Supplemental Digital Content is available in the text.
Keywords: meta-analysis, nuclear casein kinase and cyclin-dependent kinase substrate 1, Parkinson’s disease, polymorphism, Ras-related protein Rab29, solute carrier family 41 member 1
Abstract
The PARK16 locus is considered to play a protective role in Parkinson’s disease (PD). However, the epidemiological evidence on the relationships between PARK16 single-nucleotide polymorphisms (rs823128, rs1572931, and rs823156) and PD is inconsistent. Therefore, we carried out a meta-analysis to validate the relationships and performed a bioinformatic analysis to explore putative regulation mechanisms of the single-nucleotide polymorphisms in PD. Through meta-analysis, we confirmed that minor variants of rs823128A>G, rs1572931C>T, and rs823156A>G played protective roles in PD. Through bioinformatic analysis, we predicted that rs823128, rs1572931, and rs823156 as noncoding variants of NUCKS1, RAB29, and SLC41A1, respectively, might affect PD risk by altering the transcription factor-binding capability of the genes. These findings suggest new clues for PD research and potential targets for PD prevention and treatment.
Introduction
Parkinson’s disease (PD) is a common and complex neurodegenerative disorder, which is believed to be caused by the interaction of multiple genes and environmental factors. The disease affects about 1% individuals over 60 years old and the quality of life of patients with this disease is severely affected 1. However, because of the limited knowledge of the molecular mechanism in PD, effective preventive or curative strategies for the disease are still absent to date.
The etiology of most PD cases is still vague, but increasing evidence shows an important role of genetic susceptibility in PD. Therefore, studies of the relationship between genetic polymorphisms and PD susceptibility may help to elucidate the pathogenesis of the disease.
The PARK16 locus is the genetic region spanning five genes on chromosome 1. The genes are solute carrier family 45 member 3 (SLC45A3), nuclear casein kinase and cyclin-dependent kinase substrate 1 (NUCKS1), Ras-related protein Rab29 (RAB29), solute carrier family 41 member 1 (SLC41A1), and peptidase M20 domain containing 1 (PM20D1). In recent years, the PARK16 has been identified to play a protective role in PD 2,3. Rs823128, rs1572931, and rs823156 were believed to be among the most PD-associated single-nucleotide polymorphisms (SNPs) in this locus. These SNPs were identified in NUCKS1, RAB29, and SLC41A1, respectively 3. The association between rs823128 SNP and PD was identified in the White population 2, but showed no relationship in some studies in the East Asian population 4,5. Rs1572931 SNP was indicated to be associated with PD in the East Asian 6 and the Mediterranean population 7, but not in the White population 2. As for rs823156, the SNP located in SLC41A1 was considered to be associated with PD in the White population 2, but not in the Hispanic population 8. Considering the inconsistent conclusions of studies in association between these SNPs and PD, we decided to carry out a comprehensive review and meta-analysis here to further validate the association of the SNPs with PD risk. Besides these, the findings of our study might provide new insights into the pathogenesis of PD.
Methods
Publication search
Relevant literatures were searched in PubMed, Embase, the Cochrane Library, the Chinese National Knowledge Infrastructure, the China Science and Technology Journal Database (VIP), and the Wanfang database up to 4 February 2017. Inclusion and exclusion criteria were then used to screen appropriate studies for analysis. The inclusion criteria were as follows: (a) case–control studies only; (b) included an association evaluation between SNPs rs823128, rs823156 or rs1572931, and PD susceptibility; and (c) included allele or genotype frequencies for the calculation of odds ratios (ORs) and 95% confidence intervals. Articles were excluded if (a) they were case reports, reviews, or meta-analyses, and (b) they lacked the data necessary for a meta-analysis.
Statistical analysis
ORs with 95% confidence intervals were calculated to evaluate the associations between SNPs and PD susceptibility under additive models and recessive models; a P value of less than 0.05 was considered statistically significant. Heterogeneity between articles was examined by the I2 index, a quantity that indicated the consistency of data from trials 9. Fixed-effects models were used when heterogeneity across studies was low (I2<50% in meta-analysis); otherwise, random-effects models were applied. Agreement or disagreement of genotype frequencies with Hardy–Weinberg equilibrium in each study was analyzed. Publication bias was evaluated using Egger’s test and Begg’s test, with a P value of more than 0.10 considered evidence for no potential publication bias. Meta-analysis was carried out using Review Manager 5.3 and publication bias was evaluated using Stata 14 software (Stata Corporation, College Station, Texas, USA).
In silico analysis for the putative transcription factor-binding sites affected by single-nucleotide polymorphisms
Online software Gene-Regulation (http://www.gene-regulation.com) was performed to predict the possible effects of the SNPs on putative alteration of transcription factor-binding sites in the relevant genes. Parameters used for the predictions were human matrices only, with a threshold score of 75.0 points (a maximum 100.0).
Results
Study characteristics
A total of 20 eligible studies were included in this meta-analysis. The characteristics of the studies are summarized in Table 1. Among these articles, 14 articles referred to SNP rs823128, 5 referred to rs1572931, and 12 articles referred to rs823156.
Table 1.
Characteristics of the studies included in the meta-analysis

Quantitative synthesis
ORs of rs823128, rs1572931, and rs823156 in PD were evaluated; the results are shown in Table 2 and Supplementary Fig. 1 (Supplemental digital content 1, http://links.lww.com/WNR/A428). In the overall pooled analysis, PD patients showed significantly lower frequencies of the G allele and the GG genotype than control participants in SNP rs823128. The frequencies of the rs1572931T allele and the TT genotype tended to be lower in PD patients and PD patients showed rarer frequencies in the rs823156 G allele and the GG genotype compared with the controls.
Table 2.
Associations of rs823128, rs1572931, and rs823156 single-nucleotide polymorphisms with Parkinson’s disease risk in meta-analysis
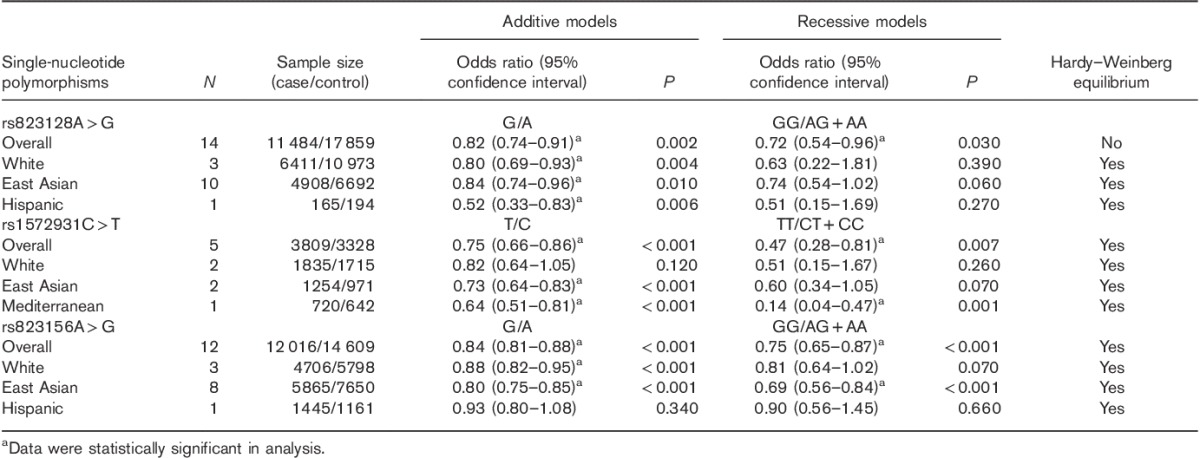
Taking the ethnic variety of association between these SNPs and PD into consideration, we carried out a subgroup analysis determined by sample ethnicity. The results indicated that, for rs823128, PD patients presented lower frequencies of the G allele in the White, East Asian, and Hispanic populations. For rs1572931, PD patients showed a significantly lower frequency of the T allele than the controls in the East Asian population, and showed the same trend of the presence of the rs1572931 T allele and the TT genotype in the Mediterranean population. However, the relationship was not identified between rs1572931 and PD risk in White patients. As for rs823156 SNP, PD patients tended to have lower G allele frequency than control participants in the White population and to have lower frequencies of the G allele and the GG genotype in the East Asian population, but it showed no link between rs823156 SNP and PD risk in the Hispanic population.
In addition, genotype frequencies of rs823128 in controls disagreed with Hardy–Weinberg equilibrium in the overall pooled analysis; thus, the result of this polymorphism should be interpreted with caution.
Publication bias
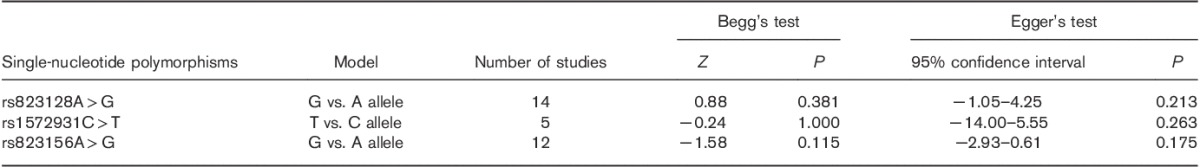
Additive models were used as representatives to be performed. As shown in Table 3 and Supplementary Fig. 2 (Supplemental digital content 2, http://links.lww.com/WNR/A429), there was no publication bias in this study.
Table 3.
Results of Egger’s and Begg’s tests for publication bias (additive models)
In-silico analysis
Through bioinformatic analysis using online software Gene-Regulation, we predicted the modified transcription factor-binding sites caused by rs823128, rs1572931, and rs823156. As shown in Fig. 1, in silico analysis, change at rs823128 was predicted to add a binding site for HOXA3 transcription factor (score, 87.5) and eliminated the sites for TSC2 (score, 100.0) and TOPORS (score, 100.0). Change at rs1572931, the transcription factor-binding sites for CTCF (score, 100.0), and estrogen receptor-β (score, 100.0) were predicted to be added and the site for PGR (score, 100.0) was predicted to be eliminated. As for change at rs823156, the SNP was identified to add putative the transcription factor-binding sites for NP-4 (score, 100.0) and RARA (score, 100.0) transcription factors and to eliminate the binding site for the NFASC (score, 100.0) transcription factor.
Fig. 1.
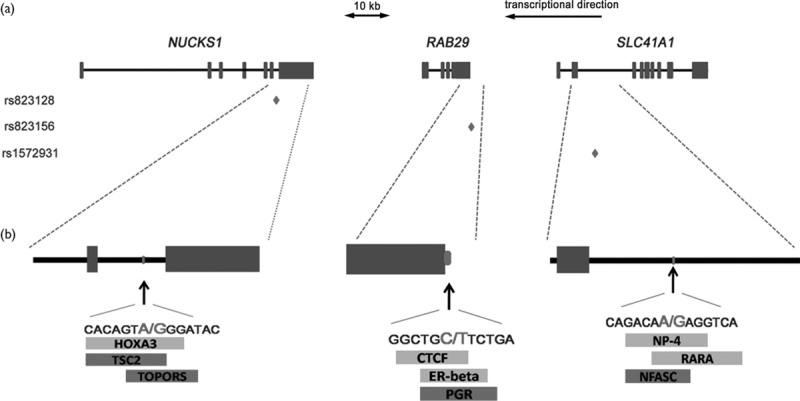
The putative effects of rs823128, rs1572931, and rs823156 on transcription factor-binding sites. (a) Scheme of the NUCKS1/RAB29/SLC41A1 (between 205 712 819 and 205 813 759 according to genome assembly GCRh37) showing the structure of NUCKS1, RAB29, and SLC41A1 genes (exons are in blue rectangles). The three diamonds represent the locations of the three SNPs analyzed in this study. (b) Enlargement of the partial regions of NUCKS1, RAB29, and SLC41A1 the SNPs is located, is conducted. Below, the positions and sequences of rs823128, rs1572931, and rs823156 are presented. The rectangles represent the length of the transcription factor-binding sites. The predicted changes associated with the different alleles are highlighted: additional transcription factor-binding sites are in green and eliminated transcription factor-binding sites are in red. NUCKS1, nuclear casein kinase and cyclin-dependent kinase substrate 1; RAB29, Ras-related protein Rab29; SLC41A1, solute carrier family 41 member 1; SNP, single-nucleotide polymorphism.
Discussion
In this meta-analysis, the results of overall pooled analysis showed that rs823128, rs1572931, and rs823156 SNPs within PARK16 were associated with PD susceptibility. The minor alleles of rs823128A>G, rs1572931C>T and rs823156A>G were associated with a reduced PD risk and polymorphisms of these three SNPs showed ethnicity-specific effects on PD, which were consistent with a previous report 24. The ethnic differences in the relationships between SNPs and PD risk were widely considered to be influenced by environmental factors, such as lifestyles of patients and the extent of pollution in the surroundings 24. However, the detailed mechanisms of these ethnic differences still need to be confirmed by further investigation.
SNPs rs823128, rs1572931, and rs823156 were identified to be located in NUCKS1, RAB29, and SLC41A1, respectively. Therefore, these genes were implicated to play protective roles in PD.
NUCKS1, the gene rs823128 SNP locates in, is a housekeeping gene expressed in various types of cells. It is a vertebrate-specific gene. Its coding protein, NUCKS1, is a chromatin-associated protein with a role in DNA damage response and homologous recombination 25. It is responsible for repairing DNA and maintaining chromosome stability, and dysfunction of the protein may lead to increasing cellular sensitivity to the harmful substance, such as reactive oxygen species (ROS) 26. Although accumulating evidence provided suggestive support that SNP rs1572931 in NUCKS1 was associated with PD risk, its molecular mechanism is still obscure to date. However, the interaction between ROS-damaged DNA and impairment of DNA repair capability in neurons was found to be an important causative factor of PD, suggesting that capability of DNA repair regulated by NUCKS1 played a critical function in PD prevention 27.
RAB29, where rs1572931 locates, is considered to exert protective effects on PD. Its coding protein, as a member of the Ras-related GTP-binding protein subfamily, is ubiquitously expressed in human tissues. The protein has been identified to cooperate with leucine-rich repeat kinase 2 (LKKR2) to reduce human PD risk 28. LKKR2, a confirmed PD-related protein, was identified to possess GTPase activity 29. It could activate the Ras signaling pathway and autophagy in neurons when formed as a protein complex with RAB29. The RAB29-LRRK2 complex, as an activator of the Ras signaling pathway, could promote the clearance of a series of PD-causing factors (such as α-synuclein and ROS) in neurons, thus preventing the development of PD 30,31. In addition, the RAB29-LRRK2 complex was also found to be able to regulate axonal elongation in neurons, contributing toward improving the function of learning and memory in PD patients 29.
SLC41A1, the gene rs823156 SNP locates in, plays a vital role in physiological function. SLC41A1 protein, as a Na+/Mg2+ exchanger in eukaryotes, is responsible for balancing magnesium homeostasis, promoting normal metabolism, and maintaining physiological function in the body 32. SLC41A1 is expressed in numerous tissues, including the kidney and the brain. The SLC41A1 protein in renal epithelial cells of distal convolution plays a crucial role in transcellular Mg2+ reabsorption in the distal convoluted tubule, contributing to magnesium homeostasis in cells, tissues, serum and cerebrospinal fluid. Decreased SLC41A1 expression/activity might decrease the SLC41A1-dependent magnesium recycle of cells, thus causing hypomagnesemia, and decreasing the free intracellular Mg2+ in cells 33. Experimental evidence based on the PD-like dopaminergic cell line PC12 showed that free intracellular Mg2+ protected cells from damage of oxidant stress, and expression deficiency of magnesium transporter protein could significantly attenuate the oxidation resistance, thus increasing the susceptibility of neurodegenerative diseases including PD 34. In all, Mg2+ homeostasis regulated by SLC41A1 may play an important role in PD prevention and treatment.
Taken together, this analysis suggests that, NUCKS1, RAB29, and SLC41A1, the gene of the PARK16 locus, might exert preventive effects on PD; minor variants of these SNPs (rs823128A>G in NUCKS1, rs1572931C>T in RAB29, and rs823156A>G in SLC41A1) were associated with reduced PD risk. Interestingly, all of these SNPs were variants in noncoding regions. To date, there are two known means for noncoding variants to alter the function of relevant genes: (a) to cause the alterative splicing of gene 35 and (b) to alter the binding of transcription factors with genes 36. To our knowledge, there are no data showing alterative splicing in NUCKS1, SLC41A1, or RAB29 caused by SNPs or noncoding variants. Thus, here, our study focused on the putative alteration of transcription factor-binding sites in these genes. Through bioinformatic analysis, we predicted that the up-regulated relevant DNA-binding capability of transcription factors HOXA3, CTCF, estrogen receptor-β, NP-4 and RARA might play protective roles in PD through regulating gene transcription, and the capability block of transcription factors TSC2, TOPORS, PGR and NFASC might have the same effects. However, the exact effects of these transcription factors on the function of the relevant genes and on the pathophysiology of PD are still obscure and need further investigation.
In summary, this study indicated that SNPs of NUCKS1, RAB29, and SLC41A1, located in PARK16, were associated with reduced PD risk. It suggested the potential roles of genes NUCKS1, RAB29, and SLC41A1 in PD, and might reveal any potential targets for PD prevention and treatment. Besides these, the findings indicated ethnicity-specific effects of rs823128, rs1572931, and rs823156 SNPs on PD, which might be useful in future genetic counseling, to assess PD susceptibility for carriers with different ethnicities. For these ethnicity-specific effects of PARK16, although the detailed mechanisms are still unclear, they showed potential population differences in PD susceptibility and PD predisposing factors across ethnicities. However, future well-designed studies, including studies with more clinical and more experimental evidence, are needed to shed more light on these findings. In addition, ethnicity-specific effects of the SNPs on PD susceptibility, even located in the same gene, are still unclear, which should be further explored as well.
Supplementary Material
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.neuroreport.com).
Acknowledgements
This work was supported by the Nature Science Foundation of Fujian Province (13161509).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Wang L, Cheng L, Li NN, Yu WJ, Sun XY, Peng R. Genetic analysis of SLC41A1 in Chinese Parkinson’s disease patients. Am J Med Genet 2015; 168:706–711. [DOI] [PubMed] [Google Scholar]
- 2.Pihlstrom L, Rengmark A, Bjornara KA, Dizdar N, Fardell C, Forsgren L, et al. Fine mapping and resequencing of the PARK16 locus in Parkinson’s disease. J Hum Genet 2015; 60:357–362. [DOI] [PubMed] [Google Scholar]
- 3.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet 2009; 41:1303–1307. [DOI] [PubMed] [Google Scholar]
- 4.Zhao YY, Lin XJ, Liu WG, Ye M, Chen JC, Wang SY, et al. Association between polymorphisms of PARK16 gene and susceptibility to Parkinson’s disease in Chinese Han population. Chinese. J Neurol 2011; 44:343–346. [Google Scholar]
- 5.Zhou LL, Zhang X, Bao QQ, Liu RP, Gong MY, Mao GY, et al. Association analysis of PARK16-18 variants and Parkinson’s disease in a Chinese population. J Clin Neurosci 2014; 21:1029–1032. [DOI] [PubMed] [Google Scholar]
- 6.Guo XY, Chen YP, Song W, Zhao B, Cao B, Wei QQ, et al. An association analysis of the rs1572931 polymorphism of the RAB7L1 gene in Parkinson’s disease, amyotrophic lateral sclerosis and multiple system atrophy in China. Eur J Neurol 2014; 21:1337–1343. [DOI] [PubMed] [Google Scholar]
- 7.Gan-Or Z, Bar-Shira A, Dahary D, Mirelman A, Kedmi M, Gurevich T, et al. Association of sequence alterations in the putative promoter of RAB7L1 with a reduced Parkinson disease risk. Arch Neurol 2012; 69:105–110. [DOI] [PubMed] [Google Scholar]
- 8.Mata IF, Yearout D, Alvarez V, Coto E, de Mena L, Ribacoba R, et al. Replication of MAPT and SNCA, but not PARK16-18, as susceptibility genes for Parkinson’s disease. Mov Disord 2011; 26:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003; 327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang XL, Mao XY, Li HH, Zhang JH, Li NN, Burgunder JM, et al. Association of GWAS loci with PD in China. Am J Med Genet B Neuropsychiatr Genet 2011; 156B:334–339. [DOI] [PubMed] [Google Scholar]
- 11.Chang KH, Chen CM, Chen YC, Lyu RK, Chang HS, Ro LS, et al. Association between PARK16 and Parkinson’s disease in the Han Chinese population: a meta-analysis. Neurobiol Aging 2013; 34:2442 e5-2442.e9. [DOI] [PubMed] [Google Scholar]
- 12.Gopalai AA, Ahmad-Annuar A, Li HH, Zhao Y, Lim SY, Tan AH, et al. PARK16 is associated with PD in the Malaysian population. Am J Med Genet B Neuropsychiatr Genet 2016; 171:839–847. [DOI] [PubMed] [Google Scholar]
- 13.Miyake Y, Tanaka K, Fukushima W, Kiyohara C, Sasaki S, Tsuboi Y, et al. PARK16 polymorphisms, interaction with smoking, and sporadic Parkinson's disease in Japan. J Neurol Sci 2016; 362:47–52. [DOI] [PubMed] [Google Scholar]
- 14.Yan YP, Mo XY, Tian J, Zhao GH, Yin XZ, Jin FY, et al. An association between the PARK16 locus and Parkinson’s disease in a cohort from eastern China. Parkinsonism Relat Disord 2011; 17:737–739. [DOI] [PubMed] [Google Scholar]
- 15.Ramirez A, Ziegler A, Winkler S, Kottwitz J, Giesen R, Diaz-Grez F, et al. Association of Parkinson disease to PARK16 in a Chilean sample. Parkinsonism Relat Disord 2011; 17:70–71. [DOI] [PubMed] [Google Scholar]
- 16.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet 2009; 41:1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer CC, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 5' of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet 2011; 20:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan EK, Kwok HH, Tan LC, Zhao WT, Prakash KM, Au WL, et al. Analysis of GWAS-linked loci in Parkinson disease reaffirms PARK16 as a susceptibility locus. Neurology 2010; 75:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilarino-Guell C, Ross OA, Aasly JO, White LR, Rajput A, Rajput AH, et al. An independent replication of PARK16 in Asian samples. Neurology 2010; 75:2248–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goudarzian M, Khaligh A, Fourozan R, Jamal Mirmoosavi S, Darvish H, Safaralizadeh T, et al. The rs1572931 polymorphism of the RAB7L1 gene promoter is associated with reduced risk of Parkinson's disease. Neurol Res 2015; 37:1029–1031. [DOI] [PubMed] [Google Scholar]
- 21.Liu QS, Zhang WL, Liu SD. [Association between RAB7L1 gene promoter polymorphism and Parkinson disease in Southwestern Chinese Han Population]. Journal of Neuroscience and Mental Health 2016; 16:297–299. [Google Scholar]
- 22.Chung SJ, Chung Y, Hong M, Kim M, You S, Kim YJ, et al. AD and PD GWAS top hits and risk of Parkinson’s disease in Korean population. Movement disorders 2014; 29 (Suppl 1):S48. [Google Scholar]
- 23.Guo JF, Li K, Yu RL, Sun QY, Wang L, Yao LY, et al. Polygenic determinants of Parkinson’s disease in a Chinese population. Neurobiol Aging 2015; 36:e1761–e1766. [Google Scholar]
- 24.Peeraully T, Tan EK. Genetic variants in sporadic Parkinson’s disease: East vs West. Parkinsonism Relat Disord 2012; 18 (Suppl 1):S63–S65. [DOI] [PubMed] [Google Scholar]
- 25.Parplys AC, Zhao W, Sharma N, Groesser T, Liang F, Maranon DG, et al. NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res 2015; 43:9817–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper SJ, Zou H, Legrand SN, Marlow LA, von Roemeling CA, Radisky DC, et al. Loss of type III transforming growth factor-beta receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene 2010; 29:2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross CA, Truant R. DNA repair: a unifying mechanism in neurodegeneration. Nature 2017; 541:34–35. [DOI] [PubMed] [Google Scholar]
- 28.Kuwahara T, Inoue K, D’Agati VD, Fujimoto T, Eguchi T, Saha S, et al. LRRK2 and RAB7L1 coordinately regulate axonal morphology and lysosome integrity in diverse cellular contexts. Sci Rep 2016; 6:29945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci 2010; 11:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrett RM, Alexopoulou Z, Tofaris GK. The endosomal pathway in Parkinson’s disease. Mol Cell Neurosci 2015; 66:21–28. [DOI] [PubMed] [Google Scholar]
- 31.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA 2009; 106:2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandt T, Song Y, Scharenberg AM, Sahni J. SLC41A1 Mg(2+) transport is regulated via Mg(2+)-dependent endosomal recycling through its N-terminal cytoplasmic domain. Biochem J 2011; 439:129–139. [DOI] [PubMed] [Google Scholar]
- 33.Goytain A, Quamme GA. Functional characterization of the mouse [corrected] solute carrier, SLC41A2. Biochem Biophys Res Commun 2005; 330:701–705. [DOI] [PubMed] [Google Scholar]
- 34.Shindo Y, Yamanaka R, Suzuki K, Hotta K, Oka K. Altered expression of Mg(2+) transport proteins during Parkinson’s disease-like dopaminergic cell degeneration in PC12 cells. Biochim Biophys Acta 2016; 1863:1979–1984. [DOI] [PubMed] [Google Scholar]
- 35.Pascale E, Di Battista ME, Rubino A, Purcaro C, Valente M, Fattapposta F, et al. Genetic architecture of MAPT gene region in Parkinson disease subtypes. Front Cell Neurosci 2016; 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu PY, Liang R, Jankovic J, Hunter C, Zeng YX, Ashizawa T, et al. Association of homozygous 7048G7049 variant in the intron six of Nurr1 gene with Parkinson’s disease. Neurology 2002; 58:881–884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.neuroreport.com).


