Abstract
Genome-wide association studies (GWASs) have identified many complex loci. However, most loci reside in noncoding regions and have unknown biological functions. Integrative analysis that incorporates known functional information into GWASs can help elucidate the underlying biological mechanisms and prioritize important functional variants. Hence, we develop a flexible Bayesian variable selection model with efficient computational techniques for such integrative analysis. Different from previous approaches, our method models the effect-size distribution and probability of causality for variants with different annotations and jointly models genome-wide variants to account for linkage disequilibrium (LD), thus prioritizing associations based on the quantification of the annotations and allowing for multiple associated variants per locus. Our method dramatically improves both computational speed and posterior sampling convergence by taking advantage of the block-wise LD structures in human genomes. In simulations, our method accurately quantifies the functional enrichment and performs more powerfully for prioritizing the true associations than alternative methods, where the power gain is especially apparent when multiple associated variants in LD reside in the same locus. We applied our method to an in-depth GWAS of age-related macular degeneration with 33,976 individuals and 9,857,286 variants. We find the strongest enrichment for causality among non-synonymous variants (54× more likely to be causal, 1.4× larger effect sizes) and variants in transcription, repressed Polycomb, and enhancer regions, as well as identify five additional candidate loci beyond the 32 known AMD risk loci. In conclusion, our method is shown to efficiently integrate functional information in GWASs, helping identify functional associated-variants and underlying biology.
Keywords: functional information, genome-wide association study, GWAS, Bayesian variable selection regression, BVSR, expectation-maximization, EM, Markov chain Monte Carlo, MCMC, age-related macular degeneration, AMD
Introduction
Genome-wide association studies (GWASs) have identified thousands of genetic loci for complex traits and diseases, providing insights into the underlying genetic architecture.1, 2, 3, 4, 5 Each associated locus typically contains hundreds of variants in linkage disequilibrium (LD),6, 7 most of which are of unknown function and located outside protein-coding regions. Unsurprisingly, the biological mechanisms underlying the identified associations are often unclear8 and pinpointing causal variants is difficult.9
Recent functional genomic studies help understand and pinpoint functional associations and mechanisms.10, 11, 12 Genetic variants can be annotated based on the genomic location (e.g., coding, intronic, and intergenic), role in determining protein structure and function (e.g., Sorting Intolerant From Tolerant [SIFT]13 and Polymorphism Phenotyping [PolyPhen]14 scores), ability to regulate gene expression (e.g., expression quantitative trait loci [eQTL] and allelic specific expression [ASE] evidence15, 16), biochemical function (e.g., DNase I hypersensitive sites [DHS], metabolomic QTL [mQTL] evidence,17 and chromatin states18, 19, 20), evolutionary significance (e.g., Genomic Evolutionary Rate Profiling [GERP] annotations21), and a combination of different types of annotation (e.g., CADD22). Many statistical methods, including stratified LD score regression23 and MQS,24 can now evaluate the role of functional annotations in GWASs through heritability analysis. Preliminary studies also show higher proportions of associated variants in protein-coding exons, regulatory regions, and cell-type-specific DHSs.25, 26, 27
Integrating functional information into GWASs is expected to help identify and prioritize true associations. However, accomplishing this goal in practice requires methods to account for both LD and computational cost. Consider two recent methods, fGWAS26 and PAINTOR,27 as examples. fGWAS assumes that variants are independent and there is at most one association signal per locus, modeling no LD, which dramatically improves computational speed and allows fGWAS to be applied at genome-wide scale; PAINTOR accounts for LD, assuming the possibility of multiple association signals per locus, but is computationally slow and can be used to fine-map small regions only (∼10 kb).
Here, we pair a flexible Bayesian method with an efficient computational algorithm. Together the two represent an attractive means to incorporate functional information into association mapping. Our model accounts for genotype correlation due to LD, allows for multiple signals per locus and, importantly, shares information genome-wide to increase association-mapping power. Our algorithm takes advantage of the local LD structure in the human genome28, 29, 30 and refines previous Markov chain Monte Carlo (MCMC) algorithms to greatly improve mixing, which is key when searching for independent signals among many associated variants in LD (but less important in other applications such as modeling total genomic heritability). We refer to our method as the Bayesian functional GWAS (bfGWAS). Below, we illustrate the benefits of our method with extensive simulations as well as real large-scale GWASs on age-related macular degeneration (AMD)31 (33,976 individuals, 9,857,286 variants) and skin cancer (17,624 individuals, 8,626,534 variants).
Material and Methods
Bayesian Variable Selection Model
Our method is based on the standard Bayesian variable selection regression (BVSR) model32 (Supplemental Note; Figure S1A),
where is the centered phenotype vector with n individuals, is the centered genotype matrix with p genetic variants, is a vector of genetic effect-sizes where each element βi follows a “spike-and-slab” variable selection prior, . Different from the standard BVSR, however, our method considers functional annotations that classify variants into K non-overlapping categories. For example, all variants could be annotated based on their most important functions in a gene, such as non-synonymous, synonymous, intronic, intergenic, or other genomic, which classifies all variants into five non-overlapping categories.
Annotation-Specific Effect-Size Priors
We assume that variants in the same annotation category q share a prior32, 33 for effect sizes, , with the same category-specific parameters . This model implies that effect sizes are normally distributed as with probability , or set to zero with probability , with denoting the point-mass function at 0. Here, represents the (unknown) causal probability for variants in the qth category and represents the (unknown) corresponding effect-size variance. An enhancement to previous Bayesian models32, 34, 35 is that we model both the proportion of associated variants and their effect-size distribution in each annotation category. Note that our model is different from simply applying BVSR on variants of each annotation, because we model the LD among variants of different annotations.
We assume independent, conjugate, and non-informative priors for , e.g., with mean 10−6 and with k1 = k2 = 0.1. Although independent and conjugate priors are assumed for the convenience of deriving closed-form expressions for the conditional posterior distributions (greatly saving computational cost), the posterior distributions of depend on each other through effect sizes and the number of signals. Non-informative priors allow the Bayesian estimates to be mainly determined by the likelihood when there exist associations in the qth category (otherwise the Bayesian estimates will be determined by the respective prior modes; see derivation details in Supplemental Note). Particularly, assuming a conservative prior mean 10−6 for (equivalent to assume one signal per 1M variants) enforces an initial sparse model, which helps control false positives and barely affects identifying real signals. Taking k1 = k2 = 0.1 makes the Inverse Gamma prior for non-informative with mode at 0.09.
Our goal is to simultaneously make inference on the category-specific parameters that represent the importance of each functional category, and on the variant-specific parameters—effect-size βi and the probability of (referred as posterior inclusion probability [PPi], representing association evidence, i.e., the probability for the variant to be associated with the phenotype). Our model shares information genome-wide to estimate the category-specific parameters, which then inform the variant-specific parameters. As a result, variant associations will be prioritized based on the inferred importance of functional categories.
Scalable EM-MCMC Algorithm
Because standard MCMC algorithms suffer from heavy computational burden and poor mixing of posterior samples for large GWASs, we develop a scalable expectation-maximization MCMC (or EM-MCMC) algorithm. Our algorithm is based on the observation that LD decays exponentially with distance and displays local block-wise structure along the human genome.28, 29, 30, 36, 37 This observation allows us to decompose the complex joint likelihood of our model into a product of block-wise likelihoods (Appendix A and Supplemental Note). Intuitively, conditional on a common set of category-specific parameters , we can infer by running the MCMC algorithm per genome block. A diagram of this EM-MCMC algorithm is shown in Figure S1B.
Running MCMC per genome-block facilitates parallel computing and reduces the search space. Unlike previous MCMC algorithms for GWASs that use proposal distributions based only on marginal association evidence (such as implemented in GEMMA38), our MCMC algorithm uses a proposal distribution that favors variants near the “causal” variants being considered in each iteration and prioritizes among these neighboring variants based on their conditional association evidence (see Supplemental Note). Our strategy dramatically improves the MCMC mixing property, encouraging our method to explore different combinations of potentially associated variants in each locus (Figure S2). In addition, we implemented memory-reduction techniques that reduce memory usage up to 97%, effectively reducing the required physical memory from 120 Gb (usage by GEMMA38) to 3.6 Gb for a GWAS with ∼33K individuals and ∼400K genotyped variants (Appendix A and Supplemental Note).
In practice, we segment the whole genome into blocks of 5,000–10,000 variants, based on marginal association evidence, genomic distance, and LD. We always ensure variants in LD (R2 > 0.1) with significant signals (p < 5 × 10−8) are in the same block (Appendix A). We first initialize the category-specific parameters , then run the MCMC algorithm per block (E-step), summarize the MCMC posterior estimates of across all blocks to update (M-step), and repeat the block-wise EM-MCMC steps until the estimates of converge (Figure S1B).
In addition, we calculate the regional posterior inclusion probability (regional-PP) per block that is the proportion of MCMC iterations with at least one signal (see Supplemental Note). Because Bayesian PP might be split among multiple variants in high LD, the threshold of regional-PP > 0.95 (conservatively analogous to false discovery rate 0.05) is used for identifying loci.
AMD and MGI GWAS Data
The GWAS data of age-related macular degeneration (AMD) consist of 33,976 unrelated European samples (16,144 advanced case subjects; 17,832 control subjects), and a total of 12,023,830 genotyped on a customized Exome-Chip and imputed against the 1000 Genomes Project phase I reference panel.31, 39 Advanced AMD case subjects include both subjects with choroidal neovascularization and subjects with geographic atrophy. Samples were aggregated across 26 studies and genotyped centrally.31
The Michigan Genomics Initiative (MGI) data are the institutional repository of DNA and electronic health records, collected from patients recruited on the day of their elective surgery or procedure at the University of Michigan Health System. DNA was extracted from blood and samples were genotyped on the Illumina HumanCoreExome v.12.1 array and then imputed against the HRC reference panel.40 The MGI GWAS data studied in this paper contain 17,624 unrelated European individuals and ∼8.7M genotyped or imputed variants with frequency > 0.5%. The phenotype of skin cancer was defined as the presence of ICD9 code 232 (carcinoma in situ of skin) on two or more visits (2,359 case subjects). The control phenotype was defined as the absence of ICD9 codes (172–173.99) on all visits (15,265 control subjects). For both MGI and the AMD genetic studies, all participants gave informed consent and the University of Michigan IRB approved our GWAS analyses.
Results
Simulation
We simulated phenotypes with the genotype data (chromosomes 18–22) from the AMD GWAS,31 including 33,976 individuals and 52,549 variants with minor allele frequency (MAF) > 0.05. We segmented this small genome into 100 × 2.5 Mb blocks, each with ∼5K variants. Within each block, we marked a 25 kb continuous region (starting 37.5 kb from the beginning of a block) as the potential locus. We randomly selected two causal SNPs per locus for ten randomly selected loci. We simulated two complementary annotations to classify variants into “coding” and “noncoding” groups, where the coding variants account for ∼1% overall variants but ∼10% variants within the causal loci (matching the pattern in the real AMD data). We simulated two scenarios: (1) coding variants ∼53× enriched among causal variants (7 coding versus 13 noncoding) and (2) no enrichment (randomly selecting causal variants in risk loci with equally distributed annotations). A total of 15% of phenotypic variance was divided equally among causal variants. We compared bfGWAS with single variant likelihood-ratio test, conditional analysis, fGWAS, and the standard Bayesian variable selection regression model (BVSR, considering no functional information). The single-variant test (also referred to as p value), conditioned p value, fGWAS posterior association probability (PP, see Appendix A), BVSR PP, and bfGWAS PP were used as criteria to identify associations. The reason that we did not include PAINTOR into comparison is because PAINTOR costs >1,000 CPU hr to finish analyzing one 2.5 Mb genome-block with ∼5K variants.
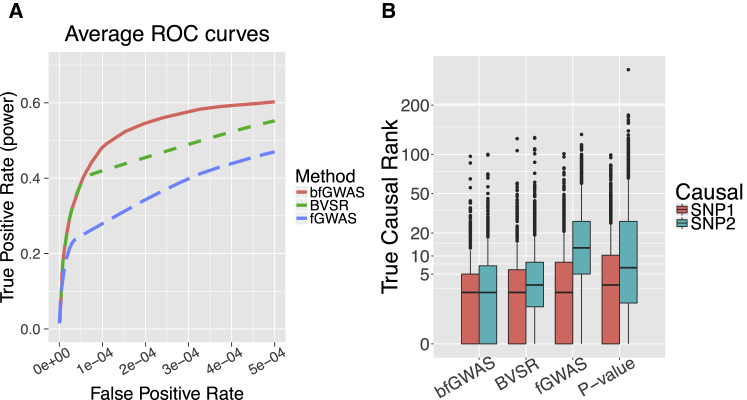
We first compared power of different methods using average ROC curves27, 32 across 100 simulation replicates. Because the p value is used differently from the other “fine-mapping” criteria (fGWAS PP, BVSR PP, bfGWAS PP), we compare only the average ROC curves of fGWAS, BVSR, and bfGWAS (Figure 1A). We found that bfGWAS (modeling LD and allowing multiple signals per locus) outperformed both fGWAS and BVSR. Specifically, with false positive rate (FPR) 2 × 10−4, the power of identifying the true associations is 0.55 by bfGWAS, 0.45 by BVSR, and 0.34 by fGWAS. In addition, for identifying associated loci with regional-PP > 0.95, bfGWAS has power 0.98 and false discovery rate (FDR) 0.005, BVSR has power 0.97 and FDR 0.006, and fGWAS has power 0.97 and FDR 0.005.
Figure 1.
Power Comparison by Simulation Studies
Compare the power of bfGWAS, the standard Bayesian variable selection regression model (BVSR), fGWAS, p value of single variant test with conditional analysis, with 100 simulation replicates and complete sample size 33,976.
(A) Average ROC curves, larger area under curve suggests higher power.
(B) Boxplot of the ranks of the true causal SNP1 (with smaller p value) and SNP2, higher rank (smaller rank value) suggests higher power.
In a typical GWAS, researchers identify a series of associated loci and then examine associated variants within each locus independently. We examined the ability of each method to prioritize the true associations in each locus. Since we simulated two causal SNPs per locus (SNP1 and SNP2), we examine the power for identifying each of these separately (Figure 1B). All methods have approximately the same median rank for causal SNP1 (typically, 2nd rank among 150 SNPs in the locus), suggesting that the strongest signal in a locus can often be identified without incorporating functional information and LD. The median rank for the second causal SNP2 was the 2nd by bfGWAS, 3rd by BVSR, 13th by fGWAS, and 6th by conditioned p value—suggesting that incorporating functional information improves power to identify multiple signals in a locus and that fGWAS is limited by the assumption of at most one signal per locus. Stratified results based on the LD between two causal variants further demonstrate that bfGWAS has the highest power for identifying the weaker signal, especially when both SNPs are in high LD (Figure S3).
Both bfGWAS and fGWAS correctly identified enrichment in scenario 1 and properly controlled for the type I error of enrichment in scenario 2, despite some numerical issues for fGWAS (Figure S4). Moreover, bfGWAS estimated the effect-size variance per annotation. For all 100 simulation replicates under both scenarios, the 95% confidence intervals of the log-ratio of estimated effect-size variances between coding and noncoding overlapped with 0 (Figure S5), suggesting that effect-size variances were similar between two annotations (matching the simulated truth).
In summary, our simulation studies show that, in comparison with competing methods, bfGWAS has highest power, especially in loci with multiple associated variants. Further, bfGWAS produces enrichment parameter estimates that can help with interpretation of association results.
GWAS of AMD
Next, we applied our method to the AMD GWAS data with 33,976 unrelated European individuals (16,144 advanced case subjects; 17,832 control subjects). We analyzed 9,866,744 (∼10M) low-frequency and common variants (MAF > 0.5%) with three types of genomic annotations: gene-based functional annotations by SeattleSeq, summarized regulatory annotations,41 and the core 15 chromatin states profiled by ChromHMM42, 43 with respect to 127 consolidated epigenomes (ROADMAP, ENCODE).44
Coding Variation and AMD
We used SeattleSeq to classify variants according to their impact on coding sequences (Table S1) and then applied our method bfGWAS and fGWAS. bfGWAS identified 37 loci out of 1,063 considered genome blocks with regional-PP > 0.95 (Tables S2, S3, and S5), including 32 among the 34 known AMD loci31 and 5 extra candidate loci. Using the threshold of Bayesian PP > 0.1068 (roughly equivalent to the p value 5 × 10−8 based on permutations of AMD data; Figure S6), we identified 150 associated variants (Figure S8A; Table S3), with 47 distributed among 42,005 non-synonymous variants, 4 among 67,165 synonymous coding variants, 54 among 3,679,235 intronic variants, 18 among 5,512,423 intergenic variants (including non-annotated variants), and 27 among 565,916 “other-genomic” variants (UTR, non-coding exons, upstream and downstream of genes). Very roughly, this corresponds to fraction of associated variants of ∼1:1,000 among non-synonymous variants, 1:15,000 among synonymous variants, 1:100,000 among intronic variants, 1:300,000 among intergenic variants, and 1:20,000 among other-genomic variants.
Similarly, fGWAS identified 39 loci by regional-PP > 0.95, including all 34 known loci and the same 5 extra candidate loci identified by bfGWAS (Tables S2, S4, and S6; Figure S9B). A total of 94 associated variants were identified by fGWAS with fGWAS PP > 0.1068, including 22 non-synonymous, 6 coding-synonymous, 28 intronic, 15 intergenic, and 23 other-genomic signals. Compared with bfGWAS, the proportion of loci that contain at least one non-synonymous variant with PP > 0.1068 is smaller (31% by fGWAS versus 49% by bfGWAS). Similarly, the proportion of non-synonymous variants prioritized by fGWAS is also smaller (30% by fGWAS versus 46% by bfGWAS), indicating that bfGWAS places greater weight on non-synonymous variants—which, as a group, appears to have both a higher prior probability of association and larger effect sizes when associated.
Besides replicating the association results within known AMD loci,31 bfGWAS identified five additional candidate loci (Table S5): missense rs7562391/PPIL3, rs61751507/CPN1, rs2232613/LBP, downstream rs114318558/ZNRD1ASP, and splice rs6496562/ABHD2. Among these five candidate loci, fGWAS identified three with the same top risk variants, a different top risk variant (coding-synonymous rs61733667) for CPN1, and a nearby locus (upstream rs116803720/HLA-K) of ZNRD1ASP (Table S6). Interestingly, there are several connections between these candidate loci and known AMD loci. Specifically, the protein encoded by LBP is part of the lipid transfer protein family (which also includes CETP among the known AMD risk loci) that promotes the exchange of neutral lipids and phospholipids between plasma lipoproteins.45 ZNRD1ASP has been associated with lipid metabolisms46 and ABHD2 has been associated with coronary artery disease,47 two other traits where the AMD loci encoding CETP, APOE, and LIPC are also involved. The gene CPN1 has been associated with age-related disease (specifically, hearing impairment48).
Multiple Signals in a Single Locus
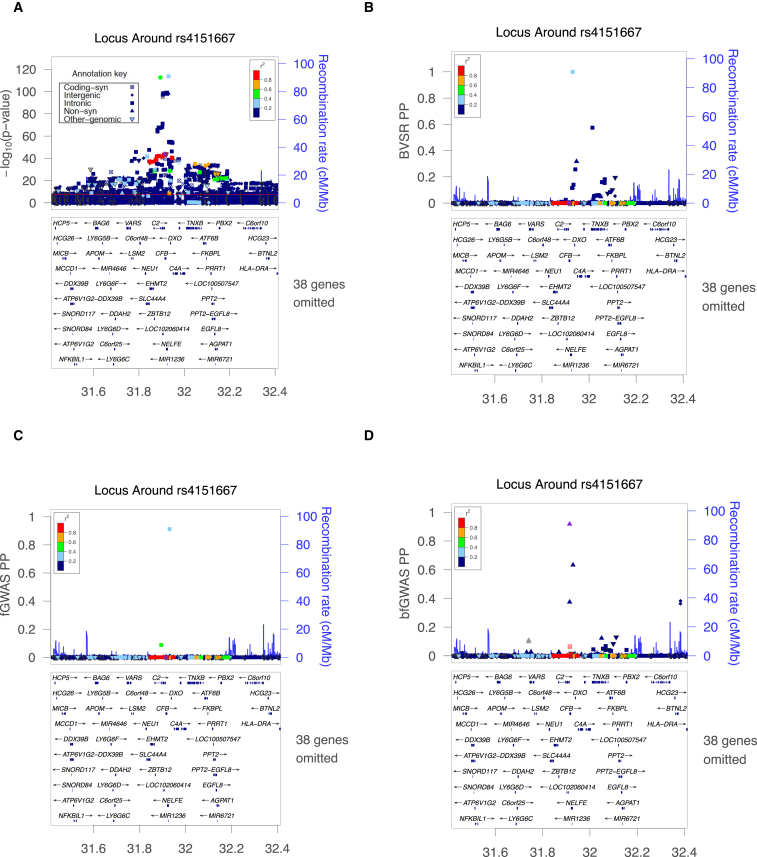
We use two examples to illustrate the importance of studying multiple signals in a single locus. Our first example focuses on a 1 Mb region around locus C2/CFB/SKIV2L on chromosome 6 where 1,862 variants have p < 5 × 10−8. There are an estimated 4 independent signals in the region by conditional analysis,31 1 variant with fGWAS PP > 0.1068, 11 with BVSR PP > 0.1068, and 8 with bfGWAS PP > 0.1068. Interestingly, the alternative methods (p value, fGWAS, and BVSR) identified intronic SNP rs116503776/SKIV2L/NELFE as the top candidates (p = 2.1 × 10−114; fGWAS PP = 0.912; BVSR PP = 1.0), while bfGWAS identified two missense SNPs, rs4151667/C2/CFB (p = 1.4 × 10−44; bfGWAS PP = 0.917) and rs115270436/SKIV2L/NELFE (p = 2.8 × 10−99; bfGWAS PP = 0.633), as the top functional candidates (Figure 2; Tables S2–S4).
Figure 2.
ZoomLocus Plots around rs4151667 in the Locus C2/CFB/SKIV2L
(A) –log(p values) by single variant tests.
(B) Posterior inclusion probabilities (PP) by the standard Bayesian variable selection regression model (BVSR).
(C) Posterior association probabilities (PP) by fGWAS.
(D) Bayesian inclusion probabilities (PP) by bfGWAS.
The top cyan squares in (A)–(C) denote the intronic variant rs116503776; the purple triangle in (D) denotes the non-synonymous variant rs4151667.
A haplotype analysis describing the odds ratios (ORs) for all possible haplotypes for SNPs rs116503776, rs4151667, and rs115270436 helps clarify the region. Intronic SNP rs116503776 with the smallest p value appears to be associated with the phenotype by tagging the other two missense SNPs (Table S15). In particular, haplotypes with rs116503776 can either increase or decrease risk, depending on alleles at the other two SNPs. To further confirm the importance of the missense SNPs rs4151667 and rs115270436, we compared the AIC/BIC/loglikelihood between two models: one model with the top two independent signals (rs116503776 and rs114254831) identified by single-variant conditional analysis,31 versus the other model with the top two signals (rs4151667 and rs115270436) identified by bfGWAS. As expected, the second model has smaller AIC/BIC and larger loglikelihood than the first one (Table S16). Thus, we can see that while alternative methods (p value, fGWAS, and BVSR) focus on the SNP with the smallest p value, our bfGWAS method finds an alternative pairing of missense signals that better accounts for all data.
Our second example focuses on a 1 Mb region around gene C3 on chromosome 19 (Figure S9) with 112 genome-wide significant variants with p < 5 × 10−8. fGWAS discovered only a single missense signal, rs2230199, with the most significant p = 1.7 × 10−77 (top blue triangle in Figures S9A and S9C). However, both BVSR and bfGWAS identified two missense variants with PPs = 1.0 and five intronic variants with 0.11 < PPs < 0.18. The top two missense signals, rs2230199 and rs147859257 (241 base pairs apart), were confirmed by conditional analysis,31 where the second signal rs147859257 has conditioned p = 6.0 × 10−33 (purple triangle in Figures S9B and S9D), overlapping with rs2230199. These two missense signals match the interpretation of previous studies.49, 50, 51 Because five other intronic variants (rs11569479, rs11569470, rs201063729, rs10408682, and rs11569466) are in high LD with R2 > 0.98 between each other, we believe this is the third independent signal whose Bayesian PP was split among five variants in high LD by bfGWAS.
Enrichment Analysis
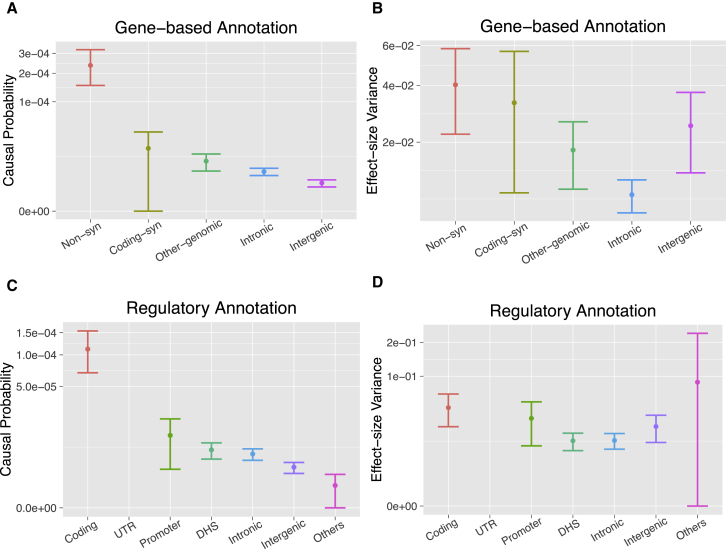
bfGWAS estimated that non-synonymous variants are 10–100 times more likely to be causal than variants in other categories and that they also have larger effect sizes (Figures 3A and 3B). To better compare enrichment among multiple categories, we define two new sets of parameters (Supplemental Note). The first set of parameters is defined to contrast the posterior association probability estimate for each category to the genome-wide average . The second set of parameters is similarly defined to contrast the effect-size variance from each category to the genome-wide average. Moreover, the square root of the effect-size variance reflects the effect-size magnitude because of the prior assumption for the effect size in our model.
Figure 3.
Category-Specific Parameter Estimates with 95% Error Bars by bfGWAS for Gene-Based Annotations and Regulatory Annotations
(A and C) Causal probabilities.
(B and D) Effect-size variances.
The estimates of UTR in (C) and (D) were estimated as their prior values due to no association was found for this annotation (hence not shown in the plots). The estimate of the effect-size variance for the “Others” category in (D) is also close to the prior because of low region-association evidence, hence it has a wide 95% error bar. The error bars denote the 95% confidence intervals for the category-specific parameter estimates.
Compared to the genome-wide average probability of causality = (Figure S12A), we found that non-synonymous category were 53× more likely to be causal (p = 7.24 × 10−84), that coding-synonymous and other variants were 4.3× and 2.2× more likely (p = 0.005, 0.003), and that intergenic variants were 0.7× less likely (p = 4.9 × 10−6), while the intronic variants matched the genome-wide average (p = 0.659). In addition, compared to the genome-wide average effect-size variance (; Figure S12B), we found that the effect size variance of was 1.9× larger for non-synonymous variants (p = 0.014; i.e., 1.4× larger effect-size), and 0.4× smaller for variants in the intronic category (p = 4.5 × 10−6); remaining categories were not significantly different (p > 0.2). The estimated enrichment parameters by fGWAS show a similar pattern, although the contrast of the estimated enrichment for non-synonymous versus other annotations is not as pronounced as by bfGWAS (Figure S12A).
Analysis with Regulatory Annotations
In addition, we analyzed the GWAS data of AMD with the summarized regulatory annotations:41 coding, UTR, promoter (defined as within 2 kb of a transcription starting site), DHS in any of 217 cell types, intronic, intergenic, and “others” (not annotated as any of the previous six categories). Overall GWAS results were similar as the ones described in previous context (Tables S7–S10). Compared to the genome-wide average association probability ( = ; Figure S12C), we found that the association probability of the coding category was 28× higher (p < 2.2 × 10−16), the promoter was 2.6× (p = 0.028) higher, and the intergenic and “others” were 0.5× and 0.9× less (p = 5.3 × 10−4, 0.033), while the DHS and intronic were not significantly different (p > 0.1). In addition, compared to the genome-wide average effect-size variance , we found that the effect-size variance of the coding category was 1.9× larger (p = 0.019; i.e., 1.4× larger effect size) and the DHS and intronic were 0.5× less (p = 0.011, 0.007), while the promoter, intergenic, and “others” were not significantly different (p > 0.1; Figure S12D). Here, fGWAS identified a slightly different enrichment pattern (Figure S12B), where UTR was identified as the second most enriched category. This is presumably because fGWAS assumes one signal per locus and tends to prioritize the variant with the smallest p value in each locus, e.g., UTR variants rs1142/KMT2E/SPRK2 and rs10422209/CNN2 have the highest fGWAS PP and the smallest p value in their respective locus (Tables S2 and S8).
Analysis with Chromatin States
Last, we considered the annotations of core 15 chromatin states profiled by ChromHMM43 with respect to 127 consolidated epigenomes (ROADMAP, ENCODE):44 active TSS (TssA), flanking active TSS (TssAFlnk), transcription at gene 5′ and 3′ (TxFlnk), strong transcription (Tx), weak transcription (TxWk), genic enhancers (EnhG), enhancers (Enh), ZNF genes & repeats (ZNF/Rpts), heterochromatin (Het), bivalent/poised TSS (TssBiv), flanking bivalent TSS/Enh (BivFlnk), bivalent enhancer (EnhBiv), repressed PolyComb (ReprPC), weak repressed PolyComb (ReprPCWk), and quiescent/low (Quies).
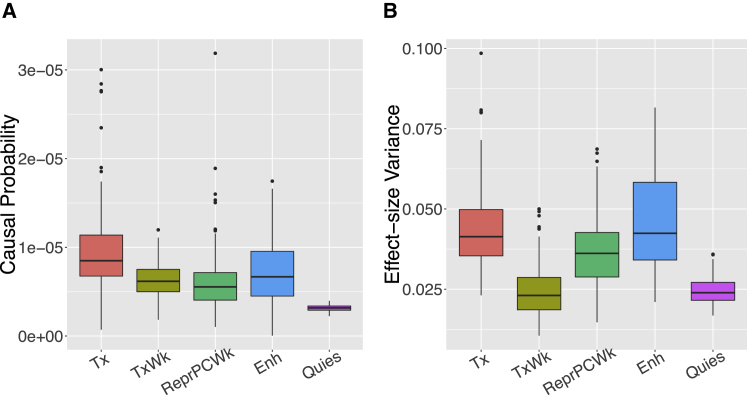
With each set of chromatin states profiled per epigenome, we applied bfGWAS on the GWAS data of AMD and then counted the frequency of the top 5 enriched chromatin states across all 127 epigenomes. We found that the associations are mostly enriched with strong transcription (Tx), weak transcription (TxWk), repressed PolyComb (ReprPC), enhancers (Enh), and Quies (Figure 4). Specifically, the highest estimates of the causal probabilities are 3.0 × 10−5 for strong transcription with respect to the fetal brain male tissue (E081), 1.2 × 10−5 for weak transcription with respect to the adipose nuclei (E063), 3.1 × 10−5 for repressed PolyComb with respect to the spleen tissue (E113), 1.7 × 10−5 for enhancers with respect to the ovary tissue (E097), and 3.9 × 10−6 for Quies with respect to the pancreatic islets.
Figure 4.
Top Five Enriched Chromatin States Identified by bfGWAS, using the AMD GWAS Data with Respect to 127 Epigenomes
(A) Boxplot of the category-specific causal probabilities for the top five enriched chromatin states.
(B) Boxplot of the effect-size variances for the top five enriched chromatin states.
We further examined the list of variants that contribute 95% posterior probabilities in the identified loci with regional-PP > 95%. We found that the results accounting for the chromatin states that are profiled with respect to the epigenome of fetal thymus (E093) gave the shortest list (average 11 variants per locus, and we present the corresponding results as an example (Figures S12E, S12F, S13A, and S13B; Tables S11–S14). For this set of enrichment analysis, we found that the repressed PolyComb had the highest causal probability (3.8× higher than the genome-wide average , p = 6.7 × 10−7; Figure S12E), and that all chromatin states have comparable effect-size variances (Figure S12F). Here, fGWAS identified transcription at gene 5′ and 3′ (TxFlnk) as the most enriched chromatin state (Figure S13C).
MGI GWAS of Skin Cancer
To illustrate the benefits of using bfGWAS for GWAS data that have relatively fewer loci, we further analyzed the MGI GWAS data with the phenotype of skin cancer, with 17,624 unrelated European samples (2,359 case subjects versus 15,265 control subjects) and ∼8.7M variants with MAF > 0.5%. We corrected the phenotype of skin cancer with respect to age, sex, PC1-4, considered the same gene-based annotations (from SeattleSeq) as for the AMD GWAS, and compared the GWAS results by p value, BVSR, fGWAS, and bfGWAS.
For this GWAS data of skin cancer, all method identified the same four loci: SLC45A2, IRF4, MC1R, and RALY (Figures S14 and S15). Both bfGWAS and fGWAS identified that non-synonymous is the most enriched annotation (Figure S16). Although BVSR, fGWAS, and bfGWAS all produced the highest PP for the leading SNP with the smallest p value, our bfGWAS method outperformed BVSR for identifying the leading SNP at locus SLC45A2, as well as produced an additional and independent non-synonymous signal in locus MC1R (missed by fGWAS) for allowing multiple signals per locus as well as accounting for functional information and LD (Figure S17). In addition, our bfGWAS method avoids the false signal on chromosome 3 by BVSR for using annotation-specific priors. Specifically, by the threshold of PP > 0.1068, bfGWAS identified 9 associated variants (3 non-synonymous, 4 intronic, and 1 other genomic), and 9 by fGWAS (2 non-synoymous, 5 intronic, and 2 intergenic).
Therefore, this set of GWAS analyses further confirmed the advantages of using our bfGWAS method for integrating functional information and fine-mapping loci with multiple signals.
Discussion
Here, we describe a scalable Bayesian hierarchical method, bfGWAS, for integrating functional information in GWASs to help prioritize functional associations and understand underlying genetic architecture. bfGWAS models both association probability and effect-size distribution as a function of annotation categories for improving fine-mapping resolution. Unlike previous methods,26, 27 bfGWAS accounts for LD and allows for the possibility of multiple signals per locus while remaining capable of genome-wide inference. Further, bfGWAS employs an improved MCMC sampling strategy to greatly improve the mixing of MCMC samples, which ensures the capability of identifying a list of independent association candidates.
By simulation studies, we demonstrated that bfGWAS had higher power than the alternative methods for identifying multiple signals in a single locus by accounting for both functional information and LD. We also showed that bfGWAS accurately estimated the enrichment patterns under scenarios with or without enrichment for one annotation in simulations. In the real GWASs of AMD and skin cancer, we further confirmed the advantages of identifying multiple independent signals per locus and prioritizing important functional associations by bfGWAS. Further, we gave two fine-mapped AMD loci, C2/CFB/SKIV2L and C3, by bfGWAS as examples with justifications by haplotype analysis, model comparison, and previous findings. Thus, we believe our method is useful for understanding the underlying genetic architecture of complex traits and diseases for efficiently integrating functional information into GWASs.
Extending bfGWAS to deal with overlapping or quantitative annotations might seem trivial in theory, by assuming a logistic model with multiple functional covariates (both categorical and quantitative) for in the BVSR model. However, the posterior estimates for the coefficients in the logistic model of no longer have analytical formulas in the M-step of the EM-MCMC algorithm (Supplemental Note). Specifically, overestimated will inflate the number of false positives. In preliminary analysis, we encountered computational challenges of controlling the false positive rate, which requires further studies.
Here, bfGWAS makes a key assumption that the variant correlation matrix has a block-wise structure, which allows us to segment the genome into approximately independent blocks, analyze variants per block by MCMC, and summarize genome-wide information by an EM algorithm. In parallel to our study, many recent studies have also explored the benefits of dividing the human genome into approximately independent LD blocks to facilitate genome-wide analyses.26, 52 Although the standard segmentation methods (e.g., based on genomic location52 as we adopted here, or the number of variants per block26) are often sufficient in practice, we expect that a better segmentation method30 based on LD blocks will further increase the association mapping power.
The biggest limitation of bfGWAS is probably computational cost, as we perform MCMC using the complete genotype data. Specifically, bfGWAS took 5,000 CPU hr (∼5 hr with parallel computations on 1,000 CPUs for the 1,063 genome blocks) to analyze the AMD GWAS data with 33,976 individuals and 9,857,286 variants. Implementing bfGWAS with summary statistics is expected to reduce the computation cost significantly, which is part of our continuing research. In addition, the variational approximation53, 54 and other approximations55, 56 of MCMC may provide an efficient alternative for posterior inference in large GWASs.
Acknowledgments
X.Z. is supported by NIH grants R01HG009124 and R01GM126553 and the National Science Foundation (NSF) grant DMS1712933. J.Y. and G.A. are supported by NIH grants R01HG007022 and R01EY022005. The authors would like to thank the Michigan Genomics Initiative (MGI) for allowing us to use the GWAS data about the skin cancer phenotype.
Published: August 24, 2017
Footnotes
Supplemental Data include 17 figures, 16 tables, and a detailed technical note and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.08.002.
Contributor Information
Xiang Zhou, Email: xzhousph@umich.edu.
Gonçalo Abecasis, Email: goncalo@umich.edu.
Appendix A
Bayesian Hierarchical Model Accounting for Functional Information
Recall the standard Bayesian variable selection regression (BVSR) model as described in the Material and Methods,
We assume that variants in the same functional category have the same spike-and-slab prior, , for the effect sizes. That is, for variants of the qth functional annotation category. Consequently, denotes the category-specific causal probability and denotes the category-specific effect-size variance (the square root of reflects the magnitude of effect size).
We further assume the following independent hyper priors:34
where follows a Beta distribution with positive shape parameters aq and bq and follows an Inverse-Gamma distribution with shape parameter k1 and scale parameter k2. In order to adjust for the unbalanced distribution of functional annotations among all variants and enforce a sparse model in our analysis, we choose values for aq and bq such that the Beta distribution has mean with (aq + bq) equal to the number of variants in category q. We set k1 = k2 = 0.1 in our analysis to induce non-informative prior for . Note that is fixed as the phenotype variance value in our Bayesian inferences (Supplemental Note).
Bayesian Inference
We introduce a latent indicator vector to facilitate computation, where each element is a binary variable and indicates whether by or by ( corresponds to the ith variant with genetic effect-size ). Equivalently,
where denotes the number of 1s in ; denotes the zero effect-size vector with ; denotes the non-zero effect-size vector with ; and denotes the diagonal covariance matrix, , corresponding to non-zero effect-sizes. Consequently, the expectation of is an estimate of the posterior inclusion probability (PP) for the ith variant, .
The posterior joint distribution of our proposed Bayesian hierarchical model is proportional to
where , is the p × Q matrix of binary annotations, and Q is the total number of annotations. The goal is to estimate the category-specific parameters and the variant-specific parameters from their posterior distributions, conditioning on the data . Here, the category-specific parameters denote the shared characteristics among all variants with the same annotation, which are also called enrichment parameters.
EM-MCMC Algorithm
The basic idea of the EM-MCMC algorithm is to segment the whole genome into approximately independent blocks each with 5,000–10,000 variants, run MCMC algorithm per block with fixed category-specific parameter values to obtain posterior estimates of (E-step), then summarize the genome-wide posterior estimates of and update values of by maximizing their posterior likelihoods (M-step). Repeat such EM-MCMC iterations for a few times until the estimates of (maximum a posteriori estimates, i.e., MAPs) converge (Figure S1).
We derive the log-posterior-likelihood functions for and the analytical formulas for their MAPs. In addition, we construct their confidence intervals using Fisher information, whose analytical forms are derived for our Bayesian hierarchical model (Supplemental Note). In our practical analyses, we find that, in general, with about 5 EM iterations and 50K MCMC iterations per block, the estimates for would achieve convergence. Our method of integrating functional information into GWAS by using the above Bayesian hierarchical model and EM-MCMC algorithm is referred as “Bayesian Functional GWAS” (bfGWAS).
Convergence Diagnosis
The MCMC algorithm implemented in bfGWAS is essentially a random walk over all possible linear regression models with combinations of variants, which can start with either a model containing multiple significant variants by sequential conditional analysis or the most significant variant by p value. In each MCMC iteration, a new model is proposed by including an additional variant, by deleting one variant from the current model, or by switching one variant within the current model with one outside; and then up to acceptation or rejection by the Metropolis-Hastings algorithm (Supplemental Note). Importantly, we refine the standard proposal strategy for the switching step by prioritizing variants in the neighborhood of the switch candidate according to their conditional association evidence (e.g., p values conditioning on variants, except the switch candidate, in the current model). As a result, this MCMC algorithm encourages our method to explore different combinations of potential signals in each locus and significantly improves the mixing property.
We used the potential scale reduction factor (PSRF)57 to quantitatively diagnose the MCMC mixing property. PSRF is essentially a ratio between the average within-chain variance of the posterior samples and the overall-chain variance with multiple MCMC chains. From the example plots of the PSRFs of Bayesian PPs (Figure S2), for 58 top marginally significant SNPs (with p < 5 × 10−8) in the WTCCC GWAS of Crohn disease,1 we can see that about half of the PSRF values by the standard MCMC algorithm (used in GEMMA35) exceed 1.2, suggesting that the standard MCMC algorithm has poor mixing property. In contrast, the PSRF values by our MCMC algorithm are within the range of (0.9, 1.2), suggesting that our MCMC algorithm has greatly improved mixing property.
Key Implementation Details
We employ two computational techniques to save memory in the bfGWAS software. One is to save all genotype data as unsigned characters in memory, because unsigned characters are equivalent to unsigned integers in (0, 256) that can be easily converted to genotype values within the range of (0.0, 2.0) by multiplying with 0.01. This technique saves up to 90% memory compared to saving genotypes in double type. Second, with an option of in-memory compression, bfGWAS will further save additional 70% memory. As a result, we can decrease the memory usage from ∼120 GB (usage by GEMMA35) to ∼3.6 GB for a typical GWAS dataset with ∼33K individuals and ∼400K variants.
The bfGWAS software wraps a C++ executable file for the E-step (MCMC algorithm) and an R script for the M-step together by a Makefile, which is generated by a Perl script and enables parallel computation through submitting jobs. Generally, 50K MCMC iterations with ∼5K variants and ∼33K individuals require about 300 MB memory and 1 hr CPU time on a 1.6 GHz core, where the computation cost is of order 0(nm2) with the sample size (n) and number of variants (m) considered in the linear models during MCMC iterations (usually m < 10). The computation cost for M-step is almost negligible due to the analytical formulas of the MAPs.
fGWAS
In this paper, the fGWAS results were generated by using summary statistics from single variant likelihood-ratio tests and the same annotation information used by bfGWAS. fGWAS26 produces variant-specific posterior association probabilities (PPs), segment-specific PPs, and enrichment estimates for all annotations. We used the same genome segmentation as used by bfGWAS for fGWAS in both simulations and real data analyses, to produce comparable results. The final fGWAS PP is given by the product of the variant-specific PP and the corresponding segment-specific PP, and the fGWAS regional-PP is given by the highest segment-specific PP in a region or genome block.
Simulation Studies
We used genotype data on chromosomes 18–22 from the AMD GWAS (33,976 individuals and 241,500 variants with MAF > 0.05) to simulate quantitative phenotypes from the standard linear regression model , where is the genotype vector of the ith individual and is the noise term generated from . We segmented the genotype data into 100× 2.5 Mb blocks each with ∼5,000 variants. Within each block, we marked a ∼25 kb continuous region (starting 37.5 kb from the beginning of a block) as the causal locus and randomly selected two causal SNPs if the genome block was selected as a risk locus. Two complementary annotations (“coding” versus “noncoding”) were simulated, where the coding variants account for ∼1% overall variants but ∼10% variants within the causal loci (matching the pattern in the real AMD analysis). We selected positive effect-size vector and noise variance such that a total of 15% phenotypic variance was equally explained by causal SNPs. We controlled the enrichment-fold of coding variants by varying the number of coding variants among the causal SNPs.
We compared bfGWAS with p value, conditioned p value, and fGWAS. In the simulation studies, p values were obtained from a series of likelihood-ratio tests based on the standard linear regression model. p values conditioning on the top significant variant per locus were used to identify the second signal by conditional analysis. fGWAS was implemented with summary statistics from single variant tests and the same genome segmentation as used by bfGWAS. We failed to include PAINTOR in the comparison, because PAINTOR cannot complete the analysis for one block in >1,000 CPU hr (on a 2.5 GHz, 64-bit CPU) and is thus expected to require >1 million CPU hr for a genome-wide analysis.
GWAS of AMD
In the GWAS data of AMD, all genotypes were generated by a customized chip that contains (1) the usual genome-wide variant content, (2) exome content comparable to the Exome chip (protein-altering variants across all exons), (3) variants in known AMD risk loci (protein-altering variants and previously associated variants), and (4) previously observed and predicted variation in TIMP3 and ABCA4 (two genes implicated in monogenic retinal dystrophies). The genotyped variants (439,350) were then imputed to the 1000 Genomes reference panel (phase I),58 resulting a total of 12,023,830 variants.
The software bfGWAS used dosage genotype data and standardized phenotypes. Phenotypes were first coded quantitatively with 1 for case subjects and 0 for control subjects; then corrected for the first and second principle components, age, gender, and source of DNA samples; and then standardized to have mean 0 and standard deviation 1. In order to make the Bayesian inferences scalable to the AMD GWAS data (33,976 individuals, 9,866,744 variants with MAF > 0.5%), we segmented the whole genome into 1,063 non-overlapped blocks, such that each block has length ∼2.5 Mb (containing ∼10,000 variants) and all previously identified loci along with variants in LD (R2 > 0.1) were not split. Then we applied the EM-MCMC algorithm with 5 EM steps and 50,000 MCMC iterations per block (including 50,000 extra burn-ins).
For comparison, p values were obtained by a series of likelihood-ratio tests, using the same “quantitative” phenotype vector as used by bfGWAS; fGWAS was implemented with the summary statistics from single variant tests and the same genome segmentation as used by bfGWAS; and a standard Bayesian variable selection regression (BVSR) method that models no functional information was also applied.
Three types of genomic annotations were considered for analyzing the AMD data: gene-based functional annotations of SNPs and small indels from SeattleSeq, summarized regulatory annotations,41 and the chromatin states profiled respectively for 127 epigenomes by ChromHMM.19, 42, 43 For variants annotated with multiple functions, we used the most severe function in the analysis: non-synonymous > coding-synonymous > other-genomic > intronic > intergenic for the gene-based annotations; coding > UTR > promoter > DHS > intronic > intergenic > “others” for the summarized regulatory annotations.
We further did sensitivity analysis using varying prior means as well as starting values (10−6, 5 × 10−6, 10−5) for , and varying starting values (10, 5, 1) for in bfGWAS with gene-based functional annotations. As expected, the results showed that the posterior inference results were not affected by various practical prior assumptions and starting values of the category-specific parameters. Specifically, all three sets of results identified the same 37 risk loci, comparable number of associated variants with Bayesian PP > 0.1068, as well as the same enrichment pattern (Figure S10).
Accession Numbers
The accession number for the AMD genotype data analyzed in this paper is dbGaP: phs001039.v1.p1.
Web Resources
bfGWAS, https://github.com/yjingj/bfGWAS
ChromHMM, http://compbio.mit.edu/ChromHMM/
Profiled chromatin states with respect to 127 epigenomes, http://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html#core_15state
SeattleSeq, http://snp.gs.washington.edu/SeattleSeqAnnotation138/
Supplemental Data
References
- 1.Wellcome Trust Case Control C., Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 7.Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 8.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carithers L.J., Moore H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015;13:307–308. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellis M., Wold B., Snyder M.P., Bernstein B.E., Kundaje A., Marinov G.K., Ward L.D., Birney E., Crawford G.E., Dekker J. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 14.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tung J., Zhou X., Alberts S.C., Stephens M., Gilad Y. The genetic architecture of gene expression levels in wild baboons. eLife. 2015;4:4. doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lea A.J., Tung J., Zhou X. A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data. PLoS Genet. 2015;11:e1005650. doi: 10.1371/journal.pgen.1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVicker G., van de Geijn B., Degner J.F., Cain C.E., Banovich N.E., Raj A., Lewellen N., Myrthil M., Gilad Y., Pritchard J.K. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X. A unified framework for variance component estimation with summary statistics in genome-wide association studies. bioaRxiv. 2017 doi: 10.1214/17-AOAS1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schork A.J., Thompson W.K., Pham P., Torkamani A., Roddey J.C., Sullivan P.F., Kelsoe J.R., O’Donovan M.C., Furberg H., Schork N.J., Tobacco and Genetics Consortium. Bipolar Disorder Psychiatric Genomics Consortium. Schizophrenia Psychiatric Genomics Consortium All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kichaev G., Yang W.Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Wall J.D., Pritchard J.K. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 30.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritsche L.G., Igl W., Bailey J.N., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2015;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan Y., Stephens M. Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann. Appl. Stat. 2011;5:1780–1815. [Google Scholar]
- 33.Chipman H., George E.I., McCulloch R.E. The Practical Implementation of Bayesian Model Selection. In: Lahiri P., editor. Model selection. Institute of Mathematical Statistics; Beachwood, OH: 2001. pp. 65–116. [Google Scholar]
- 34.Carbonetto P., Stephens M. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn’s disease. PLoS Genet. 2013;9:e1003770. doi: 10.1371/journal.pgen.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 37.Wen X., Stephens M. Bayesian Methods for Genetic Association Analysis with Heterogeneous Subgroups: From Meta-Analyses to Gene-Environment Interactions. Ann. Appl. Stat. 2014;8:176–203. doi: 10.1214/13-AOAS695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masson D., Jiang X.C., Lagrost L., Tall A.R. The role of plasma lipid transfer proteins in lipoprotein metabolism and atherogenesis. J. Lipid Res. 2009;50(Suppl):S201–S206. doi: 10.1194/jlr.R800061-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kettunen J., Tukiainen T., Sarin A.P., Ortega-Alonso A., Tikkanen E., Lyytikäinen L.P., Kangas A.J., Soininen P., Würtz P., Silander K. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fransen E., Bonneux S., Corneveaux J.J., Schrauwen I., Di Berardino F., White C.H., Ohmen J.D., Van de Heyning P., Ambrosetti U., Huentelman M.J. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur. J. Hum. Genet. 2015;23:110–115. doi: 10.1038/ejhg.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helgason H., Sulem P., Duvvari M.R., Luo H., Thorleifsson G., Stefansson H., Jonsdottir I., Masson G., Gudbjartsson D.F., Walters G.B. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat. Genet. 2013;45:1371–1374. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 50.Seddon J.M., Yu Y., Miller E.C., Reynolds R., Tan P.L., Gowrisankar S., Goldstein J.I., Triebwasser M., Anderson H.E., Zerbib J. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat. Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan X., Larson D.E., Wang C., Koboldt D.C., Sergeev Y.V., Fulton R.S., Fulton L.L., Fronick C.C., Branham K.E., Bragg-Gresham J. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat. Genet. 2013;45:1375–1379. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loh P.R., Bhatia G., Gusev A., Finucane H.K., Bulik-Sullivan B.K., Pollack S.J., de Candia T.R., Lee S.H., Wray N.R., Kendler K.S., Schizophrenia Working Group of Psychiatric Genomics Consortium Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015;47:1385–1392. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jordan M.I., Ghahramani Z., Jaakkola T.S., Saul L.K. An introduction to variational methods for graphical models. Mach. Learn. 1999;37:183–233. [Google Scholar]
- 54.Carbonetto P., Stephens M. Scalable variational inference for Bayesian variable selection in regression, and its accuracy in genetic association studies. Bayesian Analysis. 2012;7:73–108. [Google Scholar]
- 55.Rue H., Martino S., Chopin N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. Series B Stat. Methodol. 2009;71:319–392. [Google Scholar]
- 56.Singh, S.W.M., and McCallum, A. (2012). Monte Carlo MCMC: efficient inference by approximate sampling. https://ciir-publications.cs.umass.edu/getpdf.php?id=1053.
- 57.Gelman A., Rubin D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992;7:457–472. [Google Scholar]
- 58.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.