Abstract
MicroRNAs are a class of small non-coding RNAs that serve as important regulators of gene expression at the posttranscriptional level. They are stable in body fluids and pose great potential to serve as biomarkers. Here, we present a highly specific, sensitive and cost-effective system to quantify miRNA expression based on two-step RT-qPCR with SYBR-green detection chemistry called Two-tailed RT-qPCR. It takes advantage of novel, target-specific primers for reverse transcription composed of two hemiprobes complementary to two different parts of the targeted miRNA, connected by a hairpin structure. The introduction of a second probe ensures high sensitivity and enables discrimination of highly homologous miRNAs irrespectively of the position of the mismatched nucleotide. Two-tailed RT-qPCR has a dynamic range of seven logs and a sensitivity sufficient to detect down to ten target miRNA molecules. It is capable to capture the full isomiR repertoire, leading to accurate representation of the complete miRNA content in a sample. The reverse transcription step can be multiplexed and the miRNA profiles measured with Two-tailed RT-qPCR show excellent correlation with the industry standard TaqMan miRNA assays (r2 = 0.985). Moreover, Two-tailed RT-qPCR allows for rapid testing with a total analysis time of less than 2.5 hours.
INTRODUCTION
MicroRNAs (miRNAs) are short non-coding RNA molecules (∼19–24 nt long) that mediate regulation of gene expression at the post-transcriptional level (1,2). Production of miRNAs starts with the transcription of genomic DNA into long primary transcripts called pri-miRNAs. The pri-miRNAs are subsequently cleaved by RNase III Drosha into shorter precursor transcripts with hairpin structure called pre-miRNAs. The pre-miRNAs are transported into the cytoplasm where they are processed by RNase III Dicer into ∼22 nt double-stranded miRNA molecules. Both strands of this duplex may become functional mature miRNAs (3–5). MiRNAs may function as master regulators of numerous physiological and pathological processes and changes in their expression patterns are often observed in various diseases (6–10). Because of their remarkable stability in biofluids miRNAs have exciting potential to serve as minimally invasive diagnostic biomarkers (11–14).
MiRNA expression can be measured by many techniques; the three most common being microarrays, next generation sequencing (RNA-Seq), and reverse transcription quantitative PCR (RT-qPCR). In addition, non-PCR based isothermal amplification methods have also been proposed (15–18). Each of these methods has its advantages and limitations. Microarray analysis is generally more cost efficient than RNA-Seq and offers the possibility to monitor large number of targets, but, at least with conventional microarrays, specificity and dynamic range are limited. RNA-Seq is suitable for high-throughput and is the only platform capable of discovering new miRNAs. Disadvantages of RNA-Seq are the rather high cost per sample and the complexity of the workflow and data analysis. Also, the precision of quantitation is poor for the low abundant miRNAs. RT-qPCR is the method commonly used for the validation of results from screening experiments and when high accuracy and precision is required. It is also the method of choice when only a small number of targets is quantified, particularly when the amount of material is limiting. Another appealing aspect is the simple workflow easily set up in laboratories that have experience in RT-qPCR (19–22).
There are, however, significant technical challenges in miRNA expression profiling using RT-qPCR. MiRNA molecules are only 19–24 nt long, which is the length of a conventional PCR primer. The sequence of the mature miRNA is contained in its precursor molecules (pri-miRNAs and pre-miRNAs), however, only the mature miRNAs are believed to have effector functions and they usually are the targets for quantification. The GC content of miRNAs is highly variable, which complicates assay design and protocol optimization, particularly when a common protocol is sought for multiple/all miRNA targets. The sequences of miRNAs within the same family, such as let-7, may be highly similar, differing only in a single base position. MiRNAs are subject to various post-transcriptional modifications and may differ in sequence and nucleotide composition at either or both ends. This obscures specific quantification with many techniques (23–25).
Several methods to quantify miRNAs based on RT-qPCR have been developed to date, many of which are commercially available. Generally, the methods can be divided into two groups that are based on universal and specific reverse transcription, respectively. In addition, probe-ligation methods that do not require reverse transcription have also been proposed (26,27). In the universal RT approach, all miRNA molecules are elongated by an identical tail used to prime the reverse transcription into cDNA. These methods include the addition of poly-A tails to the 3′-end with the poly(A) polymerase (Exiqon′s miRCURY LNA system, Qiagen′s miScript PCR system, Quantabio′s qScript microRNA system) (28,29), polyuridylation with poly(U) polymerase (30), ligation of a universal linker with T4 RNA ligase (31), and more recently combination of linker ligation and end tailing (Thermo Fisher′s TaqMan Advanced miRNA assays). The main advantage of these approaches is that all miRNAs are converted into cDNA in the same tube. However, they may suffer from high background noise and they are often limited by the efficiency of the extra enzymatic steps needed. Some also require special reagents. Moreover, small RNAs possessing a 2′-O-methyl (2′-O-Me) modification on their 3′ terminal nucleotide, such as plant mature miRNAs and piRNAs, are resistant to polyadenylation and cannot be efficiently reverse transcribed using a polyadenylation-based cDNA synthesis approach (32). The polyadenylation and ligation steps also introduce bias (33–35). The second group of methods includes the use of linear primers (36,37), pincer probes (38), and stem–loop RT primers (39), also known as TaqMan miRNA assays (Thermo Fisher). The stem–loop method is probably the most common today and is frequently used for benchmarking and validation of other methods (40).
The stem–loop primers of the TaqMan miRNA assays are composed of a short single stranded sequence at their 3′-ends that anneals to the 3′-end of the targeted miRNA, a double-stranded segment (the stem) and a loop. The stem–loop structure shifts the equilibrium to the formation of an RNA/DNA duplex and should prevent binding of the primer to pri- and pre-miRNAs and to any dsDNA that may be present. Nevertheless, it has been reported that presence of genomic and plasmid DNA containing sequences of the corresponding pre-miRNA give rise to significant background signal leading to false positive results (41). The method uses hydrolytic probes that are costly to produce and do not allow controlling the specificity of the reaction by melting curve analysis. Notably, the TaqMan probe does not contribute to the specificity of the reaction, as it binds to a site originating from the sequence of the RT primer. Another limitation of this design is the reduced ability to reverse transcribe isomiR variants (24,41,42). Although the stem–loop approach employs target-specific RT primers, the reverse transcription step can be multiplexed using multiple RT primers in the same tube (43–45).
We have developed a novel specific and cost-effective approach to quantify miRNA expression that utilizes specific structured primers for reverse transcription and SYBR-green based qPCR named ‘Two-tailed RT-qPCR’. The Two-tailed RT primers are composed of two hemiprobes complementary to separate regions of the target miRNA and of an oligonucleotide tether folded into a hairpin. This novel design increases the binding strength of the RT primer to its template leading to increased sensitivity. The 3′-hemiprobe can be short, providing high discriminatory power to mismatches in the 3′-region and leaving enough space for the design of miRNA specific qPCR primers with sufficient melting temperature (Tm). The 5′-hemiprobe improves the discrimination between highly similar sequences, particularly when the differing nucleotide is located in the center or close to the 5′-end of the miRNA sequence. Since Two-tailed RT primers do not interact with the ends of the miRNA they are able to detect all terminal variants of any miRNA (isomiRs) and therefore accurately reflect the true miRNA content in a sample.
MATERIALS AND METHODS
Primers, templates and synthetic oligonucleotides
Sequences of the miRNA oligonucleotides were obtained from the miRBase Release 21 (www.mirbase.org) (46). Sequences of primers and targets are listed in supplementary file. Secondary structure of the Two-tailed RT primers were predicted using the UNAfold web server (http://unafold.rna.albany.edu/) (47). RNA oligonucleotides were synthesized and quantified by Integrated DNA Technologies. DNA primers were synthesized and quantified by Invitrogen. Precursor miRNAs were synthesized by in vitro transcription from corresponding PCR products using T7 RNA polymerase (New England Biolabs) according to the manufacturer′s protocol (suppl. file). Reactions were treated with the Turbo DNA-free kit (Thermo Fisher), RNA was precipitated in 3M LiCl and quantified with the Qubit 2.0 fluorometer (Thermo Fisher). Correct size of the precursor miRNA products was verified using the Fragment Analyzer (Advanced Analytical).
cDNA synthesis
RT reactions were performed with the qScript flex cDNA kit (Quantabio) in a total reaction volume of 10 μl. The reaction mixture contained either 10 ng of total RNA or synthetic miRNA template, 1× RT buffer, 0.05 μM RT primer, 1 μl GSP enhancer and 0.5 μl RT enzyme. RT reactions were incubated in a 96-well plate in a Bio-Rad CFX 1000 thermocycler for 45 min at 25°C, 5 min at 85°C and then held at 4°C. Reactions using TaqMan miRNA assays (Thermo Fisher) and Quantabio qScript microRNA system (Quantabio) were performed according to the manufacturer′s protocol except that the total reaction volume was scaled down to 10 μl. Reactions using miQPCR method were performed as described in (31) according to the protocol obtained from the corresponding author (personal communication): ligation of template miRNAs to the miLINKER adaptor was performed in a total reaction volume of 8 μl containing 0.8× T4 buffer (New England Biolabs), 5 mM MgCl2, 17% PEG 8000, 0.15 μM miLINKER adaptor, 0.1 μl RNaseOUT (40U/μl) (Thermo Fisher) and 0.18 μl T4 RNA Ligase 2, truncated K227Q (New England Biolabs). The ligation reaction was incubated for 30 min at 25°C and then placed at 4°C. The ligated miRNAs were then incubated for 2 min at 85°C with 0.5 μM dNTPs and 0.05 μM universal mQ-RT primer in a total reaction volume of 14 μl and then reverse transcribed in total reaction volume of 20 μl containing 1× RT buffer, 5 mM DTT and 1 μl SuperScript III (Thermo Fisher) for 30 min at 46°C, 5 min at 85°C and finally held at 4°C.
Quantitative PCR
qPCR was performed in a total reaction volume of 10 μl containing 1× SYBR Grandmaster Mix (TATAA Biocenter), 0.4 μM forward and reverse primer and the cDNA product diluted at least 10×. Reactions were performed in duplicates and incubated in a 96- or 384-well plate in a CFX 96 or CFX 384 Real Time Detection System (Bio-Rad) at 95°C for 30 s, followed by 45 cycles of 95°C for 5 s and 60°C for 15 s. Reaction specificity was assessed by melting curve analysis immediately after the qPCR. qPCR with TaqMan miRNA assays and Quantabio qScript microRNA system were performed according to manufacturers′ protocols in a total reaction volume of 10 μl. cDNA was diluted at least 15× or 10× for the TaqMan and Quanta reactions respectively. qPCR with the miQPCR method was performed in a total reaction volume of 10 ul containing 1x SYBR Grandmaster Mix, 0.15 μM forward and reverse primer and the cDNA product diluted 100× according to the recommended protocol (31). Reactions were incubated at 95°C for 30 s, followed by 45 cycles of 95°C for 10 s and 60°C for 35 s followed by melting curve analysis.
MiRNA profiling in mouse tissues
All procedures involving the use of laboratory animals were performed in accordance with the European Community Council Directive of 24 November 1986 (86/609/EEC) and animal care guidelines approved by the Institute of Experimental Medicine, Academy of Sciences of the Czech Republic (Animal Care Committee decision on 17 April 2009; approval number 85/2009). Mouse tissue samples from brain, cerebellum, liver, lung, kidney, heart and skeletal muscle were dissected, placed into TRI Reagent (Sigma-Aldrich) and were immediately frozen on dry ice. Before use, samples were thawed, homogenized using the TissueLyser (Qiagen) and total RNA was extracted with TRI Reagent (Sigma-Aldrich) according to the manufacturer′s protocol. RNA quantity and purity was assessed using the NanoDrop 2000 spectrophotometer (Thermo Fisher) and RNA integrity was assessed using the Fragment Analyzer (Advanced Analytical). Inhibition of the RT-qPCR workflow was tested for using an RNA spike control (Tataa Biocenter). Data were normalized to total amount of RNA. Same aliquots were used for all measurements. Cq values were transformed to quantities relative to the sample with the lowest expression for each target miRNA separately and expression values were converted to log scale. Pearson correlation coefficients were calculated based on logarithmic expression values. GenEx 6 software (MultiD) was used for data pre-processing.
RESULTS
General assay design
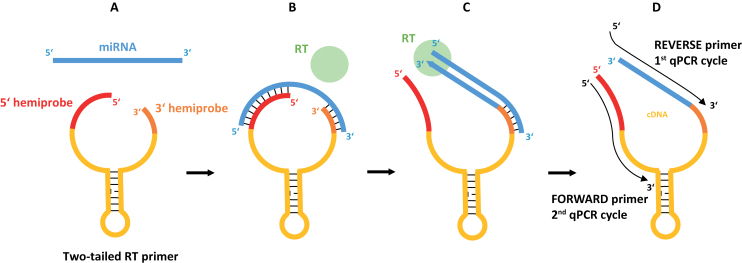
A novel two step RT-qPCR system for the quantification of microRNAs is presented (Figure 1). Reverse transcription is performed with target-specific structured primers that are about 50 nucleotides long and contain two target specific hemiprobes complementary to the miRNA. The 3′-hemiprobe is about 6 nt long and binds to the 3′-region of the target miRNA. The 5′-hemiprobe is usually longer and binds within the 5′-region of the targeted microRNA. The two hemiprobes are connected by an oligonucleotide tether designed to fold into a hairpin to prevent nonspecific interactions (Figure 1A). After hybridization, the RT reaction is primed from the 3′-hemiprobe. The 5′-hemiprobe is displaced by the RT enzyme and the Two-tailed RT primer is elongated to produce cDNA with a sequence complementary to the targeted miRNA (Figure 1B and C). The cDNA is then quantified by conventional qPCR utilizing SYBR-Green chemistry with two target-specific PCR primers. The reverse PCR primer is specific for the miRNA target sequence while the forward primer is specific for the predesigned region in the 5′-end of the Two-tailed RT primer (Figure 1D).
Figure 1.
Schematic of Two-tailed RT-qPCR. (A) Two-tailed RT primer having two hemiprobes connected by a hairpin folding sequence. (B) The hemiprobes bind cooperatively, one at each end of the target miRNA, forming a stable complex. (C) Reverse transcriptase binds the 3′-end of the hybridized Two-tailed RT primer and elongates it to form tailed cDNA. (D) The cDNA is amplified by qPCR using two target-specific primers.
The Two-tailed RT primer has 3 functions: i) it primes specifically the reverse transcription of the target miRNA template ii) it contributes with additional sequence to the cDNA making it long enough for PCR amplification iii) it contains the sequence of the forward PCR primer. We reasoned that the introduction of a second binding element, the 5′-hemiprobe that binds within the 5′-end of the miRNA, will increase the sensitivity and specificity of the RT reaction, as more nucleotides can be interrogated. Also, the 3′-hemiprobe can be made shorter (5–6 nt), providing flexibility to design the reverse PCR primer with adequate Tm without overlapping with the 3′-hemiprobe sequence, thereby avoiding the risk of undesired self-priming and primer-dimer formation. The 5′-hemiprobe also contributes to increased discriminatory power between highly similar targets that differ only by 1 nt in the center or close to the 5′-end of the miRNA sequence. The reason is that the target miRNAs are subjected to sequence interrogation twice: first in the RT and then in the qPCR.
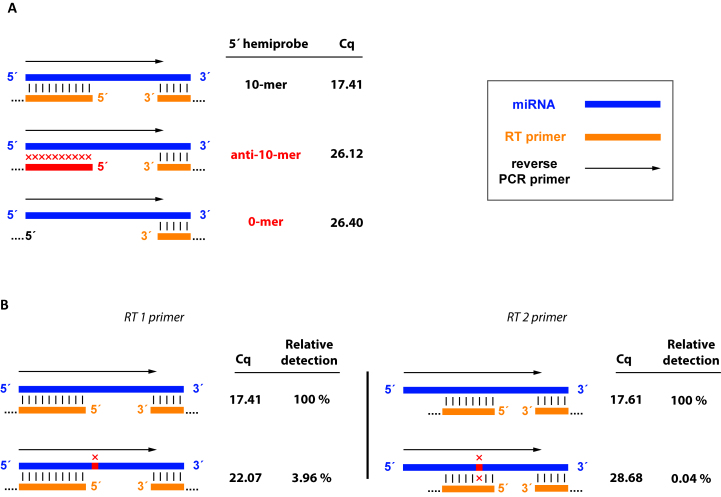
To test the concept we designed three Two-tailed RT primers with the same 3′-hemiprobe (5 nt), but different 5′-hemiprobes: one with 10 complementary nucleotides to the target, one with 10 non-complementary nucleotides, and one without 5′-hemiprobe. We found that Two-tailed RT primers that lacked complementary 5′-hemiprobe gave significantly higher qPCR Cq values demonstrating the contribution of the 5′-hemiprobe to the sensitivity of the system (Figure 2A).
Figure 2.
Importance of the 5′-hemiprobe for sensitivity and specificity. (A) Two-tailed RT primers with 5 nt complementary 3′-hemiprobe and either 10 nt complementary 5′-hemiprobe (top), 10 nt non-complementary 5′-hemiprobe (middle), or no 5′-hemiprobe at all (bottom) targeting let-7a. Cq values obtained with the Two-tailed RT primer having a complementary 5′-hemiprobe are about nine cycles lower than those obtained with the Two-tailed RT primers lacking 5′-complementarity. (B) Two-tailed RT primers used to assay targets that differ in one nucleotide. The variable nucleotide is in the non-interrogated region between the hybridization sites of the 3′- and 5′-hemiprobes (left) and the variable nucleotide is within the 5′-hemiprobe binding site (right).
To test the contribution of the 5′-hemiprobe to the specificity, we compared two Two-tailed RT primers for their ability to discriminate between two members of the Let-7 miRNA family: Let-7a and Let-7f. These differ only in one nucleotide located in the center of the miRNA sequence. RT 1 primer was designed to bind with its 5′-hemiprobe to the first ten nucleotides from the 5′-end of let-7a, while RT 2 primer had a 5′-hemiprobe that binds to 8 nucleotides in the center of the let-7a sequence (Figure 2B). With this design, the nucleotide distinguishing let-7a from let-7f is sensed by the 5′-hemiprobe of the RT 2 primer, but not by the RT 1 primer. The 5′-hemiprobe of the RT 2 primer was also shorter (8 instead of 10 nucleotides), as we reasoned the impact of a mismatch will be more prominent with a shorter probe sequence. The rest of the RT 1 and RT 2 primer sequences as well as the PCR primers used were identical. The measured ΔCq between the fully matched and the mismatched template was substantially larger when using the RT 2 primer, which overlapped the differing base with its 5′-hemiprobe (ΔCq = 11.07), than with the RT 1 primer, which did not (ΔCq = 4.66). This demonstrates the significant contribution from the 5′-hemiprobe to the specificity of the system. Notably, the Cq values for the let-7a microRNA, to which both Two-Tailed RT primers were fully complementary, were equal suggesting that the assay sensitivity remained the same even though the length of the 5′-hemiprobe differed by two bases.
Repeatability
To compare the performance of the Two-tailed RT-qPCR to other RT-qPCR methods for miRNA analysis we performed most of the experiments also with the TaqMan microRNA assays, the Quantabio qScript microRNA system, and the miQPCR method published in (31) (in the following text referred to as TaqMan, Quanta, and miQPCR). These methods use different strategies to produce cDNA for subsequent qPCR. TaqMan uses miRNA specific RT primers. Quanta uses poly(A) polymerase to add poly(A) tails to the 3′ ends of miRNAs to allow for universal reverse transcription with an oligo(dT) primer, and miQPCR ligates a defined adaptor sequence to the 3′ end of miRNAs prior to reverse transcription of ligated constructs with an universal RT primer.
We assessed the repeatability of the new Two-tailed RT-qPCR and the three known methods by measuring the imprecision, expressed as standard deviation of Cq values obtained from triplicate measurements having the same input RNA (Table 1). Three miRNAs expressed at high (let-7a), moderate (miR-21), and low levels (miR-193a) were measured in total RNA extracted from mouse cerebellum. First step in the workflow of every method was taken as the point for replication. Overall, all tested methods displayed very high repeatability as demonstrated by low standard deviations of the replicate measurements (Table 1).
Table 1. Average Cq values and standard deviations of triplicate measurements of three miRNAs quantified by four different methods. Sample was total RNA isolated from mouse cerebellum.
| Two-tailed RT-qPCR | TaqMan | Quanta | miQPCR | |||||
|---|---|---|---|---|---|---|---|---|
| Cq | St.dev | Cq | St.dev | Cq | St.dev | Cq | St.dev | |
| let-7a | 19.59 | 0.21 | 26.69 | 0.28 | 19.87 | 0.14 | 24.65 | 0.16 |
| miR-21 | 23.73 | 0.22 | 28.92 | 0.24 | 23.36 | 0.02 | 29.39 | 0.16 |
| miR-193a | 29.12 | 0.17 | 35.95 | 0.85 | 31.13 | 0.29 | 35.15 | 0.12 |
Sensitivity and dynamic range
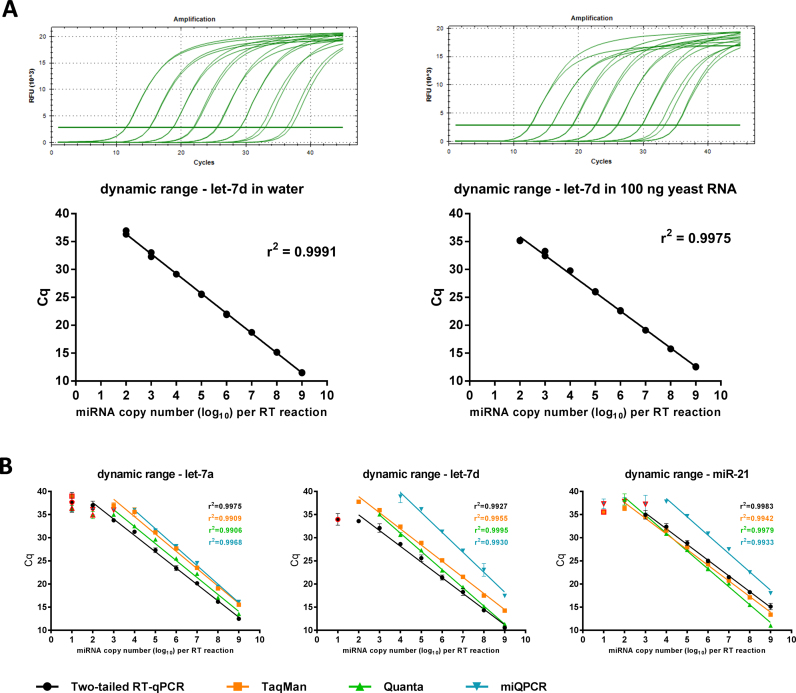
The sensitivity and dynamic range of the Two-tailed RT-qPCR were evaluated using synthetic let-7d miRNA as target. A dilution series spanning 8 orders of magnitude was prepared ranging from approx. 10 to 109 copies of let-7d miRNA molecules per RT reaction. 10% of the cDNA product was used for qPCR. The Two-tailed RT-qPCR assay exhibited excellent linearity between the log of the miRNA input and Cq values over 7 orders of magnitude and accurately quantified down to 10 cDNA copies of let-7d corresponding to 100 miRNAs in the original sample (Figure 3A). Similar results were obtained when 100 ng of yeast total RNA was added to each RT reaction to simulate the complex background in biological samples (Figure 3A). To determine the limit of detection (LoD) more precisely, a 2-fold serial dilution of let-7d approaching zero-concentration was performed with each sample analyzed in hexaplicate. LoD is estimated at the lowest concentration that produces 95% positive replicates and can be roughly estimated by fitting the fraction of positive replicates to the logarithm of the concentration using GenEx software (MultiD) (48). LoD of the two-tailed RT PCR assay for let-7d microRNA was estimated to 111 miRNA molecules, which corresponds to 11 cDNA molecules in our workflow as only 10% of the cDNA was used as template for qPCR (suppl. file). The LoD estimate relies on the miRNA stock concentration provided by the oligonucleotide manufacturer (IDT), which was determined spectroscopically. This may have overestimated the concentration of intact full length miRNA to some degree as any contaminating byproducts contribute to absorption. Hence, the LoD we estimate is an upper limit.
Figure 3.
Dynamic range and sensitivity of Two-tailed RT-qPCR, TaqMan, Quanta and miQPCR. (A) Amplification plots and standard curves of let-7d assayed in water and against a background of 100 ng yeast RNA. The dynamic range is seven logs. (B) Standard curves of let-7a, let-7d, and miR-21 assayed with Two-tailed RT-qPCR, TaqMan, Quanta, and miQPCR. Cq values outside the linear range are indicated with red border.
To further compare the sensitivity and dynamic range of the two-tailed RT-qPCR to the other methods we prepared dilution series also of let-7a, let-7d and miR-21. Three of the methods (two-tailed, TaqMan, Quanta) typically detected hundreds of miRNA copies with linear response down to at least 103 miRNAs in the original sample or 100 cDNA per RT reaction (Figure 3B). miQPCR performed less good, producing significantly higher Cq values and a narrower linear dynamic range reaching down to 104 miRNA copies (Figure 3B).
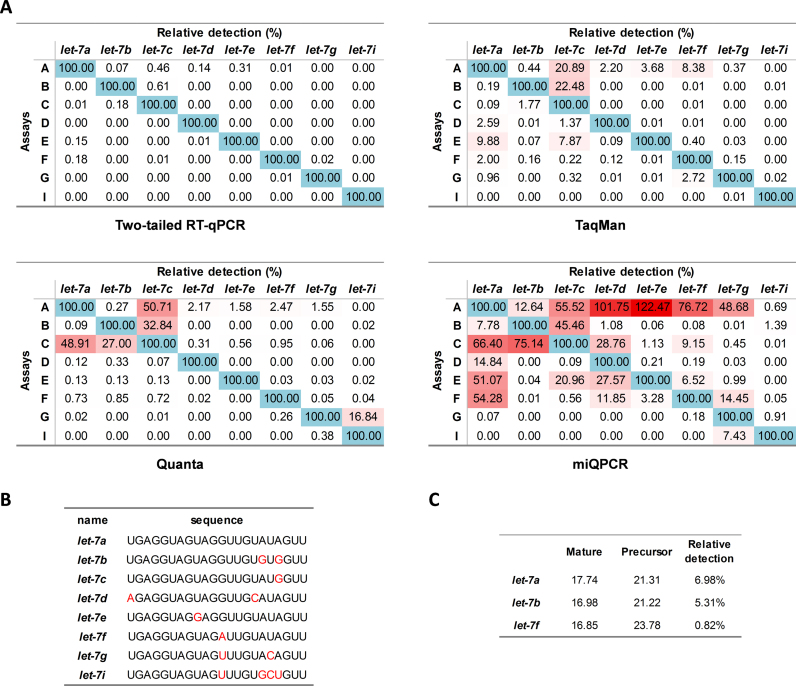
Discrimination of highly similar sequences
The capability to discriminate between highly homologous sequences with the Two-tailed RT-qPCR was tested on the let-7 miRNA family. The members of this family are highly similar as four pairs of the let-7 miRNAs differ only in a single nucleotide in different positions, posing major challenge for specific quantification (Figure 4A). We started by identifying the optimal length of the 5′-hemiprobe to maximize specificity, without compromising too much on sensitivity, and then designed assays for the let-7 family members accordingly (Supplementary Figure S1, suppl. file). We assayed approximately 2 × 108 copies of each let-7 miRNA target with each let-7 Two-tailed RT-qPCR assay. Cross-reactivity was estimated for each of the assay-target pairs based on the Cq difference between the reactions with the perfectly matched target and with the mismatched target assuming 100% efficiency for the matched target. Only negligible levels of unspecific signal were observed (<1%), and only for targets that differed from the perfect match by a single nucleotide: let-7a versus let-7c, let-7b versus let-7c, let-7a versus let-7e, let-7a versus let-7f (Figure 4B). Overall, all Two-tailed RT-qPCR assays exhibited exceptional specificity. None of the other tested methods reached similar level of specificity with false positive signals more than 200 times stronger, reaching as high as 22.48% (TaqMan), 50.71% (Quanta) and 122.47% (miQPCR) (Figure 4A).
Figure 4.
Specificity of Two-tailed RT-qPCR, TaqMan, Quanta, and miQPCR. (A) Measured false-positive levels of let-7 miRNA family members expressed relative to the level of the targeted member. (B) Sequences of eight members of the let-7 family. Nucleotide variations relative to let-7a are indicated. (C) Cq values and relative detection levels of pre-miRNAs relative to the targeted mature microRNA measured with Two-tailed RT-qPCR.
Discrimination between mature and precursor miRNAs
To test if the Two-tailed RT-qPCR assays can distinguish mature miRNAs from their precursor molecules we individually assayed the same amount of mature let-7a, let-7b and let-7f miRNAs (∼2 × 108 copies) and their corresponding precursor miRNAs. Cross-reactivity with precursor molecules was estimated from the measured ΔCq values. It ranged from 0.82% for let-7f to 6.98% for let-7a (Figure 4C), demonstrating that the Two-tailed RT-qPCR assays specifically quantify the amounts of mature miRNAs.
Performance of the system with biological samples and comparison with independent platform
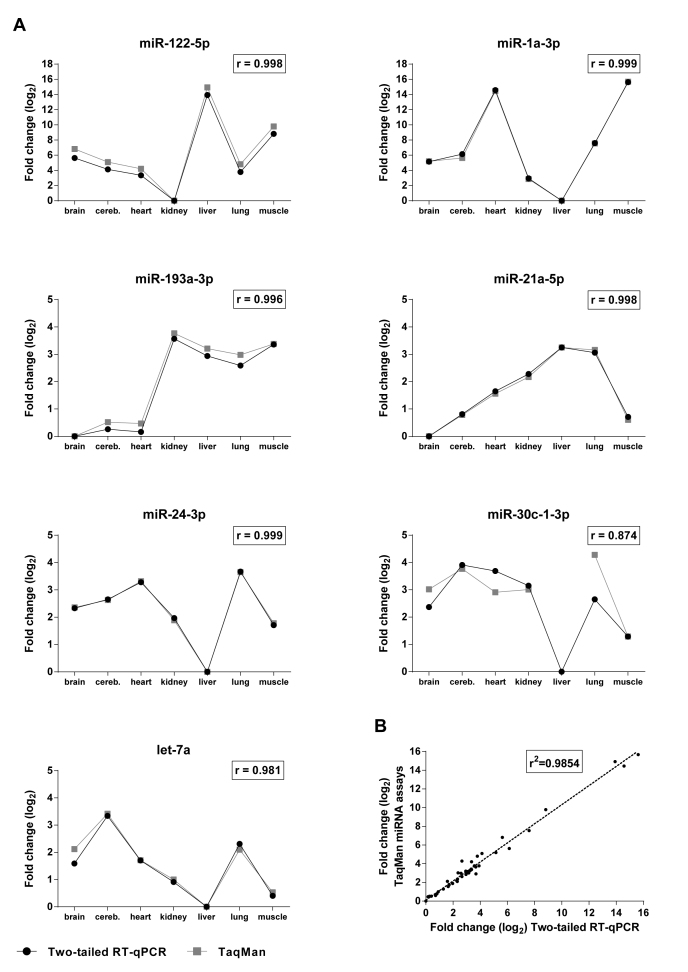
We validated the Two-tailed RT-qPCR assays on biological samples measuring the expression of 8 miRNAs across 7 mouse tissues and compared with measurements using commercially available TaqMan miRNA assays (Figure 5A). Relative expression levels across the tissues were calculated from the Cq values. The results were in agreement with previous reports (36,49) with miR-122–5p being highly expressed in liver, while miR-1a-3p having high expression in heart and muscle. The other microRNA targets exhibited lower variation in expression levels across the tissues. Let-7a and miR-21a, which are thought to have housekeeping functions, were indeed expressed at high levels in all the examined tissues. MiR-615–5p was not detected by any of the methods, suggesting it is either not present at all or only at exceedingly low levels.
Figure 5.
Comparison of expression profiles measured with Two-tailed RT-qPCR and TaqMan miRNA assays. (A) Relative fold changes of the expression of each target in seven tissues measured with Two-tailed RT-qPCR and TaqMan miRNA assays, respectively. (B) Overall correlation of the relative expression changes measured with the two methods.
The correlation between the measured ΔCq ( = Cq tissue, lowest expression – Cq tissue,x) with the Two-tailed RT-qPCR assays and with the TaqMan miRNA assays was excellent (Figure 5A). Pearson correlation coefficients (r) were 0.981 or larger for all the measured targets but miR-30c-1–3p, where r was 0.874. This lower correlation could be ascribed to the TaqMan miR-30c-1–3p assay, which generated very high and therefore uncertain Cq values (33.85 - 36.33). In the liver sample the TaqMan assay failed to detect miR-30c-1–3p, while the Two-tailed RT-qPCR assay showed clear positive signal with Cq = 32.91. Considering all the measured data the correlation between the Two-tailed RT-qPCR and the TaqMan miRNA assays was excellent (R2 = 0.985, Figure 5B).
Multiplexing of the reverse transcription
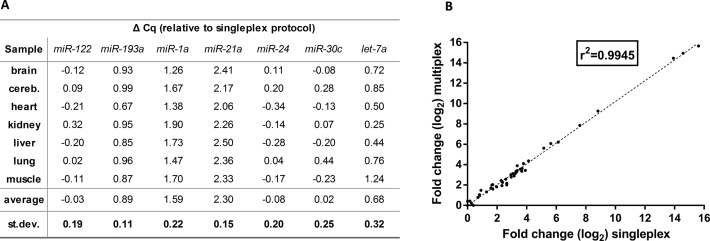
Multiplexing the reverse transcription could significantly increase the analysis throughput, save on reagents costs, and reduce the amount of material required. We tested multiplexing the RT step with the Two-tailed RT-qPCR assays by measuring expression of eight miRNA targets across seven mouse tissues. Eight Two-tailed RT primers were pooled and 10 ng of total mouse RNA was reverse transcribed in multiplex. 5% of the cDNA produced was used for each qPCR with one set of PCR primer pair. The Cq values and calculated relative expression levels from singleplex and multiplex protocols were compared. Neither protocol detected miR-615–5p, as in the previous experiment. For miR-122, miR-24, and miR-30c-1 there was no significant difference in Cq values between the singleplex and multiplex protocol. For the remaining four assays there was a shift ranging from 0.68 cycles for let-7a to 2.30 cycles for miR-21a (Figure 6A). The shift was assay specific, but constant and reproducible across samples, and therefore did not influence the calculation of relative expression levels. This is analogous to the mRNA dependent RT yields we have reported before, which do not affect calculations of relative expression levels as they too are constant and reproducible across samples (50,51). The agreement between the relative quantities measured by the multiplex and singleplex protocols was excellent (R2 = 0.995, Figure 6B).
Figure 6.
Comparison of singleplex and multiplex Two-tailed RT-qPCR. (A) ΔCq = Cqmultiplex - Cqsingleplex. (B) Overall correlation of the relative expression changes between tissues measured with the singleplex and multiplex protocol.
Detection of isomiRs
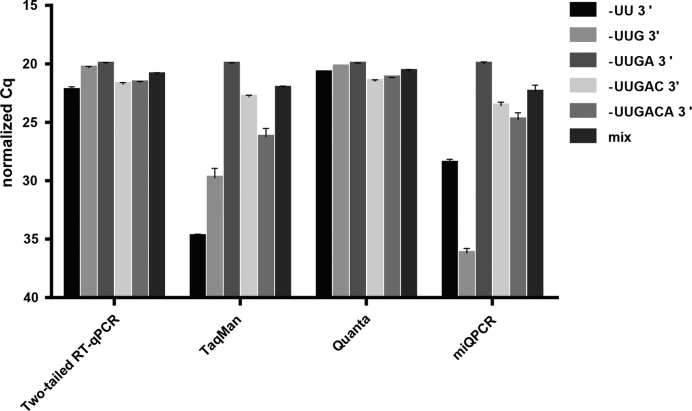
We assessed the ability of the Two-tailed RT-qPCR assays to measure isomiR variants that differ from the canonical sequence in their length and nucleotide composition in the 3′-terminus. We modified the design of the miR-21 Two-tailed assay such that the 3′-hemiprobe binding site is shifted two nucleotides upstream from the 3′-end of the miR-21 canonical sequence. This allows the detection of isomiRs that are two nucleotides shorter at the 3′-end, as well as all isomiRs with extended 3′-ends. We tested the assay analyzing equal amounts (∼2 × 108 copies) of five different synthetic variants of hsa-miR-21–5p that had 3′-terminus: (a) shortened by 2 nt, (b) shortened by 1 nt, (c) fully matching canonical sequence (d) extended by 1 nt (-C-3′) and (e) extended by 2 nt (-CA-3′) and also with the equimolar mixture of all five (suppl. file). For comparison, we also analyzed the samples with the miR-21 TaqMan, Quanta, and miQPCR assays.
We found that Two-tailed RT-qPCR along with the Quanta′s miR-21 assay reflects the amounts of different isomiRs with much better precision than the TaqMan and miQPCR miR-21 assays (Figure 7). The Cq values of the different miR-21 isomiRs measured with the Two-tailed RT-qPCR and Quanta miScript assays were similar, as expected, since the initial amount of template had been the same. This demonstrates the ability of the two-tailed RT-qPCR to detect all 3′-isomiR variants with equal efficiency. On the contrary, the TaqMan and miQPCR methods greatly underestimated the amounts of isomiRs that differ from the canonical sequence (Figure 7).
Figure 7.
Relative sensitivities of Two-tailed RT-qPCR, TaqMan, Quanta, and miQPCR to miR-21 isomiRs. Cq values are normalized such that the Cq of the canonical form is set to 20. Error bars indicate SD of two independent cDNA syntheses.
DISCUSSION
We present a new method for the quantification of miRNAs and other small RNAs by RT-qPCR (Figure 1). The new method is called Two-tailed RT-qPCR and is based on sequence specific RT primers with a novel design that allows the RT primer to hybridize to two regions of the miRNA target with separate complementary parts called hemiprobes. This design offers several advantages over existing strategies for RT-qPCR based detection of miRNAs, including high sensitivity, improved discrimination between similar miRNAs, and ability to quantify isomiRs.
It is well known that specific detection of a nucleic acid sequence requires targeting it with two probe molecules, as a single unmodified standard probe does under most conditions not confer sufficient specificity. The short length of microRNAs limits the size of probes that can be used to interrogate the sequence. For example, two regular PCR primers cannot be used to amplify a regular cDNA copy of the microRNA, as they cannot be fitted without overlap. Reducing the primers′ lengths in order to fit makes binding too weak. Binding strength can be increased by incorporation of modified bases such as the Locked Nucleic Acids (LNAs) into the primers (36). Modified primers may have higher melting temperature (Tm) and enhanced sequence discrimination (52). However, results are highly dependent on the design of the LNA oligomers, which often requires extensive trial-and-error optimization (40). LNA-containing primers are also more expensive than conventional primers and may exhibit lower amplification efficiencies (28,53).
We wanted to find a way to interrogate the sequence of a microRNA with two unmodified non-overlapping probes. To solve the thermal stability problem, we reasoned this should be possible by using two short hemiprobes that are connected. This way each hemiprobe would bind with high specificity, as a single mismatch would greatly distort the rather short hybrid it forms, while overall high thermal stability is achieved through cooperativity. Connecting the hemiprobes leads to cooperative binding as they drag each other to the binding site. As a consequence the overall binding strength is comparable to that of a long probe. Connecting the hemiprobes with an oligonucleotide stretch that forms a hairpin protects it from undesired interactions. We call this new primer ‘Two-tailed RT primer’. When used in reverse transcription, the Two-tailed RT primer is extended forming a tailed cDNA. The cDNA can then be amplified by PCR.
Another aspect of the Two-tailed RT primer is that the 3′-hemiprobe can be made rather short (5–6 nt). This leaves enough space to design an unmodified miRNA-specific qPCR primer without overlapping with the 3′-hemiprobe. Also, a short 3′-hemiprobe is more sensitive to mismatches in the target sequence, while sufficient binding strength is obtained through cooperative binding with the longer 5′-hemiprobe. Indeed, when we compared Two-tailed RT primers with the same length of the 3′-hemiprobe (5nt), but different length of the 5′-hemiprobe we found significant differences (Figure 2A).
By strategic design of the 5′-hemiprobe the specificity of the Two-tailed RT-qPCR can be optimized for different cases. For example, closely related miRNAs often differ in base positions in their 5′-regions. Those variants are poorly distinguished with current methods for miRNA analysis, which use only on one of the qPCR primers for discrimination. This holds also for the specific-primer based TaqMan miRNA assays, as this part of the microRNA is not sensed by its stem–loop primer. With the Two-tailed RT-qPCR assays, the nucleotides in the 5′-regions, such as those distinguishing let-7a, let-7e, let-7f, and let-7g, are interrogated twice: first time in the reverse transcription by the 5′-hemiprobe of the RT primer and second time in the qPCR by the reverse PCR primer (Figure 1). Specificity can be maximized using a short 5′-hemiprobe designed to sense all the sequence variants in the variable region of the miRNA (Supplementary Figure S1). Using this strategy, we designed Two-tailed RT-qPCR assays that exhibit negligible cross-reactivity between the members of the let-7 miRNA family (Figure 4B). When these members were assayed with the three other methods, substantial undesired cross-reactivity was observed (Figure 4A).
Since the sequence of the mature transcript is contained in its precursors (3), RT-qPCR assays designed to detect mature miRNAs may also amplify their precursor molecules. Although precursors are usually present in cells at much lower levels than the mature miRNAs (54–57), it may still be relevant to measure them separately. We decided to test if the two-tailed RT-qPCR can distinguish between mature miRNAs and the corresponding pre-miRNAs. Our results show that the Two-tailed RT-qPCR assays designed for mature miRNAs show minimal cross-reaction with pre-miRNAs (Figure 4C). One contributing factor is that reverse transcription is performed at rather low temperature (25°C) without any pre-heating step that would open the secondary structure of pre-miRNAs making them available for priming with the Two-tailed RT primer. We observed that even without the pre-heating step, sensitivity of the system is not influenced by potential microRNA-long RNA interactions (Supplementary Figure S2). The specificity of the two-tailed RT-qPCR assays for mature miRNAs is further confirmed by the excellent correlation with the expression profiles measured using TaqMan miRNA assays (Figure 5), which do not cross-react with precursor miRNAs because of its particular RT-priming mechanism (39,44).
Target-specific priming of the reverse transcription has several advantages including higher specificity and lower background, but a common disadvantage is that each target requires a separate RT reaction. This can be resolved by multiplexing the RT step using an RT primer pool. Multiplexing RT increases throughput, saves on reagent costs, minimizes labor, and reduces the sample amount needed. Usually, it is problematic to have many long oligomers, such as multiple RT primers, in the same reaction as it leads to unspecific amplification (58). The Two-tailed RT primers are, however, designed with a hairpin structure that prevents non-specific interactions. As demonstrated, multiplexing eight targets showed perfect agreement with the corresponding singleplex reactions (Figure 6). Importantly, the specificity of the multiplex measurement was not compromised, based on negative controls and melting curve analyses (suppl. file). This is likely due to the use of two specific PCR primers for each target in the downstream qPCR, while other methods for miRNA analysis commonly use only one miRNA specific primer combined with a universal primer. The Two-tailed RT-qPCR assays are designed with target specific PCR primers as follows: one miRNA specific reverse primer and one forward primer that is specific to an internal segment of the corresponding Two-tailed RT primer. This way both PCR primers are specific to one cDNA only. Although using a second specific PCR primer provides no advantage with the singleplex protocol, it adds specificity to the multiplex protocol, where many different cDNAs are present as qPCR templates. In our experiment, we performed reverse transcription in octaplex and then divided the cDNA into aliquots that were analyzed with target specific PCR primer pairs in singleplex qPCR using dye based detection. This workflow is, however, not limited to any particular number of targets and should be applicable to virtually any degree of multiplexing. When only a small number of microRNAs are targeted 1-step RT-qPCR can be employed, distinguishing the PCR products with fluorescent probes.
Another advantage of the Two-tailed RT-qPCR is its capability to reverse transcribe isomiRs with the same efficiency as the canonical sequence (Figure 7). IsomiRs are miRNA variants that differ in length and sequence composition in their 3′- and/or 5′-termini from the annotated canonical sequence (23,24). Growing evidence suggests isomiRs are expressed in a cell, tissue, and gender specific manner, possess relevant physiological functions, and are potential biomarkers in clinical diagnostics (59–63). In some cases, isomiRs, derived from the same precursor arm, bind to other targets than the canonical miRNA. Another function is that isomiRs cooperate with the canonical form to drive similar biology by targeting the same set of core biological networks while distributing the off-target effects and thus increasing the signal to noise ratio of gene silencing (62,64). Measuring full isomiR profiles may therefore be more valuable than targeting the canonical sequence only. This was recently demonstrated when clearly improved discrimination of cancerous and healthy tissues was obtained by inclusion of full isomiR profiles (59,65).
The heterogeneity of the miRNA sequences pose a substantial issue for many RT-qPCR methods. Those employing universal reverse transcription should generally be able to amplify all terminal sequence variants, but biochemical modifications of terminal nucleotides may interfere with the enzymatic steps upstream of the RT-qPCR. Furthermore, the design of the stem–loop primers used in the TaqMan miRNA assays renders the method less sensitive to isomiRs. Due to the stem-part, which blocks annealing to longer sequences, only the particular sequence with the defined ends is amplified with optimal efficiency, and the method may completely miss on some variants (40–42). Particularly isomiRs that differ at the 3′-end pose a problem. Notably, largest variability across isomiRs is found in their 3′-ends, and in some cases the canonical sequence represents only a small fraction of the total amount of a miRNA (24,59,62,64,66,67).
To test the ability of the Two-tailed RT-qPCR to detect different 3′-isomiRs we measured five synthetic terminal variants of miR-21. We observed that the Two-tailed RT-qPCR accurately quantifies shorter as well as longer variants of the canonical sequence, reaching the level of a poly(A)-tail based method while providing all advantages of RT-specific priming. The full isomiR repertoire of the miRNA is thus measured and no potentially important isomiRs are missed (Figure 7). This is in difference from the TaqMan and miQPCR approaches, which exhibited high variation across the miR-21 isomiRs, greatly underestimating the amounts of those that differ from the canonical sequence (Figure 7).
In some applications only a certain isomiR is of interest. Although it should be possible to target it specifically with custom-designed TaqMan miRNA assays, our results (Figure 7) as well as reports in the literature suggest this is not always the case and various degree of cross-reactivity to other isomiRs is observed (41,42,68,69). Therefore, it seems the TaqMan miRNA assays are neither specific to a single 3′-end isomiR nor do they detect all the isomiRs with equal sensitivity, which may lead to an underestimation of the total amount of a given miRNA in a sample (25,42). To our knowledge, the only qPCR based method that can be used to distinctively quantify specific isomiRs with 1 nt resolution is the ‘Dumbbell-PCR’ (69). This method exploits the properties of T4 RNA ligase 2 to ligate stem–loop adapters to the ends of the targeted isomiR. The formed dumbbell-like structure is then quantified with TaqMan qPCR. Such extreme specificity is currently not achieved with the two-tailed RT-qPCR, but the Two-tailed RT primer can be designed to reverse transcribe all isomiRs of a particular miRNA to obtain a correct quantification of the total amount.
RT-qPCR technology remains unequalled tool in small-RNA expression profiling, vital for validation of genome-wide experiments and accurate measurement of challenging samples such as liquid biopsies. However, measurements of higher number of targets significantly increases the cost of such analyses. We have developed a highly sensitive and exceedingly specific method called Two-tailed RT-qPCR, suitable for rapid and cost-effective microRNA profiling. At the same time, Two-tailed RT-qPCR reflects on the current state of microRNA field and confers several advantages over current RT-qPCR methods, including increased specificity and ability to capture the full isomiR profile (Table 2). Two-tailed RT-qPCR uses only standard oligomers, can employ either dye or probe based detection and can be used for animal and plant small RNAs alike. The whole analysis can be performed in just 2.5 h.
Table 2. Comparison of parameters of four tested RT-qPCR methods for miRNA quantification. Cost estimate per assay with 20 RT and 60 qPCR reactions is indicated (details available in suppl. file).
| cDNA synthesis strategy | Sensitivity (miRNA copies in analyzed volume) | Linear dynamic range | Unspecific cross-reaction | Accurate detection of isomiRs | Melting curve for specificity control | Detection of piRNAs and plant miRNAs | Cost | |
|---|---|---|---|---|---|---|---|---|
| Two-tailed RT-qPCR | specific RT primer | 102 - 103 | 6–7 logs | < 1% | yes | yes | yes |
|
| TaqMan | specific RT primer | 102 - 103 | 6–7 logs | < 23% | no | no | yes |
|
| Quanta | poly(A) tailing | 102 - 103 | 6–7 logs | < 51% | yes | yes | no |
|
| miQPCR | linker ligation | 104 | 5 logs | > 100% | no | yes | yes, but efficiency is reduced |
|
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pavel Honsa and Martin Valny from the Institute of Experimental Medicine CAS for providing mouse tissue samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [P303/16/10214S, P303/17/04034S]; Swedish foundation for Strategic Research [ID14–0075]; BIOCEV CZ.1.05/1.1.00/02.0109 provided by ERDF and MEYS; Operational Programme Education for Competitiveness [CZ.1.07/2.3.00/30.0045] from the European Social Fund and the state budget of the Czech Republic. Funding for open access charge: Czech Science Foundation [P303/16/10214S].
Conflict of interest statement. M.K. and R.S. have shares in TATAA Biocenter AB. Two-tailed RT-qPCR is patent applied in PCT/US15/45966.
REFERENCES
- 1. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huntzinger E., Izaurralde E.. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011; 12:99–110. [DOI] [PubMed] [Google Scholar]
- 3. Kim V.N., Han J., Siomi M.C.. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009; 10:126–139. [DOI] [PubMed] [Google Scholar]
- 4. Winter J., Jung S., Keller S., Gregory R.I., Diederichs S.. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009; 11:228–234. [DOI] [PubMed] [Google Scholar]
- 5. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 6. Tan L., Yu J.T., Tan L.. Causes and Consequences of MicroRNA Dysregulation in Neurodegenerative Diseases. Mol. Neurobiol. 2015; 51:1249–1262. [DOI] [PubMed] [Google Scholar]
- 7. Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009; 10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao C., Rajewsky K.. MicroRNA control in the immune system: basic principles. Cell. 2009; 136:26–36. [DOI] [PubMed] [Google Scholar]
- 9. Lin S., Gregory R.I.. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015; 15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hatfield S.D., Shcherbata H.R., Fischer K.A., Nakahara K., Carthew R.W., Ruohola-Baker H.. Stem cell division is regulated by the microRNA pathway. Nature. 2005; 435:974–978. [DOI] [PubMed] [Google Scholar]
- 11. He Y., Lin J., Kong D., Huang M., Xu C., Kim T.K., Etheridge A., Luo Y., Ding Y., Wang K.. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clin. Chem. 2015; 61:1138–1155. [DOI] [PubMed] [Google Scholar]
- 12. Basak I., Patil K.S., Alves G., Larsen J.P., Moller S.G.. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci.: CMLS. 2016; 73:811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moldovan L., Batte K.E., Trgovcich J., Wisler J., Marsh C.B., Piper M.. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014; 18:371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarzenbach H., Nishida N., Calin G.A., Pantel K.. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014; 11:145–156. [DOI] [PubMed] [Google Scholar]
- 15. Jiang H.X., Liang Z.Z., Ma Y.H., Kong D.M., Hong Z.Y.. G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal. Chim. Acta. 2016; 943:114–122. [DOI] [PubMed] [Google Scholar]
- 16. Liu H., Li L., Duan L., Wang X., Xie Y., Tong L., Wang Q., Tang B.. High specific and ultrasensitive isothermal detection of microRNA by padlock probe-based exponential rolling circle amplification. Anal. Chem. 2013; 85:7941–7947. [DOI] [PubMed] [Google Scholar]
- 17. Ma F., Liu M., Tang B., Zhang C.Y.. Rapid and sensitive quantification of microRNAs by isothermal helicase-dependent amplification. Anal. Chem. 2017; 89:6182–6187. [DOI] [PubMed] [Google Scholar]
- 18. Deng R., Zhang K., Li J.. Isothermal amplification for microRNA detection: from the test tube to the cell. Acc. Chem. Res. 2017; 50:1059–1068. [DOI] [PubMed] [Google Scholar]
- 19. Mestdagh P., Hartmann N., Baeriswyl L., Andreasen D., Bernard N., Chen C.F., Cheo D., D’Andrade P., DeMayo M., Dennis L. et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study (vol 11, pg 809, 2014). Nat. Methods. 2014; 11:971–971. [DOI] [PubMed] [Google Scholar]
- 20. Svoboda P. A toolbox for miRNA analysis. FEBS Lett. 2015; 589:1694–1701. [DOI] [PubMed] [Google Scholar]
- 21. Pritchard C.C., Cheng H.H., Tewari M.. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012; 13:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aldridge S., Hadfield J.. Introduction to miRNA profiling technologies and cross-platform comparison. Methods Mol. Biol. 2012; 822:19–31. [DOI] [PubMed] [Google Scholar]
- 23. Guo L., Chen F.. A challenge for miRNA: multiple isomiRs in miRNAomics. Gene. 2014; 544:1–7. [DOI] [PubMed] [Google Scholar]
- 24. Neilsen C.T., Goodall G.J., Bracken C.P.. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet.: TIG. 2012; 28:544–549. [DOI] [PubMed] [Google Scholar]
- 25. Dellett M., Simpson D.A.. Considerations for optimization of microRNA PCR assays for molecular diagnosis. Expert Rev. Mol. Diagnost. 2016; 16:407–414. [DOI] [PubMed] [Google Scholar]
- 26. Jin J., Vaud S., Zhelkovsky A.M., Posfai J., McReynolds L.A.. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res. 2016; 44:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J., Yao B., Huang H., Wang Z., Sun C., Fan Y., Chang Q., Li S., Wang X., Xi J.. Real-time polymerase chain reaction microRNA detection based on enzymatic stem–loop probes ligation. Anal. Chem. 2009; 81:5446–5451. [DOI] [PubMed] [Google Scholar]
- 28. Balcells I., Cirera S., Busk P.K.. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol. 2011; 11:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi R., Chiang V.. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. 2005; 39:519–525. [DOI] [PubMed] [Google Scholar]
- 30. Mei Q., Li X., Meng Y., Wu Z., Guo M., Zhao Y., Fu X., Han W.. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One. 2012; 7:e46890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benes V., Collier P., Kordes C., Stolte J., Rausch T., Muckentaler M.U., Haussinger D., Castoldi M.. Identification of cytokine-induced modulation of microRNA expression and secretion as measured by a novel microRNA specific qPCR assay. Scientific Rep. 2015; 5:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munafo D.B., Robb G.B.. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010; 16:2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhuang F., Fuchs R.T., Sun Z., Zheng Y., Robb G.B.. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012; 40:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sorefan K., Pais H., Hall A.E., Kozomara A., Griffiths-Jones S., Moulton V., Dalmay T.. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012; 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yehudai-Resheff S., Schuster G.. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000; 28:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raymond C.K., Roberts B.S., Garrett-Engele P., Lim L.P., Johnson J.M.. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005; 11:1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharbati-Tehrani S., Kutz-Lohroff B., Bergbauer R., Scholven J., Einspanier R.. miR-Q: a novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol. Biol. 2008; 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang T., Yang J., Liu G., Jin W., Liu Z., Zhao S., Yao M.. Quantification of mature microRNAs using pincer probes and real-time PCR amplification. PLoS One. 2015; 10:e0120160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benes V., Castoldi M.. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010; 50:244–249. [DOI] [PubMed] [Google Scholar]
- 41. Schamberger A., Orban T.I.. 3′ IsomiR species and DNA contamination influence reliable quantification of microRNAs by stem–loop quantitative PCR. PLoS One. 2014; 9:e106315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soundara Pandi S.P., Chen M., Guduric-Fuchs J., Xu H., Simpson D.A.. Extremely complex populations of small RNAs in the mouse retina and RPE/choroid. Invest. Ophthalmol. Vis. Sci. 2013; 54:8140–8151. [DOI] [PubMed] [Google Scholar]
- 43. Lao K., Xu N.L., Yeung V., Chen C., Livak K.J., Straus N.A.. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem. Biophys. Res. Commun. 2006; 343:85–89. [DOI] [PubMed] [Google Scholar]
- 44. Tang F., Hajkova P., Barton S.C., O’Carroll D., Lee C., Lao K., Surani M.A.. 220-plex microRNA expression profile of a single cell. Nat. Protoc. 2006; 1:1154–1159. [DOI] [PubMed] [Google Scholar]
- 45. Tang F., Hajkova P., Barton S.C., Lao K., Surani M.A.. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006; 34:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J.. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008; 36:D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Markham N.R., Zuker M.. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008; 453:3–31. [DOI] [PubMed] [Google Scholar]
- 48. Forootan A., Sjöback R., Björkman J., Sjögreen B., Linz L., Kubista M.. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantif. 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang Y., Ridzon D., Wong L., Chen C.. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007; 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stahlberg A., Hakansson J., Xian X., Semb H., Kubista M.. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004; 50:509–515. [DOI] [PubMed] [Google Scholar]
- 51. Stahlberg A., Kubista M., Pfaffl M.. Comparison of reverse transcriptases in gene expression analysis. Clin. Chem. 2004; 50:1678–1680. [DOI] [PubMed] [Google Scholar]
- 52. Vester B., Wengel J.. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004; 43:13233–13241. [DOI] [PubMed] [Google Scholar]
- 53. Veedu R.N., Vester B., Wengel J.. Enzymatic incorporation of LNA nucleotides into DNA strands. Chembiochem. 2007; 8:490–492. [DOI] [PubMed] [Google Scholar]
- 54. Gan L., Denecke B.. Profiling pre-MicroRNA and mature microRNA expressions using a single microarray and avoiding separate sample preparation. Microarrays. 2013; 2:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmittgen T.D., Lee E.J., Jiang J., Sarkar A., Yang L., Elton T.S., Chen C.. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008; 44:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li N., You X., Chen T., Mackowiak S.D., Friedlander M.R., Weigt M., Du H., Gogol-Doring A., Chang Z., Dieterich C. et al. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic Acids Res. 2013; 41:3619–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Faridani O.R., Abdullayev I., Hagemann-Jensen M., Schell J.P., Lanner F., Sandberg R.. Single-cell sequencing of the small-RNA transcriptome. Nat. Biotechnol. 2016; 34:1264–1266. [DOI] [PubMed] [Google Scholar]
- 58. Stahlberg A., Krzyzanowski P.M., Jackson J.B., Egyud M., Stein L., Godfrey T.E.. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016; 44:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Telonis A.G., Loher P., Jing Y., Londin E., Rigoutsos I.. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015; 43:9158–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loher P., Londin E.R., Rigoutsos I.. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014; 5:8790–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Siddle K.J., Tailleux L., Deschamps M., Loh Y.H., Deluen C., Gicquel B., Antoniewski C., Barreiro L.B., Farinelli L., Quintana-Murci L.. bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS Genet. 2015; 11:e1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cloonan N., Wani S., Xu Q., Gu J., Lea K., Heater S., Barbacioru C., Steptoe A.L., Martin H.C., Nourbakhsh E. et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011; 12:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang S., Xu Y., Li M., Tu J., Lu Z.. Dysregulation of miRNA isoform level at 5′ end in Alzheimer's disease. Gene. 2016; 584:167–172. [DOI] [PubMed] [Google Scholar]
- 64. Ahmed F., Senthil-Kumar M., Lee S., Dai X., Mysore K.S., Zhao P.X.. Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol. 2014; 11:1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koppers-Lalic D., Hackenberg M., de Menezes R., Misovic B., Wachalska M., Geldof A., Zini N., de Reijke T., Wurdinger T., Vis A. et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016; 7:22566–22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McGahon M.K., Yarham J.M., Daly A., Guduric-Fuchs J., Ferguson L.J., Simpson D.A., Collins A.. Distinctive profile of IsomiR expression and novel microRNAs in rat heart left ventricle. PLoS One. 2013; 8:e65809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Baran-Gale J., Fannin E.E., Kurtz C.L., Sethupathy P.. Beta cell 5′-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One. 2013; 8:e73240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu H., Neilson J.R., Kumar P., Manocha M., Shankar P., Sharp P.A., Manjunath N.. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007; 2:e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Honda S., Kirino Y.. Dumbbell-PCR: a method to quantify specific small RNA variants with a single nucleotide resolution at terminal sequences. Nucleic Acids Res. 2015; 43:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.