Abstract
Ski interacting protein (Skip) has been found to bind to the highly conserved region of Ski, which is required for its transforming activity. Ski is a unique oncoprotein that is involved in inducing both transformation and differentiation. At the molecular level, Ski has been shown to exhibit either co-activator or co-repressor activity depending on the cellular and promoter context. We were interested in further elucidating the biological implications of the Ski–Skip interaction. Here we have identified the SNW domain of Skip as the interaction region for Ski. This domain of Skip is highly conserved in all the Skip homologues identified from different species. Using a series of reporter plasmids, we show that Skip is a potent transcriptional activator of many different promoters, the activity of which was also mapped to the conserved core SNW domain of the protein. Addition of excess Ski further augmented the transcriptional activities of Skip, suggesting that one of the ways in which Ski brings about transformation is by binding and cooperating with the SNW domain of Skip in transcriptional activation.
INTRODUCTION
Skip was discovered as a Ski interacting protein in a two-hybrid screen, using the oncogene v-Ski as bait, and interacts with both the cellular and viral forms of Ski (1). Interestingly, Skip was found to interact with a highly conserved region of Ski required for its transforming activity (1), suggesting that this interaction was important for the ability of Ski to transform cells. v-Ski was originally identified in avian Sloan–Kettering viruses and was found to transform chicken embryo fibroblasts (2). The cellular homologue c-Ski has been identified from several species, including human, chicken and Xenopus (3–5). The v-Ski protein lacks a 292 amino acid region from the C-terminus of c-Ski, but retains the N-proximal cysteine region (6). This region is responsible for the cellular transformation and the myogenic activities of Ski (7).
Overexpression of c-Ski or v-Ski induces either transformation or muscle differentiation of quail embryo fibroblasts, depending on the growth conditions (8,9). Ski overexpression also causes postnatal hypertrophy of type II fast muscle fibres in transgenic mice (10). Moreover, germline inactivation in mice demonstrates that loss of Ski function results in decreased myofibre development in addition to other abnormalities (11). The capacity of Ski to induce both transformation (growth) and differentiation, which is usually associated with the cessation of growth, is an intriguing paradox. Because of its nuclear localisation and its ability to induce expression of muscle-specific genes in quail cells (8), Ski has been assumed to be a transcription factor. Indeed, Ski can function either as a transcriptional activator (12,13) or as a repressor (14), depending on the specific promoters involved. Recombinant c-Ski protein purified from Escherichia coli fails to bind DNA, whereas c-Ski in mammalian cell nuclear extracts retains DNA binding activity suggesting that c-Ski binds to DNA only when it is associated with other proteins (15). Ski activates transcription of muscle-specific and certain viral promoters (16), and has also been found to enhance transcriptional activation by binding nuclear factor I sequences (13). In addition, c-Ski has been observed to be a component of the histone deacetylase complex, HDAC1 (17,18), and also interacts with Smad proteins to regulate transforming growth factor-β (TGF-β) signalling (19,20). Despite these observations, the molecular mechanisms by which Ski transforms cells and regulates differentiation remain unknown, as does the role of Skip in these processes.
There is mounting evidence which suggests that Skip also has a role in the regulation of transcription. The Drosophila melanogaster homologue of Skip, Bx42, is found associated with chromatin in transcriptionally active puffs of salivary glands (21,22), suggesting a role in the regulation of transcription. Recently, Skip has been shown to have a role in the EBNA2 (the Epstein–Barr virus-encoded latency protein) activation of CBF1-repressed promoters (23). Contacts with both CBF1 and Skip were shown to be important for the effective targeting of EBNA2 to DNA. Skip was also shown to interact with the ankyrin repeat domain of NotchIC to facilitate NotchIC function in the activation of downstream target genes of the Notch signalling pathway (24). Most recently, Skip has also been shown to interact with Smad proteins to augment TGF-β-dependent transcription (25).
We were interested in further investigating the biochemical significance of the Ski–Skip interaction. Here, we have mapped the interaction site between Ski and Skip. We also show that Skip is capable of activating diverse promoters and that ectopic expression of Skip and Ski results in synergistic co-activation. This co-activation activity maps to the region of Skip required for binding Ski, which encompasses the highly conserved SNW domain. Taken together, these studies provide a biochemical basis for the transforming activity of the Ski oncoprotein.
MATERIALS AND METHODS
Plasmids
Various forms of Skip used in the study were cloned into the BamHI–EcoRI sites of pcDNA 3.1 by PCR amplification from the Skip expression plasmid, pCGSP-Skip (1). These constructs were verified by DNA sequencing using the dideoxy chain termination method (26) and were used both for in vitro translations and for expression in cells. The cellular Ski expression plasmid, pMT2-Ski, has been described previously (1) and the GST-Ski expression plasmid was a kind gift from S. Ishii (Tsukuba Institute, Ibaraki, Japan) (17). pJ4Ω.16 E2 has been described previously (27). The pTKM.32 CAT reporter plasmid has also been described previously (28). GAL4 CAT (pG5CAT), which contains five consensus GAL4 binding sites upstream of the E1B minimal promoter, was obtained from commercial sources (Clontech). The other CAT reporter constructs, p21 CAT (pWWP-CAT), pBLCAT2, pBLCAT3 and AdE2CAT have been described previously (29–31).
Cells
U2OS and Saos-2 cells were grown in DMEM supplemented with 10% foetal calf serum. Transfections were performed using calcium phosphate precipitation as described previously (32).
Transfections and CAT assays
Cells were transfected with 1 µg of reporter plasmids (unless otherwise stated) along with the indicated amounts of expression plasmids. Where titrations were performed, DNA input was balanced with empty vector DNA. After 48 h the cells were harvested in 100 µl CAT buffer (40 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA) and subjected to three cycles of freeze-thawing, followed by incubation at 65°C for 10 min. Samples were clarified by centrifugation in an eppendorf centrifuge at 14 000 r.p.m. for 2 min and the protein concentration of the supernatant was measured by the Bio-Rad protein assay. CAT assays were routinely performed with 5–10 µg of protein incubated with 2.5 µl acetyl-CoA (33.3 mg/ml) and 1.5 µl [14C]chloramphenicol (50 mCi/mol; Amersham) in a final volume of 50 µl at 37°C for 1 h. Following extraction with ethyl acetate, samples were analysed by thin layer chromatography and quantified with a Packard Instant Imager (Packard, Meriden, CT).
Expression of Skip was verified by western blotting from total cell extracts. Protein concentrations were determined using the Bradford method (Bio-Rad). Samples of total protein lysate (10 µg) were subjected to SDS–PAGE, transferred to nitrocellulose membranes (Schleicher & Schuell), and then probed with the antiserum for Skip. Skip antibodies were raised in rabbit against glutathione S-transferase (GST) fused to an N-terminal Skip fragment comprising amino acids 1–219. The haemagglutinin (HA) monoclonal antibody was obtained from Babco (16B12). Complexes of primary and horseradish peroxidase-linked secondary antibodies (Dako) were detected by enhanced chemiluminescence (ECL) (Amersham).
In vitro translations and GST pull-down assays
35[S]-labelled proteins were produced in vitro by using a coupled transcription–translation system (Promega TNT) according to the manufacturer’s instructions. For fusion protein production and purification, 100 ml of an overnight culture of E.coli strain BL-21 containing the GST–Ski expression plasmid were inoculated into 1 l of Luria broth containing ampicillin and incubated at 37°C for a further 1 h. Recombinant protein expression was induced for 3 h with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (Sigma). The cells were harvested by centrifugation, disrupted by sonication and cell debris was removed by centrifugation. The levels of protein induction were then determined by SDS–PAGE and Coomassie brilliant blue R staining.
Equal amounts of GST and GST–Ski fusion protein bound to glutathione-linked agarose (Sigma) were incubated with the in vitro translated proteins for 1 h at room temperature in a buffer containing 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 2.5 mM EDTA. Bound proteins were washed extensively in PBS containing 0.5% NP-40 before analysis by SDS–PAGE and autoradiography.
RESULTS
Identification of the Ski interacting domain on Skip
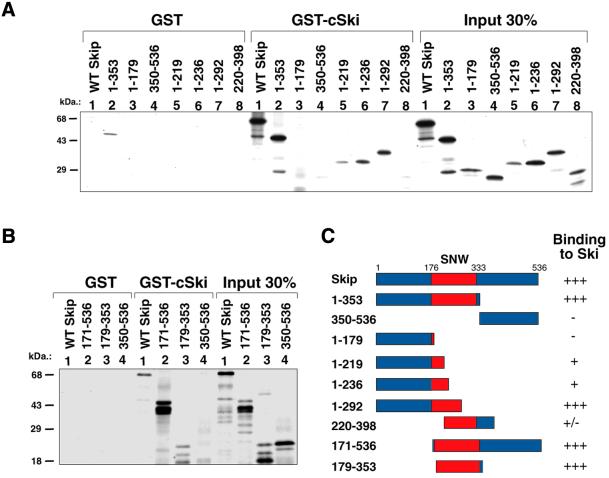
The Skip binding region of Ski has been shown to be required for its transformation activity, however the region of Skip involved in the interaction has not yet been identified. Therefore, we first made a series of Skip deletion constructs in pcDNA3.1, shown in Figure 1C. The various Skip deletion mutants were translated in vitro and GST pull-down assays were performed using GST–c-Ski fusion protein bound to glutathione-coupled agarose. The results obtained are shown in Figure 1A and B. As can be seen, the wild-type Skip showed strong binding to c-Ski, in agreement with previous studies (1). In addition, mutants 1–353 and 171–536 showed wild-type levels of interaction, whereas the C-terminal mutant containing amino acid residues 350–536 failed to bind and only weak binding was obtained with the mutant spanning residues 1–179. Interestingly, the mutants spanning residues 1–292 and 179–353 retained wild type-like binding with Ski. These results clearly show that the region of Skip required for interaction with Ski lies principally between residues 179 and 292 within the SNW domain, which is defined by amino acids from 176 to 333. Interestingly, this SNW domain is highly conserved in all the Skip homologues identified from different species (1,22,33–35), suggesting that this Ski interaction domain has been highly conserved through evolution.
Figure 1.
Mapping the c-Ski interaction domain on Skip. (A and B) Results from representative GST pull-down assays. Purified GST or GST–c-Ski on glutathione resin were incubated with the in vitro translated Skip deletion mutants for 1 h at room temperature. After extensive washing, bound proteins were analysed by SDS–PAGE and autoradiography. (C) A schematic diagram of the Skip deletion mutants used in the analysis. Numbers refer to the amino acid residues and the central red boxed region corresponds to the highly conserved SNW region, where homology with Skip proteins derived from other species is >90%. The relative strength of binding of each protein to c-Ski is also shown, together with the mean percentage binding from at least three separate assays, where +++ and +/– indicate 20–30% and 2–3% binding, respectively.
Skip activates diverse promoters
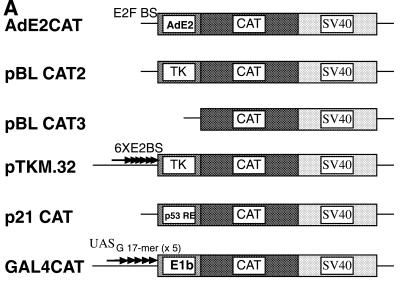
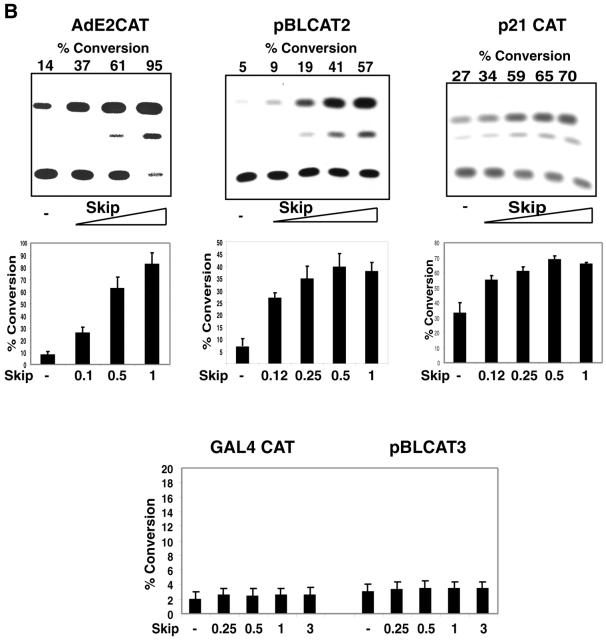
Having identified the specific interaction domain of Ski on Skip, we next investigated further characterisation of the Skip protein. There are now a number of reports that describe Skip affecting the transcriptional activities of a number of different promoters (23–25). To further investigate this, we analysed the effects of Skip on a series of different reporter constructs containing both cellular and viral promoters. These are shown schematically in Figure 2A. We investigated the effects of Skip on two viral promoters: the Adenovirus E2 (AdE2) promoter (31) and the HSV TK promoter (pBLCAT2) (30). For a cellular promoter we used the p53-responsive p21 promoter (29). U2OS cells were transfected with the different reporter constructs together with increasing amounts of Skip expression plasmid. After 48 h the cells were harvested and CAT assays were performed. The results obtained are shown in Figure 2B. It is clear from this analysis that Skip is a potent transcriptional activator of the viral and cellular promoters tested, showing strong dose-dependent activation of the AdE2 promoter, the HSV TK promoter and the p53-responsive p21 promoter. In contrast, no increase in transcription is seen using the heterologous GAL4 promoter, which cannot function without the GAL4 activation domain in mammalian cells, or when using the pBLCAT3 reporter (30), which lacks the HSV TK promoter (Fig. 2A). These results suggest that Skip may behave as a general transcriptional co-activator, since increased transcriptional activity was only obtained with Skip on already active promoters. No transcriptional activity was observed on the p53-responsive p21 promoter in p53-negative Saos-2 cells (data not shown). To further investigate the apparent general transcriptional activity of Skip, we performed promoter activation assays in the presence of the HPV-16 E2 transactivator (27,36). Cells were transfected with the pTKM.32 reporter, which contains multiple E2 binding sites upstream of the TK promoter, either alone or together with Skip and HPV-16 E2 in various combinations. After 48 h the cells were harvested and CAT assays were performed. The results obtained are shown in Figure 3. As can be seen, HPV-16 E2 transactivation activity is greatly increased in a dose-dependent manner when Skip is added in increasing concentrations (Fig. 3). These results demonstrate that Skip can act as a transcriptional co-activator with the HPV-16 E2 protein, and further supports the notion that Skip functions as a co-activator.
Figure 2.
Skip stimulates the activation of diverse viral and cellular promoters. (A) CAT reporter constructs used in the study. The promoter and SV40 polyadenylation signal is shown. BS refers to DNA binding site, RE refers to responsive element, 6XE2BS means six HPV-16 E2 binding sites. GAL4 CAT contains five consensus GAL4 binding sites, which is represented as UASG17-mer (× 5). (B) U2OS cells were transfected with the indicated reporter plasmids together with increasing amounts of the Skip expression plasmid as shown (concentrations are shown in micrograms). After 48 h the cells were harvested and CAT assays performed. In each case a representative CAT assay is shown, together with the collated results from at least three independent transfections. Standard deviations are shown.
Figure 3.
Skip is a transcriptional co-activator. U2OS cells were transfected with the HPV-16 E2-responsive pTKM.32 CAT reporter plasmid together with 0.1 or 0.25 µg of pJ4womega.16 E2, as indicated. For assessing Skip co-activation, the input of pJ4womega.16 E2 was fixed at 0.1 µg and increasing amounts of Skip expression plasmid were used as indicated (concentrations are shown in micrograms). After 48 h the cells were harvested and CAT assays performed. (A) Results from a representative assay; (B) collated results from at least three independent transfections. Standard deviations are shown.
Ski and Skip cooperate in transcriptional transactivation
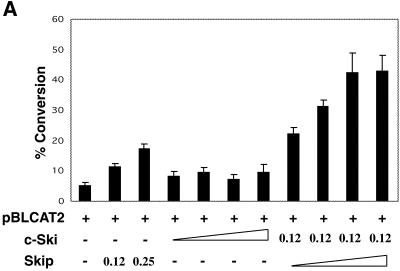
We next investigated the effect of Ski on Skip’s transcriptional activation activity. In order to do this, we chose to analyse the effects of Ski upon Skip activation of the HSV TK promoter (pBLCAT2). U2OS cells were transiently transfected with the pBLCAT2 reporter plus Skip or c-Ski, alone or in combination, and CAT assays were performed after 48 h. The results are shown in Figure 4A. As can be seen, addition of Skip alone results in a significant increase in promoter activity. In contrast, addition of Ski alone has a minimal effect. Strikingly, co-transfection of Ski and Skip results in a dramatic increase in the HSV TK promoter activity. The cooperation between Skip and Ski in transcriptional activation was also obtained in Saos-2 cells, on the AdE2 promoter, demonstrating that the synergistic activity of Skip+Ski is neither cell-type, nor promoter, specific (Fig. 4B).
Figure 4.
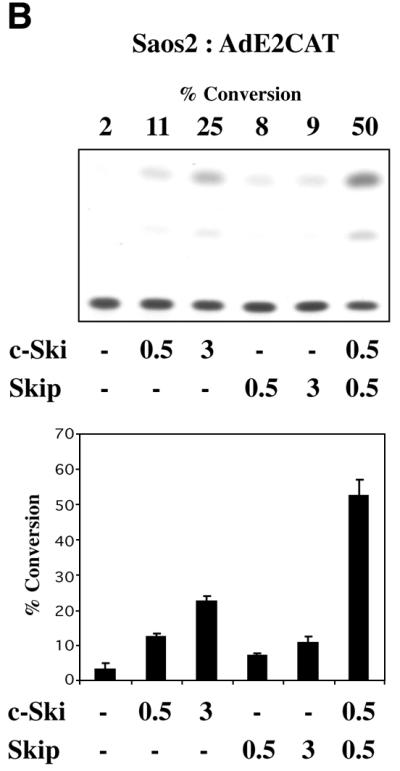
Skip and c-Ski can synergistically activate transcription. (A) HSV TK promoter. U2OS cells were transfected with the pBLCAT2 reporter plasmid together with the combinations of Skip and c-Ski expression plasmids indicated (concentrations are in micrograms). The concentrations of c-Ski and Skip expression plasmids used were 0.125, 0.25, 0.5 and 1 µg. (B) AdE2 promoter. Saos-2 cells were transfected with the AdE2 reporter plasmid together with the combinations of Skip and c-Ski expression plasmids indicated (concentrations are in micrograms). After 48 h the cells were harvested and CAT assays performed. The graph shows the collated results from at least three independent transfections. Standard deviations are shown.
The SNW domain confers transcriptional activity
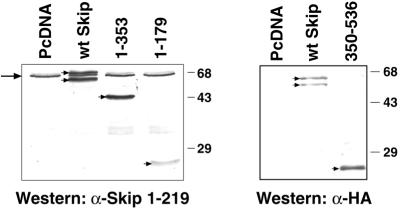
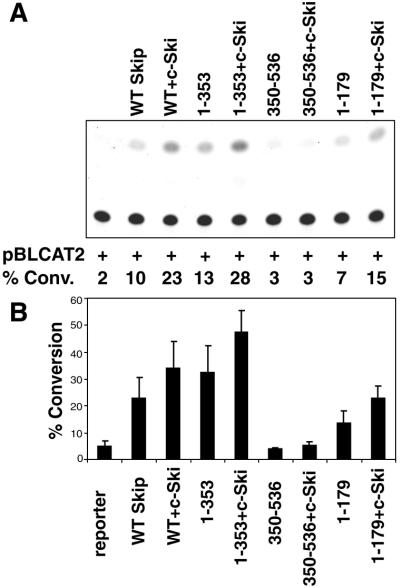
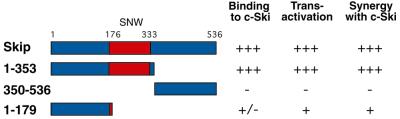
Having characterised Skip as a transcriptional activator, we next investigated whether the region of Skip required for binding Ski, the SNW domain, was also involved in transcriptional transactivation. In order to first ensure that the mutant Skip proteins were being stably expressed, U2OS cells were transfected with each of the expression plasmids, and mutant Skip expression was assessed by western blotting. The results obtained are shown in Figure 5. As can be seen, the mutants of Skip produced stable protein and comparable levels of expression. For the reporter assays, cells were transfected with the various forms of mutant Skip expression plasmids together with a pBLCAT2 reporter in the presence or absence of c-Ski. Cells were harvested after 48 h and transcriptional assays were performed: the results obtained are shown in Figure 6. Strikingly, the region of Skip found to be responsible for binding Ski is also essential for the ability of Skip to activate transcription. Thus, the construct containing amino acid residues 1–353 behaves very similarly to the wild-type Skip in both transcriptional activation and in its ability to synergise with c-Ski. In contrast, the mutant comprising amino acid residues 350–536, which is completely defective in its ability to bind c-Ski, is also defective in its ability to activate transcription and to synergise with c-Ski. Interestingly, the construct containing amino acid residues 1–179, which showed very weak binding to c-Ski, retains weak transcriptional activating activity and exhibits modest synergistic activity with c-Ski. These results are summarised in Figure 7. Taken together, they demonstrate that the transcriptional activation activity of Skip and its ability to synergise with c-Ski are largely dependent upon the ability of Skip to interact with c-Ski, which in turn is dependent upon the highly conserved SNW domain of Skip.
Figure 5.
Expression levels of Skip proteins in U2OS. Total protein extracts were prepared, transferred to nitrocellulose membrane and incubated with either anti-Skip polyclonal antibody or with an anti-HA-tag monoclonal antibody. Staining was done with the ECL detection system. Arrow indicates the position of endogenous Skip protein, arrowheads indicate position of transfected Skip protein.
Figure 6.
The c-Ski binding region of Skip is required for its ability to stimulate the HSV TK promoter. U2OS cells were transfected with the pBLCAT2 reporter construct together with 0.25 µg of the different deletion constructs of Skip. In addition, 0.5 µg of c-Ski expression plasmid was also included as indicated. (A) Results from a representative assay; (B) collated results from at least three separate transfections. Standard deviations are shown.
Figure 7.
Schematic diagram showing the domains of Skip required for binding c-Ski, activating transcription and synergising with c-Ski. Numbers refer to the amino acid residues and the central red boxed region corresponds to the evolutionarily conserved SNW region.
DISCUSSION
Ski is involved in both oncogenic transformation and terminal differentiation. The mechanism of how Ski carries out both of these diverse processes is not known. Skip was found to interact with the N-terminal region of Ski, which is essential for its transforming activity. However, the role of Skip in Ski’s transforming activity has not been elucidated. Here we focused on the specificity and biochemical significance of the Ski–Skip interaction. We provide evidence that the evolutionarily conserved SNW domain of Skip is the binding domain of Ski. A series of deletions within the Skip protein were produced and assessed for their ability to interact with c-Ski in a series of GST pull-down assays. Although a weak interaction could be mediated by amino acid residues 1–179, the principal region of Skip responsible for binding c-Ski was mapped to the portion of the protein spanning amino acid residues 179–292, which lie within the central region of the SNW domain. No binding was detected within the C-terminal half of the protein between residues 350 and 536. Strikingly, this c-Ski binding region of Skip is very highly conserved amongst all the Skip proteins that have so far been characterised from different species, including D.melanogaster (22), Dictyostelium discoideum (34) and Saccharomyces cerevisiae (33,35). This demonstrates that this interaction domain is evolutionarily very highly conserved, suggesting a central role for the SNW region in the function of these two proteins.
Bx42, the Drosophila homologue of Skip, is assumed to be involved in transcriptional regulation, since it has been found to be associated with transcriptionally active puffs of salivary glands. In support of this, studies have shown that Skip has a role in the EBNA2 and NotchIC activation of target genes (23,24). Since Skip has now been shown to play a role in the transcription of a number of different genes, we thought that this could well represent a more general transcriptional effect of Skip. Indeed, using a variety of different promoters we have been able to show that Skip is a general co-activator. We first analysed the ability of Skip to activate expression from a series of viral and cellular promoters. To do this we analysed the p53-responsive p21 promoter, the AdE2 promoter and the HSV TK promoter. In all cases strong transcriptional activation was obtained in the presence of Skip. In contrast, no transcriptional activation was obtained using the inactive GAL4 or the basal pBLCAT3, nor was there upregulation of the p21 promoter in the absence of p53, suggesting that Skip can only stimulate transcription of already active promoters. In order to address this further, we performed transactivation assays with the HPV-16 E2 transactivator on a promoter containing six E2 binding sites upstream of the HSV TK promoter. As expected, the ability of E2 to activate this promoter was greatly increased in the presence of Skip, providing additional evidence that Skip can be classed as a general transcriptional co-activator. In addition, more recently Skip was also found to activate TGF-β-dependent transcription (25), as well as muscle specific E-box mediated transcription through MyoD, which further supports our data (37).
We next proceeded to investigate the mechanisms underlying the transcriptional activation function of Skip. An obvious candidate for a role in this activity of Skip was Skip’s known interacting cellular partner, c-Ski. Indeed, addition of c-Ski to Skip transactivation assays produced a dramatic synergistic activation of diverse target promoters, indicating that Skip and c-Ski cooperate in promoter activation. Interestingly, previous studies had shown that c-Ski could also partially overcome pRb suppression of E2F-1 responsive promoters, although in our analysis we found that c-Ski could activate the E2F-1 regulated AdE2 promoter in Saos-2 cells. These cells lack functional pRb, therefore it is likely that c-Ski may also regulate transcription independently of its interaction with pRb. In addition, when mutants of Skip were assessed for their ability to activate gene expression, only those which retained the ability to interact with c-Ski could also upregulate promoter activity. Further, the synergistic transcriptional activity was only obtained when both Skip and c-Ski were present, and with Skip mutant proteins which could still interact with c-Ski. These results therefore provide a function for this highly conserved SNW region of Skip, and further demonstrate that the ability of Skip to interact with c-Ski is required both for its intrinsic transcriptional activity and for its ability to act synergistically with c-Ski. The fact that the transforming activity of Ski requires its ability to interact with Skip suggests that one of the ways in which Ski brings about transformation is through its cooperation with Skip. Taken together, these results define a function for the Skip–Ski complex and provide a molecular basis for the role of Skip’s highly conserved SNW domain.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Shunsuke Ishii for the GST–Ski expression plasmid, and to David Pim and Miranda Thomas for their comments on the manuscript. This work was supported in part by research grants from the Associazione Italiana per la Ricerca Sul Cancro and the EU Biomed 2 program to L.B. and NIH grant CA42573 to M.H.
References
- 1.Dahl R., Wani,B. and Hayman,M.J. (1998) The Ski oncoprotein interacts with Skip, the human homologue of Drosophila Bx42. Oncogene, 16, 1579–1586. [DOI] [PubMed] [Google Scholar]
- 2.Li Y., Turck,C.M., Teumer,J.K. and Stavnezer,E. (1986) Unique sequence, Ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J. Virol., 57, 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nomura N., Sasamoto,S., Ishii,S., Date,T., Matsui,M. and Ischizaki,R. (1989) Isolation of human cDNA clones of Ski and Ski-related gene Sno. Nucleic Acids Res., 17, 5489–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleeman J.P. and Laskey,R.A. (1993) Xenopus c-Ski contains a novel coiled-coil protein domain and is maternally expressed during development. Oncogene, 8, 67–77. [PubMed] [Google Scholar]
- 5.Sutrave P. and Hughes,S.H. (1989) Isolation and characterization of three distinct cDNAs for the chicken c-Ski gene. Mol. Cell. Biol., 9, 4046–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stavnezer E., Brodeur,D. and Brennan,L.A. (1989) The v-Ski oncogene encodes a truncated set of c-Ski coding exons with limited sequence and structural relatedness to v-myc. Mol. Cell. Biol., 9, 4038–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G., Teumer,J., Colmenares,C., Richmond,C. and Stavnezer,E. (1997) Identification of a core functional and structural domain of the v-Ski oncoprotein responsible for both transformation and myogenesis. Oncogene, 15, 459–471. [DOI] [PubMed] [Google Scholar]
- 8.Colmenares C. and Stavnezer,E. (1989) The Ski oncogene induces muscle differentiation in quail embryo cells. Cell, 59, 293–303. [DOI] [PubMed] [Google Scholar]
- 9.Colmenares C., Sutrave,P., Hughes,S.H. and Stavnezer,E. (1991) Activation of the c-Ski oncogene by overexpression. J. Virol., 65, 4929–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutrave P., Kelly,A.M. and Hughes,S.H. (1990) Ski can cause selective growth of skeletal muscle in transgenic mice. Genes Dev., 4, 1462–1472. [DOI] [PubMed] [Google Scholar]
- 11.Berk M., Desai,S.Y., Heyman,H.C. and Colmenares,C. (1997) Mice lacking the Ski proto-oncogene have defects in neurulation, craniofacial patterning, and skeletal muscle development. Genes Dev., 11, 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engert J.C., Servaes,S., Sutrave,P., Hughes,S.H. and Rosenthal,N. (1995) Activation of a muscle-specific enhancer by the Ski proto-oncogene. Nucleic Acids Res., 23, 2988–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarapore P., Richmond,C., Zheng,G., Cohen,S.B., Kelder,B., Kopchick,J., Kruse,U., Sippel,A.E., Colmenares,C. and Stavnezer,E. (1997) DNA binding and transcriptional activation by the Ski oncoprotein mediated by interaction with NF1. Nucleic Acids Res., 25, 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicol R. and Stavnezer,E. (1998) Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J. Biol. Chem., 273, 3588–3597. [DOI] [PubMed] [Google Scholar]
- 15.Nagase T., Mizuguchi,G., Nomura,N., Ishizaki,R., Ueno,Y. and Ishii,S. (1990) Requirement of protein co-factor for the DNA binding function of the human Ski proto-oncogene product. Nucleic Acids Res., 18, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelder B., Richmond,C., Stavnezer,E., List,E.O. and Kopchick,J.J. (1997) Production, characterization and functional activities of v-Ski in cultured cells. Gene, 202, 15–21. [DOI] [PubMed] [Google Scholar]
- 17.Nomura T., Khan,M.M., Kaul,S.C., Dona,H.-D., Wadhwa,R., Colmenares,C., Kohno,I. and Ishii,S. (1999) Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev., 13, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokitou F., Nomura,T., Khan,M.M., Kaul,S.C., Wadhwa,R., Yasukawa,T., Kohno,I. and Ishii,S. (1999) Viral Ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J. Biol. Chem., 274, 4485–4488. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y., Liu,X., Eaton,E.N., Lane,W.S., Lodish,H.F. and Weinberg,R.A. (1999) Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell, 4, 499–509. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y., Liu,X., Ng-Eaton,E., Lodish,H.F. and Weinberg,R.A. (1999) SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl Acad. Sci. USA, 96, 12442–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saumweber H., Frasch,M. and Korge,G. (1990) Two puff-specific proteins bind within the 2.5 kb upstream region of the Drosophila melanogaster Sgs-4 gene. Chromosoma, 99, 52–60. [DOI] [PubMed] [Google Scholar]
- 22.Wieland C., Mann,S., von Besser,H. and Saumweber,H. (1992) The Drosophila nuclear protein Bx42, which is found in many puffs on polytene chromosomes, is highly charged. Chromosoma, 101, 517–525. [DOI] [PubMed] [Google Scholar]
- 23.Zhou S., Fujimuro,M., Hsieh,J.J.-D., Chen,L. and Hayward,S.D. (2000) A role for Skip in EBNA2 activation of CBF1-repressed promoters. J. Virol., 74, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou S., Fujimuro,M., Hsieh,J.J.-D., Chen,L., Miyamoto,A., Weinmaster,G. and Hayward,S.D. (2000) Skip, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol., 20, 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leong G.M., Subramaniam,N., Figueroa,J., Flanagan,J.L., Hayman,M.J., Eisman,J.A. and Kouzmenko,A.P. (2001) Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-β-dependent transcription. J. Biol. Chem., 276, 18243–18248. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvard V., Storey,A., Pim,D. and Banks,L. (1994) Characterisation of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J., 13, 5451–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry F., Dostatni,N., Arnos,F. and Yaniv,M. (1990) Cooperative activation of transcription by bovine papillomavirus type 1 E2 can occur over a large distance. Mol. Cell. Biol ., 10, 4431–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Deiry W.S., Tokino,T., Velculescu,V.E., Levy,D.B., Parsons,R., Trent,J.M., Lin,D., Mercer,W.E., Kinzler,K.W. and Vogelstein,B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 30.Luckow B. and Schutz,G. (1987) CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res., 15, 5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murthy S.C., Bhat,G.P. and Thimmappaya,B. (1985) Adenovirus EIIA early promoter: transcriptional control elements and induction by the viral pre-early E1A gene, which appears to be sequence independent. Proc. Natl Acad. Sci. USA, 82, 2230–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matlashewski G., Schneider,J., Banks,L., Jones,N., Murray,A. and Crawford,L. (1987) Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J., 6, 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diehl B.R. and Pringle,J.R. (1991) Molecular analysis of Saccharomyces cerevisiae chromosome I: identification of additional transcribed regions and demonstration that some encode essential functions. Genetics, 127, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folk P., Puta,F., Krpejsova,L., Blahuskova,A., Markos,A., Rabino,M. and Dottin,R.P. (1996) The homolog of chromatin binding protein Bx42 identified in Dictyostelium. Gene, 181, 229–231. [DOI] [PubMed] [Google Scholar]
- 35.Harris S.D., Cheng,J., Pugh,T.A. and Pringle,J.R. (1992) Molecular analysis of Saccharomyces cerevisiae chromosome I on the number of genes and the identification of essential genes using temperature-sensitive-lethal mutations. J. Mol. Biol., 225, 53–65. [DOI] [PubMed] [Google Scholar]
- 36.Phelps W.C. and Howley,P.M. (1987) Transcriptional trans-activation by the human papillomavirus type 16 E2 gene product. J. Virol., 61, 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y.-J., Noguchi,S., Hayashi,Y.K., Tsukahara,T., Shimizu,T. and Arahata,K. (2001) The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum. Mol. Genet., 10, 1129–1139. [DOI] [PubMed] [Google Scholar]