Renal agenesis is a devastating birth defect, and although genes encoding retinoic acid signaling components have been shown to be important for renal...
Keywords: GREB1L, retinoic acid, renal agenesis, CAKUT, whole exome sequencing
Abstract
Renal agenesis (RA) is one of the more extreme examples of congenital anomalies of the kidney and urinary tract (CAKUT). Bilateral renal agenesis is almost invariably fatal at birth, and unilateral renal agenesis can lead to future health issues including end-stage renal disease. Genetic investigations have identified several gene variants that cause RA, including EYA1, LHX1, and WT1. However, whereas compound null mutations of genes encoding α and γ retinoic acid receptors (RARs) cause RA in mice, to date there have been no reports of variants in RAR genes causing RA in humans. In this study, we carried out whole exome sequence analysis of two families showing inheritance of an RA phenotype, and in both identified a single candidate gene, GREB1L. Analysis of a zebrafish greb1l loss-of-function mutant revealed defects in the pronephric kidney just prior to death, and F0 CRISPR/Cas9 mutagenesis of Greb1l in the mouse revealed kidney agenesis phenotypes, implicating Greb1l in this disorder. GREB1L resides in a chromatin complex with RAR members, and our data implicate GREB1L as a coactivator for RARs. This study is the first to associate a component of the RAR pathway with renal agenesis in humans.
CONGENITAL anomalies of the kidney and urinary tract (CAKUT) are one of the more common sets of birth defects noted in children and represent a significant cause of morbidity and mortality (Sanna-Cherchi et al. 2009), including end-stage renal disease (ESRD) (USRDS, 1999). Renal agenesis (RA) is defined as the complete absence of renal tissue at birth, which can be separated into unilateral and bilateral renal agenesis (Yalavarthy and Parikh 2003), and represents the most severe form of CAKUT. While unilateral renal agenesis (URA) can lead to proteinuria, hypertension, and early renal failure, it is generally compatible with life (Schreuder et al. 2008). Bilateral Renal Agenesis (BRA), in contrast, is almost invariably fatal at birth (Potter 1946, 1965). It is estimated that BRA occurs at a frequency of 1/3000–1/5000 births, while URA occurs more frequently (up to 1/1000 births), although estimating the incidence is hampered by underreporting (Norwood and Chevalier 2003; Yalavarthy and Parikh 2003). In humans, genetic etiologies for RA were first identified 30 years ago, when it was shown that relatives of a person with a nonsyndromic RA had an increased risk (from 4 to 9%) of having RA themselves (Carter et al. 1979; Roodhooft et al. 1984). At least 70 different clinical conditions or syndromes exist where RA has been identified as a component (Sanna-Cherchi et al. 2007; Kerecuk et al. 2008), including: branchio-oto-renal (Brophy et al. 2013); hypoparathyroidism, deafness, and renal dysplasia (Van Esch et al. 2000); Townes–Brocks (Kohlhase et al. 1998); and Fraser (Vrontou et al. 2003) syndromes. Additionally, variants identified in several genes (including EYA1, SIX1 and SIX2, FRAS1, GATA3, WNT4, RET, FGF20, UPK3A, and ITGA8) have been implicated in human nonsyndromic RA (Jenkins et al. 2005; Sanna-Cherchi et al. 2007; Skinner et al. 2008; Toka et al. 2010; Barak et al. 2012; Humbert et al. 2014).
In mice, variants in a variety of genes have been identified that cause RA, and several of these genes are involved in regulating developmental processes such as nephric duct formation (Pax2, Lim1) or ureter budding (GDNF, Ret, GFR alpha1) (Uetani and Bouchard 2009). Additionally, a large number of the RA-associated genes are required for the proper expression of GDNF and Ret/GFR alpha1, including Eya1, Six2, Fras1, Gata3, and Emx2 (Uetani and Bouchard 2009). However, most of the genes identified in monogenic mutant animal models have not yet been correlated with the equivalent human disease. More recently, ESRRG (Estrogen Related Receptor Gamma), a gene encoding an estrogen receptor-related nuclear hormone receptor, was implicated in RA based on chromosomal breakpoint analysis in cases affected by RA (Harewood et al. 2010), although targeted inactivation in mice only revealed agenesis of the renal papilla (Berry et al. 2011). Additionally, glomerulonephritis was observed in mice lacking the estrogen receptor alpha gene (Shim et al. 2004).
Various studies have shown that retinoic acid signaling plays a key role in kidney development. Retinoic acid [which binds a nuclear receptor highly homologous to steroid hormone receptors (Petkovich et al. 1987)], can expand the pronephric region of the kidney in animal cap assays as well as promote expression of many markers of the intermediate mesoderm and its derivatives in mouse embryonic stem cells (Osafune et al. 2002; Kim and Dressler 2005). Moreover, retinoic acid can promote ureteric bud outgrowth in the developing metanephros (Rosselot et al. 2010), which is thought to work by regulating Ret expression in the bud (Batourina et al. 2005). Furthermore, studies in Xenopus and zebrafish showed that several genes required for specification and development of the pronephros (pax8, lhx1, wt1, pteg) are under the control of retinoic acid signaling (Carroll and Vize 1999; Cartry et al. 2006; Perner et al. 2007; Bollig et al. 2009; Lee et al. 2010), and compound null mutations of genes encoding α and γ retinoic acid receptors (RARs) cause a renal agenesis phenotype (Mendelsohn et al. 1994). Finally, a recent report showed that mutation of the Nuclear Receptor Interacting Protein 1 (NRIP1) gene, encoding a transcriptional cofactor of retinoic acid receptors, caused a range of CAKUT, including renal hypo/dysplasia and vesicoureteral reflux (VUR) and/or ectopia (Vivante et al. 2017).
Here we describe identification of a novel renal agenesis locus, GREB1L, through exome sequence analysis of cases chosen from two independent RA pedigrees, and show that (1) zebrafish greb1l is required for proper specification of the pronephros and (2) F0 CRISPR mouse Greb1l mutants present with kidney agenesis phenotypes, confirming a role for GREB1L in this disorder. GREB1L was initially identified as a paralog of GREB1, and GREB1 expression was upregulated upon estrogen treatment of a human breast carcinoma cell line and shown to be highly correlated with both estrogen receptor (ER) and androgen receptor (AR) expression in breast/prostate cancer cell lines and primary tumors (Ghosh et al. 2000; Rae et al. 2005, 2006; Mohammed et al. 2013). Notably, GREB1 (which acts as a coactivator of the ER) resides in a chromatin complex with both GREB1L and Retinoic Acid Receptor components. GREB1L, on the other hand, is upregulated in a well-established cell line model of retinoic acid signaling (Laursen et al. 2012), and mutation of retinoic acid targets expressed in the developing pronephros are associated with RA in mice or humans (Kreidberg et al. 1993; Shawlot and Behringer 1995; Torres et al. 1995; Brophy et al. 2001; Bouchard et al. 2002; Meeus et al. 2004; Trueba et al. 2005). Taken together, these data strongly implicate GREB1L as a coactivator for RARs that, when reduced in dose, causes kidney agenesis phenotypes.
Materials and Methods
Case ascertainment, Iowa
The Iowa case ascertainment has been carried out on joint projects and replication efforts throughout the world. In 2005, the Brophy laboratory established an Internal Review Board (IRB) approved website for collecting RA samples (IRB # 200711705) (www.kidneygenes.com). Participants and their physicians are made aware of this study through our website, the National Center for Biotechnology Information (NCBI) web resource www.genetests.com, and our work with the National Potter’s Syndrome Support Group. From our website, appropriate consent forms and other paperwork are downloaded by the participant or their physician. This has resulted in a worldwide data and sample collection including in-depth phenotypic, clinical, and genetic material. The proband of the family included in this study was originally brought to our attention by Dr. Michael Schneider while at the University of Southern Illinois. Adult family members voluntarily filled out a health questionnaire that collected their personal health history as well as that of their extended family history in an anonymous manner. Through this method, additional potentially affected family members and their immediate relatives were identified. Members who were enrolled were asked, at their own discretion, to reach out to additional family members to inquire about participating. Members who were willing to participate contacted us and were enrolled through their local medical provider.
Case ascertainment, Denmark
The Danish cases were ascertained as part of a project on prenatally diagnosed kidney anomalies. Data on pre- and postnatal findings in the families were collected as well as DNA. The second affected fetus from family 2 was analyzed by our in-house-designed kidney-gene-targeted panel including 108 genes associated with kidney disease. This analysis did not reveal any disease-causing variants. The family was therefore selected for novel kidney-gene discovery using whole exome sequencing.
Case phenotypes and samples, Iowa
Kidney ultrasound, MRI, and intravenous pyelogram examination revealed URA as well as hypertrophy of the left kidney for II-5, and URA for II-7 (Figure 1A). Additionally, kidney ultrasound revealed URA for III-3 and III-4. In utero kidney ultrasound revealed BRA for one family member (III-6), and in utero kidney ultrasound as well as MRI revealed BRA for another (III-8). BRA was suspected in two family members (II-1, II-6) but not confirmed. Four confirmed affected family members with either URA or BRA (II-5, II-7, III-4, III-8) were selected for whole exome sequencing. The Institutional Review Board of the Carver College of Medicine, University of Iowa, approved this study, and all participants provided written consent in addition to DNA samples after being properly counseled regarding the potential of incidental findings from whole exome sequencing.
Figure 1.
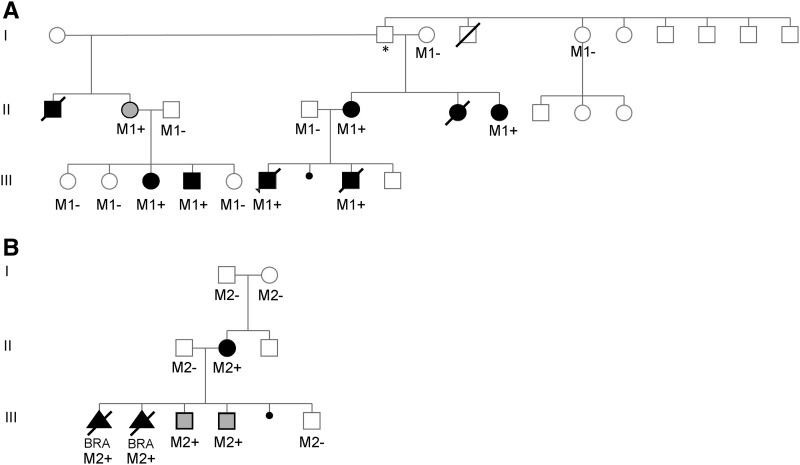
Iowa and Danish RA pedigrees. (A) Iowa pedigree showing dominant inheritance of the agenesis phenotype. (B) Danish pedigree showing transmission of the de novo GREB1L variant to both fetuses. M1, Iowa variant; M2, Danish variant; +, presence, −, absence, *, likely origin of the Iowa mutation. II-2 shaded female (Iowa) and III-3,4 shaded males (Denmark), family members with incomplete penetrance.
Case phenotypes and samples, Denmark
Two pregnancies with affected fetuses presenting with BRA (indicated in Figure 1B) were terminated following parental request and approval by the regional abortion committee. Following the second termination, the parents had a renal ultrasound, which showed left-sided URA in the mother. The father had normal kidneys. Subsequently, the mother’s parents and brother had renal ultrasound examinations, which showed normal kidneys. Prenatal ultrasound examinations of the three live-born healthy brothers were unremarkable. The second affected fetus, the affected mother, and the unaffected father as well as the unaffected maternal grandparents were selected for exome sequencing analysis. Blood samples were obtained from the adult family members. Subsequently, buccal smear samples were obtained from the live-born brothers.
The Danish National Committee of Ethics approved the whole exome sequencing study, and written informed consent was obtained for all four adult family members included in the study after being properly counseled regarding the potential of incidental findings from whole exome sequencing.
Exome sequencing analysis, Iowa
Genomic DNA was obtained from either lymphocytes isolated from whole blood samples or from tissue samples obtained during autopsy using standard laboratory methods. The genomic DNA samples from four affected individuals (II-5, II-7, III-4, III-8; Figure 1A) were prepared for WES (whole exome sequencing) using the Illumina paired-end sample prep kit (Illumina, San Diego, CA) and captured using the Nimblegen SeqCap EZ Human Exome Library v2.0 kit (Roche NimbleGen Inc, Madison, WI) following the manufacturer’s instructions. Captured samples were sequenced using Illumina HiSeq100-bp paired-end sequencing (Duke Center for Genomic and Computational Biology, Ontario Institute for Cancer Research). Next, quality control was performed using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads with average quality scores <20 were trimmed using the Burrows Wheeler Aligner (BWA) (Li and Durbin 2009) and reads <35 bp were not used for the downstream analysis. Reads were mapped to the human reference genome (version-glk_v37) using BWA. Mapping statistics of the aligned reads and coverage of exome target regions were analyzed using Qualimap software (http://qualimap.bioinfo.cipf.es/) (Garcia-Alcalde et al. 2012) and BEDtools (Quinlan and Hall 2010) (see Supplemental Material, Table S1 in File S1).
Local realignment and base quality score recalibration was performed using Genome Analysis Toolkit (GATK) (http://www.broadinstitute.org/gatk/) (McKenna et al. 2010), and fixing mate information and marking duplicates was performed using Picard tools (http://picard.sourceforge.net). Finally, Unified Genotyper was used to call genetic variants in standard Variant Call Format. Variants were annotated using SnpEff (Cingolani et al. 2012) software, the University of California, Santa Cruz human reference genome assembly hg19, and dbSNP 137. Additionally, minor allele frequencies (MAF) for all variants were generated using two databases, the 1000 Genomes Project and the National Heart Lung Blood Institute Exome Sequencing Project (NHLBI-ESP) using the wANNOVAR web server (http://wannovar.usc.edu/) (Chang and Wang 2012). We then applied GATK’s best practices of variant quality and coverage thresholds to account for false positive variant calls. A genotype filter was applied to exclude variants with diverse genotypes across all samples. Assuming that variants involved in causing Mendelian disorders would be rare in nature, we excluded variants that had an MAF ≥1% in 1000 Genomes and NHLBI-ESP. Moreover, we also excluded those variants that had an MAF tag of >5% in their dbSNP 137 annotations.
Lastly, we checked the effects of amino acid substitution on protein structure using the database of human nonsynonymous SNPs and function predictions (dbNSFP v2.0) (https://sites.google.com/site/jpopgen/dbNSFP) (Liu et al. 2013). We focused our analysis on both Polymorphism Phenotyping version 2 (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph2/) and Separating Intolerant from Tolerant (SIFT) (http://sift.jcvi.org/). The CONsensus DELeteriousness (CONDEL) program was then used to generate the weighted average of the normalized scores from PolyPhen-2 and SIFT (http://bg.upf.edu/fannsdb/) (Gonzalez-Perez and Lopez-Bigas 2011). The deleterious variants based on CONDEL predictions were retained in our final list for downstream analysis. Directed Sanger sequencing (carried out by the Iowa Institute of Human Genetics, Genomics Division, University of Iowa Carver College of Medicine) along with a TaqMan Allelic Discrimination Assay (Applied Biosystems) was then used to determine which of these variants showed the predicted segregation pattern for an etiologic variant.
Exome sequencing analysis, Denmark
Genomic DNA was extracted from cultured fetal fibroblasts, formalin-fixed paraffin-embedded fetal tissue, whole blood samples, and buccal smear samples using standard laboratory methods. DNA from two affected and three unaffected family members (Figure 1B) was prepared for WES using the KAPA HTP Library Preparation Kit (KAPA Biosystems Inc, Wilmington, MA) and captured using the SeqCap EZ MedExome Kit (Roche NimbleGen Inc, Madison, WI) according to the manufacturer’s instructions. Next, Illumina NextSequation 500 sequencing was used to generate paired-end reads (carried out by the Department of Molecular Medicine, Aarhus University Hospital, Denmark).
Reads were aligned to the human reference genome (GRCh37/hg19) and variants were called and annotated in coding exons ± 10 bp using Biomedical Genomics Workbench version 2.0 (CLC bio-Qiagen, Aarhus, Denmark). Standard settings on QIAGEN’s Ingenuity Variant Analysis (www.qiagen.com/ingenuity) software were used for data analysis. False positive variant calls were removed based on default coverage and quality thresholds. Variants with an MAF ≥0.1% in the 1000 Genomes Project, the National Heart Lung Blood Institute Exome Sequencing Project (NHLBI-ESP), the Allele Frequency Community, and the Exome Aggregation Consortium (ExAC) were excluded. Variants predicted deleterious and listed in the Human Gene Mutation Database were retained. A filter was applied to retain variants present in heterozygous form in the affected mother and the second affected fetus. Finally, a filter was applied to retrain variants present in heterozygous form only in the affected family members but not present in unaffected family members. The variants were confirmed by direct sequencing using BigDye Terminator v1.1 Cycle Sequencing Kit according to the description of the manufacturer (Applied Biosystems, Life Technology) and analyzed using ABI 3500xl Genetic Analyzer (Applied Biosystems, Foster City, CA). Additionally, the presence of the variant was tested in the female fetus and in the live-born brothers by Sanger sequencing. Primer sequences and PCR details are available upon request.
Zebrafish analyses
University of Iowa:
Zebrafish embryos and adults were reared as described previously (Westerfield 2000), in the University of Iowa Zebrafish Facility, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, following procedures approved by the University of Iowa’s Institutional Animal Care and Use Committee (IACUC). Embryos were staged by hours or days post fertilization (hpf or dpf) at 28.5° (Kimmel et al. 1995). Homozygous greb1l mutant embryos were generated from heterozygous adults of the sa1260 allele obtained from the Zebrafish International Resource Center, Eugene, OR.
To inhibit grebl1 expression we ordered an antisense morpholino oligonucleotide (MO) targeting the exon 3–intron 3 junction (sequence: 5′- TATTGGAACACCAACCTAAAAGTGC-3′) (Gene Tools, Philomath, OR). To test efficacy of the MO, we harvested RNA (separately) from embryos injected with the control MO or with the greb1l MO, generated first-strand complementary DNA (cDNA), and carried out PCR with primers flanking the splice junction on both cDNA templates. The band of expected size was found in both templates, but in the greb1l MO template an additional larger band was present. Sequence from both products confirmed the smaller band corresponded to correctly spliced RNA and the larger band to RNA in which the third intron was unspliced. To mutate the grebl1 gene with CRISPR/Cas9 we used the website https://chopchop.rc.fas.harvard.edu/index.php to identify a high-scoring guide RNA target site. An oligo specific to exon 17 of the zebrafish grebl1 gene was selected. The target site (GGTCCACACAAAAATGG) was synthesized (Integrated DNA Technologies; IDT) with the T7 promoter sequence on the 5′ end and a 20-bp overlap at the 3′ end complementary to the generic Cas9-binding scaffold oligo. The guide sequence oligo was then annealed to the generic 119-bp Cas9-binding scaffold oligo as described in the cloning-free method of generating single-stranded guide RNA (sgRNA) (Talbot and Amacher 2014). Once annealed, this product provides a DNA template complete with T7 promoter for in vitro synthesis of an sgRNA. We co-injected 1- to 2-cell-stage embryos with sgRNA (200–400 pg per embryo) and/or Cas9 protein (IDT) at 2 ng.
For in situ hybridization, 592 bp of greb1l cDNA was amplified from 24 hpf wild-type zebrafish first-strand cDNA using the following primers: forward, 5′-GTCAAGCAGGAAAAGATCTGC-3′; reverse, 5′-GGAACGATCGGTAATGTCTT-3′. The cDNA was engineered into the StrataClone vector (Agilent Technologies, Santa Clara, CA) and a DIG-labeled, antisense RNA probe was generated by in vitro transcription (Roche Diagnostics, Indianapolis, IN). Whole-mount in situ hybridization was carried out following procedures described previously (Thisse and Thisse 2008). For immunohistochemistry, a monoclonal anti-ATPase, [Na(+) K(+)] α-1 subunit antibody (a6F, Developmental Studies Hybridoma Bank at the University of Iowa), was used at a 1:100 dilution. Following primary antibody incubation for 48 hr, the embryos were blocked and then incubated with an Alexa-488 conjugated goat-anti-mouse secondary antibody for 48 hr.
Mayo Clinic:
Zebrafish procedures were approved by the Mayo Clinic’s IACUC. Embryos developed at 28.5° with 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) added at 24 hpf to prevent pigmentation for facilitating cyst visualization. Embryos were anesthetized using 0.02% tricaine (Aquatic Habitats) before observation by microscopy. Embryos were examined for cysts at 2 and 3 dpf using a Zeiss Lumar stereo fluorescence microscope and Zen software.
Generation of CRISPR mutagenized F0 embryos:
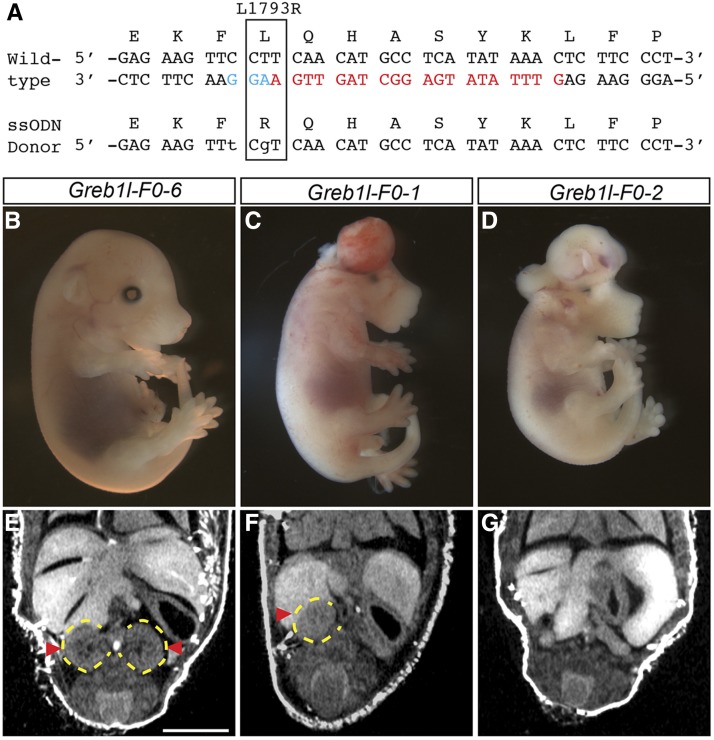
A guide sequence, GTTTATATGAGGCATGTTGA, targeting the orthologous region in mouse to the L1793R mutation was synthesized as an Ultramer (IDT) with the guide sequence embedded between the T7 promoter and portion of stem loop as described in Bassett et al. (2013). The resulting DNA template was column purified (QIAGEN) prior to in vitro transcription reaction, column purified (Zymogen), and quantified via Nanodrop before microinjection. A single-stranded donor oligonucleotide (ssODN) was designed to introduce the desired T > G point mutation to create L1793R missense mutation and also included a silent C > T substitution to ablate the PAM sequence. The ssODN was synthesized as a 125-bp Ultramer (IDT) with the introduced base pair changes underlined: ATCCTGCCCCTTCAGTACGTCTGCGCCCCTGACAGTGAACACACACTCCTGGCAGCCCCTGCACAGTTCCTCCTGGAGAAGTTTCGTCAACATGCCTCATATAAACTCTTCCCTAAAGCCATCCA. One-cell embryos were obtained from superovulated C57BL/6NJ (B6NJ; JAX stock number 5304) female donors crossed to B6NJ males. For microinjection, reagents were injected into the pronucleus at the following concentrations: 30 ng/μl Cas9 mRNA (Trilink); 15 ng/μl sgRNA; and 20 ng/μl ssODN. Embryos were collected at E15.5 and processed for microCT as described in Dickinson et al. (2016). PCR genotyping was performed on tail tip DNA using primers flanking the region of interest: Greb1l-GT-F TGACAGGCACATCTCCCATG and Greb1l-GT-R TCCAAGTCATCAAGGCAGGC that generate a 433-bp product. Individual genotypes were first assessed using Sanger sequencing and subsequently confirmed by T/A cloning and sequencing of at least eight independent clones of tail tip DNA for each putative mutant.
Data availability
File S1 compares sequences between human and zebrafish GREB1L proteins, compares kidney phenotypes in zebrafish for greb1l morpholino knockdown and CRISPR-Cas9 deletion and shows the sequences of mutagenized alleles recovered from CRISPR F0 mouse embryos. This study was approved by the University of Iowa under IRB#200711705 as well as by the Danish National Committee of Ethics. WES data is available in the Sequence Read Archive database (accession number SRP112780).
Results
Exome sequence analysis of two pedigrees reveals GREB1L as an RA gene
Four URA/BRA family members (II-5, II-7, III-4, III-8) from pedigree 1, which suggests autosomal dominant inheritance of the RA phenotype (Figure 1A), were selected for WES analysis. Across the four samples, we achieved an average targeted exome coverage of 172× with a mean mapping quality of 45.30 for calling high-quality variants (Table S1 in File S1). We focused on identifying variants shared by all four cases, and this revealed heterozygosity for novel missense variants in three genes (LHX9 c.1127 C > T, GYLTL1B c.442 T > A, GREB1L c.5378 T > G) as well as heterozygosity for a novel stop-loss variant (CLEC9A c.724 T > C) in CLEC9A (no novel variants showed homozygosity or compound heterozygosity shared by all four affected family members). PolyPhen-2, SIFT, and CONDEL analyses predicted all three missense variants to be damaging. Directed Sanger sequencing along with a TaqMan Allelic Discrimination Assay (Applied Biosystems) revealed that only the GREB1L variant showed the predicted segregation pattern for an etiologic variant: six affected family members (II-5, II-7, III-3, III-4, III-6, and III-8) all harbored the variant, while seven unaffected family members (I-3, I-5, II-3, II-4, III-1, III-2, III-5) lacked the variant; female II-2 was hypothesized to be a carrier of the GREB1L variant exhibiting incomplete penetrance, and presence of the variant was confirmed (Figure 1A). This missense variant, which was absent from the ExAC database, changes a conserved leucine to arginine in the highly conserved c-terminus of the protein (see Figure 2 and Figure S1 in File S1).
Figure 2.
Comparison of proteins encoded by Iowa, Danish, and zebrafish GREB1L mutants. The human, mouse, and fish variants (position indicated in human protein) encode proteins that are altered in the conserved c-terminus of the protein. L1793R, Iowa protein; G1870FS, Danish protein; predicted glycosyltransferase domain indicated by green lettering (Iyer et al. 2013). Iowa L–R mutation indicated in red lettering, which was recapitulated in two out of three mouse mutants along with deletion of the following Q residue (see Figure S3 in File S1); Danish frameshift amino acids indicated in blue lettering; zebrafish W to STOP mutation indicated by purple W residue. Conserved c-terminus indicated by boxed region, with asterisks denoting amino acids conserved between GREB1 and GREB1L paralogs.
Two URA/BRA affected (II-2, III-2) and three unaffected family members (I-1, I-2, II-1) from pedigree 2 (Figure 1B) were selected for WES analysis. We achieved a mean target region coverage of 119× and mapping quality of 61 for calling high-quality variants (Table S2 in File S1). Ingenuity Variant Analysis revealed two variants that were present only in affected family members, i.e., GREB1L c.5608 + 1delG and FAM21C c.1837G > C. The FAM21C missense variant was predicted to be tolerated and benign by SIFT and PolyPhen-2, respectively, with the base at that position being weakly conserved. The GREB1L variant is novel and deletes one of two G residues located at the splice donor site of the last intron (the transcribed wild-type RNA sequence reads AAAG at the 3′ end of the exon followed by GUAA at the 5′ end of the intron). Both G residues represent highly conserved nucleotides involved in splicing and therefore there are two potential effects of a single G at the splice donor site: first, a novel splice site could be created, shifted by 1 bp, resulting in the protein sequence as depicted in Figure 2; and second, splicing efficiency could be diminished, causing partial intronic read through of nonspliced messenger RNA prior to encountering a stop codon during translation. Importantly, in either scenario, the highly conserved c-terminus of the protein encoded by the last exon would be deleted.
Sanger sequencing identified the GREB1L variant in the other affected fetus as well as the two eldest healthy live-born brothers. Additionally, based on the exome sequencing data, no disease-associated copy number variants were identified.
GREB1L is a haploinsufficiency locus
Large-scale microarray and sequencing studies have helped elucidate numerous haploinsufficient or variation-intolerant regions within the genome (Petrovski et al. 2013; Zarrei et al. 2015; Ruderfer et al. 2016), and interrogation of the Database of Genomic Variants (DGV; http://dgv.tcag.ca) finds only two deletion events in control populations that involve coding portions of GREB1L. This is very significant given that as of early 2017, the DGV had identified over six million sample level CNVs from control populations of over 70 studies. Further supporting this claim is the absence of any GREB1L coding deletion CNVs within the DECIPHER database and only one within the ClinGen resource (https://decipher.sanger.ac.uk/; https://www.clinicalgenome.org/). Finally, there are no deletions involving GREB1L noted in the CNV calls from the ExAC database (http://exac.broadinstitute.org/) and the gene itself is predicted to be haploinsufficient (%HI 9.33 reported by DECIPHER) (Huang et al. 2010). Given that most of the copy number variable regions of the genome are pericentromeric, the lack of such variation within the GREB1L gene is also significant due to its proximity to the centromere of chromosome 18 (Iafrate et al. 2004; Sebat et al. 2004; Redon et al. 2006; Zarrei et al. 2015). Taken together, these data identify GREB1L as a likely haploinsufficiency locus.
GREB1L is expressed in the developing kidney
To determine whether GREB1L is expressed during genitourinary development, we accessed gene expression microarray data cataloged in GUDMAP (Genitourinary Development Molecular Anatomy Project) (McMahon et al. 2008; Harding et al. 2011). These results revealed that GREB1L is expressed primarily in the early proximal tubule as well as metanephric mesenchyme, and also in the ureteric bud (Georgas et al. 2009). However, most kidney expression studies have been performed during morphogenesis of the metanephric kidney, and thus information on factors associated with early pronephric specification is lacking. Since early events in kidney development are strongly conserved between zebrafish and mammals (Drummond 2005; Drummond and Davidson 2016), we thus turned to the zebrafish model to explore greb1l expression patterns and function during development.
Pronephric kidney development is altered in zebrafish greb1l loss-of-function mutants and greb1l-depleted embryos
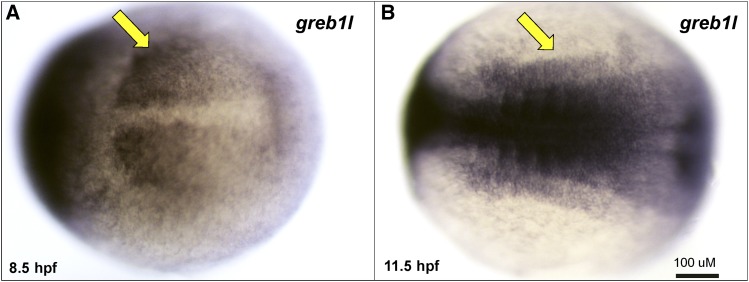
The single ortholog of GREB1L in the zebrafish genome, greb1l, is predicted to encode a protein that is 61% identical and 73% similar to the human ortholog (Figure S1 in File S1). Whole-mount in situ hybridization on wild-type embryos at 90% epiboly (8.5 hpf) and early somitogenesis stages (11.5 hpf) revealed expression of greb1l in the mesoderm, including the intermediate mesoderm, the origin of the pronephros (Figure 3, A and B).
Figure 3.
Endogenous expression of greb1l during zebrafish development. In situ hybridization of greb1l antisense probes on embryos fixed at indicated stages (panel A, 8.5 hpf; panel B, 11.5 hpf). Embryos are presented in a dorsal view with rostral to the left. Arrows indicate intermediate mesoderm signal.
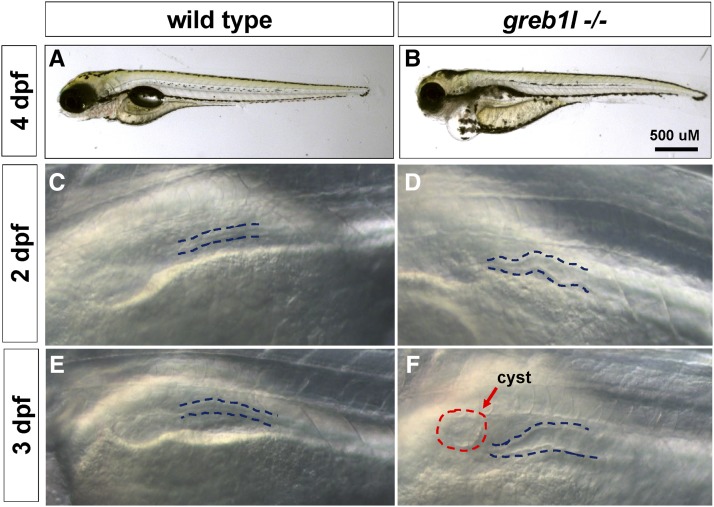
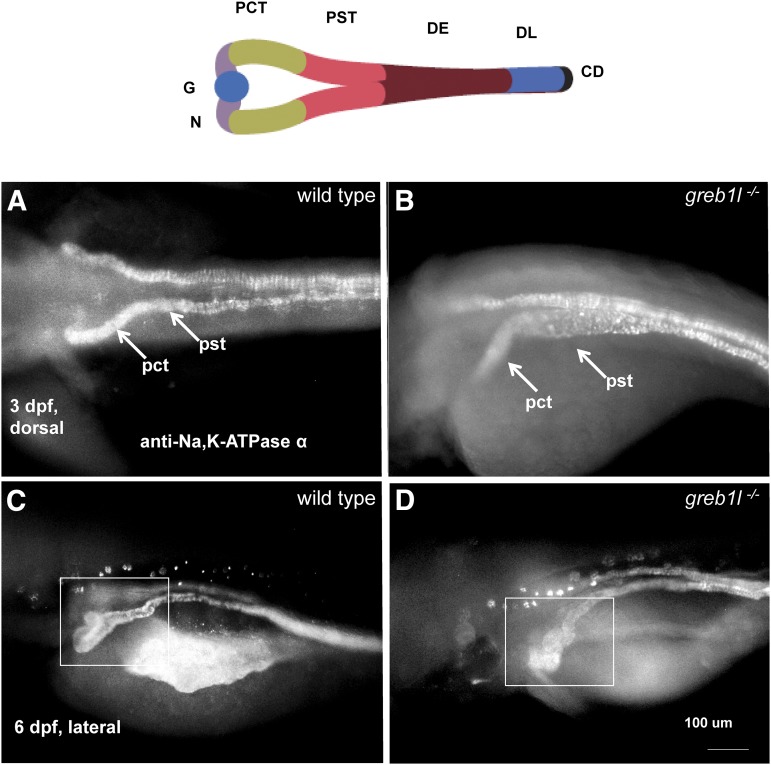
An N-ethyl-N-nitrosourea (ENU)-induced T to A substitution in the grebl1 gene, the sa1260 allele which introduces a stop codon at amino acid 1915 of the 1942 residue full-length protein, was isolated by large-scale screening (Kettleborough et al. 2013). Heterozygotes for the sa1260 allele are morphologically normal and are fertile. Homozygotes for this allele (hereafter, greb1l mutants) were readily recognized at 3 dpf by periorbital edema and pericardial effusions, consistent with a defect in ion and fluid homeostasis (16 embryos in a clutch of 109 embryos showed this phenotype and were all found to be homozygous mutants by sequencing) (Figure 4B). At 2 dpf, the developing kidney is readily discernible in normal living embryos. In most of the clutch, presumed to be wild types or heterozygous mutants, the pronephric tubule appeared normal (Figure 4C). By contrast, in greb1l mutants, tubules were dilated and kinked (n = 16) (Figure 4D). At 3 dpf, dilation was more pronounced, and the majority of the mutants had obvious cysts (15 of 16 mutants, Figure 4F). In embryos processed to reveal immunoreactivity of anti-Na/K ATPase antibody (a6f), an early marker of pronephric mesoderm, dilation of the proximal straight tubule in mutants in comparison to nonmutant siblings was evident (Figure 5, A and B). In all wild types examined, there was a characteristic hairpin turn between the proximal convoluted tubule and the neck region of the pronephros (n = 12, shown at 6 dpf, Figure 5C). In all greb1l mutants examined, this region of the pronephros is serpentine (n = 14, Figure 5D). greb1l mutants died between 10 and 12 dpf.
Figure 4.
greb1l mutants have edema and abnormal pronephros development. (A–F) Lateral views of live larvae at the indicated stage and of the indicated genotype. (B) At 4 dpf, mutants exhibit edema particularly around the heart and eyes. (D) 2 dpf mutant embryos have dilated and kinked tubules (F: dotted lines). At 3 dpf, in mutant embryos the kidney remains dilated (dotted lines) and cysts are evident in most cases (red dotted circle).
Figure 5.
Renal morphology of zebrafish greb1l mutants. In all images, rostral is to the left and embryos are processed with anti-Na,K ATPase antibody (a6F) and a fluorescent secondary antibody. (A and B) Dorsal views of representative embryos at 3 dpf. Mutants present with swelling of the proximal convoluted tubule (PCT) and proximal straight tubule (PST). (C and D) Ventral-lateral views. Mutants have a deformed junction between the PCT and neck. Top, schematic modified from Drummond and Davidson (2016).
Because the greb1l sa1260 mutant allele came from a chemical mutagenesis screen, it is conceivable that there are other mutations cosegregating with the greb1l mutation. To confirm that the abnormal kidney phenotype in sa1260 mutants results from the mutation in greb1l we reduced greb1l expression by injecting wild-type embryos with antisense MO targeting an early splice junction, or with control MO. We used RT-PCR and sequencing to confirm the MO was effective at inhibiting splicing of greb1l (see Materials and Methods). Additionally, we employed CRISPR technology by injecting wild-type embryos with Cas9 protein and a guide RNA targeting an evolutionarily conserved exon of greb1l; this is expected to yield mosaic embryos in which a variable frequency of cells experience a biallelic mutation in grebl (Talbot and Amacher 2014). In both cases we fixed injected embryos at 4 dpf and processed them to reveal anti-Na/K ATPase immunoreactivity. The large majority of embryos injected with greb1l MO exhibited the abnormal morphology of the proximal kidney seen in sa1260 mutants (Figure S2 in File S1). Moreover, ∼30% of F0 embryos injected with greb1l gRNA with Cas9 protein exhibited the proximal kidney defects, suggesting mosaic, biallelic mutation of greb1l is sufficient to yield this phenotype; notably, the efficiency of phenotypic penetrance in CRISPR/Cas9-injected F0 embryos is comparable to that seen by other groups targeting other genes (Jao et al. 2013). The convergent phenotype of greb1l mutants, embryos injected with MO targeting greb1l, and CRISPR/Cas9 reagents targeting greb1l strongly support a requirement for Greb1l in kidney morphogenesis in zebrafish and are consistent with a role for GREB1L in morphogenesis of the human kidney.
F0 CRISPR mouse Greb1l mutants display kidney agenesis phenotypes
The single ortholog of mouse GREB1L, Greb1l, is predicted to encode a protein that is 90% identical and 94% similar to the human ortholog. To test whether Greb1l plays a similar role in a mammalian model, we generated F0 mutant embryos in mice using CRISPR/Cas9, targeting mouse exon 31 and an ssODN designed to introduce the orthologous L1793R Iowa mutation (Figure 6A). Our F0 approach has previously been shown to faithfully recapitulate human disease phenotypes despite the mosaicism intrinsic to the genome editing process, and that we can establish a clear and robust genotype–phenotype relationship (Guimier et al. 2015). Our microinjections produced 56 F0 embryos that were subsequently analyzed at E15.5. Of these, we identified 9 (16%) embryos showing evidence of CRISPR/Cas9 mutagenesis with 3 (33%) phenotypically affected mutants that displayed a range of gross phenotypes including exencephaly and craniofacial dysmorphology including unilateral and bilateral cleft lip (Figure 6, B–D). For the mutant embryos, we took advantage of our high-throughput microCT imaging platform established for the Knockout Mouse Phenotyping Program (KOMP2) (Dickinson et al. 2016) to examine developmental kidney defects. In two of the affected embryos, we observed unilateral agenesis, and bilateral agenesis in the third (Figure 6, E–G). Notably, in each case of unilateral agenesis, the contralateral kidney also appears abnormal or incompletely developed. To determine the nature of the CRISPR-induced mutations, we cloned and sequenced the mutations of all embryos showing evidence of CRISPR activity and confirmed that all three embryos with RA phenotypes harbored mutations in Greb1l. Two affected embryos carried knock-in alleles harboring the L1793R mutation along with an accompanying in-frame deletion removing a conserved glutamine residue adjacent to L1793 (Figure S3 in File S1). The other affected embryo was homozygous for a 2-bp insertion resulting in a frameshift mutation and stop codon 33 amino acids downstream. The remaining six mutagenized embryos showed evidence of nonhomologous end joining (NHEJ)-induced indels but also contained wild-type alleles suggesting only partial impairment of Greb1l function. In summary, these phenotypes are consistent with an essential role for Greb1l in kidney development and suggest additional roles for Greb1l during embryonic development.
Figure 6.
Analysis of Greb1l function in CRISPR/Cas9 mutagenized F0 mouse embryos. (A) CRISPR/Cas9 strategy for introducing the L1793R mutation using an ssODN donor template. The guide sequence is colored red and the adjacent PAM sequence (AGG) is indicated in turquoise. Point mutations engineered into the donor are shown as lowercase. (B–D) Whole-mount images highlighting the observed exencephaly in two mutants (C and D) carrying KI alleles as compared to wild type (B). (E–G) Coronal sections of microCT data show unilateral and complete kidney agenesis in mutagenized F0 embryos. The position of kidneys is indicated by red arrowheads and yellow dotted circles. Bar, 2 mm.
Discussion
GREBL1 coding variants are associated with RA in two families
The Iowa pedigree structure (Figure 1A) is consistent with autosomal dominant inheritance of the RA phenotype, and the GREB1L missense variant was the only variant to cosegregate with the phenotype in all cases tested (six) but none of the unaffected family members tested (seven) except the female carrier (II-2, Figure 1A) exhibiting incomplete penetrance. The odds of such a segregation pattern occurring by chance is 1 in over 16,000. The variant was called as damaging by SIFT/PolyPhen-2/CONDEL and alters a conserved residue in a highly conserved domain in the c-terminus of the protein.
In the Danish family, both the GREB1L frameshift variant as well as a missense variant of FAM21C had arisen de novo in the affected mother and was found in one affected fetus. The FAM21C variant was predicted to be tolerated/benign, while the GREB1L variant causes a profound alteration of the c-terminus (notably, the same region affected by the Iowa variant). The GREB1L variant was also found in the second affected fetus (and thus all three affected family members), along with two unaffected brothers. Collectively, these data reveal that the GREB1L variants identified in both the Iowa and Danish families are likely the etiologic variants causing the RA phenotype with incomplete penetrance.
GREB1L expression pattern supports its role in kidney morphogenesis
Transcriptome analysis shows that GREB1L is expressed in a variety of tissues (www.genecards.org), with particularly high expression in brain, kidney, and ovary (GEO accession number GSM35549, profile GDS3052; Hildner et al. 2008). GREB1L is also robustly expressed in the early proximal tubule and metanephric mesenchyme of the metanephric kidney, with lower expression levels seen in the ureteric bud (Georgas et al. 2009). Since these expression studies were performed on the developing metanephric kidney but not on earlier morphogenetic events, we performed in situ hybridization of greb1l in the developing zebrafish embryo and found it to be expressed in the portion of the intermediate mesoderm that gives rise to the pronephros (Figure 3, A and B). Since both GREB1L human variants (as well as the variants in zebrafish and mouse Greb1l) alter the c-terminus of the protein (Figure 2 and Figures S1 and S3 in File S1), it is possible that this region may be associated with a kidney-specific function and that its alteration could produce a dominant effect. Both variants are located in one of the most highly conserved domains of the protein when comparing the GREB1 and GREB1L paralogs, or the GREB1L human and zebrafish orthologs (amino acid identity 61%). The Iowa missense variant alters a residue residing in a stretch of 24/27 (89%) conserved amino acids across paralogs as well as between human and zebrafish GREB1L, while the Danish frameshift variant deletes a region of 47/54 (87%) conserved amino acids across paralogs and 50/54 (93%) across orthologs. However, given that premature stop codons produced by both the Danish and mouse variants would probably lead to nonsense-mediated decay, it is more likely that these variants are loss-of-function, which would effectively reduce GREB1L gene dosage to half. Consistent with this idea, we have found that GREB1L is likely to be a haploinsufficiency gene. This might also explain the range of observed phenotypes (two kidneys, URA, BRA) observed in the pedigrees, with the reduced gene dosage effectively creating a “teeter-totter” scenario of stochastic developmental decisions that either result in the morphogenesis of a mature kidney, or no kidney at all.
GREB1L is a likely cofactor for steroid hormone/RARs
Although UniProt predicts GREB1L to be a single-pass membrane protein due to a predicted membrane-associated domain, we do not favor this hypothesis. Its paralog, GREB1 (54% identical and 67% similar to GREB1L), was shown to be a nuclear chromatin-bound ER coactivator that is (1) upregulated after estrogen treatment and (2) essential for ER-mediated transcription (Rae et al. 2005; Mohammed et al. 2013). Importantly, GREB1L and retinoic acid receptor members are part of the ER/GREB1 chromatin complex (Mohammed et al. 2013). Consistent with GREB1L playing a similar coactivator role, but in concert with RARs, retinoic acid treatment of F9 embryonal carcinoma stem cells (a well-established model for retinoic acid signaling) was shown to robustly upregulate GREB1L (Laursen et al. 2012), which would then be predicted to bind and activate RARs. Intriguingly, RNAi knockdown of GREB1 in cell lines was shown to block estrogen-induced growth (Rae et al. 2005), produce a G0/1 arrest with increased G1 DNA content (Kittler et al. 2007), and decrease cell viability after treatment with Paclitaxel (Whitehurst et al. 2007; Sinnott et al. 2014), suggesting that GREB proteins might be playing a role in mediating cell growth.
Zebrafish grebl1 is required for proper morphogenesis of the pronephros
Whole-mount ISH of zebrafish embryos revealed widespread labeling of the mesoderm during early somitogenesis, an area that includes the intermediate mesoderm that gives rise to the pronephros and also expresses the pronephric markers wt1a, wt1b, pax2a, pax8, and lhx1a at similar stages of development (Bollig et al. 2006; Perner et al. 2007; Drummond and Davidson 2016). It is worth noting that these genes are the orthologs of the genes that pattern the mammalian pronephros (see below), demonstrating the evolutionary conservation of pronephros specification and relevance of the zebrafish model.
Zebrafish greb1l mutants had abnormal pronephric morphology and evidence of altered function, including presence of cysts and dilated tubules evident by 2 dpf when the pronephros begins filtering (Drummond et al. 1998). Later, the mutants developed edema and disrupted proximal tubule convolution, phenotypes that could result from/contribute to defects in fluid and ion transport (Vasilyev et al. 2009, 2012) thus contributing to death of the mutants. In particular, loss of fluid flow leads to fluid accumulation and organ distension including pronephric cysts and tubule dilation (Kramer-Zucker et al. 2005). Since greb1l zebrafish mutants die just prior to the time when the mesonephros can be reliably detected (Diep et al. 2015), we were unable to assess development of the mesonephros, the mature kidney of the zebrafish. Nonetheless, these data point toward a role of greb1l in controlling early pronephros specification/morphogenesis and ultimately function.
F0 CRISPR Greb1l mouse mutants present with URA and BRA phenotypes
The high efficiency of Cas9 coupled with the short gestation period of the mouse provides a significant opportunity to functionally validate discoveries uncovered from WES efforts of human cohorts. Here, we demonstrate this powerful combination using CRISPR/Cas9-mediated genome editing to model a novel point mutation in GREB1L directly in F0 mouse embryos, thereby removing the traditional constraints of establishing animal lines and performing timed matings. The mutagenized embryos all displayed a spectrum of kidney abnormalities ranging from URA to BRA, confirming the causative etiology of the human mutations in RA. Additionally, several craniofacial abnormalities were observed in the affected embryos, highlighting a critical and more widespread role for Greb1l during embryonic development. While two of the three mutants harbored nonnull allelic combinations, the third was homozygous for a frameshift mutation, consistent with the highly conserved nature of the mutated residue and c-terminal domain of the GREB1L protein. These findings along with current advances in genome editing hold great promise for the future of performing rapid and precise modeling of human developmental disorders in a mammalian system.
GREB1L may mediate proliferation and inductive events in early kidney development
In addition to connections between estrogen/estrogen-related nuclear steroid hormone receptors and kidney morphogenesis (Shim et al. 2004; Harewood et al. 2010; Berry et al. 2011), retinoic acid also plays key roles in genitourinary system development, including promoting early pronephric kidney morphogenesis (Carroll and Vize 1999; Osafune et al. 2002; Kim and Dressler 2005; Cartry et al. 2006; Perner et al. 2007; Bollig et al. 2009; Lee et al. 2010) as well as metanephros development (Vilar et al. 1996), and absence of α and γ RARs results in murine renal agenesis (Mendelsohn et al. 1994). The hypothesis that GREB1L may be promoting kidney development through activation of RARs is particularly attractive, since several vertebrate genes required for pronephros specification and development in fish, frogs, and/or mice (Pax2, Pax8, Lhx1, Wt1, and Pteg; mouse abbreviations used for clarity) are under the control of retinoic acid signaling (Carroll and Vize 1999; Cartry et al. 2006; Perner et al. 2007; Bollig et al. 2009; Lee et al. 2010), to determine rostral/caudal and multiciliated/transporting epithelial cell fate (Wingert et al. 2007; Li et al. 2014; Cheng and Wingert 2015; Marra and Wingert 2016), and mouse or human studies have associated several of these early expressed genes (Pax2, Pax8, Lhx1, and Wt1) with RA phenotypes (Kreidberg et al. 1993; Shawlot and Behringer 1995; Torres et al. 1995; Brophy et al. 2001; Bouchard et al. 2002; Meeus et al. 2004; Trueba et al. 2005). Collectively, these data suggest a mechanism whereby retinoic acid signaling activates GREB1L expression, which in turn allows interaction of GREB1L and RARs, both of which are required for robust activation of the pronephros patterning genes. Failure to properly activate GREB1L expression, or expression of the retinoic acid-responsive PAX2/8, LHX1, and WT1 targets, could then lead to RA phenotypes.
The pronephros is formed from intermediate mesoderm (where greb1l expression is observed in the zebrafish), and although it is considered a rudimentary structure that will be temporarily replaced by the mesonephros, studies have demonstrated that the pronephric duct is essential for promoting both mesonephric as well as metanephric (adult) kidney formation via key inductive signaling events (Saxen and Sariola 1987; Vize et al. 1997; Carroll et al. 1999; Natarajan et al. 2013). Early on, the pronephric duct signals nearby intermediate mesoderm to form mesonephric tubules and these allow drainage into the mesonephric duct, the most caudal portion of the original pronephric duct. Later on, the mesonephric duct forms the ureteric bud, and mutual induction between the metanephric mesenchyme and the ureteric bud promotes mature kidney development (Piscione and Rosenblum 2002; Clarke et al. 2006; Costantini 2010). Of particular note, studies on mutants of the retinoic acid-responsive pronephros specification gene Pax2 revealed that homozygous mutant embryos were able to form both a pronephros and a mesonephros, but failed to induce the mature metanephric kidney (Torres et al. 1995; Brophy et al. 2001; Bouchard 2004). These studies underscore the importance of proper pronephros specification for mature kidney development, and our zebrafish results suggest that GREB1L might be functioning in early pronephric development to ensure that the proper downstream developmental decisions are made.
Although the RA-associated Wt1, Pax2, and Lhx1 genes are expressed in the early pronephros, they are also necessary for proper metanephric mesenchyme induction and development (Shawlot and Behringer 1995; Donovan et al. 1999; Clarke et al. 2006). Remarkably, in mice Greb1l is also expressed at high levels in the metanephric mesenchyme, suggesting that similar to pronephros development, a second retinoic acid signaling event involving Greb1l and retinoic acid signaling targets is employed. Additionally, Greb1l is expressed in ureteric buds, albeit at lower levels, and retinoic acid signaling has been shown to be required for proper expression of Ret, itself a gene associated with RA. It is thus conceivable that the agenesis phenotype seen in Greb1l mutants may also be due to alterations in specification of either the metanephric mesenchyme or ureteric bud. Further studies are needed to establish which mechanisms underlie the agenesis phenotypes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.1125/-/DC1.
Acknowledgments
The authors thank Senuri Jayatilleka for initial work on the greb1l morpholino and zebrafish in situ assays, and Caleb Heffner for assistance with mouse embryo processing. This work was funded in part by the National Institutes of Health grant R01 DE021071 (J.R.M.) as well as by the National Institutes of Health grant RC4 DK090937 (P.D.B. and J.R.M.), a National Institutes of Health grant UM1 OD023222 (S.A.M.), and a Maria Dorthea and Holger From, Haderslevs Foundation grant (M.R.).
Footnotes
Cofirst authors.
Communicating editor: J. Lupski
Literature Cited
- Barak H., Huh S. H., Chen S., Jeanpierre C., Martinovic J., et al. , 2012. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell 22: 1191–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J.-L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batourina E., Tsai S., Lambert S., Sprenkle P., Viana R., et al. , 2005. Apoptosis induced by vitamin A signaling is crucial for connecting the ureters to the bladder. Nat. Genet. 37: 1082–1089. [DOI] [PubMed] [Google Scholar]
- Berry R., Harewood L., Pei L., Fisher M., Brownstein D., et al. , 2011. Esrrg functions in early branch generation of the ureteric bud and is essential for normal development of the renal papilla. Hum. Mol. Genet. 20: 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig F., Mehringer R., Perner B., Hartung C., Schafer M., et al. , 2006. Identification and comparative expression analysis of a second wt1 gene in zebrafish. Dev. Dyn. 235: 554–561. [DOI] [PubMed] [Google Scholar]
- Bollig F., Perner B., Besenbeck B., Kothe S., Ebert C., et al. , 2009. A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros. Development 136: 2883–2892. [DOI] [PubMed] [Google Scholar]
- Bouchard M., 2004. Transcriptional control of kidney development. Differentiation 72: 295–306. [DOI] [PubMed] [Google Scholar]
- Bouchard M., Souabni A., Mandler M., Neubuser A., Busslinger M., 2002. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 16: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy P. D., Ostrom L., Lang K. M., Dressler G. R., 2001. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128: 4747–4756. [DOI] [PubMed] [Google Scholar]
- Brophy P. D., Alasti F., Darbro B. W., Clarke J., Nishimura C., et al. , 2013. Genome-wide copy number variation analysis of a Branchio-oto-renal syndrome cohort identifies a recombination hotspot and implicates new candidate genes. Hum. Genet. 132: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T. J., Vize P. D., 1999. Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev. Biol. 214: 46–59. [DOI] [PubMed] [Google Scholar]
- Carroll T., Wallingford J., Seufert D., Vize P. D., 1999. Molecular regulation of pronephric development. Curr. Top. Dev. Biol. 44: 67–100. [DOI] [PubMed] [Google Scholar]
- Carter C. O., Evans K., Pescia G., 1979. A family study of renal agenesis. J. Med. Genet. 16: 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartry J., Nichane M., Ribes V., Colas A., Riou J. F., et al. , 2006. Retinoic acid signalling is required for specification of pronephric cell fate. Dev. Biol. 299: 35–51. [DOI] [PubMed] [Google Scholar]
- Chang X., Wang K., 2012. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 49: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. N., Wingert R. A., 2015. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev. Biol. 399: 100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. C., Patel S. R., Raymond R. M., Jr, Andrew S., Robinson B. G., et al. , 2006. Regulation of c-Ret in the developing kidney is responsive to Pax2 gene dosage. Hum. Mol. Genet. 15: 3420–3428. [DOI] [PubMed] [Google Scholar]
- Costantini F., 2010. GDNF/Ret signaling and renal branching morphogenesis: from mesenchymal signals to epithelial cell behaviors. Organogenesis 6: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., et al. , 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep C. Q., Peng Z., Ukah T. K., Kelly P. M., Daigle R. V., et al. , 2015. Development of the zebrafish mesonephros. Genesis 53: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan M. J., Natoli T. A., Sainio K., Amstutz A., Jaenisch R., et al. , 1999. Initial differentiation of the metanephric mesenchyme is independent of WT1 and the ureteric bud. Dev. Genet. 24: 252–262. [DOI] [PubMed] [Google Scholar]
- Drummond I. A., 2005. Kidney development and disease in the zebrafish. J. Am. Soc. Nephrol. 16: 299–304. [DOI] [PubMed] [Google Scholar]
- Drummond I. A., Davidson A. J., 2016. Zebrafish kidney development. Methods Cell Biol. 134: 391–429. [DOI] [PubMed] [Google Scholar]
- Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., et al. , 1998. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125: 4655–4667. [DOI] [PubMed] [Google Scholar]
- Garcia-Alcalde F., Okonechnikov K., Carbonell J., Cruz L. M., Gotz S., et al. , 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28: 2678–2679. [DOI] [PubMed] [Google Scholar]
- Georgas K., Rumballe B., Valerius M. T., Chiu H. S., Thiagarajan R. D., et al. , 2009. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev. Biol. 332: 273–286. [DOI] [PubMed] [Google Scholar]
- Ghosh M. G., Thompson D. A., Weigel R. J., 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60: 6367–6375. [PubMed] [Google Scholar]
- Gonzalez-Perez A., Lopez-Bigas N., 2011. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88: 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimier A., Gabriel G. C., Bajolle F., Tsang M., Liu H., et al. , 2015. MMP21 is mutated in human heterotaxy and is required for normal left-right asymmetry in vertebrates. Nat. Genet. 47: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S. D., Armit C., Armstrong J., Brennan J., Cheng Y., et al. , 2011. The GUDMAP database – an online resource for genitourinary research. Development 138: 2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harewood L., Liu M., Keeling J., Howatson A., Whiteford M., et al. , 2010. Bilateral renal agenesis/hypoplasia/dysplasia (BRAHD): postmortem analysis of 45 cases with breakpoint mapping of two de novo translocations. PLoS One 5: e12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B. T., Purtha W. E., Diamond M., Matsushita H., et al. , 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322: 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Lee I., Marcotte E. M., Hurles M. E., 2010. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6: e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert C., Silbermann F., Morar B., Parisot M., Zarhrate M., et al. , 2014. Integrin alpha 8 recessive mutations are responsible for bilateral renal agenesis in humans. Am. J. Hum. Genet. 94: 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A. J., Feuk L., Rivera M. N., Listewnik M. L., Donahoe P. K., et al. , 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36: 949–951. [DOI] [PubMed] [Google Scholar]
- Iyer L. M., Zhang D., Burroughs A. M., Aravind L., 2013. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 41: 7635–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L. E., Wente S. R., Chen W., 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D., Bitner-Glindzicz M., Malcolm S., Hu C. C., Allison J., et al. , 2005. De novo Uroplakin IIIa heterozygous mutations cause human renal adysplasia leading to severe kidney failure. J. Am. Soc. Nephrol. 16: 2141–2149. [DOI] [PubMed] [Google Scholar]
- Kerecuk L., Schreuder M. F., Woolf A. S., 2008. Renal tract malformations: perspectives for nephrologists. Nat. Clin. Pract. Nephrol. 4: 312–325. [DOI] [PubMed] [Google Scholar]
- Kettleborough R. N., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., et al. , 2013. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496: 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Dressler G. R., 2005. Nephrogenic factors promote differentiation of mouse embryonic stem cells into renal epithelia. J. Am. Soc. Nephrol. 16: 3527–3534. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kittler R., Pelletier L., Heninger A. K., Slabicki M., Theis M., et al. , 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9: 1401–1412. [DOI] [PubMed] [Google Scholar]
- Kohlhase J., Wischermann A., Reichenbach H., Froster U., Engel W., 1998. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 18: 81–83. [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker A. G., Wiessner S., Jensen A. M., Drummond I. A., 2005. Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev. Biol. 285: 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., et al. , 1993. WT-1 is required for early kidney development. Cell 74: 679–691. [DOI] [PubMed] [Google Scholar]
- Laursen K. B., Wong P. M., Gudas L. J., 2012. Epigenetic regulation by RARalpha maintains ligand-independent transcriptional activity. Nucleic Acids Res. 40: 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Kim S., Choi S. C., Han J. K., 2010. XPteg (Xenopus proximal tubules-expressed gene) is essential for pronephric mesoderm specification and tubulogenesis. Mech. Dev. 127: 49–61. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cheng C. N., Verdun V. A., Wingert R. A., 2014. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev. Biol. 386: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jian X., Boerwinkle E., 2013. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34: E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra A. N., Wingert R. A., 2016. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev. Biol. 411: 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P., Aronow B. J., Davidson D. R., Davies J. A., Gaido K. W., et al. , 2008. GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 19: 667–671. [DOI] [PubMed] [Google Scholar]
- Meeus L., Gilbert B., Rydlewski C., Parma J., Roussie A. L., et al. , 2004. Characterization of a novel loss of function mutation of PAX8 in a familial case of congenital hypothyroidism with in-place, normal-sized thyroid. J. Clin. Endocrinol. Metab. 89: 4285–4291. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C., Lohnes D., Decimo D., Lufkin T., LeMeur M., et al. , 1994. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120: 2749–2771. [DOI] [PubMed] [Google Scholar]
- Mohammed H., D’Santos C., Serandour A. A., Ali H. R., Brown G. D., et al. , 2013. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3: 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan G., Jeyachandran D., Subramaniyan B., Thanigachalam D., Rajagopalan A., 2013. Congenital anomalies of kidney and hand: a review. Clin. Kidney J. 6: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood V. F., Chevalier R. L., 2003. Renal Developmental Disorders of the Fetus and Newborn. McGraw-Hill, New York, NY. [Google Scholar]
- Osafune K., Nishinakamura R., Komazaki S., Asashima M., 2002. In vitro induction of the pronephric duct in Xenopus explants. Dev. Growth Differ. 44: 161–167. [DOI] [PubMed] [Google Scholar]
- Perner B., Englert C., Bollig F., 2007. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 309: 87–96. [DOI] [PubMed] [Google Scholar]
- Petkovich M., Brand N. J., Krust A., Chambon P., 1987. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature 330: 444–450. [DOI] [PubMed] [Google Scholar]
- Petrovski S., Wang Q., Heinzen E. L., Allen A. S., Goldstein D. B., 2013. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscione T. D., Rosenblum N. D., 2002. The molecular control of renal branching morphogenesis: current knowledge and emerging insights. Differentiation 70: 227–246. [DOI] [PubMed] [Google Scholar]
- Potter E. L., 1946. Facial characteristics of infants with bilateral renal agenesis. Am. J. Obstet. Gynecol. 51: 885–888. [DOI] [PubMed] [Google Scholar]
- Potter E. L., 1965. Bilateral absence of ureters and kidneys: a report of 50 cases. Obstet. Gynecol. 25: 3–12. [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae J. M., Johnson M. D., Scheys J. O., Cordero K. E., Larios J. M., et al. , 2005. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat. 92: 141–149. [DOI] [PubMed] [Google Scholar]
- Rae J. M., Johnson M. D., Cordero K. E., Scheys J. O., Larios J. M., et al. , 2006. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate 66: 886–894. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., et al. , 2006. Global variation in copy number in the human genome. Nature 444: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodhooft A. M., Birnholz J. C., Holmes L. B., 1984. Familial nature of congenital absence and severe dysgenesis of both kidneys. N. Engl. J. Med. 310: 1341–1345. [DOI] [PubMed] [Google Scholar]
- Rosselot C., Spraggon L., Chia I., Batourina E., Riccio P., et al. , 2010. Non-cell-autonomous retinoid signaling is crucial for renal development. Development 137: 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer D. M., Hamamsy T., Lek M., Karczewski K. J., Kavanagh D., et al. , 2016. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 48: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna-Cherchi S., Caridi G., Weng P. L., Scolari F., Perfumo F., et al. , 2007. Genetic approaches to human renal agenesis/hypoplasia and dysplasia. Pediatr. Nephrol. 22: 1675–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna-Cherchi S., Ravani P., Corbani V., Parodi S., Haupt R., et al. , 2009. Renal outcome in patients with congenital anomalies of the kidney and urinary tract. Kidney Int. 76: 528–533. [DOI] [PubMed] [Google Scholar]
- Saxen L., Sariola H., 1987. Early organogenesis of the kidney. Pediatr. Nephrol. 1: 385–392. [DOI] [PubMed] [Google Scholar]
- Schreuder M. F., Langemeijer M. E., Bokenkamp A., Delemarre-Van de Waal H. A., Van Wijk J. A., 2008. Hypertension and microalbuminuria in children with congenital solitary kidneys. J. Paediatr. Child Health 44: 363–368. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., et al. , 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Shawlot W., Behringer R. R., 1995. Requirement for Lim1 in head-organizer function. Nature 374: 425–430. [DOI] [PubMed] [Google Scholar]
- Shim G. J., Kis L. L., Warner M., Gustafsson J. A., 2004. Autoimmune glomerulonephritis with spontaneous formation of splenic germinal centers in mice lacking the estrogen receptor alpha gene. Proc. Natl. Acad. Sci. USA 101: 1720–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott R., Winters L., Larson B., Mytsa D., Taus P., et al. , 2014. Mechanisms promoting escape from mitotic stress-induced tumor cell death. Cancer Res. 74: 3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. A., Safford S. D., Reeves J. G., Jackson M. E., Freemerman A. J., 2008. Renal aplasia in humans is associated with RET mutations. Am. J. Hum. Genet. 82: 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J. C., Amacher S. L., 2014. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11: 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B., 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3: 59–69. [DOI] [PubMed] [Google Scholar]
- Toka H. R., Toka O., Hariri A., Nguyen H. T., 2010. Congenital anomalies of kidney and urinary tract. Semin. Nephrol. 30: 374–386. [DOI] [PubMed] [Google Scholar]
- Torres M., Gomez-Pardo E., Dressler G. R., Gruss P., 1995. Pax-2 controls multiple steps of urogenital development. Development 121: 4057–4065. [DOI] [PubMed] [Google Scholar]
- Trueba S. S., Auge J., Mattei G., Etchevers H., Martinovic J., et al. , 2005. PAX8, TITF1, and FOXE1 gene expression patterns during human development: new insights into human thyroid development and thyroid dysgenesis-associated malformations. J. Clin. Endocrinol. Metab. 90: 455–462. [DOI] [PubMed] [Google Scholar]
- Uetani N., Bouchard M., 2009. Plumbing in the embryo: developmental defects of the urinary tracts. Clin. Genet. 75: 307–317. [DOI] [PubMed] [Google Scholar]
- USRDS, 1999 Excerpts from United States Renal Data System 1999 Annual Data Report. Am. J. Kidney Dis. 34 (2 Suppl 1): S1-176. [PubMed]
- Van Esch H., Groenen P., Nesbit M. A., Schuffenhauer S., Lichtner P., et al. , 2000. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 406: 419–422. [DOI] [PubMed] [Google Scholar]
- Vasilyev A., Liu Y., Mudumana S., Mangos S., Lam P. Y., et al. , 2009. Collective cell migration drives morphogenesis of the kidney nephron. PLoS Biol. 7: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilyev A., Liu Y., Hellman N., Pathak N., Drummond I. A., 2012. Mechanical stretch and PI3K signaling link cell migration and proliferation to coordinate epithelial tubule morphogenesis in the zebrafish pronephros. PLoS One 7: e39992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar J., Gilbert T., Moreau E., Merlet-Benichou C., 1996. Metanephros organogenesis is highly stimulated by vitamin A derivatives in organ culture. Kidney Int. 49: 1478–1487. [DOI] [PubMed] [Google Scholar]
- Vivante A., Mann N., Yonath H., Weiss A.-C., Getwan M., et al. , 2017. A dominant mutation in nuclear receptor interacting protein 1 causes urinary tract malformations via dysregulation of retinoic acid signaling. J. Am. Soc. Nephrol. 28: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize P. D., Seufert D. W., Carroll T. J., Wallingford J. B., 1997. Model systems for the study of kidney development: use of the pronephros in the analysis of organ induction and patterning. Dev. Biol. 188: 189–204. [DOI] [PubMed] [Google Scholar]
- Vrontou S., Petrou P., Meyer B. I., Galanopoulos V. K., Imai K., et al. , 2003. Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat. Genet. 34: 209–214. [DOI] [PubMed] [Google Scholar]
- Westerfield M., 2000. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio). University of Oregon Press, Eugene, OR. [Google Scholar]
- Whitehurst A. W., Bodemann B. O., Cardenas J., Ferguson D., Girard L., et al. , 2007. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446: 815–819. [DOI] [PubMed] [Google Scholar]
- Wingert R. A., Selleck R., Yu J., Song H. D., Chen Z., et al. , 2007. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 3: 1922–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalavarthy R., Parikh C. R., 2003. Congenital renal agenesis: a review. Saudi J. Kidney Dis. Transpl. 14: 336–341. [PubMed] [Google Scholar]
- Zarrei M., MacDonald J. R., Merico D., Scherer S. W., 2015. A copy number variation map of the human genome. Nat. Rev. Genet. 16: 172–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
File S1 compares sequences between human and zebrafish GREB1L proteins, compares kidney phenotypes in zebrafish for greb1l morpholino knockdown and CRISPR-Cas9 deletion and shows the sequences of mutagenized alleles recovered from CRISPR F0 mouse embryos. This study was approved by the University of Iowa under IRB#200711705 as well as by the Danish National Committee of Ethics. WES data is available in the Sequence Read Archive database (accession number SRP112780).