Abstract
Sm and Sm-like proteins represent an evolutionarily conserved family with key roles in RNA metabolism in Pro- and Eukaryotes. In this study, a collection of 53 mRNAs that co-purified with Sulfolobus solfataricus (Sso) SmAP2 were surveyed for a specific RNA binding motif (RBM). SmAP2 was shown to bind with high affinity to the deduced consensus RNA binding motif (SmAP2-cRBM) in vitro. Residues in SmAP2 interacting with the SmAP2-cRBM were mapped by UV-induced crosslinking in combination with mass-spectrometry, and verified by mutational analyses. The RNA-binding site on SmAP2 includes a modified uracil binding pocket containing a unique threonine (T40) located on the L3 face and a second residue, K25, located in the pore. To study the function of the SmAP2-RBM in vivo, three authentic RBMs were inserted in the 3′UTR of a lacS reporter gene. The presence of the SmAP2-RBM in the reporter-constructs resulted in decreased LacS activity and reduced steady state levels of lacS mRNA. Moreover, the presence of the SmAP2-cRBM in and the replacement of the lacS 3′UTR with that of Sso2194 encompassing a SmAP2-RBM apparently impacted on the stability of the chimeric transcripts. These results are discussed in light of the function(s) of eukaryotic Lsm proteins in RNA turnover.
INTRODUCTION
The evolutionarily conserved Sm and Sm-like (Lsm) proteins play important roles in RNA processing and degradation (1). The Sm proteins are composed of an N-terminal α-helix and five β-strands (β1–β5) connected by five loops (L1–L5) forming a continuous Sm-fold (2–6). The Sm1 motif corresponds to the β1, β2 and β3 strands, whereas the Sm2 sequence motif corresponds to the β4 and β5 strands. The Sm proteins form characteristic doughnut-shaped (torus-shaped) homomorphic or heteromorphic rings containing six or seven protein subunits. To distinguish each site of the doughnut the L3/proximal face contains loop 3, whereas the L4/distal face comprises loop 4. The homo- or heteromeric assemblies usually bind to small RNA molecules to form ribonucleoprotein complexes. The central pore on each site of the doughnut is predominantly positively charged and comprises the RNA binding site(s).
In Escherichia coli, the Hfq protein binds to and facilitates the interaction between sRNAs and target mRNAs, which frequently modulates translation and mRNA stability (7). Hfq can also associate with the 3′ end of mRNAs and inhibit 3′-5′ exonuclease activities (8). Crystal structures of Staphylococcus aureus Hfq in complex with short oligonucleotides revealed that the proximal/L3 face binds poly(U) sequences between neighboring monomers around the central pore (9). The proximal/L3 face uridine-binding pocket is conserved in Hfq proteins of Gram-negative and Gram-positive bacteria. Further studies using E. coli Hfq revealed that the distal/L4 face binds A-rich oligonucleotides with three nucleotides per subunit of Hfq (10). More recent work showed that the rim (11) and C-terminal tails of the hexamer contribute as well to Hfq RNA binding (12) and function (12,13).
In Eukaryotes the best-characterized Lsm complexes are Lsm1–7 and Lsm2–8, which are present in the cytoplasm and nucleus, respectively. Lsm1–7, in conjunction with Pat1, acts as a major regulator of mRNA stability through its association with 3′ oligoadenylated mRNAs, which protects them from 3′–5′ directional decay while simultaneously promoting decapping and decay through its interactions with the decapping machinery. Lsm2–8 is primarily involved in stabilizing the U6 snRNA and chaperoning it through the splicing process (1). RNA sequences recognized by various Sm/Lsm complexes include the Sm-class snRNA with the consensus sequence AAUUUUUGG (14). Residues of the Lsm2–8 complex involved in RNA binding have been recently identified using the penta-nucleotide GUUUU (15). Each of the four uracil bases is recognized by a conserved pattern of interactions, involving two sequence motifs: the N residue of the DxXxN motif and the R residue of the IRGX motif (15) of two neighbouring Lsm proteins. The G base is stacked by W35 from Lsm 4 and L58 from Lsm7 representing the X-residue in the DxXxN motif.
Like other Sm/Lsm proteins, the Sm-like archaeal proteins (SmAPs) are composed of an N-terminal α-helix and five β-strands forming a continuous Sm-fold (2–6). However, the homo-heptameric SmAPs display characteristic differences from bacterial and eukaryal Lsm proteins, e.g. they lack an extended C-terminus that is characteristic for some bacterial Hfq- and eukaryotic Sm proteins (16). Several studies showed that SmAPs from different Archaea bind to poly(U) stretches of different lengths with Kd's ranging from 72 nM to 10 μM depending on their origin and the length of polyU) (4,5,17–21). In SmAP1 of Archaeoglobus fulgidus (AF), Pyrococcus abyssi and Sulfolobus solfataricus the conserved poly(U) binding pocket consists of residues H40 and N42 (22) present within the conserved DxXxN motif described for the eukaryal Lsm complex (15) and residue R66 (22) within the conserved IRGX motif (15). The U base is hydrogen-bonded to residue N42 and stacked between the side-chains of H40 and R66 (22). Despite the high similarity of eukaryal Lsm complexes with SmAPs (23), the function and RNA substrates of the latter remain puzzling. In the euryarchaeaon Archaeoglobus fulgidus (Af) co-immunoprecipitation experiments revealed association of the two SmAPs with RNaseP RNA (4), which could suggest a role in tRNA processing. Another study showed that the Haloferax volcanii (Hv) Sm protein binds to several uncharacterized ncRNAs, tRNAs and C/D box snoRNAs (18), again suggesting a function in tRNA/rRNA processing. A deletion of the Sm1 motif in the Hv SmAP1 encoding gene showed a gain of function in swarming, which was paralleled by an up-regulation of transcripts encoding proteins required for motility (24).
One of the best studied crenarchaeota is S. solfataricus (Sso), which can grow chemolithotrophically and chemoorganotrophically at 80°C and at an acidic pH of 2–4. The majority of protein-coding transcripts (69%) lack a 5′UTR (25), suggesting that post-transcriptional regulation might occur at the 3′UTR (26). Alternative 3′UTRs might be formed by a second transcription termination site as observed in other Archaea, e.g. Sulfolobus acidocaladarius (27). Sso encodes three SmAP proteins (http://www-archbac.u-psud.fr/projects/sulfolobus), Sso 6454 (SmAP1), Sso 5410 (SmAP2) and Sso 0276 (SmAP3). The heptameric SmAP1 and SmAP2 share 36,4% identity, whereas they have only ∼10% identity with SmAP3. Different classes of non-coding RNAs (ncRNAs) co-purified with both, SmAP1 and SmAP2, or solely with SmAP1 or SmAP2 (6). The large number of associated intron-containing tRNAs and rRNA modifying RNAs indicated a conserved role of SmAP1 and SmAP2 in tRNA and rRNA processing (6). The comparison of the 3D structures of SmAP1 and SmAP2 revealed differences in non-conserved domains such as loop L4 and in the N- and C-terminal regions as well as in the surface charge with SmAP2 being more positively charged on the L3 face (6). Residues that are involved in RNA binding in other archaeal SmAPs (13,14,21,22) are conserved in SmAP1 but in SmAP2 H40 is replaced by T (6). In contrast to SmAP1, the co-purification study revealed a larger number of mRNAs that were exclusively enriched with SmAP2 (6), which might be attributed to the above-mentioned differences between the two proteins.
In this study, we revealed a consensus RNA binding motif for SmAP2 (SmAP2-cRBM) that is present in all 53 co-purifying mRNAs. The SmAP2-cRBM 5′-GGAUGGAUAUUAGGAAAUG-3′ was shown to bind with high affinity to SmAP2 in vitro. Moreover, UV-crosslinking in conjunction with mass-spectrometry suggested that the SmAP2-cRBM is located on the positively charged L3/proximal face of SmAP2. Moreover, the presence of SmAP2-RBMs resulted in a decrease of the steady-state levels of reporter mRNAs and apparently reduced their stability in vivo.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
The Sso strains PH1–16(pMJ05-Ara/tf55-lacS-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194-RBM-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194–3′UTR) and PH1–16(pMJ05-Ara/tf55-Δ2194-RBM-2194–3′UTR), PH1–16(pMJ05-Ara-lacS-1742-RBM-3′UTR) and PH1–16(pMJ05-Ara-lacS-2912-RBM-3′UTR) were generated as described in the Supplementary Information. All Sso strains were grown at 75°C in arabinose- or sucrose containing Brock's medium supplemented with 0.2% NZamine.
Bioinformatic analysis
The coding region and additional 100 nt of the 3′UTR of 53 mRNAs bound to SmAP2 (6) were used as input sequences and analyzed using the MEME tool (28). The default settings given in the data submission form were used as follows: Site distribution was set to zero or one occurrence per sequence; the motif width was set to a minimum of 6 and a maximum of 50; the O-order Markov model of sequence was used as background model. The SmAP2-RBM was identified in all 53 mRNAs with an E-value of 3.9 e-017. The identified motif was submitted to TomTom using the RNA (DNA-encoded) database for all species (29) and to FIMO (30) using a threshold of 1 × 10−6.
Creation of SmAP2 gene mutants and purification of the SmAP proteins
The smAP2K25A allele was created by means of recombinant DNA technology using inverse PCR with the 5′ monophosphorylated oligonucleotides 5′- CTGGTAAAACTTGCGGATGGCTCAG-3′ and 5′-CACGATTCTATTTATTGCCG-3′. The smAP2T40H allele was created likewise using the oligonucleotides 5′-CAAACTGACGGACATATGAATTTAG-3′ and 5′-TTCTAATTTTCCAATGTACTCTG-3′ and pProEx-SmAP2 (6) as template. The plasmid-template was removed by DpnI-cleavage, the PCR-products were isolated from agarose-gels, ligated, and then transformed into E. coli Top10 (Thermo Fisher scientific). The resulting constructs were validated by DNA sequencing, and then transformed into E. coli JW4130Δhfq (http://cg sc.biology.yale.edu/KeioList.php) for protein isolation. The SmAP1, the SmAP2 and the SmAP2 variants were purified by means of metal affinity chromatography, the His-tag was removed by TEV-protease, and the proteins were further purified to homogeneity by size-exclusion chromatography as described (6).
Electrophoretic mobility shift assays
The RNA nucleotides comprising the SmAP2-cRBM, the 1742-RBM and the 2194-RBM were purchased from Eurofins. 10 nM of 6-carboxyfluorescein(FAM)-labelled SmAP2-cRBM (5′-GGAUGGAUAUUAGGAAAUG-3′), FAM-labelled 2194-RBM (5′-GGAGAUAUAGUAGGAUAG-3′) and FAM-labelled 1742-RBM (5′-GGAUGGAUAUUAGGUACUGGG-3′) were incubated with 0, 50, 100, 150, 200, 250, 300 and 400 nM (SmAP2)7. Likewise, the SmAP2-cRBM was incubated either with 0, 50, 100, 200, 400, 600, 800, 1000 and 1500 nM (SmAP1)7 or 0, 200, 300, 400, 800, 1000, 2000 and 3000 nM (SmAP2)7, (SmAP2K25A)7 and (SmAP2T40H)7, respectively. For the poly(U)15 competition experiment, 10 nM of FAM-labeled SmAP2-cRBM was incubated with 200 nM (SmAP2)7 in the absence or presence of increasing amounts of poly(U)15 (50, 100, 200, 400, 800 and 1000 nM, respectively). The RNA and the respective proteins were incubated in a volume of 20 μl containing 50 mM HEPES (pH 7.0), 50 mM KCl, 5 mM MgCl2 and 1 mM β-mercaptoethanol at 65°C for 20 min. Then, 2 μl 85% glycerol was added and the samples were loaded on a 15% polyacrylamide gel. The electrophoresis was carried out at 80 V and 4°C for 90 min using 10 mM Tris, 58 mM glycine at pH 8.0 as running buffer. The RNA bands were visualized with the Typhoon Scanner using default settings for fluorescence acquisition mode.
Microscale thermophoresis
The Kd's of SmAP2 for poly(U)15 and for the SmAP2-cRBM (5-GGAUGGAUAUUAGGAAAUG-3′) were determined by microscale thermophoresis (31) using the Monolith NT.115 Green/Red or Blue/Green instrument (Nanotemper Technologies). SmAP2 was labeled with the NT-647 dye using the Monolith NT™ Protein Labeling Kit RED-NHS according to the manufacturer's instructions. 10 μl of labeled NT-647 SmAP2 protein (60 nM) was added to 16 serial dilutions (10 μl each) of either poly(U)15 or SmAP2-cRBM (final concentrations ranging from 1.53 nM to 50 μM). The reverse experiment was performed with a constant concentration of FAM-labelled SmAP2-cRBM (5′-FAM-GGAUGGAUAUUAGGAAAUG-3′), 1742-RBM (5′FAM-GGAUGGAUAUUAGGUACUGGG-3′) and 2194-RBM (5-FAM-GGAGAUAUAGUAGGAUAG-3′) (25 nM final concentration) and serial 1:1 dilutions of (SmAP2)7 (16 samples with final concentrations ranging from 0.122 nM to 2 μM). The binding reactions were carried out in MST buffer (25 mM Tris (pH 7.4), 75 mM NaCl, 5 mM MgCl2, 2.5 mM DTT, 0.05% Tween and 0.08 mg/ml BSA) at 65°C for 20 min. After centrifugation, the samples containing NT-647-labelled SmAP2 (final concentration 30 nM) were loaded onto hydrophobic capillaries (Nanotemper Technologies) and measured using red LED at 95% power and 40% MST power. The samples containing FAM-labelled SmAP2-cRBM (final concentration 25 nM) were loaded onto standard capillaries (Nanotemper Technologies) and measured using blue LED at 80% power and 20% MST power. Data analysis was performed with NTAnalysis software (Nanotemper Technologies).
UV induced protein–RNA crosslinking and mass spectrometry
The UV induced protein–RNA crosslinking and enrichment of crosslinked hetero-conjugates was performed as described (32,33). Briefly, 1 nmol of SmAP2-cRBM and 1 nmol of (SmAP2)7 protein were mixed in a 1:1 molar ratio in a total volume of 100 μl buffer containing 50 mM HEPES (pH 7.0), 50 mM KCl, 5 mM MgCl2 and 10 mM β-mercaptoethanol. The mixture was incubated at 65°C for 10 min. The sample was then transferred to black polypropylene microplates (Greiner Bio-One) and UV irradiated at 254 nm for 10 min at 4°C. The samples were precipitated with ethanol and the pellet was dissolved in 4 M urea and 50 mM Tris–HCl (pH 7.9), which was diluted 1:4 with 50 mM Tris–HCl (pH 7.9) to obtain a concentration of 1 M urea. The RNA was hydrolyzed by incubation with RNase A and RNase T1 for 2 h at 52°C followed by incubation with 1 μg Benzonase for 1 h at 37°C in the presence of 1 mM MgCl2. Following RNA digestion, the sample was treated overnight at 37°C with trypsin (Promega) with an enzyme to substrate ratio of 1:20. The non-crosslinked RNA fragments were removed by C18 desalting using in-house prepared C18 columns (Dr Maisch GmbH, Germany) and the crosslinked peptides were subjected to TiO2 enrichment using TiO2 columns (GL Sciences). The samples were finally dissolved in 12 μl 5% (v/v) ACN in water, 1% (v/v) FA in water for LC–MS/MS analysis. 8 μl were injected into a nano-LC system (Agilent 1100 series, Agilent Technologies) coupled with a Q-exactive instrument (Thermo Fisher Scientific). The ESI-MS was performed in data-dependent mode using the HCD method. The precursor ions as well as fragment ions were scanned in Orbitrap, and the resulting spectra were measured with high accuracy (<5 ppm) both at the MS1 and MS2 level. Data analysis was performed as described (33) and the high scoring crosslinked peptides were manually annotated for confirmation. The residues identified were located in the 3D structure (PDB code: 4XQ3) (6) using Swiss PDB viewer software.
β-Galactosidase assays
1 ml culture of strains PH1–16(pMJ05-Ara/tf55-lacS-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194-RBM-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194–3′UTR), PH1–16(pMJ05- Ara/tf55-Δ2194-RBM-2194–3′UTR), PH1–16(pMJ05-Ara-lacS-1742-RBM-3′UTR) and PH1–16(pMJ05-Ara-lacS-2912-RBM-3′UTR) was withdrawn at an OD600 of 0.4. The strains with plasmids containing only the arabinose-inducible promoter were grown in Brock's medium supplemented with 0.2% NZamine and 0.2% arabinose. The strains with plasmids bearing only the constitutively active tf55α promoter were grown in Brock's medium supplemented with 0.2% NZamine and 0.2% sucrose. 6 × 1 ml for ß-galactosidase assays and 10 ml for RNA-isolation were withdrawn from each biological replicate and the cells were harvested by centrifugation. The cell-pellets were snap frozen in liquid nitrogen. For the ß-galactosidase assays the cell-pellets were resuspended in 50 μl 10 mM Tris–HCl (pH 8.0). Crude cell extracts were prepared by repeated freeze and thaw cycles for 5 min at 37°C. The extracts were centrifuged for 30 min at 10 000 × g. To 20 μl of the supernatant 980 μl of pre-heated reaction buffer (50 mM sodiumphosphate (pH 6.5), 4 mg/ml ONPG) was added and the mixture was incubated for 30 min at 75°C. After incubation the absorbance was measured at 420 nM and the Miller units were calculated as described (26). Each experiment was performed with three biological and six technical replicates of the respective strain. The error bars are derived from biological and technical replicates. The statistical significance was determined using a t-test (Supplementary Table S2). The cell-pellets for RNA isolation of the three biological replicates were processed as described below.
cDNA synthesis and qPCR
Three biological replicates of strains PH1–16(pMJ05-Ara/tf55-lacS-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194-RBM-3′UTR), PH1–16(pMJ05-Ara/tf55-lacS-2194–3′UTR), PH1–16(pMJ05-Ara/tf55-Δ2194-RBM-2194–3′UTR), PH1–16(pMJ05-Ara-lacS-1742-RBM-3′UTR) and PH1–16(pMJ05-Ara-lacS-2912-RBM-3′UTR) were grown as described above. Total RNA was isolated using Trizol (Thermo Fisher Scientific) and treated with DNase I (Roche). The complete removal of DNA was confirmed by PCR using the 16S rRNA-specific oligonucleotides 5′-TTGGGATCGAGGGCTGAAAC-3′ and 5′-CTCACCCCTCTCCTACTCGG-3′. The mRNA quality was assessed by 8M Urea/8% polyacrylamide gel electrophoresis. The cDNA was synthesized using 1 μg of DNA-free total RNA using random hexamer oligonucleotides (Thermo Fisher Scientific) and SuperScript III reverse transcriptase (Thermo Fisher Scientific). For the qPCR, 10 ng cDNA of the respective strains was used together with the SSO3019/lacS-specific oligonucleotides 5′-GGAAGCGGTAGAGATGGCTG-3′ and 5′-CATCCGTGACCGTAACCTCC-3′ or together with the 16S rRNA-specific oligonucleotides (internal standard). The oligonucleotides were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3). The samples containing 5 × HOT FIREPol EvaGreen® qPCR Mix (Medibena), 10 ng cDNA and 250 nM of each oligonucleotide were placed in a real time PCR cycler (Eppendorf Mastercycler), and the reaction was started at 95°C for 15 min, followed by 40 cycles of 25 s at 95°C, 20 s at 58°C and 20 s at 72°C. All reactions including the DNA standards and the negative control (no template) were done in triplicates. The fluorescence was measured at the last step of each cycle using the automatic baseline setting to determine the Ct values. After 40 cycles, a melting curve analysis was done by raising the temperature from 45°C to 95°C every 15 s, and by measuring the fluorescence at each cycle. The melting curve analysis yielded a single peak in the melting curve. The quantification was performed with the realplex 2.2 software and the copy number was calculated by generation of a SSO3019 (lacS) standard curve from 102–107 copies of the linearized plasmid pSAV5-lacS-2194–3′UTR or 102–107 copies of a 16S rRNA standard generated by PCR with oligonucleotides 5′-TTGGGATCGAGGGCTGAAAC-3′ and 5′-GAGGTGATCCAGCCGC-3′. The lacS levels were normalized to the 16S rRNA levels. The statistical significance was determined using a t-test (Supplementary Table S2).
mRNA stability assays
The Sso strains PH1–16(pMJ05-Ara-lacS-3′UTR), PH1–16(pMJ05-Ara-lacS-2194-RBM-3′UTR), PH1–16(pMJ05-Ara-lacS-2194–3′UTR) and PH1–16(pMJ05-Ara-Δ2194-RBM-2194–3′UTR) were grown in Brock's medium at 75°C containing 0.2% arabinose. At an OD600 of 0.5, the cells were pelleted and resuspended in the same volume of Brock's medium pre-warmed to 75°C containing 0.2% sucrose. 20 ml of culture was withdrawn directly after transfer to the sucrose- containing medium (time 0) as well as after 5, 10, 15, 20, 25 and 30 min. Each experiment was performed with two biological and three technical replicates of the respective strains. After sampling the cells were immediately pelleted at 4°C and snap frozen in liquid nitrogen. Total RNA was isolated and processed for qPCR as described above.
RESULTS
Identification and validation of the SmAP2-cRBM
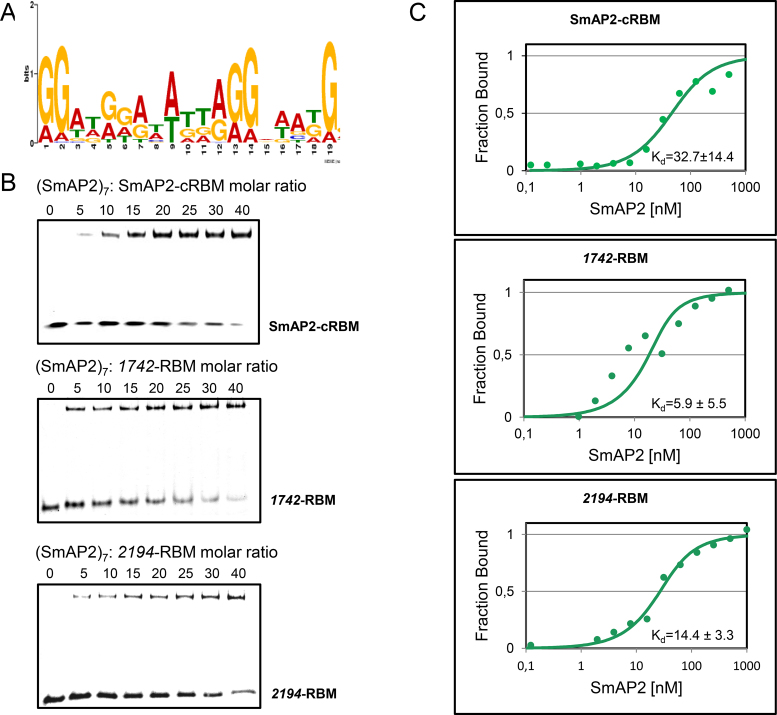
A recent study identified 53 leaderless/short-leadered mRNAs co-purifying with SmAP2 (6). To search for a SmAP2 binding motif in this population of mRNAs the MEME online tool for motif discovery was employed (28). The respective coding sequences and additionally 100 nucleotides from the 3′UTR were used as input. This survey revealed a consensus motif for SmAP2 in either of these mRNAs (Figure 1A; Supplementary Table S1). Notably, the position of the SmAP2-RBM in the 53 mRNAs was not uniformly distributed. In some mRNAs the motif is found in the coding region, whereas others bear it in the 3′UTR (Supplementary Table S1).
Figure 1.
Binding of the SmAP2-RBM to SmAP2. (A) The consensus SmAP2 RNA binding motif (SmAP2-cRBM) was deduced from 53 mRNA sequences bound to SmAP2 (6) using the MEME online tool (28). (B) EMSA assays were performed with (SmAP2)7 and the RNA substrates SmAP2-cRBM (5′-GGAUGGAUAUUAGGAAAUG-3′; top panel), the 1742-RBM (5′-GGAGAUAUAGUAGGAUAG-3′; middle panel) and the 2194-RBM 5′-GGAUGGAUAUUAGGUACUGGG-3′; bottom panel). As described in Materials and Methods, increasing concentrations of (SmAP2)7 were incubated with the SmAP2-cRBM, the 1742-RBM and the 2194-RBM in a molar ratio of 0:1, 5:1, 10:1, 15:1, 20:1, 25:1, 30:1 and 40:1, respectively. (C) Dissociation constants (Kd's) of (SmAP2)7 for the SmAP2-cRBM (top panel), for the 1742-RBM (middle panel) and for the 2194-RBM (bottom panel) revealed by microscale thermophoresis (31). 25 nM fluorescently labelled SmAP2-cRBM (top panel), 1742-RBM (middle panel) and 2194-RBM (bottom panel) were added to increasing amounts of (SmAP2)7 protein. The Kd′s were determined as described in Materials and Methods, and were expressed as mean EC50 ± EC50 confidence interval of 2 independent experiments.
To test whether SmAP2 binds to the deduced consensus motif 5′-GGAUGGAUAUUAGGAAAUG-3′ (SmAP2-cRBM), first electrophoretic mobility shift assays (EMSA) were employed. A molar ratio of 5:1 of (SmAP2)7: SmAP2-cRBM was sufficient to obtain a partial mobility shift (Figure 1B, upper panel), whereas no shift was observed with SmAP1, even when (SmAP1)7 was added in 150-fold molar excess (Supplementary Figure S1). Next, EMSA assays were performed with two authentic SmAP2 RBMs derived from the 3′UTR of Sso 1742 mRNA (5′-GGATGGATATTAGGTACTGGG-3′; 1742-RBM) and Sso 2194 mRNA (5′-GGAGAUAUAGUAGGAUAG-3′; 2194-RBM), both of which co-purified with SmAP2 (6) (Supplementary Table S1; Figure S2A/S2B). As observed for the SmAP2-cRBM, a 5-fold molar excess of (SmAP2)7 over the respective RNA was sufficient to obtain a mobility shift with the 1742-RBM (Figure 1B, middle panel) as well as with the 2194-RBM (Figure 1B, lower panel). Next, the binding affinity of SmAP2 for the SmAP2-cRBM was analyzed by microscale thermophoresis (MST) (31) and the Kd was determined with 42.1 ± 17.8 nM after incubation of increasing amounts of SmAP2-cRBM RNA with a constant concentration of NT-647 labeled SmAP2 protein (Supplementary Figure S3A).
As poly(U) is commonly used as a RNA-substrate for Sm-proteins, next the binding affinity of SmAP2 for SmAP2-cRBM was compared with the binding affinity of SmAP2 for poly(U)15. The Kd was determined with 426 ± 92 nM after incubation of increasing amounts of poly(U)15 with a constant concentration of NT-647 labelled SmAP2 protein (Supplementary Figure S3B). In agreement with the low affinity of SmAP2 for poly(U)15, a 80–100-fold molar excess of poly(U)15 was required to displace SmAP2 from the SmAP2-cRBM (Supplementary Figure S3C).
As the NT-647 labeling kit modifies K-residues, and as these residues are often involved in RNA binding the reverse experiment was performed using constant amounts of 6-carboxyfluorescein (FAM) labelled RNA and increasing amounts of non-labelled SmAP2. This experiment revealed a likewise low Kd (32.7 ± 14.4 nM) of SmAP2 for the SmAP2-cRBM (Figure 1C, upper panel). In addition, the binding affinity of SmAP2 was determined for the 1742-RBM and the 2194-RBM, respectively. The Kds were determined with 5.9 ± 5.5 nM and 14.4 ± 3.3 nM for the 1742-RBM and the 2194-RBM, respectively (Figure 1C, middle and bottom panel). The low Kd value for the 1742-RBM might be explained by its high congruence with the SmAP2-cRBM (Supplementary Table S1; Figure S2A).
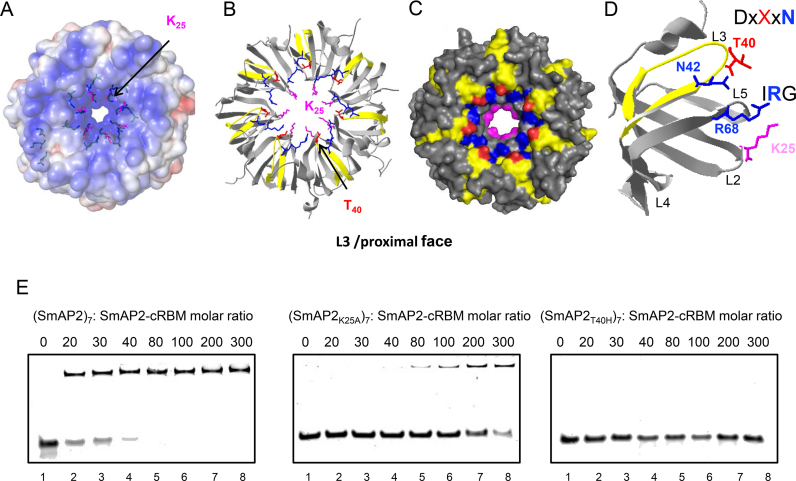
Residues located on the L3/proximal face of SmAP2 interact with the SmAP2-cRBM
Next, we aimed at identifying the binding site(s) of the SmAP2-cRBM on the surface of SmAP2 using UV-crosslinking followed by mass spectrometry. This method identified the SmAP2 residue K25 and the SmAP2-specific peptide L34–R46 as being involved in the interaction with uracil-residues of the SmAP2-cRBM (Supplementary Figure S4). The position of these residues in the 3D structure (PDB code 4XQ3) (6) of SmAP2 (Figure 2A–D) indicated that the SmAP2-cRBM is located on the positively charged L3/proximal face of SmAP2 (Figure 2A). The L34–R46 peptide encompasses the region corresponding to the above mentioned eukaryal DxXxN motif, wherein the H40 residue in SmAP1 is replaced by T40 in SmAP2 (Supplementary Figure S5). The location of T40 is highlighted in red in the heptameric structure (Figure 2B and C) and in the monomer (Figure 2D). As SmAP1 was defective in binding to the SmAP2-cRBM (Supplementary Figure S1), T40 in SmAP2 was replaced by H (SmAP2T40H) to test the requirement of T40 for SmAP2-cRBM binding. In addition, the variant SmAP2K25A was created to confirm the relevance of this crosslinked residue for SmAP2-cRBM binding. When compared with SmAP2 (Figure 2E, left panel), the SmAP2K25A (Figure 2E, middle panel) and the SmAP2T40H (Figure 2E, right panel) variants displayed a reduced- and a lack of binding to the SmAP2-cRBM, respectively. In addition, the binding stoichiometry of the SmAP2-cRBM to (SmAP2)7 was assessed using analytical size exclusion chromatography (Supplementary Figure S6). Either (SmAP2)7 or SmAP2-cRBM alone and a mix of both (1:2 molar ratio of (SmAP2)7:SmAP2-cRBM) were separated by size exclusion chromatography. When compared to the single components, only half of the RNA was bound to SmAP2, indicating that SmAP-cRBM and (SmAP2)7 form an equimolar complex.
Figure 2.
Location of residues in (SmAP2)7 and in the SmAP2 monomer interacting with uracil residue(s) of the SmAP2-cRBM revealed by UV-crosslinking and mass spectrometry. (A) Poisson–Boltzmann electrostatic potential of the solvent accessible L3/proximal face of (SmAP2)7 (15.) The location of K25 is highlighted in magenta. (B) Ribbon model of (SmAP2)7 depicting the location of K25 (magenta) and the peptide L34–R46 (yellow) involved in SmAP2-cRBM binding. The location of the T40 residue is indicated in red. (C) Space filling model of (SmAP2)7 depicting the locations of K25 (magenta), the peptide L34–R46 (yellow) and T40 (red). The SmAP2-specific T40 residue is located at the rim of the pore in the DxXxN motif, which has been described for eukaryal Lsm-complexes (10). N42 (blue) in the conserved DxXxN motif and R68 (blue) in the conserved IRG motif are involved in RNA binding in other Sm-proteins (15,22). (D) Location of residues involved in RNA-binding depicted in the SmAP2 monomer. The residues T40 (red) and N42 (blue) are located in the L3 loop in the conserved DxXxN motif, R68 (blue) is located in the L5 loop in the IRG motif and K25 (magenta) is located in the L2 domain. (E) EMSA assays performed with the (SmAP2K25A)7 and (SmAP2T40H)7 mutant proteins and the SmAP2-cRBM. Increasing concentrations of (SmAP2)7, (SmAP2K25A)7 and (SmAP2T40H)7, respectively were incubated with the SmAP2-cRBM in a molar ratio of 0:1 (lane1), 20:1 (lane 2), 30:1 (lane 3), 40:1 (lane 4), 80:1 (lane 5), 100:1 (lane 6), 200:1(lane 7) and 300:1 (lane 8), respectively.
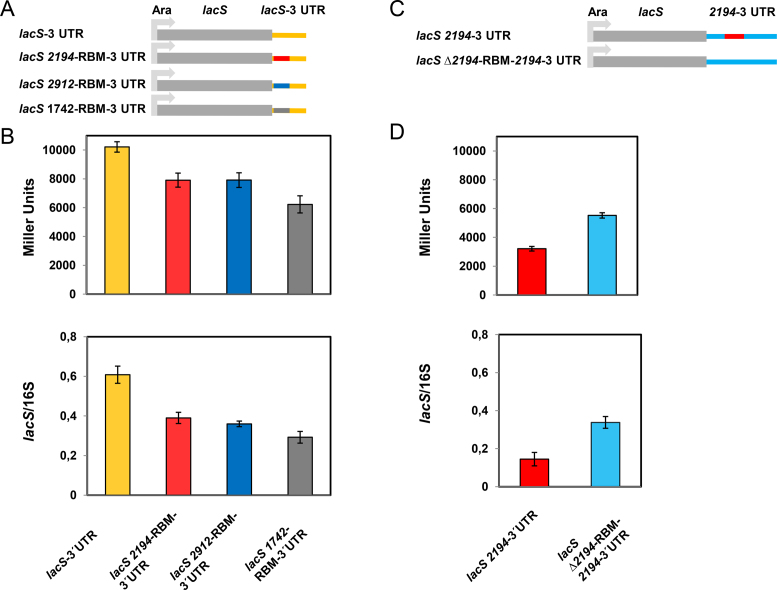
The SmAP2 RNA binding motif in the 3′UTR affects transcript abundance
To test whether the SmAP2-RBM is of functional importance in Sso, different reporter constructs were generated and placed into the Sso shuttle vector pMJ05 containing an arabinose-inducible promoter (Figure 3A and B; Supporting information). First, the SmAP2 RNA binding motifs located in the 3′ UTR of ORF 1742, 2194 and 2912 (Supplementary Table S1), were individually inserted 15 nt downstream of the stop codon of the lacS reporter gene, which is devoid of an SmAP2-RBM (Figure 3A). To test whether the presence of either SmAP2-RBM impacts on lacS expression, the β-galactosidase activities and the lacS mRNA steady state levels obtained with the lacS-1742-RBM-3′UTR (grey bars), lacS-2194-RBM-3′UTR (red bars) and lacS-2912-RBM-3′UTR (blue bars) constructs were compared with those obtained for the lacS gene comprising its authentic 3′UTR. The strains harbouring either reporter construct were grown in the presence of arabinose. When compared to the β-galactosidase activity conferred by the authentic lacS gene (orange bar) the β-galactosidase activities conferred by the lacS-2194-RBM-3′UTR (red bar) and lacS-2912-RBM-3′UTR (blue bar) constructs were ∼20% reduced, whereas that conferred by the lacS-1742-RBM-3′UTR (grey bar) construct was ∼40% reduced (Figure 3B; top panel).
Figure 3.
Regulatory function of the SmAP2-cRBM motif. (A) Schematic depiction of the SSV-1 derived Sso-shuttle plasmid pMJ05, in which the lacS gene containing its authentic 3′UTR is preceded by an arabinose-inducible promoter (Ara). In the lacS 2194-RBM-3′UTR construct, the 2194-RBM (red line) identified in the 3′ UTR of the putative thermopsine gene (Sso2194) was transplanted to the 3′UTR of the lacS gene, 14 nt downstream of the G of the TAG stop codon. The 2912-RBM identified in the 3′ UTR of a sulfate adenylyltransferase gene (Sso2912) (blue line) and the 1742-RBM identified in the 3′ UTR of the terminal quinol oxidase gene (Sso1742) (grey line) were inserted analogously. (B) β-galactosidase activities (top panel) and lacS mRNA steady state levels (bottom panel) obtained with the Sso strain PH1–16 harbouring the constructs containing the lacS 3′UTR (pMJ05-Ara-lacS-3′UTR; orange bars), the 2194-RBM inserted in the lacS 3′UTR (pMJ05-Ara-lacS-2194-RBM-3′UTR; red bars), the 2912-RBM inserted in the lacS 3′UTR (pMJ05-Ara-lacS-2912-RBM-3′UTR; blue bars), and the 1742-RBM inserted in the lacS 3′UTR (pMJ05-Ara-lacS-1742-RBM-3′UTR; gray bars) respectively. The β-galactosidase activities and the lacS mRNA levels were determined as described in Materials and Methods. Each experiment was performed with three biological replicates of the respective strains. The error bars are derived from biological and technical replicates. The statistical significance was determined using a t-test (Supplementary Table S2). (C) Schematic depiction of the SSV-1 derived Sso-shuttle plasmid pMJ05, in which the lacS 3′UTR was replaced by the 2194–3′UTR (lacS 2194–3′UTR). In the lacS Δ2194-RBM-2194–3′UTR construct, the 2194-RBM 5′-GGAGATATAGTAGGTATAGTA-3′ was replaced by the unrelated sequence 5′-TTCTGGCCCACCATACCATCGC-3′. (D) β-galactosidase activities (top panel) and lacS mRNA steady state levels (bottom panel) obtained with the Sso strain PH1–16 harbouring the constructs containing the 2194–3′UTR abutted to the lacS gene (pMJ05-Ara-lacS-2194–3′UTR; red bars) and the Δ2194-RBM-2194–3′UTR (pMJ05-Ara-Δ2194-RBM-2194–3′UTR; light blue bars), respectively. The ß-galactosidase activities and the lacS mRNA levels were determined as described in Materials and Methods. Each experiment was performed with three biological replicates of the respective strain. The error bars are derived from biological and technical replicates. The statistical significance was determined using a t-test (Supplementary Table S2).
This observation was mirrored by reduced lacS mRNA levels obtained with the lacS-2194-RBM-3′UTR (red bar), lacS-2912-RBM-3′UTR (blue bar) and the lacS-1742-RBM-3′UTR (grey bar) constructs when compared to the control (orange bar) (Figure 3B; bottom panel).
To test this further the authentic lacS 3′UTR was replaced with the 2194–3′UTR (Sso genome coordinates: 2017993–2018243) (Figure 3C). In addition, the 2194-RBM was exchanged with a random sequence (lacS-Δ2194-RBM-2194–3′UTR; Figure 3C) with the rationale that this manipulation might alleviate the negative impact of the 2194-RBM on the β-galactosidase activity and lacS transcript stability. When compared with the lacS-2194-RBM-3′UTR construct (Figure 3B; top panel; red bar), the β-galactosidase activity (Figure 3D; top panel; red bar) and the lacS transcript levels (Figure 3D, bottom panel, red bar) obtained with the lacS-2194–3′UTR construct were further reduced. However, the exchange of the 2194-RBM with an unrelated sequence (lacS-Δ2194-RBM-2194–3′UTR construct) (Figure 3D, top panel, blue bar) resulted in a ∼ 20% increase in the β-galactosidase values. The increased β-galactosidase activities (Figure 3D, top panel, blue bar) were mirrored by an increase of the lacS transcript levels (Figure 3D, bottom panel, blue bar) obtained with the lacS-Δ2194-RBM-2194–3′UTR construct. The statistical significance of the β-galactosidase assays and qPCRs were validated using a t-test (Supplementary Table S2).
To verify these results, the reporter constructs lacS-2194-RBM-3′UTR, lacS-2194–3′UTR, lacS-Δ2194-RBM-2194–3′UTR as well as the lacS gene (lacS-3′UTR) were also equipped with the constitutively active tf55α promoter (34) (Supplementary Figure S7A and B). Basically, the same results were obtained with these constructs as observed for those equipped with the arabinose-inducible promoter (Supplementary Figure S7C and D), again indicating that the presence of the 2194-RBM impacts on lacS mRNA levels.
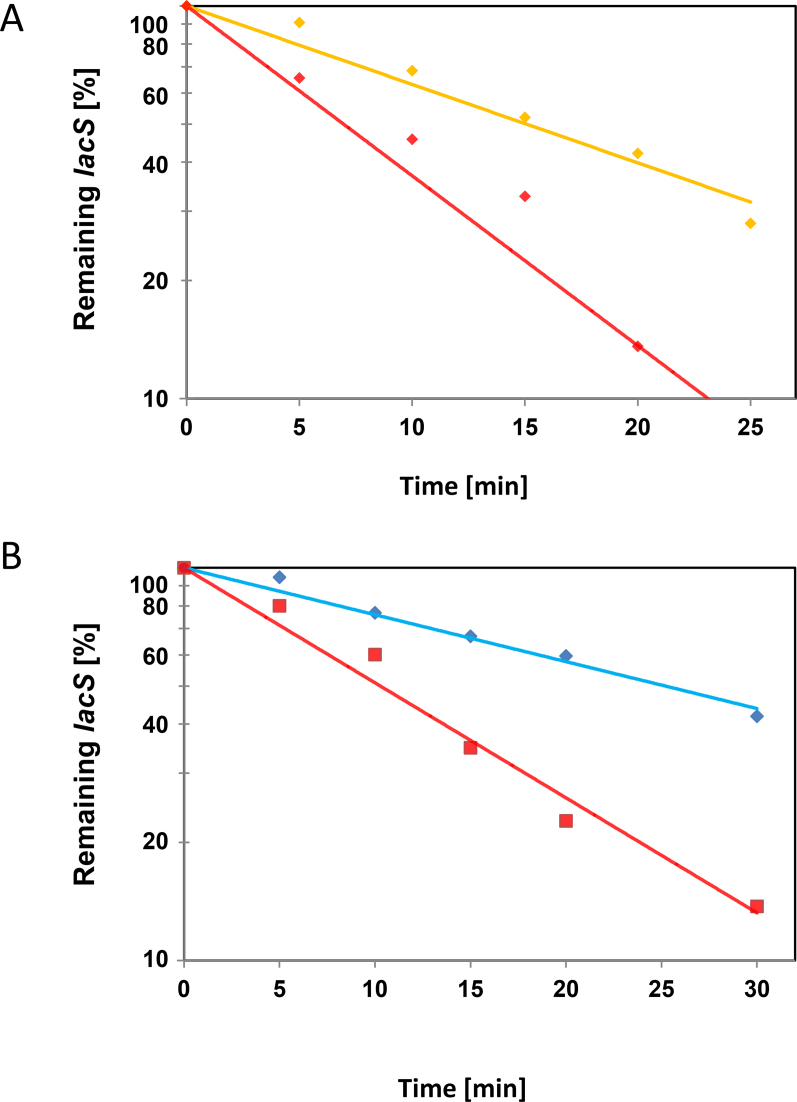
We next asked whether the presence of the 2194-RBM affects the stability of the corresponding chimeric transcripts (Figure 3A and B). Initially, actinomycin D was used to stop transcription as specified by Andersson et al. (35). However, the lacS mRNA levels from the reporter constructs increased with time showing that transcription from the plasmid borne Ara/tf55α promoters is not abolished after actinomycin D treatment, which was in contrast to chromosomally encoded control mRNAs showing a time-dependent decay (B. Märtens, unpublished). On the other hand, increased concentrations of actinomycin D resulted in cell lysis. We therefore used Ara-promoter shut-off as a means to study the remaining lacS mRNA levels in strains harboring the reporter constructs by shifting these from arabinose- to sucrose-containing medium.
To ascertain repression of the Ara-promoter by sucrose we first determined lacS mRNA levels and the β-galactosidase activities obtained with the lacS-3′UTR construct in the presence of sucrose (Supplementary Figure S8A). To test for shut-off of transcription, the lacS mRNA levels were then determined in strains harbouring the lacS-3′UTR and the lacS-2194-RBM-3′UTR constructs, respectively. The remaining lacS transcript levels were determined by qPCR at 2 h and 4 h after shift to sucrose containing medium (Supplementary Figure S8B). After 2 h the lacS mRNA levels derived from either construct were strongly decreased (0.7% remaining), indicating shut-off of the arabinose promoter in the presence of sucrose (Supplementary Figure S8B).
The remaining lacS levels in the strains harbouring the respective reporter constructs lacS-3′UTR, lacS-2194-RBM-3′UTR, lacS-2194–3′UTR, lacS-Δ2194-RBM-2194–3′UTR (see Figure 3A and B) were determined by qPCR at 0, 5, 10, 15, 20 and 25 min after shift to sucrose medium. When compared with the strain harbouring the lacS-3′UTR construct, the remaining lacS levels relative to time zero (shift from arabinose to sucrose containing growth medium) were reduced in strain carrying the lacS-2194-RBM-3′UTR construct during the time course of the experiment (Figure 4A). On the other hand, the remaining lacS levels were increased in the strain carrying the lacS-Δ2194-RBM-2194–3′UTR construct when compared with that observed in the strain bearing the lacS-2194–3′UTR construct (Figure 4B). Although the promoter shut-off approach does not preclude ongoing transcription at the beginning after the shift to sucrose, which hampers the determination of half-lifes, we interpret these experiments as showing that the presence of the 2194-RBM impacts on the stability of the respective transcripts.
Figure 4.
Reduced lacS mRNA levels upon placement of the 2194-RBM in its 3′UTR. (A) The Sso strains PH1–16(pMJ05-Ara-lacS-3′UTR) and PH1–16(pMJ05-Ara-lacS-2194-RBM-3′UTR) were grown in Brock's medium at 75°C containing 0.2% arabinose to an OD600 of 0.5. Then, the cells were harvested and resuspended in the same volume of Brock's medium containing 0.2% sucrose pre-warmed to 75°C ( = 0 min). After 5, 10, 15, 20 and 25 min, total RNA was isolated and cDNA was generated as described in Materials and Methods. The remaining lacS mRNA levels after promoter shut-off were quantified by qPCR using cDNA derived from Sso PH1–16 harboring either plasmid pMJ05-Ara-lacS-3′UTR (orange graph) or pMJ05-Ara-lacS-2194-RBM-3′UTR (red graph) and normalized to 16S rRNA levels. The experiment was performed with two biological and three technical replicates. A least square analysis of semilogarithmic plots of remaining lacS mRNA versus time is shown. The respective lacS mRNA levels at time 0 were set to 100%. (B) The experiment was performed as described in A, except that the strains PH1–16(pMJ05-Ara-lacS-2194–3′UTR) (red graph) and PH1–16(pMJ05-Ara-Δ2194-RBM-2194–3′UTR) (light blue graph) were used and a 30 min time point was included.
DISCUSSION
Hitherto only a few studies have been performed to reveal the function of SmAPs (4,6,18,24). In general archaeal Sm proteins seem to bind to different species of ncRNAs e.g. tRNAs, C/D box RNAs and other ncRNAs (6,18). We did not observe a specific motif in the ncRNAs solely enriched with Sso SmAP1 or Sso SmAP2 (6). It is conceivable that binding occurs through U-rich single stranded regions as binding of Sso SmAP1 and SmAP2 to poly(U)15 was observed (Supplementary Figure S3). In support, recent transcriptome analyses performed with two phylogenetically distant Archaea, Methanosarcina mazei and Sulfolobus acidocaldarius, revealed that many transcripts contain U-rich stretches at their 3′ ends (27).
The high affinity of Sso SmAP2 for the SmAP2-cRBM (Figure 1C) suggests that the protein binds to a population of transcripts harbouring the motif. Many of the 53 transcripts comprising the SmAP2-RBM encode transport proteins for amino acids and carbohydrates (6). However, it remains to be elucidated whether this list is exhaustive. Another issue concerns the conservation of the SmAP2-RBM in other Archaea that contain the Sm2-type Lsm protein. The motif comparison tool Tomtom (29) revealed a partial overlap of the SmAP2-cRBM with eukaryotic PCBP1–3, SRSF, PUF68, hnRNP, HRB27C and G3BP2 RNA binding motifs (Supplementary Figure S9). However, the homologues part of the SmAP2-cRBM and these eukaryotic RNA-binding motifs is rather short (≤ 7 nt). The Sm binding site in the Sm-class snRNAs consensus sequence AAUUUUUGG in Eukaryotes (14) resembles partially the middle part of the identified SmAP2-cRBM. It seems conceivable that the thermophilic lifestyle of Sso may require a longer RNA binding motif for formation of stable SmAP2-RNA complexes. We also tested whether the 1742-RBM, 2194-RBM and 2912-RBM is conserved in other crenarchaeota, which was found to be the case. In addition, we looked for the occurrence of the SmAP2-cRBM in the Sso genome using FIMO (30). Using a threshold of 1 × 10−6 we could identify 99 motifs in the entire genome (B. Märtens, unpublished).
The SmAP2-cRBM was shown to crosslink to the SmAP2 sub-sequence L34–R46 (Figure 2A–C; Supplementary Figure S4). The majority of the SmAP1 proteins in crenarchaeota and euryarchaeota contain a H40 residue within the DxXxN motif, which has been described for eukaryal Lsm-complexes (21). In contrast, the crenarchaeal SmAP2 proteins contain a T at position 40 (6), which is apparently pivotal for SmAP2-cRBM binding (Figure 2E). The uracil-binding pocket of SmAP proteins are usually formed by the N42 and H40 residues in the L3 domain and by R66 in the L5 domain (4) (Figure 2B–D; Supplementary Figure S5B). The same amino acid residues located in the DxXxN and IRG motifs of eukaryal Lsm complexes are involved in binding to 5′-GUUUU-3′ (15) (Figure 2D; Supplementary Figure S5A). When compared with the affinity of Haloferax volcanii SmAP1 for poly(U)30 (18), the Sso-SmAP2 displayed a 10–40-fold lower affinity for poly(U)15, which might be interpreted as showing that T40 actually hinders poly(U) binding.
K25, located in the L2 domain of SmAP2 (Supplementary Figure S5) and positioned in the pore (Figure 2A–C) was shown to interact with the SmAP2-cRBM (Supplementary Figure S4). K25 is a highly conserved residue within the Sm1 motif of SmAPs and is not present in eukaryal Sm/Lsm proteins (http://www.genome.jp/tools/blast). Further studies are necessary to show whether the K25-SmAP2-cRBM interaction is unique for Sso-SmAP2. However, as the K25A exchange in SmAP2 reduced binding to the SmAP2-cRBM but did not abolish it, it might be more likely that this residue contributes to overall rather than to specific binding of RNA substrates.
In Eukaryotes, deadenylation of mRNAs can either lead to 3′ to 5′ decay by the exosome or to 5′ to 3′ directional decay by Xrn1. In the latter scenario, the association of the Lsm1–7/Pat1 complex with the 3′ end of deadenylated mRNAs leads to the formation of a larger decapping complex including the decapping enzymes Dcp1/Dcp2 and the decapping activators Scd6, Edc3 and Dhh1 as well as to the recruitment of Xrn1, which in turn results in 5′ to 3′ directional decay of the mRNA (36).
The mechanisms underlying mRNA turnover in Sso are poorly understood. An exosome complex has been characterized in Sso, which catalyses 3′ to 5′ exonucleolytic decay, when the intracellular ATP level is low (37). In addition, an exoribonuclease with 5′ to 3′ directionality has been identified, which belongs to the aCPSF2 group of ß-CASP proteins. The Sso-aCPSF2 activity is affected by the phosphorylation state of the 5′ end of RNA, and was shown to impact on RNA turnover (38). How might the presence of the SmAP2-RBM affect transcript stability and turnover? A recent interactome study (39) did not reveal an interaction between SmAP2 and aCPSF2. Nevertheless, this might not exclude the possibility that aCPSF2 is recruited by SmAP2 via an adaptor protein as exemplified by Xrn1 recruitment to the Lsm1–7/Pat1 complex (36). This working hypothesis would not necessarily require a distinct location of the SmAP2-RBM, and thus would be compatible with the different locations of the SmAP2-RBM in the 53 analyzed mRNAs (Supplementary Table S1). It is further worth noting that SmAP2 was found to interact with DnaG (39), a component of the Sso exosome (37), with a putative 8.5-kDa ssh7a (Sso10610) endoribonuclease (39,40) as well as with a putative metal-dependent RNA phosphohydrolase (39). Thus, there might be other modi operandi of RNA turnover for SmAP2 than mentioned above for eukaryal Lsm proteins. In any case this awaits molecular characterization of the players involved and testing of different mRNA decay models.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. S.V. Albers for providing materials and plasmids and Dr. G. A. Bezerra for help with the analytical size exclusion chromatography. L. Rampelt is acknowledged for her technical help. The MST measurements were performed with aid of the Protein Technology Facility of the VBCF, Vienna Biocenter.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Science Fund (FWF) [22888 to U.B.]. Funding for open access charge: FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wilusz C.J., Wilusz J.. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013; 10:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collins B.M., Harrop S.J., Kornfeld G.D., Dawes I.W., Curmi P.M., Mabbutt B.C.. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001; 309:915–923. [DOI] [PubMed] [Google Scholar]
- 3. Mura C., Cascio D., Sawaya M.R., Eisenberg D.S.. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Törö I., Thore S., Mayer C., Basquin J., Seraphin B., Suck D.. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001; 20:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Törö I., Basquin J., Teo-Dreher H., Suck D.. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J. Mol. Biol. 2002; 320:129–142. [DOI] [PubMed] [Google Scholar]
- 6. Märtens B., Bezerra G.A., Kreuter M.J., Grishkovskaya I., Manica A., Arkhipova V., Djinovic-Carugo K., Bläsi U.. The heptameric SmAP1 and SmAP2 proteins of the crenarchaeon Sulfolobus solfataricus bind to common and distinct RNA targets. Life (Basel). 2015; 5:1264–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vogel J., Luisi B.F.. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011; 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Regnier P., Hajnsdorf E.. The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3′ ends of RNAs resulting from Rho-independent termination: a tentative model. RNA Biol. 2013; 10:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schumacher M.A., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G.. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002; 21:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Link T.M., Valentin-Hansen P., Brennan R.G.. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19292–19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng A., Panja S., Woodson S.A.. Arginine patch predicts the RNA annealing activity of Hfq from Gram-negative and Gram-positive bacteria. J. Mol. Biol. 2016; 428:2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schu D.J., Zhang A., Gottesman S., Storz G.. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015; 34:2557–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santiago-Frangos A., Kavita K., Schu D.J., Gottesman S., Woodson S.A.. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6089–E6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matera A.G., Terns R.M., Terns M.P.. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:209–220. [DOI] [PubMed] [Google Scholar]
- 15. Zhou L., Hang J., Zhou Y., Wan R., Lu G., Yin P., Yan C., Shi Y.. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature. 2013; 506:116–120. [DOI] [PubMed] [Google Scholar]
- 16. Mura C., Randolph P.S., Patterson J., Cozen A.E.. Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on Sm function. RNA Biol. 2013; 10:636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Achsel T., Stark H., Luhrmann R.. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:3685–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fischer S., Benz J., Spath B., Maier L.K., Straub J., Granzow M., Raabe M., Urlaub H., Hoffmann J., Brutschy B. et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010; 285:34429–34438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikulin A., Mikhailina A., Lekontseva N., Balobanov V., Nikonova E., Tishchenko S.. Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii. J. Biomol. Struct. Dyn. 2016; 1–34. [DOI] [PubMed] [Google Scholar]
- 20. Mura C., Kozhukhovsky A., Gingery M., Phillips M., Eisenberg D.. The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs). Protein Sci. 2003; 12:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thore S., Mayer C., Sauter C., Weeks S., Suck D.. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2003; 278:1239–1247. [DOI] [PubMed] [Google Scholar]
- 22. Kilic T., Thore S., Suck D.. Crystal structure of an archaeal Sm protein from Sulfolobus solfataricus. Proteins. 2005; 61:689–693. [DOI] [PubMed] [Google Scholar]
- 23. Murina V.N., Nikulin A.D.. RNA-binding Sm-like proteins of bacteria and archaea. similarity and difference in structure and function. Biochemistry (Mosc). 2012; 76:1434–1449. [DOI] [PubMed] [Google Scholar]
- 24. Maier L.K., Benz J., Fischer S., Alstetter M., Jaschinski K., Hilker R., Becker A., Allers T., Soppa J., Marchfelder A.. Deletion of the Sm1 encoding motif in the lsm gene results in distinct changes in the transcriptome and enhanced swarming activity of Haloferax cells. Biochimie. 2015; 117:129–137. [DOI] [PubMed] [Google Scholar]
- 25. Wurtzel O., Sapra R., Chen F., Zhu Y., Simmons B.A., Sorek R.. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010; 20:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Märtens B., Manoharadas S., Hasenöhrl D., Manica A., Bläsi U.. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013; 14:527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dar D., Prasse D., Schmitz R.A., Sorek R.. Widespread formation of alternative 3′ UTR isoforms via transcription termination in archaea. Nat. Microbiol. 2016; 1:16143. [DOI] [PubMed] [Google Scholar]
- 28. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics (Oxford, England). 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duhr S., Braun D.. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:19678–19682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma K., Hrle A., Kramer K., Sachsenberg T., Staals R.H., Randau L., Marchfelder A., van der Oost J., Kohlbacher O., Conti E. et al. Analysis of protein-RNA interactions in CRISPR proteins and effector complexes by UV-induced cross-linking and mass spectrometry. Methods (San Diego, Calif.). 2015; 89:138–148. [DOI] [PubMed] [Google Scholar]
- 33. Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., Urlaub H.. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014; 11:1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Albers S.V., Jonuscheit M., Dinkelaker S., Urich T., Kletzin A., Tampe R., Driessen A.J., Schleper C.. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 2006; 72:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andersson A.F., Lundgren M., Eriksson S., Rosenlund M., Bernander R., Nilsson P.. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006; 7:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nissan T., Rajyaguru P., She M., Song H., Parker R.. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 2010; 39:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evguenieva-Hackenberg E., Bläsi U.. Attack from both ends: mRNA degradation in the crenarchaeon Sulfolobus solfataricus. Biochem. Soc. Trans. 2013; 41:379–383. [DOI] [PubMed] [Google Scholar]
- 38. Hasenöhrl D., Konrat R., Bläsi U.. Identification of an RNase J ortholog in Sulfolobus solfataricus: implications for 5′-to-3′ directional decay and 5′-end protection of mRNA in Crenarchaeota. RNA (New York, N.Y.). 2011; 17:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Märtens B., Hou L., Amman F., Wolfinger M.T., Evguenieva-Hackenberg E., Bäsi U.. The SmAP1/2 proteins of the crenarchaeon Sulfolobus solfataricus interact with the exosome and stimulate A-rich tailing of transcripts. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shehi E., Serina S., Fumagalli G., Vanoni M., Consonni R., Zetta L., Deho G., Tortora P., Fusi P.. The Sso7d DNA-binding protein from Sulfolobus solfataricus has ribonuclease activity. FEBS Lett. 2001; 497:131–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.