Abstract
Identifying the sequence polymorphisms underlying complex trait variation is a key goal of genetics research, since knowing the precise causative molecular events allows insight into the pathways governing trait variation. Genetic analysis of complex traits in model systems regularly starts by constructing QTL maps, but generally fails to identify causative sequence polymorphisms. Previously we mapped a series of QTL contributing to resistance to nicotine in a Drosophila melanogaster multiparental mapping resource and here use a battery of functional tests to resolve QTL to the molecular level. One large-effect QTL resided over a cluster of UDP-glucuronosyltransferases, and quantitative complementation tests using deficiencies eliminating subsets of these detoxification genes revealed allelic variation impacting resistance. RNAseq showed that Ugt86Dd had significantly higher expression in genotypes that are more resistant to nicotine, and anterior midgut-specific RNA interference (RNAi) of this gene reduced resistance. We discovered a segregating 22-bp frameshift deletion in Ugt86Dd, and accounting for the InDel during mapping largely eliminates the QTL, implying the event explains the bulk of the effect of the mapped locus. CRISPR/Cas9 editing of a relatively resistant genotype to generate lesions in Ugt86Dd that recapitulate the naturally occurring putative loss-of-function allele, leads to a large reduction in resistance. Despite this major effect of the deletion, the allele appears to be very rare in wild-caught populations and likely explains only a small fraction of the natural variation for the trait. Nonetheless, this putatively causative coding InDel can be a launchpad for future mechanistic exploration of xenobiotic detoxification.
Keywords: xenobiotics, CRISPR, functional validation, QTL mapping, RNAi
A principal goal of research on the genetics of complex traits is to identify the precise sequence polymorphisms responsible for phenotypic variation. This is either carried out directly in natural populations or in laboratory mapping panels derived from a sample of naturally derived chromosomes. The pursuit of causative alleles is open to criticism (Rockman 2012), since with finite power it is necessarily the case that experimentally identified causative variants represent a biased set of the complete collection of sites impacting variation. Nonetheless, genomewide mapping studies have provided important contributions to our understanding of polygenic trait variation. Robust, replicable associations from unbiased genomewide association studies (GWAS) have provided considerable insight into the pathways underlying disease risk (Hirschhorn 2009; Visscher et al. 2012). In addition, the genes and molecular lesions contributing to an array of crop domestication traits have been identified (Doebley et al. 2006; Gross and Olsen 2010), providing detail on the specific molecular differences between modern crop plants and their wild progenitors and an understanding of the nature of the selection applied.
In model organisms, the hunt for causative variation commonly begins with linkage-based QTL mapping. Whether initiated with two parental strains (Lander and Botstein 1989) or, more recently, with several founders (Kover et al. 2009; Churchill et al. 2012; King et al. 2012b; Threadgill and Churchill 2012), such mapping designs have tremendous power to find QTL, and in some cases have led to the identification of specific polymorphisms contributing to trait variation (e.g., Long et al. 2000; Deutschbauer and Davis 2005; Bendesky et al. 2011; Cook et al. 2016; Linder et al. 2016). These variants facilitate a deeper understanding of specific biomedically relevant traits and collectively add to a fundamental appreciation of complex trait variation and its maintenance in populations. However, with the possible exception of yeast, where one can test vast numbers of recombinants and minimize the statistical challenges associated with finding small-effect variants at high resolution (Ehrenreich et al. 2010), it is quite clear that linkage-based genomewide mapping for most complex traits in most systems does not result in the identification of a causative mutation.
The difficulty with moving from QTL to causative site is compounded if traits are highly polygenic. Cornforth and Long (2003) have shown by simulation that standard QTL mapping approaches are unable to discriminate between a single QTL of large effect and a series of very closely linked QTL that each have independent effects. That such collections of adjacent causative sites exist is amply demonstrated by fine mapping studies that succeed in “fractioning” a single QTL into multiple causative loci (Pasyukova et al. 2000; Steinmetz et al. 2002; Kroymann and Mitchell-Olds 2005), from elegant work showing that multiple alleles combine to yield the major effect of the Adh gene on alcohol dehydrogenase expression in flies (Stam and Laurie 1996) and from recent work in multiparental mapping panels showing that QTL frequently appear to be generated by more than one segregating site (Baud et al. 2013; King et al. 2014). Thus, to follow up QTL mapping and identify functional allelic variation at the sequence level, additional molecular and functional tests are typically required.
In Marriage et al. (2014) we described initial genetic dissection of nicotine resistance in D. melanogaster larvae, expanding on studies that have identified genes and transcripts influencing related nicotine resistance traits in flies (Passador-Gurgel et al. 2007; Li et al. 2012). Nicotine is an ecologically and biomedically interesting toxin for three reasons. It is produced by a number of plants, most notably Nicotiana species, as a defense against herbivory (Glendinning 2002; Steppuhn et al. 2004). Nicotine was also commonly used as an agricultural pesticide in the middle of the 20th century (Shepard 1951; Metcalf 1955), although it has since been supplanted by other insecticides, including the chemically related neonicotinoids. Finally, nicotine continues to be responsible for large numbers of human deaths due to its addictive properties (Jha et al. 2013).
Marriage et al. (2014) used lines from the Drosophila Synthetic Population Resource (DSPR), a set of inbred strains derived from a multiparental, advanced generation intercross (King et al. 2012b), and mapped a number of nicotine resistance QTL. Two loci, Q1 on chromosome 2L and Q4 and 3R, had large effects on phenotype, explaining 8.5 and 50.3% of the broad sense heritability, respectively. Both intervals contained genes encoding detoxification enzymes, with Q1 encompassing two cytochrome P450 genes, and Q4, a set of 10 UDP-glucuronosyltransferase (or UGT) genes. These genes are strong a priori candidates to contribute to xenobiotic resistance, offering the possibility of identifying the underlying causative alleles. Our goal here was to provide evidence that one or more of these genes directly contributes to variation in resistance, employing fine mapping, expression profiling, tissue-specific RNAi, overexpression analyses, and CRISPR/Cas9 genome editing. Our data point to a single 22-bp InDel event in a coding region of Ugt86Dd as a major factor contributing to the previously mapped Q4 locus.
Materials and Methods
Larval nicotine resistance assay
We used the same assay described in Marriage et al. (2014), except here we tested larvae in narrow polystyrene vials rather than in plates. Briefly, flies were allowed to lay eggs on a cornmeal–yeast–molasses medium containing 0.5% activated charcoal, supplemented with a small amount of active yeast paste to elicit egg laying. First instar larvae were collected and placed either on no-drug media or on media containing 0.18 μl/ml nicotine (N3876; Sigma). No-drug and nicotine media were always prepared the day prior to larval collection to minimize variation due to nicotine breakdown. Replicate assay vials contained 30 first instar larvae in nearly all cases (Supplemental Material, File S1), and the phenotype for each replicate vial is given as the fraction of larvae that ultimately emerge as adults. For every genotype tested, we set up several egg-laying vials, and the mean phenotype is given as the average of multiple replicate assay vials.
All animals were reared and tested at 25° and 50% relative humidity on a 12-hr-light/12-hr-dark cycle. Those test animals that were the result of crosses were generated by pairing 10 virgin females with four to five males over several replicate vials. Prior to initiating such crosses, parental flies were allowed to recover from CO2 anesthesia for 24 hr.
Chromosome substitution
Marriage et al. (2014) identified four autosomal QTL contributing to nicotine resistance and found that for each QTL, recombinant inbred lines (RILs) harboring the A4 founder allele were on average more resistant to nicotine than those harboring the A3 allele (see figure 3 in Marriage et al. 2014). To broadly examine the phenotypic effects of the A3 and A4 alleles, we generated the chromosome substitution lines A4/A3; A4; A3 and A3/A4; A3; A4 (chromosomes are listed in the order X/Y; 2; 3). We directly compared these substitution genotypes to inbred strains A3 and A4 and carried out intercrosses among strains to explore dominance. For each genotype, we measured the phenotype using two replicate no-drug vials to test for variation in overall viability and six replicate nicotine-containing vials.
To generate chromosome substitutions, the following strains were obtained from the Bloomington Drosophila Stock Center (BDSC): the double-balancer strain 8316 (w1118; wgSp-1/CyO; sensLy-1/TM6B, Tb1), the GFP-marked chromosome 2 balancer strain 5702 [w1; snaSco/Cyo, Gal4-Hsp70, upstream activating sequence (UAS)-GFP], and the GFP-marked chromosome 3 balancer strain 5704 (w1; Sb1/TM3, Gal4-Hsp70, UAS-GFP, y+ Ser1).
Quantitative complementation tests
To test for the effects of allelic variation between strains A3 (relatively susceptible to nicotine) and A4 (relatively resistant to nicotine) at particular genomic regions and at certain genes, we carried out quantitative complementation tests. Virgin females from A3 and A4 were each crossed to males from a series of mutation-containing lines, along with their co-isogenic wild-type control lines, allowing the production of four types of F1 progeny per target mutation (A3/mutation, A3/control, A4/mutation, and A4/control). Phenotypes were based on a total of 2–8 replicate no-drug vials, and 8–18 replicate nicotine-containing vials for each genotype. We employed the following model for analysis: y = μ + S + D + (S × D), where S is the effect of founder strain (A3 or A4), D is the effect of the mutation (comparing either a deficiency or an insertion to wild type), and S × D is the interaction. A significant S × D term indicates a failure to complement (A3 and A4 functionally differ at the locus under test).
Exelixis (Parks et al. 2004) and Bloomington Stock Center (BSC) (Cook et al. 2012) deficiencies were obtained from the BDSC, specifically 7497 [w1118; Df(2L)Exel6011/CyO], 7957 [w1118; Df(3R)Exel7306/TM6B, Tb1], 7958 [w1118; Df(3R)Exel8152/TM6B, Tb1], and 26545 [w1118; Df(2L)BSC693/SM6a], along with the co-isogenic control strain 6326 (w1118). We additionally obtained the DrosDel (Ryder et al. 2004) deficiency 9083 [w1118; Df(3R)ED5506/TM6C, cu1 Sb1] and its co-isogenic control 5905 (also w1118 but not necessarily otherwise identical to 6326). All deficiencies are maintained over balancers, and since we must discriminate deficiency-containing and balancer chromosomes in larvae, prior to use we substituted all native balancers with GFP-marked versions derived from BDSC stocks 5702 and 5704 (see above). Finally, we obtained several Minos-based insertional mutants (Metaxakis et al. 2005), all of which were generated in the 5905 background: 23530 [w1118; Mi{ET1}4Cyp28d1MB03293], 23587 [w1118; Mi{ET1}Cyp28d2MB02776], 24834 [w1118; Mi{ET1}Ugt86DjMB04890], and 27861 [w1118; Mi{ET1}Ugt86DhMB11311].
RNAseq to identify nicotine resistance genes
We selected six (12) lines with relatively high (low) resistance (File S2) from the set of DSPR pB2 RILs that were previously scored for nicotine resistance (Marriage et al. 2014). Strains were raised as described above: 30 first instar larvae from each were placed on no-drug or nicotine-supplemented media for 2 hr, after which larvae were flash frozen in liquid nitrogen and stored at −80°. We isolated RNA from each sample using TRIzol reagent (15596018; Thermo Fisher Scientific, Waltham, MA) and purified using RNeasy columns (74104; Qiagen, Valencia, CA). We then pooled equal amounts of total RNA from each sample to generate four RNA pools (high/nicotine, low/nicotine, high/no-drug, low/no-drug), generated an Illumina TruSeq unstranded messenger RNA (mRNA) library from each sample, and combined libraries to sequence over a single HiSeq2500 lane and generate single-end 100-bp reads [University of Kansas (KU) Genome Sequencing Core]. Reads were quality trimmed using scythe (v0.991, github.com/vsbuffalo/scythe) and sickle (v1.200, github.com/najoshi/sickle), assembled to the D. melanogaster reference genome [National Center for Biotechnology Information (NCBI) Build 5.3, ccb.jhu.edu/software/tophat/igenomes.shtml] using TopHat v2.0.10 (Trapnell et al. 2009; Kim et al. 2013), and significant expression differences between relevant pooled samples were identified using Cufflinks v.2.2.1 (Trapnell et al. 2010, 2013).
RNAi knockdown
We employed the binary Gal4-UAS RNAi system to knock down expression of several candidate detoxification genes ubiquitously and specifically in a number of tissues. We crossed males from each Gal4 driver strain to females containing a UAS-RNAi transgene, or to appropriate control females, and assayed F1 Gal4-UAS-RNAi progeny. For each genotype we phenotyped 3–5 replicate no-drug vials and 8–10 replicate nicotine vials.
We obtained UAS-RNAi transgene-carrying stocks from the Vienna Drosophila Resource Center (VDRC) (Dietzl et al. 2007). The “GD” library strains harbor P-element-derived UAS transgenes, while the “KK” library strains harbor phiC31-based transgenes, where all transgenes reside at the same landing site. Specifically, we use VDRC UAS-RNAi strains 6016 (GD, UAS-RNAi-Ugt86Dd), 7870 (GD, UAS-RNAi-Cyp28d1), 7868 (GD, UAS-RNAi-Cyp28d2), 100353 (KK, UAS-RNAi-Ugt86Dd), 102626 (KK, UAS-RNAi-Cyp28d2), and 110259 (KK, UAS-RNAi-Cyp28d1). We directly compare genotypes containing these transgenes to the appropriate control strains 60000 (GD control strain) and 60100 (KK control strain). In addition, we obtained the Transgenic RNAi Project (TRiP) (Perkins et al. 2015) strain 53892 (UAS-RNAi-Cyp28d1) and its co-isogenic control (36304) from the BDSC.
We drove RNAi in all cells and all timepoints using Act5C-Gal4 [derived from BDSC 3954 (y1 w*; P{Act5C-GAL4}17bFO1/TM6B, Tb1)], after first substituting the third chromosome balancer in this strain for the GFP-marked version derived from BDSC 5704 (see above). To drive RNAi in specific groups of malpighian tubule cells, we employed a series of strains obtained from Julian Dow and Shireen Davies (University of Glasgow): c42-Gal4, c710-Gal4, c724-Gal4, and uro-Gal4 (Rosay et al. 1997; Sozen et al. 1997; Terhzaz et al. 2010). We obtained the anterior midgut Gal4 driver 1099 from Nicolas Buchon (flygut.epfl.ch). Finally, we obtained a number of strains expressing Gal4 in various parts of the gut from the BDSC, specifically strains 1967 [y1 w*; P{GawB}34B], 30828 [w*; P{GawB}Alp4c232], 30844 [w*; P{GawB}c601c601], and 43656 [w*; P{Scr-GAL4.4}1-3].
Overexpression of Ugt86Dd
A gain-of-function analysis was performed for Ugt86Dd by creating UAS-Ugt86Dd overexpression strains. We PCR amplified the gene from both A3 and A4, and simultaneously added attB sites, using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTACAACATGAGATTATTAACTGTGATCGCGA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAATGTTTCTTAAGCTTATCAG-3′. Following the manufacturer’s protocol (PCR Cloning System with Gateway Technology, 12535029; Thermo Fisher Scientific) we cloned the PCR products into the pDONR221 vector, creating entry clones via BP recombination reactions. Clones were Sanger sequenced using M13 primers to verify the sequence and direction of the insert. The destination vector pUASg.attB was donated by Johannes Bischof (Bischof et al. 2013) and used in combination with the LR reaction (Gateway LR Clonase, 11791020; Thermo Fisher Scientific) to generate both pUASg.Ugt86Dd-A3.attB and pUASg.Ugt86Dd-A4.attB expression clones. Expression clones were Sanger sequenced using primer hsp-GW-F (5′-GCAACTACTGAAATCTGCCAAG-3′) to verify the direction of the insert. To generate fly stocks we injected BDSC stock 24749 [y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-86Fb] with the expression plasmids at 0.510-0.515μg/μl (BestGene, Chino Hills, CA). We obtained one A3 transformant line (UAS-Ugt86DdA3), and five A4 transformants [UAS-Ugt86DdA4(1) to UAS-Ugt86DdA4(5)], that are each homozygous for the transgene-containing chromosome 3. These strains were utilized in conjunction with tissue-specific Gal4 drivers (above) to explore the effects of Ugt86Dd overexpression. For each genotype, we tested two to three replicate no-drug vials and six replicate nicotine vials.
PCR genotyping assay for Ugt86Dd InDel variant
We identified a 22-bp InDel variant in a protein-coding region of exon two of Ugt86Dd and developed a PCR assay to genotype it. Primers 5′-CGCTTTTGCTCAGCATTTTA-3′ and 5′-ATATGTGGCAGGTGAACGAA-3′ amplify 219-bp and 197-bp products in the presence of the insertion and deletion allele, respectively. PCR cycling conditions were 95° for 2 min, 35 cycles of 95° for 20 sec, 57° for 25 sec, and 72° for 30 sec, with a final 2-min extension at 72°. Products were sized on 2% gels run at low voltage for up to 2 hr.
QTL mapping
Marriage et al. (2014) detected a nicotine resistance QTL—Q4—that encompasses Ugt86Dd using the DSPR. To test whether the 22-bp InDel in Ugt86Dd might contribute to the QTL effect, we first established the allelic status of the DSPR founder lines via PCR, finding that founders A3, AB8, B6, and B7 possess the deletion (Figure S1). Next, we employed the estimated founder genotype probabilities for the DSPR RILs (King et al. 2012b) to determine the InDel status of each RIL (File S3).
Methods for QTL mapping in the DSPR are described in King et al. (2012a,b). We employed the R (r-project.org) packages DSPRqtl, DSPRqtlDataA, and DSPRqtlDataB for mapping (FlyRILs.org), analyzed the pA and pB panels of RILs separately, and—in addition to the InDel covariate—included a covariate accounting for the subpopulation from which the RIL was derived (pA1, pA2, pB1, and pB2). Note that in the analyses conducted here, we only employed RILs for which we had high confidence in the founder haplotype at the Ugt86Dd gene, so sample sizes are marginally reduced compared to those employed in Marriage et al. (2014), and estimates of the QTL effect are slightly different than previously reported. To establish significance of mapped QTL, we generated genomewide 5% significance thresholds via 1000 permutations (Churchill and Doerge 1994).
Testing the effect of the InDel variant in the Drosophila Genetic Reference Panel
Our PCR assay revealed that seven strains from the Drosophila Genetic Reference Panel (DGRP) (Mackay et al. 2012; Huang et al. 2014) were homozygous for the deletion (BDSC nos. 25176, 25177, 25206, 28185, 28199, 28213, and 37525). We also confirmed that the following seven strains were homozygous for the much more common insertion allele (25174, 25200, 28160, 28197, 28226, 28239, and 28295). We created a pair of populations, one fixed for the deletion and one fixed for the insertion, by carrying out all possible crosses between the seven founding strains (10 virgin females × 10 males), collecting 10 F1 progeny of each sex from each cross, and combining into a 1-gallon population bottle. Both populations were maintained for seven generations, at which point we phenotyped each population using 20 replicate nicotine-containing vials.
Ugt86Dd CRISPR/Cas9 editing
We used the CRISPR Optimal Target Finder (tools.flycrispr.molbio.wisc.edu/targetFinder) to identify an appropriate guide RNA (gRNA) target sequence within Ugt86Dd (Gratz et al. 2014), specifically targeting the site of the 22-bp InDel. The target sequence 5′-TCACTACGAAGTCATTGTGGAGG-3′ matches the sequence of A4 and was used to generate a gRNA that leads to double-strand breaks within the insertion sequence (Figure S1). We purchased the 5′-phosphorylated sense and antisense oligos 5′- CTTCGTCACTACGAAGTCATTGTGG-3′ and 5′-AAACCCACAATGACTTCGTAGTGAC-3′, annealed, and followed the protocol from Gratz et al. (2013) to clone the annealed oligos into the pU6–BbsI–chiRNA plasmid (Addgene, Cambridge, MA; plasmid 45946). Transformants were verified via Sanger sequencing.
Our goal was to compare the phenotypes of inbred strains differing for wild-type and mutant Ugt86Dd alleles in an otherwise genetically identical background. Preliminary sequencing revealed that chromosome 3 for the vasa-Cas9 BDSC strain 55821 [y1 M{vas-Cas9.RFP-}ZH-2A w1118] is not homozygous. Thus, we used a series of standard fly crosses with balancers to substitute chromosome 3 from founder strain A4 into 55821, creating a strain homozygous for vasa-Cas9 on the X and A4 on chromosome 3 (the state of chromosome 2 is unknown, but is likely heterozygous).
The gRNA plasmid was injected into 300 embryos from our custom vasa-Cas9; ; A4 injection strain at 250 ng/μl (BestGene) yielding 89 G0 progeny, 57 of which yielded progeny after being individually crossed to one to two animals from BDSC 5704 (see above). Where possible we collected 10 F1 balanced progeny from each G0 cross, and individually crossed to one to two animals from BDSC 5704. For crosses containing a mutation (see below), and for some that did not, we collected balanced F2 progeny of both sexes to establish stocks, removing the balancer in subsequent generations to create strains carrying homozygous third chromosomes. These strains can and do segregate on the X and chromosome 2.
After each F1 cross produced eggs, we genotyped the F1 animal for the presence of a CRISPR/Cas9-induced mutation using T7 endonuclease [New England Biolabs (NEB), Ipswich, MA; M0302L]. Briefly, following single-fly DNA isolation (Puregene Cell and Tissue Kit, 158388; Qiagen) we PCR amplified a region surrounding the gRNA target site using oligos 5′-ACGCTTTTGCTCAGCATTTT-3′ and 5′-GGCTGGGGATACCATTTCTT-3′ (95° for 2 min, 35 cycles of 95° for 20 sec, 57° for 25 sec, and 72° for 30 sec, with a final 2-min extension at 72°). Subsequently 10 μl of PCR product was mixed with 7 μl of molecular grade water and 2 μl of NEB buffer 2 (B7002S), incubated at 95° for 5 min in a heat block, and allowed to slowly cool to room temperature for ∼2 hr. We then added 1 μl of T7 endonuclease to each reaction, incubated at 37° for 15 min, added 2 μl of 0.25 mM EDTA to stop the reaction, and immediately loaded the entire reaction volume into a 1.5% agarose gel. We also ran DNA from A4 and from the F1 progeny of A3 × A4 crosses (which are heterozygous for the 22-bp InDel) through this genotyping pipeline as negative and positive controls, respectively.
Editing led to 16 strains with homozygous third chromosomes carrying independent mutations in Ugt86Dd (File S4). Simultaneously, we generated seven strains containing unedited homozygous third chromosomes. Each of the 23 strains created was phenotyped using 5–6 replicate no-drug and nicotine-containing vials. For two edited and one unedited genotype, we substituted the edited third chromosome into the A4 background, allowing a direct comparison of mutated and wild-type versions of Ugt86Dd. Each of these strains—A4-Ugt86DdDel1, A4-Ugt86DdDel11, and A4-Ugt86Ddwt (File S4)—was tested directly as an inbred genotype, and as a heterozygote following crosses to A4, using 10 replicate no-drug and nicotine-containing vials.
Estimating the frequency of the InDel in nature
We can identify the two alleles at the short InDel directly from reads resulting from next-generation sequencing of pooled population samples of D. melanogaster, thereby estimating allele frequency. We retrieved raw FASTQ files from 13 wild-caught, resequenced population samples (Bergland et al. 2014) via the NCBI Sequence Read Archive (SRA) (BioProject accession PRJNA256231), and used a Unix grep procedure to count the number of raw reads containing the insertion and deletion sequences. We ensured that instances of a sequence occurring in both reads of a single paired-end fragment were counted only once. The insertion-specific sequence was taken as the central 20-bp (i.e., ATTGTGGAGGACATTCATCG) of the 22-bp InDel and its reverse complement. The deletion-specific sequence was taken as the 10-bp either side of the 22-bp InDel (i.e., GAACGAATTCACTTCGTAGT), and its reverse complement. These query sequences match all 15 inbred DSPR founder strains (Figure S1).
Data availability
All raw phenotypes are presented in File S1. Raw RNAseq reads are available from the NCBI SRA (accession no. SRP102254).
Results
Large, dominant effect of chromosome 3 on nicotine resistance
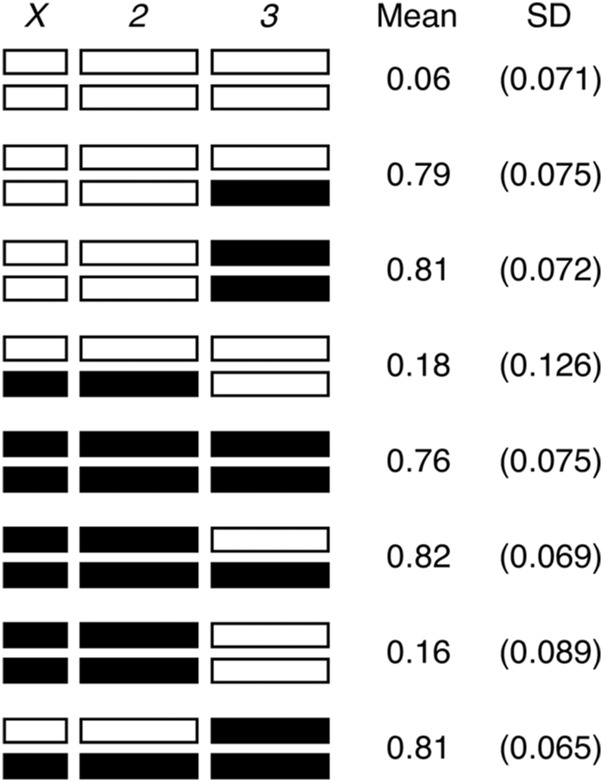
Marriage et al. (2014) identified four autosomal QTL contributing to nicotine resistance in the pA DSPR population. Three QTL on chromosome 2 (Q1–Q3) individually contributed 4.6–8.5% to the broad sense heritability of the trait, and one QTL on chromosome 3 (Q4) contributed 50.3% to the heritability. By estimating the effects associated with each founder allele at each mapped QTL, it appeared that in all cases, carrying an A4 allele at the QTL led to greater resistance than carrying an A3 allele. To confirm this, we tested various chromosome substitution genotypes and observed that moving a single third chromosome from A4 into an otherwise A3 background led to a marked increase in resistance compared to A3 (P < 10−8, Figure 1). This implies that A3 and A4 do harbor different alleles at functional variants within Q4, and that the effect of the A4 chromosome is dominant.
Figure 1.
Effects of chromosome substitution on nicotine resistance. Chromosomes derived from DSPR founder A3 are shown in white, and those from A4 are shown in black. Nicotine resistance for each genotype was tested across six replicate vials, and we performed all possible pairwise t-tests among genotypes. The three low resistance genotypes are statistically indistinguishable, as are the five high resistance genotypes, while all pairs of low and high resistance genotypes are significantly different (P < 0.0001 in all cases). The viability of each genotype was tested twice under no-drug conditions, and all genotypes showed a mean viability of >0.9.
In contrast to the large effect of chromosome 3, the combined effects of the X and chromosome 2 on nicotine resistance are relatively small; A genotype with the A3 third chromosome homozygous in an otherwise A4 background is only marginally more resistant than founder A3 (P = 0.07, Figure 1). This result confirms the smaller phenotype effects estimated at the second chromosome QTL by Marriage et al. (2014).
Fine mapping nicotine resistance QTL
One strategy to fine map QTL is to employ quantitative complementation tests, either using deletions that remove tens to hundreds of genes (Pasyukova et al. 2000), or loss-of-function mutant alleles of plausible candidate genes (Long et al. 1996). We crossed the A3 and A4 founders, which differ in nicotine resistance, to a series of mutation-carrying strains and their corresponding co-isogenic, mutation-free control strains, generating four types of progeny (A3/mutation, A3/control, A4/mutation, and A4/control). We then used a statistical test that estimates the effects of founder and mutation, and the interaction between these factors. A significant interaction—a quantitative failure to complement—implies the effects of founder alleles are different in the mutant and control backgrounds, and thus suggests the founders segregate for functional variation at the tested deficiency/gene.
We employed two small deficiencies that deleted overlapping regions of the Q1 region, one of which eliminates both of the P450 genes within the interval, Cyp28d1 and Cyp28d2, and one of which deletes only Cyp28d1. Since the deletions remove a total of 11–13 genes, including three additional P450s not within the Q1 interval, we also employed a pair of Minos insertions (Metaxakis et al. 2005) that insert within coding exons of Cyp28d1 and Cyp28d2, likely disrupting gene function. In all four cases, there was a highly significant founder × mutant interaction (P < 10−5, Figure S2) with the difference between A3 and A4 being much greater in the mutant background. Collectively these results imply that both Cyp28d1 and Cyp28d2 show functional differences between A3 and A4 that confer effects on phenotype.
We carried out a similar experiment for the Q4 region, testing three overlapping deficiencies that individually delete 11–19 genes, including variable numbers of the UGT genes resident within Q4. All three deletions exhibit a significant quantitative failure to complement (P < 10−6, Figure S2). Since Ugt86Dc is the only gene deleted by all three deletions, a parsimonious explanation is that A3 and A4 have distinct nicotine resistance alleles at this gene. However, Marriage et al. (2014) showed that expression of this gene is reduced in A4 compared to A3, which is not the pattern one might expect under the assumption that Ugt86Dc leads to the enhanced nicotine resistance of A4. Additionally, that study provided evidence that the founder allelic effects at Q4 do not fall into two groups, as would be expected if a single causative gene was responsible for the QTL. Thus, the different deletions may be uncovering variants in independent genes that affect phenotype.
Finally, we tested a Minos element in a coding exon of Ugt86Dj, and one that resides within the 3′-UTR of one of the two isoforms of Ugt86Dh. (Mutations in a known background were available only for 2/10 of the UGT genes under Q4 when this experiment was conducted.) While we observed a significant founder × mutant interaction for both genes (Ugt86Dj, P < 0.001; Ugt86Dh, P < 0.01; Figure S2), in both cases the difference between the A3 and A4 alleles was greatest in the control background. These results suggest the effects are more likely due to epistasis than allelic failure to complement (see Mackay 2001; Geiger-Thornsberry and Mackay 2004), and do not provide strong evidence for the role of functional variation at Ugt86Dh or Ugt86Dj in resistance to nicotine.
Candidate nicotine resistance genes via RNAseq
Under the assumption that some fraction of complex trait variation is regulatory in origin (Gibson and Weir 2005; Gilad et al. 2008; Gusev et al. 2014; Torres et al. 2014; Albert and Kruglyak 2015), we might expect to see changes in the expression of genes harboring causative loci. Marriage et al. (2014) used RNAseq of whole, first instar larvae from A3 and A4 tested under no-drug and nicotine-exposure conditions to attempt to resolve candidate genes. Here, we carried out a similar study employing mixed pools of RNA from relatively susceptible and relatively resistant pB DSPR RILs (File S2). Many genes showed differential expression between susceptible and resistant genotypes and/or between treatments (File S5), but those changes at loci within QTL intervals are of principal interest.
Considering those 34 genes within Q1, only Cyp28d1 was differentially expressed at a nominal 5% level, showing an induction in expression on nicotine exposure in both susceptible (P < 10−4) and resistant animals (P = 0.001), confirming the results of Marriage et al. (2014). However, unlike our previous study, we found no change in expression between susceptible and resistant pB line pools that would indicate an allelic difference between genotypes at this gene. Since Q1 was not identified in the pB DSPR panel, it is possible there is no segregating variation that leads to expression variation within this panel, although we cannot discount the possibility we failed to capture any such variation in the small number of pB lines used for the current expression study (File S2).
At Q4, 5/10 UGT genes show a change in expression in at least one contrast, with two genes showing a change between susceptible and resistant genotypes; Ugt35b shows a slight increase in expression in the resistant genotypes under no-drug conditions (P < 0.05), while Ugt86Dd increases in expression in resistant animals under both no-drug and nicotine conditions (P < 10−4 and P = 0.001, respectively). Marriage et al. (2014) found no effect at Ugt35b, but did observe expression differences between A3 and A4 in the expression of Ugt86Dd. Since Q4 was identified in both the pA and pB populations, the similar change in expression between susceptible and resistant genotypes derived from the two panels implies Ugt86Dd is a strong candidate to harbor functional genetic variation contributing to resistance.
Marriage et al. (2014) identified a small peak in the LOD score profile in pB that encompasses the Cyp6g1 gene, a gene shown to be associated with DDT (Daborn et al. 2002; Chung et al. 2007; Schmidt et al. 2010) and nicotine resistance (Li et al. 2012). However, we did not focus on the QTL in our previous study, given the modest LOD score at the peak and the relatively small number of pB RILs assayed. Here, we found a dramatic increase in expression of Cyp6g1 between susceptible and resistant pB genotypes in both no-drug and nicotine conditions (P < 10−4), providing some additional support for the effect of this gene on nicotine resistance in the DSPR.
Significant reduction in resistance following ubiquitous gene knockdown
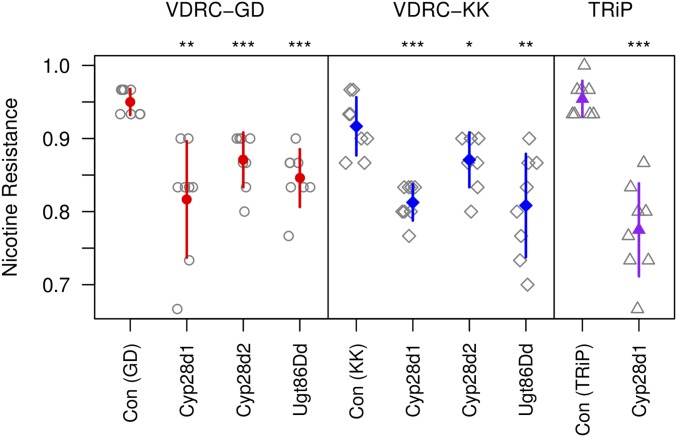
Under Q1, the genes most likely to harbor functional variation impacting nicotine resistance are Cyp28d1 and Cyp28d2. Under Q4, Ugt86Dd appears to be the strongest candidate based on RNAseq data. We knocked down the expression of these three genes using two to three different UAS transgenes per gene. In comparison with co-isogenic control strains, we see a robust reduction in resistance following knockdown of each gene (Figure 2). Notably, gene knockdown had no effect on viability under no-drug conditions; all strains showed phenotypes above 0.93 on no-drug food (compare to Figure 2), and there was no effect of gene knockdown under no-drug conditions (P > 0.4 for all tests). Thus, the effects of reducing expression of Cyp28d1, Cyp28d2, and Ugt86Dd appear to be specific to nicotine resistance.
Figure 2.
Effect of ubiquitous gene knockdown on nicotine resistance. We employed the Gal4-UAS-RNAi system to knock down the expression of three genes in all cells and at all timepoints via Act5C-Gal4. Each genotype was tested across eight replicate vials (raw data shown in gray symbols), and the mean (±1 SD) phenotype of each is shown with colored symbols/lines. Within each RNAi system (VDRC-GD, VDRC-KK, and TRiP) we tested the effect of each gene knockdown against its co-isogenic control using t-tests, and significance is highlighted by asterisks (* P < 0.05, ** P < 0.01, *** P < 0.001). All ubiquitous RNAi knockdowns reduce nicotine resistance. Note that the viability of each genotype was tested four to five times in no-drug conditions, and all genotypes showed a mean viability of >0.93.
Knockdown of Cyp28d1 and Ugt86Dd in the midgut reduces resistance
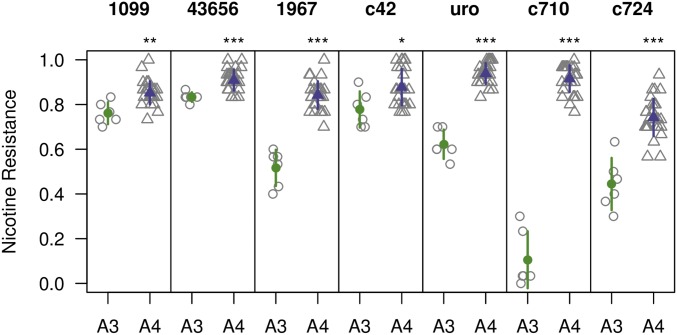
Data from FlyAtlas (Chintapalli et al. 2007; Robinson et al. 2013) indicates that Ugt86Dd expression is strongly enriched in the adult midgut, and in the adult and third instar larval malpighian tubules. In the same data set, Cyp28d1 shows considerable among-tissue variation in expression, but has highest expression in the adult midgut and larval fat body, while Cyp28d2 is strongly enriched in the larval midgut. All three genes additionally show variation in expression along the length of the midgut in third instar larvae (Harrop et al. 2014) and variation among adult midgut cell types (Dutta et al. 2015). These data show where the genes are expressed at high levels, but do not directly establish where they might act to metabolize nicotine. To help achieve this, we employed RNAi with a series of tissue-specific Gal4 drivers. Using two drivers that express Gal4 in the anterior region of the midgut, we saw that RNAi of Cyp28d1 (in 5/6 cases) and Ugt86Dd (in 4/4 cases), but not Cyp28d2 (in 0/4 cases), led to a significant reduction in resistance (Table 1). In contrast, a driver expressing Gal4 in the posterior region of the midgut showed no effect of any target gene (Table 1). These data imply that the anterior midgut is an important site of nicotine metabolism and is impacted by the action of the products of Cyp28d1 and Ugt86Dd, genes showing expression variation between resistant and susceptible genotypes (above and Marriage et al. 2014). The lack of any apparent effect of Cyp28d2 in the midgut (Table 1), in contrast to the effect observed following ubiquitous knockdown of the gene (Figure 2) might imply that Cyp28d2 is required in a different, and untested tissue. Alternatively, the ubiquitous knockdown effect described above may be a false positive, which is known to occur at low rates in RNAi screens (e.g., Mummery-Widmer et al. 2009; Schnorrer et al. 2010).
Table 1. Effect of midgut-specific gene knockdown on nicotine resistance.
| RNAi system | UAS genotype | Mean nicotine resistance (±1 SD) | ||
|---|---|---|---|---|
| Anterior midgut Gal4 (1099) | Anterior midgut Gal4 (43656) | Posterior midgut Gal4 (1967) | ||
| VDRC-GD | Control (GD) | 0.95 ± 0.032 | 0.96 ± 0.032 | 0.83 ± 0.096 |
| Cyp28d1 | 0.90 ± 0.064* | 0.92 ± 0.039* | 0.88 ± 0.047ns | |
| Cyp28d2 | 0.92 ± 0.050ns | 0.96 ± 0.033ns | 0.89 ± 0.072ns | |
| Ugt86Dd | 0.90 ± 0.070* | 0.90 ± 0.025*** | 0.89 ± 0.093ns | |
| VDRC-KK | Control (KK) | 0.84 ± 0.063 | 0.95 ± 0.032 | 0.73 ± 0.099 |
| Cyp28d1 | 0.77 ± 0.086* | 0.88 ± 0.050** | 0.75 ± 0.085ns | |
| Cyp28d2 | 0.84 ± 0.052ns | 0.93 ± 0.078ns | 0.76 ± 0.124ns | |
| Ugt86Dd | 0.77 ± 0.058* | 0.89 ± 0.053** | 0.75 ± 0.083ns | |
| TRiP | Control (TRiP) | 0.93 ± 0.060 | 0.94 ± 0.050 | 0.70 ± 0.069 |
| Cyp28d1 | 0.83 ± 0.073** | 0.91 ± 0.054ns | 0.78 ± 0.079ns | |
The Gal4-UAS-RNAi system was used to knock down expression of three genes in the midgut. The phenotype of each genotype was tested over 7–10 (mean = 9.5) replicate vials. Each gene knockdown was tested against the relevant co-isogenic control using a t-test (ns P ≥ 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001). The viability of each genotype was tested two to five times in no-drug conditions. These genotype means averaged 0.92, 24/30 of the genotypes have greater no-drug viability than nicotine viability, and for the other six genotypes no-drug viability was at most 4% less than that on nicotine food. These observations suggest the effects of these tissue-specific gene knockdowns are not a result of general defects in viability.
Malpighian tubule knockdown of target detoxification genes enhances resistance
The malpighian tubules are an important site of xenobiotic metabolism in insects (Dow and Davies 2006; Yang et al. 2007), and we tested multiple Gal4 drivers that broadly express in the malpighian tubules, and also those that target specific cell types within the organ. RNAi against three genes (Cyp28d1, Cyp28d2, and Ugt86Dd) using drivers expressing Gal4 broadly in the malpighian tubules, and in one case additionally in the hindgut and ureter, revealed no major changes in resistance (Table 2). However, in nearly all cases, knockdown of these genes in either principal or stellate cells of the malpighian tubule led to a significant increase in resistance in comparison with control genotypes (Table 2). We note that under no-drug conditions, all genotypes associated with these malpighian tubule cell-specific tests show viabilities >0.93, implying the relative increase in resistance on gene knockdown is in response to the nicotine treatment.
Table 2. Effect of malpighian tubule RNAi on nicotine resistance.
| Gal4 driver | Mean nicotine resistance (±1 SD) | |||
|---|---|---|---|---|
| Control (GD) | UAS-Cyp28d1 | UAS-Cyp28d2 | UAS-Ugt86Dd | |
| 30844 | 0.93 (0.060) | 0.94 (0.046)ns | 0.86 (0.055)* | 0.89 (0.083)ns |
| 30828 | 0.90 (0.060) | 0.92 (0.061)ns | 0.88 (0.059)ns | 0.90 (0.079)ns |
| c42 | 0.85 (0.074) | 0.92 (0.039)* | 0.91 (0.066)ns | 0.94 (0.051)** |
| uro | 0.69 (0.211) | 0.94 (0.063)** | 0.91 (0.045)** | 0.95 (0.023)** |
| c710 | 0.74 (0.079) | 0.91 (0.068)*** | 0.88 (0.061)*** | 0.92 (0.061)*** |
| c724 | 0.77 (0.083) | 0.89 (0.072)** | 0.81 (0.077)ns | 0.93 (0.054)*** |
We knocked down expression of three genes, using UAS constructs from the VDRC-GD collection, with six different Gal4 drivers. The 30844 driver expresses Gal4 in the hindgut, ureter, and malpighian tubules, 30828 expresses Gal4 in the malpighian tubules, c42-Gal4 and uro-Gal4 are specific to the principal cells of the malpighian tubules, and c710-Gal4 and c724-Gal4 are specific to tubule stellate cells. The phenotype of each genotype was tested across 10 replicate vials, and each gene knockdown was compared to the relevant co-isogenic control using a t-test (ns P ≥ 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001). The viability of each genotype was also tested two to five times under no-drug conditions. The lowest genotype mean under no-drug conditions was 0.88, and the average genotype mean was 0.95. Notably, the mean phenotypes under no-drug conditions for the non-RNAi, control genotypes were 0.91–0.98, implying that the marked reduction in viability for some of these genotypes on nicotine-supplemented media is not due to general viability defects.
This result—heightened resistance to a drug after knocking down expression of known detoxification genes in cell types known to be involved in xenobiotic metabolism—is counterintuitive. Indeed, RNAi knockdown of the P450 gene Cyp6g1 in principal cells robustly leads to a reduction in DDT resistance (Yang et al. 2007). We speculate that the presence of nicotine or its byproducts, coupled with a reduction in expression of key nicotine metabolizers in the malpighian tubule principal/stellate cells, leads to the production of additional detoxification enzymes, possibly in other tissues, that enhance resistance. Analyses of gene expression in the malpighian tubules and other gut tissues of these knockdown genotypes may provide insight into this observation.
Overexpression of functional Ugt86Dd enhances nicotine resistance
Using the Ugt86Dd sequence from the relatively susceptible A3 founder and the relatively resistant A4 founder, we made UAS-Ugt86Dd strains, allowing ectopic overexpression of the Ugt86Dd gene product. In the course of verifying clones prior to plasmid injection, we noticed that exon 2 of the A3 allele contained a 22-bp deletion relative to A4, leading to a frameshift and a premature stop codon, reducing the length of the predicted protein product from 517 to 206 amino acids. PCR and sequencing of the original founders revealed that the A3 and A4 strains differ at this variant.
Ubiquitous overexpression of UAS-Ugt86DdA4 via Act5C-Gal4 did not yield viable Gal4-UAS offspring under no-drug conditions, while similar overexpression of UAS-Ugt86DdA3 did yield Gal4-UAS progeny. These observations imply that ubiquitous overexpression of full-length Ugt86Dd is poisonous to cells and confirms that A3 carries a nonfunctional Ugt86Dd allele.
In order to test the effect of Ugt86Dd overexpression on nicotine resistance we used a series of drivers expressing Gal4 in various regions of the gut and compared a single line containing the UAS-Ugt86DdA3 transgene to five strains containing the same UAS-Ugt86DdA4 transgene. Figure 3 shows that for every Gal4 driver, overexpression of UAS-Ugt86DdA4 (containing the insertion allele of Ugt86Dd) leads to higher nicotine resistance than overexpression of UAS-Ugt86DdA3 (containing the deletion). These data suggest that the InDel variant segregating between strains A3 and A4 may be responsible for some of the difference in nicotine resistance exhibited by these strains. We note that while all six strains tested are homozygous for the same third chromosome harboring the transgene landing site, they may be variable on the second and X chromosomes due to postinjection crossing against balancers. Thus, it is conceivable that some of the variation observed between the A3 and A4 transgenes is due to differences in genetic background.
Figure 3.
Effect of tissue-specific overexpression of Ugt86Dd alleles. We overexpressed Ugt86Dd derived from strains A3 (which contains a 22-bp out-of-frame coding deletion) and A4 (which carries an allele making a full-length gene product) using seven tissue-specific Gal4 drivers. We had access to a single UAS-Ugt86DdA3 transgene line and five UAS-Ugt86DdA4 transgene lines and tested each line over six replicate nicotine vials (gray open symbols). We present the mean (±1 SD) phenotype of each genotype in colored symbols and compared the genotype means for each Gal4 driver using t-tests (* P < 0.05, ** P < 0.01, *** P < 0.001). In every case, the A4 transgene leads to higher nicotine resistance. Notably, we additionally tested all genotypes across two to three no-drug vials, and the A3 transgene showed viabilities of 0.84–0.96. This indicates that the reductions in viability under nicotine conditions are nicotine specific and likely not due to expression of the UAS-Ugt86DdA3 transgene being generally deleterious.
This overexpression experiment may not say anything about the natural site of Ugt86Dd-based nicotine detoxification, since we are ectopically expressing the gene at high levels in tissues where it may normally be expressed at more modest levels under its native promoter.
The Ugt86Dd InDel variant is associated with nicotine resistance
Four of the DSPR founder strains (A3, AB8, B6, and B7) possess the 22-bp deletion allele at Ugt86Dd (Figure S1). At the Q4 QTL identified by Marriage et al. (2014) these four founders have the lowest strain effects, suggesting the InDel may have a functional role in resistance. Furthermore, mean phenotypes of RILs carrying the insertion and deletion alleles are highly significantly different for both the pA and pB populations (t-test, P < 10−15; File S3), with the mean resistance of the deletion-containing RILs being 37% (pA) and 27% (pB) less than that of the insertion-containing RILs.
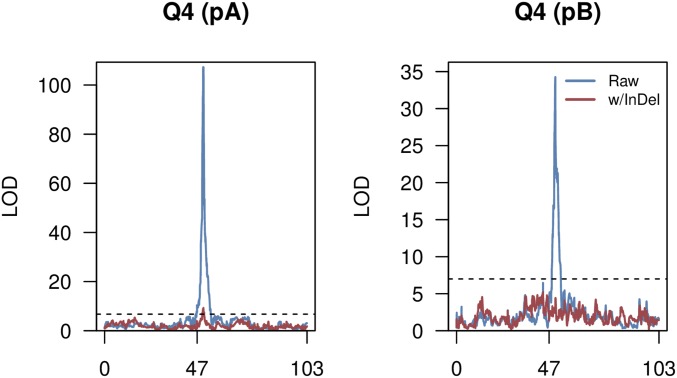
We employed InDel status as an additional covariate in the DSPR QTL mapping analysis and observed a substantial reduction in LOD score at the site of the Q4 locus (Figure 4), eliminating any above-threshold peak in pB and leaving a much more modest effect at the locus in pA; in the standard analysis, Q4 contributes 46.5% to the variation among pA lines, whereas the above-threshold peak at the same location after accounting for the InDel contributes 5.3% to the variation. These data imply that the InDel variant alone, or one or a collection of variants in linkage disequilibrium (LD) with this variant, are responsible for a large fraction of the nicotine resistance variation in the DSPR and may completely, or nearly completely, explain the major-effect QTL mapped by Marriage et al. (2014).
Figure 4.
An InDel variant in Ugt86Dd explains variation at a nicotine resistance QTL. Each plot shows the LOD curve along chromosome 3, with genetic distance along the x-axis, in the pA (left) and pB (right) DSPR populations. In blue, using a standard mapping analysis in the DSPR, one sees evidence for the major-effect QTL Q4. In red, using a similar analysis but now including a covariate accounting for the InDel status of each phenotyped RIL, one sees a peak in pA with a LOD score just surviving the threshold, and no peak in pB. These data suggest the InDel variant contributes to the effect observed at Q4.
To attempt to validate the effect of the InDel variant on nicotine resistance, we exploited the DGRP, a collection of ∼200 inbred, resequenced strains of D. melanogaster (Mackay et al. 2012; Huang et al. 2014). Seven of the DGRP strains were found to harbor the same 22-bp deletion allele as found in the DSPR. We intercrossed these lines for seven generations to generate a population fixed for the deletion allele, but otherwise outbred, and generated a similar population from seven random DGRP strains carrying the insertion allele. Comparing these populations revealed a small, yet significant reduction in resistance in the deletion population (insertion population mean = 0.94, deletion population mean = 0.89, t-test, P = 0.011). While this result does tend to validate our work in the DSPR, any effect of the Ugt86Dd InDel variant is apparently much smaller in the DGRP.
CRISPR/Cas9-derived deletion mutations at Ugt86Dd in A4 reduce resistance
The effect of the deletion allele in founder A3 is to generate a premature stop codon in Ugt86Dd. To directly test the effect of premature stop-encoding mutations in this gene, we generated a series of deletion alleles via CRISPR/Cas9 editing. We constructed a custom injection strain containing an X-linked vasa-Cas9 transgene and an A4-derived third chromosome and employed a gRNA that generates double-strand breaks at the site of InDel (Figure S1). We generated a series of 16 independent mutations (File S4), 13 of which lead to premature stop codons, and constructed strains carrying homozygous edited third chromosomes for all 16. Simultaneously, we made seven strains with homozygous unedited third chromosomes from genotypes that were passed through the CRISPR/Cas9 process (injection, balancing, and so on). Our rationale for keeping such genotypes was that during the creation of our injection strain, and while establishing homozygous edited chromosomes, we employed several generations of crossing with balancers. Since movement of genetic material via gene conversion from a balancer to the nonbalancer homolog has been inferred (Blumenstiel et al. 2009), such unedited genotypes provided valuable controls for the effects of induced mutations. All 23 strains were tested in multiple replicates for viability under no-drug and nicotine conditions, and while we observed no difference between the sets of unedited and edited genotypes in the absence of nicotine (P = 0.2), we saw a large decrease in resistance due to editing (P < 10−15, Table S1).
The fact that the three lesions that result in amino acid changes, but do not introduce a premature stop codon, still each lead to a strong reduction in nicotine resistance may suggest that the region of the protein targeted for mutation is critical to the function of Ugt86Dd.
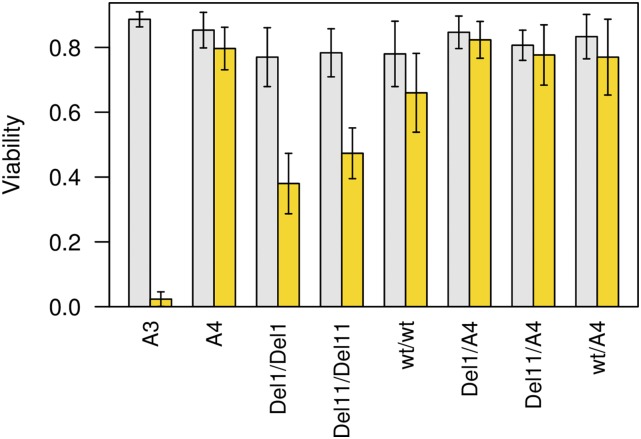
For two deletion alleles—a 1-bp and an 11-bp deletion—that both lead to premature stop codons, along with one unedited allele, we used standard fly genetics to put the third chromosome (originally derived from A4) into a complete A4 background, generating strains A4-Ugt86DdDel1, A4-Ugt86DdDel11, and A4-Ugt86Ddwt. All three have marginally reduced viability under no-drug conditions compared to the original A4 founder strain (P = 0.03–0.06), perhaps indicative of novel mutations relative to the A4 progenitor line, or slight changes in sequence due to gene conversion from balancers during stock construction. Nonetheless, Figure 5 shows that both of the deletion-carrying stocks A4-Ugt86DdDel1 and A4-Ugt86DdDel11 have significantly reduced nicotine resistance in comparison with A4-Ugt86Ddwt (P < 10−4 and P < 0.001, respectively). The induced mutations appear to behave recessively, since measuring the phenotype of their heterozygous progeny after crossing to A4 recapitulates the A4 homozygous phenotype (Figure 5).
Figure 5.
Phenotypes of background-controlled Ugt86Dd CRISPR/Cas9-derived mutations. We made a pair of Ugt86Dd mutations homozygous in the A4 background. Del1 introduces a 1-bp mutation, while Del11 introduces an 11-bp mutation, each leading to a different stop codon mutation (File S4). In addition, we passed an unedited allele (wt) through an identical crossing scheme, which should be identical to the pure A4 strain. We tested 10 replicate vials of each of the genotypes in the plot for both no-drug (gray bars) and nicotine (yellow bars) treatments and present the mean (±1 SD) viability across replicate vials. The two homozygous mutant strains (Del1/Del1 and Del11/Del11) have significantly lower nicotine resistance (t-test, P < 10−5) in comparison to the parental A4 genotype and the unedited control strain (wt/wt). Since the unedited control strain has slightly, but significantly reduced resistance in comparison to A4 (P < 0.01), the crossing scheme used to establish the homozygous CRISPR/Cas9-derived genotypes may have resulted in some additional changes in addition to the target mutation.
The difference in nicotine resistance between strains A3 and A4 is 0.69–0.77 (Figure 1 and Figure 5), while A4-Ugt86DdDel1 and A4-Ugt86DdDel11 differ from A4-Ugt86Ddwt by 0.28 and 0.19, respectively. This suggests that 25–41% of the difference between founders A3 and A4 could be due to functional variation at Ugt86Dd, with the most likely variant conferring this effect being the naturally segregating 22-bp InDel. Since the difference between the mean phenotype of RILs carrying the A3 and A4 alleles at the Q4 locus is 0.37 (Marriage et al. 2014), our gene editing data again imply that Ugt86Dd, and likely the InDel variant that results in an allele containing a similar premature stop codon to the edited mutations, confers the bulk of the effect observed at this QTL.
Discussion
Variation at Cyp28d1 may underlie nicotine resistance
Quantitative complementation tests suggest that functional allelic variation is present at Cyp28d1 (Figure S2). Previous RNAseq data (Marriage et al. 2014) suggest gene expression is higher in the more resistant A4 line than in the more susceptible A3 line, and both ubiquitous and anterior midgut-specific RNAi of Cyp28d1 led to a reduction in nicotine resistance (Figure 2 and Table 1). These results strongly implicate variation at Cyp28d1 in the genetic control of nicotine resistance. Data from Chakraborty et al. (2017), who have generated an assembly of DSPR founder A4 based on long-read sequencing data, give some indication that variation in Cyp28d1 copy number may be causative; in contrast to the D. melanogaster reference strain, founder A4 possesses two copies of Cyp28d1. The relatively high expression of Cyp28d1 in A4 (Marriage et al. 2014) supports the idea of a positive correlation between copy number and gene expression, which could in turn lead to enhanced resistance. Such positive associations between P450 copy number and xenobiotic resistance have been reported previously (Wondji et al. 2009; Schmidt et al. 2010). Nonetheless, the relationship between copy number and expression level is not straightforward (Zhou et al. 2011; Schrider et al. 2016), and while the presence of two copies of Cyp28d1 in a strain resistant to nicotine is suggestive, establishing a direct effect requires functional validation.
Evidence that Cyp28d2 also contributes to nicotine resistance, and segregates for causative variation generating the Q1 locus, is marginally more mixed. A quantitative complementation test using an insertional mutant implies the gene harbors a functional difference between strains A3 and A4 (Figure S2), strain A4 exhibits higher expression than A3 (Marriage et al. 2014), and ubiquitous knockdown of Cyp28d2 decreases resistance (Figure 2). However, knockdown of the gene in the midgut had no detectable effect (Table 1). It is possible that we did not knock down Cyp28d2 in the tissue in which it would normally act on nicotine, and failed to identify an effect. Employing an expanded catalog of Gal4 drivers (Buchon et al. 2013) may uncover the tissue in which Cyp28d2 is active against nicotine. Given our data, either Cyp28d1 or Cyp28d2, or indeed both genes, may contribute to the effect observed at the Q1 locus originally mapped by Marriage et al. (2014). Additional fine mapping in natural populations, where both genes appear to segregate for copy number variants (CNVs) (Good et al. 2014), may be profitable and allow functional effects associated with the two genes to be separated.
Ugt86Dd explains a large fraction of the variation for nicotine resistance in the DSPR
Deficiency quantitative complementation tests suggest that the set of UGT genes under Q4 may harbor functional allelic variation affecting nicotine resistance, although these tests do not directly implicate Ugt86Dd (Figure S2). RNAseq, both from the present study and Marriage et al. (2014), suggest Ugt86Dd is more highly expressed in more resistant genotypes. Knocking down the gene ubiquitously (Figure 2) and in the anterior midgut (Table 1) leads to reduced resistance, while overexpression of full-length Ugt86Dd enhances resistance compared to overexpression of a putatively null allele (Figure 3). All this evidence strongly implicates Ugt86Dd as a major source of genetic variation contributing to nicotine resistance.
The segregating 22-bp coding InDel is a compelling candidate for the actual causative site underlying much of this variation. By accounting for the polymorphism during QTL mapping the effect of the Q4 interval is markedly reduced (Figure 4), and comparison of DGRP-derived populations fixed for alternate alleles at the InDel suggest the deletion is associated with reduced resistance. Finally, while we do not specifically generate the 22-bp deletion in the A4 background, our creation and testing of other deletions at a similar location in the Ugt86Dd gene, which similarly lead to premature stop codons, provides strong evidence that the InDel is functional, and directly leads to an effect on nicotine resistance (Figure 5).
Several pieces of evidence suggest that additional polymorphisms also contribute to the effect observed at the Q4 locus. First, the strain effects at the mapped QTL do not fall into two categories, as would be expected if a biallelic causative polymorphism was solely responsible for the QTL effect (Marriage et al. 2014). Second, a quantitative complementation test using a deficiency that eliminates several UGT genes, but leaves Ugt86Dd intact, showed a significant failure to complement, implying other genes in the QTL region may segregate for functional variation (Figure S2). Third, after accounting for the InDel, QTL mapping in the pA population still yields a modest-effect QTL at the Q4 interval, suggesting factors not in perfect LD with the InDel in the DSPR are additionally involved. Such variants could impact other genes in the Q4 interval, but potentially might affect the regulation and/or coding sequence of Ugt86Dd in alleles derived from founders other than A3 and A4. Finally, the difference between the strain effects of A4 (which carries the insertion at Ugt86Dd) and A3 (which carries the deletion) at QTL Q4 is larger than the phenotypic difference between wild-type and edited mutant Ugt86Dd alleles in the same A4 background. Assuming the effects of the naturally occurring deletion at Ugt86Dd in A3 and the CRISPR/Cas9-induced deletion in A4 are the same, this observation is most likely due to variants in other genes in the QTL interval.
One factor that could contribute to the Q4 QTL in addition to Ugt86Dd is a CNV at Ugt86Dh (Chakraborty et al. 2017), where founder A4 harbors two copies of this gene relative to the D. melanogaster reference genome (although we do not yet know the status of this gene in other DSPR founders.) Notably two copies of Ugt86Dh increase expression (Chakraborty et al. 2017), implying the CNV may have some functional effect, and the gene is deleted in the one deficiency that does not eliminate Ugt86Dd (Figure S2). Given the circumstantial evidence associated with Ugt86Dh, it is a strong candidate for additional follow-up functional characterization.
Effect of the segregating InDel at Ugt86Dd
Given the apparent large effect of the InDel in the DSPR, and specifically the effect of introducing a premature stop-encoding lesion into A4, we anticipated the effect of the deletion would routinely be large. However, comparison of DGRP-derived populations fixed for alternate alleles at the InDel showed only a modest effect of the variant. Our design assumes we can estimate the effect of each allele at the InDel averaged over a similarly randomized genetic background. However, due to the paucity of DGRP lines possessing the deletion, it is likely that the insertion and deletion populations show many other fixed differences along the genome, some of which could lead to underestimating any true effect of the variant.
Outside of this technical challenge, one possibility for the very different effect estimates of the InDel in DSPR- and DGRP-derived populations is the different genetic background of the two panels. The DSPR is derived from a worldwide collection of P-element-free strains that have been present in laboratories for decades (King et al. 2012b), while the DGRP is a set of strains relatively recently collected from North Carolina (Mackay et al. 2012). One observation suggesting these populations may have different genetic architectures for nicotine resistance comes from work by Passador-Gurgel et al. (2007). This study demonstrated that flies derived from North Carolina are commonly more nicotine resistant than flies derived from California. Indeed, using our assay we find that average nicotine resistance in our DGRP-derived outbred populations is higher than all but three of the 1274 DSPR RILs assayed by Marriage et al. (2014). While we can only speculate that there is an interaction between the InDel and variation at other functional variants, we know that epistasis contributes significantly to phenotypic variation (Monnahan and Kelly 2015; Forsberg et al. 2017), and that large-effect mutations moved into a varied set of genetic backgrounds result in a spectrum of phenotypic expression (Chandler et al. 2013; Chari and Dworkin 2013; He et al. 2014; Sittig et al. 2016). Thus, there is the potential for genetic background to influence the phenotypic expression of the Ugt86Dd variant.
A way to both establish the causative effect of the 22-bp Ugt86Dd InDel in the DSPR, as well as to estimate any background-specific expression of the variant alleles in the DGRP (and other populations), is to use CRISPR/Cas9 to precisely introduce the deletion into strains carrying a wild-type Ugt86Dd sequence, and similarly “repair” the allele in strains carrying the deletion (see Gratz et al. 2013). Such reagents would additionally facilitate future work on the mechanisms underlying nicotine detoxification by Ugt86Dd, for instance, by examining the products of nicotine metabolism, and allow tests of the effect of the Ugt86Dd gene in the detoxification of other xenobiotics, perhaps neonicotinoid insecticides.
Ugt86Dd deletion is rare and recessive
Among the 15 DSPR founder strains, the 22-bp deletion allele at Ugt86Dd is fairly common (4/15 or 27%). To assess its frequency in wild-caught population samples, we searched for next-generation sequencing reads that perfectly match the deletion and insertion alleles at the variant in a series of pooled resequencing data sets derived from a number of United States populations (see Materials and Methods and Bergland et al. 2014). Averaging over these samples we roughly estimate the frequency of the deletion in nature at ∼2% (Table S2), although there is fairly large sample-to-sample variation around this value (0–11%) that could represent real biological, among-population variation, or simply be a result of sampling (of both individuals and sequencing reads.)
The deletion allele leads to a premature stop, likely ablating gene function, and purifying selection may perhaps explain the low frequency of the allele in nature. Huang et al. (2014) identified variants in the set of 200 DGRP fly strains that potentially generate a damaged protein, and found that such variants are typically less frequent than other sites (e.g., nonsynonymous variants). Similarly, studies in humans have shown that rare, protein-coding polymorphisms are enriched for putatively damaging events (MacArthur et al. 2012; Nelson et al. 2012; Tennessen et al. 2012). Since the Ugt86Dd deletion appears to be recessive (Figure 5), under the assumption there is some cost to an organism in nature carrying the deletion in homozygous form, this might explain why its frequency in nature is not even lower.
It is not clear why the deletion allele is fairly common in the DSPR founders. The founders were collected in the 1950s and 1960s from a number of different countries (King et al. 2012b), prior to the P-element sweep through D. melanogaster, and conceivably populations were enriched for the deletion during this period. However, given the small number of founders, and their arbitrary sampling from stock centers many generations after their original derivation from nature (King et al. 2012b), a deletion frequency of 4/15 may not reflect the actual worldwide population frequency of the allele at the time of collection. It is tempting to speculate that the frequency of the Ugt86Dd deletion allele has been modulated by the use of nicotine as an insecticide. Nicotine was in common use as a pesticide after World War II (Shepard 1951; Metcalf 1955), but is no longer used in this way in the United States (Environmental Protection Agency 2009), having been supplanted by other insecticides, many of which are nicotine derivatives (Goulson 2013). However, it is known that individual UGT genes can act on a range of substrates (Tukey and Strassburg 2000), so any selection acting on Ugt86Dd could be due to one or more unknown compounds to which flies can be exposed in nature.
Despite the large effect of the InDel in the DSPR, replicated by our CRISPR/Cas9 editing, the recessive nature of the deletion allele and its low frequency in nature likely means the InDel explains only a very small fraction of the variation in nicotine resistance in a natural, outbred diploid population. Using the formula appropriate for a completely recessive variant, Va = 8pq3α2 (Falconer and Mackay 1996), estimating the frequency (q) of the deletion allele at 2%, taking the effect of the deletion from the difference between the background matched wild-type Ugt86Dd genotype and the CRISPR-derived lesions (0.19–0.28), and assuming the phenotypic variance among DSPR line means (Marriage et al. 2014) is representative of that in a natural population, the InDel explains <0.01% of the phenotypic variation for the trait. This estimate is lower still if the difference between homozygous insertion- and deletion-carrying genotypes are more in line with the values observed for the DGRP populations.
Resolving causative polymorphisms underlying QTL
Several features of our experimental system facilitated the resolution of a likely causative sequence variant underlying a mapped QTL. First, our study of a xenobiotic resistance trait clearly motivated a focus on the small number of detoxification family members within QTL. This ability to home in on genes that were a priori highly likely to harbor segregating causative variation may be unavailable for many other traits, where the purpose of unbiased, genomewide mapping is to help elucidate the pathways underlying variation. Second, we were fortunate that the causative gene possessed a variant with a clear molecular signature of damage. The phenotypic effects of most polymorphisms, particularly those associated with noncoding changes, are considerably more challenging to decipher from sequence data alone. Third, despite the QTL containing multiple detoxification genes, and our prediction that the QTL was generated by the action of multiple genes/alleles (Marriage et al. 2014), the Ugt86Dd coding mutation alone has a major effect on phenotype that facilitated its detection. Other study systems will lack these advantages. Despite these factors, all studies in Drosophila can leverage the battery of functional tools available for this organism to drill down to the causative molecular lesion(s) underlying QTL and gain insight into the genetic control of trait variation.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300058/-/DC1.
Acknowledgments
We thank Tony Long for suggesting the in silico InDel frequency estimation approach, Emily Pfeifer for technical assistance, and Rob Unckless for comments on a previous version of this manuscript. We thank the TRiP [National Institutes of Health (NIH) R01-GM084947] and the Vienna Drosophila Resource Center for providing transgenic RNAi fly stocks used in this study. Various additional fly stocks, including the DGRP strains, were obtained from the Bloomington Drosophila Stock Center (NIH P40-OD018537). We also thank Julian Dow and Shireen Davies (University of Glasgow) for providing malpighian tubule Gal4 strains, and Nicolas Buchon (Cornell University) for donating a tissue-specific gut Gal4 driver (flygut.epfl.ch). Finally, we thank Johannes Bischof for donating the pUASg.attB vector used for the overexpression experiment (flyorf.ch). X.W.’s work on this project was supported via the Kansas IdeA Networks of Biomedical Research Excellence project (NIH P20-GM103418), and the University of Kansas Genome Sequencing Core is supported by an NIH Centers of Biomedical Research Excellence (P20-GM103638). This project was supported by NIH grant R01-OD010974 to S.J.M. and Anthony D. Long (University of California Irvine).
Footnotes
Communicating editor: C. Jones
Literature Cited
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. [DOI] [PubMed] [Google Scholar]
- Baud A., Hermsen R., Guryev V., Stridh P., Graham D., et al. , 2013. Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat. Genet. 45: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A., Tsunozaki M., Rockman M. V., Kruglyak L., Bargmann C. I., 2011. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., O’Brien K. R., Schmidt P. S., Petrov D. A., 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10: e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J., et al. , 2013. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140: 2434–2442. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Osman D., David F. P., Fang H. Y., Boquete J. P., et al. , 2013. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3: 1725–1738. [DOI] [PubMed] [Google Scholar]
- Chakraborty M., Zhao R., Zhang X., Kalsow S., Emerson J. J., 2017. Extensive hidden genetic variation shapes the structure of functional elements in Drosophila. bioRxiv (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler C. H., Chari S., Dworkin I., 2013. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 29: 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari S., Dworkin I., 2013. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 9: e1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., Ffrench-Constant R. H., et al. , 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Gatti D. M., Munger S. C., Svenson K. L., 2012. The diversity outbred mouse population. Mamm. Genome 23: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Tanny R. E., Seo B., Riccardi D. D., et al. , 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R. K., Christensen S. J., Deal J. A., Coburn R. A., Deal M. E., et al. , 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth T. W., Long A. D., 2003. Inferences regarding the numbers and locations of QTLs under multiple-QTL models using interval mapping and composite interval mapping. Genet. Res. 82: 139–149. [DOI] [PubMed] [Google Scholar]
- Daborn P. J., Yen J. L., Bogwitz M. R., Le Goff G., Feil E., et al. , 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D., 2006. The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dow J. A., Davies S. A., 2006. The Malpighian tubule: rapid insights from post-genomic biology. J. Insect Physiol. 52: 365–378. [DOI] [PubMed] [Google Scholar]
- Dutta D., Dobson A. J., Houtz P. L., Glasser C., Revah J., et al. , 2015. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12: 346–358. [DOI] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental Protection Agency , 2009. Nicotine; product cancellation order. Fed. Regist. 74: 26695–26696. [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Addison Wesley Longman, Essex, England. [Google Scholar]
- Forsberg S. K., Bloom J. S., Sadhu M. J., Kruglyak L., Carlborg O., 2017. Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat. Genet. 49: 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger-Thornsberry G. L., Mackay T. F., 2004. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech. Ageing Dev. 125: 179–189. [DOI] [PubMed] [Google Scholar]
- Gibson G., Weir B., 2005. The quantitative genetics of transcription. Trends Genet. 21: 616–623. [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K., 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning J. I., 2002. How do herbivorous insects cope with noxious secondary plant compounds in their diet? Entomol. Exp. Appl. 104: 15–25. [Google Scholar]
- Good R. T., Gramzow L., Battlay P., Sztal T., Batterham P., et al. , 2014. The molecular evolution of cytochrome P450 genes within and between drosophila species. Genome Biol. Evol. 6: 1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulson D., 2013. An overview of the environmental risks posed by neonicotinoid insecticides. J. Appl. Ecol. 50: 977–987. [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Olsen K. M., 2010. Genetic perspectives on crop domestication. Trends Plant Sci. 15: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A., Lee S. H., Trynka G., Finucane H., Vilhjalmsson B. J., et al. , 2014. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95: 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrop T. W., Pearce S. L., Daborn P. J., Batterham P., 2014. Whole-genome expression analysis in the third instar larval midgut of Drosophila melanogaster. G3 4: 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. Z., Ludwig M. Z., Dickerson D. A., Barse L., Arun B., et al. , 2014. Effect of genetic variation in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics 196: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., 2009. Genomewide association studies–illuminating biologic pathways. N. Engl. J. Med. 360: 1699–1701. [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 24: 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha P., Ramasundarahettige C., Landsman V., Rostron B., Thun M., et al. , 2013. 21st-century hazards of smoking and benefits of cessation in the United States. N. Engl. J. Med. 368: 341–350. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Macdonald S. J., Long A. D., 2012a Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics 191: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Merkes C. M., McNeil C. L., Hoofer S. R., Sen S., et al. , 2012b Genetic dissection of a model complex trait using the Drosophila synthetic population resource. Genome Res. 22: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. G., Sanderson B. J., McNeil C. L., Long A. D., Macdonald S. J., 2014. Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 10: e1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J., Mitchell-Olds T., 2005. Epistasis and balanced polymorphism influencing complex trait variation. Nature 435: 95–98. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bai S., Cass B. N., 2012. Accord insertion in the 5′ flanking region of CYP6G1 confers nicotine resistance in Drosophila melanogaster. Gene 502: 1–8. [DOI] [PubMed] [Google Scholar]
- Linder R. A., Seidl F., Ha K., Ehrenreich I. M., 2016. The complex genetic and molecular basis of a model quantitative trait. Mol. Biol. Cell 27: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Mullaney S. L., Mackay T. F., Langley C. H., 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics 144: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Lyman R. F., Morgan A. H., Langley C. H., Mackay T. F., 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154: 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G., Balasubramanian S., Frankish A., Huang N., Morris J., et al. , 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2001. Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2: 11–20. [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriage T. N., King E. G., Long A. D., Macdonald S. J., 2014. Fine-mapping nicotine resistance loci in Drosophila using a multiparent advanced generation inter-cross population. Genetics 198: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A., Savakis C., 2005. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf R. L., 1955. Organic Insecticides: Their Chemistry and Mode of Action. Interscience Publishers, New York. [Google Scholar]
- Monnahan P. J., Kelly J. K., 2015. Epistasis is a major determinant of the additive genetic variance in Mimulus guttatus. PLoS Genet. 11: e1005201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer J. L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., et al. , 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. R., Wegmann D., Ehm M. G., Kessner D., St Jean P., et al. , 2012. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Passador-Gurgel G., Hsieh W. P., Hunt P., Deighton N., Gibson G., 2007. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat. Genet. 39: 264–268. [DOI] [PubMed] [Google Scholar]
- Pasyukova E. G., Vieira C., Mackay T. F., 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156: 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., et al. , 2015. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. W., Herzyk P., Dow J. A., Leader D. P., 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 41: D744–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosay P., Davies S. A., Yu Y., Sozen M. A., Kaiser K., et al. , 1997. Cell-type specific calcium signalling in a Drosophila epithelium. J. Cell Sci. 110: 1683–1692. [DOI] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Schonbauer C., Langer C. C., Dietzl G., Novatchkova M., et al. , 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464: 287–291. [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Hahn M. W., Begun D. J., 2016. Parallel evolution of copy-number variation across Continents in Drosophila melanogaster. Mol. Biol. Evol. 33: 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard H. H., 1951. The Chemistry and Action of Insecticides. McGraw-Hill, New York. [Google Scholar]
- Sittig L. J., Carbonetto P., Engel K. A., Krauss K. S., Barrios-Camacho C. M., et al. , 2016. Genetic background limits generalizability of genotype-phenotype relationships. Neuron 91: 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen M. A., Armstrong J. D., Yang M., Kaiser K., Dow J. A., 1997. Functional domains are specified to single-cell resolution in a Drosophila epithelium. Proc. Natl. Acad. Sci. USA 94: 5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam L. F., Laurie C. C., 1996. Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 144: 1559–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Steppuhn A., Gase K., Krock B., Halitschke R., Baldwin I. T., 2004. Nicotine’s defensive function in nature. PLoS Biol. 2: E217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhzaz S., Finlayson A. J., Stirrat L., Yang J., Tricoire H., et al. , 2010. Cell-specific inositol 1,4,5 trisphosphate 3-kinase mediates epithelial cell apoptosis in response to oxidative stress in Drosophila. Cell. Signal. 22: 737–748. [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A., 2012. Ten years of the collaborative cross. G3 2: 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J. M., Gamazon E. R., Parra E. J., Below J. E., Valladares-Salgado A., et al. , 2014. Cross-tissue and tissue-specific eQTLs: partitioning the heritability of a complex trait. Am. J. Hum. Genet. 95: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey R. H., Strassburg C. P., 2000. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 40: 581–616. [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Brown M. A., McCarthy M. I., Yang J., 2012. Five years of GWAS discovery. Am. J. Hum. Genet. 90: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji C. S., Irving H., Morgan J., Lobo N. F., Collins F. H., et al. , 2009. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 19: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., McCart C., Woods D. J., Terhzaz S., Greenwood K. G., et al. , 2007. A Drosophila systems approach to xenobiotic metabolism. Physiol. Genomics 30: 223–231. [DOI] [PubMed] [Google Scholar]
- Zhou J., Lemos B., Dopman E. B., Hartl D. L., 2011. Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol. Evol. 3: 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw phenotypes are presented in File S1. Raw RNAseq reads are available from the NCBI SRA (accession no. SRP102254).