Abstract
Background
The TMPRSS2-ERG gene fusion is detected in approximately half of primary prostate cancers (PCa) yet the prognostic significance remains unclear. We hypothesized that ERG promotes the expression of common genes in primary PCa and metastatic castration-resistant PCa (CRPC), with the objective of identifying ERG-associated pathways, which may promote the transition from primary PCa to CRPC.
Methods
We constructed tissue microarrays (TMA) from 127 radical prostatectomy specimens, 20 LuCaP patient-derived xenografts (PDX), and 152 CRPC metastases obtained immediately at time of death. Nuclear ERG was assessed by immunohistochemistry (IHC). To characterize the molecular features of ERG-expressing PCa, a subset of IHC confirmed ERG+ or ERG-specimens including 11 radical prostatectomies, 20 LuCaP PDXs, and 45 CRPC metastases underwent gene expression analysis. Genes were ranked based on expression in primary PCa and CRPC. Common genes of interest were targeted for IHC analysis and expression compared with biochemical recurrence (BCR) status.
Results
IHC revealed that 43% of primary PCa, 35% of the LuCaP PDXs, and 18% of the CRPC metastases were ERG+ (12 of 48 patients [25%] had at least 1 ERG+ metastasis). Based on gene expression data and previous literature, two proteins involved in calcium signaling (NCALD, CACNA1D), a protein involved in inflammation (HLA-DMB), CD3 positive immune cells, and a novel ERG-associated protein, DCLK1 were evaluated in primary PCa and CRPC metastases. In ERG+ primary PCa, a weak association was seen with NCALD and CACNA1D protein expression. HLA-DMB expression and the presence of CD3 positive immune cells were decreased in CRPC metastases compared to primary PCa. DCLK1 was upregulated at the protein level in unpaired ERG+ primary PCa and CRPC metastases (p=0.0013 and p<0.0001, respectively). In primary PCa, ERG status or expression of targeted proteins was not associated with BCR-free survival. However for primary PCa, ERG+DCLK1+ patients exhibited shorter time to BCR (p=0.06) compared with ERG+DCLK1- patients.
Conclusions
This study examined ERG expression in primary PCa and CRPC. We have identified altered levels of inflammatory mediators associated with ERG expression. We determined expression of DCLK1 correlates with ERG expression and may play a role in primary PCa progression to metastatic CPRC.
Keywords: ERG, Prostate Cancer, CRPC, Metastasis, and DCLK1
Introduction
TMPRSS2 is a prostate-specific, androgen-responsive serine protease, frequently rearranged at the genomic level with members of the E26 transformation-specific (ETS) gene family [1]. The most frequent fusion involves the ‘ETS-related gene’ or ‘ERG’ occurring in approximately 40-50% of primary prostate cancer (PCa) [2-11] with slightly less prevalence in PCa metastases (∼25-40%) [6, 8, 9, 12, 13]. Interestingly, there are a number of clinical studies suggesting worsened clinical prognosis with presence of this TMPRSS2-ERG gene fusion [4, 7, 14-17] as well as studies reporting the opposite [5, 18-22]. Therefore, no overall consensus as to the clinical implication of the TMPRSS2-ERG fusion in primary PCa progression has been reached. These findings prompted us to define the role ERG in castration resistant PCa (CRPC) and examine whether ERG expression in primary PCa promotes survival, proliferation and progression to CRPC.
In this study, we analyzed ERG expression in CRPC specimens in order to define common biological pathways and downstream effectors of ERG. We have evaluated previously reported ERG-associated markers including two involved in calcium signaling NCALD [23] and CACNA1D [24, 25], two involved in inflammation HLA-DMB [25, 26] and CD3+ [27], and a novel ERG-associated protein, doublecortin-like kinase 1 (DCLK1). We found a weak association of NCALD and CACNA1D with ERG expression in primary PCa. We also found an interesting trend of decreased inflammatory mediators (HLA-DMB and CD3+ cells) in CRPC compared to primary PCa, suggesting an evolving inflammatory process with ERG+ PCa progression to CRPC. Furthermore, DCLK1 expression was associated with ERG expression in primary PCa and CRPC specimens. This is an important finding as DCLK1 is a cancer-stem cell marker thought to play a role in oncologic pluripotency and epithelial to mesenchymal transition (EMT) in several malignancies [28-33]. Influences of these genes on biochemical recurrent (BCR)-free survival in primary PCa relative to ERG status are also reported.
Materials and Methods
Tissue specimens
All samples were obtained from patients who signed written informed consent under the aegis of the Prostate Cancer Donor Program at the University of Washington, and approved by the Institutional Review Board. Primary PCa: 127 radical prostatectomy specimens were obtained from the University of Washington Medical Center. Longitudinal clinical follow up of these patients yielded 65 patients with and 62 patients without evidence of BCR. LuCaP patient-derived xenografts: 20 LuCaP patient-derived xenograft (PDX) lines derived from radical prostatectomy and CRPC specimens were used for analysis. These PDX lines are maintained by serial passage in severe combined immune deficient (SCID) male mice as previously described [34]. CRPC metastases: Bone and visceral metastases were obtained from 48 CRPC patients (152 sites total; 79 bone and 73 visceral) taken immediately at time of death as part of the University of Washington PCa tissue acquisition necropsy program described previously [35].
Tissue microarray construction
All specimens for immunohistochemistry (IHC) were formalin fixed (decalcified in formic acid for bone specimens), paraffin embedded and examined histologically for presence of nonnecrotic tumor. Tissue microarrays (TMA) were constructed with 1 mm-diameter duplicate cores from primary PCa (consisting of 127 radical prostatectomy specimens), LuCaP PDX (consisting of 20 LuCaP xenografts), and CRPC (consisting of 152 metastases; 79 bone and 73 visceral from 48 patients, up to 4 metastatic sites from each patient) specimens. Clinical characteristics of the 48 CRPC patients are shown in Supplemental Table 1.
RNA isolation
Total RNA was isolated from a subset of IHC confirmed ERG+ and ERG- specimens including 11 primary PCa (5 ERG-, 6 ERG+), 20 LuCaP PDX (13 ERG-, 7 ERG+), and 45 CRPC metastases (30 ERG-, 15 ERG+). Eight-micron thick sections from primary PCa (n=11) and visceral metastases (n=38) were cut using a Leica CM3050S cryostat, collected on PEN Membrane Frame Slides (Life Technologies) and immediately fixed in 95% ethanol. Sections were briefly stained with hematoxylin then dehydrated in 100% ethanol. 5,000-10,000 tumor cells per sample were laser capture microdissected with an Arcturus Veritas instrument and collected on CapSure® Macro LCM Caps (Life Technologies). Digital photographs were taken of tissue sections before, during, and after LCM and assessed by a pathologist to confirm the tumor content. RNA was isolated using the Arcturus PicoPure RNA Isolation Kit and the samples were DNAse treated using the Qiagen RNase-Free DNase Set. RNA was amplified for two rounds using the Ambion MessageAmp aRNA Kit. The bone metastases (n=7) frozen in OCT blocks were sampled using 1 mm diameter tissue punch in a −20°C cryostat. The sample was obtained from the region of the block where there was tumor based upon a section of an adjacent decalcified FFPE block. RNA was isolated from the tissue cores using RNeasy Plus Micro Kit (Qiagen Inc.). Tissue cores were placed in the kit's Buffer RLT Plus, to which β-mercaptoethanol had been added, and homogenized with a disposable hard tissue homogenizer tip (Omni International). Flash frozen LuCaP PDX tissue was histologically evaluated for regions of viable tumor and RNA was isolated from 15, 10-micron sections with ≥80% tumor content. For PDX tissue with <80% tumor, stroma and necrotic tissue were removed using an 18-guage needle prior to sectioning. LNCaP and VCaP human PCa cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). RNA was extracted using the Qiagen RNeasy Kit, (Qiagen Inc.), according to the manufacturer's protocol. On-column DNase digestion was performed. RNA was quantified using a Nanodrop 1000 (Thermo Scientific) and quality was assessed via Agilent Bioanalyzer 2100 (Agilent Technologies).
RNA amplification and microarray hybridization
Agilent 44K whole human genome expression oligonucleotide microarrays (Agilent Technologies, Inc.) were used to profile the PCa, LuCaP PDX, and CRPC metastases. Total RNA from PDX tissue was amplified one round; patient samples were amplified two rounds as described previously [36]. Probe labeling and hybridization against a common reference pool of prostate tumor cell lines (LNCaP, DU145, PC3, and CWR22) followed Agilent protocols. Fluorescent array images were digitized using the Agilent DNA microarray scanner G2565BA. Data were loess normalized within arrays (normexp background correction with offset 50) and quantile normalized between arrays in R using the Limma Bioconductor package. Control probes were removed, duplicate probes averaged, and spots flagged by Agilent Feature Extraction software as being foreground feature non-uniformity or population outliers were assigned a value of “NA”. Data were normalized separately for PCa, LuCaP PDXs, and CRPC metastases. CRPC metastases were subject to an additional normalization step to remove systematic batch effects by application of the ComBat function within the sva Bioconductor package to the log2 Cy3 signal intensities. Microarray data are deposited in the Gene Expression Omnibus database under the accession number GSE74367. The AR activity score was determined by the expression of a 20-gene signature and calculated as described previously [37]. Briefly, the activity score is defined as the sum of the expression Z-scores calculated across 149 CRPC metastases converted to a percent. Pearson's correlation coefficient was used to study the relationships between variables shown in scatterplots using the cor.test function in R.
Ingenuity pathway analysis (IPA)
To identify biological pathways involved in ERG regulation of CRPC metastases, Ingenuity pathway Analysis (IPA, Ingenuity Systems; https://www.ingenuity.com) was conducted on the 376 differentially expressed genes between ERG+ and ERG- groups based on SAM score ≥3 or ≤-3. Molecular and cellular functions and upstream regulator analysis was also conducted for these genes.
Immunohistochemistry
Five-micron thick sections of the TMAs were deparaffinized and rehydrated in sequential xylene and graded ethanol. Antigen retrieval was performed in 10 mM citrate buffer (pH 6.0) in a pressure cooker. Endogenous peroxidase and avidin/biotin were blocked respectively (Vector Laboratories Inc.). Sections were then incubated with 5% normal goat-horse-chicken serum, incubated with primary antibody (Supplemental Table 2), incubated with biotinylated secondary antibody (Vector Laboratories Inc.), followed by ABC reagent (Vector Laboratories Inc.), and stable DAB (Invitrogen Corp.). All sections were lightly counterstained with hematoxylin and mounted with Cytoseal XYL (Richard Allan Scientific). Mouse or rabbit IgG were used as negative controls as appropriate.
Immunohistochemical assessment
ERG immunostaining was assessed using a quasi-continuous scoring system, created by multiplying each optical density level (“0” for no brown color, “1” for faint and fine brown chromogen deposition, and “2” for clear and coarse granular chromogen clumps) multiplied by the percentage of cells at each staining level, resulting in a total score range of 0 to 200. The final score for each sample was the average of 2 duplicated tissue cores. Only nuclear positivity was evaluated in ERG IHC staining. The final scores were categorized as “none” (score range: 0), “weak” (score range: 1 to 100), and “strong” (score range: 101 to 200). HLA-DMB, DCLK1, NCALD and CACNA1D immunostaining were assessed using a four point categorical compositional scale: “0” no staining, “1” faint/equivocal or focal staining, “2” intermediate staining, and “3” strong staining, multiplied by the percentage of cells at each staining level, resulting in a total score range of 0 to 300. For CD3 we identified the number of cells staining positive in the tumor and ranked tumors as having no positive cells (score range: 0) or positive for cells with 3 different score ranges (score range: 1 to 10; 11 to 20; or above 20 cells).
Statistical analysis
Statistical Analysis of Microarray (SAM) program (http://www-stat.stanford.edu/∼tibs/SAM/) was used to analyze expression differences between the groups. Unpaired, t-tests were calculated for all probes passing filters and controlled for multiple testing by estimation of q-values using the false discovery rate (FDR) method [38]. Spearman correlations for protein validation in the respective tissues (ERG, NCALD, CACNA1D, HLA-DMB, CD3, DCLK1) were used with significance differences calculated via student's t-test (significance set at p<0.05). Kaplan-Meier curves of BCR-free survival in primary PCa patients stratified by positive or negative IHC staining of specific proteins were examined using GraphPad PRISM 6. In primary PCa specimens, Spearman correlation of BCR-free survival with proliferation (Ki-67 positivity) relative to each targeted protein was performed with student t-tests (significance set at p< 0.05). A summary of experimental materials and methods used can be found in Supplemental Figure 1.
Results
Gene expression analysis reveals heterogeneity among metastatic ERG+ CRPC
Figure 1 describes ERG gene expression in PCa lines. Non-ERG expressing LNCaP and ERG expressing VCaP cell lines are included for reference. Using ERG microarray expression, Cy3 values of primary PCa specimens were divided into those with low (<2750), medium (>2750 <20,000) and high (>20,000) levels of ERG expression. Based on these Cy3 values (arbitrary units), a smaller percentage of CRPC metastases express medium to high levels of ERG transcript relative to primary PCa (31% versus 64%). Following, there appears to be a larger subpopulation of low expressors in this CRPC cohort relative to primary PCa. The finding of a decreased number of ERG positive metastases, a larger medium ERG expressing population, and a smaller high ERG expressing population in CRPC (relative to primary PCa) suggests ERG expression is not maintained uniformly or to the same levels as in primary PCa. To determine if the change in ERG expression was due to lower overall AR expression in the CRPC cohort we used a known 20-gene expression signature of AR to compute an AR activity score [37]. ERG expression levels were compared to the AR activity score as well as AR, KLK3, HOXB13, and STEAP1. No significant correlation was made except a weak correlation with STEAP1 (p=0.005) (Supplemental Figure 2). One explanation for this result could be that adenocarcinomas with no TMPRSS2-ERG fusion typically still have high AR activity so the overall correlation is not significant. However in the medium ERG expressors a range of AR activity was observed.
Figure 1. ERG gene expression in primary PCa and CRPC metastases.
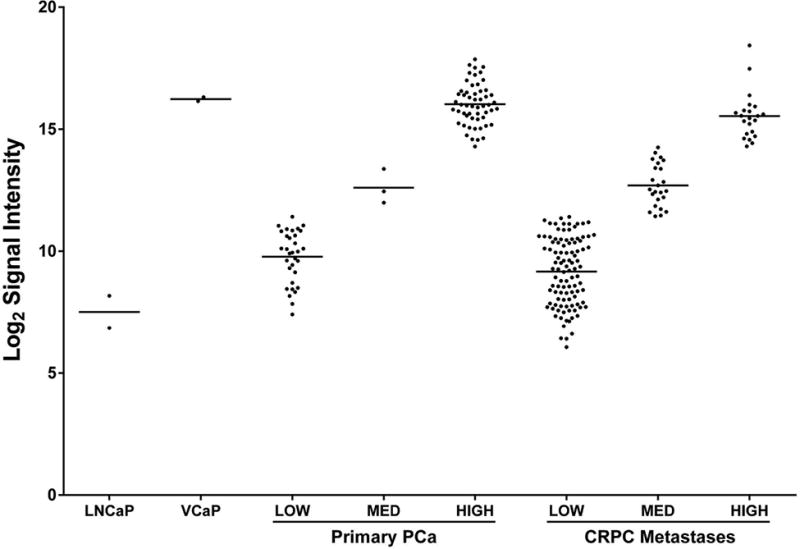
Agilent gene expression array Log2 signal intensities of LNCaP (ERG-) and VCaP (ERG+) cells, primary PCa, and CRPC metastases. Based on ERG microarray expression Cy3 values primary PCa specimens were divided into those with low (<2750), medium (>2750 <20,000) and high (>20,000) levels of ERG expression. Of note, the CRPC metastases group contains a large cohort of medium ERG expressors.
Nuclear ERG immunoreactivity is decreased in CRPC metastases when compared to primary PCa
The TMA comprising radical prostatectomy specimens revealed that 43% were ERG+ (Figure 2). Additionally, 42% of the non-recurrent and 43% of recurrent PCa patients were ERG+ showing no clear differences between these two populations. In CRPC metastases, 27 of 148 sites present on the array were positive for nuclear ERG (18%), representing 12 of 48 patients with at least 1 ERG+ metastasis (25%). These results suggest that nuclear ERG is less frequent in CRPC compared to primary PCa (Figure 2). Interestingly, we found that some patients had both ERG+ and ERG- metastases as evidenced by the presence or complete absence of ERG expression in the tumor cells by IHC analysis. The TMA comprising 20 LuCaP PDX indicated nuclear ERG positivity in 7/20 lines (35%). As a quality control measure, ERG IHC positivity was confirmed with a second antibody (Supplemental Table 2) for CRPC metastases both between patients and between metastatic sites, showing excellent correlation (r=0.85 and r=0.83 respectively, both p<0.0001).
Figure 2. ERG IHC in primary PCa and CRPC metastases.
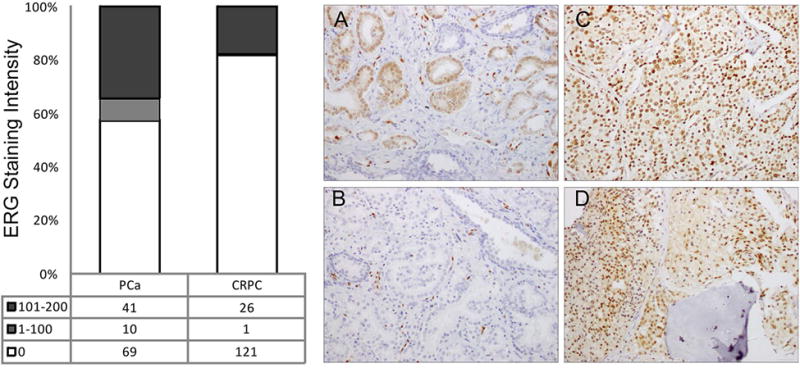
ERG+ staining was observed in 43% of primary PCa patients (bar graph, left). 18% of CRPC metastases expressed ERG+ with the majority showing high intensity staining. IHC images: A: ERG+ primary PCa with tumor cell nuclei in brown; B: ERG- PCa specimen (note ERG+ endothelial cells); C: Diffusely ERG+ visceral metastasis; D: Diffusely ERG+ bony metastasis.
Selection of target genes in ERG+ primary PCa, CRPC and LuCaP PDX using differential gene expression analysis
A subset of PCa, LuCaP PDX, and CRPC metastases annotated by their ERG IHC status underwent gene expression analysis (Figure 3). The three gene datasets were then compared for common overlapping genes based on Cy3 score (Figure 4). There were four upregulated genes and one downregulated gene common to the three groups with a complete list of the overlapping upregulated genes shown in Supplemental Table 3A. The upregulated genes were ERG, FAM19A5 (family with sequence similarity 19 [chemokine (C-C motif)-like], member A5), RORB (RAR-related orphan receptor B), and NCALD (neurocalcin delta). The one down-regulated gene common to all three datasets was WDR6 (WD repeat-containing protein 6). Since the objective of this study was to examine pathways common ERG-associated to both primary PCa and CRPC metastases, we ranked the top 100 primary PCa genes and compared these to their matched ranks in LuCaP PDX and CRPC specimens (Supplemental Table 3B). ERG ranked 2nd overall in primary PCa, 1st in CRPC and 6th in LuCaP PDXs. We selected other genes of interest based on a high rank in CRPC in relation to primary PCa, including NCALD, CACNA1D, and HLA-DMB. NCALD (a neuronal calcium sensor) and CACNA1D (a voltage-dependent, calcium channel) are both involved in calcium signaling. HLA-DMB (major histocompatibility complex, class II, DM beta) is involved in MHC assembly and antigen presentation. These three genes were all found within the top 25 ranked genes and their associations with ERG status have been reported previously [23-26]. Additionally, the 4th ranked gene in primary PCa and 51st in CRPC was DCLK1 (doublecortin kinase like 1, or CaM Kinase like 1 [DCAMKL1]). This gene has not previously been associated with ERG expression in PCa. Previous studies suggest DCLK1 is a cancer stem cell marker thought to play a role in oncologic stemness and epithelial to mesenchymal (EMT) transition in several malignancies [28-33].
Figure 3. Gene expression array of Primary PCa, LuCaP PDXs, and CRPC metastases.
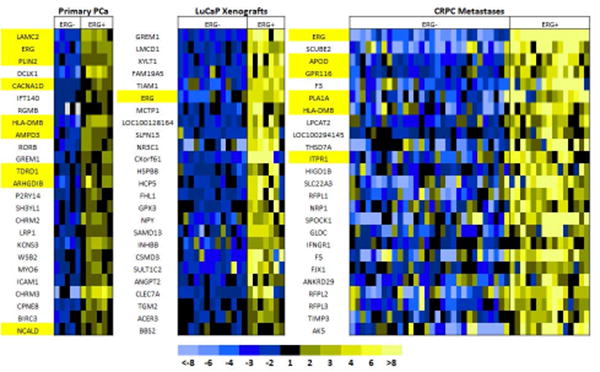
The top 25 genes found on expression analysis in 11 primary PCa (5 ERG-, 6 ERG+), 20 LuCaP PDX (13 ERG-, 7 ERG+), and 45 CRPC metastases (30 ERG-, 15 ERG+) specimens relative to ERG positivity based on IHC are shown. Genes highlighted in yellow have been previously found to correlate with ERG activity in the literature.
Figure 4. Venn diagram of overlapping up-regulated genes in ERG+ primary PCa, LuCaP PDXs, and CRPC (Score >3, Total N=16,096).
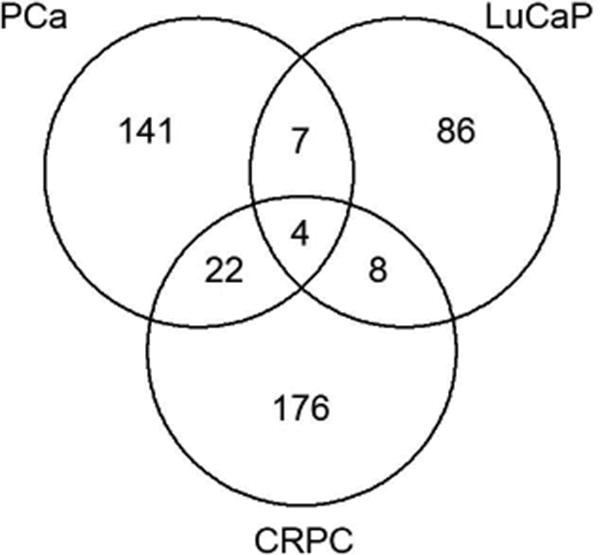
The 4 upregulated genes found on gene expression are ERG, FAM19A5 (family with sequence similarity 19 [chemokine (C-C motif)-like], member A5), RORB (RAR-related orphan receptor B), and NCALD (neurocalcin delta). A list of all upregulated, overlapping genes between any 2 groups is shown in Supplemental Table 3.
Ingenuity Pathway Analysis (IPA) reveals alterations in NF-κB and cellular movement associated networks relative to ERG gene expression
To identify ERG associated biological pathways in metastatic CRPC, a total of 376 differentially expressed genes (SAM score ≥ 3 or ≤ -3 between ERG+ and ERG- samples) were input into the IPA platform. The top two networks involved were the ERG-regulated network (p=1×10−40) and the NF-κB-associated network (p=1×10−37) with HLA-DMB seen as a direct effector of NF-κB (Supplemental Figure 3A and 3B, respectively). Furthermore, genes that regulate cellular movement were found to be the principal molecular and cellular function altered in this gene set (Supplemental Table 4) relative to ERG expression.
Verification of nuclear ERG associated protein expression in primary PCa, CRPC, and LuCaP PDX
We further evaluated four genes identified in the differential gene expression analysis (NCALD, CACNA1D, HLA-DMB, and DCLK1). An additional T-cell receptor marker (CD3) was included because of its involvement in inflammation and correlation with ERG status in PCa [27]. We performed IHC to evaluate the abundance and cellular localization of these proteins relative to ERG status with the results presented in Table 1. NCALD expression was weakly associated with ERG positivity in primary PCa (p=0.0282), a trend towards association was seen in CRPC (p=0.052) (Supplemental Figure 4). Similarly, CACNA1D was weakly associated with ERG positivity in primary PCa (p=0.0068), but not in CRPC (p=0.8746) (Supplemental Figure 5). HLA-DMB was associated with ERG positivity in primary PCa (p<0.0001) while only a trend was seen in CRPC (p= 0.0823) (Figure 5). The CD3+ cell number trend was increased in ERG+ primary PCa (p=0.0888) but decreased in CRPC (p=0.0771) (Figure 6). The novel gene, DCLK1 was associated with ERG positivity in both primary PCa (p=0.0013) and CRPC (p<0.0001) (Figure 7). No significant findings were seen in LuCaP PDXs.
Table 1. ERG-associated genes validated at the protein level in PCa, CRPC, and LuCaP PDX.
All ERG-associated proteins identified by gene expression (with addition of CD3+) are shown with Spearman correlations relative to nuclear ERG staining reported. P-values from unpaired, two tailed t-test and 95% confidence intervals shown (significance set at p<0.05, highlighted in light grey).
| PRIMARY PCa | |||||
|---|---|---|---|---|---|
| Correlation with nERG | NCALD | CACNA1D | HLA-DMB | CD3 | DCLK1* |
|
|
|||||
| Number of Specimens | 118 | 114 | 119 | 119 | 120 |
| Spearman r | 0.2021 | 0.2522 | 0.3675 | 0.1567 | 0.2896 |
| 95% confidence interval | 0.01673 to 0.3740 | 0.06612 to 0.4213 | 0.1956 to 0.5175 | −0.02942 to 0.3323 | 0.1111 to 0.4501 |
|
|
|||||
| p-value (two-tailed) | 0.0282 | 0.0068 | < 0.0001 | 0.0888 | 0.0013 |
|
|
|||||
| CRPC METASTASES | |||||
|
| |||||
| Correlation with nERG | NCALD | CACNA1D | HLA-DMB | CD3 | DCLK1* |
|
|
|||||
| Number of Specimens | 144 | 144 | 147 | 147 | 143 |
| Spearman r | 0.1623 | −0.0133 | 0.1438 | −0.1463 | 0.3910 |
| 95% confidence interval | −0.0063 to 0.3218 | −0.1812 to 0.1554 | −0.0234 to 0.3032 | −0.3055 to 0.02086 | 0.2378 to 0.5253 |
| p-value (two-tailed) | 0.0520 | 0.8746 | 0.0823 | 0.0771 | < 0.0001 |
| LuCaP XENOGRAFTS | |||||
|
| |||||
| Correlation with nERG | NCALD | CACNA1D | HLA-DMB | DCLK1* | |
|
|
|||||
| Number of Specimens | 22 | 22 | 21 | 22 | |
| Spearman r | 0.2828 | −0.1762 | −0.3422 | 0.07295 | |
| 95% confidence interval | −0.1707 to 0.6374 | −0.5657 to 0.2775 | −0.6817 to 0.1186 | −0.3713 to 0.4901 | |
| p-value (two-tailed) | 0.2022 | 0.4327 | 0.1289 | 0.747 | |
Denotes novel gene, DCLK1.
Corresponds to gene not previously reported in the literature as related to ERG/TMPRSS2 fusion. All significant correlations are highlighted in light grey.
Figure 5. HLA-DMB protein expression in ERG+ and ERG- primary PCa and CRPC.
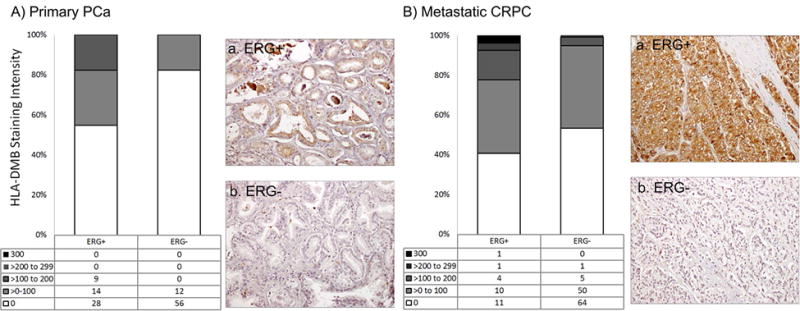
A There was a significant correlation between nuclear ERG and HLA-DMB in primary PCa (r=0.3675, p<0.0001). Representative IHC images of HLA-DMB+ in ERG+ (a) and ERG- (b) primary PCa samples are shown. B. HLA-DMB was not associated with nuclear ERG expression found in CRPC specimens (r=0.1438, p=0.082). Representative IHC images of HLA-DMB+ in ERG+ (a) and ERG- (b) CRPC samples are shown.
Figure 6. CD3 expression in ERG+ and ERG- primary PCa and CRPC.
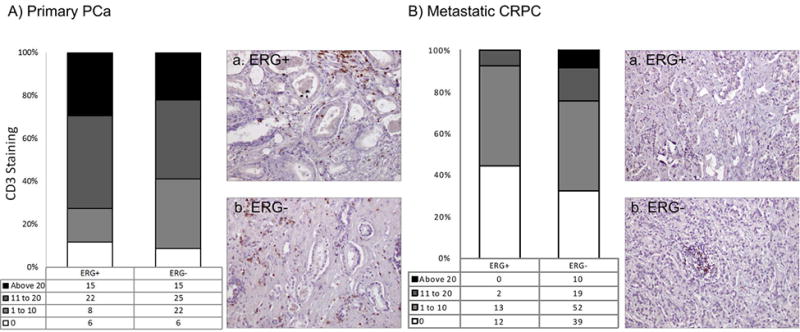
A CD3+ cells were common in primary PCa revealing a positive, non-significant correlation to ERG+ (r=0.1567, p=0.0888). Representative IHC images of CD3+ cells in ERG+ (a) and ERG- (b) primary PCa samples are shown. B. CD3+ cells were less common in CRPC and revealed a negative, non-significant correlation to ERG+ (r= -0.1463, p=0.0771). Representative IHC images of CD3+ cells in ERG+ (a) and ERG- (b) CRPC specimens are shown.
Figure 7. DCLK1 protein expression in ERG+ and ERG- primary PCa and CRPC.

A DCLK1 expression was correlated with nuclear ERG expression in primary PCa (r=0.2896, p=0.0013). Representative IHC images of DCLK1+ in ERG+ (a) and ERG- (b) primary PCa are shown. B. DCLK1 expression was also correlated with nuclear ERG expression in CRPC specimens (r=0.391, p<0.0001). Representative IHC images of DCLK1+ cells in ERG+ (a) and ERG- (b) CRPC specimens are shown.
ERG and DCLK1 influence on BCR-free survival in primary PCa
In our series, we found no difference in BCR-free survival relative to ERG expression alone (Supplemental Figure 6, p=0.41). As ERG is hypothesized to increase proliferation and confer an invasive phenotype in PCa cells [39], we also correlated proliferation (via Ki-67) with each protein relative to ERG status. No significant findings relative to ERG expression and proliferation were seen overall (p=0.6885, Supplemental Figure 6). When looking at the targeted proteins, BCR-free analysis revealed no significant differences in HLA-DMB+ vs. HLA-DMB- samples (p=0.85) or ERG+HLA-DMB+ vs. ERG+HLA-DMB- samples (p=0.10) (Supplemental Figure 7). HLA-DMB also did not show any significant correlation with proliferation alone or in ERG+ cases (r=0.051, p=0.5843; r=0.028, p=0.8451, respectively, Supplemental Figure 7). BCR-free survival for CD3 did not show any significant differences overall (CD3+ vs. CD3-, p=0.42; ERG+CD3+ vs. ERG+CD3-, p=0.5, Supplemental Figure 8). Proliferation was significantly increased in CD3+ vs. CD3- specimens (r=0.2608, p=0.0045) while this relationship only approached significance in ERG+CD3+ vs. ERG+CD3- specimens (r=0.2647, p=0.0605, Supplemental Figure 8). BCR-free survival for NCALD did not show any significant differences overall (NCALD+ vs. NCALD-, p=0.84; ERG+NCALD+ vs. ERG+NCALD-, p=0.14, Supplemental Figure 9). Proliferation was not significantly increased in NCALD+ vs. NCALD- specimens (r=0.1794, p=0.054) or ERG+NCALD+ vs. ERG+NCALD- specimens (r=0.06085, p=0.6066, Supplemental Figure 9). Similarly no significant differences were observed for BCR-free survival for CACNA1D+ vs. CACNA1D-, p=0.09. ERG+CACNA1D+ vs. ERG+CACNA1D-approached significance, but there were a limited number of patient samples in the ERG- group (p=0.05) (Supplemental Figure 10). Proliferation was weakly associated with CACNA1D+ vs. CACNA1D- specimens (r=0.1883, p=0.0468), but no association was identified in ERG+CACNA1D+ vs. ERG+CACNA1D- specimens (r=0.03380, p=0.7765, Supplemental Figure 10). For DCLK1, there was no difference in BCR in DCLK1+ vs. DCLK1- patients (p=0.11). Interestingly, when ERG status was considered, ERG+DCLK1+ patients showed a more pronounced trend towards worsened BCR-free survival (Figure 8, p=0.06) when compared to ERG+DCLK1-patients. DCLK1+ status alone was significantly correlated with proliferation (r= 0.194, p=0.0350) while in ERG+DCLK1+ vs. ERG+DCLK1- patients, this relationship was absent (r=0.110, p=0.4419).
Figure 8. Primary PCa BCR-free survival and proliferation relative to DCLK1 status.
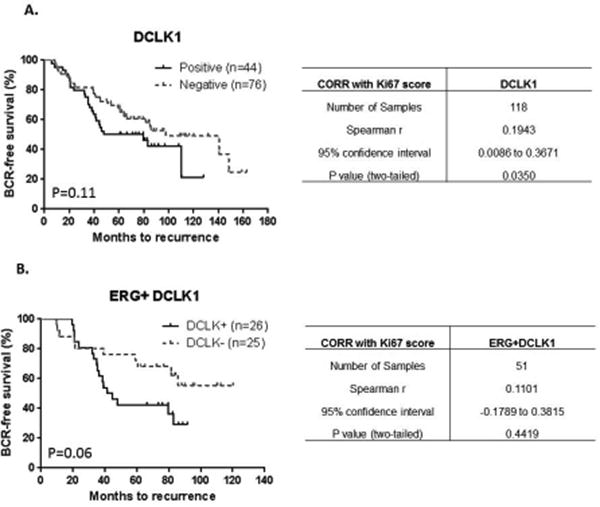
A Kaplan-Meier analysis of DCLK1+ vs. DCLK1- status did not correlate with BCR-free survival overall (p=0.11). There was a positive correlation with proliferation relative to DCLK1 status (r=0.1943, p=0.035). B. In ERG+ samples, DCLK1+ vs. DCLK1- status revealed a trend towards worsened BCR-free survival (p=0.06). DCLK1 did not correlate significantly with proliferation in this grouping (r=0.1101, p=0.4419).
Discussion
We observed ERG protein expression is decreased in CRPC patients relative to primary PCa. This finding is consistent with previous reports that the TMPRSS2-ERG fusion is frequently reported in primary PCa (40-50%) [2, 2, 4-11] but is slightly less common in PCa metastases (25-40%) [6, 8, 9, 12, 13]. This difference may indicate ERG is important in primary PCa, yet ERG expression may no longer be required in subgroups of CRPC metastases which may have acquired additional mutations/rearrangements, promoting proliferation and survival irrespective of ERG expression.
We hypothesized that ERG overexpression regulates particular genes common to primary PCa and CRPC metastases, and these genes persisting in late stage metastatic disease can identify a signature suggesting early aggressiveness. This signature could then provide clues for the translational application of anti-ERG therapy in early PCa. In our study, ERG overexpression resulted in significant associations with two mechanisms: calcium signaling and inflammatory mediators. Epidemiologic evidence suggests that modulation of calcium signaling may decrease the risk of PCa in select populations [40, 41]. CACNA1D is highly overexpressed in primary PCa, CRPC, and ERG+ tumors with calcium channel blocker experiments showing decreased androgen-mediated calcium-influx, transactivation, and cellular growth in PCa cells [24]. NCALD is a predominantly cytosolic neuronal calcium sensor binding protein, however with elevated intracellular calcium levels, can undergo a conformational change resulting in localization to the peri-nuclear trans-Golgi network [42] and protein levels have been shown to correlate significantly with ERG positivity in PCa [23]. Based on our data, the role of calcium modulation of ERG+ progression in this cohort is unclear, with significant changes seen at the gene level, a weak association observed at the protein level in primary PCa, and no significant association seen in CRPC metastases.
NF-κB a mediator of multiple inflammatory pathways has been shown to be constitutively active in several cancers and has a variety of pro-tumorigenic functions [43] including regulating epithelial to mesenchymal transition and metastases [44]. In contrast, pharmacologically immune suppressed individuals such as those with organ transplants, can also be at increased risk of malignancy [43]. Therefore, cancer surveillance by the immune system is a delicate balance in which perturbations can affect progression. ERG expression has been shown to be associated with increased NF-κB activity in PCa cells [43] and may work through toll-like receptor pathways to activate NF-κB [45]. Alternatively, it has also been shown to modulate NF-κB activity in endothelial cells by blocking IL-8 and ICAM [46]. We looked specifically at HLADMB and CD3+ IHC expression as they have been described to associate with ERG+ PCa [25-27]. HLA-DMB is a class II MHC molecule involved in antigen presentation of tumor cells. Increased mRNA and protein levels of HLA-DMB can promote cytotoxic lymphocytes in tumor tissue and has been shown to be an independent prognostic factor for survival in epithelial malignancies [47]. Through co-stimulation of the TCR (CD3) with MHC and other ligand binding (e.g. CD40L) [48], the CD3+ cell can then activate and exert its cytotoxic effects on tumor cells. Our data indicates a change in protein expression of these markers as tumors progress from primary PCa to CRPC metastases. Specifically, HLA-DMB correlates with ERG positivity significantly in primary PCa, but this association was decreased in CRPC. We found the CD3+ cell correlation changed from positive in primary PCa to negative in CRPC. Together, these findings suggest some loss of immune capability with CRPC progression. This has not been reported relative to ERG+ status in HLA-DMB, but is consistent with CD3+ findings in a study reporting low CD3+ cell numbers are associated with a worsened BCR-free survival [27]. Further, on IPA analysis we found the NF-κB pathway to be enriched with HLA-DMB as a downstream effector and movement of myeloid cells and phagocytes were predicted to be activated, suggesting immune activity involvement in ERG+ CRPC metastases. Overall, our data suggest that an imbalance of inflammatory mediators may play a role in ERG+ PCa progression.
In our study DCLK1 expression was highly correlated with nuclear ERG immunoreactivity in PCa and CRPC, a novel finding. DCLK1 was first described in the neuroscience literature as a microtubule associated kinase capable of autophosphorylation involved in neuronal migration [49] and in the developing cerebral cortex [50]. Initial work in cancer came in the gastrointestinal literature with Barrett's esophagus and esophageal carcinoma transition [51, 52]. More recent results in pancreatic and colorectal cancer (CRC) identified DCLK1 as a putative stem cell marker found in serum, circulating tumor cells, and tumor stroma [28, 30] which is associated with increased pluripotency and EMT [29, 31] with adverse prognostic implications [30, 32]. DCLK1 was recently found to be highly prevalent in renal cell carcinoma (RCC) and correlated with increasing stage of disease [33]. Further experiments with siRNA knockdown of DCLK1 resulted in decreased stemness, EMT, invasion, migration, and adhesion in RCC [33]. We found DCLK1 to be significantly upregulated in ERG+ primary PCa and CRPC metastases by gene expression analysis with IHC validation. We also saw clear separation of BCR-free Kaplan-Meier survival curves in both DCLK1+ vs. DCLK1- PCa and in ERG+ DCLK+ vs. ERG+DCLK1- PCa. Importantly, statistical significance was not reached, which is likely due to small sample size, particularly in the ERG+DCLK1+ analysis (N=26 vs. 25, Figure 8). However, in samples that are both ERG+DCLK1+, this relationship appears to be more pronounced, suggesting ERG+ may have a role as a partial driver of DCLK1 mediated PCa progression. This is consistent with our data showing that ERG status itself is not associated with BCR, but rather, may be driving progression by other means. As DCLK1 correlated with ERG+ status, it may represent a downstream effector of ERG in primary PCa and CRPC and serve as a future therapeutic target in patients with ERG+ primary and CRPC.
Conclusions
We found ERG positivity in 43% and 25% of primary PCa and CRPC patients, respectively. Gene expression analysis with IHC confirmation suggests altered inflammatory pathways may be involved in ERG+ progression to CRPC. Moreover, we identified DCLK1 as a novel ERG-associated protein. Future mechanistic studies aimed at ERG and DCLK1 are needed to define their combined role in PCa progression.
Supplementary Material
A: ERG IHC assessment was performed and TMAs created for three groups: 1. One hundred and twenty-seven primary prostatectomy specimens (65 with BCR, 62 without); 2. Twenty LuCaP PDX with primary (radical prostatectomy or transurethral resection of the prostate) or metastatic (at time of death) origin; 3. One hundred and fifty-two metastatic CRPC patient specimens (79 bone and 73 visceral metastases; up to 4 per patient). B: A subset of each IHC grouping underwent gene array analysis, sorted by ERG status. The top overexpressed genes were studied and validated at the protein level using IHC and the TMAs described in A.
A: ERG expression relative to AR activity using a known 20-gene expression signature, B: STEAP1 expression, C: HOXB13 expression, D: KLK3 expression, and E: AR expression.
Ingenuity pathway analysis of CRPC metastases is shown. ERG (A.) and NF-κβ (B.) pathways were found to be enriched suggesting involvement in ERG+ CRPC progression.
A. NCALD expression was weakly associated with nuclear ERG expression in primary PCa (r=0.2021, p=0.0282). Representative IHC images of NCALD+ in ERG+ (a) and ERG- (b) primary PCa are shown. B. NCALD expression was not associated with nuclear ERG expression in CRPC specimens (r=0.1623, p=0.052). Representative IHC images of NCALD+ in ERG+ (a) and ERG- (b) CRPC specimens are shown.
A. CACNA1D expression was weakly associated with nuclear ERG in primary PCa (r=0.2522, p=0.0068). Representative IHC images of CACNA1D+ in ERG+ (a) and ERG- (b) primary PCa are shown. B. CACNA1D expression was not associated with nuclear ERG in CRPC specimens (r=-0.0133, p=0.8746). Representative IHC images of CACNA1D+ in ERG+ (a) and ERG- (b) CRPC specimens are shown.
Kaplan-Meier analysis of ERG+ vs. ERG- status did not correlate with BCR-free survival overall (p=0.41) or proliferation via Ki-67 (r=-0.0392, p=0.6885).
A. Kaplan-Meier analysis of HLA-DMB+ vs. HLA-DMB- status did not correlate with BCR-free survival overall (p=0.85). There was also no significant correlation with proliferation (r= 0.051, p=0.5843). B. In ERG+ specimens, HLA-DMB+ vs. HLA-DMB- status did not correlate with BCR-free survival (p=0.10) or proliferation (r=0.028, p=0.8451).
A. Kaplan-Meier analysis of CD3+ vs. CD3- status did not correlate with BCR-free survival overall (p=0.42). There was a positive correlation of proliferation relative to CD3+ status (r=0.261, p=0.0045). B. In ERG+ specimens, CD3+ vs. CD3- status did not correlate with BCR-free survival (p=0.50) or proliferation (r= 0.265, p=0.0605).
A. Kaplan-Meier analysis of NCALD+ vs. NCALD- status did not correlate with BCR-free survival overall (p=0.84). There was a trend towards positive correlation of proliferation relative to NCALD+ status (r=0.1794, p=0.0540). B. In ERG+ specimens, NCALD+ vs. NCALD- status did not correlate with BCR-free survival (p=0.14) or proliferation (r= 0.06085, p=0.6066)
A. Kaplan-Meier analysis of CACNA1D+ vs. CACNA1D- status did not correlate with BCR-free survival overall (p=0.09). There was a trend towards positive correlation of proliferation relative to CACNA1D+ status (r=0.1883, p=0.0468). B. In ERG+ specimens, CACNA1D+ vs. CANAID- status just correlated with BCR-free survival (p=0.05), but with limited CACNA1D- specimens. There was no association with proliferation (r= 0.03380, p=0.7765)
Clinical data and treatment information for the tissue acquisition necropsy patients in the study are shown (SD=standard deviation). *Indicates no information available for two patients. †Indicates no information available for two patients.
A. Genes with increased expression relative to PCa, LuCaP PDXs, and CRPC specimens are shown (as described in Figure 3). B. The top 100 ranked primary PCa genes with corresponding ranks in LuCaP PDX and CRPC metastases are shown. Highlighted in grey are those genes validated by protein expression using IHC and previously reported in the literature to be associated with ERG. DCLK1 (outlined) is a novel gene not previously reported in the PCa literature or in relation to ERG expression.
Acknowledgments
We would like to thank the patients and their families who were willing to participate in the Prostate Cancer Donor Program, for without them research of this nature would not be possible. We would also like to acknowledge Drs. Dan Lin, Jonathan Wright, William Ellis, Bruce Dalkin, Sarah Holt and the tissue acquisition necropsy teams in the Urology Department at the University of Washington. This material is the result of work supported by resources from the VA Puget Sound Health Care System, Seattle, Washington (RLV is a VA Biomedical Laboratory R&D Senior Research Career Scientist, PHL is a Staff Physician), by the Department of Defense (W81XWH-12-1-0399), the Pacific Northwest Prostate Cancer SPORE (P50CA97186), the PO1 NIH grant (PO1CA085859), P01CA163227 and the Richard M. LUCAS Foundation.
Footnotes
Disclosure Statement: United States Patent and Trademark Office awarded the patent for YK-4-279 an ERG inhibitor to Georgetown University, which includes A.Ü. as one of the inventors. A license agreement has been executed between Georgetown University and Tokalas, Inc. for the patent. A.Ü. is a shareholder of Tokalas, Inc. We have no other conflicts to disclose.
References
- 1.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Albadine R, Latour M, Toubaji A, Haffner M, Isaacs WB, Platz A, Meeker AK, Demarzo AM, Epstein JI, Netto GJ. TMPRSS2-ERG gene fusion status in minute (minimal) prostatic adenocarcinoma. Mod Pathol. 2009;22:1415–1422. doi: 10.1038/modpathol.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnel AD, Lafargue CJ, Vollmer RT, Corcos J, Bismar TA. TMPRSS2-ERG fusion is frequently observed in Gleason pattern 3 prostate cancer in a Canadian cohort. Cancer Biol Ther. 2009;8:125–130. doi: 10.4161/cbt.8.2.7134. [DOI] [PubMed] [Google Scholar]
- 4.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 5.Fine SW, Gopalan A, Leversha MA, Al-Ahmadie HA, Tickoo SK, Zhou Q, Satagopan JM, Scardino PT, Gerald WL, Reuter VE. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–1333. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, Eastham JA, Scardino PT, Scher HI, Tickoo SK, Reuter VE, Gerald WL. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, Trachtenberg J, Narod SA, Seth A. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 9.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 10.Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, Zielenska M, Soares FA, Squire JA. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 12.Scheble VJ, Scharf G, Braun M, Ruiz C, Sturm S, Petersen K, Beschorner R, Bachmann A, Zellweger T, Fend F, Kristiansen G, Bubendorf L, Wernert N, Shaikhibrahim Z, Perner S. ERG rearrangement in local recurrences compared to distant metastases of castration-resistant prostate cancer. Virchows Arch. 2012;461:157–162. doi: 10.1007/s00428-012-1270-7. [DOI] [PubMed] [Google Scholar]
- 13.Teng LH, Wang C, Begin LR, Dolph M, Yilmaz A, Trpkov K, Donnelly B, Bismar TA. ERG protein expression and gene rearrangements are present at lower rates in metastatic and locally advanced castration-resistant prostate cancer compared to localized disease. Urology. 2013;82:394–399. doi: 10.1016/j.urology.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Hagen RM, Adamo P, Karamat S, Oxley J, Aning JJ, Gillatt D, Persad R, Ladomery MR, Rhodes A. Quantitative analysis of ERG expression and its splice isoforms in formalin-fixed, paraffin-embedded prostate cancer samples: association with seminal vesicle invasion and biochemical recurrence. Am J Clin Pathol. 2014;142:533–540. doi: 10.1309/AJCPH88QHXARISUP. [DOI] [PubMed] [Google Scholar]
- 15.Qi M, Yang X, Zhang F, Lin T, Sun X, Li Y, Yuan H, Ren Y, Zhang J, Qin X, Han B. ERG rearrangement is associated with prostate cancer-related death in Chinese prostate cancer patients. PLoS One. 2014;9:e84959. doi: 10.1371/journal.pone.0084959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagglof C, Hammarsten P, Stromvall K, Egevad L, Josefsson A, Stattin P, Granfors T, Bergh A. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS One. 2014;9:e86824. doi: 10.1371/journal.pone.0086824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 18.Fleischmann A, Saramaki OR, Zlobec I, Rotzer D, Genitsch V, Seiler R, Visakorpi T, Thalmann GN. Prevalence and prognostic significance of TMPRSS2-ERG gene fusion in lymph node positive prostate cancers. Prostate. 2014;74:1647–1654. doi: 10.1002/pros.22882. [DOI] [PubMed] [Google Scholar]
- 19.Taris M, Irani J, Blanchet P, Multigner L, Cathelineau X, Fromont G. ERG expression in prostate cancer: the prognostic paradox. Prostate. 2014;74:1481–1487. doi: 10.1002/pros.22863. [DOI] [PubMed] [Google Scholar]
- 20.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, Suppan C, Flavin R, Sesso HD, Rider JR, Sweeney C, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci EL, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–1509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–1036. [PubMed] [Google Scholar]
- 22.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 23.Massoner P, Kugler KG, Unterberger K, Kuner R, Mueller LA, Falth M, Schafer G, Seifarth C, Ecker S, Verdorfer I, Graber A, Sultmann H, Klocker H. Characterization of transcriptional changes in ERG rearrangement-positive prostate cancer identifies the regulation of metabolic sensors such as neuropeptide Y. PLoS One. 2013;8:e55207. doi: 10.1371/journal.pone.0055207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, Zeng X, Zhang R, Huang J, Kuang X, Yang J, Liu J, Tawfik O, Thrasher JB, Li B. Cav1.3 channel alpha1D protein is overexpressed and modulates androgen receptor transactivation in prostate cancers. Urol Oncol. 2014;32:524–536. doi: 10.1016/j.urolonc.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Paulo P, Ribeiro FR, Santos J, Mesquita D, Almeida M, Barros-Silva JD, Itkonen H, Henrique R, Jeronimo C, Sveen A, Mills IG, Skotheim RI, Lothe RA, Teixeira MR. Molecular subtyping of primary prostate cancer reveals specific and shared target genes of different ETS rearrangements. Neoplasia. 2012;14:600–611. doi: 10.1593/neo.12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boormans JL, Korsten H, Ziel-van der Made AJ, van Leenders GJ, de Vos CV, Jenster G, Trapman J. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer. 2013;133:335–345. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- 27.Flammiger A, Bayer F, Cirugeda-Kuhnert A, Huland H, Tennstedt P, Simon R, Minner S, Bokemeyer C, Sauter G, Schlomm T, Trepel M. Intratumoral T but not B lymphocytes are related to clinical outcome in prostate cancer. APMIS. 2012;120:901–908. doi: 10.1111/j.1600-0463.2012.02924.x. [DOI] [PubMed] [Google Scholar]
- 28.Qu D, Johnson J, Chandrakesan P, Weygant N, May R, Aiello N, Rhim A, Zhao L, Zheng W, Lightfoot S, Pant S, Irvan J, Postier R, Hocker J, Hanas JS, Ali N, Sureban SM, An G, Schlosser MJ, Stanger B, Houchen CW. Doublecortin-like kinase 1 is elevated serologically in pancreatic ductal adenocarcinoma and widely expressed on circulating tumor cells. PLoS One. 2015;10:e0118933. doi: 10.1371/journal.pone.0118933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sureban SM, May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Pantazis P, Rao CV, Postier RG, Houchen CW. DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS One. 2013;8:e73940. doi: 10.1371/journal.pone.0073940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirzaei A, Tavoosidana G, Modarressi MH, Rad AA, Fazeli MS, Shirkoohi R, TavakoliYaraki M, Madjd Z. Upregulation of circulating cancer stem cell marker, DCLK1 but not Lgr5, in chemoradiotherapy-treated colorectal cancer patients. Tumour Biol. 2015 doi: 10.1007/s13277-015-3132-9. [DOI] [PubMed] [Google Scholar]
- 31.Chandrakesan P, Weygant N, May R, Qu D, Chinthalapally HR, Sureban SM, Ali N, Lightfoot SA, Umar S, Houchen CW. DCLK1 facilitates intestinal tumor growth via enhancing pluripotency and epithelial mesenchymal transition. Oncotarget. 2014;5:9269–9280. doi: 10.18632/oncotarget.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagliardi G, Goswami M, Passera R, Bellows CF. DCLK1 immunoreactivity in colorectal neoplasia. Clin Exp Gastroenterol. 2012;5:35–42. doi: 10.2147/CEG.S30281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weygant N, Qu D, May R, Tierney RM, Berry WL, Zhao L, Agarwal S, Chandrakesan P, Chinthalapally HR, Murphy NT, Li JD, Sureban SM, Schlosser MJ, Tomasek JJ, Houchen CW. DCLK1 is a broadly dysregulated target against epithelial-mesenchymal transition, focal adhesion, and stemness in clear cell renal carcinoma. Oncotarget. 2015;6:2193–2205. doi: 10.18632/oncotarget.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis WJ, Vessella RL, Buhler KR, Bladou F, True LD, Bigler SA, Curtis D, Lange PH. Characterization of a novel androgen-sensitive, prostate-specific antigen-producing prostatic carcinoma xenograft: LuCaP 23. Clin Cancer Res. 1996;2:1039–1048. [PubMed] [Google Scholar]
- 35.Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, Vessella RL. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34:646–653. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 36.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, Murali R, Vickers A, Scardino PT, Sander C, Reuter V, Taylor BS, Sawyers CL. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111:11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes Dev. 2011;25:2147–2157. doi: 10.1101/gad.17546311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Debes JD, Roberts RO, Jacobson DJ, Girman CJ, Lieber MM, Tindall DJ, Jacobsen SJ. Inverse association between prostate cancer and the use of calcium channel blockers. Cancer Epidemiol Biomarkers Prev. 2004;13:255–259. doi: 10.1158/1055-9965.epi-03-0093. [DOI] [PubMed] [Google Scholar]
- 41.Fitzpatrick AL, Daling JR, Furberg CD, Kronmal RA, Weissfeld JL. Hypertension, heart rate, use of antihypertensives, and incident prostate cancer. Ann Epidemiol. 2001;11:534–542. doi: 10.1016/s1047-2797(01)00246-0. [DOI] [PubMed] [Google Scholar]
- 42.O'Callaghan DW, Ivings L, Weiss JL, Ashby MC, Tepikin AV, Burgoyne RD. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction. J Biol Chem. 2002;277:14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- 43.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R, Ren C, Ayala G, Ittmann M. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res. 2011;71:1325–1333. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dryden NH, Sperone A, Martin-Almedina S, Hannah RL, Birdsey GM, Khan ST, Layhadi JA, Mason JC, Haskard DO, Gottgens B, Randi AM. The transcription factor Erg controls endothelial cell quiescence by repressing activity of nuclear factor (NF)-kappaB p65. J Biol Chem. 2012;287:12331–12342. doi: 10.1074/jbc.M112.346791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callahan MJ, Nagymanyoki Z, Bonome T, Johnson ME, Litkouhi B, Sullivan EH, Hirsch MS, Matulonis UA, Liu J, Birrer MJ, Berkowitz RS, Mok SC. Increased HLA-DMB expression in the tumor epithelium is associated with increased CTL infiltration and improved prognosis in advanced-stage serous ovarian cancer. Clin Cancer Res. 2008;14:7667–7673. doi: 10.1158/1078-0432.CCR-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenberger SP, Toes RE, van der Voort EI, Offringa R, Melief CJ. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 49.Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess HA, Martinez S, Reiner O. KIAA0369, doublecortin-like kinase, is expressed during brain development. J Neurosci Res. 1999;58:567–575. [PubMed] [Google Scholar]
- 51.Souza RF, Krishnan K, Spechler SJ. Acid, bile, and CDX: the ABCs of making Barrett's metaplasia. Am J Physiol Gastrointest Liver Physiol. 2008;295:G211–G218. doi: 10.1152/ajpgi.90250.2008. [DOI] [PubMed] [Google Scholar]
- 52.Vega KJ, May R, Sureban SM, Lightfoot SA, Qu D, Reed A, Weygant N, Ramanujam R, Souza R, Madhoun M, Whorton J, Anant S, Meltzer SJ, Houchen CW. Identification of the putative intestinal stem cell marker doublecortin and CaM kinase-like-1 in Barrett's esophagus and esophageal adenocarcinoma. J Gastroenterol Hepatol. 2012;27:773–780. doi: 10.1111/j.1440-1746.2011.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: ERG IHC assessment was performed and TMAs created for three groups: 1. One hundred and twenty-seven primary prostatectomy specimens (65 with BCR, 62 without); 2. Twenty LuCaP PDX with primary (radical prostatectomy or transurethral resection of the prostate) or metastatic (at time of death) origin; 3. One hundred and fifty-two metastatic CRPC patient specimens (79 bone and 73 visceral metastases; up to 4 per patient). B: A subset of each IHC grouping underwent gene array analysis, sorted by ERG status. The top overexpressed genes were studied and validated at the protein level using IHC and the TMAs described in A.
A: ERG expression relative to AR activity using a known 20-gene expression signature, B: STEAP1 expression, C: HOXB13 expression, D: KLK3 expression, and E: AR expression.
Ingenuity pathway analysis of CRPC metastases is shown. ERG (A.) and NF-κβ (B.) pathways were found to be enriched suggesting involvement in ERG+ CRPC progression.
A. NCALD expression was weakly associated with nuclear ERG expression in primary PCa (r=0.2021, p=0.0282). Representative IHC images of NCALD+ in ERG+ (a) and ERG- (b) primary PCa are shown. B. NCALD expression was not associated with nuclear ERG expression in CRPC specimens (r=0.1623, p=0.052). Representative IHC images of NCALD+ in ERG+ (a) and ERG- (b) CRPC specimens are shown.
A. CACNA1D expression was weakly associated with nuclear ERG in primary PCa (r=0.2522, p=0.0068). Representative IHC images of CACNA1D+ in ERG+ (a) and ERG- (b) primary PCa are shown. B. CACNA1D expression was not associated with nuclear ERG in CRPC specimens (r=-0.0133, p=0.8746). Representative IHC images of CACNA1D+ in ERG+ (a) and ERG- (b) CRPC specimens are shown.
Kaplan-Meier analysis of ERG+ vs. ERG- status did not correlate with BCR-free survival overall (p=0.41) or proliferation via Ki-67 (r=-0.0392, p=0.6885).
A. Kaplan-Meier analysis of HLA-DMB+ vs. HLA-DMB- status did not correlate with BCR-free survival overall (p=0.85). There was also no significant correlation with proliferation (r= 0.051, p=0.5843). B. In ERG+ specimens, HLA-DMB+ vs. HLA-DMB- status did not correlate with BCR-free survival (p=0.10) or proliferation (r=0.028, p=0.8451).
A. Kaplan-Meier analysis of CD3+ vs. CD3- status did not correlate with BCR-free survival overall (p=0.42). There was a positive correlation of proliferation relative to CD3+ status (r=0.261, p=0.0045). B. In ERG+ specimens, CD3+ vs. CD3- status did not correlate with BCR-free survival (p=0.50) or proliferation (r= 0.265, p=0.0605).
A. Kaplan-Meier analysis of NCALD+ vs. NCALD- status did not correlate with BCR-free survival overall (p=0.84). There was a trend towards positive correlation of proliferation relative to NCALD+ status (r=0.1794, p=0.0540). B. In ERG+ specimens, NCALD+ vs. NCALD- status did not correlate with BCR-free survival (p=0.14) or proliferation (r= 0.06085, p=0.6066)
A. Kaplan-Meier analysis of CACNA1D+ vs. CACNA1D- status did not correlate with BCR-free survival overall (p=0.09). There was a trend towards positive correlation of proliferation relative to CACNA1D+ status (r=0.1883, p=0.0468). B. In ERG+ specimens, CACNA1D+ vs. CANAID- status just correlated with BCR-free survival (p=0.05), but with limited CACNA1D- specimens. There was no association with proliferation (r= 0.03380, p=0.7765)
Clinical data and treatment information for the tissue acquisition necropsy patients in the study are shown (SD=standard deviation). *Indicates no information available for two patients. †Indicates no information available for two patients.
A. Genes with increased expression relative to PCa, LuCaP PDXs, and CRPC specimens are shown (as described in Figure 3). B. The top 100 ranked primary PCa genes with corresponding ranks in LuCaP PDX and CRPC metastases are shown. Highlighted in grey are those genes validated by protein expression using IHC and previously reported in the literature to be associated with ERG. DCLK1 (outlined) is a novel gene not previously reported in the PCa literature or in relation to ERG expression.


