Abstract
The class II histone deacetylases HDAC4 and HDAC5 interact specifically with the myogenic MEF2 transcription factor and repress its activity. Here we show that HDAC4 is cytoplasmic during myoblast differentiation, but relocates to the nucleus once fusion has occurred. Inappropriate nuclear entry of HDAC4 following overexpression suppresses the myogenic programme as well as MEF2-dependent transcription. Activation of the Ca2+/calmodulin signalling pathway via constitutively active CaMKIV prevents nuclear entry of HDAC4 and HDAC4-mediated inhibition of differentiation. Consistent with a role of phosphorylation in HDAC4 cytoplasmic localisation, HDAC4 binds to 14-3-3 proteins in a phosphorylation-dependent manner. Together these data establish a role for HDAC4 in muscle differentiation. Recently, HDAC5 has also been implicated in muscle differentiation. However, despite the functional similarities of HDAC4 and HDAC5, their intracellular localisations are opposed, suggesting a distinct role for these enzymes during muscle differentiation.
INTRODUCTION
In recent years it has become apparent that transcriptional regulation and chromatin modification are intricately linked (1). The most intensively studied chromatin modification is nucleosomal histone acetylation, which is modulated through histone acetyltransferases and histone deacetylases. Both activities have been implicated in a variety of biological processes, including the cell cycle, differentiation, ageing and cancer (2,3).
In mammalian cells two classes of histone deacetylases have been described so far. Class I histone deacetylases HDAC1, HDAC2, HDAC3 and HDAC8 (4–8) and class II histone deacetylases HDAC4, HDAC5, HDAC6 and HDAC7 (9–11). In addition, a second family of NAD+-dependent histone deacetylases related to the yeast SIR2 protein has recently been described (12,13). Three of the class II histone deacetylases, HDAC4, HDAC5 and HDAC7, are highly homologous not only in their catalytic domains, but also in their extended N-termini. The N-termini of HDAC4 and HDAC5 specifically interact with and repress the myogenic transcription factor MEF2 (14–17). Tissue-specific expression of HDAC4 and HDAC5, with high levels in heart and skeletal muscle (9), correlates with a role of HDAC4 and HDAC5 in regulating MEF2 function. Three members of the class II family of deacetylases, HDAC4, HDAC5 and HDAC7, have also been shown to interact functionally with the nuclear hormone co-repressor N-CoR/SMRT (11,18). A surprising feature of HDAC4 is its ability to shuttle between the nucleus and cytoplasm (14). This feature could be unique to the class II enzymes since class I deacetylases do not show differential localisation (5,8).
The MEF2 transcription factors belong to the MADS-box family of DNA-binding transcription factors. In mammalian cells there are four mef2 genes, mef2a–mef2d, which share extensive sequence homology (19). Their essential role in muscle differentiation has been demonstrated in Drosophila, mouse and mammalian tissue culture cells (20–22). A variety of signalling pathways have been shown to impinge on MEF2 function. These include the mitogen-activated protein kinases (MAPKs) p38 and ERK5, which have the potential to phosphorylate MEF2 factors (23–33). In addition, Ca2+-dependent signalling through calcineurin and Ca2+/calmodulin-dependent kinases (CaMKs) has also been implicated in regulation of MEF2 activity (34–39).
In this study we demonstrate a role for the histone deacetylase HDAC4 in C2C12 muscle differentiation, namely at the step of myoblast fusion into multinucleated myotubes. HDAC4 translocates from the cytoplasm to the nucleus after myoblast fusion. Overexpression of HDAC4 in myoblasts represses both differentiation and MEF2-dependent transcription. Activation of CaMKIV and the MKK6/MAP kinase pathway overcomes HDAC4-induced inhibition of myotube fusion and activates MEF2-dependent transcription. Furthermore, CaMKIV activation, but not MKK6b activation, prevents relocation of HDAC4 to the nucleus after myoblast fusion, suggesting that CaMKIV signalling controls HDAC4 function through regulating its localisation. Furthermore, we demonstrate that HDAC4 interacts with 14-3-3 proteins in a phosphorylation-dependent manner, suggesting a mechanism for its Ca2+/calmodulin-dependent kinase-dependent localisation. Together these data identify HDAC4 localisation as a regulatory step in muscle differentiation.
MATERIALS AND METHODS
Recombinant DNA
GFP (40), RSV CaMKIV(1–313) (41), pBJ5-HDAC5-F (9) and MKK6b(E) (24) have been described previously. The HDAC5-Flag expression vector was provided by Christina Grozinger and Stuart Schreiber. The 3×MCK-MEF2-luciferase reporter construct was a generous gift from Mona Nemer. The MKK6b(E) expression plasmid was a gift from Jiahuai Han. Constitutively active CaMKIV (amino acids 1–313) was provided by Richard Maurer.
Antibodies
Two rabbit polyclonal antibodies were generated using the peptide H2N-CKPAEKRPDEEPMEEEPPL-CONH2 corresponding to amino acids 1067–1084 of HDAC4 linked to keyhole limpet haemocyanin (HD4A and HD4B). Sera were purified against immobilised peptides using affinity chromatography by standard protocols. The antibodies recognise a band of the expected molecular size that co-migrates with recombinant and in vitro translated HDAC4 on SDS–PAGE.
Cell culture and transfections
293T and C2C12 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco BRL) with 10% FBS (Gibco BRL), penicillin, streptomycin and glutamine (all Gibco BRL). Cells were grown at 37°C in an atmosphere containing 5% CO2. 293T cells were transfected using the Ca3(PO4)2 technique as described (42). C2C12 cells were transfected using lipofectamine (Gibco BRL). For differentiation assays C2C12 cells were transferred to differentiation medium containing 2% horse serum for the times indicated.
Immunoprecipitations
293T cells in culture dishes (15 cm diameter) were transfected with 30 µg expression vector. HeLa cells (10 cm dishes) were transfected with 10 µg DNA. Cells were washed in ice-cold PBS and lysed in IPH buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40) at 4°C for 30 min. Lysates were cleared by centrifugation, diluted five times in IPH buffer containing 0.1% NP-40 and incubated with 5 µg anti-Myc mouse monoclonal antibody (Boehringer Mannheim, Mannheim, Germany) for 1 h. An aliquot of 50 µl of a slurry of protein A/G–Sepharose beads (Pharmacia) was added and incubation continued for 2 h with rotation at 4°C. Precipitates were washed six times in ice-cold IPH and either resuspended in loading buffer for SDS–PAGE or used for deacetylase assays.
SDS–PAGE and western analysis
SDS–PAGE and western blotting were performed according to standard procedures (43). Anti-HA antibody and anti-Myc antibody (both Boehringer Mannheim) were used at a concentration of 1 µg/ml. The anti-MEF2A antibody (Santa Cruz) was used at 1 µg/ml.
Gene reporter assays
HeLa and 293T cells grown in culture dishes (10 cm diameter) or slide flasks were transfected at 40–60% confluency with a total of 10 µg DNA. Cells were washed 16 h after transfection and incubated for an additional 24 h, either in the presence or absence of TSA (330 nM), before harvesting. Chloramphenicol acetyltransferase and luciferase assays were performed essentially as described previously (44). Transfection efficiency was controlled through co-expression of a GFP reporter plasmid and analysis of GFP expression in each sample.
Confocal microscopy
Cells were fixed for 20 min in 4% paraformaldehyde and stained as described previously (45). Anti-HA mouse monoclonal antibody (Boehringer Mannheim) was used at 1:200 dilution. Cy5-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories Inc., USA) was diluted 1:200. Fixed and mounted cells were analysed by confocal microscopy using a Bio-Rad MRC 1024 confocal system on an upright Nikon fluorescence microscope equipped with a 60× oil lens. Using the argon ion 488 nm line, cells were scanned using 10% laser power, 2.5× zoom and 10 times Kalman averaging. Images were exported into Adobe Photoshop for processing and printing.
Microinjection, fluorescence microscopy and CCD imaging
Cells were incubated in CO2-free medium without phenol red (Gibco BRL) and incubated at 37°C using a ΔT 0.15 mm dish (Bioptechs, PA). Cells were imaged by time-lapse fluorescence microscopy using a Leica DMIRBE microscope equipped with custom filter sets (Chroma Technology, VE), a PentaMax camera (Princeton Instruments, NJ) and a Lambda 10-2 filter wheel (Sutter, CA) controlled by a PowerWave computer (PowerComputing, TX) running IP Lab Spectrum software (Scanalytics Inc., VA) as described (46). Images were exported into Adobe Photoshop and printed on a dye sublimation printer (Fuji, USA).
RESULTS
HDAC4 relocates from the cytoplasm to the nucleus during C2C12 myoblast fusion
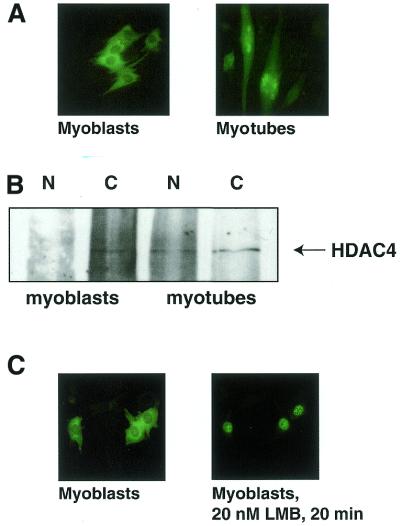
As the histone deacetylase HDAC4 is expressed in skeletal muscle (9) and has the ability to shuttle between the nucleus and cytoplasm (14), we asked whether HDAC4 localisation was regulated during C2C12 muscle differentiation. To this end we microinjected cDNA coding for a HDAC4–GFP fusion protein into C2C12 myoblasts. Using fluorescence microscopy on living cells we found that HDAC4–GFP localises to the cytoplasm when injected into undifferentiated myoblasts (Fig. 1A, left). In contrast, HDAC4–GFP injected during myoblast fusion accumulated in the nucleus after myotube formation (Fig. 1A, right). This feature of HDAC4–GFP reflects the behaviour of the endogenous protein, as demonstrated using an HDAC4-specific antibody. Figure 1B shows that in C2C12 myoblasts HDAC4 protein is localised only in the cytoplasm. In extracts prepared from differentiated cells HDAC4 is detected in both the cytoplasm and the nucleus. Considering that ∼50–70% of myoblasts undergo fusion in a differentiation assay, this observation is consistent with endogenous HDAC4 being predominantly nuclear in myotubes. These data demonstrate that endogenous HDAC4 enters the nucleus after C2C12 differentiation and that HDAC4–GFP fusions can be used as a marker for HDAC4 localisation.
Figure 1.
HDAC4 relocalises from the cytoplasm to the nucleus after myoblast fusion. (A) C2C12 myoblasts were microinjected with pcHDAC4-GFP. Four hours after injection living cells were visualised using fluorescence microscopy. To visualise HDAC4–GFP in myotubes, myoblasts were transferred to differentiation medium, incubated for 2 days and microinjected with pcHDAC4-GFP. Living myotubes were visualised using fluorescence microscopy. (B) Cytoplasmic and nuclear extracts were prepared from one 15 cm dish each of either 40–50% confluent myoblasts or myotubes after 5 days differentiation. Extracts were mixed with HDAC4-specific antibody (HD4B) and immunoprecipitated using protein A/G beads. Immunoprecipitates were separated using SDS–PAGE and western blotting was performed using a second HDAC4-specific antibody (HD4A). (C) C2C12 myoblasts were microinjected with pcHDAC4-GFP and living cells were visualised using fluorescence microscopy. Leptomycin B (LMB) was added to the medium at 20 nM and HDAC4–GFP fluorescence was monitored for 20 min.
Recently, the class II histone deacetylase HDAC5 has also been demonstrated to relocalise during C2 muscle differentiation (47). In contrast to HDAC4, however, HDAC5 was found to be nuclear in C2 myoblasts and cytoplasmic in C2 myotubes (47).
HDAC4 shuttles in C2C12 cells
Relocalisation of HDAC4 in C2C12 cells could be achieved through regulation of the rate of HDAC4 nuclear import and export. We have previously shown that HDAC4 is actively exported in HeLa cells (14). To test if HDAC4 can shuttle in C2C12 cells, myoblasts were injected with HDAC4–GFP and 3 h after injection HDAC4–GFP was visualized by live cell imaging. HDAC4–GFP was found to be predominantly cytoplasmic (Fig. 1C, left). However, after addition of leptomycin B, a crmA-specific inhibitor of nuclear export, HDAC4–GFP relocated quantitatively to the nucleus in 20 min (Fig. 1C, right). This experiment demonstrates that the predominantly cytoplasmic localisation of HDAC4 in C2C12 myoblasts is maintained by rapid nuclear export of the protein.
Overexpression of HDAC4 inhibits C2C12 myotube formation
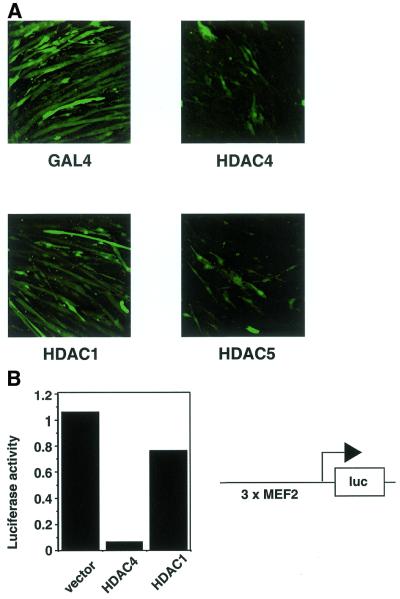
Since HDAC4 shuttles in C2C12 cells (Fig. 1C), but is quantitatively held in the cytoplasm before myoblast fusion (Fig. 1A and B), its localisation might be important for regulation of C2C12 muscle differentiation. As overexpression of HDAC4 results in partial mislocalisation of HDAC4 to the nucleus in myoblasts (Fig. 5A) we decided to use overexpression to test this hypothesis. HDAC4 was overexpressed in C2C12 cells using transient transfection and transferred to differentiation medium. Four days after transfection cells were fixed and analysed using confocal microscopy. Differentiation of transfected cells was scored using GFP co-transfection as a marker. Figure 2A (top left) shows that transfection of a control protein, the yeast transcriptional regulator GAL4, together with GFP allowed myoblast fusion to occur. In contrast, transfection of the class II histone deacetylases HDAC4 and HDAC5 (9) prevented myoblast fusion (Fig. 2A, right). This effect was specific for class II histone deacetylases, as the class I histone deacetylase HDAC1 did not show this effect (Fig. 2A, bottom left). Thus, class II histone deacetylases have the potential to block C2C12 muscle differentiation. In agreement with these data, HDAC4 and HDAC5 overexpression has been shown to deregulate myogenic markers in differentiating C2 cells (48). Furthermore, HDAC4 and HDAC5 overexpression was found to inhibit MyoD-induced differentiation of 10T1/2 cells (48).
Figure 5.
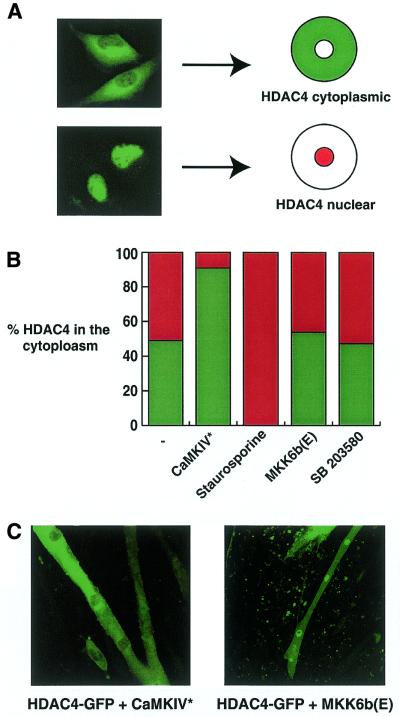
Active CaMKIV, but not MKK6b, regulates HDAC4 localisation. (A) C2C12 myoblasts were transfected with 100 ng pcHDAC4-GFP. After transfection cells were transferred to differentiation medium for 24 h before fixing. GFP staining was visualised using confocal microscopy. HDAC4-GFP localised either to the cytoplasm or the nucleus. (B) C2C12 myoblasts were transfected with 100 ng pcHDAC4-GFP either in the presence or absence of pRSV-CaMKIV* or pMKK6b(E) (500 ng each). After transfection cells were transferred to differentiation medium for 24 h before fixing. Prior to fixing staurosporine (20 µM) or SB203580 (10 µM) was added to the culture medium for 1 h, as indicated. GFP staining was visualised using confocal microscopy. Percentages of cells displaying cytoplasmic staining are given (n > 200). (C) C2C12 myoblasts were transfected with 100 ng pcHDAC4-GFP either in the presence or absence of pRSV-CaMKIV* or pMKK6b(E) (200 ng each). After transfection cells were transferred to differentiation medium for 4 days before fixing. GFP staining was visualised using confocal microscopy.
Figure 2.
HDAC4 inhibits C2C12 differentiation and MEF2 expression. (A) C2C12 myoblasts were transfected either with pcHDAC4-Flag, pBJ5-HDAC5-Flag, pcHDAC1-Flag or pcGAL4 and pEGFP as a transfection marker (100 ng each plasmid). After transfection cells were transferred to differentiation medium for 4 days before fixing. GFP staining was visualised using confocal microscopy. (B) C2C12 myoblasts were transfected with 1 µg 3×MCK-MEF2-luc and 200 ng either pcHDAC4-Flag or pcHDAC1-Flag. After transfection cells were transferred to differentiation medium for 2 days. Cell extracts were prepared and analysed for luciferase activity.
Previous studies have shown that HDAC4 has the ability to repress transcriptional activity of the MEF2 transcription factor (14–17). We therefore tested whether HDAC4 inhibited MEF2 activity during C2C12 differentiation. To this end we transfected C2C12 cells with a luciferase reporter driven by MEF2 sites (14) along with either HDAC4, HDAC1 or a vector control. Figure 2B shows that HDAC4 is able to repress activity of the MEF2-driven promoter. In contrast, HDAC1 is unable to repress MEF2 activity, which is consistent with its inability to bind MEF2A (14). Together, these observations suggest that HDAC4 represses C2C12 differentiation, at least partly, because of its ability to bind to and silence MEF2 activity, whereas HDAC1 cannot inhibit differentiation or repress MEF2 activity because it has no access to MEF2.
The MEF2-binding and deacetylase domains of HDAC4 are required for inhibition of C2C12 myotube formation
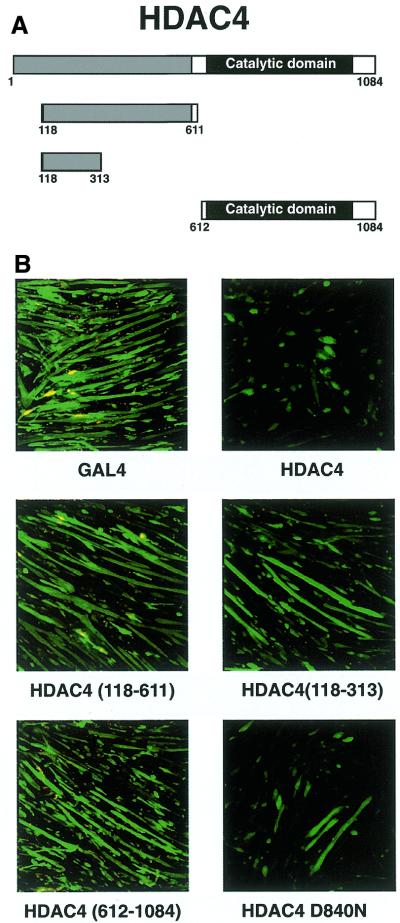
Previously we have mapped the MEF2-interaction domain on HDAC4 to its N-terminus (amino acids 118–313) (14), whereas the histone deacetylase domain is located in the C-terminal half of the protein (9,15,49). In order to establish which domain of HDAC4 is required for inhibition of myotube fusion, we transfected deletion constructs of HDAC4 along with GFP into myotubes. A diagrammatic representation of the HDAC4 constructs used is given in Figure 3A. Figure 3B (top) shows that full-length HDAC4, but not the GAL4 control, inhibits C2C12 differentiation. In contrast, two N-terminal HDAC4 constructs [HDAC4(118–611) and HDAC(118–313)], which do not possess catalytic activity but can still bind MEF2 protein, are unable to inhibit differentiation (Fig. 3B, middle two). Expression of the HDAC4 catalytic domain alone was also unable to repress differentiation (Fig. 3B, bottom left). These data suggest that both the MEF2-binding and catalytic domains of HDAC4 are required for an effect on C2C12 differentiation. To test whether the catalytic activity of HDAC4 was required in this assay, we also transfected a mutant of HDAC4 (D840N) in the same experiment. Previously we showed that HDAC4 D840N is not associated with histone deacetylase activity (14). We found that this catalytic mutant still retained considerable ability to compromise myotube formation, although its inhibitory potential was clearly compromised compared to wild-type HDAC4 (Fig. 3B, bottom right). This result might be explained through the earlier observation that HDAC4 D840N retains some repressive activity on MEF2-dependent transcription (14). This residual repressive capacity may stem from a deacetylase-independent repression domain or perhaps is an indication that the low level of deacetylase activity associated with HDAC4 D840N is sufficient for repression (14).
Figure 3.
Both the MEF2-binding and catalytic domains of HDAC4 are required for inhibition of differentiation. (A) Schematic representation of the HDAC4 deletion constructs used. (B) C2C12 myoblasts were transfected either with pcHDAC4-Myc/His, pcHDAC4(118–611)-Myc/His, pcHDAC4(118–313)-Myc/His, pcHDAC4(612–1084)-Myc/His, pcHDAC4-D840N-Myc/His or pcGAL4 and pEGFP as a tranfection marker (100 ng each plasmid). After 4 days in differentiation medium, GFP staining was visualised using confocal microscopy.
Constitutively active forms of CaMKIV and MKK6b overcome HDAC4-induced inhibition of differentiation
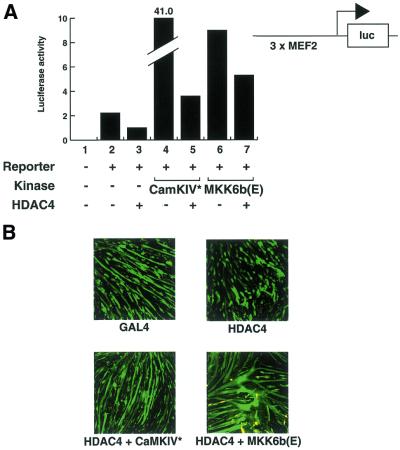
Two major pathways have been shown to positively regulate muscle differentiation: Ca2+-dependent pathways, mediated for example through Ca2+/calmodulin-dependent kinases (CaMK) (17,36,39); the p38 MAP kinase pathway (23–33). Constitutively active enzymes from each of these signalling pathways (CaMKIV and MKK6b, respectively) have been shown to stimulate MEF2-dependent transcription (17,33). We first asked if expression of either of these enzymes would overcome the ability of HDAC4 to silence MEF2 transcription. Figure 4A shows that HDAC4 can repress the activity of a MEF2-driven promoter (lanes 1–3), whereas constitutively active CaMKIV and MKK6b stimulate MEF2 activity (lanes 4 and 6). In the presence of HDAC4 and either CaMKIV* or MKK6b(E) MEF2 transcriptional activity is not decreased below endogenous levels but is instead elevated several-fold (Fig. 4A, lanes 5 and 7). These data indicate that CaMKIV, MKK6 and HDAC4 have opposing effects on MEF2-dependent transcriptional activity (as reported previously) but that in the presence of both signals the net effect is a slight increase in MEF2 activity.
Figure 4.
Activated CaMKIV and MKK6b overcome HDAC4-induced inhibition of MEF2-dependent transcription and C2C12 differentiation. (A) C2C12 myoblasts were transfected with 1 µg 3×MCK-MEF2-luc, 100 ng pcHDAC4-Myc/His, 500 ng pRSV-CaMKIV* or 500 ng pMKK6b(E) as specified for each lane. After transfection, cells were incubated in differentiation medium. After 24 h cell extracts were prepared and analysed for luciferase activity. (B) C2C12 myoblasts were transfected either with pcGAL4 or pcHDAC4-Myc/His (100 ng each), either in the presence or absence of pRSV-CaMKIV* or pMKK6b(E) (200 ng each). 100 ng pEGFP was co-transfected as a transfection marker (100 ng each plasmid). After transfection cells were transferred to differentiation medium for 4 days before fixing. GFP staining was visualised using confocal microscopy.
Since CaMKIV* and MKK6b(E) antagonise HDAC4-mediated repression of MEF2 activity, we asked if these two constitutive kinases would also override HDAC4-mediated inhibition of differentiation. Figure 4B shows that co-expression of CaMKIV* or MKK6b(E) with HDAC4 allows myotube formation (Fig. 4B, bottom) to occur to a level comparable to the GAL4 control (top left). In the same experiment transfection of HDAC4 alone inhibited myoblast differentiation (Fig. 4B, top right). Thus, the overriding effects of CaMKIV* and MKK6b(E) on HDAC4-mediated repression of MEF2-dependent transcription (Fig. 4A) are consistent with the overriding of HDAC4-mediated inhibition of differentiation (Fig. 4B).
CaMKIV but not MKK6b MAP kinase signalling regulates HDAC4 localisation
The effects of CaMKIV and MKK6/p38 signalling on MEF2-dependent transcription and C2C12 differentiation activity could be mediated either through a pathway dependent or independent of HDAC4. To distinguish between the two possibilities we decided to analyse whether either pathway had an effect on HDAC4 cellular localisation. For this we made use of the fact that overexpression of HDAC4–GFP leads either to cytoplasmic or nuclear localisation of the protein (Fig. 5A; 14). We therefore asked whether addition of CaMKIV* or MKK6b(E) affected the proportion of cells containing nuclear versus cytoplasmic HDAC4–GFP. Figure 5B summarises the results. Transfection of HDAC4–GFP alone resulted in ∼50% of the cells with nuclear GFP staining and an equal number with cytoplasmic staining. Virtually no cells showed both cytoplasmic and nuclear staining, as reported previously for HeLa cells (14). Co-transfection of CaMKIV* resulted in an increase in cells with cytoplasmic staining to >90%. In agreement with this result, treatment of the transfected cells with staurosporine, a broad specificity kinase inhibitor that also targets CaMKIV, resulted in nuclear staining of HDAC4–GFP in 100% of cells (Fig. 5B). In contrast, co-transfection of MKK6b(E) had no effect on HDAC4 cellular distribution. Similarly, a specific inhibitor of p38 MAP kinase, SB203580, did not alter HDAC4–GFP localisation (Fig. 5B).
Given that constitutively active CaMKIV increases the proportion of cytoplasmic HDAC4 in myoblasts, we asked if it could inhibit nuclear entry after myoblast fusion. As shown in Figure 1, GFP–HDAC4 is localised in the nucleus of differentiated myotubes. However, Figure 5C shows that in the presence of CaMKIV* HDAC4–GFP is predominantly cytoplasmic. In contrast, the presence of MKK6b(E) does not alter the localisation of HDAC4–GFP, which still accumulates in the nucleus. Collectively, these results indicate that constitutively active CaMKIV increases the cytoplasmic localisation of HDAC4–GFP in myoblasts and prevents its nuclear entry after myoblast fusion.
The above results suggest that the cytoplasmic localisation of HDAC4 is regulated by CaMKIV-mediated phosphorylation pathways, but not by the MKK6b/MAP kinase pathway. Thus, constitutively active CaMKIV can override HDAC4 inhibition of differentiation by excluding HDAC4 from the nucleus, whereas MKK6b(E) circumvents the inhibitory effects of HDAC4 indirectly via a distinct pathway.
HDAC4 associates with 14-3-3 proteins in a phosphorylation-dependent manner
The dramatic exclusion of HDAC4 from the nucleus mediated by the CaMKIV signalling pathway suggests that this process is mediated by phosphorylation. The family of 14-3-3 proteins share the ability to recognise phosphorylated proteins and can mediate their localisation in the cytoplasm (50–54). We therefore tested the ability of HDAC4 to bind to 14-3-3 proteins. GST–14-3-3 fusion proteins bound to glutathione beads were incubated with extracts from cells expressing HDAC4–Flag. The upper panel of Figure 6A shows that Flag–HDAC4 specifically co-precipitates with 14-3-3ɛ, 14-3-3γ and 14-3-3ζ, but not with 14-3-3ɛ carrying a point mutation in the phosphoprotein-binding domain (14-3-3ɛ mut) or beads alone. 14-3-3 proteins were expressed to approximately equal levels (Fig. 6B). Binding to 14-3-3 proteins is severely impaired after treatment of the cells with staurosporine, an inhibitor of Ca2+/calmodulin signalling pathways (Fig. 6A, bottom). The staurosporine effect was not due to changes in expression of HDAC4 (Fig. 6C). Since 14-3-3 proteins are cytoplasmic, these results suggest that a phosphorylation-dependent interaction of 14-3-3 proteins and HDAC4 may anchor the HDAC4 protein in the cytoplasm following Ca2+/calmodulin signalling. In agreement with these observations, 14-3-3 proteins have been shown to associate with HDAC4 in the yeast two-hybrid system and mammalian cell extracts (55,56).
Figure 6.
HDAC4 interacts with 14-3-3 proteins in a phosphorylation-dependent manner. 293 cells were transfected with 5 µg pcHDAC4-Flag. Thirty-six hours after transfection cells were lysed either with or without prior treatment with staurosporine (20 µM) for 60 min. (A) Cell lysates were incubated with 1 µg recombinant purified GST–14-3-3 fusion protein pre-bound to glutathione beads. Bound material was subjected to SDS–PAGE and western analysis using anti-Flag antibody. (B) Input of GST–14-3-3 fusion proteins separated using SDS–PAGE and visualised using Coomassie staining. (C) Input of cell lysates subjected to SDS–PAGE and western analysis using anti-Flag antibody.
DISCUSSION
Muscle differentiation is a complex process that involves the orchestration of a whole gene expression programme. Although a number of transcription factors and signal transduction pathways involved in this process have been identified, the picture remains far from complete. In this study we present a novel mechanism of transcriptional control during C2C12 myoblast differentiation at the step of myoblast fusion, namely regulation of HDAC4 nucleo-cytoplasmic shuttling. A model consistent with our observation is depicted in Figure 7.
Figure 7.
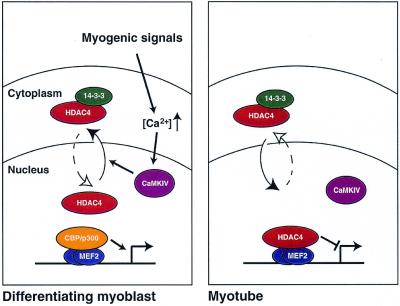
A model for the role of HDAC4 during C2C12 differentiation. In myoblasts HDAC4 is held in the cytoplasm, where it associates with 14-3-3 proteins in a phosphorylation-dependent manner. Cytoplasmic retention of HDAC4, which is stimulated by CaMKIV activity, allows MEF2-dependent transcriptional activation via CBP/p300 histone acetyltransferases. After myotube fusion HDAC4 relocates to the nucleus, where it associates with MEF2 to repress transcription.
As reported previously (14), HDAC4 has the surprising feature of shuttling between the nucleus and the cytoplasm. There are several examples where cytoplasmic retention is used to regulate the biological function of a protein, as in the case of NFκB and NFAT (57,58). Here we show that cytoplasmic retention of HDAC4 may be necessary for correct regulation of C2C12 myoblast differentiation. We have shown that inappropriate HDAC4 expression in myoblasts leads to nuclear entry (Fig. 5B) and inhibition of myoblast fusion (Fig. 2). Thus, we propose that endogenous HDAC4 is held in the cytoplasm in C2C12 myoblasts to allow myotube formation to occur (Fig. 7, left). Following myoblast fusion HDAC4 enters the nucleus, presumably because the differentiating signal, which activates the Ca2+/calmodulin signalling pathway, has ceased. Once HDAC4 enters the nucleus (Fig. 7, right) its repressive activity may serve to inhibit a subset of myogenic promoters (such as MEF2-driven ones) and thus establish a terminally differentiated state.
We speculate that MEF2 activity is an important, not unique, target for HDAC4 following its localisation to the nucleus, for several reasons: MEF2 activity is essential for C2C12 differentiation (20) and HDAC4 can repress MEF2 transcriptional activity and C2C12 differentiation (Fig. 2). In addition, a signalling pathway that stimulates MEF2 activity and induces differentiation (the Ca2+/calmodulin signalling pathway) has the ability to differentially localise HDAC4 to the cytoplasm and override HDAC4-mediated differentiation arrest (Figs 4 and 5).
If HDAC4 enters the nucleus after myoblast fusion to target MEF2-dependent transcription, MEF2-dependent target genes would be expected to be down-regulated after myoblast fusion. However, a number of MEF2 gene products have been shown to be up-regulated during myoblast fusion (19,59,60). This apparent discrepancy could be explained by the fact that the increase in RNA and protein levels seen after differentiation could be a reflection of the integrated amount of gene transcription during fusion rather than the transcriptional activity after fusion. Alternatively, HDAC4 might only regulate a subset of MEF2-responsive genes. Evidence for down-regulation of MEF2-dependent transcription during muscle development in vivo comes from transgenic mice carrying a MEF2-dependent lacZ reporter (61,62). In these mice high levels of MEF2-dependent transcription in the somites of the embryo ceases after birth.
MEF2 being a central player in the regulation of muscle differentiation, its activity is regulated through a network of signalling pathways. Although CaMKs, calcineurin, p38 and ERK5 have been implicated in regulation of MEF2 activity, integration of and potential cross-talk between these pathways is poorly understood. As both p38 and ERK5 have the ability to phosphorylate MEF2 factors within their transactivation domain, a putative mechanism of action for this pathway could be phosphorylation-dependent recruitment of co-activators, such as CBP/p300 (63). Here we propose a mechanism for CaMKIV-mediated activation of MEF2-dependent transcription, namely nuclear exclusion of the HDAC4 repressor protein.
The involvement of CaMKIV in the nuclear exclusion of HDAC4 suggests the involvement of phosphorylation in this process. The 14-3-3 proteins are good candidates to mediate such a phosphorylation-dependent localisation of HDAC4, since they are able to bind phosphorylated proteins and are localised primarily in the cytoplasm. This study suggests that phosphorylation-dependent binding of HDAC4 to 14-3-3 proteins might underlie the cytoplasmic retention of HDAC4 in C2C12 myoblasts. 14-3-3 proteins may mediate the export of HDAC4 and/or may anchor HDAC4 in the cytoplasm. It is tempting to speculate that HDAC4 itself is phosphorylated in a CaMKIV-dependent manner and directly associates with 14-3-3 proteins, although the involvement of an intermediary protein cannot be excluded at this point.
The results presented here suggest a central regulating role for HDAC4 in muscle differentiation. This unique role of HDAC4 is shared by HDAC5 and possibly other members of the class II family of histone deacetylases, such as HDAC7. These three enzymes have unique features, which may allow them to regulate myogenesis. First, they all possess a large N-terminal extension, which contains the binding site for MEF2. This N-terminus is not present in the class I enzymes (HDAC1, HDAC2, HDAC3 and HDAC8) or in the class II deacetylase HDAC6. Secondly, HDAC4, HDAC5 and HDAC7 are expressed in a tissue-specific manner with high levels of expression in muscle cells (9,10,49). In contrast, the class I enzymes are expressed ubiquitously (4,5,8), whereas HDAC6 is not expressed in muscle (9,10). The ubiquitous nature of the class I enzymes may not, however, affect the myogenic programme because these enzymes do not have access to MEF2. Indeed, we have shown that HDAC1 protein is unable to repress myogenic differentiation. Thus, the unique structural and biological features of HDAC4, HDAC5 and HDAC7 suggest that they may form part of a distinct deacetylase subgroup.
This study has identified HDAC4 as a crucial regulator of the myogenic differentiation programme. We demonstrate that HDAC4 and HDAC5 share the ability to inhibit C2C12 myoblast differentiation when overexpressed, as has also been shown in the case of the 10T1/2 cell system (48). In contrast, HDAC4 and HDAC5 localisation changes reciprocally during C2C12 myoblast differentiation (Fig. 1; 47). This might suggest that HDAC4 and HDAC5 regulate a distinct subset of MEF2-dependent and/or MEF2-independent target genes. Gene expression profiling and chromatin immunoprecipitation experiments might help us to identify those genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Paul Kemp for advice on C2C12 differentiation. We thank Michael Yaffe, Christina Grozinger, Stuart Schreiber, Richard Maurer and Jiahuai Han for reagents. This work was funded by a Cancer Research Campaign (CRC) programme grant (DR11). E.A.M. was supported by a CRC studentship and C.K. was funded by a Marie Curie fellowship from the EC.
References
- 1.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- 3.Guarente L. (2000) Sir2 links chromatin silencing, metabolism and aging. Genes Dev., 14, 1021–1026. [PubMed] [Google Scholar]
- 4.Dangond F., Hafler,D.A., Tong,J.K., Randall,J., Kojima,R., Utku,N. and Gullans,S.R. (1998) Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem. Biophys. Res. Commun., 242, 648–652. [DOI] [PubMed] [Google Scholar]
- 5.Emiliani S., Fischle,W., Van Lint,C., Al-Abed,Y. and Verdin,E. (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl Acad. Sci. USA, 95, 2795–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 7.Yang W.M., Inouye,C., Zeng,Y.Y., Bearss,D. and Seto,D. (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA, 93, 12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu E., Chen,Z., Fredrickson,T., Zhu,Y., Kirkpatrick,R., Zhang,G.F., Johanson,K., Sung,C.M., Liu,R. and Winkler,J. (2000) Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem., 275, 15254–15264. [DOI] [PubMed] [Google Scholar]
- 9.Grozinger C.M., Hassig,C.A. and Schreiber,S.L. (1999) Three proteins define a class of human histone deacetylases related to yeast hda1p. Proc. Natl Acad. Sci. USA, 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verdel A. and Khochbin,S. (1999) Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem., 274, 2440–2445. [DOI] [PubMed] [Google Scholar]
- 11.Kao H.Y., Downes,M., Ordentlich,P. and Evans,R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev., 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- 12.Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- 13.Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- 14.Miska E.A., Karlsson,C., Langley,E., Nielsen,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang A.H., Bertos,N.R., Vezmar,M., Pelletier,N., Crosato,M., Heng,H.H., Th’ng,J., Han,J. and Yang,X.J. (1999) HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol., 19, 7816–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemercier C., Verdel,A., Galloo,B., Curtet,S., Brocard,M.P. and Khochbin,S. (2000) mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem., 275, 15594–15599. [DOI] [PubMed] [Google Scholar]
- 17.Lu J., McKinsey,T.A., Nicol,R.L. and Olson,E.N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl Acad. Sci. USA, 97, 4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang E.Y., Zhang,J., Miska,E.A., Guenther,M.G., Kouzarides,T. and Lazar,M.A. (2000) Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev., 14, 45–54. [PMC free article] [PubMed] [Google Scholar]
- 19.Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol., 14, 167–196. [DOI] [PubMed] [Google Scholar]
- 20.Ornatsky O.I., Andreucci,J.J. and McDermott,J.C. (1997) A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem., 272, 33271–33278. [DOI] [PubMed] [Google Scholar]
- 21.Lilly B., Zhao,B., Ranganayakulu,G., Paterson,B.M., Schulz,R.A. and Olson,E.N. (1995) Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science, 267, 688–693. [DOI] [PubMed] [Google Scholar]
- 22.Lin Q., Schwarz,J., Bucana,C. and Olson,E.N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science, 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han T.H. and Prywes,R. (1995) Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol., 15, 2907–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J., Jiang,Y., Li,Z., Kravchenko,V.V. and Ulevitch,R.J. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature, 386, 296–299. [DOI] [PubMed] [Google Scholar]
- 25.Kato Y., Kravchenko,V.V., Tapping,R.I., Han,J., Ulevitch,R.J. and Lee,J.D. (1997) BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J., 16, 7054–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zetser A., Gredinger,E. and Bengal,E. (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem., 274, 5193–5200. [DOI] [PubMed] [Google Scholar]
- 27.Zhao M., New,L., Kravchenko,V.V., Kato,Y., Gram,H., di Padova,F., Olson,E.N., Ulevitch,R.J. and Han,J. (1999) Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol., 19, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C.C., Ornatsky,O.I., McDermott,J.C., Cruz,T.F. and Prody,C.A. (1998) Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res., 26, 4771–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke N., Arenzana,N., Hai,T., Minden,A. and Prywes,R. (1998) Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol. Cell. Biol., 18, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ornatsky O.I., Cox,D.M., Tangirala,P. andreucci,J.J., Quinn,Z.A., Wrana,J.L., Prywes,R., Yu,Y.T. and McDermott,J.C. (1999) Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res., 27, 2646–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S.H., Galanis,A. and Sharrocks,A.D. (1999) Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol., 19, 4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinissen M.J., Chiariello,M., Pallante,M. and Gutkind,J.S. (1999) A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s and extracellular signal-regulated kinase 5. Mol. Cell. Biol., 19, 4289–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puri P.L., Wu,Z., Zhang,P., Wood,L.D., Bhakta,K.S., Han,J., Feramisco,J.R., Karin,M. and Wang,J.Y. (2000) Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev., 14, 574–584. [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S., Liu,P., Borras,A., Chatila,T. and Speck,S.H. (1997) Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J., 16, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youn H.D., Sun,L., Prywes,R. and Liu,J.O. (1999) Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science, 286, 790–793. [DOI] [PubMed] [Google Scholar]
- 36.Passier R., Zeng,H., Frey,N., Naya,F.J., Nicol,R.L., McKinsey,T.A., Overbeek,P., Richardson,J.A., Grant,S.R. and Olson,E.N. (2000) CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest., 105, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H., Naya,F.J., McKinsey,T.A., Mercer,B., Shelton,J.M., Chin,E.R., Simard,A.R., Michel,R.N., Bassel-Duby,R., Olson,E.N. et al. (2000) MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J., 19, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naya F.J., Mercer,B., Shelton,J., Richardson,J.A., Williams,R.S. and Olson,E.N. (2000) Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem., 275, 4545–4548. [DOI] [PubMed] [Google Scholar]
- 39.Blaeser F., Ho,N., Prywes,R. and Chatila,T.A. (2000) Ca(2+)-dependent gene expression mediated by MEF2 transcription factors. J. Biol. Chem., 275, 197–209. [DOI] [PubMed] [Google Scholar]
- 40.Zernicka-Goetz M., Pines,J., Ryan,K., Siemering,K.R., Haseloff,J., Evans,M.J. and Gurdon,J.B. (1996) An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development, 122, 3719–3724. [DOI] [PubMed] [Google Scholar]
- 41.Sun P., Enslen,H., Myung,P.S. and Maurer,R.A. (1994) Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev., 8, 2527–2539. [DOI] [PubMed] [Google Scholar]
- 42.Hagemeier C., Bannister,A.J., Cook,A. and Kouzarides,T. (1993) The activation domain of the transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl Acad. Sci. USA, 90, 1580–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin K., Trouche,D., Hagemeier,C., Sorensen,T.S., La Thangue,N.B. and Kouzarides,T. (1995) Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature, 375, 691–694. [DOI] [PubMed] [Google Scholar]
- 44.Hagemeier C., Cook,A. and Kouzarides,T. (1993) The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res., 21, 4998–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pines J. and Hunter,T. (1991) Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol., 115, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlsson C., Pines,J. (1998) In Celis,J. (ed.), Cell Biology: A Laboratory Handbook. Academic Press, San Diego, CA, Vol. 4, pp. 246–252.
- 47.McKinsey T.A., Zhang,C.L., Lu,J. and Olson,E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J., McKinsey,T.A., Zhang,C.L. and Olson,E.N. (2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell, 6, 233–244. [DOI] [PubMed] [Google Scholar]
- 49.Fischle W., Emiliani,S., Hendzel,M.J., Nagase,T., Nomura,N., Voelter,W. and Verdin,E. (1999) A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem., 274, 11713–11720. [DOI] [PubMed] [Google Scholar]
- 50.Zeng Y., Forbes,K.C., Wu,Z., Moreno,S., Piwnica-Worms,H. and Enoch,T. (1998) Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature, 395, 507–510. [DOI] [PubMed] [Google Scholar]
- 51.Peng C.Y., Graves,P.R., Thoma,R.S., Wu,Z., Shaw,A.S. and Piwnica-Worms,H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- 52.Kumagai A. and Dunphy,W.G. (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev., 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J., Winkler,K., Yoshida,M. and Kornbluth,S. (1999) Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J., 18, 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- 55.McKinsey T.A., Zhang,C.L. and Olson,E.N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl Acad. Sci. USA, 97, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao A., Luo,C. and Hogan,P.G. (1997) Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol., 15, 707–747. [DOI] [PubMed] [Google Scholar]
- 58.Jans D.A. and Hubner,S. (1996) Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev., 76, 651–685. [DOI] [PubMed] [Google Scholar]
- 59.Gossett L.A., Kelvin,D.J., Sternberg,E.A. and Olson,E.N. (1989) A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol., 9, 5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cserjesi P. and Olson,E.N. (1991) Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol. Cell. Biol., 11, 4854–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naya F.J., Wu,C., Richardson,J.A., Overbeek,P. and Olson,E.N. (1999) Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development, 126, 2045–2052. [DOI] [PubMed] [Google Scholar]
- 62.Naya F.J. and Olson,E. (1999) MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol., 11, 683–688. [DOI] [PubMed] [Google Scholar]
- 63.Sartorelli V., Huang,J., Hamamori,Y. and Kedes,L. (1997) Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol., 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]