Abstract
Pretreatment tumor PD-L1 expression correlates with response to anti-PD-1/PD-L1 therapies. Yet, most patients with PD-L1+ tumors do not respond to treatment. The current study was undertaken to investigate mechanisms underlying the failure of PD-1–targeted therapies in patients with advanced renal cell carcinoma (RCC) whose tumors express PD-L1. Formalin-fixed, paraffin-embedded (FFPE) pretreatment tumor biopsies expressing PD-L1 were derived from 13 RCC patients. RNA was isolated from PD-L1+ regions and subjected to whole genome microarray and multiplex quantitative (q)RT-PCR gene expression analysis. A balance between gene expression profiles reflecting metabolic pathways and immune functions was associated with clinical outcomes following anti-PD-1 therapy. In particular, the expression of genes involved in metabolic and solute transport functions such as UGT1A family members, also found in kidney cancer cell lines, was associated with treatment failure in patients with PD-L1+ RCC. Conversely, tumors from responding patients overexpressed immune markers such as BACH2, a regulator of CD4+ T cell differentiation, and CCL3, involved in leukocyte migration. These findings suggest that tumor cell–intrinsic metabolic factors may contribute to treatment resistance in RCC, thus serving as predictive markers for treatment outcomes and potential new targets for combination therapy regimens with anti-PD-1.
Keywords: kidney cancer, immunotherapy, PD-1, PD-L1, tumor metabolism
INTRODUCTION
In renal cell carcinoma (RCC) and some other tumor types, monoclonal antibodies (mAbs) blocking the programmed death 1 (PD-1):PD-1 ligand (PD-L1, B7-H1) interaction, by targeting either PD-1 (e.g., nivolumab, pembrolizumab) or PD-L1 (e.g., MPDL3280A/atezolizumab, MEDI4736/durvalumab), can induce tumor regression (1–5). Approximately 15–30% of patients with advanced RCC experience durable objective tumor regressions following PD-1 pathway blockade (6–9). Attention is now focused on identifying biomarkers predicting response or resistance to anti–PD-1 treatment (10). We previously identified PD-L1 expression on the tumor cell surface as one factor associated with the clinical activity of nivolumab anti–PD-1 in solid tumors including RCC (1). This observation was supported by a subsequent study of nivolumab in RCC, showing a higher objective response rate and prolonged progression-free and overall survival in patients whose pretreatment tumor specimens were PD-L1+ (7), although this finding was not substantiated in a subsequent phase III trial of nivolumab (8). PD-L1 expression by tumor-infiltrating immune cells has been associated with an increased response rate and overall survival following atezolizumab anti-PD-L1 therapy in a nonrandomized trial in RCC (9).
Notably, a significant number of patients with PD-L1+ RCC do not respond to PD-1 pathway blockade, suggesting that additional intratumoral factors may influence treatment outcomes. The current study was undertaken to explore gene expression profiles characterizing PD-L1+ RCCs that did or did not respond to nivolumab anti–PD-1 therapy.
MATERIALS AND METHODS
Detailed information is available online in Supplementary Materials and Methods.
Tumor specimens
Consenting patients with unresectable metastatic RCC received nivolumab anti-PD-1 monotherapy at the Johns Hopkins Kimmel Cancer Center, on one of four clinical trials (NCT00441337, NCT00730639, NCT01354431, NCT01358721) approved by the Johns Hopkins Institutional Review Board. Patients were classified as responders (R) or nonresponders (NR) to anti–PD-1 therapy based on radiographic staging according to Response Evaluation Criteria in Solid Tumors (RECIST) (11). Pretreatment tumor specimens were characterized for PD-L1 expression by immunohistochemistry (IHC) as described (1, 12) (Supplementary Table S1).
Immunohistochemical (IHC) analysis
Serial 5 um-thick sections from formalin-fixed, paraffin-embedded (FFPE) PD-L1+ tumor specimens were stained for expression of selected markers with specific mAbs as previously described (13). PD-L1+ tumor specimens had ≥ 5% of tumor cells showing cell surface staining with the murine anti-human PD-L1 mAb 5H1 (from Lieping Chen, Yale University). Comparisons of IHC results from PD-L1+ tumors derived from responding vs. nonresponding patients were performed using The R Project for Statistical Computing (https://www.r-project.org/).
Multiplex quantitative (q)RT-PCR assays and statistical analysis
PD-L1+ tumor areas, identified with IHC on neighboring tissue sections, were either manually dissected by scraping with a scalpel, or were laser-capture microdissected from 5-um FFPE tissue sections as previously described (12,14) (Supplementary Fig. S1). Total RNA was isolated from PD-L1+ tumor areas and the expression of selected genes was quantified using custom-made TaqMan low-density array micro fluidic cards per protocol (TLDA; Applied Biosystems, Waltham, MA), as described (14).
Whole genome expression profiling and analysis
Global gene expression in pretreatment RCC specimens was measured by DASL (cDNA-mediated annealing, selection, extension, and ligation) assays arrayed on the Illumina Human HT-12 WG-DASL V4.0 R2 expression BeadChip, per the manufacturer's specifications (Illumina, San Diego, CA) as described (14). Gene expression data are accessible through GEO Series accession number GSE67501 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67501).
RCC cell lines
Eight RCC cell lines (786-0, A498, ACHN, Caki I, RXF-393, TK-10, SN12C, and U0-31) from the NCI-60 panel were obtained in 2009 from the NCI-Frederick Cancer Center DCTD Tumor/Cell Repository, Frederick, MD. Characterization is provided at https://dtp.cancer.gov/discovery_development/nci-60/characterization.htm. They were cryopreserved and used within 6 months of continuous culture. RCC cell lines were assessed with multiplex qRT-PCR for selected gene expression.
Cytotoxicity assays
The established melanoma cell lines 397-mel and 888-mel, generated from metastatic melanoma lesions in our laboratory and maintained as described (15,16) were transfected with the empty vector pCMV6-Entry or with pCMV6-UGT1A6 encoding human UGT1A6 (RC215957; Origene Technologies, Rockville, MD) using Lipofectamine 2000 (Life Technologies, Grand Island, NY). qRT-PCR and Western blotting revealed no UGT1A6 in these melanomas prior to transfection, and positive expression after transfection with the plasmid encoding UGT1A6, but not with the control vector. Tumor cell lysis was measured with a modified flow-cytometry-based assay detecting the disappearance of CFSE-labeled transfectants expressing UGT1A6 or not, upon incubation with nonspecific LAK cells or specific T effector cells (17).
In silico correlation of UGT1A6 expression with overall survival in RCC
RNA sequencing data from The Cancer Genome Atlas project (TCGA), including 444 clear cell RCC samples and 72 matched normal kidney samples, were used for in silico analysis (18). Data were analyzed using R/Bioconductor software.
RESULTS
Immune-related gene expression in PD-L1+ melanomas not associated with RCC outcomes
In a prior study of archival melanoma specimens, we identified immune-related genes that were coordinately overexpressed in PD-L1+ compared to PD-L1− tumors (14). They included genes associated with CD8+ T-cell activation (CD8A, IFNG, PRF1, CCL5), antigen presentation (CD163, TLR3, CXCL1, LYZ), and immunosuppression [PD1, CD274 (PD-L1), LAG3, IL10]. In the current study, multiplex qRT-PCR was used to characterize pretreatment PD-L1+ RCC specimens from 12 patients receiving anti–PD-1 (R = 4, NR = 8) for immune markers (Supplementary Table S1). Genes which were coordinately overexpressed in PD-L1+melanomas were also expressed in PD-L1+RCCs. However, none were significantly differentially expressed according to clinical outcomes after nivolumab therapy (data not shown). Similar results were obtained when immunohistochemistry (IHC) was used to examine protein expression for a more focused group of immune-related molecules in RCCs from 13 patients (R = 4, NR = 9), including PD-1, PD-L2, LAG-3, and TIM-3, and to characterize infiltrating immune cell subsets (FoxP3, CD4:CD8 ratio) (Supplementary Fig. S2). No significant differences in expression of these molecules were observed between PD-L1+RCCs from responders vs. nonresponders. Thus, all PD-L1+RCCs appeared to have an immune-reactive tumor microenvironment (TME) which, in itself, did not distinguish responders from nonresponders.
Resistance to anti–PD-1 therapy associated with increased expression of metabolic genes
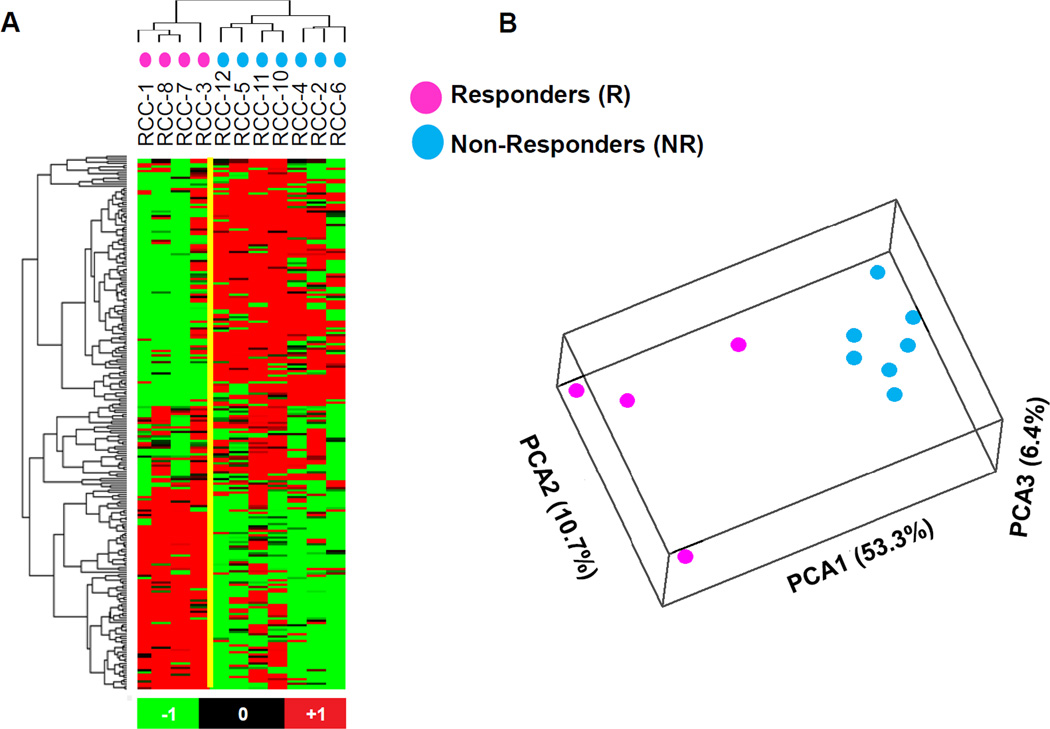
Because analysis of a selected panel of 60 immune-related genes did not reveal significant differences between PD-L1+RCCs that were responsive or resistant to anti-PD-1 therapy, we next turned to whole genome expression profiling. Eleven available RCC specimens from among the original cohort (R = 4, N = 7) (Supplementary Table S1) were analyzed for expression of 29,377 gene targets. We identified 234 probes corresponding to 226 genes that were differentially expressed between the two groups, based on a P value of ≤ 0.01 and expression fold change magnitude ≥ 1.5. Among them, 116 probes corresponding to 113 genes were upregulated in tumors from responding patients, and 118 probes corresponding to 113 genes were upregulated in tumors from nonresponding patients (Figure 1A, Supplementary Table S2). By inspection, genes upregulated in nonresponders appeared to be functionally related in metabolic pathways. In contrast, some genes that were upregulated in tumors from responding patients had immune functions. Thus there appeared to be a functional dichotomy of gene expression profiles in PD-L1+RCCs obtained from patients who responded or not to anti–PD-1 therapy. To further explore these trends, a functional annotation analysis was performed with the DAVID Bioinformatics Resources 6.7 tool, based on 1017 Illumina probes having differential expression in tumors from R vs. NR patients with fold expression change ≥ 1.5 and P ≤ 0.05. A principal component analysis (PCA) (19) of the entire set of 1017 probes further revealed the segregation of gene expression profiles in RCCs from R vs. NR patients (Fig. 1B). Among the 1017 probes, 467 and 550 were found to be overexpressed in R and NR, respectively. Analysis of the 467 probes that were overexpressed in R did not yield significant DAVID gene clusters. However, analysis of the 550 probes that were overexpressed in NR yielded 23 pathways involving mitochondrial and other metabolic functions (DAVID Benjamini FDR ≤ 0.010) (Table 1).
Figure 1. Whole genome microarray analysis of pretreatment PD-L1 + RCC specimens demonstrates differential gene expression between patients responding or not to anti–PD-1 therapy.
(A) Heat map and cluster analysis based on differentially expressed genes detected by 234 Illumina probes satisfying the criteria Student’s t test P ≤ 0.01 and fold change magnitude ≥ 1.5, comparing tumors from responders (R, n = 4) vs. nonresponders (NR, n = 7). Data were analyzed by using BRBArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html) and visualized by Stanford Cluster Program and TreeView software. Red, high gene expression; green, low gene expression. (B) Principal component analysis (PCA) based on 1017 Illumina probes having differential expression in tumors from R vs. NR patients, with P ≤ 0.05 and fold change magnitude ≥1.5. Separation of the R and NR samples is seen. The principal component axis directions are labeled, with the percent of the total variance captured by each axis in parentheses.
Table 1.
Functionally annotated gene categories from DAVID analysis of whole genome microarray results, comparing RCCs from responders to nonresponders
| Functionally-related group of genesa | Number of genes in submitted list/ total number of genes in the category (%) |
P valueb | Benjamini multiple comparison adjusted P value |
|---|---|---|---|
| GO: mitochondrion | 72/1087 (6.6) | 6.20E-13 | 2.32E-10 |
| Swiss-Prot: mitochondrion | 57/832 (6.9) | 1.02E-11 | 4.02E-09 |
| GO: coenzyme binding | 24/181 (13.3) | 1.19E-10 | 7.55E-08 |
| GO: mitochondrial part | 45/595 (7.6) | 6.51E-10 | 1.22E-07 |
| Swiss-Prot: oxidoreductase | 42/562 (7.5) | 6.89E-10 | 1.35E-07 |
| GO: oxidation reduction | 47/639 (7.4) | 8.04E-11 | 1.62E-07 |
| GO: cofactor binding | 26/249 (10.4) | 2.86E-09 | 9.08E-07 |
| GO: organelle membrane | 63/1096 (5.7) | 7.99E-09 | 9.96E-07 |
| GO: mitochondrial envelope | 34/419 (8.1) | 2.35E-08 | 2.19E-06 |
| Swiss-Prot: transit peptide | 34/476 (7.1) | 1.13E-07 | 1.11E-05 |
| GO: mitochondrial membrane | 29/394 (7.4) | 2.25E-06 | 1.69E-04 |
| GO: envelope | 38/622 (6.1) | 3.75E-06 | 2.00E-04 |
| GO: organelle envelope | 38/620 (6.1) | 3.45E-06 | 2.15E-04 |
| Swiss-Prot: endoplasmic reticulum | 40/713 (5.6) | 3.36E-06 | 2.64E-04 |
| UniProt: transit peptide:Mitochondrion | 33/467 (7.1) | 2.62E-07 | 3.06E-04 |
| Swiss-Prot: nad | 18/189 (9.5) | 4.96E-06 | 3.25E-04 |
| GO: mitochondrial inner membrane | 22/306 (7.2) | 7.10E-05 | 0.00265 |
| GO: organelle inner membrane | 23/329 (7.0) | 7.05E-05 | 0.00292 |
| GO: endoplasmic reticulum | 47/960 (4.9) | 6.68E-05 | 0.00312 |
| Swiss-Prot: mitochondrion inner membrane | 16/193 (8.3) | 9.65E-05 | 0.00540 |
| GO: acyl-CoA binding | 6/16 (37.5) | 3.17E-05 | 0.00670 |
| GO: monovalent inorganic cation transmembrane transporter activity |
12/104 (11.5) | 5.26E-05 | 0.00833 |
| GO: hydrogen ion transmembrane transporter activity |
11/90 (12.2) | 7.56E-05 | 0.00957 |
Shown are functional gene categories upregulated in tumors from nonresponders and having a Benjamini adjusted P value (FDR) from DAVID of ≤ 0.010. The list submitted to DAVID contained 550 Illumina probe IDs for which the Nonresponder/Responder expression level fold change was ≥ 1.5 and the equal variance two-sided t-test P value was ≤ 0.05. Gene groups are ordered according to the Benjamini adjusted P values.
Nomenclature per DAVID web tool (https://david.ncifcrf.gov/).
DAVID adjustment of the Fisher exact test (hypergeometric distribution) p-value.
Investigation of differentially expressed genes with multiplex qRT-PCR
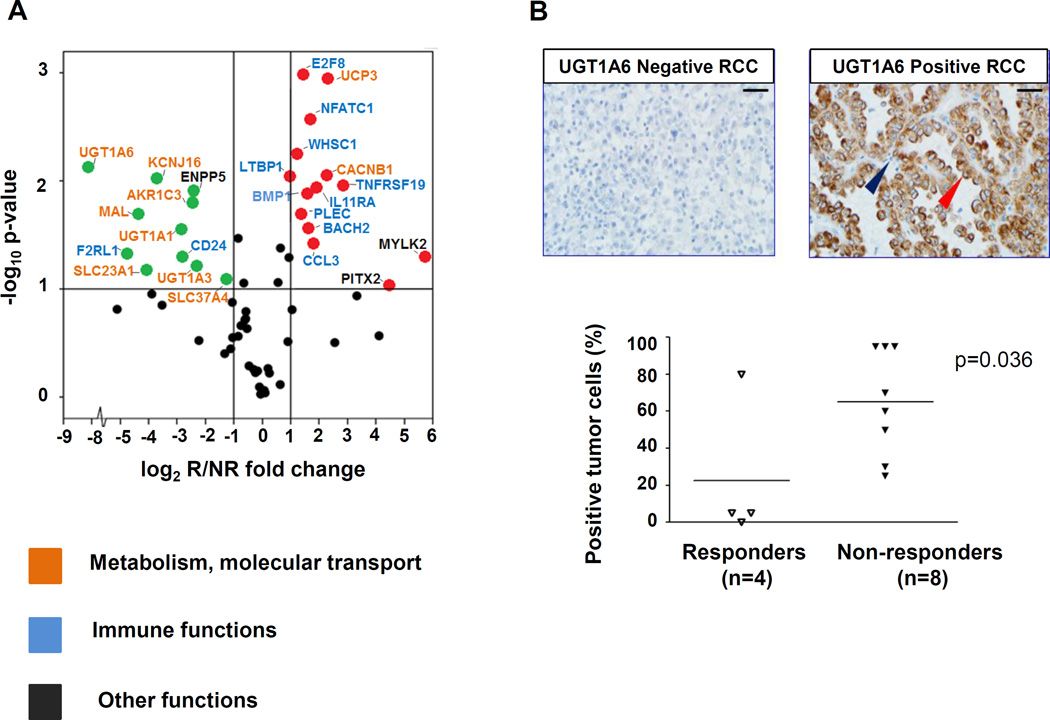
To further explore the results of global gene expression profiling, differential expression of 60 selected unique gene targets was assessed with multiplex qRT-PCR in 12 specimens (R = 4, NR = 8) (Supplementary Table S3). By considering results obtained with each of four endogenous gene controls, 25 among the 60 queried genes were confirmed to be differentially expressed in the two groups of patients with divergent clinical outcomes (Table 2). These 25 genes were not differentially expressed in RCC specimens from primary vs. metastatic sites (not shown). In particular, upregulation of genes corresponding to proteins associated with metabolic and solute transport functions was found in nonresponders (Fig. 2A). These proteins are known to have physiologic functions in normal renal epithelial cells. Among them, the UDP-glucuronosyltransferase UGT1A6 showed the greatest differential expression, being upregulated approximately 300-fold in nonresponders (P = 0.007 using GUSB endogenous control). Its family members UGT1A1 and UGT1A3 were also overexpressed in nonresponders. Additionally, genes encoding proteins involved in solute transport, such as the potassium channel rectifier KCNJ16, the glucose-6-phosphate translocase SLC37A4, the human sodium-dependent ascorbic acid (vitamin C) transporter SLC23A1, and the myelin and lymphocyte-associated protein MAL, were also significantly upregulated in RCCs from nonresponding patients. In contrast, some genes associated with immune functions were upregulated in tumors from responding patients, including the chemokine CCL3, the plectin molecule (PLEC), the nuclear factor NFATC1, the transcription regulator BACH2, and the histone methyltransferase WHSC1 (Fig. 2A). Thus qRT-PCR confirmed the dichotomous pattern of gene expression suggested by whole genome microarray in tumors from R vs. NR patients.
Table 2.
Genes differentially expressed in RCCs from responding vs. nonresponding patients, assessed by qRT-PCR
| GUSB | PTPRC | ||||
|---|---|---|---|---|---|
| Gene Symbola | Protein Functionb | FC R/NR |
P valuec | FC R/NR |
P valuec |
| AKR1C3 | Aldo-keto reductase family member, which catalyzes the conversion of aldehydes and ketones to alcohols and the reduction of prostaglandin D2 and phenanthrenequinone. |
−5.6 | 0.015 | −6.1 | 0.015 |
| BACH2 | Transcription regulator, which induces apoptosis in response to oxidative stress and represses effector programs to stabilize Treg-mediated immune homeostasis. |
3.2 | 0.027 | 2.9 | 0.150 |
| BMP1 | Bone morphogenetic protein 1, which cleaves the C-terminal propeptides of procollagen I, II and III and induces cartilage and bone formation. |
3.6 | 0.012 | 3.3 | 0.137 |
| CACNB1 | Voltage-dependent L-type calcium channel subunit beta-1, which contributes to the function of the calcium channel by increasing peak calcium current. |
4.8 | 0.009 | 4.4 | 0.017 |
| CCL3 | C-C motif chemokine 3 with inflammatory and chemokinetic properties. |
3.5 | 0.038 | 3.2 | 0.071 |
| CD24 | Signal transducer CD24, which modulates B-cell activation responses. |
−7.0 | 0.051 | −7.7 | 0.048 |
| E2F8 | Transcription factor E2F8, which participates in various processes such as angiogenesis and polyploidization of specialized cells. |
2.6 | 0.001 | 2.4 | 0.071 |
| ENPP5 | Ectonucleotide pyrophosphatase /phosphodiesterase family member, which may play a role in neuronal cell communication. |
−5.4 | 0.013 | −5.9 | 0.054 |
| F2RL1 | Proteinase-activated receptor, which mediates inhibition of tumor necrosis factor alpha (TNF). |
−26.3 | 0.047 | −28.9 | 0.047 |
| IL11RA | Receptor for interleukin-11, which might be involved in the control of proliferation and/or differentiation of skeletogenic progenitor or other mesenchymal cells. |
3.0 | 0.013 | 2.8 | 0.097 |
| KCNJ16 | Inward rectifier potassium channel, which mediates regulation of fluid and pH balance. |
−13.2 | 0.010 | −14.5 | 0.018 |
| LTBP1 | Latent-transforming growth factor beta- binding protein, which may play critical roles in controlling the activity of transforming growth factor beta 1 (TGFB) and may have a structural role in the extra cellular matrix. |
2.0 | 0.009 | 1.8 | 0.230 |
| MAL | Myelin and lymphocyte protein, which can be important component in the vesicular trafficking between the Golgi complex and the apical plasma membrane. |
−20.6 | 0.020 | −22.6 | 0.016 |
| MYLK2 | Myosin light chain kinase, implicated in the level of global muscle contraction and cardiac function. |
53.4 | 0.050 | 48.6 | 0.072 |
| NFATC1 | Nuclear factor of activated T-cells, which plays a role in the inducible expression of cytokine genes in T cells regulating their activation, proliferation but also their differentiation and programmed death. |
3.3 | 0.003 | 3.0 | 0.055 |
| PITX2 | Pituitary homeobox, which controls cell proliferation in a tissue-specific manner and is involved in morphogenesis. |
22.4 | 0.095 | 20.3 | 0.075 |
| PLEC | Plectin, which interlinks intermediate filaments with microtubules and microfilaments and anchors intermediate filaments to desmosomes. |
2.6 | 0.020 | 2.4 | 0.246 |
| SLC23A1 | Solute carrier member, which mediates electrogenic uptake of vitamin C. |
−16.6 | 0.066 | −18.2 | 0.091 |
| SLC37A4 | Glucose-6-phosphate translocase, which plays a central role in homeostatic regulation of glucose. |
−2.3 | 0.081 | −2.3 | 0.239 |
| TNFRSF19 | Tumor necrosis factor receptor family member, which mediates activation of Jun N-terminal kinase (JNK) and Nuclear Factor-kappa-B (NFKB), possible promoting caspase-independent cell death. |
7.1 | 0.011 | 6.4 | 0.112 |
| UCP3 | Mitochondrial uncoupling protein 3, involved in mitochondrial transport uncoupling oxidative phosphorylation. |
4.8 | 0.001 | 4.4 | 0.051 |
| UGT1A1 | UDP-glucuronosyltransferases, which mediate the elimination of toxic xenobiotics and endogenous compounds. |
−7.1 | 0.028 | −7.8 | 0.098 |
| UGT1A3 | −5.0 | 0.062 | −5.5 | 0.110 | |
| UGT1A6 | −287.7 | 0.007 | −316.1 | 0.012 | |
| WHSC1 | Histone-lysine N-methyltransferase, which acts as a transcription regulator of cytokines. |
2.3 | 0.006 | 2.1 | 0.293 |
Listed are genes with expression fold change magnitude (FC) ≥ 2 and P value ≤ 0.1 (Student’s t-test) when normalized to either GUSB or PTPRC (CD45) expression. Positive FC indicates genes over-expressed in tumors from responding (R) patients; negative FC indicates over-expression in tumors from nonresponding (NR) patients.
Refers to the official gene name (http://www.ncbi.nlm.nih.gov/gene/).
Obtained from HUGO Gene Nomenclature Committee website: http://www.genenames.org/.
Data were analyzed using the comparative Ct method (ΔΔCt), normalized to either GUSB (beta-glucuronidase) or PTPRC (CD45, pan immune cell marker). A 2-tailed, unpaired Student’s t-test was used to determine the statistical significance of FC values.
Figure 2. Genes overexpressed in pretreatment PD-L1+ RCC specimens from responding vs. nonresponding patients reflect immune vs. metabolic functions, respectively.
(A) Results of multiplex qRT-PCR for 60 select genes are shown, amplifying RNA isolated from pretreatment tumors in 4 responders and 8 nonresponders. Red and green dots represent genes significantly overexpressed or under-expressed, respectively, by at least 2-fold in tumors from responders compared to nonresponders, and with a 2-sided P value ≤ 0.1 (indicated by the horizontal line). Gene names are color-coded according to biological functions. The GUSB transcript was used as the internal reference. UGT1A6 was the most highly over-expressed gene associated with nonresponse to anti–PD-1. Similar results were obtained using 18S, ACTB, or PTPRC (CD45) as reference genes. Supporting information is provided in Table 2 and Supplementary Table S3. (B) UGT1A6 protein expression was evaluated by IHC in the same 12 pre-treatment PD-L1+ RCC specimens as were studied with qRT-PCR, including tumors from 4 responders and 8 nonresponders. In the top panels, representative UGT1A6 negative (left) and positive (right) RCC specimens are shown. Scale bars are equal to 25 um. Red arrow, kidney cancer cell with positive staining; blue arrow, infiltrating lymphocyte devoid of staining. In bottom panel, UGT1A6 expression is quantified by percent positive tumor cells in each specimen. Horizontal black bars indicate mean values. Enhanced UGT1A6 expression was significantly associated with nonresponse to anti–PD-1 therapy (P = 0.036, one-sided Wilcoxon rank sum test).
Genes upregulated in resistant PD-L1+RCCs expressed by kidney cancer cell lines
The RCC TME is a complex milieu containing many different cell types. To understand whether metabolic genes that were overexpressed in tumor specimens from nonresponding patients were specifically associated with renal carcinoma cells, we evaluated their expression in 8 established kidney cancer cell lines using qRT-PCR. Results showed that cultured renal carcinoma cells expressed the metabolic genes of interest (Supplementary Fig. S3).
Overexpressed UGT1A6 protein associated with nonresponse to anti–PD-1 therapy
UGT1A6 is involved in the chemical “defensome” and detoxifies exogenous and stress-related lipids (20). In whole genome expression profiling and qRT-PCR assessment, it was the most highly overexpressed gene associated with nonresponse to anti–PD-1 (~8-fold and ~300-fold, P ≤ 0.005 and P = 0.007, respectively) (Supplementary Table S2 and Fig. 2A). Therefore, the expression of UGT1A6 in RCC was further explored at the protein level with IHC, in the same 12 specimens as those used for gene expression profiling by qRT-PCR. UGT1A6 protein expression varied widely among the specimens and was associated with clinical outcomes following anti-PD-1 therapy (P = 0.036, one-sided Wilcoxon rank sum test) (Fig. 2B).
Melanoma cells transfected with UGT1A6 are not resistant to lysis by immune cells in vitro
To examine whether overexpression of UGT1A6 protein might protect tumor cells from immune cell killing, we tested the ability of nonMHC-restricted LAK cells and MHC I-restricted T cells (397 TILs or 888 TILs) to lyse 397-mel or 888-mel cells transfected with a plasmid encoding UGT1A6. No substantial difference in lysis of UGT1A6-positive or -negative melanoma cells was observed (not shown).
Expression of UGT1A6 is not associated with survival in patients with RCC
To explore whether the observed overexpression of genes such as UGT1A6 is specifically associated with adverse outcomes to anti–PD-1therapy, or generally associated with poor prognosis in patients with RCC, an in silico analysis was performed on RNA expression data obtained from 444 primary clear cell RCC specimens in The Cancer Genome Atlas project (TCGA) (18). Kaplan-Meier curves were generated using the median expression value for UGT1A6 to segregate samples into high or low expressers. There was no significant correlation between UGT1A6 mRNA expression in primary kidney cancers and overall survival in the entire cohort of 444 patients, nor with survival in a subset of 71 patients with stage IV metastatic disease (similar to patients in our study) (Supplementary Figure S4A). Furthermore, UGT1A6 expression did not correlate with RCC clinical stage when analyzing TCGA data (Supplementary Figure S4B). These findings suggest that the association of UGT1A6 expression with clinical outcomes following anti–PD-1 therapy is specifically relevant in the context of this treatment.
DISCUSSION
Expression of the immunosuppressive ligand PD-L1, detected in pretreatment tumor biopsies with IHC, has been associated with favorable clinical outcomes following PD-1 and PD-L1 blocking therapies in some studies in several types of cancer (1, 7, 9, 21). This can be understood by viewing PD-L1 as a surrogate marker for an immune-reactive TME, since inflammatory cytokines such as IFN-g are major drivers of PD-L1 expression on tumor and stromal cells. In this model, blocking the PD-1/PD-L1 interaction unleashes an immune response that is already properly trained and poised to attack cancer cells, but held in check by this immunosuppressive pathway. Despite the therapeutic impact of PD-1 blockade in many patients with certain cancer types (22), the majority of patients with PD-L1+tumors do not respond to anti–PD-1/PD-L1 drugs, suggesting the involvement of additional factors in the TME contributing to treatment resistance. The current study attempts to identify such factors by exploring the gene expression landscape of PD-L1+RCCs derived from patients with divergent clinical outcomes after anti–PD-1 therapy, and identifies groups of metabolic and immunologic factors associated with adverse or favorable clinical outcomes, respectively.
RCC has been characterized as a metabolic disease, with the signature upregulation of factors adapting to hypoxia and functioning to meet the bioenergetic demands of cellular proliferation (23). We here describe a likely metabolic shift in RCCs resistant to anti–PD-1 therapy, with overexpression of molecules associated with glucuronidation and the transport of solutes and nutrients such as glucose. Among them, UGT1A6, whose principal role is to promote cellular clearance of toxins and exogenous lipophilic chemicals (18), was the single most highly overexpressed molecule associated with anti–PD-1 treatment resistance, and other UGT1A family members and solute carriers constituting the chemical “defensome” were also upregulated. This gene expression profile may simply reflect an activated cell phenotype, however, one might hypothesize that the heightened clearance of toxins from tumor cells by molecules typified by UGT1A6 may render them more fit to outpace the immune system. Furthermore, the overexpression of metabolic genes in RCCs resistant to anti–PD-1 therapy may reflect tumor-imposed metabolic effects that can restrict the responsiveness of tumor-specific infiltrating T cells by competing for vital nutrients such as glucose within the TME (24).
Despite the limitations inherent to this retrospective study of a small number of specimens, the significantly differential gene expression profiles described here provide a basis for future exploration in larger RCC cohorts, potentially including PD-L1-positive and -negative tumors. The general approach to identifying biomarkers of clinical response to PD-1–targeted therapies has so far focused on immunologic factors in the TME. However, a deeper level of investigation may be warranted for individual tumor types, and intersections of tumor cell-intrinsic factors with immunologic factors may be particularly revealing. For instance, a recent report unexpectedly found overexpression of β-catenin to be associated with decreased infiltration of tumor-specific T cells in melanoma (25). A greater knowledge of such tumor-intrinsic mechanistic markers may reveal new therapeutic targets for combination treatment regimens based on PD-1 pathway blockade and useful markers for selecting patients most likely to respond to these therapies.
Supplementary Material
Acknowledgments
Financial Support: This study was supported by research grants from Bristol-Myers Squibb (S.L. Topalian); the National Cancer Institute, NIH [R01 CA142779 (J.M. Taube, D.M. Pardoll, S.L. Topalian) and P30 CA0006973 (all authors)]; the Bloomberg~Kimmel Institute for Cancer Immunotherapy at Johns Hopkins (R.A. Anders, C.G. Drake, D.M. Pardoll, J.M. Taube, S.L Topalian); and Stand Up To Cancer—Cancer Research Institute Cancer Immunology Translational Cancer Research Grant SU2C-AACR-DT1012 (R.A. Anders, J.M. Taube, D.M. Pardoll, S.L. Topalian). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
GJN consults for Genentech/Roche. CGD receives research grants from Bristol-Myers Squibb; consults for Bristol-Myers Squibb, Novartis, and Roche/Genentech; holds stock options in Potenza Therapeutics; and is entitled to receive patent royalties through his institution, from Bristol-Myers Squibb and Potenza. DMP receives research grants from Bristol-Myers Squibb; consults for Five Prime Therapeutics, GlaxoSmithKline, MedImmune, Pfizer, Potenza Therapeutics, and Sanofi; holds stock options in Jounce and Potenza; and is entitled to receive patent royalties through his institution, from Bristol-Myers Squibb, Potenza, and Tizona LLC. JMT receives research funding from Bristol-Myers Squibb and serves on advisory boards for Bristol-Myers Squibb. SLT receives research funding from Bristol-Myers Squibb.
The authors would like to thank Dr. Hans Hammers, Dr. Jenny Kim, Ting Wang and Yelena Millman (Johns Hopkins University) for providing clinical response data; Dr. Lieping Chen (Yale University) for providing the anti-PD-L1 mAb 5H1; Dr. Shuming Chen, George A. Crabill, and Matthew Presby (Johns Hopkins University) for helpful discussions and technical assistance; Jessica Esandrio for administrative assistance; and Dr. Mark Ratain (University of Chicago) and Dr. David Feltquate (Bristol-Myers Squibb) for helpful discussions.
Footnotes
The following authors have declared relevant financial relationships: MLA, TLM, AEB, LD, RAA, HX, TSP, JF, CC, and LC do not declare any conflicts.
REFERENCES
- 1.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med. 2016 doi: 10.1056/NEJMoa1603702. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDermott DF, Drake CG, Sznol M, Choueiri TK, Powderly JD, Smith DC, et al. Survival, durable response, and long-term safety in patients with previously treated advanced renal cell carcinoma receiving nivolumab. J Clin Oncol. 2015;33:2013–2020. doi: 10.1200/JCO.2014.58.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer RJ, Rini BI, McDermott DF, Redman BG, Kuzel TM, Harrison MR, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430–1437. doi: 10.1200/JCO.2014.59.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase Ia study. J Clin Oncol. 2016;34:833–842. doi: 10.1200/JCO.2015.63.7421. [DOI] [PubMed] [Google Scholar]
- 10.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-drivenbiomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Nat Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 12.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taube JM, Young GD, McMiller TL, Chen SM, Salas JT, Pritchard TS, et al. Differential expression of immune-regulatory genes associated with PD-L1 display in melanoma: implications for PD-1 pathway blockade. Clin Cancer Res. 2015;21:3969–3976. doi: 10.1158/1078-0432.CCR-15-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–3725. [PubMed] [Google Scholar]
- 16.Robbins PF, El-Gamil M, Kawakami Y, Stevens E, Yannelli JR, Rosenberg SA. Recognition of tyrosinase by tumor-infiltrating lymphocytes from a patient responding to immunotherapy. Cancer Res. 1994;54:3124–3126. [PubMed] [Google Scholar]
- 17.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolliffe IT. Principal Component Analysis. second. New York: Springer-Verlag New York, Inc; 2002. [Google Scholar]
- 20.Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, et al. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab Dispos. 2004;32:281–290. doi: 10.1124/dmd.32.3.281. [DOI] [PubMed] [Google Scholar]
- 21.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.