Abstract
During maturation of the phage HK97 capsid, each of the 415 capsid subunits forms covalent bonds to neighboring subunits, stabilizing the capsid. Crosslinking is catalyzed not by a separate enzyme but by subunits of the assembled capsid in response to conformational rearrangements during maturation. This report investigates the catalytic mechanism. Earlier work established that the crosslinks are isopeptide (amide) bonds between sidechains of a lysine on one subunit and an asparagine on another subunit, aided by a catalytic glutamate on a third subunit. The mature capsid structure suggests that the reaction may be facilitated by the arrival of a valine with the lysine to complete a hydrophobic pocket surrounding the glutamate, lysine and asparagine. We show that this valine has an essential role for efficient crosslinking, and that any of six other amino acids can successfully substitute for valine. Evidently none of the remaining 13 amino acids will work.
Keywords: Bacteriophage assembly, Virus capsids, Virus maturation, Capsid stabilization, Covalent cross-linking, Genetic selection
Introduction
The protein capsid (head) of bacteriophage HK97 is assembled initially into a structure known as Prohead I, which has 415 copies of the major capsid protein (MCP) arranged in T=7 icosahedral symmetry, 12 copies of the portal protein, and about 50 copies of the maturation protease (Conway et al., 2007; Duda et al., 1995a; Duda et al., 1995b). Maturation of Prohead I to the superficially similar Prohead II entails proteolysis, removing the N-terminal 102 amino acids from each copy of the MCP inside the closed shell, and autoproteolysis of the protease. The resulting peptides probably diffuse through small holes in the Prohead lattice. Expansion converts Prohead II to mature Head, the final structure. Expansion is triggered by DNA packaging in vivo or by solvent changes in vitro. It involves extensive conformational changes within and between copies of the MCP, resulting in a capsid diameter increase (Conway et al., 1995; Duda et al., 1995a). Perhaps the most striking feature of expansion is that ≈400 covalent crosslinks form between pairs of MCP subunits (Popa et al., 1991), with the result that MCP monomers are linked into covalently continuous rings of 6 or 5 subunits (Duda, 1998; Duda et al., 1995a). The rings are mutually interlinked topologically at the 3-fold positions of the capsid lattice (Helgstrand et al., 2003; Wikoff et al., 2000), forming the robust structure we have called viral chainmail. Efficient crosslinking requires that the subunits first assemble into capsids (Duda et al., 1995a; Duda et al., 1995b; Hendrix and Duda, 1998), and capsid expansion is required for crosslinking (Duda et al., 1995a; Gan et al., 2004). Efficient crosslinking in turn is essential to produce viable HK97 phage (Ross et al., 2005).
Mechanism of crosslinking
We describe here experiments to elucidate the autocatalytic chemical mechanism of crosslinking. The crosslink is an isopeptide (amide) bond between the side chains of lysine 169 (K169) of one subunit and asparagine 356 (N356) of an adjacent subunit (Duda et al., 1995a). All of the crosslinks are clearly visible in the high-resolution structure of mature Head (Helgstrand et al., 2003; Wikoff et al., 2000). The Head structure also shows that K169 is part of a β-hairpin, termed the E-loop, that extends partway over the surface of the adjacent subunit, bringing K169 near the location of N356 of the adjacent subunit to which it will crosslink (Figure 1). Structural and biochemical experiments show that the E-loops in Prohead I or Prohead II are poorly ordered and extend away from the outer surface of the capsid shell; the E-loops dock on an adjacent subunit and become ordered as part of the conformational changes that underlie capsid expansion (Conway et al., 2001; Gan et al., 2004; Gan et al., 2006; Lee et al., 2008; Li et al., 2005; Wikoff et al., 2006). The K169 and N356 residues that are destined to react are ~35 Å apart in Prohead II, but once the E-loop docks on the adjacent subunit, K169 projects into a cleft on the adjacent subunit to reach its N356 partner (Gan et al., 2006; Gertsman et al., 2009; Wikoff et al., 2006; Wikoff et al., 2000).
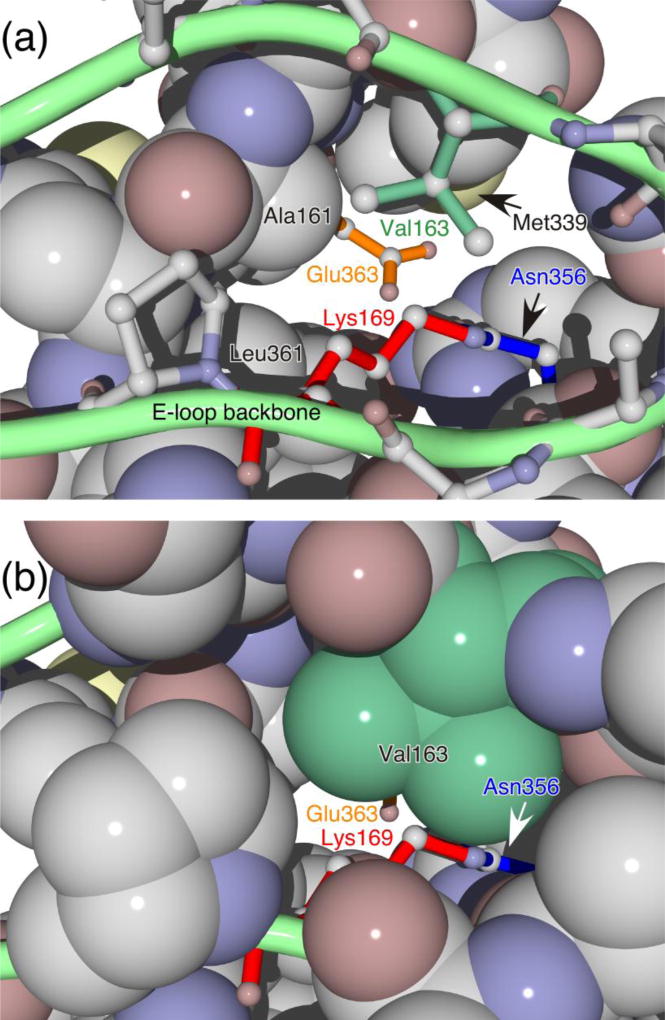
Figure 1. Structure of the crosslinking reaction site.
The view is from the outside of the capsid and represents the structure of the mature head in which the crosslink has formed. Top panel: The green loop represents the E-loop of the subunit that supplies the reactive lysine Lys 169). The side chain of Lys 169 is shown as red sticks and is attached via the crosslink to Asn 356 (blue sticks) from the adjacent subunit. The side chain of Val 163 is shown as green sticks, and the side chain of catalytic residue Glu 363, which is part of a third subunit, is shown as orange sticks. Most other side chains are represented as space-filling models, and the side chains of Met 339 and Leu 361 are indicated. Bottom panel: Same as top panel, except that Val 163 and the other E-Loop residues are shown in spacefilling form.
The high-resolution structure of mature Head shows an unexpected feature that may provide insights into the catalytic mechanism of crosslinking: the carboxylate of glutamate 363 (E363) from a third subunit is located ~2Å from the crosslink (Helgstrand et al., 2003; Wikoff et al., 2000). E363 is in a comparable position in Prohead II, where the crosslink has not yet formed (Gertsman et al., 2009). We proposed that E363 may play a catalytic role in crosslinking (Helgstrand et al., 2003; Wikoff et al., 2000). Subsequent experiments support this hypothesis (Dierkes et al., 2009) and further suggest that an important property of E363 for performing this catalytic role is its ability to accept a proton from K169—that is, to act as a general base.
We proposed the model shown in Figure 2 from the facts outlined above. We suggest that E363, the catalytic glutamate, accepts a proton from the ε-amino group of K169, converting the lysine ε-amino group into a good nucleophile that attacks the γ-carbon of N356 to form the observed isopeptide crosslink. The side chains of lysine and glutamate have very different pKA’s in aqueous solvent and thus a proton would not be expected to pass readily from lysine to glutamate. However, examination of the high-resolution structure of Head shows that the crosslink and the catalytic glutamate – which mark the presumed catalytic site – are enclosed within a hydrophobic environment bordered by the side chains of three other amino acids: Leucine 361, Methionine 339, and Valine 163 (Figure 1). If the positive charge of K169 and the negative charge of E363 are indeed enclosed together in a hydrophobic pocket prior to crosslinking, the hypothesized transfer of a proton from K169 to E363 is expected to be energetically more favorable. Two of the three hydrophobic amino acid side chains that line the interior of the hydrophobic pocket occupy essentially the same positions in Prohead II as in Head. However the third hydrophobic residue, Valine 163 (V163), is part of the E-loop that also contains K169. The side chains of K169 and V163 extend from the same side of the E-loop with the consequence that when the E-loop docks on the neighboring subunit and positions K169 for crosslinking, it simultaneously positions V163 to complete the hydrophobic cage surrounding the crosslinking reaction site. The experiments reported here test various aspects of the reaction mechanism proposed in Figure 2, particularly the role of V163.
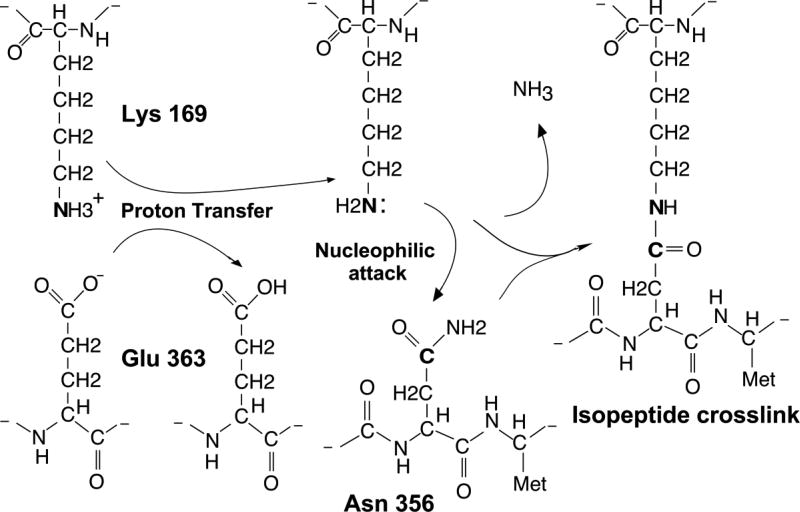
Figure 2. Proposed reaction mechanism.
The catalytic residue Glu 363 is proposed to accept a proton from the epsilon amino group of reactant Lys 169, rendering Lys 169 a good nucleophile that attacks the gamma carbon of the crosslinking partner, Asn 356. The proton transfer from Lys 169 to Glu 363 is proposed to be facilitated by a hydrophobic environment around the reaction site, of which a crucial component is supplied by Val 163.
Results
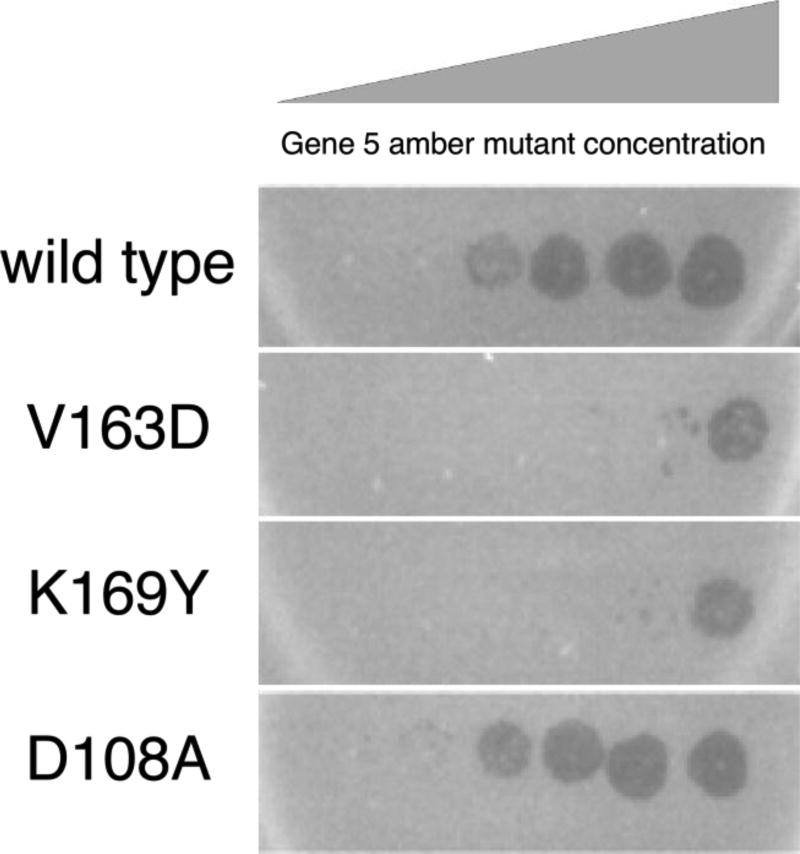
We wished to determine whether V163 plays an essential role in assembly and head maturation, and particularly whether V163 has a direct role in crosslinking. We changed the V163 codon (GTG) to an aspartate codon (GAT) in the context of plasmid pV0. This plasmid has HK97 genes 4 (protease) and 5 (MCP) under the control of a phage T7 promoter and an inducible T7 RNA polymerase (Duda et al., 1995b). This V163D mutant MCP is unable to support phage growth, as judged by its failure to complement an infecting phage bearing an amber mutation in gene 5, the MCP gene (Figure 3).
Figure 3. Complementation assay for functionality of mutant forms of the major capsid protein gp5.
The photographs show bacterial lawns of amber non-suppressing strains expressing gp5 of the indicated genotype. Each lawn was spotted with a dilution series of HK97 carrying an amber mutation in gene 5. Clearing of the lawn at higher dilutions of the infecting phage (more leftward spots) indicates positive complementation. Mutant D108A complements as well as wild type, but mutants V163D (the focus of this study) and K169Y (missing the crosslink reactant lysine) both fail to complement.
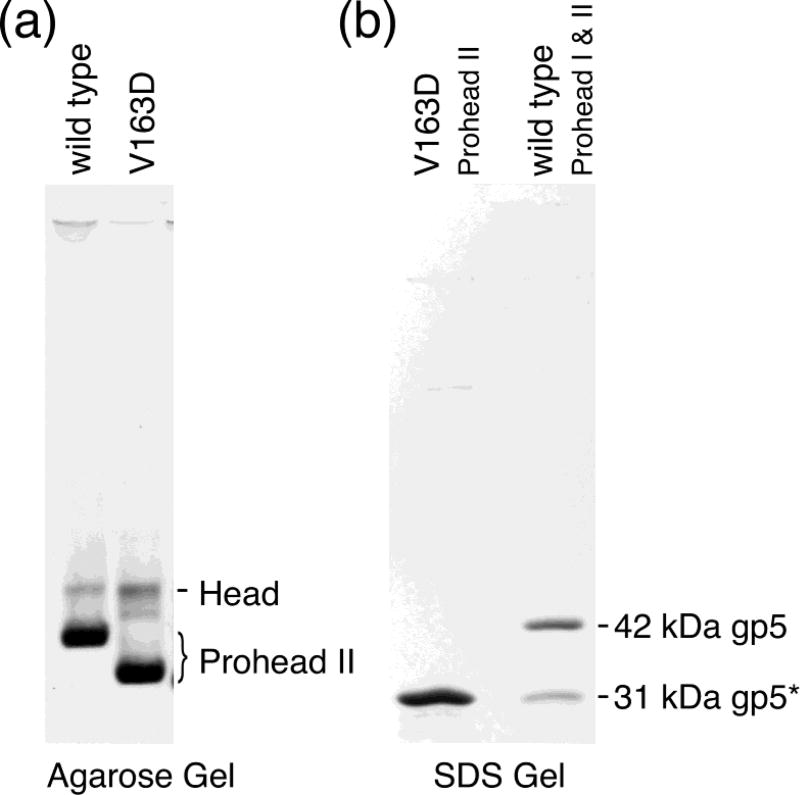
We expressed the V163D mutant MCP from the plasmid that also carries the protease to ask whether V163D can complete capsid assembly and maturation steps. With wild-type genes, this construct abundantly produces Prohead II that can crosslink efficiently in response to certain changes in solvent composition. The V163D mutant likewise produces Prohead II-like particles at the same high level seen for the wild type. We characterized these particles first by agarose gel electrophoresis (Figure 4a). The mutant particles migrate as a band characteristic of Prohead II except faster migrating than wild-type Prohead II, a property we ascribe to the 420 extra negative charges on the outside of these Proheads due to the mutation. We also analyzed the V163D particles on an SDS polyacrylamide gel, which showed that all of the mutant MCP, like the wild type, had been processed to the 31 kDa cleaved form (Duda et al., 1995a; Duda et al., 1995b) (Figure 4b). We conclude that mutant V163D MCP can fold, assemble, and undergo cleavage indistinguishably from wild-type MCP.
Figure 4. Gel analysis of wild-type and V163 mutant proheads.
(a) Agarose gel of Prohead II of wild-type and V163D mutant capsids. Each capsid protein was co-expressed with protease. After incubation, cells were lysed, and the lysate was processed through our prohead purification protocol. The resulting particles were analyzed on a 0.8% agarose gel. The heavy bands near the bottom of the gel are the proheads. The slower migrating lighter bands are spontaneously expanded proheads (i.e., heads). (b) SDS-polyacrylamide gel of the V163D mutant Proheads. The sample in the left lane is the V163D mutant Prohead preparation shown in lane 2 of the agarose gel in Figure 4. It displays a single subunit size. The right lane has a mixture of wild-type Prohead I and Prohead II, which are made of 42 kDa unprocessed gp5 or 31 kDa proteolytically processed gp5*, respectively. They serve as size standards and identify the subunits of the mutant prohead as the 31 kDa processed form of gp5.
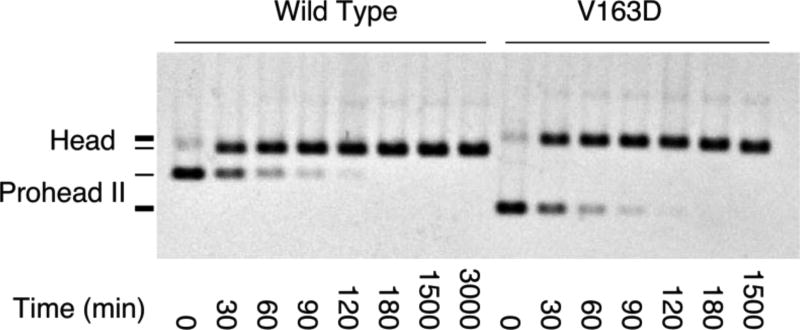
The next step in head maturation is expansion, leading to crosslink formation. We treated V163D and wild-type Prohead II with high-salt buffer at low pH, one of the treatments that induces expansion of Prohead II (Dierkes et al., 2009; Duda et al., 1995a; Gan et al., 2004), and analyzed the particles on an agarose gel to separate expanded from unexpanded particles (Figure 5). Both wild-type and mutant proheads expanded under this treatment. Expansion appears to follow the same kinetics for the two particles, as verified by quantifying all the bands on the gel (data not shown).
Figure 5. Expansion of wild-type and V163 mutant Prohead II.
Proheads were treated with DMF expansion buffer for the indicated times and analyzed on a 0.8% agarose gel. The positions of Prohead II and expanded Heads are indicated in the figure. The V163D mutant particle positions are indicated by thicker lines on the left. The mutant proheads migrate faster than wild-type proheads, as also seen in Figure 4(a), presumably because of the increased negative charge from the V163D mutation. This added negative charge is apparently buried during expansion, so the mutant heads no longer migrate faster than wild type.
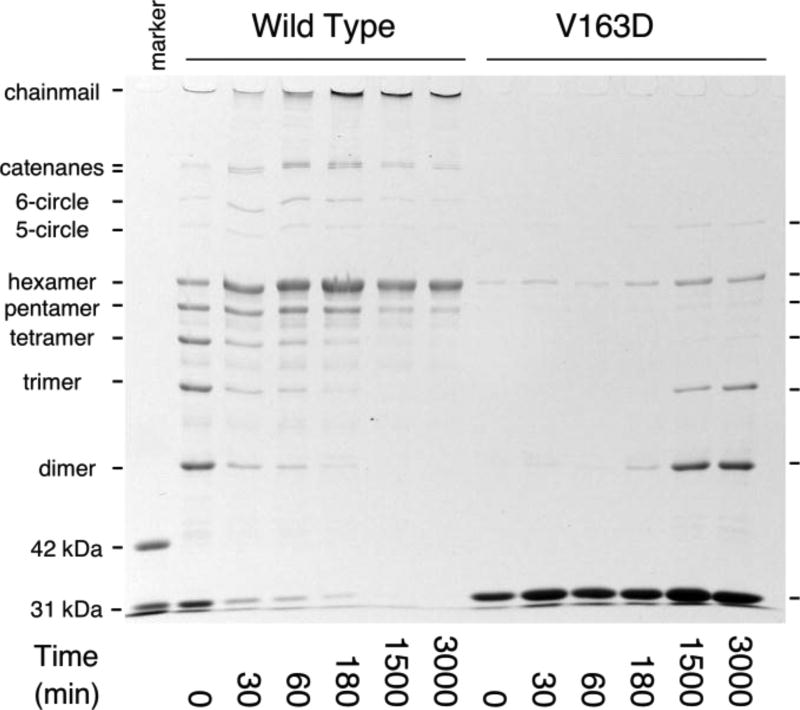
The conformational changes of expansion facilitate crosslinking for wild type, and we asked whether the same could be true for V163D by inducing expansion and assaying for the appearance of crosslinked species by SDS polyacrylamide gel electrophoresis (Figure 6). We used three different methods for expansion, and all three gave essentially the same results. Figure 6 compares wild-type and V163D Prohead II expansion induced by treatment with DMF buffer. For wild-type Prohead II, the non-crosslinked (monomer) form of the subunit is consumed by crosslinking by the end of the time course. Over time, the protein progresses through a series of crosslinked forms, from dimer to linear hexamer plus topologically circular pentamer and hexamer and eventually to the fully crosslinked network that is trapped in the well of the SDS gel (Duda, 1998; Duda et al., 1995a; Gan et al., 2004). In contrast, the monomer band intensity for V163D Prohead II does not decrease appreciably over the course of the experiment. Small amounts of dimer, trimer and linear hexamer appear, but only very late in the reaction, and no highly crosslinked mature chainmail network is detected.
Figure 6. Expansion-induced crosslinking by wild-type or V163D mutant Prohead II.
Proheads were treated with DMF expansion buffer for an hour and neutralized to initiate crosslinking. The “marker” lane has a mixture of wild-type Prohead I and Prohead II, as in Figure 4(b), to show the migration of unprocessed (42 kDa) and processed (31 kDa) gp5 monomers.
To summarize, the V163D mutant protein folds and assembles into proheads as well as the wild-type protein; it is cleaved normally by the maturation protease; the resulting Prohead II expands with normal efficiency and kinetics; however, crosslinking is severely defective. Possibly owing to crosslinking failure, the V163D protein is not functional, as judged by its inability to complement plaque formation for a phage bearing an amber mutation in the MCP gene.
To learn what other amino acids could substitute for V163 and support plaque formation – and by inference efficient crosslinking – a mutant V163D (codon GAT) was introduced by recombineering into an otherwise wild-type HK97 prophage. We also included a silent substitution at V162 (GTG to GTC) to mark all revertants and thus exclude contaminating wild-type HK97 phage from scoring as revertants. UV-light induction of the mutant prophage gave plaque-forming phages about 109-fold less frequently than induction of a wild-type prophage. After viable revertant plaque recovery and purification, the MCP gene was sequenced from four plaque-forming phages recovered independently (Table 1). Codon 163 was altered in each phage, once to CTT – a leucine codon – and three times to GTT – a valine codon – while the linked silent substitution at codon 162 was retained in all four. One of the GTT revertants had an additional substitution K166R (AAA to AGA), which must be a tolerated irrelevant change. This V163D reversion analysis shows that leucine can replace valine for phage growth. Our interpretation is that valine or leucine arriving with the E-loop can serve as a lid to close the hydrophobic cage around the crosslinking reaction site.
Table 1.
Spontaneous Revertants of Lethal Mutation V163D.
| Revertant | Residue (Codon) | Mutation (Codon) | Revertant (Codon)* |
|---|---|---|---|
| 1 | V163 (GTG) | V163D (GAT) | V163L (CTT) |
| 2◊ | V163 (GTG) | V163D (GAT) | V163V (GTT) |
| 3◊ | V163 (GTG) | V163D (GAT) | V163V (GTT)# |
| 4 | V163 (GTG) | V163D (GAT) | V163V (GTT) |
All revertants retained the linked codon change at codon 162 (GTG to GTC).
Two revertant plaques were recovered from the same induction experiment.
An additional change was found nearby, K166R (AAA to AGA).
Owing to the technical challenge of recovering a large number of rare revertants, a different approach was implemented to streamline testing the complete set of natural amino acids at position 163. PCR products were made with a randomized library of triplets at codon 163; these libraries were recombineered into a lysogen. Two independent versions of this randomized library were made: the first had 32 codons as NNW, while the second had 32 codons as NNS. (N means that all four bases were included during synthesis at that position, while W means A or T and S means G or C.) Only viable plaque-forming phages should be recovered after recombineering and prophage induction. Since every possible codon should be represented at about 3% in one library or the other, codons for every functional amino acid replacement at codon 163 should be recovered repeatedly from a sufficient number of plaque-forming phages. We recovered and characterized 68 individuals with the marker substitution but with no other changes besides codon 163. Sequencing showed that codons for six different amino acids were found at position 163 – Cys, Gln, Ile, Leu, Thr, and of course Val (Table 2). Transformation and induction was done multiple times for each library to reduce the recovery of siblings. An alternative NNW library had a non-silent linked marker D161E and produced both Gln and Ala revertants. The Ala revertant was only found in this context, so we independently created the V163A mutation on a plasmid without the linked D161E mutation and found that V163A was viable by complementation, so Ala is also included in Table 2. Because rapid crosslinking is apparently required for HK97 phage viability (Dierkes et al., 2009), we interpret plaque formation to indicate a sufficiently rapid rate of crosslinking leading to a fully crosslinked capsid.g.
Table 2.
Functional Replacements for V163 from Library Selections.
| Amino Acid | Codon | Number | Library |
|---|---|---|---|
| 1. Ile | ATC, ATT, ATA | 19 | NNW, NNS |
| 2. Val | GTG, GTA | 18 | NNS, NNW |
| 3. Thr◊ | ACG, ACC, ACT | 18 | NNS, NNW |
| 4. Gln | CAG | 8 (+ 1) | NNS, NNW-1 |
| 5. Cys | TGT | 4 | NNW |
| 6. Leu | TTG | 1 | NNS |
| 7. Ala# | GCA | 1 | NNW-1 |
All revertants retained a linked codon change at codon 161 (GAC to GAT for libraries NNW and NNS, and GAC to GAG for libary NNW-1).
This mutation was only isolated in the presence of a D161E mutation, so we independently created the V163A mutation in a wild-type context and found that it was viable using plasmid complementation.
The recombinants with randomized sequences at codon 163 should include multiple examples of all possible codons, so we conclude that only those seven amino acids that were actually recovered at codon 163 in plaque-forming phages are able to support efficient crosslinking. By this argument, the other thirteen amino acids – Asp, Asn, Arg, Glu, Gly, His, Lys, Met, Phe, Pro, Ser, Trp, Tyr – that were not recovered at position 163 in plaque-forming phages probably do not support efficient crosslinking or fail at some prior step in maturation.
A potential flaw in this argument, pointed out by a reviewer, is that failure to recover a particular amino acid could be due to bias in the “randomized” codon sequences rather than failure of that amino acid to be functional at position 163. To address this concern, we constructed plasmids carrying codons at position 163 specifying six of the thirteen amino acids (Asn, Glu, Gly, Met, Phe, Ser) that we failed to recover in the experiments described above. These all failed to complement. As controls, we included the previously characterized plasmids encoding Asp, Ala, and Val (wild type) and plasmids encoding the other five amino acids previously recovered in plaque-forming phages. These all complemented well, except for Asp which of course did not complement. The complementation results are summarized in Table 3. For all of the variants whose complementation properties are reported in Table 3, we also assessed their ability to assemble proheads. All of the variants, both complementing and non-complementing, made proheads (Table 3).
Table 3.
Phenotypes of thirteen V163 mutants when expressed from plasmids.
| Position 163 amino acid |
Complementation efficiency* |
Complementation score# |
Recovered as viable phage |
Prohead production§ |
|---|---|---|---|---|
| Val | = 1.0 | + | (wt) | + |
| Ile | 1.0 | + | + | + |
| Thr | 1.0 | + | + | + |
| Gln | 1.0 | + | + | + |
| Cys | 1.0 | + | + | + |
| Ala | 1.0 | + | + | + |
| Leu | 0.1 | + | + | + |
| Met | 0.03 | − | − | + |
| Asn | 0.001 | − | − | + |
| Ser | 0.001 | − | − | + |
| Phe | 0.00003 | − | − | + |
| Gly | 0.00001 | − | − | + |
| Glu | 0.00001 | − | − | + |
| Asp | 0.00001 | − | − | + |
Complementation was done with HK97 phage bearing an amber mutation in gene 5 on a lawn of bacteria with a plasmid expressing the named V163X variant. The efficiency reported is calculated relative to the complementation achieved when using the wild-type plasmid, which is set to “1”.
Complementation of 0.1 or higher was judged to be “+”, lower than 0.1 as “–”.
Prohead production was determined in small scale cultures after induction of gene expression from a plasmid carrying the wild-type gene 4 and the named V163X variant of gene 5.
Discussion
Determining the chemical mechanism of the HK97 crosslinking reaction presents some interesting experimental problems. For example, attempts to synchronize all of the individual reactions in the structure suffer from the fact that seven “quasi-equivalent” geometrically distinct groups of reaction sites exist in the T=7 Prohead II, and apparently these do not all react with the same kinetics (Duda, 1998; Duda et al., 1995a; Gan et al., 2004; Gan et al., 2006; Lee et al., 2008). The genetic approach we have taken here for testing the other natural amino acids in place of V163 ties the available biochemical and structural information about the crosslinking reaction to the biological outcome of forming viable phage and gives results that support the idea that V163 (or its functional replacement) helps to form an appropriate environment for crosslinking. The set of functional replacements provides new information about the nature of that appropriate reaction environment.
With the exception of Gln and Thr, which will be discussed below, the functional amino acids at position 163 are hydrophobic and roughly the same size as Val. This is consistent with the hypothesis that Val (or its successful substitute) completes the hydrophobic pocket enclosing the reaction site. This is further supported by our observation that Asp fails to support crosslinking and probably none of the four charged amino acid side chains supports crosslinking, since they were never recovered from the randomized libraries, and the two that were tested explicitly, Asp and Glu, do not support crosslinking.
The size of the side chain at position 163 also appears to be significant. Despite their hydrophobicity, none of the aromatic side chains was recovered, so we conclude that none of these supports crosslinking, and we showed this explicitly for Phenylalanine. We suggest that this may indicate an inability of the rigid and bulky aromatic side chains to fit accurately into the cleft in the neighboring subunit normally occupied by V163. Two distinct consequences of failure of aromatic side chains to fit accurately might prevent efficient crosslinking. First, failure of the side chain to dock fully would prevent the E-loop from fully descending and consequently prevent the reactant K169 that is part of the E-loop from reaching the reaction site. Second, failure of the side chain to descend to the level of the reaction site might result in failure to close the hydrophobic pocket and thus failure to displace polar solvent from the hydrophobic pocket.
Our tentative conclusion is that the amino acid at position 163 must have two features to support crosslink formation: 1) it must offer a surface with enough hydrophobic character to close a hydrophobic pocket around the reaction site, and 2) it must exclude polar molecules, either bulk solvent or polar parts of the side chain itself, from the vicinity of the hydrophobic pocket.
The role of Thr or Gln, which apparently support crosslinking when at position 163, seems less clear. Both Thr and Gln side chains offer hydrophobic and polar parts. Compared to Ser and Asn, which do not support crosslinking, Thr and Gln, respectively, have one additional carbon atom (methyl- and methylene-, respectively). While we have no direct information about how these side chains are positioned in the cleft where the wild-type V163 normally rests, we speculate that the additional carbon groups may give Thr and Gln sufficient hydrophobic character to contribute to the necessary environment of the reaction site.
The cases of Ser and Cys are interesting in this context. The side chains of these two amino acids are essentially the same size and shape, yet Cys supports crosslinking while Ser does not. We speculate that the difference may lie in the fact that Cys is substantially more hydropathic (hydrophobic) than Ser.
The two smallest side chains, those for Ala and Gly, gave different results - only Ala was able to support crosslinking. Unlike the aromatic side chains, these both should fit into the valine cleft and so allow full descent of the E-loop and the reactant K169. We suggest that the failure of Gly, but not Ala to support effective crosslinking implies that there are subtle aspects of the reaction environment that we fail to appreciate. It could be that only Ala, but not Gly can effectively seal the reaction pocket, suggesting the possibility that solvent exchange is inhibitory and must be prevented to allow the reaction to occur. It may also be that the methyl side chain of Ala but not the hydrogen side chain of Gly provides sufficient hydrophobic surface to support the reaction.
This view of the role for V163 in supporting crosslinking is consistent with and so provides support for the reaction scheme illustrated in Figure 2. Several previously derived results similarly provide support for the reaction scheme. These include 1) identification of the two reactant amino acids K169 and N356 that join to form the crosslinked bond (Duda et al., 1995a), as well as characterizing mutants of those residues (Duda et al., 1995b), 2) structural analysis of proheads and mature heads (Conway et al., 1995; Conway et al., 2001; Gan et al., 2004; Gan et al., 2006; Gertsman et al., 2009; Helgstrand et al., 2003; Lee et al., 2008; Wikoff et al., 2006; Wikoff et al., 2000), including the positions of the E-loop, the two reacting residues and the catalytic E363 residue, before and after the reaction, 3) characterizing E363 as the catalytic residue (Dierkes et al., 2009), including evidence for it acting as a general base. While these results, even in aggregate, cannot be said to provide rigorous proof of the proposed reaction scheme, we suggest that the scheme is both chemically plausible and provides an excellent explanation for the experimental results.
When we first reported that the MCP of HK97 is covalently crosslinked to other MCP subunits (Popa et al., 1991), there were already hints that this phenomenon might occur beyond phage HK97, because two mycobacteriophages (Ford et al., 1998; Hatfull and Sarkis, 1993) as well as HK97’s close relative, HK022, showed the same unusual pattern of MCP bands in an SDS gel of virions as had been characterized for HK97. Several more tailed phages (Borriss et al., 2007; Gilakjan and Kropinski, 1999; Lubbers et al., 1995; van Sinderen et al., 1996) have subsequently been shown to have the SDS gel pattern indicative of crosslinking. We speculate that MCP crosslinking may be moderately common among the tailed phages; if so, it is an intriguing but as yet unanswered question whether they all accomplish crosslinking by using the same chemical mechanism that we outlined here for HK97. It should be noted, however, that not all tailed phages crosslink their MCP subunits. Most of the phages studied in detail over the past 60 years do not crosslink their MCP, though many of them have alternative means to stabilize their capsids, such as accessory proteins that bind to the outside of the capsid. We regard MCP crosslinking as a phenotypic feature that some tailed phages employ and others do not, in the same way that some tailed phages have contractile tails and others do not.
An interesting but distinct example of spontaneous post-translational formation of an isopeptide crosslink bond has been documented in the pilus structures of Gram-positive bacteria (Hendrickx et al., 2011; Hendrickx et al., 2012; Kang et al., 2007; Wang et al., 2013; Zahner et al., 2011). Some examples of these crosslinks join adjacent subunits of the pilus structure, while others occur within individual subunits. The crosslinks between subunits are catalyzed by transpeptidase enzymes called sortases (Ton-That and Schneewind, 2004), while the crosslinks within subunits are self-catalyzed via a mechanism that appears to closely follow the bacteriophage capsid paradigm (Kang et al., 2007). Although the structures of the phage proteins and the pilus proteins are quite different, as are the geometries of the two types of reaction sites, it appears there may be much in common between the two reaction mechanisms. The pilus reaction has a catalytic residue, which is either glutamate or aspartate in different instances, the isopeptide bond forms between side chains of lysine and asparagine (or aspartate in one instance), and the hydrophobic environment for the reaction appears to be supplied by the side chains of flanking aromatic amino acids (Kang et al., 2007).
Materials & Methods
Bacteria, plasmids, and lysogens
E. coli strains Ymel (mel-1 supF58) and LE392 (F-, metB1, trpR55, lacY1, galK2, galK22, supE44, supF58, hsdR514) were used for propagating HK97 wild-type and amber mutants. T7 promoter expression strain E. coli BL21(DE3)/pLysS (Studier et al., 1990) was used for plasmid complementation tests and for production of proheads from plasmids. E. coli DH10B was used for molecular cloning steps. Plasmid pV0 expresses HK97 protease gp4 and major capsid protein gp5 and efficiently produces HK97 Prohead II after induction (Duda et al., 1995a; Duda et al., 1995b). Mutation V163D in gene 5 was created in plasmid pV0 by site-directed mutagenesis using overlapping primers GCCGACGTCGATGCAGAGAA and TTCTCTGCATCGACGTCGGC using the QuickChange protocol (Stratagene). Candidate plasmids were sequenced to ensure that only the desired changes were present. Plasmids containing all of the mutations in Table 3 except V163D were created by PCR based methods that produced a DNA fragment with 25 bp overlapping ends suitable for assembly into a SacI-DraIII-cleaved wild-type plasmid (pV0SacI) backbone using the NEBuilder enzyme mix (New England Biolabs, Ipswich, MA). For the mutants isolated as revertants in phages, a phage stock was added directly to a PCR reaction to generate an appropriate fragment containing the mutation and used for assembly. For the rest, a four-primer site-directed mutagenesis method (Adereth et al., 2005) was used to create the desired mutation in a PCR fragment suitable for NEBuilder assembly. Plasmid clones were sequenced to ensure that the sequences of all incorporated DNA fragments were correct. All HK97 mutant lysogens were derived from the recombineering strain BW25113 (Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, rrnB-3, hsdR514 ) containing the plasmid pKD46, which expresses the phage λ Red recombination proteins under arabinose control (Datsenko and Wanner, 2000). The galK positive/negative selection system was used to facilitate selecting recombinants (Warming et al., 2005). A galK deletion of BW25113 that is also a HK97 lysogen called DT002 (Tso et al., 2014) was used to create strain DT005 in which the HK97 DNA from codon 224 in gene 4 through codon 264 in gene 5 was replaced by galK (Tso et al., 2014) using recombineering. This strain was subsequently used for introducing mutations from plasmids into the HK97 lysogen via further recombineering steps as described below.
Generating dsDNA fragments with libraries of substitutions at residue 163
Libraries of double-stranded DNA recombineering substrates with a randomized codon near their centers were made using a PCR-based method described previously (Adereth et al., 2005), but using Phusion polymerase (Finnzymes Oy) instead of Pfu (Stratagene). The technique entails creating two PCR products that are each phosphorylated on one end, which are subsequently joined by blunt-end ligation and re-amplified. PCR primers were designed in which the nucleotide sequence NNS or NNW (N: any nucleotide; S: C or G; W: A or T) replaced the normal V163 codon (GTG) in order to produce randomized codons at position 163. Production of the DNA libraries was done in two steps. First, DNA fragments which contain the first half and the second half of gene 5 were made by PCR; the first half using primers LP1 = 5´GGTGACGGGAAGCAGGGG with V163XR = 5´P-SNNCACATCGGCGTTATTGGTAAACACC or V163X2RS = 5´P-WNNCACATCGGCGTTATTGGTAAACACC or V163X-1R WNNCACCTCGGCGTTATTGGTAAACACC; the second half using primers gp5f1 = 5’P-GCAGAGAAAGCACTGAAGCCAG and gp6-r1 = 5´GTCGTCCCTGTCGTCTTCCTC. The two first-round PCR products were gel purified (Gubin and Kincaid, 1998), ligated, and re-amplified with primers LP1 and gp6-r1 to synthesize full-length gene 5. The final products that contain a randomized codon 163 in HK97 gene 5 were gel purified and used for recombineering as described below. A silent mutation (GAC to GAT at codon 161) was included in primers V163X2RS (for library NNW) and V163XR (for library NNS) to use as a tag indicating incorporation and to detect contaminating wild-type phage or wild-type DNA during recombineering. A third library (NNW-1 ) used a primer V163XR1 with a D161E linked mutation (GAC to GAG at codon 161), but most of this library was discarded after the first few revertants were sequenced.
Constructing mutant HK97 lysogens and selection of surviving phages
Recombineering techniques (Court et al., 2002; Datsenko and Wanner, 2000) were used to transfer the mutation by homologous recombination into the HK97 lysogen DT005 (above and (Tso et al., 2014)) in which the HK97 DNA from codon 224 in gene 4 through codon 264 in gene 5 is replaced by galK. The galK gene in HK97 gene 5 was replaced by DNA fragments which contain the desired mutations and recombinants were selected on M63 minimal salts plates containing 2-deoxy-galactose (DOG) (Henderson and Giddens, 1977), which counterselects strains expressing the galK gene.
HK97 lysogen DT005 was prepared for recombineering by growth in LB medium with 100 µg/ml ampicillin and 10 mM L-arabinose at 30°C to a density of ~4 × 108 cells/ml to accumulate lambda Red proteins. Cells were chilled on ice for 10 min, centrifuged at 6700 rpm for 10 min at 4°C, washed three times with ice-cold sterile water and suspended in a small volume of ice-cold water. The dsDNA recombineering substrates were then introduced into the lysogens via electroporation. Cells were diluted into LB with 10 mM L-arabinose to recover and to allow further expression of functional lambda Red proteins. After incubation at 30°C for 3 hours, cells were washed three times with 1x minimal salts (M63 or M9) to remove LB medium and spread on the DOG-containing selection plates. Alternatively, for screening of cells recombined with DNA containing randomized-codon libraries, after 3 hours of growth the cells were washed in UV induction buffer and immediately induced, as described below.
Controls included were 1) HK97 lysogen without L-arabinose induction but with transformation by DNA substrates, 2) HK97 lysogen with L-arabinose induction but without transformation by DNA substrates. No colonies or very few colonies would normally appear on these control plates. Recombination efficiency was one recombinant in ~106 surviving cells after transformation. To quickly screen revertants and to rule out contamination by wild-type HK97, a silent mutation was generated next to the designed mutation in gp5 to create a restriction polymorphism. Transfer of mutations into lysogens was confirmed by amplifying segments of the gene 5 DNA from candidate clones by PCR, followed by testing for restriction polymorphisms and sequencing. Plasmid pKD46 was sometimes evicted from the lysogens by growth at 42°C, followed by testing for antibiotic resistance before being used for further experiments.
Induction of HK97 lysogens by ultraviolet irradiation
Mutant lysogen DT-V163D was induced by ultraviolet-light (UV) irradiation, and any resulting revertant phages were selected by their ability to plaque on E. coli host Ymel. Pools of transformed cells from recombineering experiments with random libraries at codon 163 were induced by UV-irradiation immediately after the 3 hr post-transformation growth step. Lysogens to be tested were normally grown until early exponential phase, pelleted by centrifugation and resuspended in UV induction buffer (10 mM Tris-Cl pH 7.5, 5mM MgSO4). A control strain DT-HK97 (HK97 lysogen without pKD46), was treated with various doses of UV light to test for optimal induction conditions for HK97, usually 5 to 10 seconds. After UV exposure, lysogens were pelleted immediately, suspended in LB medium, and incubated at 37°C. Chloroform was added after 1.5 hours to aid in lysis. After 2 hours, the lysate was cleared by centrifugation, and the supernatant was used for plaque assays. For all mutant lysogens, a set of candidate revertants which were able to plaque on Ymel were randomly selected for further analysis. Segments of HK97 gene 5 DNA from each candidate plaque were amplified by PCR, and the amplified DNA was tested for any restriction polymorphisms, and sequenced.
Complementation tests
Spot-complementation tests were done to compare the efficiency of plating (EOP) of amber mutant phage on a strain which produces mutant capsid proteins from plasmids to the EOP when the wild-type gp5 is provided from a control plasmid (Dierkes et al., 2009; Ross et al., 2005). Overnight cultures of BL21(DE3)/pLysS containing plasmids were grown in LB medium with 50 µg/ml ampicillin, 25 µg/ml chloramphenicol, and 0.4% maltose; 0.2 ml of each culture to be tested was mixed with 2.5 ml soft agar and poured onto an LB plate containing the same antibiotics. Plasmid gene expression was induced by supplementing the soft agar with 1% lactose. Stocks of wild-type HK97 and HK97 amber mutants in genes 4 and 5 at 109 pfu/ml were serially diluted in five 10-fold steps with TMG buffer (Tris-HCl pH 7.5, 10 mM MgSO4, 0.01% gelatin). After the soft agar solidified, 0.004 ml of each phage dilution was spotted onto lawns seeded with mutant or control strains. Plates were examined for phage growth after overnight incubation at 37°C. A score of 10 was given for each fully cleared spot and a score of 3 given for a partially cleared spot and the scores multiplied to approximate the plaquing ability. For relative complementation efficiency calculations, the score for a mutant plasmid was divided by the score obtained for the wild-type plasmid.
Preparation of HK97 Prohead II V163D
Strain BL21(DE3)pLysS carrying plasmid pV0-V163D was grown in 1 L LB medium at 28°C, induced by adding 0.4 mM IPTG, and incubated for 16 hrs. Proheads were purified by differential centrifugation, polyethylene glycol precipitation, velocity sedimentation in glycerol gradients, and ion-exchange chromatography as described previously (Dierkes et al., 2009; Duda et al., 1995a; Li et al., 2005). The proheads in peak fractions from the ion-exchange step were pelleted at 35,000 rpm in a Beckman Type 45Ti rotor, suspended in a small volume of buffer (20 mM Tris-HCL, 40 mM NaCl), and stored at 4°C.
Small-scale (mini-prohead) expression tests for mutant function
High-level expression of the cloned HK97 genes was done using 1.5 mL cultures of strain BL21(DE3)pLysS carrying wild-type or mutant plasmids (pV0-V163X) for 24 hrs under vigorous aeration conditions using the TYM5052 autoinduction media developed by Studier (Studier, 2005). Under these conditions, which have been described in detail (Oh et al., 2014; Tso et al., 2014), HK97 capsid proteins are expressed at levels high enough to be clearly visible in crude extracts using stained SDS-polyacrylamide gels and assembled capsomers, proheads, or heads are easily detected using native agarose gels of the same extracts as described below.
Electrophoresis in agarose and polyacrylamide gels
Native agarose gels were run in TAMg buffer (40 mM Tris base, 20 mM acetic acid, pH 8.0, 1 mM MgSO4) to detect the presence of assembled proteins, such as HK97 capsomers and proheads (Duda et al., 1995a). Mobility shifts were observed in the agarose gels when the size or the surface charge of proheads or capsomers were changed by mutation. After electrophoresis, gels were stained with Coomassie Brilliant Blue R250. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate (SDS-PAGE) (Laemmli, 1970) was used to detect HK97 gp5 production and to monitor proteolytic cleavage of gp5 (Duda et al., 1995a). Samples were precipitated in cold 10% trichloroacetic acid (TCA) for 10 min at 4°C and washed three times with acetone to remove TCA. The protein pellets were dried under vacuum followed by suspending samples with 1xSDS loading buffer. Samples were boiled for 2–3 min to denature proteins before loading on SDS-PAGE. After electrophoresis, gels were fixed and stained by Coomassie Brilliant Blue R250.
Crosslinking assays
HK97 capsid protein crosslinking is a consequence of prohead expansion. Three different conditions to induce expansion for testing the ability of proheads to crosslink are described separately (Dierkes et al., 2009; Duda et al., 1995a). 1) SDS-glycerol expansion was initiated by mixing proheads with 1xSDS sample buffer (15 mg/ml Prohead II was diluted 1:4 into SDS sample buffer) followed by incubation at room temperature. Reactions were stopped at time points 0 min, 30 min, 2 hours, or 24 hours by heating in boiling water for 2.5 min. Samples were analyzed on SDS-PAGE. 2) Low-pH expansion was initiated by diluting proheads into low-pH expansion buffer (20 mg/ml Prohead II diluted 1:10 into 50 mM sodium acetate, pH 4.1, 400 mM KCl) for an hour, followed by diluting samples into neutralizing buffer (100 mM Tris-HCl pH 8.0) to allow full expansion and crosslinking. After neutralizing, crosslinking was terminated at 0 min, 30 min, 60 min, 120 min, 1 day, 4 days, or 7 days by precipitation with TCA for analysis on SDS-PAGE. 3) Dimethyl formamide (DMF) expansion was initiated by diluting proheads into DMF buffer (20 mg/ml Prohead II diluted 1:10 into 40% (v/v) DMF in TAMg gel buffer (pH 8.0)) After an hour, samples were diluted 10-fold into TAMg buffer without DMF and incubated for 0 min, 30 min, 60 min, 180 min, 1 day, or 2 days before precipitation with TCA for analysis on SDS-PAGE. All expansion/crosslinking experiments were repeated to confirm the results.
Structural analysis
The structure of HK97 Head II (PDB ID: 1OHG) was visualized using the Swiss-PDB viewer (Guex and Peitsch, 1997) and rendered using the POV ray tracer (http://www.povray.org).
Construction of an HK97 lysogen recombineering strain
Recombineering techniques were used to create the strain needed to introduce lethal missense mutations into a HK97 lysogen by homologous recombination (Court et al., 2002; Datsenko and Wanner, 2000). The strain used in the experiments described here, DT005, is derived from BW25113 and has a galK deletion and is lysogenic for a HK97 variant that has a region of HK97 head genes from codon 224 of gene 4 to codon 264 of gene 5 replaced by galK, as described previously (Tso et al., 2014). The host for all recombineering experiments was E. coli strain BW25113 containing the plasmid pKD46, which expresses the phage λ Red recombination proteins under arabinose control (Datsenko and Wanner, 2000). The galK positive/negative selection system was used to facilitate selecting recombinants (Warming et al., 2005).
Highlights.
The protein subunits of bacteriophage HK97 capsid are locked together by covalent bonds between subunits.
We propose a model for the chemical mechanism of crosslink formation.
During capsid maturation a valine moves to close a hydrophobic pocket enclosing the reaction site.
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM47795 to R.W.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adereth Y, Champion KJ, Hsu T, Dammai V. Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. BioTechniques. 2005;38:864, 866, 868. doi: 10.2144/05386BM03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriss M, Lombardot T, Glockner FO, Becher D, Albrecht D, Schweder T. Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles : life under extreme conditions. 2007;11:95–104. doi: 10.1007/s00792-006-0014-5. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. Journal of structural biology. 2007;158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annual review of genetics. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL. Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J Virol. 2009;83:2088–2098. doi: 10.1128/JVI.02000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995a;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995b;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- Ford ME, Sarkis GJ, Belanger AE, Hendrix RW, Hatfull GF. Genome structure of mycobacteriophage D29: implications for phage evolution. J Mol Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Molecular cell. 2004;14:559–569. doi: 10.1016/j.molcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, Hendrix RW, Duda RL, Liljas L, Johnson JE. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–1665. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilakjan ZA, Kropinski AM. Cloning and analysis of the capsid morphogenesis genes of Pseudomonas aeruginosa bacteriophage D3: another example of protein chain mail? Journal of bacteriology. 1999;181:7221–7227. doi: 10.1128/jb.181.23.7221-7227.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin AN, Kincaid RL. A pressure-extrusion method for DNA extraction from agarose gels. Analytical biochemistry. 1998;258:150–152. doi: 10.1006/abio.1998.2581. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Molecular microbiology. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Henderson PJ, Giddens RA. 2-Deoxy-D-galactose, a substrate for the galactose-transport system of Escherichia coli. The Biochemical journal. 1977;168:15–22. doi: 10.1042/bj1680015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nature reviews Microbiology. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, Poor CB, Jureller JE, Budzik JM, He C, Schneewind O. Isopeptide bonds of the major pilin protein BcpA influence pilus structure and bundle formation on the surface of Bacillus cereus. Molecular microbiology. 2012;85:152–163. doi: 10.1111/j.1365-2958.2012.08098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gan L, Tsuruta H, Moyer C, Conway JF, Duda RL, Hendrix RW, Steven AC, Johnson JE. Virus capsid expansion driven by the capture of mobile surface loops. Structure. 2008;16:1491–1502. doi: 10.1016/j.str.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Conway JF, Cheng N, Steven AC, Hendrix RW, Duda RL. Control of Virus Assembly: HK97 "Whiffleball" Mutant Capsids Without Pentons. J Mol Biol. 2005;348:167–182. doi: 10.1016/j.jmb.2005.02.045. [DOI] [PubMed] [Google Scholar]
- Lubbers MW, Waterfield NR, Beresford TP, Le Page RW, Jarvis AW. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Applied and environmental microbiology. 1995;61:4348–4356. doi: 10.1128/aem.61.12.4348-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B, Moyer CL, Hendrix RW, Duda RL. The delta domain of the HK97 major capsid protein is essential for assembly. Virology. 2014;456–457:171–178. doi: 10.1016/j.virol.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa MP, McKelvey TA, Hempel J, Hendrix RW. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J Virol. 1991;65:3227–3237. doi: 10.1128/jvi.65.6.3227-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. Embo J. 2005;24:1352–1363. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods in enzymology. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends in microbiology. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Tso D, Hendrix RW, Duda RL. Transient contacts on the exterior of the HK97 prohead that are essential for capsid assembly. Journal of Molecular Biology. 2014;426:2112–2129. doi: 10.1016/j.jmb.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters MH, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Molecular microbiology. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao S, Edwards SA, Grater F. Isopeptide bonds mechanically stabilize spy0128 in bacterial pili. Biophysical journal. 2013;104:2051–2057. doi: 10.1016/j.bpj.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, Duda RL, Hendrix RW, Steven AC, Johnson JE. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. Journal of structural biology. 2006;153:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Zahner D, Gandhi AR, Stuchlik O, Reed M, Pohl J, Stephens DS. Pilus backbone protein PitB of Streptococcus pneumoniae contains stabilizing intramolecular isopeptide bonds. Biochemical and biophysical research communications. 2011;409:526–531. doi: 10.1016/j.bbrc.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]