Abstract
Cytochrome P450 enzymes (P450s) are some of the most exquisite and versatile biocatalysts found in nature. In addition to their well-known roles in steroid biosynthesis and drug metabolism in humans, P450s are key players in natural product biosynthetic pathways. Natural products, the most chemically and structurally diverse small molecules known, require an extensive collection of P450s to accept and functionalize their unique scaffolds. In this review, we survey the current catalytic landscape of P450s within the Streptomyces genus, one of the most prolific producers of natural products, and comprehensively summarize the functionally characterized P450s from Streptomyces. A sequence similarity network of >8500 P450s revealed insights into the sequence–function relationships of these oxygen-dependent metalloenzymes. Although only ~2.4% and <0.4% of streptomycete P450s have been functionally and structurally characterized, respectively, the study of streptomycete P450s involved in the biosynthesis of natural products has revealed their diverse roles in nature, expanded their catalytic repertoire, created structural and mechanistic paradigms, and exposed their potential for biomedical and biotechnological applications. Continued study of these remarkable enzymes will undoubtedly expose their true complement of chemical and biological capabilities.
Graphical abstract
1 Cytochromes P450
Cytochromes P450 (P450s or CYPs) form a superfamily of ubiquitous heme-dependent enzymes that catalyze a diverse array of reactions via a complex multistep mechanism. They are most well known for their roles in xenobiotic detoxification, steroid biosynthesis, and drug metabolism in humans,1,2 but also play key roles in the biosynthesis of natural products.3–5 The heme (iron protoporphyrin IX) prosthetic group, which is coordinated on the proximal side by a thiolate ion, plays a key role in the reductive activation of molecular oxygen, a defining feature of P450 enzymes. P450s (P = pigment) were named as such due to their distinctive spectroscopic absorption maximum at 450 nm when the thiolate–ferrous–CO complex is reduced.6,7 P450s are also commonly known as monooxygenases, due to the insertion of only one of the oxygen atoms from molecular oxygen into the substrate; the other is reduced to water. Correspondingly, hydroxylation of a carbon atom on a hydrophobic organic substrate is considered the archetypal P450 reaction. However, labeling a P450 as “just another hydroxylase” is a gross misrepresentation of their catalytic capabilities. The diversity and versatility of these natural biocatalysts highlights nature’s ability to evolve enzymes for chemical reactions.
P450s are of special interest to a variety of scientific disciplines. These oxygen-dependent metalloenzymes, with their fascinating chemical and physical properties, ability to catalyze C–H activation reactions, and diverse roles in human health, are relevant to pharmacologists and medicinal chemists, enzymologists and biochemists, bioinorganic chemists and biophysicists, natural product chemists, and biomimetic and synthetic chemists. Consequently, P450s have been thoroughly reviewed from a variety of perspectives.3,8–19 In this review, we aim to highlight the sequences, structures, and functions of P450s involved in the natural product biosynthetic pathways found in Streptomyces and how the study of these microbial enzymes has advanced the field of cytochrome P450 chemistry and enzymology.
2 Function
This review is not intended to extensively discuss the catalytic mechanism of P450s; it has been excellently covered elsewhere.9,12,17,20–22 However, a description of the sophisticated P450 catalytic cycle is needed to discuss the ability of P450s to catalyze diverse chemical transformations.
The currently accepted P450 catalytic cycle, shown here as resulting in monooxygenation, is depicted in Fig. 1. In the resting state of the low-spin ferric (FeIII) enzyme, a water molecule coordinates to the heme iron as the sixth ligand. Upon substrate binding, the coordinated water molecule is displaced, resulting in a change of the ferric iron spin state to high-spin. The iron, with its more positive reductive potential, is first reduced to the ferrous (FeII) state, molecular oxygen then binds, followed by a second reduction event resulting in the formation of a peroxo-ferric (FeIII–OO2−) intermediate. Two successive protonations of the distal oxygen, the first yielding the hydroperoxo-ferric intermediate Compound 0 (Cpd 0, FeIII–OOH−) and the second generating a transient intermediate (FeIII–OOH2), is followed by heterolytic fission of the O–O bond to yield the high valent oxo-ferryl (FeIV=O) π-cation porphyrin radical generally referred to as Compound I (Cpd I), while concomitantly releasing a water molecule. Cpd I then abstracts a hydrogen from the substrate forming a substrate radical species, which then, in the prototypical hydroxylation reaction, rebounds with the hydroxyl radical to form the hydroxylated product. Disassociation of the product from the enzyme allows water to return as the sixth heme ligand and finalizes the regeneration of the resting state of the hemoprotein. While Cpd I is accepted as the primary active species in heme oxygenase chemistry,9 other intermediates are also capable of catalyzing oxidative reactions.20,22,23 It should also be mentioned that several of the steps are likely in equilibrium and the rate-limiting step may vary, supporting a more dynamic view of the P450 catalytic cycle.24
Fig. 1.
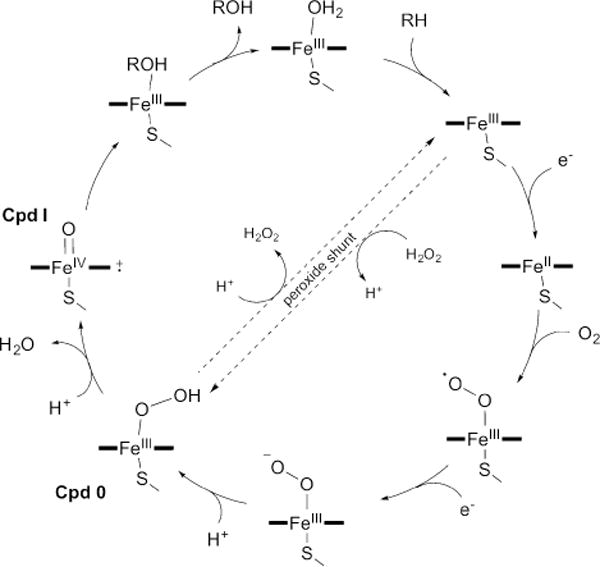
The cytochrome P450 catalytic cycle. The cycle, shown here depicting hydroxylation of the substrate RH to yield the product ROH, is described in the text. The peroxide shunt pathway can directly form Cpd 0 from the substrate-bound high-spin FeIII state using H2O2.
The electrons required for heme-FeIII reduction are generally supplied by NAD(P)H through redox protein partners.10,25 There are two main classes of P450 redox systems: the bacterial and mitochondrial soluble class I and the eukaryotic microsomal membrane-bound class II. Typically, class I consists of a three-component system in which an NAD(P)H-dependent and FAD-containing reductase shuttles electrons to a [2Fe-2S] ferredoxin, which in turn shuttles the electrons to the P450 protein. The membrane-bound class II system is generally a two-component system that utilizes an NAD(P)H-dependent FAD- and FMN-containing reductase for electron supply. While most redox systems fit into either class I or II, there is a growing trend of exceptions, indicating there is still much to be uncovered regarding the diversity of P450 redox partners.10 The most well-known and biotechnological useful examples include the fused multiprotein single-component systems P450BM3 from Bacillus megaterium26,27 and P450RhF from Rhodococcus sp.28 The recent discoveries of the self-sufficient P450 CYP102D1 from Streptomyces avermitilis29 and the redox-independent P450 CYP154A1 from Streptomyces coelicolor30 continue to amend the paradigms of electron transfer systems for P450s.
One significant challenge of the bacterial three-component systems is to identify the native ferredoxin reductase and ferredoxin partners for each P450, and much work has focused on understanding these relationships in Streptomyces.31–35 One advantage of bacterial P450s, and those from Streptomyces in particular, is the inherent flexibility of the P450s to accept electrons from heterologous redox partners. While each P450 behaves differently, streptomycete P450s have been shown to accept redox proteins from other Streptomyces species,36–38 other bacterial genera including putidaredoxin reductase and putidaredoxin from the P450cam system in Pseudomonas putida39,40 and flavodoxin reductase and flavodoxin from Escherichia coli,41,42 and eukaryotes such as the commercially available spinach ferredoxin reductase and ferredoxin.43,44 Inspired by the one-component systems, Streptomyces P450s have also been engineered into redox self-sufficient enzymes by the fusion of the P450 protein to the reductase domains of P450RhF45–47 or CYP102D1.48 Finally, some P450s, are able to utilize H2O2 as a surrogate for the oxygens, electrons, and proton needed to directly generate Cpd 0 from the substrate-bound high-spin ferric P450 (i.e., peroxide shunt or peroxygenation; Fig. 1).9,49,50
2.1 P450s in natural product biosynthesis
Natural products are the most chemically and structurally diverse class of small molecules and consequently possess an extraordinarily wide range of biological activities.51 The complexity of natural products arises from nature’s ability to take simple building blocks and form complex and highly functionalized scaffolds. There are many strategies to biosynthesize natural products, but one common theme is the initial construction of a chemical scaffold followed by a set of reactions to functionalize the (typically) inert skeleton. Whether it be a linear or macrocyclic polyketide or a polycyclic terpenoid carbon skeleton, enzymes that can functionalize unactivated C–H bonds are vital to the production of biologically active natural products. The newly introduced functional groups can act as polar handles for substrate recognition, be used as attachment points for other moieties, or provide the chemical properties necessary for its mode of action or target interaction. Derivatization of the building blocks that nature uses adds another layer of complexity to the biosynthesis of natural products.
P450s are one of the most utilized enzymes that functionalize natural product scaffolds. This becomes anecdotally apparent when one considers two facts: (i) the sheer numbers of P450s found in the most prevalent producers of natural products, including plants, fungi, and the actinomycetes (see online databases http://drnelson.uthsc.edu/cytochromeP450.html and http://p450.riceblast.snu.ac.kr),4,5,31,52,53 and (ii) the frequency of P450-encoding genes found within known and putative secondary metabolite gene clusters. The tremendous diversity in natural product structures requires diversity in the enzymes that act on them, and P450s are no exception. P450s that act in natural product biosynthetic pathways have some of the most diverse sequences, structures, functions, and mechanisms. It is because of these characteristics that natural product-related P450s have been crucial in revealing their diverse roles in nature, expanding their catalytic repertoire, creating structural and mechanistic paradigms, and exposing their potential for biomedical and biotechnological applications.
2.2 P450s in Streptomyces
After the genomes of S. avermitilis, the avermectin industrial strain, and S. coelicolor, the actinorhodin-producing model strain, were sequenced,54–56 it was clear that the Streptomyces genus harbored a significant number of P450s. S. coelicolor and S. avermitilis contained 18 and 33 P450s, respectively, accounting for 0.2% and 0.4% of all coding sequences.31,53 In comparison, many prokaryotes have only a few P450s encoded within their genomes; Escherichia coli and Salmonella typhimurium have none.8,57 Since P450s are usually associated with either secondary metabolism or xenobiotic transformations, the number of P450s in Streptomyces and their diversity may reflect the extraordinary biosynthetic potential of these strains to produce diverse natural products, their ability to detoxify chemicals they come into contact with, or both.
Actinomycetes are generally considered as the workhorse producers of natural products, with Streptomyces being particularly proficient.58 The recent emphasis on bacterial sequencing projects has continued to support these ideas, and it is now believed that we may have missed ~90% of the natural product biosynthetic potential of actinomycetes.58 These sequencing efforts and the subsequent explosion of new sequence data have also assembled an enormous library of orphan enzymes, i.e., enzymes with unknown functions and/or unknown endogenous substrates.11
This review is limited to P450s from the Streptomyces genus for several reasons. Streptomycetes are one of the greatest reservoirs of natural products and the enzymes that catalyse their biosynthesis. Natural variants of enzymes found within natural product biosynthetic pathways have specialized functions that have evolved over millions of years to be catalytically efficient with regio- and stereoselectivities, and secondary metabolism-based P450s possess immense capacity for diverse chemical reactions. Streptomycetes are also traditionally more amenable to genetic manipulations, which allows the elucidation of the physiological functions of enzymes on endogenous substrates. The clustering of biosynthetic-related genes into operons or clusters, a common paradigm in microbial genomes,59 also facilitates the identification of the endogenous natural product the P450 is involved in its biosynthesis. For these reasons, we considered Streptomyces to be an appropriate starting point to discuss the sequence–structure–function aspects of P450s involved in natural product biosyntheses.
There are 184 functionally characterized P450s of streptomycete origin (Table 1, access to the comprehensive P450 Excel spreadsheet is available online at www.scripps.edu/shen/NPLI/database.html). P450s were regarded as “functionally characterized” in this review if they met one or more of the following conditions: (i) the P450-encoding gene was cloned, the P450 protein was produced, and an in vitro experiment was conducted, or alternatively the protein was heterologously produced for biotransformation reactions (i.e., in vitro characterization); (ii) the P450-encoding gene was inactivated in the native Streptomyces strain or was transferred to another host for genetic complementation (i.e., in vivo characterization); or (iii) a crystal structure was solved (i.e., structural characterization). Although there are many additional genes that encode P450s found in genomes and gene clusters or published in annotated gene cluster tables, mere functional prediction, comparison with highly homologous P450s, or having been named with the systematic nomenclature was not sufficient for inclusion in this “functionally characterized” collection. These sequences, however, were included in the P450 sequence similarity network (SSN), as discussed below.
Table 1.
Experimentally characterized P450s of streptomycete origin
| P450a | CYP nameb | Biosynthetic pathway | Function(s) | In vitroc/in vivo/structure | PDB ID(s) | Ref. |
|---|---|---|---|---|---|---|
| AcmG8 | – | Actinomycin G | Hydroxylation (PCP-tethered)d |
–/Y/– | 172 | |
| AknT | – | Aclacinomycin A | Glycosyltransferase (GTase) activator | Y/–/– | 202,203 | |
| AmphL | 161A3 | Amphotericin B | Hydroxylation | –/Y/– | 204 | |
| AmphN | 105H4 | Amphotericin B | Hydroxylation/oxidation to acid | –/Y/– | 178,179 | |
| AryC | – | Arylomycin | Biaryl ring coupling (C–C) | –/Y/– | 205 | |
| AurH | 151Ae | Aureothin | Hydroxylation/ether formation | Y/Y/Y | 3P3L, 3P3O, 3P3X, 3P3Z | 115–118 |
| AveE | 171A1 | Avermectin | Ether formation | –/Y/– | 119 | |
| AziB1 | – | Azinomycin B | Hydroxylation | Y/–/– | 74 | |
| BecO | 1045A3 | BE-14106 | Hydroxylation | –/Y/– | 206 | |
| BorI | – | Borrelidin | Hydroxylation/oxidation to aldehyde/oxidation to nitrile | Y/Y/– | 90 | |
| BoxA | 105Ae | Xenobiotics | Hydroxylation | Y/–/– | 131,207 | |
| CanCf | 105H5 | Candicidin | Hydroxylation | –/Y/– | 208–211 | |
| ChoP | 105C1 | Unknown | Unknown | Y/–/– | 212 | |
| ChryOIII | – | Chrysomycin | Desaturation | –/Y/– | 213 | |
| CldC | – | Cyslabdan A | Hydroxylation/epoxidation | –/Y/– | 214 | |
| ComI | 165E1 | Complestatin | Biaryl ring coupling (C–C) | –/Y/– | 165 | |
| ComJ | 165B5 | Complestatin | Biaryl ring coupling (C–O) | –/Y/– | 165 | |
| CotB3 | – | Cyclooctatin | Hydroxylation | –/Y/– | 215 | |
| CotB4 | – | Cyclooctatin | Hydroxylation | –/Y/– | 215 | |
| CSP4 | 107P3 | Xenobiotics | Dealkylation | Y/–/– | 216 | |
| CYP102B1 | 102B1 | Fatty acids | Hydroxylation/epoxidation | Y/Y/– | 31,217 | |
| CYP102D1 | 102D1 | Fatty acids | Hydroxylation | Y/–/– | 29 | |
| CYP105D4 | 105D4 | Xenobiotics | Hydroxylation | Y/–/– | 131 | |
| CYP105D5 | 105D5 | Xenobiotics | Hydroxylation | Y/Y/– | 27,31,32,34,131 | |
| CYP105D6 | 105D6 | Filipin | Hydroxylation | Y/–/Y | 3ABB | 218 |
| CYP105D7 | 105D7 | Pentalenolactone/xenobiotics | Hydroxylation | Y/Y/Y | 4UBS | 48,219–224 |
| CYP105F2 | 105F2 | Oleandomycin | Hydroxylation | Y/–/– | 225 | |
| CYP105N1 | 105N1 | Coelibactin | Unknown | Y/–/Y | 3TYW, 4FXB | 31,226,227 |
| CYP105P1 | 105P1 | Filipin | Hydroxylation | Y/–/Y | 3ABA, 3E5J, 3E5K, 3E5L | 218,228 |
| CYP105P2 | 105P2 | Flavones | Hydroxylation | Y/–/Y | 5IT1 | 40,190,229 |
| CYP107AJ1 | 107AJ1 | Xenobiotics | Dealkylation | Y/–/– | 154 | |
| CYP107L2 | 107L2 | Fatty acids | Unknown | Y/–/Y | 5CJE, 5CWE | 230 |
| CYP107P1 | 107P1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP107P2 | 107P2 | Xenobiotics | Hydroxylation | Y/–/– | 171 | |
| CYP107T1 | 107T1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP107U1 | 107U1 | Xenobiotics | Oxidation to ketone | Y/Y/– | 31,231 | |
| CYP107W1 | 107W1 | Oligomycin A | Hydroxylation | Y/–/Y | 4WPZ, 4WQ0 | 232,233 |
| CYP107Y1 | 107Y1 | Xenobiotics | Hydroxylation | Y/–/– | 171 | |
| CYP107Z13 | 107Z13 | Avermectin | Oxidation to ketone | Y/–/– | 133,234 | |
| CYP125A2 | 125A2 | Xenobiotics | Hydroxylation | Y/–/– | 171 | |
| CYP147F1 | 147F1 | Fatty acids | Hydroxylation | Y/–/– | 235–237 | |
| CYP154A1 | 154A1 | Dipentaenone/xenobiotics | Cycloaddition/dealkylation | Y/Y/Y | 1ODO | 30–32,188,207,238 |
| CYP154C1 | 154C1 | Pikromycin/methymycin/neomethymycin | Hydroxylation | Y/–/Y | 1GWI | 31,32,239 |
| CYP154C3 | 154C3 | Xenobiotics | Hydroxylation | Y/–/– | 45 | |
| CYP155A1 | 155A1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP156A1 | 156A1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP156B1 | 156B1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP157A1 | 157A1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP157B1 | 157B1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP157C1 | 157C1 | Unknown | Unknown | Y/–/– | 31,151 | |
| CYP157C4 | 157C4 | Xenobiotics | Dealkylation | Y/–/– | 240 | |
| CYP158A1 | 158A1 | Flaviolin | Biaryl ring coupling (C–C) | Y/–/Y | 2DKK, 2NZ5, 2NZA | 31,32,42 |
| CYP158A2 | 158A2 | Flaviolin | Biaryl ring coupling (C–C) | Y/–/Y | 1SE6, 1S1F, 1T93, 2D0E, 2D09, 3TZO, 5DE9 | 31,32,42,102,111,112 |
| CYP159A1 | 159A1 | Unknown | Unknown | Y/–/– | 31 | |
| CYP170A1f | 170A1 | Albaflavenone | Hydroxylation/oxidation to ketone/diphosphate ionization | Y/Y/Y | 3DBG, 3EL3 | 145–147,241 |
| CYP170A2 | 170A2 | Albaflavenone | Hydroxylation/oxidation to ketone | –/Y/– | 242 | |
| CYP170B1 | 170B1 | Albaflavenone | Hydroxylation/oxidation to ketone | Y/–/– | 147 | |
| CYP450Y110 | – | Xenobiotics | Hydroxylation | Y/–/– | 243 | |
| CYP51f | 170A1 | Xenobiotics | Dealkylation | Y/Y/– | 244 | |
| CYPSvh01f | 105C1 | Xenobiotics | Hydroxylation | Y/–/– | 245 | |
| CYPSvu022 | 154He | Xenobiotics | Hydroxylation | Y/–/– | 245 | |
| DesVIII | – | Pikromycin | GTase activator | Y/Y/– | 141–143,246,247 | |
| DnrQ | 131A1 | Duanorubicin | GTase activator | –/Y/– | 143,246,248 | |
| DoxA27952 | 129A2 | Doxorubicin | Hydroxylation/oxidation to ketone | Y/–/– | 35 | |
| DoxA29050 | 129A2 | Doxorubicin | Hydroxylation/oxidation to ketone | Y/Y/– | 86,249,250 | |
| DoxAC5 | 129A1 | Doxorubicin | Hydroxylation/oxidation to ketone | Y/–/– | 251,252 | |
| Ema1 | 107Z12 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema2f | 107Z10 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema3 | 107Z2v2 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema4f | 107Z5v3 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema5 | 107Z6 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema6 | 107Z5v2 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema7 | 107Z3 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema8 | 107Z2v1 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema9 | 107Z11 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema10f | 107Z5v3 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema11 | 107Z1 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema12 | 107Z9 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema13 | 107Z8 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema14f | 107Z10 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema15 | 107Z5v1 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema16 | 107Z4 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| Ema17 | 107Z7 | Xenobiotics | Hydroxylation/oxidation to ketone | Y/–/– | 132 | |
| EncR | 107R1 | Enterocin | Hydroxylation | Y/Y/– | 253,254 | |
| FcpCf | 105e | Xenobiotics | Hydroxylation | Y/–/– | 38 | |
| FilC | – | Filipin | Hydroxylation | –/Y/– | 255 | |
| FilD | – | Filipin | Hydroxylation | –/Y/– | 255 | |
| FosK | – | Fostriecin | Hydroxylation | –/Y/– | 256 | |
| FscPf | 105H5 | Candicidin | Hydroxylation/oxidation to acid | –/Y/– | 209–211 | |
| GalD | – | Galbonolide | Hydroxylation or epoxidation | –/Y/– | 257 | |
| GbnD | – | Galbonolide | Hydroxylation or epoxidation | Y/–/– | 258 | |
| GdmP | 105U1 | Geldanamycin | Desaturation | –/Y/– | 92–94,259 | |
| GerPI | – | Dihydrochalcomycin | Epoxidation | –/Y/– | 260 | |
| GerPII | – | Dihydrochalcomycin | Hydroxylation | –/Y/– | 260 | |
| GfsF | 105e | FD-891 | Hydroxylation/epoxidation | Y/Y/– | 261,262 | |
| GilOIII | – | Gilvocarcin | Desaturation | –/Y/– | 213,263 | |
| GrhO3 | 105D9 | Griseorhodin | Epoxidation | –/Y/– | 264 | |
| HerG | – | Herboxidiene | Hydroxylation | Y/–/– | 265,266 | |
| HerO | – | Heronamide | Hydroxylation | Y/Y/– | 46 | |
| HlsH | 107e | Halstoctacoanolide | Hydroxylation/oxidation to ketone | –/Y/– | 164 | |
| HlsI | 107e | Halstoctacoanolide | Hydroxylation | –/Y/– | 164 | |
| HmtN | – | Himastatin | Hydroxylation | Y/Y/Y | 4E2P | 95,126 |
| HmtS | – | Himastatin | Biaryl ring coupling (C–C) | –/Y/– | 95 | |
| HmtT | – | Himastatin | C–N bond formation | Y/Y/Y | 4GGV | 95,126 |
| JulI | – | Julichrome | Biaryl ring coupling (C–C) | Y/Y/– | 96 | |
| LkmF | 107A2 | Lankomycin | Hydroxylation | –/Y/– | 267,268 | |
| LkmK | 107AP1 | Lankomycin | Hydroxylation | –/Y/– | 267,268 | |
| LnmA | 107AC1 | Leinamycin | Hydroxylation | Y/Y/Y | 4Z5P | 269 |
| LnmZ | 107AG1 | Leinamycin | Hydroxylation | Y/Y/Y | 4Z5Q | 269 |
| LtmK | – | Lactimidomycin | Desaturation | –/Y/– | 91 | |
| MeiE | 171A2 | Meilingmycin | Ether formation | –/Y/– | 173,174 | |
| MfnN | – | Marfomycin | Hydroxylation | –/Y/– | 270 | |
| MgsK | – | iso-Migrastatin | Hydroxylation | –/Y/– | 271 | |
| MonD | 124B1 | Monensin | Hydroxylation (ACP-tethered)d |
–/Y/– | 79 | |
| NcsB3 | 154J1 | Neocarzinostatin | Hydroxylation | Y /–/– | 272,273 | |
| NikF | 105K1 | Nikkomycin | Hydroxylation | –/Y/– | 167,274 | |
| NikQ | 162A1 | Nikkomycin | Hydroxylation (PCP-tethered) |
Y/Y/– | 167,168 | |
| NovI | 163A1 | Novobiocin | Hydroxylation (PCP-tethered)d |
Y/–/– | 166 | |
| NysL | 161A1 | Nystatin | Hydroxylation | Y/Y/– | 180,181,275 | |
| NysN | 105H1 | Nystatin | Hydroxylation/oxidation to acid | –/Y/– | 180,181 | |
| NzsA | – | Neocarazostatin | Hydroxylation | Y/Y/– | 276 | |
| OleP | 107D1 | Oleandomycin/xenobiotics | Epoxidation/hydroxylation | Y/–/Y | 4XE3 | 72,73,131,277,278 |
| OleP1 | 235A1 | Oleandomycin | GTase activator | –/Y/– | 248,279 | |
| ORF-A | 107C1 | Carbomycin | Epoxidation | Y/–/– | 280 | |
| P450CLA | 105M1 | Clavulanic acid | Unknown | Y/–/– | 281 | |
| P450mel | 107F1 | Melanin | Biaryl ring coupling (C–C) | Y/Y/– | 103 | |
| P450sca-2 | 105A3 | Xenobiotics | Hydroxylation | Y/–/– | 43,282–285 | |
| P450sky | 163B3 | Skyllamycin | Hydroxylation (PCP-tethered)d |
Y/Y/Y | 4L0E, 4L0F, 4PWV, 4PXH | 76–78 |
| P450SU-1 | 105A1 | Xenobiotics | Hydroxylation/epoxidation/dealkylation | Y/Y/Y | 2ZBX, 2ZBY, 2ZBZ, 3CV8, 3CV9 | 36,131,134–137,139,286–294 |
| P450SU-2 | 105B1 | Xenobiotics | Hydroxylation/dealkylation | Y/Y/– | 36,138,286–289,295 | |
| P450terf | 107Le | Xenobiotics | Hydroxylation | Y/Y/– | 296,297 | |
| PenM | 161C3 | Pentalenolactone | Oxidative rearrangement | Y/Y/– | 113 | |
| PikC | 107L1 | Pikromycin/methymycin/neomethymycin/xenobiotics | Hydroxylation/oxidation to ketone | Y/Y/Y | 2BVJ, 2CA0, 2CD8, 2C6H, 2C7X, 2VZM, 2VZ7, 2WH2, 2WI9, 3ZK5, 3ZPI, 4B7D, 4B7S, 4BF4, 4UMZ | 47,65–69,298–306 |
| PimD | 161A2 | Pimaricin | Epoxidation | Y/Y/Y | 2XBK, 2X9P | 50,70,71 |
| PlaO2 | – | Phenalinolactone | Nonfunctional | –/Y/– | 83 | |
| PlaO3 | – | Phenalinolactone | Hydroxylation | –/Y/– | 82 | |
| PlaO4 | – | Phenalinolactone | Hydroxylation | –/Y/– | 83 | |
| PlaO5 | – | Phenalinolactone | Hydroxylation | Y/Y/– | 83 | |
| PldB | 107e | Pladienolide | Hydroxylation | Y/Y/– | 307 | |
| PlmS2 | 107L8 | Phoslactomycin | Hydroxylation | Y/Y/– | 308 | |
| PntM | 161C2 | Pentalenolactone | Oxidative rearrangement | Y/Y/Y | 5L1O, 5L1P, 5L1Q, 5L1R, 5L1S, 5L1T, 5L1U, 5L1V, 5L1W | 113,114 |
| ProP450 | – | Xenobiotics | Hydroxylation | Y/–/– | 49,309 | |
| PsmA | 105e | Pladienolide | Hydroxylation | Y/Y/– | 310,311 | |
| PtlI | 183A1 | Pentalenolactone | Hydroxylation/oxidation to aldehyde | Y/–/– | 312 | |
| PtmO5 | – | Platensimycin | Ether formation | –/Y/– | 120 | |
| Qui15 | – | Echinomycin | Hydroxylation (PCP-tethered)d |
–/Y/– | 170 | |
| RapJ | 122A2 | Rapamycin | Hydroxylation | –/Y/– | 313,314 | |
| RapN | 107G1 | Rapamycin | Hydroxylation | –/Y/– | 313,314 | |
| RavOIII | – | Ravidomycin | Desaturation | –/Y/– | 213 | |
| RevI | – | Reveromycin | Hydroxylation | Y/Y/Y | 3WVS | 315 |
| RmnC | – | Raimonol | Hydroxylation | –/Y/– | 214 | |
| SamR0478 | – | Stambomycin | Hydroxylation | –/Y/– | 316 | |
| SamR0479 | – | Stambomycin | Hydroxylation | –/Y/– | 316 | |
| SanH | 105K2 | Nikkomycin | Hydroxylation | –/Y/– | 317,318 | |
| SanQ | 162A2 | Nikkomycin | Hydroxylation (PCP-tethered) |
–/Y/– | 169 | |
| Sclav_p0067 | – | (–)-Drimenol | Unknowng | –/Y/– | 319 | |
| ScnD | – | Pimaricin | Epoxidation | Y/Y/– | 182,183 | |
| ScnG | – | Pimaricin | Hydroxylation/oxidation acid | –/Y/– | 182,183 | |
| SgcD3 | 211A1 | C-1027 | Hydroxylation | –/Y/– | 320 | |
| SgvP | 107e | Griseoviridin | C–S bond formation | Y/Y/Y | 4MM0 | 129,130,189 |
| SKCTCFkbDf | – | FK506/FK520 | Hydroxylation/oxidation to ketone | Y/Y/– | 321 | |
| Sl5NT | – | Unknown | Nitration | Y/–/– | 125 | |
| SlgO2 | – | Streptolydigin | Hydroxylation/oxidation to ketone | –/Y/– | 322,323 | |
| SlStaN | 244A1 | Staurosporine | C–N bond formation | Y/–/– | 127 | |
| SMAFkbD | – | FK506/FK520 | Hydroxylation/oxidation to ketone | –/Y/– | 324 | |
| SMg15NT | – | Unknown | Nitration | Y/–/– | 125 | |
| SoCYP158A2 | 158A2 | Phenol/indole | Hydroxylation | Y/–/– | 325 | |
| SoyC | 105D1 | Xenobiotics | Hydroxylation/epoxidation/dealkylation/desaturation | Y/–/– | 37,326–333 | |
| Srm13 | – | Spiramycin | Hydroxylation/oxidation to aldehyde | –/Y/– | 334,335 | |
| StaF | 165A4 | A47934 | Biaryl ring coupling (C–O) (PCP-tethered) |
Y/Y/Y | 5EX8, 5EX9 | 99,106,107 |
| StaG | 165D1 | A47934 | Biaryl ring coupling (C–O) | –/Y/– | 99 | |
| StaH | 165B4 | A47934 | Biaryl ring coupling (C–O) (PCP-tethered) |
Y/Y/Y | 5EX6 | 99,106,107 |
| StaJ | 165C5 | A47934 | Biaryl ring coupling (C–C) | –/Y/– | 99 | |
| StaP | 245A1 | Staurosporine | Biaryl ring coupling (C–C)/oxidative decarboxylation | Y/–/Y | 2Z3T, 2Z3U, 3A1L | 100,101,108–110,336 |
| StFkbDf | – | FK506/FK520 | Hydroxylation/oxidation to ketone | Y/Y/– | 337,338 | |
| StStaN | 244A1 | Staurosporine | C–N bond formation | –/Y/– | 100,128 | |
| StuD1 | – | Thiolactomycin/thiotetronate | Epoxidation (ACP-tethered)d |
–/Y/– | 80 | |
| StuD2 | – | Thiolactomycin/thiotetronate | Hydroxylation/oxidation to acid | –/Y/– | 80 | |
| Sv5NT | – | Unknown | Nitration | Y/–/– | 125 | |
| TamIf | – | Tirandamycin | Hydroxylation/oxidation to ketone/epoxidation | Y/Y/– | 84,85 | |
| TetrKf | – | Tetramycin | Hydroxylation | Y/Y/– | 339 | |
| ThnC | – | Thienodolin | C–S bond formation | Y/Y/– | 340 | |
| TmcR | – | Tautomycetin | Hydroxylation/oxidation to ketone | –/Y/– | 341,342 | |
| TrdIf | – | Tirandamycin | Hydroxylation/oxidation to ketone/epoxidation | –/Y/– | 343 | |
| TtmDf | – | Tetramycin | Hydroyxlation | Y/Y/–g | 339,344 | |
| TtnI | – | Tautomycetin | Hydroxylation/oxidation to ketone | –/Y/– | 345 | |
| TxtC | 246A1 | Thaxtomin | Hydroxylation | Y/Y/– | 346 | |
| TxtE | – | Thaxtomin | Nitration | Y/Y/Y | 4L36, 4TPO, 4TPN, 5D3U, 5D40 | 122–125,347 |
| TylHI | 105L1 | Tylosin | Hydroxylation | –/Y/– | 348–350 | |
| TylMIII | – | Tylosin | GTase activator | –/Y/– | 143,246,351 | |
| Unk. P450 | – | Xenobiotics | Dealkylation | –/Y/– | 352 | |
| XiaM | – | Xiamycin | Hydroxylation/oxidation to acid | Y/Y/– | 89 |
Commonly used P450 names. Duplicated names are differentiated by adding the first letter of the genus and species, e.g., SlStaN = Streptomyces longisporoflavus StaN.
CYP names identified from reference or the Cytochrome P450 Homepage (drnelson.uthsc.edu).163
In this study, heterologous P450 biotransformations are considered in vitro experiments.
P450 acts on a PCP- or ACP-tethered substrate.
Only CYP family or subfamily was given.
CanC = FscP; CYP170A1 = CYP51; CYPSvh01 = FcpC; Ema2 = Ema14; Ema4 = Ema10; StFkbD = SKCTCFkbD; TamI = TrdI; TetrK = TtmD.
The function of Sclav_p0067 is still unknown, but in vivo results support alteration of FPP cyclization by the sesquiterpene synthase Sclav_p0068.319
2.3 Diverse functions
P450s are commonly associated with the hydroxylation, epoxidation, and dealkylation of xenobiotics found in human drug metabolism.1 Yet, P450s are extremely versatile and their unique catalytic cycle plays a key role in bestowing these hemoproteins with a wide variety of functional capabilities.16,21,23,24 These enzymes, although generally considered to show substrate promiscuity, can also be substrate specific, showing regio- and stereoselectivity, particularly when involved in the biosynthesis of natural products. As described in section 2, many P450 functions can be attributed to the use of Cpd I as the oxidizing species. It is thought that the structural diversity of natural products, translating into diverse electronic properties, may allow P450 catalytic intermediates that are less active than Cpd I to act as alternative oxidants.3 This section aims at highlighting selected examples of Streptomyces P450 to give an overview of the types of P450 transformations that are known to occur on the structurally diverse skeletons of natural products (Fig. 2).
Fig. 2.
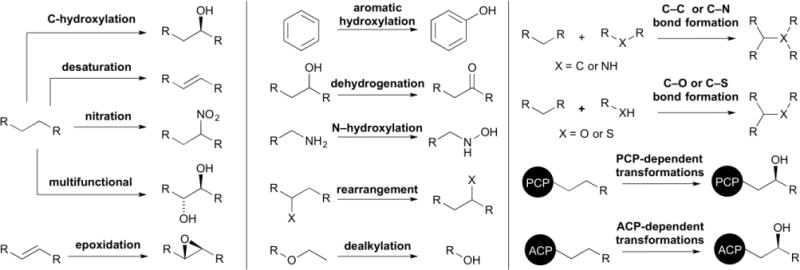
The functional diversity of P450s in Streptomyces. P450s catalyze a wide variety of functionalizations (selected examples are shown) in natural product biosynthetic pathways and in xenobiotic degradation.
2.3.1 Oxygenation
Hydroxylation of an aliphatic carbon is the prototypical P450 transformation, but P450s can also oxygenate carbons by epoxidation and aromatic hydroxylation. When involved in the functionalization of natural products, these P450 oxidative reactions are very often stereo- and regioselective, resulting in retention of configuration.12 C–H activation, one of the most difficult and most sought after synthetic and biosynthetic reactions,60–62 is a trademark of P450s. It is this ability, to insert an oxygen into an unactivated C–H bond, that piques the interest of synthetic and biosynthetic chemists, making P450s one of the most targeted types of enzymes for biotechnological applications.13,14,19,63,64
The majority of P450s in natural product biosynthetic pathways catalyze hydroxylation. While much of this review focuses on the sequence, structure, and function of P450s that catalyze other types of reactions, it should be noted that more than two-thirds of the characterized P450s from Streptomyces catalyze hydroxylations (Table 1). The most well known and studied P450 from Streptomyces, PikC (CYP107L1), catalyzes the aliphatic hydroxylation of several different macrolide antibiotics. PikC hydroxylates 12-membered macrolides to form methymycin and neomethymycin, and can hydroxylate twice at different positions to yield novamethymycin (Fig. 3).65,66 PikC also hydroxylates the 14-membered macrolide narbomycin to give pikromycin.67–69
Fig. 3.
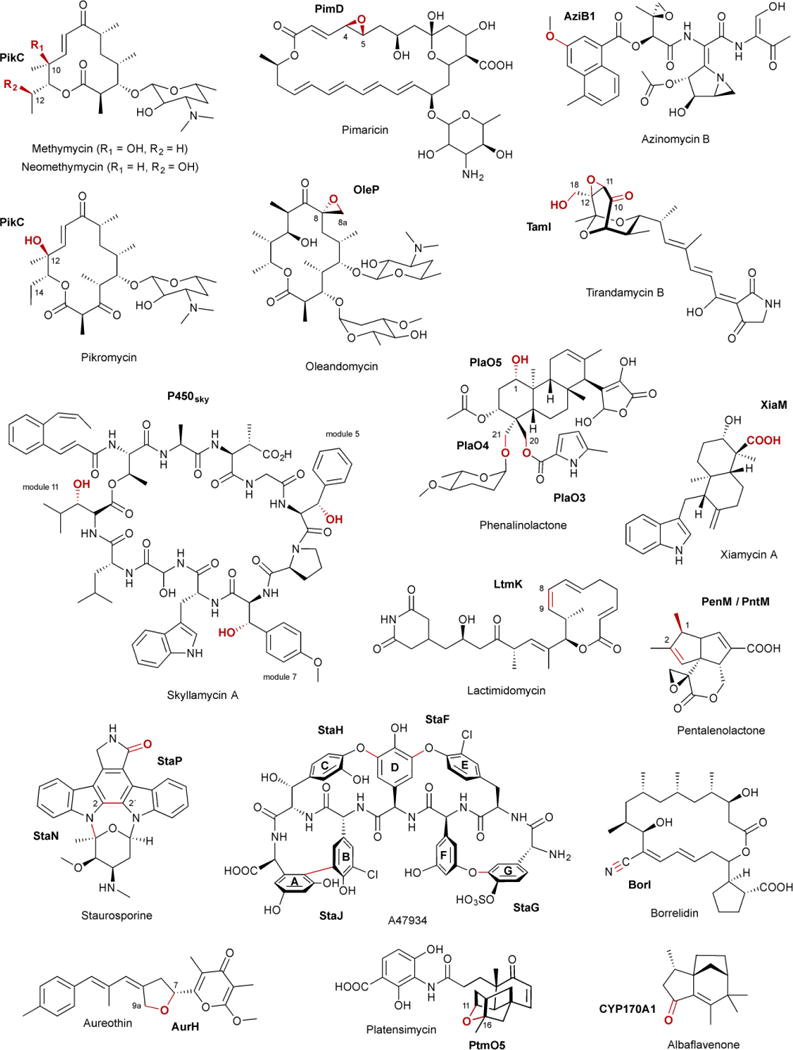
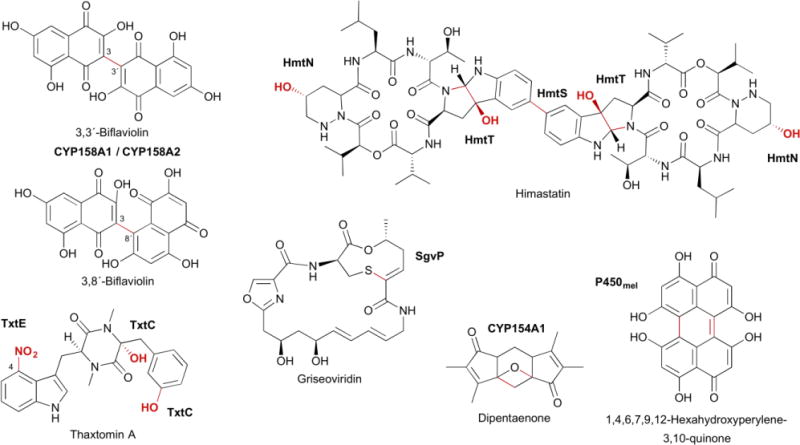
Selected natural products, P450s, and the biosynthetic transformations they catalyze, as discussed in the text. Functional groups and bonds colored in red are catalyzed by the P450s labeled in bold.
In addition to hydroxylation, P450s also commonly convert C–C double bonds into epoxides with retention of configuration.22 P450 epoxidations are frequently seen on polyketides due to the prevalence of olefins. PimD (CYP161A2), which epoxidizes the C-4–C-5 conjugated olefin in the polyene macrolide pimaricin (Fig. 3), is likely catalyzed by Cpd 0 (discussed in more detail in section 4.3.1).50,70,71. OleP (CYP107D1), a P450 epoxidase responsible for the C-8–C-8a epoxide in oleandomycin (Fig. 3), is unique in that it catalyzes epoxidation of two aliphatic carbons, although there is evidence that this transformation proceeds through an OleP-generated olefinic intermediate.72,73
P450s can also hydroxylate aromatic carbons, with the hydroxylation of naphthoic acid by AziB1 in the biosynthesis of azinomycin B as one example from Streptomyces (Fig. 3).74 The commonly accepted mechanism for P450 aromatic hydroxylation is initial epoxidation of the aromatic ring, followed by epoxide opening and rearomatization via hydride migration.8,75
Most P450s bind free substrates, but some achieve substrate selectivity by directly interacting with the peptidyl-carrier protein (PCP) of a nonribosomal peptide synthetase (NRPS) or the acyl-carrier protein (ACP) of a polyketide synthase (PKS), respectively. P450sky (CYP163B3) was demonstrated to utilize PCP-tethered amino acids as substrates in the modular biosynthesis of the cyclic depsipeptide skyllamycin.76–78 P450sky β-hydroxylates L-Phe, OMe-L-Tyr, and L-Leu in modules 5, 7, and 11, respectively (Fig. 3), but does not hydroxylate the PCP-tethered amino acids in the other eight modules of the NRPS, indicating that P450sky harbors innate PCP domain selectivity.77 P450s that utilize ACP-tethered substrates are also involved in natural product biosynthetic pathways, including monensin79 and thiolactomycin,80 although direct evidence is yet to be established. P450–ACP complexes, however, have been reported in other systems.81
A common theme throughout natural product biosynthesis is the late-stage functionalization at various positions on the scaffold. Many biosynthetic gene clusters contain multiple P450s to functionalize one scaffold, either sequentially or with additional modification steps in between each P450-catalyzed step. There are four P450-encoding genes, plaO2–plaO5, within the phenalinolactone gene cluster.82 With the exception of PlaO2, each P450 catalyzes the hydroxylation of a different carbon on the diterpene scaffold. PlaO3 and PlaO4 hydroxylate the C-20 and C-21 methyl groups, respectively, and PlaO5 stereoselectively hydroxylates C-1 (Fig. 3).82,83 PlaO2 was determined to be nonessential in phenalinolactone formation,83 although it is not clear whether it is nonfunctional, can be complemented by another P450, or has another biological role in the organism. An alternative strategy to functionalize one scaffold multiple times is the utilization of a multifunctional P450, with TamI being the quintessential example. Acting through repeated exchange with the FAD-dependent oxidase TamL, TamI catalyzes an oxidative cascade on the tirandamycin sketeton, beginning with stereoselective hydroxylation at C-10, and followed by successive epoxidation at C-11–C-12 and hydroxylation of the C-18 methyl (Fig. 3).84,85 TamI, in the absence of TamL, can also bypass the TamL dehydrogenation reaction and form minor amounts of the C-10 ketone.85
2.3.2 Dehydrogenation
P450s are also known to catalyze two or more electron dehydrogenations, converting an sp3 hybridized carbon into an sp2 or sp hybridization state. The classification of P450 “dehydrogenations” can sometimes be a misnomer, however, due to ketones, aldehydes, and carboxylic acids forming through second or third hydroxylations with subsequent (likely) nonenzymatic dehydrations.21,75,86–88 In Table 1, these “dehydrogenations” are simply labeled as “oxidations.” For example, XiaM, which converts a methyl into a carboxylic acid in the biosynthesis of the indolosesquiterpene alkaloid xiamycin A biosynthesis (Fig. 3), was shown to have hydroxyl, geminol diol, and aldehyde intermediates.89 Another extraordinary example is BorI.90 BorI first produces an aldehyde group from a methyl in a four electron oxidative process. After the aldehyde undergoes BorJ-mediated transamination, BorI completes the conversion of the amine into a nitrile, presumably through N,N-dihydroxy and aldoxamine intermediates (Fig. 3).90
The other subset of dehydrogenation reactions are performed by the P450 desaturases. When Cpd I creates the substrate radical, there is a competition between hydrogen atom abstraction and hydroxylation. Most often, the hydroxylated compound is the major product, and the hydroxylated and desaturated congeners can not be interconverted.21 In the biosynthesis of some natural products, desaturation is the predominant P450 product, supporting that desaturation can be enzymatically controlled. In the final step of lactimidomycin, LtmK converts 8,9-dihydro-LTM into LTM at >95% efficiency (Fig. 3).91 The presence of <5% of both the 8S- and 9R-hydroxy-LTM analogues (hydroxyl groups are on the same face), suggests that LtmK can generate a radical on either the C-8 or C-9 position. GdmP (CYP105U1), a desaturase in geldanamycin biosynthesis,92,93 may also allow radical rebound to the heme-bound oxygen in a non-stereoselective manner as both stereochemistries of the C-4 hydroxylated congener were isolated from the producing strain.94
2.3.3 Biaryl ring coupling
The coupling of aromatic rings can create structural diversity (e.g., himastatin95 and julichromes96), give flexible backbones rigid conformations [e.g., glycopeptide antibiotics (GPA)97–99 and staurosporine100,101], and polymerize monomers to yield natural sunscreens (e.g., melanins102,103). The signature structural feature in the GPA family, the most well known being vancomycin, is the heptapeptide backbone with multiple biaryl (C–C) or biaryl ether (C–O–C) bridges.104 Linear and incompletely processed intermediates or analogues do not retain the strong antibacterial activities associated with these natural products.97 In Streptomyces, the GPA A47934 has four of these aromatic bridges;105 the vancomycin-type only has three.104 Four CYP165 members, StaH, StaG, StaF, and StaJ, sequentially install the C–O–D, F–O–G, D–O–E, and A–B ring, respectively (Fig. 3).99,106 Adding to the complexity and interest of this family of P450s, they utilize PCP-tethered substrates and interact with the so-called X-domain, found in the last NRPS module, for P450 recruitment and cyclization efficiency.97,98,107
StaP (CYP245A1) is responsible for intramolecular C–C bond formation in staurosporine biosynthesis. StaP was proposed and theoretically supported to delocalize the π-cation porphyrin radical of Cpd I over the indole moieties of chromopyrrolic acid (CPA).108,109 CPA, upon losing an electron and proton, forms a C–C bond between the two indole rings at C-2 with a coupled second electron transfer, and goes through a series of protonations and tautomerizations to yield the six-ring indolocarbazole (Fig. 3).108,109 After biaryl coupling of CPA, StaP also catalyzes oxidative decarboxylation, mediated by the FAD-dependent enzyme StaC or RebC (from the rebeccamycin pathway).101,110
CYP158A1 and CYP158A2 from S. coelicolor polymerize flaviolins to form a variety of bi- and tri-flaviolins that may protect against UV radiation.42,102,111,112 Although these two P450s share 61% sequence identity and bind the same substrate, CYP158A1 only forms two biflaviolins, whereas CYP158A2 forms at least four polymers (Fig. 3). Similarly, P450mel (CYP107F1) in Streptomyces griseus catalyzes sequential oxidations of 1,3,6,8-tetrahydroxynaphthalene (THN), first through an intermolecular aryl coupling and then through an intramolecular aryl coupling, to form the 1,4,6,7,9,12-hexahydroperylene-3,10-quinone (HPQ) melanin (Fig. 3).103 For many of these biaryl ring couplings, it is still unclear whether they utilize radical or cationic mechanisms and whether both “substrates” form radicals before coupling or one attacks the neutral distal substrate.
2.3.4 Other transformations
The versatility of P450s is on display in Streptomyces natural product biosynthetic pathways. PenM (CYP161C3) and PntM (CYP161C2) from Streptomyces exfoliatus and arenae, respectively, catalyze a rare oxidative rearrangement in pentalenolactone biosynthesis (Fig. 3).113,114 After initial substrate radical formation, steric hindrance of the C-1 radical by substituents on the substrate itself severely suppresses oxygen rebound. This reduced rebound rate allows the oxidative formation of a neopentyl carbocation intermediate, which undergoes a 1,2-methyl shift followed by deprotonation to yield pentalenolactone.114
P450s can install oxygens on unactivated carbons and form ether linkages with pre-installed oxygens. Understandably, some P450s are able to do both. AurH, a P450 from Streptomyces thioluteus, catalyzes stereoselective tetrahydrofuran ring formation in aureothin biosynthesis (Fig. 3).115–118 Two C–O bonds are sequentially formed: the first at the methylene C-7 to yield a 7R-hydroxy intermediate and the second between the newly installed hydroxyl and methyl C-9a.116,117 It is unknown whether the heterocyclization steps utilize a transient dihydroxylated or radical intermediate. A similar mechanism may be used by AveE (CYP171A1) to form the tetrahydrofuran ring in avermectin.119 PtmO5 also catalyzes tetrahydrofuran ring formation in platensimycin biosynthesis (Fig. 3).120 PtmO5 likely stereoselectively hydroxylates the C-11 position (11S) on the ent-kauranol scaffold, then uses the pre-installed R-hydroxyl group at the C-16 position as a leaving group to form the 11S,16S ether ring.120
P450s are also able to form C–N bonds using at least two different strategies. Hemoproteins such as P450s can generate reactive nitrogen species, although reaction of molecular oxygen with heme-bound nitric oxide (NO) can cause irreversible enzyme inhibition.121 TxtE, a P450 found in several plant-pathogenic Streptomyces spp. responsible for the production of thaxtomins, nitrates free L-Trp at the C-4 position (Fig. 3).122–125 The proposed mechanism differs from typical Cpd I chemistry diverting after ferric superoxide (FeIII–OO−) is formed. Reaction of NO with ferric superoxide forms ferric peroxynitrite (FeIII–OONO), which then undergoes heterolytic cleavage to yield NO2 and FeIV=O followed by nitration and a FeIV=O-mediated hydrogen abstraction.122 Nitration may also occur via electrophilic aromatic substitution if heterolytic cleavage is achieved through protonation of the ferric peroxynitrite complex.122 The second strategy for C–N bond formation is the oxidative formation of an electrophilic functional group, which can subsequently be attacked by a nearby nitrogen atom. HmtT is responsible for hexahydropyrroloindole formation in himastatin going through a putative 2,3-epoxyindoline intermediate.95,126 The nitrogen atom then attacks C-2 forming a C–N bond and leaving the hydroxyl group on C-3 (Fig. 3). Similarly, StaN (CYP244A1) may form the glycosidic bond in staurosporine via hydroxylation at C–5 of the sugar, followed by protonation and dehydration to yield the electrophilic oxonium cation (Fig. 3).127,128
In a unique C–S bond forming reaction, SgvP, from the griseoviridin biosynthetic pathway, forms a nine-membered thio-ene-containing lactone ring (Fig. 3).129,130 Although only speculated at this time, the epoxidation of an α,β-unsaturated amide may lead to sulfur nucleophilic attack at the α carbon,130 mirroring the proposals for C–N bond formation described above.
2.3.5 Xenobiotic transformations
Although this review mainly focuses on P450s in natural product biosynthesis, we would be remiss to not discuss the inherent role that P450s play in the oxidative transformations of xenobiotics. Given their associations with compound degradation and steroid biosynthesis, researchers have utilized P450s from Streptomyces, as well as other sources, as a tool to convert target compounds into compounds of interest. Notable examples include (i) a systematic screening of P450s for the regio- and stereoselective hydroxylation of testosterone,131 (ii) the biosynthetic conversion of avermectin to 4´-oxo-avermectin for the agriculturally important insecticide emamectin,44,132,133 and (iii) the multipurpose P450SU-1 (CYP105A1), which, among other reactions, catalyzes the conversion of vitamin D3 into its active form 1α,25-dihydroxyvitamin D3,134–136 O- and N-dealkylations of coumarin and sulfonylurea herbicides,36,37,137,138 and the hydroxylation and epoxidation of diterpenoid-based resin acids.139 In many cases, P450s with “xenobiotic” activities have unknown endogenous substrates and functions (i.e., orphan P450s).
2.3.6 Non-P450 transformations
It is not unusual for proteins of similar sequence and/or function to perform vastly different types of chemistries with the triosephosphate isomerase (TIM) barrel fold being the classic example.140 Proteins that are homologous to P450s in sequence and structure but catalyze unrelated reactions are beginning to be discovered. It has yet to be determined how prevalent these enzymes are in nature and in natural product biosynthesis.
In the biosynthesis of the methymycins and pikromycin in Streptomyces venezuelae, the glycosyltransferase (GTase) DesVII attaches TDP-desoamine to 10-deoxymethynolide or narbonolide.141 However, DesVII requires a protein partner for proper activity. DesVIII, a protein with a P450-like sequence minus the conserved Cys, was shown to have protein-protein interactions with DesVII and improve DesVIII activity at least 1000-fold.141–143 EryCII, a homologue of DesVII from the erythromycin biosynthetic pathway in Saccharopolyspora erythraea, was recently confirmed to have the P450-like fold and allosterically activates the GTase EryCIII by forming a homotetramer.144 By analogy, DesVIII and its homologous non-heme containing P450-like proteins are allosteric activators of macrolide GTases.
CYP170A1 catalyzes two sequential and nonstereoselective allylic oxidations in the conversion of the sesquiterpene epi-isozizaene into the antibiotic albaflavenone in S. coelicolor (Fig. 3).145 Interestingly, CYP170A1 generated isomers of farnesene, farnesol, and nerolidol in a redox-independent manner when incubated with farnesyl diphosphate (FPP).146 A second active site on the P450 scaffold was found, mimicking a terpene synthase in both sequence and structure.146 Homologues of CYP170A1, such as CYP170B1 from Streptomyces albus, do not have the terpene synthase functionality, likely due to the inability to bind Mg2+ given its DGXXR motif, instead of the canonical DDXXD terpene synthase motif.147 It is unclear if the moonlighting terpene synthase function of CYP170A1 is biologically relevant.
3 Sequence
It is well known that the P450 superfamily exhibits significant sequence heterogeneity and lacks conserved sequence motifs. Until 2006, there were only three residues that were thought to be strictly conserved among all of the P450 sequences in the public databases (Fig. 4). The first invariant residue, and catalytically most important, was a Cys, the fifth axial ligand of the heme iron.148 The other two invariant residues were the Glu and Arg that composed the EXXR motif found in the K-helix.149 These two charged residues form ionic interactions with the region prior to the Cys-ligand loop (meander region) to assure correct tertiary structure and stabilization of the heme within the protein.150 Once the genomes of S. avermitilis and S. coelicolor were available, it was clear that there were P450s in nature that did not possess the EXXR motif.31,53 CYP157C1 from S. coelicolor was later confirmed to be a legitimate heme-containing P450, indicating that only the iron-binding cysteine is strictly conserved.151 There are two other highly conserved, but nonessential, residues found in P450s: a Thr in the I-helix, which has been proposed to be involved in oxygen activation152 and the prevention of auto-inactivation,153 and a Gly four residues upstream of the conserved Cys, which allows the formation of the β-hairpin turn.150 Sequence alignment of the 184 (minus CYP102D1 due to its significantly longer sequence) functionally characterized P450s from Streptomyces reveals the highly conserved nature of the EXXR motif and the heme-binding Cys, Thr, and Gly residues (Fig. 4). Although no residues are completely conserved within these P450s, there are indeed several highly conserved motifs. HXXXR in the C-helix, Arg in β5, and Asp, Arg, and Phe in the meander region. Many of these residues comprise the heme-binding pocket; however, the roles of other conserved motifs outside of the active site is unknown. These residues may contribute to the structural integrity of the P450 fold or protein-protein interactions with redox partners.
Fig. 4.
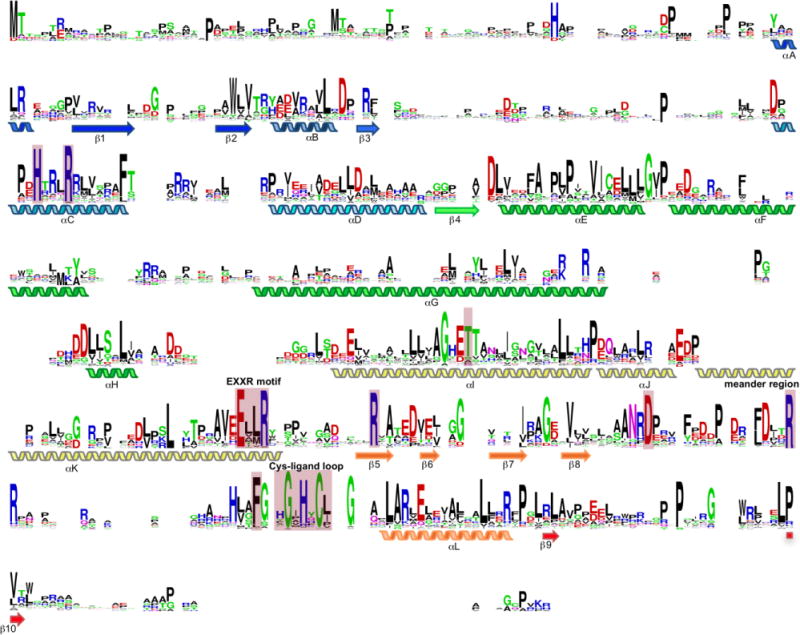
Sequence alignment of the 184 functionally characterized P450s from Streptomyces (minus CYP102D1, for simplicity, given its extended length). The heme-binding Cys, EXXR motif in the K-helix, and Thr in the I-helix are highly conserved. In addition, there are other highly conserved motifs and residues, both within and outside of the active site, in Streptomyces P450s. Residues discussed in the text are highlighted by pink boxes. Positions with no residues represent gaps in the alignment due to sequence length differences.
Interestingly, as more P450 sequences become available, even the heme-binding Cys paradigm is being challenged. CYP107AJ1 from Streptomyces peucetius does not contain the expected Cys, yet was shown to be a legitimate P450 that retains peroxygenase activity.154 Other natural variants of P450s without the axial Cys have also been identified in other organisms, although their functions are still unknown.155 Engineered P450s with Cys-to-Ser or Cys-to-His mutations have also been shown to promote unnatural carbene- and nitrene-transfers, as well as peroxidase activity,156–159 suggesting that natural variants without the conserved Cys may still in fact be functional P450s. Finally, as described in section 2.3.6, there are P450-like proteins, e.g., GTase activator DesVIII and its homologues, that lack both the axial Cys and heme,143,144 indicating that P450s are evolving to utilize the P450 scaffolding for alternate functions.
Given the amino acid sequence disparity among P450s, as well as our limited understanding of P450 sequence–structure–function relationships, it is currently impossible to predict P450 function or substrate preference based on sequence alone. However, with the enormous influx of bacterial genomes, we set out to map the current catalytic landscape of P450 enzymes of streptomycete origin.
3.1 Network generation
P450 protein sequences, limited to the taxonomic order of Streptomycetales, were obtained from the InterPro database.160 As of January 3, 2017, there were 8578 P450 sequences; of these, 7818 were unique (i.e., <100% identity). To emphasize the rapid rate at which the number of P450s in the database are increasing, there were 6722 P450 sequences of streptomycete origin on September 15, 2016, corresponding to an increase of >18 new P450s per day! Given this extraordinary rate of growth, a systematic approach to prioritizing P450s is necessary, along with innovative high-throughput methods, for focused and informative future studies.
We used 8579 P450 sequences (the 8578 sequences from InterPro plus SoCYP158A2, as it was not found in the InterPro database) and generated a P450 sequence similarity network (SSN)161 using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST).162 After using a minimum length cutoff of 200 amino acids and a representative node network of 100% identity, there were 7579 unique P450 sequences. Of these, 184 (~2.4%) have been functionally characterized (Table 1). To select an E value threshold for the network, we attempted to parallel the systematic CYP nomenclature (i.e., family and subfamily >45% and >55% identity, respectively)163 to give the community a valid point of reference. As the networks are based on E values, which take sequence length into consideration, but the CYP nomenclature are based on sequence-independent percent identities, we used the functionally characterized P450s that have corresponding CYP names as a guide for threshold selection. For the CYP family level, we selected a threshold of 10−85 to prevent HlsI164 from separating from CYP107 family. For the CYP subfamily level, we selected a threshold of 10−124 to keep ComJ165 within the CYP165B subfamily. The E values of 10−85 and 10−124 are approximately equivalent to median values of 45% and 58%, respectively, for P450s with 400 residues. Access to the P450 SSN is available at www.scripps.edu/shen/NPLI/database.html.
3.2 Pre-CYP family network
Before separating the P450s into their approximate CYP family and subfamily networks, we evaluated the global organization of streptomycete P450s. At an E value of 10−35 (low stringency threshold), the two main classes of P450s can already be distinguished. These two major clusters of proteins represent the bacterial and mitochondrial soluble class I and the eukaryotic microsomal membrane-bound class II (described in section 2). The larger class I subtype contains >2300 unique P450 sequences and includes the vast majority of the functionally characterized enzymes. The smaller class II subtype, which contains the self-sufficient CYP102D1 and the CYP170 family, is comprised of <400 unique P450s. In addition to the two major clusters, there are a handful of smaller clusters revealing P450 or P450-like proteins that are significantly different in sequence from the two main classes. All but one of these small clusters only contain uncharacterized members. The only known P450 included in this group is CYP155A1, a legitimate P450 with an unknown endogenous function.31 Conspicuously, the P450-like GTase activators already show signs of separation from the class I cluster at this threshold.
As in all SSNs, as the similarity threshold is increased, differences in P450 sequences becomes more apparent. In general, before the CYP family cutoff of 10−85 is reached, the large clusters begin to separate into what will become the CYP family clusters. One of the most diverse P450s that resolves into an independent cluster early is the C–N bond forming StaN,100,127,128 perhaps indicative of its CYP244 designation. Interestingly, a cluster of sequences that starts to splinter off the class I subtype at 10−45 and has clear separation at 10−55 contains only P450s that work on ACP- or PCP-tethered substrates including MonD,79 NovI,166 NikQ,167,168 SanQ,169 Qui15,170 and P450sky.76–78 This finding suggests that this cluster of P450s (>180 members) may all work on ACP- or PCP-tethered substrates and gives insight into the putative endogenous function of CYP125A2, a P450 shown to hydroxylate a xenobiotic flavonoid.171 Not all characterized P450s that act on ACP- or PCP-tethered substrates are found within this cluster, with AcmG8,172 StaF, and StaH being notable exceptions,99,106 suggesting there is more than one type of sequence that can utilize protein-protein interactions for substrate selectivity.
3.3 CYP family and subfamily networks
While we used the systematically named and functionally characterized P450s from Streptomyces as a guide for determining the CYP family network found in this review, it is clear that the determined E value of 10−85 is not perfect in separating members of different CYP families from each other. There are >100 clusters with at least five unique P450 sequences and >300 smaller clusters or singletons (Fig. 5). Parenthetically, bacterial P450s encompass CYP100–CYP2999;57 although it is not clear how many are found within Streptomyces. While many of these clusters are separated into different CYP families, there are exceptions. In addition, the diversity and growing number of new P450 sequences results in common neighbors of CYP families, linking these clusters together. As with the CYP nomenclature, one SSN threshold will never be perfect; however, utilizing a range of thresholds likely gives the best opportunity to systematically categorize such a large number of protein sequences.
Fig. 5.
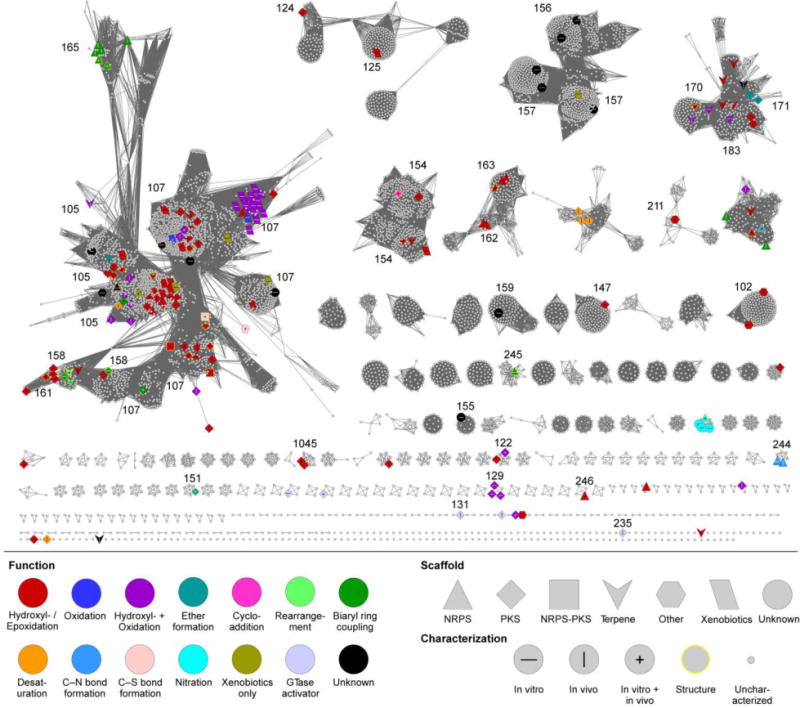
CYP family SSN of Streptomyces P450s. The SSN is shown at a BLAST E value cutoff = 10−85 (median 45% identity over 400 residues). Larger nodes are functionally characterized P450s with node labels describing how it was characterized. Colors and shapes of nodes represent P450 function and substrate type (type of natural product scaffold). See inset legend for details. CYP families of functionally characterized P450s are labeled.
After the CYP family network was annotated, depicting P450 function, substrate (type of natural product scaffold), and functional characterization by color, shape, and label of each node, respectively, several conclusions are immediately apparent. (i) Streptomycetes possess a great diversity of P450s, most of which have not been studied. More than 75 clusters with at least five unique members do not have a functionally characterized P450, and that does not include the >50 P450s with unknown endogenous functions. (ii) Large CYP family clusters give an impression of conservativity across different Streptomyces species, suggesting these may be involved in much less specialized roles, e.g., detoxification. The largest supercluster is composed of both the CYP105 and CYP107 families, the two largest CYP families in bacteria.4 Smaller clusters and singletons, P450s that are not present in many different species, likely have evolved specific functions for less common biosynthetic pathways or substrates. (iii) At this similarity level, there are no overall tendencies of P450s of similar function or preferred substrate to cluster together. For example, not all ether-forming P450s cluster together, with AurH,115–118 AveE/MeiE,119,173,174 and PtmO5120 existing in three different clusters. This corresponds to the difficulty in predicting P450 function based on overall P450 sequence.
(iv) From a more local perspective, there are some clusters that show similarities in function and/or substrate preference. As described above, a group of P450s that act on ACP- and PCP-tethered substrates showed early separation from the larger class I cluster. At an E value of 10−85, the ACP-dependent MonD79 separated from a group of 145 P450s containing only known enzymes that hydroxylate PCP-tethered amino acids at the β position, i.e., NovI,166 NikQ,167,168 SanQ,169 Qui15,170 and P450sky.76–78 Their separation from other P450s, combined with their sequence similarities suggest that they possess sequence–function or sequence–substrate relationships. Although the amino acid specificities differ amongst these P450s, the P450–PCP (or P450–phosphopantetheine) interactions are likely responsible for substrate recognition. Several predicted, but uncharacterized, P450s involved in the biosyntheses of β-amino acid containing natural products such as simocyclinone,175 clorobiocin,176 and courmermycin,177 also cluster in this group. Finally, (v) the biaryl ring coupling-catalyzing P450s involved in glycopeptide biosynthesis, are splintering off of the CYP105 family. These include the aforementioned StaH, StaG, StaF, and StaJ99,106 and ComI and ComJ from complestatin biosynthesis.165 Although only StaH and StaF were characterized in vitro showing a requirement for the PCP-tethered substrate and the X-domain,107 it is plausible that the other four enzymes, along with the other unidentified members in this splintered group follow the same reactivity requirements.
The CYP subfamily network, with a E value of 10−124, significantly increases the level of complexity within the P450 network (Fig. 6). The number of clusters with at least five unique P450 sequences, clusters with less than five, and singletons inflated to >200, >300, and >500 respectively. As with the lower threshold network, there are a few exceptions to our attempt to limit one subfamily per cluster. Importantly, and in opposition to the CYP family network, nodes of similar colors and/or shapes, i.e., functionally characterized P450s with similar functions and/or substrates, cluster together more frequently. For example, AmphN,178,179 NysN,180,181 and ScnG,182,183 P450s that all convert a methyl group into a carboxylic acid in the biosynthesis of polyenes amphotericin B, nystatin, and pimaricin, respectively, cluster together into one subfamily (CYP105H). TxtE, the tryptophan-nitrating enzyme122–124 clusters together with seven other P450s, including the three P450s that were identified to nitrate the C-5 position on the indole ring of tryptophan.125 Even NikQ167,168 and SanQ,169 the PCP-dependent β-hydroxylating P450s separate from the other group of PCP-dependent β-hydroxylating enzymes. This may indicate sequence differences that may account for PCP specificity, or alternatively, a difference in amino acid preference. The most striking revelation in this CYP subfamily network is the immensity of unknown and uncharacterized P450s found within both large and small clusters.
Fig. 6.
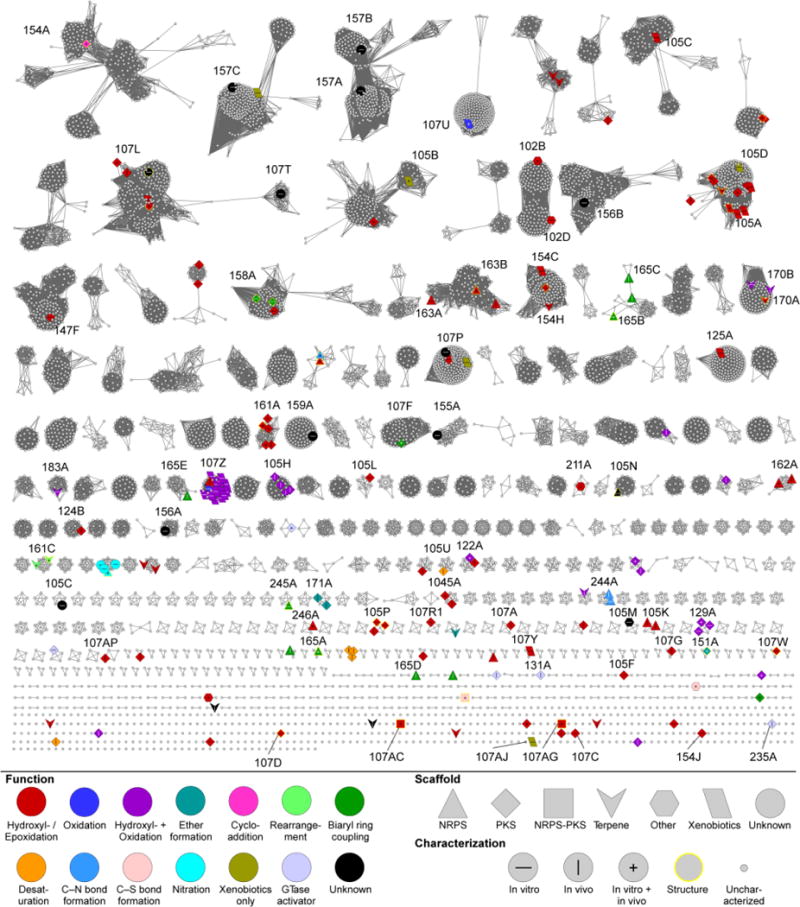
CYP subfamily SSN of Streptomyces P450s. The SSN is shown at a BLAST E value cutoff = 10−124 (median 58% identity over 400 residues). Larger nodes are functionally characterized P450s with node labels describing how it was characterized. Colors and shapes of nodes represent P450 function and substrate type (type of natural product scaffold). See inset legend for details. CYP subfamilies of functionally characterized P450s are labeled.
Overall, the P450 SSN clearly emphasizes (i) the sequence diversity of P450s found within Streptomyces, (ii) that higher levels of sequence similarity, in general, can be used to cluster P450s of similar tendencies, and thus identify clusters of novel, uncharacterized P450s, and (iii) the majority of these clusters contain no functionally characterized members. The identification of the most promising P450s and their functional and structural characterizations will surely expand the chemical reaction space of P450s found in the biosynthesis of natural products.
4 Structure
Since the first crystal structure of a P450, P450cam (CYP101A1) from P. putida, was determined in 1985,184 the rate of P450 structures has dramatically increased every year. At the time of writing this review, there are around 750 P450 structures deposited within the Protein Data Bank (PDB). Twenty-nine of these are from Streptomyces, with 13, eight, three, and four coming from polyketide, nonribosomal peptide, hybrid polyketide–nonribosomal peptide, and terpene biosynthesis, respectively; one is still functionally unknown. In the CYP family SSN, less than 15 clusters with at least five unique members have at least one PDB entry, revealing a significant lack of CYP families and subfamilies that have been structurally characterized. In this section, we describe the general structure of P450s, structures from Streptomyces that catalyze interesting chemistries or give mechanistic insight into this family of enzymes, and examples showing the potential of structure-based engineering of P450s. While other reviews have covered the topic of P450 structure,3,16,185 we aim here to highlight those from Streptomyces.
4.1 Common structural aspects
There are now a sufficient number of P450 structures from eukaryotes, bacteria, and archaea sources to verify that all P450s share similar two- and three-dimensional structures, even in spite of low sequence identities between some members.185 The fold and overall structures of the 29 P450s from Streptomyces are nearly identical, particularly the protein architecture surrounding the heme binding site. The root-mean-square deviations (RMSD) of the 29 structurally aligned P450s ranged from 1.3 Å to 2.6 Å, with an average value of 1.8 Å. The P450s from Streptomyces mainly consist of 12 α-helices (αA–αL) and 10 β-strands (β1–β10) and fold into prism-like structures (Figs. 4 and 7A). As in all P450s, the P450s from Streptomyces feature an extended I-helix passing through the length of the protein. The binding environment of the heme prosthetic group is quite conserved, being sandwiched between the I-helix and Cys-ligand loop. The Cys-ligand loop forms a rigid architecture via two main chain H-bonds, which positions the conserved Cys at an ideal place for coordination with the heme iron. A highly conserved Phe (Fig. 4), seven amino acids prior to the conserved Cys, interacts with the heme via T-shaped π-π stacking. Two His (C-helix and Cys-ligand loop) and two Arg (C-helix and β5 helix), both highly conserved residues, (Fig. 4) bind to the two propionate groups of heme via electrostatics interactions. Superposition of the 29 P450 structures revealed both regions of strict alignment and variability (Fig. 7C). Regions that were most structurally conserved were the I- and L-helices and the Cys-ligand loop; regions showing subtle differences between structures were mainly in the N-terminal loop, A–B and B–C loops, and F–G region (including the F–G loop and the F- and G-helices). In general, the substrate binding site of P450s is located near the B–C and F–G loops, helices F, G, and I, and the heme. The B–C loop (in some P450s, contains 1–3 small helices) and F–G region form the active site entrance and are responsible for substrate recognition. The B–C loop and F–G region undergo a conformational change, from an open to closed state, upon substrate binding (Fig. 7B).185 However, the conformations of some P450s are not controlled by substrate binding. For example, the crystal structure of substrate-free PikC (PDB ID: 2BVJ) was shown to exist in both open and closed conformations within the two polypeptide chains.186
Fig. 7.
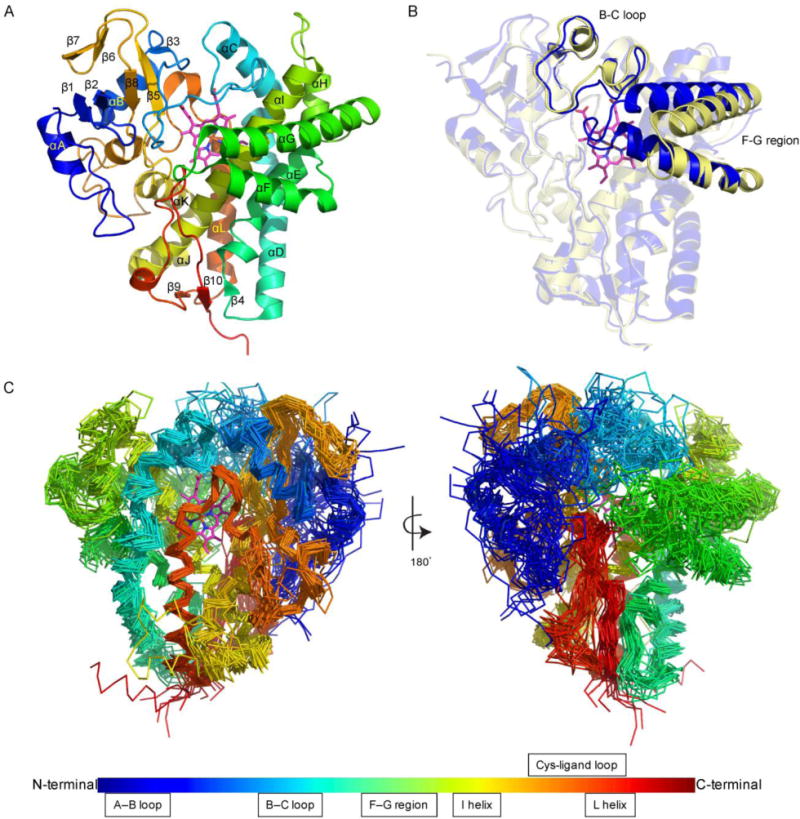
Common structural aspects of P450s from Streptomyces. (A) The overall structure of P450s (exemplified by PDB ID: 3ABA). (B) The open and closed states of the P450 active site are facilitated by conformational changes in the B–C loop and the F–G region [PDB IDs: 1SE6 (open) and 2D09 (closed)]. (C) Structural superposition of the 29 structurally characterized P450s from Streptomyces highlighting the regions of high, moderate, and low structural conservativity.
4.2 Variations in structure
As described above, the 29 P450 structures from Streptomyces are highly conserved. Given the inherent ability of P450s to perform diverse chemistries on diverse substrates, there must be structural variations between P450s that account for these differences. The same concept is found in the SSN, where there is not a universal pattern for sequence–function relationships, but local trends do exist. Since structure is typically determined based on sequence (with the exception of convergent evolution of protein folds), the P450 SSN gives insight into finding P450s with unique structural properties. For example, the PCP-dependent P450s, which show sequence similarity, likely have sequence-associated structural similarities due to their requirement for PCP-P450 protein-protein interactions. Another interesting observation is that none of the P450s found within the cluster containing the EXXR motif-less CYP157C1 (264 members in total) possess the EXXR motif, supporting that P450s with similar sequences have similar structural motifs. No members within this cluster have been structurally characterized yet. The structures of selected P450s from Streptomyces with unusual biochemical properties are presented in this section.
4.2.1 Heme group inversions
Almost all P450s incorporate the prosthetic heme group in the same orientation, based on the positions of the two protoheme vinyl groups. A crystal structure of CYP121A1 (PDB ID: 1N40) from Mycobacterium tuberculosis revealed that the heme group can be in two distinct orientations, the normal orientation and one in which the heme is flipped 180°.187 The crystal structure of CYP154A1 (PDB ID: 1ODO) from S. coelicolor revealed that its heme group only resides in the 180°-flipped orientation (Fig. 8A).188 CYP154A1 catalyzes an unusual redox-independent Paternò–Büchi-like [2+2] cycloaddition of a dipentaenone into a oxetane-containing product.30 The role of this natural product or intermediate is not clear, but deletion of the CYP154A1-encoding gene compromised the stability of the bacterial spores of the producing strain.30 Three additional structures of P450s from Streptomyces, StaF (PDB ID: 5EX8),106 SgvP (4MM0),189 and CYP105P2 (5IT1),190 revealed that their heme groups also adopt this unusual flipped orientation (Fig. 8A). It remains unclear whether heme group orientation is the consequence of protein sequence or a distinct incorporation process during P450 folding, and how, if at all, it affects the structure or catalytic mechanism of the P450. The four P450s containing this unusual flipped heme group did not cluster together in the SSN at either the CYP subfamily or family level.
Fig. 8.
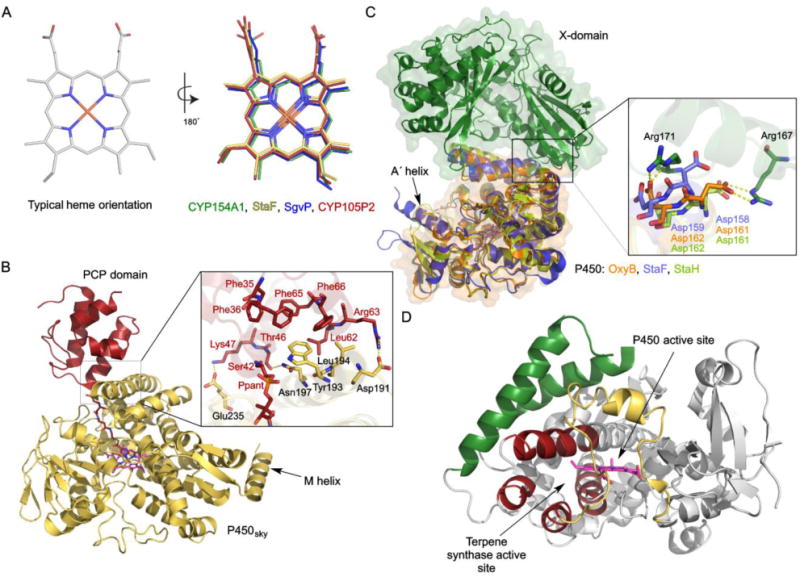
Selected variations in the structures of P450s from Streptomyces. (A) Typical heme orientations and inversed orientations in four different P450s (PDB IDs: 1ODO, 5EX8, 4MM0, 5IT1). (B) The crystal structure of P450sky in complex with an inhibitor-tethered PCP domain (PDB ID: 4PXH). The inset shows a zoomed-in look at the interface of P450sky and the PCP domain highlighting the hydrophobic and electrostatic interactions. The unusual C-terminal M-helix is labeled. (C) Structural superposition of StaF (PDB ID: 5EX8) and StaH (PDB ID: 5EX6) with OxyB in complex with the X-domain (PDB ID: 4TX3). The inset shows the two Asp residues from the conserved PRDD motif, which are proposed to be involved in recruitment by the X-domain via interaction with two Arg residues from the X-domain. (D) The crystal structure of CYP170A1 (PDB ID: 3EL3). The B–C loop and F–G region, colored in yellow and green, respectively, form the P450 active site. The four helices, colored in red, form the terpene synthase active site.
4.2.2 Carrier protein recognition
In some instances, P450s recognize substrates that are tethered to carrier proteins found in natural product biosynthetic assembly lines. P450sky catalyzes β-hydroxylation of three PCP-tethered amino acids in the biosynthesis of skyllamycin.76–78 The crystal structure of P450sky revealed a typical P450 fold (PDB ID: 4L0E, 4L0F, 4PWV, 4PXH).77,78 The complex structure of P450sky and an inhibitor-tethered PCP domain from module 7 revealed that the interfaces of P450sky and PCP form several hydrophobic interactions and a few electrostatics interactions (Fig. 8B).78 In contrast to other typical P450s, the structure of P450sky in the P450–PCP complex only shows differences to that of unbound P450sky in the interface of the F- and G-helices; the B–C loop of P450sky does not rearrange upon binding to the PCP. Furthermore, the structure of P450sky exhibits an unusual additional C-terminal M-helix (Fig. 8B), which may serve to mediate interactions with either the redox partner proteins of P450sky or other NRPS domains in close spatial proximity to the PCP domain (Fig. 8B).77 P450sky, which hydroxylates PCP-tethered amino acids in modules 5, 7, and 11 of the skyllamycin NRPS, did not show any interaction with either the PCP domain from module 10 of the skyllamycin NRPS or a heterologous ACP from E. coli,78 indicating that P450sky recognizes specific carrier proteins.
4.2.3 P450 recruitment by an NRPS X-domain
The teicoplanin-like GPA A47934 involves four P450s that not only accept PCP-tethered substrates, but also interact with the X-domain for P450 recruitment and cyclization efficiency.97,98,107 The biosynthetic gene cluster of A47934 possesses four genes encoding P450s, StaH, StaG, StaF and StaJ, which were demonstrated to participate in C–O–D, F–O–G, D–O–E, and A–B ring formations, respectively.99,106 The X-domain was demonstrated to recruit StaF and StaH to catalyze moderate levels of PCP-tethered peptide cyclization.106,107 The interaction between the P450 and X-domain was first identified based on the crystal structure of OxyB in complex with the X-domain in the teicoplanin system (PDB ID: 4TX3) (Fig. 8C).191 OxyB binds to the X-domain through interaction with the D-, E-, F- and G-helices. Two Asp residues in a PRDD motif, which is conserved in the F-helix within all GPA crosslinking P450s, contact two conserved Arg residues from the X-domain. The crystal structures of StaF (PDB ID: 5EX8) and StaH (PDB ID: 5EX6) were also recently solved, revealing typical cytochrome P450 structures and high structural homology with OxyB (RMSD of 0.7 Å and 1.2 Å, respectively).106,107 As in OxyB, the F helix PRDD motifs of StaF and StaH are proposed to be involved in recruitment by the X-domain (Fig. 8C). Furthermore, StaF, like its orthologue OxyA in teicoplanin biosynthesis, features a long A´ helix at the N-terminus, which is unique for D–O–E ring-catalyzing P450s in GPA biosynthesis (Fig. 8C).106,192 However, the function of this extra helix remains unclear.
4.2.4 Moonlighting active site
CYP170A1 is an extremely unique P450. Not only is it a multifunctional monooxygenase that catalyzes two sequential allylic oxidations in the biosynthesis of albaflavenone,145 it also has terpene synthase activity that converts FPP into a mixture of farnesene isomers. Within its normal prism-like P450 structure, CYP170A1 possesses a novel terpene synthase active site (PDB ID: 3EL3, 3DBG).146 This active site contains the Mg2+ binding motifs DDXXD and DTE that are typical of canonical terpene synthases. The terpene synthase active site is formed by a four helix α-helical barrel (helices C, H, I, and L), which is unusual compared to the six α-helices found in all other terpene synthases (Fig. 8D).146 This is the first functional P450 that possesses another non-P450 active site that is moonlighting on the basic P450 structure. It is unknown if the other members of the CYP170 family, those without the DDXXD and DTE motifs and thus the terpene synthase activity, have the same four α-helical barrel. CYP170A1 appears to be an example of evolution caught-in-action, supporting that structural elements, beyond the heme binding site, in the P450-fold are conducive for evolving into other functions.
4.3 Structure-based mechanistic studies
The functional versatility of P450s is clearly evident. However, except for the commonly accepted mechanism for hydroxylation, the catalytic mechanisms of many of the unusual functions of P450s remain unclear. Crystal structures from natural product P450 enzymes, particularly those in complex with their endogenous substrates, help to investigate and understand substrate binding modes, protein conformational changes, and enzyme catalytic mechanisms. Here we present some examples of P450 crystal structures from natural product biosynthetic pathways in Streptomyces that shed light on some of the uncommon P450 reactions.
4.3.1 Epoxidation via the hydroperoxyferric intermediate
PimD catalyzes epoxidation of the C-4–C-5 double bond in the polyene macrolide pimaricin.50,70,71 The crystal structure of PimD in complex with its substrate, 4,5-desepoxypimaricin (PDB ID: 2XBK), revealed that the π-orbitals of the C-4–C-5 double bond of 4,5-desepoxypimaricin point away from the iron at an angle of ~125° and are too distant for synchronous oxygen insertion by Cpd I (Fig. 9A).50 In fact, the bond angle and distance (3.7 Å) appear to be favorable for a typical abstraction and rebound hydroxylation mechanism, but the high dissociation energy of abstracting a hydrogen from a vinyl carbon likely precludes it (Fig. 9A).50 Alternatively, epoxidation by PimD may occur via a concerted substrate-assisted mechanism in which the hydroperoxoferric intermediate, Cpd 0, acts as the oxidant. The highly nucleophilic peroxoferric intermediate first abstracts a hydrogen from the C-7 hydroxyl of the substrate, then a concerted and cyclic six-electron rearrangement of Cpd 0 with the C-4–C-5 double bond inserts the distal oxygen atom yielding the epoxy product (Fig. 9A).50 Evidence for Cpd 0 oxidation for the PimD reaction mechanism was further confirmed as the formation of pimaricin was detected when incubated with hydrogen peroxide, but not with organic peroxides that do not form Cpd 0.50 If steric and/or electronic factors prevent interaction of Cpd I with the substrate, as in the case of PimD, the reactivity of other intermediates in the P450 catalytic cycle can be invoked to catalyze the desired reaction.
Fig. 9.
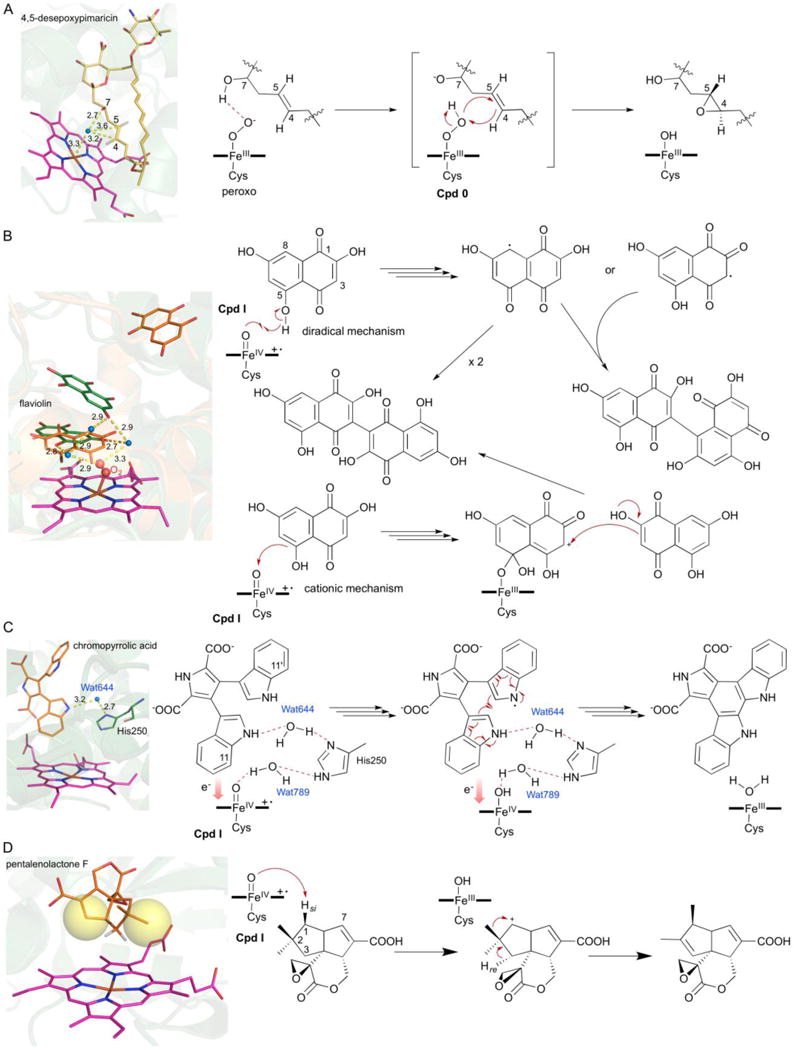
Structure-based mechanistic studies of P450s from Streptomyces. (A) Epoxidation by PimD via the hydroperoxyferric intermediate, Cpd 0 (PDB ID: 2XBK). (B) Substrate-assisted biaryl ring coupling by CYP158A1 and CYP158A2 (PDB IDs: 2NZ5 and 2D09). The two different binding modes of the two biflaviolin substrates are shown in orange and green. (C) Intramolecular biaryl ring coupling by StaP (PDB IDs: 2Z3U). Wat644 and His250 are shown in the active site; Wat789 is liberated during the formation of Cpd I (Wang 2009). (D) Oxidative rearrangement by PntM via a carbocation intermediate (PDB IDs: 5L1O). Blue dots in each figure represent water molecules; yellow spheres depict steric hindrance.
4.3.2 Substrate-assisted biaryl ring coupling
CYP158A2 catalyzes C–C bond formation to polymerize flaviolins.42,102,111,112 CYP158A2 is a substrate-assisted enzyme, requiring its substrate to contain a proton donor or acceptor to stabilize water molecules as it does not contain the highly conserved Thr in the I-helix. The crystal structure of CYP158A2 contains two molecules of flaviolin in the active site cavity (PDB ID: 1T93, 2D09) (Fig. 9B).102 The C-5 and C-7 hydroxyl groups of one molecule of flaviolin stabilize catalytically important water molecules that are involved in dioxygen activation (Fig. 9B).111 Both diradical and cationic mechanisms have been proposed for this reaction. In the diradical mechanism, initial hydrogen abstraction is postulated to occur from the C-5 hydroxyl group (Fig. 9B). The radicals localize at C-3 or C-8 and subsequently undergo C–C coupling reaction. In the cationic mechanism, an initial ipso attack at C-5 yields a flaviolin-iron covalent intermediate and generates a positive charge at C-3. The carbon cation is then nucleophilically attacked by C-3 of the adjacent flaviolin molecule to from the C–C bond (Fig. 9B).102,111
CYP158A1 shares high sequence identity and structure similarity to CYP158A2. CYP158A1 also catalyzes the biaryl ring coupling reaction between flaviolin molecules, albeit with a different preferred regioselectivity compared to that of CYP158A2.42 The major biflaviolin products of CYP158A1 and CYP158A2 are the C-3–C-3 and C-3–C-8 coupled products, respectively.42 As in the CYP158A2 structure, two flaviolin molecules were found in the crystal structure of CYP158A1 (PDB ID: 2NZ5).42 One flaviolin molecule is positioned over the heme group, similar to that found in CYP158A2; the second flaviolin molecule is bound at the entrance to the substrate access channel, which is too far from the first flaviolin for dimerization (Fig. 9B). Conformational changes were proposed to facilitate movement of the second flaviolin molecule towards the heme-adjacent flaviolin molecule.42 Although it is still unclear whether CYP158A1 and CYP158A2 utilize diradical or cationic mechanisms, the structures of these two P450s provide insights into the binding of multiple substrates in one P450 active site and reveals important mechanistic features applicable to P450 intermolecular biaryl ring coupling enzymes.
4.3.3 Intramolecular biaryl ring coupling
StaP catalyzes intramolecular C–C bond formation in the biosynthesis of staurosporine.108,109 The crystal structure of StaP in complex with its substrate CPA (PDB ID: 2Z3U) revealed that the two carboxyl groups of CPA are bound in the active site via several hydrogen bonds and electrostatics interactions. The two indole rings are held in place by intramolecular T-shaped π–π interactions (Fig. 9C).108 Combined with structural analysis, theoretical QM/MM calculations, along with mutagenesis studies, the catalytic mechanism of StaP was proposed.109 The substrate CPA loses two protons via assistance of a Wat–His–Wat triad and subsequently transfers two electrons to Cpd I (Fig. 9C). The C–C bond formation was formed during the second electron transfer.109 StaP dramatically lost enzymatic activity when incubated with a (C-11, C11´)-chloro derivative of CPA (CCA). The crystal structure of the StaP–CCA complex (PDB ID: 3A1L) revealed the absence of one of the water molecules in the Wat–His–Wat triad,109 supporting the essential role of the Wat–His–Wat triad for the StaP-catalyzed coupling reaction. Furthermore, the activities of two StaP mutants, the His of the Wat–His–Wat triad replaced by Ala and Phe, decreased to 25% and 72%, respectively, compared to that of wild-type StaP. These mutagenesis studies indicate that the His is not absolutely required for the proton relay pathway, and that a Wat–Wat diad, in absence of the His, may adequately relay protons for the StaP reaction.109
4.3.4 Oxidative rearrangement via a carbocation intermediate
PntM catalyzes a unique oxidative rearrangement in pentalenolactone biosynthesis.113,114 The crystal structure of substrate-free PntM (PDB ID: 5L10) and in complex with various ligands (PDB ID: 5L1P, 5L1Q, 5L1R, 5L1S, 5L1T, 5L1U, 5L1V), including its substrate (pentalenolactone F), product (pentalenolactone), and substrate analogue (6,7-dihydropentalenolactone F), revealed that the three residues, Phe232, Met77, and Met81, which bind to the ligands, are unique in PntM and its orthologues.114 The crystal structures support that C-1 of pentalenolactone F is sufficiently sterically hindered by its own axial C-2si-methyl and the C-7-vinylidene substituents, that oxygen rebound does not occur after initial radical formation (Fig. 9D). Instead, electron transfer outcompetes the reduced oxygen rebound rate, resulting in carbocation formation at C-1. Subsequent C-2 methyl migration and C-3 deprotonation form pentalenolactone (Fig. 9D). The unusual carbocation intermediate is proposed to result from an outer shell electron transfer from the transiently generated C-1 radical to the heme-FeIII–OH radical species.114
4.4 Structure-based engineering of P450s
P450s are well regarded as versatile biocatalysts that activate C–H bonds and perform a vast variety of chemistries. Crystal structures of P450s reveal insights into the shapes and sizes of active site cavities, how substrates bind and are oriented inside these typically hydrophobic pockets, and what controls regio- and stereoselectivity. Using details gleaned from these studies, structure-based enzymes or substrate engineering can be and have been carried out to rationally augment P450s as biocatalysts and generate novel natural product analogues.
4.4.1 Functional switch
AurH is a multifunctional enzyme involved in the biosynthesis of the polyketide antibiotic aureothin.115–118 AurH catalyzes a tandem oxygenation of C-7 and C-9a resulting in tetrahydrofuran formation. The crystal structure of AurH is the first structure of the CYP151A subfamily (PDB ID: 3P3L, 3P3O, 3P3X, 3P3Z),117 a relatively small group of P450s. A protein-ligand docking model revealed that the pyrone ring of the substrate occupies a hydrophobic pocket built by Phe85, Phe89, Leu175, Val242, Ala243, and Thr239.117 The Phe89Trp and Thr239Phe mutants significantly decreased enzymatic performance and transformed the substrate into new aureothin derivatives (Fig. 10A).117 The bulky Trp and Phe residues alter the binding mode of the pyrone entity via steric effects. Both engineered mutants catalyze the regioselective six-electron transfer of the nonactivated C-9a methyl group into a carboxylic acid via hydroxyl and aldehyde intermediates. New aureothin derivatives were isolated by fermentation of the aurH (Phe89Trp)- and aurH (Thr239Phe)-complemented ΔaurH variant strains, respectively.117 This work provides new insights into the finesse of enzyme-mediated oxygenation reactions, but also sets the stage for enzyme engineering in the field of synthetically useful biotransformations.
Fig. 10.
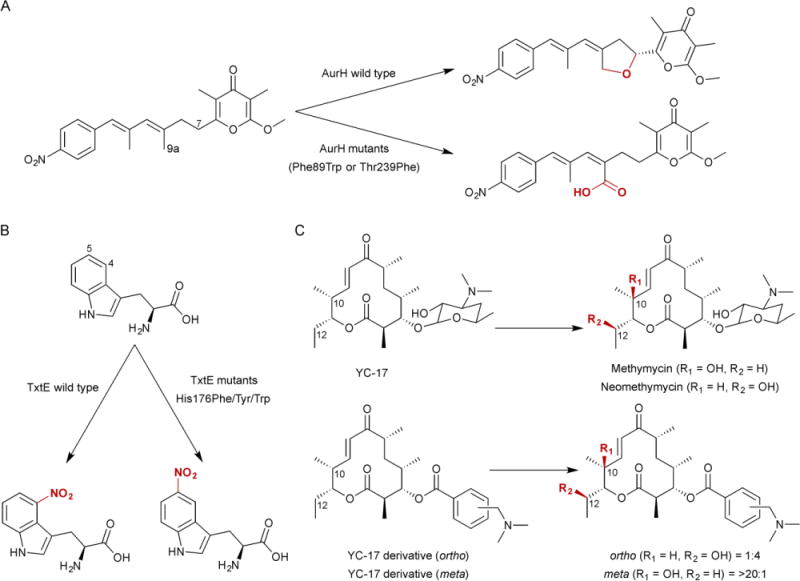
Structure-based engineering of P450s in Streptomyces. (A) Functional switch, from hydroxylation and ether formation to hydroxylation and oxidation to carboxylic acid, of AurH via site-directed mutagenesis. (B) Nitration regioselectivity switch, from C-4 to C-5, of TxtE via site-directed mutagenesis. (C) Substrate engineering of PikC shifting the natural abundance of regioisomers. The ratios representing the C-10:C-12 hydroxylated products changes from 1:1 with the natural desosamine anchoring group to 1:4 or >20:1 depending on the synthetic anchoring variant used.
4.4.2 Regioselectivity switch
TxtE was the first enzyme reported to efficiently catalyze regioselective nitration of L-tryptophan.122 The crystal structures of the substrate-free TxtE and in complex with L-tryptophan were reported (PDB ID: 4L36, 4TPN, 4TPO);123,124 however, information about the interactions between L-tryptophan and the F–G loop was missing due to the disordered nature of the F–G loop. The F–G loop of TxtE was rebuilt via computing using Morkov state model molecular dynamics (MD) simulations, and three enzyme states, open lid, closed lid and transition, were proposed.125 His176, from the F–G loop, plays a role in controlling the enzyme state (open, closed, and transition states) and forms edge-to-face π–π interactions with the substrate. Interestingly, His176Phe, His176Tyr, and His176Trp variants all resulted in nitration exclusively at the C-5 position instead of the C-4 position (Fig. 10B).125 Computational models revealed that the increased steric demand of Phe/Tyr/Trp at position 176 shifts the substrate alignment relative to the heme group, placing the C-5 of indole closest to the nitrogen of the ferric peroxynitrite. This enzyme engineering shows a rare example of how a single residue in the P450 F–G loop can interact directly with the substrate to contribute to active-site organization and control regiochemistry.125
4.4.3 Substrate engineering
PikC performs multiple hydroxylation reactions on structurally diverse macrolides. The key to this significant substrate tolerance is the presence of the deoxyamino sugar desosamine.186 The crystal structures of PikC in complex with its substrates, YC-17 and narbomycin (PDB ID: 2C6H, 2C7X), showed the desosamine moiety of the substrates is bound in two distinct binding pockets, one buried and the other surface-exposed.186 Elimination of the surface-exposed negative charge at Asp50 results in a significant improvement in catalytic activity.193 In the crystal structure of the PikC mutant Asp50Asn, the desosamine moiety of both YC-17 and narbomycin was bound in a catalytically productive “buried site” (PDB ID: 2WHW, 2WI9).194 A two-step substrate binding mechanism was proposed with desosamine recognition in the two subsites to allow the macrolide substrate to sequentially progress toward a catalytically favorable orientation.194 By harnessing its unique desosamine-anchoring functionality via a “substrate engineering” strategy, PikC is able to hydroxylate a series of carbocyclic rings linked to the desosamine glycoside via an acetal linkage in a regioselective manner.195 Furthermore, the size, stereochemistry, and rigidity of the anchoring group influence the regioselectivity of enzymatic hydroxylation.195 The natural anchoring group desosamine affords a 1:1 mixture of regioisomers, while synthetic anchors shift the ratio of C-10/C-12 hydroxylation of the YC-17 analogue from 1:4 to 20:1 (Fig. 10C).195 Substrate engineering has provided new insights into the structural parameters that govern productive substrate binding and conversion of potential P450 substrates.
5 Conclusions and future perspectives
Genomics has changed the way scientists think about and study enzymes. Traditional enzymology begins with the observation of a phenomenon, and leads to the identification of the responsible protein followed by the encoding gene. With the advent and widespread use of DNA and whole genome sequencing, genes and their encoding proteins are known before any biochemistry is attempted. This leads to unlimited opportunities to discover new transformations, investigate enzyme mechanisms, and find patterns in sequence-structure-function relationships. However, with the rate of sequencing easily outpacing enzyme characterization, large-scale analysis and prioritization is needed to efficiently and effectively select the most promising candidates for further study. Analogous to strain prioritization for natural product discovery,196–199 SSNs can be utilized to facilitate enzyme differentiation and prioritization.161,200,201
Cytochromes P450 are one of most exquisite and versatile biocatalysts found in nature. Although most of the research in P450 enzymology has focused on their direct relevance to human health through steroid biosynthesis and drug metabolism, P450s are key players in the biosynthesis of biologically active natural products. Due to their almost ubiquitous nature in secondary metabolite biosynthetic pathways, P450s possess tremendous diversity in sequence, substrate preference, and function. The P450s found within one of the most prolific producers of natural products, the Streptomyces genus (previously termed the CYPome31), and the SSNs described in this review, effectively confirms this premise. With only ~2.4% (even less with known physiological functions) and <0.4% of streptomycete P450s functionally and structurally characterized, respectively, it is clear that the true chemical and biological capabilities of these remarkable enzymes, in Streptomyces and all organisms, is not fully understood.
It is universally accepted that protein functions are directly related to their primary amino acid sequences and structures. The diversity and versatility, both in function and substrate preference, of P450s makes it extremely difficult to find a generalized rule-of-thumb for sequence–structure–function relationships. While the community does not currently have the capacity to correctly predict function or structure solely based on P450 sequence, efforts are continuing to understand these connections. Structural-based engineering and directed evolution of P450 enzymes for biotechnology applications have succeeded, but only on a case-by-case basis. Expanding the reaction space of microbial P450s, either through engineering or natural P450 variant discovery, will continue to improve the understanding and predictive power of sequence–structure–function relationships of P450s.
Natural P450 variants possess an immense capacity for chemical reactions and will dramatically advance the field of P450 enzymology, chemistry, and biotechnological applications. The discovery of natural TxtE variants that selectivity nitrate the C-5 position of tryptophan, after engineering a TxtE mutant to have the exact same regioselectivity,125 perfectly highlights this idea. Nature, with its use of evolution and consequential diversity, has already developed many of the reactions and selectivities that scientists desire, and even ones that are still unimaginable. P450s are already used as an alternative to synthetic approaches to C–H functionalization. Discovery of new P450s, repurposing of known P450s, and taking inspiration from the natural product biosynthetic strategy of late-stage functionalization using regio- and stereoselective P450s will advance the field of biocatalysis and become a valuable technology.
Nature possesses the greatest diversity of chemical space. Enzymes are the natural catalysts that both biosynthesize and modify these complex scaffolds. New enzyme functions and selectivities found within natural product biosynthesis translate into novel natural products with unique chemical and biological characteristics. The prevalence of P450s to functionalize natural products, combined with the total number of uncharacterized enzymes, reinforces the idea of the vastness of natural product diversity that has yet to be discovered. Given the striking diversity of sequence and function that are found in P450s, the tremendous amount of available genomics data, and a precedence of P450s to catalyze unique or multifunctional transformations in natural product biosynthetic pathways, study of streptomycete P450s will undoubtedly continue to make fundamental contributions to enzymology, structural biology, natural products chemistry and biosynthesis.
Supplementary Material
Acknowledgments
Studies on natural product biosynthesis, engineering, and drug discovery in the Shen laboratory are currently supported in part by National Institutes of Health grants CA078747, CA106150, GM114353, GM115575, and CA204484. JDR and CYC are supported in part by an Arnold O. Beckman Postdoctoral Fellowship and the Fellowship of Academia Sinica–The Scripps Research Institute Postdoctoral Talent Development Program, respectively.
Footnotes
Conflicts of Interest
There are no conflicts of interest to declare.
References
- 1.Guengerich FP. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 523–785. [Google Scholar]
- 2.Auchus RJ, Miller WL. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 851–879. [Google Scholar]
- 3.Podust LM, Sherman DH. Nat Prod Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLean KJ, Leys D, Munro AW. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 261–407. [Google Scholar]
- 5.Schuler MA. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 409–449. [Google Scholar]
- 6.Klingenberg M. Arch Biochem Biophys. 1958;75:376–386. doi: 10.1016/0003-9861(58)90436-3. [DOI] [PubMed] [Google Scholar]
- 7.Omura T, Sato R. J Biol Chem. 1962;237:1375–1376. [PubMed] [Google Scholar]
- 8.Cryle MJ, Stok JE, De Voss JJ. Aust J Chem. 2003;56:749–762. [Google Scholar]
- 9.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 10.Munro AW, Girvan HM, McLean KJ. Nat Prod Rep. 2007;24:585–609. doi: 10.1039/b604190f. [DOI] [PubMed] [Google Scholar]
- 11.Guengerich FP, Tang Z, Salamanca-Pinzon SG, Cheng Q. Mol Interv. 2010;10:153–163. doi: 10.1124/mi.10.3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortiz de Montellano PR. Chem Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasan R. ACS Cat. 2012;2:647–666. [Google Scholar]
- 14.Kelly SL, Kelly DE. Philos Trans R Soc, B: Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0476. 20120476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb DC, Waterman MR, Zhao B. Exp Opin Drug Metabol Toxicol. 2013;9:1279–1294. doi: 10.1517/17425255.2013.806485. [DOI] [PubMed] [Google Scholar]
- 16.Lamb DC, Waterman MR. Philos Trans R Soc, B: Biol Sci. 2013;368:20120434. doi: 10.1098/rstb.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. Trends Biochem Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Cochrane RVK, Vederas JC. Acc Chem Res. 2014;47:3148–3161. doi: 10.1021/ar500242c. [DOI] [PubMed] [Google Scholar]
- 19.Girvan HM, Munro AW. Curr Opin Chem Biol. 2016;31:136–145. doi: 10.1016/j.cbpa.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Denisov IG, Sligar SG. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 69–109. [Google Scholar]
- 21.Guengerich FP. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz De Montellano PR, De Voss JJ. Nat Prod Rep. 2002;19:477–493. doi: 10.1039/b101297p. [DOI] [PubMed] [Google Scholar]
- 23.Guengerich FP, Munro AW. J Biol Chem. 2013;288:17065–17073. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guengerich FP. J Biochem Mol Toxicol. 2007;21:163–168. doi: 10.1002/jbt.20174. [DOI] [PubMed] [Google Scholar]
- 25.Waskell L, Kim J-JP. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 33–68. [Google Scholar]
- 26.Narhi LO, Fulco AJ. J Biol Chem. 1982;257:2147–2150. [PubMed] [Google Scholar]
- 27.Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TWB, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, Page CC, Dutton PL. Trends Biochem Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 28.Roberts GA, Grogan G, Greter A, Flitsch SL, Turner NJ. J Bacteriol. 2002;184:3898–3908. doi: 10.1128/JB.184.14.3898-3908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi KY, Jung EO, Jung DH, Pandey BP, Yun H, Park HY, Kazlauskas RJ, Kim BG. FEBS J. 2012;279:1650–1662. doi: 10.1111/j.1742-4658.2011.08462.x. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Q, Lamb DC, Kelly SL, Lei L, Guengerich FP. J Am Chem Soc. 2010;132:15173–15175. doi: 10.1021/ja107801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb DC, Skaug T, Song HL, Jackson CJ, Podust LM, Waterman MR, Kell DB, Kelly DE, Kelly SL. J Biol Chem. 2002;277:24000–24005. doi: 10.1074/jbc.M111109200. [DOI] [PubMed] [Google Scholar]
- 32.Lei L, Waterman MR, Fulco AJ, Kelly SL, Lamb DC. Proc Natl Acad Sci USA. 2004;101:494–499. doi: 10.1073/pnas.2435922100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun YJ, Shimada T, Waterman MR, Guengerich FP. Biochem Soc Trans. 2006;34:1183–1185. doi: 10.1042/BST0341183. [DOI] [PubMed] [Google Scholar]
- 34.Chun YJ, Shimada T, Sanchez-Ponce R, Martin MV, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC, Guengerich FP. J Biol Chem. 2007;282:17486–17500. doi: 10.1074/jbc.M700863200. [DOI] [PubMed] [Google Scholar]
- 35.Rimal H, Lee SW, Lee JH, Oh TJ. Arch Biochem Biophys. 2015;585:64–74. doi: 10.1016/j.abb.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Hussain HA, Ward JM. Appl Environ Microbiol. 2003;69:373–382. doi: 10.1128/AEM.69.1.373-382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain HA, Ward JM. Enz Microb Technol. 2003;32:790–800. [Google Scholar]
- 38.Wang W, Wang FQ, Wei DZ. Appl Environ Microbiol. 2009;75:4202–4205. doi: 10.1128/AEM.02606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson JA, Lorence MC, Amarneh B. J Biol Chem. 1990;265:6066–6073. [PubMed] [Google Scholar]
- 40.Niraula NP, Bhattarai S, Lee NR, Sohng JK, Oh TJ. J Microbiol Biotechnol. 2012;22:1059–1065. doi: 10.4014/jmb.1201.01037. [DOI] [PubMed] [Google Scholar]
- 41.Taylor M, Lamb DC, Cannell R, Dawson M, Kelly SL. Biochem Biophys Res Commun. 1999;263:838–842. doi: 10.1006/bbrc.1999.1427. [DOI] [PubMed] [Google Scholar]
- 42.Zhao B, Lamb DC, Lei L, Kelly SL, Yuan H, Hachey DL, Waterman MR. Biochemistry. 2007;46:8725–8733. doi: 10.1021/bi7006959. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka T, Miyakoshi S, Tanzawa K, Nakahara K, Hosobuchi M, Serizawa N. Eur J Biochem. 1989;184:707–713. doi: 10.1111/j.1432-1033.1989.tb15070.x. [DOI] [PubMed] [Google Scholar]
- 44.Jungmann V, Molnar I, Hammer PE, Hill DS, Zirkle R, Buckel TG, Buckel D, Ligon JM, Pachlatko JP. Appl Environ Microbiol. 2005;71:6968–6976. doi: 10.1128/AEM.71.11.6968-6976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makino T, Katsuyama Y, Otomatsu T, Misawa N, Ohnishi Y. Appl Environ Microbiol. 2014;80:1371–1379. doi: 10.1128/AEM.03504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, Zhang W, Chen Y, Yuan C, Zhang H, Zhang G, Ma L, Zhang Q, Tian X, Zhang S, Zhang C. ChemBioChem. 2015;16:2086–2093. doi: 10.1002/cbic.201500281. [DOI] [PubMed] [Google Scholar]
- 47.Narayan ARH, Jimenez-Oses G, Liu P, Negretti S, Zhao W, Gilbert MM, Ramabhadran RO, Yang YF, Furan LR, Li Z, Podust LM, Montgomery J, Houk KN, Sherman DH. Nature Chem. 2015;7:653–660. doi: 10.1038/nchem.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi KY, Jung E, Jung DH, An BR, Pandey BP, Yun H, Sung C, Park HY, Kim BG. Microb Cell Fact. 2012;11:81. doi: 10.1186/1475-2859-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berrie JR, Williams RAD, Smith KE. Biochem Soc Trans. 1997;25:18S. doi: 10.1042/bst025018s. [DOI] [PubMed] [Google Scholar]
- 50.Kells PM, Ouellet H, Santos-Aberturas J, Aparicio JF, Podust LM. Chem Biol. 2010;17:841–851. doi: 10.1016/j.chembiol.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman DJ, Cragg GM. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 52.Chen W, Lee MK, Jefcoate C, Kim SC, Chen F, Yu JH. Genome Biol Evol. 2014;6:1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Biochem Biophys Res Commun. 2003;307:610–619. doi: 10.1016/s0006-291x(03)01231-2. [DOI] [PubMed] [Google Scholar]
- 54.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 56.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nature Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 57.Nelson DR. Human Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nett M, Ikeda H, Moore BS. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh CT, Fischbach MA. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crabtree RH. Chem Rev. 2010;110:575. doi: 10.1021/cr900388d. [DOI] [PubMed] [Google Scholar]
- 61.Lewis JC, Coelho PS, Arnold FH. Chem Soc Rev. 2011;40:2003–2021. doi: 10.1039/c0cs00067a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davies HML, Morton D. J Org Chem. 2016;81:343–350. doi: 10.1021/acs.joc.5b02818. [DOI] [PubMed] [Google Scholar]
- 63.Girhard M, Bakkes PJ, Mahmoud O, Urlacher VB. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 451–520. [Google Scholar]
- 64.Sakaki T. Biol Pharm Bull. 2012;35:844–849. doi: 10.1248/bpb.35.844. [DOI] [PubMed] [Google Scholar]
- 65.Graziani EI, Cane DE, Betlach MC, Kealey JT, McDaniel R. Bioorg Med Chem Lett. 1998;8:3117–3120. doi: 10.1016/s0960-894x(98)00553-8. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Q, Sherman DH. J Nat Prod. 2001;64:1447–1450. doi: 10.1021/np010146r. [DOI] [PubMed] [Google Scholar]
- 67.Betlach MC, Kealey JT, Betlach MC, Ashley GW, McDaniel R. Biochemistry. 1998;37:14937–14942. doi: 10.1021/bi981699c. [DOI] [PubMed] [Google Scholar]
- 68.Xue Y, Wilson D, Zhao L, Liu HW, Sherman DH. Chem Biol. 1998;5:661–667. doi: 10.1016/s1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 69.Lee SK, Park JW, Kim JW, Jung WS, Park SR, Choi CY, Kim ES, Kim BS, Ahn JS, Sherman DH, Yoon YJ. J Nat Prod. 2006;69:847–849. doi: 10.1021/np060026p. [DOI] [PubMed] [Google Scholar]
- 70.Mendes MV, Recio E, Fouces R, Luiten R, Martin JF, Aparicio JF. Chem Biol. 2001;8:635–644. doi: 10.1016/s1074-5521(01)00033-3. [DOI] [PubMed] [Google Scholar]
- 71.Mendes MV, Anton N, Martin JF, Aparicio JF. Biochem J. 2005;386:57–62. doi: 10.1042/BJ20040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaisser S, Lill R, Staunton J, Mendez C, Salas J, Leadlay PF. Mol Microbiol. 2002;44:771–781. doi: 10.1046/j.1365-2958.2002.02910.x. [DOI] [PubMed] [Google Scholar]
- 73.Montemiglio LC, Parisi G, Scaglione A, Sciara G, Savino C, Vallone B. Biochim Biophys Acta. 2016;1860:465–475. doi: 10.1016/j.bbagen.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 74.Ding W, Deng W, Tang MC, Zhang Q, Tang GL, Bi YR, Liu W. Mol BioSyst. 2010;6:1071–1081. doi: 10.1039/b926358f. [DOI] [PubMed] [Google Scholar]
- 75.Ortiz de Montellano PR. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 111–176. [Google Scholar]
- 76.Pohle S, Appelt C, Roux M, Fiedler HP, Suessmuth RD. J Am Chem Soc. 2011;133:6194–6205. doi: 10.1021/ja108971p. [DOI] [PubMed] [Google Scholar]
- 77.Uhlmann S, Suessmuth RD, Cryle MJ. ACS Chem Biol. 2013;8:2586–2596. doi: 10.1021/cb400555e. [DOI] [PubMed] [Google Scholar]
- 78.Haslinger K, Brieke C, Uhlmann S, Sieverling L, Suessmuth RD, Cryle MJ. Angew Chem, Int Ed. 2014;53:8518–8522. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 79.Huttel W, Spencer JB, Leadlay PF. Beilstein J Org Chem. 2014;10:361–368. doi: 10.3762/bjoc.10.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tao W, Yurkovich ME, Wen S, Lebe KE, Samborskyy M, Liu Y, Yang A, Ju Y, Deng Z, Tosin M, Sun Y, Leadlay PF. Chem Sci. 2016;7:376–385. doi: 10.1039/c5sc03059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cryle MJ. Biochem Soc Trans. 2010;38:934–939. doi: 10.1042/BST0380934. [DOI] [PubMed] [Google Scholar]
- 82.Duerr C, Schnell HJ, Luzhetskyy A, Murillo R, Weber M, Welzel K, Vente A, Bechthold A. Chem Biol. 2006;13:365–377. doi: 10.1016/j.chembiol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 83.Daum M, Schnell HJ, Herrmann S, Guenther A, Murillo R, Mueller R, Bisel P, Mueller M, Bechthold A. ChemBioChem. 2010;11:1383–1391. doi: 10.1002/cbic.201000117. [DOI] [PubMed] [Google Scholar]
- 84.Carlson JC, Fortman JL, Anzai Y, Li S, Burr DA, Sherman DH. ChemBioChem. 2010;11:564–572. doi: 10.1002/cbic.200900658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlson JC, Li SY, Gunatilleke SS, Anzai Y, Burr DA, Podust LM, Sherman DH. Nature Chem. 2011;3:628–633. doi: 10.1038/nchem.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dickens ML, Priestley ND, Strohl WR. J Bacteriol. 1997;179:2641–2650. doi: 10.1128/jb.179.8.2641-2650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morrone D, Chen X, Coates RM, Peters RJ. Biochem J. 2010;431:337–344. doi: 10.1042/BJ20100597. [DOI] [PubMed] [Google Scholar]
- 88.Guengerich FP, Sohl CD, Chowdhury G. Arch Biochem Biophys. 2011;507:126–134. doi: 10.1016/j.abb.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Q, Li H, Li S, Zhu Y, Zhang G, Zhang H, Zhang W, Shi R, Zhang C. Org Lett. 2012;14:6142–6145. doi: 10.1021/ol302782u. [DOI] [PubMed] [Google Scholar]
- 90.Olano C, Moss SJ, Brana AF, Sheridan RM, Math V, Weston AJ, Mendez C, Leadlay PF, Wilkinson B, Salas JA. Mol Microbiol. 2004;52:1745–1756. doi: 10.1111/j.1365-2958.2004.04090.x. [DOI] [PubMed] [Google Scholar]
- 91.Seo JW, Ma M, Kwong T, Ju J, Lim SK, Jiang H, Lohman JR, Yang C, Cleveland J, Zazopoulos E, Farnet CM, Shen B. Biochemistry. 2014;53:7854–7865. doi: 10.1021/bi501396v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shin JC, Na Z, Lee D, Kim W, Lee K, Shen YM, Paik SG, Hong YS, Lee JJ. J Microbiol Biotechnol. 2008;18:1101–1108. [PubMed] [Google Scholar]
- 93.Lin L, Ni SY, Wu LZ, Wang YG, Wang YC, Tao PZ, He WQ, Wang XJ. Biosci, Biotechnol, Biochem. 2011;75:2042–2045. doi: 10.1271/bbb.110361. [DOI] [PubMed] [Google Scholar]
- 94.Li T, Ni S, Jia C, Wang H, Sun G, Wu L, Gan M, Shan G, He W, Lin L, Zhou H, Wang Y. J Nat Prod. 2012;75:1480–1484. doi: 10.1021/np3001738. [DOI] [PubMed] [Google Scholar]
- 95.Ma J, Wang Z, Huang H, Luo M, Zuo D, Wang B, Sun A, Cheng YQ, Zhang C, Ju J. Angew Chem, Int Ed. 2011;50:7797–7802. doi: 10.1002/anie.201102305. [DOI] [PubMed] [Google Scholar]
- 96.Praeg A, Gruening BA, Haeckh M, Luedeke S, Wilde M, Luzhetskyy A, Richter M, Luzhetska M, Guenther S, Mueller M. J Am Chem Soc. 2014;136:6195–6198. doi: 10.1021/ja501630w. [DOI] [PubMed] [Google Scholar]
- 97.Bischoff D, Pelzer S, Holtzel A, Nicholson GJ, Stockert S, Wohlleben W, Jung G, Sussmuth RD. Angew Chem, Int Ed. 2001;40:1693–1696. doi: 10.1002/1521-3773(20010504)40:9<1693::aid-anie16930>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 98.Zerbe K, Woithe K, Li DB, Vitali F, Bigler L, Robinson JA. Angew Chem, Int Ed. 2004;43:6709–6713. doi: 10.1002/anie.200461278. [DOI] [PubMed] [Google Scholar]
- 99.Hadatsch B, Butz D, Schmiederer T, Steudle J, Wohlleben W, Suessmuth R, Stegmann E. Chem Biol. 2007;14:1078–1089. doi: 10.1016/j.chembiol.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 100.Onaka H, Taniguchi SI, Igarashi Y, Furumai T. J Antibiot. 2002;55:1063–1071. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- 101.Howard-Jones AR, Walsh CT. J Am Chem Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 102.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 103.Funa N, Funabashi M, Ohnishi Y, Horinouchi S. J Bacteriol. 2005;187:8149–8155. doi: 10.1128/JB.187.23.8149-8155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicolaou KC, Boddy CNC, Brase S, Winssinger N. Angew Chem, Int Ed. 1999;38:2096–2152. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 105.Boeck LD, Mertz FP. J Antibiot. 1986;39:1533–1540. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- 106.Ulrich V, Brieke C, Cryle MJ. Beilstein J Org Chem. 2016;12:2849–2864. doi: 10.3762/bjoc.12.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ulrich V, Peschke M, Brieke C, Cryle MJ. Mol BioSyst. 2016;12:2992–3004. doi: 10.1039/c6mb00373g. [DOI] [PubMed] [Google Scholar]
- 108.Makino M, Sugimoto H, Shiro Y, Asamizu S, Onaka H, Nagano S. Proc Natl Acad Sci USA. 2007;104:11591–11596. doi: 10.1073/pnas.0702946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Chen H, Makino M, Shiro Y, Nagano S, Asamizu S, Onaka H, Shaik S. J Am Chem Soc. 2009;131:6748–6762. doi: 10.1021/ja9003365. [DOI] [PubMed] [Google Scholar]
- 110.Goldman PJ, Ryan KS, Hamill MJ, Howard-Jones AR, Walsh CT, Elliott SJ, Drennan CL. Chem Biol. 2012;19:855–865. doi: 10.1016/j.chembiol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao B, Guengerich FP, Voehler M, Waterman MR. J Biol Chem. 2005;280:42188–42197. doi: 10.1074/jbc.M509220200. [DOI] [PubMed] [Google Scholar]
- 112.Zhao B, Bellamine A, Lei L, Waterman MR. Arch Biochem Biophys. 2012;518:127–132. doi: 10.1016/j.abb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu DQ, Seo MJ, Ikeda H, Cane DE. J Am Chem Soc. 2011;133:2128–2131. doi: 10.1021/ja111279h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Duan L, Jogl G, Cane DE. J Am Chem Soc. 2016;138:12678–12689. doi: 10.1021/jacs.6b08610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.He J, Mueller M, Hertweck C. J Am Chem Soc. 2004;126:16742–16743. doi: 10.1021/ja046104h. [DOI] [PubMed] [Google Scholar]
- 116.Richter MEA, Traitcheva N, Knuepfer U, Hertweck C. Angew Chem, Int Ed. 2008;47:8872–8875. doi: 10.1002/anie.200803714. [DOI] [PubMed] [Google Scholar]
- 117.Zocher G, Richter MEA, Mueller U, Hertweck C. J Am Chem Soc. 2011;133:2292–2302. doi: 10.1021/ja110146z. [DOI] [PubMed] [Google Scholar]
- 118.Richter M, Busch B, Ishida K, Moore BS, Hertweck C. ChemBioChem. 2012;13:2196–2199. doi: 10.1002/cbic.201200406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pang CH, Matsuzaki K, Ikeda H, Tanaka H, Omura S. J Antibiot. 1995;48:59–66. doi: 10.7164/antibiotics.48.59. [DOI] [PubMed] [Google Scholar]
- 120.Rudolf JD, Dong LB, Manoogian K, Shen B. J Am Chem Soc. 2016;138:16711–16721. doi: 10.1021/jacs.6b09818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roncone R, Barbieri M, Monzani E, Casella L. Coord Chem Rev. 2006;250:1286–1293. [Google Scholar]
- 122.Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, Challis GL. Nature Chem Biol. 2012;8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu F, Li M, Xu C, Wang Z, Zhou H, Yang M, Chen Y, Tang L, He J. PloS one. 2013;8:e81526. doi: 10.1371/journal.pone.0081526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dodani SC, Cahn JKB, Heinisch T, Brinkmann-Chen S, McIntosh JA, Arnold FH. ChemBioChem. 2014;15:2259–2267. doi: 10.1002/cbic.201402241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dodani SC, Kiss G, Cahn JKB, Su Y, Pande VS, Arnold FH. Nature Chem. 2016;8:419–425. doi: 10.1038/nchem.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang H, Chen J, Wang H, Xie Y, Ju J, Yan Y, Zhang H. FEBS Lett. 2013;587:1675–1680. doi: 10.1016/j.febslet.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 127.Salas AP, Zhu L, Sanchez C, Brana AF, Rohr J, Mendez C, Salas JA. Mol Microbiol. 2005;58:17–27. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Onaka H, Asamizu S, Igarashi Y, Yoshida R, Furumai T. Biosci, Biotechnol, Biochem. 2005;69:1753–1759. doi: 10.1271/bbb.69.1753. [DOI] [PubMed] [Google Scholar]
- 129.Xie Y, Wang B, Liu J, Zhou J, Ma J, Huang H, Ju J. ChemBioChem. 2012;13:2745–2757. doi: 10.1002/cbic.201200584. [DOI] [PubMed] [Google Scholar]
- 130.Xie Y, Li Q, Song Y, Ma J, Ju J. ChemBioChem. 2014;15:1183–1189. doi: 10.1002/cbic.201400062. [DOI] [PubMed] [Google Scholar]
- 131.Agematu H, Matsumoto N, Fujii Y, Kabumoto H, Doi S, Machida K, Ishikawa J, Arisawa A. Biosci, Biotechnol, Biochem. 2006;70:307–311. doi: 10.1271/bbb.70.307. [DOI] [PubMed] [Google Scholar]
- 132.Molnar I, Hill DS, Zirkle R, Hammer PE, Gross F, Buckel TG, Jungmann V, Pachlatko JP, Ligon JM. Appl Environ Microbiol. 2005;71:6977–6985. doi: 10.1128/AEM.71.11.6977-6985.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang X, Liu W, Ji Y, Niu J, Li M. Appl Microbiol Biotechnol. 2012;93:1957–1963. doi: 10.1007/s00253-011-3490-1. [DOI] [PubMed] [Google Scholar]
- 134.Sawada N, Sakaki T, Yoneda S, Kusudo T, Shinkyo R, Ohta M, Inouye K. Biochem Biophys Res Commun. 2004;320:156–164. doi: 10.1016/j.bbrc.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 135.Sugimoto H, Shinkyo R, Hayashi K, Yoneda S, Yamada M, Kamakura M, Ikushiro S-i, Shiro Y, Sakaki T. Biochemistry. 2008;47:4017–4027. doi: 10.1021/bi7023767. [DOI] [PubMed] [Google Scholar]
- 136.Hayashi K, Sugimoto H, Shinkyo R, Yamada M, Ikeda S, Ikushiro S, Kamakura M, Shiro Y, Sakaki T. Biochemistry. 2008;47:11964–11972. doi: 10.1021/bi801222d. [DOI] [PubMed] [Google Scholar]
- 137.O’Keefe DP, Tepperman JM, Dean C, Leto KJ, Erbes DL, Odell JT. Plant Physiol. 1994;105:473–482. doi: 10.1104/pp.105.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Braatz JA, Bass MB, Ornstein RL. J Comput-Aided Mol Des. 1994;8:607–622. doi: 10.1007/BF00123668. [DOI] [PubMed] [Google Scholar]
- 139.Janocha S, Zapp J, Hutter M, Kleser M, Bohlmann J, Bernhardt R. ChemBioChem. 2013;14:467–473. doi: 10.1002/cbic.201200729. [DOI] [PubMed] [Google Scholar]
- 140.Nagano N, Orengo CA, Thornton JM. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 141.Borisova SA, Zhao L, Melancon CE, III, Kao C-L, Liu H-w. J Am Chem Soc. 2004;126:6534–6535. doi: 10.1021/ja049967j. [DOI] [PubMed] [Google Scholar]
- 142.Borisova SA, Zhang C, Takahashi H, Zhang H, Wong AW, Thorson JS, Liu H-w. Angew Chem, Int Ed. 2006;45:2748–2753. doi: 10.1002/anie.200503195. [DOI] [PubMed] [Google Scholar]
- 143.Borisova SA, Liu H-w. Biochemistry. 2010;49:8071–8084. doi: 10.1021/bi1007657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moncrieffe MC, Fernandez MJ, Spiteller D, Matsumura H, Gay NJ, Luisi BF, Leadlay PF. J Mol Biol. 2012;415:92–101. doi: 10.1016/j.jmb.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. J Biol Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR. J Biol Chem. 2009;284:36711–36719. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moody SC, Zhao B, Lei L, Nelson DR, Mullins JGL, Waterman MR, Kelly SL, Lamb DC. FEBS J. 2012;279:1640–1649. doi: 10.1111/j.1742-4658.2011.08447.x. [DOI] [PubMed] [Google Scholar]
- 148.Kalb VF, Loper JC. Proc Natl Acad Sci USA. 1988;85:7221–7225. doi: 10.1073/pnas.85.19.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson JA, Deisenhofer J. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 150.Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 151.Rupasinghe S, Schuler MA, Kagawa N, Yuan H, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC. FEBS Lett. 2006;580:6338–6342. doi: 10.1016/j.febslet.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 152.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 153.Yoshigae Y, Kent UM, Hollenberg PF. Biochemistry. 2013;52:4636–4647. doi: 10.1021/bi4004843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Niraula NP, Kanth BK, Sohng JK, Oh TJ. Enz Microb Technol. 2011;48:181–186. doi: 10.1016/j.enzmictec.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 155.Sezutsu H, Le Goff G, Feyereisen R. Philos Trans R Soc, B: Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0428. 20120428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Coelho PS, Wang ZJ, Ener ME, Baril SA, Kannan A, Arnold FH, Brustad EM. Nat Chem Biol. 2013;9:485–487. doi: 10.1038/nchembio.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McIntosh JA, Coelho PS, Farwell CC, Wang ZJ, Lewis JC, Brown TR, Arnold FH. Angew Chem, Int Ed. 2013;52:9309–9312. doi: 10.1002/anie.201304401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McIntosh JA, Heel T, Buller AR, Chio L, Arnold FH. J Am Chem Soc. 2015;137:13861–13865. doi: 10.1021/jacs.5b07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang ZJ, Renata H, Peck NE, Farwell CC, Coelho PS, Arnold FH. Angew Chem, Int Ed. 2014;53:6810–6813. doi: 10.1002/anie.201402809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios SYI, Yeh LS, Young SY, Mitchell AL. Nucl Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. PloS one. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, Okuda K, NebertP DW. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 164.Tohyama S, Kakinuma K, Eguchi T. J Antibiot. 2006;59:44–52. doi: 10.1038/ja.2006.7. [DOI] [PubMed] [Google Scholar]
- 165.Park OK, Choi HY, Kim GW, Kim WG. ChemBioChem. 2016;17:1725–1731. doi: 10.1002/cbic.201600241. [DOI] [PubMed] [Google Scholar]
- 166.Chen H, Walsh CT. Chem Biol. 2001;8:301–312. doi: 10.1016/s1074-5521(01)00009-6. [DOI] [PubMed] [Google Scholar]
- 167.Lauer B, Russwurm R, Bormann C. Eur J Biochem. 2000;267:1698–1706. doi: 10.1046/j.1432-1327.2000.01162.x. [DOI] [PubMed] [Google Scholar]
- 168.Chen H, Hubbard BK, O’Connor SE, Walsh CT. Chem Biol. 2002;9:103–112. doi: 10.1016/s1074-5521(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 169.Zeng H, Tan H, Li J. Curr Microbiol. 2002;45:175–179. doi: 10.1007/s00284-001-0115-4. [DOI] [PubMed] [Google Scholar]
- 170.Zhang C, Kong L, Liu Q, Lei X, Zhu T, Yin J, Lin B, Deng Z, You D. PloS one. 2013;8:e56772. doi: 10.1371/journal.pone.0056772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pandey BP, Lee N, Choi K-Y, Jung E, Jeong D-h, Kim B-G. Enz Microb Technol. 2011;48:386–392. doi: 10.1016/j.enzmictec.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 172.Wang X, Tabudravu J, Rateb ME, Annand KJ, Qin Z, Jaspars M, Deng Z, Yu Y, Deng H. Mol BioSyst. 2013;9:1286–1289. doi: 10.1039/c3mb70081j. [DOI] [PubMed] [Google Scholar]
- 173.Sun Y, Zhou X, Tu G, Deng Z. Arch Microbiol. 2003;180:101–107. doi: 10.1007/s00203-003-0564-1. [DOI] [PubMed] [Google Scholar]
- 174.He Y, Sun Y, Liu T, Zhou X, Bai L, Deng Z. Appl Environ Microbiol. 2010;76:3283–3292. doi: 10.1128/AEM.02262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Galm U, Schimana J, Fiedler HP, Schmidt J, Li SM, Heide L. Arch Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 176.Pojer F, Li SM, Heide L. Microbiology. 2002;148:3901–3911. doi: 10.1099/00221287-148-12-3901. [DOI] [PubMed] [Google Scholar]
- 177.Wang ZX, Li SM, Heide L. Antimicrob Agents Chemother. 2000;44:3040–3048. doi: 10.1128/aac.44.11.3040-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Carmody M, Murphy B, Byrne B, Power P, Rai D, Rawlings B, Caffrey P. J Biol Chem. 2005;280:34420–34426. doi: 10.1074/jbc.M506689200. [DOI] [PubMed] [Google Scholar]
- 179.Stephens N, Rawlings B, Caffrey P. Biosci, Biotechnol, Biochem. 2012;76:384–387. doi: 10.1271/bbb.110673. [DOI] [PubMed] [Google Scholar]
- 180.Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, Strom AR, Valla S, Zotchev SB. Chem Biol. 2000;7:395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 181.Brautaset T, Sletta H, Degnes KF, Sekurova ON, Bakke I, Volokhan O, Andreassen T, Ellingsen TE, Zotchev SB. Appl Environ Microbiol. 2011;77:6636–6643. doi: 10.1128/AEM.05780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Qi Z, Kang Q, Jiang C, Han M, Bai L. Appl Microbiol Biotechnol. 2015;99:6745–6752. doi: 10.1007/s00253-015-6635-9. [DOI] [PubMed] [Google Scholar]
- 183.Liu S-P, Yuan P-H, Wang Y-Y, Liu X-F, Zhou Z-X, Bu Q-t, Yu P, Jiang H, Li Y-Q. Microbiol Res. 2015;173:25–33. doi: 10.1016/j.micres.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 184.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 185.Poulos TL, Johnson EF. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th. Ortiz de Montellano PR, editor. Springer International Publishing; Switzerland: 2015. pp. 3–32. [Google Scholar]
- 186.Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. J Biol Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Leys D, Mowat CG, McLean KJ, Richmond A, Chapman SK, Walkinshaw MD, Munro AW. J Biol Chem. 2003;278:5141–5147. doi: 10.1074/jbc.M209928200. [DOI] [PubMed] [Google Scholar]
- 188.Podust LM, Bach H, Kim Y, Lamb DC, Arase M, Sherman DH, Kelly SL, Waterman MR. Protein Sci. 2004;13:255–268. doi: 10.1110/ps.03384804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li Q, Li Q, Zhang H, Chen Y, Zhang G. FEBS Lett. 2017;591:1295–1304. doi: 10.1002/1873-3468.12643. [DOI] [PubMed] [Google Scholar]
- 190.Lee Chang W, Park H, Lee Jun H, Lee Chang W, Park H, Lee Jun H, Lee J-H, Rimal H, Oh T-J. Int J Mol Sci. 2016;17 [Google Scholar]
- 191.Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ. Nature. 2015;521:105–109. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 192.Haslinger K, Cryle MJ. FEBS Lett. 2016;590:571–581. doi: 10.1002/1873-3468.12081. [DOI] [PubMed] [Google Scholar]
- 193.Li S, Ouellet H, Sherman DH, Podust LM. J Biol Chem. 2009;284:5723–5730. doi: 10.1074/jbc.M807592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li S, Chaulagain MR, Knauff AR, Podust LM, Montgomery J, Sherman DH. Proc Natl Acad Sci USA. 2009;106:18463–18468. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Negretti S, Narayan ARH, Chiou KC, Kells PM, Stachowski JL, Hansen DA, Podust LM, Montgomery J, Sherman DH. J Am Chem Soc. 2014;136:4901–4904. doi: 10.1021/ja5016052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xie P, Ma M, Rateb ME, Shaaban KA, Yu Z, Huang SX, Zhao LX, Zhu X, Yan Y, Peterson RM, Lohman JR, Yang D, Yin M, Rudolf JD, Jiang Y, Duan Y, Shen B. J Nat Prod. 2014;77:377–387. doi: 10.1021/np401063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hindra, Huang T, Yang D, Rudolf JD, Xie P, Xie G, Teng Q, Lohman JR, Zhu X, Huang Y, Zhao L-X, Jiang Y, Duan Y, Shen B. J Nat Prod. 2014;77:2296–2303. doi: 10.1021/np5006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ju KS, Gao J, Doroghazi JR, Wang KKA, Thibodeaux CJ, Li S, Metzger E, Fudala J, Su J, Zhang JK, Lee J, Cioni JP, Evans BS, Hirota R, Labeda DP, van der Donk WA, Metcalf WW. Proc Natl Acad Sci USA. 2015;112:12175–12180. doi: 10.1073/pnas.1500873112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rudolf JD, Yan X, Shen B. J Ind Microbiol Biotechnol. 2016;43:261–276. doi: 10.1007/s10295-015-1671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Brown SD, Babbitt PC. J Biol Chem. 2012;287:35–42. doi: 10.1074/jbc.R111.283408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhao S, Jacobson MP, Sakai A, Zhang X, Kumar R, San Francisco B, Solbiati J, Gerlt JA, Vetting MW, Hillerich B, Seidel RD, Almo SC, Steves A, Brown S, Akiva E, Barber A, Babbitt PC. eLife. 2014;3:e03275. doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lu W, Leimkuhler C, Gatto GJ, Kruger RG, Oberthur M, Kahne D, Walsh CT. Chem Biol. 2005;12:527–534. doi: 10.1016/j.chembiol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 203.Yuan Y, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. J Am Chem Soc. 2005;127:14128–14129. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Byrne B, Carmody M, Gibson E, Rawlings B, Caffrey P. Chem Biol. 2003;10:1215–1224. doi: 10.1016/j.chembiol.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 205.Jin X, Rao M, Wei W, Ge M, Liu J, Chen D, Liang Y. Biotechnol Lett. 2012;34:2283–2289. doi: 10.1007/s10529-012-1032-2. [DOI] [PubMed] [Google Scholar]
- 206.Joergensen H, Degnes KF, Sletta H, Fjaervik E, Dikiy A, Herfindal L, Bruheim P, Klinkenberg G, Bredholt H, Nygaard G, Doeskeland SO, Ellingsen TE, Zotchev SB. Chem Biol. 2009;16:1109–1121. doi: 10.1016/j.chembiol.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 207.Zhou Y, Minami T, Honda K, Omasa T, Ohtake H. J Biosci Bioeng. 2010;110:403–407. doi: 10.1016/j.jbiosc.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 208.Campelo AB, Gil JA. Microbiology. 2002;148:51–59. doi: 10.1099/00221287-148-1-51. [DOI] [PubMed] [Google Scholar]
- 209.Chen S, Mao X, Shen Y, Zhou Y, Li J, Wang L, Tao X, Yang L, Wang Y, Zhou X, Deng Z, Wei D. Appl Environ Microbiol. 2009;75:1778–1781. doi: 10.1128/AEM.00859-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mao X, Wang F, Zhang J, Chen S, Deng Z, Shen Y, Wei D. Appl Biochem Biotechnol. 2009;159:673–686. doi: 10.1007/s12010-008-8502-y. [DOI] [PubMed] [Google Scholar]
- 211.Martin JF, Aparicio JF. Meth Enzymol. 2009;459:215–242. doi: 10.1016/S0076-6879(09)04610-2. [DOI] [PubMed] [Google Scholar]
- 212.Horii M, Ishizaki T, Paik SY, Manome T, Murooka Y. J Bacteriol. 1990;172:3644–3653. doi: 10.1128/jb.172.7.3644-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kharel MK, Nybo SE, Shepherd MD, Rohr J. ChemBioChem. 2010;11:523–532. doi: 10.1002/cbic.200900673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ikeda H, Shin-Ya K, Nagamitsu T, Tomoda H. J Ind Microbiol Biotechnol. 2016;43:325–342. doi: 10.1007/s10295-015-1694-6. [DOI] [PubMed] [Google Scholar]
- 215.Kim SY, Zhao P, Igarashi M, Sawa R, Tomita T, Nishiyama M, Kuzuyama T. Chem Biol. 2009;16:736–743. doi: 10.1016/j.chembiol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 216.Shrestha P, Oh TJ, Sohng JK. Biotechnol Lett. 2008;30:1101–1106. doi: 10.1007/s10529-008-9654-0. [DOI] [PubMed] [Google Scholar]
- 217.Lamb DC, Lei L, Zhao B, Yuan H, Jackson CJ, Warrilow AGS, Skaug T, Dyson PJ, Dawson ES, Kelly SL, Hachey DL, Waterman MR. Appl Environ Microbiol. 2010;76:1975–1980. doi: 10.1128/AEM.03000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Xu LH, Fushinobu S, Takamatsu S, Wakagi T, Ikeda H, Shoun H. J Biol Chem. 2010;285:16844–16853. doi: 10.1074/jbc.M109.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pandey BP, Roh C, Choi KY, Lee N, Kim EJ, Ko S, Kim T, Yun H, Kim BG. Biotechnol Bioeng. 2009;105:697–704. doi: 10.1002/bit.22582. [DOI] [PubMed] [Google Scholar]
- 220.Takamatsu S, Xu LH, Fushinobu S, Shoun H, Komatsu M, Cane DE, Ikeda H. J Antibiot. 2011;64:65–71. doi: 10.1038/ja.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Roh C. Molecules. 2013;18:3028–3040. doi: 10.3390/molecules18033028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pandey BP, Lee N, Choi KY, Kim JN, Kim EJ, Kim BG. Appl Microbiol Biotechnol. 2014;98:5009–5017. doi: 10.1007/s00253-014-5525-x. [DOI] [PubMed] [Google Scholar]
- 223.Xu LH, Ikeda H, Liu L, Arakawa T, Wakagi T, Shoun H, Fushinobu S. Appl Microbiol Biotechnol. 2015;99:3081–3091. doi: 10.1007/s00253-014-6148-y. [DOI] [PubMed] [Google Scholar]
- 224.Liu L, Yao Q, Ma Z, Ikeda H, Fushinobu S, Xu LH. J Mol Cat B: Enzymatic. 2016;132:91–97. [Google Scholar]
- 225.Shrestha P, Oh TJ, Liou K, Sohng JK. Appl Microbiol Biotechnol. 2008;79:555–562. doi: 10.1007/s00253-008-1455-9. [DOI] [PubMed] [Google Scholar]
- 226.Zhao B, Moody SC, Hider RC, Lei L, Kelly SL, Waterman MR, Lamb DC. Int J Mol Sci. 2012;13:8500–8513. doi: 10.3390/ijms13078500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lim YR, Hong MK, Kim JK, Doan TTN, Kim DH, Yun CH, Chun YJ, Kang LW, Kim D. Arch Biochem Biophys. 2012;528:111–117. doi: 10.1016/j.abb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 228.Xu LH, Fushinobu S, Ikeda H, Wakagi T, Shoun H. J Bacteriol. 2009;191:1211–1219. doi: 10.1128/JB.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kanth BK, Liou K, Sohng JK. Comput Biol Chem. 2010;34:226–231. doi: 10.1016/j.compbiolchem.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 230.Han S, Pham TV, Kim JH, Lim YR, Park HG, Jeong D, Yun CH, Chun YJ, Kang LW, Kim D. Biochem Biophys Res Commun. 2017;482:902–908. doi: 10.1016/j.bbrc.2016.11.131. [DOI] [PubMed] [Google Scholar]
- 231.Tian Z, Cheng Q, Yoshimoto FK, Lei L, Lamb DC, Guengerich FP. Arch Biochem Biophys. 2013;530:101–107. doi: 10.1016/j.abb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Han S, Pham TV, Kim JH, Lim YR, Park HG, Cha GS, Yun CH, Chun YJ, Kang LW, Kim D. Arch Biochem Biophys. 2015;575:1–7. doi: 10.1016/j.abb.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 233.Han S, Pham TV, Kim JH, Lim YR, Park HG, Cha GS, Yun CH, Chun YJ, Kang LW, Kim D. Molecules Cells. 2016;39:211–216. doi: 10.14348/molcells.2016.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Liu W, Jiang X, Ji Y, Niu J, Li M. Weishengwu Xuebao. 2011;51:410–416. [Google Scholar]
- 235.Parajuli N, Basnet DB, Lee HC, Sohng JK, Liou K. Arch Biochem Biophys. 2004;425:233–241. doi: 10.1016/j.abb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 236.Bhattarai S, Liou K, Oh TJ. J Microbiol Biotechnol. 2012;22:917–922. doi: 10.4014/jmb.1112.12053. [DOI] [PubMed] [Google Scholar]
- 237.Bhattarai S, Liou K, Oh TJ. Arch Biochem Biophys. 2013;539:63–69. doi: 10.1016/j.abb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 238.Zhou Y, Minami T, Honda K, Omasa T, Ohtake H. Appl Microbiol Biotechnol. 2010;87:647–655. doi: 10.1007/s00253-010-2505-7. [DOI] [PubMed] [Google Scholar]
- 239.Podust LM, Kim Y, Arase M, Neely BA, Beck BJ, Bach H, Sherman DH, Lamb DC, Kelly SL, Waterman MR. J Biol Chem. 2003;278:12214–12221. doi: 10.1074/jbc.M212210200. [DOI] [PubMed] [Google Scholar]
- 240.Rimal H, Yu SC, Jang JH, Oh TJ. J Microbiol Biotechnol. 2015;25:1417–1424. doi: 10.4014/jmb.1504.04057. [DOI] [PubMed] [Google Scholar]
- 241.Zhao B, Waterman MR. IUBMB Life. 2011;63:473–477. doi: 10.1002/iub.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Takamatsu S, Lin X, Nara A, Komatsu M, Cane DE, Ikeda H. Microb Biotechnol. 2011;4:184–191. doi: 10.1111/j.1751-7915.2010.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Park JW, Lee JK, Kwon TJ, Yi DI, Park YI, Kang SM. J Microbiol Biotechnol. 2001;11:1011–1017. [Google Scholar]
- 244.Lamb DC, Fowler K, Kieser T, Manning N, Podust Larissa M, Waterman Michael R, Kelly DE, Kelly SL. Biochem J. 2002;364:555–562. doi: 10.1042/BJ20011380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Li ZZ, Li XF, Yang W, Dong X, Yu J, Zhu SL, Li M, Xie L, Tong WY. BMC Genomics. 2013;14:130. doi: 10.1186/1471-2164-14-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hong JSJ, Park SJ, Parajuli N, Park SR, Koh HS, Jung WS, Choi CY, Yoon YJ. Gene. 2007;386:123–130. doi: 10.1016/j.gene.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 247.Wu H, Li W, Xin C, Zhang C, Wang Y, Ren S, Ren M, Zhao W, Yuan L, Xu Z, Yuan H, Geng M, Zhang L, Weaver DT, Zhang B. Appl Microbiol Biotechnol. 2016;100:2257–2266. doi: 10.1007/s00253-015-7036-9. [DOI] [PubMed] [Google Scholar]
- 248.Otten SL, Liu X, Ferguson J, Hutchinson CR. J Bacteriol. 1995;177:6688–6692. doi: 10.1128/jb.177.22.6688-6692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dickens ML, Strohl WR. J Bacteriol. 1996;178:3389–3395. doi: 10.1128/jb.178.11.3389-3395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lomovskaya N, Otten SL, Doi-Katayama Y, Fonstein L, Liu XC, Takatsu T, Inventi-Solari A, Filippini S, Torti F, Colombo AL, Hutchinson CR. J Bacteriol. 1999;181:305–318. doi: 10.1128/jb.181.1.305-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Walczak RJ, Dickens ML, Priestley ND, Strohl WR. J Bacteriol. 1999;181:298–304. doi: 10.1128/jb.181.1.298-304.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Walczak RJ, Hines JV, Strohl WR, Priestley ND. Org Lett. 2001;3:2277–2279. doi: 10.1021/ol015998x. [DOI] [PubMed] [Google Scholar]
- 253.Piel J, Hertweck C, Shipley PR, Hunt DM, Newman MS, Moore BS. Chem Biol. 2000;7:943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 254.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Nat Chem Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 255.Payero TD, Vicente CM, Rumbero A, Barreales EG, Santos-Aberturas J, de Pedro A, Aparicio JF. Microb Cell Fact. 2015;14:1–14. doi: 10.1186/s12934-015-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu X-j, Kong R-x, Niu M-s, Qiu R-g, Tang L. J Nat Prod. 2013;76:524–529. doi: 10.1021/np300667r. [DOI] [PubMed] [Google Scholar]
- 257.Kim HJ, Karki S, Kwon SY, Park SH, Nahm BH, Kim YK, Kwon HJ. J Biol Chem. 2014;289:34557–34568. doi: 10.1074/jbc.M114.602334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liu C, Zhang J, Lu C, Shen Y. Antonie van Leeuwenhoek. 2015;107:1359–1366. doi: 10.1007/s10482-015-0415-5. [DOI] [PubMed] [Google Scholar]
- 259.Li S, Ni S, Wu L, Li L, Jiang B, Wang H, Sun G, Gan M, Li J, He W, Lin L, Wang Y, Bai S, Si S. J Nat Prod. 2013;76:969–973. doi: 10.1021/np4000679. [DOI] [PubMed] [Google Scholar]
- 260.Malla S, Thuy Ta Thi T, Oh Tae J, Sohng Jae K. Arch Microbiol. 2011;193:95–103. doi: 10.1007/s00203-010-0648-7. [DOI] [PubMed] [Google Scholar]
- 261.Kudo F, Motegi A, Mizoue K, Eguchi T. ChemBioChem. 2010;11:1574–1582. doi: 10.1002/cbic.201000214. [DOI] [PubMed] [Google Scholar]
- 262.Kudo F, Kawamura K, Furuya T, Yamanishi H, Motegi A, Komatsubara A, Numakura M, Miyanaga A, Eguchi T. ChemBioChem. 2016;17:233–238. doi: 10.1002/cbic.201500533. [DOI] [PubMed] [Google Scholar]
- 263.Liu T, Kharel MK, Fischer C, McCormick A, Rohr J. ChemBioChem. 2006;7:1070–1077. doi: 10.1002/cbic.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yunt Z, Reinhardt K, Li A, Engeser M, Dahse HM, Guetschow M, Bruhn T, Bringmann G, Piel J. J Am Chem Soc. 2009;131:2297–2305. doi: 10.1021/ja807827k. [DOI] [PubMed] [Google Scholar]
- 265.Yu D, Xu F, Shao L, Zhan J. Bioorg Med Chem Lett. 2014;24:4511–4514. doi: 10.1016/j.bmcl.2014.07.078. [DOI] [PubMed] [Google Scholar]
- 266.Pokhrel AR, Dhakal D, Jha AK, Sohng JK. Appl Microbiol Biotechnol. 2015;99:8351–8362. doi: 10.1007/s00253-015-6860-2. [DOI] [PubMed] [Google Scholar]
- 267.Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. Mol Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 268.Arakawa K, Kodama K, Tatsuno S, Ide S, Kinashi H. Antimicrob Agents Chemother. 2006;50:1946–1952. doi: 10.1128/AAC.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.B. Shen, Unpublished data.
- 270.Liu J, Wang B, Li H, Xie Y, Li Q, Qin X, Zhang X, Ju J. Org Lett. 2015;17:1509–1512. doi: 10.1021/acs.orglett.5b00389. [DOI] [PubMed] [Google Scholar]
- 271.Ma M, Kwong T, Lim SK, Ju J, Lohman JR, Shen B. J Am Chem Soc. 2013;135:2489–2492. doi: 10.1021/ja4002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. Chem Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 273.Hang VTT, Oh TJ, Yamaguchi T, Sohng JK. FEMS Microbiol Lett. 2010;311:119–125. doi: 10.1111/j.1574-6968.2010.02075.x. [DOI] [PubMed] [Google Scholar]
- 274.Bruntner C, Lauer B, Schwarz W, Mohrle V, Bormann C. Mol Gen Genet. 1999;262:102–114. doi: 10.1007/pl00008637. [DOI] [PubMed] [Google Scholar]
- 275.Volokhan O, Sletta H, Ellingsen TE, Zotchev SB. Appl Environ Microbiol. 2006;72:2514–2519. doi: 10.1128/AEM.72.4.2514-2519.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Huang S, Elsayed SS, Lv M, Tabudravu J, Rateb ME, Gyampoh R, Kyeremeh K, Ebel R, Jaspars M, Deng Z, Yu Y, Deng H. Chem Biol. 2015;22:1633–1642. doi: 10.1016/j.chembiol.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 277.Rodriguez AM, Olano C, Mendez C, Hutchinson CR, Salas JA. FEMS Microbiol Lett. 1995;127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- 278.Shah S, Xue Q, Tang L, Carney JR, Betlach M, McDaniel R. J Antibiot. 2000;53:502–508. doi: 10.7164/antibiotics.53.502. [DOI] [PubMed] [Google Scholar]
- 279.Olano C, Rodriguez AM, Michel JM, Mendez C, Raynal MC, Salas JA. Mol Gen Genet. 1998;259:299–308. doi: 10.1007/s004380050816. [DOI] [PubMed] [Google Scholar]
- 280.Arisawa A, Tsunekawa H, Okamura K, Okamoto R. Biosci, Biotechnol, Biochem. 1995;59:582–588. doi: 10.1271/bbb.59.582. [DOI] [PubMed] [Google Scholar]
- 281.Goto LS, Hokka CO, Lima JF, Prieto T, Araujo APU, Nantes IL, Nascimento OR. J Braz Chem Soc. 2012;23:913–920. [Google Scholar]
- 282.Serizawa N, Matsuoka T. Biosci, Biotechnol, Biochem. 1993;57:1777–1778. doi: 10.1271/bbb.57.1686. [DOI] [PubMed] [Google Scholar]
- 283.Ito S, Matsuoka T, Watanabe I, Kagasaki T, Serizawa N, Hata T. Acta Crystallogr, Sect D: Biol Crystallogr. 1999;D55:1209–1211. doi: 10.1107/s0907444999003716. [DOI] [PubMed] [Google Scholar]
- 284.Ba L, Li P, Zhang H, Duan Y, Lin Z. Biotechnol Bioeng. 2013;110:2815–2825. doi: 10.1002/bit.24960. [DOI] [PubMed] [Google Scholar]
- 285.Ba L, Li P, Zhang H, Duan Y, Lin Z. Biotechnol J. 2013;8:785–793. doi: 10.1002/biot.201200097. [DOI] [PubMed] [Google Scholar]
- 286.Romesser JA, O’Keefe DP. Biochem Biophys Res Commun. 1986;140:650–659. doi: 10.1016/0006-291x(86)90781-3. [DOI] [PubMed] [Google Scholar]
- 287.O’Keefe DP, Romesser JA, Leto KJ. Arch Microbiol. 1988;149:406–412. [Google Scholar]
- 288.Omer CA, Lenstra R, Little PJ, Dean C, Tepperman JM, Leto KJ, Romesser JA, O’Keefe DP. J Bacteriol. 1990;172:3335–3345. doi: 10.1128/jb.172.6.3335-3345.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Harder PA, O’Keefe DP, Romesser JA, Leto KJ, Omer CA. Mol Gen Genet. 1991;227:238–244. doi: 10.1007/BF00259676. [DOI] [PubMed] [Google Scholar]
- 290.Hayashi K, Yasuda K, Sugimoto H, Ikushiro S, Kamakura M, Kittaka A, Horst RL, Chen TC, Ohta M, Shiro Y, Sakaki T. FEBS J. 2010;277:3999–4009. doi: 10.1111/j.1742-4658.2010.07791.x. [DOI] [PubMed] [Google Scholar]
- 291.Sakaki T, Sugimoto H, Hayashi K, Yasuda K, Munetsuna E, Kamakura M, Ikushiro S, Shiro Y. Biochim Biophys Acta. 2011;1814:249–256. doi: 10.1016/j.bbapap.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 292.Dasgupta K, Ganesan S, Manivasagam S, Ayre BG. BMC Plant Biol. 2011;11:67. doi: 10.1186/1471-2229-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kleser M, Hannemann F, Hutter M, Zapp J, Bernhardt R. J Biotechnol. 2012;157:405–412. doi: 10.1016/j.jbiotec.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 294.Janocha S, Bernhardt R. Appl Microbiol Biotechnol. 2013;97:7639–7649. doi: 10.1007/s00253-013-5008-5. [DOI] [PubMed] [Google Scholar]
- 295.O’Keefe DP, Gibson KJ, Emptage MH, Lenstra R, Romesser JA, Litle PJ, Omer CA. Biochemistry. 1991;30:447–455. doi: 10.1021/bi00216a021. [DOI] [PubMed] [Google Scholar]
- 296.El Ouarradi A, Lombard M, Buisson D. J Mol Cat B: Enzymatic. 2010;67:172–178. [Google Scholar]
- 297.Lombard M, Salard I, Sari MA, Mansuy D, Buisson D. Arch Biochem Biophys. 2011;508:54–63. doi: 10.1016/j.abb.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 298.Xue Y, Zhao L, Liu HW, Sherman DH. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Cane DE, Graziani EI. J Am Chem Soc. 1998;120:2682–2683. [Google Scholar]
- 300.Yoon YJ, Beck BJ, Kim BS, Kang HY, Reynolds KA, Sherman DH. Chem Biol. 2002;9:203–214. doi: 10.1016/s1074-5521(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 301.Srinivasan A, Bach H, Sherman DH, Dordick JS. Biotechnol Bioeng. 2004;88:528–535. doi: 10.1002/bit.20285. [DOI] [PubMed] [Google Scholar]
- 302.Kim BG, Lee SK, Liou K, Sohng JK, Yoon YJ, Lee HC. J Ind Eng Chem. 2004;10:759–765. [Google Scholar]
- 303.Li S, Podust LM, Sherman DH. J Am Chem Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Basnet DB, Park JW, Yoon YJ. J Biotechnol. 2008;135:92–96. doi: 10.1016/j.jbiotec.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 305.Jha AK, Dhakal D, Pham TTV, Pokhrel AR, Yamaguchi T, Jung HJ, Yoon YJ, Sohng JK. Appl Microbiol Biotechnol. 2015;99:3421–3431. doi: 10.1007/s00253-015-6431-6. [DOI] [PubMed] [Google Scholar]
- 306.Lee SK, Park JW, Park SR, Ahn JS, Choi CY, Yoon YJ. J Microbiol Biotechnol. 2006;16:974–978. [Google Scholar]
- 307.Machida K, Arisawa A, Takeda S, Tsuchida T, Aritoku Y, Yoshida M, Ikeda H. Biosci, Biotechnol, Biochem. 2008;72:2946–2952. doi: 10.1271/bbb.80425. [DOI] [PubMed] [Google Scholar]
- 308.Ghatge MS, Reynolds KA. J Bacteriol. 2005;187:7970–7976. doi: 10.1128/JB.187.23.7970-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Berrie JR, Williams RAD, Smith KE. J Steroid Biochem Mol Biol. 1999;71:153–165. doi: 10.1016/s0960-0760(99)00132-6. [DOI] [PubMed] [Google Scholar]
- 310.Machida K, Aritoku Y, Nakashima T, Arisawa A, Tsuchida T. J Biosci Bioeng. 2008;105:649–654. doi: 10.1263/jbb.105.649. [DOI] [PubMed] [Google Scholar]
- 311.Machida K, Aritoku Y, Tsuchida T. J Biosci Bioeng. 2009;107:596–598. doi: 10.1016/j.jbiosc.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 312.Quaderer R, Omura S, Ikeda H, Cane DE. J Am Chem Soc. 2006;128:13036–13037. doi: 10.1021/ja0639214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Molnar I, Aparicio JF, Haydock SF, Khaw LE, Schwecke T, Koenig A, Staunton J, Leadlay PF. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 314.Chung L, Liu L, Patel S, Carney JR, Reeves CD. J Antibiot. 2001;54:250–256. doi: 10.7164/antibiotics.54.250. [DOI] [PubMed] [Google Scholar]
- 315.Takahashi S, Nagano S, Nogawa T, Kanoh N, Uramoto M, Kawatani M, Shimizu T, Miyazawa T, Shiro Y, Osada H. J Biol Chem. 2014;289:32446–32458. doi: 10.1074/jbc.M114.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Song L, Laureti L, Corre C, Leblond P, Aigle B, Challis GL. J Antibiot. 2014;67:71–76. doi: 10.1038/ja.2013.119. [DOI] [PubMed] [Google Scholar]
- 317.Chen W, Tian Y, Yang H, Tan H. Weishengwu Xuebao. 2000;40:598–604. [PubMed] [Google Scholar]
- 318.Xie Z, Niu G, Li R, Liu G, Tan H. Curr Microbiol. 2007;55:537–542. doi: 10.1007/s00284-007-9028-1. [DOI] [PubMed] [Google Scholar]
- 319.Yamada Y, Kuzuyama T, Komatsu M, Shin-Ya K, Omura S, Cane DE, Ikeda H. Proc Natl Acad Sci USA. 2015;112:857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.B. Shen, Unpublished data.
- 321.Ban YH, Shinde PB, Hwang J-y, Song M-C, Kim DH, Lim S-K, Sohng JK, Yoon YJ. J Nat Prod. 2013;76:1091–1098. doi: 10.1021/np4001224. [DOI] [PubMed] [Google Scholar]
- 322.Olano C, Gomez C, Perez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Brana AF, Mendez C, Salas JA. Chem Biol. 2009;16:1031–1044. doi: 10.1016/j.chembiol.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 323.Gomez C, Olano C, Palomino-Schaetzlein M, Pineda-Lucena A, Carbajo RJ, Brana AF, Mendez C, Salas JA. J Antibiot. 2012;65:341–348. doi: 10.1038/ja.2012.37. [DOI] [PubMed] [Google Scholar]
- 324.Motamedi H, Shafiee A, Cai SJ, Streicher SL, Arison BH, Miller RR. J Bacteriol. 1996;178:5243–5248. doi: 10.1128/jb.178.17.5243-5248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yue C, Liu N, Liu M, Lu Y, Shao M, Wang M, Ai G, Huang Y. World J Microbiol Biotechnol. 2015;31:541–548. doi: 10.1007/s11274-015-1825-2. [DOI] [PubMed] [Google Scholar]
- 326.Sariaslani FS, Kunz DA. Biochem Biophys Res Commun. 1986;141:405–410. doi: 10.1016/s0006-291x(86)80187-5. [DOI] [PubMed] [Google Scholar]
- 327.Trower MK, Sariaslani FS, Kitson FG. Biochem Biophys Res Commun. 1988;157:1417–1422. doi: 10.1016/s0006-291x(88)81033-7. [DOI] [PubMed] [Google Scholar]
- 328.Sariaslani FS, Trower MK, Kunz DA. Biocatalysis. 1989;2:151–160. [Google Scholar]
- 329.Sariaslani FS, Stahl RG., Jr Biochem Biophys Res Commun. 1990;166:743–749. doi: 10.1016/0006-291x(90)90872-k. [DOI] [PubMed] [Google Scholar]
- 330.Trower MK, Lenstra R, Omer C, Buchholz SE, Sariaslani FS. Mol Microbiol. 1992;6:2125–2134. doi: 10.1111/j.1365-2958.1992.tb01386.x. [DOI] [PubMed] [Google Scholar]
- 331.Lamb DC, Kelly DE, Masaphy S, Jones GL, Kelly SL. Biochem Biophys Res Commun. 2000;276:797–802. doi: 10.1006/bbrc.2000.3541. [DOI] [PubMed] [Google Scholar]
- 332.Kaderbhai MA, Ugochukwu CC, Kelly SL, Lamb DC. Appl Environ Microbiol. 2001;67:2136–2138. doi: 10.1128/AEM.67.5.2136-2138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Akhtar MK, Kaderbhai NN, Hopper DJ, Kelly SL, Kaderbhai MA. J Biol Chem. 2003;278:45555–45562. doi: 10.1074/jbc.M212685200. [DOI] [PubMed] [Google Scholar]
- 334.Karray F, Darbon E, Oestreicher N, Dominguez H, Tuphile K, Gagnat J, Blondelet-Rouault MH, Gerbaud C, Pernodet JL. Microbiology. 2007;153:4111–4122. doi: 10.1099/mic.0.2007/009746-0. [DOI] [PubMed] [Google Scholar]
- 335.Nguyen HC, Darbon E, Thai R, Pernodet JL, Lautru S. Antimicrob Agents Chemother. 2013;57:3836–3842. doi: 10.1128/AAC.00512-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Onaka H, Taniguchi S-i, Igarashi Y, Furumai T. Biosci, Biotechnol, Biochem. 2003;67:127–138. doi: 10.1271/bbb.67.127. [DOI] [PubMed] [Google Scholar]
- 337.Chen D, Zhang L, Pang B, Chen J, Xu Z, Abe I, Liu W. J Bacteriol. 2013;195:1931–1939. doi: 10.1128/JB.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Huang D, Xia M, Li S, Wen J, Jia X. J Ind Microbiol Biotechnol. 2013;40:1023–1037. doi: 10.1007/s10295-013-1301-7. [DOI] [PubMed] [Google Scholar]
- 339.Cao B, Yao F, Zheng X, Cui D, Shao Y, Zhu C, Deng Z, You D. ChemBioChem. 2012;13:2234–2242. doi: 10.1002/cbic.201200402. [DOI] [PubMed] [Google Scholar]
- 340.Wang Y, Wang J, Yu S, Wang F, Ma H, Yue C, Liu M, Deng Z, Huang Y, Qu X. ChemBioChem. 2016;17:799–803. doi: 10.1002/cbic.201500670. [DOI] [PubMed] [Google Scholar]
- 341.Choi SS, Hur YA, Sherman DH, Kim ES. Microbiology. 2007;153:1095–1102. doi: 10.1099/mic.0.2006/003194-0. [DOI] [PubMed] [Google Scholar]
- 342.Kim D, Nah JH, Choi SS, Shin HS, Sherman DH, Kim ES. J Ind Microbiol Biotechnol. 2012;39:1563–1568. doi: 10.1007/s10295-012-1157-2. [DOI] [PubMed] [Google Scholar]
- 343.Mo X, Wang Z, Wang B, Ma J, Huang H, Tian X, Zhang S, Zhang C, Ju J. Biochem Biophys Res Commun. 2011;406:341–347. doi: 10.1016/j.bbrc.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 344.Ren J, Cui Y, Zhang F, Cui H, Ni X, Chen F, Li L, Xia H. Microbiol Res. 2014;169:602–608. doi: 10.1016/j.micres.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 345.Yang D, Li W, Huang SX, Shen B. Org Lett. 2012;14:1302–1305. doi: 10.1021/ol300187p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. J Bacteriol. 2002;184:2019–2029. doi: 10.1128/JB.184.7.2019-2029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Barry SM, Challis GL. Meth Enzymol. 2012;516:171–194. doi: 10.1016/B978-0-12-394291-3.00001-0. [DOI] [PubMed] [Google Scholar]
- 348.Fouces R, Mellado E, Diez B, Barredo JL. Microbiology. 1999;145:855–868. doi: 10.1099/13500872-145-4-855. [DOI] [PubMed] [Google Scholar]
- 349.Bate N, Cundliffe E. J Ind Microbiol Biotechnol. 1999;23:118–122. doi: 10.1038/sj.jim.2900707. [DOI] [PubMed] [Google Scholar]
- 350.DeMars MD, Sheng F, Park SR, Lowell AN, Podust LM, Sherman DH. ACS Chem Biol. 2016;11:2642–2654. doi: 10.1021/acschembio.6b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Gandecha AR, Large SL, Cundliffe E. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 352.Sutherland JB. Appl Environ Microbiol. 1986;52:98–100. doi: 10.1128/aem.52.1.98-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


