MA, Gumundsdottir PF, Erlendsdottir H, Kristinsson KG, Haraldsson G. 2016. Cocolonization of pneumococcal serotypes in healthy children attending day care centers: molecular versus conventional methods. Pediatr Infect Dis J 35:477–480. doi: 10.1097/INF.0000000000001059. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. 2008. Estimates of disease burden and cost-effectiveness. World Health Organization, Geneva, Switzerland. http://www.who.int/immunization/monitoring_surveillance/burden/estimates/en/ Accessed 9 April 2015.
- 5.Kim JO, Romero-Steiner S, Sorensen UB, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun 67:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avery OT, Dubos R. 1931. The protective action of a specific enzyme against type III pneumococcus infection in mice. J Exp Med 54:73–89. doi: 10.1084/jem.54.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Hardy GG, Magee AD, Ventura CL, Caimano MJ, Yother J. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect Immun 69:2309–2317. doi: 10.1128/IAI.69.4.2309-2317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyams C, Yuste J, Bax K, Camberlein E, Weiser JN, Brown JS. 2010. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect Immun 78:716–725. doi: 10.1128/IAI.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melin M, Jarva H, Siira L, Meri S, Kayhty H, Vakevainen M. 2009. Streptococcus pneumoniae capsular serotype 19F is more resistant to C3 deposition and less sensitive to opsonophagocytosis than serotype 6B. Infect Immun 77:676–684. doi: 10.1128/IAI.01186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady AM, Calix JJ, Yu J, Geno KA, Cutter GR, Nahm MH. 2014. Low invasiveness of pneumococcal serotype 11A is linked to ficolin-2 recognition of O-acetylated capsule epitopes and lectin complement pathway activation. J Infect Dis 210:1155–1165. doi: 10.1093/infdis/jiu195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer BL, Saad JS, Shenoy AT, Orihuela CJ, Nahm MH. 24 April 2017. Position of O-acetylation within the capsular repeat unit impacts the biological properties of pneumococcal serotypes 33A and 33F. Infect Immun doi: 10.1128/IAI.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fusco PC, Farley EK, Huang CH, Moore S, Michon F. 2007. Protective meningococcal capsular polysaccharide epitopes and the role of O acetylation. Clin Vaccine Immunol 14:577–584. doi: 10.1128/CVI.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tielen P, Strathmann M, Jaeger KE, Flemming HC, Wingender J. 2005. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res 160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Slauch JM, Mahan MJ, Michetti P, Neutra MR, Mekalanos JJ. 1995. Acetylation (O-factor 5) affects the structural and immunological properties of Salmonella typhimurium lipopolysaccharide O antigen. Infect Immun 63:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas EL, Jefferson MM, Bennett JJ, Learn DB. 1987. Mutagenic activity of chloramines. Mutat Res 188:35–43. doi: 10.1016/0165-1218(87)90112-1. [DOI] [PubMed] [Google Scholar]
- 17.Cuzzi B, Cescutti P, Furlanis L, Lagatolla C, Sturiale L, Garozzo D, Rizzo R. 2012. Investigation of bacterial resistance to the immune system response: cepacian depolymerisation by reactive oxygen species. Innate Immun 18:661–671. doi: 10.1177/1753425911435954. [DOI] [PubMed] [Google Scholar]
- 18.Rosenthal RS, Blundell JK, Perkins HR. 1982. Strain-related differences in lysozyme sensitivity and extent of O-acetylation of gonococcal peptidoglycan. Infect Immun 37:826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crisostomo MI, Vollmer W, Kharat AS, Inhulsen S, Gehre F, Buckenmaier S, Tomasz A. 2006. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol Microbiol 61:1497–1509. doi: 10.1111/j.1365-2958.2006.05340.x. [DOI] [PubMed] [Google Scholar]
- 20.Davis KM, Akinbi HT, Standish AJ, Weiser JN. 2008. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog 4:e1000241. doi: 10.1371/journal.ppat.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox KL, Yildirim HH, Deadman ME, Schweda EK, Moxon ER, Hood DW. 2005. Novel lipopolysaccharide biosynthetic genes containing tetranucleotide repeats in Haemophilus influenzae, identification of a gene for adding O-acetyl groups. Mol Microbiol 58:207–216. doi: 10.1111/j.1365-2958.2005.04814.x. [DOI] [PubMed] [Google Scholar]
- 22.Vann WF, Liu TY, Robbins JB. 1978. Cell-free biosynthesis of the O-acetylated N-acetylneuraminic acid capsular polysaccharide of group C meningococci. J Bacteriol 133:1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudlavalleti SK, Datta AK, Tzeng YL, Noble C, Carlson RW, Stephens DS. 2004. The Neisseria meningitidis serogroup A capsular polysaccharide O-3 and O-4 acetyltransferase. J Biol Chem 279:42765–42773. doi: 10.1074/jbc.M313552200. [DOI] [PubMed] [Google Scholar]
- 24.Orskov F, Orskov I, Sutton A, Schneerson R, Lin W, Egan W, Hoff GE, Robbins JB. 1979. Form variation in Escherichia coli K1: determined by O-acetylation of the capsular polysaccharide. J Exp Med 149:669–685. doi: 10.1084/jem.149.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calix JJ, Brady AM, Du VY, Saad JS, Nahm MH. 2014. Spectrum of pneumococcal serotype 11A variants results from incomplete loss of capsule O-acetylation. J Clin Microbiol 52:758–765. doi: 10.1128/JCM.02695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calix JJ, Saad JS, Brady AM, Nahm MH. 2012. Structural characterization of Streptococcus pneumoniae serotype 9A capsule polysaccharide reveals role of glycosyl 6-O-acetyltransferase wcjE in serotype 9V capsule biosynthesis and immunogenicity. J Biol Chem 287:13996–14003. doi: 10.1074/jbc.M112.346924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geno KA, Saad JS, Nahm MH. 15 February 2017. Discovery of novel pneumococcal serotype, 35D: a natural WciG-deficient variant of serotype 35B. J Clin Microbiol doi: 10.1128/JCM.00054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, van Putten JP. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun 71:6192–6198. doi: 10.1128/IAI.71.11.6192-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkateswaran PS, Stanton N, Austrian R. 1983. Type variation of strains of Streptococcus pneumoniae in capsular serogroup 15. J Infect Dis 147:1041–1054. doi: 10.1093/infdis/147.6.1041. [DOI] [PubMed] [Google Scholar]
- 30.Jansson PE, Lindberg B, Lindquist U, Ljungberg J. 1987. Structural studies of the capsular polysaccharide from Streptococcus pneumoniae types 15B and 15C. Carbohydr Res 162:111–116. doi: 10.1016/0008-6215(87)80205-7. [DOI] [PubMed] [Google Scholar]
- 31.Rajam G, Carlone GM, Romero-Steiner S. 2007. Functional antibodies to the O-acetylated pneumococcal serotype 15B capsular polysaccharide have low cross-reactivities with serotype 15C. Clin Vaccine Immunol 14:1223–1227. doi: 10.1128/CVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Lin J, Kim KH, Benjamin WH Jr, Nahm MH. 2011. Development of an automated and multiplexed serotyping assay for Streptococcus pneumoniae. Clin Vaccine Immunol 18:1900–1907. doi: 10.1128/CVI.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee LH, Gu XX, Nahm MH. 2014. Towards new broader spectrum pneumococcal vaccines: the future of pneumococcal disease prevention. Vaccines (Basel) 2:112–128. doi: 10.3390/vaccines2010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper D, Emini EA, Gu J, Han M, Jansen KU, Kainthan RK, Kim JH, Prasad AK, Pride MW, Watson WJ. July 2015. Immunogenic compositions comprising conjugated capsular saccharide antigens and uses thereof. US patent 20,150,202,309 A1.
- 35.Beall B, McEllistrem MC, Gertz RE Jr, Wedel S, Boxrud DJ, Gonzalez AL, Medina MJ, Pai R, Thompson TA, Harrison LH, McGee L, Whitney CG, Active Bacterial Core Surveillance Team. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol 44:999–1017. doi: 10.1128/JCM.44.3.999-1017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez BE, Hulten KG, Lamberth L, Kaplan SL, Mason EO Jr. 2006. Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr Infect Dis J 25:301–305. doi: 10.1097/01.inf.0000207484.52850.38. [DOI] [PubMed] [Google Scholar]
- 37.Gherardi G, Fallico L, Del Grosso M, Bonanni F, D'Ambrosio F, Manganelli R, Palu G, Dicuonzo G, Pantosti A. 2007. Antibiotic-resistant invasive pneumococcal clones in Italy. J Clin Microbiol 45:306–312. doi: 10.1128/JCM.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laufer AS, Thomas JC, Figueira M, Gent JF, Pelton SI, Pettigrew MM. 2010. Capacity of serotype 19A and 15B/C Streptococcus pneumoniae isolates for experimental otitis media: implications for the conjugate vaccine. Vaccine 28:2450–2457. doi: 10.1016/j.vaccine.2009.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarewicz O, Lucas M, Brandt C, Herrmann L, Albersmeier A, Ruckert C, Blom J, Goesmann A, van der Linden M, Kalinowski J, Pletz MW. 2017. Whole genome sequencing of 39 invasive Streptococcus pneumoniae sequence type 199 isolates revealed switches from serotype 19A to 15B. PLoS One 12:e0169370. doi: 10.1371/journal.pone.0169370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz-Elias EJ, Marcano J, Camilli A. 2008. Isolation of Streptococcus pneumoniae biofilm mutants and their characterization during nasopharyngeal colonization. Infect Immun 76:5049–5061. doi: 10.1128/IAI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS Jr, Bakaletz LO. 2007. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol 65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 42.Trappetti C, Ogunniyi AD, Oggioni MR, Paton JC. 2011. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One 6:e19844. doi: 10.1371/journal.pone.0019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keller LE, Jones CV, Thornton JA, Sanders ME, Swiatlo E, Nahm MH, Park IH, McDaniel LS. 2013. PspK of Streptococcus pneumoniae increases adherence to epithelial cells and enhances nasopharyngeal colonization. Infect Immun 81:173–181. doi: 10.1128/IAI.00755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh RL, Camilli A. 2011. Streptococcus pneumoniae is desiccation tolerant and infectious upon rehydration. mBio 2:e00092-11. doi: 10.1128/mBio.00092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajam G, Phillips DJ, White E, Anderton J, Hooper CW, Sampson JS, Carlone GM, Ades EW, Romero-Steiner S. 2008. A functional epitope of the pneumococcal surface adhesin A activates nasopharyngeal cells and increases bacterial internalization. Microb Pathog 44:186–196. doi: 10.1016/j.micpath.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Gladstone RA, Jefferies JM, Tocheva AS, Beard KR, Garley D, Chong WW, Bentley SD, Faust SN, Clarke SC. 2015. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine 33:2015–2021. doi: 10.1016/j.vaccine.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun 69:3755–3761. doi: 10.1128/IAI.69.6.3755-3761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun 75:83–90. doi: 10.1128/IAI.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park IH, Kim KH, Andrade AL, Briles DE, McDaniel LS, Nahm MH. 2012. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. mBio 3:e00035-12. doi: 10.1128/mBio.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins JB, Austrian R, Lee CJ, Rastogi SC, Schiffman G, Henrichsen J, Makela PH, Broome CV, Facklam RR, Tiesjema RH, Parke JC Jr. 1983. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis 148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 51.Burton RL, Nahm MH. 2006. Development and validation of a fourfold multiplexed opsonization assay (MOPA4) for pneumococcal antibodies. Clin Vaccine Immunol 13:1004–1009. doi: 10.1128/CVI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardy GG, Caimano MJ, Yother J. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J Bacteriol 182:1854–1863. doi: 10.1128/JB.182.7.1854-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calix JJ, Oliver MB, Sherwood LK, Beall BW, Hollingshead SK, Nahm MH. 2011. Streptococcus pneumoniae serotype 9A isolates contain diverse mutations to wcjE that result in variable expression of serotype 9V-specific epitope. J Infect Dis 204:1585–1595. doi: 10.1093/infdis/jir593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xayarath B, Yother J. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol 189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trzcinski K, Thompson CM, Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol 69:7364–7370. doi: 10.1128/AEM.69.12.7364-7370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burton RL, Geno KA, Saad JS, Nahm MH. 2016. Pneumococcus with the “6E” cps locus produces serotype 6B capsular polysaccharide. J Clin Microbiol 54:967–971. doi: 10.1128/JCM.03194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camilli R, Spencer BL, Moschioni M, Pinto V, Berti F, Nahm MH, Pantosti A. 2014. Identification of Streptococcus pneumoniae serotype 11E, serovariant 11Av and mixed populations by high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and flow cytometric serotyping assay (FCSA). PLoS One 9:e100722. doi: 10.1371/journal.pone.0100722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanchette KA, Shenoy AT, Milner J II, Gilley RP, McClure E, Hinojosa CA, Kumar N, Daugherty SC, Tallon LJ, Ott S, King SJ, Ferreira DM, Gordon SB, Tettelin H, Orihuela CJ. 2016. Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect Immun 84:2922–2932. doi: 10.1128/IAI.00277-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez CJ, Hinojosa CA, Shivshankar P, Hyams C, Camberlein E, Brown JS, Orihuela CJ. 2011. Changes in capsular serotype alter the surface exposure of pneumococcal adhesins and impact virulence. PLoS One 6:e26587. doi: 10.1371/journal.pone.0026587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marks LR, Reddinger RM, Hakansson AP. 2014. Biofilm formation enhances fomite survival of Streptococcus pneumoniae and Streptococcus pyogenes. Infect Immun 82:1141–1146. doi: 10.1128/IAI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burton RL, Nahm MH. 2012. Development of a fourfold multiplexed opsonophagocytosis assay for pneumococcal antibodies against additional serotypes and discovery of serological subtypes in Streptococcus pneumoniae serotype 20. Clin Vaccine Immunol 19:835–841. doi: 10.1128/CVI.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke ML, Burton RL, Hill AN, Litorja M, Nahm MH, Hwang J. 2010. Low-cost, high-throughput, automated counting of bacterial colonies. Cytometry A 77:790–797. doi: 10.1002/cyto.a.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Part 11):3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 65.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.