Abstract
Objective
Healthcare communities have identified a significant need for disease-specific information. Disease-specific ontologies are useful in assisting the retrieval of disease-relevant information from various sources. However, building these ontologies is labor intensive. Our goal is to develop a system for an automated generation of disease-pertinent concepts from a popular knowledge resource for the building of disease-specific ontologies.
Methods
A pipeline system was developed with an initial focus of generating disease-specific treatment vocabularies. It was comprised of the components of disease-specific citation retrieval, predication extraction, treatment predication extraction, treatment concept extraction, and relevance ranking. A semantic schema was developed to support the extraction of treatment predications and concepts. Four ranking approaches (i.e., occurrence, interest, degree centrality, and weighted degree centrality) were proposed to measure the relevance of treatment concepts to the disease of interest. We measured the performance of four ranks in terms of the mean precision at the top 100 concepts with five diseases, as well as the precision-recall curves against two reference vocabularies. The performance of the system was also compared to two baseline approaches.
Results
The pipeline system achieved a mean precision of 0.80 for the top 100 concepts with the ranking by interest. There were no significant different among the four ranks (p = 0.53). However, the pipeline-based system had significantly better performance than the two baselines.
Conclusions
The pipeline system can be useful for an automated generation of disease-relevant treatment concepts from the biomedical literature.
Keywords: Information extraction, Data mining, Ontology, MEDLINE citations, SemMedDB, Treatment
1. Introduction
Disease-specific ontologies are knowledge bases intended to structure and represent disease-relevant information including disease etiology, diagnostic characteristics, treatments and prognosis [1]. By providing rich domain knowledge, they can be very useful in assisting the retrieval of disease-relevant information from sources like clinical data repositories, biomedical literature, and online health resources, which therefore can better meet the information needs of various healthcare communities.
Building and maintaining such ontologies is labor-intensive. One major challenge is to identify disease-specific vocabularies that form the core of disease-specific ontologies. For example, in a previous study [1] we asked medical experts to manually develop reference vocabularies for three diseases from selected biomedical literature sources. The annotation of selected documents took around 100 man-hours, not counting the document preparation, guideline development, experts training, adjudication, and concept mapping. From the same study, we also found that existing literature sources were sufficient to provide disease-specific vocabulary. Therefore, there is an opportunity for the development of algorithms that can automatically extract vocabulary components from these sources.
In the present study, we address the challenge described above by developing a set of knowledge extraction techniques that automatically generate disease-pertinent vocabulary from existing sources. We chose the MEDLINE database as our knowledge source because it contains a large collection of published journal citations and covers a variety of diseases. We have focused on treatment concepts associated with the disease of interest, including direct treatment and prevention of the problem or complications caused by the problem; however, the method can also be adapted to other disease domains (e.g., signs, symptoms, diagnostic tests).
2. Background
2.1. Disease-specific information needs and barriers
Disease-specific information is frequently sought by people in the healthcare communities, including clinicians, healthcare consumers, clinical researchers and medical knowledge engineers. The types of information that have been sought include medical knowledge (information that is understood to be generalizable to the care of all patients), patient data (information about a specific person), and population statistics (aggregated data about groups or populations of patients) [2]. For example, a variety of published studies investigated physicians’ information needs by analyzing their clinical questions raised in the course of patient care [2–4]. A large proportion of the questions were related to disease-specific medical knowledge, such as “what is the drug of choice for condition x?”, “what test is indicated in situation x?”, and “how should I treat condition x?” [5]. Clinicians also frequently seek disease-specific patient information (e.g., medical history, physical exam) from clinical data repositories. With the wide adoption of electronic medical records, available patient data has been shown a marked increase. Thus, it is important to be able to distil and filter medical records to show the patient information that is relevant to a specific problem of interest.
Healthcare consumers also frequently seek health information online to better understand and manage their own health [6]. Research shows that the top two major health topics searched online are related to the personal medical problems and the treatment for these problems [7]. Clinical researchers and medical knowledge engineers also demand disease-specific information in order to understand, model, and analyze clinical data. For example, when conducting a retrospective clinical study, clinical researchers need to understand the details of the clinical problem (e.g., disease-specific signs and symptoms, diagnostic tests, comorbidities) in order to properly identify “research subjects” from an EHR.
2.2. Disease-specific ontologies
Ontologies are explicit and formal representations of domain knowledge, which enable the management, sharing, and reuse of domain knowledge [8,9]. Disease-specific ontologies intend to integrate vocabularies of different aspects of the disease, such as signs and symptoms, medications, therapeutic procedures, diagnostic procedures, and laboratory tests and imaging. To minimize information overload, it is crucial to develop effective information retrieval systems capable of retrieving relevant information to meet different information needs. For healthcare consumers, who are likely to have low health literacy [10], it is important to assist them forming optimal queries to retrieve relevant information from online health sources [11]. We anticipate that these ontologies will facilitate the retrieval of specific information from a variety of sources, such as websites [12], biomedical literature [13], and clinical data repositories [14–16]. Disease-specific ontologies can support information retrieval systems by providing domain-specific concepts and relations necessary to direct the formulation or expansion of initially simple queries tied to clinical concepts. In addition, the medical knowledge contained in disease-specific ontologies could be used by clinical researchers and medical knowledge engineers to understand the diseases, and the vocabularies in the ontologies may further assist their research or engineering work (e.g., cohort selection, text annotations).
2.3. Relation extraction in biomedical domain
Domain experts can develop disease-specific ontologies, but individuals with the required expertise are scarce and expensive. A long-term goal of our research is to create a platform to facilitate large-scale development of these ontologies. One of the critical tasks in building disease-specific ontologies is to acquire medical knowledge like concepts and relationships related to the disease of interest [8,17]. This kind of medical knowledge has been substantially documented in sources like the biomedical literature, web documents, and clinical data repositories, although most of it is represented in unstructured and narrative format. We therefore hope to take advantage of these sources and investigate automatic techniques to extract disease-specific medical knowledge from them.
Automatic extraction of relational medical knowledge from the biomedical literature is an active subject of research interest [18–22]. Researchers have attempted to extract disease-specific medical knowledge from the biomedical literature ever since the 1990s [23,24]. In the earliest stage, the methods merely relied on co-occurrence-based statistics. For examples, Zeng and Cimino used MeSH co-occurrence information from the UMLS to obtain disease-chemical associations [24]. Chen et al. used co-occurrence statistics to extract disease-drugs relations from MEDLINE abstracts [18].
Along with the advanced development of NLP techniques, a variety of rule-based and machine-learning-based methods have been used for relation extraction. A typical example of rule-based system is SemRep [25,26] which is built upon UMLS and MetaMap. It interprets the biomedical knowledge presented in a given sentence from the scientific literature in the form of predications {subject PREDICATE object}, where the subject and object are biomedical concepts from the UMLS Metathesaurus and the PREDICATE is a semantic relation from the UMLS Semantic Network [25–27]. For example, from the sentence “this paper will review the earlier and present studies in the development of rasagiline for treatment of PD and discuss its pharmacology and applicable mechanism of action”, SemRep extracts the predication {Rasagiline TREATS Parkinson’s disease}. Based on a preliminary evaluation, the precision and recall of SemRep are 78% and 49% respectively [26]. More recently, Xu and Wang applied a pattern-based approach to extract disease-drug and disease-disease risk relationships from MEDLINE citations [21,22].
Machine learning techniques have also been successfully applied to relation extraction. From one standpoint, relation extraction is a classification problem which is to predict semantic relations held between two identified entities in a given sentence [28]. Researchers have employed different classification models using diverse lexical, syntactic and semantic features derived from the text to make predication on the relations. For example, Rosario and Hearst compared graphical models and neural network using lexical, syntactic, and semantic features to distinguish among seven relation types that can occur between the entities “treatment” and “disease” in bioscience texts [29]. Zeng et al. exploited a convolutional deep neural network to extract lexical and sentence level features which were fed into a softmax classifier to predict the relationships between two marked nouns [30]. From another standpoint, relation extraction is a sequence labeling problem, for which researchers have applied kernel-based approaches to label the relationships between two entities. For example, Bundschus et al. used conditional random field technologies to extract disease-treatment associations from PubMed abstracts [31]. Giuliano et al. investigated a kernel-based approach based on shallow linguistic processing for extracting relations between entities from biomedical literature [32].
In the present work, we intend to develop an automated approach to extract treatment vocabularies from the biomedical literature for a given disease of interest. Unlike previous studies which worked on semantic interpretation of the relationships from the biomedical literature, we focused on filtering and ranking disease-specific concepts for a given disease of interest. In addition, our work builds on previous tools and methods, in particular SemRep.
2.4. SemMedDB
SemRep is routinely used to process the entire set of MEDLINE citations (i.e., the titles and abstracts) to extract structured predications, which are then stored in a repository called SemMedDB [33]. There are currently over 83 million semantic predications in this database version June 30, 2015, approximately 93% of which are associative (or, non- “IS-A” predication). Although SemMedDB provides structured predications that could facilitate the acquisition of medical knowledge from the biomedical literature, further inference is needed to filter noisy data and to retrieve information that is most useful for a disease-specific ontology. For example, a query in SemMedDB for a collection of predications that include congestive heart failure retrieves thousands of predications. Within these predications, concepts may range widely from pharmaceutical substances to signs and symptoms and related genes. Therefore, many retrieved concepts and predications could be outside scope for a disease-specific ontology. In addition, concepts that are irrelevant to the main search topic may be retrieved due to errors in the underlying SemRep NLP process and inaccurate or outdated information presented in MEDLINE abstracts. We addressed these issues in the development of our automatic knowledge extraction system from SemMedDB.
3. Materials and methods
The study method is comprised of two parts: (1) the development of a pipeline-based process to extract disease-specific, treatment-related information from biomedical literature; and (2) an experiment to compare the pipeline-based process to extract disease-specific treatment vocabulary with two baseline approaches in terms of precision-recall curves and mean average precision.
3.1. Pipeline-based process
The pipeline-based process developed in the present study consists of the following steps (see Fig. 1): (1) retrieval of therapeutic citations from MEDLINE for the disease of interest using a search strategy that aims to retrieve scientifically sound studies; (2) retrieval of all predications and their corresponding sentences from SemMedDB for the citations retrieved in Step 1; (3) development of a semantic schema from the UMLS and existing disease-specific ontologies to identify treatment-related predications from this list; (4) retrieval of treatment-related predications from the predications in Step 2 using the semantic schema from Step 3; (5) extraction of treatment concepts from the treatment predications extracted in Step 4 from the list generated in Step 3; (6) ranking of the treatment concepts extracted in Step 5 using four ranking algorithms.
Fig. 1.
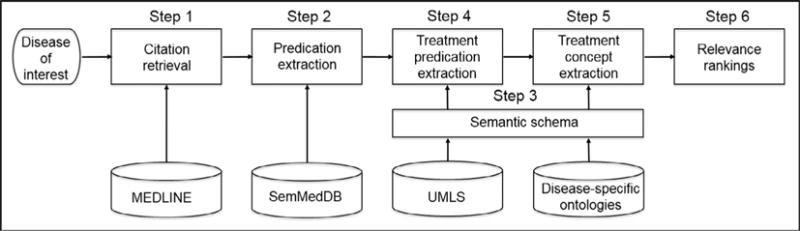
Flowchart of automatically extracting disease-specific, treatment vocabulary from the biomedical literature and the ranking of treatment concepts.
3.1.1. Step 1: Retrieval of disease-pertinent MEDLINE citations
The first step retrieves biomedical citations from MEDLINE database regarding the therapy of a given disease. We built a search strategy based on the PubMed Clinical Queries, which is a set of filters that are tuned to retrieve scientifically sound clinical studies in topics such as treatment, diagnosis, and prognosis [34–36]. The Clinical Query filters provide two modes: broad and narrow. The broad treatment filter has shown a sensitivity of 99% and a specificity of 70%, while the narrow treatment has shown a sensitivity of 93% and specificity of 97%. In the present study, we focused on sensitivity and used the broad filter.
Although Clinical Query filters perform well in retrieving clinical trial studies, the query does not cover other types of study design, such as systematic reviews, which would also be useful for retrieving disease-specific medical knowledge. Hence, we extended the Clinical Query treatment filter to retrieve systematic review articles (see Fig. 2). In addition, we added the following restrictions: English language, abstract available, human subjects, and core clinical journals. We obtained the list of clinical journals by combining the PubMed core clinical journals (http://www.nlm.nih.gov/bsd/aim.html) with a list of journals categorized under “clinical medicine” in Web of Science (http://ip-science.thomsonreuters.com/mjl/). For each disease of interest, we added a MeSH term for the disease as a major topic. The modified Clinical Query filter can also be extended to retrieve articles for other disease-associated concepts, such as etiology, diagnosis, and prognosis.
Fig. 2.

Modified Clinical Query for retrieving treatment-related citations for the disease of interest from MEDLINE. In the query, “QUERY_TERM” is the MeSH term for the disease of interest. “JOURNALLIST” is a list of clinical journals, e.g., “CA-CANCER J CLIN”, “NEW ENGL J MED”.
3.1.2. Step 2: Predication extraction with SemRep
In this step, the input is all the PMIDs that were assigned to those MEDLINE citations retrieved from step 1. The output is the predications generated by the SemRep from those MEDLINE citations as well as the sentences where the predications came from. More specifically, we took all the PMIDs to form SQL scripts to query the SemMedDB [33] to retrieve all the predications and sentences. The version of SemMedDB we used was updated with citations published through June 30, 2015. Citations published after this date were not yet available in SemMedDB, therefore we excluded those citations from the study.
3.1.3. Step 3: Development of semantic schema
The semantic schema consisted of a set of metapredications whose arguments are defined based on high-level domains based on UMLS semantic groups [37]; for example, {Procedures TREATS Disorders}. The development of a semantic schema is a one-time process that supports knowledge extraction of treatment concepts for any disease of interest. The development of the semantic schema was performed in two steps: (1) selection of relevant semantic groups to filter treatment concepts (Step 4), and (2) definition of relevant metapredications to filter treatment predications (Step 5).
To select relevant semantic groups, we analyzed the semantic groups and types that were present in the heart failure reference vocabulary (http://bioportal.bioontology.org/ontologies/HFO) that had been manually created in a previous study [1]. The rationale for this approach is the assumption that the majority of semantic groups and types covered in disease treatment vocabularies would also be covered in the heart failure reference vocabulary.
A total of 413 treatment concepts were retrieved, from 38 semantic types and 9 semantic groups (i.e., Chemicals & Drugs, Procedures, Physiology, Devices, Activities & Behaviors, Concepts & Ideas, Objects, Disorders, and Organizations). The majority of the heart failure treatment concepts belonged to two semantic groups: Chemicals & Drugs and Procedures. We manually reviewed the other seven semantic groups and, based on domain knowledge, decided to include only four semantic groups: Chemicals & Drugs, Procedures, Devices, and Activities & Behaviors (Table 1). We also excluded a subset of the semantic types from the Procedures and Devices semantic groups. For example, from Procedures, we excluded Diagnostic Procedure, Laboratory Procedure, Molecular Biology Research Technique, and Research Activity.
Table 1.
The semantic types and groups of treatment concepts.
| Semantic groups | Included semantic types |
|---|---|
| Procedures | Educational Activity, Health Care Activity, Therapeutic or Preventive Procedure |
| Chemicals & Drugs | Alla |
| Activities & Behaviors | Alla |
| Devices | Medical Device |
Refer to http://semanticnetwork.nlm.nih.gov/download/SemGroups.txt for detailed semantic types included by a specific semantic group.
We followed a similar process for metapredications, also using the heart failure vocabulary.
We retrieved a total of 54,991 predications from SemMedDB from 15,994 citations. Forty percent (N = 22,019) of the predications contained treatment concepts from the heart failure vocabulary. We then generated 205 unique metapredications based on the retrieved predications, such as {Chemicals & Drugs, ADMINISTERED_TO, Living Beings}. Next, we removed the metapredications that did not contain any of the four semantic groups selected in the previous step. In addition, we excluded metapredications whose predicate was not treatment-related predicates, such as DIAGNOSES, CAUSES, STIMULATES, PRODUCES, PREDISPOSES, as well as negation predications. The remaining metapredications were grouped into four categories (Table 2). For each category, we identified the predication arguments that were most relevant for extracting treatment concepts. However, we noted some exceptions. For example, in category 3, for metapredications where the arguments are Chemical & Drugs and Devices, their corresponded predications are usually about the comparison or co-occurrence of a treatment (Chemical & Drugs) with a “placebo” (Devices), therefore, only the concepts from the position of Chemical & Drugs will be retrieved.
Table 2.
Semantic schema for classifying treatment predications. The predication arguments in underline are the ones from which treatment concepts are extracted.
| Category | Subject | Relation | Object |
|---|---|---|---|
| 1 | Chemicals & Drugs/Procedures/Devices/Activities & Behaviors | ADMINISTERED_TO/AUGMENTS/AFFECTS/ASSOCIATED_WITH/DISRUPTS/INHIBITS/TREATS/PREVENTS | ANY semantic groups |
| 2 | ANY semantic groups | USES | Chemical & Drugs/Procedures/Devices/Activities & Behaviors |
| 3a | Chemicals & Drugs/Procedures/Devices/Activities & Behaviors | COEXISTS_WITH/compared_with/same_as/INTERACTS_WITH/METHOD_OF/lower_than/higher_than | Activities & Behaviors/Drugs/Procedures/Devices |
| 4 | Chemicals & Drugs/Procedures/Devices/Activities & Behaviors | ISA | Activities & Behaviors/Drugs/Procedures/Devices |
For metapredications where the subject is Chemical & Drugs and the object is Devices, and vice-versa, only Chemical & Drugs concepts are extracted.
3.1.4. Step 4: Extraction of relevant treatment predications
Many predications retrieved in Step 2 could be not related to the treatment (e.g., a predication {congestive heart failure CAUSES cardiomyopathy, dilated}), or were generic and of little interest (e.g., {pharmaceutical preparations TREATS pneumonia}). To filter out generic predications, we adopted the novelty approach proposed by Fiszman et al. [38]. A predication is considered as generic when it has a generic concepts which is determined by whether the hierarchical depth in the Metathesaurus is less than an empirical distance. Each concept of the predications has the attribute of novelty in the SemMedDB. We exclude predications that contain non-novel concepts.
We then used the semantic schema to separate the treatment predications from irrelevant predications. To do so, we excluded predications that did not match one of the metapredications. For example, the predication {Adrenergic beta-Antagonists PREVENTS heart failure} matches the metapredication {Chemicals & Drugs PREVENTS Disorders}, while predication {congestive heart failure CAUSES cardiomyopathy, dilated} does not match any metapredications in the semantic schema.
3.1.5. Step 5: Extraction of disease-specific treatment concepts
After obtaining treatment predications, we extracted the concepts in the subject or object according to the semantic schema in Table 2. However, these extracted concepts could still be too general for the disease of interest. To exclude general concepts, we used an approach based on the assumption that concepts associated with a large number of diseases (i.e., common concepts) are likely to be general.
In order to identify common concepts, we took all MeSH terms (from UMLS Version 2014AB) with the semantic type of disorders (N = 5109), and repeated Steps 1, 2, and 4 above to generate disease-treatment pairs. A subset of 2683 MeSH terms were associated with disease-treatment pairs. Then, we analyzed the retrieved treatment concepts and the number of associated disorders for each treatment concept. If a treatment concept was associated with more than an arbitrary threshold of 20% of disease MeSH terms (N = 536), the concept was considered to be a common concept. Applying this criterion, we generated a set of 69 common concepts. Table 3 shows examples of common concepts.
Table 3.
Sampled common concepts.
| CUI | UMLS concept | # of co-occurred diseases |
|---|---|---|
| C0040808 | Treatment Protocols | 1445 |
| C1273870 | Management procedure | 1418 |
| C1273869 | Intervention regimes | 1361 |
| C0011900 | Diagnosis | 1326 |
| C1533685 | Injection procedure | 1265 |
| C0543467 | Operative Surgical Procedures | 1248 |
| C0184661 | Procedures | 1201 |
| C0032042 | Placebos | 1193 |
| C0001617 | Adrenal Cortex Hormones | 1172 |
| C0728940 | Excision | 1091 |
| C1522577 | Follow-up | 1083 |
| C0185125 | Application procedure | 1064 |
| C0023977 | Long-term care | 1041 |
| C0220908 | Screening procedure | 989 |
3.1.6. Step 6: Concept ranking
Ranking concepts has three purposes. First, the ranking might convey the information of the strength of the association. As we know, some treatment concepts might have stronger association with the disease of interest. For example, both “carvedilol” and “fish oil” are retrieved as treatment of heart failure, however, “carvedilol” is mentioned much more frequently in the literature than fish oil as a treatment of heart failure. Second, ranking concepts could make the true relevant concepts appear earlier in the result list than the noise. Although the semantic schema are able to filter some treatment-irrelevant information, noisy information can still be introduced because the semantic schema was focused on sensitivity. For example, given a disease of interest (i.e., heart failure), we extracted a treatment predication {Trastuzumab TREATS Breast cancer metastatic}, where the concept “Trastuzumab” was discussed as a cause of heart failure rather a treatment. Last but not least, a ranked list could speed up the review of automatically extracted concepts. The knowledge authors could prioritize their work with the ranked output.
We explored four approaches to rank the concepts: occurrence, interest, degree centrality, and weighted degree centrality.
- Occurrence: the frequency of the occurrence of a treatment concept in the retrieved treatment predications for a given disease of interest (Formula (1)). The assumption is that the more often a concept is mentioned in the context of disease-specific treatment predications, the stronger the confidence that it is as a treatment for the disease of interest.
where ai is the frequency of the occurrence of a concept ti in the treatment predications.(1) - Interest: A treatment concept may have a high occurrence score among the other extracted treatment concepts simply because it frequently occurs in the entire database. However, the relation between the concept and the disease of interest can still be weak. Interest is a measure that attempts to correct this weakness of occurrence, the idea of which is very similar to the TF-IDF (term frequency inverse document frequency) – a statistic that is intended to reflect how important a word is to a document in a collection of corpus [39]. We define the interest is the ratio of the occurrence of a treatment concept to the sum of the occurrence of all treatment concepts retrieved for a given disease of interest divided by logarithm of the ratio of the occurrence of a treatment of interest to all treatment concepts in the database (see Formula (2)). The denominator is a simple way of measuring the commonality of a concept.
where ai is the frequency of the occurrence of a concept ti in the treatment predications, Ai is the total frequency of the occurrence of the concept ti in the entire database, while M is the total number of retrieved treatment concepts.(2) - Degree centrality: Occurrence-based statistics ignore the linkage between concepts. Since the treatment predications extracted in step 4 can form a graph, we analyzed the formed network and use the centrality to identify important vertices (i.e., treatment concepts) within the graph. Degree centrality is the simplest of many centrality approaches, which measures the significance of the concepts in the graph by counting their connectivity to other concepts. We do not look at whether a concept is directly connected to the disease of interest or not; rather, we assess whether concepts are in the center of the graph. The following formula was used to calculate the degree centrality of a given concept in the graph:
where i is the focal node, j represents all other nodes, N is the total number of nodes, and x is the adjacency matrix, in which the cell xij is defined as 1 if node i is connected to node j, and 0 otherwise. Zhang et al. have used degree centrality for semantic abstraction summarization of therapeutic studies, in which degree centrality was used to select important nodes from a graph [37]. Özgür et al. also used degree centrality for mining gene-disease association from biomedical literature [40].(3) - Weighted degree centrality: Weighted degree centrality is a harmonization between the frequency of occurrence and degree centrality [41].
where ki is the degree centrality score of node i, or CD(i) as described in Formula (3). Si is the sum of weighted adjacency matrix in which wij is the value that represents the weight of the edge (i.e., the occurrence of a predication) between node i and node j. α is a positive tuning parameter that can be set according to the research setting and data. We used α = 0.5 in this study to harmonize the occurrence and the degree centrality in one ranking.(4)
3.2. Experiment
We conducted an experiment to test the following null hypotheses: there is no difference in precision at top 100 extracted concepts among the rankings produced by the four ranking approaches in the pipeline-based algorithms (H1); and there is no difference in precision at top 100 extracted concepts among the rankings produced by the pipeline, predication, and MeSH-based extraction methods (H2). In addition, we also evaluated the performance of the system against the manually extracted treatment vocabulary with precision-recall curves.
3.2.1. Baseline approaches
We compared our approach with two baselines in terms of extracting disease-specific treatment concepts from MEDLINE citations.
Baseline 1
The Medical Subject Headings (MeSH) vocabulary is used to index and catalog articles in MEDLINE. MeSH qualifier terms, in conjunction with the MeSH main headings, offer a convenience to group citations together when they are related to a particular aspect of a subject. For example, Platelet Aggregation Inhibitors/therapeutic use indicates that the citation is about the use of the drug class platelet aggregation inhibitors in the treatment of a disease. After reviewing the qualifiers defined in the MeSH Topical Qualifiers [42] and examples in the MEDLINE database of how those qualifiers were used with the MeSH headings, we selected the following qualifiers: “methods”, “instrumentation”, “therapeutic use”, “pharmacology”, and/or “administration & dosage”. For example, the qualifier “administration & dosage” is defined as “used with drugs for dosage forms, routes of administration, frequency and duration of administration, quantity of medication, and the effects of these factors.”, a drug MeSH term could be possibly assigned with the qualifier “administration & dosage”. Based in their definition, the qualifiers “methods” and “instrumentation” were used with procedures and techniques, including diagnostic procedures and therapeutic procedures. The qualifiers “therapeutic use”, “pharmacology”, and/or “administration & dosage” were used with drugs or chemical substances.
From the articles retrieved by Step 1, we were able to extract a collection of MeSH terms associated with the therapeutic qualifiers of interest. We then obtained the UMLS concepts for these MeSH terms using the mappings established in the UMLS Metathesaurus. Next, the resulting UMLS concepts were restricted using the same semantic types and groups described in Table 1 in order to avoid the inclusion of concepts not related to treatment. The remaining concepts were ranked based on their frequency of occurrence.
Baseline 2
This baseline approach simply used the predications to obtain disease-specific treatment concepts. We first extracted the predications with the pattern of {Subject TREATS/PREVENTS Object}, where the object is the disease of interest. We then extracted all the concepts in the subject position. Thereafter, we ranked the concepts based on their frequency of the occurrence in the retrieved predications.
3.2.2. Validation of extracted concepts
We selected five diseases cases for hypothesis testing. Two diseases, pulmonary embolism (PE) and rheumatoid arthritis (RA), were chosen from a previous study, for which we have developed reference treatment vocabularies with 80 and 232 concepts respectively. The reference vocabularies are available in BioPortal as rheumatoid arthritis ontology (https://bioportal.bioontology.org/ontologies/RAO) and pulmonary embolism ontology (https://bioportal.bioontology.org/ontologies/PE). The other three diseases (diabetes mellitus, asthma, and schizophrenia) were chosen from a previous publication on knowledge extraction from existing knowledge resources [18].
In order to measure the performance of different knowledge extraction approaches, we validated the extracted concepts for the selected diseases. This was done by comparing to reference standards (for the two diseases with reference standards) and manual review.
For automated comparison to reference standards, we used exact matching and one-way hierarchical matching where any extracted concepts that were children of reference concepts were considered as positive. The hierarchical relationships were obtained from the UMLS Metathesaurus MRREL and MRHIER tables.
For manual review, the goal was to verify if false-positive concepts according to the reference standard were indeed true-positives or just gaps in the reference standard. For example, “tumor necrosis factor-alpha inhibitor” (a drug class used to treat rheumatoid arthritis) was extracted by our system as a treatment for rheumatoid arthritis. However, this drug class was not present in the reference standard. Upon review one of the source sentences: “Tumour necrosis factor-alpha (TNFalpha) inhibitors are effective agents in treating RA; however, their cost effectiveness as first-line agents has not been investigated”, we confirmed that “tumor necrosis factor-alpha inhibitor” is indeed a treatment for rheumatoid arthritis. This review was done by one of the authors (LW) with additional clinician review if such judgement could not be made directly based on the source sentences.
3.2.3. Outcome measures
The primary outcome for the two hypotheses was precision at K and secondary outcomes were the overall precision and recall. Precision at K was the ratio of the number of “true positive” concepts among the top K ranked concepts divided by K. We calculated the precision at K for five testing diseases for different rankings and algorithms. We choose the parameter K = 100, believing that as knowledge engineers, it is a fair amount of concepts that they would go through. When calculating the precision at K, for diseases having reference standards, we not only validated the extracted concepts with the reference standards, but also manually verified false positive concepts in case they were in fact correct concepts, but missing in the reference standard. For three diseases without reference standards, the top 100 concepts of each disease were manually validated.
To evaluate ranked results, interpolated precision-recall curves were plotted to visualize the trade-off between precision and recall, where the precision and recall were calculated based on the reference standards. The precision-recall curves also provided a visual comparison among the ranks in the pipeline-based approach and between the pipeline-based approach and the baselines. We plotted the interpolated precision-recall curves only for the two diseases with reference vocabularies. An error analysis were also conducted based on manual inspection of false-positive and false-negative concepts.
3.2.4. Statistical analysis
To test the difference among the different rankings in the pipeline-based system (H1), we first measured the top 100 precision obtained by four different rankings for five diseases. We then calculated the mean precision for each ranking. We used analysis of variance (ANOVA) to test the significance of the difference. For pairwise comparisons, we used the Tukey honest significant difference (HSD) post-hoc test.
To test the difference between the pipeline-based system vs. predication-based system and the pipeline-based system vs. the MeSH-based approach (H2), we calculated the mean top 100 precision for the two baselines across the same five diseases. We used ANOVA to test the significance of the differences between pipeline-based system and predication-based approach, followed by the Dunnett post-hoc test for comparisons between the four ranks in the pipeline-based system with the control (or the baseline). In the same way, we tested the significance of difference between the pipeline-based system and the MeSH-based approach. All statistical analyses were based on a significance level of 0.05 and were performed with R version 3.2.5.
4. Results
4.1. System outputs on five diseases
Table 4 shows the number of citations, predications, treatment predications, and treatment concepts retrieved from each step for the five test diseases. The number of retrieved citations varied by disease. On average, each citation was able to generate 4–5 predications, and less than half of those predications were treatment predications. The number of candidate treatment concepts also varied based on the disease of interest.
Table 4.
The numbers of retrieved citations, predications, treatment predication, and treatment concepts for five testing diseases.
| Test cases | Citations | Predications | Treatment predications | Candidate treatment concepts |
|---|---|---|---|---|
| Rheumatoid arthritis | 11,263 | 53,039 | 26,914 | 1984 |
| Pulmonary embolism | 3031 | 12,820 | 5101 | 706 |
| Diabetes mellitus | 32,552 | 166,140 | 72,730 | 3873 |
| Asthma | 17,286 | 94,001 | 39,189 | 2385 |
| Schizophrenia | 6910 | 25,086 | 14,701 | 1018 |
Table 5 shows sample output from the pipeline-based system for rheumatoid arthritis. The output consists of the following attributes: UMLS CUI, concept name, semantic type, four ranking scores (occurrence, interest, degree centrality, and weighted degree centrality), and sentences extracted from the abstract and titles of the published articles.
Table 5.
Exampled output for rheumatoid arthritis with ranking scores and sample source sentences.
| CUI | Concept | Semantic type | Occurrence | Interest | DC | WDC | Source sentences |
|---|---|---|---|---|---|---|---|
| C0025677 | Methotrexate | Phsu | 4102 | 0.3095 | 394 | 1271.29 | CONCLUSIONS: This study confirms previous observations from a dose-ranging study showing that anakinra, in combination with MTX, is an effective and safe treatment for patients with RA who have inadequate responses to MTX alone |
| C0666743 | Infliximab | Phsu | 1974 | 0.1724 | 212 | 646.91 | Infliximab therapy was also associated with improvements in health-related quality of life in patients with Crohn’s disease or rheumatoid arthritis |
| C0717758 | Etanercept | Aapp | 1313 | 0.1263 | 148 | 440.82 | CONCLUSION: Etanercept as monotherapy was safe and was superior to MTX in reducing disease activity, arresting structural damage, and decreasing disability over 2 years in patients with early, aggressive RA |
| C0242708 | Antirheumatic Drugs, Disease-Modifying | Phsu | 940 | 0.1002 | 158 | 385.38 | Early diagnosis and treatment with disease-modifying antirheumatic drugs (DMARDs) are necessary to reduce early joint damage, functional loss, and mortality |
| C0393022 | Rituximab | Aapp | 1004 | 0.0844 | 125 | 354.26 | CONCLUSIONS: Evidence from RCTs suggests that RTX and ABT are more effective than supportive care |
DC = degree centrality; WDC = weighted degree centrality; phsu = pharmaceutical substance; aapp = amino acid, peptide, or protein; MTX = methotrexate; RA = rheumatoid arthritis; RTX = rituximab; ABT = abatacept; RCTs = randomised controlled trials.
4.2. Performance of pipeline-based algorithms versus baselines
Table 6 shows the precision of the top 100 treatment concepts extracted by the pipeline system and baselines on five diseases: rheumatoid arthritis, pulmonary embolism, diabetes mellitus, Alzheimer’s disease, and asthma.
Table 6.
Top 100 precision for treatment concepts extracted for five diseases.
| Diseases | Top 100 precision
|
|||||
|---|---|---|---|---|---|---|
| Pipeline-based
|
B1 | B2 | ||||
| Occurrence | Interest | DC | WDC | |||
| Rheumatoid arthritis | 0.87 | 0.89 | 0.82 | 0.84 | 0.56 | 0.76 |
| Pulmonary embolism | 0.63 | 0.66 | 0.65 | 0.63 | 0.31 | 0.38 |
| Diabetes mellitus | 0.78 | 0.82 | 0.66 | 0.75 | 0.46 | 0.54 |
| Asthma | 0.8 | 0.81 | 0.76 | 0.81 | 0.54 | 0.66 |
| Schizophrenia | 0.81 | 0.83 | 0.74 | 0.77 | 0.31 | 0.62 |
| Mean precision | 0.78 | 0.80 | 0.73 | 0.76 | 0.44 | 0.59 |
| Std. deviation | 0.089 | 0.085 | 0.071 | 0.081 | 0.121 | 0.142 |
| 95% Confidence interval | (0.67, 0.89) | (0.70, 0.91) | (0.638, 0.814) | (0.66, 0.86) | (0.29, 0.59) | (0.41, 0.77) |
DC = degree centrality; WDC = weighted degree centrality; B1 = MeSH-based baseline; B2 = predication-based baseline.
In the pipeline-based approaches, the difference among occurrence, interest, degree centrality, and weighted degree centrality was not significant (mean top 100 precision = 0.78 vs. 0.80 vs. 0.73 vs. 0.76; p = 0.53).
According to the ANOVA test, there was a significant difference in mean precision at top 100 among the pipeline-based and predication-based approaches (occurrence 0.78 vs. interest 0.80 vs. degree centrality 0.73 vs. weighted degree centrality 0.76 vs. predication-based 0.59; p = 0.022). With the HSD post-hoc test, three ranks (i.e., interest, occurrence, and weighted degree centrality) in the pipeline-based system significantly outperformed the predication-based baseline (see Fig. 3), while no significant difference was found between the degree centrality and the predication-based baseline. According to the ANOVA test, there was a significant difference in mean precision at top 100 among the pipeline-based and the MeSH-based baseline (occurrence 0.78 vs. interest 0.80 vs. degree centrality 0.73 vs. weighted degree centrality 0.76vs. MeSH-based 0.44; p < 0.0001). With the HSD post-hoc test, the pipeline-based approach with all four ranks significantly outperformed the MeSH-based approach (see Fig. 4).
Fig. 3.
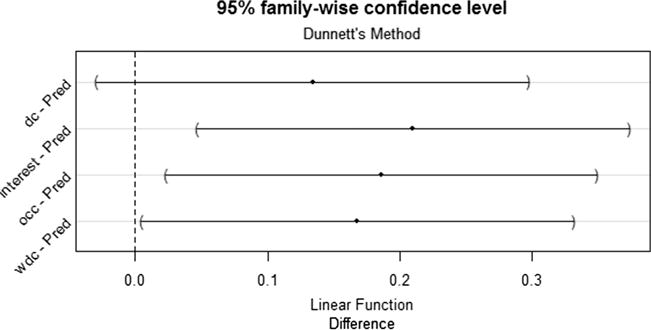
95% family-wise confidence level for the difference of the precision of top 100 concepts between the pipeline-based system and the Predication-based system.
Fig. 4.
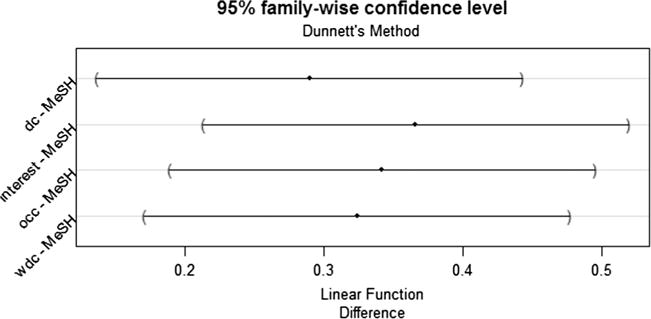
95% family-wise confidence level for the difference of the precision of top 100 concepts between the pipeline-based system and the MeSH-based system.
Figs. 5 and 6 provide a visualization of the treatment vocabularies generated by the pipeline-based system for asthma and diabetes.
Fig. 5.
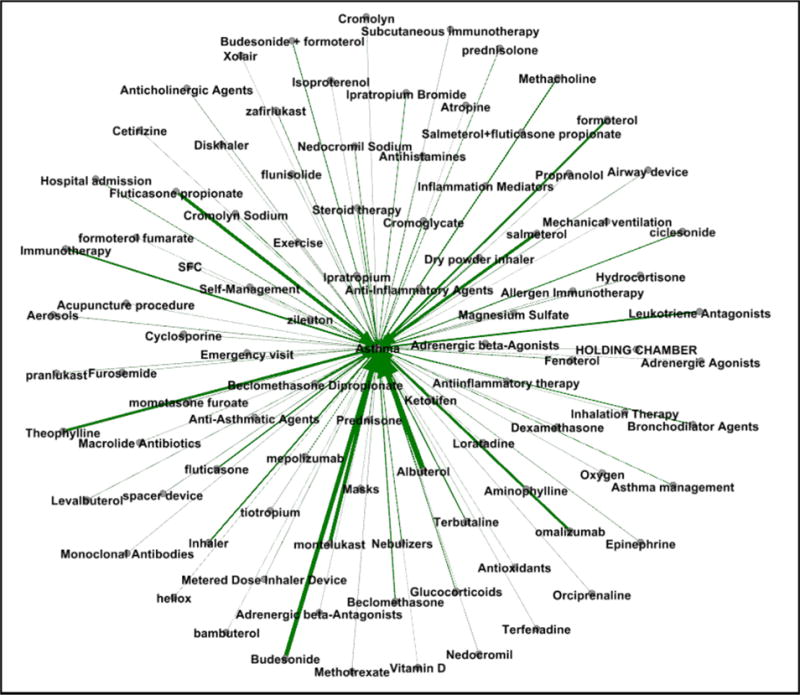
Weighted graph of exampled treatment concepts for asthma.
Fig. 6.
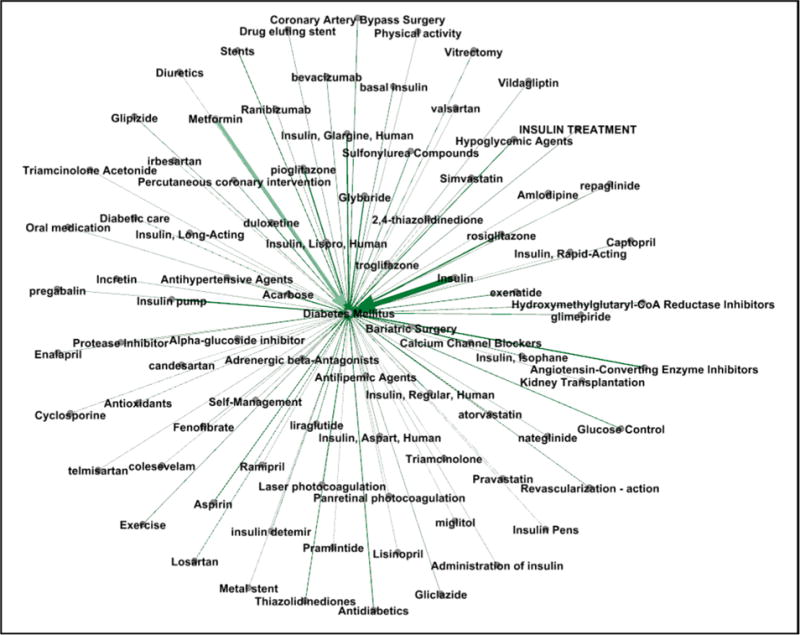
Weighted graph of exampled treatment concepts for diabetes mellitus.
4.3. Precision-recall curves
The precision-recall curves compared the performance of the different approaches against the manually developed reference vocabularies. Fig. 7 shows the interpolated precision-recall curves on rheumatoid arthritis and pulmonary embolism. By including all extracted concepts, the recall of rheumatoid arthritis was 0.59, and the recall of pulmonary embolism was 0.66. Recall for the pipeline based approach was less than 1 for both diseases, indicating that the automated system captured only a subset of the concepts in the gold standard. The predication-based baseline approach reached a recall of 0.58 for rheumatoid arthritis and 0.56 for pulmonary embolism while, the MeSH-based baseline reached a recall of 0.34 for both pulmonary embolism and rheumatoid arthritis.
Fig. 7.
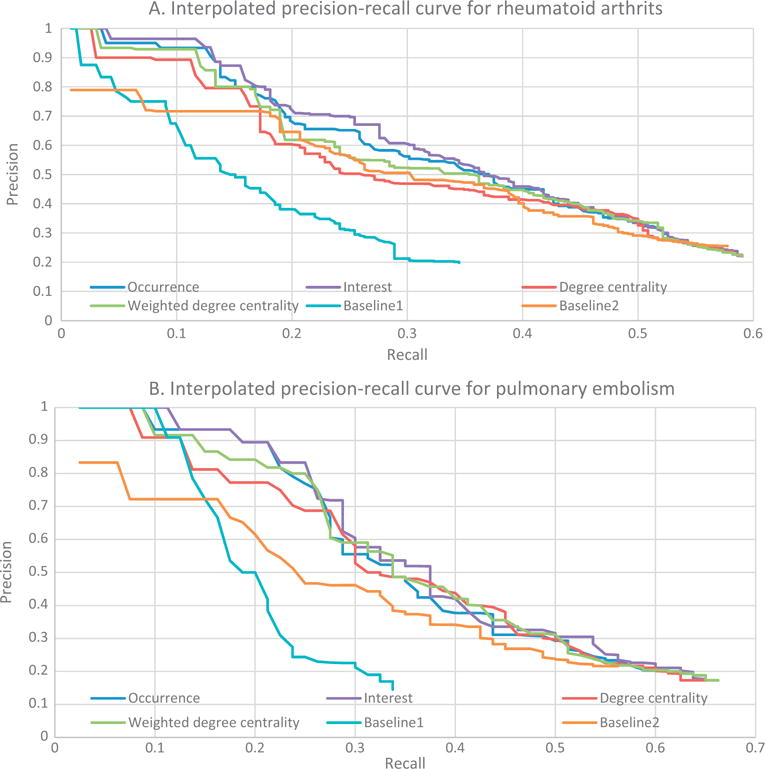
(A) Interpolated precision-recall curves for rheumatoid arthritis; (B) Interpolated precision-recall curves for pulmonary embolism.
4.4. Error analysis
We identified 143 false negative concepts for rheumatoid arthritis, and 43 false negative concepts for pulmonary embolism. All these false negative concepts were included in the error analysis. We identified over two thousand false positive concepts for these two diseases and analyzed the false positive concepts among the top 100 ranked concepts of each disease retrieved by any of the ranks, which resulted in 47 false positive concepts for rheumatoid arthritis and 76 for pulmonary embolism.
Three main reasons could be attributed to false negative concepts or lowered recall: (1) about one third of the reference concepts were not present in the extracted sentences and predications (e.g., “fluindione” and “lanoteplase” for pulmonary embolism). A few false negative concepts were missed because their semantic types were not included in the semantic schema of the automated system, such as ‘systemic’ and ‘nutritional’. (2) One third of the reference concepts existed in the extracted citations and sentences, however were missed because they were not captured by SemRep. For example, in “Tai Chi and yoga are complementary therapies which have, during the last few decades, emerged as popular treatments for rheumatologic and musculoskeletal diseases” two predications were extracted: {Complementary therapies TREATS Rheumatologist} and {Complementary therapies TREATS Musculoskeletal Diseases}; however, none of the predications included the relevant concepts “Tai Chi” and “yoga”. (3) One third of reference concepts were missed because equivalent annotations were mapped to UMLS CUIs with different granularity in the reference vocabulary. For example, ‘resistance training’ was mapped to C0872279 (Resistance Training) in the reference standard, but was mapped to C0814409 (Resistance education) in SemMedDB. The reference was more likely to include the entire annotation as a concept while SemRep mapped more granular fragments to UMLS concepts. For example, from the sentence “in this systematic review, outcomes for total wrist fusion were comparable and possibly better than those for total wrist arthroplasty in rheumatoid patients”, SemRep extracted the predication {Arthroplasty TREATS Patients}, while in the reference the “total wrist arthroplasty” was mapped to C0408314 (total wrist arthroplasty).
Several reasons were attributed to false positive concepts or lowered precision. (1) Among the analyzed false positive concepts, 40% were correct disease-specific treatments that were missing in the reference vocabularies. Examples include “methotrexate treatment”, “tumor necrosis factor therapy”, and “Hip Replacement, Total” for rheumatoid arthritis; and “Prescription of prophylactic anticoagulant”, “Prescription of prophylactic anticoagulant”, “Compression Stockings”, and “Angioplasty, Balloon” for pulmonary embolism. (2) Many false positive concepts were biomarkers of tests and assessments for treatment monitoring, usually with the semantic type of “amino acid, peptide, or protein”. Examples include “neurohormonal factor”, “N-terminal pro-B-type natriuretic peptide”. (3) The false positive concepts could be studied as adverse events or risk factors for the disease of interest. Especially for pulmonary embolism, many false positive concepts were related to complications of certain procedures or medications that increase the risk of pulmonary embolism, such as “Arthroplasty”, “Repair of hip”, “Splenectomy”. (4) False positive concepts were also caused by errors introduced by NLP tools. For example, from the sentence “this indicates that the MHAQ and RA-HAQ generally fail to identify appropriately the extent of functional loss in RA”, the predication {Ametantrone TREATS Rheumatoid Arthritis} was extracted, where “HAQ” (Health Assessment Questionnaire) was incorrectly mapped to “ametantrone”.
5. Discussion
In this study, we developed a pipeline-based knowledge extraction system to automatically generate disease-specific treatment vocabularies from the biomedical literature. The system is designed to retrieve disease-specific treatment-related articles, predications, and a ranked list of concepts. Comparing to a MeSH-based and a predication-based concept extraction approaches, our system had significantly higher precision for extracting the top 100 concepts. We also compared different algorithms ranking the extracted concepts; there was no significant difference among four ranks. Our system achieved an average precision of 0.8 for the top 100 concepts. We conclude that this pipeline-based system could be useful in generating disease-specific treatment vocabulary from the biomedical literature for building disease-specific ontologies. Besides, manual review of the system output would be necessary in order to generate a high-quality treatment vocabulary from these automated generated concepts. As an individual without much clinical background, we estimated the time for judging the relevance of the treatment concepts to the disease of interest by reading the origin sentences and citations, which is about one minute per concept. Comparing to manually acquisition, this could be much more efficient.
We reported that the pipeline system has achieved an average precision of 0.80 ranked by interest based on five test diseases. However, as the results show, for well-studied diseases (e.g., rheumatoid arthritis) with many associated biomedical articles, the system would have higher precision, while for those with less articles (e.g., pulmonary embolism), their precision is relatively lower. Therefore, the reported performance would not reflect the system’s performance on diseases that have not been extensively investigated, such as new or rare diseases.
Our system has achieved a relatively low recall based on two test diseases (i.e., pulmonary embolism and rheumatoid arthritis). Based on the error analysis in Section 4.4, approximately two thirds of the false negative concepts were probably attributed to the relation extraction tool we have used. However, there exist many other approaches aimed at extracting semantic relations from the biomedical literature or web documents, and some of them were also used UMLS and/or MetaMap [43]. Therefore, our system may gain further recall by incorporating the output of other relation extraction approaches or tools as secondary knowledge sources in addition to the SemMedDB to our proposed pipeline process.
Although the automated generated vocabulary was not able to identify 100% of the concepts in our manually generated reference vocabularies, the automated approach was able to extract some relevant treatment concepts that were missing in these reference vocabulary. This included cases of concepts with finer granularity or new information that was not included in the guidelines, textbooks, or online documents used to build the reference vocabularies. What’s more, rather than starting from scratch, we build our system upon publically available resources, such as PubMed Clinical Queries, MEDLINE citations, and SemMedDB. In addition, we developed semantic schemas for treatment from an existing disease-specific treatment vocabulary to filter treatment predications rather simply relying on predicates such as “TREATS” or “PREVENTS”. In this way, more information could be captured, for example, the evidence about the comparison between two medications can also be identified.
The main contribution of our study lies in three areas; the tuned selection of articles, the filtering of predications from millions of predications in the SemMedDB, and the ranking of concepts specific to the disease of interest. As Fig. 7 shows, predication-based approach has lower precision comparing to the pipeline system, which indicates that purely using SemRep predications would require much more review effort. In addition, the MeSH-based approach have lower recall comparing to the pipeline system, which indicates that using MeSH heading in the MEDLINE citations would not result as good coverage of the treatment vocabulary as using the pipeline system.
Our approach is innovative in two ways. First, compared to previous studies [18,22], we not only retrieve disease-specific pharmaceutical substances, but also other types of treatment, such as procedures, devices, and activities. In terms of disease-drug pairs, it is interesting to compare the results with previous studies [18,22]. However, we found such comparisons to be difficult since there were substantial differences in study goals, evaluation methods, and reference standards. In a simple comparison to the work of Chen et al. [18], our study found a greater number of disease-relevant citations and disease-drug pairs. Comparing to Xu’s work [22], we have achieved a similar recall at a precision of 0.80, with the caveat that the reference standards used in both studies were different. Second, we were able to collect the source sentences and PubMed citations related to the disease-specific treatments. This could be useful for anyone who are interested in expanding their knowledge on a specific treatment. The extracted concepts also provide an index for over thousands of disease-specific treatment-related citations and sentences from MEDLINE. Researchers or clinicians can use this index to trace the evidence in the biomedical literature of a specific treatment for the disease of interest.
Our proposed approach was designed to be generalizable to other disease domains, such as diagnostic tests, signs, and symptoms. Yet, some adaptation is necessary including developing specific semantic schemas and defining common concepts for other disease domains. The same approach used to develop the semantic schema and define common concepts in the present study can be followed to adapt the algorithms to other disease domains.
The study has several limitations. First, the semantic schema for extracting treatment predications and concepts were developed based on a reference vocabulary of one disease (i.e., heart failure), and might not be generalize to some types of disease. Second, we defined a list of common concepts to be filtered from extracted treatment concepts in Section 3.1.4. The selection of common concepts is based on an arbitrary cut-off threshold. Third, as the algorithm evaluation demonstrated, our reference standards had gaps in coverage and therefore were not perfect. Last the approach to judging the correctness of extracted concepts for diseases without a reference vocabulary was not as rigorous as the approach used to develop the reference vocabularies.
6. Conclusions
We investigated a pipeline-based approach to extract disease-specific treatment concepts from the biomedical literature to assist the development of disease-specific vocabularies. The pipeline-based approach obtained a mean precision of 0.8 for the top 100 retrieved concepts, which was significantly higher than two baselines. The performance of four ranking strategies (e.g., occurrence, degree centrality, weighted degree centrality, and interest) was not statistically significant different. In the future, we intend to extend the system to extract concepts on other disease aspects, including signs, symptoms, and diagnostic tests.
Acknowledgments
The authors thank Thomas Rindflesch and Marcelo Fiszman for providing access to Semantic MEDLINE and useful input related to the database. The authors also thank Olivier Bodenreider for inputs on using the UMLS and MeSH. This work was supported in part by Grants LM010482 and 1R01LM011416 from the National Library of Medicine.
References
- 1.Wang L, Bray BE, Shi J, Del Fiol G, Haug PJ. A method for the development of disease-specific reference standards vocabularies from textual biomedical literature resources. Artif Intell Med. 2016 doi: 10.1016/j.artmed.2016.02.003. http://dx.doi.org/10.1016/j.artmed.2016.02.003. [DOI] [PMC free article] [PubMed]
- 2.Gorman PN. Information needs of physicians. J Am Soc Inform Sci. 1995;46:729–736. http://dx.doi.org/10.1002/(SICI)1097-4571(199512)46:10<729::AID-ASI3>3.0.CO;2-2. [Google Scholar]
- 3.Covell D, Uman G, Manning PR. Information needs in office practice: are they being met? Ann Intern Med. 1985;103:596–599. doi: 10.7326/0003-4819-103-4-596. [DOI] [PubMed] [Google Scholar]
- 4.Del Fiol G, Workman TE, Gorman PN. Clinical questions raised by clinicians at the point of care. JAMA Int Med. 2014;174:710. doi: 10.1001/jamainternmed.2014.368. http://dx.doi.org/10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 5.Ely JW, Osheroff JA, Gorman PN, Ebell MH, Chambliss ML, Pifer EA. A taxonomy of generic clinical questions: classification study. BMJ. 2000;321:429–432. doi: 10.1136/bmj.321.7258.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline RJ, Haynes KM. Consumer health information seeking on the Internet: the state of the art. Health Educ Res. 2001;16:671–692. doi: 10.1093/her/16.6.671. http://dx.doi.org/10.1093/her/16.6.671. [DOI] [PubMed] [Google Scholar]
- 7.Fox S, Fallows D. Internet Health Resources. PewResearchCenter. 2003 < http://www.pewinternet.org/2003/07/16/internet-health-resources/>.
- 8.Noy NF, Mcguinness DL. Ontology development 101: a guide to creating your first ontology. Stanford Knowl Syst Lab Tech Rep KSL-01-05 Stanford Med Informatics Tech Rep SMI-2001-0880. 2001 [Google Scholar]
- 9.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. IMIA Yearb Med Inform. 2008;47:67–79. [PMC free article] [PubMed] [Google Scholar]
- 10.Eysenbach G, Jadad AR. Evidence-based patient choice and consumer health informatics in the internet age. J Med Internet Res. 2001;3:e19. doi: 10.2196/jmir.3.2.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng QT, Crowell J, Plovnick RM, Kim E, Ngo L, Dibble E. Assisting consumer health information retrieval with query recommendations. J Am Med Infor Assoc. 2006;13:80–90. doi: 10.1197/jamia.M1820. http://dx.doi.org/10.1197/jamia.M1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu G, Jones CB, Abdelmoty AI. Ontology based spatial query expansion in information retrieval. Lect Notes Comput Sci – ODBASE2005. 2005;3761:11466–11482. [Google Scholar]
- 13.Lowe HJ, Barnett GO. Understanding and using the medical subject headings (MeSH) vocabulary to perform literature searches. JAMA. 1994;271:1103–1108. http://dx.doi.org/10.1001/jama.271.14.1103. [PubMed] [Google Scholar]
- 14.Haug PJ, Ferraro JP, Holmen J, Wu X, Mynam K, Ebert M, et al. An ontology-driven, diagnostic modeling system. J Am Med Inform Assoc. 2013;20:e102–e110. doi: 10.1136/amiajnl-2012-001376. http://dx.doi.org/10.1136/amiajnl-2012-001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malhotra A, Younesi E, Gündel M, Müller B, Heneka MT, Hofmann-Apitius M. ADO: a disease ontology representing the domain knowledge specific to Alzheimer’s disease. Alzheimer’s Dement. 2014;10:238–246. doi: 10.1016/j.jalz.2013.02.009. http://dx.doi.org/10.1016/j.jalz.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Chalortham N, Buranarach M, Supnithi T. Ontology development for type II diabetes mellitus clinical support system. Proc 4th Int Conf Knowl Inform Creat Support Syst. 2009 [Google Scholar]
- 17.Buitelaar P, Cimiano P, Magnini B. Ontology Learning From Text: Methods, Evaluation and Applications. IOS Press; Amsterdam, The Netherlands: 2005. doi: 10.1.1.70.3041. [Google Scholar]
- 18.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease-drug knowledge from biomedical and clinical documents: an initial study. J Am Med Infor Assoc. 2008;15:87–98. doi: 10.1197/jamia.M2401. http://dx.doi.org/10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Chused A, Elhadad N, Friedman C, Markatou M. Automated knowledge acquisition from clinical narrative reports. AMIA Annu Sympos Proc. 2008:783–787. [PMC free article] [PubMed] [Google Scholar]
- 20.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform. 2010;43:891–901. doi: 10.1016/j.jbi.2010.09.009. http://dx.doi.org/10.1016/j.jbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Xu R, Li L, Wang Q. DRiskKB: a large-scale disease-disease risk relationship knowledge base constructed from biomedical text. BMC Bioinform. 2014;15:105. doi: 10.1186/1471-2105-15-105. http://dx.doi.org/10.1186/1471-2105-15-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu R, Wang Q. Large-scale extraction of accurate drug-disease treatment pairs from biomedical literature for drug repurposing. BMC Bioinform. 2013;14:181. doi: 10.1186/1471-2105-14-181. http://dx.doi.org/10.1186/1471-2105-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimino JJ, Barnett GO. Automatic knowledge acquisition from MEDLINE. Methods Inf Med. 1993;32:120–130. [PubMed] [Google Scholar]
- 24.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS. AMIA Annu Sympos Proc. 1998:568–572. [PMC free article] [PubMed] [Google Scholar]
- 25.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–477. doi: 10.1016/j.jbi.2003.11.003. http://dx.doi.org/10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Rindflesch TC, Fiszman M, Libbus B. Semantic interpretation for the biomedical research literature. In: Fuller S, Hersh W, Friedman C, Chen H, editors. Med Infor Knowl Manage Data Min Biomed. Springer; 2005. pp. 399–422. [Google Scholar]
- 27.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGAR: extraction of drugs, genes and relations from the biomedical literature. Pac Sympos Biocomput. 2000:517–528. doi: 10.1142/9789814447331_0049. http://dx.doi.org/10.1016/j.bbi.2008.05.010. [DOI] [PMC free article] [PubMed]
- 28.Hendrickx I, Kim SN, Kozareva Z, Nakov P, Romano L, Szpakowicz S. SemEval-2010 Task 8: Multi-Way Classification of Semantic Relations Between Pairs of Nominals. 5th Int Work Semant Eval ACL 2010. 2010:33–8. [Google Scholar]
- 29.Rosario B, Hearst Ma. Classifying semantic relations in bioscience texts. Proc 42nd Annu Meet Assoc Comput Linguist. 2004:430. http://dx.doi.org/10.3115/1218955.1219010.
- 30.Zeng D, Liu K, Lai S, Zhou G, Zhao J. Relation classification via convolutional deep neural network. COLING. 2014:2335–2344. [Google Scholar]
- 31.Bundschus M, Dejori M, Stetter M, Tresp V, Kriegel HP. Extraction of semantic biomedical relations from text using conditional random fields. BMC Bioinfor. 2008;9:207. doi: 10.1186/1471-2105-9-207. http://dx.doi.org/10.1186/1471-2105-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giuliano C, Lavelli A, Romano L, Sommarive V. Exploiting shallow linguistic information for relation extraction from biomedical literature. EACL. 2006 [Google Scholar]
- 33.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28:3158–3160. doi: 10.1093/bioinformatics/bts591. http://dx.doi.org/10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes RB, Wilczynski NL, McKibbon KA, Walker JC, Sinclair CJ. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Infor Assoc. 1994;1:447–458. doi: 10.1136/jamia.1994.95153434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haynes RB, Wilczynski NL. Optimal search strategies for retrieving scientifically strong studies of diagnosis from Medline: analytical survey. BMJ. 2004;328:1040. doi: 10.1136/bmj.38068.557998.EE. http://dx.doi.org/10.1136/bmj.38068.557998.EE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haynes RB, McKibbon KA, Wilczynski NL, Walter SD, Werre SR. Optimal search strategies for retrieving scientifically strong studies of treatment from medline: analytical survey. BMJ. 2005;330:1179. doi: 10.1136/bmj.38446.498542.8F. http://dx.doi.org/10.1136/bmj.38446.498542.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Fiszman M, Shin D, Miller CM, Rosemblat G, Rindflesch TC. Degree centrality for semantic abstraction summarization of therapeutic studies. J Biomed Inform. 2011:830–838. doi: 10.1016/j.jbi.2011.05.001. http://dx.doi.org/10.1016/j.jbi.2011.05.00 (in press) [DOI] [PMC free article] [PubMed]
- 38.Fiszman M, Rindflesch TC, Kilicoglu H. Abstraction summarization for managing the biomedical research literature, in. Proc HLT-NAACL Work Comput Lex Semant. 2003:76–83. [Google Scholar]
- 39.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Inform Process Manage. 1988;24:513–523. [Google Scholar]
- 40.Ozgür A, Vu T, Erkan G, Radev DR. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24:i277–i285. doi: 10.1093/bioinformatics/btn182. http://dx.doi.org/10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Opsahl T, Agneessens F, Skvoretz J. Node centrality in weighted networks: generalizing degree and shortest paths. Soc Netw. 2010;32:245–251. http://dx.doi.org/10.1016/j.socnet.2010.03.006. [Google Scholar]
- 42.https://www.nlm.nih.gov/mesh/topsubscope.html
- 43.Nebot V, Berlanga R. Exploiting semantic annotations for open information extraction: an experience in the biomedical domain. Knowl Inf Syst. 2014;38:365–389. http://dx.doi.org/10.1007/s10115-012-0590-x. [Google Scholar]


