Abstract
Enhancers are regulatory DNA sequences that can activate transcription over large distances. Recent studies have revealed the widespread role of distant activation in eukaryotic gene regulation and in the development of various human diseases, including cancer. Here we review recent progress in the field, focusing on new experimental and computational approaches that quantify the role of chromatin structure and dynamics during enhancer–promoter interactions in vitro and in vivo.
Keywords: : activation, distant action, enhancers, mechanisms, promoters
Transcription of the vast majority of eukaryotic genes is regulated by enhancers [1] – DNA sequences that bind transcription factors and activate transcription at promoter sites over variable distances (up to more than a million base pairs [1 Mb] away) [2]. The enhancers work in concert with other distantly placed regulatory elements (e.g., silencers [3] and insulators [4]) on the core promoters [5]. Genomic studies have identified specific ‘signatures’ of enhancers and promoters, such as histone modifications, associated proteins and RNA [6]. There are about a million human enhancers [7,8], with thousands active in particular cell types [9,10]. However, the majority of enhancers are described based only on evolutionary sequence conservation, without corresponding functional characterization (e.g., see [11]). The poor sequence similarity between different enhancers makes bioinformatics analysis challenging (reviewed in [12]). Most promoters associate with a single enhancer, but about 25% associate with two or more enhancers at any given time [13]. Recent genomic studies using various versions of the chromatin conformation capture (3C) method, in other words, combining chemical cross-linking with high-throughput sequencing, have revealed widespread use of enhancers in gene regulation [6,14]. In fact, enhancers are the regulatory elements that mediate the transcriptional activation of the vast majority of human genes [15] and largely dictate the state of chromatin near promoter regions [16]. Changes in enhancer-dependent gene expression can lead to the development of many types of cancers as well as to heart and autoimmune diseases in humans [17–21].
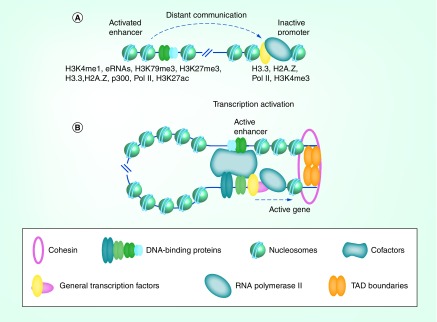
To be activated, a gene must be positioned within a more open chromatin region within nuclei (euchromatin) [22–24]. These changes in chromatin structure can affect the transcription of genes [25,26]: that is, the same genes can show different levels of expression in euchromatin as compared with heterochromatin, where the DNA is more tightly packed. In mammals, heterochromatin domains spread to cover developmentally regulated genes during the differentiation of stem cells [27]. The activation of genes starts with the labeling of inactive enhancers by ‘bivalent’ marks (H3K4me1 and H3K27me3), histone variants (H3.3 and H2A.Z) and/or elongating (paused) RNA polymerase II (Pol II) complexes with short aborted transcripts [28–31]. The enhancers and promoters in different cell types are then activated by cell-specific transcription factors and become associated with short enhancer RNAs, p300, H3K79me3, H3K27ac [32,33] and various coactivators, including histone modifiers and ATP-dependent chromatin remodelers [34–36] (Figure 1A). Note that the model described above has been developed for re-activated enhancers already carrying epigenetic marks; it is not clear whether the model is applicable to enhancers activated de novo during development. The activated enhancers subsequently communicate with their target promoters. Once the enhancer-bound protein(s) reach the target promoter(s), they establish direct interaction with promoter-bound protein(s), accompanied by formation of a loop containing the intervening chromatin [37] (Figure 1B). Formation of the loop is facilitated by CTCF/cohesin interactions, which form a fixed ring likely encompassing the base of the loop [38], and by various cofactors including Mediator that serve as a bridge between promoters and enhancers (Figure 1B) ([39,40], reviewed in [41]). After formation of the enhancer–promoter loop, activation of the promoter can occur either by recruiting of general transcription factor TFIID to the promoter and its activation through interaction with general transcription factor TFIIA or by activating Pol II paused on poised promoters [31,42–46].
Figure 1. . Proposed mechanism of action of eukaryotic enhancers.
(A) Prior to enhancer activation, enhancers and target promoters are marked by histone variants (H3.3, H2A.Z), pioneering transcription factors, various histone modifications (e.g. H3K4me1, H3K79me3, H3K27me3, H3K27ac), eRNAs, p300, Pol II and specific transcription factors. (B) Activation of the enhancer is followed by enhancer–promoter communication, chromatin looping (assisted by CTCF/cohesin bound at the TAD boundaries and by various cofactors including Mediator) and by promoter activation.
eRNA: Enhancer RNA; TAD: Topologically associating domain.
Super-enhancers constitute a specific group of enhancers, which drive the expression of genes that determine cell fate [47,48]. Super-enhancers are large clusters of transcriptional enhancers characterized by a high density of transcription factor binding sites (more than ten-times that of a typical enhancer), specific histone marks (H3K4me1 or H3K27ac) and the mediator complex Med1 [47,49]. Malfunctioning super-enhancers are associated with many human diseases, including cancer and neurodegenerative disorders (see [50] for review).
Although various factors participating in enhancer action have been identified, understanding their interplay and the mechanistic aspects of enhancer action trails behind, in part because of the limitations of current experimental approaches. Below we review some new experimental techniques that are making it possible to dissect the mechanism of enhancer action and recent progress in the field; other aspects of enhancer action have been covered in several excellent reviews [51–54].
Approaches for analysis of enhancer–promoter interactions
Experimental approaches for analysis of enhancer–promoter communication in vitro
Long-range communication between sequentially distant elements on DNA was initially quantified through assays of the ligation-circularization propensities of ‘sticky’ complementary, single-stranded oligonucleotide extensions at the ends of long DNA molecules [55–57] and of nucleosome-bound chromatin constructs in vitro [58]. However, extensive internucleosomal interactions strongly complicate interpretation of the experiments involving chromatin [e. nizovtseva, u npublished Data]. The more recent development of a novel fluorescence-based, protein-free approach now makes it possible to measure the looping of single DNA molecules in real time [59] (Figure 2A). The method detects the association/dissociation of dyes at the ends of 8- to 10-nucleotide overhangs on the analyzed DNA fragments. Although this approach could prove useful in the future for analysis of communication in chromatin, neither it nor the classic ligation assay offers insight into enhancer-specific mechanisms of communication with promoters (i.e., the role of protein–protein or DNA–protein interactions facilitating communication).
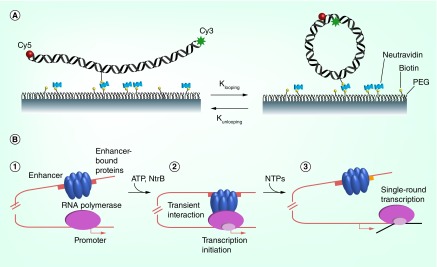
Figure 2. . Experimental approaches for analysis of enhancer-promoter communication in vitro.
(A) Looping of single DNA molecules in real time. Donor (Cy3) and acceptor (Cy5) labeled DNA molecules are immobilized on a surface via biotin–neutrAvidin interaction and the rate of dye association/dissociation is monitored by analysis of the efficiency of Förster resonance energy transfer. (B) Measuring enhancer–promoter communication [60,63]. 1. Before transcription is induced, RNA polymerase is bound at the promoter but cannot initiate transcription, and activator proteins are bound to the enhancer but cannot communicate with the promoter. 2. Enhancer interacts with RNA polymerase after fast phosphorylation of the activator proteins by protein kinase NtrB, causing looping of the intervening DNA and initiation of transcription. 3. In the presence of NTPs RNA polymerase completes transcription, and the yield of transcripts that correlates with the efficiency of enhancer–promoter communication is measured.
NTP: Nucleotide triphosphate.
(A) Reproduced with permission from [59].
A new quantitative assay of the rates of distant communication between a bacterial transcription regulatory element (enhancer) and its target (promoter) on DNA versus chromatin is providing fresh insights into the contributions of proteins and DNA to long-range transcriptional regulation [60–63] (Figure 2B). In this experimental system, enhancer–promoter communication (EPC) occurs within the same DNA molecule, communication is the rate-limiting step in the entire transcription reaction, and the yield of transcripts is directly proportional to the rate of communication on both DNA and chromatin templates [62]. The nucleosomes on the chromatin constructs lie between the enhancer and promoter, with the communicating regions being nucleosome-free [60]. The technique has been used to study the mechanisms of distant communication on histone-free DNA [62,64] and in chromatin [37,65,66]. In the case of histone-free DNA, the experimental results quantitatively mirror the expected end-to-end communication properties of DNA [67]. The approach has been used to measure communication rates on relatively short chromatin fragments containing up to 25 nucleosomes [37]. Since this approach provides quantitative information on the rates of EPC in chromatin, it was also used to evaluate predictions of the computational approaches described below [65,68].
Experimental approaches for analysis of EPC in vivo
Approaches currently in use for the analysis of enhancer–promoter interaction in vivo are far less quantitative than those used in vitro. The growing set of in vivo approaches now includes: chromatin immunoprecipitation (ChIP) and DNase I treatments combined with DNA sequencing (ChIP-seq and DNAse-seq, respectively) to identify enhancers; chromosome conformation capture methods to detect enhancers and promoters in direct physical proximity; and functional assays such as clustered regularly interspaced short palindromic repeats and transcription activator-like effectors (TALEs) to evaluate activation of transcription [69]. Note that these tools provide information about average genome organization in a population of cells, as opposed to that in individual cells which could differ dramatically. Newly developed single-cell combinatorial indexed high-throughput sequencing data hint of the potential variation of 3D genome structure in single cells [12,70].
The ChIP-seq approach involves the cross-linking of transcription factors to their DNA binding sites in vivo. The resulting cross-linked complexes are isolated via immunoprecipitation using an antibody against the transcription factor, RNA polymerase or a histone mark. Subsequent genomic sequencing makes it possible to identify the enhancers that are active in a specific chromatin environment, such at sites of lysine 27 acetylation on histone H3 (H3K27ac), lysine 4 monomethylation on histone H3 (H3K4me1) or other modifications detected in cell culture and in human tissues (Figure 1A; for review see [69,71,72]). The ChIP-seq method makes it possible to identify active and potentially active enhancers and promoters and is very useful for analysis of enhancer activation in response to drugs and other changes in the cellular environment. The approach, however, does not provide information about chromatin topology and does not identify the target promoters activated by a particular enhancer. Different variants of the 3C methods are used for this purpose.
Chromosome conformation capture is based on the idea that closely localized genomic regions can be cross-linked in vivo. The in vivo cross-linked DNA fragments are then digested with restriction enzymes or sonicated and re-ligated to one another. The newly ligated DNA fragments are identified using DNA sequencing or the PCR with primers to the regions of interest (3C [73]). Different modifications of the 3C approach employ different methods to separate the DNA fragments after cross-linking (4C, 5C [74,75]) and allow for high-throughput data analysis and generation of genome-wide contact maps (Hi-C [76,77]). A combination of Hi-C and ChIP, so-called ChIA-PET [78], allows for genome-wide detection of the protein-mediated loops captured by the various 3C methods described above. Although the ligation frequencies can be quantified, the data obtained by any of the currently available 3C methods cannot be directly translated into contact frequencies in vivo. Indeed, the efficiency of cross-linking utilized by these methods is apt to vary dramatically and to depend on the relative distances between the analyzed genomic regions, the accessibility of the cross-linker and the availability of the DNA ends for ligation (reviewed in [77]). The rates of EPC are also extremely difficult to analyze by current 3C techniques.
A variety of functional assays can be applied to investigate minimal enhancer sequences and the dependence of enhancer action on the distance from the target promoter, the chromatin environment and the positioning of the enhancers within the topologically associating domains (TADs) of chromatin (reviewed in [79]). Other new-generation functional assays include clustered regularly interspaced short palindromic repeats/cas9 editing machinery that allows for enhancer activation (CRISPR; a segment of DNA containing short repetitions of base sequences, involved in the defense mechanisms of prokaryotic organisms to viruses [80]) or inhibition (CRISPi; CRISPR-inhibiting enhancers [81]) and TALEs, which target specific DNA sequences after fusion of the effector DNA-interacting proteins to a repressor or activator domain. TALE has been used to study protein domains involved in activation or repression of transcription after binding to a target DNA sequence [82].
Computational approaches for analysis of EPC
Computational treatment of the arrays of nucleosomes in native chromatin fibers lies beyond the scope of conventional atomic-level macromolecular methods [83–86]. Computer resources preclude detailed study of the atoms in such large assemblies. The supporting low-resolution experimental data also call for interpretations that conform to the characteristics of the molecular system. An understanding of the long-range properties of chromatin requires knowledge of the contributions of whole nucleosomes, the interspersed protein-free DNA linkers and the surrounding molecular environment. These molecular components (Figure 3) provide a simple means to interpret various mesoscale properties of chromatin fibers, such as the strong internucleosomal associations detected with force-extension technology [87] and electric dichroism measurements [88]; the effects of nucleosome composition and number of the global dimensions of chromatin fibers [89]; the packing of nucleosomes in solid-state structures [81,90,91]; the contacts of nucleosomes deduced through chemical cross-linking [92]; and the ability of chromatin fibers to support EPC [37,65].
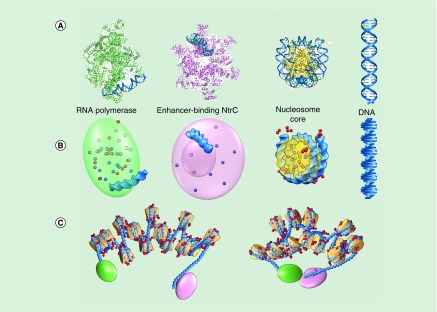
Figure 3. . Modeling long-range communication between regulatory elements on chromatin arrays.
(A) Protein-bound DNA segments – the glnAp2 promoter recognized by RNA polymerase, the enhancer site bound by the bacterial nitrogen regulatory protein C and the DNA on the nucleosome core particle – follow pathways, highlighted in deep blue, from known high-resolution structures [99,100,129–134]. The DNA linkers between successive protein–DNA complexes undergo small elastic deformations about the ideal double-helical structure. (B) A reduced depiction of protein and DNA facilitates computational treatment of the chromatin assembly. The simplification of DNA to a series of base pairs (blue blocks) and the cationic/anionic amino-acid residues to a set of point charges (small red/blue spheres at positive/negative sites) make it possible to bridge the gap between the size of systems that can be studied in full atomic detail and events, such as enhancer–promoter communication, that takes place on a larger molecular scale. The representation of the DNA-bound proteins as simple geometric objects enclosing the folded polypeptide chains expedites detection of molecular collisions. (C) The simulated changes in large-scale structure, depicted in open (left) and successfully contacted (right) states, arise from the deformations of the protein-free DNA linker segments guided by collision avoidance and electrostatic interactions between protein and DNA. Note the contacts of the mobile charges on the histone tail (red spheres) with the exposed faces of nearby nucleosomes in the more compact state.
Recent computational treatments of long-range communication on nucleosome arrays [68,93,94] build upon earlier coarse-grained models of deformable chromatin fibers [95,96], which combine a static representation of the nucleosome with known mechanical properties of protein-free DNA. The DNA on both the nucleosomes and the regulatory proteins adopts the precise 3D pathways found in known high-resolution structures (Figure 3A) [97–100] while the protein-free DNA linkers undergo elastic deformations, in other words, small fluctuations in bending and twisting, consistent with the solution properties of double-helical DNA [101]. The treatment of DNA as a sequence of base pairs rather than a series of longer helical elements [96,102] makes it possible to capture the ‘high-resolution’ information contained in the experiments, such as the effect of small shifts in nucleosome positioning on long-range communication. Representation of the proteins as rigid, impenetrable objects, for example, ellipsoids or cylinders (Figure 3B), with shapes compatible with their known 3D folds facilitates identification of contact-free arrangements of protein and DNA. The added consideration of atomic charges helps to distinguish the more likely configurations. A small set of point charges, extracted from the spatial locations of cationic and anionic amino-acid residues, preserves key features of the folded histone core, for example, the so-called acidic patches on the exposed faces of the nucleosome core particle [103]. Contacts of the acidic patch on one nucleosome with mobile point charges on the exposed histone tails of nearby nucleosomes [65,94] give rise to attractive interactions that enhance chain compaction (Figure 3C). The charges on DNA and the terminal regulatory proteins similarly contribute to the formation of closed loops. Models of rigid, fully extended fibers, such as those used to identify the optimal arrangements of nucleosomes in chromatin [104–107], do not capture the variations in communication levels with the numbers, composition and spacing of nucleosomes. The rigid structures, nevertheless, serve as critical reference points for simulations of deformable oligonucleosome chains.
Importantly, the combination of these computational approaches with experimental measurements in several recent studies has identified critical factors contributing to the formation of chromatin fibers and helped to unravel mechanisms of EPC. For example, trends in the predicted interactions of nearby nucleosomes in regular nucleosome arrays compared with those detected through a combination of electron microscopy and chemical footprinting have pointed to various factors that may promote the higher-order structure of chromatin, such as enhanced bending of the protein-free DNA linkers between successive nucleosomes [92]. Recent comparisons of computer-simulated models with the observed associations of nucleosomes in the chromatin from HeLa cells at two stages of the cell cycle suggest how the level of linker histone uptake may contribute to the deformability and looping propensities of nucleosome-decorated DNA chains [108].
Mechanisms of EPC in chromatin
Most enhancers activate transcription in cis, over DNA stretches with an average length of approximately 100 kb [109]; the actual contour length between the enhancer and target promoter on a chromatin fiber is considerably shorter (equivalent to ∼3 kb of stretched linear DNA), considering the approximately 30-fold DNA compaction associated with chromatin folding [110]. Given that the enhancer must physically reach the promoter, in most cases over a large distance in cis (Figure 1B), long-range communication is the principal aspect of enhancer action. In fact, this early step could be rate-limiting in the regulation of at least some genes (reviewed in [37]). Therefore, it is essential to identify chromatin properties that allow for efficient enhancer action.
The need for close association raises the question of how an enhancer can efficiently reach a sequentially distant promoter. Chromatin structure per se is a highly efficient communication device [58], mediating long-range interactions by looping the chromatin positioned between a promoter and an enhancer in vitro [66,111]. Importantly, the high efficiency of EPC cannot be explained by chromatin-induced DNA compaction alone [66,68]. Furthermore, modulation of chromatin architecture is one of the key factors in dynamic transcriptional regulation [112] (see [52,113] for review). These observations raise a number of important questions, such as: What elements of chromatin structure mediate efficient EPC? What are the mechanisms of efficient EPC in chromatin? How can EPC be inhibited or facilitated by various factors? Some of these questions have been answered in recent studies involving the combination of experimental and computational approaches described above.
Recent data obtained using well-defined chromatin arrays and an enhancer–promoter system assembled from purified components suggest that the presence of nucleosomes along with DNA facilitates communication between transcriptional proteins [68]. Here the experimentally measured signals, expressed in terms of the levels of enhancer-dependent RNA transcripts produced on nucleosome-decorated versus nucleosome-free DNA of the same length, provide critical benchmarks for evaluating mesoscale models of chromatin. Correspondence between the predicted likelihood of contact between the ends of computer-simulated oligonucleosome arrays and the experimental data lends credence to the models and points to details of macromolecular structure and deformation potentially responsible for the observed behavior of the chromatin constructs.
The data further show that internucleosomal interactions involving the histone tails are essential for highly efficient, long-range EPC in chromatin in vitro [68]. The molecular simulations of the communicating chromatin fibers suggest that transient binary internucleosomal interactions can mediate distant communication between the enhancer and promoter in chromatin [68]. In particular, electrostatic interactions between the N-terminal tails of the core histones and nearby nucleosomal DNA (Figure 3C) enhance the computed probability of interaction between DNA sites that lie far apart along the sequence. Importantly, the histone tails not only contribute to the high efficiency of EPC, but also allow for nearly distance-independent communication [68].
It has been proposed that long-range, transient internucleosomal interactions through the histone N-terminal tails [114,115] could keep chromatin fibers in close proximity while at the same time allow the relocation of interacting fibers relative to one another [68]. These relocations could occur by a mechanism involving sliding of interacting chromatin fibers or by re-establishing internucleosomal interactions de novo [37,116]. Although internucleosomal interactions themselves are not sufficient to account for action over a distance, these transient nonspecific interactions within the chromatin fiber strongly enhance long-distance regulatory enhancer–promoter interactions [68,116]. The subdivision of a loop into two topologically independent loops by insulators inhibits interdomain interactions and is expected to strongly inhibit enhancer action over a distance [116]. Thus, nonspecific internucleosomal interactions are likely important for both enhancer action over a distance and insulator activity of the boundary elements.
It has been shown that nucleosome spacing and the presence of nucleosome-free DNA regions can considerably affect the structure/dynamics of chromatin fibers and, in turn, affect the efficiency of enhancer–promoter interactions in vitro [65]. In particular, on nucleosomal arrays containing more than seven nucleosomes in the presence of a single nucleosome-free DNA region can positively affect the rate of EPC [65]. The presence of nucleosome-free DNA regions could be essential for communication since the chromatin fiber is a rigid polymer that otherwise does not support efficient formation of loops in vivo [117,118].
The studies described above are currently focused on the analysis of enhancer–promoter interactions over relatively short distances (1–5 kb). However, knowledge of the spatial arrangements of interacting nucleosomes in regular nucleosome arrays provides a basis for linking the ‘local’ mesoscale features of chromatin to higher-order macromolecular structures, such as the long, approximately 100 kb loops of chromatin, detected within the TADs of chromosomes [119,120]. The nucleosome–nucleosome interactions extracted from base-pair-level simulations of short oligonucleosome arrays that account for experimental data provide the necessary input for direct, nucleosome-level simulations of the closure propensities and structural features of chromosomal loops of the same chemical make-up [94,121]. Moreover, given the known sensitivity of nucleosome positioning to DNA base sequence [122–124], a nucleosome-level depiction of chromatin provides a direct link from primary nucleotide sequence, double-helical structure and nucleosome-decorated DNA to looped architectures detected at the chromosomal level.
As emphasized in this review, the configurational properties of nucleosome-decorated DNA reflect the spacing and composition of the constituent nucleosomes. The precise positions and chemical make-up of nucleosomes modulate the communication between regulatory proteins at the ends of well-defined oligonucleosome arrays. Ensembles of modeled structures consistent with these observations exhibit very different 3D features upon repositioning or modification of nucleosomes (Figure 4). The local structures and deformabilities of the nucleosomes can, in turn, influence the folding and interactions of longer stretches of chromatin [94,121]. Other factors known to modulate the local spatial arrangements of nucleosomes can similarly contribute to the long-range properties of chromatin. For example, changes in DNA composition that perturb the ‘breathing’ propensities of nucleosomes, such as replacement of the 601 sequence used in recent studies [37,65] by a less perfect nucleosome-positioning sequence, could affect the average local spatial arrangements of nucleosomes and thereby influence the likelihood of contact between terminal regulatory proteins. The introduction of chromosomal factors that recognize specific features of the nucleosome core particle may not only perturb local nucleosome structure but also contribute to the relative positions of nucleosomes on DNA and the consequent remodeling of chromatin. Proteins known to recognize the acidic patch, such as the polycomb repressive complex [125], can disrupt the stacking of nucleosomes found in regular oligonucleosome arrays, while the uptake of linker histones in the vicinity of the nucleosomal dyad [126,127] can perturb and potentially stiffen the pathways of DNA entering and exiting the nucleosome.
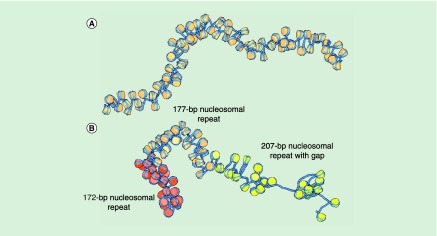
Figure 4. . Molecular snapshots illustrating the potential effects of nucleosome positioning on large-scale chromatin folding.
(A) The smoothly deformed pathway of a regularly spaced construct, depicted for a stretch of 78 intact nucleosomes with uniform 177-bp spacing, undergoes compression/extension and large-scale bending/twisting upon changes in nucleosome positioning. (B) The 26 nucleosomes (deep brown) positioned at increments of 172 bp at the beginning of a 77-nucleosome construct rearrange and form a more compact ‘secondary structure’, as compared with the 26 nucleosomes in the middle and the 25 nucleosomes at the end of the same chain, with respective nucleosome repeats of 177 bp (light brown) and 207 bp (yellow). Note that the junction between ‘helical’ stretches in the simulated construct becomes more pronounced when the spacing changes by roughly a half turn of DNA (172 - 177 = -5 bp) than when altered by a multiple of the tenfold double-helical repeat (207 - 177 = 30 bp) [65]. The presence of a nucleosome-free gap, here located within the stretch of nucleosomes spaced at 207-bp intervals, enhances the flexibility of the modeled structure, allowing the chain to bend in various directions.
Making the connection between the sequence-dependent information in short chromatin constructs and the in vivo looping of DNA captured with assorted new technologies require extension of present nucleosome level studies of regular chromatin arrays to oligonucleosomes with the mixed spacings mapped at base-pair resolution on genomic sequences [122,123,128]. The loops of chromatin detected in genomic studies contain hundreds of nucleosomes and, as emphasized here (Figure 4), the likelihood of their formation depends upon the spacing and composition of the constituent nucleosomes. The predicted properties of chromatin constructs with nucleosomes positioned at the sites mapped in a given genome can potentially be compared with the known intrachromosomal contacts along the same DNA. Characterization of the long-range interactions between the unevenly spaced nucleosomes on chromatin, however, requires knowledge of the configurational properties of a very large number of well-defined oligonucleosomes. The interpretation of chromosomal looping also requires knowledge of the proteins that mediate these structures at the genomic level and the interactions, or bridging, of nearby loops are believed to occur within TADs.
Future perspective
Although the recent studies described above document considerable progress in mechanistic understanding of enhancer action in chromatin, the in vitro and in vivo work remains largely disconnected. Development of more quantitatively tractable in vivo approaches and in vitro chromatin-covered templates supporting enhancer-promoter interaction on larger nucleosomal arrays would fill in the gap and allow for development of authentic systems recapitulating physiologically relevant aspects of enhancer action and analysis of enhancer malfunction in human diseases.
In spite of the rapid progress in understanding of the principles of enhancer-dependent transcription, many aspects of this process and its role in human diseases remain to be studied. Main remaining challenges include functional characterization of enhancers, identification of their target genes and trancription factor partners, mechanistic understanding of the impact of the changes in chromatin structure on enhancer action and analysis of the mechanisms of enhancer action in single cells.
Executive summary.
Enhancers play widespread role in eukaryotic gene regulation and in the development of various human diseases.
The enhancer-promoter interaction is an early and likely regulated step in gene activation.
The efficiency of enhancer-promoter interaction are largely dictated by the structure/dynamics of intervening chromatin.
New experimental approaches allow quantitative analysis of gene activation over a distance in vitro.
In vitro and in vivo studies are currently largely disconnected.
Acknowledgements
We thank R Young for helpful comments on the work.
Footnotes
Financial & competing interests disclosure
This work was supported by NIH research grants RO1-GM34809 to WK Olson and RO1-GM119398 to VM Studitsky, by Fox Chase Cancer Center start-up funds to VM Studitsky, and by Russian Science Foundation grant 14–24–00031. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Wasylyk B, Wasylyk C, Chambon P. Short and long range activation by the SV40 enhancer. Nucleic Acids Res. 1984;12(14):5589–5608. doi: 10.1093/nar/12.14.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vokes SA, Ji H, Wong WH, Mcmahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22(19):2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghirlando R, Giles K, Gowher H, et al. Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta. 2012;1819(7):644–651. doi: 10.1016/j.bbagrm.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13(4):233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 6.Maston GA, Landt SG, Snyder M, Green MR. Characterization of enhancer function from genome-wide analyses. Annu. Rev. Genomics Hum. Genet. 2012;13:29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- 7.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennacchio LA, Ahituv N, Moses AM, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 12.Slattery M, White KP. Enhanced dissection of the regulatory genome. Nat. Methods. 2013;10(8):710–712. doi: 10.1038/nmeth.2577. [DOI] [PubMed] [Google Scholar]
- 13.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22(3):490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15(4):272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 15.Li G, Ruan X, Auerbach RK, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014;21(3):210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- 18.Wamstad JA, Wang X, Demuren OO, Boyer LA. Distal enhancers: new insights into heart development and disease. Trends Cell Biol. 2014;24(5):294–302. doi: 10.1016/j.tcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Peeters JG, Vervoort SJ, Tan SC, et al. Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep. 2015;12(12):1986–1996. doi: 10.1016/j.celrep.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 20.Sur I, Taipale J. The role of enhancers in cancer. Nat. Rev. Cancer. 2016;16(8):483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 21.Abraham BJ, Hnisz D, Weintraub AS, et al. Small genomic insertions form enhancers that misregulate oncogenes. Nat. Commun. 2017;8:14385. doi: 10.1038/ncomms14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morey C, Da Silva NR, Kmita M, Duboule D, Bickmore WA. Ectopic nuclear reorganisation driven by a Hoxb1 transgene transposed into Hoxd. J. Cell Sci. 2008;121(Pt 5):571–577. doi: 10.1242/jcs.023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118(5):555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Naughton C, Sproul D, Hamilton C, Gilbert N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol. Cell. 2010;40(3):397–409. doi: 10.1016/j.molcel.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila . Cold Spring Harb. Perspect. Biol. 2013;5(8):a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins RD, Hon GC, Lee LK, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellner WA, Ramos E, Van Bortle K, Takenaka N, Corces VG. Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters. Genome Res. 2012;22(6):1081–1088. doi: 10.1101/gr.136929.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin C, Zang C, Wei G, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009;41(8):941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golob JL, Kumar RM, Guenther MG, et al. Evidence that gene activation and silencing during stem cell differentiation requires a transcriptionally paused intermediate state. PLoS ONE. 2011;6(8):e22416. doi: 10.1371/journal.pone.0022416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonn S, Zinzen RP, Girardot C, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44(2):148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 34.D'alessio JA, Wright KJ, Tjian R. Shifting players and paradigms in cell-specific transcription. Mol. Cell. 2009;36(6):924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11(6):426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 36.Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011;22(7):759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 37.Kulaeva OI, Nizovtseva EV, Polikanov YS, Ulianov SV, Studitsky VM. Distant activation of transcription: mechanisms of enhancer action. Mol. Cell. Biol. 2012;32(24):4892–4897. doi: 10.1128/MCB.01127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haarhuis JHI, Van Der Weide RH, Blomen VA, et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169(4):693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uthe H, Vanselow JT, Schlosser A. Proteomic analysis of the Mediator complex interactome in Saccharomyces cerevisiae . Sci. Rep. 2017;7:43584. doi: 10.1038/srep43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrenko N, Jin Y, Wong KH, Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell. 2016;64(3):443–454. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16(3):155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 43.Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature. 2010;465(7300):956–960. doi: 10.1038/nature09080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilchrist DA, Dos Santos G, Fargo DC, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng B, Li T, Rahl PB, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell. 2012;45(1):38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loven J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niederriter AR, Varshney A, Parker SC, Martin DM. Super enhancers in cancers, complex disease, and developmental disorders. Genes. 2015;6(4):1183–1200. doi: 10.3390/genes6041183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169(1):13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson LA, Tsai LH. In the loop: how chromatin topology links genome structure to function in mechanisms underlying learning and memory. Curr. Opin. Neurobiol. 2016;43:48–55. doi: 10.1016/j.conb.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adam RC, Fuchs E. The yin and yang of chromatin dynamics in stem cell fate selection. Trends Genet. 2016;32(2):89–100. doi: 10.1016/j.tig.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shore D, Langowski J, Baldwin RL. DNA flexibility studied by covalent closure of short fragments into circles. Proc. Natl Acad. Sci. USA. 1981;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell. 2004;14(3):355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 57.Crothers DM, Drak J, Kahn JD, Levene SD. DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- 58.Stein A, Dalal Y, Fleury TJ. Circle ligation of in vitro assembled chromatin indicates a highly flexible structure. Nucleic Acids Res. 2002;30(23):5103–5109. doi: 10.1093/nar/gkf671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vafabakhsh R, Ha T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science. 2012;337(6098):1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polikanov YS, Studitsky VM. Analysis of distant communication on defined chromatin templates in vitro . Methods Mol. Biol. 2009;543:563–576. doi: 10.1007/978-1-60327-015-1_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polikanov YS, Rubtsov MA, Studitsky VM. Biochemical analysis of enhancer-promoter communication in chromatin. Methods. 2007;41(3):250–258. doi: 10.1016/j.ymeth.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Bondarenko V, Ninfa A, Studitsky VM. DNA supercoiling allows enhancer action over a large distance. Proc. Natl Acad. Sci. USA. 2001;98(26):14883–14888. doi: 10.1073/pnas.261477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bondarenko VA, Liu YV, Jiang YI, Studitsky VM. Communication over a large distance: enhancers and insulators. Biochem. Cell Biol. 2003;81(3):241–251. doi: 10.1139/o03-051. [DOI] [PubMed] [Google Scholar]
- 64.Bondarenko VA, Jiang YI, Studitsky VM. Rationally designed insulator-like elements can block enhancer action in vitro . EMBO J. 2003;22(18):4728–4737. doi: 10.1093/emboj/cdg468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nizovtseva EV, Clauvelin N, Todolli S, et al. Nucleosome-free DNA regions differentially affect distant communication in chromatin. Nucleic Acids Res. 2017;45(6):3059–3067. doi: 10.1093/nar/gkw1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rubtsov MA, Polikanov YS, Bondarenko VA, Wang YH, Studitsky VM. Chromatin structure can strongly facilitate enhancer action over a distance. Proc. Natl Acad. Sci. USA. 2006;103(47):17690–17695. doi: 10.1073/pnas.0603819103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polikanov YS, Bondarenko VA, Tchernaenko V, et al. Probability of the site juxtaposition determines the rate of protein-mediated DNA looping. Biophys. J. 2007;93(8):2726–2731. doi: 10.1529/biophysj.107.111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulaeva OI, Zheng G, Polikanov YS, et al. Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J. Biol. Chem. 2012;287(24):20248–20257. doi: 10.1074/jbc.M111.333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buffry AD, Mendes CC, Mcgregor AP. The functionality and evolution of eukaryotic transcriptional enhancers. Adv. Genet. 2016;96:143–206. doi: 10.1016/bs.adgen.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Ramani V, Deng X, Qiu R, et al. Massively multiplex single-cell Hi-C. Nat. Methods. 2017;14(3):263–266. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle. 2014;13(18):2847–2852. doi: 10.4161/15384101.2014.949201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowman SK. Discovering enhancers by mapping chromatin features in primary tissue. Genomics. 2015;106(3):140–144. doi: 10.1016/j.ygeno.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 74.Simonis M, Klous P, Splinter E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38(11):1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 75.Zhao Z, Tavoosidana G, Sjolinder M, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38(11):1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 76.Ferraiuolo MA, Sanyal A, Naumova N, Dekker J, Dostie J. From cells to chromatin: capturing snapshots of genome organization with 5C technology. Methods. 2012;58(3):255–267. doi: 10.1016/j.ymeth.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denker A, De Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30(12):1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kvon EZ. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106(3):185–192. doi: 10.1016/j.ygeno.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Horlbeck MA, Gilbert LA, Villalta JE, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5:e19760. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thakore PI, D'ippolito AM, Song L, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12(12):1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crocker J, Stern DL. TALE-mediated modulation of transcriptional enhancers in vivo . Nat. Methods. 2013;10(8):762–767. doi: 10.1038/nmeth.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruscio JZ, Onufriev A. A computational study of nucleosomal DNA flexibility. Biophys. J. 2006;91(11):4121–4132. doi: 10.1529/biophysj.106.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Materese CK, Savelyev A, Papoian GA. Counterion atmosphere and hydration patterns near a nucleosome core particle. J. Am. Chem. Soc. 2009;131(41):15005–15013. doi: 10.1021/ja905376q. [DOI] [PubMed] [Google Scholar]
- 85.Biswas M, Langowski J, Bishop TC. Atomistic simulations of nucleosomes. WIREs Comput. Mol. Sci. 2013;3:378–392. [Google Scholar]
- 86.Shaytan AK, Armeev GA, Goncearenco A, Zhurkin VB, Landsman D, Panchenko AR. Coupling between histone conformations and DNA geometry in nucleosomes on a microsecond timescale: atomistic insights into nucleosome functions. J. Mol. Biol. 2016;428(1):221–237. doi: 10.1016/j.jmb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chien F-T, Van Noort J. 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr. Pharm. Biotechnol. 2009;10(5):474–485. doi: 10.2174/138920109788922128. [DOI] [PubMed] [Google Scholar]
- 88.Mcghee JD, Rau DC, Charney E, Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- 89.Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 90.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436(7047):138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 91.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30 nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl Acad. Sci. USA. 2006;103(17):6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl Acad. Sci. USA. 2009;106(32):13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olson WK, Clauvelin N, Colasanti AV, Singh G, Zheng G. Insights into gene expression and packaging from computer simulations. Biophys. Revs. 2012;4(3):171–178. doi: 10.1007/s12551-012-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clauvelin N, Lo P, Kulaeva OI, et al. Nucleosome positioning and composition modulate in silico chromatin flexibility. J. Phys. Condens. Matter. 2015;27(6):064112. doi: 10.1088/0953-8984/27/6/064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katritch V, Bustamante C, Olson WK. Pulling chromatin fibers: computer simulations of direct physical micromanipulations. J. Mol. Biol. 2000;295(1):29–40. doi: 10.1006/jmbi.1999.3021. [DOI] [PubMed] [Google Scholar]
- 96.Wedemann G, Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys. J. 2002;82(6):2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 98.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296(5571):1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 99.Lee S-Y, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17(20):2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon T. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20(11):1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Czapla L, Swigon D, Olson WK. Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theor. Comput. 2006;2(3):685–695. doi: 10.1021/ct060025+. [DOI] [PubMed] [Google Scholar]
- 102.Arya G, Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 104.Ben-Haïm E, Lesne A, Victor J-M. Chromatin: a tunable spring at work inside chromosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001;64(5 Pt. 1):051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 105.Koslover EF, Fuller CJ, Straight AF, Spakowitz AJ. Local geometry and elasticity in compact chromatin structure. Biophys J. 2010;99(12):3941–3950. doi: 10.1016/j.bpj.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scipioni A, Turchetti G, Morosetti S, De Santis P. Geometrical, conformational and topological restraints in regular nucleosome compaction in chromatin. Biophys Chem. 2010;148(1–3):56–67. doi: 10.1016/j.bpc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 107.Norouzi D, Zhurkin VB. Topological polymorphism of the two-start chromatin fiber. Biophys J. 2015;108(10):2591–2600. doi: 10.1016/j.bpj.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl Acad. Sci. USA. 2016;113(5):1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spitz F. Gene regulation at a distance: from remote enhancers to 3D regulatory ensembles. Semin. Cell Dev. Biol. 2016;57:57–67. doi: 10.1016/j.semcdb.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 110.Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2015;589(20 Pt A):2914–2922. doi: 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laybourn PJ, Kadonaga JT. Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science. 1992;257(5077):1682–1685. doi: 10.1126/science.1388287. [DOI] [PubMed] [Google Scholar]
- 112.Won H, De La Torre-Ubieta L, Stein JL, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538(7626):523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vermunt MW, Creyghton MP. Transcriptional dynamics at brain enhancers: from functional specialization to neurodegeneration. Curr. Neurol. Neurosci. Rep. 2016;16(10):94. doi: 10.1007/s11910-016-0689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 2005;280(40):33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 115.Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 2007;27(6):2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mukhopadhyay S, Schedl P, Studitsky VM, Sengupta AM. Theoretical analysis of the role of chromatin interactions in long-range action of enhancers and insulators. Proc. Natl Acad. Sci. USA. 2011;108(50):19919–19924. doi: 10.1073/pnas.1103845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boedicker JQ, Garcia HG, Johnson S, Phillips R. DNA sequence-dependent mechanics and protein-assisted bending in repressor-mediated loop formation. Phys. Biol. 2013;10(6):066005. doi: 10.1088/1478-3975/10/6/066005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diesinger PM, Kunkel S, Langowski J, Heermann DW. Histone depletion facilitates chromatin loops on the kilobasepair scale. Biophys. J. 2010;99(9):2995–3001. doi: 10.1016/j.bpj.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rao SSP, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanborn AL, Rao SSP, Huang S-C, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA. 2015;112(47):E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Todolli S, Perez PJ, Clauvelin N, Olson WK. Contributions of sequence to the higher-order structures of DNA. Biophys. J. 2017;112(3):416–426. doi: 10.1016/j.bpj.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brogaard K, Xi L, Wang J-P, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nat. Protoc. 2012;486(7404):496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moyle-Heyrman G, Zaichuk T, Xi L, et al. Chemical map of Schizosaccharomyces pombe reveals species-specific features in nucleosome positioning. Proc. Natl Acad. Sci. USA. 2013;110(50):20158–20163. doi: 10.1073/pnas.1315809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cui F, Chen L, Loverso PR, Zhurkin VB. Prediction of nucleosome rotational positioning in yeast and human genomes based on sequence-dependent DNA anisotropy. BMC Bioinformatics. 2014;15:313. doi: 10.1186/1471-2105-15-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mcginty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514(7524):591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou B-R, Feng H, Kato H, et al. Structural insights into the histone H1-nucleosome complex. Proc. Natl Acad. Sci. USA. 2013;110(48):19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou B-R, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell. 2015;59(4):628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl Acad. Sci. USA. 2011;108(31):12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296(5571):1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 130.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. 2015;4:e08504. doi: 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pelton JG, Kustu S, Wemmer DE. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 1999;292(5):1095–1110. doi: 10.1006/jmbi.1999.3140. [DOI] [PubMed] [Google Scholar]
- 132.Batchelor JD, Doucleff M, Lee C-J, et al. Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation. J. Mol. Biol. 2008;384(5):1058–1075. doi: 10.1016/j.jmb.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 133.Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24(8):814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Davey CA, Sargent DF, Luger K, Mäder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1087–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]