Summary
Listeria monocytogenes, the causative agent of listeriosis, is an intracellular pathogen that is exquisitely evolved to survive and replicate in the cytosol of eukaryotic cells. Eukaryotic cells typically restrict bacteria from colonizing the cytosol, likely through a combination of cell autonomous defenses, nutritional immunity, and innate immune responses including induction of programmed cell death. This suggests that L. monocytogenes and other professional cytosolic pathogens possess unique metabolic adaptations, not only to support replication, but also to facilitate resistance to host-derived stresses/defenses and avoidance of innate immune activation. In this review, we outline our current understanding of L. monocytogenes metabolism in the host cytosol and highlight major metabolic processes which promote intracellular replication and survival.
Access to the cytosol is essential for virulence
L. monocytogenes is a Gram-positive facultative intracytosolic pathogen and the causative agent of listeriosis in humans and livestock (Freitag et al., 2009). Ingestion of L. monocytogenes-laden food products and entry into the host via the gastrointestinal tract is responsible for periodic outbreaks of listeriosis (Lecuit, 2007). Infections with L. monocytogenes normally results in mild gastroenteritis, however severe L. monocytogenes infection, especially in at-risk populations (immunocompromised, elderly or pregnant individuals) may result in complications such as septicemia, meningitis, endocarditis, or spontaneous abortion. Disseminated infections with L. monocytogenes are deadly, with a lethality rate of up to ~30% even with antibiotic treatment (Swaminathan and Gerner-Smidt, 2007).
L. monocytogenes enters host cells either via phagocytosis or through receptor-mediated endocytosis facilitated by the bacterial surface proteins internalin A (InlA) and internalin B (InlB) (Mengaud et al., 1996; Shen et al., 2000). L. monocytogenes is initially encapsulated in a host vacuole and subsequently secretes the cholesterol-dependent pore-forming toxin listeriolysin O (LLO, encoded by hly) and the phospholipases PlcA and PlcB to promote escape into the cytosol (Portnoy et al., 1988; Camilli et al., 1989; Mengaud et al., 1991; Vazquez-Boland et al., 1992; Marquis et al., 1995). Once in the cytosol, ActA promotes L. monocytogenes actin-based motility and cell-to-cell spread (Kocks et al., 1992). Genetic deletion of any of these virulence factors or their master regulator, PrfA (Freitag et al., 2009), attenuates L. monocytogenes virulence, signifying necessity for L. monocytogenes to access the cytosol to cause disease. Importantly, not only access to, but maintenance of the cytosolic niche is essential for L. monocytogenes virulence as mutants that trigger host cell death, either due to LLO toxicity or activation of innate immune cell death pathways, are highly attenuated in vivo (Glomski et al., 2003; Sauer et al., 2010).
Metabolic adaptions to the cytosolic environment are crucial for L. monocytogenes intracellular replication and survival. Although our understanding of L. monocytogenes physiology and metabolism in the cytosol is currently incomplete, robust genetic tools coupled with exciting new approaches to transcriptomics, metabolomics and proteomics are leading to a renaissance in our molecular understanding of L. monocytogenes intracellular life. A complete and comprehensive discussion of L. monocytogenes’ metabolic potential is beyond the scope of this review, instead, we will focus on major metabolic processes critical for L. monocytogenes replication and survival in the eukaryotic cytosol.
The inhospitable cytosolic environment
Listeria monocytogenes inhabits the host cytosol, the dense (400 mg/ml of macromolecules) (Guigas et al., 2007) gel-like fluid surrounding the organelles in the cytoplasm that is the major site for glycolysis, gluconeogenesis and the pentose phosphate pathway. Indirect measurements determine that the cytosol of various eukaryotic cells is approximately 1- to 10-fold greater viscosity than bulk water (Luby-Phelps, 2000). Additionally, the cytosol is typically a pH neutral environment (~7.4) (Llopis et al., 1998) that is highly reducing (oxidized:reduced glutathione is approximately 30:1) (Hwang et al., 1992) and contains low concentrations of free amino acids (Piez and Eagle, 1957) and free metal ions such as magnesium, sodium, calcium, potassium, and iron (Ray et al., 2009).
Despite limited availability of amino acids and metals, it was originally thought that the cytosol was a nutrient replete environment, where bacteria should thrive if they had access. This idea was supported by the observation that Bacillus subtilis engineered to express LLO were capable of replicating in the cytosol of host cells following phagosomal escape (Bielecki et al., 1990). However, subsequent studies by Goebel and colleagues challenged this idea and demonstrate that multiple different non-cytosol-adapted bacteria, microinjected or engineered to enter the host cytosol, will not replicate in the cytosol and appear to be cleared (Goetz et al., 2001; Slaghuis et al., 2004). They further demonstrated that rare replication of cytosolic B. subtilis occurred only in dead or dying cells, potentially explaining the original observations by Bielecki et al. Even more recently it has become clear that some vacuolar pathogens that accidently escape into the cytosol of macrophages are also unable to replicate and appear to be cleared (Beuzon et al., 2000; Beuzón et al., 2002; Creasey and Isberg, 2012; Ge et al., 2012). In the case of Salmonella typhimurium this appears to be cell-type specific, as S. typhimurium are restricted in the cytosol of macrophages but replicate in the cytosol of epithelial cells, suggesting that certain host cells are better able to prevent bacterial cytosolic replication and survival (Beuzón et al., 2002; Knodler et al., 2010).
Finally, several metabolic pathways in L. monocytogenes and Francisella spp. have been reported to promote not just cytosolic replication but survival, as disruptions in these pathways lead to cytosolic bacteriolysis, in some cases in a cell-type specific manner (Sauer et al., 2010; Peng et al., 2011; Pensinger et al., 2016; Chen et al., 2017). Importantly, bacteriolysis in the cytosol can potently activate innate immunity through a series of cytosol specific DNA sensing pathways including cGAS/STING and AIM2, leading to type I interferon induction and inflammasome activation, respectively (Sauer et al., 2010; Woodward et al., 2010; Sauer, Sotelo-Troha, et al., 2011; Rae et al., 2011; Hansen et al., 2014). As discussed above, maintenance of the cytosolic niche is essential for L. monocytogenes and other cytosolic pathogens to causes disease and as such, activation of inflammasome attenuates virulence (Warren et al., 2011; Sauer, Pereyre, et al., 2011). Together these data suggest through unknown mechanisms the cytosol of various cells vary in their ability to restrict bacterial replication and survival. To promote its virulence, L. monocytogenes must modulate its metabolism not only to acquire host nutrients but also to tolerate cytosolic stresses and evade host immune defenses.
Host-derived metabolites required for virulence
Like all intracellular pathogens, L. monocytogenes steals nutrients from its host cell. To facilitate this, the L. monocytogenes genome encodes over 330 putative transporters which is 2 and 3 times more transport genes than either Escherichia coli or Bacillus subtilis, respectively (Glaser et al., 2001). Of these transporters, 84 assemble into 29 complete phosphoenolpyruvate-dependent phosphotransferase systems (PTS) (Stoll and Goebel, 2010). Interestingly despite this abundance of PTSs, they are downregulated during infection and are dispensable for intracellular replication in macrophages (Stoll and Goebel, 2010; Aké et al., 2011). During intracellular replication, L. monocytogenes transports host-derived hexose-phosphates, such as glucose-1-phosphate and glucose-6-phosphate, through Hpt (gene uhpT), a permease required for virulence (Chico-Calero et al., 2002) whose expression is tightly controlled by the master virulence regulator, PrfA (Ripio et al., 1997). A second major intracellular carbon source for L. monocytogenes is glycerol. Glycerol utilization mutants (glp and dha genes) are defective for intracellular replication in both epithelial cells and macrophages (Joseph et al., 2006; Joseph et al., 2008). 13C isotopologue studies suggest that host derived glucose-6-phosphate and glycerol are diverted to separate metabolic pathways in L. monocytogenes such that glucose-6-phosphate is oxidized by the pentose phosphate pathway and likely used as precursors for nucleotide biosynthesis and aromatic compounds, whereas glycerol feeds lower glycolysis for energy and amino acids biosynthesis (Grubmüller et al., 2014). Surprisingly a combinatorial mutant that can neither use glucose-6-phosphate or glycerol (ΔC3ΔuhpT) was still partially able to replicate in macrophages (Grubmüller et al., 2014), suggesting that alternative carbon sources may still be available during intracellular replication. Although it is unlikely that lipids act as the direct alternative carbon source due to a lack of genes required for beta-oxidation of fatty acids (Glaser et al., 2001), breakdown of phosphatidylethanolamine (PE) in mammalian membranes (Tsoy et al., 2009), possibly by the L. monocytogenes phosphatidylcholine phospholipase C (PlcB), may liberate both ethanolamine and glycerol (Geoffroy et al., 1991). Ethanolamine can be further degraded into acetaldehyde and ammonia by L. monocytogenes’ adenosylcobalamin-dependent ethanolamine lyase (encoded by eut genes) (Joseph and Goebel, 2007). In support of this hypothesis eut mutants are defective for intracellular replication in epithelial cells (Joseph et al., 2006). Whether ethanolamine and/or other host metabolites are used by L. monocytogenes as carbon sources during intracellular replication remains an important unanswered question. Furthermore, it is likely that diversification of carbon sources would help L. monocytogenes fulfill its nutritional requirements in the cytosol while minimizing metabolic perturbations that can be detected by the host cell (Grubmüller et al., 2014).
During intracellular replication L. monocytogenes likely assimilates nitrogen from various host sources, such as glutamine. Indeed genes for glutamate synthase, used to assimilate nitrogen from glutamine (Schreier, 1993), are up-regulated during infection (Joseph et al., 2006). Furthermore, the glutamine ABC transporter (GlnPQ) is required for optimal intracellular replication and virulence suggesting that L. monocytogenes needs to scavenge glutamine from the host (Haber et al., 2017). In the absence of glutamine, other nitrogen sources such as ammonia, ethanolamine and arginine may sustain growth of L. monocytogenes in the cytosol (Tsai and Hodgson, 2003; Kutzner et al., 2016). Inorganic ammonia transporters (encoded by nrgAB) are upregulated in epithelial cells (Joseph et al., 2006) but downregulated in macrophages (Chatterjee et al., 2006), possibly signifying differences in nitrogen source availability between these two cell types. Ammonia may also be acquired through ethanolamine degradation (Joseph et al., 2006). Finally, genes for the arginine ABC transporter appear to be induced intracellularly (Klarsfeld et al., 1994; Joseph et al., 2006). As with carbon acquisition, nitrogen assimilation from different host sources may benefit L. monocytogenes replication but also facilitate avoidance of host defense pathways as arginine metabolism plays a critical role in nitric oxide synthase and polyamine synthesis. Indeed, several pathogens are known to deplete arginine levels to avoid killing by host cells (Gobert et al., 2001; Abu-Lubad et al., 2014; Goldman-Pinkovich et al., 2016).
Although L. monocytogenes possesses all the metabolic pathways required for de novo synthesis of both essential and non-essential amino acids (Glaser et al., 2001) during infection, host-derived amino acids serve as building blocks for listerial proteins and other macromolecules, as well as for assimilation of nitrogen as discussed above. Mutants in cysteine, arginine and glutamine transport systems are each partially impaired for intracellular replication and attenuated for virulence (Klarsfeld et al., 1994; Schauer et al., 2010; Haber et al., 2017), while other amino acid transporters have yet to be characterized. 13C isotopologue profile analysis demonstrates that host-derived amino acids are efficiently taken up by cytosolic L. monocytogenes and quickly incorporated into protein with little to no detectable catabolism of amino acids (Eylert et al., 2008; Grubmüller et al., 2014). Amino acids may also be acquired in the form of host-derived oligopeptides (Marquis et al., 1993), possibly through an ATP-dependent oligopeptide transporter, OppABCDF (Verheul et al., 1998) or the di/tripeptide transporter, DtpT (Wouters et al., 2005) where they are then degraded by aminopeptidases (Perry and Higgins, 2013; Cheng et al., 2015). Assimilation of host amino acids and de novo synthesis of certain amino acids (discussed later) are crucial for L. monocytogenes replication in the cytosol, however the relative contribution of amino acid scavenging versus synthesis is not clear. This is important since nutritional immunity through amino acid depletion is a well-defined host defense mechanism which L. monocytogenes likely must overcome (Appelberg, 2006).
L. monocytogenes does not possess complete pathways required to de novo synthesize a variety of cofactors including thiamine, lipoate, biotin, and riboflavin (Premaratne et al, 1991; Phan-Thanh and Gormon, 1997), suggesting L. monocytogenes must scavenge these cofactors from the host cytosol. Transport of biotin and riboflavin from the host cytosol likely occurs through the putative biotin (BioMNY) and riboflavin transporters (Lmo1945, Ecf and RibU), respectively (Dowd et al, 2011; Karpowich et al, 2015; Matern et al, 2016). L. monocytogenes is unable to synthesize thiamine de novo, due to absence of ThiC, a HMP-P synthase, however, supplementation of HMP-P/HMP in vitro facilitates growth of L. monocytogenes in the minimal media lacking thiamine, suggesting that latter steps in the thiamine biosynthesis pathway are intact (Schauer et al, 2009). Interestingly growth of L monocytogenes in epithelial cells requires both the thiamine transporter (ThiT) and enzymes for latter de novo thiamine biosynthesis (ThiD) (Schauer et al, 2009), suggesting that thiamine concentrations in the host are scarce and L. monocytogenes must concurrently scavenge host derived thiamine and synthesize additional thiamine from HMP to support robust growth of L. monocytogenes in the cytosol. Finally, L. monocytogenes possesses two lipoate ligase enzymes, LplA1 and LplA2 which are required for lipoylation of dehydrogenase complexes (Keeney et al, 2007) including the pyruvate dehydrogenase complex (PDH), branched-chain α-keto acid dehydrogenase (BKD) and glycine cleavage pathway (GCV). Although L. monocytogenes possess two of these enzymes their functions are not fully overlapping or redundant; LplA1 is the major lipoate host scavenging enzyme active in the cytosol as evidenced by its higher affinity for lipoate (Christensen et al, 2011) and its essentiality for virulence (O’Riordan et al., 2003; Keeney et al., 2007). Interestingly, both PDH and BKD mutants are significantly attenuated for virulence and may explain the necessity for lipoate scavenging (Sun and O’Riordan, 2010; Chen et al., 2017). These host-derived cofactors are responsible for enzymatic activity in numerous metabolic pathway, some of which may be required for intracellular replication and/or survival, however, in many cases the specific metabolic pathways impaired when L. monocytogenes is unable to scavenge these cofactors from the host have not been characterized.
Undoubtedly nutrient acquisition from the host is essential for replication of L. monocytogenes, though, why L. monocytogenes has evolved redundant mechanisms to acquire diverse and sometimes overlapping host nutrients remains unclear. One possibility is that overlap of carbon and nitrogen utilization pathways may be important to overcome host nutritional immunity (Appelberg, 2006). Additionally, diversification of nutrient sources may reduce metabolic burdens on the host and prevent detection by the host immune system. Multiple recent studies have highlighted a variety of mechanisms by which host cell sense disruptions of metabolic flux and/or bacterial metabolites directly as indicators of infection to trigger innate immune responses (Wynosky-Dolfi et al., 2014; Sanman et al., 2015; Gaudet et al., 2015; Wolf et al., 2016). Interestingly, although recent host metabolomics studies clearly indicate that theft of nutrients from the host cytosol by L. monocytogenes and Shigella flexneri influences host metabolism (Gillmaier et al., 2012; Kentner et al., 2014; Grubmüller et al., 2014), these pathogens effectively evade known metabolically triggered host innate immune responses. Together these findings suggest that L. monocytogenes metabolism is not only important for replication but also countering host immunity, though the mechanisms by which this occurs remain to be elucidated. Importantly, the intersection of metabolism and innate immunity in the context of L. monocytogenes infection in an intact animal has also not been addressed.
Bacterial metabolic programs required for virulence
Although L. monocytogenes acquires many essential nutrients directly from the host, there are also many intrinsic metabolic pathways essential for cytosolic replication, survival and ultimately virulence. Genetic screens have uncovered an exhaustive list of metabolic pathways vital to L. monocytogenes intracellular replication (Camilli et al., 1989; Joseph et al., 2006; Schauer et al., 2010), though in many cases these pathways’ contributions to intracellular survival, innate immune evasion and virulence of L. monocytogenes have not been thoroughly examined.
Although most amino acids utilized in L. monocytogenes protein synthesis appear to be host-derived, pathways for biosynthesis of Thr, His, Arg, Ser, Met, the branched chain amino acids (BCAA), and the aromatic amino acids are induced during intracellular replication and are necessary for efficient replication during infection (Marquis et al., 1993; Joseph et al., 2006; Chatterjee et al., 2006; Camejo et al., 2009; Schauer et al., 2010; Lobel et al., 2012; Chen et al., 2017). These data suggest that although host derived amino acids are transported and used directly in protein synthesis, certain amino acids are inadequately supplied in the host cell cytosol. In some cases, inefficient replication and/or defective stress responses are not due to defects in protein synthesis, but instead are due to the roles that amino acids or their precursors play in non-protein synthesis processes. For example, BCAAs are necessary not only for protein synthesis, but are also a major precursor of branched chain fatty acid biosynthesis and are necessary for proper L. monocytogenes lipid/membrane homeostasis and resistance to intracellular stresses (Sun and O’Riordan, 2010). BCAAs levels also signal the global metabolic regulator CodY, which represses and activates genes for amino acid biosynthesis, nutrient transport, stress response and virulence (Bennett et al., 2007; Lobel et al., 2012; Lobel and Herskovits, 2016). Moreover, L. monocytogenes codY mutants are partially attenuated for virulence both ex vivo and in vivo (Bennett et al., 2007; Lobel et al., 2012; Whiteley et al., 2015). Alternatively, the aro genes, required for chorismate biosynthesis are upregulated during infection and are essential for virulence. However, chorismate is a precursor not only for aromatic amino acids (Tyr, Trp, Phe) but also menaquinone (MK) and folate, and the attenuation of the aro mutants is primarily due to disruption of MK and/or 1,4-dihydroxy-2-naphthoate (DHNA) biosynthesis (Stritzker et al., 2004; Chen et al., 2017). Taken together, these observations demonstrate that virulence defects associated with amino acid biosynthesis mutants may be mistakenly attributed to protein synthesis and that the functions of amino acids/precursors in other metabolic pathways should be carefully evaluated to fully understand L. monocytogenes’ intracellular metabolic requirements.
Lipids play a critical role in L. monocytogenes replication and survival during infection. L. monocytogenes and other Gram-positive bacterial membranes are highly enriched for branched chain fatty acids (Whittaker et al., 2005). Lipoate-dependent BKD synthesizes constituents for branched chain fatty acids from BCAAs (Massey et al., 1976) and BKD mutants are severely attenuated ex vivo and in vivo (Sun and O’Riordan, 2010) due to killing by lysozyme and cathelicidin-related antimicrobial peptides (CRAMP) (Sun et al., 2012). Finally, modulation of lipid composition between anteiso-, iso-, and straight-chain fatty acids drastically alters virulence phenotypes of L. monocytogenes within macrophages (Sun and O’Riordan, 2010; Sun et al., 2012). These data together highlight the importance for proper membrane homeostasis and biogenesis during intracellular replication of L. monocytogenes.
Likewise, connections between central metabolism and cell wall homeostasis are likely important for L. monocytogenes survival and replication within host cells. For example, L-rhamnose utilization genes are active during intracellular replication of serogroup 1/2 L. monocytogenes strains that decorate their wall teichoic acids (WTA) with rhamnose (Uchikawa et al., 1986; Lobel et al., 2012). As WTA modifications have been implicated in resistance to cell wall stress, this could indicate a critical role for WTA modification or other L-rhamnose dependent processes during infection. Indeed, mutants defective for L-rhamnosylation of their WTA are attenuated during infection in a cationic peptide-dependent manner (Carvalho et al., 2015). Similarly, the bacterial second messenger cyclic di-adenosine monophosphate (c-di-AMP) is emerging as a critical signaling molecule in L. monocytogenes that integrates regulation of central metabolism with cell wall homeostasis and osmoregulation (Kaplan Zeevi et al., 2013; Huynh et al., 2014; Tadmor et al., 2014; Sureka et al., 2014; Whiteley et al., 2015; Whiteley et al., 2017). Mutants lacking c-di-AMP (ΔdacA) are prone to bacteriolysis in the cytosol of macrophages leading to hyperactivation of DNA-sensing innate immune signaling pathways. Not surprisingly given the central role of c-di-AMP in regulating metabolism and cell-envelope stress ΔdacA mutants are highly attenuated for virulence (Witte et al., 2013). Finally, the PASTA (penicillin-binding-protein and serine/threonine associated) kinase PrkA and its associated protein of unknown function YvcK have been shown to be important for glycerol metabolism in L monocytogenes (Görke et al., 2005; Mir et al., 2014; Pensinger et al., 2016). Through some unknown mechanism these proteins also direct cell wall homeostasis, cytosolic survival, and virulence of L. monocytogenes (Sauer et al., 2010; Pensinger et al., 2016). The molecular mechanisms by which L. monocytogenes intracellular metabolism facilitates proper cell wall maintenance and resistance against cytosolic stresses remains to be elucidated.
L. monocytogenes is a facultative aerobe that can respire on oxygen but not on nitrate, nitrite or other terminal electron acceptors since it is missing genes for these reductases (Glaser et al., 2001). While it is clear that L. monocytogenes possesses the capacity for both aerobic and fermentative growth (Romick et al., 1996), which mode of growth is dominant during intracellular replication has not been resolved, though several lines of evidence point towards aerobic respiration. L. monocytogenes genes for the electron transport chain are induced in cell culture and in vivo, including genes for MK biosynthesis genes (men) and L. monocytogenes’ cytochrome bd and aa3 oxidases in vivo (Chatterjee et al., 2006; Camejo et al., 2009). These data suggest that the electron transport chain and generation of a membrane potential is necessary for infection, and indeed MK deficient mutants are attenuated in vivo (Stritzker et al., 2004; Perry and Higgins, 2013; Chen et al., 2017). Additionally, the fact that the non-fermentable substrate, glycerol, is a primary carbon and energy source during intracellular replication (Grubmüller et al., 2014) suggests that respiration is required for intracellular replication. Likewise, computational modeling of L. monocytogenes metabolic flux also predict that oxygen consumption and CO2 efflux occur in the cytosol (Lobel et al., 2012). In contrast anaerobic growth likely occurs during transit through the gastrointestinal tracts and plays a role in priming L. monocytogenes for intracellular invasion (Wallace et al., 2017). Understanding the relative contribution of aerobic and anaerobic metabolism and its temporal and spatial regulation during infection is a key next step in understanding the progression of listeriosis.
While a key role for electron transport is to facilitate ATP synthesis through oxidative phosphorylation, the electron transport chain also governs reduction-oxidation (redox) balance (Bueno et al., 2012), detoxification of oxidative/nitrosative stress (Giuffrè et al., 2014), protein localization during cell division (Strahl and Hamoen, 2010), nucleotide biosynthesis (Kilstrup et al., 2005), and solute transport of betaine/carnitine (Wargo and Meadows, 2015) in bacteria. As such, it is difficult to know which of the key functions of the electron transport chain are essential for virulence. Even more confounding is a recent study demonstrating that MK biosynthetic mutants are killed in the macrophage cytosol independent of respiration defects or even the synthesis of full length MK. Instead, the cytosolic survival defects and at least some of the virulence defect of the MK biosynthetic mutants is due to lack of the MK biosynthetic intermediate, 1,4-dihydroxynaphthoate (DHNA) (Chen et al., 2017). It is likely that DHNA/MK, a functioning electron chain and oxidative phosphorylation all contribute to the virulence of L. monocytogenes in vivo, however, since these processes are intimately linked, studies to tease apart their relative contributions will be necessary in the future.
Integration of metabolism and virulence gene expression
PrfA is a Crp/Fnr family transcription factor that is responsible for regulating the entire virulence regulon in L. monocytogenes, and not surprisingly it is both regulated by and regulates metabolism (de las Heras et al., 2011) (Milohanic et al., 2003). Most notably, while PrfA activity is necessary to fully activate expression of uhpT as discussed above, PrfA function is inhibited by glucose and other phospho-transfer-system (PTS) substrates not present or utilized by L. monocytogenes in the host cytoplasm (Milenbachs et al., 1997; Mertins et al., 2007). The inhibition of PrfA is not mediated directly by the sugars, instead, uptake and metabolism of these sugars affects the serine phosphorylation (Ser-P) of the phosphocarrier protein Hpr (Deutscher et al., 1995) and it is hypothesized that PrfA inhibition is directly mediated by Ser-P Hpr (Herro et al., 2006) or other components of the PTS (Joseph et al., 2008; Vu-Khac and Miller, 2009). PrfA in turn likely regulates the PTS system since Hpr levels are lowered in vivo (Fuchs et al., 2012) and a constitutively active PrfA mutant (PrfA*) has impaired growth on glucose (Eylert et al., 2008), although the mechanism by which this is regulated is not defined. Consistent with the regulation of PrfA activity by carbon source availability, glycerol metabolism also increases PrfA activity, again through an unknown mechanism (Mertins et al., 2007). Through this regulation, L. monocytogenes ensures that virulence factors required for colonizing a host are only turned on once the bacteria gain access to a host. Indeed, loss of this regulation through genetic manipulation leads to loss of fitness during L. monocytogenes’ environmental lifestyle (Bruno and Freitag, 2010), demonstrating the importance of tying metabolism to virulence factor regulation.
In addition to regulation of PrfA function through sensing of carbon metabolism, several studies have investigated other metabolic regulators of PrfA function. For example, in response to low levels of BCAAs L. monocytogenes upregulates BCAA synthesis and the transcriptional regulator CodY which promotes transcription of prfA (Bennett et al., 2007; Lobel et al., 2012; Lobel et al., 2014). Additionally, translation of PrfA is repressed in trans by S-adenosyl methionine dependent riboswitches which bind the prfA UTR (Loh et al., 2009). Finally, PrfA is post-translationally regulated allosterically by glutathione (Reniere et al., 2015), an abundant peptide which maintains redox balance in the host cytosol (Meister and Anderson, 1983) such that high exogenous and endogenous glutathione increase PrfA activity as much as 150 fold (Reniere et al., 2015). Ultimately, defining additional molecular mechanisms by which metabolism regulates PrfA activity transcriptionally, translationally or post-translationally will be key to further understanding how L. monocytogenes rapidly adapts to its intracellular niche.
Perspective
Despite previous assumptions about the bountiful and protected nature of the cytosol, it is now clear that specific adaptations are required to colonize this niche. Increasing evidence suggests that different cell types vary in their ability to restrict bacteria from colonization, highlighting the need for L. monocytogenes to have metabolic flexibility (Beuzón et al., 2002; Chen et al., 2017). Much work is still required to characterize the unique host cytosolic environment and understand what metabolic adaptations are necessary to live in this restrictive environment (Fig. 1). While historically L. monocytogenes metabolic adaptations have been studied in the context of nutrients required for replication, it is likely that metabolic adaptations are also necessary to facilitate stress responses as L. monocytogenes encounters cytosolic antimicrobial effectors such as ubiquicidin (Hiemstra et al., 1999), interferon-inducible guanylate binding proteins (GBP) (Man et al., 2016), lysozyme (Rae et al., 2011) autophagy (Knodler and Celli, 2011) and other yet to be defined cell autonomous defenses. In the future, it will be important to clearly delineate functions of metabolism related to stress responses versus replication, what host cell processes impose these selective pressures and how the physiochemical, nutritional, and immunological characteristics of the cytosol differ between cell types.
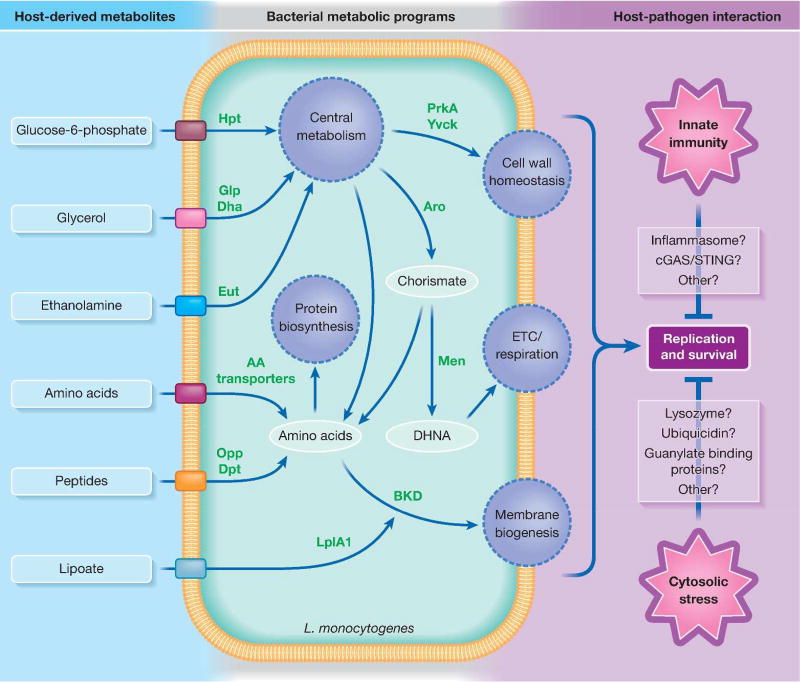
Fig 1.
Overview of select L. monocytogenes metabolic pathways that determine intracellular replication and survival. During colonization of host cells, L. monocytogenes transports host-derived metabolites through various nutrient transport systems. These metabolites are used by L. monocytogenes both for catabolic and anabolic pathways to replicate in the host cytosol and resist cytosolic stresses. Green text denotes L. monocytogenes enzymes crucial for intracellular replication and/or survival. Abbreviations: ETC - electron transport chain, DHNA - 1,4-dihydroxy-2-naphthoate, BKD - branched-chain α-keto acid dehydrogenase complex.
Finally, infection by intracellular pathogens could impose a tremendous metabolic burden on the host cell, and it is likely that some pathogens, including L. monocytogenes, modulate their metabolism to minimize this impact and promote survival of the host cell (Gillmaier et al., 2012; Kentner et al., 2014). Furthermore, it is becoming increasing clear that host cells monitor metabolic activity such that both host and pathogen metabolites can be potent activators of the host immune system (Wynosky-Dolfi et al., 2014; Sanman et al., 2015; Gaudet et al., 2015; Wolf et al., 2016). This further highlights the need for pathogens to tightly regulate their metabolism to promote survival and replication without alerting the innate immune system to their presence, and points to our need to further understand pathogen metabolism during infection as a potential option for therapeutic interventions. Using a combination of newly developed metabolomics, transcriptomics and genetic approaches we can start probing both the host and L. monocytogenes metabolism simultaneously during infection to get a systematic picture of host-pathogen interactions in the cytosol.
Acknowledgments
We are grateful for Adam Schaenzer for critical reading of this manuscript. This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases (T32AI055397 [G.Y.C.]) and National Cancer Institute (R01CA188034 [J.-D.S.]).
Footnotes
These authors declare no conflict of interests.
References
- Abu-Lubad M, Meyer TF, Al-Zeer MA. Chlamydia trachomatis inhibits inducible NO synthase in human mesenchymal stem cells by stimulating polyamine synthesis. J Immunol. 2014;193:2941–2951. doi: 10.4049/jimmunol.1400377. [DOI] [PubMed] [Google Scholar]
- Aké FMD, Joyet P, Deutscher J, Milohanic E. Mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol. 2011;81:274–293. doi: 10.1111/j.1365-2958.2011.07692.x. [DOI] [PubMed] [Google Scholar]
- Appelberg R. Macrophage nutriprive antimicrobial mechanisms. J Leukoc Biol. 2006;79:1117–28. doi: 10.1189/jlb.0206079. [DOI] [PubMed] [Google Scholar]
- Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, et al. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63:1453–67. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. Embo J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- Bielecki J, Youngman P, Connelly P, Portnoy D. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990;345:175–176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- Bruno JC, Freitag NE. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One. 2010;5:1–12. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno E, Mesa S, Bedmar EJ, Richardson DJ, Delgado MJ. Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal. 2012;16:819–852. doi: 10.1089/ars.2011.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camejo A, Buchrieser C, Couvé E, Carvalho F, Reis O, Ferreira P, et al. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009;5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A, Paynton CR, Portnoy DA. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc Natl Acad Sci U S A. 1989;86:5522–5526. doi: 10.1073/pnas.86.14.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F, Atilano ML, Pombinho R, Covas G, Gallo RL, Filipe SR, et al. L-Rhamnosylation of Listeria monocytogenes Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane. PLOS Pathog. 2015;11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, et al. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, McDougal CE, D’Antonio MA, Portman JL, Sauer J-D. A genetic screen reveals that synthesis of 1,4-dihydroxy-2-naphthoate (DHNA), but not full-length menaquinone, is required for Listeria monocytogenes cytosolic survival. MBio. 2017;8:e00119–17. doi: 10.1128/mBio.00119-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Wang X, Dong Z, Shao C, Yang Y, Fang W, et al. Aminopeptidase T of M29 family acts as a novel intracellular virulence factor for Listeria monocytogenes infection. Sci Rep. 2015;5:17370. doi: 10.1038/srep17370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH, Hagar Ja, O’Riordan MXD, Cronan JE. A complex lipoate utilization pathway in Listeria monocytogenes. J Biol Chem. 2011;286:31447–31456. doi: 10.1074/jbc.M111.273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci U S A. 2012;109:3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Dowd GC, Joyce SA, Hill C, Gahan CGM. Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect Immun. 2011;79:369–379. doi: 10.1128/IAI.00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eylert E, Schär J, Mertins S, Stoll R, Bacher A, Goebel W, Eisenreich W. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol Microbiol. 2008;69:1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- Freitag NE, Port GC, Miner MD. Listeria monocytogenes - from saprophyte to intracellular pathogen. Nat Rev Microbiol. 2009;7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs TM, Eisenreich W, Kern T, Dandekar T. Toward a systemic understanding of Listeria monocytogenes metabolism during infection. Front Microbiol. 2012;3:23. doi: 10.3389/fmicb.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet RG, Sintsova A, Buckwalter CM, Leung N, Cochrane A, Li J, et al. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science. 2015;348:1251–1255. doi: 10.1126/science.aaa4921. [DOI] [PubMed] [Google Scholar]
- Ge J, Gong Y-N, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc Natl Acad Sci U S A. 2012;109:6193–6198. doi: 10.1073/pnas.1117490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy C, Raveneau J, Beretti J, Lecroisey A, Alouf JE, Berche P, Leon U, De Purification and characterization of an extracellular 29-kilodalton phospholipase C from Listeria monocytogenes. Infect Immun. 1991;59:2382–2388. doi: 10.1128/iai.59.7.2382-2388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmaier N, Götz A, Schulz A, Eisenreich W, Goebel W. Metabolic responses of primary and transformed cells to intracellular Listeria monocytogenes. PLoS One. 2012;7:e52378. doi: 10.1371/journal.pone.0052378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrè A, Borisov VB, Arese M, Sarti P, Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta. 2014;1837:1178–1187. doi: 10.1016/j.bbabio.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- Glomski IJ, Decatur AL, Portnoy DA. Listeria monocytogenes mutants that fail to compartmentalize listeriolysin O activity are cytotoxic, avirulent, and unable to evade host extracellular defenses. Infect Immun. 2003;71:6754–6765. doi: 10.1128/IAI.71.12.6754-6765.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, et al. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: A strategy for bacterial survival. Proc Natl Acad Sci. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Bubert A, Wang G, Chico-Calero I, Vazquez-Boland JA, Beck M, et al. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc Natl Acad Sci. 2001;98:12221–12226. doi: 10.1073/pnas.211106398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Pinkovich A, Balno C, Strasser R, Zeituni-Molad M, Bendelak K, Rentsch D, et al. An arginine deprivation response pathway Is induced in Leishmania during macrophage invasion. PLOS Pathog. 2016;12:e1005494. doi: 10.1371/journal.ppat.1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Foulquier E, Galinier A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology. 2005;151:3777–91. doi: 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- Grubmüller S, Schauer K, Goebel W, Fuchs TM, Eisenreich W. Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front Cell Infect Microbiol. 2014;4:156. doi: 10.3389/fcimb.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigas G, Kalla C, Weiss M. Probing the nanoscale viscoelasticity of intracellular fluids in living cells. Biophys J. 2007;93:316–323. doi: 10.1529/biophysj.106.099267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A, Friedman S, Lobel L, Burg-Golani T, Sigal N, Rose J, et al. L-glutamine induces expression of Listeria monocytogenes virulence genes. PLOS Pathog. 2017;13:e1006161. doi: 10.1371/journal.ppat.1006161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jørgensen SE, Rahbæk SH, Jensen SB, et al. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014:1–13. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herro R, Poncet S, Cossart P, Buchrieser C, Gouin E, Glaser P, Deutscher J. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J Mol Microbiol Biotechnol. 2006;9:224–234. doi: 10.1159/000089650. [DOI] [PubMed] [Google Scholar]
- Hiemstra PS, Barselaar MT, van den, Roest M, Nibbering PH, Furth R, van Ubiquicidin, a novel murine microbicidal protein present in the cytosolic fraction of macrophages. J Leukoc Biol. 1999;66:423–428. doi: 10.1002/jlb.66.3.423. [DOI] [PubMed] [Google Scholar]
- Huynh TN, Luo S, Pensinger D, Sauer J, Tong L, Huynh TN, et al. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A. 2014;112:1–10. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Joseph B, Goebel W. Life of Listeria monocytogenes in the host cells’ cytosol. Microbes Infect. 2007;9:1188–1195. doi: 10.1016/j.micinf.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, et al. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stu C, Schauer K, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan Zeevi M, Shafir NS, Shaham S, Friedman S, Sigal N, Nir-Paz R, et al. Listeria monocytogenes MDR transporters and c-di-AMP that contribute to Type I interferons induction, play a role in cell wall stress. J Bacteriol. 2013 doi: 10.1128/JB.00794-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowich NK, Song JM, Cocco N, Wang D-N. ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism. Nat Struct Mol Biol. 2015;22:565–571. doi: 10.1038/nsmb.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Stuckey Ja, O’Riordan MXD. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol. 2007;66:758–770. doi: 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner D, Martano G, Callon M, Chiquet P, Brodmann M, Burton O, et al. Shigella reroutes host cell central metabolism to obtain high-flux nutrient supply for vigorous intracellular growth. Proc Natl Acad Sci U S A. 2014;111:9929–34. doi: 10.1073/pnas.1406694111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilstrup M, Hammer K, Ruhdal Jensen P, Martinussen J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev. 2005;29:555–590. doi: 10.1016/j.femsre.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Klarsfeld AD, Goossens PL, Cossart P. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol Microbiol. 1994;13:585–597. doi: 10.1111/j.1365-2958.1994.tb00453.x. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler La, Vallance Ba, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A. 2010;107:17733–8. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kutzner E, Kern T, Felsl A, Eisenreich W, Fuchs TM. Isotopologue profiling of the listerial N-metabolism. Mol Microbiol. 2016;100:315–327. doi: 10.1111/mmi.13318. [DOI] [PubMed] [Google Scholar]
- de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol. 2011;14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Lecuit M. Human listeriosis and animal models. Microbes Infect. 2007;9:1216–1225. doi: 10.1016/j.micinf.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci U S A. 1998;95:6803–8. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Herskovits AA. Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes. PLoS Genet. 2016;12:1–27. doi: 10.1371/journal.pgen.1005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Sigal N, Borovok I, Belitsky BR, Sonenshein AL, Herskovits AA. The metabolic regulator CodY links L. monocytogenes metabolism to virulence by directly activating the virulence regulatory gene, prfA. Mol Microbiol. 2014;95:624–644. doi: 10.1111/mmi.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Listeria monocytogenes Virulence. PLoS Genet. 2012;8:e1002887. doi: 10.1371/journal.pgen.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Man SM, Place DE, Kuriakose T, Kanneganti T-D. Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J Leukoc Biol. 2016;100 doi: 10.1189/jlb.4MR0516-223R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis H, Bouwer HG, Hinrichs DJ, Portnoy DA. Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect Immun. 1993;61:3756–3760. doi: 10.1128/iai.61.9.3756-3760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis H, Doshi V, Portnoy DA. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey LK, Sokatch JR, Conrad RS. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976;40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern A, Pedrolli D, Groβhennig S, Johansson J, Mack M. Uptake and metabolism of antibiotics roseoflavin and 8-demethyl-8-aminoriboflavin in riboflavin-auxotrophic Listeria monocytogenes. J Bacteriol. 2016;198:3233–3243. doi: 10.1128/JB.00388-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Mengaud J, Braun-Breton C, Cossart P. Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol Microbiol. 1991;5:367–372. doi: 10.1111/j.1365-2958.1991.tb02118.x. [DOI] [PubMed] [Google Scholar]
- Mengaud J, Ohayon H, Gounon P, Mege R-M, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–32. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- Mertins S, Joseph B, Goetz M, Ecke R, Seidel G, Sprehe M, et al. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J Bacteriol. 2007;189:473–490. doi: 10.1128/JB.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenbachs AA, Brown DP, Moors M, Youngman P. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol Microbiol. 1997;23:1075–1085. doi: 10.1046/j.1365-2958.1997.2711634.x. [DOI] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppée J-Y, Frangeul L, Vega Y, Vázquez-Boland Ja, et al. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- Mir M, Prisic S, Kang C-M, Lun S, Guo H, Murry JP, et al. Mycobacterial gene cuvA is required for optimal nutrient utilization and virulence. Infect Immun. 2014;82:4104–4117. doi: 10.1128/IAI.02207-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensinger DA, Boldon KM, Chen GY, Vincent WJB, Sherman K, Xiong M, et al. The Listeria monocytogenes PASTA kinase PrkA and its substrate YvcK are required for cell wall homeostasis, metabolism, and virulence. PLOS Pathog. 2016;12:e1006001. doi: 10.1371/journal.ppat.1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KJ, Higgins DE. A differential fluorescence-based genetic screen identifies Listeria monocytogenes determinants required for intracellular replication. J Bacteriol. 2013;195:3331–3340. doi: 10.1128/JB.00210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan-Thanh L, Gormon T. A chemically defined minimal medium for the optimal culture of Listeria. Int J Food Microbiol. 1997;35:91–95. doi: 10.1016/s0168-1605(96)01205-6. [DOI] [PubMed] [Google Scholar]
- Piez KA, Eagle H. The free amino acid pool of cultured human cells. J Biol Chem. 1957;231:533–45. [PubMed] [Google Scholar]
- Portnoy D, Jacks P, Hinrichs D. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne RJ, Lin WJ, Johnson Ea. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 1991;57:3046–8. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae CS, Geissler A, Adamson PC, Portnoy DA. Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect Immun. 2011;79:3596–3606. doi: 10.1128/IAI.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. Glutathione activates virulence gene expression of an intracellular pathogen. Nature. 2015;517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripio MT, Brehm K, Lara M, Suarez M, Vazquez-Boland JA. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romick TL, Fleming HP, Mcfeeters RF. Aerobic and anaerobic metabolism of Listeria monocytogenes in defined glucose medium. Appl Environ Microbiol. 1996;62:304–307. doi: 10.1128/aem.62.1.304-307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman LE, Qian Y, Eisele NA, Ng TM, Linden WA, van der, Monack DM, et al. Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. Elife. 2015;53:1689–1699. doi: 10.7554/eLife.13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J-D, Pereyre S, Archer KA, Burke TP, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci U S A. 2011;108:12419–12424. doi: 10.1073/pnas.1019041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J-D, Sotelo-Troha K, Moltke von J, Monroe KM, Rae CS, Brubaker SW, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–94. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J-D, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Geginat G, Liang C, Goebel W, Dandekar T, Fuchs TM. Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling. BMC Genomics. 2010;11:573. doi: 10.1186/1471-2164-11-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Stolz J, Scherer S, Fuchs TM. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol. 2009;191:2218–2227. doi: 10.1128/JB.01636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier HJ. Bacillus subtilis and Other Gram-Positive Bacteria. American Society of Microbiology; 1993. Biosynthesis of glutamine and glutamate and the assimilation of ammonia; pp. 281–298. [Google Scholar]
- Shen Y, Naujokas M, Park M, Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103:501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- Slaghuis J, Goetz M, Engelbrecht F, Goebel W. Inefficient replication of Listeria innocua in the cytosol of mammalian cells. J Infect Dis. 2004;189:393–401. doi: 10.1086/381206. [DOI] [PubMed] [Google Scholar]
- Stoll R, Goebel W. The major PEP-phosphotransferase systems (PTSs) for glucose, mannose and cellobiose of Listeria monocytogenes, and their significance for extra- and intracellular growth. Microbiology. 2010;156:1069–1083. doi: 10.1099/mic.0.034934-0. [DOI] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritzker J, Janda J, Schoen C. Growth, virulence, and immunogenicity of Listeria monocytogenes aro mutants. Infect Immun. 2004;72:5622–5629. doi: 10.1128/IAI.72.10.5622-5629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, O’Riordan MXD. Branched-chain fatty acids promote Listeria monocytogenes intracellular infection and virulence. Infect Immun. 2010;78:4667–4673. doi: 10.1128/IAI.00546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O’Riordan MXD. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J Bacteriol. 2012;194:5274–84. doi: 10.1128/JB.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Choi PH, Precit M, Delince M, Pensinger Da, Huynh TN, et al. The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function. Cell. 2014;158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Tadmor K, Pozniak Y, Burg Golani T, Lobel L, Brenner M, Sigal N, Herskovits Aa. Listeria monocytogenes MDR transporters are involved in LTA synthesis and triggering of innate immunity during infection. Front Cell Infect Microbiol. 2014;4:1–14. doi: 10.3389/fcimb.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H-N, Hodgson DA. Development of a synthetic minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 2003;69:6943–6945. doi: 10.1128/AEM.69.11.6943-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J Bacteriol. 2009;191:7157–64. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa K, Sekikawa I, Azuma I. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J Biochem. 1986;99:315–27. doi: 10.1093/oxfordjournals.jbchem.a135486. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland J-A, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheul A, Rombouts FM, Abee T. Utilization of oligopeptides by Listeria monocytogenes Scott A. Appl Environ Microbiol. 1998;64:1059–1065. doi: 10.1128/aem.64.3.1059-1065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu-Khac H, Miller KW. Regulation of mannose phosphotransferase system permease and virulence gene expression in Listeria monocytogenes by the EIItMan transporter. Appl Environ Microbiol. 2009;75:6671–6678. doi: 10.1128/AEM.01104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace N, Newton E, Abrams E, Zani A, Sun Y. Metabolic determinants in Listeria monocytogenes anaerobic listeriolysin O production. Arch Microbiol. 2017;0:0. doi: 10.1007/s00203-017-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Meadows JA. Carnitine in bacterial physiology and metabolism. Microbiology. 2015;161:1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SE, Duong H, Mao DP, Armstrong A, Rajan J, Miao EA, Aderem A. Generation of a Listeria vaccine strain by enhanced caspase-1 activation. Eur J Immunol. 2011;41:1934–1940. doi: 10.1002/eji.201041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Garelis NE, Peterson BN, Choi PH, Tong L, Woodward JJ, Portnoy DA. c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance, and osmoregulation. Mol Microbiol. 2017:1–23. doi: 10.1111/mmi.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Pollock AJ, Portnoy DA. The PAMP c-di-AMP Is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe. 2015;17:788–98. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker P, Fry FS, Curtis SK, Al-Khaldi SF, Mossoba MM, Yurawecz MP, Dunkel VC. Use of fatty acid profiles to identify food-borne bacterial pathogens and aerobic endospore-forming bacilli. J Agric Food Chem. 2005;53:3735–3742. doi: 10.1021/jf040458a. [DOI] [PubMed] [Google Scholar]
- Witte CE, Whiteley AT, Burke TP. Cyclic di-AMP Is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio. 2013;4:1–10. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, et al. Hexokinase Is an innate immune receptor for the detection of bacterial peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters JA, Hain T, Darji A, Hufner E, Wemekamp-Kamphuis H, Chakraborty T, Abee T. Identification and characterization of di- and tripeptide transporter DtpT of Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2005;71:5771–5778. doi: 10.1128/AEM.71.10.5771-5778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynosky-Dolfi MA, Snyder AG, Philip NH, Doonan PJ, Poffenberger MC, Avizonis D, et al. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]