Abstract
Background:
Phenotyping is an automated technique that can be used to distinguish patients based on electronic health records. To improve the quality of medical care and advance type 2 diabetes mellitus (T2DM) research, the demand for T2DM phenotyping has been increasing. Some existing phenotyping algorithms are not sufficiently accurate for screening or identifying clinical research subjects.
Objective:
We propose a practical phenotyping framework using both expert knowledge and a machine learning approach to develop 2 phenotyping algorithms: one is for screening; the other is for identifying research subjects.
Methods:
We employ expert knowledge as rules to exclude obvious control patients and machine learning to increase accuracy for complicated patients. We developed phenotyping algorithms on the basis of our framework and performed binary classification to determine whether a patient has T2DM. To facilitate development of practical phenotyping algorithms, this study introduces new evaluation metrics: area under the precision-sensitivity curve (AUPS) with a high sensitivity and AUPS with a high positive predictive value.
Results:
The proposed phenotyping algorithms based on our framework show higher performance than baseline algorithms. Our proposed framework can be used to develop 2 types of phenotyping algorithms depending on the tuning approach: one for screening, the other for identifying research subjects.
Conclusions:
We develop a novel phenotyping framework that can be easily implemented on the basis of proper evaluation metrics, which are in accordance with users’ objectives. The phenotyping algorithms based on our framework are useful for extraction of T2DM patients in retrospective studies.
Keywords: phenotyping, positive predictive value (PPV), sensitivity, support vector machine (SVM), type 2 diabetes mellitus (T2DM)
An estimated 415 million people worldwide have diabetes mellitus. Approximately 87% to 91% of these people have type 2 diabetes mellitus (T2DM).1 Therefore, there is an increasing need for T2DM clinical research, clinical decision support systems, and clinical trial recruitments;2,3 the identification of T2DM patients is crucial to fulfill these purposes. EHR data are expected to be useful for improving the efficiency of cohort research and clinical trial recruitments and for improving the overall quality of medical care.4-7 The diagnoses in EHRs are often used for the identification of patients; however, to date, the diagnoses in EHRs have been limited in terms of accuracy and completeness.4,8-11 Moreover, manually distinguishing patients on the basis of EHRs can be extremely time consuming. Thus, the demand for automated techniques for distinguishing patients based on EHRs, so-called phenotyping, has been increasing. Some published studies, such as eMERGE projects,5,6,11,12 describe automated phenotyping techniques employed in the United States. Many T2DM phenotyping algorithms exist;13,14 however, they have some limitations.
The first limitation is that high performance is incompatible with the readability regarding the techniques used to develop phenotyping algorithms; the rule-based methods are dominant although there is a growing trend of data-driven methods.5 Rule-based methods are easy for users to understand and the performance in detecting T2DM is 65-70% sensitivity and 88-95% positive predictive value (PPV).5,13,15,16 However, even complicated algorithm that combines 9 rules tend to miss some patients who have incomplete data.17 Classification of such patients requires other techniques to interpret EHR data deeply, thus we cannot classify these patients completely only with rules. Meanwhile, data-driven methods classify even such patients by patterns learned from data and sometimes produce higher performance than rule-based methods; the performance for T2DM is 80-90% sensitivity and ~89% PPV.18 However, they tend to be the “black-box” type and lack readability.19 If we apply some existing data-driven methods to other data sets and find new specific error cases, we cannot understand why the patients were not classified correctly, and tuning the method in accordance with these cases is nearly impossible.18,20 Therefore, we believe that a phenotyping algorithm that combines rule-based and data-driven methods has strength. Second, the appropriateness of the evaluation metrics may also present a limitation. Appropriate evaluation metrics can differ depending on their objectives. Accordingly, high sensitivity—which means that a phenotyping algorithm can identify almost all T2DM patients—should be given priority to if users intend to use the algorithm for screening. Moreover, high PPV—which means that almost all patients identified by a phenotyping algorithm are T2DM patients—should be prioritized if users intend to use the patients identified by the algorithm as clinical research subjects without later screening. In previous studies, phenotyping algorithms have been proposed on the basis of developers’ individual objectives. These algorithms are applicable for only a particular objective and are independent of other algorithms. Therefore, we believe that a phenotyping framework that can be adjusted based on users’ objectives is practical because each user can apply it on the basis of appropriate evaluation metrics appropriate for his/her objective. Nevertheless, such a framework has not been studied.
We thus propose a practical phenotyping framework that consists of expert knowledge and a machine learning approach to take advantage of each. Accordingly, we develop 2 types of phenotyping algorithms: one has high sensitivity and the other has high PPV. In addition, this paper introduces 2 evaluation metrics for detecting whether our proposed algorithm can have high sensitivity while retaining high PPV, and vice versa. The present study used EHR data in Japan and the first automated T2DM phenotyping algorithm in Asia.
Methods
This section describes the proposed framework and its evaluation. We will describe the materials used for evaluation, the proposed framework, and the method to evaluate it.
Eligible Subjects and EHR Data
The subjects employed in our study were patients with at least 2 visits to the University of Tokyo (UT) Hospital between January 1, 2009, and December 31, 2014, and with at least 1 visit between January 1, 2012, and December 31, 2012. We obtained EHR data for eligible patients over 6 years (2009 to 2014). For the application of algorithms, we used diagnoses, medications, and laboratory tests stored based on HL7 version 2.5 (ISO 27931:2009).21
Proposed Algorithms
We propose a hybrid phenotyping framework that consists of 2 steps: the first step uses expert knowledge as rules; the second uses a machine learning approach (Figure 1(i)). Our framework first applies rules to exclude the obvious non-T2DM patients. A subsequent machine learning approach correctly selects T2DM patients. Because only certain types of diagnoses, medications, and laboratory tests are related to T2DM, we supposed that a rule-based algorithm based on practice guidelines for treatment of T2DM22 could easily include almost all T2DM patients and exclude the obvious non-T2DM patients.
Figure 1.
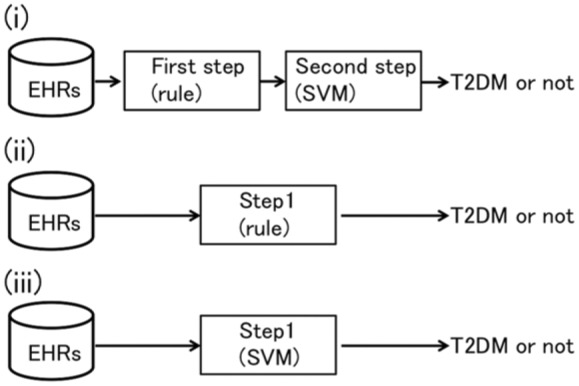
Framework of proposed algorithms. (i) Phenotyping algorithms (A) and (B). (ii) Algorithms (D), (F), and (G). (iii) Algorithms (C) and (E).
We chose to use a support vector machine (SVM) for our machine learning approach. Regarding T2DM, even without considering disease names, a few studies have compared the phenotyping performance of machine learning methods and no consensus has been reached on the best performing method.18,20 Nevertheless, for this limited scenario, SVMs exhibited good performance in all those studies. SVM is a multidimensional and nonlinear classifier that can specify the best classification boundary to classify even complicated patients that rule-based algorithms cannot classify accurately. SVM is appropriate for one of our objectives; combining rule- and machine-learning-based methods. SVMs also have the general advantage of being known as one of the best classifiers in binary classification tasks like the one in this study.19 In addition, there are many SVM studies using cost-sensitive learning,23-25 which is suitable for the other objective; developing 2 types of algorithms based on this phenotyping framework. One is tuned for high sensitivity; the other is tuned for high PPV. Cost-sensitive learning can tune it for the 2 algorithms by changing the costs; for example, we can tune it for high sensitivity by increasing the cost of false negatives, which results in a narrow non-T2DM area.
(A) Rule + SVM (designed for high sensitivity)
<First step (rule) > (1) Patients with T2DM related ICD-10 codes (E11x or E14x) who are treated with insulin or T2DM medications OR (2) Patients with random glucose ≥ 200 mg/dL OR (3) Patients with HbA1c ≥ 6.5%.
<Second step (SVM) > Features and the maximum-margin hyperplane are determined following Table 1.
(B) Rule + SVM (designed for high PPV)
<First step (rule) > The same rules as those of the first step of (A).
<Second step (SVM) > See Table 1.
Table 1.
Details of Tuning of SVM.
| Cost-sensitive learning |
||
|---|---|---|
| Algorithm | Maximum-margin hyperplane for the highest sensitivity | Maximum-margin hyperplane for the highest PPV |
| (A) | Yes | No |
| (B) | No | Yes |
| (C) | Yes | No |
| (E) | No | Yes |
To consider definite T2DM patients as cases, we scaled each feature and excluded features with zero and near-zero-variance features as SVM preprocessing, to improve data quality. We performed cost-sensitive learning, which can decrease the proportion of FN cases or FP cases by changing the cost for these cases. The costs were determined for constructing the maximum-margin hyperplane for the highest PPV or sensitivity.
Table 2.
Nineteen Features Used for SVM.
| Features | Definition | |
|---|---|---|
| Disease name (frequency) | T2DM codes as main diagnosis | E11x (ICD-10 code) |
| T1DM codes | Disease control number,34 including T1DM | |
| Malnutrition-related-DM | E12x (ICD-10 code) | |
| Secondary DM | E13x (ICD-10 code) | |
| Unspecified DM as main diagnosis | E14x (ICD-10 code) | |
| Slowly progressive insulin-dependent diabetes mellitus (SPIDDM) | Disease control number,34 including SPIDDM. (If one ICD-10 code corresponds to different disease names in Japanese, each disease name has its own disease control number. We could have performed a more detailed coding using the disease control number than by using ICD-10 alone.) | |
| Medication (frequency) | Insulin | 24924x, 24925x, 24926x (National Health Insurance Drug List) |
| T2DM medication | 396x (National Health Insurance Drug List) | |
| Medication with a function of rise in blood glucose | The medications or injections that have descriptions about hyperglycemia as a side effect in the package inserts. (The National Health Insurance Drug List provides the codes of drugs used in Japan.) | |
| Injection with a function of rise in blood glucose | ||
| Laboratory tests (maximum value) | Random glucose | There were no criteria for distinguishing random blood glucose levels and fasting blood glucose levels in HL7 storage at UT Hospital. We thus assumed that all blood glucose levels were random blood glucose levels. |
| Glycoalbumin | ||
| HbA1c (J) | HbA1c (J) was the HbA1c standard used in Japan until March 2012, and HbA1c (J) 6.1% is equivalent to HbA1c 6.5%. | |
| HbA1c | ||
| GAD antibody | ||
| IA2 antibody | ||
| 125I-insulin biding ratio | ||
| (minimum value) | C-peptide |
We selected features that are correlated with T2DM and other subtypes of DM.
Experimental Evaluation: Five Phenotyping Algorithms for Comparison
We present SVM (C) and rule (D) algorithms, which are tuned for high sensitivity.
We also present SVM algorithm (E), which is tuned for high PPV. We propose a modified PheKB algorithm (F), which is also tuned for high PPV, for comparison. We selected PheKB algorithm (G)14,17 as a solely rules-based baseline algorithm tuned for high PPV because it is one of the most popular PPV-maximizing T2DM phenotyping algorithms. However, the diagnosis criteria of T2DM in Japan differ from those in the United States.22 We thus expected that some negative cases, such as secondary DM or temporary hyperglycemia, would not be fully excluded by (G). Accordingly, we propose the modified PheKB algorithm (F).
(C) SVM (Designed for high sensitivity)
See Table 1.
(D) Rule (Designed for high sensitivity)
The same rules as those of the first step of (A).
(E) SVM (Tuned for high PPV)
See Table 1.
(F) Rule (Designed for high PPV)
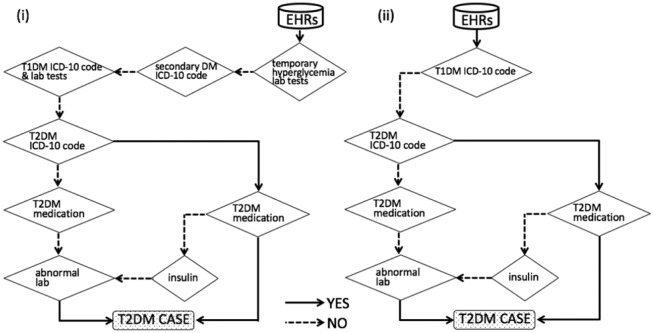
We modified algorithm (G) using data from UT Hospital in Japan (Figure 2(i)).
Figure 2.
(i) Modified PheKB algorithm (F), which is designed for high PPV. Algorithm (D) could include the obvious T2DM patients in accordance with the diagnosis criteria in Japan. This is also an aggressive strategy to specifically exclude T1DM, secondary DM, and temporary hyperglycemia. These points greatly differentiate (D) from (C). Abnormal lab indicates the following: a maximum value of random glucose ≥ 200 mg/dL and (1) a maximum value of HbA1c ≥ 6.5% or (2) a maximum value of HbA1c (J) ≥ 6.1%. T1DM lab tests are one of the following: (1) a maximum GAD-antibody value of ≥ 1.5 U/mL or (2) a maximum 125I-insulin biding ratio of ≥ 7% or (3) a maximum IA2-antibody value of ≥ 0.4 U/mL. The details of other steps are shown in Table 2. (ii) PheKB algorithm (G). Steps of T2DM Dx by physician ≥ 2, T2DM Rx precedes T1DM Rx, and diabetes medical supplies were excluded because they were data that were not included in the UT Hospital HL7 data. Abnormal lab means one of the following: (1) a maximum random glucose value of ≥ 200 mg/dL, (2) a maximum HbA1c of ≥ 6.5%, or (3) a maximum HbA1c (J) of ≥ 6.1%. The details of other steps are shown in Table 2.
(G) PheKB algorithm
We translated the PheKB algorithm to apply it to the HL7 data from UT Hospital (Figure 2(ii)).
Annotation of EHRs
Billing codes are not sufficient for use as a gold standard; we thus annotated EHRs manually. Two medical doctors and 1 non-medical-doctor researcher (1 of the authors) annotated randomly selected EHRs of 510 patients for algorithms (C) to (G). For algorithms (A) and (B), 3 annotators checked randomly selected EHRs of 850 patients, who were selected in the first step of either algorithm (A) or (B). This is because the number of patients remaining after the first step of algorithm (A) or (B) would be insufficient for SVM if 510 randomly selected patients were considered for algorithm (A) or (B). The number of false negative cases excluded by the first step of (A) or (B) was calculated using the proportion of false negative cases of algorithm (D). Two annotators checked each EHR. Two medical doctors discussed and made final decisions regarding patients with mismatched annotations. We classified the EHRs into 2 subtypes: T2DM and non-T2DM.
Evaluation Metrics
We employed 4 evaluation metrics in this study. Sensitivity is True Positive (TP) / (TP + False Negative (FN)). We used the sensitivity that should be prioritized if algorithms are to be used for screening. PPV is TP / (TP + False Positive (FP)). We used PPV, which should be prioritized if users intend to use the patients identified by the algorithm as clinical research subjects without later screening. We are also interested in the extent to which sensitivity can be increased while maintaining a high PPV, and vice versa. We thus introduce the following metrics: area under the precision-sensitivity curve (AUPS) with high sensitivity and AUPS with high PPV. In AUPS with a high sensitivity, the sensitivity ranged from 0.9 to 1. In AUPS with a high PPV, the PPV ranged from 0.9 to 1.
The Software Used for Developing the Algorithms
Rule algorithms were developed using the Konstanz Information Miner (KNIME).26 SVM analyses and validations were performed using R-3.1.3,27 package kernlab, version 0.9-23,28 and package ROCR, version 1.0-7.29 We used 20% of the data to decide the type of kernel to use and the value of hyper-parameters; these were determined for the highest accuracy based on a grid search using package caret, version 6.0-64.30 A Gaussian kernel was selected. We conducted 5-fold cross-validation using the remaining 80% of the data. We performed cost-sensitive learning based on a grid search using package kernlab.
Results
Subjects
Table 3 lists the 104,522 patients who were eligible for this study. 11.4% of the subjects were annotated as T2DM patients and the rest were non-T2DM patients.
Table 3.
Demographic Characteristics of Eligible Patients.
| Characteristic | Value |
|---|---|
| Age (years), mean (SD) | 52.6 (26.5) |
| Female sex | 57.2% |
| Blood glucose test | 58.1% |
| HbA1c test | 44.3% |
| E14x (DM, ICD-10 code) | 36.2% |
| E11x (T2DM, ICD-10 code) | 22.6% |
| T2DM medication | 7.2% |
| GAD antibody | 2.6% |
| IA2 antibody | 0.1% |
Results of Seven Phenotyping Algorithms
Table 4 shows the performance of each algorithm. Our proposed algorithm, (B), demonstrates in the highest PPV, highest AUPS with high sensitivity, and highest AUPS with high PPV. Proposed algorithm (A) demonstrates the second-highest sensitivity and second-highest AUPS with high sensitivity. Proposed algorithms (A) and (B) are both based on our proposed framework. The only difference is the tuning at each second step. These results show that our proposed framework can be used to develop 2 types of phenotyping algorithms by changing the SVM tuning.
Table 4.
Performance of Each Algorithm.
| Designed for | Sensitivity | PPV | AUPS with high sensitivity | AUPS with high PPV | ||
|---|---|---|---|---|---|---|
| Hybrid | (A) Rule + SVM | High sensitivity | 90.9% | 80.0% | 0.72 | 0.52 |
| (B) Rule + SVM | High PPV | 51.8% | 98.3% | 0.80 | 0.83 | |
| Simple | (C) SVM | High sensitivity | 85.7% | 37.5% | 0.37 | 0.42 |
| (D) Rule | High sensitivity | 92.2% | 72.3% | — | — | |
| (E) SVM | High PPV | 57.1% | 66.7% | 0.43 | 0.42 | |
| (F) Modified PheKB algorithm | High PPV | 51.0% | 96.2% | — | — | |
| (G) PheKB algorithm | High PPV | 78.0% | 63.9% | — | — |
Each SVM has a different hyperplane and rule-based algorithms, and the same rules are used for algorithms (A), (B), and (D) (see the Proposed Algorithms and Experimental Evaluation: Five Phenotyping Algorithms for Comparison sections).
In addition, rule-based algorithm (D) produced the highest sensitivity, but algorithm (A) produced the second-highest sensitivity in addition to high PPV; thus, algorithm (A) is more practical for use in screening than algorithm (D).
Algorithm Validation Using ROC Curves and PS Curves
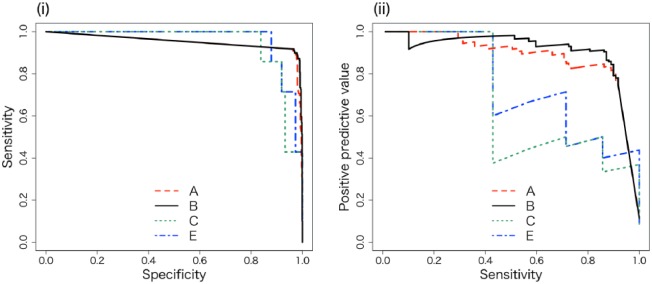
We drew the ROC curves and PS curves of algorithms (A), (B), (C), and (E). The ROC curves show that proposed algorithms (A) and (B) exhibit high sensitivity with high specificity (Figure 3(i)). The PS curves show that proposed algorithms (A) and (B) exhibit high PPV and high sensitivity (Figure 3(ii)).
Figure 3.
(i) ROC curves of algorithms (A), (B), (C), and (E). (ii) The PS curves of the algorithms. We drew PS curves and evaluated whether the PPV maintained a high score while retaining a high sensitivity. The axes of the ROC curve consist of specificity = (1 – (TN / (FP + TN))) and sensitivity = (TP / (TP + FN)). The axes of the PS curve consist of precision = PPV and sensitivity, where TN = true negative, FP = false positive, TP = true positive, and FN = false negative.
Error Analysis
Table 5 shows the false positive (FP) and false negative (FN) cases of each algorithm. Proposed algorithm (B) greatly decreases the proportion of FP cases. Proposed algorithm (A) decreases the proportion of FN cases more than that of FP cases. These results show that algorithms (A) and (B) each offer different advantages in accordance with each tuning characteristic. FN cases of algorithms (A) and (B) consist of cases classified as non-T2DM by both the SVM in the second step and the rules in the first step. Therefore, regarding the proportion of FN cases classified by the SVM, algorithm (A) performed better than algorithm (C), whereas algorithms (A) and (C) decreases the proportion of FN cases classified by both whole algorithms equally well. The inclusion of rules influences the performance of the subsequent SVM and corresponds with one of our theses; a hybrid framework could take advantage of rules and SVM.
Table 5.
FP and FN Cases of Phenotyping Algorithms.
| Patterns of cases | (A) | (B) | (C) | (D) | (E) | (D) | (G) | |
|---|---|---|---|---|---|---|---|---|
| FP | Temporary glycemia | 5 | 0 | 5 | 17 | 0 | 0 | 3 |
| Secondary DM | 5 | 0 | 1 | 0 | 0 | 0 | 2 | |
| T1DM | 2 | 1 | 0 | 1 | 0 | 1 | 0 | |
| Non-T2DM (except above cases) | 13 | 0 | 4 | 0 | 2 | 0 | 17 | |
| FN | Regarded as T1DM | 0 | 0 | 0 | 0 | 0 | 2 (one case overlaps case with T2DM and T1DM) | 4 |
| No E11x (T2DM) ICD-10 code | 1 | 11 | 1 | 0 | 1 | 3 | 2 | |
| Regarded as temporary hyperglycemia | 0 | 0 | 0 | 0 | 1 | 10 | 0 | |
| No abnormal lab owing to good control of T2DM | 0 | 3 | 0 | 4 | 1 | 4 | 3 | |
| No test of HbA1c and glycoalbumin | 0 | 2 | 0 | 0 | 0 | 1 | 1 | |
| No glucose test | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Treated with exercise or diet therapy | 0 | 14 | 0 | 0 | 0 | 5 | 0 | |
| No T2DM medication (treated in other hospitals) | 0 | 12 | 0 | 0 | 0 | 0 | 0 | |
| Others | 9 | 11 | 0 | 0 | 0 | 0 | 0 |
Some overlapping cases exist between patterns in FN cases of proposed algorithm (B). Nine FN cases of algorithms (A) or (B) were classified as non-T2DM by the rule in the first step.
Discussion
The Importance of Phenotyping Algorithms and Evaluation Metrics Depending on Application Purposes
As mentioned, the proposed hybrid phenotyping framework consists of a rule-based algorithm and an SVM machine learning algorithm. Proposed algorithms (A) and (B) show higher performance compared to algorithms that use only rules or only an SVM. We assume that this framework uses expert knowledge and an SVM complementarily, resulting in higher performance.
Moreover, algorithm (B), which is based on our framework, shows the highest PPV, the highest AUPS with high sensitivity, and the highest AUPS with high PPV; algorithm (A) has the second-highest sensitivity and second-highest AUPS with high sensitivity. Proposed algorithm (A) is therefore appropriate for screening, whereas proposed algorithm (B) is thus the most appropriate for researchers who intend to use output cases identified by algorithms as subjects of clinical research without later screening. This framework is user-friendly and practical because users can develop appropriate phenotyping algorithms in accordance with their objectives simply by changing the tuning at the second step of each algorithm. Outside of our work, such a framework has not yet been discussed. We believe that it is important that users be able to easily develop appropriate phenotyping algorithms in accordance with their objectives.
Comparison With PheKB
According to the results, algorithm (G) does not produce a PPV equivalent to that of PheKB (sensitivity 67.1%, PPV 95.0%).14 It is difficult to compare the performance of algorithm (G) with that of PheKB because of the differences in patient demographics and clinical criteria of T2DM between the United States and Japan.31 To overcome these problems, we propose algorithm (F), which is modified based on data from UT Hospital in Japan. This algorithm excludes T1DM, secondary DM, and temporary hyperglycemia. Approximately 25% of FP cases in the results of algorithm (G) are secondary DM and temporary hyperglycemia, and algorithm (F) achieves a 33% higher PPV than algorithm (G). Algorithm (F) produced a PPV equivalent to that of PheKB. Proposed algorithm (B) resulted in higher PPV than algorithm (F), the modified PheKB algorithm, meaning that proposed algorithm (B) is more appropriate for research subjects.
Analysis of FP and FN Cases and Structured Data Limitations
The error analysis of FP and FN cases is outlined in Table 5. In this section, we discuss the limitations of our data based on these results.
We used only structured data, with which it is difficult to determine the names of patients’ diseases. Physicians in Japan must register diagnoses as billing codes; such diagnostic information is registered for insurance claim, and can differ from patients’ actual clinical condition. Conversely, accurate diagnoses sometimes are not registered when billing codes are not required.
Excluding specific disease, for example T1DM, based on laboratory tests is similarly difficult. T1DM is known to have a low C-peptide; however, T2DM patients with a decrease in insulin secretion can also have a low C-peptide. GAD antibodies, IA2 antibodies or 125I-insulin binding ratios are generally performed only when patients are initially diagnosed. To overcome this problem, the disease names in EHR text or the descriptions of clinical situations from EHR text could help in developing phenotyping algorithms with a higher performance.32,33
Moreover, if a patient has abnormal blood glucose levels a few times in a specific clinical situation, such as postsurgical recovery or shock, abnormal glucose levels are due not to DM, but to temporary hyperglycemia. Such clinical contexts cannot be fully determined using only structured data. The technique for interpretation of clinical text in EHR is required. However, the standardization of natural language processing techniques and the equalization of their accuracy across all languages are very difficult tasks and are currently impractical.
We wanted to determine whether T2DM medication preceded insulin treatment, because this information implies a diagnosis of T2DM as opposed to T1DM. However, we could not use the data because only electronic data from approximately last 6 years from UT Hospital could be used.
The Possibility of Other Machine Learning Methods
The possibility that other machine learning methods outperform SVM is relatively low based on existing studies.18,20 However, our results cannot deny the possibility that other methods would lead to different conclusion (that is, the hybrid algorithm would not outperform the nonhybrid algorithm). We will apply another method to our hybrid framework in the future. The progress of machine learning algorithms is remarkable and machine learning alone may soon show significantly higher performance. If that is the case, however, humans can easily modify rules-based approaches, which is a strength of the hybrid method that will not change.
General Applicability
Our framework was evaluated with patients from 1 university hospital in Japan. We must evaluate this framework with patients at other hospitals in several different countries with different demographics and regional characteristics to assess its general applicability. We also intend to evaluate our framework with other diseases.
Conclusion
We propose a practical T2DM phenotyping framework using expert knowledge and an SVM to develop an algorithm that can be used for both screening and identifying research subjects. Our proposed framework can be used to develop 2 types of phenotyping algorithms by changing the method of tuning SVM: one is tuned for a high sensitivity for screening; the other is tuned for a high PPV for detecting research subject. Both algorithms showed a higher performance than baseline algorithms. The proposed framework is a user-friendly and novel method that each user can employ evaluation metrics appropriate for their objectives.
Footnotes
Abbreviations: AUC, area under the ROC curve; AUPS, area under precision-sensitivity curve; EHR, electronic health record; FN, false negative; FP, false positive; PPV, positive predictive value; ROC, receiver operating characteristic; SVM, support vector machine; T2DM, type 2 diabetes mellitus.
Authors’ Note: This research was approved by the Research Ethics Committee of the Graduate School of Medicine and Faculty of Medicine, University of Tokyo (permission number 10733 [2015]).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by JSPS KAKENHI Grant Number 16J05555.
References
- 1. International Diabetes Federation. IDF diabetes atlas 2015. http://www.diabetesatlas.org/resources/2015-atlas.html. Accessed July 4, 2016.
- 2. Bernard Z, Christoph W, John ML, et al. Empagliflozin, cardiovascular outcomes, and mortality in type 2 diabetes. New Eng J Med. 2015;373(22):2117-2128. [DOI] [PubMed] [Google Scholar]
- 3. Tai ES, Goh SY, Lee JJ, et al. Lowering the criterion for impaired fasting glucose: impact on disease prevalence and associated risk of diabetes and ischemic heart disease. Diabetes Care. 2004;27(7):1728-1734. [DOI] [PubMed] [Google Scholar]
- 4. Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. JAMIA. 2013;20(1):117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. JAMIA. 2014;21(2):221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jie X, Luke VR, Pamela LS, et al. Review and evaluation of electronic health records-driven phenotype algorithm authoring tool for clinical and translational research. JAMIA. 2015;22(6):1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albers DJ, Elhadad N, Tabak E, Perotte A, Hripcsak G. Dynamical phenotyping: using temporal analysis of clinically collected physiologic data to stratify populations. PLOS ONE. 2014;9(6):e96443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCormick N, Lacaille D, Bhole V, Avina-Zubieta JA. Validity of heart failure diagnoses in administrative databases: a systematic review and meta-analysis. PLOS ONE. 2014;9(8):e104519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hogan WR, Wagner MM. Data accuracy in computer-based patient records. JAMIA. 1997;4(5):342-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ariana EA, Wesley TK, April T, Tong L, Jiayang X, Mark SC. Electronic health record phenotyping improves detection and screening of type 2 diabetes in the general United States population: a cross-sectional, unselected, retrospective study. J Biomed Inform. 2016;60:162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rachel LR, Shelley AR, Douglas W, et al. A comparison of phenotype definitions for diabetes mellitus. JAMIA. 2013;20(e2):e319-e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type2 diabetes mellitus. JAMIA. 2012;19(2):219-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nichols GA, Desai J, Elston Lafata J, et al. Construction of a multisite DataLink using electronic health records for the identification, surveillance, prevention, and management of diabetes mellitus: the SUPREME-DM project. Prev Chronic Dis. 2012;9:E110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hebert PL, Geiss LS, Tierney EF, Engelgau MM, Yawn BP, McBean AM. Identifying persons with diabetes using Medicare claims data. Am J Med Qual. 1999;14(6):270-277. [DOI] [PubMed] [Google Scholar]
- 17. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. JAMIA. 2012;19(2):212-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li D, Simon G, Chute CG, Pathak J. Using association rule mining for phenotype extraction from electronic health records. AMIA Annu Symp Proc. 2013;2013;142-146. [PMC free article] [PubMed] [Google Scholar]
- 19. Wu X, Kumar V, Quinlan JR, et al. Top 10 algorithm in data mining. Knowl Inf Syst. 2008;14(1):1-37. [Google Scholar]
- 20. Kawaler E, Cobian A, Peissig P, Cross D, Yale S, Craven M. Learning to predict post-hospitalization VTE risk from EHR data. AMIA Annu Symp Proc. 2012;2012:436-445. [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell SL. HL7 (health level 7)—the future becomes a reality. Health Inform. 1990;7(5):24-26. [PubMed] [Google Scholar]
- 22. Japan Diabetes Society. Evidence-based practice guideline for treatment of diabetes in Japan. 2013. http://www.jds.or.jp/modules/en/index.php?content_id=44. Accessed February 29, 2016.
- 23. Zadrozny B, Langford J, Abe N. Cost-sensitive learning by cost-proportionate example weighting. Third IEEE International Conference on Data Mining 2003;435-442. [Google Scholar]
- 24. Batuwita R, Palade V. Class imbalance learning methods for support vector machines. In: He H, Ma Y, eds. Imbalanced Learning: Foundations, Algorithms, and Applications. New York, NY: John Wiley; 2013:83-100. [Google Scholar]
- 25. Veropoulos K, Campbell C, Cristianini N. Controlling the sensitivity of support vector machines. IJCAI Proc. 1999;55-60. [Google Scholar]
- 26. Berthold MR, Cebron N, Dill F, et al. KNIME: the Konstanz Information Miner. 4th Annual Industrial Simulation Conference 2006;58-61. [Google Scholar]
- 27. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 28. Alexandros K, Alex S, Kurt H. kernlab—an S4 package for kernel methods in R. https://cran.r-project.org/web/packages/kernlab/vignettes/kernlab.pdf. Accessed February 29, 2016.
- 29. Tobias S, Oliver S, Niko B, Thomas L. Visualizing the performance of scoring classifiers. http://rocr.bioinf.mpi-sb.mpg.de/. Accessed February 29, 2016.
- 30. Max K, Jed W, Steve W, et al. Package caret. https://github.com/topepo/caret/. Accessed February 29, 2016.
- 31. Slivio EI, Richard MB, John BB, et al. ADA/EASD: management of hyperglycemia in type 2 diabetes: a patient-centered approach. Diabetic Care. 2012;35(6):1364-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medication from electronic health records provides superior phenotyping performance. JAMIA. 2016;23(e1):e20-e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLOS ONE. 2015;10(8):e136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Information Retrieval System of Japanese Standard Disease-Code Master [in Japanese]. http://www.dis.h.u-tokyo.ac.jp/byomei/. Accessed February 29, 2016. [PMC free article] [PubMed]