Karmaus et al. show that loss of mechanistic target of rapamycin complex 1 (mTORC1) signaling impairs myelopoiesis. M-CSF–dependent myelopoiesis requires mTORC1 signaling and anabolic metabolism, which in turn promote expression of M-CSFR and transcription factors PU.1 and IRF8, thereby constituting a feed-forward loop for myelopoiesis.
Abstract
Myelopoiesis is necessary for the generation of mature myeloid cells during homeostatic turnover and immunological insults; however, the metabolic requirements for this process remain poorly defined. Here, we demonstrate that myelopoiesis, including monocyte and macrophage differentiation, requires mechanistic target of rapamycin complex 1 (mTORC1) signaling and anabolic metabolism. Loss of mTORC1 impaired myelopoiesis under steady state and dampened innate immune responses against Listeria monocytogenes infection. Stimulation of hematopoietic progenitors with macrophage colony-stimulating factor (M-CSF) resulted in mTORC1-dependent anabolic metabolism, which in turn promoted expression of M-CSF receptor and transcription factors PU.1 and IRF8, thereby constituting a feed-forward loop for myelopoiesis. Mechanistically, mTORC1 engaged glucose metabolism and initiated a transcriptional program involving Myc activation and sterol biosynthesis after M-CSF stimulation. Perturbation of glucose metabolism or disruption of Myc function or sterol biosynthesis impaired myeloid differentiation. Integrative metabolomic and genomic profiling further identified one-carbon metabolism as a central node in mTORC1-dependent myelopoiesis. Therefore, the interplay between mTORC1 signaling and metabolic reprogramming underlies M-CSF–induced myelopoiesis.
Introduction
Myeloid cells, including monocytes, neutrophils, and eosinophils, make up the majority of blood leukocytes, yet are among the cells with the shortest life spans in the body (Ginhoux and Jung, 2014; Manz and Boettcher, 2014; Kotzin et al., 2016). The generation of mature myeloid cells during myelopoiesis requires sequential progression from hematopoietic stem cells (HSCs) to precursor populations before terminal differentiation. The rate of progression increases during immunological insults to meet the demand for greater myeloid cell numbers (Manz and Boettcher, 2014; Varol et al., 2015). For example, in response to Listeria monocytogenes infection, inflammatory monocytes are generated from BM precursors and play crucial roles in clearance of bacterial infection (Shi and Pamer, 2011). The generation of myeloid cells during hematopoiesis requires myelopoietic cytokines, including G-CSF, M-CSF, and GM-CSF (Ginhoux and Jung, 2014; Manz and Boettcher, 2014), which are up-regulated in infection, inflammation, and cancer (Hamilton, 2008). In addition, Toll-like receptor–mediated signaling in myeloid progenitors stimulates myelopoiesis in response to pathogens (Nagai et al., 2006). M-CSF (encoded by Csf1) and M-CSF receptor (M-CSFR, also known as CD115; encoded by Csf1r) are particularly important for the development of the monocytic lineage, as indicated by the loss of monocytes and macrophages in Csf1r−/− (Dai et al., 2002) and Csf1op/op mice (Wiktor-Jedrzejczak et al., 1990; Yoshida et al., 1990). In addition, monocytes and macrophages share a committed myeloid progenitor, which is distinct from dendritic cells and other myeloid cells (Hettinger et al., 2013). In summary, M-CSF–mediated myelopoiesis induces differentiation of the monocytic lineage from BM precursors.
The differentiation of hematopoietic progenitors into mature myeloid cells is contingent on the activation of gene expression programs under the control of lineage-defining transcription factors (Orkin and Zon, 2008; Moignard et al., 2013). In particular, PU.1 is essential for the development of the monocytic lineage. High PU.1 expression levels relative to other lineage-defining transcription factors support monocytic lineage development (DeKoter and Singh, 2000; Nutt et al., 2005), and loss of PU.1 abrogates common myeloid progenitor (CMP) and granulocyte-macrophage progenitor (GMP) differentiation but spares megakaryocyte-erythroid progenitors (Scott et al., 1994; Dakic et al., 2005; Iwasaki et al., 2005). PU.1 functions in part by forming a heterodimer with interferon regulatory factor 8 (IRF8), another critical transcription factor for myelopoiesis (Kurotaki et al., 2014). Both PU.1 and IRF8 bind to the M-CSFR promoter to drive gene transcription (Kurotaki et al., 2014; Satoh et al., 2014). Moreover, Krüppel-like factor 4 (KLF4) can partially rescue monocyte differentiation in the absence of IRF8 (Kurotaki et al., 2013). Despite our knowledge of the roles of cytokines and transcription factors in myelopoiesis, mechanisms connecting extrinsic signals to transcriptional responses and cell fate decisions remain poorly defined.
Emerging studies highlight the critical roles of metabolic reprogramming in innate and adaptive immunity. Studies on the metabolic regulation of myeloid cells are largely restricted to innate immune responses (O’Neill and Pearce, 2016) and myeloid leukemia (Galluzzi et al., 2013), whereas little is known about the metabolic processes driving nonmalignant myelopoiesis. One common denominator among normal myelopoiesis and leukemic and other pathological conditions is the preference for glucose as a fuel source (Akers et al., 2011; Nagareddy et al., 2013; Sarrazy et al., 2016). Further, leukemia cells and hematopoietic progenitors are sensitive to perturbations in aerobic glycolysis, whereas HSCs are less sensitive to such stress (Wang et al., 2014). Among the regulators of immune and cancer metabolism is signaling via mechanistic target of rapamycin (mTOR), a serine/threonine protein kinase that controls multiple cellular processes including protein translation, cell growth, and metabolism. mTOR forms two complexes of discrete functions, which are defined by the obligate adapter proteins Raptor (encoded by Rptor; mTOR complex 1; mTORC1) and Rictor (encoded by Rictor; mTORC2; Laplante and Sabatini, 2012). The mTOR pathway coordinates immune signaling and cell metabolism to determine cell fate decisions in immune responses (Chi, 2012), although how immune signals are sensed and integrated to drive metabolic changes for cell fate decisions during myelopoiesis is poorly understood.
In this study, we show that mTORC1 signaling, but not mTORC2, is a crucial positive determinant of myelopoiesis during infection of L. monocytogenes, under homeostatic conditions and after M-CSF stimulation. M-CSF stimulation activates mTORC1, which engages a positive feed-forward loop by promoting glucose uptake and anabolic metabolism to subsequently up-regulate expression of M-CSFR in developing myeloid cells. We further identify Myc and sterol biosynthesis as mTORC1 downstream effector pathways important for metabolic reprogramming and M-CSFR–dependent functions during myelopoiesis. Finally, integrative profiling of the metabolome and transcriptome reveals one-carbon metabolism as a critical node of mTORC1-dependent myeloid differentiation. These results provide novel insights into how metabolic reprogramming of M-CSF–instructed precursor cells drives myelopoiesis to mediate antibacterial immunity and homeostatic control of hematopoiesis.
Results
Deletion of Rptor but not Rictor depletes myeloid cells and impairs host resistance to L. monocytogenes
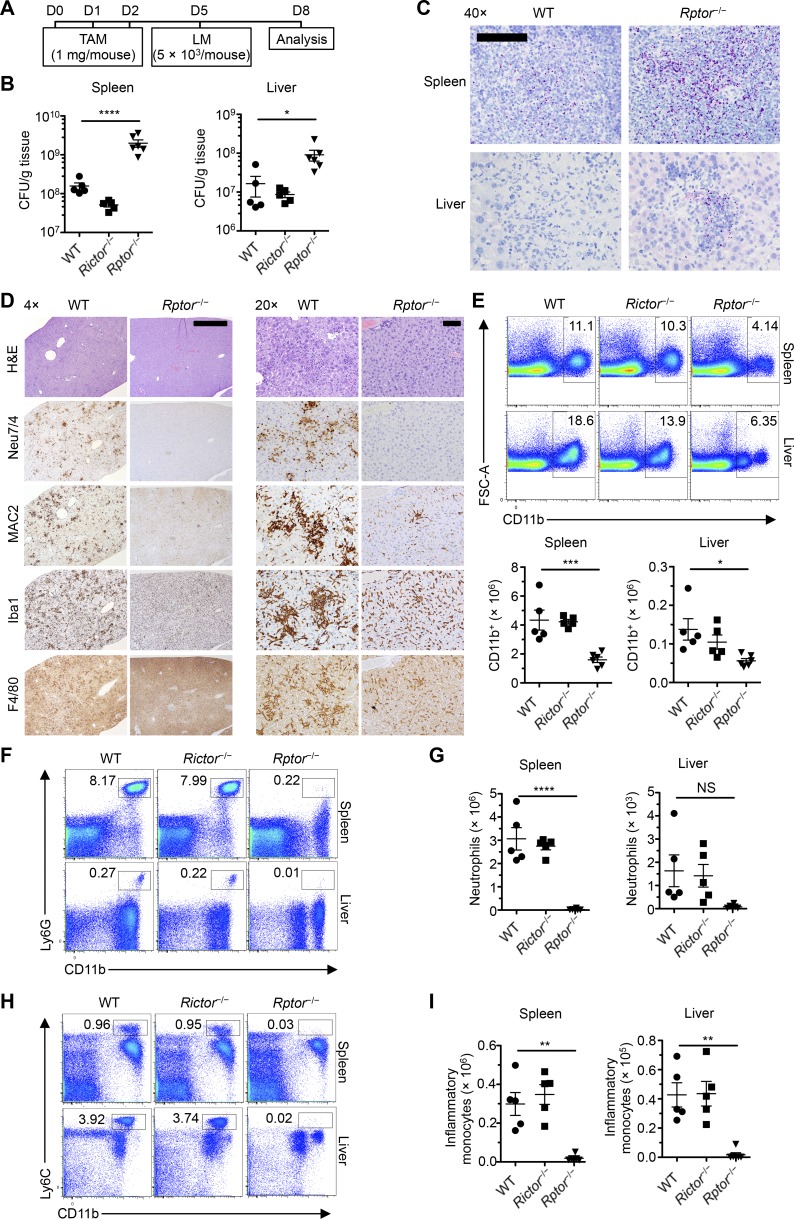
To investigate the roles of mTORC1 and mTORC2 in myeloid cell development, we used mice harboring loxP-flanked alleles of Rptor and Rictor, respectively. We were unable to obtain viable mice when Rptor was constitutively deleted in hematopoietic cells via the Vav-icre system (de Boer et al., 2003). To overcome such early lethality, we developed inducible deletion systems by breeding Rptorfl/fl and Rictorfl/fl alleles with Rosa26-CreERT2 mice expressing a Cre-ERT2 fusion gene in the ubiquitously expressed Rosa26 locus (called Rptor−/− and Rictor−/− mice, respectively). To bypass any potential effects of nonhematopoietic loss of mTOR signaling, we used BM cells from these mice or Rosa26-CreERT2 control mice (called WT mice) to reconstitute lethally irradiated recipient mice and generated chimeras, wherein administration of tamoxifen induced acute deletion of Rptor or Rictor in hematopoietic cells selectively. At day 5 after initial tamoxifen treatment, we challenged WT, Rptor−/−, and Rictor−/− BM chimeras with L. monocytogenes to determine whether loss of either of these genes affected the antibacterial immune response (Fig. 1 A). Rptor−/−, but not Rictor−/−, BM chimeras had markedly increased bacterial burden versus WT BM chimeras in the spleen and liver, the primary target organs of L. monocytogenes infection (Fig. 1 B), which was further verified by immunohistochemical detection of L. monocytogenes (Fig. 1 C). Thus, Raptor deficiency impairs antibacterial immunity.
Figure 1.
Hematopoietic Rptor ablation renders mice susceptible to L. monocytogenes infection. (A) Scheme of experimental design of tamoxifen (TAM) treatment and L. monocytogenes infection. (B) Total colony-forming units of L. monocytogenes per gram of tissue in WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). (C) Immunohistochemistry for L. monocytogenes antigen in the spleen and liver tissues from WT and Rptor−/− mice (n = 4–5 mice per group). Bar, 0.1 mm. (D) Hematoxylin and eosin staining for general pathology and immunohistochemical detection of neutrophils (Neu7/4) and macrophages (MAC2, Iba1, and F4/80) in the liver of WT and Rptor−/− mice (n = 4–5 mice per group). Bars: (left) 1.0 mm; (right) 400 µm. (E) Flow cytometry analysis and numbers of CD11b+ myeloid cells in the spleen and liver of WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). (F) Flow cytometry analysis of neutrophils (CD11b+Ly6G+) in spleen and liver of WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). (G) Number of neutrophils in spleen and liver of WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). (H) Flow cytometry analysis of inflammatory monocytes (CD11b+Ly6C+) in spleen and liver of WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). (I) Number of inflammatory monocytes in spleen and liver of WT, Rictor−/−, and Rptor−/− mice (n = 5 mice per group). Numbers indicate percentages of cells in gates. Data are mean ± SEM and representative of four (E–I) or two (B–D) independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant; one-way ANOVA with Dunnet’s post hoc test.
Detailed histological analysis also revealed that the liver of WT chimeras contained widespread inflammation characterized by myriad coalescing microgranulomas composed of clusters of neutrophils (positive for Neu7/4) and macrophages (positive for MAC2, Iba1, and F4/80), and often associated with apoptotic hepatocytes (Fig. 1 D), but Listeria bacteria and antigens were not detected in any of these lesions. In contrast, the liver of Rptor−/− mice contained much fewer inflammatory foci, which were typically composed of smaller numbers of macrophages and neutrophils and sometimes included numerous intact Listeria organisms. In addition, although there was a diffuse increase in neutrophils both in periportal areas and diffusely in sinusoids of WT mice, there were only rare neutrophils present in hepatic sinusoids and blood vessels from Rptor−/− mice (Fig. 1 D).
Consistent with the histological analysis, Rptor−/− mice had markedly reduced percentages and numbers of CD11b+ myeloid cells in the spleen and liver (Fig. 1 E). To obtain an unbiased view of the CD11b+ population in these organs, we performed t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction on the flow cytometry data. Of total CD11b+ cells, neutrophils (Ly6G+) and inflammatory monocytes (Ly6C+) were markedly reduced in Rptor-deleted mice (Fig. S1, A and B). Detailed flow cytometry analysis verified the tSNE dimensionality reduction and showed markedly reduced neutrophils (CD11b+Ly6G+; Fig. 1, F and G) and inflammatory monocytes (CD11b+Ly6C+; Fig. 1, H and I) in these organs. Because of the drastic decreases in monocytes, we also analyzed their precursor populations in the BM (see gating scheme in Fig. S1 C according to Hettinger et al. [2013] and Chong et al. [2016]). There were drastic decreases in total monocytes, especially the CXCR4+ (premonocytes) compartment, as well as common monocyte progenitors (cMoPs; Fig. S1 D), whereas monocyte-macrophage DC progenitors (MDPs) showed a small increase, probably as a result of the blocked differentiation of these progeny populations (Fig. S1 D). Thus, Raptor is important for host defense against L. monocytogenes, and its deficiency disrupts BM monocyte development and prevents accumulation of myeloid cells at sites of infection.
We next analyzed the myeloid cell compartment in the spleen after tamoxifen treatment of WT, Rptor−/−, and Rictor−/− mice under homeostatic conditions. At day 10 after tamoxifen treatment, Raptor-deficient mice showed reduction of myeloid populations compared with WT mice (Fig. S1, E and F), with the loss of such cells varying in the different lineages. Specifically, Rptor ablation reduced monocytes (CD11b+CD115+) by ∼90%, with a preferential reduction of the Ly6Chi subset (Fig. S1 E), and neutrophils were largely depleted in Rptor−/− mice (Fig. S1 F). To determine the kinetics by which myeloid cells disappeared in the absence of Rptor, we analyzed myeloid populations in the peripheral blood of WT or Rptor−/− BM chimeras at different time points after tamoxifen treatment (Fig. S1 G). Monocytes and neutrophils rapidly disappeared in the blood of Rptor−/− mice and became essentially undetectable at days 5 and 7 after tamoxifen treatment, whereas T and B cells were largely maintained in the time points examined (Fig. S1 H). At day 4 after the onset of tamoxifen treatment, monocytes and neutrophils were modestly reduced in Rptor−/− mice compared with WT mice (Fig. S1 I), so we measured mTORC1 activity in these cells. The phosphorylated forms of the canonical mTORC1 downstream effectors, 4E-BP1 (p-4E-BP1) and S6 (p-S6), were markedly curtailed in monocytes and neutrophils (Fig. S1 J). The mTORC1 activity in B and T cells was much lower than that in CD11b+ cells, and thus basal mTORC1 activity was not altered by Raptor deficiency (Fig. S1 K). These results indicate that Raptor and mTORC1 signaling are required to maintain peripheral myeloid populations under inflammatory and homeostatic conditions.
Diminished response to hematopoietic growth factors in the absence of Raptor
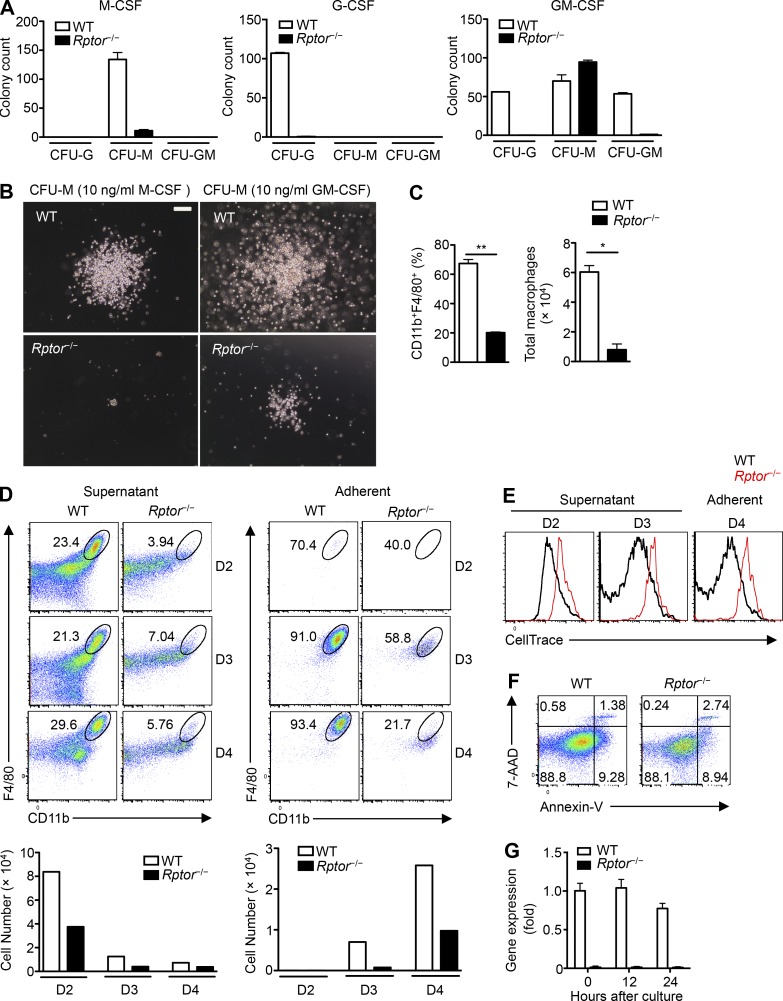
Although Rptor deletion impairs myeloid cells in vivo, it remained unclear whether these effects are caused by reduced myelopoiesis from precursors. We therefore examined the ability of BM cells from WT and Rptor−/− mice to generate CFU in response to different myeloid growth factors: M-CSF, G-CSF, and GM-CSF. Compared with BM cells from WT mice, Rptor−/− cells had severely impaired CFU generation in response to stimulation with M-CSF (CFU-M) and G-CSF (CFU-G; Fig. 2 A). In addition to fewer colonies, the colony size of Rptor−/− M-CSF–derived CFU-M was greatly decreased in comparison to WT controls (Fig. 2 B). In response to GM-CSF stimulation, WT BM cells formed discrete CFU-G, CFU-M, and CFU-GM colonies, whereas Rptor−/− BM cells formed only CFU-M colonies (Fig. 2 A), which were also smaller in size compared with WT controls (Fig. 2 B).
Figure 2.
Impaired M-CSF–induced myeloid development after Raptor deletion. (A) Number of colony-forming units (CFU-G, CFU-M, or CFU-GM) for BM cells of WT and Rptor−/− mice incubated with M-CSF, G-CSF, or GM-CSF in methylcellulose. (B) Representative images of CFU-M 7 d after total BM cell culture from WT and Rptor−/− mice. Bar, 100 µm. (C) Frequency (left) and number (right) of macrophages (CD11b+F4/80+) after liquid culture of WT and Rptor−/− BM cells with M-CSF (10 ng/ml) for 5 d. (D–F) Lin– BM cells from WT and Rptor−/− mice were under liquid culture with M-CSF (10 ng/ml) for indicated times. (D) Flow cytometry analysis of F4/80 and CD11b (top) and number (bottom) of CD11b+F4/80+ of supernatant (left) and adherent cell fractions (right) at day 2 (D2), D3, or D4. (E) CellTrace dilution of supernatant cell fraction (left and middle) at D2 and D3 and adherent cell fraction (right) at D4. (F) Flow cytometry analysis of 7-AAD and Annexin-V (n = 1 or 2 mice per group) of Lin– BM cells from WT and Rptor−/− mice from liquid culture with M-CSF (10 ng/ml) at day 2. (G) Gene expression of Rptor in Lin– BM cells stimulated with M-CSF (10 ng/ml) for the indicated times. Numbers indicate percentages of cells in gates or quadrants. Data are mean ± SEM and representative of four (A–E) or two (F and G) independent experiments. *, P < 0.05; **, P < 0.01; Student’s t test for parametric data or Mann–Whitney test for nonparametric data.
Under in vitro liquid cultures, BM cells respond to M-CSF by differentiating along the monocytic lineage, culminating in the generation of macrophages (CD11b+F4/80+). Consistent with the CFU findings, Rptor−/− BM cells were impaired in the generation of CD11b+F4/80+ macrophages in response to M-CSF (Fig. 2 C). To circumvent any potential contributions of existing myeloid populations in the BM, we purified Lin– cells from WT and Rptor−/− BM and cultured them with M-CSF in vitro. The generation of macrophages from Rptor−/− Lin– progenitors was reduced throughout the culture period, as indicated by lower percentages and numbers of CD11b+F4/80+ cells (Fig. 2 D). Moreover, this effect was observed in both the adherent and suspension cell fractions, excluding a possible involvement of adhesion in this process (Fig. 2 D). The defect was associated with impaired proliferation of Rptor−/− cells, as measured by CellTrace dilution (Fig. 2 E), but not excessive cell death, as measured by Annexin-V/7-AAD staining (Fig. 2 F). Throughout the culture conditions, Rptor deletion was complete in Rptor−/− cells (Fig. 2 G). These results indicate that Raptor is required for M-CSF–mediated differentiation of the monocyte-macrophage lineage.
Raptor is necessary for M-CSFR (CD115) expression
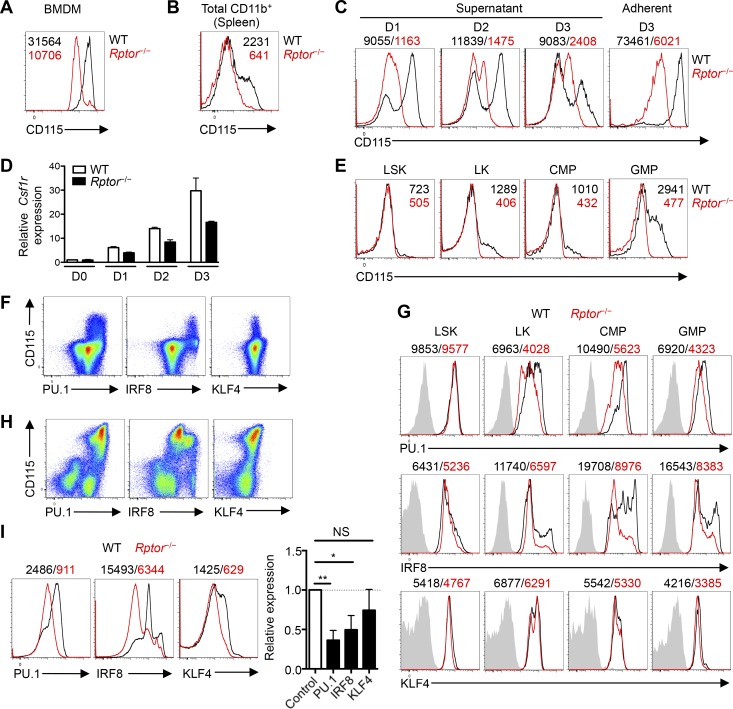
M-CSFR expression is crucial to mediate M-CSF–induced myeloid differentiation (Stanley and Chitu, 2014). We therefore analyzed CD115 expression on myeloid cells from different sources in WT and Rptor−/− mice. Macrophages generated from WT BM cells expressed approximately threefold higher levels of CD115 than Raptor-deficient cells (Fig. 3 A), and CD115 expression was practically absent in CD11b+ cells from the spleen of Rptor−/− mice in vivo (Fig. 3 B). Also, the expression of CD115 within CD11b+CD115+ monocytes was reduced on a per-cell basis in Rptor−/− mice at an early time after tamoxifen treatment (day 4; Fig. S2 A). Further, after M-CSF stimulation in vitro, Lin– cells from Rptor−/− mice showed markedly impaired ability to up-regulate surface CD115 expression compared with Lin– cells from WT mice, and the defects persisted at later time points (Fig. 3 C). Csf1r mRNA expression was also reduced in M-CSF–stimulated Lin– cells from Rptor−/− mice (Fig. 3 D). Hence, Raptor is required for CD115 expression on mature and developing myeloid cells.
Figure 3.
M-CSFR expression is reduced after Raptor deletion in developing myeloid precursors. (A) Expression of CD115 on WT and Rptor−/− macrophages after liquid culture of BM cells with M-CSF (10 ng/ml) for 5 d, with mean fluorescence intensity (MFI) plotted within graph. (B) Expression of CD115 on CD11b+ cells in the spleen of WT and Rptor−/− mice. (C) Expression of CD115 on supernatant and adherent cell fractions of WT and Rptor−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 1, 2, or 3 d, with MFI plotted above graphs. (D) Analysis of Csf1r mRNA in WT and Rptor−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 0, 1, 2, and 3 d. (E) Expression of CD115 on WT and Rptor−/− myeloid progenitor cell populations, with MFI plotted within graphs. (F) Flow cytometry analysis of CD115 together with PU.1, IRF8, or KLF4 in WT Lin– BM cells. (G) Expression of PU.1, IRF8, or KLF4 in various myeloid progenitor cell populations of WT and Rptor−/− mice, with MFI plotted above graphs. (H) Flow cytometry analysis of PU.1, IRF8, or KLF4 with CD115 in WT Lin– BM cells 2 d after liquid culture with M-CSF (10 ng/ml). (I) Flow cytometry analysis of PU.1, IRF8, or KLF4 in WT Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 2 d. Right, summary plot of PU.1, IRF8, or KLF4 in Rptor−/− cells with expression relative to WT controls. Data are mean ± SEM and representative of four (A and C), three (B and F–I), two (D), or six (E) independent experiments. *, P < 0.05; **, P < 0.01; NS, not significant; Student’s t test.
We next determined the effect of Rptor deletion on the homeostasis of hematopoietic progenitors and the expression of CD115, which is known to be progressively up-regulated during myelopoiesis (Stanley and Chitu, 2014). The frequencies of various progenitor populations (within the CD127– fraction) such as Lin–Sca-1+c-Kit+ (LSK) stem cells, Lin–c-Kit+ cells (LK; myeloid progenitors), CMPs (Lin–Sca-1–c-Kit+CD34+FcγRII/IIImid), and GMPs (Lin–Sca-1–c-Kit+CD34+FcγRII/IIIhigh; see Fig. S2 B for gating schemes) were largely comparable between WT and Rptor−/− mice, with the exception of a modest increase in the CMP population in Rptor−/− mice (Fig. S2 B), which likely reflects a blocked differentiation into the downstream granulocyte-macrophage lineage. We also determined the monocyte precursor populations under homeostasis (Fig. S2 C) and found trends similar to those we observed in mice infected with L. monocytogenes. We next used BrdU incorporation and active caspase-3 staining to measure cell proliferation and death, respectively. CMP cells from Rptor−/− mice had a small decrease in proliferation, whereas other precursor populations showed a normal extent of BrdU incorporation (Fig. S2 D). Also, apoptotic cell death was unchanged in cells from Rptor−/− mice (Fig. S2 E). Consistent with the observations we have described, Rptor−/− mice showed reduced CD115 expression on LK cells and the two LK subpopulations, CMPs and GMPs, whereas CD115 was expressed at a low and similar level on LSK cells between WT and Rptor−/− mice (Fig. 3 E). In contrast, expression of CD116 (GM-CSFR, encoded in part by Csf2ra) surface protein and Csf2ra transcript levels were largely normal or only slightly altered in Rptor−/− Lin– cells after stimulation with M-CSF in vitro (Fig. S2, F and G), whereas in freshly isolated BM progenitor populations, the expression of CD116 was slightly enhanced or unchanged in Rptor−/− cells (Fig. S2 H). Similarly, expression levels of CD114 (G-CSFR), CD123 (IL-3Rα), and CD135 (FLT-3) on various hematopoietic progenitors were largely comparable between WT and Rptor−/− mice (Fig. S2 H). Thus, Rptor−/− progenitors show a selective reduction of CD115 expression.
PU.1, IRF8, and KLF4 orchestrate the differentiation of monocytes/macrophages from myeloid progenitors (Scott et al., 1994; Kurotaki et al., 2013). We observed a positive correlation between PU.1 and CD115 expression, and between IRF8 and CD115 expression in total Lin– cells, whereas KLF4 expression did not show a strong correlation with CD115 (Fig. 3 F). We next examined the effects of Rptor deletion on the expression of these transcription factors. Progenitor cells from Rptor−/− mice had reduced PU.1 and IRF8 expression compared with WT cells, but similar KLF4 levels (Fig. 3 G). After culturing Lin– cells from WT mice with M-CSF for 2 d, the increased CD115 expression correlated with up-regulation of PU.1, IRF8, and to a lesser extent, KLF4 (Fig. 3 H). M-CSF–stimulated Rptor−/− cells showed significantly reduced levels of PU.1 and IRF8, whereas the reduction of KLF4 was rather small (Fig. 3 I). Collectively, among cytokine receptors responsible for myelopoiesis, expression of M-CSFR is particularly sensitive to Rptor ablation and the resulting impairment of mTORC1 signaling. The decrease in M-CSFR expression is associated with lower levels of PU.1 and IRF8 in Rptor−/− cells, especially after stimulation with M-CSF, which may collectively contribute to impairment of M-CSF–induced myeloid development.
Raptor ablation reduces anabolic metabolism in developing myeloid cells
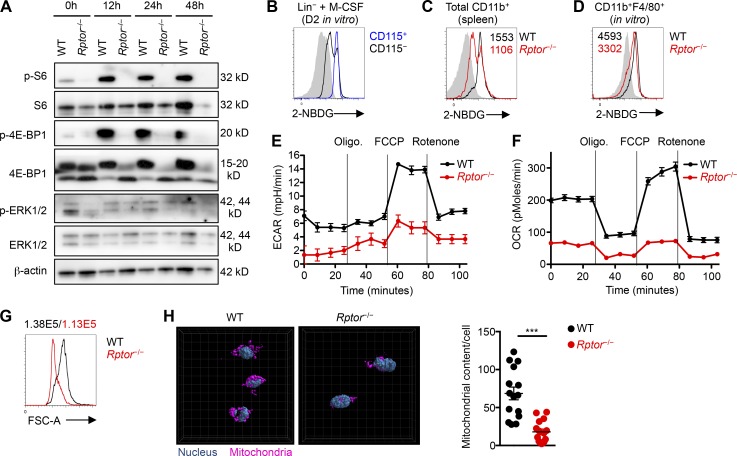
HSCs are quiescent but undergo extensive metabolic reprogramming during myeloid differentiation (Shyh-Chang et al., 2013); however, specific metabolic pathways and molecular regulators in this differentiation process remain poorly understood. We therefore investigated multiple metabolic features in developing myeloid cells and the effects of Rptor deletion in this context. First, M-CSF stimulation of WT Lin– cells up-regulated mTORC1 activity, as measured by phosphorylation of S6 and 4E-BP1, but only modest changes were observed in p-ERK1/2 after stimulation (Fig. 4 A). Deletion of Rptor abrogated S6 and 4E-BP1 phosphorylation but did not have strong effects on ERK1/2 phosphorylation (Fig. 4 A). Phosphorylation of AKT (S473), indicative of mTORC2 activity, was modestly increased in Rptor−/− cells at 48 h (Fig. S3 A), most likely as a compensatory response to the loss of mTORC1 activity (Zeng et al., 2013). Second, we used a fluorescently labeled glucose analog, 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG), to measure glucose uptake. Neutrophils—and monocytes in particular—readily incorporated 2-NBDG (Fig. S3 B), indicating active glucose uptake (Hard, 1970; Newsholme et al., 1986, 1987). Further, upon M-CSF stimulation of Lin– cells, the CD115+ subset exhibited higher uptake of 2-NBDG than the CD115– counterpart (Fig. 4 B), suggesting robust glucose uptake during M-CSF–induced myeloid differentiation. Rptor deletion diminished the uptake of 2-NBDG by splenic CD11b+ cells in vivo (Fig. 4 C), and M-CSF–stimulated cells in vitro (Fig. 4 D). Thus, Raptor loss reduces glucose uptake in a controlled, nutrient-replete environment.
Figure 4.
Reduced anabolic metabolism in M-CSF–stimulated Rptor−/− myeloid cells. (A) Immunoblot analysis of phosphorylated and total proteins of S6, 4E-BP1, ERK1/2, and β-actin in fresh (0 h) or M-CSF–stimulated (10 ng/ml, for the indicated times) Lin– cells from WT and Rptor−/− mice. (B) 2-NBDG staining of CD115+ and CD115– cells in WT mice. (C) 2-NBDG staining of CD11b+ cells from the spleen of WT and Rptor−/− mice, with mean fluorescence intensity (MFI) plotted within graph. (D) 2-NBDG staining of CD11b+F4/80+ cells from Lin– cells of WT and Rptor−/− mice stimulated with M-CSF (10 ng/ml) for 3 d, with MFI plotted within graph. (E) Measurement of ECAR in developing myeloid cells (stimulated with 10 ng/ml M-CSF for 2 d) in response to the indicated mitochondrial inhibitors. Oligo, Oligomycin; FCCP, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone. (F) Measurement of OCR in Lin– cells from WT and Rptor−/− mice stimulated with M-CSF (10 ng/ml) for 2 d. (G) Cell size of CD11b+F4/80+ cells from WT and Rptor−/− mice, with MFI plotted above graph. (H) Representative images (left) and quantification of mitochondrial content per cell (right) of Lin– cells from WT and Rptor−/− mice stimulated with M-CSF (10 ng/ml) for 2 d. Data are mean ± SEM and representative of two (A and B), six (D), three (C and F), or four (E, G, and H) independent experiments. ***, P < 0.001; Student’s t test.
Third, to determine metabolic reprogramming in response to M-CSF stimulation, we measured extracellular acidification rate (ECAR) and mitochondrial oxygen consumption rate (OCR), which are indicative of activities of glycolysis and mitochondrial oxidative phosphorylation, respectively. M-CSF stimulation up-regulated ECAR and OCR, indicating metabolic activation (Fig. S3, C and D). Rptor deletion markedly reduced basal ECAR and OCR, as well as maximal glycolytic capacity and glycolytic reserve, and maximal and spare respiratory capacity (Fig. 4, E and F). In accordance with these observations, Rptor deletion reduced cell growth and mitochondrial biogenesis, as indicated by smaller cell size (Fig. 4 G), and reduced mitochondrial content compared with WT cells (Fig. 4 H). These results reveal that Raptor loss in developing myeloid cells impairs both glycolytic and oxidative metabolism.
Glucose is required for M-CSF–dependent myeloid differentiation and CD115 expression
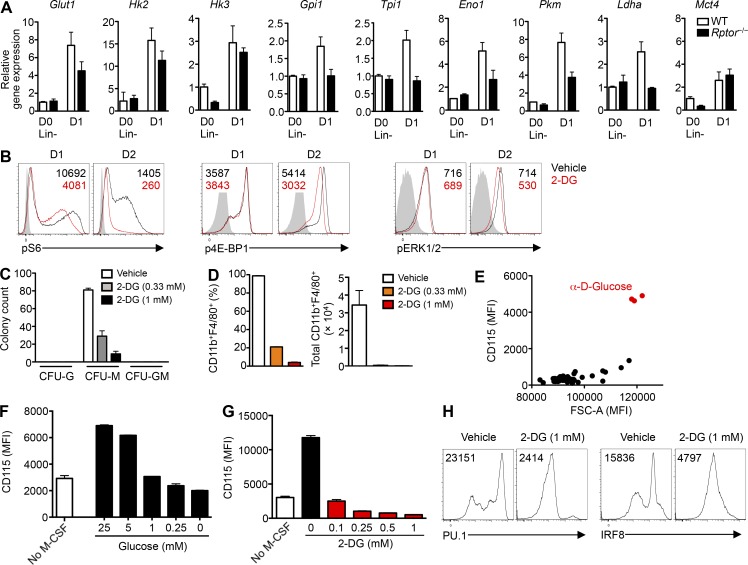
The impairments in glucose uptake and metabolism in Rptor−/− cells prompted us to determine whether the availability of glucose contributes to the development of macrophages and the underlying cellular mechanisms of myeloid differentiation. First, we stimulated Lin– cells with M-CSF and observed increased gene expression of multiple glycolytic enzymes (Fig. 5 A). Rptor deletion impeded the up-regulation of many of these enzymes, including Glut1, Hk2, Gpi1, Tpi1, Eno1, Pkm, and Ldha (Fig. 5 A). To test whether there is reciprocal regulation between mTORC1 and glycolysis, we used 2-deoxy-d-glucose (2-DG), a nonhydrolyzable glucose analog and hexokinase inhibitor, and examined its effects on mTORC1 activity. We found that glucose inhibition impaired mTORC1 activity, but pERK1/2 levels were mostly unchanged (Fig. 5 B). Additionally, 2-DG reduced CFU-M counts in response to M-CSF (Fig. 5 C) and generation of macrophages from BM progenitors in vitro (Fig. 5 D), suggesting the presence of a feed-forward loop between mTORC1 and glucose metabolism in driving myeloid differentiation.
Figure 5.
Glucose is necessary for M-CSF–induced myelopoiesis. (A) mRNA analysis of Glut1, Hk2, Hk3, Gpi1, Tpi1, Eno1, Pkm, Ldha, or Mct4 in freshly isolated or M-CSF–stimulated (10 ng/ml, for 1 d) Lin– cells from WT and Rptor−/− mice. (B) Flow cytometry analysis of phosphorylation of S6 (left), 4E-BP1 (middle), and ERK1/2 (right) of Lin– cells stimulated with M-CSF (10 ng/ml) for 1 or 2 d in the presence of PBS (vehicle) or 2-DG (1 mM), with mean fluorescence intensity (MFI) plotted within graphs. (C) Number of CFU (CFU-G, CFU-M, and CFU-GM) for BM cells of WT mice incubated with M-CSF containing methylcellulose in the presence of 2-DG (0.33 or 1.0 mM) or PBS (vehicle). (D) Frequency (left) and number (right) of CD11b+F4/80+ macrophages after liquid culture of BM cells with M-CSF (10 ng/ml) for 5 d in the presence of 2-DG (0.33 or 1.0 mM) or PBS (vehicle). (E) MFI of cell size (FSC-A) and CD115 of Lin– cells stimulated with M-CSF (10 ng/ml) for 2 d in Phenotype Microarray (carbon and energy sources; PM-M1) plates. (F) MFI of CD115 on Lin– cells stimulated without or with M-CSF (10 ng/ml) for 2 d in the presence of varying concentrations of glucose. (G) MFI of CD115 on Lin– cells stimulated without or with M-CSF (10 ng/ml) for 2 d in the presence of varying 2-DG concentrations. (H) Expression of PU.1 (left) or IRF8 (right) in WT Lin– cells stimulated with M-CSF (10 ng/ml) for 2 d in the presence of PBS (vehicle) or 2-DG (1 mM), with MFI plotted within graphs. Data are mean ± SEM and representative of three (A–D and H), two (E), or five (F and G) independent experiments.
Next, we adopted a carbon energy source screening method to determine the ability of different carbon sources to up-regulate CD115 expression on Lin– cells stimulated with M-CSF in glucose-free medium. Not only was glucose necessary for maximal CD115 expression induced by M-CSF treatment, but more importantly, it could not be substituted by any other common carbon sources (Fig. 5 E). We then determined the effect of varying glucose concentrations in the culture medium on CD115 expression. Indeed, decreasing levels of glucose (Fig. 5 F) or increasing amounts of 2-DG (Fig. 5 G) reduced CD115 surface expression on developing myeloid cells stimulated with M-CSF in a concentration-dependent manner. Finally, to examine whether glycolysis is necessary for the transcriptional changes related to CD115 expression, we stimulated Lin– cells with M-CSF in the presence of 2-DG. We found that 2-DG decreased levels of PU.1 and IRF8 in M-CSF–stimulated cells (Fig. 5 H). The attenuating effect of 2-DG on the generation of CFU-M and macrophages or expression of CD115 and transcription factors mimicked the phenotypes observed in myeloid cells lacking Rptor. These results point to a self-amplifying loop during myelopoiesis, involving glucose metabolism, M-CSFR signaling, mTORC1 activation, and transcriptional activation.
Sterol biosynthesis and Myc activity mediate mTORC1 signaling after M-CSF stimulation
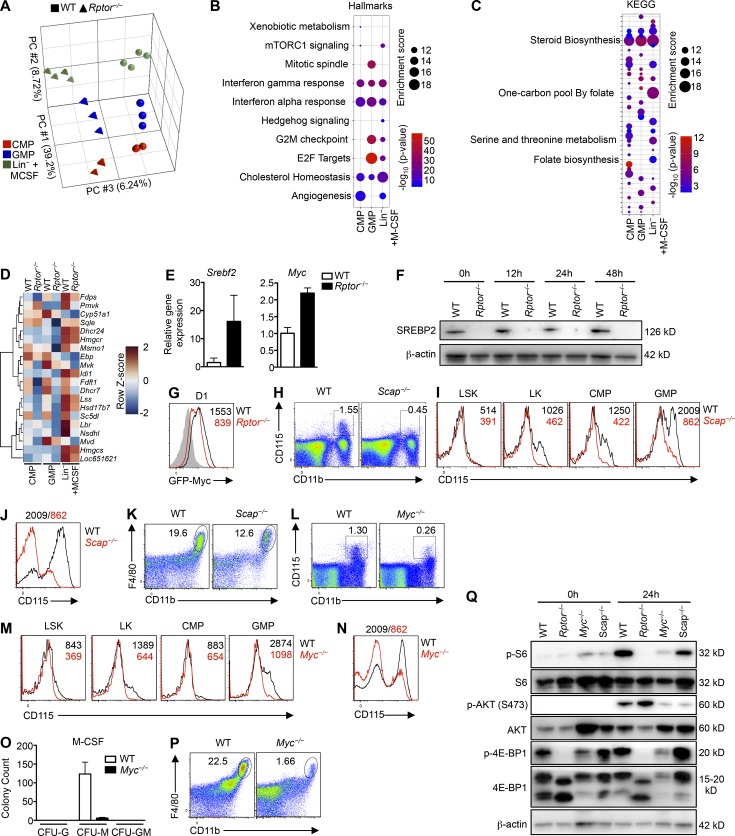
To understand the molecular basis of Raptor in myeloid development, we performed transcriptome profiling to compare CMP, GMP and M-CSF–stimulated Lin– cells from WT and Rptor−/− mice. Principal component analysis showed the separation of different populations based on cell types (principal component 1 [PC1] and PC2) and genotypes (PC3; Fig. 6 A). For the differentially expressed transcripts between WT and Rptor−/− mice (with false discovery rate [FDR] <0.05 and >1.5-fold difference), the Venn diagram showed distinct and overlapping genes regulated by Raptor in these three populations (Fig. S4 A). As expected, Csf1r expression was reduced in cells from Rptor−/− compared with WT controls (Fig. S4 B). To identify pathways underlying Raptor-dependent myelopoiesis in an unbiased manner, we performed gene set enrichment analysis (GSEA) using the Hallmark and KEGG gene sets. Among the strongest enriched gene sets in WT versus Rptor−/− cells (i.e., down-regulated in Rptor−/− cells) were those related to cholesterol or steroid biosynthesis (Fig. 6, B and C), with two genes, Idi1 and Sqle, validated by real-time PCR analysis (Fig. S4 C). In addition to cholesterol biosynthesis, we observed a profound reduction of the KEGG gene sets related to one-carbon metabolism in Rptor−/− cells (Fig. 6 C). The heat map in Fig. 6 D showed that multiple genes involved in cholesterol biosynthesis were increased after M-CSF treatment in WT cells, whereas Raptor deficiency lowered their expression in different populations, especially in M-CSF–stimulated cells. Using network analysis, we further identified the transcription factor Myc as the most centrally regulated node in the top differentially regulated gene network between M-CSF–stimulated WT and Rptor−/− cells (Fig. S4 D). Interestingly, although the mRNA levels for sterol regulatory element binding protein 2 (SREBP2; a critical transcription factor for cholesterol synthesis; encoded by Srebf2) and Myc were up-regulated in Raptor-deficient cells (Fig. 6 E), their protein levels were dampened by Raptor deletion (Fig. 6, F and G), suggesting posttranscriptional regulation mediated by mTORC1.
Figure 6.
Loss of sterol biosynthesis or Myc impairs M-CSF–induced myeloid development. (A) Principal component analysis of WT and Rptor−/− CMP, GMP, or Lin– cells stimulated with M-CSF (10 ng/ml) for 12 h (n = 3–4 mice per group). The top three PCs are shown. (B) GSEA enrichment plot of the top Hallmark gene sets enriched in CMP, GMP, or M-CSF–stimulated Lin– cells from WT vs Rptor−/− mice (n = 3–4 mice per group). The thresholds for Hallmark pathways were set at FDR <5% and enrichment score ≥10. (C) GSEA enrichment plot of the top KEGG gene sets enriched in CMP, GMP, or M-CSF–stimulated Lin– cells from WT vs Rptor−/− mice (n = 3–4 mice per group). The thresholds for KEGG pathways were set at FDR <5% and enrichment score ≥10. (D) Heatmap of the “reactome cholesterol biosynthesis” gene set for indicated CMP, GMP, or M-CSF–stimulated Lin– cells from WT and Rptor−/− mice (n = 3–4 mice per group). Colorization depicts row z-scores as indicated in the scale (right). (E) Gene expression analysis of freshly isolated Lin– cells of WT and Rptor−/− mice. (F) Immunoblot for SREBP2 protein in Lin– cells of WT and Rptor−/− mice stimulated with M-CSF (10 ng/ml) for the indicated time periods. (G) GFP-Myc analysis for Lin– cells of WT and Rptor−/− mice stimulated with M-CSF for 24 h. (H) Flow cytometry analysis of monocytes (CD11b+CD115+) in spleen of WT and Scap−/− mice. (I) Expression of CD115 on WT and Scap−/− myeloid progenitor cell populations, with mean fluorescence intensity (MFI) plotted within graphs. (J) Expression of CD115 on WT and Scap−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 2 d, with MFI plotted above graph. (K) Flow cytometry analysis of F4/80 and CD11b on WT and Scap−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 2 d. (L) Flow cytometry analysis of monocytes (CD11b+CD115+) in spleen of WT and Myc−/− mice. (M) Expression of CD115 on WT and Myc−/− myeloid progenitor cell populations, with MFI plotted within graphs. (N) Expression of CD115 on WT and Myc−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 2 d, with MFI plotted above graph. (O) Number of CFU (CFU-G, CFU-M, CFU-GM) for BM cells of WT and Myc−/− mice incubated with M-CSF containing methylcellulose. (P) Flow cytometry analysis of F4/80 and CD11b on WT and Myc−/− Lin– BM cells after liquid culture with M-CSF (10 ng/ml) for 2 d. (Q) Immunoblot analysis of freshly isolated (0 h) or M-CSF (10 ng/ml)–stimulated (24 h) Lin– BM cells from WT, Rptor−/−, Myc−/−, and Scap−/− mice. Shown are total and phosphorylated S6, AKT, and 4E-BP1, as well as β-actin. Numbers indicate percentages of cells in gates. Data are mean ± SEM. Data are one experiment (A–D) or representative of two (E–G and Q) or four (H–P) independent experiments.
Scap serves as the intracellular sensor for cholesterol and allows for SREBP activation in response to low cholesterol levels for increased cholesterol biosynthesis (Cyster et al., 2014). To determine the role of sterol biosynthesis in myelopoiesis, we crossed mice expressing loxP-flanked Scap alleles (Scapfl/fl) with Rosa26-CreERT2 mice (Scap−/−), allowing for the tamoxifen-inducible deletion of Scap. Similar to Rptor deletion, Scap deletion reduced splenic monocytes (Fig. 6 H). Further, Scap deletion reduced the levels of CD115 expressed on myeloid progenitor populations including LK, CMP, and GMP cells (Fig. 6 I) and M-CSF–stimulated Lin– cells in vitro (Fig. 6 J). Also, the percentages of CD11b+F4/80+ cells were reduced at day 2 in Scap-deleted Lin– cells during in vitro cultures with M-CSF (Fig. 6 K).
Using GFP-Myc knock-in mice (the endogenous Myc locus is modified by a GFP-Myc fusion protein; Huang et al., 2008) to track Myc expression, we found that Myc expression was readily detectable in hematopoietic progenitor populations (Fig. S4 E) but not in mature myeloid populations (Fig. S4 F), indicating active regulation of Myc expression in myelopoiesis. To directly measure Myc function, we crossed mice expressing loxP-flanked Myc alleles (Mycfl/fl) with Rosa26-CreERT2 mice (Myc−/−), allowing for tamoxifen-mediated deletion of Myc. Myc deletion reduced splenic monocytes (Fig. 6 L), associated with reduced CD115 expression on LK, CMP, and GMP progenitor populations (Fig. 6 M) and M-CSF–stimulated Lin– cells (Fig. 6 N). Compared with WT cells, Myc−/− cells had lower CFU-M colony counts and reduced CD11b+F4/80+ percentages (Fig. 6, O and P).
Next we assessed whether Scap or Myc deletion impairs mTOR signaling. We observed reduced signal for the phosphorylation of 4E-BP1, S6, and AKT, especially in Myc-deficient cells (Fig. 6 Q), consistent with our hypothesis for an M-CSF–induced mTORC1-dependent feed-forward loop of myelopoiesis. Overall, these results identify the requirement of both sterol biosynthesis and Myc transcriptional activity for CD115 up-regulation and myeloid differentiation. Therefore, mTORC1 relays myelopoiesis-inducing signals by increasing sterol biosynthesis and Myc activation.
M-CSF–induced myelopoiesis requires one-carbon metabolism
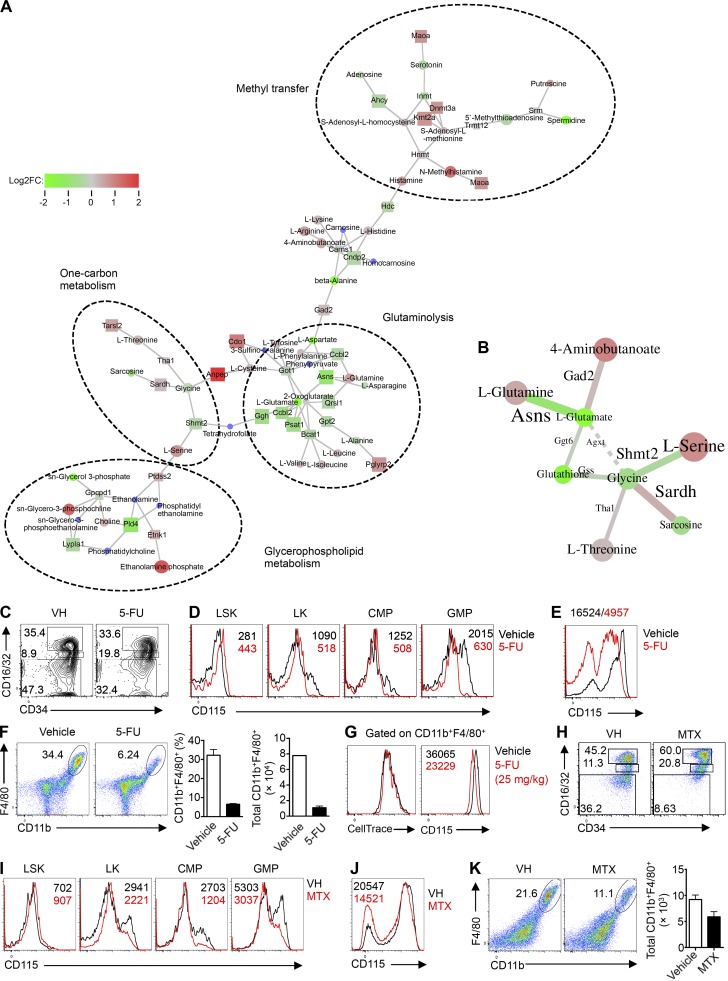
Our data indicate that Rptor deletion impairs anabolic and glucose metabolism and the overall metabolic fitness of these cells. To gain a better understanding of the metabolic dysregulation, we performed unbiased metabolomic analysis of Rptor−/− cells. Specifically, Lin– cells from WT and Rptor−/− mice were stimulated with M-CSF for 1 d, and metabolite profiles were analyzed using high-resolution liquid chromatography-mass spectrometry (LC-MS). Next, combining both the microarray and metabolomics data, we generated enriched metabolic networks using genes and metabolites (GAM) analysis (Sergushichev et al., 2016). These enriched networks include glutaminolysis, which is related to Myc activity (Wise et al., 2008); glycerophospholipid metabolism; methyl transfer; and one-carbon metabolism (Fig. 7 A). When solving for optimality to generate a maximum-weight connected subgraph, glutamate, glycine, and glutathione were among the most decreased metabolites in Rptor−/− cells (Fig. 7 B). These metabolites participate in one-carbon metabolism, which integrates the availability of biosynthetic fuel sources, including glucose and amino acids, to drive anabolic metabolism and cell growth (Locasale, 2013). Furthermore, Rptor−/− cells showed a pronounced increase of serine (Fig. 7 B), the catabolism of which requires the one-carbon pathway (Bao et al., 2016). To ascertain the functional effect of one-carbon metabolism on myelopoiesis, we used 5-flourouracil (5-FU), an inhibitor of one-carbon metabolism (Locasale, 2013). Specifically, we treated mice with 5-FU and isolated BM cells 2 d after treatment. Similar to Rptor−/− mice, BM cells from 5-FU–treated mice showed a slight increase in the CMP population compared with control mice (Fig. 7 C). More importantly, 5-FU treatment resulted in a pronounced reduction in CD115 expression in LK, CMP, and GMP progenitor populations (Fig. 7 D) and M-CSF–stimulated Lin– cells in vitro (Fig. 7 E). Further, BM cells from 5-FU–treated mice were less efficient in developing into CD11b+F4/80+ macrophages upon M-CSF stimulation (Fig. 7 F). Further, the CellTrace dilution of cultured CD11b+F4/80+ macrophages was not affected by 5-FU, but the surface levels of CD115 were decreased by 5-FU (Fig. 7 G). To ascertain our findings supporting a role for one-carbon metabolism in myelopoiesis, we tested another one-carbon pathway inhibitor, methotrexate (MTX), which targets dihydrofolate reductase (Locasale, 2013). MTX inhibited myelopoiesis and largely phenocopied 5-FU treatment and Rptor deletion, including an increased CMP population (Fig. 7 H), impaired CD115 expression on myeloid precursors (Fig. 7 I) and M-CSF–stimulated Lin– cells in vitro (Fig. 7 J), and reduced numbers of CD11b+F4/80+ macrophages (Fig. 7 K). In summary, inhibition of one-carbon metabolism by 5-FU phenocopied the defects observed in Rptor−/− cells, indicating that one-carbon metabolism supports M-CSF–induced mTORC1 activation during myelopoiesis (Fig. S5).
Figure 7.
mTORC1 regulates one-carbon metabolism to support myelopoiesis. (A) GAM analysis of nodes (n = 3–4 mice per group). (B) GAM analysis of optimally solved edges (n = 3–4 mice per group). (C) Flow cytometry analysis of CD16/32 and CD34 on BM Lin– LK cells from mice treated with vehicle (PBS) and 5-FU (25 mg/kg). (D) Expression of CD115 on myeloid progenitor cell populations from mice treated with vehicle (PBS) and 5-FU, with mean fluorescence intensity (MFI) plotted within graphs. (E) Expression of CD115 on Lin– BM cells from mice treated with vehicle (PBS) and 5-FU after liquid culture with M-CSF (10 ng/ml) for 2 d, with MFI plotted above graph. (F) Flow cytometry analysis (left) of F4/80 and CD11b, frequency (middle), and number (right) of CD11b+F4/80+ in Lin– BM cells from mice treated with vehicle (PBS) and 5-FU after liquid culture with M-CSF (10 ng/ml) for 2 d. (G) Flow cytometry analysis of CellTrace (left) and CD115 (right) within the gated CD11b+F4/80+ cells from cultured (M-CSF, 10 ng/ml, 2 d) Lin– BM cells from mice treated with vehicle (PBS) and 5-FU, with MFI plotted within graphs. (H) Flow cytometry analysis of CD16/32 and CD34 on BM Lin– LK cells from mice treated with vehicle (DMSO) and MTX (32 mg/kg). (I) Expression of CD115 on myeloid progenitor cell populations from mice treated with vehicle (DMSO) and MTX, with MFI plotted within graphs. (J) Expression of CD115 on Lin– BM cells from mice treated with vehicle (DMSO) and MTX after culture with M-CSF (10 ng/ml) for 2 d, with MFI plotted within graph. (K) Flow cytometry analysis (left) of F4/80 and CD11b and number (right) of CD11b+F4/80+ cells in Lin– BM cells from mice treated with vehicle (DMSO) and MTX after culture with M-CSF (10 ng/ml) for 2 d. Numbers indicate percentage of cells in gates. Data are mean ± SEM. Data are one experiment (A and B) or representative of three experiments (C–K).
Discussion
In this study, we describe that M-CSF–mediated myelopoiesis depends on the interplay between mTORC1-mediated signaling and metabolic reprogramming. Specifically, mTORC1 signaling promotes expression of M-CSFR and selective lineage-specific transcription factors, and loss of Rptor results in defective myelopoiesis under homeostasis and impaired response to bacterial infection. Additionally, mTORC1 impinges on metabolic programs in myelopoiesis, including sterol biosynthesis, Myc activation, and one-carbon metabolism. Disruption of each metabolic program highlights critical roles of cellular metabolism in supporting M-CSF–mediated myelopoiesis. Further, metabolic reprogramming by M-CSF requires glucose without possible substitution by any other carbon sources tested. These results point to a previously unappreciated feed-forward loop with mTORC1 as a signaling node to integrate cytokine signaling, metabolic reprogramming, and transcription factor activation in developing myeloid cells.
Cell fate determination in hematopoiesis requires signaling via cytokine receptors and sequential engagement of transcription factors, but what connects cell surface and nuclear events remains much less understood. In myelopoiesis, signaling by M-CSF and M-CSFR interaction engages the transcription factors PU.1, IRF8, and KLF4, which are established regulators of hematopoietic development, including monocyte-macrophage differentiation (Scott et al., 1994; Kurotaki et al., 2013). Here we show that PU.1 and IRF8 expression in myeloid precursors correlates with M-CSFR expression, and more importantly, loss of Raptor impairs expression of M-CSFR, PU.1, and IRF8, suggesting specific effects of mTORC1 signaling on myelopoiesis. We further establish the dependence of M-CSFR expression and myelopoiesis on mTORC1-dependent anabolic metabolism. Although previous studies linked mTORC1 signaling to HSC regeneration and leukemogenesis (Hoshii et al., 2012; Kalaitzidis et al., 2012), our studies unveil novel roles of mTORC1 in mediating M-CSF–dependent myelopoiesis and connecting M-CSFR expression and metabolic reprogramming at the mechanistic level. Our findings further highlight the physiological importance of this pathway in the generation of inflammatory monocytes and neutrophils in bacterial infection, as indicated by the marked susceptibility of Rptor−/− mice to L. monocytogenes infection.
To mount a sustained cellular response to a weak signal, positive transcriptional feed-forward loops are frequently engaged to potentiate such effects. For example, in a positive interaction between PU.1 and M-CSFR, PU.1 activates the transcription of Csf1r, leading to higher receptor expression and ligand sensitivity, which in turn induce more PU.1 (Sarrazin et al., 2009). Moreover, PU.1 directly binds its own genomic control elements and induces its own transcription in a mode of autoregulation (Chen et al., 1995; Okuno et al., 2005; Staber et al., 2013). As a result of these amplification steps, HSCs, which express low levels of M-CSFR, can be directly instructed toward myeloid lineage fate in response to M-CSF (Mossadegh-Keller et al., 2013). Our studies highlight that mTORC1 signaling, mTORC1-dependent metabolic and transcriptional activation, and M-CSFR constitute a novel feed-forward loop in M-CSF–dependent myelopoiesis. M-CSF stimulation activates mTORC1, which then further promotes expression of M-CSFR, as well as PU.1 and IRF8, for enhanced responsiveness to M-CSF. Moreover, this amplification loop requires activation of mTORC1-dependent anabolic metabolism, as deletion of Myc and Scap causes drastic reductions in M-CSFR levels on myeloid progenitors. Interestingly, compared with M-CSF–stimulated Lin– progenitors in which Raptor deficiency greatly dampens metabolic activities, CMP and GMP cells also show dependence on Raptor for their metabolic gene expression programs, albeit to a lesser extent. Because the development of CMP and GMP cells is largely undisturbed in Rptor−/− mice, these results suggest that defective metabolism is intrinsic to mTORC1 deficiency, not simply a consequence of impaired differentiation. Further, the more profound defects in M-CSF–stimulated Rptor−/− Lin– cells suggest that strong activation of mTORC1 and metabolism in response to M-CSF stimulation underlies robust myelopoiesis. In contrast, deletion of Raptor in mature myeloid cells using LysM-Cre does not result in decreased myeloid cell populations in immune organs (unpublished data). Collectively, mTORC1-dependent metabolic programs are differentially adopted by myeloid precursors to meet energy and biosynthetic needs, and these serve as an important mechanism to amplify signals from M-CSF, thereby forming a feed-forward loop to drive cell differentiation.
Glucose is a ubiquitous energy source, and many cellular decisions are centered on its availability. In response to growth factors, cells increase glucose uptake and metabolism to support bioenergetic and biosynthetic needs (Ward and Thompson, 2012). M-CSF is detectable in the serum during homeostasis, and its levels increase in response to infection (Wing et al., 1985), inflammation, and cancer (Hamilton, 2008). Our data here suggest that regulation of glucose availability offers an independent route to control cellular responses to M-CSF. Specifically, we found that glucose is the required carbon source for anabolic metabolism in developing myeloid cells, and that M-CSFR expression and M-CSF–mediated myelopoiesis are very sensitive to glucose availability. Further, mTORC1 activity is required for glucose uptake, glycolysis, and downstream anabolic metabolism. In addition, sustained mTORC1 signaling induced by M-CSF requires the presence of glucose, and blockade of glucose metabolism by 2-DG impairs M-CSF–mediated myelopoiesis, suggesting that metabolic reprogramming is a prerequisite in this process. Therefore, mTORC1 bridges M-CSFR signaling and glucose-dependent anabolic metabolism. We noted that increased glucose availability in vivo caused by hyperglycemia drives myelopoiesis above homeostatic levels (Nagareddy et al., 2013), and defective uptake of glucose via Glut1 deletion directly impairs myelopoiesis (Sarrazy et al., 2016). Further, AML cells have a high glycolytic metabolism but, unlike developing myeloid cells, can adapt to metabolic stress, such as the reduced presence of glucose (Saito et al., 2015). Whether mTORC1-mediated glucose and anabolic metabolism in myeloid cells contribute to these pathological conditions warrants further investigation.
Although several pathways are involved in the generation of macromolecules necessary for growth and proliferation, one-carbon metabolism serves as an allocation node for anabolic metabolism. One-carbon metabolism incorporates cellular availability of “building blocks,” such as glucose and glutamine, and diverts their metabolism through methionine and folate cycles for various cellular end points (Locasale, 2013; Yang and Vousden, 2016). Our microarray, GSEA, and unbiased network analysis using GAM (Sergushichev et al., 2016) reveal one-carbon metabolism as a critical pathway regulated by mTORC1 after M-CSF stimulation, and pharmacological inhibition of one-carbon metabolism drastically impairs M-CSF–mediated myelopoiesis. It is noteworthy that although mTORC1 deficiency impairs the gene expression program of cholesterol biosynthesis in CMP and GMP populations, the transcriptional program of one-carbon metabolism is largely unchanged in these precursor populations from Rptor−/− mice. In contrast, M-CSF–mediated up-regulation of both metabolic programs depends on mTORC1. Furthermore, expression of methylenetetrahydrofolate dehydrogenase 2 (Mthfd2), an enzyme in one-carbon metabolism and arguably the most up-regulated metabolic enzyme in certain cancer cells versus normal cells (Nilsson et al., 2014), is reduced in Rptor−/− cells in our microarray analysis. Although part of one-carbon metabolism occurs in the cytosol, serine catabolism occurs primarily in the mitochondria (Yang and Vousden, 2016). Thus, in the case of mitochondrial failure, cells accumulate serine because of feedback mechanisms, leading to higher production of serine and the inability to consume serine by mitochondria (Bao et al., 2016). Consistent with this notion, we observed a drastic reduction in mitochondrial biogenesis but increased serine levels in Rptor−/− cells (Fig. 7 B), further supporting a role of mTORC1 in activating one-carbon metabolism in myelopoiesis.
In summary, our study has revealed a crucial role for mTORC1 signaling and downstream transcriptional and metabolic pathways in M-CSF–instructed myelopoiesis. Our results suggest a metabolism-centric model for myelopoiesis, in addition to the previously discovered transcription factor–dependent model, and the critical roles of mTORC1 signaling in integrating these pathways. Given the importance of mTORC1 in myeloid cell development, the identification of the signaling network and metabolic dependencies provides new insight into therapeutic intervention of aberrant myeloid development observed in cancer and immune-mediated diseases. From this perspective, one-carbon metabolism is an attractive target for cancer therapy because of its central role in cell growth and proliferation (Locasale, 2013; Yang and Vousden, 2016). Our data suggest that targeting mTORC1 signaling or downstream metabolic pathways such as one-carbon metabolism in either tumor cells or immunosuppressive myeloid cells, such as M-CSFR–expressing myeloid-derived suppressor cells (Kumar et al., 2016), could produce synergistic effects on cancer therapies.
Materials and methods
Mice
Mice were housed and bred at the St. Jude Children’s Research Hospital animal care facilities in specific pathogen–free conditions. C57BL/6 and Scapfl mice were purchased from the Jackson Laboratory. Rosa26-CreERT2 (which express a Cre-ERT2 fusion gene under the control of the ubiquitously expressed Rosa26 locus), Rptorfl, Rictorfl, and Mycfl mice have been described (Liu et al., 2010; Wang et al., 2011; Yang et al., 2011; Zeng et al., 2013). GFP-Myc knock-in mice were kindly provided by B. Sleckman (Weill Cornell Medical College, New York, NY; Huang et al., 2008). Cre-expressing mice were used as controls, and littermates were used whenever possible. Mice were backcrossed for at least 10 generations to the C57BL/6 background strain. For the generation of BM chimeras, host mice were lethally irradiated for a total of 1,100 rads before receiving 3 million BM donor cells retroorbitally. Mice remained on antibiotic (Baytril) water for 2–3 wks, and after 6 wks, reconstitution was determined by flow cytometry analysis of blood samples. Mice were used 6–8 wks after chimera generation for experiments. All experiments with mice were conducted in accordance with the St. Jude Children’s Research Hospital institutional policies, and animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
L. monocytogenes infection
L. monocytogenes bacterial cultures were grown overnight from glycerol stocks at 30°C in brain–heart infusion (BHI) medium under constant mixing. On the day of infection, overnight cultures were subcultured in BHI at 37°C under constant mixing until logarithmic growth was achieved. Subcultures were diluted in ice-cold PBS, and 5 × 103 CFU of L. monocytogenes were injected into mice using 100 µl PBS retroorbitally. Titers were confirmed by plating on BHI agar plates overnight and CFU counting. Mice were killed 3 d after infection, and spleens and livers were harvested, weighed, and homogenized for further analysis.
Tamoxifen treatment
Mice were injected with tamoxifen (1 mg/mouse) dissolved in 0.2 ml corn oil (Sigma) into the intraperitoneal cavity for four consecutive days or as described. This treatment regimen resulted in complete deletion of the target genes in multiple cell types. Rptor−/− and Rictor−/− mice were killed 10 d after the initial tamoxifen treatment unless otherwise noted. Because of issues with viability of long-term treatment, Myc−/− and Scap−/− mice were killed 5 d after the initial tamoxifen treatment.
Cell purification and culture
Cells isolated from spleen or BM were treated with ACK lysis buffer (Gibco) for 3 min to remove red blood cells. Whole or sorted Lin– BM cells were cultured in DMEM (Gibco) medium (plus β-mercaptoethanol) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin in the presence of M-CSF (10 ng/ml) for the indicated times. Unless otherwise stated, cells were cultured at 106 cells/ml in 0.5 ml (48-well plate) or 1 ml (24-well plate) for the indicated times in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 40 µM β-mercaptoethanol. G-CSF (10 ng/ml), M-CSF (10 ng/ml), or GM-CSF (10 ng/ml) was added to the cultures, and cells were incubated for the indicated times. Nonadherent cells were removed from the wells by pipetting, and adherent cells were removed after incubation with Versene (Gibco) according to the manufacturer’s instructions. In glucose titration experiments, glucose-free DMEM (Gibco) and dialyzed FBS (Gibco) were used. For carbon energy source screening, we used Phenotype MicroArrays PM-M1 plates (Biolog) and cultured Lin– cells with M-CSF (10 ng/ml) for 48 h in 50 µl and glucose-free DMEM.
Growth factor cultures in methylcellulose
BM samples were mixed with methylcellulose (M3134) and IMDM (both from Stem Cell Technologies) according to the manufacturer’s instructions in the presence of 10% FBS and cytokines: G-CSF (10 ng/ml), GM-CSF (10 ng/ml), or M-CSF (10 ng/ml). Each methylcellulose dish contained 2 × 104 cells, and samples were plated in replicate. 7 d after the start of culture, CFUs were enumerated according to colony appearance and descriptions supplied by the manufacturer using an optical microscope and a grid Petri dish.
Chemicals, inhibitors, and other reagents
2-DG and 5-FU (both from Sigma-Aldrich) were dissolved in aqueous media, and MTX (Sigma) was dissolved in DMSO initially and further dissolved in PBS for injection. 2-NBDG (Thermo Fisher Scientific) was dissolved in 100% ethanol and used at 10 µM in complete DMEM. Recombinant mouse G-CSF, GM-CSF, or M-CSF (all from Peprotech) were dissolved in PBS containing 0.1% (wt/vol) BSA. Aliquots of 1,000× were stored at −80°C and used within fewer than three freeze-thaw cycles.
Flow cytometry
For analysis of surface markers, cells were stained in PBS (Gibco) containing 2% (wt/vol) BSA. Surface receptors were stained for 30 min on ice. Annexin V staining was performed per manufacturer’s instructions (BD Biosciences). CellTrace labeling was performed according to manufacturer’s instructions (Thermo Fisher Scientific). Intracellular staining was performed with eBioscience Foxp3/Transcription Factor Staining Buffer Set according to the manufacturer’s instructions. The Lin– cocktail kit (B220, CD3, Gr-1, CD11b, TER-119) was purchased from BD Biosciences. Streptavidin-conjugated fluorophores were from BioLegend. Myeloid precursor populations were defined by the following markers: LSK (Lin–Sca-1+c-Kit+CD127–), LK (Lin–Sca-1–c-Kit+CD127–), CMP (Lin–Sca-1–c-Kit+CD127–CD34+CD16/32mid), GMP (Lin–Sca-1–c-Kit+CD127–CD34+CD16/32hi), MDP (Lin– [in this case B220, Ly6G, CD3, Ter-119], CD115+CD135+), and cMoPs (Lin–CD115+CD135–). For in vivo BrdU labeling, mice were injected intraperitoneally with BrdU (1 mg) and analyzed 1 h later according to the manufacturer’s instructions (BD Biosciences; 552598). Caspase-3 staining was performed according to the manufacturer’s instructions (BD Biosciences). The following antibodies were used with alternate names, clone numbers, phosphorylation sites (if applicable) and sources in parentheses: anti-CD135 (Flt3; A2F10.1; BD Biosciences); anti-Sca-1 (D7), anti-Ly6C (AL-21), anti-CD115 (AFS98), anti-F4/80 (BM8), anti-CD11b (M1/70), anti-B220 (RA3-6B2), anti-TCRβ (H57-597; all from BioLegend); anti-KLF4 (polyclonal), anti-GM-CSFR (CD116; 698423), anti-CD114 (polyclonal; all from R&D Systems); anti-Ly6G (1A8), anti-CD127 (A7R34; both from Tonbo Biosciences); anti–c-Kit (CD117; 2B8), anti-CD16/32 (93), anti-CD34 (RAM34), anti-CD123 (5B11), anti-IRF8 (V3GYWCH; all from eBioscience); and anti-PU.1 (9G7), anti-phosphorylated (T37/46) 4E-BP1 (236B4), anti-phosphorylated (S235/236) S6 (DX57.2.2E; all from Cell Signaling Technology). Certain flow cytometry data were analyzed using the R library “flowCore,” subjected to dimensionality reduction by tSNE with at least 8,000 cells using a Barnes–Hut implementation with the R library “Rtsne,” and visualized using ggplot2.
RNA and immunoblot analyses
Real-time PCR analysis was performed with primers and probe sets from Applied Biosystems, as described (Liu et al., 2009). Immunoblots were performed as described previously (Liu et al., 2010), using the primary antibodies anti-CD115, anti-phosphorylated (T37/46) 4E-BP1 (236B4), anti-phosphorylated (S235/236) S6 (DX57.2.2E), anti-phosphorylated ERK1/2 (D13.14.4E), anti-phosphorylated (S473) AKT (D9E), anti-S6 (5G10), anti-4E-BP-1 (53H11), anti-ERK1/2 (137F5), anti Akt (polyclonal; all from Cell Signaling Technology), anti-SREBP2 (ab30682; Abcam), and anti-β-actin (AC-74; Sigma-Aldrich) and the secondary horseradish peroxidase–conjugated antibodies anti–rabbit IgG (W401B) and anti–mouse IgG (W402B; both from Promega).
Microarray analysis
RNA samples of CMP (Lin–Sca-1–c-Kit+CD34+FcγRII/IIImid), GMP (Lin–Sca-1–c-Kit+CD34+FcγRII/IIIhi) from WT and Rptor−/−, and cultures of BM Lin– cells from WT and Rptor−/− mice stimulated with M-CSF (10 ng/ml) for 12 h were analyzed with the Affymetrix Mouse Gene 2.0 ST GeneChip array, and expression signals were summarized with the robust multiarray average algorithm (Affymetrix Expression Console v1.1) or by fitting a linear model implemented in the R package “limma” (Ritchie et al., 2015). Lists of differentially expressed genes by 1.5-fold or more were analyzed for functional enrichment using the Ingenuity Pathways (www.ingenuity.com). Up-regulated and down-regulated genes under each condition were annotated using Hallmark, KEGG, and Gene Ontology gene sets v6.0 downloaded from MsigDB (Subramanian et al., 2005), and functional enrichment of each pathway in the gene set was performed using Fisher’s exact test. Fisher’s exact p-value was corrected for multiple testing using the Benjamini–Hochberg method. Pathways were deemed significantly enriched at FDR <5% and enrichment score ≥10.
Gene expression data generated from CMP, GMP, and Lin− cells treated with M-CSF (10 ng/ml) isolated from WT and Rptor−/− mice are available in the GEO database under accession no. GSE100256.
Metabolic assay
ECAR and OCR were measured using the Seahorse XF24-3 Extracellular Flux Analyzer per the manufacturer’s instructions (Seahorse Bioscience) in response to 1 µM oligomycin, 2 µM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone, and 1 µM rotenone.
Microscopy
Live cells were stained using MitoTracker Deep Red (M22426; Thermo Fisher Scientific) and Hoechst 33342 (62249; Thermo Fisher Scientific) for 20 min. Images were acquired using a Zeiss Axio ObserverZ.1 microscope equipped with a CSU-22 spinning disk (Yokagawa Electric), Delta Evole EMCCD camera (Photometrics), 100×/1.45-NA oil objective, and Slidebook imaging software (Intelligent Imaging Innovations). Frequency and volume of MitoTracker+ puncta (mitochondrial content) per cell was determined for each sample.
Pathology and immunohistochemistry
Formalin-fixed liver and spleen were processed and embedded in paraffin using standard techniques, sectioned at 4 µm, mounted on positively charged glass slides (Superfrost Plus; Thermo Fisher Scientific), and dried at 60°C for 20 min. Neutrophils were detected with anti-Neu7/4 antibody (1:500 dilution, NBP2-13077; Novus Biologicals). The three antibodies used to detect macrophages were anti-Iba1 (1:300 dilution, CP290A; Biocare Medical), anti-F4/80 (1:500, clone BM8; Thermo Fisher Scientific), and anti-MAC2 (1:500, ACL-8942AP; Accurate Chemical and Scientific). A rabbit polyclonal antibody was used to detect L. monocytogenes in tissue sections (1:1,000, NB100-65667; Novus Biologicals). All sections were examined by a pathologist blinded to the experimental group assignments.
Metabolomics analysis
WT and Rptor−/− BM Lin– cells were cultured at 106 cells/ml in 1 ml (24-well plate) in the presence of M-CSF (10 ng/ml) for 24 h, followed by three PBS washes. Metabolites were extracted from cell pellets and culture medium using 80% methanol containing 0.05 ng/µl inosine-15N4 and 0.05 ng/µl thymine-d4 as internal standards (Cambridge Isotope Laboratories). The samples were centrifuged (10 min, 9,000 g, 4°C), and the supernatants were collected. Negative-ion targeted profiling of polar metabolites was performed using an Acquity UPLC (Waters Corporation) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (SCIEX). Extracts (10 µl) were injected directly onto a 150 × 2.0-mm Luna NH2 column (Phenomenex). The column was eluted at a flow rate of 400 µl/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide [Sigma-Aldrich] in water [VWR International]) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 vol/vol acetonitrile/methanol [VWR International]) followed by a 10-min linear gradient to 100% mobile phase A. The ion spray voltage was −4.5 kV, and the source temperature was 500°C. Positive ionization mode profiling of polar metabolites was performed using a Nexera ×2 U-HPLC (Shimadzu Corporation) Q Exactive Orbitrap (Thermo Fisher Scientific) LC-MS system. The 80% methanol extracts (100 µl) were dried using a nitrogen evaporator (TurboVap LV; Biotage) and resuspended in 10 µl water and 90 µl of 74.9:24.9:0.2 vol/vol/vol acetonitrile/methanol/formic acid containing stable isotope–labeled internal standards (valine-d8, Isotec; and phenylalanine-d8, Cambridge Isotope Laboratories). The samples were centrifuged (10 min, 9,000 g, 4°C), and the supernatants were injected directly onto a 150 × 2-mm Atlantis HILIC column (Waters Corporation). The column was eluted isocratically at a flow rate of 250 µl/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 min followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. The electrospray ionization voltage was 3.5 kV, and data were acquired using full-scan analysis over m/z 70–800 at 70,000 resolution. Reversed-phase C18 chromatography/negative ion mode MS analyses of free fatty acids and bile acids were conducted using an LC-MS system comprised of a Shimadzu Nexera X2 UHPLC (Shimadzu Corporation) coupled to a Q Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific). Medium (30 µl) was extracted using 90 µl methanol containing PGE2-d4 (Cayman Chemical) and centrifuged (10 min, 9,000 g, 4°C), and the cell extracts were analyzed directly. The samples were injected onto a 150 × 2.1-mm Acquity BEH C18 column (Waters Corporation). The column was eluted isocratically at a flow rate of 450 µl/min with 80% mobile phase A (0.01% formic acid in water) for 3 min followed by a linear gradient to 100% mobile phase B (acetonitrile with 0.01% acetic acid) over 12 min. MS analyses were performed in the negative ion mode using electrospray ionization, full-scan MS acquisition over 70–850 m/z, and a resolution setting of 70,000. Metabolite identities were confirmed using authentic reference standards. Other MS settings were as follows: spray voltage, −3.5 kV; capillary temperature, 320°C; and heater temperature, 300°C. LC-MS data were processed and visually inspected using MultiQuant 2.1 (SCIEX) and TraceFinder 3.1 software (Thermo Fisher Scientific).
Genes and metabolite analysis
Analysis of GAM datasets was performed according to instructions available online (https://artyomovlab.wustl.edu/shiny/gam/; Sergushichev et al., 2016).
Statistical analysis
P-values were calculated by Student’s t test for parametric data or Mann–Whitney test for nonparametric data when comparing two samples, as well as one-way ANOVA with Dunnet’s post hoc test for parametric data or Kruskal–Wallis for nonparametric data when comparing more than two samples. Differences between groups were considered statistically significant with a p-value cutoff of 0.05. Data are represented as mean ± SEM.
Online supplemental material
Fig. S1 shows the homeostatic loss of myeloid cells in Rptor−/− mice. Fig. S2 shows gating of myeloid precursor populations, proliferation and cell death analysis, and expression of myelopoietic cytokine receptors. Fig. S3 shows AKT phosphorylation, glucose uptake in myeloid cells, and M-CSF–stimulated metabolism. Fig. S4 shows a summary of differentially expressed genes between cells from WT and Rptor−/− mice in CMP, GMP, and M-CSF–stimulated Lin– cells, the Myc network from transcriptome analysis, and GFP-Myc expression in myeloid cells. Fig. S5 shows a summary of the present study depicting a metabolism-centric model of myelopoiesis.
Supplementary Material
Acknowledgments
The authors acknowledge B. Sleckman for GFP-Myc mice; Y. Wang and N. Chapman for editing of the manuscript; C. Cloer, M. Hendren, A. KC, S. Rankin, and B. Rhode for animal colony management; and St. Jude Immunology FACS core facility for cell sorting.
This work was supported by the National Institutes of Health (AI105887, AI101407, CA176624, and NS064599 to H. Chi) and the American Asthma Foundation (to H. Chi).
Author contributions: P.W.F. Karmaus conceived and designed the study, performed cellular, molecular, and biochemical experiments, analyzed data, and wrote the manuscript; A.A. Herrada and L. Long helped perform cellular, molecular, and biochemical experiments; C. Guy performed microscopy; G. Neale and Y. Dhungana helped with bioinformatic analysis of the microarrays; P. Vogel performed histopathology analysis; J. Avila and C.B. Clish performed metabolomic analyses; and H. Chi helped conceive and design experiments, wrote the manuscript, and provided overall direction. The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 2-DG
- 2-deoxy-d-glucose
- 2-NBDG
- 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose
- 5-FU
- 5-fluorouracil
- BHI
- brain–heart infusion
- CMP
- common myeloid progenitor
- CSFR
- CSF receptor
- ECAR
- extracellular acidification rate
- GAM
- genes and metabolites
- GMP
- granulocyte-macrophage progenitor
- HSC
- hematopoietic stem cell
- MDP
- monocyte-macrophage progenitor
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- MTX
- methotrexate
- OCR
- oxygen consumption rate
- tSNE
- t-distributed stochastic neighbor embedding
References
- Akers L.J., Fang W., Levy A.G., Franklin A.R., Huang P., and Zweidler-McKay P.A.. 2011. Targeting glycolysis in leukemia: A novel inhibitor 3-BrOP in combination with rapamycin. Leuk. Res. 35:814–820. 10.1016/j.leukres.2010.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X.R., Ong S.E., Goldberger O., Peng J., Sharma R., Thompson D.A., Vafai S.B., Cox A.G., Marutani E., Ichinose F., et al. . 2016. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 5:e10575 10.7554/eLife.10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ray-Gallet D., Zhang P., Hetherington C.J., Gonzalez D.A., Zhang D.E., Moreau-Gachelin F., and Tenen D.G.. 1995. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 11:1549–1560. [PubMed] [Google Scholar]
- Chi H. 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12:325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.Z., Evrard M., Devi S., Chen J., Lim J.Y., See P., Zhang Y., Adrover J.M., Lee B., Tan L., et al. . 2016. CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J. Exp. Med. 213:2293–2314. 10.1084/jem.20160800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Dang E.V., Reboldi A., and Yi T.. 2014. 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 14:731–743. 10.1038/nri3755 [DOI] [PubMed] [Google Scholar]
- Dai X.M., Ryan G.R., Hapel A.J., Dominguez M.G., Russell R.G., Kapp S., Sylvestre V., and Stanley E.R.. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 99:111–120. 10.1182/blood.V99.1.111 [DOI] [PubMed] [Google Scholar]
- Dakic A., Metcalf D., Di Rago L., Mifsud S., Wu L., and Nutt S.L.. 2005. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 201:1487–1502. 10.1084/jem.20050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., and Kioussis D.. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325. 10.1002/immu.200310005 [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., and Singh H.. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 288:1439–1441. 10.1126/science.288.5470.1439 [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Vander Heiden M.G., and Kroemer G.. 2013. Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 12:829–846. 10.1038/nrd4145 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., and Jung S.. 2014. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392–404. 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- Hamilton J.A. 2008. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 8:533–544. 10.1038/nri2356 [DOI] [PubMed] [Google Scholar]
- Hard G.C. 1970. Some biochemical aspects of the immune macrophage. Br. J. Exp. Pathol. 51:97–105. [PMC free article] [PubMed] [Google Scholar]
- Hettinger J., Richards D.M., Hansson J., Barra M.M., Joschko A.C., Krijgsveld J., and Feuerer M.. 2013. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14:821–830. 10.1038/ni.2638 [DOI] [PubMed] [Google Scholar]
- Hoshii T., Tadokoro Y., Naka K., Ooshio T., Muraguchi T., Sugiyama N., Soga T., Araki K., Yamamura K., and Hirao A.. 2012. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J. Clin. Invest. 122:2114–2129. 10.1172/JCI62279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Bredemeyer A.L., Walker L.M., Bassing C.H., and Sleckman B.P.. 2008. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38:342–349. 10.1002/eji.200737972 [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Somoza C., Shigematsu H., Duprez E.A., Iwasaki-Arai J., Mizuno S., Arinobu Y., Geary K., Zhang P., Dayaram T., et al. . 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 106:1590–1600. 10.1182/blood-2005-03-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D., Sykes S.M., Wang Z., Punt N., Tang Y., Ragu C., Sinha A.U., Lane S.W., Souza A.L., Clish C.B., et al. . 2012. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell. 11:429–439. 10.1016/j.stem.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin J.J., Spencer S.P., McCright S.J., Kumar D.B.U., Collet M.A., Mowel W.K., Elliott E.N., Uyar A., Makiya M.A., Dunagin M.C., et al. . 2016. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 537:239–243. 10.1038/nature19346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Patel S., Tcyganov E., and Gabrilovich D.I.. 2016. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 37:208–220. 10.1016/j.it.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D., Osato N., Nishiyama A., Yamamoto M., Ban T., Sato H., Nakabayashi J., Umehara M., Miyake N., Matsumoto N., et al. . 2013. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 121:1839–1849. 10.1182/blood-2012-06-437863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D., Yamamoto M., Nishiyama A., Uno K., Ban T., Ichino M., Sasaki H., Matsunaga S., Yoshinari M., Ryo A., et al. . 2014. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 5:4978 10.1038/ncomms5978 [DOI] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Burns S., Huang G., Boyd K., Proia R.L., Flavell R.A., and Chi H.. 2009. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat. Immunol. 10:769–777. 10.1038/ni.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yang K., Burns S., Shrestha S., and Chi H.. 2010. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat. Immunol. 11:1047–1056. 10.1038/ni.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale J.W. 2013. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer. 13:572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz M.G., and Boettcher S.. 2014. Emergency granulopoiesis. Nat. Rev. Immunol. 14:302–314. 10.1038/nri3660 [DOI] [PubMed] [Google Scholar]
- Moignard V., Macaulay I.C., Swiers G., Buettner F., Schütte J., Calero-Nieto F.J., Kinston S., Joshi A., Hannah R., Theis F.J., et al. . 2013. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 15:363–372. 10.1038/ncb2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., and Sieweke M.H.. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 497:239–243. 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., and Kincade P.W.. 2006. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 24:801–812. 10.1016/j.immuni.2006.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagareddy P.R., Murphy A.J., Stirzaker R.A., Hu Y., Yu S., Miller R.G., Ramkhelawon B., Distel E., Westerterp M., Huang L.S., et al. . 2013. Hyperglycemia promotes myelopoiesis and impairs the resolution of atherosclerosis. Cell Metab. 17:695–708. 10.1016/j.cmet.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P., Curi R., Gordon S., and Newsholme E.A.. 1986. Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem. J. 239:121–125. 10.1042/bj2390121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P., Gordon S., and Newsholme E.A.. 1987. Rates of utilization and fates of glucose, glutamine, pyruvate, fatty acids and ketone bodies by mouse macrophages. Biochem. J. 242:631–636. 10.1042/bj2420631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., and Mootha V.K.. 2014. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5:3128 10.1038/ncomms4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Metcalf D., D’Amico A., Polli M., and Wu L.. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 201:221–231. 10.1084/jem.20041535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A., and Pearce E.J.. 2016. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213:15–23. 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Huang G., Rosenbauer F., Evans E.K., Radomska H.S., Iwasaki H., Akashi K., Moreau-Gachelin F., Li Y., Zhang P., et al. . 2005. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25:2832–2845. 10.1128/MCB.25.7.2832-2845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I.. 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 132:631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Chapple R.H., Lin A., Kitano A., and Nakada D.. 2015. AMPK protects leukemia-initiating cells in myeloid leukemias from metabolic stress in the bone marrow. Cell Stem Cell. 17:585–596. 10.1016/j.stem.2015.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S., Mossadegh-Keller N., Fukao T., Aziz A., Mourcin F., Vanhille L., Kelly Modis L., Kastner P., Chan S., Duprez E., et al. . 2009. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 138:300–313. 10.1016/j.cell.2009.04.057 [DOI] [PubMed] [Google Scholar]
- Sarrazy V., Viaud M., Westerterp M., Ivanov S., Giorgetti-Peraldi S., Guinamard R., Gautier E.L., Thorp E.B., De Vivo D.C., and Yvan-Charvet L.. 2016. Disruption of Glut1 in hematopoietic stem cells prevents myelopoiesis and enhanced glucose flux in atheromatous plaques of ApoE−/− Mice. Circ. Res. 118:1062–1077. 10.1161/CIRCRESAHA.115.307599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J., Asahina N., Kitano S., and Kino Y.. 2014. A comprehensive profile of ChIP-Seq-based PU.1/Spi1 target genes in microglia. Gene Regul. Syst. Bio. 8:127–139. 10.4137/GRSB.S19711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.W., Simon M.C., Anastasi J., and Singh H.. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 265:1573–1577. 10.1126/science.8079170 [DOI] [PubMed] [Google Scholar]
- Sergushichev A.A., Loboda A.A., Jha A.K., Vincent E.E., Driggers E.M., Jones R.G., Pearce E.J., and Artyomov M.N.. 2016. GAM: A web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 44:W194–W200. 10.1093/nar/gkw266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., and Pamer E.G.. 2011. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11:762–774. 10.1038/nri3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G.Q., and Cantley L.C.. 2013. Stem cell metabolism in tissue development and aging. Development. 140:2535–2547. 10.1242/dev.091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber P.B., Zhang P., Ye M., Welner R.S., Nombela-Arrieta C., Bach C., Kerenyi M., Bartholdy B.A., Zhang H., Alberich-Jordà M., et al. . 2013. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell. 49:934–946. 10.1016/j.molcel.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E.R., and Chitu V.. 2014. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 6:a021857 10.1101/cshperspect.a021857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Mildner A., and Jung S.. 2015. Macrophages: Development and tissue specialization. Annu. Rev. Immunol. 33:643–675. 10.1146/annurev-immunol-032414-112220 [DOI] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., and Green D.R.. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882. 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Israelsen W.J., Lee D., Yu V.W., Jeanson N.T., Clish C.B., Cantley L.C., Vander Heiden M.G., and Scadden D.T.. 2014. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 158:1309–1323. 10.1016/j.cell.2014.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P.S., and Thompson C.B.. 2012. Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect. Biol. 4:a006783 10.1101/cshperspect.a006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiktor-Jedrzejczak W., Bartocci A., Ferrante A.W. Jr., Ahmed-Ansari A., Sell K.W., Pollard J.W., and Stanley E.R.. 1990. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc. Natl. Acad. Sci. USA. 87:4828–4832. 10.1073/pnas.87.12.4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing E.J., Barczynski L.C., Waheed A., and Shadduck R.K.. 1985. Effect of Listeria monocytogenes infection on serum levels of colony-stimulating factor and number of progenitor cells in immune and nonimmune mice. Infect. Immun. 49:325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., and Thompson C.B.. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 105:18782–18787. 10.1073/pnas.0810199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., and Vousden K.H.. 2016. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 16:650–662. 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- Yang K., Neale G., Green D.R., He W., and Chi H.. 2011. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 12:888–897. 10.1038/ni.2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Hayashi S., Kunisada T., Ogawa M., Nishikawa S., Okamura H., Sudo T., Shultz L.D., and Nishikawa S.. 1990. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 345:442–444. 10.1038/345442a0 [DOI] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499:485–490. 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.