Abstract
We describe a reflection imaging system which consists of a plasmonic crystal, a common laboratory microscope, and bandpass filters for use in the quantitative imaging and in-situ monitoring of live cells and their substrate interactions. Surface plasmon resonance (SPR) provides a highly sensitive method to monitor changes in physicochemical properties occurring at metal-dielectric interfaces. Polyelectrolyte thin films deposited using the layer-by-layer (LBL) self-assembly method provide a reference system to calibrate the reflection contrast changes that occur when the polyelectrolyte film thickness changes, as well as provide insight into the optical responses that originate from the multiple plasmonic features supported by this imaging system. Finite-difference time-domain (FDTD) simulations of the optical responses measured experimentally from the polyelectrolyte reference system are used to provide a calibration of the optical system for subsequent use in quantitative studies investigating live cell dynamics in cultures supported on a plasmonic crystal substrate. Live Aplysia californica pedal ganglion neurons cultured in artificial sea water were used as a model system through which to explore the utility of this plasmonic imaging technique. Here, the morphology of cellular peripheral structures < ~80 nm in thickness were quantitatively analyzed and the dynamics of their trypsin-induced surface detachment visualized. These results illustrate capacities of this system for use in investigations of the dynamics of ultrathin cellular structures within complex bioanalytical environments.
Keywords: surface plasmon resonance (SPR), optical cell imaging, neurons, layer-by-layer (LBL) assembly, nanoimprint soft lithography, Finite-Difference Time-Domain (FDTD), plasmonics
TOC image
1. INTRODUCTION
Cell morphology and the processes mediating its physiological and pathological dynamics are essential characteristics of the mechanisms underlying cellular differentiation, growth, apoptosis, and autophagy. Better understandings of their complex nature are needed to sustain progress in medical sciences and biotechnology.1–6 Time-resolved imaging of living systems can provide insights into an individual cell’s organization and function that static observations of fixed cells cannot.7–12 Because most cells are optically transparent, live cell imaging at high spatial resolution is difficult using conventional optics without staining or tagging structures of interest in the cell.13–15 Regardless of the imaging technique, good cellular health is crucial to preserve a normal metabolic state of the studied cells throughout the entire duration of the imaging experiment.16 When imaging cellular dynamics, experimental conditions such as pH17, 18, oxygenation19, temperature20 and osmolality21 need to be sustained to maintain viability of cells. In addition, high intensity illumination may induce temperature fluctuations and/or damage of cells and cellular components such as DNA (via UV light absorption), which may lead to cell death.22 It is critical, therefore, that live cell imaging techniques be designed so as to minimize physical and chemical damage to the cell and retain the ability to conduct measurements in its physiological environment.
A number of optics-based analytical tools have been developed for spatiotemporal studies of structure and function in living cells. Fluorescence microscopy is the most widely used technique for live cell imaging because, by tagging cell components with fluorophores (such as small organic dyes, fluorescent proteins, or quantum dots), it is capable of probing specific structures including organelles, multimolecular complexes, and individual molecules inside the cell.23–25 Fluorescence imaging, however, can be complicated by the toxicity associated with introducing foreign fluorescent labels into the system and the susceptibility of fluorophores to photobleaching.9, 26, 27 Phase contrast microscopy28, 29 and differential interference contrast microscopy30, 31 make it possible to visualize unstained biological specimens with enhanced contrast and resolution. These imaging methods provide a useful means to study cellular structures and processes, albeit with lesser quantitative capabilities for analyzing live cell morphology. Three-dimensional (3D) images of live cells can be obtained using confocal microscopy – wherein the resultant images are generated from multiple 2D optical slices measured at different depths within a sample.32–35 Confocal microscopy, typically, requires incorporation of fluorescent labels in the cells, which introduces prospects for the aforementioned problems and increases the overall complexity of the experiment.36 Digital holographic microscopy,12 interference reflection microscopy,37 and spatial light interference microscopy (SLIM)38 are extremely powerful analytical methods that provide label-free forms of quantitative spatiotemporal live cell imaging, producing information on morphology and dynamics of the cell. These optical techniques do require complex/demanding instrumental setups for enhanced contrast and/or significant resources for data processing. Atomic force microscopy (AFM) stands as one notable alternative to optical imaging techniques that can be used to generate topographical maps of specimens with high resolution in the perpendicular direction.39–42 While AFM affords height data at nanometer resolution, it is more limited in its utility for studying live cell dynamics due in part to long scan times and physical impacts arising from contact of the AFM tip with the cell surface.43
The utility of plasmonic imaging as an analytical tool in biology, in that it can be performed on samples located in a complex environment (i.e. in the liquid cell-culture media), is well appreciated and a subject treated in some depth in the literature.44–48 We recently described a label-free reflection imaging technique using a plasmonic crystal that in principle makes it possible to quantitatively map the index-convolved thicknesses of complex biological specimens placed in contact with the crystal surface. We explored this in the earlier work using dried samples of fixed Aplysia californica pedal neurons.49 This model was chosen due to its robustness in culture on a variety of surfaces, and for its thorough characterization in the neuroscience literature.50,51 This imaging method is label-free and highly sensitive to changes in refractive index within the near-surface region of the plasmonic crystal. We show in the present work that nanoimprinted optics of this form can be used as substrates for cell culture, and allow a simple and quantitative way of imaging live cells, including their morphological dynamics. There are a number of important biological phenomena that are expected to yield perturbations within the limiting volume element probed in this experiment, ones that might be probed given suitable means to separate those responses from other interfering optical effects (e.g. cell-substrate interactions, mechanical parameters of cells including rigidity/flexibility, extracellular matrix chemical and physical properties, etc.).Using measurements made of polyelectrolyte layer-by-layer (LBL) assemblies as a reference calibration in conjunction with theoretical simulations performed utilizing finite-difference time-domain (FDTD) methods, a multispectral reflection contrast calibration was applied to representative reflection images of live Aplysia pedal neurons cultured on the surface of a gold-coated plasmonic crystal substrate. The results demonstrate that reflection-mode imaging can quantitatively map the morphological parameters of peripheral regions of live cells in a growth-permitting environment. We further show this imaging method can be utilized to investigate dynamic changes, as illustrated in quantitative analyses made of the trypsin-mediated detachment of live Aplysia pedal neurons from their supporting substrate.
2. EXPERIMENTAL METHODS
Reagents were used as received without further purification. Commercially available chemical reagents were purchased from Norland Products, Gelest, and Sigma-Aldrich. A complete reagent list is given in Supplementary Methods (Supporting Information).
The 3D plasmonic crystals were fabricated using soft nanoimprint lithography technique as previously reported.9, 52–56 To improve cell outgrowth on the plasmonic crystal surface and prevent water penetration at the interface between the metal film and the photocured polyurethane (NOA) nanostructures, a ~6 nm thin layer of aluminum oxide (Al2O3) was deposited via ALD (atomic layer deposition, Cambridge Nanotech). Full details of the 3D plasmonic crystal fabrication process are given in Supplementary Methods (Supporting Information).
Polyelectrolyte assemblies were created on the surfaces of both Au-coated plasmonic crystals and reference Au thin films supported by a silicon wafer using a LBL method. The thicknesses of polyelectrolyte LBL assemblies on the Au-coated silicon substrate in artificial sea water were measured using an MFP-3D atomic force microscope (AFM, Asylum Research, Santa Barbara, CA) and the refractive indices of the polyelectrolyte films under the same conditions determined using a Woollam VASE spectroscopic ellipsometer with a home-built liquid cell (shown in Figure S1). Analysis of the AFM data was performed using Igor Pro (WaveMetrics, wavemetrics.com). The thickness data from the AFM measurement was used in calculations made to determine the refractive indices of the polyelectrolyte films based on ellipsometry data. Full details of the polyelectrolyte LBL growth process and AFM measurements are given in Supplementary Methods (Supporting Information).
Reflection mode images of both the polyelectrolyte films and live cells in culture on the plasmonic crystal surface were obtained using an AxioScope A1 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) operated with a halogen light source and a 20X and 0.40 NA objective lens. Grayscale reflection images were processed and analyzed using Matlab (MathWorks Inc., www.mathworks.com) and ImageJ (National Institutes of Health, Bethesda, MD). To calibrate the normalized reflection contrast changes as a function of the thickness of the polyelectrolyte assemblies, a portion of the 16-mercaptohexadecanoic acid (MHDA)-coated nanohole array (Figure 1a) was covered with a cured NOA film, creating a region at which the surface refractive index remained unchanged during the polyelectrolyte film deposition. The reflection images of this plasmonic crystal and coating layer were taken at every second round of the polyelectrolyte deposition. The normalized reflection contrasts for the coated regions were calculated using a method reported previously.49 Full details of the acquisition parameters and calibration procedures are included in the Supplementary Methods (Supporting Information).
Figure 1.
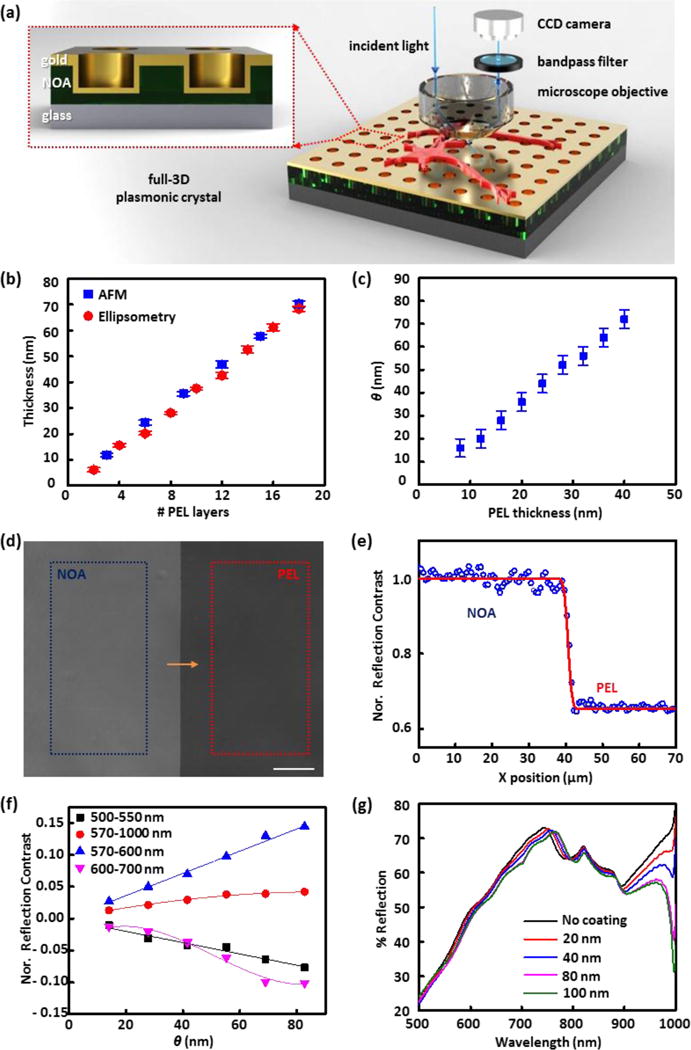
(a) Schematic illustration of the plasmonic reflection imaging system setup with a laboratory microscope, bandpass filters, CCD camera, and a full 3D plasmonic crystal. (Inset) A close-up illustration of the cross-section of the full 3D plasmonic crystal. (b) Thickness values of polyelectrolyte assemblies measured at increasing numbers of polyelectrolyte layer (PEL) depositions using AFM and ellipsometry. (c) Corresponding pairs of thicknesses of polyelectrolyte films (n = 1.48) and index corrected mass coverages (θ) of live cells (n = 1.42). (d) Reflection image (using a 500–550 nm bandpass filter) of a polyelectrolyte layer-by-layer assembly (PEL) and photocured polyurethane film (NOA) on a plasmonic crystal used for reflection contrast calibration. Average pixel signal intensities were calculated in the regions marked with the blue (NOA region) and red box (PEL region), and used for reflection contrast calibration. The scale bar corresponds to 100 μm. (e) Step-edge profile of normalized reflection contrast (blue circles) along the yellow arrow in (d) and fitted red curve with a Gaussian width of 0.84 μm. (f) Normalized reflection contrast (NRC) as a function of index corrected thickness (θ) determined using different bandpass filters: 500–550 nm (black squares), 570–1000 nm (red circles), 570–600 nm (blue triangles), and 600–700 nm (pink inverted triangles). The error bars are obscured by the data markers themselves as a result of the large number of pixels (more than 30,000) used to determine the value. (g) FDTD calculated reflection spectra for varying thickness of index corrected material on a nanostructured plasmonic crystal.
Aplysia Californica (100–300 g), supplied by the National Resource for Aplysia (Miami, FL), were kept in circulated, aerated seawater at 14 °C. Prior to dissection, the animals were anesthetized by injection of isotonic magnesium chloride solution into the body cavity (~30–50 % of body weight). Individual Aplysia pedal neurons were isolated as described previously57, 58 and cultured in artificial sea water overnight. Normal outgrowth of cellular processes was observed for most of the cultured cells. Cell detachment from the plasmonic crystal surface was induced by injection of trypsin into the culture media. AFM measurements of fixed Aplysia neurons on plasmonic crystals were performed. The AFM data was analyzed with Igor Pro. Full details regarding neuron isolation protocols and cell culture on the plasmonic crystal, as well as imaging and dissociation protocol are given in Supplementary Methods (Supporting Information).
Several 3D FDTD simulations were carried out to model the zero-order reflection/transmission spectra and optical behavior of the plasmonic crystal as a function of thin film thickness using an FDTD software package (Lumerical Solutions Inc., Vancouver, Canada). Details regarding the unit cell geometry and models used are given in Supplementary Methods (Supporting Information).
3. RESULTS AND DISCUSSION
3.1. Plasmonic Crystals and Computational Models
The optical setup of the system used in this work is shown in Figure 1a. Reflection images of live Aplysia neurons on a plasmonic crystal immersed in liquid media (artificial sea water) were acquired with a laboratory microscope with specific bandpass filters (500–550 nm, 570–1000 nm, 570–600 nm, or 600–700 nm) inserted in front of a CCD camera, as described in the Experimental and Supplementary Methods. A series of 3D plasmonic crystals similar to those used in the earlier study with fixed cells44 was employed as a substrate for live-cell culturing and imaging. The inset of Figure 1a shows an illustration of the cross section of a full 3D plasmonic crystal consisting of a square array of embossed nanoholes (540 nm diameter, 280 nm depth, 740 nm pitch) conformally coated with a thin film of Au. This optical system generates multiple plasmonic features that respond with varying sensitivities to changes occurring at the metal/dielectric interface, responses that for this design rule span visible to near-infrared wavelengths49, 52–56 The optical features of the experimental spectra are well modeled by the FDTD calculations (Figure 1g and Figure S2b) using the appropriate boundary conditions and geometrical model for the periodic nanohole array. Figure S2a presents the structural model of the plasmonic crystal used in these calculations and illustrates the strong agreement it affords between the experimentally measured and calculated transmission spectra.
3.2. Index Corrected Mass Coverage
The polyelectrolyte LBL assemblies of poly(allylamine hydrochloride) (PAH) and poly(sodium 4-styrenesulfonate) (PSS) were used as a reference system to calibrate the reflection contrast changes seen as a function of thin film thickness on the plasmonic crystal. For further theoretical analysis using FDTD calculations, accurate values for the thickness and refractive indices of the polyelectrolyte layers in artificial sea water are required. The thickness of one layer of PAH/PSS and its refractive index in dry condition were determined as described in previous studies.49, 55, 59 These values are altered in the artificial sea water environment requiring corrected values calculated using immersed AFM and ellipsometry data (Figure 1b) as measured on patterned substrates (illustrated in Figure S3) hydrated in artificial sea water.
Figure S4a shows a comparison of the ellipsometric responses of the polyelectrolyte film measured before and after removal of the resist layer, demonstrating that the polyelectrolyte film is not affected by the stripping step. The steep step edge of the polyelectrolyte pattern (Figure S4b) is easily quantified using AFM measurements. These AFM data reveal that each PAH/PSS deposition step increases the thickness of the polyelectrolyte assembly by 3.85 ± 0.043 nm when measured in artificial sea water.
Using the AFM thickness data and a Cauchy model, the refractive indices were then calculated from the spectroscopic ellipsometry data. The analysis yielded a calculated refractive index at 600 nm of 1.484, in agreement with previous reports.60 The thickness increase due to each PAH/PSS deposition step, as determined by AFM measurement and ellipsometric analysis, is plotted in Figure 1b. As the volume change of the polyelectrolyte assemblies in artificial sea water originates from osmotic swelling due to artificial sea water penetration into the thin film, the refractive index of polyelectrolyte in artificial sea water can be approximated using the Arago-Biot equation, n = φ1n1 + φ2n2 where n is the refractive index of polyelectrolyte assemblies in artificial sea water, n1 is the refractive index of dried polyelectrolyte assemblies (1.64)55, 59, n2 is the refractive index of artificial sea water (1.34)61, φ1 is the volume fraction of the polyelectrolyte assemblies, and φ2 is the volume fraction of artificial sea water. According to this relationship, the calculated refractive index of the polyelectrolyte film is 1.485, which is in good agreement with the experimentally measured refractive index of polyelectrolyte assemblies in artificial sea water using spectroscopic ellipsometry.
The plasmonic response of the system is dependent on both the local refractive index and changes in the thickness of any dielectric material present on the surface of the plasmonic crystal. A living cell is an unquestionably complex dielectric structure, one comprised of components with very different refractive indices. For example, the refractive index of even the relatively uniform component, the cell membrane (1.46–1.53), is higher than that of the highly structurally complex cytoplasm (1.35–1.38).62, 63 Since the optical properties in this range differ sufficiently from the polyelectrolyte calibration system, the imaging mode contrasts must be modified to account for the local refractive indices of the cellular structures in contact with the substrate. The useful quantitative reflection imaging range of this technique is limited to a thickness range of approximately 80 nm, due to the exponential decay of the evanescent electric field associated with plasmon resonances (although other optical effects can and do contribute to sensitivities at greater distances).49 At least a quarter of the cellular component is expected to be the cell membrane (for reference, the thickness of the cell membrane is ~ 10 nm59, 64). Therefore, using the Arago-Biot approximation, we estimated an effective refractive index for regions of the cell in contact of 1.42 as a starting point for the analyses of sensitivity discussed below. A detailed analysis of the limitations and sensitivity of this system has been provided in previous studies.49,53,55 The calculations that follow aim to optimize and calibrate this system for performing measurements in the complex optical/fluid environment of a living cell. Finite-difference time-domain (FDTD) calculations were used to determine the index corrected mass coverage (θ) for a biological material (n ~ 1.42) that exhibits the same optical response as the polyelectrolyte assemblies (n ~ 1.48) at different thicknesses. Figure S5 presents the calculated reflection spectra for plasmonic crystals immersed in artificial sea water and coated with a conformal layer of either polyelectrolyte or the equivalent index corrected material for live cells. The small discrepancies between the spectra at higher thicknesses (40 nm polyelectrolyte and 72 nm index-corrected material) likely originate from the limitations in the grid spacing (4 nm) used for the structural modeling. The corresponding pairs for FDTD calculated polyelectrolyte film thicknesses and index corrected mass coverages are presented in Figure 1c.
The cell membrane outer lipid layer is usually separated from the substrate involved in cell adhesion to the surface by an extracellular matrix, a collection of fibrous proteins and glycosaminoglycans. These interlayer structures can be reasonably large, as noted in a previous study, where the distance to the substrate at a peripheral lamellipodia contact was determined to be 25 ± 10 nm.64–66 We used several models evaluating either 0 nm, 12 nm, or 24 nm artificial sea water gaps between the plasmonic crystal substrate and the index corrected material to assess the effects of the extracellular matrix layer on the outcome of plasmonic measurements. The calculated reflection spectra presented in Figure S6, show a negligible effect of a thin artificial sea water gap (~ 24 nm) on plasmonic crystal reflectance. For this reason, we did not consider the presence of gaps between the cell and substrate for further quantitative analyses of live cell imaging, noting the important caveat that the presence of a low index boundary layer may contribute to the composite changes in dielectric structure evidenced in the data.
3.3. Reflection Imaging Contrast Calibration
Reflection images of polyelectrolyte thin films deposited on the plasmonic crystal substrate were acquired in artificial sea water using a series of bandpass filters inserted in front of the camera (Figure 1a). A representative reflection image used for contrast calibration is presented in Figure 1d. The NOA covered region, where the refractive index is held constant, provides a means to correct variations in illumination intensity between different images (as discussed in the S.I.).49
Figure 1e shows the step-edge profile of the normalized reflection contrast at the boundary (measured along the yellow arrow shown in Figure 1d) and the contrast difference between the polyelectrolyte and NOA-coated regions. This step-edge profile was fit using a cumulative Gaussian curve67 (red line, as presented in Figure 1e) to estimate the lateral resolution of the plasmonic imaging sensor. The resultant fit (a Gaussian width of 0.84 μm) gives a value that is slightly larger than the resolution limit of the CCD camera (~0.65 μm). As discussed in a previous report49, any single pixel in the reflection image overlaps part of one or several nanoholes, given that the pixel size (~0.65 μm) and periodicity of the nanoholes (~0.74 μm) are comparable.
Using a series of bandpass filters, the relation of the normalized reflection contrast (NRC) to θ was calculated from the calibration data (Figure 1f). Because of the large number of pixels (~ 30,000) used in this calculation, the errors bars are smaller than the points used in the plot. A variety of trends are noticed in these data. The reflection contrast, for example, increases with θ in the wavelength ranges of 570–600 nm and 570–1000 nm, but decreases and becomes negative at the wavelength ranges of 500–550 nm and 600–700 nm. The changes in reflection contrast also vary in functional form, varying non-linearly over the wavelength number range of 600–700 and 570–1000 nm, but linearly over 500–550 and 570–600 nm. These complex behaviors result as a consequence of the multiple plasmonic modes generated by the nanohole array. To simplify the analysis, a linear approximation for the normalized reflection contrast was used for both the 500–550 and 570–600 nm wavelength ranges using specific values for the data shown as given in the Supporting Information (Supplementary Methods).
The reflection spectra of a plasmonic crystal (Figure 1g) and a flat Au substrate (Figure S7) in artificial sea water with increasing θ were calculated using the FDTD computational method, simulations that theoretically affirm the optical responses seen experimentally. The FDTD calculated reflectance of the flat Au surface decreases over the wavelength range of 400–1000 nm as θ increases, whereas the reflection spectra of plasmonic crystal under the same conditions shows a more complex and wavelength-dependent optical response. For example, wavelengths ranging between 600–740 nm exhibit a decrease in reflectance with thicker θ while the opposite trend was observed in the 770–800 nm range. These trends support the experimentally observed, wavelength-dependent, contrast inversion of the reflection images shown in Figure 1f. Using the FDTD calculations, we also estimated the z (surface-normal) dependence of the sensing volume of the plasmonic optics system. The results presented in Figure 1g show that the quantitatively measurable range for thin films supported on the plasmonic crystal lies in the region extending to ~80 nm from the surface.
3.4. Reflection Imaging of Live Aplysia Pedal Ganglion Neurons Cultured on Plasmonic Crystals
To explore the quantitative analytical capabilities of plasmonic crystal-based live-cell imaging, isolated Aplysia pedal ganglion neurons were cultured on the surface of a device immersed in artificial sea water and examined using an optical microscope. Figures 2a and 2b show reflection images taken in the outgrowth region of a representative live Aplysia neuron, as acquired using different bandpass filters (full images are shown in Figure S8). The image shown in Figure 2a was collected using a 500–550 nm bandpass filter, and that in Figure 2b using a 570–600 nm bandpass filter. The peripheral regions of the live neurons appear darker than the surrounding area in the 500–550 nm image (Figure 2a) while those in the 570–600 nm image (Figure 2b) appear brighter. The contrast inversion seen here can be used to develop a more quantitative analysis of features appearing in the outgrowth region using appropriate reference data (Figure 1f) as is discussed in the sections below.
Figure 2.
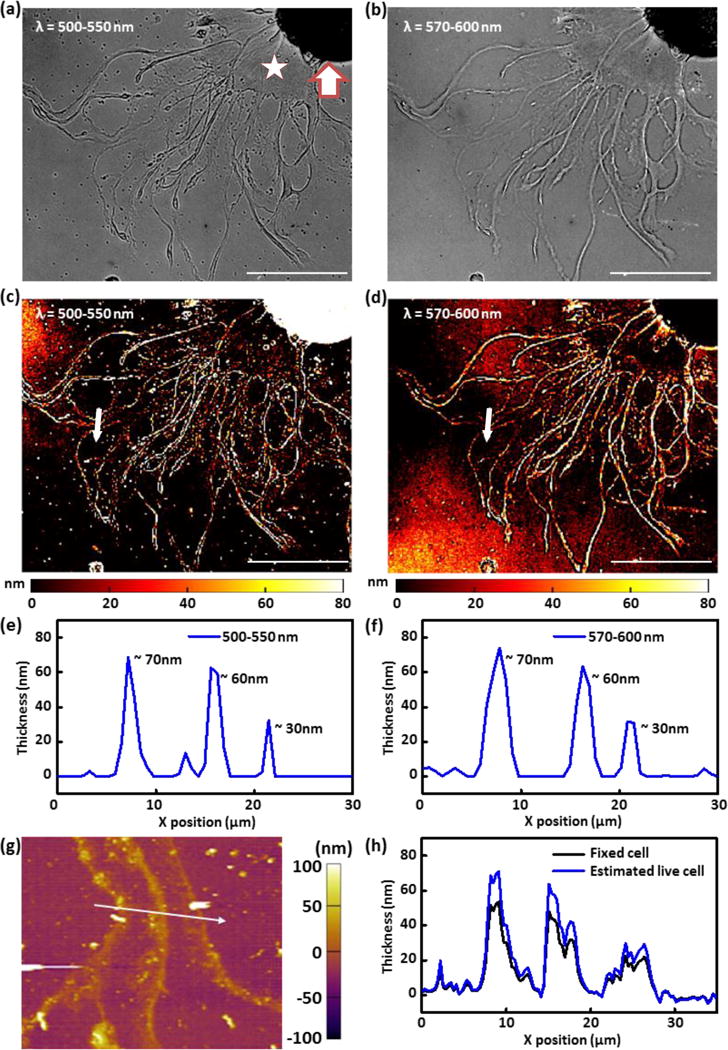
(a, b) Reflection images of a live Aplysia neuron cultured on a plasmonic crystal surface acquired using (a) 500–550 nm bandpass filter and (b) 570–600 nm bandpass filter. (c, d) Mass coverage transformed images generated by applying normalized reflection contrast calibrations to images of a live Aplysia neuron cultured on a plasmonic crystal acquired using (c) 500–550 nm bandpass filter and (d) 570–600 nm bandpass filter. The z-scale for the image is restricted to the plasmonic sensing volume (0–80 nm). (e, f) Height profiles (along the white arrow) of the quantitative reflection image acquired using (e) 500–550 nm bandpass filter and (f) 570–600 nm bandpass filter. (g) AFM image of a fixed Aplysia neuron after FFT filtering to remove plasmonic nanostructure contribution. The imaged neuronal cell is identical with the one presented in Figure 2a–d. (h) AFM height profile (fixed cell) along the white arrow drawn on Figure 2g and estimated thickness of the live cell by consideration of volume changes resulting from water loss during sample fixation and drying. The scale bars in (a–d) correspond to 100 μm. Thick arrow marks cell body. Star is placed on representative examined thin region near the cell body.
Although Aplysia neurons have many structural features that range in thickness from tens of nanometers to tens of microns, only the peripheral area is targeted for quantitative imaging using this SPR-based technique, given the 80 nm limit to the plasmonic sensitivity (while many optical effects are seen throughout these images, our current analytical models do not allow their assessment in more rigorous form). Therefore, we focus here only on thin features with dimensions within the 80 nm limit using a linear regression of optical response as a function of θ observed in the 500–550 nm and 570–600 nm wavelength ranges as a basis for analysis.
The linear contrast calibration data presented in Figure 1f was applied to the reflection images shown in Figures 2a and 2b to analyze the thickness of subregions of live Aplysia pedal neurons. The normalized reflection contrast was calculated by mathematically dividing each pixel in the cell image by the average pixel value of the reference area, as described in the Supplementary methods section. The reference area was selected to be adjacent to the cell, but where cell features were absent. Even though the “empty” reference regions in the image lacked cell features, they do present a thin film of poly-L-lysine, which was applied to the surface prior to cell culture to improve the biocompatibility of the plasmonic crystal and help neuronal cells attach and grow on the surface of the substrate. The contributions of this uniform layer on the reflection contrast can be neglected during normalization for this reason. Figures 2c and 2d show the resultant thickness-transformed cell images using the 500–550 nm and 570–600 nm bandpass filters, respectively. In the latter images, the calculated thicknesses of cellular features were restricted to a range of 0 – 80 nm due to the limitation of sensing volume of SPR-based imaging. Pixels with values corresponding to cell thicknesses outside of this range were truncated and assigned to minimum and maximum contrast values. The cell body was not analyzed due to its overall thickness exceeding that of the plasmonic sensing volume and contrast changes as are seen result as a convolution of various optical phenomena (such as light absorption, reflection, and scattering by cellular structures, interference effects, as well as SPR). In their total, these effects cause the thicker regions of the soma to appear dark in both of the images shown in Figures 2a and 2b. The thinner outgrowths (terminals) and growth cones do show contrast sensitivities whose form depends on the wavelength ranges used to collect the image. From the thickness-transformed images obtained using both wavelength regions (Figures 2c and 2d), the thickness of thin features of live Aplysia pedal neurons can be roughly identified; the thickness of thin regions near the cell body is ~10–20 nm and that of string-like processes is ~60 nm or more.
The self-consistency of measured thicknesses determined using different bandpass filters (Figures 2c and 2d) illustrates the inter-reliability of this thickness evaluating protocol. Figures 2e and 2f show almost identical thickness profiles along the white arrows on the cell images in Figure 2c (using a 500–550 nm bandpass filter) and Figure 2d (using a 570–600 nm bandpass filter). The thickness values of the filament features located along the white arrows are in good agreement (Figures 2e and f): thickness of ~70 nm, ~60 nm, and ~30 nm at X ~8 μm, ~16 μm, and ~20 μm, respectively. The correspondence of the thickness data becomes poorer with very small features: for example, the small feature seen at X ~ 13 μm in the 500–550 nm image (Figure 2e), but not in the 570–600 nm image (Figure 2f). The discrepancies in those two images likely originate from the relatively coarse linear regressions used in the reference contrast calculations. We note that, as shown in previous work49, a combination of multispectral reflection images can be used to increase the sensitivity for measurements made of small features.
Atomic force microscopy (AFM) was used to evaluate the reliability of the SPR-based method for quantifying the thickness of small features present in the live neuronal cultures. We found, however, that it was difficult to measure the heights of specific parts of live Aplysia pedal neurons without damaging the cell without fixation. For this reason, the neuronal cells were fixed on the plasmonic crystals for AFM scanning after the SPR imaging experiments. The height of features in a fixed cell should be reduced compared to that in a live cell due to dehydration (about 70% of the cell volume is composed of water) and changes in neuronal cytoskeleton. This suggests that fixation should lead to a ~33% decrease in cell height. The FFT filtered (to remove the periodic modulation of the substrate) AFM image of the fixed neuronal cell measured in the region along the white arrow in the live-cell reflection images (Figures 2c and 2d) is presented in Figure 2g. The measured height profile of the fixed cell along the white arrow in the AFM image (Figure 2g) is shown in Figure 2h; the height profile estimated from these data for a live cell (adjusted for water loss, Figure 2h) reveals good agreement with height profiles calculated from the reflection images presented in Figures 2e and 2f. These findings are in good agreement with our previously reported results on fixed-cell imaging.49
3.5. Plasmonic Imaging Analysis of the Effect of Trypsin Treatment on Cell Position
Trypsin, a serine protease with high specificity for cleavage of peptide chains at the carboxyl- sides of lysine and arginine residues, is widely used to detach adherent cells from supporting substrate surfaces (Figure S9).68–70 Here we examined the process of cell detachment as a model for testing the quantitative capabilities of this analytical platform for characterizing dynamic transformations of live cells.
Figure S10 shows the changes in the reflection contrasts that result due to the supplementation of the culturing media with trypsin. We utilized the 570–600 nm bandpass filter to collect these data because of the benefits for sensitivity of detection that result from its steep and linear response. Key features of the acquired temporal data, analyzed as mass coverage transformed images, are presented in Figure 3. Surprisingly, distinguishable changes in θ are not observed during the first ~30 min following the addition of trypsin to the culture media, indicating a large number of molecules involved in the cell attachment to the surface and/or restricted access of the enzyme molecules to these molecular anchors and/or rigidity of the cell structure which changes only at some areas after detachment. Progressive changes in the reflection contrasts are noted only after a 30 min incubation. For example, an outgrowth filament readily distinguishable in the t = 0 min image (labeled ‘i’ in Figures 3a and 3b) is not observed in images acquired 50 min after applying trypsin. This exemplifies changes seen in the contrast profiles of many other outgrowth filaments (e.g. labeled ‘ii’ and ‘iii’ in Figures 3a and 3b), where changes from white-yellow to yellow-red denote changes in index as might result due to a progressive form of cell detachment in these regions.
Figure 3.
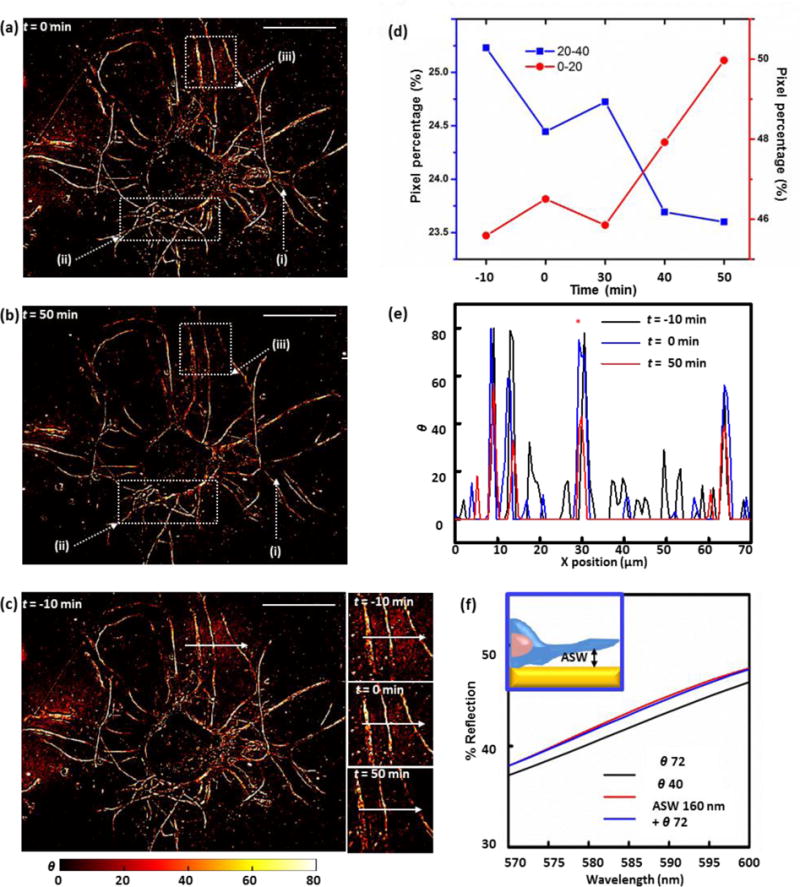
(a, b) Mass coverage transformed images of the live cultured Aplysia neuron acquired for time-lapse analysis of trypsin-induced cell detachment at t=0 (a) and t=50 min (b) after trypsin addition. Dotted-line boxes mark outgrowth filaments with distinguishable changes in contrast profiles over time. The scale bars correspond to 100 μm. (c) Mass coverage transformed images of a live Aplysia neuron acquired 10 min before the addition of trypsin. Insets: neuron extensions of focus during the time-lapse detachment analysis 10 min before trypsin addition, at the time of trypsin addition, and 50 min after the trypsin addition. Scale bar: 100 μm. (d) Pixel percentage distributions across two representative ranges of θ values of mass coverage transformed images of the live Aplysia neuron acquired during time-lapse analysis of cell detachment induced by trypsinization, showing an inversion in the trend of pixel percentage change with time. (e) Line profiles of index corrected mass coverage along the white arrows in Figure 3a at t= −10 min (black line), t= 0 min (blue line), and t=60 min (red line). (f) Comparison between three FDTD simulated reflection spectra acquired in the 570–600 nm range: plasmonic crystals covered with 72 of index corrected mass coverage (black line), 160 nm artificial sea water (ASW) gap between plasmonic crystal and 72 of index corrected mass coverage (blue line), and 40 of index corrected mass coverage, within the optimal 570–600 nm range (red line).
We find that the effects of the dynamic environment on the measured contrast changes (which reflect essentially near surface contact regions of the cell-plasmonic crystal interface) are generally small as compared to those induced by the trypsin treatment, as seen in an exemplary comparison of reflection live-cell images of an outgrowth region obtained 10 min before (t = −10 min, Fig. 3c, inset) and right at the point of trypsin application (t = 0 min, Fig. 3c, inset). After the addition of trypsin, changes progressively increasing in intensity are observed, ones denoting the likely increase in the thickness of artificial sea water layer separating the cell membrane and the substrate surface (t = 50 min, Figure 3c, inset). This analysis can be extended to a full image format, following temporal changes on a pixel-by-pixel basis. Figure S11 shows the results of this analysis in terms of distributions measured across different ranges of index-corrected mass coverage in time-lapse live-cell images. These data demonstrate that the number of pixels in the full frame image having higher index corrected mass coverage values (θ = 40–80) decreases markedly after the trypsin addition, while those with lower values (θ = 0–20) increase in number with time. Figure 3d shows the time-lapse profile of two representative index corrected mass coverage categories (θ = 0–20 and θ = 20–40) that illustrate the inversion of this trend of decrease in reflection contrast as the apparent artificial sea water content present at the cell/plasmonic crystal interface increases. The pixel percentage corresponding to the lower index corrected mass coverage range (θ = 0–20) increases with time after trypsin application, whereas that corresponding to θ = 20–40 decreases. Comparisons made between two exemplary mass-coverage transformed images of a live Aplysia pedal neuron (t = 0 min, Figure 3c, inset; t = 50 min, Figure 3c, inset) show the expected trends in reflection contrast. Figure 3e shows the quantitative changes in mass coverage profiles measured in a specific line scan along the white arrows in the cell images due to trypsinization (Figure 3c, insets). The smaller θ profile values obtained from the t = 50 min reflection image validate the progressive increase in thickness of a low index (presumably artificial sea water) material layer between the cell and the plasmonic crystal, suggesting that cell detachment can be monitored in real time using 2D reflection images of this type.
Additional FDTD calculations were performed to quantitatively investigate the nature of the low index layer formed between the cell and the plasmonic crystal during cell detachment. In Figure 3e, the θ value of the outgrowth filament (indicated with a red asterisk) is reduced from ~70 to ~40 as a result of trypsin treatment for 50 min. This suggests a limiting thickness for an artificial sea water layer present at this interface. It is known that average cell-substrate distances at the peripheral region of the growth cone are of the order of 10–30 nm, with the cell membrane of other regions of the cell separated from the substrate by 100–150 nm.57 The FDTD simulations (Figure 3f and Figure S12a) suggest an interphase having a gap of artificial sea water as large as 160 nm between itself and the initial 72 nm of index corrected contacting process material has formed in this region after 50 minutes (i.e. a 160 nm artificial sea water gap + a 72 θ process contact yields a contrast matching the experimentally measured 40 θ value). We caution that these compositional attributes should not be construed as being stiffly determined by these analyses, rather that they illustrate a likely general trend of artificial sea water layer thickness chance during the detachment of the cell process from the surface. While promising, additional work is needed to firmly establish the quantitative attributes of the imaging as an analytical technique through which to explore cell-substrate dynamics.
CONCLUSIONS
We describe a plasmonic imaging technique that is capable of quantitatively characterizing different attributes of the morphologies and dynamics of live cells and cell-substrate contacts, using it to investigate cultured pedal ganglion neurons of Aplysia californica. Polyelectrolyte films deposited via layer-by-layer assembly provide a reference system to quantitatively evaluate the reflection contrast changes evidence as a function of the thickness of a contacting thin film layer. FDTD simulations extend these calibrations to support imaging-mode analyses of biological specimens, explored explicitly in this work in quantitative analyses made of thin outgrowth regions of live cells and the supporting interactions occurring between them and their substrate. Trypsin-mediated cell detachment was further visualized and quantified using label-free plasmonic reflection imaging without negatively affecting cell viability. The data suggest that reflection imaging techniques of this type represent a potentially useful analytical method for studying the morphologies and dynamics of live ultrathin biological structures at interfaces, providing a reliable, user-friendly, fast, and cost-effective alternative to other live imaging techniques with similar quantitative capabilities.
Supplementary Material
Acknowledgments
The authors thank Xiying Wang for preparing the Aplysia cell cultures. This work was supported by the U.S. Department of Energy under prime Award No. DE-SC0001293 via subcontract 67N-1087758 from California Institute of Technology as part of the DOE Energy Frontier Research Center on Light-Material Interactions in Energy Conversion and was performed using resources at the Frederick Seitz Materials Research Laboratory Central Facilities at the University of Illinois, including the Center for Microanalysis of Materials; the Micro/Nanofabrication Facility; and the Laser and Spectroscopy Facility. SRR and JVS were partially supported by the National Institute on Drug Abuse by Award No. P30 DA018310. A. californica were partially provided by the National Resource for Aplysia funded by PHS grant P40 OD010952.
Footnotes
Supporting Information
Supplementary methods; Scheme of the cross section of the full 3D plasmonic crystal; Experimental transmission spectra and FDTD calculated transmission spectra of full 3D plasmonic crystal; Schemes of polyelectrolyte film pattering process using permanent marker; Ellipsometric spectra of Au-coated silicon pieces to verify no effect of acetone on quality of polyelectrolyte film; 3D AFM image of the step edge of a polyelectrolyte pattern; Digital picture of a home-built liquid cell for ellipsometry; Comparison of FDTD calculated reflection spectra for pairs of optically equivalent polyelectrolyte and index-corrected material; Comparison of FDTD calculated reflection spectra of plasmonic crystals with/without artificial sea water gap between cell and substrate; FDTD calculated reflection spectra for varying thicknesses of index-corrected material on flat Au film; Raw reflection image of live Aplysia neuronal cells; Mass coverage transformed images of the live Aplysia neuronal cell acquired for time-lapse analysis of cell detachment induced by trypsinization; Pixel percentage distributions across various ranges of θ values of mass coverage transformed images of the live Aplysia neuron acquired for time-lapse analysis of cell detachment induced by trypsinization; Comparison between the three FDTD simulated reflection spectra in Fig. 3f acquired in the 450–650 nm range.
Notes
The authors declare no competing financial interest.
References
- 1.Clarke PGH. Developmental Cell-Death - Morphological Diversity and Multiple Mechanisms. Anat Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 2.Hogeweg P. Evolving mechanisms of morphogenesis: on the interplay between differential adhesion and cell differentiation. J Theor Biol. 2000;203:317–333. doi: 10.1006/jtbi.2000.1087. [DOI] [PubMed] [Google Scholar]
- 3.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Bio. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 4.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 5.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 6.Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Bio. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 7.Conn PM. Imaging and spectroscopic Analysis of Living Cells: Optical and Spectroscopic Techniques. Method Enzymol. 2012;504:1–506. [Google Scholar]
- 8.Jaiswal JK, Goldman ER, Mattoussi H, Simon SM. Use of quantum dots for live cell imaging. Nat Methods. 2004;1:73–78. doi: 10.1038/nmeth1004-73. [DOI] [PubMed] [Google Scholar]
- 9.Stephens DJ, Allan VJ. Light microscopy techniques for live cell Imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 10.Marquet P, Depeursinge C, Magistretti PJ. Exploring Neural Cell Dynamics with Digital Holographic Microscopy. Annu Rev Biomed Eng. 2013;15:407–431. doi: 10.1146/annurev-bioeng-071812-152356. [DOI] [PubMed] [Google Scholar]
- 11.Kimata Y, Higaki T, Kawashima T, Kurihara D, Sato Y, Yamada T, Hasezawa S, Berger F, Higashiyama T, Ueda M. Cytoskeleton dynamics control the first asymmetric cell division in Arabidopsis zygote. P Natl Acad Sci. 2016;113:14157–14162. doi: 10.1073/pnas.1613979113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees MD, Thomas SR. Using Cell-substrate Impedance and Live Cell Imaging to Measure Real-time Changes in Cellular Adhesion and De-adhesion Induced by Matrix Modification. J Vis Exp. 2015;96:e52423. doi: 10.3791/52423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung YJ, Choi W, Fang-Yen C, Badizadegan K, Dasari RR, Feld MS. Optical diffraction tomography for high resolution live cell imaging. Opt Express. 2009;17:266–277. doi: 10.1364/oe.17.000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquet P, Rappaz B, Magistretti PJ, Cuche E, Emery Y, Colomb T, Depeursinge C. Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt Lett. 2005;30:468–470. doi: 10.1364/ol.30.000468. [DOI] [PubMed] [Google Scholar]
- 15.Downes A. Raman Microscopy and Associated techniques for label-free imaging of cancer tissue. Appl Spectrosc Rev. 2015;50:641–653. doi: 10.1080/05704928.2015.1052817. [DOI] [Google Scholar]
- 16.Frigault MM, Lacoste J, Swift JL, Brown CM. Live-cell microscopy - tips and tools. J Cell Sci. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- 17.Valiahdi SM, Egger AE, Miklos W, Jungwirth U, Meelich K, Nock P, Berger W, Hartinger CG, Galanski M, Jakupec MA, Keppler BK. Influence of extracellular pH on the cytotoxicity, cellular accumulation, and DNA interaction of novel pH-sensitive 2-aminoalcoholatoplatinum(II) complexes. J Biol Inorg Chem. 2013;18:249–260. doi: 10.1007/s00775-012-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busa WB. Mechanisms and Consequences of Ph-Mediated Cell Regulation. Annu Rev Physiol. 1986;48:389–402. doi: 10.1146/annurev.ph.48.030186.002133. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths L, Dachs GU, Bicknell R, Harris AL, Stratford IJ. The influence of oxygen tension and pH on the expression of platelet-derived endothelial cell growth factor thymidine phosphorylase in human breast tumor cells grown in vitro and in vivo. Cancer Res. 1997;57:570–572. [PubMed] [Google Scholar]
- 20.Davidson JF, Whyte B, Bissinger PH, Schiestl RH. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. P Natl Acad Sci USA. 1996;93:5116–5121. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasantes-Morales H, Lezama RA, Ramos-Mandujano G, Tuz KL. Mechanisms of cell volume regulation in hypo-osmolality. Am J Med. 2006;119:S4–S11. doi: 10.1016/j.amjmed.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 26.Ovecka M, Vaskebova L, Komis G, Luptovciak I, Smertenko A, Samaj J. Preparation of plants for developmental and cellular imaging by light-sheet microscopy. Nat Protocols. 2015;10:1234–1247. doi: 10.1038/nprot.2015.081. [DOI] [PubMed] [Google Scholar]
- 27.Hoebe RA, Van Oven CH, Gadella TWJ, Dhonukshe PB, Van Noorden CJF, Manders EMM. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat Biotechnol. 2007;25:249–253. doi: 10.1038/nbt1278. [DOI] [PubMed] [Google Scholar]
- 28.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 29.Barty A, Nugent KA, Paganin D, Roberts A. Quantitative optical phase microscopy. Opt Lett. 1998;23:817–819. doi: 10.1364/ol.23.000817. [DOI] [PubMed] [Google Scholar]
- 30.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 31.Allen RD, Allen NS, Travis JL. Video-Enhanced Contrast, Differential Interference Contrast (Avec-Dic) Microscopy - a New Method Capable of Analyzing Microtubule-Related Motility in the Reticulopodial Network of Allogromia Laticollaris. Cell Motil Cytoskel. 1981;1:291–302. doi: 10.1002/cm.970010303. [DOI] [PubMed] [Google Scholar]
- 32.Dodt HU, Zieglgansberger W. Visualizing Unstained Neurons in Living Brain-Slices by Infrared Dic-Videomicroscopy. Brain Res. 1990;537:333–336. doi: 10.1016/0006-8993(90)90380-t. [DOI] [PubMed] [Google Scholar]
- 29.Shepherd JNH, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG. 3D Microperiodic Hydrogel Scaffolds for Robust Neuronal Cultures. Adv Funct Mater. 2011;21:47–54. doi: 10.1002/adfm.201001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaami T, Otsuki S, Tomosada N, Kosugi Y, Shimizu M, Ishida H. High-speed 1-frame/ms scanning confocal microscope with a microlens and Nipkow disks. Appl Optics. 2002;41:4704–4708. doi: 10.1364/ao.41.004704. [DOI] [PubMed] [Google Scholar]
- 34.Nakano A. Spinning-disk confocal microscopy - A cutting-edge tool for imaging of membrane traffic. Cell Struct Funct. 2002;27:349–355. doi: 10.1247/csf.27.349. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 36.Pawley J. Fundamental Limits in Confocal Microscopy. Handbook of Biological Confocal Microscopy. 1989:15–26. Revised Edition. [Google Scholar]
- 37.Barr VA, Bunnell SC. Interference Reflectance Microscopy. In: Bonifacino SA, editor. Current protocols in cell biology. Wiley; New York: 2009. Chapter:Unit-4.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Millet L, Mir M, Ding HF, Unarunotai S, Rogers J, Gillette MU, Popescu G. Spatial light interference microscopy (SLIM) Opt Express. 2011;19:1016–1026. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotsch C, Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: An atomic force microscopy study. Biophys J. 2000;78:520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys J. 1996;70:556–567. doi: 10.1016/S0006-3495(96)79602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis MW, Russell B. Micromechanical regulation in cardiac myocytes and fibroblasts: implications for tissue remodeling. Pflug Arch Eur J Phy. 2011;462:105–117. doi: 10.1007/s00424-011-0931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radmacher M, Tillmann RW, Fritz M, Gaub HE. From Molecules to Cells - Imaging Soft Samples with the Atomic Force Microscope. Science. 1992;257:1900–1905. doi: 10.1126/science.1411505. [DOI] [PubMed] [Google Scholar]
- 43.Heinisch JJ, Lipke PN, Beaussart A, Chatel SE, Dupres V, Alsteens D, Dufrene YF. Atomic force microscopy - looking at mechanosensors on the cell surface. J Cell Sci. 2012;125:4189–4195. doi: 10.1242/jcs.106005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quidant R. Plasmon Nano-Optics: Designing Novel Nano-Tools for Biology and Medicine. In: Enoch S, Bonod N, editors. Plasmonics. Springer-Verlag; Berlin Heidelberg: 2012. pp. 201–222. (Springer Series in Optical Sciences 167). [Google Scholar]
- 45.Leroy L, Maillart E, Livache T. Biological Applications of Surface Plasmon Resonance Imaging. In: Fritzsche W, Popp J, editors. Optical Nano- and Microsystems for Bioanalytics. Springer-Verlag; Berlin Heidelberg: 2012. pp. 211–226. (Springer Series on Chemical Sensors and Biosensors 10). [Google Scholar]
- 46.Xiao L, Yeung ES. Optical imaging of individual plasmonic nanoparticles in biological samples. Annu Rev Anal Chem. 2014;7:89–111. doi: 10.1146/annurev-anchem-071213-020125. [DOI] [PubMed] [Google Scholar]
- 47.Wong CL, Olivo M. Surface Plasmon Resonance Imaging Sensors: A Review. Plasmonics. 2014;9:809–824. [Google Scholar]
- 48.Lee SE, Lee LP. Biomolecular plasmonics for quantitative biology and nanomedicine. Curr Opin Biotechnol. 2010;21:489–97. doi: 10.1016/j.copbio.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le AP, Kang S, Thompson LB, Rubakhin SS, Sweedler JV, Rogers JA, Nuzzo RG. Quantitative Reflection Imaging of Fixed Aplysia californica Pedal Ganglion Neurons on Nanostructured Plasmonic Crystals. J Phys Chem B. 2013;117:13069–13081. doi: 10.1021/jp402731f. [DOI] [PubMed] [Google Scholar]
- 50.Moroz LL. Aplysia. Current Biology. 2011;21(2):R60–R61. doi: 10.1016/j.cub.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Hla T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127(7):1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart ME, Mack NH, Malyarchuk V, Soares JANT, Lee TW, Gray SK, Nuzzo RG, Rogers JA. Quantitative multispectral biosensing and 1D imaging using quasi-3D plasmonic crystals. P Natl Acad Sci USA. 2006;103:17143–17148. doi: 10.1073/pnas.0606216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao JM, Stewart ME, Maria J, Lee TW, Gray SK, Rogers JA, Nuzzo RG. Seeing molecules by eye: Surface plasmon resonance imaging at visible wavelengths with high spatial resolution and submonolayer sensitivity. Angew Chem Int Edit. 2008;47:5013–5017. doi: 10.1002/anie.200800501. [DOI] [PubMed] [Google Scholar]
- 54.Stewart ME, Anderton CR, Thompson LB, Maria J, Gray SK, Rogers JA, Nuzzo RG. Nanostructured plasmonic sensors. Chem Rev. 2008;108:494–521. doi: 10.1021/cr068126n. [DOI] [PubMed] [Google Scholar]
- 55.Stewart ME, Yao JM, Maria J, Gray SK, Rogers JA, Nuzzo RG. Multispectral Thin Film Biosensing and Quantitative Imaging Using 3D Plasmonic Crystals. Anal Chem. 2009;81:5980–5989. doi: 10.1021/ac900819j. [DOI] [PubMed] [Google Scholar]
- 56.Yao JM, Le AP, Gray SK, Moore JS, Rogers JA, Nuzzo RG. Functional Nanostructured Plasmonic Materials. Adv Mater. 2010;22:1102–1110. doi: 10.1002/adma.200904097. [DOI] [PubMed] [Google Scholar]
- 57.Ye XY, Xie F, Romanova EV, Rubakhin SS, Sweedler JV. Production of Nitric Oxide within the Aplysia californica Nervous System. ACS Chem Neurosci. 2010;1:182–193. doi: 10.1021/cn900016z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang LP, Ota N, Romanova EV, Sweedler JV. A Novel Pyridoxal 5′-Phosphate-dependent Amino Acid Racemase in the Aplysia californica Central Nervous System. J Biol Chem. 2011;286:13765–13774. doi: 10.1074/jbc.M110.178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caruso F, Niikura K, Furlong DN, Okahata Y. Ultrathin multilayer polyelectrolyte films on gold: Construction and thickness determination. Langmuir. 1997;13:3422–3426. [Google Scholar]
- 60.Richert L, Arntz Y, Schaaf P, Voegel JC, Picart C. pH dependent growth of poly(L-lysine)/poly(L-glutamic) acid multilayer films and their cell adhesion properties. Surf Sci. 2004;570:13–29. [Google Scholar]
- 61.Jonasz M, Fournier GR. Light scattering by particles in water: Theoretical and experimental constraints. Academic Press; San Diego, CA: 2007. p. 714. [Google Scholar]
- 62.Voros J. The density and refractive index of adsorbing protein layers. Biophys J. 2004;87(1):553–561. doi: 10.1529/biophysj.103.030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benesch J, Askendal A, Tengvall P. The determination of thickness and surface mass density of mesothick immunoprecipitate layers by null ellipsometry and protein 125Iodine labeling. J Colloid Interface Sci. 2002;249(1):84–90. doi: 10.1006/jcis.2002.8247. [DOI] [PubMed] [Google Scholar]
- 64.Curtis H, Barnes NS. Biology. 5th. W. H. Freeman; 1989. [Google Scholar]
- 65.Hine R. The Facts on File Dictionary of Biology. 3rd. Checkmark Books; 1999. [Google Scholar]
- 66.Giebel KF, Bechinger C, Herminghaus S, Riedel M, Leiderer P, Weiland U, Bastmeyer M. Imaging of cell/substrate contacts of living cells with surface plasmon resonance microscopy. Biophys J. 1999;76:509–516. doi: 10.1016/s0006-3495(99)77219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang D, Tamburo R, Stetten G. Cumulative Gaussian Curve Fitter for Boundary Parameterization. Insight Journal, Special Issue on MICCAI Workshop on Open-Source Software. 2005 [Google Scholar]
- 68.Oyama Y, Hori N, Allen CN, Carpenter DO. Influences of Trypsin and Collagenase on Acetylcholine Responses of Physically Isolated Single Neurons of Aplysia-Californica. Cell Mol Neurobiol. 1990;10(2):193–205. doi: 10.1007/BF00734573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao B, Li L, Wang L, Wang CY, Yu JD, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Gene Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasparakis G, Manouras T, Selimis A, Vamvakaki M, Argitis P. Laser-Induced Cell Detachment and Patterning with Photodegradable Polymer Substrates. Angew Chem Int Edit. 2011;50:4142–4145. doi: 10.1002/anie.201007310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


