Abstract
Analogues of the mRNA 5′-cap are useful tools for studying mRNA translation and degradation, with emerging potential applications in novel therapeutic interventions including gene therapy. We report the synthesis of novel mono- and dinucleotide cap analogues containing dihalogenmethylenebisphosphonate moiety (i.e. one of the bridging O atom substituted with CCl2 or CF2) and their properties in the context of cellular translational and decapping machineries, compared to phosphate-unmodified and previously reported CH2-substituted caps. The analogues were bound tightly to eukaryotic translation initiation factor 4E (eIF4E), with CCl2-substituted analogues having the highest affinity. When incorporated into mRNA, the CCl2-substituted dinucleotide most efficiently promoted cap-dependent translation. Moreover, the CCl2-analogues were potent inhibitors of translation in rabbit reticulocyte lysate. The crystal structure of eIF4E in complex with the CCl2-analogue revealed a significantly different ligand conformation compared to that of the unmodified cap analogue, which likely contributes to the improved binding. Both CCl2- and CF2- analogues showed lower susceptibility to hydrolysis by the decapping scavenger enzyme (DcpS) and, when incorporated into RNA, conferred stability against major cellular decapping enzyme (Dcp2) to transcripts. Furthermore, the use of difluoromethylene cap analogues was exemplified by the development of 19F NMR assays for DcpS activity and eIF4E binding.
INTRODUCTION
The cap is the characteristic feature present on the 5′ end of eukaryotic mRNAs. It consists of 7-methylguanosine connected via 5′-5′ triphosphate linkage to the first nucleotide of the mRNA (Figure 1A) (1). The cap structure is involved in mRNA recognition and metabolism including synthesis, transport, translation and turnover (2,3). Therefore, synthetic cap analogues have found a wide range of applications in biological studies, biotechnology and medicine, either as small molecule inhibitors of cap-dependent processes or as reagents for the modification of 5′ end of mRNA (4,5). Among variety of cap binding proteins, eIF4E (eukaryotic translation initiation factor) appears to be especially promising candidate for inhibition studies. eIF4E is a constituent of translation initiation complex and its binding to mRNA cap is the first event in protein biosynthesis. Several studies have shown that targeting translation initiation machinery is one of possible strategies for development of novel anti-cancer therapies (6–8). Although eIF4E is involved in general cap-dependent translation mechanism in eukaryotic cells, eIF4E overexpression leads to translational upregulation only of a subset of oncogenic transcripts (so called ‘weak’ mRNAs). Consequently, targeting eIF4E by various approaches has been shown to impede tumor growth with minimal or without any toxic effect on healthy cells (9–11).
Figure 1.
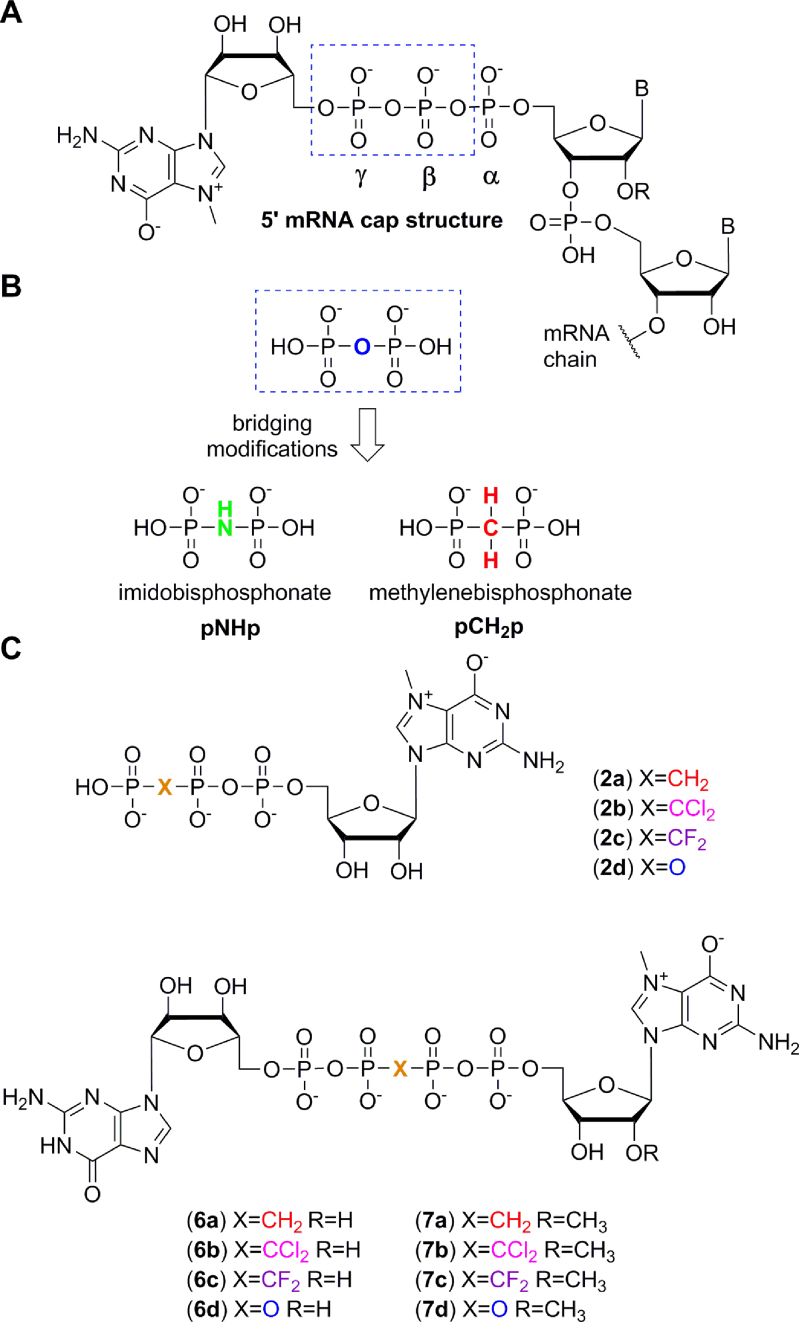
(A) Schematic structure of the mRNA 5′-cap. (B) Imidodiphosphonate (pNHp) and methylenebisphosphonate (pCH2p) are previously reported substitutions of bridging oxygen in the mRNA 5′-cap. (C) Structures of the mono- and dinucleotide cap analogues used in this study, including newly synthesized pCCl2p and pCF2p analogues.
On the other hand, capped mRNAs have been recently intensively investigated in the context of gene therapy applications and have already entered clinical trials (12–14). For example, it has been shown that dendritic cells can be targeted in vivo with intravenously administered RNA-lipoplexes to trigger release of interferon-α (14). Chemically modified cap analogues have been shown to increase mRNA half-life and translation levels in vivo, thereby improving the pharmacological properties of recombinant antigen-encoding RNAs (15). In particular, chemical modifications that stabilize cap analogues against specific decapping pyrophosphatases are of interest (5). Two major decapping enzymes have been identified in the cells: DcpS (decapping scavenger) and decapping complex Dcp1–Dcp2 (16–18). DcpS acts in the 3′-to-5′ decay by removing cap structures released after mRNA degradation by the exosome. In contrast, the Dcp1-Dcp2 complex, in which Dcp1 is the regulatory and Dcp2 the catalytic subunit, targets only capped transcripts thereby directing them to the 5′-to-3′ decay pathway. However, new enzymes displaying, at least in vitro, similar decapping activities are being continuously reported and their biological roles investigated (19–22). The modifications of the triphosphate chain that have been previously employed to prevent decapping include substitutions of non-bridging oxygen atoms with other heteroatoms (S, Se, BH3) or substitutions of bridging oxygen atoms with methylene (CH2) or imido (NH) groups (23–26) (Figure 1B). The substitutions in the bridging positions lack the P-stereocenter, which is created in the case of non-bridging substitutions, and thus are easier to produce and purify. Methylenebisphosphonate- (pCH2p) and imidodiphosphonate (pNHp) isosteres of pyrophosphate are widely used as tools in various binding and enzymatic activity studies (27–33). In general, pNHp is a closer mimic of pyrophosphate, both in respect to the pKa of phosphate groups and to geometry. However, it suffers from hydrolytic instability, especially in acidic conditions (6,34). On the other hand, the pCH2p moiety is hydrolytically stable but to larger extent affects the pKa of neighboring phosphates and cannot be engaged in H-bond interactions (31). This may result in disruption of the polar interactions within protein binding sites (35,36). The previously reported cap analogues containing methylenebisphosphonate (pCH2p) moiety were characterized by diminished affinity to cap-binding translation initiation factor 4E (eIF4E) (36,37). Consequently, these analogues were relatively poor inhibitors of cap-dependent translation and when incorporated into the 5′ end of mRNA, led to poorly translated transcripts. Therefore, in this work investigated another type of pyrophosphate isosters, dihalogenated bisphosphonates, i.e. difluoro- (pCF2p) and dichlorobisphosphonate (pCCl2p). We expected that the electronic effects associated with the electronegativity of fluorine and chlorine, which include lowering the pKa of phosphonates (30), might improve the protein binding properties of such cap analogues while maintaining their stability toward enzymatic decapping.
Two series of cap analogues were designed: mononucleoside triphosphates (2b–c) and dinucleoside tetraphosphates (6b–c, 7b–c, Figure 1C). In the dinucleotide series, the phosphate linkage was extended from three to four phosphate units in order to increase the binding affinities for eIF4E, as previously demonstrated (35,38). In both series, β/γ-bridging oxygen was chosen as the modification site to yield cap analogues with dichloromethylene and difluoromethylene linkers. The choice of modification site was dictated by synthetic availability and assumption that it could confer stability towards both cap-specific pyrophosphatases DcpS and Dcp2. In the dinucleotide series the ‘Anti-Reverse Cap Analogs’’ (ARCA) (39,40) containing an additional 2′-O-methylation in the 7-methylguanosine moiety (7b–c) were also synthesized. These cap analogues were dedicated for incorporation into mRNA 5′ ends by in vitro transcription. For comparison, previously reported pCH2p cap analogues (2a, 6a, 7a) and cap analogues with unmodified phosphate chain (2d, 6d, 7d) were used (Figure 1C).
MATERIALS AND METHODS
Synthesis
Procedures for the synthesis and characterization of the cap analogues are given in the Supporting Information.
pKa measurments
The nucleotides were dissolved in a mixture of H2O and D2O (86:14) at concentrations ranging from 3 to 20 mM. The pH of the samples was adjusted in steps of ∼0.5 pH units with 100 mM aqueous solution of HCl (containing 16% D2O) or 100 mM NaOH (16% D2O). The pH was measured using the HANNA HI1093B pH electrode at 20°C. NMR spectra were obtained on Bruker Avance III 500 MHz spectrometer equipped with a high stability temperature unit using 5 mm PABBO BB/19F-1H/D Z-GRD probe at 471 MHz (19F NMR) or 202.50 MHz (31P NMR). Both 19F and 31P NMR spectra were measured at 25°C. The 31P NMR chemical shifts were referenced to 20% phosphorus acid in D2O (δP = 0 ppm) as an external standard. The 19F NMR chemical shifts were reported to external 10 mM NaF in D2O (δF = −121.5 ppm). The obtained data were processed using OriginLab software and the pKa values were obtained by fitting to the experimental points a curve described by the following equation:
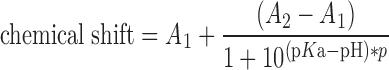 |
Protein preparation
Mouse eukaryotic initiation factor eIF4E (residues 28–217) and human DcpS (41) were expressed as described before (42). Human Dcp2 was expressed in Escherichia coli and purified as described previously (17).
DcpS enzymatic stability assays
A standard sample contained 40 μM cap analogue, 62.5 nM hDcpS in 0.4 ml of 50 mM Tris–HCl pH 7.6 containing 0.2 M KCl, 0.5 mM EDTA, 1.2 mM DTT at 37°C. 100 μl aliquots of the reaction mixture were collected after 15, 30, 60, 120 min and thermally deactivated at 100°C for 3 min. Mixtures containing no hDcpS were used as controls. Control samples were incubated for 120 min at 37°C and then incubated for 3 min at 100°C. All cap analogues in control samples (without DcpS added) were found to be stable after 120 min incubation in the experimental conditions. The samples were stored on ice and analyzed by RP-HPLC using a linear gradient from 0 to 50% of methanol in 0.1 M KH2PO4 pH 6.0 within 15 min (UV-detection at 260 nm). The experiment was performed in duplicate for each analogue.
Percentage hydrolysis was calculated from the following equation:
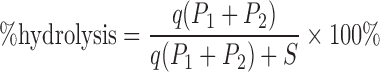 |
where S is absorbance of a substrate, P the absorbance of a cleavage product and q = ϵS/(ϵP1 + ϵP2), where ϵS is extinction coefficient for a substrate, ϵP the extinction coefficient for a product. The extinction coefficients used for calculations were as follows: ϵ = 22 600 M−1 cm−1 is (dinucleotide analogues containing guanosine and 7-methylguanosine), ϵ = 11 400 M−1 cm−1 is (mononucleotide analogues containing 7-methylguanosine), ϵ = 12 080 M−1 cm−1 is (mononucleotide analogues containing non-methylated guanosine). In the case of hydrolysis of dinucleotide analogues q = 0.9, for hydrolysis of mononucleotide analogues q = 1 and the equation is simplified to % hydrolysis = P/(P + S) × 100%.
Fluorescence titration experiments
The affinities of cap analogues for eIF4E was studied by time-synchronized titration method as described earlier (43). Fluorescence data were collected using an LS-55 spectrofluorometer (Perkin-Elmer Co.) and quartz cuvette (Hellma) with optical path length of 4 mm for absorption and 10 mm for emission. The experiments were performed at 20°C either in 50 mM HEPES/KOH buffer pH 7.20 containing 100 mM KCl, 0.5 mM EDTA and 1 mM DTT. Titration measurements were carried out by adding 1 μl aliquots of tested ligand to 1400 μl solution of 0.1 μM eIF4E. eIF4E fluorescence was excited at 280 nm (slit 2.5 nm, auto cut-off filter) and monitored at 340 nm (slit 4 nm, 290 nm cut-off filter), respectively. Data correction for sample dilution and inner filter effect were applied. Association equilibrium constants (KAS) were determined by fitting the theoretical dependence of fluorescence intensity in the function of total concentration of tested ligand, according to the equation described earlier (43). Each experiment was repeated from three to six times, the final association constants KAS were calculated as weighted average, with the weights taken from reciprocal standard deviations squared. Data were analyzed using Origin 6.0 software.
Inhibition of translation assays
Inhibition of cap-dependent translation in RRL (Flexi Rabbit Reticulocyte Lysate,Promega) by cap analogues, analysis of their stability in RRL and calculation of IC50 values were performed as described previously. Briefly, the in vitro translation reaction was performed in 12.5 μl volume for 60 min at 30°C, in conditions determined for cap-dependent translation. The reaction mixture was pre-incubated for 60 min at 30°C prior to addition of dinucleotide cap analogue (inhibitor) and m27,3′-OGpppG-capped luciferase mRNA to start the translation reaction. To analyse stability of studied cap analogues in rabbit reticulocyte lysate translation system, each cap analogue was incubated in a translation mixture for 60 min at 30°C and then luciferase mRNA was added to start translation. Reactions were stopped by chilling on ice and the luciferase activity was measured in a luminometer (Glomax, Promega).
Translation efficiency in RRL system
Capped, polyadenylated luciferase mRNAs were synthesized in vitro on a dsDNA template (amplified by PCR reaction) that contained: SP6 promoter sequence of DNA-dependent RNA polymerase, 5′-UTR sequence of rabbit β-globin, the entire firefly luciferase ORF and a string of 31 adenosines. A typical in vitro transcription reaction mixture (40 μl) contained: SP6 transcription buffer (ThermoFisher Scientific), 0.7 μg of DNA template, 1 U/μl RiboLock Ribonuclease Inhibitor (ThermoFisher Scientific), 0.5 mM ATP/CTP/UTP and 0.1 mM GTP and 0.5 mM dinucleotide cap analogue (molar ratio cap analog:GTP 5:1). The reaction mixture was preincubated at 37°C for 5 min before addition of SP6 RNA polymerase (Fermentas) to final concentration 1 U/μl and reaction was continued for 45 min at 37°C. After incubation, reaction mixtures were treated with DNase RQ1 (Promega), in transcription buffer, for 20 min at 37°C at concentration 1 U per 1 μg of template DNA. RNA transcripts were purified using NucAway Spin Columns (Ambion), integrity of transcripts was checked on a non-denaturating 1% agarose gel and concentrations were determined spectrophotometrically. A translation reaction in RRL was performed in 10 μl volume for 60 min at 30°C, in conditions determined for cap-dependent translation. A typical reaction mixture contained: 40% RRL lysate, mixture of amino acids (0.01 mM), MgCl2 (1.2 mM), potassium acetate (170 mM) and 5′-capped mRNA. Four different concentrations of each analysed transcript were tested in in vitro translation reaction. Activity of synthesized luciferase was measured in a luminometer.
In vitro RNA decapping assay with hDcp2
RNAs initiated with cap analogues were synthesized by in vitro transcription with SP6 polymerase and were generated on template of annealed oligonucleotides (ATACGATTTAGGTGACACTATAGAAGAAGCGGGCATGCGGCCAGCCATAGCCGATCA and TGATCGGCTATGGCTGGCCGCATGCCCGCTTCTTCTATAGTGTCACCTAAATCGTAT), which contain SP6 promoter sequence (ATTTAGGTGACACTATAGA) and encodes modified 35-nt sequence (GAAGAAGCGGGCATGCGGCCAGCCATAGCCGATCA) (44). Typical in vitro transcription reaction (20 μl) was incubated in 40°C for 3 h and contained: RNA Pol Buffer (New England BioLabs),1 U/μl SP6 polymerase, 1 U/μl RiboLock RNase Inhibitor, 0.5 mM ATP/CTP/UTP, 0.125 mM GTP, 1.25 mM cap analogue and 0.1 μM annealed oligonucleotides as a template. To prepare uncapped RNAs, in vitro transcription reaction was performed as before, but with 0.5 mM GTP. Following 3 h incubation, 1 U/μl DNase I (ThermoFisher Scientific) was added and incubation was continued for 30 min at 37°C, after that EDTA was added to 25 μM final concentration. RNAs were purified using RNA Clean & Concentrator-25 (Zymo Research) and their quality was checked on 15% acrylamide/7 M urea 1× TBE gels, whereas concentrations were determined spectrophotometrically.
Moreover, to obtain homogenous 3′ end of synthetized RNAs, obtained 35-nt long transcripts (1 μM) (capped RNAs were 36-nt long) were incubated with 1 μM DNAzyme 10–23 (TGATCGGCTAGGCTAGCTACAACGAGGCTGGCCGC) in 50 mM MgCl2 and 50 mM Tris–HCl pH 8.0 for 1 h at 37°C (44), what allow to produce a 3′ homogenous 25-nt RNAs (capped RNAs were 26-nt long). Prepared transcripts were purified with RNA Clean & Concentrator-25 (Zymo Research) and DNase on-column. Their quality and concentration were checked as above.
In vitro decapping activity was measured with obtained transcripts after DNazyme 10–23 treatment. 100 ng of capped RNAs were subjected to digestion with 630 nM hDcp2 in decapping buffer (50 mM Tris–HCl pH 8.0, 50 mM NH4Cl, 0.01% NP-40, 1 mM DTT, 5 mM MgCl2 and 2 mM MnCl2). Reactions were performed at 37°C for the indicated times and terminated by adding equal volume of loading dye (5 M urea, 44% formamide, 20 mM EDTA, 0.03% bromophenol blue, 0.03% xylene cyanol). RNAs after hDcp2 treatment were resolved electrophoretically on denaturing 15% acrylamide / 7 M urea 1× TBE gel and were stained with SYBR Gold (Invitrogen) and visualized using a Typhoon FLA 9500 (GE Healthcare).
Cell culture and in vivo measurements of translation efficiency
mRNAs encoding firefly luciferase for measurement of translational efficiency in HeLa cells were synthetized by in vitro transcription with SP6 polymerase using pJET_luc_128A plasmid digested with AarI (ThermoFisher Scientifics) as a template. This plasmid was obtained by cloning sequence encoding firefly luciferase, two repeats of β-globin 3′UTR and 128 adenines from hRLuc-pRNA2(A)128 plasmid (45) into pJET1.2 vector (ThermoFisher Scientific). Typical in vitro transcription reaction (20 μl) was incubated in 40°C for 3 h and contained: RNA Pol Buffer,1 U/μl SP6 polymerase, 1 U/μl RiboLock RNase Inhibitor, 0.5 mM ATP/CTP/UTP, 0.125 mM GTP, 1.25 mM cap analogue and 0.1 μg of digested plasmid as a template. Following 3 h incubation, 1 U/μl DNase I (ThermoFisher Scientific) was added and incubation was continued for 30 min at 37°C, after that EDTA was added to 25 μM final concentration. Obtained mRNAs were purified with NucleoSpin RNA Clean-up XS (Macherey-Nagel) and their quality was checked on native 1.2% 1× TBE agarose gel, whereas concentrations were determined spectrophotometrically.
Human cervical carcinoma HeLa cells were grown in DMEM (Gibco) (with 2 mM l-glutamine (Gibco)) supplemented with 10% FBS (Sigma-Aldrich) and 1% penicillin/streptomycin (Gibco) at 5% CO2 and 37°C. In a typical experiment 24 h before transfection, 104 cells were seeded in 100 μl medium without antibiotics per well of 96-well plate. Cells in each well were transfected for 1 h using 0.3 μl Lipofectamine MessengerMAX Transfection Reagent (Invitrogen), 0.1 μg mRNA and 10 μl Opti-MEM (Gibco). After transfection, cells were washed with PBS and supplemented with fresh medium without antibiotics, and grown for the indicated times. Cell lysis was performed using Luciferase Assay System (Promega) and bioluminescence flux was measured with the Synergy H1 microplate reader (BioTek). Obtained firefly luminescence was depicted as a function of time. Total protein expression was calculated as integral of the line segments connecting obtained data points for firefly luminescence.
Crystallography
Murine eukaryotic translation initiation factor 4E (eIF4E, residues 28–217) was expressed and purified as described previously (42) with an additional gel filtration purification step. The protein was concentrated to 4.75 mg/ml (Amicon 10 000 molecular weight Ultra Centrifugal Filter Device) in a gel filtration buffer (20 mM HEPES pH 7.2, 100 mM KCl, 0.5 mM EDTA, 2 mM DTT) and incubated with 0.5 mM cap analogues at room temperature for 15 min. Thin plate-like crystals were obtained using hanging drop vapor diffusion method at 18°C. Diffraction-quality crystals were formed in 13% PEG 5000 monomethyl ether, 0.1 M Tris–HCl pH 8.0 solution for eIF4E-m7GppppG complex and in 15% PEG 3350, 2% 1,4-dioxane, 0.1 M Tris–HCl pH 8.0 for eIF4E-m27,2’OGppCCl2ppG within 1 day. Harvested crystals were transferred to the cryoprotectant solution (reservoir solution and glycerol 2:1 v/v) and flash frozen in liquid nitrogen. The X-Ray diffraction data sets were collected at 100 K at Bessy II (Helmholtz-Zentrum Berlin, Germany) Beamline 14.1 using a Dectris PILATUS detector and processed using XDS (46) with XDSAPP GUI (47). Structures were solved by Molecular Replacement using Phaser (48) and chain B of eIF4E-m7GpppG complex structure (PDB id: 1L8B) as a search model. Ligand dictionaries were generated using ProDRG (49) and JLigand (50). The model building and ligand fitting was performed in Coot (51), and the structures were refined using phenix.refine (52). The structures contain four copies of the protein in the asymmetric unit (chains A-D). High R-factor values result from significant disorder of two outermost protein molecules (chain C and D) as indicated by the wwPDB Validation report. Nonetheless, the electron density maps are very well defined for the majority of chain A and chain B residues and for the cores of chain C and D (Supplementary Figure S10). Data collection and refinement statistics are summarized in Supplementary Table S4.
19F NMR binding assays
Decoupled 19F{31P} NMR experiments were recorded using a Bruker AVIII 600 with BB-F/1H Prodigy N2 cryoprobe operating at 298 K using 5 mm diameter NMR tubes (Norell). 19F 90o pulse lengths were 15 μs and spectra were typically obtained using 128 scans for displacement experiments and a recovery delay of 2 s. Data were processed with 5 Hz Lorentzian line broadening using TopSpin 3.1 software (Bruker) and were referenced to the internal trifluoroacetic acid (TFA) standard (0.025 mM, –75.45 ppm). Samples typically contained 0.015 mM eIF4E and 0.025 mM m7GppCF2ppG in HEPES (50 mM, pH 7.2) containing 100 mM KCl, 0.5 mM EDTA, 1 mM DTT and 10% D2O. Increasing amounts of displacing ligand (m7GppCCl2p and m7GppCCl2ppG) were added from stock solutions. The protein–ligand complex concentration [PL], which is equal to the concentration of m7GppCF2ppG displaced from the protein, was then plotted against ligand concentration (L0). The apparent association constant values Kapp (defined as the inverse concentration of the free ligand (1/L) when 50% of the protein is bound to the ligand in the presence of the reporter ligand) were determined by fitting the experimental points to a theoretical curve [PL] = P0L0/(1/Kapp + L0).
The actual association constants were calculated using equations derived from equations previously described by Nikolovska-Coleska et al. (53) and Leung et al. (54). When concentration of the reporter ligand R0 and its association constant (Krep) are known, the correlation between apparent association constant Kapp of the ligand and the actual association constant KAS is given by equation: KAS = Kapp([R50]Krep + [P]Krep+1), where [R50] is the free reporter ligand concentration when 50% of the ligand is displaced from the enzyme and [P] is the free enzyme concentration in the presence of reporter ligand. [R50] is described as: [R50] = R0 – 0.5(P0 – [P]), where P0 is the initial (i.e. total) concentration of enzyme and R0 is the total concentration of the reporter ligand. [P] is given by equation: [P] = -0.5(1/Krep + R0 – P0) + 0.5 ((1/Krep + R0 – P0)2 + 4P0/Krep)0.5. For calculations following values were used: P0 = 15 μM, R0 = 25 μM, Krep = 57.6 μM−1.
Monitoring enzymatic reactions by 19F NMR
The reactions were run in standard 5 mm NMR tubes and contained 2 mM m7GppCF2ppG (6c) in 600 μl of DcpS buffer (50 mM Tris–HCl pH 7.9 containing 150 mM NaCl, 2 mM DTT and 15% D2O). Just before placing in the NMR apparatus 20 μl of 5 μM hDcpS enzyme were added to the samples (to a final concentration of 160 nM). The reactions were monitored at 30°C using a standard 19F NMR pulse sequence (16 transients) in 2.7 min intervals. The spectra were recorded on 400 MHz spectrometer using 5 mm 4NUC probe. The resonances were assigned based on reference 19F NMR spectra of pure substrate (m7GppCF2ppG, 6c) and product (GppCF2p, 4c), respectively.
RESULTS
Synthesis
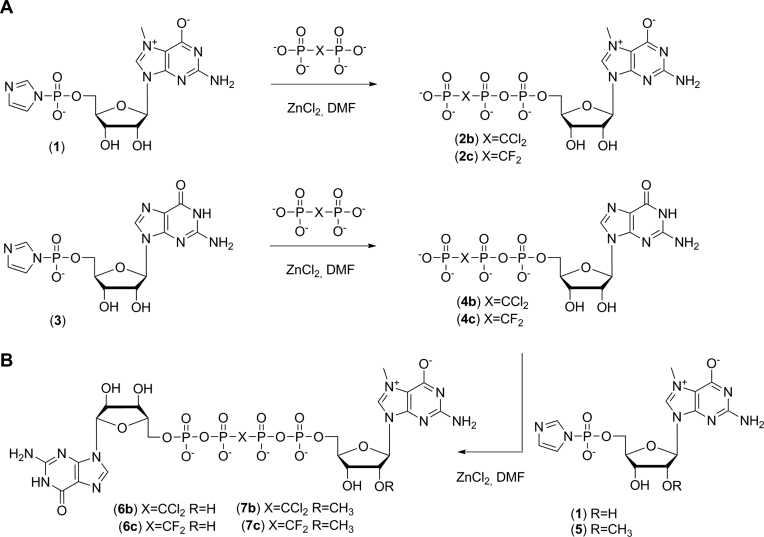
The mononucleotide cap analogues 2b-c were synthesized by the coupling of an appropriate triethylammonium dihalomethylenebisphosphonate with an imidazole derivative of a 7-methylguanosine monophosphate 1 in DMF in the presence of ZnCl2 (40,55). Similarly, coupling of an appropriate dihalomethylenebisphosphonate (pCF2p or pCCl2p) with an imidazole derivative of a guanosine monophosphate 3 yielded modified guanosine triphosphates 4b–c, which were then further coupled with an imidazole derivative of either 7-methyl (1) or 7,2’-O-dimethylguanosine monophosphate (5) to yield dinucleotide cap analogues 6b–c and 7b–c, respectively (Scheme 1).
Scheme 1.
Synthesis of dihalogenated cap analogues.
Biophysical and spectroscopic characterization
To assess the influence of various bridging oxygen substitutions on the electronic properties of the tri- or tetraphosphate bridge within the cap structure, we first compared the 31P NMR chemical shifts (δP) for cap analogues 2a–d and 6a–d (Table 1, values measured in D2O). The δP values for difluoromethylene, followed by dichloromethylene-containing caps were closer to the δP values of the pyrophosphate-containing analogues than to those of methylene-containing caps. This observation confirmed our suggestion that, the introduction of halogen substituents at the methylene moiety might lead to closer mimics of pyrophosphate. Since the chemical shifts of ionizable groups may significantly depend on the pH, we also investigated the pH dependence of δP for mononucleotides 2a–d (Supplementary Figure S1). The chemical shifts for β and γ phosphates were highly dependent on the pH in the range from 5 to 9, but the general tendency observed for the samples prepared in D2O (initial measurements of 31P NMR spectra) was preserved. Furthermore, this experiment enabled the determination of pKa4 values for analogues 2a–d (Table 1, Supplementary Table S1). The pKa value was the highest for methylene analogue 2a (pKa4 = 8.6) and decreased with the addition of chlorine and fluorine substituents (pKa4 = 7.4 and 6.4 for 2b and 2c, respectively). The unmodified analogue 2d was characterized by a pKa4 value of 6.9, placing it between dichloromethylene- and difluoromethylene analogues 2b and 2c. These data were in good agreement with the previously reported values for adenosine 5′-triphosphate analogues (14) (Supplementary Table S2, Figure S3).
Table 1. 31P NMR chemical shifts and pKa4 values for modified cap analogues.
| Cap analogue | 31P NMR chemical shifts—δP [ppm] | pKa4 | |||
|---|---|---|---|---|---|
| Pα | Pβ | Pγ | Pδ | ||
| m7GppCH2p (2a) | −10.16 | 10.21 | 15.03 | − | 8.63 ± 0.01 |
| m7GppCCl2p (2b) | −10.58 | 0.88 | 7.90 | − | 7.32 ± 0.07 |
| m7GppCF2p (2c) | −10.90 | −3.51 | 3.49 | − | 6.28 ± 0.07 |
| m7Gppp (2d) | −10.56 | −22.22 | −9.83 | − | 6.56 ± 0.03 |
| m7GppCH2ppG (6a) | −10.76 | 7.75 | 7.75 | −10.76 | − |
| m7GppCCl2ppG (6b) | −10.81 | −1.41 | −1.41 | −10.81 | − |
| m7GppCF2ppG (6c) | −11.08 | −6.37 | −6.37 | −11.08 | − |
| m7GppppG (6d) | −10.47 | −22.04 | −22.04 | −10.47 | − |
Binding to eIF4E
Initiation of translation is the rate limiting step of protein biosynthesis and involves binding of the mRNA cap by the eIF4F pre-initiation complex. Specifically, the cap structure is recognized by eIF4E, which is a part of the eIF4F assembly, therefore high affinity for eIF4E is a desirable feature that could indicate an increase in translational potency of mRNAs with chemically modified cap analogues at their 5′ ends. On the other hand, eIF4E is often overexpressed in cancer cells and is a target of interest in anti-cancer therapy (6–8,56), thus high-affinity ligand binding to eIF4E is also required for obtaining small molecule inhibitors of translation. Targeting eIF4E with cap analogues is one of the potential strategies to counteract its overexpression in cancer cells. Therefore, eIF4E-cap interaction is one of the most important properties determined for novel cap analogues. The dihalomethylene cap analogues (2b–c, 6b–c and 7b–c) were evaluated as eIF4E binders by determining equilibrium association constants (KAS) using a fluorescence titration method (Table 2, Supplementary Figure S3) (43). In the case of mononucleotide triphosphates (2a–d), the dihalomethylene cap analogues 2b and 2c were characterized by KAS values similar and 1.5-fold higher compared to the unmodified parent analogue 2d (KAS of 188.1 and 115 μM−1 for 2b and 2c, respectively, and 108.7 μM−1 for 2d). In contrast, the methylene analogue 2a exhibited a KAS 6-fold lower than 2d (KAS = 19.5 μM−1). The binding affinities for the dinucleotide tetraphosphates 6a–d and 7a–d revealed the same general tendency however, with smaller differences between the methylene and dihalogenomethylene modifications. As in the case of mononucleotides, the highest affinity among the phosphate-modified dinucleotides was observed for the dichloromethylene compounds 6b and 7b, which had KAS values only slightly lower than those of the phosphate-unmodified parent dinucleotides 6d and 7d (KAS of 77.9 and 82.8 μM−1 for 6b and 7b, respectively; 102.8 μM−1 and 99.8 μM−1 for 6d and 7d, respectively). The difluormethylene analogues 6c and 7c were weaker binders (KAS of 57.6 and 50.0 μM−1 for 6c and 7c, respectively), with KAS values almost 2-fold lower compared to those of the unmodified parent dinucleotides (6d and 7d, respectively) and only slightly higher than the affinities of the methylene analogues 6a and 7a (KAS of 48.6 μM−1 and 44.0 μM−1 for 6a and 7a, respectively).
Table 2. Equilibrium association constants (KAS) for cap analogue—eIF4E complexes as measured by fluorescence quenching titration experiments.
| K AS [μM−1] | |||
|---|---|---|---|
| X | m7GppXp | m7GppXppG | m27,2′-OGppXppG |
| O | 108.7 ± 4.0a | 102.8 ± 4.4a | 99.8 ± 6.0b |
| CH2 | 19.5 ± 1.4 | 48.6 ± 2.1c | 44.0 ± 3.2c |
| CCl2 | 188.1 ± 7.4 | 77.9 ± 1.7 | 82.8 ± 1.1 |
| CF2 | 115.2 ± 5.8 | 57.6 ± 0.8 | 50.0 ± 1.8 |
Crystallography
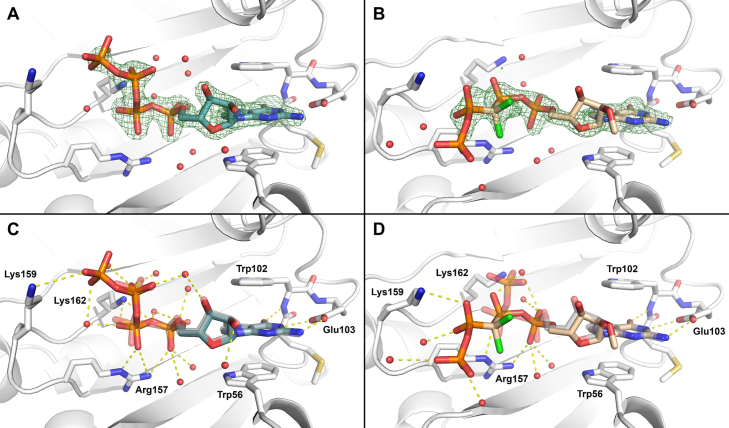
To further investigate the role of a pCCl2p moiety in the stabilization of cap-eIF4E interactions, we obtained co-crystal structures of a murine eIF4E (residues 28−217) in a complex with the dichloromethylene analogue 7b (m27,2’-OGppCCl2ppG) (PDB id: 5J5Y) and a cap analogue 6d unmodified in the tetraphosphate bridge (m7GppppG) (PDB id: 5J5O) determined to 1.75 Å and 1.87 Å resolution, respectively. In both complexes the 7-methylguanine is stacked between two tryptophan sidechains (Trp56 and Trp102) and hydrogen bonded to the carboxylic group of Glu103. The ribose ring adopts a C3’-endo conformation so that the phosphorus atoms of the two downstream phosphates (δ and γ for tetraphosphates) lay nearly coplanar with the 7-methyguanine, participating in ionic interactions with the positively charged side chains of Lys162 and Arg157 and an extensive net of hydrogen bonds mediated by water molecules (Figure 2A and B).
Figure 2.
(A) Close-up view of the ligand binding in the crystal structure of eIF4E in complex with m7GppppG (6d) (PDB id: 5J5O, including simulated annealing omit Fo − Fc electron density map contoured at 3σ). (B) Close-up view of the ligand binding in the crystal structure of eIF4E in complex with m27,2’OGppCCl2ppG (7b) (PDB id: 5J5Y, including a simulated annealing omit Fo − Fc electron density map contoured at 3σ). (C) Superposition of crystal structures of eIF4E in complex with m7GppppG (6d) (PDB id: 5J5O) and m7GpppG (PDB id: 1L8B) reveal similar binding modes for both ligands. (D) Superposition of crystal structures of eIF4E in complex with m27,2’OGppCCl2ppG (7b) (PDB id: 5J5Y) and m7GpppG (PDB id: 1L8B) reveal different conformation of the tetraphosphate bridge of analogue 7b in comparison to m7GpppG.
The structure of eIF4E in a complex with m7GppppG (6d) reveals a very similar ligand binding mode as in the previously reported eIF4E-m7GpppG complex (PDB id: 1L8B, overlay in Figure 2C) (43). For 6d, similar to m7GpppG, the oligophosphate bridge continues in a perpendicular direction toward the C-terminal loop, resulting in strong ionic contacts between a β-phosphate and Lys162 as well as the formation of several hydrogen bonds with water molecules. An additional phosphate in 6d (α-phosphate) forms an ionic contact with Lys159, which is probably responsible for the increase of the association constant, compared to m7GpppG.
On the other hand, in the structure of eIF4E in complex with m27,2’OGppCCl2ppG (7b), the tetraphosphate bridge of 7b adopts an essentially different conformation (Figure 2D). The conformation of analogue 7b is likely forced by steric hindrance between the chlorine atom and C5’ of 7-methylguanosine. To avoid a clash, the bridging β/γ-CCl2- carbon atom of 7b is located approximately coplanar with γ and δ phosphates and the β-phosphate continues outside of the binding pocket, impairing ionic interactions with Lys162. However, the β-phosphate moiety forms ionic contact with Lys159, thus compensating the energetic loss to some extent. Finally, in all the 6d, 7b, and m7GpppG co-crystal structures the second nucleoside is disordered, so that the electron density was not defined for that part of the ligands, suggesting that they do not form any specific and strong interactions with the protein.
Susceptibility to enzymatic hydrolysis by hDcpS
The utility of cap analogues for studies in complex biological mixtures and in vivo is quite often hampered by their poor stability arising from hydrolysis by cellular pyrophosphatases (38). In humans, short capped RNA oligonucleotides and mono- and dinucleotide caps are targets for a human decapping scavenger enzyme (hDcpS), which is a member of HIT family pyrophosphatases (57) and cleaves the triphosphate chain of a dinucleotide cap between the γ and β phosphates (Supplementary Figure S4) (58). The influence of a dihalomethylene moiety on the enzymatic susceptibility of analogues 2b,c, 6b,c and 7b,c to hDcpS was studied using an HPLC assay (37,38,59). The enzymatic reaction progress was analyzed at four different time points (15, 30, 60 and 120 min). The previously studied unmodified parent compounds and their corresponding methylene analogues were included in the assay as references. The results after 1h are shown in Table 3, whereas data for all the time points are summarized in the Supporting Information (Supplementary Figure S5, Table S3).
Table 3. Enzymatic hydrolysis of cap analogues by hDcpS.
| % hydrolysis after 1 h incubation with hDcpSb | |||
|---|---|---|---|
| X | m7GppXp | m7GppXppG | m27,2′-OGppXppG |
| O | 43.9 ± 3.5 | 90.0 ± 2.3 | – |
| CH2a | 0 | 0 | 0 |
| CCl2 | 1.1 ± 0.3 | 0 | 0 |
| CF2 | 6.9 ± 0.0 | 50.1 ± 1.8 | 8.1 ± 0.7 |
aData from Rydzik et al. (20).
bReactions were carried out using 0.04 mM cap analogue and 62.5 nM hDcpS in 50 mM TRIS buffer pH = 7.5 containing 0.2 M KCl, 0.5 mM EDTA and 1.2 mM DTT.
In agreement with previous reports (37,38,59), we found that dinucleotide analogues are generally more susceptible to hydrolysis by hDcpS than mononucleotides. While the unmodified analogue 44% of m7Gppp (2d) was hydrolyzed after 1 h incubation with hDcpS, the dinucleotide cap m7GppppG (6d) was hydrolyzed to ∼90%. Under the same conditions, the modified mononucleotides were hydrolyzed significantly slower: difluoromethylene analogue (2c) was hydrolyzed in 7%, whereas dichloromethylene (2b) and methylene (2a) analogues were virtually resistant toward enzymatic hydrolysis (Table 3). In the dinucleotide series, the methylene (6a) and dichloromethylene (6b) analogues were also resistant to hDcpS, whereas the difluoromethylene compound (6c) was a fairly good substrate for hDcpS (approximately 50% cleavage after 1 h).
The ARCA compounds, i.e. those bearing an additional methyl group in the 2′-O position of the ribose ring, are known to be less susceptible to hDcpS than their unmethylated counterparts (35–37). Accordingly, compound 7c was hydrolyzed slower than 6c (8% and 50%, after 1 h, respectively).
Susceptibility to decapping enzyme hDcp2
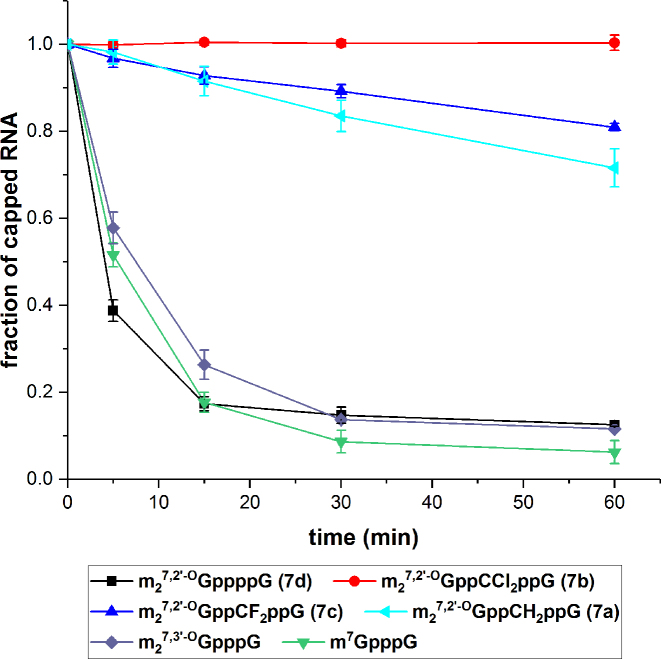
Decapping is the first irreversible event in 5′-to-3′ mRNA degradation pathway. mRNA decapping is executed by Dcp1-Dcp2 complex, in which Dcp2 is the catalytically active subunit and a member of Nudix (nucleoside diphosphate linked to another moiety X) hydrolase superfamily of proteins. In contrast to DcpS, hDcp2 catalyzes cleavage between α and β phosphates in the 5′–5′-triphosphate chain, releasing 7-methylguanosine diphosphate and 5′ monophosphorylated mRNA (Supplementary Figure S4) (60). The decapped mRNA is no longer available for translational machinery and is subjected to further degradation by Xrn1, main cytoplasmic 5′-3′ exonuclease (3). Increased resistance of mRNA cap to hDcp2 has been shown to extend mRNA half-life in cells, thereby prolonging and increasing overall protein expression. In vitro susceptibility to decapping of transcripts capped with analogs 7a-d was assayed by incubation of short capped transcripts with hDcp2, followed by resolution of the reaction mixtures on high resolution polyacrylamide gel in order to separate capped and decapped RNAs (Supplementary Figure S6). Gels were stained with fluorescent dye, visualized and intensity of bands corresponding to both versions of RNA was determined densitometrically. Figure 3 depicts the fraction of capped RNA remaining at various timepoints, while Table 4 shows RNA decapping susceptibilities, defined as a percentage of decapped RNA to total RNA at a timepoint 15 min. As expected, the presence of methylene or dihalomethylene moiety in the cap's tetraphosphate chain stabilized the transcripts against Dcp2 (Table 4, Figure 3). The decapping susceptibilities for RNA capped with compounds 7a, 7b and 7c were 8 ± 3, 0 ± 1 and 7 ± 2, respectively, compared to 74 ± 3, and 83 ± 2 for reference analogs m27,3′-OGpppG and m27,2′-OGppppG (7d), respectively. In particular, RNA capped with the dichloro-containing analogue 7b was fully resistant against hDcp2 under the assay conditions.
Figure 3.
Susceptibility of differently capped RNAs to hDcp2. The data represent quantified results from duplicate experiments (gels from the decapping assay are shown in Supplementary Figure S6). Fraction of capped RNA was calculated as a ratio of capped RNA band intensity to total RNA at a given time. All data were normalized to time 0’.
Table 4. Enzymatic hydrolysis of capped RNAs by hDcp2.
| Cap analogue | Decapping susceptibility (%)a,b |
|---|---|
| m27,2′-OGppppG (7d) | 83 ± 2 |
| m27,2′-OGppCH2ppG (7a) | 8 ± 3 |
| m27,2′-OGppCCl2ppG (7b) | 0 ± 1 |
| m27,2′-OGppCF2ppG (7c) | 7 ± 2 |
| m27,3′-OGpppG | 74 ± 3 |
| m7GpppG | 82 ± 2 |
a% of RNA decapping after 15 min incubation with hDcp2.
bReactions were carried out using 100 ng of capped RNA and 630 nM hDcp2 50 mM Tris-HCl pH 8.0, 50 mM NH4Cl, 0.01% NP-40, 1 mM DTT, 5 mM MgCl2 and 2 mM MnCl2.
Inhibition of translation in RRL system
Following the promising results from eIF4E binding and enzymatic stability experiments (susceptibility to hDcpS and to hDcp2) we investigated the modified cap analogues as inhibitors of cap-dependent translation. The IC50 values for cap analogues 2a–c, 6a–c and 7a–c were determined in the rabbit reticulocyte lysate (RRL) system utilizing a luciferase reporter gene based assay (38) (Table 5). Two set of experiments were conducted. In the first, the cap analogue of interest and a reporter mRNA were added simultaneously to the RRL system and the mixture was incubated for 60 min at 30°C prior to the measurement (set A). In the second, the cap analogue was preincubated in the RRL system for 60 min prior to the addition of a reporter mRNA and then mixture was incubated for a further 60 min at 30°C before the measurement (set B) (Supplementary Figures S7 and S8). The second set of experiments took into account enzymatic stability of the studied compounds through their extended exposure to cellular lysate of reticulocytes (38). The pCH2p analogues (2a, 6a and 7a) were characterized by the highest IC50 values, which is in agreement with previous findings (61) and likely results from their low binding affinities to eIF4E. A clear improvement in the IC50 values was observed in the case of pCCl2p analogues compared to their pCH2p and unmodified counterparts. Consistent with the highest binding affinities for eIF4E, the pCCl2p-containing caps (2b, 6b and 7b) were characterized by the lowest IC50 values. Unlike analogues unmodified in the phosphate bridge (2d, 6d and 7d), the pCH2p and pCCl2p analogues (2a–b, 6a–b and 7a–b) were relatively stable in RRL since they retained a similar activity both with and without pre-incubation. In particular, the pCCl2p analogues 2b and 7b were identified as potent inhibitors of translation in the RRL system (IC50 of 0.97 and 0.95 μM for 2b and 7b respectively).
Table 5. IC50 values for the inhibition of translation in the rabbit reticulocyte lysate system by cap analogues.
| IC50 [μM] | ||||||
|---|---|---|---|---|---|---|
| X | m7GppXp | m7GppXppG | m27,2′-OGppXppG | |||
| Inhibitiona | Stabilitya | Inhibitiona | Stabilitya | Inhibitiona | Stabilitya | |
| O | 3.52 ± 0.25 | 7.57 ± 0.27 | 5.0 ± 0.5b | > 20b | 2.8 ± 0.2b | 4.2 ± 0.1b |
| CH2 | 8.09 ± 1.18 | 9.87 ± 0.84 | 4.6 ± 0.6b | 3.8 ± 0.2b | 2.8 ± 0.2b | 2.9 ± 0.3b |
| CCl2 | 0.97 ± 0.13 | 1.37 ± 0.01 | 1.34 ± 0.15 | 1.52 ± 0.10 | 0.95 ± 0.10 | 1.23 ± 0.11 |
| CF2 | 1.81 ± 0.13 | 2.43 ± 0.07 | 2.95 ± 0.32 | 4.99 ± 0.41 | 2.16 ± 0.23 | 3.59 ± 0.28 |
aSet A and B refer to the experiments conducted without (A) and with (B) cap analogue preincubation in RRL system.
bData from Rydzik et al. (20).
Synthesis and translation efficiency of 5′-capped mRNAs
Cap is an integral part of mRNA and therefore we were interested in evaluating how introduction of dichloro- and difluoromethylene modifications into phosphate chain would influence the efficiency of translation of mRNAs capped with such analogues. For these experiments, transcripts encoding firefly luciferase and capped with ARCAs 7a–d or a commercially available ARCA (m27,3′-OGpppG) were synthesized by in vitro transcription using SP6 polymerase. The translation efficiencies in rabbit reticulocyte lysate system (RRL) for differently capped RNAs were measured by luminometry after 1 h and referred to the control mRNA capped with m7GpppG (Table 6, Supplementary Figure S9). Additionally, the level of cap-independent translation was estimated using transcripts capped with the non-functional analogue ApppG. The translation efficiencies of m27,3’-OGpppG-capped mRNAs were 1.6-fold higher than that of m7GpppG-capped RNA, which is in agreement with previously published work and attributed to the ‘‘anti-reverse’’ properties of m27,3’-OGpppG (40). The unmodified tetraphosphate ARCA analogue 7d had slightly higher translation efficiency than m27,3’-OGpppG, which also agrees with previous observations and has been explained by an increased affinity for eIF4E conferred by the additional phosphate group (40,56). Despite higher affinity to eIF4E, transcripts capped with the pCH2p analogue 7a were translated less efficiently than mRNA capped with m27,3’-OGpppG however, mRNAs capped with the dihalomethylene analogues 7b and 7c both had higher translational efficiencies than the methylene analogue 7a. Notably, the pCCl2p analogue 7b outperformed other modified analogues and conferred an mRNA translational efficiency similar to that of the unmodified teraphosphate 7d.
Table 6. In vitro translation efficiency of mRNA capped with different cap analogues.
| Cap analogue | Relative translation efficiency |
|---|---|
| m7GpppG | 1.00 ± 0.02 |
| m27,3’-OGpppG | 1.60 ± 0.16 |
| m27,2’-OGppppG (7d) | 1.77 ± 0.18 |
| m27,2’-OGppCH2ppG (7a) | 1.39 ± 0.33 |
| m27,2’-OGppCCl2ppG (7b) | 1.71 ± 0.25 |
| m27,2’-OGppCF2ppG (7c) | 1.57 ± 0.10 |
| ApppG | 0.19 ± 0.10 |
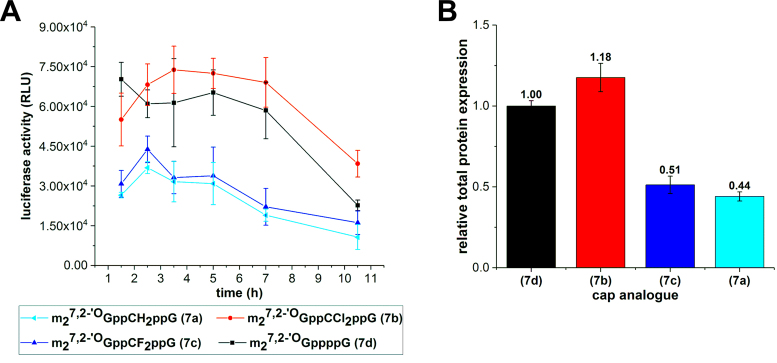
Next, we investigated whether mRNAs capped with the dihalomethylene analogues 7b and 7c would yield more protein than transcript capped with analogue 7a in a more complex system, i.e. living cells. Taking into account lower susceptibility to hDcp2 of transcripts capped with 7b and 7c compared to transcripts capped with 7d (Table 4, Figure 3) we envisaged that mRNAs carrying analogues 7b and 7c could be more stable in vivo than transcripts capped with 7d, thereby giving higher overall protein yield. Translation efficiencies were determined in HeLa cells transfected with equal amounts of RNA encoding firefly luciferase and capped with analogues 7a–d. After cell lysis, luciferase activity was measured and plotted as a function of time (Figure 4A). Moreover, the relative total protein expression, defined as the integral under expression curve normalized to the expression of RNA capped with 7d, was calculated for each transcript (Figure 4B). We found that the total protein expression was higher for mRNA capped with the pCCl2p analogue 7b than with unmodified tetraphosphate ARCA 7d (relative total protein expression equal 1.18), which is expressed about two fold more efficiently than transcripts capped with analogues 7a and 7c.
Figure 4.
Expression of capped-mRNA in HeLa cell line. (A) Firefly luciferase activity depicted as function of time. (B) Total protein expression of firefly luciferase normalized to m27,2’-OGppppG-mRNA. Values above each bar represent relative protein expression.
Applications of fluorinated cap analogues in 19F NMR studies
19F NMR is gaining increasing attention as a useful tool for studying protein-ligand interactions (62–65). In particular, the virtual absence of fluorine in biological material allows the measurement of specific interactions between fluorinated reporter tags and proteins even in complex samples including cells (66,67). Therefore, we tested the fluorinated cap analogue 6c as a reporter ligand in binding assays as well as an activity-based probe for monitoring enzymatic hydrolysis by 19F NMR.
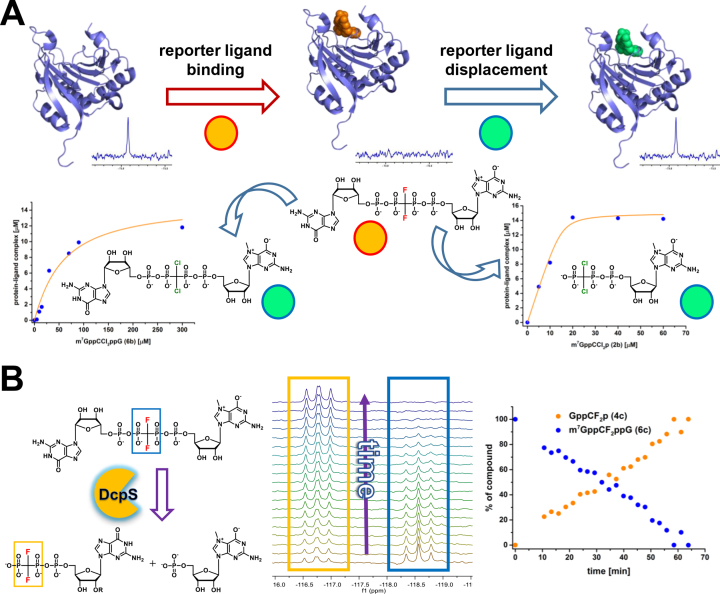
eIF4E is overexpressed in many types of cancer and constitutes an important molecular target for anti-cancer therapy (6). Therefore, methods for studying binding to the eIf4E active site are of interest, in particular from the inhibitor discovery perspective. Having established that the difluoromethylene-containing cap analogues are good eIF4E binders, we decided to evaluate their use as reporter molecules in the 19F NMR binding assay. The assay is based on the initial binding of a fluorinated reporter molecule to eIF4E, which typically results in the significant broadening of the 19F NMR signal of a ligand owing to increased transverse relaxation rates. Subsequently, the reporter ligand is displaced from complex with eIF4E, which results in a recovery of the 19F signal of the unbound reporter (Figure 5A) (68). Here, we exemplify the method using the dinucleotide analogue 6c (m7GppCF2ppG), as it was characterized by a lower affinity for eIF4E in comparison to mononucleotide analogue 2c. This might be of advantage in the displacement assays, especially when testing ligands with low binding affinities. In our case, the two pCCl2p analogues 2b and 6b, which exhibit different affinities to eIF4E, were used as model ligands (Figure 5A). Using the 19F NMR observed method, we were able to determine apparent association constant values (KAS,app = 0.164 and 0.017 μM−1 for 2b and 6b respectively), which led to actual KAS values of 166 and 17.2 μM−1 for the respective analogues. This is in a good agreement with the data obtained from fluorescence titration experiments (188 and 78 μM−1 for analogues 2b and 6b respectively, Table 2).
Figure 5.
Applications of fluorinated cap analogues. (A) Basis of the displacement assay followed by the 19F NMR signal of the reporter molecule m7GppCF2ppG (6c). The displacement of 6c by m7GppCCl2p (2b) and m7GppCCl2ppG (6b) yields binding curves typical for strong and weaker binders, respectively. (B) Hydrolysis of m7GppCF2ppG (6c) by the DcpS enzyme followed by 19F NMR.
Another therapeutically relevant cap-binding protein is hDcpS, which was identified as a molecular target in spinal muscular atrophy (34,69). However, the same approach as used for eIF4E cannot be applied to search for hDcpS binders, since all difluoromethylene cap analogues are susceptible to degradation by hDcpS. Instead, we applied the cap analogue m7GppCF2ppG (6c) as an activity-based probe to monitor DcpS-mediated hydrolysis leading to the production of m7GMP and GppCF2p (4c) (Figure 5B). The fluorinated analogue 6c was observed to be a relatively good substrate for hDcpS and therefore might find use as a reporter ligand for hDcpS inhibition studies.
DISCUSSION
We described the synthesis and properties of novel mRNA 5′-cap analogues bearing the dichloromethylene- or difluoromethylenebisphosphonate moiety as a bridging modification of the oligophosphate chain. 31P NMR and pKa measurements confirmed that introduction of an electronegative substituent into the methylene moiety influenced the electronic properties of neighboring phosphates to more closely mimic the parent pyrophosphate containing cap structure.
High affinity of ligand binding to eIF4E is required for obtaining small molecule inhibitors of translation or, in the case of mRNA modification, high translational efficiencies. Therefore, eIF4E-cap interaction is one of the most important properties determined for novel cap analogues. Generally, the introduction of both pCCl2p and pCF2p moieties stabilized the cap-eIF4E complex compared to the pCH2p-containing analogues, however, the effect was more pronounced for mononucleotides. The significantly lower affinity of m7GppCH2p (2a) might be partially explained by a smaller negative net charge compared to other mononucleotides. It has been shown that electrostatic interactions between the negatively charged phosphate chain and basic amino acids in the cap-binding pocket play an important role in complex stabilization, and that the affinity of cap analogues for eIF4E increases with the increasing negative net charge of the polyphosphate chain (43,56). The pKa value for the compound 2a determined by 31P NMR was above 8 whereas for the other analogues it was below 7.5. Therefore, at the pH of the binding experiment (pH 7.2) compound 2a might have a notably smaller negative charge compared to the other mononucleotides. However, this observation does not account for the general stabilization of cap-eIF4E complexes observed for dichloromethylene analogues compared to methylene and difluoromethylene substitutions. This was observed both in the mono- and dinucleotides series, and despite the lower pKa of pCF2p analogue 2c compared to pCCl2p analogue 2b, suggests that an additional stabilizing effect is provided exclusively by the pCCl2p group.
To obtain a deeper insight into the interaction of chlorinated cap analogue with the eIF4E protein, we obtained crystal structures of m7GppCCl2ppG (6c) and m7GppppG (6d) in complex with eIF4E. Notably, the dichloromethylene moiety in analogue 6c appears to alter the ligand binding mode compared to a cap analogue unmodified in the tetraphosphate bridge (6d). It indicates, that apart from the electronic effects, the geometry of a polyphosphate moiety might make a major contribution to the binding affinity.
The utility of cap analogues as potential inhibitors of eIF4E is also determined by their hydrolytic stability in a cell environment. We have evaluated the susceptibility of cap analogues 2a–d, 6a–d and 7a–d to cap specific pyrophosphatase DcpS, which in human targets short capped RNA oligonucleotides and mono- and dinucleotide caps (57,58). In the experimental conditions, apart from the unmodified nucleotides, only difluoromethylene-containing analogues were significantly susceptible to hDcpS-mediated hydrolysis. This indicates that the pCF2p moiety is the closest mimic of a pyrophosphate in respect to recognition by hDcpS. This might result from the pCF2p moiety being a better leaving group compared to pCH2p, combined with its small size allowing the avoidance of steric hindrance in the cap binding pocket. Such steric hindrance might also be a reason for the resistance of the pCCl2p-containing analogues toward hDcpS. This feature, combined with their high affinity toward eIF4E, makes them attractive candidates for stable small molecule inhibitors of translation.
Further studies in the rabbit reticulocyte lysate system have shown that pCCl2p analogues 2b and 7b were potent and stable inhibitors of translation (IC50 of 0.97 and 0.95 μM for 2b and 7b respectively). The in vivo use of nucleoside triphosphates is hampered by their low cell-penetrability associated with the electronegativity of their phosphate units. However, methods for delivery systems/ modification strategies have been developed (70–73), which could subsequently be applied for testing the potency of novel cap analogues in cell systems.
The binding and inhibition experiments allowed us to characterize properties of cap analogues as small molecule ligands interacting with eIF4E. However, since the cap is an integral part of an mRNA chain in vivo, and in vitro transcribed mRNAs gain increasing attention as potential vaccines and gene replacement therapeutics (74), we were also interested in evaluating the novel cap analogues as reagents for modification of 5′ end of mRNA, and in the determining the translation efficiencies of mRNAs capped with various modified analogues. Our results are in general agreement with previous observations that high affinity for eIF4E is a necessary but not sufficient condition for efficient translation (40,75), indicating that other factors play a role in defining mRNA translational properties. For instance, the previously described imidodiphosphate modification was found to generally stabilize the eIF4E-cap complex, but produced rather poorly translated mRNAs (75). In fact, the pCCl2p analogue 7b described here is the first bridging-oxygen-substitution analogue that does not impair the translational efficiency of mRNA. This signifies the pCCl2p analogues as potentially useful reagents for the synthesis of in vitro transcribed mRNAs with applications in exogenous gene delivery. Moreover, to fully evaluate the usefulness of the halomethylene-containing analogues for mRNA modification, further studies on the susceptibility to hDcp2 pyrophosphatase of transcripts capped with these analogues were performed. It occurred that RNAs capped with cap analogues carrying β/γ-bridging substitution for methylene or halomethylene group are less susceptible to hDcp2 than transcripts capped with tri- or tetraphosphate ARCA. Notably, under the conditions of our assay, RNAs capped with CCl2-modified analogue were completely resistant to decapping. Such exceptionally high resistance to decapping by Dcp2 has been previously observed in the case of cap analogs modified with double O-to-S substitutions at the non-bridging positions (2S-analogs) (26). However, the synthesis and purification of 2S analogs is much more challenging, especially because, in contrast to CCl2-analogues, they exist in four P-diastereomeric forms. Interestingly, compounds carrying the CCl2-modification were also the most resistant to hDcpS activity.
Next, we determined the translational properties of mRNAs capped with dihalomethylene analogues in living cells. The translation efficiencies determined in HeLa cells revealed that mRNA capped with pCCl2p analogue 7b is expressed over 2-fold more efficiently than mRNAs caped with other methylene analogues (7a and 7c), and slightly more efficiently than RNA capped with unmodified tetraphosphate analog, 7d. However, in contrast to 7d compound 7b was resistant do Dcp2, and why this difference appears to little have only influence on mRNA expression is currently unclear to us. Possible explanations require further investigation and may include some HeLa cells-specific effects or interference from other cap-binding proteins or decapping enzymes.
Taken altogether, the dichloromethylene substitution distinguished itself as superior in the context of mRNA properties relevant to therapeutic applications, when compared to all other bridging cap modifications tested so far. This fact, combined with synthetic availability and lack of stereogenic center in the oligophosphate chain modified with pCCl2p moiety (in contrast to all non-bridging modifications), makes the analogue 7b a promising capping reagent for mRNAs produced for in vivo applications.
Finally, we have also demonstrated that despite not optimal biochemical properties, the difluoromethylene cap analogues can be applied as molecular tools in cap-related biochemical and biophysical assays. The difluoromethylene analogue 6c was successfully used as a reporter ligand in 19F NMR based assays, allowing observation of ligand binding to eIF4E as well as the monitoring of DcpS-mediated hydrolysis. We anticipate that in the future these assays might facilitate inhibitor discovery for cap-dependent proteins.
ACCESSION NUMBERS
Structures of eIF4E in complex with m7GppppG (6d) (PDB id: 5J5O ) and with m27,2’OGppCCl2ppG (7b) (PDB id: 5J5Y).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Mike Kiledjian (Rutgers University) for hDcpS and hDcp2 encoding plasmids and Prof. Stephen R. Ikeda (The National Institute on Alcohol Abuse and Alcoholism) for providing hRLuc-pRNA2(A)128 plasmid.
Footnotes
Present address: Anna M. Rydzik, Department of Chemistry, Ludwig-Maximilians University Munich, Butenandtstr. 5-13, 81377 Munich, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Center, Poland [UMO-2012/05/E/ST5/03893, UMO-2016/21/B/ST5/02556, UMO-2013/08/A/NZ1/00866]; Ministry of Science and Higher Education, Poland [DI2012 024842]. HPLC and MS measurements were performed in the Biopolymers Laboratory, Division of Biophysics, Institute of Experimental Physics, Faculty of Physics, University of Warsaw, supported by the ERDF within the Innovation Economy Operational Programme [POIG.02.01.00-14-122/09]. Funding for open access charge: National Science Center, Poland [UMO-2012/05/E/ST5/03893].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shatkin A.J. mRNA caps-old and newer hats. Bioessays. 1987; 7:275–277. [DOI] [PubMed] [Google Scholar]
- 2. Furuichi Y., Shatkin A.J.. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000; 55:135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Houseley J., Tollervey D.. The Many Pathways of RNA Degradation. Cell. 2009; 136:763–776. [DOI] [PubMed] [Google Scholar]
- 4. Ziemniak M., Strenkowska M., Kowalska J., Jemielity J.. Potential therapeutic applications of RNA cap analogs. Future Med. Chem. 2013; 5:1141–1172. [DOI] [PubMed] [Google Scholar]
- 5. Jemielity J., Kowalska J., Rydzik A., Darzynkiewicz E.. Synthetic mRNA cap analogs with a modified triphosphate bridge–synthesis, applications and prospects. N. J. Chem. 2010; 34:829–844. [Google Scholar]
- 6. Karaki S., Andrieu C., Ziouziou H., Rocchi P., Donev R.. The eukaryotic translation initiation factor 4E (eIF4E) as a therapeutic target for cancer. Adv. Prot. Chem. Struct. Biol. 2015; 101:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siddiqui N., Sonenberg N.. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 2015; 43:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I.. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015; 14:261–278. [DOI] [PubMed] [Google Scholar]
- 9. Graff J., Konicek B., Vincent T., Lynch R., Monteith D., Weir S., Schwier P., Capen A., Goode R., Dowless M. et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 2007; 117:2638–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li S., Jia Y., Jacobson B., McCauley J., Kratzke R., Bitterman P., Wagner C.. Treatment of breast and lung cancer cells with a N-7 benzyl guanosine monophosphate tryptamine phosphoramidate pronucleotide (4Ei-1) results in chemosensitization to gemcitabine and induced elF4E proteasomal degradation. Mol. Pharm. 2013; 10:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong D.S., Kurzrock R., Oh Y., Wheler J., Naing A., Brail L., Callies S., Andre V., Kadam S.K., Nasir A. et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin. Cancer Res. 2011; 17:6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kranz L., Diken M., Haas H., Kreiter S., Loquai C., Reuter K., Meng M., Fritz D., Vascotto F., Hefesha H. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016; 534:396–401. [DOI] [PubMed] [Google Scholar]
- 13. Diken M., Kreiter S., Selmi A., Britten C., Huber C., Tureci O., Sahin U.. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011; 18:702–708. [DOI] [PubMed] [Google Scholar]
- 14. Sahin U., Kariko K., Tureci O.. mRNA-based therapeutics - developing a new class of drugs. Nat. Rev. Drug Discov. 2014; 13:759–780. [DOI] [PubMed] [Google Scholar]
- 15. Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U.. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010; 17:961–971. [DOI] [PubMed] [Google Scholar]
- 16. van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B.. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002; 21:6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Jiao X., Carr-Schmid A., Kiledjian M.. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S.W., Jiao X., Liu H., Gu M., Lima C.D., Kiledjian M.. Functional analysis of mRNA scavenger decapping enzymes. RNA. 2004; 10:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taverniti V., Seraphin B.. Elimination of cap structures generated by mRNA decay involves the new scavenger mRNA decapping enzyme Aph1/FHIT together with DcpS. Nucleic Acids Res. 2015; 43:482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grudzien-Nogalska E., Kiledjian M.. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscipl. Rev.-RNA. 2017; 8, doi:10.1002/wrna.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang J.H., Jiao X., Chiba K., Oh C., Martin C.E., Kiledjian M., Tong L.. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5΄-3΄ exoribonuclease activity. Nat. Struct. Mol. Biol. 2012; 19:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiao X., Doamekpor S.K., Bird J.G., Nickels B.E., Tong L., Hart R.P., Kiledjian M.. 5′ End nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell. 2017; 168:1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalska J., Lewdorowicz M., Zuberek J., Grudzien-Nogalska E., Bojarska E., Stepinski J., Rhoads R.E., Darzynkiewicz E., Davis R.E., Jemielity J.. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA. 2008; 14:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kowalska J., Lukaszewicz M., Zuberek J., Darzynkiewicz E., Jemielity J.. Phosphoroselenoate dinucleotides for modification of mRNA 5΄ end. Chembiochem. 2009; 10:2469–2473. [DOI] [PubMed] [Google Scholar]
- 25. Kowalska J., Wypijewska del Nogal A., Darzynkiewicz Z.M., Buck J., Nicola C., Kuhn A.N., Lukaszewicz M., Zuberek J., Strenkowska M., Ziemniak M. et al. Synthesis, properties, and biological activity of boranophosphate analogs of the mRNA cap: versatile tools for manipulation of therapeutically relevant cap-dependent processes. Nucleic Acids Res. 2014; 42:10245–10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strenkowska M., Grzela R., Majewski M., Wnek K., Kowalska J., Lukaszewicz M., Zuberek J., Darzynkiewicz E., Kuhn A.N., Sahin U. et al. Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential. Nucleic Acids Res. 2016; 44:9578–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blackburn G., Guo M.. Synthesis, physical, chemical, and enzyme studies on bis-2,6-diaminopurine beta-d-ribofuranoside p1,p4-tetraphosphate. Nucleosides Nucleotides. 1991; 10:549–551. [Google Scholar]
- 28. Blackburn G., Langston S.. Novel p1,p2-substituted phosphonate analogs of 2΄-deoxyadenosine and 2΄-deoxythymidine 5΄-triphosphates. Tetrahedron Lett. 1991; 32:6425–6428. [Google Scholar]
- 29. Blackburn G., Guo M., Langston S., Taylor G.. Novel phosphonate and thiophosphate analogs of ap3a, diadenosine 5΄,5΄''-p1,p3-triphosphate. Tetrahedron Lett. 1990; 31:5637–5640. [Google Scholar]
- 30. Blackburn G., Eckstein F., Kent D., Perree T.. I sopolar vs isosteric phosphonate analogs of nucleotides. Nucleosides Nucleotides. 1985; 4:165–167. [Google Scholar]
- 31. Blackburn G., Kent D., Kolkmann F.. 3 new beta,gamma-methylene analogs of adenosine-triphosphate. J. Chem. Soc.-Chem. Commun. 1981; 1188–1190. [Google Scholar]
- 32. Oertell K., Wu Y., Zakharova V., Kashemirov B., Shock D., Beard W., Wilson S., McKenna C., Goodman M.. Effect of beta,gamma-CHF- and beta,gamma-CHCl-dGTP halogen atom stereochemistry on the transition state of DNA polymerase beta. Biochemistry. 2012; 51:8491–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Upton T., Kashemirov B., McKenna C., Goodman M., Prakash G., Kultyshev R., Batra V., Shock D., Pedersen L., Beard W. et al. alpha,beta-Difluoromethylene deoxynucleoside 5΄-triphosphates: a convenient synthesis of useful probes for DNA polymerase beta structure and function. Org. Lett. 2009; 11:1883–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh J., Salcius M., Liu S.W., Staker B.L., Mishra R., Thurmond J., Michaud G., Mattoon D.R., Printen J., Christensen J. et al. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem. Biol. 2008; 3:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wypijewska A., Bojarska E., Stepinski J., Jankowska-Anyszka M., Jemielity J., Davis R.E., Darzynkiewicz E.. Structural requirements for Caenorhabditis elegans DcpS substrates based on fluorescence and HPLC enzyme kinetic studies. FEBS J. 2010; 277:3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalek M., Jemielity J., Darzynkiewicz Z.M., Bojarska E., Stepinski J., Stolarski R., Davis R.E., Darzynkiewicz E.. Enzymatically stable 5΄ mRNA cap analogs: synthesis and binding studies with human DcpS decapping enzyme. Bioorg. Med. Chem. 2006; 14:3223–3230. [DOI] [PubMed] [Google Scholar]
- 37. Rydzik A.M., Lukaszewicz M., Zuberek J., Kowalska J., Darzynkiewicz Z.M., Darzynkiewicz E., Jemielity J.. Synthetic dinucleotide mRNA cap analogs with tetraphosphate 5΄,5΄ bridge containing methylenebis(phosphonate) modification. Org. Biomol. Chem. 2009; 7:4763–4776. [DOI] [PubMed] [Google Scholar]
- 38. Kowalska J., Lukaszewicz M., Zuberek J., Ziemniak M., Darzynkiewicz E., Jemielity J.. Phosphorothioate analogs of m7GTP are enzymatically stable inhibitors of cap-dependent translation. Bioorg. Med. Chem. Lett. 2009; 19:1921–1925. [DOI] [PubMed] [Google Scholar]
- 39. Grudzien-Nogalska E., Stepinski J., Jemielity J., Zuberek J., Stolarski R., Rhoads R.E., Darzynkiewicz E.. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymol. 2007; 431:203–227. [DOI] [PubMed] [Google Scholar]
- 40. Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E.. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003; 9:1108–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cohen L., Mikhli C., Friedman C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Davis R.. Nematode M(7)GpppG and m(3)(2,2,7)GpppG decapping: activities in Ascaris embryos and characterization of C-elegans scavenger DcpS. RNA. 2004; 10:1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuberek J., Wyslouch-Cieszynska A., Niedzwiecka A., Dadlez M., Stepinski J., Augustyniak W., Gingras A., Zhang Z., Burley S., Sonenberg N. et al. Phosphorylation of eIF4E attenuates its interaction with mRNA 5΄ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA. 2003; 9:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niedzwiecka A., Marcotrigiano J., Stepinski J., Jankowska-Anyszka M., Wyslouch-Cieszynska A., Dadlez M., Gingras A., Mak P., Darzynkiewicz E., Sonenberg N. et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5΄ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 2002; 319:615–635. [DOI] [PubMed] [Google Scholar]
- 44. Coleman T., Wang G., Huang F.. Superior 5΄ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004; 32:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Williams D.J., Puhl H.L., Ikeda S.R.. A simple, highly efficient method for heterologous expression in mammalian primary neurons using cationic lipid-mediated mRNA transfection. Front. Neurosci. 2010; 4:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krug M., Weiss M., Heinemann U., Mueller U.. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 2012; 45:568–572. [Google Scholar]
- 48. Mccoy A., Grosse-Kunstleve R., Adams P., Winn M., Storoni L., Read R.. Phaser crystallographic software. J. Appl. Cryst. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schuttelkopf A., van Aalten D.. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Cryst. D. 2004; 60:1355–1363. [DOI] [PubMed] [Google Scholar]
- 50. Lebedev A., Young P., Isupov M., Moroz O., Vagin A., Murshudov G.. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Cryst. D. 2012; 68:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 52. Adams P., Afonine P., Bunkoczi G., Chen V., Davis I., Echols N., Headd J., Hung L., Kapral G., Grosse-Kunstleve R. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nikolovska-Coleska Z., Wang R., Fang X., Pan H., Tomita Y., Li P., Roller P., Krajewski K., Saito N., Stuckey J. et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004; 332:261–273. [DOI] [PubMed] [Google Scholar]
- 54. Leung I., Demetriades M., Hardy A., Lejeune C., Smart T., Szollossi A., Kawamura A., Schofield C., Claridge T.. Reporter ligand NMR screening method for 2-oxoglutarate oxygenase inhibitors. J. Med. Chem. 2013; 56:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E.. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3΄-O-methyl)GpppG and 7-methyl (3΄-deoxy)GpppG. RNA. 2001; 7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 56. Zuberek J., Jemielity J., Jablonowska A., Stepinski J., Dadlez M., Stolarski R., Darzynkiewicz E.. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5΄ terminus. Biochemistry. 2004; 43:5370–5379. [DOI] [PubMed] [Google Scholar]
- 57. Liu H., Rodgers N.D., Jiao X., Kiledjian M.. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002; 21:4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gu M., Fabrega C., Liu S.W., Liu H., Kiledjian M., Lima C.D.. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol. Cell. 2004; 14:67–80. [DOI] [PubMed] [Google Scholar]
- 59. Wypijewska A., Bojarska E., Lukaszewicz M., Stepinski J., Jemielity J., Davis R.E., Darzynkiewicz E.. 7-methylguanosine diphosphate (m(7)GDP) is not hydrolyzed but strongly bound by decapping scavenger (DcpS) enzymes and potently inhibits their activity. Biochemistry. 2012; 51:8003–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. She M., Decker C.J., Chen N., Tumati S., Parker R., Song H.. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006; 13:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kalek M., Jemielity J., Grudzien E., Zuberek J., Bojarska E., Cohen L.S., Stepinski J., Stolarski R., Davis R.E., Rhoads R.E. et al. Synthesis and biochemical properties of novel mRNA 5΄ cap analogs resistant to enzymatic hydrolysis. Nucleos. Nucleot. Nucleic Acids. 2005; 24:615–621. [DOI] [PubMed] [Google Scholar]
- 62. Kitevski-LeBlanc J., Prosser R.. Current applications of F-19 NMR to studies of protein structure and dynamics. Progr. Nucl. Magn. Reson. Spectr. 2012; 62:1–33. [DOI] [PubMed] [Google Scholar]
- 63. Dalvit C., Fagerness P., Hadden D., Sarver R., Stockman B.. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J. Am. Chem. Soc. 2003; 125:7696–7703. [DOI] [PubMed] [Google Scholar]
- 64. Chen H., Viel S., Ziarelli F., Peng L.. F-19 NMR: a valuable tool for studying biological events. Chem. Soc. Rev. 2013; 42:7971–7982. [DOI] [PubMed] [Google Scholar]
- 65. Lindon J.C., Wilson I.D.. eMagRes. 2015; 4:John Wiley & Sons, Ltd. [Google Scholar]
- 66. Smith A., Zhang Z., Pielak G., Li C.. NMR studies of protein folding and binding in cells and cell-like environments. Curr. Opin. Struct. Biol. 2015; 30:7–16. [DOI] [PubMed] [Google Scholar]
- 67. Veronesi M., Giacomina F., Romeo E., Castellani B., Ottonello G., Lambruschini C., Garau G., Scarpelli R., Bandiera T., Piomelli D. et al. Fluorine nuclear magnetic resonance-based assay in living mammalian cells. Anal. Biochem. 2016; 495:52–59. [DOI] [PubMed] [Google Scholar]
- 68. Baranowski M.R., Nowicka A., Rydzik A.M., Warminski M., Kasprzyk R., Wojtczak B.A., Wojcik J., Claridge T.D., Kowalska J., Jemielity J.. Synthesis of fluorophosphate nucleotide analogues and their characterization as tools for 19F NMR studies. J. Org. Chem. 2015; 80:3982–3997. [DOI] [PubMed] [Google Scholar]
- 69. Howell M., Singh N., Singh R.. Advances in therapeutic development for spinal muscular atrophy. Future Med. Chem. 2014; 6:1081–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005; 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 71. Zochowska M., Piguet A., Jemielity J., Kowalska J., Szolajska E., Dufour J., Chroboczek J.. Virus-like particle-mediated intracellular delivery of mRNA cap analog with in vivo activity against hepatocellular carcinoma. Nanomed.-Nanotechnol. Biol. Med. 2015; 11:67–76. [DOI] [PubMed] [Google Scholar]
- 72. Gollnest T., de Oliveira T., Schols D., Balzarini J., Meier C.. Lipophilic prodrugs of nucleoside triphosphates as biochemical probes and potential antivirals. Nat. Commun. 2015; 6:8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wagner C., Iyer V., McIntee E.. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med. Res. Rev. 2000; 20:417–451. [DOI] [PubMed] [Google Scholar]
- 74. Vallazza B., Petri S., Poleganov M., Eberle F., Kuhn A., Sahin U.. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. Wiley Interdiscipl. Rev.-RNA. 2015; 6:471–499. [DOI] [PubMed] [Google Scholar]
- 75. Rydzik A.M., Kulis M., Lukaszewicz M., Kowalska J., Zuberek J., Darzynkiewicz Z.M., Darzynkiewicz E., Jemielity J.. Synthesis and properties of mRNA cap analogs containing imidodiphosphate moiety-fairly mimicking natural cap structure, yet resistant to enzymatic hydrolysis. Bioorg. Med. Chem. 2012; 20:1699–1710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.