ABSTRACT
Segmented filamentous bacteria (SFB) are known modulators of the mammalian immune system. Currently, the technology for investigating SFB culture in vitro is immature, and as a result, the mechanisms of SFB colonization and immune regulation are not yet fully elucidated. In this study, we investigated the gene diversity and host specificity of SFB flagellin genes. The fliC1 and fliC2 genes are relatively conserved, while the fliC3 and fliC4 genes are more variable, especially at the central and C-terminal regions. Host specificity analysis demonstrated that the fliC1 genes do not cluster together based on the host organism, whereas the fliC3 and fliC4 genes were host specific at the nucleotide and deduced amino acid levels. SFB flagellin protein expression in the ileum mucosa and cecal contents was detected by using fluorescence in situ hybridization (FISH) combined with immunohistochemical (IHC) analysis, immunoblotting, and liquid chromatography-tandem mass spectrometry (LC-MS/MS). Although the purified SFB FliC3 protein originating from both mouse and rat was able to activate Toll-like receptor 5 (TLR5)-linked NF-κB signaling, no host specificity was observed. Interestingly, the patterns of interaction with mouse ileum mucosal proteins were different for mouse FliC3 (mFliC3) and rat FliC3 (rFliC3). Gene Ontology (GO) and KEGG analyses indicated that more adherence-related proteins interacted with mFliC3, while more lysosome- and proteolysis-related proteins interacted with rFliC3. In vitro degradation experiments indicated that the stability of rFliC3 was lower than that of mFliC3 when they were incubated with mouse ileum mucosal proteins. In summary, the gene diversity and host specificity of SFB flagellin genes were investigated, and SFB flagellin expression was detected in gut samples.
IMPORTANCE Since SFB genomes contain only one copy of each FliC gene, the diversity of FliC is representative of SFB strain diversity. Currently, little is known regarding the diversity and specificity of members of the group of SFB. The work presented herein demonstrates that select SFB strains, exhibiting unique FliC patterns, are present in a variety of mammalian hosts. SFB fliC genes were found to interact with a number of unique targets, providing further evidence for SFB host selection. Together, this work represents a major advancement in identifying SFB and delineating how members of the group of SFB interact with the host. Future examination of FliC genes will likely enhance our knowledge of intestinal colonization by the gut microbiota.
KEYWORDS: segmented filamentous bacteria, flagellin, host specificity, gene diversity, ileum mucosal proteins
INTRODUCTION
Recently, segmented filamentous bacteria (SFB) have received attention due to their roles in regulating host immune system development (1, 2), especially in relation to stimulating Th17 cell differentiation (2–4). SFB are gut commensal bacteria that colonize the ileum mucosa of various vertebrates, including humans, mice, rats, chickens, and pigs (5–8). However, SFB colonization is host specific (9, 10), and a direct interaction with the ileum mucosa may be required for regulating the differentiation of Th17 cells (2, 4, 10). SFB genes related to colonization and immune regulation could be important for this process, and some SFB peptides have been found to promote Th17 cell differentiation (11). Intestinal dendritic cells and major histocompatibility complex (MHC) class II cells also take part in this process (4, 12), although the antigen molecules and mechanisms for initial Th17 cell differentiation are still unresolved. Mechanisms that are involved in SFB regulation of the T cell response and IgA secretion need to be investigated further.
Ericsson et al. and Schnupf et al. established culturing methods for mouse SFB in vitro (13, 14), although passaging of SFB in vitro is still problematic (13). Culture-independent bioinformatic analysis could be used as a tool in finding candidate SFB functional genes. There are a large number of publications regarding flagellum-mediated adherence (15), and some studies demonstrated that flagellin has a role in regulating Th17 cell differentiation (16, 17), IgA plasma cell generation, and Th1 functions (18). Comparative genomic analyses indicated that there is a set of genes that encode flagellin proteins within SFB genomes (19, 20) and that the homology between SFB flagellin proteins is conserved in comparison to the most closely related Clostridium species (19). Thus, it is reasonable to assume that SFB flagellin may play very important roles in colonization and immune regulation.
To date, researchers have not observed SFB flagellin via electron microscopy (5, 20–22). In this study, we first investigated the gene diversity and host specificity of the SFB functional genes fliC1, fliC2, fliC3, and fliC4. The expression of these SFB flagellin genes in mouse gut samples was measured by using fluorescence in situ hybridization (FISH) combined with immunohistochemical (IHC) analysis, immunoblotting, and liquid chromatography-tandem mass spectrometry (LC-MS/MS). In addition, the function of flagellins cloned from mouse and rat SFB (i.e., mouse and rat SFB flagellins) in inducing the Toll-like receptor 5 (TLR5)-linked NF-κB signal response was assessed. By utilizing LC-MS/MS to identify interacting proteins, we found that mouse and rat SFB flagellins have different patterns of interactions with mouse ileum mucosal proteins. KEGG analysis indicated that pathways for adherens junctions and oxidative phosphorylation were significantly enriched by mouse SFB FliC3. In contrast, pathways related to tight junctions and lysosomes were significantly enriched by rat SFB FliC3.
RESULTS
Diversity analysis of SFB flagellin genes.
PCR primers targeted to SFB 16S rRNA genes, 779F and 1380R, were used to detect the presence of SFB in the collected mouse and rat gut samples. All ileum and cecal samples collected from six mice and six rats were positive for SFB (see Table S1 in the supplemental material). PCR primers for SFB fliC genes (fliC1, fliC2, fliC3 and fliC4) were then designed by using previously reported SFB fliC gene sequences and used to detect the SFB flagellin genes (Table 1). All SFB fliC1, fliC3, and fliC4 genes were detected in mouse and rat samples (Table S1). However, SFB fliC2 genes were detected only in mouse samples. No rat SFB fliC2 genes were obtained even after PCR amplification and electrophoresis (data not shown), even though several primers that targeted different regions of the rat SFB fliC2 gene were utilized (Table 1).
TABLE 1.
Primers used for PCR analysis of flagellin genes from SFB
| Gene | Primer directionc | Sequence (5′→3′) | Location (positions) |
|---|---|---|---|
| Mouse SFB fliC1 | FW | ATGAAGTTGACTTTTAATTCTAAT | 1033492–1033515a (complement) |
| RV | TTTATATAATTTACCCAAAACATC | 1032688–1032702a | |
| Mouse SFB fliC2 | FW | ATGATAATAAATMACAATATTAATGCGATG | 692490–692519a |
| RV | TTTTAAAATAGAAATAACTTGTTGCGGTGAT | 693647–693677a (complement) | |
| Mouse and rat SFB fliC3 | FW | ATGATAATTAAYCACAATATGAATG | 637535–637559a |
| RV | TCTYAAKATWGAAAGWACTTGTTGT | 637363–637387a (complement) | |
| Mouse and rat SFB fliC4 | FW | ATGATAATTAAYCACAATATGAATGCGATG | 637535–637564a |
| RV | TTTYAATAATGAAAGKACTTGTTGTGGTGC | 638696–638725a (complement) | |
| Rat SFB fliC1 | FW | GCTATGAAAACATTTATGACT | 1033471–1033491a (complement) |
| RV | ACCCAAAACATCTATCAAATC | 1032691–1032711a | |
| Rat SFB fliC2-1 | FW | ATGATAATAAATMACAATATTAATGCGATG | 626924–626953b |
| RV | TTTTAAAATCGACATAACTTGTTGAGGCAA | 628079–628108b (complement) | |
| Rat SFB fliC2-2 | FW | ATTAATGCGATGAATGCACATA | 626942–626963b |
| RV | TAACTTGTTGAGGCAACTGATT | 628073–628094b (complement) | |
| Rat SFB fliC2-3 | FW | TTGAGTTCGGGATTAAGGATA | 627011–627031b |
| RV | TAACCTTTCAATCGCCTCT | 627841–627859b (complement) | |
| Rat SFB fliC2-4 | FW | AATCTAAAGTCTCGCACAT | 626843–626861b |
| RV | ACTTGTTGAGGCAACTGAT | 628074–628092b (complement) |
Position in the full genome sequence of SFB-mouse-Japan (BioProject accession no. PRJDA66727).
Position in the full genome sequence of SFB-rat-Yat (BioProject accession no. PRJDA67837).
FW, forward; RV, reverse.
To investigate the gene diversity of SFB flagellin genes, PCR-amplified products were gel extracted and ligated into a PMD-18T vector. The ligation products were then transformed into chemically competent Escherichia coli DH5α cells. Clones grown on plates containing LB (Luria-Bertani) plus carbenicillin were then randomly selected for sequencing using primers M13F and M13R. In total, 66 rat fliC1 sequences, 52 rat fliC3 sequences, 42 rat fliC4 sequences, 49 mouse fliC1 sequences, 61 mouse fliC2 sequences, 66 mouse fliC3 sequences, and 54 mouse fliC4 sequences were obtained by clone library sequencing (Tables S2 and S3). The sequences of the SFB fliC1 and fliC2 genes are relatively conserved, and the sequence identities between the SFB fliC1 and fliC2 genes are high at both the nucleotide and deduced amino acid levels. The lowest identities for fliC1 were 86.6% and 96.3% at the nucleotide and deduced amino acid levels, respectively. The lowest similarities for fliC2 were 99.2% and 89% at the nucleotide and deduced amino acid levels, respectively. However, the sequence identities among the SFB fliC3 and fliC4 genes were more variable, as the lowest similarities for the fliC3 and fliC4 genes from mice were 81.6% at the nucleotide level and 66.7% at the deduced amino acid level. The lowest similarities among fliC3 and fliC4 genes from rats were 74.9% at the nucleotide level and 50% at the deduced amino acid level. The presence of an early stop codon was detected more frequently in the fliC3 and fliC4 genes than in the fliC1 and fliC2 genes.
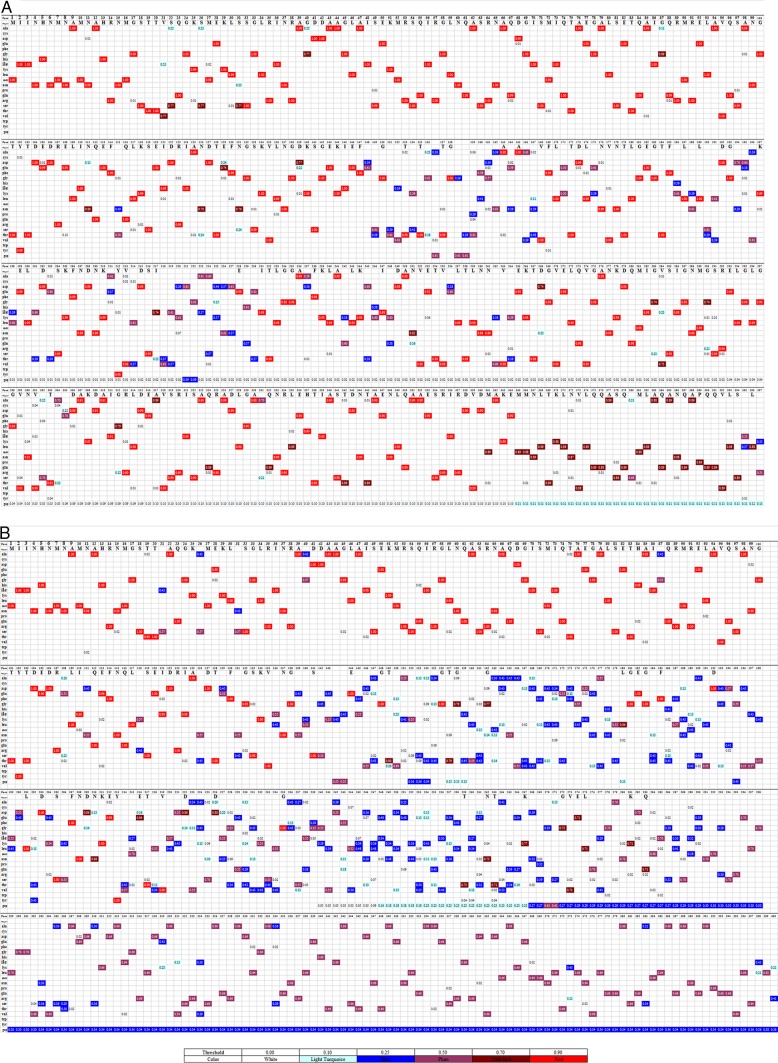
The highly variable regions of SFB FliC3 and FliC4 at the amino acid level were further analyzed. Due to the high level of homology between the fliC3 and fliC4 genes, both the fliC3 and fliC4 sequences were combined for further analysis. The N-terminal and TLR5-binding regions of the SFB FliC3 and FliC4 proteins are well conserved (Fig. 1). The N-terminal regions of most deduced sequences shared identical amino acids at a few positions; however, the central regions of these sequences were quite variable. In addition, the level of variation was higher for rat SFB FliC3 and FliC4 sequences (Fig. 1).
FIG 1.
Visualization of the mutant variable regions of the deduced SFB FliC3 and FliC4 amino acid sequences. Fifty-two rat SFB fliC3 gene sequences, 42 rat SFB fliC4 gene sequences, 66 mouse SFB fliC3 gene sequences, and 51 mouse SFB fliC4 gene sequences were obtained by clone library sequencing. For each sample, duplicate sequences were deleted, and one sequence was preserved for further analysis. Deduced amino acid sequences that were shorter than half the length of the SFB FliC genes were removed. (A) In total, 76 mouse SFB FliC3 and FliC4 sequences obtained in this study and 6 mouse SFB FliC3 and FliC4 sequences downloaded from the NCBI database were used for alignment. (B) Fifty-four rat SFB FliC3 and FliC4 sequences obtained in this study and 2 rat SFB FliC3 and FliC4 sequences downloaded from the NCBI database were used for alignment. JProfileGrid software (33) was used for multiple-sequence alignment visualization. Each row represents different amino acids. Different colors represent homology at the site for each amino acid. The numbers represent the residue frequency occurring at each column position.
Host specificity analysis of SFB flagellin genes.
Before investigation of the host specificity of mouse and rat SFB fliC genes, rat SFB 16S rRNA genes were analyzed. In total, 65 rat SFB 16S rRNA gene sequences, obtained from six rat ileum and cecal samples, were obtained by random clone library sequencing. The sequence identities of the rat SFB 16S rRNA genes in each sample were analyzed by using DNAStar software (see Table S3 in the supplemental material). One predominant sequence was noted for each sample, and the similarity among these sequences was >99.4%. The rat SFB 16S rRNA gene sequences were compared with previously reported human, mouse, and chicken SFB 16S rRNA gene sequences (6), which demonstrated that rat SFB clustered together and were separated from other host SFB sequences (Fig. S1).
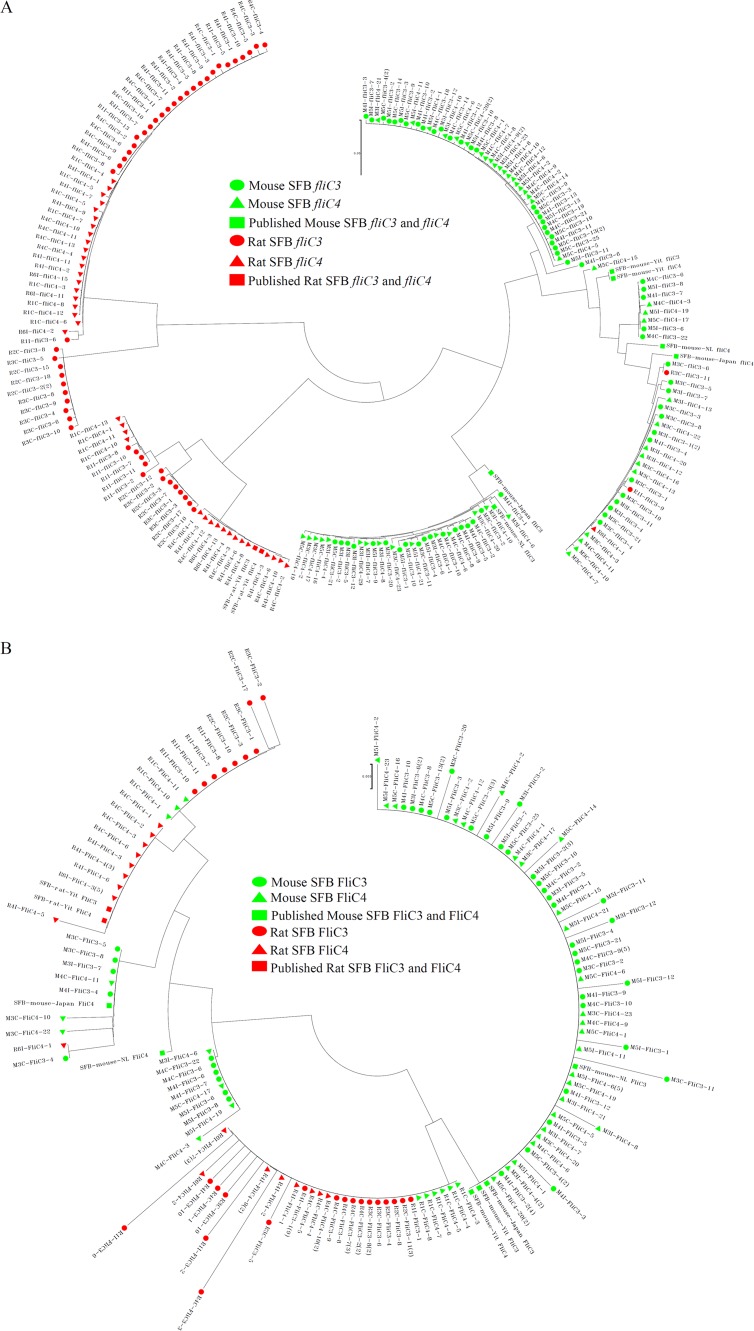
For host specificity analysis of SFB flagellin genes, all flagellin nucleotide sequences obtained in this study or downloaded from the NCBI database were analyzed. Deduced amino acid sequences that were shorter than half the length of the reported amino acid sequences for SFB fliC genes were removed. Phylogenetic analysis of these genes was performed by using MEGA 6. The fliC1 nucleotide sequences (Fig. S2A) and the deduced amino acid sequences (Fig. S2B) did not cluster together with the host organism. However, the fliC3 and fliC4 nucleotide sequences (Fig. 2A) and deduced amino acid sequences (Fig. 2B) clustered together according to the host organism. At the nucleotide level, each clade clustered together for the mouse and rat fliC3 and fliC4 sequences. Although two clades clustered together for the mouse and rat FliC3 and FliC4 amino acid sequences, mouse host sequences were separated from rat host sequences. The homology between SFB FliC3 and FliC4 was much higher and could not be separated on the phylogenetic trees (Fig. 2).
FIG 2.
Phylogenetic analyses of SFB FliC3 and FliC4 genes at the nucleotide and deduced amino acid levels. Fifty-two rat SFB fliC3 gene sequences, 42 rat SFB fliC4 gene sequences, 66 mouse SFB fliC3 gene sequences, and 51 mouse SFB fliC4 gene sequences were obtained by clone library sequencing. For each sample, duplicate sequences were deleted, and one sequence was preserved for further analysis. Deduced amino acid sequences shorter than half the length of the previously reported SFB fliC genes were removed. In total, 201 fliC3 and fliC4 nucleotide sequences and 130 deduced FliC3 and FliC4 amino acid sequences were used for phylogenetic analysis. Three mouse SFB fliC3 sequences, 3 mouse SFB fliC4 sequences, 1 rat SFB fliC3 sequence, and 1 rat SFB fliC4 sequence were downloaded from the NCBI GenBank database. Finally, 209 fliC3 and fliC4 nucleotide sequences (A) and 138 deduced amino acid sequences (B) were aligned and then used to construct a phylogenetic tree by using MEGA 6 with the neighbor-joining algorithm.
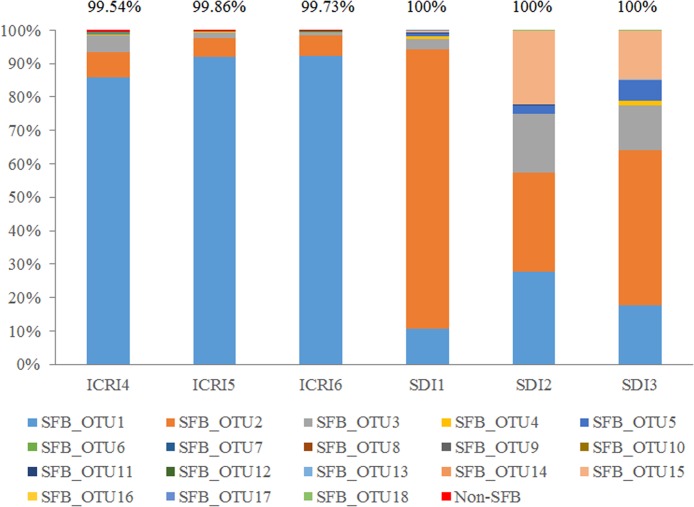
High-throughput sequencing was further used to detect the host specificity of SFB FliC3 and FliC4, and a new pair of primers was specifically designed for the SFB fliC3 and fliC4 genes according to the conserved region. The specificity of this primer was verified by alignment with >99.5% of the obtained sequences that were identified as being SFB sequences (Fig. 3). The sequencing results also demonstrated that host specificity of SFB exists between mouse and rat. SFB operational taxonomic unit (OTU) 1 (SFB_OTU1) was predominant in mouse samples, and SFB_OTU2 was the main species in rat samples.
FIG 3.
Diversity and host specificity analyses of the SFB fliC3 and fliC4 genes using high-throughput sequencing. A new pair of primers, fliC3/4 F and fliC3/4 R, was designed according to the conserved regions of the fliC3 and fliC4 genes. The PCR amplicons of 3 mouse and rat ileal samples were sent for high-throughput sequencing using Illumina MiSeq. A 97% similarity cutoff was used to define OTUs by using Mothur. One sequence from each OTU was selected and considered representative. These sequences were classified by using the rdp classifier method and the NCBI nucleotide database. The values at the top represent the SFB ratio identified for each sample.
Expression of FliC3 in mouse gut samples.
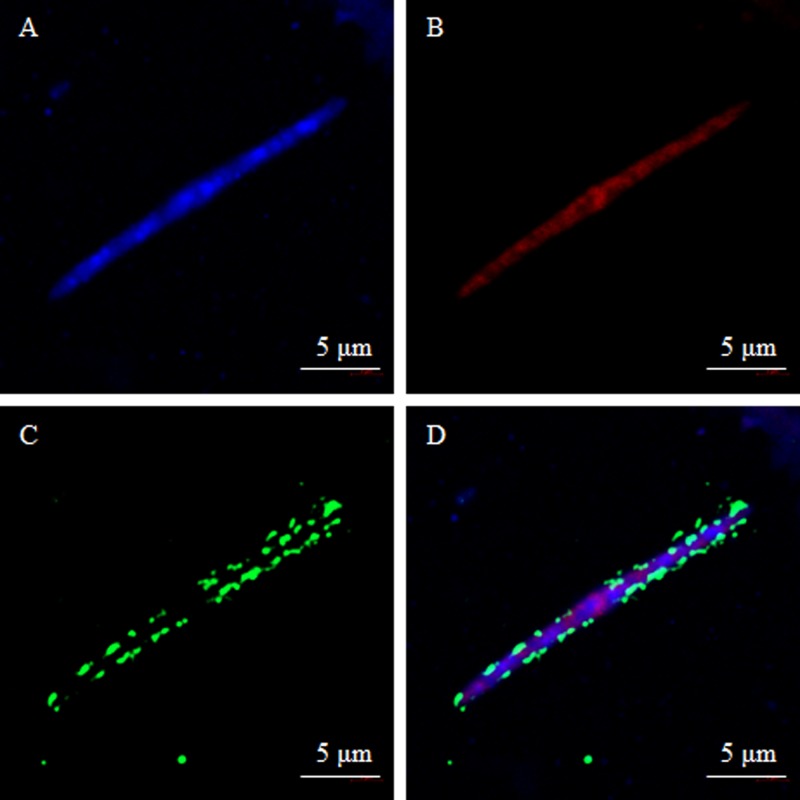
The predominant mouse SFB FliC3 sequence was subcloned into the expression vector pET-28a. The FliC3 protein was then purified from an E. coli expression system after the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) for induction. To detect the expression of FliC3 proteins in gut samples, an anti-mouse FliC3 (mFliC3) polyclonal antibody was obtained by injecting the purified mFliC3 protein into rabbits. The anti-mFliC3 polyclonal antibody was collected and separated from rabbit serum 20 days later. The specificity of this antibody was detected by staining with E. coli BL21 and Salmonella strain CVCC519. As shown in Fig. S3 in the supplemental material, SFB FliC3 antibody did not interact with these bacteria. The expression of SFB FliC3 was then detected by using FISH and immunohistochemical analysis. FliC3 expression in mouse ileum and cecal samples was detected by using a fluorescence microscope (Fig. S4). The nucleotide probe signal was red and was specifically targeted to the 16S rRNA gene of SFB (Fig. S4B and S4F), which was hybridized to the bacteria. The green signal indicated proteins hybridized to the anti-mFliC3 polyclonal antibody (Fig. S4C and S4G). The FISH and IHC signals overlapped very well (Fig. S4D and S4H). SFB FliC3 protein expression was further verified by imaging under a copolymerization microscope. SFB flagellin was expressed mainly at the bacterial surface (Fig. 4D). To explore where SFB is localized in vivo, a copolymerization microscope was used to observe mouse ileum and colon tissues. SFB flagellin was attached mainly to the epithelium of the ileum but not the colon (Fig. S5).
FIG 4.
Detection of FliC3 expression in mouse ileum samples using FISH and IHC analysis. Ileal contents were fixed in 4% paraformaldehyde and sequentially hybridized with the SFB-specific oligonucleotide probe, rabbit anti-FliC3 primary antibody, and FITC goat anti-rabbit IgG(H+L) antibody. DAPI was used to stain nuclei. A two-photon confocal microscope (LSM 800; Zeiss) was used to observe the cells. (A) DAPI stain results; (B) hybridization results with the SFB-specific oligonucleotide probe; (C) hybridization results with rabbit anti-FliC3 primary antibody and FITC goat anti-rabbit IgG(H+L) antibody; (D) overlapping signal between panels B and C.
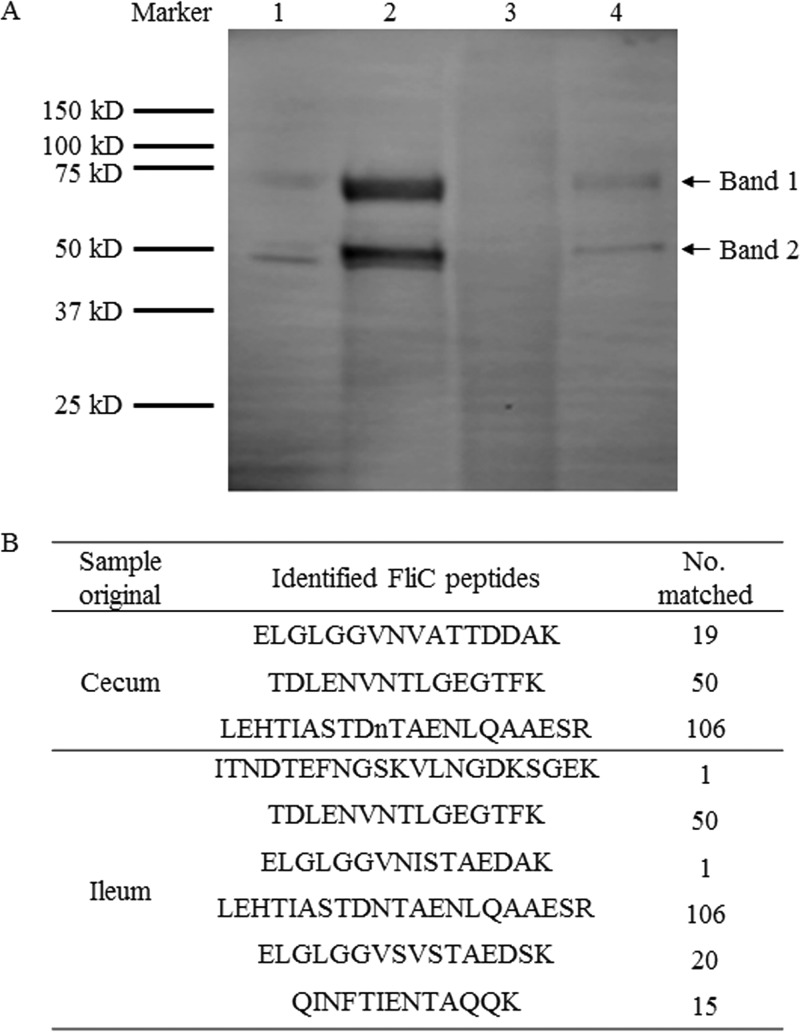
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotting, and LC-MS/MS were used to further verify fliC3 gene expression in gut samples. After the proteins were extracted from mouse ileum tissue and cecal contents, immunoblotting was performed by using anti-mFliC3 polyclonal antibody and chemiluminescence assay kits, and two bands corresponded to purified mFliC3 proteins in mouse ileum and cecal samples (Fig. 5A). Proteins on the SDS-PAGE gel that corresponded to the region of the two bands were extracted and digested for LC-MS/MS analysis. The peptides identified by LC-MS/MS were then confirmed by using the deduced FliC proteins generated in this study and downloaded from the SFB UniProt database. Three and six SFB flagellin peptides were identified from cecal and ileum samples, respectively (Fig. 5B). Other flagellar assembly-related proteins, such as Flip, FliH, FlgC, FlgG, FliK, and FliD, were also identified from the LC-MS/MS data (Fig. S6). To confirm that both bands were SFB FliC3 proteins, bands 1 and 2 of purified mFliC3 (as illustrated in Fig. 5) were sent for LC-MS/MS analysis. Peptides corresponding to the SFB FliC3 protein were identified from both bands, although more SFB FliC3 peptides were observed in band 1 (Fig. S7).
FIG 5.
Analysis of expression of FliC3 in mouse gut samples using SDS-PAGE, immunoblotting, and LC-MS/MS. Mouse ileal mucosal and cecal content proteins were extracted and analyzed by using SDS-PAGE and immunoblotting. (A) Rabbit anti-FliC3 antibody and goat anti-rabbit immunoglobulin G(H+L) were used to visualize the proteins. Lane 1, purified mFliC3 protein; lane 2, protein extracted from cecal contents; lane 4, protein extracted from the ileum mucosa. The SDS-PAGE gel of an intermediate region containing the two bands was cut and removed for trypsin digestion. The digested protein peptides were then sent for LC-MS/MS analysis. The deduced SFB FliC3 proteins were generated as a database for LC-MS/MS identity, and the matched peptides were then used for BLAST analysis. (B) Peptides identified as unique SFB flagellin fragments. The numbers represent the number of times that the peptides were present in the LC-MS/MS data.
Immunostimulation activity of mouse and rat FliC3 proteins.
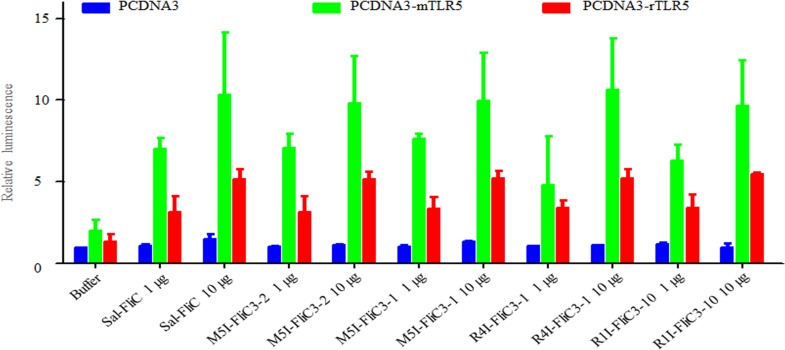
Previously, SFB FliC3 was shown to activate TLR5 and induce the NF-κB signal response. Both TLR5 (see Fig. S8 in the supplemental material) and SFB FliC3 (Fig. 2) have host specificity at the nucleotide and amino acid levels. The interaction of host specificity between TLR5 and FliC3 needs to be investigated further. However, both mouse and rat SFB FliC3 proteins were able to induce the NF-κB signal pathway when mouse or rat TLR5 was present (Fig. 6). Although the immunostimulation of rat SFB FliC3 activity was lower than that of the mouse protein, the diversity between mouse and rat SFB FliC3 proteins does not affect the activation of the TLR5-linked NF-κB signal. Differences between mouse and rat FliC3 proteins in TLR5 activation were not significant (Table S4).
FIG 6.
Activity of SFB FliC3 proteins on the TLR5-linked NF-κB signaling pathway. Luciferase activities in HeLa cells (expressing either mouse TLR5, rat TLR5, or no TLR5) after stimulation with recombinant Salmonella flagellin or mouse and rat SFB flagellins were measured. Results are presented as fold increases relative to the control value. The identity between M5I–FliC3-2 and M5I–FliC3-1 is 86.6%. The identity between R4I–FliC3-1 and R4I–FliC3-10 is 69.2%. The designations M5I, R1I, and R4I indicate strains originating from the ileum sample from mouse 5, the ileum sample from rat 1, and the ileum sample from rat 4, respectively.
Patterns of interaction of mouse and rat SFB FliC3 proteins with mouse ileum mucosal proteins.
The interactions between extracted mouse ileum mucosal proteins and purified mouse or rat SFB FliC3 proteins were investigated by using a pulldown assay and LC-MS/MS. Mouse mucosal proteins that were collected by pulldown with mouse SFB FliC3 proteins, rat SFB FliC3 proteins, or the empty control were sent for LC-MS/MS analysis. In all, two independent pulldown experiments were performed to collect interacting proteins, and LC-MS/MS analysis was performed in duplicate for each experiment. In total, four LC-MS/MS analyses were performed for each group. In sum, 487 and 482 proteins enriched 2-fold above the set threshold using mouse SFB FliC3 and rat SFB FliC3 were identified, respectively. Among the enriched mouse and rat SFB FliC3 proteins, 209 were found to overlap between the two groups, and 278 and 273 proteins were selectively enriched by using mFliC3 and rat FliC3 (rFliC3), respectively.
The functions of these enriched proteins were then further analyzed by using the DAVID bioinformatics database (https://david.ncifcrf.gov/). Although there were some overlapping Gene Ontology (GO) terms between the enriched mFliC3 and rFliC3 proteins (Table S5), more adherens- and binding-related proteins were enriched by using mFliC3, and more exopeptidase activity-, lysosome-, and proteolysis-related proteins were enriched by using rFliC3 (Table 2). Similar correlations were noted using KEGG pathway analysis (Table 3). Several proteins related to adherens junction, oxidative phosphorylation, and tight junction pathways were enriched by using both mFliC3 and rFliC3. Adherens junction-related genes such as the Rous sarcoma virus oncogene, actinin alpha 1, cadherin 1, casein kinase 2, catenin alpha 1, and vinculin interacted with both mFliC3 and rFliC3. However, pulldown proteins related to some unique pathways interacted with mFliC3 or rFliC3. Proteins related to antigen processing and presentation, endocytosis, and NOD-like receptor signaling pathways were isolated only by using mFliC3, whereas proteins related to the lysosome, substance metabolism, and degradation were isolated only by using rFliC3. For lysosome pathways, three proteins related to proteolytic cleavage (cathepsin B, cathepsin C, and cathepsin H) and five proteins related to hydrolysis (N-acetyl galactosaminidase, arylsulfatase B, hexosaminidase A, iduronidase, and mannosidase 2) were significantly enriched by using rFliC3 (Fig. S9). However, no proteins related to the lysosome pathway were enriched by using mFliC3 (Table 3).
TABLE 2.
Unique significantly enriched Gene Ontology terms for mFliC3 and rFliC3
| Categorya | Unique terms |
|
|---|---|---|
| mFliC3 | rFliC3 | |
| MF | Cofactor binding, unfolded protein binding | Exopeptidase activity |
| CC | Adherens junction, anchoring junction, basolateral plasma membrane, endoplasmic reticulum lumen, envelope, F-actin capping protein complex, I band, organelle envelope, Z disc | Actin filament bundle, actomyosin, cell cortex, cytosol, keratin filament, lysosome, lytic vacuole, mitochondrial lumen, mitochondrial matrix, soluble fraction, stress fiber, vacuole |
| BP | Electron transport chain, negative regulation of cellular component organization, negative regulation of cytoskeleton organization, negative regulation of organelle organization, negative regulation of protein complex disassembly, oxidation reduction, protein folding, regulation of actin cytoskeleton organization, regulation of actin filament length, regulation of actin filament polymerization, regulation of actin filament-based process, regulation of actin polymerization or depolymerization, regulation of cytoskeleton organization, regulation of organelle organization, regulation of protein complex disassembly | Cellular macromolecular complex subunit organization, homeostasis of no. of cells, macromolecular complex assembly, macromolecular complex subunit organization, protein complex assembly, protein complex biogenesis, proteolysis |
MF, molecular function; CC, cell component; BP, biology process.
TABLE 3.
KEGG pathway analysis of mouse ileum tissue proteins that interact with mouse or rat SFB FliC3a
| KEGG pathway term | mFliC3 |
rFliC3 |
||
|---|---|---|---|---|
| No. of genes | Benjamini P value | No. of genes | Benjamini P value | |
| Adherens junction | 9 | 0.025 | 7 | 0.120 |
| Alzheimer's disease | 20 | 0.000 | 13 | 0.071 |
| Antigen processing and presentation | 7 | 0.300 | UD | UD |
| Arginine and proline metabolism | 6 | 0.240 | 6 | 0.100 |
| Ascorbate and aldarate metabolism | UD | UD | 3 | 0.260 |
| Beta-alanine metabolism | UD | UD | 4 | 0.120 |
| Biosynthesis of unsaturated fatty acids | UD | UD | 4 | 0.190 |
| Butanoate metabolism | 4 | 0.450 | 6 | 0.065 |
| Cardiac muscle contraction | UD | UD | 7 | 0.130 |
| Citrate cycle (TCA cycle) | 4 | 0.360 | 5 | 0.085 |
| Dilated cardiomyopathy | UD | UD | 7 | 0.200 |
| Endocytosis | 10 | 0.450 | UD | UD |
| Fatty acid metabolism | 5 | 0.340 | 6 | 0.083 |
| Gap junction | 6 | 0.430 | UD | UD |
| Glutathione metabolism | UD | UD | 5 | 0.240 |
| Glycine, serine, and threonine metabolism | 4 | 0.370 | UD | UD |
| Histidine metabolism | 4 | 0.330 | UD | UD |
| Huntington's disease | 22 | 0.000 | 14 | 0.059 |
| Hypertrophic cardiomyopathy | UD | UD | 8 | 0.081 |
| Limonene and pinene degradation | UD | UD | 4 | 0.089 |
| Lipoic acid metabolism | 2 | 0.440 | UD | UD |
| Lysine degradation | UD | UD | 5 | 0.140 |
| Lysosome | UD | UD | 11 | 0.046 |
| NOD-like receptor signaling pathway | 5 | 0.430 | UD | UD |
| Oxidative phosphorylation | 16 | 0.000 | 11 | 0.071 |
| Parkinson's disease | 16 | 0.000 | 13 | 0.032 |
| Propanoate metabolism | 4 | 0.360 | 4 | 0.220 |
| Prostate cancer | 7 | 0.330 | UD | UD |
| Pyruvate metabolism | 5 | 0.310 | 6 | 0.068 |
| Ribosome | UD | UD | 8 | 0.088 |
| Spliceosome | 9 | 0.270 | 9 | 0.120 |
| TGF-β signaling pathway | UD | UD | 6 | 0.350 |
| Tight junction | 8 | 0.440 | 13 | 0.019 |
| Tryptophan metabolism | 6 | 0.091 | 6 | 0.069 |
| Valine, leucine, and isoleucine degradation | 5 | 0.310 | 6 | 0.077 |
| Viral myocarditis | UD | UD | 8 | 0.110 |
UD, undetected; TCA, tricarboxylic acid; TGF-β, transforming growth factor β.
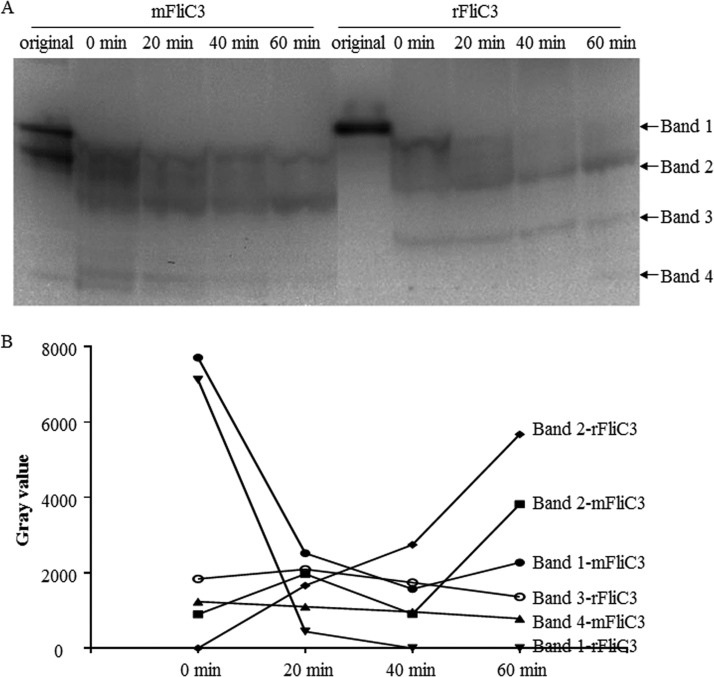
To compare the stabilities of mFliC3 and rFliC3 with mouse ileum tissue proteins, incubated samples were collected for immunoblot detection with anti-His antibodies. The initial concentrations were comparable for mFliC3 and rFliC3 (Fig. 7). Although degradation of both mFliC3 and rFliC3 was detected after mixing these proteins with mouse ileum tissue proteins, long-fragment band 1 of mFliC3 (corresponding to the predicted full length of mFliC3) was still detectable after a 60-min incubation. However, long-fragment band 1 of rFliC3 could not be detected after a 20-min incubation. The degradation rate of band 1 of rFliC3 was higher than that of mFliC3, and the short-fragment band increased more for rFliC3 (Fig. 7B). Similar trends were found when mFliC3 and rFliC3 were incubated with rat ileum tissue proteins. As illustrated in Fig. S10 in the supplemental material, long-fragment band 1 of rFliC3 was more stable than that of mFliC3 when incubated with rat ileum tissue proteins.
FIG 7.
Degradation of mFliC3 and rFliC3 with mouse ileum tissue proteins. (A) Purified mFliC3 and rFliC3 were interacted in vitro with mouse mucosal proteins. Interacted samples were collected at different time points and detected by using SDS-PAGE and immunoblotting with an anti-His antibody. (B) The concentrations of the protein bands were quantified by using ImageJ.
DISCUSSION
Although diversity among SFB 16S rRNA genes has been reported (6), and there are multiple copies of 16S rRNA genes within SFB genomes (19, 20), methods to evaluate the diversity of SFB strains have not been developed. Single-cell sequencing demonstrated that single nucleotide polymorphism (SNP) mutations exist within disparate SFB genomes isolated from the same host (23). Current technology for SFB culture and passage in vitro is relatively unrefined (13, 14). There are no simple single-cell sequencing methods available for detecting various SFB strains. Different strains may have various functions, providing a rationale for investigating the strain diversity of SFB. A connection between the functional diversity of SFB and different SFB strains could provide a foundation for further functional studies of SFB and the development of related products. In the present study, potential functional SFB flagellin genes (20) were selected to investigate their diversity. Clone library sequencing and alignment analysis demonstrated that the fliC1 and fliC2 genes are relatively conserved at both the nucleotide and deduced amino acid levels (see Tables S2 and S3 in the supplemental material). However, the sequences of the fliC3 and fliC4 genes are very diverse (Fig. 1 and Tables S2 and S3). High-throughput sequencing further verified the diversity of SFB fliC3 and fliC4 genes in mouse and rat samples, with larger variation being observed between rat samples. OTU2 is the dominant strain in the rat ileum sample SDI1, while OTU1, OTU2, OTU3, and OTU15 are the main strains in the rat ileum samples SDI2 and SDI3 (where 1, 2, and 3 in the designations represent samples from different rats). Because SFB can exist as either vegetative cells or spores, and the genomic DNA of spores is generally hard to extract, the diversity of SFB could be richer than we detected. There is only one copy of each of the FliC1 to FliC4 genes carried by each SFB genome (19); thus, the diversity of the FliC1 to FliC4 genes is reflected the diversity of SFB strains in each host. Previous studies indicated that flagellin gene diversity is related to flagellar function (24) and that SFB fliC2 to fliC4 can stimulate TLR5-linked NF-κB signaling (20). We compared the activation of SFB FliC3 in TLR5-linked NF-κB signaling. As the TLR5-binding region is highly conserved (Fig. S11), amino acid diversity should not affect activity against TLR5-linked NF-κB signaling (Fig. 6). However, other functions affected by gene diversity need further study. The highly variable region of SFB FliC3 was located mainly along the central and C-terminal regions. Although no specific function has been assigned to the central region, its deletion leads to a reduction in filament stability (25), and previous studies demonstrated that the central region of flagellin is the surface-exposed antigenic portion of flagellar filaments (24, 26). KEGG pathway analysis results also indicated that antigen processing- and presentation-related proteins were uniquely enriched with mFliC3, and similar results were observed uniquely for lysosome-related proteins with rFliC3 (Table 3). The amino acid differences between mFliC3 and rFliC3 are concentrated between the central and C-terminal regions (Fig. S11). The effects of the variations among SFB FliC3 orthologs on protein stability and immune regulation require additional investigation.
The prokaryotic flagellum is a motility organelle that is responsible for bacterial movement and is necessary for chemotaxis (27). Recently, a role for flagellum-mediated adherence was demonstrated for both pathogenic and opportunistic bacteria (e.g., Salmonella enterica, Campylobacter jejuni, Pseudomonas aeruginosa, and E. coli) in many different animal infection models (28). Potential roles for flagella in immune response regulation, including promoting Th17 cell differentiation and T cell balance, have also been reported (16–18). Full-genome sequencing has been used to determine that only one set of flagellin genes is carried by SFB genomes. Thus, functional studies of SFB flagellin may be useful for understanding the molecular mechanism of SFB colonization and immune response regulation. Unfortunately, there are currently no methods that allow SFB flagellin observation by microcopy, complicating research into these unique cellular structures. In this study, rabbit anit-mFliC3 polyclonal antibodies were prepared and used for FISH combined with IHC analysis (Fig. 4), immunoblotting, and LC-MS/MS (Fig. 5) to detect SFB flagellin expression. These experiments revealed that the fliC3 gene is expressed in the gut, in addition to other flagellar assembly-related proteins (Fig. S6). Confocal microscopy was used to observe the attachment of SFB flagellin to the mouse ileum epithelium (Fig. S5). Previous studies indicated that actin accumulation within the host cytoplasm is associated with SFB attachment to the mouse and rat ileum (29), and in this study, many actin-binding or polymerization-related proteins were enriched during pulldown with mFliC3 and rFliC3 (Table 2). It is reasonable to infer that SFB FliC3 may be related to SFB attachment and colonization.
Although cross-colonization experiments indicated that rat SFB cannot colonize the mouse ileum (9, 10) and that the SFB fliC3 genes are host specific, there was no specificity for actin binding with mFliC3 and rFliC3 (Table S6). This suggests that the host specificity of the SFB mFliC3 and rFliC3 proteins is more important for associations with other reported interaction proteins than for actin binding. The pulldown assays resulted in 209 enriched proteins that were shared for mFliC3 and rFliC3. In contrast, 278 and 273 proteins were specifically enriched with mFliC3 and rFliC3, respectively. The majority of the mFliC3-interacting proteins were related to binding, while most of the rFliC3-interacting proteins were related to lysosomal and proteolysis GO terms (Table 2). Eleven rFliC3-associated proteins were components of the lysosomal pathway. This is in contrast to mFliC3, which did not associate with lysosomal proteins (Table 3). Lysosomes contain a variety of enzymes that degrade biomolecules that are engulfed by the cell, including peptides, nucleic acids, carbohydrates, and lipids (30). Our in vitro degradation experiments verified that rFliC3 was degraded faster than mFliC3 with mouse ileum tissue proteins (Fig. 7). Rat flagellin could be more sensitive to degradation in the mouse ileum, which may explain why rat SFB cannot colonize the ilea of mice.
In conclusion, multiple distinct SFB strains were identified in several host organisms. The expression of SFB flagellin was detected in the mouse intestine. The host specificity of SFB flagellin did not affect functions related to the activation of TLR5-linked NF-κB signaling and actin binding. Interestingly, patterns of interaction with ileum mucosal proteins were different between mFliC3 and rFliC3; more proteins related to the degradation of exogenous macromolecules interacted with rFliC3. However, the functions of SFB flagellin related to colonization and immune response regulation require further elucidation using SFB isolates with microbial genetic methods and animal experiments.
MATERIALS AND METHODS
Animals and sample preparation.
Eight-week-old male ICR mice and Sprague-Dawley (SD) rats were obtained from the SLRC Laboratory Animal Center (Shanghai, China). For bacterial genome DNA and protein extraction, ileum mucosa at the distal ends of the small intestine (approximately 5 cm anterior to the ileocecal junction) and cecal contents were collected after animal sacrifice. All samples were stored at −80°C for further analysis. For FISH and IHC analysis, ileal tissue (approximately 5 cm anterior to the ileocecal junction) and cecal contents were collected and then fixed with 4% paraformaldehyde. All animals used in these experiments were handled in strict compliance with current regulations and guidelines concerning the use of laboratory animals in China. The procedures were approved by the Laboratory Animal Care and Usage Committee of Zhejiang University.
DNA extraction and PCR amplification.
Bacterial genomic DNA was extracted by using a QIAamp DNA stool minikit according to the manufacturer's instructions (Qiagen, Germany). The extracted DNA concentration was determined by using a NanoDrop ND-2000 instrument (NanoDrop Technologies, USA), and its integrity and size were confirmed by agar gel (1.0%) electrophoresis. To determine the quality of the DNA, primers 341F (5′-ATT ACC GCG GCT GCT GG-3′) and 534R (5′-CCT ACG GGA GGC AGC AG-3′) were used to amplify the V3 region of the bacterial 16S rRNA gene. The existence of SFB in these samples was detected by using SFB-specific PCR primers 779F (5′-TGT GGG TTG TGA ATA ACA AT-3′) and 1008R (5′-GCG GGC TTC CCT CAT TAC AAG G-3′) (31, 32). SFB-positive DNA samples were then used to detect SFB flagellin genes. SFB flagellin gene primers were designed by using Oligo 6 software, using the previously reported SFB flagellin sequences. The primers used are listed in Table 1. Standard PCR was performed with a total volume of 25 μl containing 2.5 μl of deoxynucleoside triphosphate (dNTP) (2.5 mmol), 2.5 μl of 10× Ex Taq buffer, 1 μl of each primer (10 μmol), 0.3 μl of Ex Taq polymerase (5 U/μl; TaKaRa), and 100 ng of prepared nucleic acids. The reaction program was performed with a DNA thermal cycler (Bio-Rad) as follows: 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 45 s. This was followed by incubation at 72°C for 5 min. Amplification products were separated by using Tris-acetate-EDTA (TAE)-agarose gel (1.0%) electrophoresis.
Cloning and sequencing.
PCR amplification products were purified by using a Biospin gel extraction kit (Bioer Technology Co., Ltd.) and were then cloned into the pMD18-T vector (TaKaRa Biotechnology [Dalian] Co., Ltd., China) according to the manufacturer's instructions, ultimately producing a shotgun library. Clones from each library were selected randomly, and sequencing with primers M13F and M13R was performed by Sangon Biotech (Shanghai, China). Ten high-quality sequences were obtained per library.
Similarity and phylogenetic analyses of SFB fliC gene and 16S rRNA gene sequences.
Totals of 49 fliC1, 61 fliC2, 66 fliC3, and 51 fliC4 gene sequences were obtained from mouse samples, and totals of 66 fliC1, 52 fliC3, 42 fliC4, and 65 16S rRNA gene sequences were obtained from rat samples. Similarities at the nucleotide and deduced amino acid levels were analyzed by using the MegAlign program included in the DNAStar software package. A phylogenetic tree was constructed by using the MEGA 6 neighbor-joining algorithm.
Primers specifically for SFB fliC3 and fliC4 gene amplification.
A new pair of primers, fliC3/4 F (5′-CAC AAT ATG AAT GCG ATG AAT G-3) and fliC3/4 R (5-GCT GTT TGA ATC ATT GAA AT-3′), was designed according to the conserved region in the SFB fliC3 and fliC4 gene sequences that were obtained in this study. The specificity of these primers was verified by using high-throughput sequencing.
Heterologous expression, extraction, and purification of the SFB FliC3 protein.
Mouse and rat SFB fliC3 genes were subcloned into the pET-28a vector by using double-digestion and ligation methods. FliC3 proteins were overexpressed in chemically competent BL21(DE3) cells [F− ompT hsdS(rB− mB−) gal dcm (DE3); Transgen Biotech, Beijing, China] by adding IPTG as an inducer. Total bacterial proteins were extracted by using BugBuster master mix (Merck Millipore, Germany). SFB FliC3 proteins were purified by using a His Bind purification kit (Merck Millipore, Germany). The protein concentration in purified samples was determined by using a bicinchoninic acid (BCA) protein assay kit (Sangon Biotech, China), and the SFB FliC3 proteins were then stored at −80°C for further analysis.
Preparation of anti-FliC3 polyclonal antibody.
An anti-FliC3 polyclonal antibody was prepared by injecting mFliC3 proteins into rabbits, which were raised by Abgent Biotechnology Co., Ltd. (Suzhou, China). After inoculation, rabbit serum was collected for affinity purification and polyclonal antibody generation.
FISH and IHC analysis.
For cecal contents, samples were washed three times by using phosphate-buffered saline (PBS). Pellets were collected and then centrifuged at 12,000 × g for 3 min. For ileum tissue samples, the ileum tissue was suspended in PBS and then fixed in a 4% paraformaldehyde solution. Fixed cecal bacteria were spotted onto glass slides and allowed to air dry for 1 h. Fixed ileum tissue samples were prepared as paraffin or frozen sections. A 20-μl aliquot of hybridization solution (0.9 M NaCl, 20 mM Tris-HCl, 0.01% SDS [pH 7.2]) containing 5 ng of a Cy3-labeled SFB-specific oligonucleotide probe (GGG TAC TTA TTG CGT TTG CGA CGG CAC) (32) was spotted onto the slides and incubated overnight at 50°C. After the slides had been immersed in washing solution (0.9 M NaCl, 20 mM Tris-HCl, 0.01% SDS [pH 7.2]) at 48°C for 15 min, the slides were briefly rinsed with sterile water. For immunostaining, bacteria were incubated sequentially with a primary antibody (1:200 mix of the rabbit anti-mFliC3 or mouse anti-muc2 antibody, catalog no. MA5-12345; Thermo Fisher Scientific) and a secondary antibody [fluorescein isothiocyanate (FITC) goat anti-rabbit IgG(H+L) or donkey anti-mouse IgG(H+L) antibody]. 4′,6-Diamidino-2-phenylindole (DAPI) was then used to stain nuclei for 15 min. After washing with 80% ethanol and sterilized water, air-dried bacterial cells were observed and photographed by using a DM2500 fluorescence microscope (Leica, Germany). The ileum tissue samples were also observed by using a two-photon confocal microscope (LSM 800; Zeiss, Germany).
SDS-PAGE and immunoblotting.
The extracted protein was mixed with 2× sample loading buffer, analyzed by using SDS-PAGE, and then transferred to a polyvinylidene difluoride (PVDF) membrane for immunodetection. Anti-FliC3 antibody (1:1,000 dilution) was used as the primary antibody, while a goat anti-rabbit IgG(H+L) antibody that was conjugated with horseradish peroxidase (1:5,000; Abcam, USA) was used as the secondary antibody. Finally, the proteins were visualized by using enhanced chemiluminescence detection reagents (Pierce, USA).
Pulldown assay.
Mouse ileum tissue proteins were extracted by using a protein lysis solution (Pierce, USA) that was supplemented with a protease inhibitor (Roche, USA). The interaction between SFB FliC3 and mouse tissue proteins was detected by using a His protein interaction pulldown kit (Pierce). A 16-tube SureBeads magnetic rack (Bio-Rad) was used for interactive protein collection.
LC-MS/MS.
The isolated interacting proteins and trypsin-digested protein peptides were sent for LC-MS/MS analysis at Zhejiang University (Hangzhou, China) and Bio-Fly Bioscience Co. (Beijing, China). A mass spectrometer (LTQ Orbitrap Elite; Thermo Fisher) and SEQUEST software were used in this study.
Detection of activation of Toll-like receptor 5-linked NF-κB signaling.
The function of purified mouse and rat SFB FliC3 proteins on Toll-like receptor 5-linked NF-κB signaling was determined by a luciferase reporter assay system as described previously (20). Briefly, HeLa cells were transformed with a pGMNF-κB luciferase reporter plasmid (Qcbio Science & Technologies Co., Ltd., Shanghai, China) and pGMLR-TK (Qcbio Science & Technologies Co., Ltd., Shanghai, China), in addition to the mouse or rat TLR5-encoding pCDNA3.1 vector or the empty vector. Twenty hours after transfection, the cells were stimulated with 1 or 10 μg of purified recombinant mouse or rat SFB flagellin FliC3 for 24 h. After stimulation, the cells were harvested, and the luciferase activities in the cell lysates were measured by using a dual-luciferase reporter assay system (Promega), using a multifunctional microplate reader (SpectraMax M5; Molecular Devices, USA). Luciferase activity was expressed as the ratio of NF-κB-dependent firefly luciferase activity to the control Renilla luciferase activity (relative luciferase units). These data were further statistically analyzed by using SPSS 20.0 software.
Degradation of mFliC3 and rFliC3 in vitro with mouse ileum tissue proteins.
Purified mFliC3 and rFliC3 were incubated with mouse and rat mucosal proteins, respectively, on ice. Interacted samples were collected at different time points and detected by using SDS-PAGE and immunoblotting with anti-His antibodies.
Bioinformatic analysis.
Mouse ileum tissue proteins that were isolated during pulldown with mouse or rat SFB FliC3 were recognized as enriched proteins if the fold change was >2 in comparison to the control. The enriched proteins were then used for GO and KEGG pathway analyses using DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/).
Accession number(s).
The clone sequencing data in the present study were deposited in the National Center for Biotechnology Information (NCBI) database under accession no. KX658686 to KX659137. The sequence data have been deposited in the NCBI SRA database under accession no. SRR5341187 to SRR5341192.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (973 Program; no. 2013CB531404), the National High Technology Research and Development Program of China (863 Program; no. 2015AA020701), the National Nature Science Foundation (NSFC) (no. 31370156), and the State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (no. 2010DS700124-ZZ1604).
We thank Weizhong Gu from Zhejiang University for help in the IHC staining. We thank LetPub for its linguistic assistance during the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01061-17.
REFERENCES
- 1.Ericsson AC, Hagan CE, Davis DJ, Franklin CL. 2014. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med 64:90–98. [PMC free article] [PubMed] [Google Scholar]
- 2.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. 2013. Host interactions with segmented filamentous bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin Immunol 25:342–351. doi: 10.1016/j.smim.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. 2014. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klaasen HL, Koopman JP, Poelma FG, Beynen AC. 1992. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev 8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 6.Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, Zhu B, Yu HD, Xiang C, Wang X. 2013. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonsson H. 2013. Segmented filamentous bacteria in human ileostomy samples after high-fiber intake. FEMS Microbiol Lett 342:24–29. doi: 10.1111/1574-6968.12103. [DOI] [PubMed] [Google Scholar]
- 8.Caselli M, Tosini D, Gafa R, Gasbarrini A, Lanza G. 2013. Segmented filamentous bacteria-like organisms in histological slides of ileo-cecal valves in patients with ulcerative colitis. Am J Gastroenterol 108:860–861. doi: 10.1038/ajg.2013.61. [DOI] [PubMed] [Google Scholar]
- 9.Tannock GW, Miller JR, Savage DC. 1984. Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl Environ Microbiol 47:441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K. 2015. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR. 2014. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geem D, Medina-Contreras O, McBride M, Newberry RD, Koni PA, Denning TL. 2014. Specific microbiota-induced intestinal Th17 differentiation requires MHC class II but not GALT and mesenteric lymph nodes. J Immunol 193:431–438. doi: 10.4049/jimmunol.1303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ericsson AC, Turner G, Montoya L, Wolfe A, Meeker S, Hsu C, Maggio-Price L, Franklin CL. 2015. Isolation of segmented filamentous bacteria from complex gut microbiota. Biotechniques 59:94–98. doi: 10.2144/000114319. [DOI] [PubMed] [Google Scholar]
- 14.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. 2015. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. 2015. Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog 11:e1004483. doi: 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, Kiyono H, Miyasaka M, Ishii KJ, Akira S. 2008. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 17.Van Maele L, Carnoy C, Cayet D, Songhet P, Dumoutier L, Ferrero I, Janot L, Erard F, Bertout J, Leger H, Sebbane F, Benecke A, Renauld JC, Hardt WD, Ryffel B, Sirard JC. 2010. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J Immunol 185:1177–1185. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uematsu S, Akira S. 2009. Immune responses of TLR5(+) lamina propria dendritic cells in enterobacterial infection. J Gastroenterol 44:803–811. doi: 10.1007/s00535-009-0094-y. [DOI] [PubMed] [Google Scholar]
- 19.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, Taylor TD, Ohno H, Umesaki Y, Hattori M. 2011. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe 10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, Ishifune C, Maekawa Y, Yasutomo K, Hattori M, Hayashi T. 2011. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res 18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyerholz DK, Stabel TJ, Cheville NF. 2002. Segmented filamentous bacteria interact with intraepithelial mononuclear cells. Infect Immun 70:3277–3280. doi: 10.1128/IAI.70.6.3277-3280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi KE, Snel J. 2000. Transmission electron microscopic demonstration of phagocytosis and intracellular processing of segmented filamentous bacteria by intestinal epithelial cells of the chick ileum. Infect Immun 68:6496–6504. doi: 10.1128/IAI.68.11.6496-6504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. 2012. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res 22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid SD, Selander RK, Whittam TS. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol 181:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muskotal A, Seregelyes C, Sebestyen A, Vonderviszt F. 2010. Structural basis for stabilization of the hypervariable D3 domain of Salmonella flagellin upon filament formation. J Mol Biol 403:607–615. doi: 10.1016/j.jmb.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Winstanley C, Morgan JA. 1997. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology 143(Part 10):3071–3084. doi: 10.1099/00221287-143-10-3071. [DOI] [PubMed] [Google Scholar]
- 27.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haiko J, Westerlund-Wikstrom B. 2013. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jepson MA, Clark MA, Simmons NL, Hirst BH. 1993. Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infect Immun 61:4001–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Settembre C, Fraldi A, Medina DL, Ballabio A. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int J Syst Bacteriol 45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 32.Urdaci MC, Regnault B, Grimont PA. 2001. Identification by in situ hybridization of segmented filamentous bacteria in the intestine of diarrheic rainbow trout (Oncorhynchus mykiss). Res Microbiol 152:67–73. doi: 10.1016/S0923-2508(00)01169-4. [DOI] [PubMed] [Google Scholar]
- 33.Roca AI. 2014. ProfileGrids: a sequence alignment visualization paradigm that avoids the limitations of sequence logos. BMC Proc 8:S6. doi: 10.1186/1753-6561-8-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.