Abstract
The eukaryal single-stranded DNA binding protein replication protein A (RPA) binds short oligonucleotides with high affinity but exhibits low cooperativity in binding longer templates, opposite to prokaryal counterparts. This discrepancy could reflect the smaller size of the replicative template portion availed to RPA. According to current models, this portion accommodates an RNA–DNA primer (RDP) of <40 nt (nested discontinuity) or a several-fold longer Okazaki fragment (initiation zone). Previous in situ UV-crosslinking revealed that RPA also interacts with nascent DNA, especially growing RDPs. Here we compare nascent SV40 DNA chains UV-crosslinked to the middle and large RPA subunits and use the data to re-examine the two models. The middle subunit interacted with the nascent chains after a few DNA residues were added to the RNA primer while the large subunit became accessible after extension by several more. Upon RDP maturation, the middle subunit disengaged while the large subunit remained accessible during further limited extension. A corresponding shift in preference in favor of the large subunit has been reported for purified RPA and synthetic gapped duplexes upon reduction of the gap from 19 to 9 nt. Combined, these facts support the proposal that the mature RDP faces downstream a correspondingly small gap, possibly created by removal of the RNA primer moiety from an adjacent, previously synthesized RDP (nested discontinuity) but insufficient for continuous elongation of the RDP into an Okazaki fragment (initiation zone).
INTRODUCTION
Single-stranded (ss)DNA binding proteins (SSB) stabilize exposed portions of the parental DNA template and mobilize other proteins to this site (1). In replicating prokaryal DNA, the exposed ssDNA portions reach ∼1 kb and accommodate numerous SSB molecules. A single SSB molecule binds its isolated target site weakly but high positive cooperativity ensures processive coating of the entire replicative template. In contrast, the generic eukaryal SSB, heterotrimeric replication protein A (RPA; subunits of 70, 34 and 14 kDa in mammals, Rpa1, 2 and 3, respectively) exhibits high affinity (Kd ∼10–10 M) to target sites reaching ∼30 nt and weak cooperativity in binding longer ligands (2,3). The discrepant behaviors of the two SSB types may reflect the differences between the replicative templates they encounter. Although the eukaryal template is likely shorter, its actual size remains uncertain. Models of eukaryal lagging strand DNA synthesis termed ‘initiation zone’ and ‘nested discontinuity’ have addressed this issue, albeit indirectly (reviewed in 4). Both models suggest, based on the SV40 paradigm, that DNA polymerase (pol) α-primase initiates the DNA chain by synthesizing a mixed RNA–DNA primer (RDP) <40 nt. RDPs are converted into the several-fold larger Okazaki fragments in a process mediated by the PCNA-dependent pol δ (5–8). However, in the first model, an Okazaki fragment arises by continuous elongation of a single RDP; in the second, it is assembled from juxtaposed RDPs in a repair-like manner. The first model resembles the prokaryal paradigm. The second, ‘counter-intuitive’ model was invoked by the bimodal decay of RNA primers observed during SV40 chromosome replication. That is, while the bulk of RNA primers are removed early in Okazaki fragment growth, a minor fraction persists and is carried intact onto the long nascent chain (9–11). According to the nested discontinuity model, the short-lived fraction originates from internal units of an RDP array destined to become an Okazaki fragment, the more persistent fraction from a 5′-terminal unit. Near contiguous RDP arrays have been visualized by simulated syntheses of Okazaki fragments in isolated SV40 replicating intermediate DNA (11,12). These experiments have suggested that a newly synthesized RDP is separated from downstream chains by a gap of ∼12 nt. Filling this gap permits stepwise ligation of the newly made RDP to downstream chains, eventually yielding Okazaki fragment-sized products. The physiological significance of the RDP array has been reinforced by recent data showing that human DNA ligase I is involved not only in conversion of Okazaki fragments into long nascent DNA chains (13,14) but also in processing of RNA-primed DNA intermediates of ∼20 nt (T.Nethanel, A.Simons, D.Canaani, C.Cardoso and G.Kaufmann, unpublished results). Successive deposition of RDPs by pol α-primase is also consistent with the high frequencies of DNA chain start sites in lagging template portions flanking the origins of bi-directional replication of SV40 (15), Saccharomyces cerevisiae (16) and human replicons (17). Finally, the ability to UV-crosslink Rpa2 to growing SV40 RDPs but not more advanced intermediates is in keeping with the nested discontinuity model (18).
Here we show that Rpa2 interacts with the emerging RDP earlier than Rpa1 and that the preference is reversed upon RDP maturation. Correlated with reported patterns of UV-crosslinking of RPA to synthetic primed templates (19–22), RPA’s structural attributes (23,24) and the ligand-binding properties (2,3), the data lead to the following suggestions. First, the interface of the eukaryal SSB with the replicative template is adjusted to the extension of the primer and contraction of the gap downstream during the RDP growth cycle. Secondly, they confirm the proposal that a mature RDP, inaccessible to Rpa2, faces a short gap, possibly formed by removal of the RNA moiety from an adjacent RDP and, accordingly, insufficient for continuous elongation of the RDP into an Okazaki fragment (9,11).
MATERIALS AND METHODS
Materials
Purified recombinant RPA was prepared as described (25). Monoclonal antibodies (MAbs) specific to Rpa1 and Rpa2, anti-SSB70A and B, and anti-SSB34A, respectively, and corresponding hybridomas (26) were received from Dr Jerard Hurwitz (Memorial Sloan Kettering Cancer Center, New York, NY). The synthesis of 5-[N-(2-nitro-5-azidobenzoyl)-trans-3-aminopropenyl]dUTP (NAB-dUTP) (27) was adapted from a reported procedure (28). TBE-urea (TBE: 90 mM Tris–borate buffer, pH 8.3; 2 mM Na2EDTA) nucleic acid separation gels, Nu-Page protein separation gels, LDS sample buffer [40% (w/v) glycerol, 0.1 M Tris–HCl buffer, pH 8.5, 8% (w/v) lithium dodecyl sulfate, 0.075% (w/v) Serva Blue G250 and 0.025% (w/v) Phenol Red] and SeeBlue Plus2 protein size markers were purchased from NOVEX. Protease inhibitor cocktail tablets (Complete Mini) were purchased from Boehringer Mannheim. Radioactive nucleotides were purchased from NEN and Amersham. The DNA oligomer 5′-GACTGACTGA CTGACTGACTG ACTACACGA TCCCCCGGGA CAGAAGGTGT CCCGGGGGAT CGTG(BrdU)-3′ was custom synthesized by Genosys-Sigma. Microcones-10 ultrafiltration device was purchased from Millipore S.A. T4 polynucleotide kinase and the large (Klenow) fragment of Escherichia coli DNA polymerase I lacking 3′-exonuclease were purchased from New England BioLabs. Other materials have been previously described (18,29).
DNA–protein UV-crosslinking in isolated replicating SV40 chromatin
Pulse labeling of nascent SV40 DNA in isolated replicating SV40 chromatin and subsequent DNA–protein UV-crosslinking were carried out essentially as described (18). Briefly, replication mixtures (70 µl) contained 20–30 µl aliquots of the SV40 replicating chromatin preparation (corresponding to 106 SV40-infected CV-1 monkey kidney cells), 2 µM each of dGTP and dCTP, 20 µM BrdUTP or NAB-dUTP, as indicated, 0.5 µM [α-32P]dATP (300–1000 Ci/mmol), 2 mM ATP and 20 µM of the other rNTPs, 40 mM HEPES-Na, pH 7.8, 5 mM MgCl2, 30 mM potassium acetate, 250 mM ethylene glycol, 0.25 mM dithiothreitol (DTT) and 60 mM sucrose and the protease inhibitor (Complete Mini). The protease inhibitor was included in all solutions described next except those containing SDS. The DNA was pulse-labeled by incubating the mixtures at 30°C (or as indicated) for the specified time. In pulse-chase experiments, the radioactive and photoreactive dNTPs were replaced during the chase by 100 µM dATP and 2 mM dTTP, respectively, followed by further incubation as indicated. To radiolabel the nascent DNA in the RNA moiety, the replication mixture was made to 0.6–1.2 µM [α-32P]UTP (6000 Ci/mmol), 2 mM ATP, 20 µM of the three other rNTPs, 20 µM BrdUTP or NAB-dUTP, 2 µM each of the other dNTPs and α-amanitin at 200 µg/ml (10).
At the end of the incubation, the mixtures were chilled on ice and transferred to a polystyrene culture plate and placed on a TFX-20M transilluminator providing radiation spectrum with a peak at 312 nm. The transilluminator was pre-chilled to 4°C. After irradiation for 5–8 min at 4°C, the sample was collected from the plate. The plate was washed three times with 150 µl of ice-cold low salt buffer (20 mM HEPES-Na, pH 7.8, 5 mM K-acetate, 0.5 mM MgCl2, 0.5 mM DTT) and the fractions were combined. The SV40 minichromatin was then sedimented at 220 000 g for 90 min through a 200 µl cushion of 10% glycerol in low salt buffer. Where indicated, the chromatin pellet was suspended in 80 µl DNase I buffer and digested with DNase I (18). To prepare the sample for native immunoprecipitation (IP), it was diluted 10-fold by adding Nonidet P-40 lysis buffer, yielding final concentrations of 50 mM Tris–HCl buffer, pH 7.5, 300 mM NaCl, 1% Nonidet P-40. The diluted sample was further incubated for 10 min at 4°C and the suspension clarified then by centrifugation at 22 000 g and 4°C for 10 min. Alternatively, the irradiated sample was collected and DNase I digested in DNase I buffer and then concentrated to 10–20 µl by ultrafiltration followed by IP as above.
To characterize the crosslinked DNA, the chromatin pellet was pre-incubated in 100 µl Nonidet P-40 lysis buffer containing 2 M NaCl. The high salt concentration helped remove non-crosslinked proteins from the irradiated SV40 chromatin. To minimize rebinding of RPA upon dilution, the sample was supplemented with 10–20 µg of salmon sperm ssDNA and diluted with Nonidet P-40 low salt lysis buffer to the final concentration described above and then treated like the DNase I digested sample.
Immunochemical detection of photolabeled RPA subunits
Normal mouse IgG (10 µg) was adsorbed to 20 µl of Protein G–Sepharose beads in phosphate-buffered saline (10 mM potassium phosphate buffer, pH 7.4, 0.15 M NaCl) containing 1% bovine serum albumin. The washed beads were used to pre-clear the photolabeled suspensions by incubation on a rotator for 4–8 h at 4°C. When the DNA was left intact, the pre-clearing procedure was repeated. Specific IP was performed with anti-Rpa1 MAbs (anti-SSB70A or B) or with anti-SSB34A. Anti-SSB34A and anti-SSB70A or B hybridoma supernatants were used in amounts of 1–1.5 ml per sample (20 µl of Protein G–Sepharose beads). The pre-cleared sample was added to the Protein G–antibody beads and the suspension rotated for 6–16 h at 4°C. The beads were rinsed four times with washing buffer containing 0.2% Nonidet P-40 (29). The immunoprecipitates were released from the Protein G beads by heating in 150 µl denaturation buffer (50 mM Tris–HCl buffer, pH 7.5, 0.5% SDS, 70 mM β-mercaptoethanol). The beads were washed four times with denaturation buffer. The supernatants were combined, extracted with phenol and proteins were precipitated from the phenol phase and interphase with acetone. The acetone precipitates were dissolved by heating for 10 min at 100°C in 20 µl SDS–PAGE sample buffer or LDS loading buffer (Novex) and separated then on SDS–10% polyacrylamide gel or Nu-PAGE 4–12% gradient gel (Novex). The gels were blotted onto a nitrocellulose membrane or dried as such. Radioactivity was monitored and quantified by autoradiography and densitometry (UMAX Astra 1220s) using TINA software (Raytest Isotopenmessgeräte Gmbh). These tools were also used for the densitometry of the enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech) images of the immunoblots.
UV-crosslinking of purified RPA to a synthetic oligonucleotide
The oligonucleotide 65mer was labeled either at the 5′-end with T4 polynucleotide kinase using [γ-32P]ATP or at the 3′-end by addition of [α-32P]dAMP in a reaction catalyzed by Klenow DNA polymerase lacking 3′-exonuclease. About 15 pmol of the radiolabeled oligonucleotide was incubated with 15 pmol of recombinant RPA in 30 µl of HI buffer [30 mM Na–HEPES buffer, pH 7.8; 0.25 mM EDTA, 0.5% (w/v) inositol, 0.01% (v/v) NP-40] for 30 min at 0°C (25). The mixture was UV-irradiated, digested with DNase I and the photolabeled protein was immunopurified as described above for the in situ photolabeled counterpart.
Sizing crosslinked DNA chains
Crosslinked DNA was monitored essentially as described (18). Briefly, gel portions containing photolabeled proteins of interest were excised, swollen in 10 mM Tris–HCl buffer, pH 7.5, 0.1 mM EDTA, and ground. The slurry was suspended in 300 µl of a similar buffer also containing 1% SDS and 100 µg/ml proteinase K and incubated for 2 h at 37°C. In certain experiments, a comparable gel portion was extracted without protease treatment to provide the background of free DNA migrating on SDS–PAGE with the photolabeled proteins. The released DNA was mixed with 5–10 µg of carrier nucleic acid such as salmon sperm ssDNA, extracted with phenol and separated on denaturing 12% polyacrylamide–7 M urea gel in 25 mM Tris–borate buffer, pH 8.3, or TBU 15% (Novex; pre-cast 15% polyacrylamide gel in 89 mM Tris–borate buffer, pH 8.3; 2 mM Na2EDTA, 7 M urea). After autoradiography and densitometric scanning, the profile of the non-proteolyzed control was subtracted from that of the proteolyzed sample to provide the net profile of the released crosslinked DNA. The upper and lower borders of the sized distributions were set at points of half maximal value.
RESULTS
Size difference between nascent DNA chains crosslinked to Rpa1 and Rpa2
Mammalian replication proteins can be functionally characterized by in situ UV-crosslinking to SV40 or cellular nascent DNA and immunopurification (29). The nascent DNA is pulse-labeled with a radioactive rNTP or dNTP precursor as well as a photoreactive dNTP precursor. After UV-irradiation, proteins are separated from the free nascent DNA and photolabeled proteins of interest are immunopurified. The crosslinked DNA may be characterized by size, RNA primer content and, in the case of SV40, replication fork polarity. In addition, the protein–nascent DNA interactions can be followed dynamically by performing the crosslinking during continuous labeling and pulse-chase regimens.
Using this protocol, Rpa2 and Rpa1 have been shown to interact with nascent SV40 DNA. In the case of Rpa2, the crosslinked nascent DNA has been characterized and shown to comprise growing RDPs but not more advanced precursors of Okazaki fragments (18). Here we compared the sizes and dynamics of the nascent SV40 DNA populations interacting with two RPA subunits. To avoid selective loss of Rpa1, which is less soluble in its isolated form (2), the two subunits were immunopurified within the native photolabeled heterotrimer. The Rpa2-specific MAb anti-SSB34A that yields the same Rpa1/Rpa2 photolabeling ratio as three anti-Rpa1 MAbs (18) was used routinely for this purpose. For mere detection of the photolabeled subunits, non-covalently linked proteins were removed by exhaustive digestion of the irradiated SV40 chromatin with DNase I before immunoprecipitation. However, when characterization of the crosslinked nascent DNA chains was also intended, the bulk proteins were separated from the photolabeled conjugates by pre-incubating the irradiated SV40 chromatin in 2 M NaCl. The immunopurified RPA exhibited the same ratio of Rpa1/Rpa2 photolabeling regardless of the salt concentration in the pre-incubation mixture or whether anti-SSB34A or anti-SSB70B was the immunoprecipitating MAb (18 and data not shown).
Separation of the native, DNase I-treated photolabeled RPA by SDS–PAGE yielded two major radioactive bands (Fig. 1, lane 3). They migrated as expected of photolabeled derivatives of Rpa1 and Rpa2 containing the residual DNase I resistant DNA adduct (18). Their identity was ascertained by the similar electrophoretic mobility of corresponding markers prepared by UV-crosslinking purified RPA to the radiolabeled 3′-end of the DNA hairpin oligonucleotide 66mer followed by digestion with DNase I and immunopurification with anti-SSB34A under native conditions (lane 1). IP with anti-SSB34A under denaturing/partially renaturing conditions yielded only the Rpa2 derivative, both with the marker (lane 2) and in situ photolabeled RPA (lane 4). The retardation of the latter by the equivalent of ∼4 kDa is attributed to lack of phosphorylation of the recombinant subunit and presence of RNA primer moieties in part of the in situ photolabeled Rpa2. Attempts to immunoprecipitate the dissociated photolabeled Rpa1 by the cognate MAb anti-SSB70A yielded no detectable signal, both with the marker and in situ photolabeled Rpa1 (not shown), probably due to the poor solubility of free Rpa1 (2). The minor in situ photolabeled products found in the native immunoprecipitate, which migrated at the high molecular weight range and between the two subunits (lane 3), were probably derived from other photolabeled replication proteins associated with the native RPA.
Figure 1.
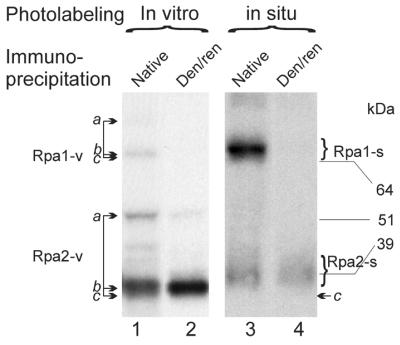
Comparison of in situ and in vitro photolabeled RPA subunits. Purified recombinant RPA (25) was UV-crosslinked to the hairpin oligonucleotide 66mer containing 3′-penultimate BrdU and 3′-terminal [α-32P]dAMP residues (Materials and Methods). The conjugate was digested with DNase I, leaving a residual DNA adduct on the major products (marked by arrows b). A minor fraction (marked as bands a and visible in the exposure shown with Rpa2) remained protected from digestion probably by binding of a second RPA molecule. The products of bands a migrated similarly to the respective RPA subunits crosslinked to the undigested DNA oligonucleotide (not shown). The photolabeled RPA was immunoprecipitated by anti-SSB34 in native form (lane 1) or under denaturing partially/renaturing conditions (lane 2) and separated by SDS–PAGE. The in situ photolabeled RPA was isolated from replicating SV40 chromatin pulse-labeled by BrdUTP and [α-32P]dATP for 90 s. After DNase I digestion, an aliquot was immunopurified using anti-SSB34A under native (lane 3) or denaturing/partially renaturing conditions (lane 4). The positions of unmodified Rpa1 and Rpa2 (indicated by arrows c) were determined by western blotting as previously described (18). kDa, protein size markers. Rpa1-v and Rpa2-v, in vitro photolabeled forms of the respective subunits; Rpa1-s and Rpa2-s indicate the corresponding in situ photolabeled forms. Native and Den/ren indicate IP under native and denaturing/partially renaturing conditions, respectively.
To size the DNA crosslinked to the two RPA subunits, the photolabeled native heterotrimer was immunopurified without prior DNase I digestion. This broadened the bands of the photolabeled subunits and enhanced the signals of the non-specific co-precipitating products (Fig. 2A, lane 2). The photolabeled RPA subunits were extracted from the indicated gel areas and treated with proteinase K. The released DNA populations were purified by phenol extraction and sized by denaturing polyacrylamide–urea gel electrophoresis (Fig. 2B, lanes 1 and 2). Controls not treated with the protease indicated that little, if any, free nascent DNA had migrated with the photolabeled protein conjugates on SDS–PAGE (18 and data not shown). The photolabeled Rpa1 and Rpa2 conjugates yielded partially overlapping size distributions, peaking in respective order at 29 and 36 nt in the experiment shown (Fig. 2B and C). By setting the lower and upper borders of distributions at half peak values, respective increments of 3.3 and 11.1 nt in favor of Rpa1 were obtained in the experiment shown and 6.7 and 10.3 nt in another. These differences were attributed to the crosslinked DNA itself rather than residual peptide adducts left by proteinase K. Namely, proteolysis of photolabeled Rpa1 and Rpa2 markers obtained by UV-crosslinking purified RPA to the 3′-end of a 5′-32P-labeled oligonucleotide 65mer (Materials and Methods) released in each case a product indistinguishable in electrophoretic mobility from the original oligonucleotide (not shown).
Figure 2.
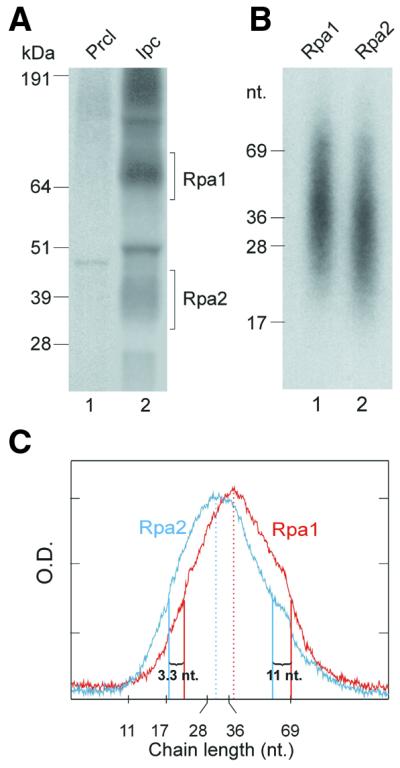
Sizing nascent SV40 DNA chains radiolabeled in the DNA moiety and crosslinked to RPA subunits. (A) Photolabeled RPA was isolated from the pulse-labeled replication mixture without DNase I treatment, as detailed in Figure 1 and Materials and Methods. Precipitates of pre-clearing (lane 1) and IP of RPA under native conditions (lane 2) were resolved by SDS–PAGE. (B) The Rpa1 and Rpa2 bands were excised from the indicated gel sections. The photolabeled proteins were extracted and treated with proteinase K. After phenol extraction, the released nascent DNA was separated by electrophoresis on 15% TBU gel (Novex). (C) Densitometric profiles of (B), lane 1 (red) and lane 2 (cyan). Peak values and half peak values marking distributions borders are indicated by respective full or dashed vertical lines in corresponding color. kDa, protein size markers; nt, nucleotide size markers; Prcl, pre-clearing immunoprecipitate; Ipc, specific immunoprecipitate.
Rpa2 selectively UV-crosslinks a distinct subpopulation of short RDPs
Radiolabeling the RNA moiety accentuates the signal of the shortest nascent DNA chains UV-crosslinked to Rpa2, revealing a distinct subpopulation of ∼15 nt. This effect has been attributed mainly to a shift in representation from weight to number distribution (18), although additional contributions due to different dynamics of the incorporation and turn over of the RNA and DNA precursors cannot be excluded. UV-crosslinking of RPA to nascent DNA radiolabeled in the RNA moiety was performed here using the potent photoreactive precursor NAB-dUTP (20) instead of BrdUTP used routinely. In the presence of NAB-dUTP, Rpa2 yielded a pattern of two distinct subpopulations crosslinked to Rpa2 similar to that seen with the BrdU-containing nascent DNA (18). However, the NAB-dU-containing DNA released from the RPA subunits was retarded by an equivalent of 5–10 nt, probably due to the bulkier substituent (20 and data not shown). The Rpa1 profile overlapped the longer subpopulation released from Rpa2 (Fig. 3B, compare lanes 1 and 2). Interestingly, the longer chains seen specifically in the Rpa1 profile when the DNA moiety was radioactively labeled (Fig. 2B and C) could not be detected when the RNA moiety was radiolabeled (Fig. 3B and C). Hence, these longer, Rpa1-specific products were either devoid of RNA or had been primed before the onset of the in vitro incubation with the radiolabeled rNTP.
Figure 3.
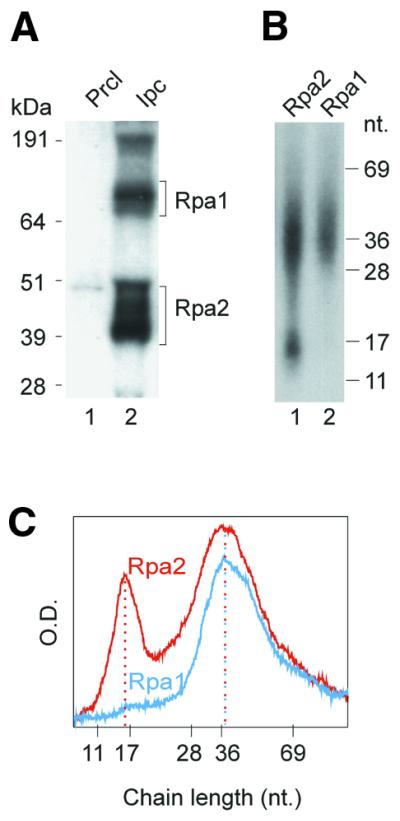
Sizing nascent SV40 DNA chains radiolabeled in the RNA moiety and crosslinked to RPA subunits. (A) SDS–PAGE pattern of photolabeled RPA subunits crosslinked to nascent SV40 DNA pulse-labeled with NAB-dUTP and [α-32P]UTP. (B) Size distribution of the DNA chains released by proteinase K from the photolabeled RPA subunits indicated in (A). (C) Densitometric profiles of (B), lanes 1 (red) and 2 (cyan). kDa, protein size markers; nt, nucleotide size markers; Prcl, pre-clearing immuno-precipitate; Ipc, specific immunoprecipitate.
Different interaction schedules of the two RPA subunits with nascent DNA
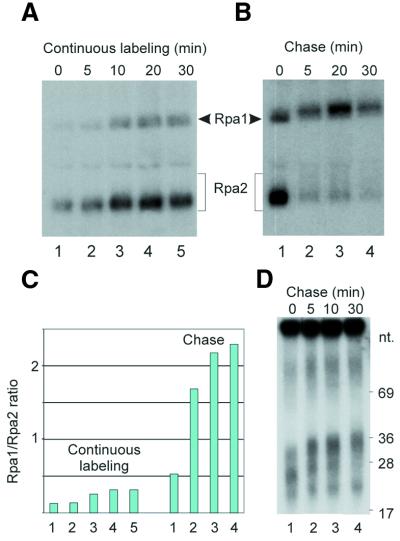
The different size distributions of nascent DNA chains crosslinked to the two RPA subunits (Figs 2 and 3) suggested that Rpa2 encounters growing RDPs earlier than Rpa1 and that the crosslinking preference is reversed with RDP maturation and further processing. To examine this possibility, the photolabeling efficiencies of the two subunits were compared during RDP growth and further processing. Significant differences between the dynamics of photolabeling of the two subunits were not detected when the UV-crosslinking was performed during incubation in the standard replication mixture at 30°C (not shown). However, distinction could be made by slowing down or arresting RDP elongation or allowing it to proceed without priming new chains. Reducing the incubation temperature of the replication mixture to 0°C delayed RDP maturation considerably. Under these conditions, the Rpa1/Rpa2 photolabeling ratio increased moderately with RDP elongation during the continuous-labeling period and, more dramatically, during the subsequent chase period (Fig. 4A–C). The growth of the crosslinked DNA chain itself was indicated by the slight increase in the retardation of the photolabeled proteins on SDS–PAGE (Fig. 4A and B). It is noteworthy that the residual adduct left by DNase I reached ∼20 nt. Hence, within this range, limited RDP elongation could be visualized by the retardation effect.
Figure 4.
Rpa1/Rpa2 photolabeling ratio during RDP growth at 0°C. Replicating SV40 chromosomes incubated at 0°C were pulse-labeled with BrdUTP and [α-32P]dATP and chased with excess dTTP and non-labeled dATP. Aliquots withdrawn at the indicated time points were UV-irradiated and the photolabeled proteins digested with DNase I and further processed as in the Materials and Methods. (A and B) SDS–PAGE patterns of RPA subunits photolabeled during pulse-labeling or subsequent chase, respectively. The 5 min interval of UV-irradiation at 4°C is not included in the incubation times shown. (C) Rpa1/Rpa2 photolabeling ratios of (A and B). (D) Size distribution of nascent DNA chains of the indicated chase times.
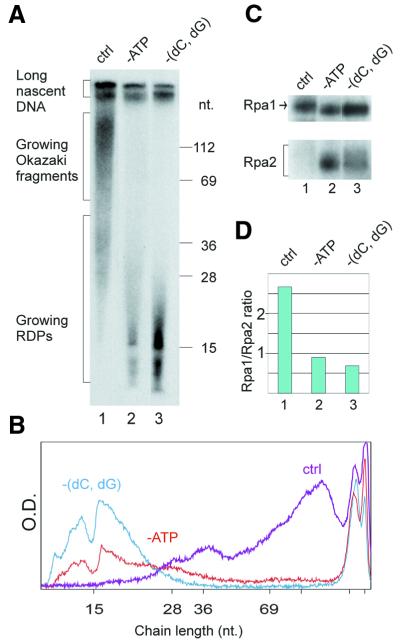
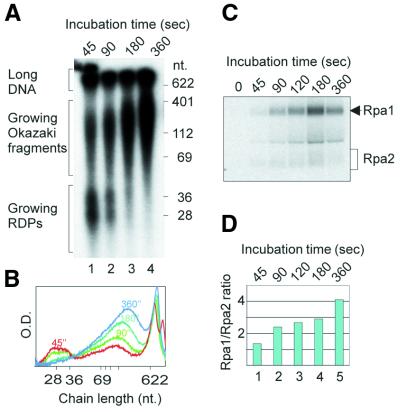
RDP maturation and conversion into Okazaki fragments are inhibited by depletion of ATP from the replication mixture (12), probably due to the failure to load PCNA on the mature RDP (7). The inhibition of RDP elongation and further processing elicited by ATP depletion (Fig. 5A, lane 2, and B) were accompanied by preferential photolabeling of Rpa2 (Fig. 5C, lane 2, and D). Omission of dCTP and dGTP, which arrested RDP elongation, likewise shifted the preference in favor of Rpa2 (Fig. 5A and C, compare lanes 1 and 3 in each, and D). In contrast, formation of longer nascent DNA chains in the control mixture (Fig. 5A, lane 1, and B) was associated with preferential crosslinking to Rpa1 (Fig. 5C, lane 1, and D). When UTP was omitted, priming of new chains was inhibited while pre-existing RDPs were elongated and further processed (Fig. 6A and B). This was accompanied by a progressive increase in the Rpa1/Rpa2 photolabeling ratio (Fig. 6C and D).
Figure 5.
Effect of arresting RDP growth on Rpa1/Rpa2 photolabeling ratio. Replicating SV40 chromosomes were pulse-labeled with BrdUTP and [α-32P]dATP as in the standard mixture or in the absence of the indicated nucleotides. (A) Size distributions of nascent DNA chains synthesized in the control (ctrl), ATP-deficient (-ATP) or dCTP- and dGTP-deficient [-(dC,dG)] replication mixtures. (B) Densitometric tracing of DNA size distributions of (A) lanes 1 (purple), 2 (red) and 3 (cyan). (C) Separation of photolabeled RPA subunits from the indicated reaction mixtures. (D) Rpa1/Rpa2 photolabeling ratios.
Figure 6.
Change in Rpa1/Rpa2 photolabeling ratio during RDP elongation and further processing. Replicating SV40 chromosomes were pulse-labeled with BrdUTP and [α-32P]dATP in the absence of UTP. (A) Size distributions of nascent DNA chains at different incubation times. (B) Densitometric tracing of nascent DNA populations of the incubation times shown in (A). (C) Separation of RPA subunits photolabeled at the indicated incubation times. (D) Rpa1/Rpa2 photolabeling ratios at incubation times shown in (C).
DISCUSSION
Modulation of RPA interface with the primed template during RDP growth
The data shown indicate that Rpa2 is poised to interact with the emerging RDP as soon as a few dNMPs are added to the RNA primer while Rpa1 is availed after extension by several more. Both subunits remain accessible during RDP maturation. Subsequently, Rpa2 disengages while Rpa1 stays put during further, limiting DNA chain growth. These switches, correlated with the reported crystal structures of RPA, ligand-binding properties and patterns of crosslinking to model oligonucleotides, suggest that RPA modulates its interface with the replicative DNA template during DNA chain growth, as discussed below and depicted schematically in Figure 7.
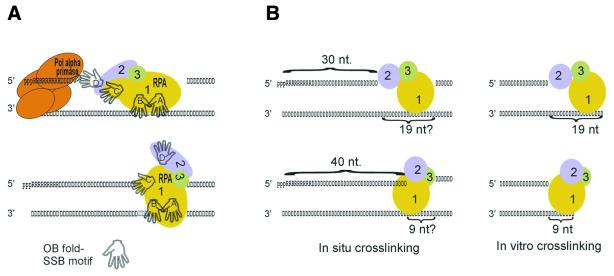
Figure 7.
Hypothetical changes in RPA interface with the primed template during RDP growth. (A) Presumptive switch of individual SSB motifs of RPA from template binding to potential interaction with the primer end during elongation of the DNA moiety of an RDP and contraction of the gap downstream. (B) The correlation of the Rpa2 → Rpa1 switch in crosslinking preference upon RDP maturation in situ (18 and this work) with that due to diminution of the synthetic template overhang or gap of in vitro reconstituted complexes (21,22) suggests that the ssDNA gap 3′ to the mature RDP is <20 nt. pppRRRRRRRRRR, RNA primer; DDDDDDDD . . . DDD, DNA strand. A–D indicate respective SSB motifs of RPA (24,35).
The central portion of Rpa1 contains two structurally homologous SSB motifs termed A and B. They interact with an octanucleotide in a co-crystal structure in the 5′ A → B 3′ polarity (23). The RPA heterotrimer has greater affinity for ssDNA and occludes up to 30 nt (30–33). The enhanced binding of the heterotrimer is attributed to two additional lone SSB motifs, motif C of the C-terminal domain of Rpa1 and motif D of Rpa2 (24,34–38). Originally, the SSB activity of RPA was implicated with template strand binding only (2). However, UV-crosslinking of RPA to synthetic primed templates (19) and nascent SV40 DNA (18) has indicated that RPA is also poised to contact the growing primer strand. The UV-crosslinking data, combined with RPA’s ligand-binding properties (30,31) and crystal structures (23,24), suggest specific assignments of individual SSB motifs in the dynamic interaction with the replicating DNA template. Thus, at the onset of RDP synthesis, the free ssDNA template downstream may be occupied by all four SSB motifs oriented, perhaps 5′ A → B → C → D 3′. This arrangement agrees with the proximity of motifs C and D in the Rpa2/Rpa3 subassembly and their upstream position relative to the template portion occupied by motifs A and B (2,23). With elongation of the DNA chains and contraction of the ssDNA gap downstream, motifs D and C may be gradually displaced from the template and availed instead to the growing primer strand. This scenario is hinted at by the ability of Rpa2 to crosslink young RDPs as short as ∼15 nt and of Rpa1 to follow suit after addition of a few more (18) (Figs 2 and 3). Presumably, the ∼15 nt fraction contains the minimal number of dNMP residues needed to bind the lone SSB motif of Rpa2 (24). It is noteworthy that neither RPA subunit crosslinks nascent DNA pulse-labeled with a photoreactive RNA precursor (G.Mass, T.Nethanel and G.Kaufmann, unpublished results), in keeping with their specificity for DNA (2). It is noteworthy that the 15 nt sized chains, which abounded among those crosslinked to Rpa2, constitute only a small fraction of the nascent DNA chains. However, as only <1% of the nascent DNA was crosslinked to Rpa2 and the other replication proteins, this bias may reflect a greater tendency of these early products to interact with Rpa2 due, perhaps, to greater propensity of pol α to abandon the growing RDP at this early stage. Further extension of the RDP also permitted crosslinking of Rpa1. This change could reflect the later displacement of motif C from the template by the growing primer end. An alternative possibility that a longer DNA portion is needed to accommodate the two RPA subunits is deemed less likely, as either subunit can interact with the 3′-end of the synthetic primer strand (19–22). In other words, the interaction of the two subunits with the primer strand may be mutually exclusive. Interaction of motifs A and B with the growing RDP also seems unlikely due to the downstream position they occupy (23) and as the crosslinking of RPA to the primer end depends on a minimal template overhang, sufficient for binding motifs A and B (19).
Upon RDP maturation, Rpa2 motif D disengages from the primer end, judged from the failure of Rpa2 to crosslink longer products (18) (Figs 2 and 3). Rpa2 also fails to access the 3′-end of the primer in synthetic primed templates or gapped duplexes when the ssDNA overhang or gap is reduced below a critical threshold, between 19 and 9 nt. Under these conditions, only Rpa1 crosslinks the primer end (19,21,22). The coincident failure of Rpa2 to interact with mature RDPs in replicating SV40 chromosomes (Figs 2–6) and with the 3′-end of the synthetic primer when the gap is reduced below the critical threshold (19,21,22) suggests that the mature RDP faces a short gap close in size to the threshold value. Such a gap could arise by removal of an RNA moiety between adjacent RDPs (nested discontinuity model) but would be insufficient for continuous elongation of the RDP to form an Okazaki fragment (initiation zone model).
Continued interaction of Rpa1 with the growing primer strand after the ssDNA gap contracts below the threshold value may also apply to gaps separating more advanced precursors including Okazaki fragments about to be joined to the long nascent chain. However, the corresponding crosslinking products were absent or under-represented in the profile of nascent DNA chains crosslinked to Rpa1, which tapered off at >80 nt (compare Figs 2 and 5B). According to the nested discontinuity model, 3′ portions of the more advanced products are also the oldest. Hence, they could be largely synthesized before the onset of in vitro pulse labeling and, hence, be deficient in the photoreactive precursor. The interaction of Rpa1 with intermediates facing short gaps could occur during their extension by pol δ. Persistence of RPA on the template after the pol α → pol δ switch (5), i.e. after RDP maturation (7,12,39), has been inferred from in vitro reconstitution studies (40).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jerard Hurwitz of the Memorial Sloan Kettering Cancer Research Institute for anti-RPA antibodies and hybridomas and Yaron Daniely for critical reading. G.K. is an incumbent of the Louise and Nahum Barag Chair in Molecular Genetics and Cancer Biology. This work was supported by grants from the US–Israel Binational Science Foundation (to G.K. and M.S.W.), the German Isareli Foundation of Scientific Research and Development and Israel Cancer Society (to G.K.) and from the Russian Foundation for Basic Research (grant 99-04-49277 to O.I.L.).
References
- 1.Lohman T.M. and Ferrari,M.E. (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem., 63, 527–570. [DOI] [PubMed] [Google Scholar]
- 2.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 3.Iftode C., Daniely,Y. and Borowiec,J.A. (1999) Replication protein A (RPA): the eukaryotic SS. Crit. Rev. Biochem. Mol. Biol., 34, 141–180. [DOI] [PubMed] [Google Scholar]
- 4.Salas M., Miller,T.J., Leis,J. and DePamphilis,M.L. (1996) Mechanisms for priming DNA synthesis. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 131–176.
- 5.Tsurimoto T., Melendy,T. and Stillman,B. (1990) Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature, 346, 534–539. [DOI] [PubMed] [Google Scholar]
- 6.Nethanel T. and Kaufmann,G. (1990) Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J. Virol., 64, 5912–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock P.A., Seo,Y.S. and Hurwitz,J. (1991) Initiation of simian virus 40 DNA synthesis in vitro. Mol. Cell. Biol., 11, 2350–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waga S., Bauer,G. and Stillman,B. (1994) Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. J. Biol. Chem., 269, 10923–10934. [PubMed] [Google Scholar]
- 9.Kaufmann G., Anderson,S. and DePamphilis,M.L. (1977) RNA primers in simian virus 40 DNA replication: distribution of 5′ terminal oligoribonucleotides in nascent DNA. J. Mol. Biol., 111, 549–568. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann G. (1981) Characterization of initiator RNA from replicating simian virus 40 DNA synthesized in isolated nuclei. J. Mol. Biol., 147, 25–39. [DOI] [PubMed] [Google Scholar]
- 11.Nethanel T., Reisfeld,S., Dinter-Gottlieb,G. and Kaufmann,G. (1988) An Okazaki piece of simian virus 40 may be synthesized by ligation of shorter precursor chains. J. Virol., 62, 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nethanel T., Zlotkin,T. and Kaufmann,G. (1992) Assembly of simian virus 40 Okazaki pieces from DNA primers is reversibly arrested by ATP depletion. J. Virol., 66, 6634–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prigent C., Satoh,M.S., Daly,G., Barnes,D.E. and Lindahl,T. (1994) Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol., 14, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenney V.J., Barnes,D.E. and Lindahl,T. (1997) Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J. Biol. Chem., 272, 11550–11556. [DOI] [PubMed] [Google Scholar]
- 15.Hay R. and DePamphilis,M.L. (1982) Initiation of SV40 DNA replication in vivo: location and structure of 5′ ends of DNA synthesized in the ori region. Cell, 28, 767–779. [DOI] [PubMed] [Google Scholar]
- 16.Bielinsky A.K. and Gerbi,S.A. (1998) Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science, 279, 95–98. [DOI] [PubMed] [Google Scholar]
- 17.Abdurashidova G., Deganuto,M., Klima,R., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science, 287, 2023–2026. [DOI] [PubMed] [Google Scholar]
- 18.Mass G., Nethanel,T. and Kaufmann,G. (1998). The middle subunit of replication protein A contacts RNA–DNA primers within replicating SV40 chromosomes. Mol. Cell. Biol., 18, 6399–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavrik O.I., Nasheuer,H.P., Weisshart,K., Wold,M.S., Prasad,R., Beard,W.A., Wilson,S.H. and Favre,A. (1998) Subunits of human replication protein A are crosslinked by photoreactive primers synthesized by DNA polymerases. Nucleic Acids Res., 26, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavrik O.I., Kolpashchikov,D.M., Nasheuer,H.P., Weisshart,K. and Favre,A. (1998) Alternative conformations of human replication protein A are detected by crosslinks with primers carrying a photoreactive group at the 3′-end. FEBS Lett., 441, 186–190. [DOI] [PubMed] [Google Scholar]
- 21.Kolpashchikov D.M., Weisshart,K., Nasheuer,H.P., Khodyreva,S.N., Fanning,E., Favre,A. and Lavrik,O.I. (1999) Interaction of the p70 subunit of RPA with a DNA template directs p32 to the 3′-end of nascent DNA. FEBS Lett., 450, 131–134. [DOI] [PubMed] [Google Scholar]
- 22.Kolpashchikov D.M., Khodyreva,S.N., Khlimankov,D.Y., Wold,M.S., Favre,A. and Lavrik,O.I. (2001) Polarity of human replication protein A binding to DNA. Nucleic Acids Res., 29, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkarev A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- 24.Bochkarev A., Bochkareva,E., Frappier,L. and Edwards,A.M. (1999) The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J., 18, 4498–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henricksen L.A., Umbricht,C. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 26.Kenny M.K., Schlegel,U., Furneaux,H. and Hurwitz,J. (1990) The role of human single stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem., 13, 7693–7700. [PubMed] [Google Scholar]
- 27.Kolpashchikov D.M., Ivanova,T.M., Boghachev,V.S., Nasheuer,H.P., Weisshart,K., Favre,A., Pestryakov,P.E. and Lavrik,O.I. (2000) Synthesis of base-substituted dUTP analogues carrying a photoreactive group and their application to study human replication protein A. Bioconjug. Chem., 11, 445–451. [DOI] [PubMed] [Google Scholar]
- 28.Bartholomew B., Tinker,R.L., Kassavetis,G.A. and Geiduschek,E.P. (1995) Photochemical cross-linking assay for DNA tracking by replication proteins. Methods Enzymol., 262, 476–494. [DOI] [PubMed] [Google Scholar]
- 29.Zlotkin T., Kaufmann,G., Jiang,Y., Lee,M.Y.W.T., Uitto,L., Syvaoja,J., Dornreiter,I., Fanning,E. and Nethanel,T. (1996) DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J., 15, 2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 30.Blackwell L.J. and Borowiec,J.A. (1994) Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol., 14, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C., Paulus,B.F. and Wold,M.S. (1994) Interactions of human replication protein A with oligonucleotides. Biochemistry, 33, 14197–14206. [DOI] [PubMed] [Google Scholar]
- 32.Pfuetzner R.A., Bochkarev,A., Frappier,L. and Edwards,A.M. (1997) Replication protein A. Characterization and crystallization of the DNA binding domain. J. Biol. Chem., 272, 430–434. [DOI] [PubMed] [Google Scholar]
- 33.Kim C. and Wold,M.S. (1995) Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry, 34, 2058–2064. [DOI] [PubMed] [Google Scholar]
- 34.Philipova D., Mullen,J.R., Maniar,H.S., Lu,J., Gu,C. and Brill,S.J. (1996) A hierarchy of SSB protomers in replication protein A. Genes Dev., 10, 2222–2233. [DOI] [PubMed] [Google Scholar]
- 35.Brill S.J. and Bastin-Shanower,S. (1998) Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol., 18, 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochkareva E., Frappier,L., Edwards,A.M. and Bochkarev,A. (1998) The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem., 273, 3932–3936. [DOI] [PubMed] [Google Scholar]
- 37.Sibenaller Z.A. and Wold,M.S. (1998) The 32- and 14-kDa subunits of replication protein are responsible for species-specific interactions with ssDNA. Biochemistry, 37, 12496–12506. [DOI] [PubMed] [Google Scholar]
- 38.Walther A.P., Bjerke,M.P. and Wold,M.S. (1999) A novel assay for examining the molecular reactions at the eukaryotic replication fork: activities of replication protein A required during elongation. Nucleic Acids Res., 27, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waga S. and Stillman,B. (1994) Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature, 369, 207–212. [DOI] [PubMed] [Google Scholar]
- 40.Yuzhakov A. (1999) Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J., 18, 6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]