Abstract
Strigolactones (SLs), a group of carotenoid derived terpenoid lactones, are root-to-shoot phytohormones suppressing shoot branching by inhibiting the outgrowth of axillary buds. DWARF 53 (D53), the key repressor of the SL signaling pathway, is speculated to regulate the downstream transcriptional network of the SL response. However, no downstream transcription factor targeted by D53 has yet been reported. Here we report that Ideal Plant Architecture 1 (IPA1), a key regulator of the plant architecture in rice, functions as a direct downstream component of D53 in regulating tiller number and SL-induced gene expression. We showed that D53 interacts with IPA1 in vivo and in vitro and suppresses the transcriptional activation activity of IPA1. We further showed that IPA1 could directly bind to the D53 promoter and plays a critical role in the feedback regulation of SL-induced D53 expression. These findings reveal that IPA1 is likely one of the long-speculated transcription factors that act with D53 to mediate the SL-regulated tiller development in rice.
Keywords: tillering, plant architecture, phytohormone, protein interaction, Oryza Sativa L.
Introduction
Strigolactones (SLs), a collection of terpenoid-derived compounds produced by plants, were firstly identified as the host-derived germination signals of root parasitic plants1, and later as stimulants of hyphal branching of arbuscular mycorrhizal fungi2. The characterizion of more axillary growth (max) mutants in Arabidopsis thaliana3,4,5,6,7,8,9, dwarf (d) mutants in rice (Oryza SativaL.)10,11,12,13,14,15,16, ramosus (rms) mutants in pea (Pisum sativum)4,17, and decreased apical dominance (dad) mutants in petunia (Petunia hybrida)18,19,20 have indicated that SLs function as root-to-shoot phytohormones that regulate shoot branching21,22 and also regulate leaf senescence and root development23,24,25,26,27,28. Subsequently, the elucidation of the biosynthetic and signaling pathways of SLs has attracted great attention. Carlactone is the conserved endogenous precursor of SLs, which is derived from all-trans-β-carotene through DWARF27 (D27)-mediated isomerization and subsequent cleavage by the carotenoid cleavage dioxygenases 7 and 8 (CCD7 and CCD8)14,29,30. In Arabidopsis thaliana, cytochrome P450 MAX1 has been shown to oxidize carlactone to carlactonoic acid (CLA)31, which is then converted into methyl carlactonoate (MeCLA) and further catalyzed to an unidentified strigolactone-like compound by an oxidoreductase-like enzyme, Lateral Branching Oxidoreductase (LBO)32. However, in rice, carlactone is first converted into ent-2′-epi-5-deoxy-strigol by one MAX1 homolog and further into orobanchol by a second MAX1 homolog33. Highly branched phenotypes that can be rescued by SL treatment are displayed by SL biosynthesis deficient mutants that include d27, d10, and d17 in rice; rms1 in pea; and max1, max3, max4, and lbo in Arabidopsis thaliana14,21,22,31,32.
Compared with the SL biosynthetic pathways, the knowledge of SL-mediated signaling is still obscure. Through studying SL insensitive mutants in rice, three components have been identified involved in SL perception and signaling. DWARF14 (D14) encodes a member of the α/β-hydrolase fold family protein, which binds and hydrolyses SL to form a covalently linked intermediate molecule (CLIM)11,12,15,34,35,36,37. This reaction triggers a conformational change to form a complex with DWARF3 (D3), which encodes an F-box protein13,38,39. D14 could interact with D3 and DWARF53 (D53) in the presence of SLs, leading to the ubiquitination and degradation of the nuclear-localized repressor D5338,39. D53 contains three ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs, which are essential to recruit the transcriptional co-repressor TOPLESS (TPL) proteins and induce their oligomerization to form a repressor-corepressor-nucleosome complex40. In accord, D53 was shown to interact with a TPL-related protein, TPR2, in rice38. However, whether D53 regulates the transcription of genes in the endogenous SL signaling pathway and what are its direct downstream transcription factors remain to be determined. In other angiosperms such as Arabidopsis thaliana and pea, the orthologs of D14, D3 and D53 have similar functions6,7,9,41,42,43,44.
Ideal Plant Architecture1 (IPA1) is a key regulator in determining plant architecture, which encodes a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family transcription factors, SPL1445,46. Two microRNAs, miRNA156 and miRNA529, regulate IPA1 expression, and point mutations in the miRNA156 recognition site perturb miRNA156-regulated degradation of IPA1 mRNA, leading to decreased tiller number and increased plant height and panicle branches45,46,47. In contrast, the CRISPR/Cas9-generated ipa1 loss-of-function mutants exhibited opposite phenotypes48, reminiscent of the dwarf and high tillering phenotype of SL-deficient or -signaling mutant plants. IPA1 directly binds to the promoter of a negative regulator of tiller bud outgrowth, TEOSINTE BRANCHED1 (OsTB1), to suppress rice tillering47. The ortholog of OsTB1 in Arabidopsis thaliana, BRANCHED1 (BRC1), is a key regulator of branch outgrowth and one of the SL-responsive genes49,50. Previous studies showed that SL-induced bud growth inhibition occurs at least partially through the regulation of BRC1 transcription in Arabidopsis thaliana and pea41,51,52,53. These results lead to the hypothesis that D53 sequesters transcriptional co-repressor TPL proteins to suppress IPA1, a key transcription factor regulating tillering. In this study, we reported that IPA1 is the direct downstream component targeted by D53 to regulate the SL response and SL-induced gene expression in rice. D53 interacts physically with IPA1 and inhibits its transcriptional activation activity. Furthermore, IPA1 binds directly to the promoter of D53 and plays a critical role in the feedback regulation of SL-induced D53 expression. Together, these findings provide new insights into understanding the function of D53, and identify IPA1 as one of the D53-targeted downstream transcription factors in the SL signaling pathway in rice.
Results
Generation and phenotypes of IPA1 loss-of-function mutants
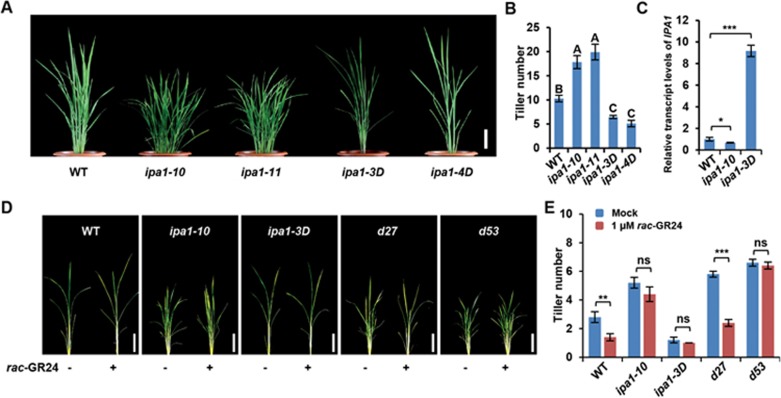
Our previous study has shown that IPA1 functions as a transcriptional activator in regulating tiller number and that its expression is regulated at both mRNA and protein levels; mRNA-level regulation was demonstrated by isolating and characterizing the two gain-of-function mutants, ipa1-1D and ipa1-2D45,54,55. To understand whether loss-of-function mutations of IPA1 could affect rice plant architecture, especially tillering, we used a targeting induced local lesions in genomes (TILLING) method to generate point mutations in IPA1, which led to amino acid substitutions in the gene product. However, none of the mutant lines exhibited obvious phenotypes, suggesting that these amino acids are not essential for the function of IPA1 (Supplementary information, Figure S1). By applying a genome editing approach, four lines of ipa1 mutants were generated, including two loss-of-function mutants, ipa1-10 and ipa1-11, and two gain-of-function mutants, ipa1-3D and ipa1-4D48. The ipa1-10 mutant resulted from a 5-bp deletion in the coding sequence; the ipa1-11 mutant arose from a 57-bp insertion in the replacement of 102-bp deletion in the coding sequence; ipa1-3D and ipa1-4D were gain-of-function mutants resulting from 12-bp and 21-bp in-frame deletions in the miRNA156/529 target sites respectively (Supplementary information, Figure S2), which abolish the miRNA regulation without interfering the normal function of IPA1. The transcript and phenotypic analyses showed that the ipa1-10 and ipa1-11 loss-of-function mutants exhibited high tillering and dwarf phenotypes (Figure 1A-1C), indicating that IPA1 is a negative regulator of rice tillering. In contrast, both ipa1-3D and ipa1-4D homozygous plants have high levels of IPA1 mRNA accumulation and low tillering (Figure 1A-1C).
Figure 1.
Phenotypic characterization of IPA1 loss-of-function mutants. (A) Gross morphologies of wild-type (WT), ipa1-10, ipa1-11, ipa1-3D, and ipa1-4D plants before the heading stage. Bar = 10 cm. (B) Statistical analysis of tiller number in (A). Values are means ± sem (n = 11). Different letters at top of each column indicate a significant difference at P < 0.05 determined by Tukey's HSD test. (C) IPA1 expression levels in WT, ipa1-10, and ipa1-3D. Values are means ± SD (n = 3). The asterisks represent significant differences determined by Student's t test. *P < 0.05, **P < 0.001. (D) Gross morphologies of wild-type (WT), ipa1-10, ipa1-3D, d27, and d53 seedlings with or without rac-GR24 treatment. Seedlings were treated with 1.0 μM rac-GR24 (+) or mock (–). Bar = 5 cm. (E) Statistical analysis of tiller number in (D). Values are means ± SD (n = 5). The asterisks represent significant differences determined by Student's t test. **P < 0.01, **P < 0.001. ns, no significant difference.
Based on previous findings that the phenotypes of several rice tillering mutants are subject to SL regulation14,22,38, we therefore investigated whether IPA1 functions in the SL signaling pathway by examining the SL sensitivity of the respective loss- and gain-of-function mutants, ipa1-10 and ipa1-3D, in response to rac-GR24, a synthetic SL analogue. The mutant d27 showed a high tillering phenotype, due to a SL biosynthesis defect, which could be rescued by exogenously applied SL14. In the d53 plant, the gain-of-function mutant D53 protein is resistant to ubiquitination and degradation. This blocks SL signaling and results in dwarf plants with high tillering phenotypes, which could not be rescued by exogenously applied SL38,39. Hence d27 and d53 mutants were used as the respective, negative and positive controls in this assay. Upon SL treatment, tiller number is unchanged in ipa1-10 and ipa1-3D mutants, suggesting that they are insensitive to SL (Figure 1D and 1E). These results demonstrate that IPA1 plays an important role in the SL signaling pathway.
IPA1 interacts with D53 protein in vivo and in vitro
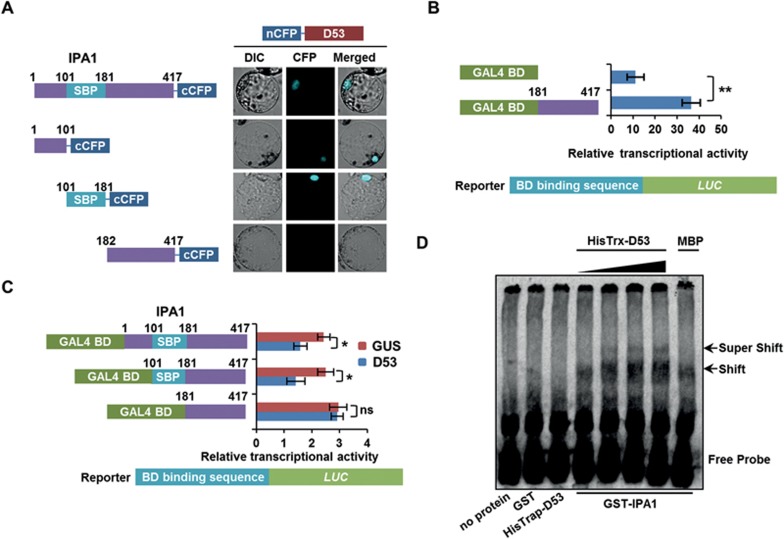
To further study the roles of IPA1 in the SL signaling pathway, we first tested whether mRNA or protein levels of IPA1 are regulated by SLs and found that they showed no significant changes after GR24 treatment (Supplementary information, Figure S3). Similarly, we also found that the protein levels of IPA1 showed no differences between wild-type Nipponbare and SL-related mutants (Supplementary information, Figure S4). These results suggest that SLs do not affect the transcript and protein levels of IPA1. We then asked whether IPA1 could physically interact with D53, the repressor in the SL signaling pathway, in several ways. We first carried out a yeast two-hybrid assay in which we fused IPA1 to GAL4 DNA binding domain (BD) and D53 to GAL4 activation domain (AD) to form BD-IPA1 and AD-D53. We found that expression of BD-IPA1 and AD-D53 in co-transformed yeast cells could activate the expression of the ADE reporter gene to allow the transformed yeast cells to grow on the SD medium without adenine, indicating an interaction between IPA1 and D53 (Figure 2A). We confirmed an interaction between IPA1 and D53 by bimolecular fluorescence complementation (BiFC), GST-pull down using rice calli, and co-immunoprecipitation (Co-IP) assays (Figure 2B-2D). Finally, the direct interaction between the two proteins was further demonstrated in an in vitro pull down assay using proteins purified from a prokaryotic expression system (Figure 2E), indicating that no other plant protein is required for the interaction between IPA1 and D53. Taken together, these results allow us to conclude that IPA1 directly interacts with D53 in rice.
Figure 2.
Interaction of IPA1 with D53 in vitro and in vivo. (A) Interaction between IPA1 and D53 revealed by yeast two-hybrid. IPA1 was fused with the GAL4 binding domain (BD) and D53 with the GAL4 activation domain (AD). The yeast clones were grown on the SD medium without leucine, tryptophan, histone and adenine (SD-L-W-H-A) with dilutions to 10−1 and 10−2. Yeast grown on the SD medium without leucine and tryptophan (SD-L-W) were used as a loading control. (B) Interaction between IPA1 and D53 revealed by the BiFC assay in rice protoplasts. IPA1 was fused with the N-terminal of CFP and D53 with the C-terminal of CFP. (C) Interaction between IPA1 and D53 revealed by GST pull-down with GST-IPA1 purified from bacteria and D53 extracted from rice calli. D53 was detected by rabbit polyclonal antibodies anti-D53 and GST by mouse monoclonal antibody anti-GST. (D) In vivo interaction between 7mIPA1-GFP and D53 revealed by the Co-IP assay in rice protoplasts. Proteins were extracted from ProIPA1:7mIPA1-GFP or ProUB:GFP. D53 was detected by rabbit polyclonal antibodies anti-D53 and GFP by mouse monoclonal antibody anti-GFP. (E) Interaction between IPA1 and D53 revealed by the GST pull-down assay with HisTrx-D53 and GST-IPA1 purified from bacteria. D53 was detected by rabbit polyclonal antibodies anti-D53 and GST by Ponceau S staining.
D53 inhibits the transcriptional activator function of IPA1
D53 contains three EAR motifs in its C-terminal part and interacts with the transcriptional co-repressor TPL/TPR proteins38,40. However, the molecular mechanisms downstream of D53 remain elusive. The interaction between D53 and IPA1 raises the possibility that D53 could suppress the transcriptional activation activity of IPA1. We therefore investigated the effect of D53 upon the transcriptional activity of IPA1 using a reporting system in tobacco (Nicotiana benthamiana). In this system, the LUC (Luciferase) reporter gene was driven by a DNA sequence containing the SQUAMOSA PROMOTER BINDING PROTEIN (SBP) binding element GTAC. When co-infiltrating Agrobacterium hosts expressing IPA1-MYC together with the reporter construct, the expression of LUC was dramatically enhanced (Figure 3A and 3B; Supplementary information, Figure S5A). This indicates that IPA1 can activate the transcription of LUC. We then co-expressed FLAG-D53 and IPA1-MYC in tobacco leaves where we found these two proteins could interact with each other (Supplementary information, Figure S5B). Finally, we infiltrated Agrobacterium hosts expressing IPA1-MYC with varying ratios of GFP to FLAG-D53 into tobacco leaves and found that the activity of LUC gradually decreased in accordance with the amount of added FLAG-D53 proteins (Figure 3C and 3D; Supplementary information, Figure S5C). This was further confirmed by the western blotting analysis (Figure 3E), thus demonstrating that D53 could inhibit the transcriptional activation activities of IPA1.
Figure 3.
D53 represses the transcriptional activation activity of IPA1. (A) Transcriptional activity assay in tobacco leaves, showing that IPA1 promotes the expression of the reporter gene Luciferase (LUC) driven by the GTAC-containing promoter. A. tumefaciens transformed with Pro35S:IPA1-MYC, Pro35S:GFP, Pro35S:GUS, and ProGTAC:LUC were mixed and injected into tobacco leaves. D-luciferin was used as the substrate of LUC. (B) Statistical analysis of (A). Values are means ± SD (n = 3). The asterisks represent significant difference determined by Student's t test. **P < 0.01. (C) Transcriptional activity assay in tobacco, showing that D53 represses the transcriptional activation activity of IPA1. A. tumefaciens transformed with Pro35S:IPA1-MYC, Pro35S:GFP, ProGTAC:LUC, Pro35S:GUS, and Pro35S:FLAG-D53 were mixed and injected into tobacco leaves. D-luciferin was used as the substrate of LUC. (D) Statistical analysis of (C). Values are means ± SD (n = 3). Different letters at top of each column indicate a significant difference at P < 0.05 determined by Tukey's HSD test. (E) Protein levels in different infiltration combinations in (C). D53 was detected by rabbit polyclonal antibodies anti-D53, GFP by mouse monoclonal antibody anti-GFP, and IPA1 was detected by rabbit polyclonal antibodies anti-IPA1. Ponceau S staining was used as loading control. See also Supplementary information, Figure S5.
Requirement of D53-IPA1 interaction for D53 repression
To investigate which domains of the two protein molecules are required for the direct interaction between IPA1 and D53, we first dissected their interaction domains through a domain mapping approach. Since the SBP domain of IPA1 is responsible for DNA binding activity and the C-terminal region functions as the activation domain47, we assayed the binding ability of the N-terminal, SBP-box and C-terminal regions of IPA1 with D53 by the BiFC assay. We found that both the N-terminal and the SBP-box of IPA1 could interact with D53 in rice protoplasts, but its C-terminal could not (Figure 4A). This is consistent with our previous finding that the activation domain of IPA1 is located at the C-terminal using the X-gal assay in yeast cells47. Here, we further found that the IPA1 C-terminal domain is sufficient to activate the reporter gene in the rice protoplast system (Figure 4B) and that D53 could repress the activation activity of the full length IPA1 and IPA1-ΔN but not that of IPA1-ΔN-ΔSBP (Figure 4C). Thus, the direct interaction between D53 and IPA1 was required for the suppression of IPA1.
Figure 4.
Interaction of IPA1 and D53 inhibits IPA1 transcriptional activation activity. (A) Interaction between the N-terminal, the SBP domain or the C-terminal of IPA1 and D53 revealed by the BiFC assay in rice protoplasts. The N-terminal, SBP domain or C-terminal of IPA1 were fused with cCFP and D53 was fused with nCFP. (B) The C-terminal of IPA1 was sufficient for the transcriptional activation activity of IPA1. Values are means ± sem (n = 3). The double asterisks represent significant difference determined by Student's t test at P < 0.01. (C) Transcriptional activity assay in rice protoplasts, showing that the N-terminal and the SBP domain are necessary for the D53-mediated repression of IPA1 transcriptional activation activity. Values are means ± SD (n = 3). The asterisk represents significant differences determined by Student's t test at P < 0.05. ns, no significant difference. GUS was used as a control. The transcriptional activation activity of full length IPA1 or IPA1-ΔN, which could bind to D53, was repressed by added D53 compared with the control. However, the activity of IPA1-ΔN-ΔSBP, which could not bind to D53, was not influenced by added D53 compared with control. (D) EMSA assay, showing that D53 does not affect the DNA binding activity of IPA1. The shift bands indicated the binding of GST-IPA1 to the probe containing GTAC element, and the super shift bands indicated the binding of GST-IPA1 together with HisTrx-D53, which was enhanced by adding HisTrx-D53, but not by MBP.
Considering that D53 might inhibit the function of IPA1 through affecting the DNA binding activity or the transcriptional activation activity of IPA1, we wanted to investigate whether D53 could repress the DNA binding activity of IPA1 in an electrophoretic mobility shift assay (EMSA). In the presence of the HisTrx-D53 fusion protein, the DNA binding activity of IPA1 was not affected and a super-shifted band could be observed (Figure 4D), indicating that the interaction between D53 and IPA1 did not affect the DNA binding activity of IPA1. Taken together, these results demonstrate that D53 forms a complex with IPA1 and inhibits the transcriptional activation activity of IPA1 without affecting the DNA binding activity of IPA1.
IPA1 binds to the promoter of D53 and regulates its transcription
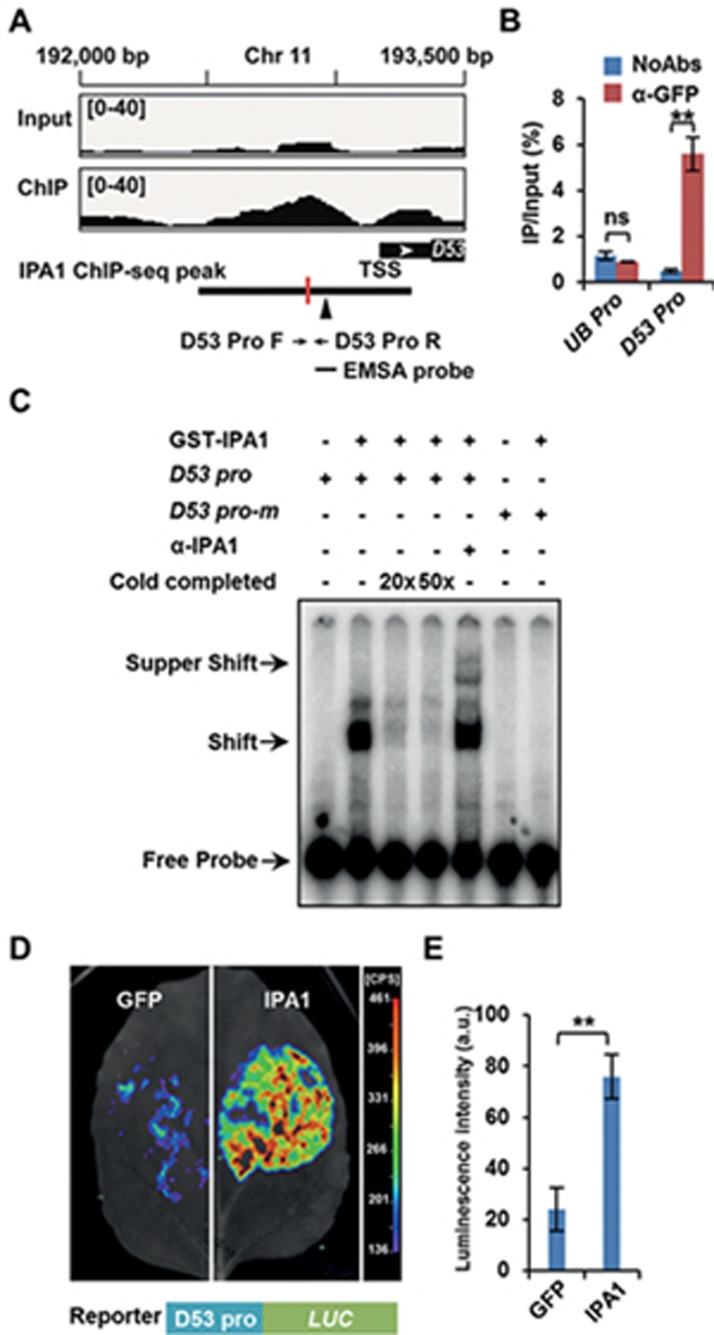
Although SLs play key roles in the regulation of shoot branching and other biological functions, only a few early SL-responsive genes have been identified39,50. In rice, the D53 transcript is rapidly induced upon GR24 treatment38,39. By analyzing IPA1 Chromatin Immunoprecipitation (ChIP)-seq data47, we found one IPA1 binding peak on the promoter region of D53 in shoot apexes with the peak summit 286 bp upstream of the D53 transcription start site (TSS). This peak contains a GTAC element which may be responsible for IPA1 binding (Figure 5A). Thus, we conducted ChIP-qPCR assays using specific primers and an anti-GFP antibody in the shoot base of ProIPA1:7mIPA1-GFP transgenic plants, and found that the IPA1 enrichment levels at the D53 promoter were significantly higher than at the Ubiquitin promoter (Figure 5B). To test whether IPA1 could directly bind to the GTAC motif in the D53 promoter, we performed an EMSA and found that GST-IPA1 could dramatically reduce the migration of the 59-bp probe from the D53 promoter and the added IPA1 antibodies could intensify the retardation, indicating that IPA1 could directly bind to the D53 promoter (Figure 5C).
Figure 5.
IPA1 binds to the D53 promoter and regulates D53 expression. (A) IPA1 binding profile in the promoter of D53. The solid arrowhead refers to the GTAC around the peak summit, and red vertical line to peak summit. Primer pairs D53-ProF and D53-ProR (Supplementary information, Table S1) were used for ChIP-qPCR. The probe was used in EMSA. TSS, transcription start site. (B) Validation of IPA1 direct binding sites in the D53 promoter by ChIP-qPCR analysis. Values are means ± sem (n = 3). The double asterisk represents significant difference determined by the Student's t test at P < 0.01; ns, no significant difference. (C) Direct binding of IPA1 to the D53 promoter in the EMSA assay. The 20- and 50-fold excess non-labeled probes were used for competition. The D53 pro-m is a mutated version of D53 pro probe with the SBP binding motif GTAC changing to ATAC. (D) Transcriptional activity assay in tobacco leaf, showing that IPA1 could enhance the expression of the D53 promoter-drived LUC reporter. A. tumefaciens transformed with Pro35S:IPA1-MYC, Pro35S:GFP, and ProD53:LUC were mixed and injected into tobacco leaves. D-luciferin was used as the substrate of Luciferase. (E) Statistical analysis of (D). Values are means ± SD (n = 3). The asterisk represents significant difference determined by Student's t test. **P < 0.01.
Our findings that the D53 promoter is the target of IPA1 suggest that IPA1 might promote the expression of D53. We tested this in a transient expression assay in N. benthamiana using the D53 promoter fused to the LUC reporter gene. We co-infiltrated N. benthamiana leaves with the 35S:IPA1-MYC construct and the ProD53:LUC reporter and found that the luminescence intensity was dramatically elevated (Figure 5D and 5E), indicating that IPA1 could directly bind to the D53 promoter and activate its expression.
Induction of D53 transcript by SL depends on IPA1 and is part of a feedback loop
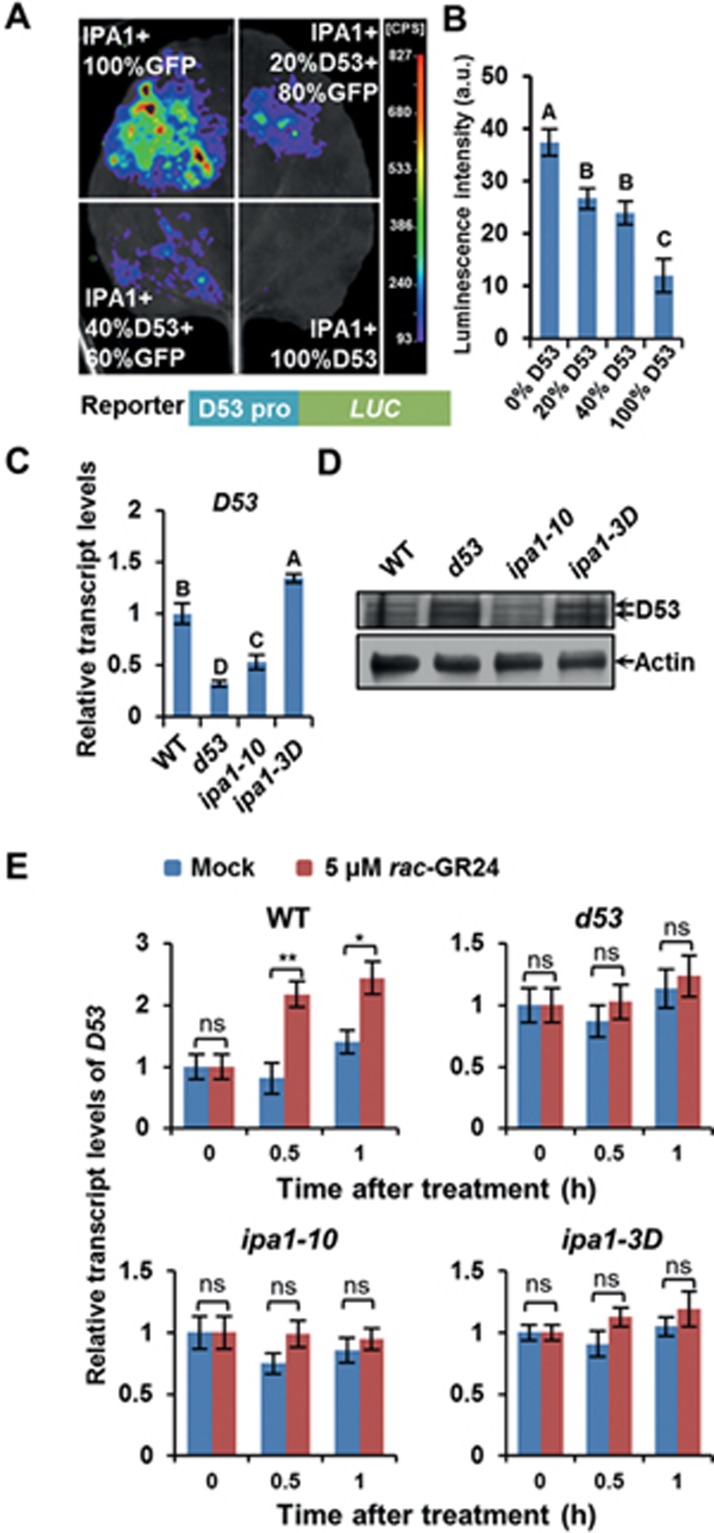
Based on the findings that D53 could inhibit the transcriptional activation activity of IPA1 and that IPA1 could directly regulate D53 expression, we explored the possibility of whether the D53 protein could inhibit its own transcription through IPA1. Thus using the transient expression system expressing the ProD53:LUC reporter construct in N. benthamiana, we co-infiltrated Agrobacterium hosts expressing IPA1-MYC together with varying ratios of Agrobacterium hosts expressing GFP and FLAG-D53. This revealed that the activity of LUC gradually decreased when more FLAG-D53 was added (Figure 6A and 6B). In accord with the result of the transient assay, disruption of IPA1 in ipa1-10 leads to downregulated transcript and protein levels of D53 compared with that in the wild-type plant, whereas ipa1-3D, the gain-of-function mutant, displays elevated levels (Figure 6C and 6D). These results demonstrate that IPA1 can promote the expression of D53, which in turn inhibits the transcriptional activation activity of IPA1, thus forming a negative feedback loop.
Figure 6.
D53 and IPA1 form a feedback regulation loop in SL signaling. (A) Transcriptional activity assay in tobacco, showing that D53 represses the IPA1-drived activation of the D53 promoter. A. tumefaciens transformed with Pro35S:IPA1-MYC, Pro35S:GFP, ProD53:LUC, and Pro35S:FLAG-D53 were mixed and injected into tobacco leaves. D-luciferin was used as the substrate of LUC. (B) Statistical analysis of (A). Values are means ± SD (n = 3). Different letters at top of each column indicate a significant difference at P < 0.05 determined by Tukey's HSD test. (C) D53 transcript levels in WT, d53 and ipa1 mutants. Values are means ± sem (n = 3). Different letters at top of each column indicate a significant difference at P < 0.05 determined by Tukey's HSD test. (D) D53 protein levels in WT, d53 and ipa1 mutants. D53 was detected by rabbit polyclonal antibodies anti-D53. Actin was used as the loading control. (E) Mutations in IPA1 disrupt SL-induced D53 transcription after rac-GR24 treatment. Values are means ± sem (n = 3). Statistical differences between mock and treatment at same time points were determined by Student's t test. *P < 0.05, **P < 0.01; ns, no significant difference.
We further investigated whether IPA1 is responsible for the SL-induction of D53 transcripts by treating wild-type, d53, ipa1-10, and ipa1-3D seedlings with rac-GR24. As shown in Figure 6E, upon the rac-GR24 treatment the D53 transcripts were significantly increased in WT plants but they showed no significant difference to the mock treated d53, ipa1-10, and ipa1-3D plants, indicating that the induction of the D53 transcription by SL depends on normal expression of IPA1. This result is consistent with the fact that the shoot branching phenotypes of ipa1-10 and ipa1-3D plants are both insensitive to an exogenous rac-GR24 treatment. Taken together, these results show that in the rice SL signaling pathway D53 represses the transcriptional activation activity of IPA1 to down regulate its downstream genes, and this in turn affects the D53 transcription, forming a feedback-loop in response to SLs.
Cross talk between miRNA156 and the SL signaling pathway
IPA1 is post-transcriptionally controlled by miRNA156 and miRNA52945,46. In miRNA156 overexpressing (miR156OE) plants, the tillering number is dramatically increased (Supplementary information, Figure S6) and SPL genes including IPA1 are remarkably down-regulated56,57. When treated with rac-GR24, the inhibition of bud outgrowth in miR156OE plants was considerably weaker than in the wild type (Figure 7A, 7B). Moreover, the induction of the D53 transcription by rac-GR24 was strongly impaired in miR156OE plants (Figure 7C). These data suggest that IPA1 may be the common target of miRNA156 and the SL signaling pathway, and that the impairment of the SL signaling in miR156OE plants is probably due to the diminished expression of IPA1.
Figure 7.
Overexpression of miRNA156 compromises SL response. (A) Gross morphologies of wild-type and miRNA156 overexpression (miR156OE) plants with or without rac-GR24 treatment. Seedlings were treated with 1 μM rac-GR24 (+) or mock (–). Bar = 5 cm. (B) Statistical analysis of tiller number in (A). Values are means ± sem (n = 5). The asterisks represent significant difference determined by Student's t test. ***P < 0.001; ns, no significant difference. (C) Overexpression of miRNA156 disrupts SL-induced D53 transcription after rac-GR24 treatment. Values are means ± sem (n = 3). Statistical differences between mock and treatment at the same time points were determined by Student's t test. **P < 0.01; ns, no significant difference. (D) Gross morphologies of d53, ipa1-1D, and d53 ipa1-1D double mutant plants. Bar = 20 cm. (E) Statistical analysis of (D). Values are means ± sem (n = 8). Different letters at top of each column indicate a significant difference at P < 0.05 determined by Tukey's HSD test. (F) A proposed model of the IPA1-mediated SL signaling pathway. In the absence of SLs, the D53 protein binds to IPA1, and together with TPL/TPR proteins represses the transcriptional activity of IPA1. In the presence of SLs, perception of SL leads to degradation of D53 by the proteasome system, which in turn releases the repression of IPA1-regulated gene expression and leads to SL response.
To further investigate the role of IPA1 in SL signaling, we constructed the ipa1-1D d53 double mutant by crossing Ri22, an ipa1-1D allele, with d53. Genetic analysis showed that ipa1-1D could significantly suppress the high tillering phenotype of d53 (Figure 7D and 7E). Similarly, we generated the ipa1-1D d27 and ipa1-1D d10 double mutants, and found that ipa1-1D could repress the high tillering phenotype of both d27 and d10 (Supplementary information, Figure S7), consistent with the previous observation that the overexpression of IPA1 could suppress the high-tillering phenotype of d1058. These genetic data substantiate the finding that IPA1 works downstream of D53 in the SL signaling pathway, where D53 functions as a transcriptional repressor; IPA1 functions in downstream of D53 as a transcription factor to regulate rice tillering as well as enhance D53 expression as a feedback mechanism (Figure 7F).
Discussion
D53 has been identified as a key transcriptional repressor in SL signaling based upon the finding that D53 can be ubiquitinated and degraded by the SCFD3 complex in an SL-dependent manner and that D53 contains three EAR motifs and physically interact with transcriptional co-repressor TPL/TRP proteins38,39,40,59,60. However, the transcription factors targeted by D53 remain elusive and the molecular mechanism of the transcriptional regulation by D53 are still in debate44,61. One major concern is that although SLs regulate multiple developmental processes, few genes have been identified to respond to SL treatment50. Significantly, the transcription of D53 is induced within 1 h of SL treatment and D53 expression levels are greatly repressed in a series of d mutants38. In this study, we demonstrate that D53 can physically interact with IPA1, a member of the SPL family transcription factors, and inhibit its transcriptional activation activity. In the ipa1 loss-of-function and overexpression plants, GR24 treatment could neither inhibit bud outgrowth nor induce the expression of D53 transcripts, indicating that IPA1 functions downstream of D53 in the SL signaling pathway. Therefore, we propose a working model, in which IPA1 functions as the transcription factor that works directly downstream of D53 in the SL signaling pathway (Figure 7F). In the absence of SLs, D53 protein binds to IPA1, and together with TPL/TPR proteins represses the transcriptional activity of IPA1. In response to SLs, D53 is degraded by the proteasome system, which in turn releases the repression of IPA1-regulated gene expression resulting in the SL response. Moreover, we found that IPA1 can directly bind to the D53 promoter and upregulate D53 expression, forming a negative feedback regulation loop.
Feedback loops are considered to play important roles in different signaling pathways62,63,64. In the SL biosynthetic and signaling pathways, several key components are tightly regulated through negative feedback loops10,38,39,41,50,65. In Arabidopsis thaliana, the key biosynthetic genes MAX3 and MAX4 are repressed by GR24 treatment but show elevated expression in max mutants41,50. AtD14 has been shown to undergo 26S-proteasome-dependent degradation after GR24 treatment, which would effectively limit the duration and intensity of SL signaling65. In rice, the transcript levels of D53 in d27, d17, d10, d3, and d14 are lower than that in wild type, in contrast, the D53 protein accumulates in all d mutants whereas the D53 transcript level increases but D53 protein is degraded rapidly after the SL treatment38. This opposite phenomenon raises the possibility that a negative feedback loop may exist to regulate the activity of D53 in planta. However, the specific mechanism of these feedback regulations in SL biosynthesis and signaling is still elusive. Here we found that IPA1 can directly bind to the promoter of D53 and activate D53 expression and that the D53 protein can form a complex with IPA1 and repress transcriptional activation of D53. Thus, the D53 protein may directly inhibit D53 mRNA transcription through interacting with IPA1 and repressing its activity. This feedback regulation would cause a substantial drop in SL signal transduction and would be important for SLs to precisely regulate plant development to enable a rapid response to environmental stimulants.
The mechanism of SL perception and signaling mediated by the SL receptor D14, the F-box protein D3 and the repressor D53 are conserved across monocotyledonous and dicotyledonous species6,9,13,34,37,38,41,43. In Arabidopsis thaliana and pea, BRC1 is considered as one important downstream gene in the SL signaling pathway, which is strongly induced after GR24 treatment to negatively regulate the development of rosette and cauline branches49,51,52. Meanwhile, BRC1 transcripts are down-regulated in max2 and max3 mutants, but up-regulated in smxl6/7/8 triple mutants41. In rice, it has been reported that mutants of OsTB1, the rice ortholog of BRC1, are insensitive to GR24 for tiller outgrowth66, leading to the assumption that the downstream SL signaling pathway may also be conserved. However, in the 2-week-old rice seedling, OsTB1 expression shows no induction following 2 μM rac-GR24 treatment in shoot bases containing the shoot apical meristem (SAM), axillary buds, young leaves and nodes66. Further understanding is needed of transcription regulation of OsTB1 in a more specific tissue, especially in the bud whose elongation is regulated by SL. In our previous work, we showed that IPA1 could bind to the promoter of OsTB1 and regulate its expression46. But in Arabidopsis thaliana, loss-of-function mutants of different SPL genes are sensitive to SL treatment, and no evidence has been found of BRC1's regulation by SPL proteins, suggesting that AtSPLs are probably not involved in BRC1 induction by SLs67. It seems that the downstream SL signaling pathway is not fully conserved between monocotyledonous and dicotyledonous plants, and that OsSPL genes and OsTB1 may have diverse functions with AtSPL genes and BRC1 in response to SLs. However, further evidence is required to support this hypothesis. Moreover, in Arabidopsis thaliana, it is reported that SLs modulate polar auxin transport through the endocytosis of PIN-FORMED1 (PIN1)68,69. PIN1 is localized to the plasma membrane and it is rapidly depleted after SL treatment, independently of protein synthesis69. In rice, it is reported that basipetal polar auxin transport is elevated in d2714, and that IAA level is increased in d328, suggesting that the crosstalk between SL and auxin, two key plant hormones for plant development, is of great importance in both species. However, the mechanism of this crosstalk is still obscure. From IPA1 ChIP-seq data, we found that IPA1 binding sites are located on the promoter of PIN1b47, but whether and how IPA1 and PIN1b are involved in the crosstalk between auxin and SL needs further study.
The high tillering phenotypes of d mutants have been well studied, but there are other significant phenotypes such as dwarfism and low fertility that need further investigation12,13,14. As a pleiotropic regulator, IPA1 regulates many important phenotypes, such as tiller number, plant height and panicle morphology in rice45,46. More importantly, IPA1 has been widely used in breeding elite rice varieties due to its great potential in improving rice yield45,54,55. It will be impoartant to have an in-depth characterization of the regulatory network of IPA1 and D53 to reach a better understanding of rice architecture and provide a powerful tool and new targets for molecular breeding in rice.
Materials and Methods
Plant materials
Rice (Oryza sativa L.) ssp. japonica Nipponbare, ProIPA1:7mIPA1-GFP transgenic line, ZH11, ipa1-10, ipa1-11, ipa1-3D, ipa1-4D, miR156OE, Ri22, d53 and other mutants were grown either in the green house under 16 h light and 8 h dark at 28 °C or the experimental field of the Institute of Genetics and Developmental Biology. Double mutants of ipa1-1D d53, ipa1-1D d10, and ipa1-1D d27 were generated from crosses between Ri22 (japonica), an ipa1-1D cultivar, and d53, d10, and d27 mutants, respectively.
TILLING
TILLING was performed in a population of 6 000 EMS-mutagenized M2 rice plants in the ZH11 background (Oryza sativa L. japonica) using PCR primer pairs targeted to a conserved domain of IPA1. Crude CEL1 endonuclease was extracted from celery70,71, and used for the cleavage of PCR products at mismatched positions of re-annealed heteroduplexes. Point mutations were detected by capillary electrophoresis using AdvanCEFS96 (Advanced Analytical Technologies, USA).
Protein interaction analysis
The primer pairs used in protein interaction analysis are listed in Supplementary information, Table S1. For the yeast two-hybrid assay, the IPA1 coding sequence (CDS) and D53 CDS were inserted into pGBKT7 and pGADT7 respectively. The Matchmaker® Gold Yeast Two-Hybrid System (Clontech) was used to test the interaction between IPA1 and D53 following the manufacturer's instructions. For the BiFC assay, the D53 CDS and different truncations of the IPA1 CDS were amplified and cloned into pUC-SCYNE(R) and pUC-SCYCE respectively72. The BiFC assay was performed as described previously47. For the Co-IP assay, total proteins were extracted from ProIPA1:7mIPA1-GFP or ProUB:GFP transgenic calli according to the method described previously38. Following the manufacturer's instructions, 30 μl of the agarose-conjugated anti-GFP monoclonal antibody (MBL, D153-8) were added into 500 μl total extracted proteins and incubated at 4 °C for 3 h with gentle rotation. The beads were washed three times with 350 μl extraction buffer, and eluted with 30 μl SDS-PAGE sample buffer. Immunoblotting was performed as described38,47. For the GST pull-down, purified GST, GST-IPA1, HisTrx-D53 and proteins extracted from rice calli were used in the assay, which was performed as described38. HisTrx-D53 fusion protein and D53 protein from rice calli were detected by rabbit polyclonal antibodies anti-D5338 and GST fusion proteins by Ponceau S or a monoclonal mouse antibody anti-GST (Abmart, M20007).
RNA isolation and quantitative real-time PCR
RNA isolation, reverse transcription and real-time PCR were performed as described previously47. Primer pairs used in real-time PCR were listed in Supplementary information, Table S1.
Expression and purification of fusion proteins
The full-length CDS of IPA1 or D53 was amplified by the primer pairs listed in Supplementary information, Table S1 and cloned into the Escherichia coli expression vector pGEX-6p-1 (GE Healthcare) or pDEST51. Expression and purification of fusion proteins were conducted as described38,47.
ChIP-qPCR assay
The ChIP-qPCR using the ProIPA1:7mIPA1-GFP transgenic seedlings was performed according to the method described previously47,73. PCR reactions were performed in triplicate for each sample, and expression levels were normalized to the input sample for enrichment detection. The fold enrichment was calculated against the Ubiquitin promoter. No addition of antibodies (NoAbs) was served as a negative control.
EMSA
The probe sequences are listed in Supplementary information, Table S1. Probe labelling and EMSA were performed as described previously47.
Transcriptional activity assay in tobacco leaf
To generate the ProGTAC:LUC and ProD53:LUC, synthesized GTAC sequence and D53 promoter were cloned into the pCAMBIA1301-LUC vector. To generate the Pro35S:IPA1-MYC and Pro35S:FLAG-D53, CDS of IPA1 and D53 were cloned to the 1300-MYC and pDEST1300-FLAG vectors, respectively. The plasmids used in this study were transformed to Agrobacterium tumefaciens strain EHA105. p19 was used to suppress RNA silencing, pCAMBIA1301 for GUS expression as an internal control. Agroinfiltration and luciferase imaging were performed as described previously74. GUS activities were measured with 4-methylumbelliferyl-β-D-glucuronide (Sigma) according to the manufacturer's instructions.
Transcriptional activity assay in rice protoplasts
The plasmids containing GAL4BD-IPA1, 35sLUC and pRTL or GAL4BD-IPA1-ΔN, 35sLUC and pRTL or GAL4BD-IPA1-ΔN-ΔSBP, 35sLUC and pRTL were introduced into rice leaf protoplasts as described75, with plasmids containing GAL4BD, pRTL and 35sLUC as a negative control. The assay was performed as described47.
GR24 treatment
Two-week-old hydroponically cultured rice seedlings were grown in the climatic cabinet at 80% humidity, under 16 h light at 25 °C and 8 h dark at 16 °C. The seedlings were treated with 5 μM rac-GR24 (StrigoLab) or equal volume of acetone, then 0.5 cm shoot base samples were harvested at indicated time points. RNA isolation, cDNA synthesis, qRT-PCR, protein extraction, and immunoblotting were performed as described above.
Accession numbers
Gene sequence used in this study can be found in the Rice Genome Annotation Project under accession numbers: LOC_Os08g39890 (IPA1) and LOC_Os11g01330 (D53).
Author Contributions
XS, ZL, HY and JL conceived this project and designed all experiments. XS, ZL, HY, GS, JX, XM, YJ, GL, JD, XFY, CML and HL performed experiments. XS, ZL, HY, GX, YW and JL analyzed data. XS, ZL, HY and JL wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Prof Shouyi Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for providing the plasmids GAL4BD, 35sLUC and pRTL, Prof Qian Qian and Dr Xingming Hu (State Key Laboratory of Rice Biology, China National Rice Research Institute) for providing the rice miRNA156 overexpressing lines, Prof Chuanyou Li for sharing the Plant Imaging System (Berthold Technologies), and lab members (Drs Lei Wang, Liang Jiang, Xue Liu, Huihui Liu) for their technical assistance, and Dr Bing Wang for comments on the manuscript. We are grateful to the Genomics and Bioinformatics Facility of State Key Laboratory of Plant Genomics, Institute of Genetic and Developmental Biology for phosphor imaging. This work was supported by grants from the National Key Research and Development Program of China (Grant 2016YFD0101800), National Natural Science Foundation of China (Grant 91635301), and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” (Grant XDPB0401).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
TILLING lines of IPA1 have no obvious phenotype.
Genomic structure and mutant alleles of IPA1.
Expression and protein levels of IPA1 after rac-GR24 treatment.
Protein levels of IPA1 in various dwarf mutants.
Expression of the fusion proteins and interaction between IPA1 and D53 in tobacco leaves.
High tillering phenotype of miRNA156-overexpressing transgenic plants.
ipa1-1D represses the high tillering phenotype of d10 and d27.
Primers used in this study
References
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 1966; 154:1189–1190. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005; 435:824–827. [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 2004; 14:1232–1238. [DOI] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 2003; 17:1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 2005; 8:443–449. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HMO. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 2007; 50:80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van de Sande K, Leyser HMO. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002; 129:1131–1141. [DOI] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 2012; 159:1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012; 139:1285–1295. [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 2007; 51:1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 2009; 50:1416–1424. [DOI] [PubMed] [Google Scholar]
- Gao Z, Qian Q, Liu X, et al. Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol Biol 2009; 71:265–276. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 2005; 46:79–86. [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009; 21:1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wu C, Fu Y, et al. Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 2009; 230:649–658. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, et al. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 2006; 48:687–698. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, et al. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 2006; 142:1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Janssen BJ, et al. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 2012; 22:2032–2036. [DOI] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 2007; 143:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, et al. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 2005; 17:746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature 2008; 455:189–194. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008; 455:195–200. [DOI] [PubMed] [Google Scholar]
- Koltai H. Strigolactones are regulators of root development. New Phytol 2011; 190:545–549. [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, et al. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 2014; 65:6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Umehara M. Possible roles of strigolactones during leaf senescence. Plants (Basel) 2015; 4:664–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant 2013; 6:18–28. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Ligerot Y, Dun EA, et al. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol 2013; 163:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang D, Chen D, Liu G, et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc Natl Acad Sci USA 2014; 111:11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Sado A, Asami K, et al. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc Natl Acad Sci USA 2014; 111:1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, et al. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012; 335:1348–1351. [DOI] [PubMed] [Google Scholar]
- Abe S, Sado A, Tanaka K, et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc Natl Acad Sci USA 2014; 111:18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Yoneyama K, Filardo F, et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 2016; 113:6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, van Dijk ADJ, Scaffidi A, et al. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 2014; 10:1028–1033. [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016; 536:469–473. [DOI] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Yi W, et al. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res 2015; 25:1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 2013; 18:147–160. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, et al. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 2013; 4:2613. [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013; 504:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013; 504:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Duan J, Ke J, et al. A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 2017; 3:e1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, et al. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 2015; 27:3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, et al. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 2015; 27:3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 2011; 108:8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ward S, Li P, Bennett T, Leyser O. SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 2016; 28:1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 2010; 42:541–544. [DOI] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 2010; 42:545–549. [DOI] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, et al. Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 2013; 25:3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li X, Zhou Z, et al. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 2016; 7:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007; 19:458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, et al. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 2009; 73:2460–2465. [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 2012; 158:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 2012; 158:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale M, Bennett T, Leyser O. BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 2017; 144:1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu H, Xiong G, et al. Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 2017; 29:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu H, Ma B, et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat Commun 2017; 8: 14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Shen J, Hou X, et al. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 2012; 158:1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 2006; 142:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Li W, Miura K, Ashikari M, Kyozuka J. Control of tiller growth of rice by OsSPL14 and strigolactones, which work in two independent pathways. Plant Cell Physiol 2012; 53:1793–1801. [DOI] [PubMed] [Google Scholar]
- Smith SM, Li J. Signalling and responses to strigolactones and karrikins. Curr Opin Plant Biol 2014; 21:23–29. [DOI] [PubMed] [Google Scholar]
- Xiong G, Wang Y, Li J. Action of strigolactones in plants. Enzymes 2014; 35:57–84. [DOI] [PubMed] [Google Scholar]
- Waldie T, McCulloch H, Leyser O. Strigolactones and the control of plant development: lessons from shoot branching. Plant J 2014; 79:607–622. [DOI] [PubMed] [Google Scholar]
- Charest PG, Firtel RA. Feedback signaling controls leading-edge formation during chemotaxis. Curr Opin Genet Dev 2006; 16:339–347. [DOI] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016; 143:3238–3248. [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 2012; 22:1732–1737. [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sanchez-Ferrero JC, et al. Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014; 26:1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 2010; 51:1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Liang Y, Seale M, Ward S, Muller D, Leyser O. Strigolactone regulates shoot development through a core signalling pathway. Biol Open 2016; 5:1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, et al. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 2010; 137:2905–2913. [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 2013; 11:e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang GQ, Yao XF, Liu CM. A simple CELI endonuclease-based protocol for genotyping both SNPs and InDels. Plant Mol Biol Rep 2013; 31:1325–1335. [Google Scholar]
- Till BJ, Zerr T, Comai L, Henikoff S. A protocol for TILLING and Ecotilling in plants and animals. Nat Protoc 2006; 1:2465–2477. [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 2008; 56:505–516. [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 2008; 3:1018–1025. [DOI] [PubMed] [Google Scholar]
- Zhang G, Song X, Guo H, Wu Y, Chen X, Fang R. A small G protein as a novel component of the rice brassinosteroid signal transduction. Mol Plant 2016; 9:1260–1271. [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2006; 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TILLING lines of IPA1 have no obvious phenotype.
Genomic structure and mutant alleles of IPA1.
Expression and protein levels of IPA1 after rac-GR24 treatment.
Protein levels of IPA1 in various dwarf mutants.
Expression of the fusion proteins and interaction between IPA1 and D53 in tobacco leaves.
High tillering phenotype of miRNA156-overexpressing transgenic plants.
ipa1-1D represses the high tillering phenotype of d10 and d27.
Primers used in this study