Abstract
Delayed or impaired wound healing is a major health issue worldwide, especially in patients with diabetes and atherosclerosis. Here we show that expression of the circular RNA circ-Amotl1 accelerated healing process in a mouse excisional wound model. Further studies showed that ectopic circ-Amotl1 increased protein levels of Stat3 and Dnmt3a. The increased Dnmt3a then methylated the promoter of microRNA miR-17, decreasing miR-17-5p levels but increasing fibronectin expression. We found that Stat3, similar to Dnmt3a and fibronectin, was a target of miR-17-5p. Decreased miR-17-5p levels would increase expression of fibronectin, Dnmt3a, and Stat3. All of these led to increased cell adhesion, migration, proliferation, survival, and wound repair. Furthermore, we found that circ-Amotl1 not only increased Stat3 expression but also facilitated Stat3 nuclear translocation. Thus, the ectopic expressed circ-Amotl1 and Stat3 were mainly translocated to nucleus. In the presence of circ-Amotl1, Stat3 interacted with Dnmt3a promoter with increased affinity, facilitating Dnmt3a transcription. Ectopic application of circ-Amotl1 accelerating wound repair may shed light on skin wound healing clinically.
Keywords: circular RNA, circ-Amotl1, Stat3, Dnmt3a, wound healing, circRNA
Yang et al. show that expression of the circular RNA circ-Amotl1 accelerated wound healing and increased levels of Stat3 and Dnmt3a. The increased Dnmt3a methylated miR-17 promoter, decreasing miR-17-5p levels but increasing fibronectin expression. Furthermore, circ-Amotl1 facilitated Stat3 nuclear translocation to promote cell activities and wound repair.
Introduction
As the largest organ of human body, the skin acts as the first line of protection against environmental hazards. Dysfunctions of the skin’s wound-healing process can result in cosmetic problems, metabolic disorders, and lethal infection. Cutaneous wound healing is a complex biological process that consists of hemostasis, inflammation, re-epithelization, vascularization, and tissue remolding. Delayed or impaired wound healing has been a major public health issue worldwide, especially in patients with diabetes mellitus and vascular atherosclerosis. We recently found that a newly detected class of genetic material circular RNAs (circRNAs) may be crucial in tissue remodeling, because the circRNA circ-Foxo3 plays roles in regulating cell cycle progression, cell senescence, cardiovascular protection, and tumor formation.1, 2, 3, 4 Recent studies have shown that a wide array of endogenous circRNAs are expressed in animal cells, while certain circRNAs are highly specific to cell type and/or developmental stage, suggesting potential roles in developmental regulation.5, 6, 7, 8, 9 Genome-wide analyses have revealed high levels of abundance and evolutionary conservation of circRNAs across species, suggesting specific roles in cellular physiology.9, 10, 11, 12
One mode of action found on some circRNAs is the sponging activity of this class of molecules to bind miRNAs, allowing them to arrest miRNA activity.13, 14, 15 The circRNA CiRS-7 or CDR1as, which is highly expressed in neuronal tissues, possesses many microRNA (miRNA)-binding sites and has been shown to sponge miRNA functions.7 The circRNA SRY, which is highly expressed in murine testes, functions as miR-138 sponge.7, 16 We have recently found that circ-Foxo3, along with the pseudogene of Foxo3, can sponge a number of miRNAs and repress breast cancer development.1 In the present report, we show that the circRNA circ-Amotl1 can accelerate wound healing by binding to Stat3. circ-Amotl1 then facilitated Stat3 nuclear translocation and binding to Dnmt3a promoter, which enhanced Dnmt3a expression and modulated miR-17 function.
Results
Enhanced Wound Healing in Mice Delivered with circ-Amotl1
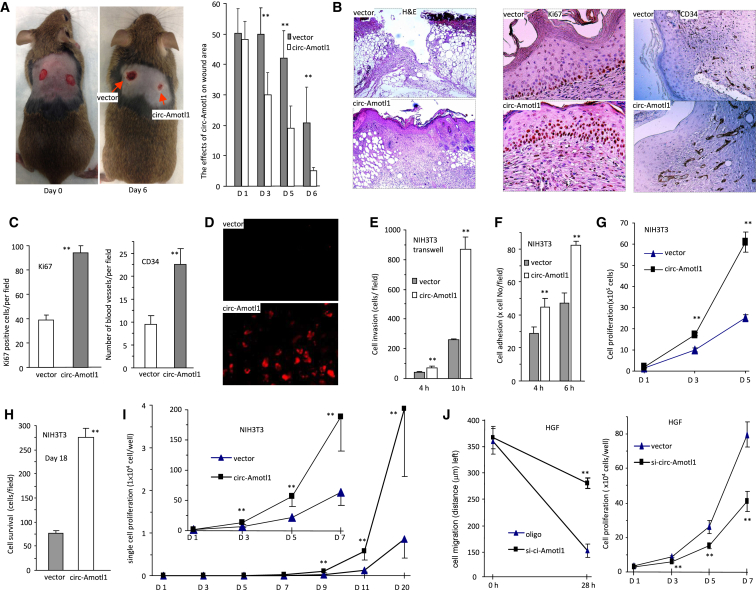
In this study, we explored the potential involvement of circ-Amotl1 in wound repair. C57BL/6xCBA mice were subject to a cervical dermal punch biopsy, which left full-thickness excisional wounds of about 5 mm on both sides of the back. The next day, the wound areas were injected with circ-Amotl1 expression plasmids (Figure S1A) or a control vector at a volume of 100 μL, containing 50 μg plasmids per site. The injection was repeated every other day. The sizes of the wound areas were measured every other day. Six days after wounding, the wounds injected with circ-Amotl1 expression plasmids showed enhanced healing compared with the wounds injected with the vector (Figure 1A, left). Studies have shown that genders and sex steroids might affect tissue repair and regeneration.17 In our studies, both male and female mice injected with circ-Amotl1 displayed accelerated wound healing. The difference in wound area between two groups was statistically significant after 6 days (Figure 1A, right). Measurements of wound area revealed that the ratios of unhealed space (day 6/day 1) were significantly smaller in the group injected with circ-Amotl1 than that in the control (Figure S1B).
Figure 1.
circ-Amotl1 Enhanced Wound Healing, Proliferation, and Migration
(A) Left: wild-type mice were subjected to wound healing assay (n = 10). Pictures were taken from the sixth day after wounding, showing that injection with circ-Amotl1 plasmids enhanced wound healing. Right: graph representing wound sizes during the 6-day healing process, which were measured by multiplying longest length by greatest width (n = 10). (B) Sections of wound tissues from circ-Amotl1- and vector-injected mice were subjected to H&E staining (left) and immunostaining for Ki67 and CD34 expression (right). (C) The numbers of Ki67-positive cells and stained blood vessels were counted and quantified. circ-Amotl1 injection enhanced Ki67 and CD34 expression in the wound tissues (n = 5). (D) In situ hybridization showing expression of circ-Amotl1 in the wound tissues. (E) Transwell migration assay showed that more circ-Amotl1-transfected cells migrated through the membrane than control groups after 4 and 10 hr (n = 5). (F) The cells were incubated in a Petri dish for 4 or 6 hr. More circ-Amotl1-transfected cells adhered to the plates than control (n = 5). (G) Cell proliferation was assayed. Cells transfected with circ-Amotl1 displayed increased proliferation compared with control (n = 5). (H) The cells were maintained in serum-free medium for survival assay. circ-Amotl1 expression increased survival rates after 18 days (n = 5). (I) In single-cell culturing assay, the number of cells per well was counted as indicated for up to 20 days. Cells transfected with circ-Amotl1 displayed increased cell proliferation compared with control (n = 20). (J) Left: wound-healing assay showed that silencing circ-Amotl1 decreased cell migration in HGF cells transfected with circ-Amotl1 siRNA. Right: silencing circ-Amotl1 repressed cell proliferation in HGF cells transfected with siRNA. **p < 0.01. Error bars denote SD (n = 5).
On the seventh day, tissues of the wound were biopsied for histological examination and immunohistochemistry staining. Wound healing is driven by proliferation and migration of cells in the wound, including fibroblasts and myofibroblasts. H&E staining showed that there was increased cell density in the wounds injected with circ-Amotl1, compared with the control wounds (Figure 1B, left). We also found that there were more new blood vessels and Ki67-positive cells generated in the wounds injected with circ-Amotl1, as evaluated by CD34 levels (Figure 1B, right). Quantitation of the numbers of stained blood vessels and Ki67-positive cells showed significant increases in the circ-Amotl1-injected mice (Figure 1C). We confirmed that the circ-Amotl1 plasmids were expressed in the wound by in situ hybridization using a probe specifically recognizing circ-Amolt1 (Figure 1D).
circ-Amotl1 Expression Accelerates Fibroblast Proliferation and Migration
Fibroblasts are known to play essential roles in tissue repair. They move to wound areas upon wound formation and synthesize collagen and fibronectin together with other extracellular matrix (ECM) molecules, generating the force required to contract the wound. To study the function of circ-Amotl1 on fibroblast activities, NIH 3T3 fibroblasts were stably transfected with the plasmids. Real-time PCR was used to confirm the expression of circ-Amotl1 in the transfected cells. There was a significant elevation of circ-Amotl1 in cells transfected with circ-Amotl1 expression plasmids (Figure S1C).
We performed a number of cell activity assays to test the effects of circ-Amotl1 on cell biology associated with wound repair. In cell migration assay, the circ-Amotl1-transfected cells showed a greater ability to migrate compared with the vector-transfected cells (Figure S1D). The locomotion of fibroblasts during wound healing includes migration as well as deformation. Thus, transwell migration assay was performed to test both functions. After being placed above a cell-permeable membrane for 10 hr, more circ-Amotl1-transfected cells migrated through microspores of the membrane (Figures 1E and S1E). In cell adhesion assay, NIH 3T3 cells were incubated in a Petri dish for 4–6 hr. It was found that more circ-Amotl1-transfected cells were able to attach to the surface of the Petri dish (Figure 1F). In proliferation and survival assays, we found that circ-Amotl1 expression increased cell proliferation (Figure 1G) and survival (Figure 1H). We examined the effect of circ-Amolt1 on each cell by performing single-cell culture assay and found that circ-Amotl1 expression greatly promoted cell proliferation (Figure 1I).
The effect of circ-Amotl1 was also performed in the human gingival fibroblast (HGF; named CRL-2014, from ATCC) cell line, which allowed us to obtain stably transfected cell lines and could be maintained for long-term experiments for mechanistic studies. After confirming upregulation of circ-Amotl1 (Figure S2A), we detected increased cell migration (Figure S2B), adhesion (Figure S2C), proliferation (Figure S2D), and survival (Figure S2E). We examined the effect of endogenous circ-Amotl1 by silencing circ-Amotl1 using a small interfering RNA (siRNA) approach (Figure S2F). Silencing endogenous circ-Amotl1 decreased HGF cell migration and proliferation (Figure 1J). It also decreased cell adhesion (Figure S2G) and survival (Figure S2H).
Interaction of circ-Amotl1 with Stat3
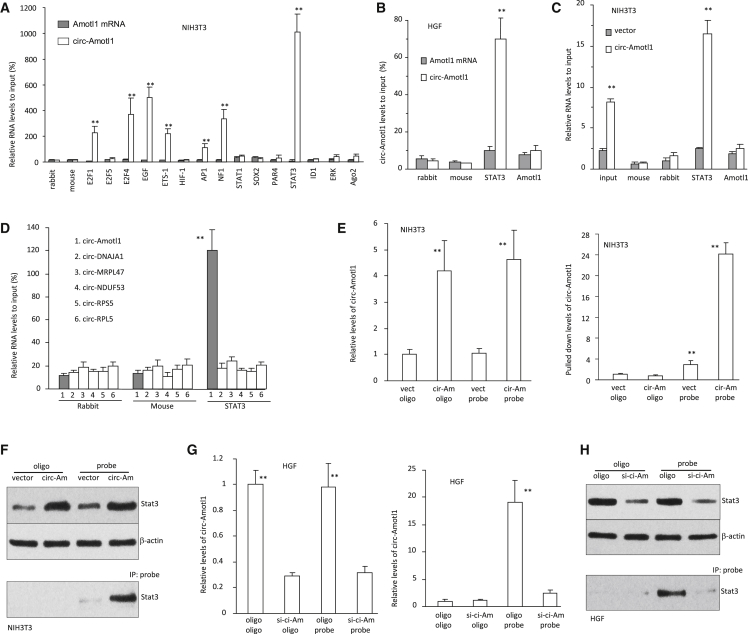
Because circ-Amotl1 greatly promoted cell proliferation, we anticipated that circ-Amotl1 might interact with mitosis-associated proteins. Cell lysis prepared from NIH 3T3 cells were subject to immunoprecipitation for pre-screening with anti-rabbit IgG, mouse IgG, E2F1, E2F5, E2F4, EGF, ETS-1, HIF-1a, AP1, NF1, Stat1, Sox2, Par4, Stat3, ID1, Erk, and Ago2 antibodies, followed by real-time PCR with primers specific for the linear Amotl1 mRNA or circ-Amotl1. The experiment showed that circ-Amotl1 was pulled down by immunoprecipitation experiments with antibodies against E2F1, E2F4, EGF, ETS-1, AP1, NF1, and Stat3, which did not pull down linear Amotl1 mRNA (Figure 2A). The anti-Stat3 antibody pulled down the highest level of circ-Amotl1. Stat3 is a transcription factor-mediating signal transducer and activator of transcription 3. We confirmed this result by including antibody against AMOTL1, which was shown not to bind circ-Amotl1, and found that anti-Stat3 antibody bound only circ-Amotl1 (Figure 2B). We validated these results by preparing RNAs from the vector- and circ-Amotl1-transfected NIH 3T3 fibroblasts and performing pull-down assay. Anti-Stat3 antibody was found to pull down significantly higher levels of circ-Amotl1 from the circ-Amotl1-transfected cells than those from the vector-transfected cells (Figure 2C). To analyze the specificity of the interaction, we examined the levels of other circRNAs, including circ-DNSJA1, circ-MRPL47, circ-NDUF53, circ-RPS5, and circ-PRL5. We found that antibodies against Stat3 did not pull down these circRNAs (Figure 2D), suggesting specific interaction between circ-Amotl1 and Stat3.
Figure 2.
Interaction of circ-Amotl1 with Stat3
(A) Cell lysis prepared from NIH 3T3 cells was subjected to immunoprecipitation with rabbit IgG, mouse IgG, and antibodies as indicated, followed by real-time PCR with primers specific for linear Amotl1 mRNA or circ-Amotl1. Anti-Stat3 antibody pulled down the highest levels of circ-Amotl1 (n = 4). (B) RNAs isolated from HGF cells incubated with rabbit and mouse IgG, or antibody against STAT3 or AMOTL1, were subjected to real-time PCR. Anti-Stat3 antibody pulled down circ-Amotl1 but not Amotl1 mRNA (n = 4). (C) RNAs isolated from NIH 3T3 cells transfected with vector or circ-Amotl1 were subjected to real-time PCR. Anti-Stat3 antibody pulled down more circ-Amotl1 from the circ-Amotl1-transfected cells. (D) RNAs isolated from NIH 3T3 cells were subjected to real-time PCR with primers specific for circular RNAs as indicated. Antibodies against Stat3 pulled down circ-Amotl1 but not the other circular RNAs (n = 4). (E) Left: lysates prepared from NIH 3T3 transfected with circ-Amotl1 or mock control were mixed with biotinylated probes or a control oligo and then subjected to RNA pull-down assays. circ-Amotl1 transfection produced higher levels of circ-Amotl1 relative to control (n = 4). Right: the mixture was subjected to pull-down assays. circ-Amotl1 probe pulled down higher levels of circ-Amotl1 in the cells transfected with circ-Amotl1 (n = 4). (F) Protein lysates prepared from mock- and circ-Amotl1-transfected NIH 3T3 cells were subject to western blot probed with antibody against STAT3. Top: increased STAT3 levels in the circ-Amotl1-transfected cell lysate. Bottom: pull-down of Stat3 by the probe in lysates of the circ-Amotl1-tranbsfected cells. (G) Left: transfection with circ-Amotl1 siRNA silenced circ-Amotl1 expression. Right: real-time PCR showed that circ-Amotl1 probe pulled down less circ-Amotl1 in the cells transfected with circ-Amotl1 siRNA. **p < 0.01. Error bars denote SD (n = 4). (H) The probe pulled down less Stat3 in the siRNA-transfected cells compared with control.
We further tested the interaction between circ-Amotl1 and Stat3. Lysates from NIH 3T3 and HGF cells transfected with circ-Amotl1 or vector were mixed with a biotinylated probe specifically recognizing circ-Amotl1. The mixture was subject to real-time PCR, and the experiments showed that circ-Amotl1-transfected NIH 3T3 cells expressed high levels of circ-Amotl1 (Figure 2E, left). The mixture was subject to a pull-down assay by incubation with streptavidin beads, followed by real-time PCR, and we found that significantly higher levels of circ-Amotl1 were pulled down by the probe relative to the control (Figure 2E, right).
We tested whether the circ-Amotl1 probe was able to pull down Stat3. The probe was added to the lysates prepared from circ-Amotl1-transfected cells. Western blot analysis confirmed that transfection with circ-Amotl1 increased Stat3 levels (Figure 2F). The mixtures were incubated with streptavidin beads followed by western blotting, which showed that pulling down circ-Amotl1 precipitated Stat3 from cells transfected with circ-Amotl1 (Figure 2F).
To corroborate the interaction, we used a siRNA approach to knock down circ-Amotl1. Lysates prepared from HGFs transfected with circ-Amotl1 siRNA or a control oligo were mixed with the probes. Real-time PCR confirmed that transfection with circ-Amotl1 siRNA decreased circ-Amotl1 levels (Figure 2G, left). In the presence of the probe, significantly lower levels of circ-Amotl1 were pulled down (Figure 2G, right). In co-immunoprecipitation assay, pulling down circ-Amotl1 precipitated decreased levels of Stat3 from the circ-Amotl1 siRNA-transfected cells (Figure 2H).
Cross-Talk between circ-Amotl1 and Stat3
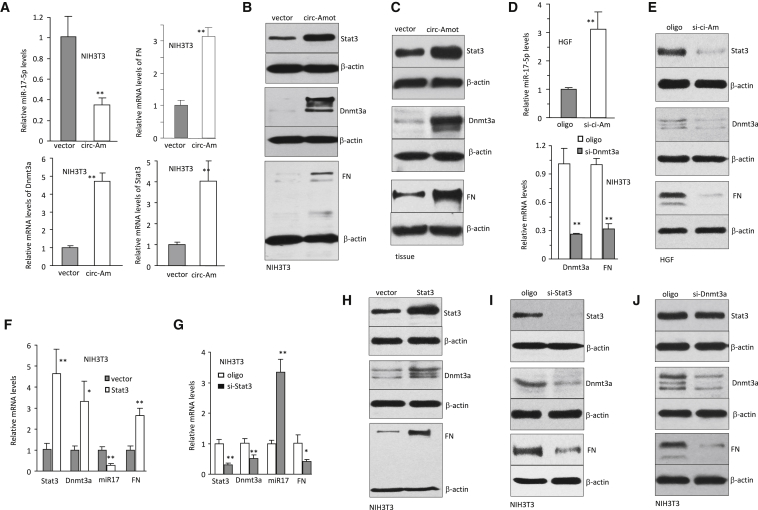
We explored how Stat3 might be mediating circ-Amotl1 function in cell migration and proliferation by screening potential targets of Stat3. Computational algorithm (http://bio.sz.tsinghua.edu.cn) prediction indicated that miR-17-5p was an excellent candidate in regulating Stat3 expression (Figure S2I). Our previous studies also showed that miR-17-5p could target fibronectin18 and Dnmt3a.19 We analyzed expression of miR-17-5p, Stat3, Dnmt3a, and fibronectin in the circ-Amotl1-transfected cells. We found that although miR-17-5p levels decreased, expression of Stat3, Dnmt3a, and fibronectin was upregulated in mRNA (Figure 3A) and protein levels (Figure 3B). In the wounds injected with circ-Amotl1, expression of Stat3, Dnmt3a, and fibronectin was also upregulated (Figures 3C and S2J). Consistently, silencing circ-Amotl1 increased miR-17-5p, and silencing Dnmt3a decreased fibronectin mRNA levels (Figure 3D). Silencing circ-Amotl1 decreased protein levels of Stat3, Dnmt3a, and fibronectin (Figure 3E).
Figure 3.
Crosstalk between circ-Amotl1 and Stat3
(A) In real-time PCR, downregulation of miR-17-5p and upregulation of fibronectin, Dnmt3a, and Stat3 levels were detected in the circ-Amotl1-transfected cells. **p < 0.01. Error bars denote SD (n = 4). (B) Transfected with circ-Amotl1 increased the expression of Stat3, Dnmt3a, and fibronectin (FN). (C) circ-Amotl1 injection increased the expression of Dnmt3a, fibronectin, and Stat3 in the wound tissues. (D) Top: real-time PCR showed that silencing circ-Amotl1 increased miR-17-5p expression. Bottom: silencing Dnmt3a decreased expression of Dnmt3a and fibronectin. (E) Western blotting confirmed that silencing circ-Amotl1 repressed expression of Stat3, Dnmt3a, and fibronectin. (F) Real-time PCR showed that ectopic expression of Stat3 increased the levels of Stat3, Dnmt3a, and fibronectin but decreased the levels of miR-17-5p. (G) Real-time PCR showed that silencing Stat3 decreased levels of Stat3, Dnmt3a, and fibronectin but increased levels of miR-17-5p. (H) Western blotting confirmed that ectopic expression of Stat3 increased expression of Dnmt3a and fibronectin. (I) Western blotting confirmed that silencing Stat3 decreased expression of Dnmt3a and fibronectin. (J) Western blotting confirmed that silencing Dnmt3a repressed expression of fibronectin but had no effect on Stat3 expression.
We examined whether Dnmt3a and fibronectin were downstream molecules of Stat3. Real-time PCR analysis indicated that ectopic expression of Stat3 increased levels of Dnmt3a and fibronectin but decreased miR-17-5p expression (Figure 3F). Silencing endogenous Stat3 produced the opposite effect (Figure 3G). Western blot analysis confirmed that transfection with Stat3 increased Dnmt3a and fibronectin expression (Figure 3H), but silencing Stat3 decreased these protein levels (Figure 3I). Silencing Dnmt3a could clearly downregulated fibronectin expression (Figure 3J).
Effects of miR-17-5p on Wound Repair and Stat3 Expression
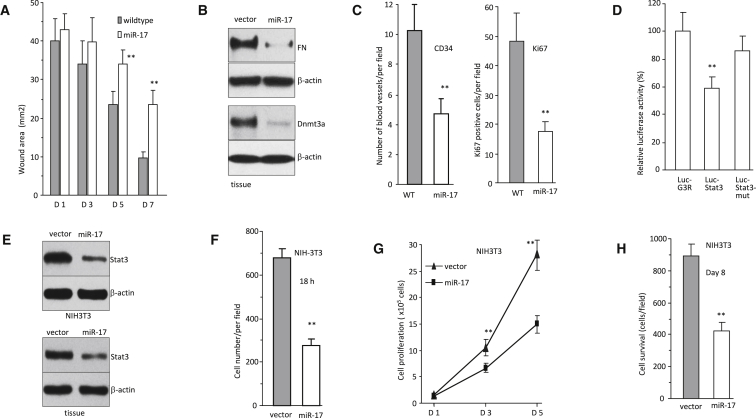
Because we have developed miR-17-transgenic mice in C57BL/6xCBA,18, 19 we used these mice, miR-17-transgenic and wild-type from the same litters, to explore the involvement of miR-17-5p in Stat3-mediated circ-Amotl1 function. Both male and female mice injected with circ-Amotl1 displayed accelerated wound healing. After confirmation of miR-17-5p upregulation (Figure S3A), the mice were subjected to a cervical dermal punch wounds of about 5 mm on both sides of the back. The sizes of the wound areas were measured as above. It appeared that the miR-17-transgenic mice showed delay in wound healing (Figure S3B). The difference in wound area between two groups was statistically significant (Figure 4A). The ratios of unhealed space (day 5/day 1 and day 7/day 1) were significantly greater in the miR-17-transgenic mice than that in the wild-type mice 5 days after wounding (Figure S3C).
Figure 4.
Effects of miR-17-5p on Wound Healing and Stat3 Expression
(A) The miR-17-transgenic and wild-type (from the same letters) mice were subject to wound healing assay (n = 10). Graphical representation of wound sizes showing that miR-17 transgenic mice had larger wounds than wild-type mice after 5 and 7 days. (B) The wound tissues were subject to western blotting to confirm repression of fibronectin and Dnmt3a in the miR-17 transgenic mice. (C) Quantitation analysis showed decreased levels of CD34 and Ki67 in the miR-17 transgenic mice. **p < 0.01. Error bars denote SD (n = 5). (D) Decreased luciferase activities were observed in cells co-transfected with miR-17-5p and Luc-stat3, which was reversed when the miR-17-5p binding site was mutated (Luc-stat3-mut) (n = 4). (E) Protein lysates from NIH 3T3 cells transfected with miR-17-5p and control oligo (top) or the wound tissues of miR-17 transgenic mice and wild-type mice (bottom) were subjected to immunoblotting. MiR-17 expression decreased Stat3 levels. (F) MiR-17 expression decreased cell migration in the trans-well assay compared with control (n = 5). (G) Ectopic expression of miR-17-5p repressed cell proliferation (n = 5). (H) In serum-free medium, miR-17 expression decreased cell survival (n = 5).
On the seventh day, the wounded tissues were biopsied for western blotting, histological examination, and immunohistochemistry staining. H&E staining showed that there was decreased cell density in the wounds of miR-17-transgenic mice, compared with the wild-type (Figure S3D). Western blot analysis confirmed repression of fibronectin and Dnmt3a (Figure 4B). We also found that there were fewer CD34- and Ki67-positive cells in the miR-17 transgenic mice (Figure S3E). Quantitation of the numbers of stained blood vessels and Ki67-positive cells showed significant decrease in the miR-17-transgenic mice (Figure 4C).
In our previous studies, we showed that expression of miR-17-5p repressed expression of Dnmt3a and fibronectin.18, 19 Our results reported here suggested that Stat3 was a target of miR-17-5p. We tested this by generating luciferase constructs harboring the 3′UTR of Stat3. Site-directed mutagenesis was also performed in the miR-17-5p binding site. Luciferase assay indicated that luciferase activity was repressed when the cells were co-transfected with the luciferase construct and miR-17-5p (Figure 4D). The inhibitory effect was abolished when the miR-17-5p binding site was mutated. Western blotting showed that in the cells stably transfected with miR-17 (Figure S3F, confirming upregulation of miR-17-5p), levels of Stat3 were repressed (Figure 4E). The effects of miR-17 transfection were found to decrease 2D (Figure S3G) and 3D (Figures 4F and S3H) cell migration, cell proliferation (Figure 4G), and survival (Figure 4H). We confirmed that expression of Stat3, Dnmt3a, and fibronectin was downregulated in the miR-17-transgenic mice (Figure S3I).
Mutual Regulation of circ-Amotl1, Stat3, Dnmt3a, Fibronectin, and miR-17-5p
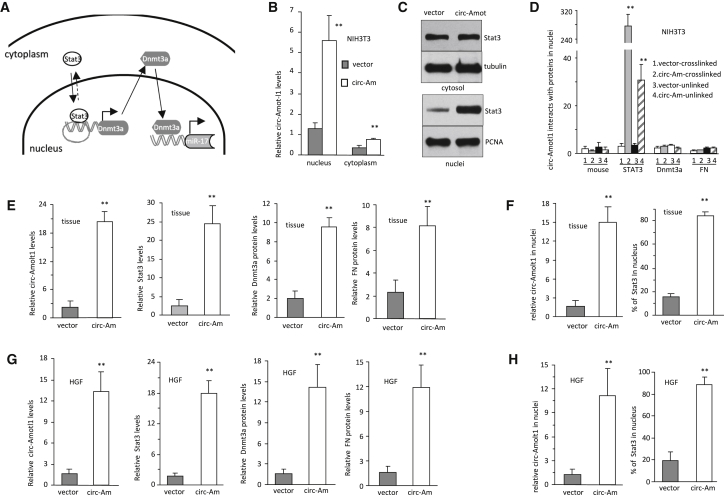
We then designed experiments to address how circ-Amotl1 increased Dnmt3a expression. Because Stat3 is a transcription factor, we proposed that circ-Amotl1 bound to Stat3 and Dnmt3a promoter, facilitating Dnmt3a transcription (Figure 5A). Experiments were conducted to validate this proposal. We found that the expressed circ-Amotl1 was translocated mainly to the nucleus (Figure 5B), and the upregulated Stat3 was seen mainly in the nucleus (Figure 5C). In the crosslinking assays, we found that circ-Amotl1 was precipitated by anti-Stat3 antibody but not the other antibodies (Figure 5D).
Figure 5.
Effects of circ-Amotl1 on Stat3 Nuclear Translocation
(A) A diagram showing that expression of circ-Amotl1 enhanced Stat3 nuclear translocation, which promoted Dnmt3a transcription. Increased Dnmt3a expression repressed miR-17 transcription. (B) Real-time PCR showed that significantly higher levels of circ-Amotl1 were detected in the nuclei compared with the cytoplasm (n = 4). (C) Western blotting showed that ectopic expression of circ-Amotl1 increased Stat3 levels in the nucleus but not in the cytoplasm. (D) Cell nuclear lysis prepared from NIH 3T3 cells was subjected to immunoprecipitation, followed by real-time PCR. Anti-Stat3 antibody pulled down significantly higher levels of circ-Amotl1 when the samples were pre-treated with crosslinking (n = 6). (E) Quantitation of circ-Amotl1, Stat3, Dnmt3a, and fibronectin in wound tissues injected with circ-Amotl1 or the control plasmid. (F) Quantitation of nuclear translocation of circ-Amotl1 and Stat3 in the wound tissues. (G) Quantitation of circ-Amotl1, Stat3, Dnmt3a, and fibronectin in HGF cells transfected with circ-Amotl1 or the vector. (H) Quantitation of nuclear translocation of circ-Amotl1 and Stat3 in HGF cells.
Confocal microscopic analysis detected mainly nuclear distribution of Stat3 in the circ-Amotl1-injected wound tissues (Figure S4A). Using a probe specifically recognizing circ-Amotl1, we detected co-localization of circ-Amotl1 and Stat3 in the nuclei of the circ-Amotl1-injected tissues. Expression of Dnmt3a and fibronectin increased in the wound tissues delivered with circ-Amotl1 (Figure S4B). Quantification analysis showed that expression of circ-Amotl1, Stat3, Dnmt3a, and fibronectin increased in the wound tissues delivered with circ-Amotl1 (Figure 5E). The levels of circ-Amotl1 and Stat3 were significantly higher in the nuclei of the circ-Amotl1-treated tissues relative to the vector-treated tissues (Figure 5F). In the circ-Amotl1-transfected cells, we detected increased expression of circ-Amotl1, Stat3, Dnmt3a, and fibronectin (Figures 5G and S5A), and nuclear translocation of circ-Amotl1 and Stat3 (Figures 5H and S5B).
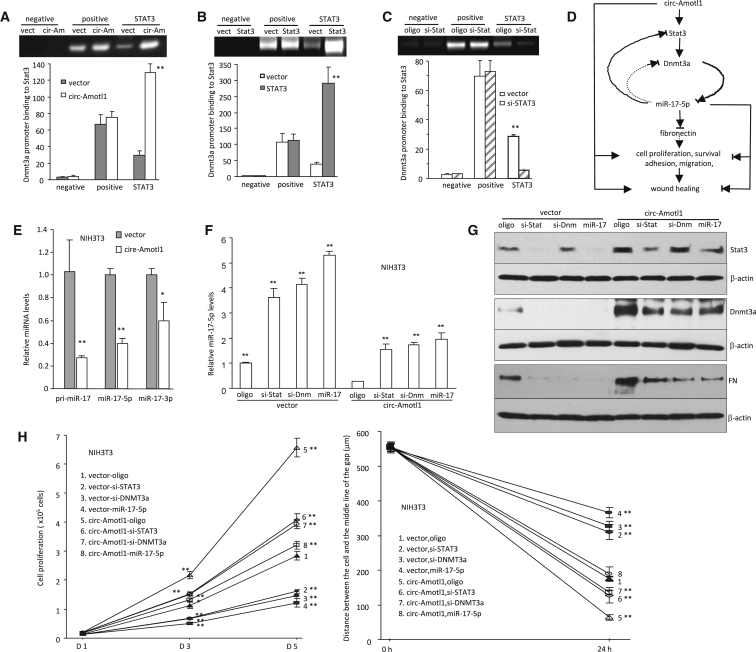
We tested the interaction between Stat3 and Dnmt3a promoter in the cells transfected with circ-Amotl1 or the vector. Chromatin was isolated, digested, and immunoprecipitated with antibodies against Stat3, Histone H3 (serving as a positive control), and rabbit IgG (serving as a negative control), followed by PCR with primers flanking a fragment of DNA in the Dnmt3a promoter. It showed that antibody against Stat3 pulled down significantly higher levels of Dnmt3a promoter in the presence of circ-Amotl1 than in the control (Figure 6A). We further examined the effect of Stat3 on the interaction with Dnmt3a promoter by ectopic expression of Stat3 expression construct. Ectopic expressed Stat3 increased pulling down the DNA promoter (Figure 6B). Silencing endogenous Stat3 decreased pulling down the promoter (Figure 6C).
Figure 6.
Effect of circ-Amotl1 on Wound Healing
(A) Top: increased interaction of Stat3 with Dnmt3a promoter in the circ-Amotl1-transfected cells. Bottom: quantitation analysis (n = 4). (B) Increased interaction of Stat3 with Dnmt3a promoter in the Stat3-transfected cells. (C) Decreased interaction of Stat3 with Dnmt3a promoter in the cells transfected with Stat3 siRNA (n = 4). (D) The pathway by which circ-Amotl1 affected wound healing. (E) Real-time PCR showed that expression of circ-Amotl1 decreased levels of pri-miR-17, miR-17-5p, and miR-17-3p compared with the vector. *p < 0.05, **p < 0.01. Error bars denote SD (n = 5). (F) circ-Amotl1- and vector-transfected NIH 3T3 cells were transfected with control oligo, miR-17-5p, Stat3 siRNA, and Dnmt3a siRNA. Transfection with miR-17-5p or silencing Stat3 and Dnmt3a repressed expression of miR-17-5p in the circ-Amotl1-transfected cells. (G) Western blot showed that transfection with miR-17-5p or silencing Stat3 and Dnmt3a increased expression of Stat3, Dnmt3a, and fibronectin in the circ-Amotl1-transfected cells relative to the vector-transfected cells. (H) Upregulation of Stat3, Dnmt3a, and fibronectin in the cells transfected with circ-Amotl1 displayed higher rates of proliferation (left) and migration (right).
On the basis of the above results, we proposed that ectopic expression of circ-Amotl1 increased Stat3 expression. Increased Stat3 levels enhanced Dnmt3a expression. Dnmt3a could then methylate miR-17 gene promoter. Because Stat3, Dnmt3a, and fibronectin are the targets of miR-17-5p, decreased miR-17-5p levels would increase expression of fibronectin, Dnmt3a, and Stat3. All of these activities led to increased cell proliferation, survival, migration, and wound repair (Figure 6D). A few key experiments were performed to further validate this proposed mode of action. We examined whether ectopic expression of circ-Amotl1 would decrease primary miR-17, miR-17-5p, and miR-17-3p, and our experiment confirmed this (Figure 6E). We transfected the circ-Amotl1- and vector-stable cell lines with Dnmt3a siRNA, Stat3 siRNA, miR-17-5p, or a control oligo. Real-time PCR analysis indicated that the miR-17-5p levels were upregulated in the cells transfected with siRNAs targeting Stat3 or Dnmt3a (Figure 6F). We also found upregulation of Stat3, Dnmt3a, and fibronectin in the cells transfected with circ-Amotl1 (Figure 6G). These cells also displayed higher rates of proliferation and migration relative to the controls (Figure 6H).
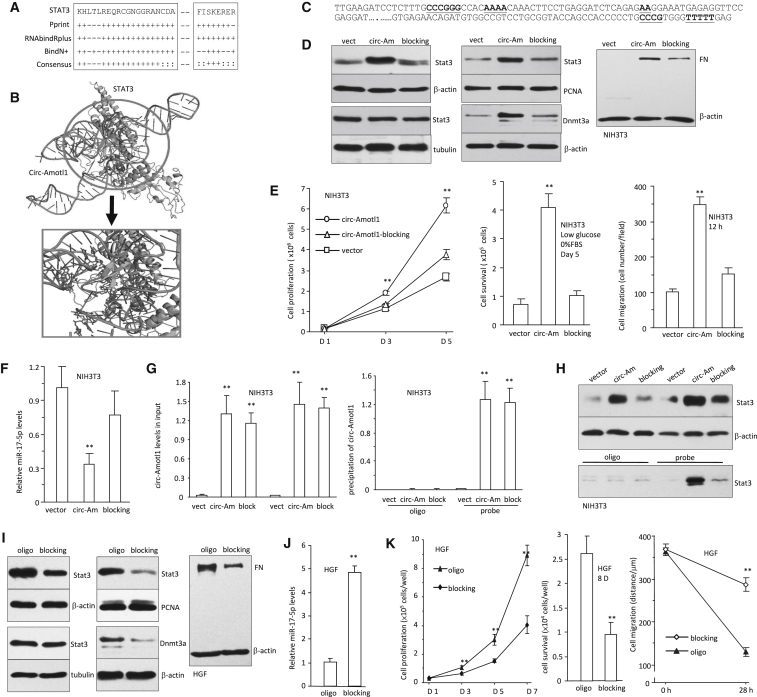
Computational Analysis of circ-Amotl1 Interacting with Stat3
In silico, we found that Stat3 shared consensus sequences with other RNA-binding proteins (Figure 7A). The predicted secondary structure of circ-Amotl1 was obtained by analyzing its thermodynamic properties using the formula ΔG = ΔH − TΔS, where ΔG = −14.70 kcal/mol at 37°C, ΔH = −380.00 kcal/mol, and ΔS = −1,177.8 cal/(K·mol), T (K) is the absolute temperature, and ΔG, ΔH, and ΔS denote the changes in free energy, enthalpy, and entropy, respectively. The secondary structure delineated in dot bracket notation was then analyzed using the software RNAComposer for tertiary structure prediction. Finally, NPDock was used to carry out the in silico molecular docking of circ-Amotl1 with Stat3 (Figures 7B and S6A). The Stat3 used in the docking procedure was derived from Protein Data Bank (PDB) entry 1BG1. The molecular simulation result supports that circ-Amotl1 could perfectly dock Stat3 and predicts a minimal binding region of circ-Amotl1 for Stat3: ‘AACCTTCAC’‘AAC’‘AGGAAGAA’. The contact map (Figure S6B), the residue-level resolution contact maps (Figure S6C), the MC score (Figure S6D), the contact distance (Table S1), the accessible surface area (Table S2), and the interaction overview (Table S3) all supported the conclusion that circ-Amotl1 sufficiently docked Stat3.
Figure 7.
circ-Amotl1 Interacting with Stat3
(A) Prediction of probable RNA-binding residues of Stat3 was carried out by submitting the Stat3 sequence to Pprint, RNABindRPlus, and BindN+ servers. Plus signs indicate the predicted RNA-binding residues. (B) Graphical representation of 3D structures of the docking models of circ-Amotl1 with Stat3 and zoomed-in images of the binding interface done by NPDcok (color graphic provided in Figure S6A). (C) circ-Amotl1 sequence labeling Stat3 binding site (red). (D) Delivery of the blocking oligo repressed expression of Stat3, Dnmt3a, and fibronectin and nuclear translocation of Stat3. (E) Delivery of the blocking oligo decreased cell proliferation (left), survival (middle), and migration (right). (F) Expression of miR-17-5p was enhanced by the blocking oligo. (G) In the presence of the blocking oligo, expression of circ-Amotl1 was not affected (left), and the probe could still precipitate equal amounts of circ-Amotl1 (right). (H) In the presence of the blocking oligo, the probe pulled down decreased levels of Stat3. (I) The blocking oligo inhibited expression of Stat3, Dnmt3a, and fibronectin and nuclear translocation of Stat3. (J) The blocking oligo increased miR-17-5p expression. (K) Delivery of the blocking oligo decreased cell proliferation (left), survival (middle), and migration (right).
To evaluate the effects of the binding sites on mediating circ-Amotl1 functions, we designed blocking oligo complementary to the binding sites (Figure 7C). NIH 3T3 cells expressing circ-Amotl1 were transfected with blocking oligo inhibiting circ-Amotl1-Stat3 interaction, followed by western blotting. We detected decreased levels and nuclear translocation of Stat3 (Figure 7D). As well, expression of Dnmt3a and fibronectin decreased. The cells transfected with the blocking oligo also displayed decreased proliferation, survival, and migration (Figure 7E) but increased levels of miR-17-5p (Figure 7F) relative to circ-Amotl1-transfected cells. We confirmed that the probe pulled down equal amounts of circ-Amotl1 (Figure 7G), but it pulled down a decreased amount of Stat3 (Figure 7H) in the presence of the blocking oligo.
To corroborate these results, we delivered the blocking oligo or the control oligo into HGF cells to test the effect of endogenous circ-Amotl1. We found that transfection with the blocking oligo decreased levels and nuclear translocation of Stat3 and decreased expression of Dnmt3a and fibronectin (Figure 7I) but increased miR-17-5p expression (Figure 7J). Delivery of the blocking oligo decreased proliferation, survival, and migration (Figure 7K).
Discussion
In this study, we demonstrated that delivery of a circRNA circ-Amotl1 enhanced wound repair by promoting cell proliferation, survival, adhesion, and migration. The increased cell activities occurred through circ-Amotl1 enhancing nuclear translocation of Stat3. Nuclear Stat3 could then bind to Dnmt3a promoter, facilitating transcription and translation of Dnmt3a. Dnmt3a induced methylation of miR-17 promoter, decreasing miR-17-5p expression. We have previously identified Dnmt3a and fibronectin as the targets of miR-17-5p.1, 18 In the present study, we identified Stat3 as the miR-17-5p target. Thus, decreased expression of miR-17-5p promoted protein expression of these targets, which was validated by a number of experiments including western blotting, luciferase assay, and immunostaining. Increased expression of these proteins led to promotion of cell proliferation, survival, adhesion, and migration, which led to acceleration of wound repair.
circRNAs have recently emerged as a novel class of non-coding RNAs. Their biological functions are largely unknown. Several studies have shown that circRNAs can function as sponges to regulate miRNA activities.1, 7, 13, 15 We have also found that circRNA circ-Foxo3 repressed cell cycle progression by binding to p21 and CDK24 and enhanced cell senescence by binding to a number of proteins associated with stress and senescent responding factors.2 Although circRNAs have been detected in mammalian cells for a few years, the number of circRNAs has been found to be more than that of mRNAs. It is conceivable that circRNAs may play roles in regulating a variety of physiological activities. Our study reported here shows that delivery of circ-Amotl1 to wound tissues accelerated wound repair. Our mouse wound healing model did not contain the splinting step, compared with the mouse excisional wound splinting model. This would facilitate wound healing faster, allowing us to measure the effect of circ-Amotl1 more effectively. In the mouse excisional wound splinting model, much more time is needed to heal the wound, which would require many more injections of the plasmids and could generate greater deviation technically. In future studies using transgenic mice overexpressing circRNAs, the mouse excisional wound splinting model would better mimic human skin wound healing.
Wound repair is achieved by complex and active physiological processes.20, 21, 22 In the wound, leukocytes are responsible for immune response, while keratinocytes generate epithelial cover, and fibroblasts produce contractile forces between cell-cell and cell-ECM conjunctions.23, 24 These cells are actively taking up nutrition from the environment and producing building blocks for the healing process.25 This allows effective uptake of the delivered circRNA-expressing plasmids, and we did detect extensive expression of circ-Amotl1 in the wound tissues. Our model appears to be an ideal one for tissue regeneration, especially for topical application to the wound. Because the expressed product is a natural component of the cell, this approach may have few side effects on the body.
One important aspect of our study is that each circ-Amotl1-transfected NIH 3T3 fibroblast could grow well in single-cell culture relative to the vector control. We developed a single-cell culture experiment to mimic the early stage of wound repair, when the cells are sparse and most of the cells are far away from one another. We found that the vector-transfected fibroblasts either died or grew slowly when they were maintained individually one cell per well. However, most of the circ-Amotl1-transfected cells could survive and grew fast. This acquired property allowed the wound to heal effectively.
In looking for protein that might be mediating circ-Amotl1 effects, we performed a binding assay with antibodies against a number proteins involved in the pathways of cell proliferation, survival, adhesion, and migration. It was found that Stat3 appeared to bind circ-Amotl1 with highest activity, as Stat3 plays important role in regulating cell activity.26, 27, 28 Because several other proteins, including E2F1, E2F4, FGF, ETS-1, and NF1, could also bind to circ-Amotl1 significantly, we cannot exclude the possibility that some of these proteins may also play roles in mediating the role of circ-Amotl1 in wound healing. This awaits future investigation. Nevertheless, the interaction of circ-Amotl1 with Stat3 appeared to be specific, as other circRNAs tested could not bind to Stat3.
Our results that circ-Amotl1 could regulate miR-17-5p expression and that miR-17-5p could repress Stat3 levels, resulting in downregulation of circ-Amotl1 function, have added an additional layer to understanding of the mutual regulation of these two classes of non-coding RNAs. Previous studies have shown that circRNAs can sponge miRNAs and decrease their activities, but it is not known whether miRNAs can regulate circRNA activities. Our results show a feedback loop in the mutual regulation of Stat3 and miR-17-5p expression, suggesting a mutual regulation of circ-Amotl1 and miR-17-5p functions.
Materials and Methods
Materials
The monoclonal or polyclonal antibodies against Ki67 (550609; BD Pharmingen), CD34 (ab81289; Abcam), Dnmt3a (ab23565; Abcam), fibronectin (ab2413; Abcam) were obtained from different sources as indicated. Horseradish peroxidase-conjugated goat anti-mouse IgG and horseradish peroxidase-conjugated goat anti-rabbit IgG were obtained from Bio-Rad. RNA and DNA extraction kits and RNA RT and PCR kits were obtained from QIAGEN. Immunoblotting was performed using the ECL western blot detection kit (Amersham Biosciences). Protein A-Sepharose 4B Conjugate and Dynabeads MyOne Streptavidin C1 magnetic beads were obtained from Invitrogen.
In Vivo Wound Healing Experiment with circ-Amotl1
A circ-Amotl1 expression construct was generated by our lab. The plasmids contained a Bluescript backbone, a CMV promoter driving circ-Amotl1 or a non-related sequence serving as a control, and the GFP expression unit. All primer sequences used are listed in Figure S6E.
At 4 weeks of age, 10 miR-17 transgenic and 10 wild-type mice of both genders were subject to skin biopsy using a punch (Miltex) of 5 mm in diameter. The transgenic mice were generated by our lab as previously described.18 A pair of full-thickness, excisional wounds were created on the dorsal region of each mouse. Wound size was measured by multiplying longest length by greatest width, and all mice were sacrificed 1 week afterward. Tissue samples were collected for further study. Using similar procedure, we also created wounds followed by injection of circ-Amotl1 plasmids or a control vector at a volume of 100 μL containing 50 μg plasmids per site. All experimental procedures were approved by the Animal Care Committee of Sunnybrook Research Institute and followed current law regarding animal protection.
Synthesis of the deliver complexes (plasmid-PEG-Au NP) was performed as previously described.29, 30, 31, 32 Briefly, 500 μg circ-Amotl1 plasmid was dissolved in 800 μL of RNase-free water. mPEGSH (PG1-TH-2k; Nanocs) was mixed with plasmids (1:20 molar ratio). Then, 10 nm gold nanoparticles (AuNP; Cytodiagnostics) were mixed with plasmid-PEG at a weight ratio of 1:1 for conjugation. The mixture was gently shaken at 60°C for 30 min and then transferred into a syringe for local injection into the wound tissues.
Cell Adhesion and Migration Assays
In adhesion assays, 1 × 106 cells were seeded in Petri dishes with 10% or 25% FBS/DMEM and maintained at 37°C. Pictures were taken in randomly selected high-power fields under an inverted light microscope (Carl Zeiss Microscopy), and cell numbers were counted at 4 or 6 hr. Cell migration was performed by wound healing and transwell assays. In wound healing assays, NIH 3T3 fibroblasts were seeded in 6-well plates at a density of 1 × 106 cells/well for 12 hr. To diminish the influence of proliferation, the cells were treated with Mitomycin C (Sigma-Aldrich) at 10 μg/mL for 2 hr before being changed to serum-free media. The cultures were then scraped linearly with 200 μL micropipette tips (BioMart). At 0, 18, and 24 or 30 hr, cell migration patterns were recorded by inverted light microscopy, and the migration distance was measured and quantified. In order to test cell motility in a 3D way, PET track-membrane (Coster; Sigma-Aldrich) was placed in 24-well tissue culture dishes, and 1 × 105 cells in 100 μL media without FBS were loaded into the upper part of the membrane. The lower portions of the wells were filled with 800 μl DMEM containing 10% FBS. After incubation at 37°C for 4, 10, 12, or 18 hr, nonmigrated cells were removed with a cotton swab, and invaded cells were fixed by 100% methanol for 30 min, followed by staining with Coomassie Brilliant Blue (Bio-Rad) for 10 min. Photographs were taken under an inverted light microscope.
Cell Proliferation and Survival Assays
Cells (NIH 3T3, 1 × 105 cells; HGF, 2 × 104 cells) were seeded in 6-well or 12-well cell culture plates with 10% or 25% FBS/DMEM and maintained at 37°C for 5 or 7 days. Cell number was counted every other day.33 Single-cell proliferation was performed as previously described.34 To test cell survival, 1 × 105 cells were seeded in 6-well dishes, cultured in 10% (NIH 3T3 cells) or 25% (HGF cells) FBS/DMEM, and maintained at 37°C. After 12 hr of culture, the medium was removed, and the cultures were washed with PBS two times, followed by addition of serum-free DMEM. Pictures were taken in randomly selected high-power fields under an inverted light microscope, and cell number was counted on day 5, 8, or 18.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was performed using the SimpleChIP chromatin IP kit (Cell Signaling Technology) according to the manufacturer’s instructions. Briefly, cells were treated with formaldehyde solution, and the chromatin was isolated, digested, and immunoprecipitated with antibody against Stat3. A total of 10% of the inputs were used for immunoblotting. The captured chromatin was eluted, crosslinking was reversed, and the DNA was recovered. ChIP DNA was subject to PCR using specific primers flanking a DNA sequence in the promoter region of Dnmt3a.
Protein and RNA Analyses
For preparation of nuclei, cells were lysed in 1 mL nuclear extraction buffer (20 mM HEPES, pH 7.2, 10 mM KCl, 2 mM MgCl2, and 10 μL PI) and homogenized with a prechilled Dounce homogenizer with 20 strokes. The lysis was centrifuged at 4,200 rpm for 5 min. The pellet was washed with PBS five times and resuspended in 100 μL lysis buffer containing 0.5 M NaCl. After centrifugation at 13,000 rpm for 10 min, the supernatants containing nuclear extract were collected for analysis.
Protein analysis on western blotting was performed as previously described.35 Immunohistochemistry was performed in tissue sections as previously described.30, 36 Real-time PCR analysis of RNAs was performed as previously described.1, 37 The procedures for protein precipitating RNA and RNA pull-down proteins were performed as previously described.19 Fluorescence in situ hybridization (FISH) was performed as previously described.38 NIH 3T3 cells were seeded in 12-well tissue culture dishes at a density of 1 × 105 cells/well. The cells were co-transfected with the luciferase reporter constructs and miR-17-5p duplex, using Lipofectamine 2000. Luciferase activity was performed as previously described.35
Statistical Analysis
All experiments were performed in triplicate, and numerical data were subject to independent-samples t tests. Levels of significance were set at *p < 0.05 and **p < 0.01.
Author Contributions
Z.-G.Y., W.W.D., Y.Z., J.L., and S.G. performed the experiments. F.M.A. performed computational analysis. Z.-G.Y., W.W.D., W.Y., and B.B.Y. designed the project and experiments. Z.-G.Y., W.Y., and B.B.Y. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (grant PJT-149083) to B.B.Y., who is the recipient of a Career Investigator Award (CI7418) from the Heart and Stroke Foundation of Ontario. Z.-G.Y. is supported by a scholarship from the China Scholarship Council. W.W.D. is supported by a postdoctoral fellowship from the Breast Cancer Foundation of Ontario. F.M.A. is supported by the Higher Education Commission of Pakistan.
Footnotes
Supplemental Information includes six figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.05.022.
Supplemental Information
References
- 1.Yang W., Du W.W., Li X., Yee A.J., Yang B.B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 2.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 3.Xie Y.Z., Yang F., Tan W., Li X., Jiao C., Huang R., Yang B.B. The anti-cancer components of Ganoderma lucidum possesses cardiovascular protective effect by regulating circular RNA expression. Oncoscience. 2016;3:203–207. doi: 10.18632/oncoscience.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobi T., Czaja-Hasse L.F., Reinhardt R., Dieterich C. Profiling and validation of the Circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J., Ao Y. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci. Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 17.Shu Y.Y., Maibach H.I. Estrogen and skin: therapeutic options. Am. J. Clin. Dermatol. 2011;12:297–311. doi: 10.2165/11589180-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 18.Shan S.W., Lee D.Y., Deng Z., Shatseva T., Jeyapalan Z., Du W.W., Zhang Y., Xuan J.W., Yee S.P., Siragam V., Yang B.B. MicroRNA miR-17 retards tissue growth and represses fibronectin expression. Nat. Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 19.Du W.W., Yang W., Xuan J., Gupta S., Krylov S.N., Ma X., Yang Q., Yang B.B. Reciprocal regulation of miRNAs and piRNAs in embryonic development. Cell Death Differ. 2016;23:1458–1470. doi: 10.1038/cdd.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al Haj Zen A., Nawrot D.A., Howarth A., Caporali A., Ebner D., Vernet A., Schneider J.E., Bhattacharya S. The retinoid agonist tazarotene promotes angiogenesis and wound healing. Mol. Ther. 2016;24:1745–1759. doi: 10.1038/mt.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühl T., Mezger M., Hausser I., Handgretinger R., Bruckner-Tuderman L., Nyström A. High local concentrations of intradermal MSCs restore skin integrity and facilitate wound healing in dystrophic epidermolysis bullosa. Mol. Ther. 2015;23:1368–1379. doi: 10.1038/mt.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Xing Q., Zhai Q., Tahtinen M., Zhou F., Chen L., Xu Y., Qi S., Zhao F. Pre-vascularization enhances therapeutic effects of human mesenchymal stem cell sheets in full thickness skin wound repair. Theranostics. 2017;7:117–131. doi: 10.7150/thno.17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghatak S., Chan Y.C., Khanna S., Banerjee J., Weist J., Roy S., Sen C.K. Barrier function of the repaired skin is disrupted following arrest of Dicer in keratinocytes. Mol. Ther. 2015;23:1201–1210. doi: 10.1038/mt.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H.J., Jin S.E., Park J.S., Ahn W.S., Kim C.K. Accelerated wound healing by smad3 antisense oligonucleotides-impregnated chitosan/alginate polyelectrolyte complex. Biomaterials. 2008;29:4831–4837. doi: 10.1016/j.biomaterials.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Frauenstein K., Sydlik U., Tigges J., Majora M., Wiek C., Hanenberg H., Abel J., Esser C., Fritsche E., Krutmann J., Haarmann-Stemmann T. Evidence for a novel anti-apoptotic pathway in human keratinocytes involving the aryl hydrocarbon receptor, E2F1, and checkpoint kinase 1. Cell Death Differ. 2013;20:1425–1434. doi: 10.1038/cdd.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y., Zhou W., Cheng L., Chen C., Huang Z., Fang X., Wu Q., He Z., Xu S., Lathia J.D. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 2017;24:167–180. doi: 10.1038/cdd.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J., Liu X., Wang P., Xue Y., Ma J., Qu C., Liu Y. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol. Ther. 2016;24:1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Tsuta K., Nokihara H., Tamura T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai X.Y., Liu H.J., Lu C., Liang C.L., Sun Y., Chen S., Sun B., Li Y., Liu Y.R., Zhang Q. Phenytoin silver: a new nanocompound for promoting dermal wound healing via comprehensive pharmacological action. Theranostics. 2017;7:425–435. doi: 10.7150/thno.17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Chang L., Du W.W., Gupta S., Khorshidi A., Sefton M., Yang B.B. Anti-microRNA-378a enhances wound healing process by upregulating integrin beta-3 and vimentin. Mol. Ther. 2014;22:1839–1850. doi: 10.1038/mt.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deveza L., Choi J., Lee J., Huang N., Cooke J., Yang F. Polymer-DNA nanoparticle-induced CXCR4 overexpression improves stem cell engraftment and tissue regeneration in a mouse hindlimb ischemia model. Theranostics. 2016;6:1176–1189. doi: 10.7150/thno.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura N., Shaheen S.M., Akita H., Nakamura T., Harashima H. A KALA-modified lipid nanoparticle containing CpG-free plasmid DNA as a potential DNA vaccine carrier for antigen presentation and as an immune-stimulative adjuvant. Nucleic Acids Res. 2015;43:1317–1331. doi: 10.1093/nar/gkv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeyapalan Z., Deng Z., Shatseva T., Fang L., He C., Yang B.B. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Q. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017 doi: 10.1038/cdd.2017.86. Published online June 5, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutnam Z.J., Du W.W., Yang W., Yang X., Yang B.B. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 2014;5:2914. doi: 10.1038/ncomms3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W.W., Liu F., Shan S.W., Ma X.C., Gupta S., Jin T., Spaner D., Krylov S.N., Zhang Y., Ling W., Yang B.B. Inhibition of dexamethasone-induced fatty liver development by reducing miR-17-5p levels. Mol. Ther. 2015;23:1222–1233. doi: 10.1038/mt.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Wu Q., Bu M., Hu L., Du W.W., Jiao C., Pan H., Sdiri M., Wu N., Xie Y., Yang B.B. Ergosterol peroxide activates Foxo3-mediated cell death signaling by inhibiting AKT and c-Myc in human hepatocellular carcinoma cells. Oncotarget. 2016;7:33948–33959. doi: 10.18632/oncotarget.8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.