Abstract
In this review, we detail our current knowledge of PrPSc structure on the basis of structural and computational studies. We discuss the progress toward an atomic resolution description of PrPSc and results from the broader field of amyloid studies that may further inform our knowledge of this structure. Moreover, we summarize work that investigates the role of PrPSc structure in its toxicity, transmissibility, and species specificity. We look forward to an atomic model of PrPSc, which is expected to bring diagnostics and/or therapeutics to the field of prion disease.
The misfolded prion protein, PrPSc, is responsible for the transmission of prion diseases. Although the detailed structure of PrPSc has been elusive, several models have been proposed.
NATIVE STRUCTURE OF THE MAMMALIAN PRION PROTEIN
Structure of PrPC
Before considering the structure of the infectious (scrapie) form of the prion protein (PrPSc), it is instructive to review the studies that inform us of the structure of the benign, natively folded cellular form (PrPC). For consistency, we quote residue numbers based on the human PrP sequence. In humans, the PRNP gene encodes a 231-residue protein after removal of its 22-residue signal peptide (Bamborough et al. 1996). A description of the PrP sequence beginning near its N terminus includes five conserved octapeptide repeats (residues 51–90) followed by a glycine-alanine-rich (GA-rich) segment (residues 113–127); a short β-strand (residues 128–132) that leads to a first α-helix (residues 144–156) and a second short strand (residues 160–164); a turn (residues 165–173) that leads to a second α-helix (residues 174–193); and a final helix (residues 199–229) that is linked to the membrane by a glycosylphosphatidylinositol (GPI) anchor (Fig. 1). Together these create the native fold of PrPC, a protein with a flexible N-terminal tail and a GPI anchor at its C terminus. This structure provides an important reference point to the initiation of PrPc aggregation during conversion to its pathogenic form. A number of nuclear magnetic resonance (NMR) and crystallographic experiments illuminated the structure of PrPC and provided us with several atomic models from a variety of species and sequence variants.
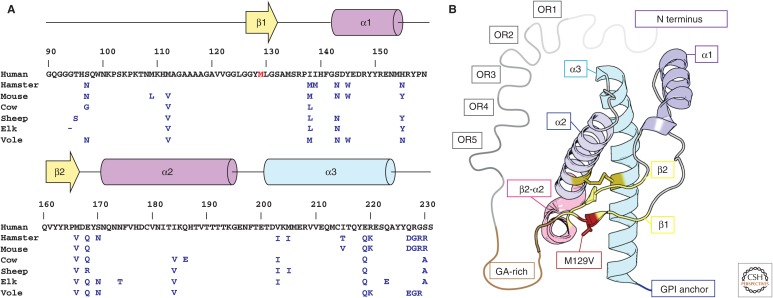
Figure 1.
Sequence and structure of human PrPC. (A) Primary sequence and secondary structure of human PrPC residues 90–230. Residues that differ between hamster, mouse, cow, sheep, elk, and bank vole sequences are indicated in blue, and those of the native human sequence are shown in black. The genetic variant M129V is shown in red. (B) Model of the crystal structure of the M129V variant of human PrPC. Secondary structure elements are labeled using a color scheme that follows A; these include five octapeptide repeats (OR1–5, gray), a glycine-alanine-rich stretch (GA-rich, brown), two β-strands (β1, β2, yellow), and three α-helices (α1–α3, magenta-blue), variant M129V (red), the β2–α2 turn (pink), and a C-terminal glycosylphosphatidylinositol (GPI) anchor (blue).
The structure of PrPC is central to the protein-only hypothesis of prions. It is this structure that first unfolds and later refolds into an infectious and toxic form. Using high-resolution protein NMR, Riek, Wüthrich, and coworkers were the first to obtain a model of the solution structure of the globular portion of PrPC. Their model is a recombinant form of the mouse protein containing residues 121–231 (Fig. 1) (Riek et al. 1996) and shows three α-helices and two β-strands—a compact globular fold that includes many residues, which when mutated, play a role in prion disease. The high sequence similarity between mouse and other mammalian prion proteins suggested they might have similar overall folds. Several other solution structures of the recombinant globular domains of PrPC from mouse and Syrian hamster soon followed (Donne et al. 1997; James et al. 1997; Riek et al. 1997). Some of the models that followed also included the GA-rich segment of the protein and its flexible N-terminal tail, although these flexible regions appeared largely unstructured (Donne et al. 1997; Riek et al. 1997). It would be four years from the first solution NMR structure of mouse PrPC until a model for human PrPC would be obtained (Zahn et al. 2000). The human model, recombinantly expressed and also determined by NMR, conserved the previously reported fold for mouse and hamster, but differed in the backbone flexibility of a few regions, including the terminal portions of the protein and several of its turns (Zahn et al. 2000). A wide variety of other NMR models have now been determined for PrPC, including a model of elk PrPC (Gossert et al. 2005). Models of the elk PrPC structure are distinct compared with models from other species because the region linking the second β-sheet and α-helix of the globular domain in the elk protein is well ordered (Gossert et al. 2005). Collectively, these models present a conserved fold for the highly homologous structures of PrPC, with some differences in the flexibility of certain regions.
High-resolution atomic structures of PrPC have also been obtained by crystallographic studies. The first of these studies determined the structure of recombinant human PrPC as a domain-swapped dimer (Knaus et al. 2001). This structure is the first and only atomic resolution structure of a putative PrP oligomer, formed by the exchange and interlocking of the third α-helix between the globular domains of two PrPC molecules. The disulfide bond in this structure is intermolecular, rather than intramolecular, as in monomeric PrPC, but each functional unit within the dimer retains its overall monomeric fold (Liu and Eisenberg 2002). A comparison between the NMR model of human PrPC and the domain-swapped model shows a small root-mean-square deviation in the swapped helix, indicating that the overall structure for this swapped segment is similar to that previously reported, despite the different topology (Knaus et al. 2001). A crystal structure of the globular domain of sheep PrPC also shows a similar overall structure to its corresponding NMR model and offers a near atomic view of native side-chain packing in PrPC, particularly of its β2–α2 loop (Haire et al. 2004). An atomic structure of the native fold of the human PrPC monomer would later emerge bound by a therapeutic antibody (Antonyuk et al. 2009). Although the packing of this structure within the crystal lattice differed from that observed in the sheep PrPC structure, possibly because of the presence of the bound antibody, no pronounced differences in the backbone structure were visible between monomers (Antonyuk et al. 2009). This set of NMR and crystal atomic structures provides a template from which new models, both computational and structural, can be established to better understand prion conversion.
The Role of PrPC Structure and Sequence in Conversion to PrPSc
The conversion of PrP from its native (PrPC) to its scrapie (PrPSc) or infectious state remains the subject of active investigation. The structural differences between these two states serve as constraints on the conversion process. For example, PrP in its scrapie form is β-sheet-rich (Pan et al. 1993), contains a protease-resistant core (Meyer et al. 1986), assembles into higher-order aggregates, including fibrils (Merz et al. 1983; Meyer et al. 1986), and contains epitopes that differ from those recognized by antibodies against PrPC (Korth et al. 1997). The Fourier-transform infrared spectra of PrPC correlate well with its known structure, which is largely α-helical. In contrast, the spectra of PrPSc and its proteolyzed ∼30-kDa fragment (residues 90–231 of the PrP sequence) show a predominantly β-sheet character (Pan et al. 1993). This suggests that PrPSc fibrils are β-sheet-rich amyloid rods (Prusiner et al. 1983), a finding corroborated by cross-β diffraction obtained from infectious prion aggregates (Nguyen et al. 1995; Inouye and Kirschner 1997). Although the atomic resolution structure of PrPSc remains unknown, these biochemical properties place specific demands on such a structure. They suggest that PrPSc is a well-ordered fibrillar amyloid formed primarily of tightly packed β-sheets.
The conversion from PrPC to PrPSc can be influenced by several factors including overabundance of PrPC and genetic mutation. High expression of wild-type PrPC alone can, in principle, drive disease by promoting aggregation of PrPC and, ultimately, its conversion to PrPSc. However, prion disease caused by PrP overexpression does not always correlate with accumulation of PrPSc (Westaway et al. 1994; Hegde et al. 1998). Prion disease caused by familial mutants of PrP varies based on the nature of the mutation. Mutations in PrP can affect both disease phenotype and its degree of severity (Mead 2006). The mutations that elicit prion disease are scattered across a large region of the protein and predominantly occur in its globular domain (residues 20–231) (Mead 2006). Explanations for why mutations lead to disease include the possibility that an interaction between a prion and an unknown protein is affected by mutation (Kaneko et al. 1997), that mutations influence the structural stability of the prion protein (Riek et al. 1998; Swietnicki et al. 1998), or that mutations directly stabilize the structure of PrPSc (Surewicz and Apostol 2011). However, not all variants of PrPC lead to disease. For example, heterozygosity of the M129V polymorphism is protective against prion disease (Palmer et al. 1991).
In addition to genetic mutation, differences between species can also influence disease progression by affecting the conversion of PrPC to PrPSc (Telling et al. 1994). Differences in the PrP sequence of only one or a few amino acids between species can prevent or diminish transmission of prion disease (Carlson et al. 1989; Kocisko et al. 1995; Moore et al. 2005). A comparison of PrP sequences from multiple species shows a highly conserved sequence, with few residue differences scattered throughout the protein (Fig. 1). Of special interest are those that occur near or in the β2–α2 loop, residues 165–176 in human PrP (Kurt et al. 2014a,b). Specific residues within this loop can significantly affect the level of PrPSc transmission from sheep, cow, deer, and elk prions to mouse PrPC (Kurt et al. 2014a,b). The structural properties of this region also differ between species, potentially driving its role in transmissibility (Gossert et al. 2005). Further study of the structure–function relationships between PrP variants and their corresponding phenotypes could elucidate the mechanism behind PrPSc transmission.
STRUCTURAL REQUIREMENTS FOR PrPSc FUNCTION
PrPSc Infectivity and Species Specificity
Three molecular events that influence the conversion of PrPC into PrPSc include unfolding of PrPC, nucleation of PrPSc aggregates, and growth of PrPSc fibrils. We can learn how each of these events might affect PrPSc by investigating other amyloid proteins, including yeast prions (Eisenberg and Jucker 2012).
Infectivity and species specificity are tied to the growth of amyloid fibrils. This growth is governed by the processes of primary nucleation, elongation of existing fibrils, and secondary nucleation (Ferrone et al. 1985; Collins et al. 2004; Cohen et al. 2013). To distinguish among these processes, we can consider the conversion of unfolded monomer into amyloid fibril. Whereas for primary nucleation the growth of fibrils depends only on the concentration of free monomer (Collins et al. 2004; Knowles et al. 2009), secondary nucleation depends on the concentration of both free monomer and fibrils (Ferrone et al. 1985; Knowles et al. 2009).
The addition of monomers onto a growing fibril requires stacking of a β-strand from a partially folded monomer onto the fibril. The strand becomes a new rung in the fibril and hydrogen-bonds with the rung ∼4.8 Å below it (Eisenberg and Jucker 2012). This periodic spacing of β-strands along the fibril is one of the structural hallmarks of amyloid. A second hallmark is the spacing between β-sheets in a fibril, which is typically ∼10 Å and is determined by the molecular specificity between the strands in the sheet (Eisenberg and Jucker 2012). In the case of steric zippers, a high shape complementarity creates tight interfaces between the strands in a fibril, which creates a typically dry interface (Fig. 2). This type of interaction is highly specific and could be a strong determinant of species or strain specificity.
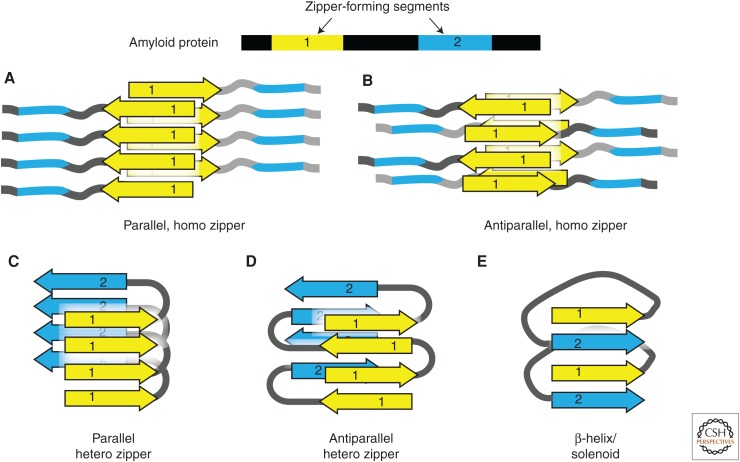
Figure 2.
Steric zipper hypothesis of amyloid fibril structure. Amyloid proteins often contain two or more amyloidogenic segments (steric zippers). The homo or hetero association of zipper segments results in five types of β-sheet architecture, giving rise to a large variety of possible assemblies of amyloid molecules. (A,B) The homotypic association between identical zipper segments in parallel (A) and antiparallel (B) arrangement. (C,D) The hetero association between different zipper segments in parallel (C) and antiparallel (D) arrangement. (E) “Hetero” interaction is built from the parallel stacking of different segments in the right-hand β-helix or solenoid.
A fibril can also experience breaking or epitaxial growth. If a fibril breaks, a molecule can be added to the newly exposed ends. Because fibrils are brittle (Tanaka et al. 2006), the formation of new ends could happen often and significantly grow the number of new fibrils as well as the overall fibril mass. Alternatively, a newly added molecule can adhere to a static part of the fibril (e.g., along its side). This molecule can template an entirely new fibril that grows alongside the first. This type of growth requires different molecular specificity. When a molecule adheres to the side of the fibril, it must do so in a way that complements the surface of that fibril. The inherent periodicity in the fibril could facilitate these interactions, offering an array of charges or hydrophobic patches for molecules to nucleate.
PrPSc Toxicity
Before discussing the structures that might give rise to PrPSc toxicity, it is beneficial to review the potential toxic elements present in PrPSc and their mechanisms of action. It is clear that prion disease cannot propagate without the presence of PrP (Sailer et al. 1994; Mallucci et al. 2003), but which of the two principal structural components of PrPSc—fibrils or oligomers—are responsible for its toxic action? Studies with some amyloid proteins favor the hypothesis that toxicity originates primarily from fibrils, other experiments favor oligomers, and yet others suggest that both elements play a role (Hill and Collinge 2003; Silveira et al. 2005; Novitskaya et al. 2006; Simoneau et al. 2007).
Recent studies suggest that infectivity and toxicity may be distinct processes in prion transmission. Although infectivity correlates with overall fibril load, toxicity appears to be induced by the presence of PrPSc fibrils but not in proportion to their abundance (Sandberg et al. 2011). One interpretation of this phenomenon is that different PrPSc functions could be assigned to different structural states within the same pathway, as in oligomers versus fibrils; to different structural polymorphs, as in different types of fibrils; or to the requirement for another agent, as in a PrPSc receptor. Because high titers of PrPSc do not always induce toxicity, PrPSc fibrils alone may be necessary but not sufficient for disease progression (Hill and Collinge 2003). These findings support the notion that the bulk of PrPSc fibrils present in an infected brain are not themselves the toxic species, but that their presence and structure are catalysts for the formation of toxic species. These toxic species are perhaps transient or less abundant polymorphs (either oligomers or fibrils).
STRUCTURAL MODELS OF PrPSc AMYLOID
What structures can we envision for amyloid fibrils of PrPSc? Although PrPSc could exhibit an entirely unknown fold, it is more likely that it resembles known amyloid structures. Several amyloid structures have been determined, both by crystallographic means and NMR, including structures made up of short segments and full proteins. These structures can serve as reference examples in the discussion of potential structures formed by PrPSc. In the following sections, we review possible structural models of PrPSc based on crystallographic structures of PrP segments, X-ray and electron diffraction of PrPSc fibrils, and imaging of PrPSc aggregates.
Steric Zipper Hypothesis of Amyloid
The steric zipper hypothesis of amyloid explains the structural properties of amyloid fibrils. These properties include β-sheet character, templated fibrillar growth, and thermodynamic stability. The zipper hypothesis also explains an important biophysical property of amyloid fibrils: cross-β diffraction (Nelson and Eisenberg 2006), which is of the type first observed by William Astbury in 1935 (Astbury et al. 1935). When an amyloid fibril or aligned fibril bundle is illuminated by an X-ray beam, it produces a characteristic diffraction pattern with a strong reflection at 4.8 Å positioned on the axis parallel to the fibril axis and another reflection at 6–11 Å positioned on the axis perpendicular to the fibril axis (Eisenberg and Jucker 2012). These two reflections can be explained by a zipper model in which an amyloid fibril is composed of closely packed, complementary β-sheets, where amino-acid side chains of two β-strands interdigitate like the teeth of a zipper. An extended network of hydrogen bonds holds the amyloid fibril together. Some of these hydrogen bonds dictate the spacing between adjacent fibril strands, producing the 4.8-Å reflection. The interdigitation of side chains between opposing sheets determines their complementarity, which in turn dictates their closeness. This closeness is reflected in the 6- to 11-Å reflection of the cross-β pattern. Together, these structural properties confer the fibril’s stability and can confer specificity in that fibril for particular amyloid segments.
The first atomic structure of steric zipper was determined from crystals of a seven-residue peptide segment, GNNQQNY, from the yeast prion protein Sup35 (Nelson et al. 2005). The structure of this zipper reveals parallel in-register β-strands and represents the cross-β spine of the Sup35 amyloid fibril. Based on this structure, the propensity for a given segment to form a steric zipper can be predicted (Goldschmidt et al. 2010). Moreover, using the X-ray crystallographic methods that helped determine this structure from micron-sized crystals, other atomic structures of amyloid-forming segments have been obtained (Sawaya et al. 2007). These segments, which belong to a small number of amyloidogenic proteins, all reveal a common steric zipper motif. However, despite sharing a common motif, the zipper structures are not identical; they exhibit polymorphisms (Wiltzius et al. 2009). The classification of zipper structures is based on the symmetry that governs the packing of β-strands in the fibril spine (Stroud 2013).
The longest zipper segment whose atomic structure has been determined is the 11-residue peptide expected to form the toxic core of α-synuclein fibrils, the cause of Parkinson’s disease. This structure was solved by electron diffraction from crystals only hundreds of nanometers thick (Rodriguez et al. 2015). Because the structures of longer zipper segments are less likely to produce large well-ordered crystals, the diffraction-based atomic structure of an entire amyloid fibril remains elusive. However, a collection of high-resolution structures from amyloid-forming segments can be used to piece together the core of an amyloid fibril (Nelson and Eisenberg 2006; Rodriguez et al. 2015). This may require knowledge of both homo zippers (segments that are complementary to their own sequence) and hetero zippers (those that complement a different sequence). Successful models of the amyloid fibril core may provide a foundation for structure-based design of drugs that target the fibril (Sievers et al. 2011; Jiang et al. 2013).
Evidence Supporting a Steric Zipper Model of PrPSc
Based on our knowledge of zipper formation by peptide segments, the propensity for any given segment of a prion protein to form a steric zipper can be estimated using the database ZipperDB (http://services.mbi.ucla.edu/zipperdb/) (Fig. 3) (Goldschmidt et al. 2010). This database provides a guide to putative high-propensity segments within the PrP sequence that might drive its assembly into fibrils through the formation of steric zippers. Although a single fragment from a loop, helix, or sheet within a large globular protein may not be informative of its overall topology, the influence of zipper segments on the core of amyloid fibrils is great. Zippers exist in a fibril as stacks of identical segments. Therefore, even a single zipper segment could comprise the entire core of a fibril. Atomic structures of peptide segments from mammalian prions have already revealed a number of steric zippers. These structures have been obtained from segments spanning several regions within the prion protein; their sequences are derived from several species. These regions include the GA-rich segment of the protein, residues in and around both the loop that links β1 and α1 and the loop between β2 and α2. Here, we group these structures by region and discuss their implication on the form and function of PrPSc.
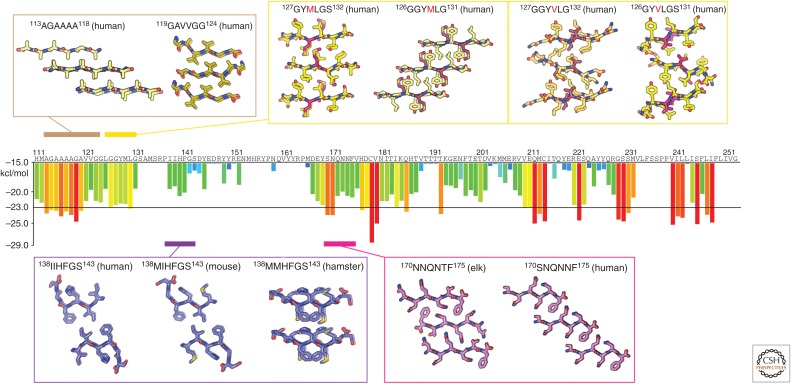
Figure 3.
Atomic structures of PrP steric zippers. The propensity for any given six-residue segment in the sequence of human PrP to form a steric zipper was estimated using the ZipperDB database (http://services.mbi.ucla.edu/zipperdb/). The evaluation of a single six-residue segment gives an energy score shown in kcal/mol on the bar graph axis (left). A colored bar representing each score is assigned to the first residue of each six-residue window throughout the PrP sequence. More negative scores are more favorable, and segments with a score of less than −23 kcal/mol are the most likely to form zippers. Atomic structures of zipper-forming segments from four regions of the sequence are shown. Two structures of the human sequence cover the region residues 113–124. Several structures cover residues 126–132 and contain either a methionine or valine at position 129. Structures from human, mouse, and hamster sequences span residues 138–143. Two structures—one with an elk sequence and the other human—span residues 170–175. As indicated in Table 1, numbers correspond to the human sequence.
We focus on PrP residues 78–226, which make up the protease-resistant core of PrPSc, abbreviated PrP 27-30 (Oesch et al. 1985). The 27-30 notation represents the molecular weight of the core fragment after digestion with proteinase K. At the N-terminal region of PrP 27-30 is the GA-rich stretch of PrP, a flexible segment populated by small hydrophobic residues. Two atomic resolution structures of zipper-forming segments have been determined from this region (Table 1) (M Apostol, HT McFarlane, DS Eisenberg, et al., unpubl.). The structures were obtained from peptides spanning residues 113–124 of human PrP, the first spanning residues 113–118, and the second spanning 119–124 (Fig. 3). Although these two structures show different packing arrangements for the β-strands that make up their fibril spines, both structures show tightly packed β-sheets that come together to form a hydrophobic interface. Other hydrophobic stretches in amyloidogenic proteins also show zippers with tightly packed hydrophobic interfaces, including the toxic core of prion-like amyloid from α-synuclein, although that zipper structure contains coordinated water molecules at its core (Rodriguez et al. 2015).
Table 1.
Atomic structures of PrP steric zippers
| Residues | Sequence | Zipper classa | Mutation | Species | PDB ID | Reference |
|---|---|---|---|---|---|---|
| 113–118 | AGAAAA | 7 | Human | M Apostol, HT McFarlane, DS Eisenberg, et al., unpubl. | ||
| 119–124 | GAVVGG | 4 | Human | M Apostol, HT McFarlane, DS Eisenberg, et al., unpubl. | ||
| 126–131 | GGYMLG | 7 | Human | 4tut | Yu et al. 2015 | |
| 127–132 | GYMLGS | 8 | Human | 3nhc | Apostol et al. 2010 | |
| 126–131 | GGYVLG | 8 | M129V | Human | 4uby | Yu et al. 2015 |
| 127–132 | GYVLGS | 8 | M129V | Human | 3nhd | Apostol et al. 2010 |
| 138–143 | MMHFGN | 6 | Hamster | 3nve | Apostol et al. 2011 | |
| 138–143 | IIHFGS | 1 | Human | 3nvf | Apostol et al. 2011 | |
| 138–143 | MIHFGN | 1 | Mouse | 3nvg | Apostol et al. 2011 | |
| 170–175 | NNQNTF | 1 | Elk | 3fva | Wiltzius et al. 2009 | |
| 170–175 | SNQNNF | 2 | Human | 2ol9 | Sawaya et al. 2007 |
All numbers correspond to the human sequence.
PDB, Protein Data Bank.
aZipper classes are defined in Sawaya et al. 2007.
Structural studies suggest that heterozygosity at residue 129, in the first β-strand of the globular domain, confers protection from prion disease. Residue 129 is a natural genetic variant with polymorphism M129V that resides within the first β-strand of PrPC. To investigate this genetic polymorphism, atomic resolution structures were obtained from six residue peptides, encoding residues 127–132 of human PrP, and containing the polymorphic residue at their core (Table 1) (Apostol et al. 2010). Although the zipper structures of these segments reveal extended β-strands, with a similar space group and unit cell dimensions, differences between their respective interfaces suggest ways in which genetic polymorphisms could affect disease susceptibility. A look down the spine of the fibril formed by each of these structures reveals differences in the orientation of their β-sheet pairs, where sheets of GYMLGS are parallel to each other, and sheets of GYVLGS are skewed (Apostol et al. 2010). This difference in packing is supported by differences in the relative buried surface area (135 Å2 for GYMLGS and 119 Å2 for GYVLGS) but not the surface complementarity score (0.74 for GYMLGS and 0.75 for GYVLGS) of each structure (Apostol et al. 2010). Because of these differences, when one sequence is modeled into the structure determined for the other, steric clashes appear. This observation has been confirmed by structures of segments from human PrP spanning residues 126–131, GGYMLG and GGYVLG (Yu et al. 2015). The segment containing the valine residue forms slanted sheets that do not overlay onto the structure containing the methionine residue. This means that a mixture of segments with different sequences may not form a steric zipper like those formed by homogeneous assemblies of either segment. In this case, structural polymorphism could explain why heterozygosity for this genetic polymorphism appears protective in humans.
A similar logic offers an explanation of the species barrier, the relative propensity of PrPSc from one species to infect another species. A comparison of the atomic structures of corresponding segments from human, mouse, and hamster PrP, spanning residues 138–144, shows nearly identical structures for human and mouse segments, but a different structure for Syrian hamster (Apostol et al. 2011). Structures of the human and mouse segments show zippers whose strands run parallel to each other when looking down the fibril axis. In this geometry, the residues in each strand that face the fibril core are the same. In contrast, the sheets in the structure from the hamster segment show adjacent β-strands running antiparallel to each other when looking down the fibril axis. This geometry has different residues facing the fibril core from each of the two sheets that make up the fibril. In categorizing these structures, we would consider the mouse and human structures to be commensurate and therefore belong to the same strain, whereas the hamster structure belongs to a different strain (Apostol et al. 2011). The differences in this collection of structures illustrate a possible explanation for how structure may dictate strain specificity and place barriers for transmission of PrPSc between species. That is, transmission may require formation of a hetero zipper between PrP of the donor and the recipient.
Structural polymorphism in another region of PrP is also thought to contribute to the species barrier and differences between strains of PrPSc. Structural differences have been observed for the loop between β2 and α2 in NMR models of PrPC (Gossert et al. 2005). These differences are expected to influence the susceptibility of certain species to prion disease (Kurt et al. 2014a). Two atomic resolution structures of zipper-forming segments from this region have been determined from analogous elk and human sequences (Table 1). Comparing the structures from this region, we can discern polymorphic differences that highlight the potential incompatibilities between the zipper polymorphs from different species. The structure of the segment spanning residues 170–175 of human PrP is composed of parallel strands running down each sheet in the zipper but different residues facing the fibril core from each of the two sheets that make up the fibril (Sawaya et al. 2007). The corresponding segment in elk PrP, residues 173–178, shows high sequence homology with the human segment but is structurally polymorphic. The elk zipper structure also reveals parallel strands in each sheet, but in this case, the same residues in each of the sheets face the fibril core (Wiltzius et al. 2009). Both of these structures have the residues xNxNxF at the fibril core, yet their interfaces differ, likely because residues that face away from the core can influence packing of those buried within the core (Apostol et al. 2011). A greater breadth of structures from various species within this region of PrP could help further classify species-specific polymorphisms and to provide a basis for deciphering their contribution to PrPSc transmissibility between species.
In Table 1 and Figure 3, we summarize 11 atomic resolution structures of steric zippers from PrP. These structures inform us of the structural differences that, when present within zipper-forming segments, could explain the behavior of PrPSc. These differences, sometimes induced by changes to a single residue, can have profound structural and phenotypic effects, including species specificity and protection against disease.
β-Solenoid Model of Yeast Prions
Although the functions of fungal and mammalian prions differ, fungal prions are also a proteinaceous infectious particle. This similarity presents an opportunity to glean from structural features of microbial prions the features that facilitate structural conversion. A number of fungal prion proteins have been identified and investigated, including Sup35 and HET-s (Alberti et al. 2009), and a structural model of the HET-s fibril core has been determined (Wasmer et al. 2008). However, sequences of microbial prions are not homologous to mammalian prions (Alberti et al. 2009), and we thus face limits in extending their structures to PrPSc.
Despite these limits, two structures of yeast prion could serve as templates for determining the structure of PrPSc. The first is a zipper structure like those reviewed in the preceding section, and the second is a model of the fibril core of the HET-s prion, which shows a β-solenoid with a triangular geometry and a hydrophobic core (Wasmer et al. 2008). HET-s is a yeast prion involved in heterokaryon incompatibility, a condition under which yeast fusions are lethal between strains expressing two different alleles (HET-s and HET-S) only when HET-s exists as a prion (Saupe and Daskalov 2012). (For more on the function of HET-s, see Riek and Saupe 2016.) The model of a HET-s fibril was obtained by solid-state NMR, revealing a rigid core of roughly 70 residues (218–289) that forms a left-handed β-solenoid of parallel strands stacked into three sheets, one of short strands and two of longer strands (Fig. 4). In the model, interchain interactions are limited to the flat surfaces above and below each chain (Fig. 4). This model resembles other left-handed β-helices, including those in antifreeze proteins and some lyases, although the latter do not always show a triangular topology (Yoder and Jurnak 1995). The HET-s model has similarities to the cell-puncturing phage protein gp27, which has a triple-stranded β-helix that forms an equilateral triangle (Kanamaru et al. 2002), and the p22 tailspike protein, which is hypothesized to exist as a protease- and detergent-resistant triple β-helix (Kreisberg et al. 2002). Importantly, in the case of p22, only the triple β-helix is resistant to protease and detergent, but not monomers, dimers, or trimers. This highlights the importance of intermolecular contacts to the β-helix fold.
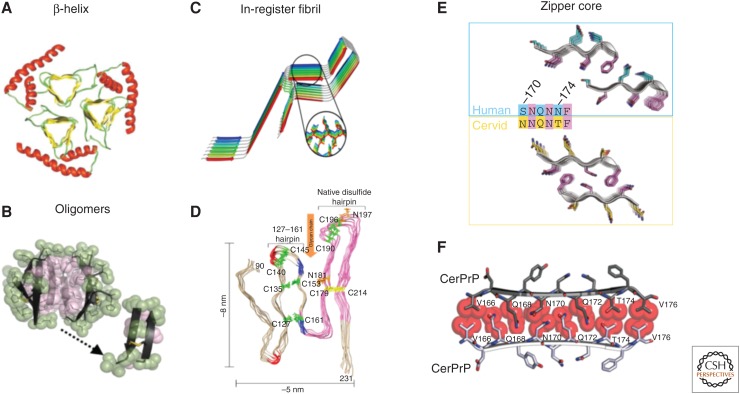
Figure 4.
Proposed models for PrPSc. (A) A triangular β-helix model of PrPSc was based on a transmission electron microscopy (TEM) image. (Courtesy of Govaerts et al. 2004; reprinted, with permission, from the National Academy of Sciences, U.S.A. © 2001.) (B) An atomic structure for a PrP oligomer was determined from pairs of naturally disulfide-linked segments with PrP sequences. (Courtesy of Apostol et al. 2013; reprinted, with permission, from the American Chemical Society © 2013.) (C,D) In-register fibril models have been obtained from different experiments. (C) An in-register stacking model was proposed based on electron paramagnetic resonance (EPR) of human PrP fibrils converted in vitro. (Courtesy of Cobb et al. 2007; reprinted, with permission, from the National Academy of Sciences, U.S.A. © 2007.) (D) Another parallel in-register β-sheet architecture using solid-state NMR spectroscopy of recombinant PrP amyloid fibrils that were seeded with PrPSc. (Courtesy of Groveman et al. 2014; adapted, with permission, © 2014, by the American Society for Biochemistry and Molecular Biology.) (E,F) Zipper structures of segments in or near the β2–α2 loop highlight its importance in conversion and transmission. (E) Microcrystal structures of human (cyan, top) and cervid (yellow, lower) β2–α2 loop segments belong to different classes of steric zippers, supporting the idea that side-chain mismatches may account for species barriers in prion disease. (Courtesy of Apostol et al. 2011; adapted, with permission, © 2011 American Chemical Society.) (F) Zipper model of the β2–α2 loop highlights tight, complementary side-chain interactions that are required for efficient prion conversion. (Courtesy of Kurt et al. 2014b; adapted, with permission, © 2014, by the American Society for Biochemistry and Molecular Biology.)
Evidence Supporting the β-Solenoid/Helix Model of PrPSc
Heterogeneity in PrPSc structure makes its crystallization a challenge and has limited the structural information available from which to build models of its fibril core. However, using imaging or diffraction methods to examine PrPSc fibrils has inspired models that include a β-helix or a β-solenoid fold (Wille et al. 2002, 2009; Govaerts et al. 2004).
The first β-helical model of PrPSc was proposed on the basis of electron diffraction experiments from negatively stained two-dimensional crystals of N-terminally truncated PrPSc (PrP 27-30) and a mini-prion construct (PrPSc106) (Wille et al. 2002). PrPSc106 differs from PrP 27-30 in that it lacks residues 23–88 and 141–176; it contains only 106 residues. Both of these preparations support the propagation of prions and therefore encode the structural information required for infection by PrPSc. Because crystals of these preparations were found to be isomorphous at a resolution of near 7 Å, a projection difference map revealed a 36-residue internal deletion within the mini-prion (Wille et al. 2002). Together with the expected location of sugars and the native sequence of a PrP monomer, models were created that satisfied the projected density. A parallel β-helix model was identified as the most likely model for PrPSc. In this model, the PrPSc subunits are assembled into a trimer by using the sidewall of the β-helix as an interface, and the trimers are stacked into fibrils by forming an inter-subunit β-helix. The model does not explain what accounts for robust aggregation beyond the trimer level.
Further refinement of the initial β-helix model was achieved by computational threading of the PrP sequence onto known β-helix folds. The computational experiment indicated PrP may exist as a parallel left-handed β-helix (Govaerts et al. 2004). The refined model distinguished between two potential parallel β-helix models that both appeared to fit the crystallographic data. The two models in question were a trimer or hexamer of either right-handed or left-handed β-helices. A PrPsc model was proposed with β-sheets forming a parallel left-handed helix. In addition, because left-handed β-helices have been shown to form trimers, a trimeric model was assumed.
Further experimental evidence in support of a β-helix model for PrPSc was obtained through X-ray fiber diffraction experiments (Wille et al. 2009). These experiments showed that diffraction of X-rays by recombinantly expressed PrP fibrils was different from that obtained with brain-derived PrPSc. The recombinant fibrils matched patterns expected for steric zippers, including a strong meridional reflection at 4.8 Å and an equatorial reflection at 10.5 Å—classic cross-β diffraction (Wille et al. 2009). Because brain-derived PrPSc fibrils produced a different diffraction pattern, synthetic diffraction data were generated from a trimeric β-helix model and fit against the measured pattern. For comparison, synthetic patterns from steric zipper models based on the structure of the GNNQQNY peptide were also evaluated. These modeling experiments suggest that brain-derived PrPSc does not exist as a simple steric zipper, such as GNNQQNY, and does not exhibit cross-β diffraction. Instead, the collective experiments in support of the β-helix model of PrPSc indicate that infectious fibrils of PrP have a complex structure. The experiments do not ensure that the structure is a left-handed β-helix, but neither do they disprove this model. Ultimately, a near atomic resolution model of PrPSc fibrils may be required to reveal the true topology of its amyloid core.
Evidence Supporting Oligomer and Other Models of PrPSc
The bulk of studies searching for the high-resolution structure of PrPSc have focused on fibrillar forms, as outlined above. The search for other species of PrPSc includes investigations of the structure of prion oligomers. Oligomers of PrPSc are expected to be both toxic and infectious (Silveira et al. 2005; Simoneau et al. 2007), with some studies suggesting that infectivity peaks with particles 17–27 nm in size (300–600 kDa; about 10 to 20 copies of PrP 27–30) (Silveira et al. 2005). The structure of an oligomeric species would illuminate the nature of polymorphs and strains.
With an approach similar to that applied to determining atomic resolution structures from steric-zipper-forming segments, a proposed atomic structure for a PrP oligomer was obtained (Apostol et al. 2013). The approach relies on selecting minimal fragments that may represent the core of such an oligomer and applying crystallographic tools to determine its atomic structure. Similar approaches have been successful in solving the atomic structure of β-sheet-rich oligomers of other amyloid-forming proteins, such as α-B crystallin. The atomic structure of an oligomer from α-B crystallin revealed a cylindrical barrel, composed of six antiparallel β-strands of identical sequence, and was termed a cylindrin (Laganowsky et al. 2012). In contrast, the PrP oligomer structure was determined from pairs of disulfide-linked segments with different sequences, belonging to the regions of α2 and α3 that form a natural disulfide bridge within the globular domain of PrPC. The atomic structure of these linked segments shows a hexameric oligomer of antiparallel β-sheets where a pair of disulfide-linked chains makes one face of a triangular prism (Apostol et al. 2013). This structural motif is different from other structures determined from amyloid-forming segments and could represent the core of a PrP oligomer. However, given the uniqueness of the structure, it is difficult to predict whether this structure is on pathway to the expected structures for PrPSc fibrils. If the oligomers discovered are not on pathway, what do their structures tell us about infectivity and toxicity?
Some models of PrP toxicity include the flexible N-terminal tail of PrP (residues 23–123) (Sonati et al. 2013). Antibody-based targeting of PrPC has been shown to elicit toxicity to primary brain cultures. Biochemical studies coupled with the structure of an antibody fragment bound to PrPC reveal a potential interaction between the N-terminal tail of PrP and the native globular domain of PrPC (Sonati et al. 2013). The N-terminal tail consists primarily of four to five 8-amino-acid-long repeats that are unusually glycine-rich. The structure of an antibody fragment bound to one of the octapeptide repeats has been obtained (Swayampakula et al. 2013) and reveals that binding of the antibody fragment to this octapeptide-repeat segment forces it to adopt an extended conformation, which is different from the expected β-turn topology for this sequence. This is important for the design of ligands that target these repeats in efforts to reduce the toxicity that results from the N-terminal region of PrP (Swayampakula et al. 2013). Active investigation continues into the roles of the N-terminal tail of PrP and other ligands and their structural influence on PrPSc.
STRUCTURAL POLYMORPHISM IN PrPSc
Structural polymorphs can affect all aspects of PrPSc function, and we can think of PrPSc strains themselves as structural polymorphs. In this sense, polymorph compatibility likely defines infectivity and transmissibility. In addition, only certain polymorphs may be toxic. Structural polymorphisms can be subtle, with a single residue taking on a different conformation, but even these polymorphisms can lead to substantial phenotypic differences. A single heterologous residue at a pivotal position could block prion infection. Several studies have aimed to map the key amino acid positions and substitutions that govern cross-species prion conversion. Residue substitutions at particular positions have been identified that restrict transmission of PrPSc, including residues in the β2–α2 loop region of PrP. A series of in vitro prion conversion experiments have investigated amino acid substitution within the β2–α2 loop in PrPC. Collectively, these studies have shown that the amyloid-forming propensity of key segments of PrPC can dictate prion conversion, more so than simple sequence differences (Kurt et al. 2014a,b). Because a catalog of structural polymorphs may be necessary to fully understand how form takes function in PrPSc, this task remains elusive for now. Instead, a link between changes to the primary sequence and their structural outcomes may help clarify the roles of particular segments in the context of various polymorphs.
How do certain residue positions or short PrP segments control polymorph compatibility and therefore prion conversion? Microcrystal structures of steric zipper segments from amyloid-like fibrils may offer clues (Sawaya et al. 2007). The steric zipper hypothesis of amyloid structure provides a theoretical foundation for prion conversion and transmissibility. Transmission of a PrPSc zipper requires a donor zipper, and a recipient PrP loop segments to form a tight zipper interface. Because side-chain mismatches lead to steric clashes and cavities in the zipper model, these would prevent conversion and thereby account for species barriers in prion disease (Apostol et al. 2010, 2011). Consistent with this hypothesis, microcrystal structures of human and cervid β2–α2 loop segments belong to different classes of steric zippers with variation in the arrangement of β-strands and β-sheets (Sawaya et al. 2007; Apostol et al. 2011). Moreover, computational analyses suggest that human and elk loop segments do not form complementary steric zippers.
SUMMARY OF STRUCTURAL MODELS OF PrPSc
Several distinct structural models of PrPSc have been proposed. We summarize them here, noting key features that may contribute to PrPSc structure (Fig. 4).
Short, Compact Fibrils with a Potential β-Helix Fold
Prusiner and colleagues proposed an experimentally based molecular model of PrPSc from electron microscopic (EM) images of a two-dimensional crystal in 2004 (Wille et al. 2002; Govaerts et al. 2004). As described above, their model shows PrPSc as a triangular β-helix containing four turns and an intact C-terminal α2–α3 helix bundle. Kunes and colleagues proposed another model based on a ladder-shaped three-dimensional EM image of a PrPSc fibril. In this model, the N-terminal (residues G89–D143) and the C-terminal (residues Y154–G227) regions of core PrP were independently converted into β-helices. The β-helices were stacked into rails, and the linker peptide between the β-helices formed the steps of the ladder (Kunes et al. 2008). Similarly, Langedijk and colleagues proposed a β-helix model that assumed conversion of the N-terminal region of core PrP (residues P104–D143) into a β-helix, according to sequence comparisons and molecular dynamics (MD) simulations (Langedijk et al. 2006).
Parallel In-Register β-Sheets
Based on the three-dimensional reconstruction of an EM image of a mouse PrP fibril converted in vitro, Tattum and colleagues proposed a ladder-shaped fibril structure (Tattum et al. 2006). Meanwhile, Cobb et al. (2007) measured electron paramagnetic resonance (EPR) of human PrP fibrils converted in vitro and proposed an in-register-stacking model based on the inter-residue distances. A more recent study using solid-state NMR spectroscopy (ssNMR) to analyze the formation of recombinant PrP amyloid fibrils that were seeded with PrPSc proposed that the C-terminal, cysteine-flanked core that contains α-helices 2 and 3 also adopts a parallel in-register β-sheet architecture (Groveman et al. 2014). The investigators determined the space between labeled isoleucine (Ile-1-13C), phenylalanine (Phe-1-13C), and leucine (Leu-1-13C) residues in recombinant PrP amyloid and observed a distance of ∼0.5 nm for four Ile residues (182, 184, 203, and 205). Based on this observation, the investigators concluded that these residues refold from an α-helical structure into a parallel in-register intermolecular β-sheet. As we have reviewed, atomic structures of a number of PrP segments show steric zippers with tight interfaces, the type of structures that would hold a fibril core of parallel β-sheets tightly clasped (Nelson and Eisenberg 2006; Sawaya et al. 2007; Wiltzius et al. 2009; Apostol et al. 2010, 2011).
Involvement of the β2 Strand and the β2–α2 Loop
DeMarco and Daggett (2004) observed extension of a native β-sheet consisting of β1 and β2 strands and recruitment of a nascent β-strand to the β-sheet in an MD simulation, and proposed a model in which subunits were assembled into a spiral fibril by forming intersubunit β-sheets. Mutations and zipper structures of segments in or near the β2–α2 loop also highlight its importance in conversion and transmission (Apostol et al. 2011; Kurt et al. 2014a,b).
CONCLUSION
The studies we summarize here offer a variety of experimental and computational approaches to understanding the structural nature of the infective and toxic form of PrP or PrPSc. Yet despite much effort, the structure of PrPSc remains elusive. At present, it is not certain that the infective and toxic structures are the same.
Although it remains elusive, the goal of determining the structure of PrPSc is of great biological importance. Knowing this structure will surely suggest paths toward effective diagnostics and therapeutics for prion disease. Moreover, the structure of PrPSc will likely tell us much about the mechanism of the species barrier and also illuminate the nature of conformational prion strains, which has long been a mystery. We hope our review will be helpful to those who are working on this important goal.
ACKNOWLEDGMENTS
We thank Drs. Heather Macfarlan, Marcin Apostol, Michael Sawaya, Timothy Hurt, Christina Sigurdson, and Sangho Lee for discussions and the Howard Hughes Medical Institute Department of Energy-OBER and the National Institutes of Health for support.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonyuk SV, Trevitt CR, Strange RW, Jackson GS, Sangar D, Batchelor M, Cooper S, Fraser C, Jones S, Georgiou T, et al. 2009. Crystal structure of human prion protein bound to a therapeutic antibody. Proc Natl Acad Sci 106: 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol MI, Sawaya MR, Cascio D, Eisenberg D. 2010. Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. J Biol Chem 285: 29671–29675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol MI, Wiltzius JJW, Sawaya MR, Cascio D, Eisenberg D. 2011. Atomic structures suggest determinants of transmission barriers in mammalian prion disease. Biochemistry 50: 2456–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol MI, Perry K, Surewicz WK. 2013. Crystal structure of human prion protein fragment reveals a motif for oligomer formation. J Am Chem Sci 135: 10202–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astbury WT, Dickinson S, Bailey K. 1935. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J 29: 2351–2360.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamborough P, Wille H, Telling GC, Yehiely F, Prusiner SB, Cohen FE. 1996. Prion protein structure and scrapie replication: Theoretical, spectroscopic, and genetic investigations. Cold Spring Harb Symp Quant Biol 61: 495–509. [PubMed] [Google Scholar]
- Carlson GA, Westaway D, DeArmond SJ, Peterson-Torchia M, Prusiner SB. 1989. Primary structure of prion protein may modify scrapie isolate properties. Proc Natl Acad Sci 86: 7475–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. 2007. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci 104: 18946–18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SI, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TP. 2013. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci 110: 9758–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Douglass A, Vale RD, Weissman JS. 2004. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol 2: e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco ML, Daggett V. 2004. From conversion to aggregation: Protofibril formation of the prion protein. Proc Natl Acad Sci 101: 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. 1997. Structure of the recombinant full-length hamster prion protein PrP(29–231): The N terminus is highly flexible. Proc Natl Acad Sci 94: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. 2012. The amyloid state of proteins in human diseases. Cell 148: 1188–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrone FA, Hofrichter J, Eaton WA. 1985. Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J Mol Biol 183: 611–631. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. 2010. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci 107: 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wüthrich K. 2005. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci 102: 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts C, Wille H, Prusiner SB, Cohen FE. 2004. Evidence for assembly of prions with left-handed β-helices into trimers. Proc Natl Acad Sci 101: 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, Caughey B. 2014. Parallel in-register intermolecular β-sheet architectures for prion-seeded prion protein (PrP) amyloids. J Biol Chem 289: 24129–24142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire LF, Whyte SM, Vasisht N, Gill AC, Verma C, Dodson EJ, Dodson GG, Bayley PM. 2004. The crystal structure of the globular domain of sheep prion protein. J Mol Biol 336: 1175–1183. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science 279: 827–834. [DOI] [PubMed] [Google Scholar]
- Hill AF, Collinge J. 2003. Subclinical prion infection. Trends Microbiol 11: 578–584. [DOI] [PubMed] [Google Scholar]
- Inouye H, Kirschner DA. 1997. X-ray diffraction analysis of scrapie prion: Intermediate and folded structures in a peptide containing two putative α-helices. J Mol Biol 268: 375–389. [DOI] [PubMed] [Google Scholar]
- James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, Kaneko K, Groth D, Mehlhorn I, Prusiner SB, et al. 1997. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci 94: 10086–10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu C, Leibly D, Landau M, Zhao M, Hughes MP, Eisenberg DS. 2013. Structure-based discovery of fiber-binding compounds that reduce the cytotoxicity of amyloid β. Elife 2: e00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415: 553–557. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, James TL, Cohen FE, Prusiner SB. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci 94: 10069–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. 2001. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol 8: 770–774. [DOI] [PubMed] [Google Scholar]
- Knowles TP, Waudby CA, Devlin GL, Cohen SI, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. 2009. An analytical solution to the kinetics of breakable filament assembly. Science 326: 1533–1537. [DOI] [PubMed] [Google Scholar]
- Kocisko DA, Priola SA, Raymond GJ, Chesebro B, Lansbury PT, Caughey B. 1995. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: A model for the scrapie species barrier. Proc Natl Acad Sci 92: 3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, et al. 1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 390: 74–77. [DOI] [PubMed] [Google Scholar]
- Kreisberg JF, Betts SD, Haase-Pettingell C, King J. 2002. The interdigitated β-helix domain of the P22 tailspike protein acts as a molecular clamp in trimer stabilization. Protein Sci 11: 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes KC, Clark SC, Cox DL, Singh RR. 2008. Left handed β helix models for mammalian prion fibrils. Prion 2: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt TD, Bett C, Fernández-Borges N, Joshi-Barr S, Hornemann S, Rülicke T, Castilla J, Wüthrich K, Aguzzi A, Sigurdson CJ. 2014a. Prion transmission prevented by modifying the β2-α2 loop structure of host PrPC. J Neurosci 34: 1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt TD, Jiang L, Bett C, Eisenberg D, Sigurdson CJ. 2014b. A proposed mechanism for the promotion of prion conversion involving a strictly conserved tyrosine residue in the β2-α2 loop of PrPC. J Biol Chem 289: 10660–10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, et al. 2012. Atomic view of a toxic amyloid small oligomer. Science 335: 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langedijk JP, Fuentes G, Boshuizen R, Bonvin AM. 2006. Two-rung model of a left-handed β-helix for prions explains species barrier and strain variation in transmissible spongiform encephalopathies. J Mol Biol 360: 907–920. [DOI] [PubMed] [Google Scholar]
- Liu Y, Eisenberg D. 2002. 3D domain swapping: As domains continue to swap. Protein Sci 11: 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallucci G, Dickinson A, Linehan J, Klöhn PC, Brandner S, Collinge J. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302: 871–874. [DOI] [PubMed] [Google Scholar]
- Mead S. 2006. Prion disease genetics. Eur J Hum Genet 14: 273–281. [DOI] [PubMed] [Google Scholar]
- Merz PA, Somerville RA, Wisniewski HM, Manuelidis L, Manuelidis EE. 1983. Scrapie-associated fibrils in Creutzfeldt–Jakob disease. Nature 306: 474–476. [DOI] [PubMed] [Google Scholar]
- Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. 1986. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci 83: 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Vorberg I, Priola SA. 2005. Species barriers in prion diseases—Brief review. In Infectious diseases from nature: Mechanisms of viral emergence and persistence (ed. Peters PD, Calisher PC), pp. 187–202. Springer, Vienna. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. 2006. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol 16: 260–265. [DOI] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Inouye H, Baldwin MA, Fletterick RJ, Cohen FE, Prusiner SB, Kirschner DA. 1995. X-ray diffraction of scrapie prion rods and PrP peptides. J Mol Biol 252: 412–422. [DOI] [PubMed] [Google Scholar]
- Novitskaya V, Bocharova OV, Bronstein I, Baskakov IV. 2006. Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons. J Biol Chem 281: 13828–13836. [DOI] [PubMed] [Google Scholar]
- Oesch B, Westaway D, Wälchli M, McKinley MP, Kent SBH, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE, et al. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell 40: 735–746. [DOI] [PubMed] [Google Scholar]
- Palmer MS, Dryden AJ, Hughes JT, Collinge J. 1991. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt–Jakob disease. Nature 352: 340–342. [DOI] [PubMed] [Google Scholar]
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. 1993. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci 90: 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35: 349–358. [DOI] [PubMed] [Google Scholar]
- *.Riek R, Saupe SJ. 2016. The HET-S/s prion motif in the control of programmed cell death. Cold Spring Harb Perspect Biol 8: a023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. 1996. NMR structure of the mouse prion protein domain PrP(121–231). Nature 382: 180–182. [DOI] [PubMed] [Google Scholar]
- Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. 1997. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett 413: 282–288. [DOI] [PubMed] [Google Scholar]
- Riek R, Wider G, Billeter M, Hornemann S, Glockshuber R, Wüthrich K. 1998. Prion protein NMR structure and familial human spongiform encephalopathies. Proc Natl Acad Sci 95, 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, Sangwan S, Guenther EL, Johnson LM, Zhang M, et al. 2015. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer A, Büeler H, Fischer M, Aguzzi A, Weissmann C. 1994. No propagation of prions in mice devoid of PrP. Cell 77: 967–968. [DOI] [PubMed] [Google Scholar]
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. 2011. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 470: 540–542. [DOI] [PubMed] [Google Scholar]
- Saupe SJ, Daskalov A. 2012. The [Het-s] prion, an amyloid fold as a cell death activation trigger. PLoS Pathog 8: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447: 453–457. [DOI] [PubMed] [Google Scholar]
- Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Münch J, Baker D, Eisenberg D. 2011. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau S, Rezaei H, Salès N, Kaiser-Schulz G, Lefebvre-Roque M, Vidal C, Fournier JG, Comte J, Wopfner F, Grosclaude J, et al. 2007. In vitro and in vivo neurotoxicity of prion protein oligomers. PLoS Pathog 3: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonati T, Reimann RR, Falsig J, Baral PK, O’Connor T, Hornemann S, Yaganoglu S, Li B, Herrmann US, Wieland B, et al. 2013. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 501: 102–106. [DOI] [PubMed] [Google Scholar]
- Stroud JC. 2013. The zipper groups of the amyloid state of proteins. Acta Crystallogr D Biol Crystallogr 69: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewicz WK, Apostol MI. 2011. Prion protein and its conformational conversion: A structural perspective. In Prion proteins (ed. Tatzelt J), pp. 135–167. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Swayampakula M, Baral PK, Aguzzi A, Kav NNV, James MNG. 2013. The crystal structure of an octapeptide repeat of the prion protein in complex with a Fab fragment of the POM2 antibody. Protein Sci 22: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swietnicki W, Petersen RB, Gambetti P, Surewicz WK. 1998. Familial mutations and the thermodynamic stability of the recombinant human prion protein. J Biol Chem 273: 31048–31052. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. 2006. The physical basis of how prion conformations determine strain phenotypes. Nature 442: 585–589. [DOI] [PubMed] [Google Scholar]
- Tattum MH, Cohen-Krausz S, Thumanu K, Wharton CW, Khalili-Shirazi A, Jackson GS, Orlova EV, Collinge J, Clarke AR, Saibil HR. 2006. Elongated oligomers assemble into mammalian PrP amyloid fibrils. J Mol Biol 357: 975–985. [DOI] [PubMed] [Google Scholar]
- Telling GC, Scott M, Hsiao KK, Foster D, Yang SL, Torchia M, Sidle KC, Collinge J, DeArmond SJ, Prusiner SB. 1994. Transmission of Creutzfeldt–Jakob disease from humans to transgenic mice expressing chimeric human–mouse prion protein. Proc Natl Acad Sci 91: 9936–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Melckebeke HV, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science 319: 1523–1526. [DOI] [PubMed] [Google Scholar]
- Westaway D, DeArmond SJ, Cayetano-Canlas J, Groth D, Foster D, Yang SL, Torchia M, Carlson GA, Prusiner SB. 1994. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell 76: 117–129. [DOI] [PubMed] [Google Scholar]
- Wille H, Michelitsch MD, Guénebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB. 2002. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci 99: 3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, et al. 2009. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci 106: 16990–16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. 2009. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol 16: 973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Jurnak F. 1995. Protein motifs. 3. The parallel β helix and other coiled folds. FASEB J 9: 335–342. [DOI] [PubMed] [Google Scholar]
- Yu L, Lee SJ, Yee VC. 2015. Crystal structures of polymorphic prion protein β1 peptides reveal variable steric zipper conformations. Biochemistry 54: 3640–3648. [DOI] [PubMed] [Google Scholar]
- Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López García F, Billeter M, Calzolai L, Wider G, Wüthrich K. 2000. NMR solution structure of the human prion protein. Proc Natl Acad Sci 97: 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]