Abstract
More than 3000 type II restriction endonucleases have been discovered. They recognize short, usually palindromic, sequences of 4–8 bp and, in the presence of Mg2+, cleave the DNA within or in close proximity to the recognition sequence. The orthodox type II enzymes are homodimers which recognize palindromic sites. Depending on particular features subtypes are classified. All structures of restriction enzymes show a common structural core comprising four β-strands and one α-helix. Furthermore, two families of enzymes can be distinguished which are structurally very similar (EcoRI-like enzymes and EcoRV-like enzymes). Like other DNA binding proteins, restriction enzymes are capable of non-specific DNA binding, which is the prerequisite for efficient target site location by facilitated diffusion. Non-specific binding usually does not involve interactions with the bases but only with the DNA backbone. In contrast, specific binding is characterized by an intimate interplay between direct (interaction with the bases) and indirect (interaction with the backbone) readout. Typically ∼15–20 hydrogen bonds are formed between a dimeric restriction enzyme and the bases of the recognition sequence, in addition to numerous van der Waals contacts to the bases and hydrogen bonds to the backbone, which may also be water mediated. The recognition process triggers large conformational changes of the enzyme and the DNA, which lead to the activation of the catalytic centers. In many restriction enzymes the catalytic centers, one in each subunit, are represented by the PD . . . D/EXK motif, in which the two carboxylates are responsible for Mg2+ binding, the essential cofactor for the great majority of enzymes. The precise mechanism of cleavage has not yet been established for any enzyme, the main uncertainty concerns the number of Mg2+ ions directly involved in cleavage. Cleavage in the two strands usually occurs in a concerted fashion and leads to inversion of configuration at the phosphorus. The products of the reaction are DNA fragments with a 3′-OH and a 5′-phosphate.
INTRODUCTION
Restriction endonucleases occur ubiquitously among prokaryotic organisms (1,2). Their principal biological function is the protection of the host genome against foreign DNA, in particular bacteriophage DNA (3). Other functions are still being discussed, such as an involvement in recombination and transposition (4–7). In addition, there is evidence that the genes for restriction and modification enzymes may act together as selfish elements (8).
By definition, restriction endonucleases are parts of restriction–modification (RM) systems, which comprise an endonuclease and a methyltransferase activity. Whereas the substrate of the restriction enzyme is foreign DNA, which is cleaved in response to defined recognition sites, that of the modification enzyme is the DNA of the host which is modified at the recognition sequence and, thereby, protected against attack by the restriction endonuclease. Three types of RM systems have been found and were classified according to their subunit composition, cofactor requirement and mode of action (9). The distinction between type I, II and III systems is still useful, but it is becoming apparent that there are intermediate cases (vide infra).
The present review will deal with the type II restriction endonucleases, which, because of their extraordinary importance for gene analysis and cloning work, have been studied in great detail. Moreover, they have proven to be excellent model systems to study highly specific protein–nucleic acid interactions, to investigate structure–function relationships and, last but not least, to understand the mechanisms of evolution within a large family of functionally related enzymes.
The last comprehensive reviews on the structure and function of type II restriction endonucleases appeared in 1993 (1) and 1997 (10). Since then about 1000 new type II restriction enzymes [compare entry numbers in (11) and (12)] were identified, eight more crystal structures determined (giving a total of 12 structures) and many biochemical studies published (http://rebase.neb.com).
From these structural and functional studies it is clear that the family of type II restriction endonucleases is more heterogeneous than originally thought. To point out the common features of these enzymes and the peculiarities of some of them will be the main focus of this review.
THE DIVERSITY OF RESTRICTION ENDONUCLEASES
The main criterion for classifying a restriction endonuclease as a type II enzyme is that it cleaves specifically within or close to its recognition site and that it does not require ATP hydrolysis for its nucleolytic activity.
The orthodox type II restriction endonuclease is a homodimer of ∼2 × 30 kDa molecular mass, which recognizes a palindromic sequence 4–8 bp in length, and in the presence of Mg2+ cleaves the two strands of the DNA within or immediately adjacent to the recognition site to give a 5′-phosphate and a 3′-OH end. Typical representatives (Table 1) are EcoRI (which produces sticky ends with 5′-overhangs) (13), EcoRV (which produces blunt ends) (14) and BglI (which produces sticky ends with 3′-overhangs) (15).
Table 1. Nomenclature of type II restriction endonucleases.
| Subtype |
Characteristic feature |
Examplea |
Recognition sequenceb |
||||||||||||
| Orthodox | Palindromic recognition site, which is recognized by a homodimeric enzyme, cleavage occurs within or adjacent to the recognition site | EcoRI | G↓C | A T | A T | T A | T A↑ | C G | |||||||
| EcoRV | G C | A T | T↓ A↑ | A T | T A | C G | |||||||||
| BglI | GC | C G | C G | N N↑ | N N | N N | N↓ N | N N | G C | G C | C G | ||||
| Type IIS | Asymmetric recognition site with cleavage occuring at a defined distance | FokI | G C | G C | A T | T A | G C | N9↓ N9 | N N | N N | N N | N N↑ | |||
| Type IIE | Two sites required for cleavage, one serving as allosteric effector | NaeI | G C | C G | G↓ C↑ | C G | G C | C G | |||||||
| Type IIF | Two sites required for cleavage, both sites are cleaved in a concerted reaction by a homotetrameric enzyme | NgoMIV | G↓ C | C G | C G | G C | G C↑ | C G | |||||||
| Type IIT | Different subunits with restriction and modification activity | Bpu10I | C G | C↓ G | T A | N N | A T↑ | G C | C G | ||||||
| BslI | C G | C G | N N | N N↑ | N N | N N | N↓ N | N N | N N | G C | G C | ||||
| Type IIG | One polypeptide chain with restriction and modification activity | Eco57I | C G | T A | G C | A T | A T | G C | N14 N14↑ | N N | N↓ N | ||||
| Type IIB | Cleavage on both sides of the recognition site | BcgI | N ↑N | N↓ N | N10 N10 | C G | G C | A T | N6 N6 | T A | G C | C G | N10 N10↑ | N N | N↓ N |
| BplI | N ↑N | N4↓ N4 | N8 N8 | G C | A T | G C | N5 N5 | C G | T A | C G | N8 N8↑ | N4 N4 | N↓ N | ||
| Type IIM | Methylated recognition site | DpnI | G C | mA↓ T ↑ | T µA | C G | |||||||||
aRestriction endonucleases whose crystal structure is known are depicted in bold letters.
bThe site of cleavage is indicated by ↓.
Many type II restriction endonucleases do not conform to this narrow definition, making it necessary to define subdivisions. A new nomenclature for these heterodox type II restriction endonucleases (Table 1) has recently been proposed (R.Roberts, personal communication).
Type IIS restriction endonucleases recognize asymmetric sequences and cleave these sequences at a defined distance (reviewed in 16), for example FokI. Until recently it was believed that these enzymes function as monomers. However, it is now clear from studies on FokI that it dimerizes on the DNA and this may be a more general phenomenon (17).
Type IIE restriction endonucleases interact with two copies of their recognition sequence, one being the target for cleavage, the other serving as allosteric effector (18), for example NaeI.
Type IIF restriction endonucleases are similar to type IIE enzymes, in as much as they interact with two copies of their recognition sequence. They differ from the type IIE enzymes in that they cleave both sequences in a concerted reaction (19), for example NgoMIV.
Type IIT restriction endonucleases are composed of two different subunits, for example Bpu10I and BslI. Bpu10I recognizes an asymmetric sequence and functions as a heterodimer (αβ) in which both subunits presumably have one active site (20). BslI recognizes a palindromic sequence and functions as a heterotetramer (α2β2) (21).
Type IIB restriction endonucleases cleave DNA at both sides of the recognition sequence, for example BcgI which recognizes an asymmetric sequence, or BplI which recognizes a symmetric sequence. These enzymes are composed of different subunits (BcgI, α2β; BplI, αβ) and have restriction and modification activity. They require the presence of AdoMet for restriction (22,23). For BcgI, it was shown that the catalytic centers for restriction and modification are located in the α-subunit, whereas the β-subunit harbors the target recognition domain (24).
Type IIG restriction endonucleases like IIB enzymes are stimulated by AdoMet but have both restriction and modification activity present in a single polypeptide chain (25), for example Eco57I.
Type IIM restriction endonucleases recognize methylated DNA (26), for example DpnI.
Restriction endonucleases, like McrBC, also require a methylated DNA substrate. They resemble type I and type III enzymes in as much as they are dependent on nucleoside triphosphate hydrolysis (GTP in the case of McrBC) for DNA cleavage. Escherichia coli McrBC for cleavage requires two C5- or N4-methylated (or C5-hydroxymethylated) PuC sites (Pu = A or G), carrying at least one methyl group per half-site, at a distance of 40 to ∼2000 bp (27). Cleavage occurs somewhere between the two sites (28). Whereas the McrB subunit is responsible for DNA recognition (29) and GTP cleavage (30), the McrC subunit harbors the catalytic center for phosphodiester bond hydrolysis (U.Pieper and A.Pingoud, submitted). The fact that McrBC requires GTP hydrolysis for cleavage would also justify classifying it as a variant of the type III enzymes. These restriction endonucleases have not been included in Table 1 because they are dependent on nucleoside triphosphate hydrolysis.
It is clear that this nomenclature does not do justice to borderline cases. Consider for example FokI, the archetypal IIS enzyme, which according to recent investigations could also be considered a type IIE enzyme (192), as it requires binding to a second recognition sequence. The recently discovered restriction enzyme HaeIV like BcgI cleaves double-stranded DNA on both sides of its recognition sequence, which means that it should be classified as a type IIB enzyme. On the other hand it harbors restriction and modification activity in one polypeptide chain, making it similar to type IIG enzymes but, in contrast, is not stimulated by AdoMet (31). Type IIT enzymes were originally classified as similar to type IIS enzymes that recognize an asymmetric sequence, but consist of two different subunits. Only last year a type IIT enzyme was discovered, BslI (21), that recognizes a palindromic sequence. Of course, restriction endonucleases that do not fit into any of these subdivisions will continue to be discovered. Eventually this will lead to new subdivisions.
THE SIMILARITY OF RESTRICTION ENDONUCLEASE STRUCTURES
With a few obvious exceptions of closely related isoschizomers, like EcoRI and RsrI (recognizing G↓AATTC), MthT1, FnuDI and NgoPII (recognizing GG↓CC), XmaI and Cfr9I (recognizing CCC↓GGG), BanI and HgiCI (recognizing G↓GYRCC), TaqI/TtHB8I (recognizing T↓CGA), BsoBI and AvaII (recognizing C↓YCRG), to name a few that share between 50 and 80% identical amino acid residues, type II restriction enzymes display little, if any, sequence homology, which had been interpreted to mean that these enzymes are evolutionarily unrelated (4,32). This conviction began to lose credibility by the observation that there is a statistically highly significant correlation between the genotype (amino acid sequence) and the phenotype (recognition sequence, site of cleavage) of restriction enzymes (33).
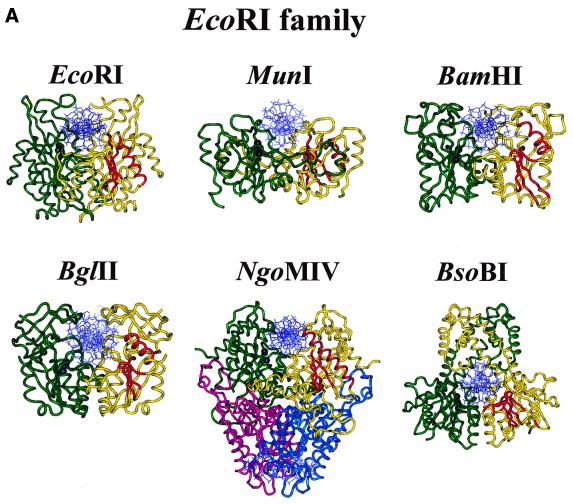
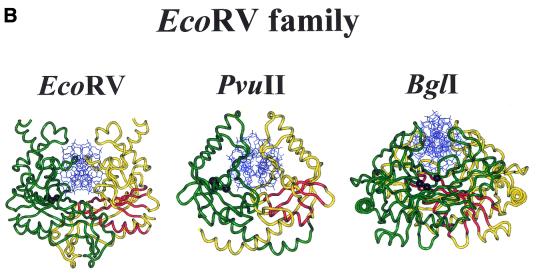
With the determination of more crystal structures it became clear that all restriction endonuclease structures so far known (Fig. 1) have a very similar core (34), including orthodox restriction enzymes producing sticky ends with a 5′-overhang (BamHI, BglII, EcoRI, MunI, BsoBI), sticky ends with a 3′-overhang (BglI) or blunt ends (EcoRV, PvuII), as well as members of the type IIS (FokI), type IIE (NaeI) and type IIF (Cfr10I, NgoMIV) subdivisions (Fig. 1). This core consists of a five-stranded mixed β-sheet flanked by α-helices, as first recognized by a comparison of the structures of EcoRI and EcoRV (35). Intriguingly, this core is also present in four other proteins with an endonuclease function, namely λ-exonuclease (34,36), MutH (37) which is involved in methyl-directed mismatch repair, Vsr endonuclease (38) which is involved in the repair of TG mismatches, and TnsA (39), one of two subunits of the Tn7 transposase. The conserved core harbors the catalytic center: it brings into spatial proximity two carboxylates, typically one aspartate and one glutamate or aspartate residue, and one lysine residue.
Figure 1.
Crystal structures of specific restriction endonuclease–DNA complexes. The two subunits of the homodimeric restriction endonucleases are shown in yellow and green (for the homotetrameric NgoMIV the individual subunits are colored yellow and green, purple and cyan), the DNA is shown in blue. In one subunit the four strictly conserved β-strands (EcoRI: β1, β2, β3 and β4; EcoRV: βc, βd, βe and βg) and one α-helix (EcoRI: α2; EcoRV: αB) of the common core are shown in red, in the other subunit the Cα-positions of the three essential amino acid residues of the PD . . . D/EXK motif are depicted as black spheres. For PDB codes see Table 2.
The structural similarity of the type II restriction endonucleases suggests that they indeed have a common, although distant, ancestor. On the basis of a comparison of protein structures a phylogeny of the restriction endonuclease superfamily was proposed (40), with two main branches, one comprising BglI, EcoRV and PvuII (as well as MutH and λ-exonuclease), the other BamHI, Cfr10I, EcoRI and FokI. The distinction between an EcoRI-like family and EcoRV-like family had been made before and not only associated with similarities in structure but also with similarities in function: EcoRI, like BamHI, binds the DNA from the major groove side and produces sticky ends with 5′-overhangs, whereas EcoRV, like PvuII, approaches the DNA from the minor groove side and produces blunt ends. This has consequences for the positioning of the two active sites and, therefore, for the arrangement of the two subunits in the homodimer. Thus, the nature of the cleavage pattern rather than the DNA sequence recognized, appears to be the most important constraint on the mode of dimerization of restriction endonucleases (41).
Within the common core, characteristic for type II restriction endonucleases, only four β-strands are absolutely conserved, two of these strands (β2 and β3 in EcoRI; βd and βe in EcoRV) contain the amino acid residues directly involved in catalysis, the remaining ones may be critical for formation of the β-sheet and the hydrophobic core. The other secondary structure elements of the common core could have been altered during divergent evolution (42). In this context, it was also observed that the EcoRI and EcoRV families differ in the orientation of a β-strand (β5 in EcoRI; βh in EcoRV), as noted before based on a smaller data set (43).
Several restriction enzymes function as homotetramers. The crystal structures of two of them, Cfr10I and NgoMIV, the latter as an enzyme–product complex, were determined (44,45). As expected from their function as type IIF enzymes, they can be considered as dimers of dimers, with a back-to-back orientation, which puts the DNA binding sites of the primary dimers at opposite ends of the tetramer. In the NgoMIV tetramer, the dimers are rotated relative to each other by ∼60° around their 2-fold axis, in Cfr10I the angle is more like 90°. In both cases, dimer–dimer contacts are extensive, the total contact surface area between primary dimers being 3200 Å2 (NgoMIV) and 2300 Å2 (Cfr10I), respectively. As shown for Crf10I, tetramerization nevertheless can be easily disrupted by a single amino acid substitution at a strategic position in a loop at the tetramer interface (44). This argues for a continuous transition between type IIF enzymes and orthodox type II enzymes, some of which, e.g. EcoRI (46), also tend to be homotetramers at higher concentrations.
All restriction enzymes are composed of subdomains, one of which constitutes the common core with the catalytic center. The other subdomains, which are in part responsible for DNA binding and dimerization, are more diverse in structure than the catalytic core. Consider for example the related proteins EcoRV (47) and PvuII (48). Both have an N-terminal dimerization subdomain which in EcoRV is formed by a short α-helix, a two-stranded antiparallel β-sheet, followed by a long α-helix, while in PvuII it consists of a long α-helix connected via a loop to a shorter α-helix. In spite of the difference in size of EcoRV and PvuII, the dimerization interface is of similar size (2300 Å2). BglI, which belongs to the same family as EcoRV and PvuII, but recognizes an interrupted sequence (GCCNNNN↓NGGC) and cleaves the DNA to produce sticky ends with 3′-overhangs, has an usually large dimer interface (3100 Å2) in which one ‘side’ of each subunit is involved (49). In contrast, EcoRI (50) and BamHI (51) have a very similar dimerization module, two α-helices which in the dimer form a four-helix bundle. EcoRI in addition has a small two-stranded antiparallel β-sheet, which interacts with the symmetry related β-sheet of the other subunit. Altogether, BamHI has a considerably smaller subunit interface than EcoRI (800 versus 2600 Å2). BsoBI, which is closely related to BamHI and EcoRI (as well as the tetrameric Crf10I) but with a molecular mass of 36.7 kDa per subunit the largest one of the three, has a large all helical subdomain fused to the cleavage domain. This subdomain is closely associated with the symmetry related other subdomain: between them 3500 Å2 of surface area is buried, whereas between the pair of catalytic subdomains only 1000 Å2 is buried. With 4800 Å2 BsoBI has the largest subunit–subunit interface among the dimeric restriction enzymes whose structure is known so far (52). It must be emphasized that the principal functions of restriction enzymes, namely dimerization, DNA binding and DNA cleavage, are interwoven, which means that regions involved in one function are often also of importance for another function (see also Fig. 4).
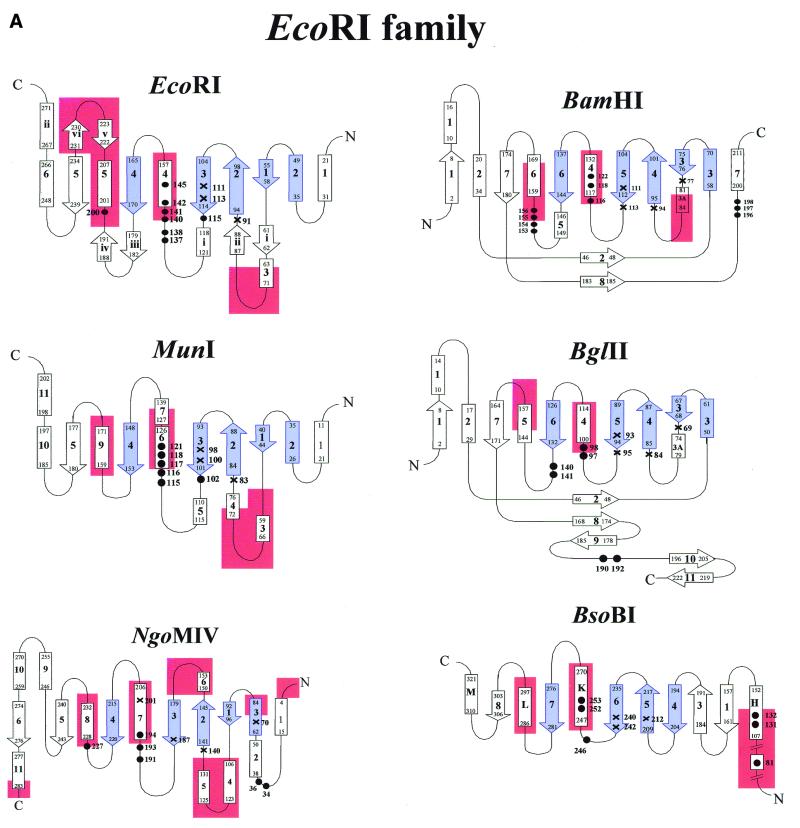
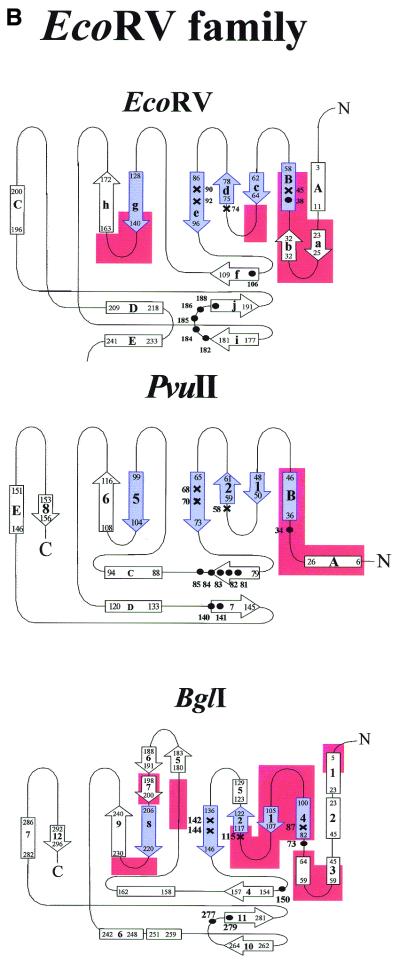
Figure 4.
(Opposite and above) The topologies of selected type II restriction endonucleases to illustrate similarities of architecture and to identify functionally important regions. Shown are the secondary structure diagrams for those restriction enzymes for which crystal structures of specific enzyme–DNA complexes have been determined (see Fig. 1). The elements comprising the common core are indicated in blue. Catalytically important amino acid residues are marked with a cross and those involved in contacts to the bases are marked with black filled circles. Regions involved in dimerization contacts are colored red. The numbering scheme is that of the respective crystallographers. The large helical N-terminal domain of BsoBI is not included in the diagram.
The type IIS restriction endonuclease FokI has a two-domain structure (53), a recognition domain comprising three smaller subdomains which are structurally related to the helix–turn–helix motif containing DNA binding domain of the catabolite gene activator protein, and a cleavage domain which is similar to a BamHI monomer. In the crystal, FokI is a dimer (54), in which dimerization is mediated by the cleavage domain. The total surface area buried in the dimer interface is unusually small (800 Å2) which may explain why FokI is a monomer in solution. Dimerization is required for DNA cleavage: presumably, a FokI monomer binds DNA at its recognition site and then recruits a second FokI monomer bound to another recognition site to form a dimer which catalyzes cleavage at the first site (17,54).
Another restriction endonuclease with a two-domain structure is the homodimeric type IIE enzyme NaeI (42). One domain (‘Endo’ domain) is structurally very similar to other type II restriction endonucleases and is responsible for substrate binding and cleavage as well as for dimerization, the other domain (‘Topo’ domain) contains a helix–turn–helix motif, similar to the catabolite gene activator protein, and presumably harbors the effector DNA binding site of NaeI (42). It is likely that this domain is also responsible for the topoisomerase activity of the L43K variant of NaeI (55).
The unusually large amino acid sequence of some type II restriction endonucleases suggest that they are composed of more than one domain. EcoRII, for example, is a homodimer with a subunit molecular mass of 45.6 kDa (56). Its enzymatic activity depends on the simultaneous binding of two copies of the recognition sequence (57), which means that it must have two DNA binding sites: indeed, it was shown recently that EcoRII like NaeI (58) induces loops in DNA containing two recognition sites (59). This could be interpreted to mean that EcoRII has a similar structural organization to NaeI, with one active site and one allosteric site (60) or, although less likely, two tightly coupled active sites as normally observed with type IIF enzymes (61,62), which are homotetramers. Another example of a large restriction endonuclease is Sau3AI, which is a monomer with a molecular mass of 56.5 kDa (63). Biochemical experiments demonstrate that it dimerizes on the DNA and like a type IIE or F enzyme, requires two recognition sites for efficient DNA cleavage (193). A remote sequence similarity between the N- and C-terminal halves of Sau3AI suggests that Sau3AI is a pseudodimer which dimerizes in the presence of DNA and thus could be considered to be a pseudotetramer in its active form. The gene for HgiDII codes for a protein of 68 kDa (64); inspection of the sequence revealed that it contains in its N-terminal half all consensus elements typically found in the GHKL family of ATPases, the significance of this observation being unclear (P.Friedhoff, personal communication).
THE INTERACTION OF RESTRICTION ENDONUCLEASES WITH DNA
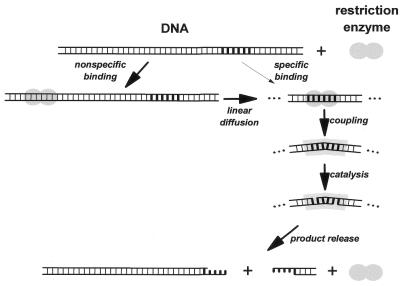
Restriction endonucleases interact with DNA in a complex manner. Because of the large size of a normal DNA substrate the reaction of a restriction enzyme with DNA cannot be simply formulated as a sequence of two or three steps. Figure 2 presents a minimal scheme for the individual steps involved in DNA cleavage by a type II restriction endonuclease. The reaction cycle starts with non-specific binding to the macromolecular DNA, which is followed by a random diffusional walk of the restriction endonuclease on the DNA. If a recognition site is not too far away from the initial site of contact it will most likely be located within one binding event. At the recognition site, conformational changes take place that constitute the recognition process and lead to the activation of the catalytic centers. After phosphodiester bond cleavage in both strands the product is released, either by direct dissociation of the enzyme–product complex or by a transfer of the enzyme to non-specific sites on the same DNA molecule. Often this step is rate limiting for DNA cleavage by restriction enzymes under multiple turnover conditions. In the following sections we will deal with the individual steps of this reaction cycle.
Figure 2.
Schematic illustration of the steps involved in DNA binding and cleavage by type II restriction endonucleases.
DNA BINDING AND TARGET SITE LOCATION
All restriction endonucleases bind DNA not only specifically but also, with considerably weaker affinity, non-specifically, similar to other proteins that recognize a specific DNA sequence (65). Upon non-specific complex formation, counterions and water molecules are released from the protein–DNA interface (66), which because of the associated favorable entropy changes balances the unfavorable loss of translational and rotational entropies of the protein and DNA upon complex formation. Protein–phosphate contacts on the other hand will lead to positive enthalpy changes. For EcoRI (67) and EcoRV (68) it has been shown by analyzing osmotic pressure effects on DNA binding that non-specific complex formation is accompanied by a release of 70–80 water molecules.
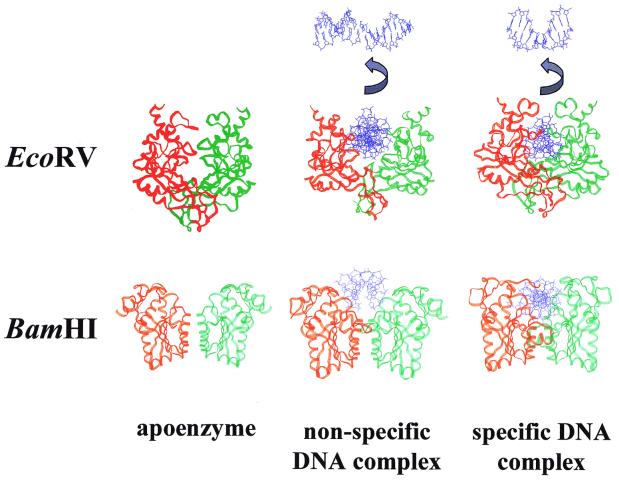
It is likely that upon non-specific DNA binding conformational changes occur, mainly in the protein, which will lead to an adaptation of the surface of the two macromolecules as is apparent from structural studies on EcoRV (47) and BamHI (69). Figure 3 shows the structures of EcoRV and BamHI together with the structures of their non-specific and specific DNA complexes. For EcoRV it is obvious that the enzyme has to open its DNA-binding site, which requires a conformational transition that presumably is triggered by a transient contact between the outer sides of the C-terminal arms of EcoRV and the DNA (70) and that allows the DNA (non-specific or specific) to enter the binding cleft. A similar mechanism of DNA binding has been discovered recently for the T7 helicase–primase protein (71). A region at the floor of the DNA binding site of EcoRV (the Q-loop), which is disordered in the free enzyme, becomes ordered in the non-specific complex. The stable non-specific complex differs from the specific complex by being less compact and having a much smaller protein–DNA contact surface: 1370 versus 2173 Å (47). No base contacts are seen in the non-specific complex; DNA backbone contacts are fewer in numbers and differ substantially from those observed for the specific complex. For BamHI, it seems as if major conformational transitions are not required to allow access of the DNA to the DNA binding cleft. Nevertheless, DNA binding is accompanied by an induced fit, as a large segment at the floor of the DNA binding site (residues 79–92), which is disordered in the free enzyme, becomes ordered in the non-specific complex. It is intriguing to note that no base-specific contacts and no direct DNA backbone contacts are seen in the non-specific complex, only a few water-mediated contacts even though the non-specific DNA used in the co-crystallization experiment differed only in one base pair from the specific DNA sequence (69). As observed with EcoRV, the non-specific BamHI–DNA complex is more open and less compact than the specific complex.
Figure 3.
Comparison of the crystal structures of the free enzyme, the non-specific and the specific DNA complex for the restriction endonucleases EcoRV and BamHI. The two subunits are shown in red and green, the DNA in blue. For EcoRV, the DNA is also shown viewed along the dyad axis to demonstrate the absence of bending in the non-specific complex and the presence of bending in the specific complex. PDB code numbers are: EcoRV (1RVE), non-specific complex (2RVE), specific complex (4RVE); BamHI (1BAM), non-specific complex (1ESG), specific complex (1BHM). It is apparent that large conformational changes take place when the free enzymes bind to non-specific DNA and to specific DNA, respectively. In the case of EcoRV, these conformational changes include the DNA which is bent by 50° in the specific complex. It is noteworthy that in the specific complex the DNA is almost completely encircled which gives the specific complex a more compact appearance compared to the non-specific one.
Non-specific DNA binding is the prerequisite for one-dimensional diffusion of proteins along DNA (72). The structures of the non-specific complexes of EcoRV and BamHI ‘provide remarkable snapshots of enzymes poised for linear diffusion (rather than cleavage)’ (69), the enzymes being only loosely bound to the DNA and their catalytic centers at a safe distance from the phosphodiester backbone. One-dimensional diffusion is defined as translocation along a DNA molecule, which does not involve a true free state of the protein: it includes sliding (i.e. a helical movement due to tracking along a groove of the DNA), hopping (i.e. a movement more or less parallel to the DNA, during which the protein does not leave the ‘DNA domain’) as well as intersegment transfer (which requires two DNA binding sites on the protein) (72–74). It has been shown for EcoRI and EcoRV that sliding is the most important process in target site location (75,76). Leaving the target site after DNA cleavage might involve either sliding or hopping (77,78). The biological significance of linear diffusion is obvious. It can accelerate target site location, as shown for EcoRI (75,79,80), BamHI (80,81), HindIII (80), EcoRV (76,82,83) and BssHII (84), it can increase processivity as for example shown for EcoRI (85) or EcoRV (78) and it can accelerate the dissociation from the specific site after cleavage, as is the case for EcoRI (79). Under optimum conditions, restriction endonucleases can scan ∼106 bp in one binding event. As this scan is a random walk, the effective sliding distance is much shorter, ∼1000 bp, as shown for EcoRI and EcoRV (75,76,80). During linear diffusion, EcoRI follows the helical pitch of the DNA, does not overlook any recognition site on its route and pauses at sites that resemble the recognition site; proteins firmly bound to DNA or unusual DNA structures constitute ‘road blocks’ (75). The ionic milieu, in particular the Mg2+ concentration, has a strong influence on the effective sliding distance, as shown for EcoRI (85) and EcoRV (76). It must be emphasized that linear diffusion is not just a test tube curiosity but a process of importance in vivo (83,86), because the biological function of many enzymes acting on DNA requires fast target site location.
DNA RECOGNITION
Restriction endonucleases while linearly diffusing along the DNA must constantly scan the major groove, possibly also the minor groove, for recognition elements at the edges of the bases. Coming into contact with some idiosyncratic features of the DNA backbone and the bases, characteristic for the recognition sequence, triggers the highly cooperative conversion of a non-specific to a specific complex, which requires major conformational changes of both the protein and the DNA, as well as the expulsion of solvent molecules from the interface to allow for more intimate contacts. For EcoRI it was shown that altogether about 150 water molecules are released upon specific DNA binding (67), much more than upon non-specific binding (87). Interestingly, binding of BamHI to its cognate sequence is accompanied by the release of a somewhat smaller number of solvent molecules (88). Whereas non-specific DNA binding by EcoRI and BamHI has a ΔCp° ≈ 0 and is enthalpy driven, specific DNA binding by these enzyme has a ΔCp° < 0. Depending on temperature, specific binding is enthalpy or entropy driven (89).
The EcoRV system provides an excellent and so far unique example among type II restriction endonuclease of a major protein-induced conformational change of the DNA. In the specific complex the DNA is bent by ∼50° (compared with little if any bending in the non-specific complex), as determined both in the crystal (47) and in solution (90–92). This angle varies somewhat, depending on the crystal form, the particular oligodeoxynucleotide and EcoRV variant used for the co-crystallization (93) (see also Fig. 6). Bending of the DNA is largest at the central TA step, which leads to an unstacking of the bases, widening of the minor groove with a concomitant compression of the major groove, which most importantly brings the scissile phosphates deeper into the active site. It is interesting that the DNA bend is preserved in the product complex (94) as well as in a quasi-product complex in which the 5′-phosphate is missing at the site of cleavage (95), indicating that the continuity of the phosphodiester bond is not required for bending. In this context it is worth mentioning that chemically modified oligodeoxynucleotide substrates (in which G is replaced by inosine and C by 5-methyl cytosine) are bent to a similar extent as the corresponding unmodified oligodeoxynucleotide (96) and that in the crystal, bending is also observed in the absence of divalent cations (47). This means that bending is required but not sufficient for DNA recognition.
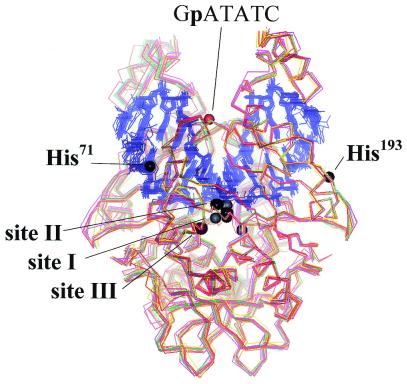
Figure 6.
Superposition of all solved structures of EcoRV and metal-ion binding site in EcoRV. The figure shows how similar these structures are although they are determined from crystals in different crystal lattices, using different substrates and metal ions. In addition all divalent metal ion binding sites as seen in different co-crystal structures and as deduced from biochemical experiments are indicated. The metal-ion binding sites near the active site are shown for both subunits, the additional sites (near His71, near His193 and near the GpATATC phosphate) are shown only for one subunit.
Another example of DNA bending in the specific complex, although not as pronounced as with EcoRV, is provided by the EcoRI (97) and MunI systems (98). For both enzymes, which recognize the same AATT core sequence in their hexanucleotide recognition sequence (G↓AATTC and C↓AATTG, respectively), a central kink is observed, accompanied by unwinding of the DNA. A similar but more localized unwinding and a similar overall bending but without a central kink of the DNA has been observed in the specific BglII–DNA complex (99). In contrast, BamHI, which recognizes the same GATC core sequence within its hexanucleotide recognition sequence as BglII (G↓GATCT and A↓GATCT, respectively), does not bend, kink or unwind the DNA significantly (100). Whereas no major DNA distortion is observed for PvuII (43) which like EcoRV is a blunt end cutter, BglI (49) which is a sticky end cutter leaving 3′-overhangs, bends the DNA by ∼20°, more or less smoothly without major kinks, the largest deviations from B-form DNA being seen in the two recognition half-sites of the interrupted GCCN4↓NGGC recognition sequence. Also, for the most recently reported structure of a specific restriction enzyme–DNA complex, BsoBI (52), no pronounced DNA bending is observed; however, slight deviations from canonical B-form exist: the DNA is extended and undertwisted, making the major and minor groove wider and more shallow.
Taken together, no generalization can be made for the kind and extent of distortion type II restriction enzymes induce in their DNA substrate. In general, however, the local helical parameters of the DNA in the specific complex differ from ideal B-DNA parameters (or where it is known from the helical parameters of the specific oligodeoxynucleotide used in the co-crystallization). It is important to note that distortions are an intimate part of the recognition process. In a few instances this has been experimentally verified by facilitating such a distortion using chemically modified substrates or substrate analogs, for example EcoRV (101), and demonstrating that they are bound more firmly than the natural (undistorted in the free state) substrate.
For EcoRV (47,93–95,102,103) and BamHI (69,100) the structural changes occurring in the protein during the transition from the non-specific complex to the specific complex are known from detailed crystallographic analyses (Fig. 3). In EcoRV correlated movements of the protein occur in concert with the binding and unwinding of the DNA (93). These movements are characterized by a translation and rotation of the long B-helices as well as a rotation of the DNA binding domains by ∼25°; they lead to an induced fit of the protein around the DNA substrate. Of particular importance is the ordering of the recognition loop, which appears to be largely unstructured in the non-specific complex and becomes structured only in the specific complex. It is responsible for making all base-specific contacts in the major groove and presumably is part of a communication network between the two identical subunits which have to act in concert to achieve double-strand cleavage in one binding event (104). The specific complex is more compact than the non-specific one, mainly because of the rotation of the DNA binding domains which brings these two domains closer together and allows them to encircle the DNA almost completely (Fig. 3).
The conformational changes that BamHI undergoes in the transition from non-specific to specific DNA binding are very different from those observed for EcoRV, in spite of the fact that in both cases the binding cleft is wider in the non-specific complex than in the specific complex and that the specific complex is more compact than the non-specific complex. The more compact structure of the specific BamHI–DNA complex is in part due to the fact that a segment of the protein is ‘pushed back’ into the protein core by the specific DNA, while the same segment is located in the binding cleft in the non-specific DNA complex. Whereas the non-specific BamHI–DNA complex (69) preserves the 2-fold symmetry of the free enzyme (51), the specific complex is characterized by a pronounced asymmetry (100), produced by the unfolding of the C-terminal α-helix in both subunits and the insertion of the unfolded polypeptide segment in one subunit only in an extended conformation into the minor groove of the DNA, while in the other subunit the unfolded polypeptide makes a side-by-side contact with the phosphodiester backbone (Fig. 3). Furthermore, whereas in the non-specific complex the DNA is only loosely bound within the cleft formed by the two subunits such that it protrudes out of the cleft (more so than with EcoRV), in the specific complex it is almost surrounded by the enzyme (like in EcoRV). Another remarkable difference between the specific and the non-specific complex concerns the orientation of the DNA relative to the two subunits of BamHI. In both complexes, the 2-fold axis of the dimeric protein coincides with the 2-fold axis of the DNA. However, compared with the non-specific complex, the enzyme is tilted about this axis by ∼20°, resulting in a different contact area at the periphery of the DNA binding site in the non-specific and the specific complex (69).
Given the fact that at present only in two systems, EcoRV and BamHI, can a comparison be made between the non-specific and the specific complex, only very general statements can be made regarding the structural changes accompanying the transition from non-specific to specific binding. Because of the similarities in function it is likely for all type II restriction endonucleases that the specific complex will be more compact than the non-specific one, in order to allow for a tighter contact between enzyme and substrate. Presumably, this will be achieved by a reorientation of the two subunits towards each other and the DNA, which will lead to a compaction of the DNA binding site and a more or less complete encircling of the DNA. The re-orientation can be substantial, as is apparent from the comparison of the structures of BglII in the free (105) and bound state (98): to bind DNA, the enzyme has to open by a ‘scissor-like’ motion of the subunits parallel to the DNA helix axis, which is accompanied by a complete rearrangement of the α-helices at the dimer interface. In contrast, in EcoRV and BamHI opening the binding cleft is achieved essentially by a motion of the subunits in a direction perpendicular to the helix axis.
There is an interesting difference between EcoRV on one side and BamHI as well as most of the other restriction endonucleases on the other side, including EcoRI. EcoRV requires the presence of Mg2+ (106) or Ca2+ (91) for specific binding. In the presence of EDTA, EcoRV in a gel electrophoretic mobility shift assay produces multiple bands, whose concentration-dependent distribution demonstrates that this enzyme binds all DNA sequences with similar affinity (82), a conclusion that was challenged (107), and then confirmed by binding studies with oligodeoxynucleotides in solution (108). In a more recent study, the preference of EcoRV for its cognate sequence in the absence of divalent cations was shown to be within a factor of 10 at neutral pH (109), in agreement with results obtained previously for wild-type EcoRV (110) and an EcoRV variant (111). Similar results were reported for PaeR7 (112), TaqI (113), Cfr9I (114), BcgI (115), MunI (116), Cfr10I (117) and BglI (118), which also require Ca2+ (as a substitute for Mg2+) for specific binding. For MunI it was shown that this requirement could be relaxed by protonation or substitution of the active site carboxylates (119), indicating that the divalent cation is required to decrease the electrostatic repulsion between the protein and the DNA at the active center. For EcoRV, additional Mg2+ binding sites outside the catalytic center are required for specific binding, as the substitution of the active site carboxylates does not alleviate the Mg2+-dependence of specific binding (111). We suggest, therefore, that for some restriction endonucleases Mg2+ (or other divalent cations) is involved in the recognition process, not only in the transition state, where its contribution is obvious, but also for preferential and strong (i.e. specific) binding of the recognition sequence. That restriction endonucleases have additional divalent metal ion binding sites already in the absence of DNA has been shown by metal ion mapping experiments for TaqI (120) and by crystallography for PvuII near Tyr94 (M.Kokkinidis, personal communication). This residue has been discussed previously as being involved in metal ion positioning on the basis of a PvuII mutant–DNA co-crystal structure (121). The Tyr94 site of PvuII is only seen with Mg2+ soaked into the crystals and not with Mn2+, which may explain why this site was not seen in the metal ion mapping experiments carried out with Fe2+ (122). In the presence of the DNA substrate more divalent metal ion binding sites may appear, as has been shown by crystallography and biochemical studies for EcoRV at position His71, His193 and a phosphodiester group within the recognition site (GpATATC), respectively (93,111) (F.Winkler, personal communication). Figure 6 gives a compilation of all metal ion binding sites observed in EcoRV so far illustrating that the interaction of a restriction enzyme with metal ions must be considered a very complicated issue. It is interesting to note that restriction enzymes that do not require divalent cations for specific DNA binding, like EcoRI, can be made dependent on divalent cations by introducing amino acid substitutions in critical positions. The EcoRI K130A or E and R131E variants behave like EcoRV in requiring Ca2+ for specific binding (123). This argues against a fundamental difference between enzymes that achieve specificity already at the binding step (in the absence of Mg2+) or only in the catalytic step (in the presence of Mg2+).
Since 1997, when we discussed the recognition process for EcoRI, EcoRV, BamHI and PvuII (for details see 10), six more co-crystal structures of specific restriction endonuclease–DNA complexes were determined: FokI (53), BglI (49), MunI (98), BglII (99), NgoMIV (45) and BsoBI (52).
FokI
FokI, a type IIS enzyme recognizes the asymmetric sequence GGATG and makes a staggered cut 9 and 13 nt, respectively, downstream of the recognition sequence, after dimerization on the DNA via its cleavage domain (17,54). FokI approaches the DNA from the major groove side and appears to surround the DNA. The recognition domain consists of three subdomains (D1, D2 and D3), which all contain a helix–turn–helix motif and are similar to the DNA binding domain of the catabolite gene activator protein. DNA recognition is based on two modules: subdomain D1, which covers the major groove at the 3′-end of the recognition sequence (GGATG), and subdomain D2, which contacts the 5′-end of the recognition sequence (GGATC). Subdomain D3 is not involved in protein–DNA but rather in protein–protein interactions. FokI, like all other restriction endonucleases, makes extensive interactions to all bases of the recognition sequence: almost all hydrogen-bond acceptors and donors at the edges of the bases in the major groove are involved in direct contacts with the protein.
BglI
BglI, an orthodox type II enzyme, recognizes the sequence GCCN5GGC and cleaves between the fourth and fifth unspecified nucleotide to produce 3′-overhanging ends. BglI approaches the DNA from the minor groove side (Fig. 1), similarly to EcoRV and PvuII with which it shares many structural features, in spite of the fact that the two subunits are arranged differently than in these two proteins in order to accommodate the unspecified sequence between the two recognition half-sites and to produce the different cleavage pattern (3′-overhangs versus blunt ends). Due to the long distance between both recognition half-sites, each subunit of BglI contacts only one half-site and cleaves close to it: there is no cross-over mode of recognition, as observed for most of the other type II restriction endonucleases and argued to be beneficial for concerted double-strand cleavage (10). This might be achieved in this case solely by the extensive hydrogen-bonding network that connects the catalytic centers of the two subunits. BglI makes base contacts predominantly in the major groove. The unspecified 5 bp between the two half-sites are contacted at the sugar–phosphate backbone. The base contacts in the major groove involve amino acid residues located on or near to a small three-stranded β-sheet (‘recognition sheet’), in a topologically similar location as observed for EcoRV and also PvuII (Fig. 4). Per recognition site there are 16 direct hydrogen bonds and two water-mediated ones, which saturates the hydrogen-bonding potential in the major groove. Moreover, there is one direct and several indirect, i.e. water-mediated, contacts to the bases from the minor groove side. In addition to these base contacts (direct readout), there are numerous backbone contacts (indirect readout); altogether 17 direct and 21 water-mediated hydrogen bonds per subunit to the DNA phosphates.
MunI
MunI recognizes the sequence C↓AATTG. The core sequence AATT as well as the cleavage pattern is the same as that for EcoRI. This and the identification of local sequence similarities, which concern structural elements of EcoRI involved in recognition and cleavage, led to the suggestion that MunI might employ a similar mechanism for DNA recognition and cleavage (124). The determination of the co-crystal structure of the specific MunI–DNA complex confirmed this proposition (Figs 1 and 4) and thereby provides the first example in which two restriction enzymes contact common parts of their recognition sequence by homologous structural elements. MunI, like EcoRI, approaches the DNA from the major groove side and distorts the DNA in a similar manner as EcoRI. MunI makes base contacts only in the major groove. There are altogether 16 hydrogen bonds to the edges of the bases and six van der Waals contacts per hexanucleotide recognition site. The outer GC base pair is contacted by Arg115, which has no counterpart in EcoRI. The AATT core sequence is recognized by amino acid residues located on one segment (Arg115 to Arg121), which in its topological location and function has a correlate in EcoRI, where it is responsible for the recognition of the same core sequence (AATT). In addition to base-specific contacts, numerous contacts exist between the sugar–phosphate backbone of the DNA and the protein, extending to two phosphate residues outside of the recognition sequence. These contacts come from several regions of the protein, which in part are also involved in base contacts. Thus, direct and indirect readout are interwoven. Some of these contacts are very similar to those observed previously in the EcoRI–DNA complex and they are considered to stabilize the distorted DNA conformation (50,125). Thus, not only are there common features in base recognition between EcoRI and MunI, but also in backbone recognition (see also 126). Deibert et al. (98) suggest that this finding may eventually be extended to ApoI, which recognizes and cleaves the sequence Pu↓AATTPy.
BglII
BglII recognizes and cleaves the sequence A↓GATCT, which closely resembles the recognition sequence of BamHI (G↓GATCC). The determination of the co-crystal structure (Fig. 1) of a specific BglII–DNA complex (99) allowed for a comparison of the strategies employed for recognition (see also 126). Although the enzymes have a similar core structure, there are remarkable differences in the way these two enzymes interact with their substrate. The most obvious difference regarding the mode of recognition is that in BglII the core structure is augmented by a β-sandwich subdomain that fully encircles the DNA and is responsible for the minor groove contacts as well as some of the backbone contacts. Different from the EcoRI/MunI systems, BamHI and BglII, which also share a common tetranucleotide in their respective hexanucleotide recognition sequences, interact with this tetranucleotide sequence differently and—with one exception (Asn140 and Ser141 in BglII correspond to Asp154 and Asp155 in BamHI, both recognizing the respective outer base pairs)—use different structural elements for recognition (Fig. 4). Although BglII (like BamHI) approaches the DNA from the major groove side, contacts are also made to the edges of the bases in the minor groove. Three loops are responsible for all base contacts: Asn140 and Ser141 (loop C) recognize via their side chain functions the first TA base pair and the C of the second CG base pair. The G of the second base pair is contacted by water-mediated bidentate hydrogen bonds from Asn98 (loop B). The T of the third TA base pair is recognized by Tyr190 of one subunit (loop D), and the A by Ser97 of the other subunit (loop B). There are in addition four more water-mediated hydrogen bonds between the minor groove face of the bases and Tyr190 and Arg192. Altogether there are 14 hydrogen bonds to the major groove and five hydrogen bonds to the minor groove. There is a pronounced intertwining of the recognition of the two strands/two halves of the recognition sequence on one side and the two subunits on the other side. Numerous interactions exist between the protein and the DNA; they extend by two phosphate residues beyond the recognition sequence. Altogether there are 28 backbone contacts, 20 of them are water-mediated.
BsoBI
BsoBI recognizes the degenerate sequence C↓PyCGPuG. A remarkable feature of the co-crystal structure of the specific BsoBI–DNA complex is the complete encirclement of the DNA by the protein to form a 20 Å long ‘tunnel’ (52). Approximately 3800 Å2 of the solvent accessible surface of the enzyme and the DNA are buried in the protein–DNA interface (Fig. 1 and Table 2). As expected from its mode of cleavage, BsoBI approaches the DNA from the major groove side. Each subunit interacts with each recognition half-site and makes base-specific contacts in the major and minor grooves. The outer and inner CG base pairs are involved in several hydrogen bonds to the protein. Of particular interest is how this enzyme manages to accept a CG or TA and GC or AT base pair in the second and fourth position of the recognition sequence. This is now understood in structural terms because this PyPu base pair is involved in only one direct hydrogen bond between Lys81 and the N7 of the purine (A in the co-crystal structure), and in one water-mediated bidentate hydrogen bond between Asp246 and N7 of the purine as well as the substituent in position 6 of the purine (N6 of A in the co-crystal structure). In addition to these 22 hydrogen bonds to the bases, several van der Waals contacts to the bases exist as well as 64 hydrogen bonds (24 water-mediated) to the backbone per site. The backbone contacts extend to two residues to the left and right of the recognition sequence.
Table 2. Overview of co-crystal structures determined for enzyme–substrate complexes of orthodox type II restriction endonucleases.
| |
EcoRI |
EcoRVa |
BamHI |
PvuIIa |
BglI |
MunI |
BglII |
NgoMIV |
BsoBI |
| Recognition sequence | G↓AATTC | GAT↓ATC | G↓GATCC | CAG↓CTG | GCCN4↓NGGC | C↓AATTG | A↓GATCT | G↓CCGGC | C↓PyCGPuG |
| PBD code | 1ERI | 1RVB | 1BHM | 3PVI | 1DMU | 1DO2 | 1DFM | 1FIU | – |
| Length of DNA (bp) | 13 | 11 | 12 | 13 | 17 | 10 | 16 | 11 | 12 |
| R-factor (%) | 17.0 | 16.5 | 18.9 | 20.5 | 17.7 | 18.0 | 19.2 | 17.4 | 19.0 |
| Resolution (Å) | 2.7 | 2.1 | 2.2 | 1.6 | 2.2 | 1.7 | 1.5 | 1.6 | 1.7 |
| Protein–DNA interface (Å2)b | 2200 | 2400 | 2000 | 1800 | 3300 | 1300 | 2800 | 2100 | 3800 |
| Dimerization interface (Å2)b | 2600 | 2300 | 800 | 2300 | 3800 | 2500 | 780 | 1000 (10 800c) | 2800 |
aWhere more than one co-crystal structure has been determined only the first high resolution structure is given.
bDetermined by the Connolly method (188) using a probe with a radius of 1.4 Å. Note that this method yields different results from other methods used in the literature. The dimerization interface is defined by: 2 × surface monomer – surface dimer. The protein–DNA interface is defined by: surface dimer + surface DNA – surface enzyme–DNA complex.
cTetramerization interface (A2). Tetramerization interface is defined by: 4 × surface monomer – surface tetramer.
NgoMIV
For NgoMIV only the structure of the enzyme–product complex has been determined (45). It is likely that many of the sequence-specific contacts required for the recognition of the substrate are preserved in the enzyme–product complex (as is the case in the EcoRV and BamHI systems). NgoMIV approaches the DNA from the major groove side and makes most of the base-specific contacts in the major groove of the target sequence recognized (G↓CCGGC). One subunit forms hydrogen bonds to the GCC half-site in the major groove, while the neighbor subunit forms a hydrogen bond to the C of the outer GC base pair in the minor groove. Base-specific contacts come from three structural elements, namely loops preceding α-helix 2, 7 and 8 (Fig. 4). It is noteworthy that three neighboring amino acids (Arg191, Asp193, Arg194) make all possible hydrogen bonds to the two adjacent GC base pairs in the major groove. Altogether there are 18 direct and two water-mediated hydrogen bonds to the bases of the NgoMIV recognition sequence. Interestingly, there is a hydrogen bond contact from Ser36 to the C on the 5′-side of the sequence, which may explain the flanking sequence preference of NgoMIV. In addition to the base-specific contacts, numerous contacts to the sugar–phosphate backbone exist, mainly from the other subunit, such that direct readout for which one subunit is responsible is interwoven with indirect readout for which the other subunit is responsible. Altogether, there are six direct and eight water-mediated contacts to the DNA–phosphates.
The region from Arg191–Arg194 (RSDR) has a structural equivalent in Cfr10I (RPDR), which recognizes a similar recognition sequence as NgoMIV (compare Pu↓CCGGPy with G↓CCGGC) and also has a Glu residue six residues away which is part of the catalytic center of NgoMIV (45). It is very likely that the recognition of the adjacent GG sequence is done by Crf10I using the equivalent residues as in NgoMIV. One possibly could extend this suggestion to other restriction endonucleases that recognize adjacent GC base pairs (Table 3). In the lack of structural data or a detailed mutational analysis this is speculative. For some of these enzymes, for example SsoII (V.Pingoud, personal communication), biochemical evidence exists that the RXXR motif plays an important role in DNA binding.
Table 3. Type II restriction endonucleases having recognition sequences with adjacent G residues and harboring a RXXR motif.
| Enzyme |
RXXR |
Recognition sequence |
| NgoMIV | K187X3RSDRX6E | G/CCGGC |
| Cfr10I | K190X3RPDRX6E | R/CCGGY |
| BsrFI | K198X3RPDRX6E | R/CCGGY |
| SgrAI | K242X3RSDRX6E | CR/CCGGYG |
| EcoRII | K324X3K-DRX6E | /CCWGG |
| PspGI | K160X3R-ERX6E | /CCWGG |
| SsoII | K182X3R-ERX6E | /CCNGG |
| AccI | K196X3K-DRX6E | T/CCGGA |
The increasing numbers of co-crystal structures available for specific restriction endonuclease–DNA complexes and complementary biochemical studies allows us to make generalizations regarding the mechanism of DNA recognition. (i) Enzymes that produce blunt ends or sticky ends with 3′-overhangs approach the DNA from the minor groove side, whereas enzymes that produce sticky ends with 5′-overhangs contact the DNA from the major groove side. (ii) DNA binding is accompanied by more or less pronounced distortions of the DNA and conformational adaptations of the enzyme, which in many cases lead to a partial encircling of the DNA by the protein. (iii) Specific DNA binding is accompanied by the release of counter ions and partial dehydration of the enzyme and the DNA at the protein–DNA interface. (iv) Enzymes that produce blunt ends or sticky ends with 3′-overhangs mainly use a β-strand and β-like turn for DNA recognition. In contrast, enzymes that produce sticky ends with 5′-overhangs mainly use an α-helix and a loop. (v) Recognition is achieved by direct and indirect readout, i.e. base contacts, and backbone contacts, respectively. Contacts to the bases are predominantly in the major groove and usually exhaust the hydrogen bonding potential in the major groove. This means that a hexanucleotide sequence is recognized by ∼20 hydrogen bonds to the bases of the recognition sequence. Interactions with the backbone are often water-mediated. (vi) Individual recognition modules (short sequence motifs) begin to show up that are used by different restriction endonucleases to recognize common parts in similar recognition sites.
COUPLING BETWEEN RECOGNITION AND CATALYSIS
Specific DNA binding by restriction endonucleases is defined as strong and, more importantly, preferential binding to the recognition site. Its outcome is what we usually see in the co-crystal structures or what we measure in binding experiments (including footprinting and crosslinking experiments). Specific binding does not necessarily mean recognition that is defined operationally, i.e. by the reaction that follows. By this definition the co-crystal structures of the specific restriction endonuclease–DNA complexes only mimic the recognition complex. In a similar argument, the results of binding experiments only address the mechanism of specific binding and not in the strictest sense the mechanism of recognition. Nevertheless, there is no doubt that the investigation of specific binding helps to understand recognition, presumably because the enzyme–substrate complex (studied in the absence of Mg2+ or in the presence of Ca2+) as well as the enzyme–product complex (studied in the presence of Mg2+, after turnover) is very similar to the ground state complex in the presence of Mg2+. In this context it is important to note that the discrimination between specific and non-specific sites requires multiple contacts to be formed between enzyme and substrate. In order to prevent these contacts, formed in the ground state complex, from impairing the catalytic efficiency (given by the difference in the Gibb’s free energy of the transition state complex and the ground state complex), it is necessary that these interactions must also stabilize the transition state (127). Therefore, the ground state complex is likely to resemble the transition state complex very much, differences being localized to the site of phosphodiester bond cleavage.
The coupling of specific binding, recognition and catalysis is ill understood. There have been many attempts to understand how the catalytic machinery is activated during the recognition process. Ideally one would like to ‘see’ this by time-resolved crystallography or NMR. This, however, has not yet been achieved. Instead, crystallographic studies of wild-type and mutant enzymes with canonical and chemically modified substrates, in the absence and presence of divalent metal ion cofactors, have been carried out and their results interpreted together with the results of single and multiple turnover cleavage studies. The best studied system in this respect is the EcoRV system, for which the structures of different enzyme–substrate complexes in different crystal lattices were determined and for which detailed biochemical data were obtained. Recognition can be formally divided into direct and indirect readout. In EcoRV the recognition (R)-loop, comprising residues 182–187, whose importance for recognition has been confirmed by a mutional analysis (128), makes all the base-specific contacts in the major groove. A hydrogen bond network links the R-loops to the scissile phosphates and the catalytic centers via Asn188 and Lys92 (94). Base recognition in the minor groove is accomplished by the glutamine (Q)-loop, comprising residues 68–70. Gln69 is in close proximity to one catalytic center and via Thr37 also to the other catalytic center (94); these two residues are very important for catalysis (128–132). Thr37 is also one of the key amino residues involved in indirect readout (94,96,130,131,133). The results of these investigations concerning coupling of recognition and catalysis assign a critical role to the symmetry related B-helices and Q-loops, which connect β-strands c and d. This region is located at the floor of the DNA binding site, vis à vis the phosphodiester bonds to be cleaved. It is known that this region adopts slightly different conformations in various co-crystal structures of EcoRV (93), which makes it likely that it has sufficient conformational freedom to be involved in activation of the catalytic centers. Residues whose position is affected by these conformational changes include Asp36 and Lys38, which also have been shown by mutational analyses to be essential for EcoRV (134,135).
A major aspect of the mechanism of activation of the catalytic centers of restriction enzymes concerns the positioning of the divalent metal ion cofactors and the water molecules, one of which in each catalytic center must take up a position in-line with the phosphodiester bond to be cleaved. For EcoRV it has been shown that Asp74 and Asp90, as well as the scissile phosphate and its 3′-neighbor, which all cooperate in Mg2+ and water binding at the catalytic centers, take up slightly different positions in different co-crystal structures of EcoRV (93). This is not unexpected for catalytically relevant residues (128,129,136) in a complex that is not active in the crystal. Of course, one would like to know how all amino acid residues in specific contact with the bases and the backbone communicate with the catalytic centers. The fact that this communication must be highly cooperative will make it very difficult to identify an intramolecular signal transduction pathway. It must be admitted, therefore, that at present, it is at best partially understood how the catalytic centers of EcoRV or any other restriction endonuclease are activated during the recognition process. Probably this is the main reason why all efforts to change or expand the specificity of restriction endonucleases by rational, i.e. structure-guided, design failed so far (137,138) or were not as successful as one had hoped (139).
Coupling of recognition to catalysis not only concerns intrasubunit but also intersubunit communication, as restriction endonucleases in general catalyze a concerted double-strand cut. This means that the information of ‘recognition’ must be passed on from one subunit to the other. As pointed out above, with few exceptions each subunit of a restriction enzyme makes contacts to both halves of the recognition sequence, which integrates the recognition process. This has been demonstrated directly for EcoRV, using artificial heterodimers (104,131,140) and for PvuII using a single chain variant (194): substitution of a residue involved in base-specific contacts in only one subunit affects cleavage in both strands, whereas substitution of a catalytic residue in one active center does not affect the other catalytic center and allows for cleavage in one strand (nicking).
THE MECHANISM OF CATALYSIS OF PHOSPHODIESTER BOND CLEAVAGE BY RESTRICTION ENDONUCLEASES
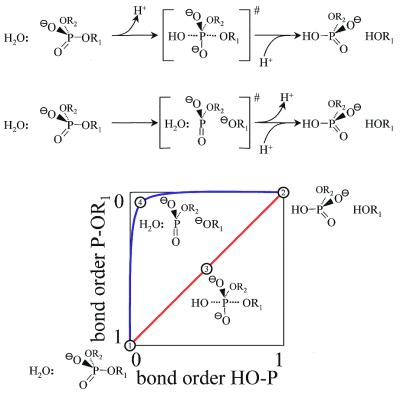
The catalysis of phosphodiester bond cleavage by restriction endonucleases can be considered as a phosphoryl transfer to water. For such a reaction two principal mechanisms may be operative, an associative and a dissociative mechanism (reviewed in 141). Both result in an inversion of configuration at phosphorus, as shown experimentally for EcoRI (142), EcoRV (143), HpaIII and SfiI (144). The associative mechanism (Fig. 5, top) requires a general base to generate the active nucleophile, a hydroxide ion, which attacks the phosphorus of the scissile phosphodiester bond, a Lewis acid that stabilizes the extra negative charge at the pentacoordinated phosphorus during the transition state, and a general acid that stabilizes the leaving group. This can be a Brønsted acid that protonates the leaving group (the fragment with a 3′-O–) or a metal ion that associates with the alcoholate. It is this mechanism that usually is explicitly or implicitly assumed to be operative in DNA cleavage by restriction enzymes. The dissociative mechanism (Fig. 5, middle) is not so much dependent on a general base to generate the active nucleophile, as there is only a small amount of bond formation to the incoming nucleophile, water, but a large amount of bond cleavage to the outgoing leaving group, the fragment with a 3′-O–. Therefore, in this mechanism stabilization of the leaving group is the most important aspect of catalysis. In addition, for this mechanism a metaphosphate-like species has to be stabilized in the transition state. This could be done by positively charged amino acid side chains, amino acid residues with hydrogen-bond donor functions (including protonated carboxylate side chains) or divalent metal ions (Mg2+), which all must preferentially interact with the trigonal bipyramidal transition state relative to the tetrahedral ground state (145).
Figure 5.
Principle mechanisms of phosphoryl transfer reactions. On top the mechanism of phosphodiester bond cleavage following an associative or a dissociative route is shown. These mechanisms differ in the amount of bond formation and bond breakage in the transition state. This is illustrated in the graph on the bottom in which the bond order of the bond to be cleaved is plotted against the bond order of the bond being formed. For an associative mechanism (red line) both bond orders change proportionally, whereas for a dissociative mechanism (blue line), the bond to the leaving group is largely cleaved while the bond to the attacking nucleophile is not yet formed. 1, Enzyme–substrate commplex; 2, enzyme–product complex; 3, transition state of the associative mechanism; 4, transition state of the dissociative mechanism.
It must be emphasized that the associative and dissociative mechanisms represent two extremes of possible mechanisms. The actual mechanisms used by a restriction endonuclease is likely not to follow any of these extremes (in Fig. 5, bottom, which depicts the change in bond order for the bond being made and the bond being broken, the reaction pathway may follow any line between the red line characteristic for an associative mechanism and the blue line characteristic for the dissociative nechanism). For alkaline phosphatase, for example, it has been shown that this enzyme can achieve substantial catalysis via a transition state with dissociative character (146).
All type II restriction endonucleases, whose crystal structures have been determined, have a catalytic sequence motif in common, the PD . . . D/EXK motif (41,128). As shown in Table 4 the consensus is somewhat relaxed which makes it hard in some cases to locate the catalytic sequence motif by an inspection of the sequence only. Nevertheless, it serves as a guideline to identify the presumptive active center of a type II restriction endonuclease, which then must be confirmed by a mutional analysis. Examples of such an identification include BsoBI (VD212 . . . E240LK) (147), Eco57I (PD78 . . . E92AK) (148), BcgI (PE53–E66DK) (24), EcoRII (PD77 . . . E96KR) (149) and HindIII (PE51 . . . D108AK) (150). It must be noted, however, that the identification of the catalytic sequence motif by a mutational analysis only is not fail-safe. An example is provided by the homing endonuclease I-PpoI for which a presumptive active site apparently was identified by a mutational analysis (PD109 . . . D140NK), which could not be confirmed by the crystal structure analysis (151,152).
Table 4. Catalytic sequence motifs in type II restriction endonucleases.
| Enzyme |
PD motif |
D/EXK motif |
Reference |
| EcoRI | PD91 | E111AK | 189 |
| EcoRV | PD74 | D90IK | 47 |
| BamHI | ID74 | E111FE | 100 |
| PvuII | ND58 | E68LK | 43,48 |
| Crf10Ia | PD134 | (E204) S188VK | 190,191 |
| FokI | PD450 | D467TK | 53 |
| BglI | PD116 | D142IK | 49 |
| MunI | PD83 | E98IK | 98 |
| NaeI | TD86 | D95CK | 42 |
| BglII | ID84 | E93VQ | 99 |
| NgoMIVa | PD140 | (E201) S185CK | 45 |
| BsoBI | VD212 | E240LK | 52 |
aThe crystal structure analysis has shown that the second acidic residue of the PD . . . D/EXK motif is recruited for the catalytic center from a distal part of the structure.
The PD . . . D/EXK motif has also been found in other nucleases as shown by crystal structure analyses, for example λ-exonuclease (PD119 . . . E129LK) (34), E.coli MutH (QD70 . . . E77LK) (37) and E.coli TnsA [D114 . . . (E149)Q130VK] (39) or tentatively assigned by mutational analyses, for example Sulfolobus solfataricus (PD42 . . . E55MK) (153) and Pyrococcus furiosus Holliday junction resolvase (VD36 . . . E46VK) (154), as well as E.coli McrBC (McrC: TD243 . . . D256AK; U.Pieper and A.Pingoud, submitted). In E.coli Vsr the PD . . . D/EXK motif is only partially present, although the endonuclease fold is conserved (38)
The fact that so many restriction endonucleases have a common catalytic sequence motif could suggest that they follow basically the same mechanism, with at least one obvious exception: BfiI, a type IIS restriction enzyme, that does not require a divalent metal ion cofactor, and in this respect differs from all type II restriction endonucleases known (155). This enzyme in its C-terminal part shows sequence homology with the Salmonella typhimurium NucA, a member of the HXKX4DX6GSXN superfamily of phosphodiesterases, whose crystal structure is known (156). It is likely that BfiI like FokI has a two-domain structure, one domain being responsible for DNA recognition and the other for cleavage. As NucA is a homodimer with one catalytic center which is formed by the two subunits, it might well be that BfiI has to form a tetramer to afford double-strand cleavage. More of these exceptions are likely to follow, as can be expected on the basis of recent sequence comparisons, which suggest that some type II restriction enzymes belong to the HNH- and GIY-YIG-families of endonucleases (157,158).
For the great majority of type II restriction endonucleases Mg2+ is an essential cofactor which can be substituted with Mn2+ (Fe2+, Co2+, Ni2+, Zn2+, Cd2+, depending on the enzyme) but not, however, by Ca2+. There is no doubt that Mg2+ is intimately associated with the catalytic process, as in nearly all cases where divalent cations were soaked into crystals of type II restriction endonucleases or into co-crystals of enzyme–DNA complexes, a divalent cation was found associated with the carboxylates of the catalytic center, with interesting variations: Mg2+, Mn2+, Ca2+, the divalent metal ions that are usually used in soaking experiments, do not necessarily occupy the same site. For example, a soaking experiment with PvuII and 50 mM Mg2+ had shown that Mg2+ is bound in one subunit to Asp58 and Glu68, i.e. at the catalytic center, while in the other subunit Mg2+ is bound in the immediate vicinity of Tyr94. This site seems to be of physiological importance, because when it is destroyed by a Tyr→Phe exchange, the DNA cleavage is affected and not concerted anymore (A.Spyridaki, C.Matzen, T.Lanio, A.Jeltsch, A.Simoncsits, A.Athanasiadis, E.Scheuring-Vanamee, M.Kokkinidis and A.Pingoud, submitted). If a soaking experiment is carried out with 50 mM Mn2+, a site close to Leu69 and Ser71 is occupied. In contrast, when co-crystals of a PvuII–DNA complex are grown at pH 4.5 in the presence of 50 mM Ca2+, then crosslinked with glutaraldehyde and then step-wise transferred to pH 6.5 conditions, co-crystals are obtained that contain two Ca2+ ions in each subunit associated with Asp58 and Glu68 (159). Table 5 gives a compilation of all published data concerning divalent metal binding sites in restriction endonuclease–DNA substrate complexes, which demonstrates the variety encountered among divalent metal ions in binding sites of different restriction endonucleases under different conditions.
Table 5. Divalent metal ion binding sites at the catalytic centers of specific restriction enzyme–DNA complexes.
| Protein |
Metal ion |
pH |
Subunit 1 |
Subunit 2 |
Reference |
| EcoRIa | 5 mM Mn2+ (soaking) | 7.0 | Asp90, Glu111 | Asp90, Glu111 | 166 |
| EcoRV | 30 mM Mg2+ (soaking) | 6.8 | Asp74, Asp90, Glu45, Asp74 | – | 94 |
| EcoRV | 30 mM Mn2+ (soaking) | 6.8 | Asp74, Asp90 | – | 94 |
| EcoRV | 30 mM Co2+ (soaking) | 6.8 | Asp74, Asp90, Glu45, Asp74 | Asp74, Asp90, Glu45, Asp74 | 94 |
| EcoRV | 30 mM Ca2+ (soaking) | 6.8 | Asp74, Asp90 | Asp74, Asp90 | 94 |
| EcoRV | 15 mM Mg2+ + 15 mM Ca2+ (soaking) | 6.8 | Asp74, Asp90 | Asp74, Asp90 | 94 |
| EcoRV | 1 mM Mn2+ + 20 mM Ca2+ (soaking) | 6.8 | Asp74, Asp90, Glu45, Asp74 | Asp74, Asp90, Glu45, Asp74 | 94 |
| EcoRV | 40 mM Ca2+ (co-crystallizition) | 6.8 | Asp74, Asp90 | Asp74, Asp90 | 102 |
| EcoRV T93A | 10 mM Ca2+ (co-crystallization) | 7.5 | Asp74, Ile91, Glu45, Asp74 | – | 103 |
| EcoRVb (GACITC) | 25 mM Mn2+ (co-crystallization) | 7.5 | – | – | 96 |
| EcoRVb,c (GAMITC) | 30 mM Mn2+ (co-crystallization) | 7.5 | Asp74, Asp90 | Asp74, Asp90 | 96 |
| EcoRVd (3′S) | 50 mM Mg2+ | 7.5 | Asp74, Ile91, Glu45, Asp74 | Asp74, Ile91 Glu45, Asp74 | 93 |
| EcoRVd (3′S) | 50 mM Mn2+ | 7.5 | Glu45, Asp74, Ile91, Glu45, Asp74, His71 | His71, His193 | 93 |
| EcoRVd (3′S) | 50 mM Ca2+ | 7.5 | Asp74, Ile91 Glu45, Asp74 | Asp74, Ile91, Glu45 | 93 |
| BamHIa | 10 mM Mn2+ (co-crystallization) | 5.3 | Asp94, Glu111, Phe112, Glu77, Asp94 | – | 178 |
| BglI | 125–150 mM Ca2+ (soaking) | 6.5 | Asp116, Asp142, Ile143, Asp116 | Asp116, Asp142, Ile143 Asp116 | 49 |
| MunI | 50 mM Ca2+ | 6.0 | – | – | 98 |
| BglII | Unidentified metal ion (co-crystallization) | 5.2 | Asp84, Val94 | Asp84, Val94 | 99 |
| PvuII | 50 mM Ca2+ (soaking) | 4.5–6.5 | Asp58, Glu68, Leu69, Asp58, Glu68 | Asp58, Glu68, Leu69, Asp58, Glu68 | 159 |
aActive in crystallo.
bI = inosin.
cM = 5-methyl cytosine.
dDeoxyribo-3′-S-phosphorothiolate at scissile phosphate.
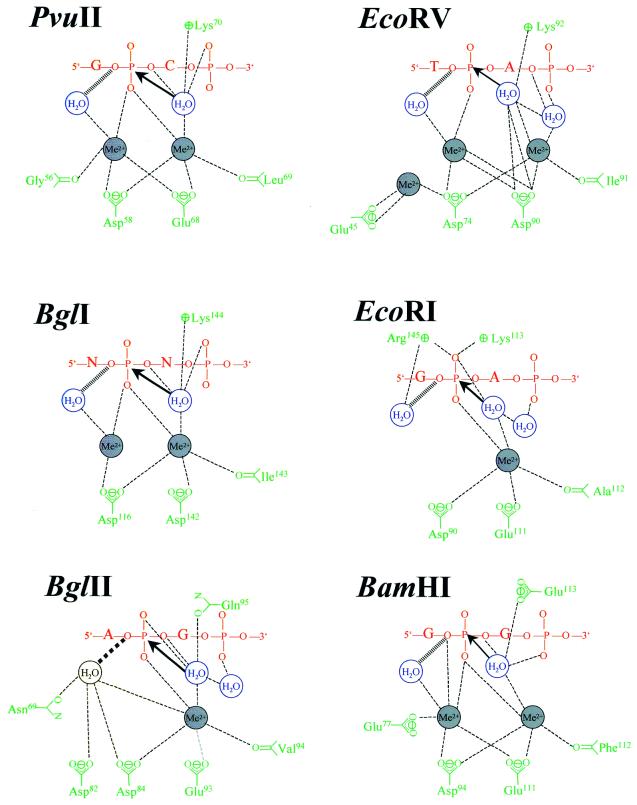
Table 5 contains 13 entries for EcoRV, which show that depending on the divalent metal ion, the substrate (or substrate analog) and the variant, different binding sites can be occupied, most of them comprising carboxylates from the catalytic centers. For EcoRV three sites are available for Ca2+, but only two are occupied simultaneously: site I comprises Asp74 (Asp90) and Ile91 (main chain carbonyl), site II Asp74 and Asp90, site III Glu45 and Asp74 (the roman numerals are used to compare these sites with analogous divalent metal binding sites in other restriction endonucleases, see also Fig. 7). It is noteworthy that in several instances only one binding site per subunit is occupied and/or that only one subunit has divalent metal ions bound, in spite of the fact that very high concentrations of divalent metal ions were used for the soaking, but also for the co-crystallization experiments. The fact that high concentrations are being used for these experiments is a point of concern, as they might lead to occupation of unphysiological binding sites (160,161). The occupation of such unphysiological sites may be the reason why only in two cases was activity demonstrated in crystallo (Table 5), an argument that is supported by biochemical data, which clearly demonstrate inhibition of DNA cleavage by EcoRV at higher concentrations of divalent cations (162). Mg2+ seems to have the same options as Ca2+ in occupying sites associated with the catalytic centers of EcoRV. Mn2+, however, can occupy additional sites outside of the catalytic centers (Fig. 6). Mn2+ binding to one of these sites modulates the catalytic efficiency and sequence selectivity of the enzyme (163). To further demonstrate the complexity of the situation, biochemical experiments suggest that there must be additional divalent metal ion binding sites in the EcoRV–DNA complex, not yet seen in the co-crystals, which are involved in recognition (111). Figure 6 shows a superposition of the divalent metal ion binding sites so far identified in EcoRV.
Figure 7.
Comparison of the active sites of restriction enzymes as deduced from co-crystal structures of specific protein–DNA substrate complexes obtained in the presence of divalent metal ions (based on 159). In EcoRV the sites formed by Asp90 and Ile91, Asp74 and Asp90 as well as Glu45 and Asp74 are numbered I, II and III, respectively, in this review. The same numbering scheme is applied to all other enzymes.
The situation seems to be not as complicated with the other restriction enzymes, possibly only because fewer structures are available. For two enzymes, EcoRI and BglII, only one metal ion per subunit (Mn2+ in the case of EcoRI, an unidentified metal ion or Ca2+, respectively, in the case of BglII) was seen in the co-crystal structures. It may be a coincidence, but these sites were found occupied at 5 mM Mn2+ and without added divalent metal ion, respectively (Table 5). The Ca2+ binding sites of BamHI, BglI and PvuII are homologous: in all three enzymes two binding sites per subunit are present, corresponding to binding sites I and II of EcoRV (Table 5). Figure 7 shows a comparison of the Ca2+ (Mn2+ in the case of EcoRI) binding sites observed in the co-crystals of BamHI, BglI, BglII, EcoRI, EcoRV and PvuII (based on ref. 159). This comparison reveals some intriguing similarities: all these restriction enzymes have a binding site I which is formed by the absolutely conserved first and second acidic amino acid residues of the PD . . . D/EXK motif and by a main chain carbonyl of the non-polar amino acid residue of this motif, PD . . . D/EXK. The divalent metal ion occupying binding site I is coordinated to a water molecule which is directly or indirectly (via a second water molecule) associated with the proRp-oxygen of the phosphate 3′ to the scissile phosphodiester bond. This water molecule is in a position for an in-line attack on the bond to be cleaved. It is held there by the last residue of the PD . . . D/EXK motif, usually a Lys, but in BamHI a Glu and in BglII a Gln residue, which all (Glu should be protonated for that purpose) are capable of forming a hydrogen bond to the attacking water molecule. The divalent metal ion in binding site II which is formed by the first and in some cases also by the second acidic amino acid residue of the PD . . . D/EXK motif, could serve to polarize the P–O bond and thereby to make the phosphorus more susceptible for a nucleophilic attack. It could also help to coordinate a water molecule, which could be used to protonate the leaving group, the 3′-O– or directly associate with this group. For EcoRV, the direct association has been ruled out (164).
The elucidation of the mechanism (or mechanisms) of DNA cleavage by type II restriction enzymes is critically dependent on the knowledge of how many Mg2+ ions are directly involved in catalysis. Crystal structure analyses are certainly very helpful to answer this question, but they can be also very misleading, because they do not tell us whether the configuration seen in the structure is productive. For EcoRV at least three different mechanisms were proposed, all based on structural information but differing in the numbers and sites of Mg2+ ions involved (‘one, two or three metals’) (36). The first mechanism was based on the comparison of the co-crystal structures of EcoRI and EcoRV as well as on molecular modeling (136,165). Based on the finding that a water molecule modeled into the structures at the position of the attacking nucleophile forms a hydrogen bond to the phosphodiester group 3′ to the scissile phosphodiester bond, it has been suggested that the phosphate 3′ to the scissile phosphodiester bond serves as the general base. Leaving group protonation would be afforded by a water molecule from the hydration sphere of a Mg2+ ion bound to Asp74 and Asp90 (site I/II). This aspect of the mechanism has become commonly accepted because, in all structures determined so far, a water in the hydration sphere of a metal ion is hydrogen bonded to the leaving group making it an obvious candidate for leaving group protonation. The characteristic feature of this mechanism is the substrate assisted catalysis, i.e. a direct role of the DNA substrate itself in the catalytic mechanism. A variant of this mechanism has been proposed for EcoRI recently (166). Although recruiting a phosphodiester group as the catalytic base might be considered surprising, this function is not uncommon. It has been proposed for other enzymes involved in nucleotidyl- or phosphoryl transfer: p21ras and other G-proteins (167), aminoacyl-tRNA synthetases (168,169) and acylphosphatase (170). Recently, it has been shown that the catalytic mechanism of peptidyl transfer in the prokaryotic ribosome also might involve a phosphodiester group of the rRNA as final proton acceptor in a relay that starts at the N3 of an adenine residue and involves also the O6 and N2 of a guanine residue (171,172). An important point in favor of the 3′-phosphate being the general base is the finding that in several high resolution structures of specific restriction enzyme–DNA complex a water molecule is seen hydrogen bonded to the 3′-phosphate and in a position ready for an in-line attack on the scissile phosphodiester bond.
Two alternative mechanisms for DNA cleavage by EcoRV were based on the Kostrewa and Winkler structure (94). They consider two Mg2+ ions as essential, but differ in an important detail, the identification of the general base. Kostrewa and Winkler (94) suggested that a water molecule in the hydration sphere of Mg2+ ion bound to Asp74 and Asp90 (site II) provides the attacking nucleophile and a water molecule in the hydration sphere of Mg2+ ion bound to Glu45 and Asp74 (site III) serves to protonate the leaving group. Vipond et al. (173) in contrast proposed that Mg2+ in site III functions as the general base. Later, Halford and coworkers (132,174) modified details of the stereochemistry of their mechanism to take into account the results of recent kinetic data for the Co2+ supported DNA cleavage by EcoRV and the results of a molecular dynamics simulation. In an accompanying study (135) it was suggested that the water molecule in the hydration sphere of Mg2+ in site III is deprotonated with the help of Glu45 (of one subunit) and Asp36 (of the other subunit). The most recent addition to the list of mechanisms proposed for DNA cleavage by EcoRV has been provided by Perona and coworkers (93,103). They had, as mentioned above, identified a new divalent metal binding site (site I) and intergrated this site into a three-metal ion mechanism, which they called a metal ion-mediated substrate-assisted catalysis mechanism to account for the fact that the metal ion in site I is coordinated to two water molecules, one that bridges to the proSp-oxygen of the 3′-phosphate, and the other attacks the scissile phosphate. This metal ion is bound to Asp74, Asp90 and Ile91 [site I; note that Horton et al. (103) use a different numbering system]. A second metal ion is bound to Asp74 and Asp90 (site II) as well as to the scissile phosphate; in its coordination sphere there is a water molecule that is close to the 3′-O of the leaving group. A third divalent metal ion, bound by Glu45 and Asp74 (site III), is not seen in any structure together with the divalent metal ions occupying site I and II, but in principle could take up this position. It is proposed to have a structural role, which is in agreement with the results of biochemical studies (162).
We have described the various proposals for the mechanism of DNA cleavage by EcoRV in detail, to make clear that even with detailed structural information it is not straightforward to derive the mechanism of an enzymatic reaction. A critical issue in the one-metal ion substrate-assisted catalysis mechanism and also, albeit to a lesser degree, in the two- and three-metal ion-mechanisms, is the pKa of the general base. The pKa of a phosphodiester (pKa < 2) or of a Mg2+-aquo complex (pKa ≈ 11–12) clearly is far off to explain the pH-optimum of the EcoRV-catalyzed reaction (pH ≈ 8.5) (135,175). There are four possible ways out of this dilemma: (i) an efficient general base is not needed, if the transition state has dissociative character; (ii) the attacking nucleophile is not generated in situ but comes from bulk solvent and is only stabilized in the catalytic center, as also suggested for DNA polymerase I (176); (iii) amino acid residues close to the active center provide assistance in lowering the pKa of the Mg2+-aquo complex, like Asp36 (135) [for a different view (134)]; and (iv) the pH profile of the cleavage reaction reflects a deprotonation of a residue (or of residues) which are not part of the catalytic center. In this context it is worthwhile to note that restriction enzymes are among the best regulated enzymes as they have to ensure that the catalytic centers are only activated if the correct sequence is bound. Any residue important in DNA recognition and coupling of recognition and catalysis could, therefore, dominate the apparent pH-dependence. As noted by Horton et al. (93), ‘establishing the structural basis for DNA cleavage beyond reasonable doubt requires visualization of essential catalytic elements within the confines of a single high-resolution crystal structure of a ternary complex. Clearly this elusive goal is yet to be attained’. Time-resolved spectroscopy using NMR (177) and EPR should be particularly suited to clarify how many divalent metal ions are required for catalysis and to elucidate their particular roles.
For EcoRV (94), as for BamHI (178) and NgoMIV (45), the structure of an enzyme–product complex in the presence of Mg2+ or Mn2+ were determined. In the EcoRV–product complex structure (obtained by co-crystallization) two Mg2+ ions were seen in both subunits, associated with the 5′-phosphate of one product as well as liganded to Glu45 and Asp74 (Mg2+ no. 1) and Gln69 (Mg2+ no. 2), respectively. In the BamHI–product complex structure (obtained by soaking), one Mn2+ ion is associated with the cleaved phosphate, Asp94 and Phe112, while the other is coordinated to Asp94, but not anymore to Glu111. In the NgoMIV–product complex structure (obtained by co-crystallization) two Mg2+ ions are seen in each active site, both coordinated by the cleaved phosphate, Asp140 and an acetate ion, one Mg2+ ion in addition by Cys186 (main chain carbonyl). The presence of two divalent metal ions at the catalytic centers of restriction endonuclease–product complexes should not be taken as evidence for their participation in catalysis, but rather as a reflection of the need to have a balance of charges. The acetate ion seen in the NgoMIV–product structure associated with both Mg2+ ions illustrates this requirement.
Two co-crystal structures of specific restriction enzyme–DNA complexes were determined without divalent metal ions: MunI (98) and BsoBI (52). The pronounced similarity between MunI and EcoRI suggests that MunI follows a similar mechanism as EcoRI. BsoBI, however, has particular features that merit a special discussion. This enzyme has a normal PD . . . D/EXK motif (Asp212, Glu240, Lys242) which means that it could follow a similar mechanism as EcoRI. BsoBI shares with other restriction endonucleases the position of the attacking water molecule which is located 3.7 Å away from the phosphorus atom of the scissile phosphodiester bond and in hydrogen bonding distance of the pro-Rp oxygen of the phosphate 3′ to the scissile phosphodiester bond. In addition, this water molecule is only 3.8 Å away from Lys243, which in turn contacts the pro-Sp oxygen of the phosphorus atom to be attacked. These features are similar in all other restriction enzymes (see Fig. 7). BsoBI, on the other hand, is the only restriction enzyme, which has an obvious general base close to the active center, His253 (from the other subunit), which may also couple recognition to catalysis, as it is involved in a hydrogen bond to N7 of the inner G (C↓PyCGPuG). It is not clear, however, how Mg2+ binding will affect these relationships. A two-metal ion mechanism is discussed by van der Woerd et al. (52), which is different from the other two-metal ion mechanisms discussed above, as the role of His253 has to be considered. One Mg2+ is suggested to bind to Asp212 and Glu240 as well as the pro-Sp oxygen of the scissile phosphodiester bond, a second Mg2+ to the proRp-oxygen and Glu252 of the other subunit.
The mechanism by which restriction endonucleases achieve DNA cleavage has not yet been proven for any restriction enzyme. For the advocates of the two-metal ion mechanism often the mechanism by which the 5′–3′ exonuclease domain of DNA polymerase I hydrolyzes DNA is taken as the paradigm (179), but in principle there the same uncertainty as to the participation of one- or two-metal ions in catalysis exists (160,161,180). A similar controversy as for EcoRV is going on for other nucleases. For example, for RNaseH two alternative mechanisms have been proposed. One is a two-metal ion mechanism (181), where one of the two metal ions activates the attacking nucleophile, and the other is a one-metal ion mechanism (182), where an amino acid residue is considered to be responsible for generating the hydroxide ion, faciliated by a neighboring phospodiester group of the substrate (183). Another example is provided by the transposases and integrases, which share a common catalytic triad, the DDE motif. Most of the structures reported for these enzymes show the binding of one Mn2+ ion at a site located between two of the three carboxylates, for example in the Tn5 transposase synaptic complex (184), but crystals for ASV integrase core soaked with Zn2+ or Cd2+ exhibit metal ion binding at two adjacent sites (185).
The BglII and BamHI restriction enzymes which cleave very similar sequences, A↓GATCT and G↓GATCC, have been discussed as representatives of type II restriction endonucleases which follow a one- and two-metal ion mechanism, respectively, and which have diverged from a common ancestor (186). The active site of BglII, on the other hand, has a similar make-up to the evolutionarily unrelated homing endonuclease I-CreI, which has one-metal ion bound per catalytic center (187). This has been considered as evidence for convergent evolution of these two enzymes. This conclusion certainly is valid concerning the amino acid residues involved in catalysis, but must be considered as tentative only concerning the number of metal ions involved.
It must not only be emphasized that at present the mechanism of DNA cleavage has not yet been elucidated for any restriction endonuclease, but also that it is not even known whether all type II restriction endonucleases with a bona fide PD . . . D/EXK motif follow the same mechanism. We tend to believe that this is the case, the argument being one of parsimony. Why else should this motif be conserved, in particular as it is possible to explain the deviations. For example, the substitution of the D/E by a S in Cfr10I and NgoMIV (the crystal structure shows that the E is supplied from another structural element, but takes up the required position in the three-dimensional structure). Likewise, the K is substituted by an E and Q in BamHI and BglII, respectively, but the hydrogen-bond donor function is preserved (when the E is protonated in BamHI). With these considerations in mind, the following features appear as invariant among all type II restriction endonucleases whose co-crystal structure has been determined. (i) The essential divalent metal ion cofactor is bound to the two carboxylates and the main chain carbonyl of the hydrophobic residue X of the PD . . . D/EXK motif as well as one of the non-bridging oxygens of the phosphate to be attacked. Whether a second (or third) divalent metal ion is required for cleavage is still a matter of debate. (ii) The phosphorus atom of the scissile phosphodiester bond is attacked in-line with the bond to be cleaved by a water molecule, held in position by the essential divalent metal ion, the K of the PD . . . D/EXK motif and one of the non-bridging oxygens of the phosphate 3′-adjacent to the phosphate to be attacked. (iii) Whether cleavage of the phosphodiester bond follows a more associative or a more dissociative mechanism is not known. Accordingly, the structure of the transition state is a matter of speculation. For its stabilization, the divalent metal ion cofactor, positively charged amino acid residues as well as amino acid residues with hydrogen bond donor functions are required. (iv) The function of the general base that has to abstract a proton from the attacking water (not very important if a dissociative mechanism is operative) could be taken over by the divalent metal ion, the K of the PD . . . D/EXK motif and/or the 3′-adjacent phosphate. All these chemical entities require a considerable shift in pKa (from their unperturbed value in the free state) to function as the general base. Whether additional amino acid residues are recruited for this purpose may differ from system to system. (v) Protonation of the leaving group or its transient stabilization, respectively, could be performed at the expense of a water molecule bound to the divalent metal ion or the divalent metal ion itself, respectively. (vi) Whatever the precise mechanism of phosphodiester bond cleavage by type II restriction endonucleases is, it involves acid–base catalysis. For this type of catalysis a proton relay has to be considered involving chemical entities not only at but also close to the site of cleavage.
CODA
Up to 1% of the genome of prokaryotic organisms is taken up by the genes for RM systems. Although it is not clear whether the majority of RM systems are required for the maintenance of the integrity of the genome or whether they are spreading as selfish genetic elements, they are key players in the ‘genomic metabolism’ of prokaryotic organisms. As such they deserve the attention of biologists in general.
Restriction enzymes (as well as their companion modification enzymes) constitute a huge family of enzymes with similar functions, the essential features of which are present also in other proteins interacting specifically with DNA, in particular proteins with nuclease function (recombinases, resolvases, transposases, integrases, repair enzymes). Elucidating the mechanism of action of restriction enzymes, therefore, is of interest also to those interested in the mechanism of DNA recognition and cleavage in general.
Finally, restriction enzymes are the work horses of molecular biology. Understanding their enzymology will be advantageous to those who use these enzymes, and essential for those who are devoted to the ambitious goal of changing the properties of these enzymes, and thereby make them even more useful.
Acknowledgments
ACKNOWLEDGEMENTS
We are obliged to all colleagues who have sent us reprints, preprints and manuscripts. Thanks are in particular due to Drs M. Deibert, V. Siksnys and A. Friedman for providing coordinates before their official release. We thank Drs R. Roberts, V. Siksnys, F. Winkler and X. Cheng for comments on the manuscript. We would also like to express our appreciation to Drs R. Roberts and D. Macelis for making available REBASE, an invaluable tool for those who work with restriction enzymes. We thank all our colleagues at Giessen, in particular Drs P. Friedhoff, T. Lanio, V. Pingoud and W. Wende, for numerous discussions and useful information, as well as Mrs K. Urbach for typing the manuscript. Work in the authors’ laboratory has been supported by grants from the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie, and the European Union.
References
- 1.Roberts R.J. and Halford,S.E. (1993) Type II restriction endonucleases. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, pp. 35–88.
- 2.Raleigh E.A. and Brooks,J.E. (1998) In De Bruijn,F.J., Lupski,J.R. and Weinstock,G.M. (eds), Bacterial Genomes. Chapman & Hall, New York, pp. 78–92.
- 3.Arber W. (1979) Promotion and limitation of genetic exchange. Science, 205, 361–365. [DOI] [PubMed] [Google Scholar]
- 4.Heitman J. (1993) On the origins, structures and functions of restriction-modification enzymes. In Setlow,J.K. (ed.), Genetic Engineering. Plenum Press, New York, Vol. 15, pp. 57–108. [DOI] [PubMed]
- 5.McKane M. and Milkman,R. (1995) Transduction, restriction and recombination patterns in Escherichia coli. Genetics, 139, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson K. and Kosturko,L.D. (1998) Endonuclease II of coliphage T4: a recombinase disguised as a restriction endonuclease? Mol. Microbiol., 27, 671–676. [DOI] [PubMed] [Google Scholar]
- 7.Bilcock D.T., Daniels,L.E., Bath,A.J. and Halford,S.E. (1999) Reactions of type II restriction endonucleases with 8-base pair recognition sites. J. Biol. Chem., 274, 36379–36386. [DOI] [PubMed] [Google Scholar]
- 8.Naito T., Kusano,K. and Kobayashi,I. (1995) Selfish behavior of restriction-modification systems. Science, 267, 897–899. [DOI] [PubMed] [Google Scholar]
- 9.Smith H.O. and Nathans,D. (1973) A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J. Mol. Biol., 81, 419–423. [DOI] [PubMed] [Google Scholar]
- 10.Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R.J. and Macelis,D. (1997) REBASE-restriction enzymes and methylases. Nucleic Acids Res., 25, 248–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts R.J. and Macelis,D. (2000) REBASE – restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedgpeth J., Goodman,H.M. and Boyer,H.W. (1972) DNA nucleotide sequence restricted by the RI endonuclease. Proc. Natl Acad. Sci. USA, 69, 3448–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schildkraut I., Banner,C.D.B., Rhodes,C.S. and Parekh,S. (1984) The cleavage site for the restriction endonuclease EcoRV is 5′-GAT↓ATC-3′. Gene, 27, 327–329. [DOI] [PubMed] [Google Scholar]
- 15.Van Heuverswyn H. and Fiers,W. (1980) Recognition sequence for the restriction endonuclease BglI from Bacillus globigii. Gene, 9, 195–203. [DOI] [PubMed] [Google Scholar]
- 16.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes – a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 17.Bitinaite J., Wah,D.A., Aggarwal,A.K. and Schildkraut,I. (1998) FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger D.H., Kupper,D., Meisel,A., Reuter,M. and Schroeder,C. (1995) The significance of distance and orientation of restriction endonuclease recognition sites in viral DNA genomes. FEMS Microbiol. Rev., 17, 177–184. [DOI] [PubMed] [Google Scholar]
- 19.Halford S.E., Bilcock,D.T., Stanford,N.P., Williams,S.A., Milsom,S.E., Gormley,N.A., Watson,M.A., Bath,A.J., Embleton,M.L., Gowers,D.M. et al. (1999) Restriction endonuclease reactions requiring two recognition sites. Biochem. Soc. Trans, 27, 696–699. [DOI] [PubMed] [Google Scholar]
- 20.Stankevicius K., Lubys,A., Timinskas,A., Vaitkevicius,D. and Janulaitis,A. (1998) Cloning and analysis of the four genes coding for Bpu10I restriction-modification enzymes. Nucleic Acids Res., 26, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh P.-C., Xiao,J.-P., O’Loane,D. and Xu,S.-Y. (2000) Cloning, expression and purification of a thermostable nonhomodimeric restriction enzyme, BslI. J. Bacteriol ., 182, 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong H. and Smith,C.L. (1997) Substrate DNA and cofactor regulate the activities of a multi-functional restriction-modification enzyme, BcgI. Nucleic Acids Res., 25, 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitkute J., Maneliene,Z., Petrusyte,M. and Janulaitis,A. (1997) BplI, a new BcgI-like restriction endonuclease, which recognizes a symmetric sequence. Nucleic Acids Res., 25, 4444–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong H. (1998) Analyzing the functional organization of a novel restriction modification system, the BcgI system. J. Mol. Biol., 279, 823–832. [DOI] [PubMed] [Google Scholar]
- 25.Janulaitis A., Vaisvila,R., Timinskas,A., Klimasauskas,S. and Butkus,V. (1992) Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res., 20, 6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacks S. and Greenberg,B. (1975) A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem., 250, 4060–4066. [PubMed] [Google Scholar]
- 27.Stewart F.J. and Raleigh,E.A. (1998) Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem., 379, 611–616. [PubMed] [Google Scholar]
- 28.Panne D., Raleigh,E.A. and Bickle,T.A. (1999) The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol., 290, 49–60. [DOI] [PubMed] [Google Scholar]
- 29.Krüger T., Wild,C. and Noyer-Weidner,M. (1995) McrB: a prokaryotic protein specifically recognizing DNA containing modified cytosine residues. EMBO J., 14, 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieper U., Schweitzer,T., Groll,D.H. and Pingoud,A. (1999) Defining the location and function of domains of McrB by deletion mutagenesis. Biol. Chem., 380, 1225–1230. [DOI] [PubMed] [Google Scholar]
- 31.Piekarowicz A., Golaszewska,M., Sunday,A.O., Siwinska,M. and Stein,D.C. (1999) The HaeIV restriction modification system of Haemophilus aegyptius is encoded by a single polypeptide. J. Mol. Biol., 293, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 32.Wilson G.G. and Murray,N.E. (1991) Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. [DOI] [PubMed] [Google Scholar]
- 33.Jeltsch A., Kröger,M. and Pingoud,A. (1995) Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene, 160, 7–16. [DOI] [PubMed] [Google Scholar]
- 34.Kovall R.A. and Matthews,B.W. (1998) Structural, functional and evolutionary relationships between lambda-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venclovas C., Timinskas,A. and Siksnys,V. (1994) Five-stranded β-sheet sandwiched with two α-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins, 20, 279–282. [DOI] [PubMed] [Google Scholar]
- 36.Kovall R.A. and Matthews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol., 3, 578–583. [DOI] [PubMed] [Google Scholar]
- 37.Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutakawa S.E., Jingami,H. and Morikawa,K. (1999) Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell, 99, 615–623. [DOI] [PubMed] [Google Scholar]
- 39.Hickman A.B., Li,Y., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- 40.Bujnicki J.M. (2000) Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol., 50, 39–44. [DOI] [PubMed] [Google Scholar]
- 41.Anderson J.E. (1993) Restriction endonucleases and modification methylases. Curr. Opin. Struct. Biol., 3, 24–30. [Google Scholar]
- 42.Huai Q., Colandene,J.D., Chen,Y., Luo,F., Zhao,Y., Topal,M.D. and Ke,H. (2000) Crystal structure of NaeI – an evolutionary bridge between DNA endonuclease and topoisomerase. EMBO J., 19, 3110–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X., Balendiran,K., Schildkraut,I. and Anderson,J.E. (1994) Structure of PvuII endonuclease with cognate DNA. EMBO J., 13, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siksnys V., Skirgaila,R., Sasnauskas,G., Urbanke,C., Cherny,D., Grazulis,S. and Huber,R. (1999) The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol., 291, 1105–1118. [DOI] [PubMed] [Google Scholar]
- 45.Deibert M., Grazulis,S., Sasnauskas,G., Siksnys,V. and Huber,R. (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol., 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 46.Modrich P. and Zabel,D. (1976) EcoRI endonuclease. Physical and catalytic properties of the homogeneous enzyme. J. Biol. Chem., 251, 5866–5874. [PubMed] [Google Scholar]
- 47.Winkler F.K., Banner,D.W., Oefner,C., Tsernoglou,D., Brown,R.S., Heathman,S.P., Bryan,R.K., Martin,P.D., Petratos,K. and Wilson,K.S. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Athanasiadis A., Vlassi,M., Kotsifaki,D., Tucker,P.A., Wilson,K.S. and Kokkinidis,M. (1994) Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nature Struct. Biol., 1, 469–475. [DOI] [PubMed] [Google Scholar]
- 49.Newman M., Lunnen,K., Wilson,G., Greci,J., Schildkraut,I. and Phillips,S.E.V. (1998) Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J., 17, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y., Grable,J.C., Love,R., Greene,P.J. and Rosenberg,J.M. (1990) Refinement of EcoRI endonuclease crystal structure: A revised protein chain tracing. Science, 249, 1307–1309. [DOI] [PubMed] [Google Scholar]
- 51.Newman M., Strzelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1994) Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature, 368, 660–664. [DOI] [PubMed] [Google Scholar]
- 52.van der Woerd M.J., Pelletier,J.J., Xu,S.-y. and Friedman,A.M. (2001) Restriction enzyme BsoBI-DNA complex: A tunnel for recognition of degenerate DNA sequences and potential histidine catalysis. Structure, 9, 133–144. [DOI] [PubMed] [Google Scholar]
- 53.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Structure of the multimodular endonuclease FokI bound to DNA. Nature, 388, 97–100. [DOI] [PubMed] [Google Scholar]
- 54.Wah D.A., Bitinaite,J., Schildkraut,I. and Aggarwal,A.K. (1998) Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jo K. and Topal,M.D. (1995) DNA topoisomerase and recombinase activities in NaeI restriction endonuclease. Science, 267, 1817–1820. [DOI] [PubMed] [Google Scholar]
- 56.Kossykh V., Repyk,A., Kaliman,A. and Buryanov,Y. (1989) Nucleotide sequence of the EcoRII restriction endonuclease gene. Biochim. Biophys. Acta, 1009, 290–292. [DOI] [PubMed] [Google Scholar]
- 57.Krüger D.H., Barcak,G.J., Reuter,M. and Smith,H.O. (1988) EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res., 16, 3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Topal M.D., Thresher,R.J., Conrad,M. and Griffith,J. (1991) NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry, 30, 2006–2010. [DOI] [PubMed] [Google Scholar]
- 59.Mücke M., Lurz,R., Mackeldanz,P., Behlke,J., Krüger,D.H. and Reuter,M. (2000) Imaging DNA loops induced by restriction endonuclease EcoRII. A single amino acid substitution uncouples target recognition from cooperative DNA interaction and cleavage. J. Biol. Chem., 275, 30631–30637. [DOI] [PubMed] [Google Scholar]
- 60.Colandene J.D. and Topal,M.D. (1998) The domain organization of NaeI endonuclease: Separation of binding and catalysis. Proc. Natl Acad. Sci. USA, 95, 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wentzell L.M., Nobbs,T.J. and Halford,S.E. (1995) The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol., 248, 581–595. [DOI] [PubMed] [Google Scholar]
- 62.Nobbs T.J., Szczelkun,M.D., Wentzell,L.M. and Halford,S.E. (1998) DNA excision by the SfiI restriction endonuclease. J. Mol. Biol., 281, 419–432. [DOI] [PubMed] [Google Scholar]
- 63.Seeber S., Kessler,C. and Gotz,F. (1990) Cloning, expression and characterization of the Sau3AI restriction and modification genes in Staphylococcus carnosus TM300. Gene, 94, 37–43. [DOI] [PubMed] [Google Scholar]
- 64.Düsterhoft A. and Kroger,M. (1991) Cloning, sequence and characterization of m5C-methyltransferase-encoding gene, hgiDIIM (GTCGAC), from Herpetosiphon giganteus strain Hpa2. Gene, 106, 87–92. [DOI] [PubMed] [Google Scholar]
- 65.von Hippel P.H. (1994) Protein–DNA recognition: new perspectives and underlying themes. Science, 263, 769–770. [DOI] [PubMed] [Google Scholar]
- 66.Parsegian V.A., Rand,R.P. and Rau,D.C. (1995) Macromolecules and water: probing with osmotic stress. Methods Enzymol., 259, 43–94. [DOI] [PubMed] [Google Scholar]
- 67.Robinson C.R. and Sligar,S.G. (1998) Changes in solvation during DNA binding and cleavage are critical to altered specificity of the EcoRI endonuclease. Proc. Natl Acad. Sci. USA, 95, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wenner J.R. and Bloomfield,V.A. (1999) Buffer effects on EcoRV kinetics as measured by fluorescent staining and digital imaging of plasmid cleavage. Anal. Biochem., 268, 201–212. [DOI] [PubMed] [Google Scholar]
- 69.Viadiu H. and Aggarwal,A.K. (2000) Structure of BamHI bound to nonspecific DNA: A model for DNA sliding. Mol. Cell, 5, 889–895. [DOI] [PubMed] [Google Scholar]
- 70.Schulze C., Jeltsch,A., Franke,I., Urbanke,C. and Pingoud,A. (1998) Crosslinking the EcoRV restriction endonuclease across the DNA-binding site reveals transient intermediates and conformational changes of the enzyme during DNA binding and catalytic turnover. EMBO J., 17, 6757–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahnert P., Picha,K.M. and Patel,S.S. (2000) A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J., 19, 3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Hippel P.H. and Berg,O.G. (1989) Facilitated target location in biological systems. J. Biol. Chem., 264, 675–678. [PubMed] [Google Scholar]
- 73.Berg O.G., Winter,R.B. and von Hippel,P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry, 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- 74.Shimamoto N. (1999) One-dimensional diffusion of proteins along DNA. Its biological and chemical significance revealed by single-molecule measurements. J. Biol. Chem., 274, 15293–15296. [DOI] [PubMed] [Google Scholar]
- 75.Jeltsch A., Alves,J., Wolfes,H., Maass,G. and Pingoud,A. (1994) Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry, 33, 10215–10219. [DOI] [PubMed] [Google Scholar]
- 76.Jeltsch A. and Pingoud,A. (1998) Kinetic characterization of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry, 37, 2160–2169. [DOI] [PubMed] [Google Scholar]
- 77.Terry B.J., Jack,W.E., Rubin,R.A. and Modrich,P. (1983) Thermodynamic parameters governing interaction of EcoRI endonuclease with specific and nonspecific DNA sequences. J. Biol. Chem., 258, 9820–9825. [PubMed] [Google Scholar]
- 78.Stanford N.P., Szczelkun,M.D., Marko,J.F. and Halford,S.E. (2000) One and three dimensional pathways for proteins to reach specific DNA sites. EMBO J., 19, 6546–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jack W.E., Terry,B.J. and Modrich,P. (1982) Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl Acad. Sci. USA, 79, 4010–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehbrecht H.-J., Pingoud,A., Urbanke,C., Maass,G. and Gualerzi,C. (1985) Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem., 260, 6160–6166. [PubMed] [Google Scholar]
- 81.Nardone G., George,J. and Chirikjian,J.G. (1986) Differences in the kinetic properties of BamHI endonuclease and methylase with linear DNA substrates. J. Biol. Chem., 261, 12128–12133. [PubMed] [Google Scholar]
- 82.Taylor J.D., Badcoe,I.G., Clarke,A.R. and Halford,S.E. (1991) EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry, 30, 8743–8753. [DOI] [PubMed] [Google Scholar]
- 83.Jeltsch A., Wenz,C., Stahl,F. and Pingoud,A. (1996) Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. EMBO J., 15, 5104–5111. [PMC free article] [PubMed] [Google Scholar]
- 84.Berkhout B. and van Wamel,J. (1996) Accurate scanning of the BssHII endonuclease in search for its DNA cleavage site. J. Biol. Chem., 271, 1837–1840. [DOI] [PubMed] [Google Scholar]
- 85.Terry B.J., Jack,W.E. and Modrich,P. (1985) Facilitated diffusion during catalysis by EcoRI endonuclease: nonspecific interactions in EcoRI catalysis. J. Biol. Chem., 260, 13130–13137. [PubMed] [Google Scholar]
- 86.Dowd D.R. and Lloyd,R.S. (1990) Biological significance of facilitated diffusion in protein–DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J. Biol. Chem., 265, 3424–3431. [PubMed] [Google Scholar]
- 87.Sidorova N.Y. and Rau,D.C. (1996) Differences in water release for the binding of EcoRI to specific and nonspecific DNA sequences. Proc. Natl Acad. Sci. USA, 93, 12272–12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lynch T.W. and Sligar,S.G. (2000) Macromolecular hydration changes associated with BamHI binding and catalysis. J. Biol. Chem., 275, 30561–30565. [DOI] [PubMed] [Google Scholar]
- 89.Jen-Jacobson L., Engler,L.E. and Jacobson,L.A. (2000) Structural and thermodynamic strategies for site-specific DNA binding proteins. Struct. Fold. Des., 8, 1015–1023. [DOI] [PubMed] [Google Scholar]
- 90.Stöver T., Köhler,E., Fagin,U., Wende,W., Wolfes,H. and Pingoud,A. (1993) Determination of the DNA bend angle induced by the restriction endonuclease EcoRV in the presence of Mg2+. J. Biol. Chem., 268, 8645–8650. [PubMed] [Google Scholar]
- 91.Vipond I.B. and Halford,S.E. (1995) Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry, 34, 1113–1119. [DOI] [PubMed] [Google Scholar]
- 92.Garcia R.A., Bustamante,C.J. and Reich,N.O. (1996) Sequence-specific recognition by cytosine C5 and adenine N6 DNA methyltransferases requires different deformations of DNA. Proc. Natl Acad. Sci. USA, 93, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horton N.C. and Perona,J.J. (2000) Crystallographic snapshots along a protein-induced DNA-bending pathway. Proc. Natl Acad. Sci. USA, 97, 5729–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kostrewa D. and Winkler,F.K. (1995) Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Angstrom resolution. Biochemistry, 34, 683–696. [DOI] [PubMed] [Google Scholar]
- 95.Horton N.C. and Perona,J.J. (1998) Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexes with d(AAAGAT) + d(ATCTT). J. Mol. Biol., 277, 779–787. [DOI] [PubMed] [Google Scholar]
- 96.Martin A.M., Sam,M.D., Reich,N.O. and Perona,J.J. (1999) Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nature Struct. Biol., 6, 269–277. [DOI] [PubMed] [Google Scholar]
- 97.Kim Y., Choi,J., Grable,J.C., Greene,P., Hager,P. and Rosenberg,J.M. (1994) In Sarma,R.H. and Sarma,M.H. (eds.), Structural Biology: The State of the Art. Adenine Press, New York, NY, pp. 225–246.
- 98.Deibert M., Grazulis,S., Janulaitis,A., Siksnys,V. and Huber,R. (1999) Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 Å resolution. EMBO J., 18, 5805–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution Nature Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 100.Newman M., Strzelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1995) Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science, 269, 656–663. [DOI] [PubMed] [Google Scholar]
- 101.Blättler M.O., Wenz,C., Pingoud,A. and Benner,S.A. (1998) Distorting duplex DNA by dimethylenesulfone substitution: a new class of ‘transition state analog’ inhibitors for restriction enzymes. J. Am. Chem. Soc., 120, 2674–2675. [Google Scholar]
- 102.Perona J.J. and Martin,A.M. (1997) Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis. J. Mol. Biol., 273, 207–225. [DOI] [PubMed] [Google Scholar]
- 103.Horton N.C., Newberry,K.J. and Perona,J.J. (1998) Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 13489–13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stahl F., Wende,W., Jeltsch,A. and Pingoud,A. (1996) Introduction of asymmetry in the naturally symmetric restriction endonuclease EcoRV to investigate intersubunit communication in the homodimeric protein. Proc. Natl Acad. Sci. USA, 93, 6175–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2001) Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease. Nature Struct. Biol., 8, 126–130. [DOI] [PubMed] [Google Scholar]
- 106.Thielking V., Selent,U., Köhler,E., Landgraf,Z., Wolfes,H., Alves,J. and Pingoud,A. (1992) Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry, 31, 3727–3732. [DOI] [PubMed] [Google Scholar]
- 107.Engler L.E., Welch,K.K. and Jen-Jacobson,L. (1997) Specific binding by EcoRV endonuclease to its DNA recognition site GATATC. J. Mol. Biol., 269, 82–101. [DOI] [PubMed] [Google Scholar]
- 108.Erskine S.G. and Halford,S.E. (1998) Reactions of the EcoRV restriction endonuclease with fluorescent oligodeoxynucleotides: identical equilibrium constants for binding to specific and non-specific DNA. J. Mol. Biol., 275, 759–772. [DOI] [PubMed] [Google Scholar]
- 109.Reid S.L., Parry,D., Liu,H.-H. and Connolly,B.A. (2001) Binding and recognition of VATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarisation. Biochemistry, 40, 2484–2494. [DOI] [PubMed] [Google Scholar]
- 110.Alves J., Selent,U. and Wolfes,H. (1995) Accuracy of the EcoRV restriction endonuclease: binding and cleavage studies with oligodeoxynucleotide substrates containing degenerate recognition sequences. Biochemistry, 34, 11191–11197. [DOI] [PubMed] [Google Scholar]
- 111.Jeltsch A., Maschke,H., Selent,U., Wenz,C., Köhler,E., Connolly,B.A., Thorogood,H. and Pingoud,A. (1995) DNA binding specificity of the EcoRV restriction endonuclease is increased by Mg2+ binding to a metal ion binding site distinct from the catalytic center of the enzyme. Biochemistry, 34, 6239–6246. [DOI] [PubMed] [Google Scholar]
- 112.Ghosh S.S., Obermiller,P.S., Kwoh,T.J. and Gingeras,T.R. (1990) Analysis of substrate specificity of the PaeR7I endonuclease: effect of base methylation on the kinetics of cleavage. Nucleic Acids Res., 18, 5063–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zebala J., Choi,J. and Barany,F. (1992) Characterization of steady state, single-turnover and binding kinetics of the TaqI restriction endonuclease. J. Biol. Chem., 267, 8097–8105. [PubMed] [Google Scholar]
- 114.Siksnys V. and Pleckaityte,M. (1993) Catalytic and binding properties of restriction endonuclease Cfr9I. Eur. J. Biochem., 217, 411–419. [DOI] [PubMed] [Google Scholar]
- 115.Kong H., Roemer,S.E., Waite-Rees,P.A., Benner,J.S., Wilson,G.G. and Nwankwo,D.O. (1994) Characterization of BcgI, a new kind of restriction-modification system. J. Biol. Chem., 269, 683–690. [PubMed] [Google Scholar]
- 116.Lagunavicius A., Grazulis,S., Balciunaite,E., Vainius,D. and Siksnys,V. (1997) DNA binding specificity of MunI restriction endonuclease is controlled by pH and calcium ions: Involvement of active site carboxylate residues. Biochemistry, 36, 11093–11099. [DOI] [PubMed] [Google Scholar]
- 117.Skirgaila R. and Siksnys,V. (1998) Ca2+-ions stimulate DNA binding specificity of Cfr10I restriction enzyme. Biol. Chem., 379, 595–598. [PubMed] [Google Scholar]
- 118.Gormley N.A., Bath,A.J. and Halford,S.E. (2000) Reactions of BglI and other type II restriction endonucleases with discontinuous recognition sites. J. Biol. Chem., 275, 6928–6936. [DOI] [PubMed] [Google Scholar]
- 119.Lagunavicius A. and Siksnys,V. (1997) Site-directed mutagenesis of putative active site residues of MunI restriction endonuclease: replacement of catalytically essential carboxylate residues triggers DNA binding specificity. Biochemistry, 36, 11086–11092. [DOI] [PubMed] [Google Scholar]
- 120.Cao W., Lu,J., Welch,S.G., Williams,R.A.D. and Barany,F. (1998) Cloning and thermostability of TaqI endonuclease isoschizomers from Thermus species SM32 and Thermus filiformis Tok6A1. Biochem. J., 333, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Horton J.R., Nastri,H.G., Riggs,P.D. and Cheng,X. (1998) Asp34 of PvuII endonuclease is directly involved in DNA minor groove recognition and indirectly involved in catalysis. J. Mol. Biol., 284, 1491–1504. [DOI] [PubMed] [Google Scholar]
- 122.Hlavaty J.J., Benner,J.S., Hornstra,L.J. and Schildkraut,I. (2000) Identification of the metal-binding sites of restriction endonucleases by Fe2+-mediated oxidative cleavage. Biochemistry, 39, 3097–3105. [DOI] [PubMed] [Google Scholar]
- 123.Windolph S. and Alves,J. (1997) Influence of divalent cations on inner-arm mutants of restriction endonuclease EcoRI. Eur. J. Biochem., 244, 134–139. [DOI] [PubMed] [Google Scholar]
- 124.Siksnys V., Zareckaja,N., Vaisvila,R., Timinskas,A., Stakenas,P., Butkus,V. and Janulaitis,A. (1994) CAATTG-specific restriction-modification munI genes from Mycoplasma: sequence similarities between R.MunI and R.EcoRI. Gene, 142, 1–8. [DOI] [PubMed] [Google Scholar]
- 125.Jen-Jacobson L., Engler,L.E., Lesser,D.R., Kurpiewski,M.R., Yee,C. and McVerry,B. (1996) Structural adaptations in the interaction of EcoRI endonuclease with methylated GAATTC sites. EMBO J., 15, 2870–2882. [PMC free article] [PubMed] [Google Scholar]
- 126.Lukacs C.M. and Aggarwal,A.K. (2001) BglII and MunI: what a difference a base makes. Curr. Opin. Struct. Biol., 11, 14–18. [DOI] [PubMed] [Google Scholar]
- 127.Jen-Jacobson L. (1997) Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers, 44, 153–180. [DOI] [PubMed] [Google Scholar]
- 128.Thielking V., Selent,U., Köhler,E., Wolfes,H., Pieper,U., Geiger,R., Urbanke,C., Winkler,F.K. and Pingoud,A. (1991) Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry, 30, 6416–6422. [DOI] [PubMed] [Google Scholar]
- 129.Selent U., Rüter,T., Köhler,E., Liedtke,M., Thielking,V., Alves,J., Oelgeschlager,T., Wolfes,H., Peters,F. and Pingoud,A. (1992) A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry, 31, 4808–4815. [DOI] [PubMed] [Google Scholar]
- 130.Wenz C., Jeltsch,A. and Pingoud,A. (1996) Probing the indirect readout of the restriction enzyme EcoRV. J. Biol. Chem., 271, 5565–5573. [DOI] [PubMed] [Google Scholar]
- 131.Stahl F., Wende,W., Wenz,C., Jeltsch,A. and Pingoud,A. (1998) Intra- vs intersubunit communication in the homodimeric restriction enzyme EcoRV: Thr37 and Lys38 involved in indirect readout are only important for the catalytic activity of their own subunit. Biochemistry, 37, 5682–5688. [DOI] [PubMed] [Google Scholar]
- 132.Thomas M.P., Brady,R.L., Halford,S.E., Sessions,R.B. and Baldwin,G.S. (1999) Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group. Nucleic Acids Res., 27, 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thorogood H., Grasby,J.A. and Connolly,B.A. (1996) Influence of the phosphate backbone on the restriction and hydrolysis of DNA by the EcoRV restriction endonuclease. J. Biol. Chem., 271, 8855–8862. [DOI] [PubMed] [Google Scholar]
- 134.Stahl F., Wende,W., Jeltsch,A. and Pingoud,A. (1998) The mechanism of DNA cleavage by the type II restriction enzyme EcoRV: Asp36 is not directly involved in DNA cleavage but serves to couple indirect readout to catalysis. Biol. Chem., 379, 467–473. [DOI] [PubMed] [Google Scholar]
- 135.Stanford N.P., Halford,S.E. and Baldwin,G.S. (1999) DNA cleavage by the EcoRV restriction endonuclease: pH dependence and proton transfers in catalysis. J. Mol. Biol., 288, 105–116. [DOI] [PubMed] [Google Scholar]
- 136.Jeltsch A., Alves,J., Wolfes,H., Maass,G. and Pingoud,A. (1993) Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA, 90, 8499–8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jeltsch A., Wenz,C., Wende,W., Selent,U. and Pingoud,A. (1996) Engineering novel restriction endonucleases: principles and applications. Trends Biotechnol., 14, 235–238. [DOI] [PubMed] [Google Scholar]
- 138.Lanio T., Jeltsch,A. and Pingoud,A. (2000) On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: a case study employing EcoRV as the target. Protein Eng., 13, 275–281. [DOI] [PubMed] [Google Scholar]
- 139.Schöttler S., Wenz,C., Lanio,T., Jeltsch,A. and Pingoud,A. (1998) Protein engineering of the restriction endonuclease EcoRV: structure-guided design of enzyme variants that recognize the base pairs flanking the recognition site. Eur. J. Biochem., 258, 184–191. [DOI] [PubMed] [Google Scholar]
- 140.Wende W., Stahl,F. and Pingoud,A. (1996) The production and characterization of artificial heterodimers of the restriction endonuclease EcoRV. Biol. Chem. Hoppe Seyler, 377, 625–632. [DOI] [PubMed] [Google Scholar]
- 141.Benkovic S.J. and Schray,K.J. (1973) Chemical basis of biological phosphoryl transfer. In Boyer, P.D. (ed.), The Enzymes. Academic Press, New York, NY, Vol. VIII, pp. 201–238.
- 142.Connolly B.A., Eckstein,F. and Pingoud,A. (1984) The stereochemical course of the restriction endonuclease EcoRI-catalyzed reaction. J. Biol. Chem., 259, 10760–10763. [PubMed] [Google Scholar]
- 143.Grasby J.A. and Connolly,B.A. (1992) Stereochemical outcome of the hydrolysis reaction catalyzed by the EcoRV restriction endonuclease. Biochemistry, 31, 7855–7861. [DOI] [PubMed] [Google Scholar]
- 144.Mizuuchi K., Nobbs,T.J., Halford,S.E., Adzuma,K. and Qin,J. (1999) A new method for determining the stereochemistry of DNA cleavage reactions: application to the SfiI and HpaII restriction endonucleases and to the MuA transposase. Biochemistry, 38, 4640–4648. [DOI] [PubMed] [Google Scholar]
- 145.Admiraal S.J. and Herschlag,D. (1995) Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. Chem. Biol., 2, 729–739. [DOI] [PubMed] [Google Scholar]
- 146.Hollfelder F. and Herschlag,D. (1995) The nature of the transition state for enzyme-catalyzed phosphoryl transfer. Hydrolysis of O-aryl phosphorothioates by alkaline phosphatase. Biochemistry, 34, 12255–12264. [DOI] [PubMed] [Google Scholar]
- 147.Ruan H., Lunnen,K.D., Pelletier,J.J. and Xu,S.-Y. (1997) Overexpression of BsoBI restriction endonuclease in E.coli, purification of the recombinant BsoBI and identification of catalytic residues of BsoBI by random mutagenesis. Gene, 188, 35–39. [DOI] [PubMed] [Google Scholar]
- 148.Rimseliene R. and Timinskas,A. (1997) Site-directed mutagenesis of type IV restriction endonuclease Eco57I. Biologija, 1, 31–33. [Google Scholar]
- 149.Reuter M., Schneider-Mergener,J., Kupper,D., Meisel,A., Mackeldanz,P., Krueger,D.H. and Schroeder,C. (1999) Regions of endonuclease EcoRII involved in DNA target recognition identified by membrane-bound peptide repertoires. J. Biol. Chem., 274, 5213–5221. [DOI] [PubMed] [Google Scholar]
- 150.Dahai T., Ando,S., Takasaki,Y. and Tadano,J. (1999) Site-directed mutagenesis of restriction endonuclease HindIII. Biosci. Biotechnol. Biochem., 63, 1703–1707. [DOI] [PubMed] [Google Scholar]
- 151.Wittmayer P.K. and Raines,R.T. (1996) Substrate binding and turnover by the highly specific I-PpoI endonuclease. Biochemistry, 35, 1076–1083. [DOI] [PubMed] [Google Scholar]
- 152.Galburt,E.A., Chevalier,B., Tang,W., Jurica,M.S., Flick,K.E., Monnat,R.J.J. and Stoddard,B.L. (1999) A novel endonuclease mechanism directly visualized for I-PpoI. Nature Struct. Biol., 6, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 153.Wardleworth B.N., Kvaratskhelia,M. and White,M.F. (2000) Site-directed mutagenesis of the yeast resolving enzyme Cce1 reveals catalytic residues and relationship with the intron-splicing factor Mrs1. J. Biol. Chem., 275, 23725–23728. [DOI] [PubMed] [Google Scholar]
- 154.Komori K., Sakae,S., Daiyasu,H., Toh,H., Morikawa,K., Shinagawa,H. and Ishino,Y. (2000) Mutational analysis of the Pyrococcus furiosus Holliday junction resolvase Hjc revealed functionally important residues for dimer formation, junction DNA binding and cleavage activities. J. Biol. Chem., 275, 40385–40391. [DOI] [PubMed] [Google Scholar]
- 155.Sapranauskas R., Sasnauskas,G., Lagunavicius,A., Vilkaitis,G., Lubys,A. and Siksnys,V. (2000) Novel subtype of type IIs restriction enzymes. J. Biol. Chem., 275, 30878–30885. [DOI] [PubMed] [Google Scholar]
- 156.Stuckey J.A. and Dixon,J.E. (1999) Crystal structure of a phospholipase D family member. Nature Struct. Biol., 6, 278–284. [DOI] [PubMed] [Google Scholar]
- 157.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Survey and summary: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bujnicki J.M., Rychlewski,L. and Radlinska,M. (2001) Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci., 26, 9–11. [DOI] [PubMed] [Google Scholar]
- 159.Horton J.R. and Cheng,X. (2000) PvuII endonuclease contains two calcium ions in active sites. J. Mol. Biol., 300, 1049–1056. [DOI] [PubMed] [Google Scholar]
- 160.Cowan J.A. (1997) Metal mediated hydrolysis of biological phosphate esters: a critical analysis of the essential metal ion stoichiometry for magnesium dependent nuclease activation. J. Biol. Inorg. Chem., 2, 168–176. [Google Scholar]
- 161.Cowan J.A. (1998) Metal activation of enzymes in nucleic acid biochemstry. Chem. Rev., 98, 1067–1087. [DOI] [PubMed] [Google Scholar]
- 162.Groll D.H., Jeltsch,A., Selent,U. and Pingoud,A. (1997) Does the restriction endonuclease EcoRV employ a two-metal ion mechanism for DNA cleavage? Biochemistry, 36, 11389–11401. [DOI] [PubMed] [Google Scholar]
- 163.Sam M.D., Horton,N.C., Nissan,T.A. and Perona,J.J. (2001) Catalytic efficiency and sequence selectivity of a restriction endonuclease modulated by a distal manganese ion binding site. J. Mol. Biol., 306, 851–861. [DOI] [PubMed] [Google Scholar]
- 164.Vyle J.S., Connolly,B.A., Kemp,D. and Cosstick,R. (1992) Sequence- and strand-specific cleavage in oligodeoxyribonucleotides and DNA containing 3′-thiothymidine. Biochemistry, 31, 3012–3018. [DOI] [PubMed] [Google Scholar]
- 165.Jeltsch A., Alves,J., Maass,G. and Pingoud,A. (1992) On the catalytic mechanism of EcoRI and EcoRV. A detailed proposal based on biochemical results, structural data and molecular modelling. FEBS Lett., 304, 4–8. [DOI] [PubMed] [Google Scholar]
- 166.Horvath M., Choi,J., Kim,Y., Wilkosz,P.A., Chandrasekhar,K. and Rosenberg,J.M. (2001) The integration of recognition and cleavage: X-ray structures of pre-transition state and post-reactive DNA EcoRI endonuclease complexes. Protein Sci., in press. [Google Scholar]
- 167.Schweins T., Langen,R. and Warshel,A. (1994) Why have mutagenesis studies not located the general base in ras p21. Nature Struct. Biol., 1, 476–484. [DOI] [PubMed] [Google Scholar]
- 168.Perona J.J., Rould,M.A. and Steitz,T.A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry, 32, 8758–8771. [DOI] [PubMed] [Google Scholar]
- 169.Cavarelli J., Eriani,G., Rees,B., Ruff,M., Boeglin,M., Mitschler,A., Martin,F., Gangloff,J., Thierry,J.C. and Moras,D. (1994) The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J., 13, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Thunnissen M.M., Taddei,N., Liguri,G., Ramponi,G. and Nordlund,P. (1997) Crystal structure of common type acylphosphatase from bovine testis. Structure, 5, 69–79. [DOI] [PubMed] [Google Scholar]
- 171.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 172.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 173.Vipond I.B., Baldwin,G.S. and Halford,S.E. (1995) Divalent metal ions at the active sites of the EcoRV and EcoRI restriction endonucleases. Biochemistry, 34, 697–704. [DOI] [PubMed] [Google Scholar]
- 174.Baldwin G.S., Sessions,R.B., Erskine,S.G. and Halford,S.E. (1999) DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J. Mol. Biol., 288, 87–103. [DOI] [PubMed] [Google Scholar]
- 175.Sam M.D. and Perona,J.J. (1999) Catalytic roles of divalent metal ions in phosphoryl transfer by EcoRV endonuclease. Biochemistry, 38, 6576–6586. [DOI] [PubMed] [Google Scholar]
- 176.Fothergill M., Goodman,M.F., Petruska,J. and Warshel,A. (1995) Structure-energy analysis of the role of metal ions in phosphodiester bond hydrolysis by DNA polymerase I. J. Am. Chem. Soc., 117, 11619–11627. [Google Scholar]
- 177.Dupureur C.M. and Conlan,L.H. (2000) A catalytically deficient active site variant of PvuII endonuclease binds Mg(II) ions. Biochemistry, 39, 10921–10927. [DOI] [PubMed] [Google Scholar]
- 178.Viadiu H. and Aggarwal,A.K. (1998) The role of metals in catalysis by the restriction endonuclease BamHI. Nature Struct. Biol., 5, 910–916. [DOI] [PubMed] [Google Scholar]
- 179.Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two-metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Aaqvist J. and Warshel,A.J. (1990) Free energy relationships in metalloenzyme-catalyzed reactions. Calculations of the effects of metal ion substitutions in Staphylococcal nuclease. J. Am. Chem. Soc., 112, 2860–2868. [Google Scholar]
- 181.Yang W., Hendrickson,W.A., Crouch,R.J. and Satow,Y. (1990) Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science, 249, 1398–1405. [DOI] [PubMed] [Google Scholar]
- 182.Nakamura H., Oda,Y., Iwai,S., Inoue,H., Ohtsuka,E., Kanaya,S., Kimura,S., Katsuda,C., Katayanagi,K., Morikawa,K. et al. (1991) How does RNase H recognize a DNA.RNA hybrid? Proc. Natl Acad. Sci. USA, 88, 11535–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Haruki M., Tsunaka,Y., Morikawa,M., Iwai,S. and Kanaya,S. (2000) Catalysis by Escherichia coli ribonuclease HI is facilitated by a phosphate group of the substrate. Biochemistry, 39, 13939–13944. [DOI] [PubMed] [Google Scholar]
- 184.Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- 185.Bujacz G., Alexandratos,J., Wlodawer,A., Merkel,G., Andrake,M., Katz,R.A. and Skalka,A.M. (1997) Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem., 272, 18161–18168. [DOI] [PubMed] [Google Scholar]
- 186.Galburt E.A. and Stoddard,B.L. (2000) Restriction endonucleases: one of these things is not like the others. Nature Struct. Biol., 7, 89–91. [DOI] [PubMed] [Google Scholar]
- 187.Jurica M.S., Monnat,R.J.,Jr and Stoddard,B.L. (1998) DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell, 2, 469–476. [DOI] [PubMed] [Google Scholar]
- 188.Connolly M.L. (1993) The molecular surface package. J. Mol. Graph., 11, 139–141. [DOI] [PubMed] [Google Scholar]
- 189.Rosenberg J.M. (1991) Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol., 1, 104–113. [DOI] [PubMed] [Google Scholar]
- 190.Bozic D., Grazulis,S., Siksnys,V. and Huber,R. (1996) Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 Å resolution. J. Mol. Biol., 255, 176–186. [DOI] [PubMed] [Google Scholar]
- 191.Skirgaila R., Grazulis,S., Bozic,D., Huber,R. and Siksnys,V. (1998) Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues. J. Mol. Biol., 279, 473–481. [DOI] [PubMed] [Google Scholar]
- 192.Vanamee E.S. Santagata,S., and Aggarwal,A.K. (2001) FokI requires two specific DNA sites for cleavage. J. Mol. Biol., 309, 69–78. [DOI] [PubMed] [Google Scholar]
- 193..Friedhoff P., Lurz,R., Lüder,G. and Pingoud,A. (2001) Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. J. Biol. Chem., 276, 23581–23588. [DOI] [PubMed] [Google Scholar]
- 194.. Simoncsits A.,Tjörnhammar,M.-L., Raskó,T., Kiss,A. and Pongor,S. (2001) Covalent joining of the subunits of a homodimeric type II restriction endonuclease: single-chain PvuII endonuclease. J. Mol. Biol., 309, 89–97. [DOI] [PubMed] [Google Scholar]