Abstract
Protein turnover rates severely decline in aging organisms, including C. elegans. However, limited information is available on turnover dynamics at the individual protein level during aging. We followed changes in protein turnover at one-day resolution using a multiple-pulse 15N-labeling and accurate mass spectrometry approach. Forty percent of the proteome shows gradual slowdown in turnover with age, whereas only few proteins show increased turnover. Decrease in protein turnover was consistent for only a minority of functionally related protein subsets, including tubulins and vitellogenins, whereas randomly diverging turnover patterns with age were the norm. Our data suggests increased heterogeneity of protein turnover of the translation machinery, whereas protein turnover of ubiquitin-proteasome and antioxidant systems are well-preserved over time. Hence, we presume that maintenance of quality control mechanisms is a protective strategy in aging worms, although the ultimate proteome collapse is inescapable.
The downturn of protein homeostasis (proteostasis), including the commonly observed slowdown of protein synthesis and degradation (protein turnover), is a major hallmark of aging (1, 2). Proteome mismanagement along with age-dependent protein aggregation, oxidation and misallocation, likely leads to the overall functional decline in senescent organisms (3).
The nematode Caenorhabditis elegans (C. elegans)1 is one of the best-studied model organisms in aging research because of its relatively short lifespan and the extensive molecular toolbox available for this organism. Microarray experiments (4, 5) and proteomic analyses (6–10) have been used to profile the changes in gene expression and protein abundance levels of aging C. elegans, respectively. Overall, these studies report an age-dependent decline in ribosomal proteins and an increase of proteasome complexes and small heat shock proteins (6, 9), corroborating the link between the aging process and proteostasis.
Recently, we found a drastic decrease of protein turnover rates in aging C. elegans using a classical 35S pulse-chase labeling (11). Liang et al. (2014) profiled changes in the pool of de novo synthesized proteins in old versus young worms, whereas another recent study has used a single pulse-chase SILAC technique to estimate protein appearance at proteomic scale during adult lifespan (12). A similar approach was used by another group, which reports protein half-lives for developing L4 and day 5 old worms (13). Although these studies point out shifts in protein synthesis rates in old worms, the gradual change in turnover of individual proteins with increasing chronological age has not been investigated previously. Do aging worms display a heterogeneous decline in proteome turnover or do specific subsets of proteins exhibit distinct trends in protein turnover with age?
To address this question, we analyzed the change of individual protein half-lives in aging worms using a multiple-pulse metabolic 15N-labeling method. Hereto, subsamples of an aging C. elegans cohort were taken daily and pulse-chased individually. These series of timed samples were subsequently analyzed with high-resolution mass spectrometry to estimate protein half-lives. Our data indicates that turnover of aging proteome slows down partially, although this pattern cannot be generalized for all proteins. Instead, it seems that most protein turnover rates are affected in a heterogeneous way as clear patterns are absent within functionally or spatially related protein groups, which indicates a nonorchestrated collapse of proteome management in aging worms. On the other hand, some distinct protein pools do show specific tendencies in protein turnover with age, such as the proteasomal proteins which maintain their fast turnover rates and the consistent slowdown observed for all tubulins and vitellogenins. In summary, increased heterogeneity in protein turnover occur during normal aging in C. elegans.
EXPERIMENTAL PROCEDURES
Strains
In this study, we used the strain GA153 glp-4(bn2ts)I daf-16(mgDf50)I; daf-2(e1370)III which was kindly provided by David Gems at the University College of London. Worms were maintained as described previously (14).
Metabolic 15N- and 14N-labeling of the Escherichia coli Bacteria
E. coli K12 was freshly grown overnight at 37 °C at shaking at 120 rpm in minimal medium, containing 42 mm Na2HPO4, 22 mm KHPO4, 9 mm NaCl, trace elements (17 mm EDTA, 3.1 mm FeCl3·6H2O, 0.62 mm ZnCl2, 0.076 mm CuCl2·2H2O, 0.042 mm CoCl2·6H2O, 0.16 mm H3BO3, 0.0068 mm MnCl2·6H2O), glucose 20% (w/v), 1 mm MgSO4, 0.3 mm CaCl2, 0.0041 mm biotin, 0.0033 mm thiamin, and 93 mm 15N- or 14N-containing NH4Cl (sterilized over 0.22 μm filter). Overnight cultures were concentrated 50-fold.
Culturing and Sampling of C. elegans
Synchronized L1 nematodes were plated on nitrogen-free agarose (1.2%) plates containing NaCl (0.3%), cholesterol (0.0005%), 1 mm CaCl2, 1 mm MgSO4 and 25 mm K2HPO4/KH2PO4 (pH 6.0) and a lawn of freshly grown 14N-labeled E. coli K12 cells. Worms were grown at 17 °C until the third larval stage (L3) and then switched to 24 °C for the remainder of the experiment. Worms that had freshly molted to adults (adult day 0) were washed three times and transferred into a Fernbach flask (not exceeding 1500 worms/ml) containing S-basal (100 mm NaCl, 50 mm potassium phosphate, pH 6.0), 12.93 μm cholesterol, 75 μm 5-fluoro-2′-deoxyuridine (Acros Organics, Geel, Belgium), and 14N-labeled E. coli K12 cells (A550 = 1.0). Starting from day one of adulthood, daily subcultures were collected and washed thoroughly (2× S-basal, 1× S-basal containing 2.5 mm EDTA, 2× S-basal) to remove bacteria. Next, worms were pulsed in new culture flasks containing S-basal, 12.93 μm cholesterol, 75 μm 5-fluoro-2′-deoxyuridine, and 15N-labeled E. coli K12 cells (A550 = 1.0). Samples were taken at 20 and 40 h labeling for three independent aging cohorts. Worms were washed thoroughly (2× S-basal, 1× S-basal containing 2.5 mm EDTA, 2× S-basal) to remove bacteria, after which dead worms were removed via Percoll (Sigma-Aldrich, St. Louis, MO) washing. The worm pellet was resuspended in 200 μl denaturing solution (8 m urea, 1 mm EDTA, 10 mm DTT, 50 mm TrisHCl, pH 8.0) and immediately dropwise frozen in liquid nitrogen and stored at −80 °C. The sample size was chosen based on a previously published study (14).
Experimental Design and Statistical Rationale
Samples (54 in total) were processed in blocked groups comprising all daily samples (day 1 till day 7) from one specific post-labeling time point (0 h, 20 h, or 40 h). Per time point, three biological replicates were used to achieve statistically valid results. Samples within a batch were analyzed blindly and in random order (Fig. 1B).
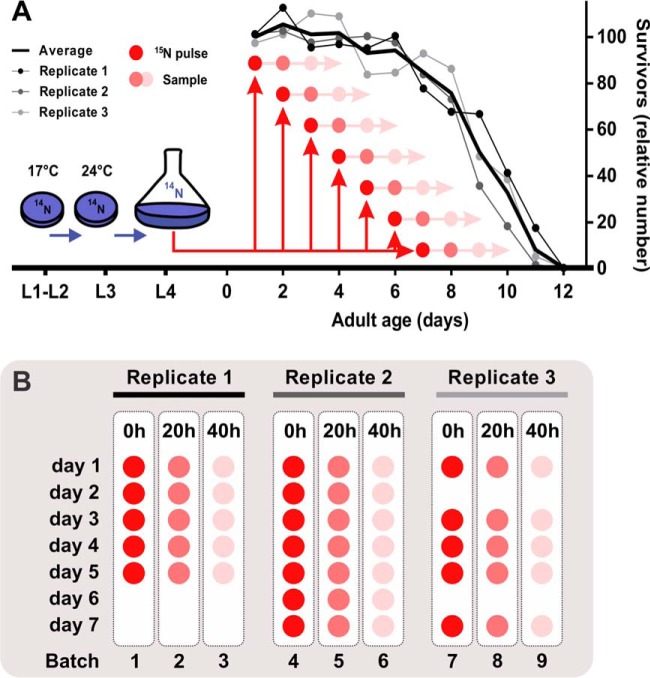
Fig. 1.
Experimental study design. A multiple-pulse 15N-labeling approach was performed to analyze the daily changes of individual protein half-lives in aging C. elegans. A, Subsamples of three independent aging cohorts were individually pulse-chased starting from day 1 until day 7 of adulthood (old worms). Survival was estimated daily by manual counting of live worms in culture subsamples and expressed as the percentage of the initial population. B, Samples were blocked based on the pulse-time, randomized and blindly analyzed using LC-MS/MS.
Preparation of the Tryptic Peptide Samples
The frozen worm beads (∼1500 worms in denaturing solution) were homogenized using a BioPulverizer (BioSpec, Bartlesville, OK) pre-conditioned in liquid nitrogen. The fine powder was recovered into 1.5 ml tube, thawed, centrifuged (2 min, 5000 rpm), and sonicated for 30 s in a 5510 Branson ultrasonic water bath (Branson Ultrasonics, Soest, Netherlands). Protein concentrations were determined using Coomassie assay. For further processing steps we took the aliquots containing 150 μg of protein and readjusted the volume with denaturation buffer. The samples were subjected to cysteine alkylation by 40 mm iodoacetamide for 1 h at 37 °C in dark shaking conditions (900 rpm), 4-fold dilution with 50 mm ammonium bicarbonate (pH 7.8), and digestion with trypsin (3 μg per sample, Promega, Madison, WI) for 12 h at 37 °C shaking conditions (400 rpm). Next, tryptic peptide samples were cleaned-up via C-18 SPE columns (Discovery DSC-18, SUPELCO, 52601-U) and concentrations were adjusted to 0.2 μg/μl (14, 15).
LC/MS Analysis and Peptide Identification
Each individual sample was analyzed with a constant-pressure capillary HPLC system coupled online to an LTQ Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) using an electrospray ionization interface. Instrument settings were described previously by Depuydt et al. (2013). The resulting 54 data sets were converted to the mzXML format using MSConvert (3.0.7680) (settings: -mzXML -32 -z), a part of the ProteoWizard software suite (16). Peptide identification was performed by preprocessing data sets with DeconMSn (2.3.1.3) (17), DtaRefinery (v1.2.0.1) (18) followed by the search tool MS-GF+ (v 1.1.0) (19) against the WormBase V238 database (20) containing 26248 WormPep entries with added human keratins and pig trypsin as contaminants. The search was aimed at only light 14N-peptides as we did not anticipate more than 50% label incorporation in such a short labeling pulse. The key MS-GF+ settings are: PMTolerance = 10 ppm; NumMods = 3; StaticMod = C2H3N1O1, C, fix, any, Carbamidomethyl; DynamicMod = None; EnzymeID = 1 (trypsin); IsotopeError = 0.1; NTT = 2 (no nonspecific cleavages allowed); minLenght = 6; maxLenght = 50; minCharge = 1, maxCharge = 4. The results of the MS/MS searches were saved as mzIdentML files. Peptide and protein identification information is listed in supplemental Table S1.
Processing 15N/14N LC-MS/MS Data
The analysis of the LC-MS/MS data sets and the estimation of protein half-lives is described in recently accepted work by our group (21). In brief, the processing of the data was carried out in the R computing environment (version 3.0.2), using a package N14N15 for the analysis of the 15N/14N isotopic data (available from GitHub, https://github.com/vladpetyuk/N14N15). Two files were required as an input: (1) mzXML with raw MS spectra and (2) mzIdentML containing peptide identifications. The peptide-to-spectrum matches were considered confident if they passed 5 ppm parent ion mass measurement tolerance and SpecEvalue < 10−9 thresholds. Peptide identification false discovery rate (FDR) was estimated using a reverse protein sequence approach (22). The FDR was estimated to be 0.11% on average per data set or 1.7% in total at unique peptide sequence across all 110 LC-MS/MS data sets. Extracted ion chromatograms were generated for the parent ions in the vicinity of the MS2 spectrum that yielded identification. After chromatographic peak detection, MS1 spectra within full width half maximum intensity were summed. Further, given the known elemental composition of the peptide, the summed MS1 spectrum intensities were fitted with the mixed isotopic distribution assuming one natural and one nonnatural proportion of the 15N isotope. The peptides that fitted to the 14N/15N isotopic distribution were filtered based on the following criteria: (1) the proportion of 15N isotope in the heavy component was constrained from 55% to 90% and (2) the R2 value between the theoretical and experimental intensities in the isotopic envelopes (that primarily result from ∼1% natural presence of 13C) for light and heavy components had to be at least 0.9 and 0.5, correspondingly. Deviation of peptide mass derived from the experimental data from the theoretical was limited with 5 ppm (after zero-centering the mass measurement error histogram to account for imperfect mass spectrometer instrument calibration). Peak picking was performed using wavelet-based approach (23). The signal-to-noise ratio of detected peaks was required to be at least 3.
The proportion of 15N-labeled peptide was log-transformed for convenience of visualization (log2, further denoted as x). Calculation of the protein half-lives included following steps:
We assumed that, during the two-day pulse labeling, the total protein abundance did not change significantly in the sterile adult worms, whereby protein synthesis equals degradation, collectively designated as protein turnover:
The change in 14N-labeled proteins with time is:
or
In this equation, P0 is the total protein concentration right before the labeling pulse, kdp is the protein degradation kinetic constant and Δt is pulse length. According to the previous assumption, protein concentration does not change during pulse labeling. Therefore:
Plugging-in the proportion of 15N as x and log-transforming the equations results in:
Protein half-life could be estimated using following equation:
Protein half-life was calculated using the weighted average from 20- and 40- hour incorporation values, in which the 40 h incorporation value was assigned double the weight as its protein half-life estimation is more accurate.
Pavlidis Template Matching and Functional Analysis
Protein turnover patterns were analyzed with Pavlidis template matching (PTM), implemented in the MultiExperiment Viewer, part of the TM4 microarray software suit (24, 25). The data set was searched for proteins with increasing or decreasing protein half-lives with age using a defined template profile. The PTM algorithm is based on the Pearson correlation between the template and the proteins in the data set and protein turnover patterns were allowed to match one of the defined templates when p values were lower than 0.05 (corresponding to a regression coefficient R of more than 0.75). Patterns that did not match both templates were considered not unchanged over age. p values were adjusted for multiple testing using FDR-based (False Discovery Rate) correction. The Database for Annotation, Visualization, and Integrated Discovery (DAVID 2015) (26) and Wormbase (Version WS246) were used for protein annotation and evaluation of functionally and spatially related proteins (supplemental Table S5). In addition, tissue specificity of expression was predicted using an online bioinformatic tool (http://worm-tissue.princeton.edu) (27). Graphs and statistical tests (including tests for normality) were performed with Graphpad Prism version 6.05 for Windows (Graphpad Software, La Jolla, CA).
Label-free Quantification
Along with the 15N-labeled worm samples, we collected aliquots of worm cultures prior to labeling. This allowed us to estimate relative changes in protein abundance over lifetime. Quantification was performed using a label-free LFQ approach (28). The MS/MS searches and quantitative analysis was performed using the MaxQuant tool (29) with default settings with exception that trypsin was not allowed to cleave after proline and the number of missed cleavages was restricted to two.
RESULTS
Experimental Design to Monitor Progressive Changes in Protein Turnover
To investigate the change of protein half-lives in the aging proteome, we used the normal-lived glp-4(bn2) daf-16(mgDf50); daf-2(e1370) mutant strain. We chose this reference strain in line with our previous proteomic studies (14, 21, 30), to enable convenient comparison between the resulting data sets. The DAF-16 dependent lifespan-extending daf-2 mutation is completely nullified by the daf-16(mgDf50) knockout allele present in this worm. In addition, we took advantage of working with a temperature-sensitive sterile mutant background (glp-4) to avoid the elimination of 15N label via egg laying and to focus on the somatic cells of the aging worms. It has been shown that germline-ablated animals increase lifespan via DAF-16 activation (31–33). Hence, effects of glp-4 will be minimal because these worms carry the daf-16(mgDf50) mutation. To monitor age-dependent changes in protein turnover, we used Stable Isotope Labeling by Nitrogen in Caenorhabditis elegans (SILeNCe), a metabolic labeling approach that has been shown to be efficient in C. elegans (21, 34, 35). The experimental study design is shown in Fig. 1. Synchronized L1 worms were grown at 17 °C on nitrogen free agar plates seeded with 14N-labeled E. coli bacteria. At the third larval stage, worms were switched to 24 °C in liquid Fernbach culture, shaken at 120 rpm. From the first day of adulthood daily subcultures were taken and pulsed with 15N-labeled E. coli after which samples were taken at 20 and 40 h of incubation. Pulsing of new subcultures was continued until day 7 and the last sample was collected at day 9, which coincides with mean lifespan of the population under these culture conditions (Fig. 1A, supplemental Table S2). Blocked groups based on sampling time were analyzed using accurate mass spectrometry (MS)-based quantitative proteomics in a randomized and blind manner (Fig. 1B). Molecular proportions of heavy (15N) and light (14N) isotope peaks of the peptides were extracted using a custom R package and the estimated protein half-lives represent the weighted average from 20- and 40-hour incorporation values.
Diverse Impact of Aging on Protein Turnover
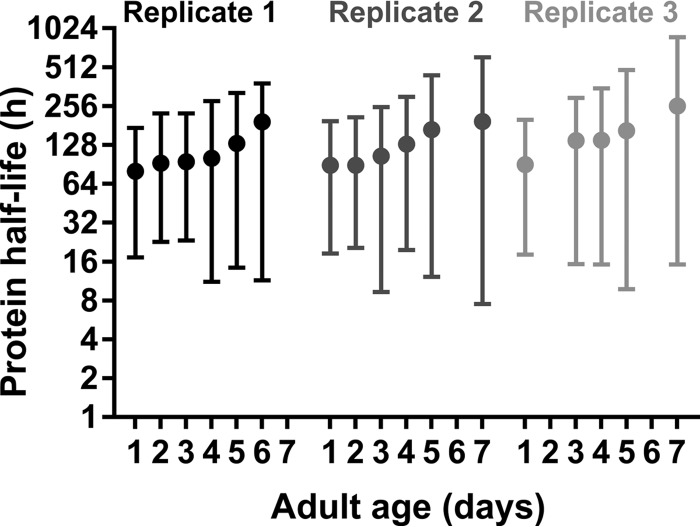
We were able to monitor the age-dependent changes in turnover of 878 peptides, corresponding to 546 unique proteins, of three independent aging cohorts. All estimated protein half-lives are listed in supplemental Table S3. Median protein half-life increases gradually with chronological age, starting from 69 h at the first day of adulthood and reaching 132 h at day 7 (Fig. 2). These findings are in line with previous results from classical 35S pulse-chase labeling, showing decreased protein turnover in old worms (11). In addition, variation of protein half-lives escalates with age as is reflected by the expansion of the interquartile ranges over time.
Fig. 2.
Increasing variability of protein turnover in aging C. elegans. Three independently sampled replicate cultures showing gradual increase in median protein half-life over time.
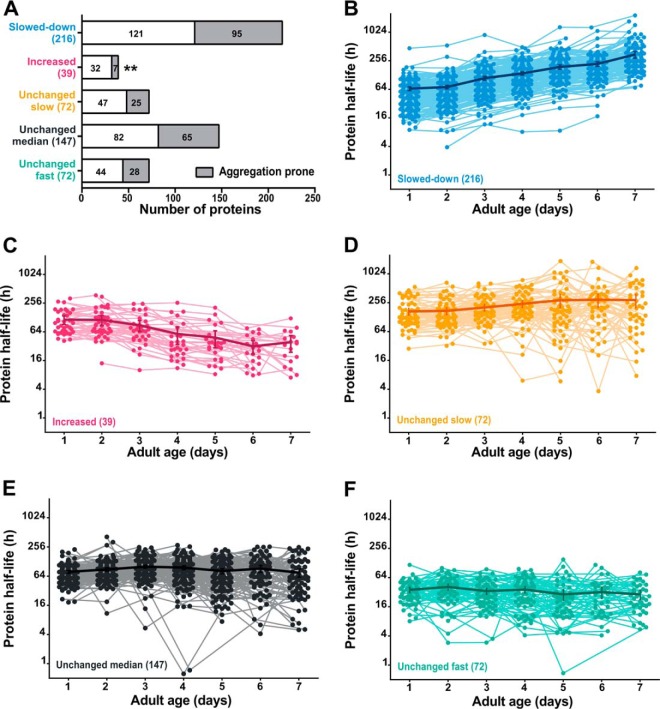
To investigate whether turnover rates are affected differently in several subsets of proteins according to their function or subcellular location, we discriminated proteins of which turnover was decreased or increased via Pavlidis Template Matching (PTM) (36). Turnover rate of 40% of the identified proteins gradually slows down with age (Fig. 3A, 3B), whereas only 7% shows an increase in turnover (Fig. 3A, 3C). Unmatched protein turnover trends (291 proteins) were further divided into three subgroups using the quartiles of the overall average half-life over age as cutoff values, representing proteins that have a steady slow (average half-life > 129 h, Fig. 3A, 3D), median (average half-life between 55 and 129 h, Fig. 3A, 3E) or fast (average half-life < 55 h, Fig. 3A, 3F) turnover during aging (supplemental Table S4 for a detailed overview).
Fig. 3.
Protein turnover patterns with age. (A) Overview of proteins per trend based on PTM-analysis, with indication of aggregation propensity (38). Detailed overview of proteins with slower (B), faster (C), unchanged slow (D), unchanged median (E) and unchanged fast (F) protein turnover with advancing age. Each dot represents a half-life estimation based on the weighted average of 20- and 40-hour incorporation values. Thick line with error bars (indicating S.E.) visualizes the overall tendency per group. (log2-y axis) ** p = 0.003. Summary of the data from replicate experiments is included in supplemental Table S4.
It has been proposed that protein aggregation actively contributes to a decrease in proteolysis with age (37). We therefore considered protein aggregation as a possible driving force in the slowdown of protein turnover. To evaluate this hypothesis, we compared our data set with a list of C. elegans proteins prone to aggregation at advanced age (38). We found comparable aggregation propensity for proteins with unchanged and decreased protein turnover during aging, independent of their absolute turnover rates. This finding contradicts our proposition that fast turnover is sufficient to counter late onset aggregation. However, in the small group of proteins of which turnover rate increases with age, aggregation-prone proteins are significantly underrepresented (Fig. 3A, Fisher's test, p value = 0.003). This observation suggests that, in some proteins, either turnover is actively increased at advanced age to avoid aggregation, or that decreased aggregation is a side effect of these increased protein dynamics.
Proteostasis-related Proteins Display Disparate Turnover Trends with Age
Proteome management relies on a complex network of biological processes, including mRNA translation, folding and degradation by the ubiquitin/proteasomal and autophagosomal pathways (39). Recent work showed increased proteome imbalance with age in C. elegans (9), possibly resulting from impaired turnover of proteostasis machinery components. To verify this idea, we performed a functional annotation analysis (supplemental Table S5). Aggregation propensity (38) of these groups was also monitored (supplemental Fig. S1).
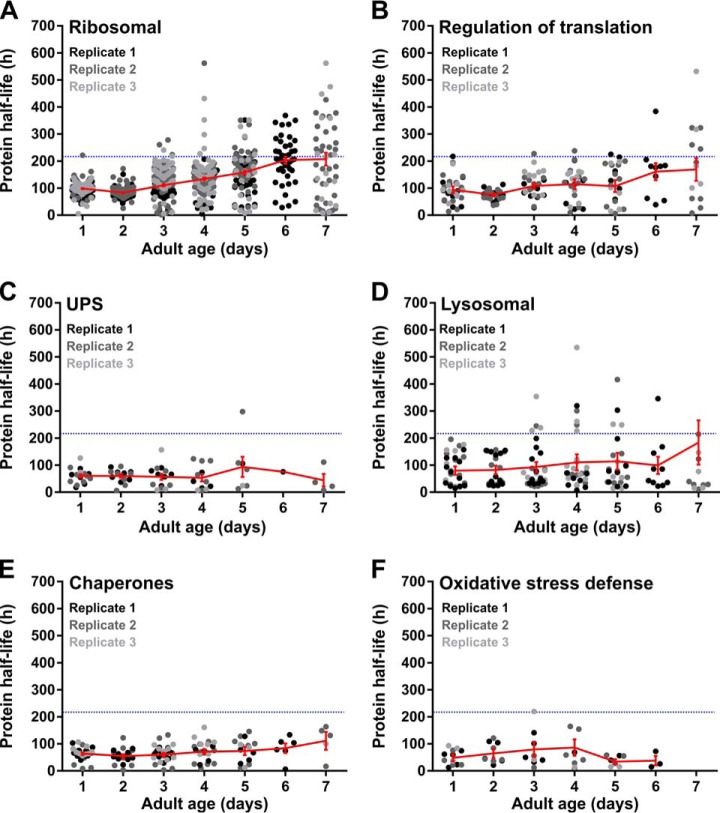
The protein synthesis machinery, including ribosomal subunits and initiation and elongation factors, display very heterogeneous patterns of turnover with age. Turnover of half of the identified ribosomal proteins slows down with age, whereas the other half remains unchanged (Fig. 4A, 4B). Half-lives of all identified ribosomal proteins in young worms are all close to 100 h, whereas over about 1 week, these figures drastically fan out over a range of about 600 h (Fig. 4A). The fact that 63% of these proteins tend to aggregate with age (supplemental Fig. S1), supports the notion of an overall dysregulation of the translation machinery in old worms.
Fig. 4.
Proteins of the proteostasis network show divergent protein turnover patterns with age. Patterns are shown for mRNA translation components, including ribosomal subunits (A) and translation regulation proteins (B); degradation-associated proteins including UPS-machinery (C) and lysosomal-related proteins (D); chaperones responsible for protein folding (E) and proteins associated with oxidative stress defense (F). Each dot represents a half-life estimation based on the weighted average of 20- and 40-hour incorporation values. Protein half-lives of three independent replicate samples are shown in different gray tones. The average trend (± S.E.) is indicated in red. The blue dotted line indicates the average lifespan of the experimental population. Summary of the data is included in supplemental Table S5.
A completely contrasting scenario is observed for proteins involved in proteolytic processes. The ubiquitin-proteasome system (UPS) machinery for protein degradation retains its high turnover rate, even in old worms (Fig. 4C). The same pattern is observed for two lysosomal proteases, ASP-1 and ASP-4, although turnover of most lysosomal enzymes does slowdown when worms get older (Fig. 4D). Subunits of the V-ATPases, proton pumps responsible for lysosomal acidification, display a clear shortening in protein half-life in old animals (day 7) despite considerable variation among biological replicates. Surprisingly, more than half of the detected UPS components are consistently found in protein aggregates in aging worms, regardless of their maintained fast turnover rates. Lysosome-related proteins show less aggregation propensity, with only ASP-4 and two V-ATPase subunits being consistently found in aggregates (supplemental Fig. S1). Taken together, these observations raise the possibility that aging-induced proteome perturbations trigger increased protein turnover of proteolytic key players to maintain a functionally intact degradation machinery with age.
Another important group of proteins implicated in proteome management comprises the chaperones, as they support protein folding and refolding under normal and stressful conditions (39). Protein turnover of folding chaperones, such as the cytosolic chaperonin CCT (Chaperonin Containing TCP-1 or TRiC homolog) proteins and endoplasmic protein disulfide isomerases, resembles the pattern of the translation machinery (Fig. 4E), although with milder fanning. Likewise, 75% of these assisted protein folding components and all detected HSP70/HSP90 molecular chaperones tend to aggregate with age (supplemental Fig. S1).
Interestingly, we found that SOD-1, a superoxide dismutase known to protect cells from oxidative damage, shows increased protein turnover with age. A similar trend was detected for the peroxidase PRDX-3 in 5-day old worms. Catalases, with exception of CTL-2, display fast turnover rates, which remain steady over age (Fig. 4F). Moreover, these antioxidant enzymes are barely found in protein aggregates (supplemental Fig. S1). This suggests an overall attempt to cope with the age-associated oxidative damage accumulation.
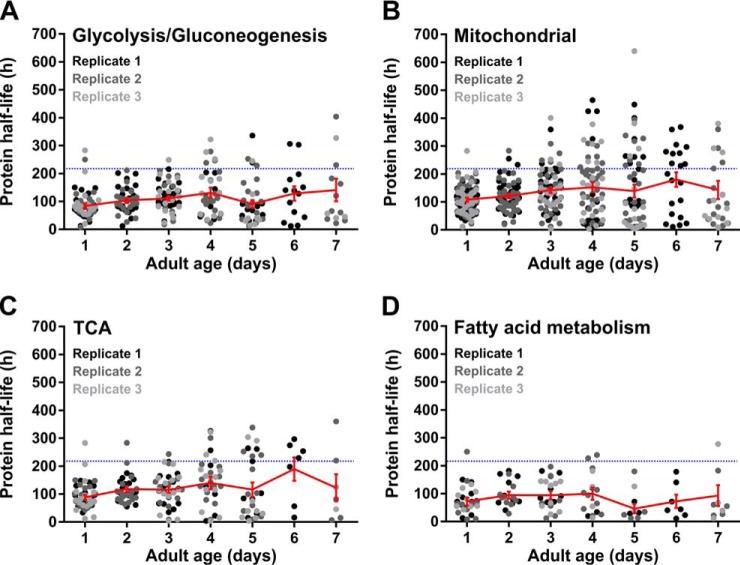
No Consistent Turnover Pattern of Proteins Involved in Energy Metabolism
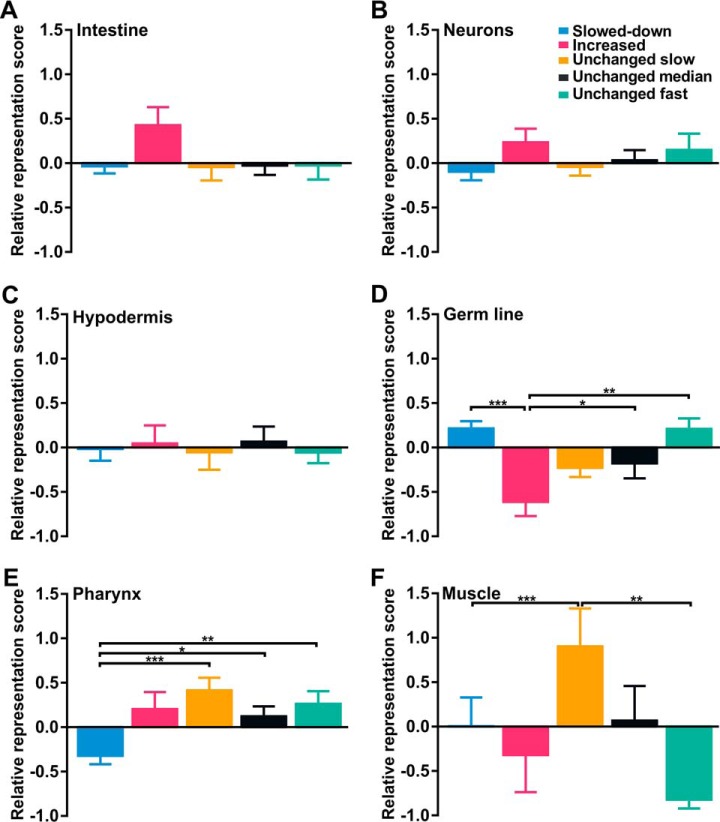
In C. elegans, metabolic rate declines exponentially with age (40, 41). Earlier, it was suggested that slowdown of protein turnover causes conformational changes in metabolic enzymes, resulting in an overall progressive decline in metabolic performance with age (42, 43). The relatively high aggregation propensity of these proteins (supplemental Fig. S1) points in the same direction. Nevertheless, no consistent pattern in the turnover rates of these proteins could be observed in aging worms, with highly varying half-lives with age (Fig. 5). Nevertheless, we found turnover of some proteins involved in glycolysis and the TCA cycle tend to slow down with age (Fig. 5A, 5C). In contrast, we found unchanged and increased protein turnover tendencies for fatty acid metabolic proteins (Fig. 5D). In line, analysis of tissue-specific expression scores (27) shows an overrepresentation of proteins with increased turnover rates for the intestine, the primary site of fat metabolism (Fig. 6A).
Fig. 5.
One-quarter of proteins involved in energy metabolism shows slowdown in protein turnover in aging worms. Patterns are shown for enzymes involved in the glycolysis/gluconeogenesis (A), mitochondrial proteins (B), enzymes of the TCA-cycle (C) and proteins associated with fatty acid metabolism (D). Color schemes and indications are identical as in Fig. 4. Summary of the data is included in supplemental Table S5.
Fig. 6.
Worm-tissues show specific patterns of protein turnover with age. Relative representation scores of identified proteins per trend based on PTM-analysis are shown for intestine (A), neurons (B), hypodermis (C), germ line (D), pharynx (E) and muscle (F) (27). Statistical significance was tested with Kruskal-Wallis test and Dunn's multiple comparison test. Statistical significance is indicated as * p < 0.05, ** p < 0.01, *** p < 0.001. Color scheme is identical to that of Fig. 3.
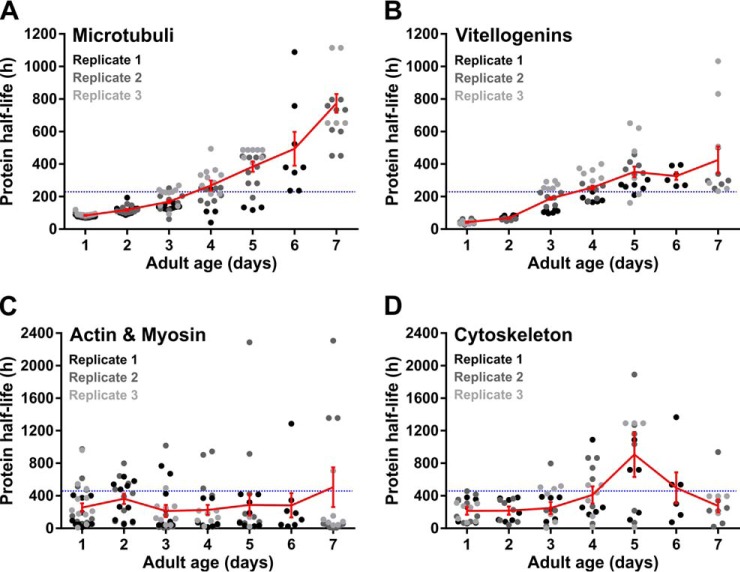
Strong and Consistent Slowdown of Tubulins and Vitellogenin Turnover
Although proteins of many functional groups show mixed protein turnover patterns with age, some functional groups show very consistent patterns. The basic structural units of microtubules, α- and β-tubulins, display a 10-fold increase in protein half-life in old compared with young worms (Fig. 7A). Remarkably, none of these proteins are consistently found in protein aggregates (supplemental Fig. S1). Vitellogenins (yolk proteins) show the same pattern (Fig. 7B). Although we used a germ-line deficient strain, these yolk proteins are detected by the mass spectrometer as they are produced in the intestine. As in fertile worms vitellogenins are translocated to oocytes, and thus considered germ line proteins by the tissue prediction tool, this results in a relative over-representation of slowed-down proteins in the germ line (Fig. 6D). In contrast to the microtubular proteins, we found that all detected vitellogenins are likely to aggregate with age (supplemental Fig. S1). Based on these two contrasting examples, it can be argued that protein aggregation is not the determining factor for protein turnover decline during aging.
Fig. 7.
Specific subsets of proteins show consistent patterns in protein turnover in aging worms. Patterns are shown for tubulins (A), vitellogenins (B), muscle-specific proteins (C) and other cytoskeletal proteins (D). Color schemes and indications are identical as in Fig. 4. Summary of the data is included in supplemental Table S5.
Body Wall Muscle and the Pharyngeal Proteins Follow Distinct Turnover Tendencies with Age
Aging worms, like humans, suffer from the progressive age-related muscle deterioration (44). During aging, it might be possible that protein degradation rates exceed protein synthesis, leading to sarcopenia in old worms. Yet, no extensive alterations in protein half-lives could be observed for muscle-specific proteins (Fig. 7C), as most of them rarely turn over during lifespan (Fig. 6F), which is consistent with earlier findings (21). Interestingly, we noticed strong representation of proteins with increased or fast protein turnover rates in the pharynx, whereas these groups are under-represented in body wall muscle (Fig. 6E, 6F).
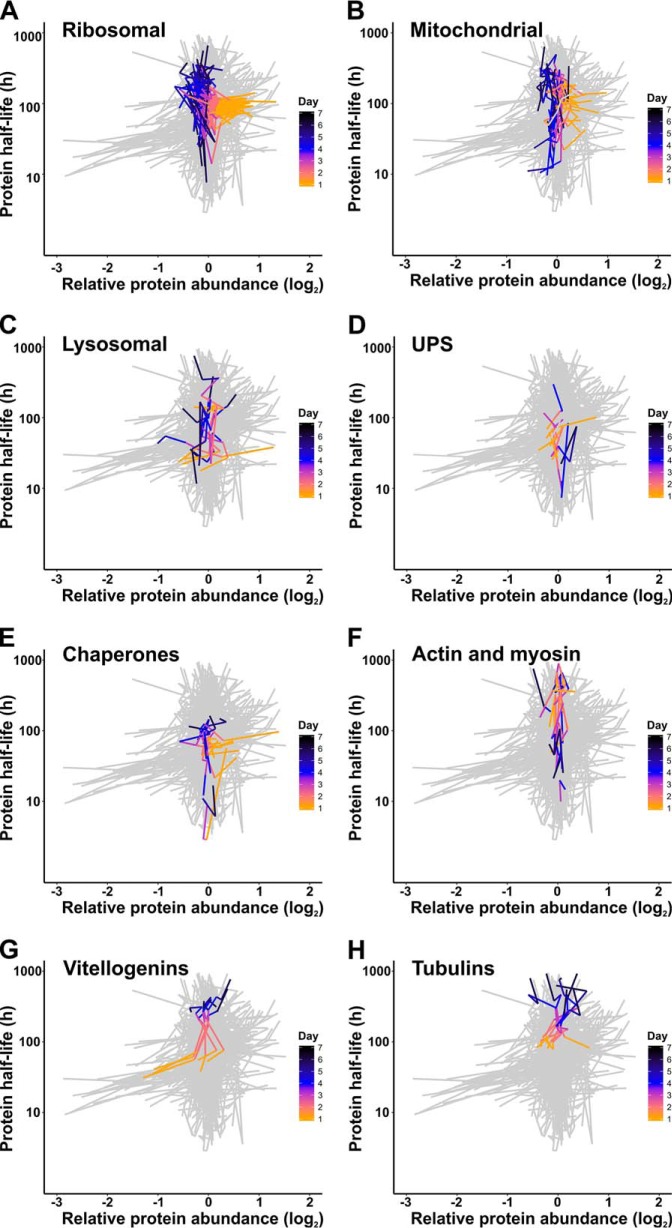
Changes in Protein Half-lives Do Not Consistently Relate to Protein Abundance Over Time
The balance between protein synthesis and degradation likely dictates protein expression changes (45). We wondered what contribution changing protein half-lives make to protein abundance changes in aging worms. Therefore, we compared the relative protein abundance with protein half-lives over time (Fig. 8, supplemental Table S6). Ribosomal protein levels consistently decrease in aging worms, whereas both increasing and decreasing trends in protein turnover occur (Fig. 8A). A similar pattern was observed for mitochondrial proteins (Fig. 8B). In contrast, proteolytic proteins, show no clear time-dependent relation between relative protein abundance and turnover (Fig. 8C, 8D). The protein levels of chaperones seem to be lower in older worms, whereas their protein half-lives remain unchanged or even decrease (Fig. 8E). In accordance with previous findings (14, 46), it is not surprising to find stable levels of cytoskeletal and muscle related proteins over age. Increased turnover is only observed for pharynx-associated muscle proteins, whereas the majority of other proteins in this group shows minor changes in protein turnover (Fig. 8F). Protein half-lives consistently increase for vitellogenins and tubulins in aging worms. The increase of vitellogenin abundance seems to be tightly linked to a general slowdown of their turnover rates (Fig. 8G). In contrast, slowdown of tubulin turnover does not result in clear alterations in its abundance (Fig. 8H). Although several groups of functionally related proteins show consistent patterns, there seems no general trend in the relation between protein abundance and turnover in aging C. elegans.
Fig. 8.
The relation between abundance and turnover rates of several protein subsets in aging worms. Patterns are shown for ribosomal proteins (A), mitochondrial proteins (B), lysosomal proteins (C), proteins of the UPS machinery (D), chaperones (E), actins and myosins (F), vitellogenins (G), and tubulins (H). Lines represent the changes of protein abundance (horizontal axis) and turnover (vertical axis) as a function of adult age (orange-to-black color gradient). A background scatter plot of all proteins detected in the experiment is added as a reference in light gray. Summary of the data is included in supplemental Table S6.
DISCUSSION
The overall downturn in protein turnover is a common observation in senescent organisms, including nematodes (2, 3, 42). Because of increased protein dwell times with age, it is likely that proteins have ample time to undergo oxidation, aggregation and misallocation. It is still unclear whether the age-related decrease in protein turnover is consistent over the entire proteome or whether groups of functionally related proteins show specific patterns. Using a pulse labeling method, we have monitored the change in individual protein turnover in aging C. elegans. Our findings are consistent with a previous study applying a single SILAC-based label-chase approach in aging worms over time (12) and a second study testing protein half-lives of young developing animals and day 5 old worms (13). However, our experimental setup with daily pulses and chases provided much more detail on individual protein turnover dynamics over adult age. We report the gradual slowdown of the turnover of a considerable part of the aging proteome, whereas only a minor fraction shows increased turnover with age. One may assume that worms tend to eat less E. coli with increasing age, resulting in a reduced uptake of 15N. Because differences in feeding rate and/or label uptake can confound the turnover estimates, we checked the atomic proportion of 15N in newly synthesized peptides over time. Overall, we observed a slight, but very slow decline of 15N proportion (70% at day 1, 65% at day 7), which indicates no substantial confounding effect of label incorporation in aging worms (supplemental Fig. S2). Disparate protein turnover changes within the majority of functionally and spatially related protein groups indicate the heterogeneous impact of aging on protein turnover. Notably, these trends are consistently observed across the studied aging cohorts, indicating that functionally or spatially related proteins become less stable with advancing age. Therefore, it is likely that the variable change of protein turnover might be one of the important components underlying the age-dependent deterioration. Conceivably, the protein synthesis machinery itself might be a key player driving this dysregulation, as it shows an escalation in variation of turnover with age. This idea is further corroborated in overall decline in ribosomal protein abundances with age, with a prominent imbalance in the relative ribosomal subunit stoichiometry (9). Thus, dysregulation of protein turnover of the translation machinery in addition with slower turnover of the aggregation-prone folding chaperones (9), likely promote the ultimate collapse of the aging proteome.
Nevertheless, consistent trends over time within functional subsets of proteins do exist. Our data shows high maintenance of the degradation apparatus in aged worms, which highly contrasts the hampered turnover of the translation machinery. Moreover, an up-regulation in proteasomal subunits has been observed earlier in aging C. elegans (9). Notwithstanding their fast turnover rates, these components likely get trapped into aggregates with age. Thus, the sustained protein turnover of proteasomal subunits at advanced age is likely the ongoing endeavor to keep up the degradation machinery to further deal with the proteome imbalance. However, chronic proteotoxicity will eventually exceed the proteasomal capacity (47).
No uniform slowdown in turnover of enzymes involved in energy metabolism could be observed, because only one-fourth shows a significant decline in protein turnover with age. As mitochondria are main producers of ROS, proteins with slow turnover are liable to oxidative modification, resulting in the accumulation of altered enzymes. Although the turnover of only minor fraction of mitochondrial proteins slows down with age, these damage-prone enzymes may be the driving force of the functional decline in metabolic performance with age (40, 41). Interestingly, fatty-acid β-oxidation proteins, that primarily reside in the intestine (48), exhibit increased turnover with age, which is also reflected in the tissue enrichment analysis. These findings may point to a shift from carbohydrate metabolism to fatty-acid β-oxidation in the intestine of old worms. Alternatively, the fatty-acid β-oxidation machinery may need improved turnover compared with enzymes involved in carbohydrate metabolism because of its specific location in the mitochondria and peroxisomes, which are well-known ROS generation sites that may impose excessive oxidative damage to residing proteins (49, 50).
Tubulins and vitellogenins show consistent slowdown of turnover rates with age. Because the latter groups have opposite aggregation propensities, protein aggregation cannot be the sole underlying mechanism responsible for the deceleration of protein turnover with age and vice versa. Microtubules are responsible for a variety of functions, including cellular transport (51). Aged cells frequently display the accumulation of cell organelles probably because of insufficient organelle transport along disrupted microtubules (52). Hence, we presume that attenuation of tubulin turnover may underlie the changing microtubule organization and dynamics during aging. On the other hand, protein aggregation might be a driving force in slowing down the turnover of vitellogenins, as these macromolecules, irrelevant to post-reproductive worms, accumulate throughout the body cavity in old worms (44). Vitellogenins, trapped into accumulating protein aggregates over time, likely show increased protein half-lives with age as they become less susceptible for protein degradation in aging worms.
Aging nematodes are characterized by a progressive locomotory decline (53). Muscle-specific proteins are very stable proteins, showing barely any turnover during a worm's lifespan (this study and previous findings (21)). Therefore, the lack of protein turnover may be associated with the age-related structural loss of sarcomere integrity (44). Interestingly, in pharyngeal muscle cells, proteins with increased and fast turnover rates are relatively over-represented, compared with other muscle tissues. One possibility is that the pharyngeal muscle cells are, besides general deterioration, more susceptible to microbial attacks than body wall muscles (54). Hence, proteins with higher turnover rates might be more flexible to cope with this additional stress.
In conclusion, our data suggests that senescent C. elegans is characterized by a variety of changes in protein turnover rates. Diverging dwell times of proteins involved in translation, consistently observed over three independent biological replicates, are likely responsible for a collapse of the translation machinery over time. Intriguingly, we found that aging worms seem to preserve their (protein) quality control mechanisms, especially the UPS and antioxidant machinery, possibly to cope with the increasing proteotoxic and oxidative stress with age. However, this maintenance fights a losing battle, eventually resulting in the ultimate collapse of the proteome.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (55) via the PRIDE partner repository with the data set identifier PXD002901 and 10.6019/PXD002901.
Supplementary Material
Acknowledgments
We thank the Gems laboratory for providing the strain GA153. We are grateful to Caroline Vlaeminck and Renata Coopman for technical support with the SILeNCe experiments. We thank Andy Vierstraete for his technical assistance in data-processing. Preliminary experiments were performed by Dr. Filip Mathijssens, who was a postdoctoral member of the Braeckman lab.
Footnotes
Author contributions: I.D., V.P., and B.P.B. designed the experiments. I.D. and S.B. carried out the experiments. I.D. and H.M.B. performed the sample preparation and V.P. carried out the instrumental analysis. I.D., V.P., and B.P.B. analyzed and interpreted the data. I.D. and B.P.B. wrote the manuscript and this was further edited by G.D., V.P., R.D.S., and B.P.B.
* ID acknowledges a PhD grant from the fund for Scientific Research-Flanders, Belgium (FWO11/ASP/031). Portions of this work were supported by NIH P41GM103493 (to R.D.S). The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
 This article contains supplemental material.
This article contains supplemental material.
E-mail authors: Ineke.Dhondt@ugent.be; Vladislav.Petyuk@pnnl.gov; sophie_bauer@gmx.at; rds@pnl.gov; geert.depuydt@bio.kuleuven.be; Bart.Braeckman@ugent.be.
1 The abbreviations used are:
- C. elegans
- Caenorhabditis elegans
- E. coli
- Escherichia coli
- FDR
- false discovery rate
- PTM
- Pavlidis Template Matching
- ROS
- reactive oxygen species
- SILAC
- stable isotope labeling with amino acids in cell culture
- SILeNCe
- Stable Isotope Labeling by Nitrogen in Caenorhabditis elegans
- TCA
- tricarboxylic acid cycle
- UPS
- ubiquitin-proteasome system.
REFERENCES
- 1. Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., and Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ryazanov A. G., and Nefsky B. S. (2002) Protein turnover plays a key role in aging. Mech. Ageing Dev. 123, 207–213 [DOI] [PubMed] [Google Scholar]
- 3. Rattan S. I. (1996) Synthesis, modifications, and turnover of proteins during aging. Exp Gerontol. 31, 33–47 [DOI] [PubMed] [Google Scholar]
- 4. Golden T. R., and Melov S. (2007) Gene expression changes associated with aging in C. elegans. WormBook. 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lund J., Tedesco P., Duke K., Wang J., Kim S. K., and Johnson T. E. (2002) Transcriptional profile of aging in C. elegans. Curr. Biol. 12, 1566–1573 [DOI] [PubMed] [Google Scholar]
- 6. Liang V., Ullrich M., Lam H., Chew Y. L., Banister S., Song X., Zaw T., Kassiou M., Gotz J., and Nicholas H. R. (2014) Altered proteostasis in aging and heat shock response in C. elegans revealed by analysis of the global and de novo synthesized proteome. Cell Mol. Life Sci. 71, 3339–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Copes N., Edwards C., Chaput D., Saifee M., Barjuca I., Nelson D., Paraggio A., Saad P., Lipps D., Stevens S. M. Jr, and Bradshaw P. C. (2015) Metabolome and proteome changes with aging in Caenorhabditis elegans. Exp. Gerontol. 72, 67–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., and Yates J. R. 3rd. (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- 9. Walther D. M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R. I., Dobson C. M., Vendruscolo M., Mann M., and Hartl F. U. (2015) Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narayan V., Ly T., Pourkarimi E., Murillo A. B., Gartner A., Lamond A. I., and Kenyon C. (2016) Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 3, 144–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Depuydt G., Shanmugam N., Rasulova M., Dhondt I., and Braeckman B. P. (2016) Increased protein stability and decreased protein turnover in the Caenorhabditis elegans Ins/IGF-1 daf-2 mutant. J. Gerontol. A Biol. Sci. Med. Sci. 71, 1553–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vukoti K., Yu X., Sheng Q., Saha S., Feng Z., Hsu A. L., and Miyagi M. (2015) Monitoring newly synthesized proteins over the adult life span of Caenorhabditis elegans. J Proteome Res. 14, 1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Visscher M., De Henau S., Wildschut M. H., van Es R. M., Dhondt I., Michels H., Kemmeren P., Nollen E. A., Braeckman B. P., Burgering B. M., Vos H. R., and Dansen T. B. (2016) Proteome-wide changes in protein turnover rates in C. elegans models of longevity and age-related disease. Cell Rep. 16, 3041–3051 [DOI] [PubMed] [Google Scholar]
- 14. Depuydt G., Xie F., Petyuk V. A., Shanmugam N., Smolders A., Dhondt I., Brewer H. M., Camp D. G., Smith R. D., and Braeckman B. P. (2013) Reduced insulin/IGF-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans. Mol. Cell. Proteomics. 12, 3624–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petyuk V. A., Qian W. J., Smith R. D., and Smith D. J. (2010) Mapping protein abundance patterns in the brain using voxelation combined with liquid chromatography and mass spectrometry. Methods 50, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kessner D., Chambers M., Burke R., Agus D., and Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mayampurath A. M., Jaitly N., Purvine S. O., Monroe M. E., Auberry K. J., Adkins J. N., and Smith R. D. (2008) DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 24, 1021–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petyuk V. A., Mayampurath A. M., Monroe M. E., Polpitiya A. D., Purvine S. O., Anderson G. A., Camp D. G. 2nd, and Smith R. D. (2010) DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol. Cell. Proteomics 9, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howe K. L., Bolt B. J., Cain S., Chan J., Chen W. J., Davis P., Done J., Down T., Gao S., Grove C., Harris T. W., Kishore R., Lee R., Lomax J., Li Y., Muller H. M., Nakamura C., Nuin P., Paulini M., Raciti D., Schindelman G., Stanley E., Tuli M. A., Van Auken K., Wang D., Wang X., Williams G., Wright A., Yook K., Berriman M., Kersey P., Schedl T., Stein L., and Sternberg P. W. (2016) WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774–D780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dhondt I., Petyuk V.A., Cai H., Vandemeulebroucke L., Vierstraete A., Smith R.D., Depuydt G., and Braeckman B.P., (2016) FOXO/DAF-16 activation slows down turnover of the majority of proteins in C. elegans. Cell Rep. 16, 3028–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elias J. E., and Gygi S. P. (2009) Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du P., Kibbe W. A., and Lin S. M. (2006) Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics 22, 2059–2065 [DOI] [PubMed] [Google Scholar]
- 24. Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., Li J., Thiagarajan M., White J. A., and Quackenbush J. (2006) TM4 microarray software suite. Methods Enzymol. 411, 134–193 [DOI] [PubMed] [Google Scholar]
- 25. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., and Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 26. Dennis G. Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., and Lempicki R. A. (2003) DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 27. Chikina M. D., Huttenhower C., Murphy C. T., and Troyanskaya O. G. (2009) Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comput. Biol. 5, e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 30. Depuydt G., Xie F., Petyuk V. A., Smolders A., Brewer H. M., Camp D. G. 2nd, Smith R. D., and Braeckman B. P. (2014) LC-MS proteomics analysis of the insulin/IGF-1-deficient Caenorhabditis elegans daf-2(e1370) mutant reveals extensive restructuring of intermediary metabolism. J. Proteome Res. 13, 1938–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arantes-Oliveira N., Apfeld J., Dillin A., and Kenyon C. (2002) Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505 [DOI] [PubMed] [Google Scholar]
- 32. Hsin H., and Kenyon C. (1999) Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 [DOI] [PubMed] [Google Scholar]
- 33. Lin K., Hsin H., Libina N., and Kenyon C. (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28, 139–145 [DOI] [PubMed] [Google Scholar]
- 34. Geillinger K. E., Kuhlmann K., Eisenacher M., Meyer H. E., Daniel H., and Spanier B. (2012) Dynamic changes of the Caenorhabditis elegans proteome during ontogenesis assessed by quantitative analysis with 15N metabolic labeling. J. Proteome Res. 11, 4594–4604 [DOI] [PubMed] [Google Scholar]
- 35. Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., and Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 36. Pavlidis P., and Noble W. S. (2001) Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2, RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grune T., Jung T., Merker K., and Davies K. J. (2004) Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 36, 2519–2530 [DOI] [PubMed] [Google Scholar]
- 38. David D. C., Ollikainen N., Trinidad J. C., Cary M. P., Burlingame A. L., and Kenyon C. (2010) Widespread protein aggregation as an inherent part of aging in C. elegans. PLos Biol. 8, e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 40. Braeckman B. P., Houthoofd K., De Vreese A., and Vanfleteren J. R. (1999) Apparent uncoupling of energy production and consumption in long-lived Clk mutants of Caenorhabditis elegans. Curr. Biol. 9, 493–496 [DOI] [PubMed] [Google Scholar]
- 41. Shoyama T., Ozaki T., Ishii N., Yokota S., and Suda H. (2007) Basic principle of the lifespan in the nematode C. elegans. Mech. Ageing Dev. 128, 529–537 [DOI] [PubMed] [Google Scholar]
- 42. Rothstein M., and Sharma H. K. (1978) Altered enzymes in the free-living nematode, Turbatrix aceti, aged in the absence of fluorodeoxyuridine. Mech. Ageing Dev. 8, 175–180 [DOI] [PubMed] [Google Scholar]
- 43. Braeckman B. P., Houthoofd K., and Vanfleteren J. R. (2000) Patterns of metabolic activity during aging of the wild type and longevity mutants of Caenorhabditis elegans. J. Am. Aging Assoc. 23, 55–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herndon L. A., Schmeissner P. J., Dudaronek J. M., Brown P. A., Listner K. M., Sakano Y., Paupard M. C., Hall D. H., and Driscoll M. (2002) Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814 [DOI] [PubMed] [Google Scholar]
- 45. Kristensen A. R., Gsponer J., and Foster L. J. (2013) Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 9, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dhondt I., Petyuk V. A., Cai H., Vandemeulebroucke L., Vierstraete A., Smith R. D., Depuydt G., and Braeckman B. P. (2016) FOXO/DAF-16 Activation Slows Down Turnover of the Majority of Proteins in C. elegans. Cell Rep. 16, 3028–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hipp M. S., Park S. H., and Hartl F. U. (2014) Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 [DOI] [PubMed] [Google Scholar]
- 48. Ashrafi K. (2007) Obesity and the regulation of fat metabolism. WormBook. 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adachi H., Fujiwara Y., and Ishii N. (1998) Effects of oxygen on protein carbonyl and aging in Caenorhabditis elegans mutants with long (age-1) and short (mev-1) life spans. J. Gerontol. A Biol. Sci. Med. Sci. 53, B240–B244 [DOI] [PubMed] [Google Scholar]
- 50. Nguyen A. T., and Donaldson R. P. (2005) Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch. Biochem. Biophys. 439, 25–31 [DOI] [PubMed] [Google Scholar]
- 51. Cooper G.M., The Cell: A Molecular Approach. 2000: ASM Press, Washington, D.C. [Google Scholar]
- 52. Schatten H., Chakrabarti A., and Hedrick J. (1999) Centrosome and microtubule instability in aging Drosophila cells. J. Cell. Biochem. 74, 229–241 [PubMed] [Google Scholar]
- 53. Bolanowski M. A., Russell R. L., and Jacobson L. A. (1981) Quantitative measures of aging in the nematode Caenorhabditis elegans. I. Population and longitudinal studies of two behavioral parameters. Mech. Ageing Dev. 15, 279–295 [DOI] [PubMed] [Google Scholar]
- 54. Chow D. K., Glenn C. F., Johnston J. L., Goldberg I. G., and Wolkow C. A. (2006) Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Exp. Gerontol. 41, 252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (55) via the PRIDE partner repository with the data set identifier PXD002901 and 10.6019/PXD002901.