Abstract
The evolutionary processes that shape patterns of diversity in highly mobile marine species are poorly understood, but important towards transferable inference on their effective conservation. In this study, bottlenose dolphins (Tursiops sp.) are studied to address this broader question. They exhibit remarkable geographical variation for morphology, life history, and genetic diversity, and this high level of variation has made the taxonomy of the genus controversial. A significant population structure has been reported for the most widely distributed species, the common bottlenose dolphin (T. truncatus), in almost all ocean basins, though no data have been available for the western North Pacific Ocean (WNP). The genetic diversity of bottlenose dolphins in the WNP was investigated based on 20 microsatellite and one mitochondrial DNA markers for samples collected from Taiwanese, Japanese, and Philippine waters (9°–39°N, 120°–140°E) during 1986–2012. The results indicated that there are at least four genetically differentiated populations of common bottlenose dolphins in the western and central North Pacific Ocean. The pattern of differentiation appears to correspond to habitat types, resembling results seen in other populations of the same species. Our analyses also showed that there was no evident gene flow between the two “sister species”, the common bottlenose dolphins, and the Indo-Pacific bottlenose dolphins (T. aduncus) occurring sympatrically in our study region.
Electronic supplementary material
The online version of this article (doi:10.1007/s00227-017-3232-8) contains supplementary material, which is available to authorized users.
Introduction
A wildlife management unit is usually defined by the significance of morphological, genetic, or demographic differences among populations, often associated with geographic barriers or distance (e.g. Allendorf and Luikart 2006). Identifying management units is imperative in wildlife conservation, as it assists the preservation of intra-species diversity and the species’ future adaptive potential. Some oceanic dolphin species show an unexpected level of population structure, given their capacity for extensive dispersion and the lack of obvious geographic barriers (Hoelzel 2009). The bottlenose dolphin (Tursiops sp.) has provided a number of classic examples regarding the parapatric or sympatric distribution of differentiated populations or species (e.g., see Moura et al. 2013). Here, we investigate bottlenose dolphin populations in a region where there is ongoing impact due to bycatch and drive fisheries, and, therefore, a pressing need for conservation management.
Bottlenose dolphins are widely distributed in the world’s tropical-to-temperate marine environment, including along the coasts of all major continents and many oceanic islands, over shallow offshore banks or sandbars, and in pelagic open waters (Rice 1998). There is considerable geographical variation in bottlenose dolphin skeletal morphology, life history, and genetic diversity, which makes the taxonomy of the genus controversial (Rice 1998; Wells and Scott 2009; Charlton-Robb et al. 2015). In our study region in the western North Pacific Ocean (WNP), two species of dolphins in the genus Tursiops have been recognized: the Indo-Pacific bottlenose dolphin (T. aduncus; hereinafter IPBD) and the common bottlenose dolphin (T. truncatus; hereinafter CBD). These two species are distributed parapatrically, or even sympatrically in particular areas. The distribution of IPBD is chiefly in the coastal waters of warm-temperate-to-tropical Indo-Pacific regions from southern Japan to western South Africa and southeastern Australia, where the water depth is always less than 200 m (Wang and Yang 2009). The distribution of CBD in the WNP, on the other hand, ranges from the southern Okhotsk Sea to the South China Sea and to Hawaiian waters, in both coastal and pelagic habitats (Miyashita 1993; Rice 1998; Wells and Scott 2009). The distribution range of these two species overlaps from the East China Sea and Taiwan Strait to the South China Sea (Zhou and Qian 1985; Wang et al. 1999, 2000; Yang et al. 2005). Although the broader taxonomy of the genus remains unresolved, the alpha taxonomy of IPBD and CBD is well supported (LeDuc et al. 1999; Wang et al. 1999, 2000; Hale et al. 2000; Kemper 2004; Natoli et al. 2004; Yang et al. 2005; Kurihara and Oda 2007; Moura et al. 2013).
Within the CBD species, a significant differentiation between coastal and offshore populations has been reported from various locations, including the western North Atlantic Ocean (Hoelzel et al. 1998; Kingston and Rosel 2004), the eastern North Atlantic Ocean (Louis et al. 2014a), and the eastern North Pacific Ocean (Lowther-Thieleking et al. 2015). The population structure of CBD can also be defined on a finer regional scale, such as within the Gulf of Mexico (Sellas et al. 2005; Richards et al. 2013), Northern Bahamas (Parsons et al. 2006), west coasts of the United States (Rosel et al. 2009), the waters around New Zealand (Tezanos-Pinto et al. 2009), Ireland (Mirimin et al. 2011), Hawaiian Islands (Martien et al. 2012), and the Adriatic Sea (Gaspari et al. 2015a).
Two recent papers analysed mitochondrial DNA (mtDNA) control region sequence data for CBD and IPBD with samples from the western South Pacific Ocean and hypothesised that the coastal ecotype of CBD is lacking in the Indo-western Pacific Ocean and that this is because the coastal habitat has been occupied by IPBD (Tezanos-Pinto et al. 2009; Oremus et al. 2015a). However, not all coastal and pelagic CBD lineages are reciprocally monophyletic (Moura et al. 2013), sometimes likely due to incomplete lineage sorting (Segura et al. 2006; Lowther-Thieleking et al. 2015). Therefore, this assessment should be confirmed using nuclear markers.
Miyashita (1993) proposed a three-stock structure for CBD in the WNP (for the waters off eastern Japan) based on 8-year transect line survey data: a Japanese coastal population (from the east coasts of Japan to the west of 142°E), a Japanese offshore population (between 30° and 42°N and from the east of 145°E to the antimeridian), and a southern offshore population (between 23° and 30°N, and between 127°E and the antimeridian). However, this three-stock hypothesis has yet to be tested using molecular markers. Kita et al. (2013) sequenced a group of 165 CBD culled in a drive fishery hunt in Japan for 402 bp mtDNA control region and compared these against published sequences worldwide (using 290 bp). They report that these dolphins were “related more closely to oceanic types from Chinese waters than other geographic regions” (p. 476). The study was unfortunately unable to provide further insights into the population structure of CBD in the WNP.
For IPBD, it has been proposed that there are at least six populations in Japanese waters (Amano 2007; Brownell and Funahashi 2013). Kakuda et al. (2002) studied the genetic structure of IPBD from Mikura Island (about 200 km south of Tokyo) using mtDNA control region sequences and concluded that the dolphins were genetically similar to the IPBD in Taiwanese waters. Hayano (2013) used the same genetic marker and reported a clear population differentiation among Mikura, Amakusa, Amami, and Ogasawa Islands. The residency of Amakusa, Mikura Island, and Kagoshima Bay populations has been proposed based on photo-identification records (Shirakihara et al. 2002; Kogi et al. 2004; Nanbu et al. 2006). A significant geographic vocalization variation is found among dolphin populations around Amakusa, Mikura, and Ogasawa Islands (Morisaka et al. 2005). In Taiwanese waters, the distribution of IPBD is seemly discontinuous: current field observations and records of fishery interactions showed that this species aggregates around the Penghu archipelago (in the Taiwan Strait, west of Taiwan), and the coastal waters off Kengting, southeast of Taiwan (Wang et al. 1999; Wang 2000).
Both CBD and IPBD are affected by multiple anthropogenic threats, such as small-scale whaling and negative fishery interactions in this WNP study region (Perrin et al. 2005; Kasuya 2007; Young and Iudicello 2007; Robards and Reeves 2011). There were more than 26,000 bottlenose dolphins caught in Japanese waters during 1972–2008 (Kasuya 2011), and about 1700 bottlenose dolphins are incidentally killed in human fisheries in the western-central Pacific Ocean every year (Young and Iudicello 2007). The aim of this study is to promote the more effective conservation of these highly mobile marine species through a better understanding of the pattern and origin of population structure, and the relevant processes. We focus on the CBD species in the WNP to help assess the impact of human disturbance, since this species is a common target in the dolphin drive fishery (Kasuya 2007; Oremus et al. 2015b).
Materials and methods
Tissue sample collection and genomic DNA preparation
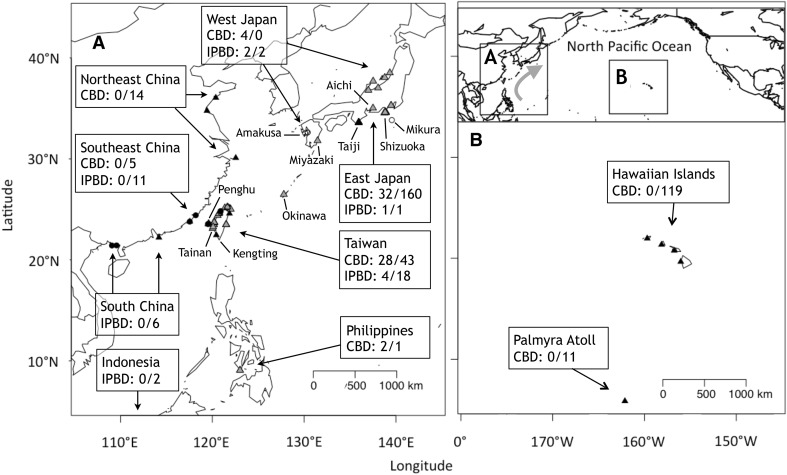
Sixty-six CBD and seven IPBD tissue samples collected from various locations in Japan, Taiwan, and the Philippines were included in this study (detailed locations and numbers shown in Fig. 1 with further details provided in supplementary Table S1). Species identity was acquired from the archives and verified by our genetic assessments. For CBD samples, each was assigned to one of the four putative populations based on its sampling location (i.e., West Japan, East Japan, Taiwan, and the Philippines; Fig. 1a; Table S1). The origin of the 14 samples collected from Japanese aquariums was unknown, but assigned to the East Japan group based on our factorial correspondence analysis (FCA) result (see below). For IPBD, the a prior population assignment was based on sampling location (Fig. 1a; Table S1). For a set of 15 samples from a drive fishery in eastern Japan, we note that none of these samples were collected to support this work. Our use of archived materials derived from those activities is not meant as an endorsement, but rather as a means to contribute to the provision of data critical to the effective conservation of these populations.
Fig. 1.
Sampling locations are provided for the Indo-Pacific bottlenose dolphins (IPBD; open circle) and common bottlenose dolphins (CBD; grey triangle) examined in this study, and the IPBD (solid circle) and CBD (solid triangle) mitochondrial DNA sequences acquired from GenBank. The numbers indicate the sample size for microsatellite/mtDNA data. Note that the sampling locations of the two IPBD samples from Indonesia are unknown, therefore, not indicated (see Wang et al. 1999). The grey arrow in upper right panel indicates the flow of Kuroshio current
Genomic DNA was isolated and purified by a standard proteinase-K digestion/phenol–chloroform extraction protocol (Sambrook et al. 1989), and preserved in TE buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH7.4). The Philippine samples were provided as extracted genomic DNA by the Southwest Fisheries Science Center, National Oceanic and Atmospheric Administration (USA).
DNA fragment amplification and genotyping
We examined 20 microsatellite loci and a 388 bp mtDNA control region sequence that have been conventionally used and validated in other bottlenose dolphin genetic studies (Shinohara et al. 1997; Hoelzel et al. 1998; Krützen et al. 2001; Natoli et al. 2004; Mirimin et al. 2011). The microsatellite loci and associated annealing temperatures are given in Table S2. Polymerase chain reaction (PCR) reagents were in 20 or 10 μl using GoTaq® Taq DNA polymerase (Promega) or multiplex polymerase (Multiplex PCR Kit, Qiagen), respectively, cycled at 95 °C for 120 s (15 m for multiplex polymerase), followed by 35 cycles of 40 s at 94 °C, 40 s at the best annealing temperature of the locus, and 70 s at 72 °C, and a post-extension at 72 °C for 10 min. Fragments were visualised on an Applied Biosystems 3730 DNA Analyser, and the allele size was determined by an internal standard marker (Genescan-500 ROX, Applied Biosystems) and visualised in Peak Scanner v.1 (Applied Biosystems). Every locus in each sample was examined at least twice and the scores blind confirmed by a second person.
The mtDNA sequences were amplified using primers designed to amplify cetacean mtDNA control region (Thr and Phe primers from Hoelzel et al. 1991). The PCR reactions were in 20 μl cycling at 95 °C for 120 s, followed by 35 cycles of 40 s at 94 °C, 40 s at 50 °C, and 70 s at 72 °C, and a post-extension at 72 °C for 10 min. The amplified mtDNA fragments were purified using QIAquick® PCR Purification Kit (Qiagen) and then sequenced on an Applied Biosystems 3730 DNA Analyser. All sequencing results were visualised in FinchTV (PerkinElmer) and manually corrected using MEGA 5.05 (Tamura et al. 2011).
Microsatellite data analysis
Genetic diversity and differentiation
We used Arlequin 3.5.1 (Excoffier and Lischer 2010) to examine linkage disequilibrium (LD), observed heterozygosity (HO), and expected heterozygosity (HE), and to assess the significance of any deviation from Hardy–Weinberg equilibrium (HWE) using Fisher’s exact test and Markov chain method (number of steps in Markov chain, 1000,000; number of dememorization steps, 100,000). The inbreeding coefficient (FIS) for each locus in each putative population was estimated using FSTAT 2.9.3.2 (Goudet 2002). We also used Arlequin to assess the degree of population differentiation by F ST (Wright 1951) and R ST (Slatkin 1995), only for populations with a sufficient sample size (East Japan; n = 32 and Taiwan; n = 28). We used a non-parametric permutation approach with 10,000 permutations to assess the statistical significance (p < 0.05 after Bonferonni correction).
Genetic structure was also investigated by FCA implemented in Genetix 4.0 (Belkhir et al. 2004), and using the Bayesian assignment method implemented in STRUCTURE (Pritchard et al. 2000). FCA was run both with and without referencing individuals to a population centre (the “sur population” option). In STRUCTURE, we used six independent runs for each value of K assuming admixture and applying a burn-in length of 100,000 and a length of simulation of 1,000,000 repeats. The analysis was undertaken with and without using the “LOCPRIOR” function. Delta K (ΔK) reflecting the highest hierarchical level was determined by the Evanno method implemented in Structure Harvester (Earl and von Holdt 2012), and the result was optimized using CLUMPP (Jakobsson and Rosenberg 2007) and DISTRUCT (Rosenberg 2004). Once K was determined, we used the USEPOPINFO option to search for potential hybrids or descents of hybrids following the method described in Martien et al. (2012).
We also used the R package Geneland (Guillot et al. 2005) to assess the population structure in a spatial context. Because the program requires information of precise spatial coordinates for each genotyped individual, those CBD samples with ambiguous sampling locations were excluded for this analysis. In particular, the Japanese samples collected from the aquariums and the Taiwanese samples confiscated in the fish markets were excluded. We conducted the analysis using the procedure described in Fontaine et al. (2007), but the number of clusters (K) was set to vary from 1 to 6 clusters, and the maximum rate of Poisson process fixed to 41 (number of samples), uncertainty attached to spatial coordinates was fixed to 100 km, maximum number of nuclei in the Poisson–Voronoi tessellation was fixed to 123, and the posterior probabilities of population membership for each individual and each pixel of the spatial domain were calculated with a burn-in of 100 iterations and a spatial domain of 151 pixels along the X-axis and 250 along the Y-axis.
CBD population dynamics in the WNP
The effective population size (N e) and long-term gene flow (the number of migrants per generation; N e m) were estimated using maximum-likelihood coalescent methods implemented in MIGRATE version 3.6.6 (Beerli and Felsenstein 1999, 2001). The settings were after Martien et al. (2012), but we used a heating scheme and repeated the analysis five times. An approximate N e was calculated as the N e μ divided by an average expected microsatellite mutation rate, μ = 5 × 10−4 (Whitaker et al. 2003; Hoelzel et al. 2007; Hollatz et al. 2011). The ratio of effective to census population size (N e N −1; Frankham 1995) was calculated using published estimates of the census population sizes for CBD populations found in Japanese waters. We used the census population size (N) estimated for the “Japanese Coastal” population (N = 37,000; Miyashita 1993) for the “East Coast Cluster” (see below), and the N estimated for the CBD in southwestern Japanese waters (N = 35,000; Kasuya 2011) for the “West Coast Cluster”. We used GeneClass2 to search for potential first-generation migrants (Piry et al. 2004) and computed the likelihood using the algorithm described in Paetkau et al. (2004), with a frequency-based method (Paetkau et al. 1995). The probability was estimated using MCMC resampling of 1000 individuals and the type I error was set to 0.01. Tests for sex-biased dispersal were implemented in FSTAT (Goudet et al. 2002). This is based on differences between the sexes for statistics associated with mean and variance of assignment indices, F IS, F ST, relatedness, H O, and within-group gene diversity (H S) with t tests using 1000 permutations.
Mitochondrial DNA data analyses
Multi-region network
Published mtDNA control region sequences for both CBD and IPBD from the same or adjacent regions, i.e., Taiwan and southeastern China (Wang et al. 1999; Yang et al. 2005), Japan (Kita et al. 2013), northeastern China (Yang et al. 2005), and Hawaiian Islands and Palmyra Atoll (Martien et al. 2012), were acquired from GenBank (see Table S3). To address strong kin bias for our samples, one individual from all recognized parent–offspring pairs was discarded. In the published sequences, the pedigree relationship among individuals for Japanese, Hawaiian, and Palmyra Atoll samples was well documented (Martien et al. 2012; Kita et al. 2013), but the kinship information for Chinese samples was not available (Wang et al. 1999; Yang et al. 2005). We assumed that there was no parent–offspring pair sampled in the Chinese samples, because (1) they were collected in independent strandings or occasional fishery interaction events, (2) only a few individuals shared the same haplotype, and more importantly, (3) those samples sharing the same haplotype were not collected at the same time or location. We aligned the sequences together with ours in MEGA and obtained a consensus sequence (388 bp) from which we generated a median joining network using the program POPART (Leigh and Bryant 2015). We then assigned these published mtDNA sequences together with ours to six putative populations based on their sampling geography; that is, Japan, Northeast China (including Zhoushan, Qingdao, and Lianyunggang), Southeast China (including Dongshan, Taiwan, Hong Kong, the Philippines), South China (Beihai), Indonesia, Hawaiian Islands, and Palmyra Atoll (Fig. 1; Table S3).
Genetic diversity and tests for population expansion history
We used DnaSP v5 (Librado and Rozas 2009) to identify the haplotypes and estimate the nucleotide diversity (π) and haplotype diversity (h) for each putative population, as well as for the overall species. The neutrality tests Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) were estimated using DnaSP to look for signals that could be interpreted as either selection or population expansion. Mismatch distributions generated in Arlequin were produced to test for population expansion signals (Rogers and Harpending 1992; Schneider and Excoffier 1999; Excoffier 2004; Ray et al. 2003). The confidence intervals of the estimates were obtained under 10,000 bootstrap simulations of an instantaneous expansion under a coalescent framework. The sum of square deviations (SSD) between the observed and the expected mismatch and the raggedness index (r) of the observed distribution were calculated and tested to evaluate fit to models (Harpending 1994; Schneider and Excoffier 1999).
CBD population structure in the western and central North Pacific Ocean
Global tests of genetic differentiation among samples, as well as a differentiation test between all pairs of putative populations, were assessed using a Fisher’s exact test (Raymond and Rousset 1995) implemented in Arlequin, using 10,000 permutations. Pairwise F ST and ΦST between all pairs of putative populations were calculated and tested for significance using Arlequin. The significance level was set as p < 0.05.
We used MrBayes 3.2 (Ronquist et al. 2012) to reconstruct the phylogeny of all CBD haplotypes using a Bayesian Markov Chain Monte Carlo (MCMC) analysis. The evolutionary model for the test was determined by jModelTest 2.1.5 (Darriba et al. 2012); the sampling increment was set at 100 and diagnostics at every 1000 generations; at least 900,000 generations were simulated to generate the consensus tree. The final consensus tree was visualized and edited for optimal display in FigTree v.1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
CBD microsatellite locus diversity and differentiation
All samples were successfully genotyped, and the missing data rate was less than 5% for all loci and samples. No significant LD or deviation from HWE was detected. Summary statistics are shown in Table 1 for each putative population (see Table S4 for the information by locus). The F ST between East Japan and Taiwan was 0.013 and significantly greater than zero (p < 0.01); while R ST was 0.055, but not significant (p = 0.068 ± 0.002). As expected based on simulation studies (e.g., Latch et al. 2006), the magnitude of differentiation was too small for clear detection by STRUCTURE. However, although LnP(K) indicated K = 1, ΔK = 2 when the LOCPRIOR function is applied, and then differentiation between East Japan and Taiwan/West Japan could be detected (Fig. S1).
Table 1.
For common bottlenose dolphins (CBD) and Indo-Pacific bottlenose dolphins (IPBD), the number of alleles, expected heterozygosity (HE), observed heterozygosity (HO), allelic richness (AR), and inbreeding coefficient (F IS) averaged across loci within populations (Mean ± SD)
| Population | n | No. of alleles | H E | H O | AR | F IS |
|---|---|---|---|---|---|---|
| CBD | ||||||
| Taiwan | 28 | 7.100 ± 2.882 | 0.715 ± 0.193 | 0.697 ± 0.192 | 1.715 | 0.025 |
| East Japan | 32 | 7.350 ± 3.558 | 0.715 ± 0.172 | 0.702 ± 0.189 | 1.715 | 0.019 |
| West Japan | 4 | 3.750 ± 1.517 | 0.695 ± 0.225 | 0.684 ± 0.248 | 1.661 | 0.019 |
| Philippines | 2 | 2.650 ± 0.875 | 0.769 ± 0.173 | 0.694 ± 0.349 | 1.692 | 0.138 |
| All samples | 66 | 8.400 ± 3.789 | 0.717 ± 0.179 | 0.695 ± 0.176 | 1.717 | |
| IPBD | ||||||
| Taiwan | 4 | 3.111 ± 0.963 | 0.651 ± 0.134 | 0.625 ± 0.196 | 1.586 | 0.046 |
| Amakusa | 2 | 2.385 ± 0.506 | 0.641 ± 0.165 | 0.615 ± 0.300 | 1.417 | 0.059 |
| Mikura Island | 1 | NA | NA | NA | NA | NA |
| All samples | 7 | 3.350 ± 1.137 | 0.601 ± 0.190 | 0.549 ± 0.180 | 1.601 | |
See Table S4 for the estimates by locus within each population
For comparisons among CBD populations using the “sur population” option, all population-specific clusters could be identified and FC1 and FC2 accounted for 82.95% of the variance (Fig. 2a). Without using the option, the genotypes of the Philippine samples remained highly distinct, but other putative populations were less well resolved (Fig. 2b). The 14 captive dolphin samples provided by Japanese aquariums grouped with samples from East Japan rather than West Japan, while West Japan samples grouped with samples from Taiwan for FC1 vs. FC2. It is noteworthy that an East Japan sample, EW4842, was clustered with the Taiwan–West Japan samples, and the same clustering pattern was also found in the Geneland analysis (see below). This young male dolphin was stranded at the coast of Miyazaki, which was the most southerly sampling site for the putative East Japan population (Fig. 1). This “mis-grouping” could reflect limitations to the resolution of the analysis, evidence of direct migration between populations, or the result of a carcass drifting between regions (Bilgmann et al. 2011). The West Japan samples were segregated from the Taiwan samples and became an independent cluster by the third factor, FC3. This factor explained the remaining 17.05% of the variance (Fig. S2).
Fig. 2.
Results of the FCA for the CBD: a using the “sur population” option; b without using the “sur population” option. The two most informative factors (FC1 and FC2) were assigned as X and Y axes in the plot, and the numbers in parentheses in each axis indicate the percentage of the variance explained by the factor
The ten simulations in the first step of the Geneland analysis all indicated the most likely number of populations for our sample set was K = 3. With the K fixed to K = 3 in the second step, the analysis suggested eight variations of population distribution patterns for CBD among the 10 runs with the highest LPP in 100 simulations. These eight variations all showed approximately the same clustering pattern, with a few samples showing inconsistent population membership (Fig. 3). The basic pattern was a cluster grouping samples from the west coast of Japan, west and north coast of Taiwan, and the sample collected in Miyazaki, Japan (“the West Coast Cluster”); a cluster for the samples from the east coast of Taiwan and from Taiji, Japan (“the East Coast Cluster”); and a cluster for the samples from the Philippines (“the South Tropical Cluster”). The samples collected from Tainan (southwestern Taiwan) and Shizuoka (eastern Japan) overlapped with all three clusters, but usually grouped with the South Tropical Cluster (Fig. 3a, b, f, h). The sample collected from Aichi (eastern Japan) grouped with the East Coast Cluster (Fig. 3a–e, g) and the West Coast Cluster (Fig. 3f, h).
Fig. 3.
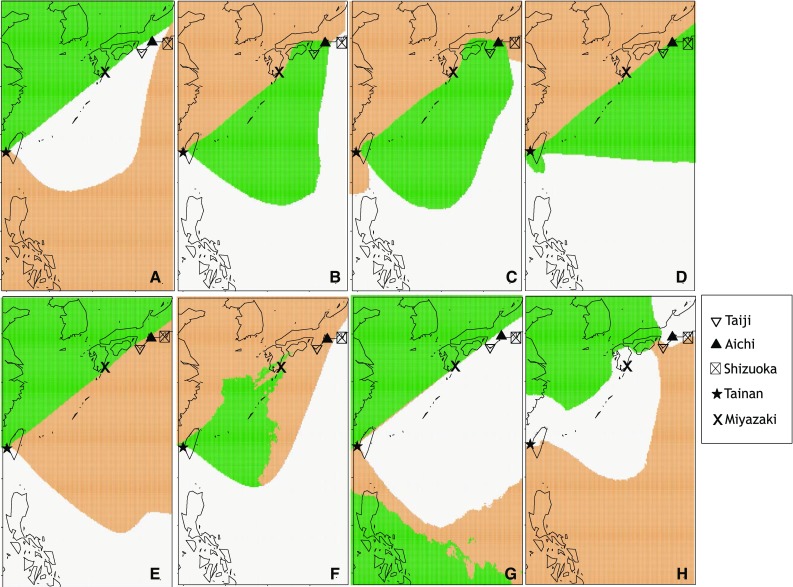
Eight variations of the individual population membership assignment patterns shown in the 10 runs with the highest LPP for K = 3 in Geneland analysis. The colours indicate the distribution of K clusters based on the mode of simulated posterior probability for each pixel. The landmarks mentioned in the text are shown
Population dynamics for CBD inferred from microsatellite analyses
To evaluate the population dynamics for CBD populations, we regrouped the samples into two clusters (for which sample sizes were deemed sufficient) based on the result of Geneland analysis: a West Coast Cluster (the samples from Miyazaki and the west coasts of Taiwan and Japan), and an East Coast Cluster (the samples from the east coast of Taiwan and Taiji). The samples from Tainan, Aichi, and Shizuoka were excluded due to the uncertainty of their population identity. N e estimated from Migrate for the East Coast Cluster was slightly larger than for the West Coast Cluster (Table 2). N e N −1 for both populations was similar in magnitude, ranging from 0.042 to 0.059, or from 0.084 to 0.118, depending on what microsatellite mutation rate was used, and migration estimates suggested a higher rate from the West to the East Coast cluster (Table 2). The GeneClass analysis identified three potential first-generation migrants (Table S5). There was no support for sex-biased dispersal (Table S6).
Table 2.
Estimates of effective population size times mutation rate (N e μ) and number of migrants per generation (N e m) from the two CBD populations recognized in Geneland analysis
| Source population | Host population | ||||
|---|---|---|---|---|---|
| East coast | West coast | ||||
| N e μ | 0.400 | (0.367–0.437) | 0.321 | (0.293–0.353) | |
| N e (low) | 2000 | (1836–2186) | 1605 | (1465–1766) | |
| N e (high) | 4001 | (3671–4371) | 3211 | (2930–3532) | |
| N e (low)/N | 0.054 | (0.05–0.059) | 0.046 | (0.042–0.05) | |
| N e (high)/N | 0.108 | (0.099–0.118) | 0.092 | (0.084–0.101) | |
| N e m | East coast | 0.057 | (0.046–0.070) | ||
| West coast | 0.106 | (0.089–0.125) | |||
The N e is calculated assuming that the average microsatellite mutation rate (μ) is 0.01% for N e (high) and 0.02% for N e (low). The ratio of effective to census population size (N e/N) is calculated using the census population size (N) estimated for the “Japanese Coastal” population (N = 37,000; Miyashita 1993) for the East Coast Cluster, and the N for the CBD in the southwestern Japanese waters (N = 35,000; Kasuya 2011) for West Coast Cluster. The 2.5th and 97.5th profile likelihood estimates are given in parentheses
MtDNA genetic diversity of CBD and IPBD in the western and central North Pacific Ocean
We sequenced 42 CBD samples from Taiwan, East Japan, and Philippines, and seven IPBD samples from Taiwan and Japan. Together with the published sequences acquired from GenBank (n = 344), we used a 388 bp consensus mtDNA sequence from a total of 393 sequences and reconstructed five putative CBD populations (East Japan, Northeast China, Southeast China, Hawaiian Islands, and Palmyra Atoll) and four IPBD populations (Japan, Southeast China, South China, and Indonesia) (Fig. 1; Table S3). According to the AIC and BIC indices calculated by jModelTest, the best model for reconstructing a phylogenetic tree for the genus using our mtDNA sequences was HKY + I+G.
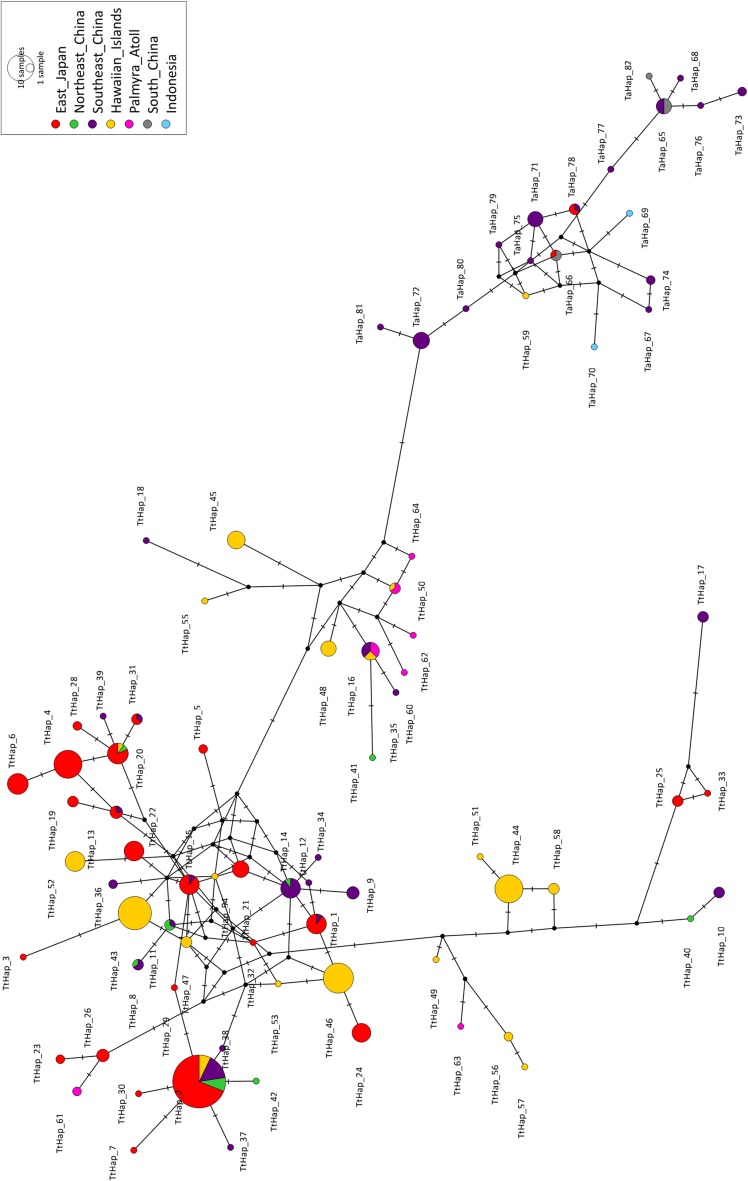
For CBD, we examined 353 sequences and identified 64 haplotypes defined by 82 variable sites, including two deletion gaps (Table S7). The overall haplotype diversity (h) was 0.935 and nucleotide diversity (π) was 0.0197. The h and π for each population are shown in Table 3. TtHap_2 was the most widespread haplotype, found in all populations except Palmyra Atoll (Fig. 4; Fig. S3; Table S8). It was also the dominant haplotype in the WNP (28.3% of all samples), where it was most common in Northeast China (42.9%, accession number AF459509-15), and in the school of dolphins culled in the drive fishery in 2005 (30.4%, previously published as Haplotype Ttr06, GenBank accession number AB303159). TtHap_16 was the only haplotype shared between the WNP (in Southeast China) and the tropical central Pacific (in Palmyra Atoll). It is noteworthy that in the phylogenetic tree, TtHap_17 (from southeast China), 25, and 33 (from eastern Japan) were isolated from the major CBD-IPBD lineage, potentially indicating that a lineage sorting process is still ongoing in CBD in the WNP populations (Fig. S3).
Table 3.
Summary of the mtDNA haplotype diversity, nucleotide diversity, and indices for testing locus neutrality for the CBD and IPBD populations
| n | No. of haplotypes | Haplotype diversity (h) | Nucleotide diversity (π) | Tajima’s D | Fu’s Fs | |
|---|---|---|---|---|---|---|
| CBD | ||||||
| East Japan | 160 | 23 | 0.870 (0.019) | 0.01368 (0.00103) | −0.81835 | −1.788 |
| SE China | 49 | 20 | 0.908 (0.025) | 0.02193 (0.00314) | −0.74485 | −1.607 |
| NE China | 14 | 8 | 0.824 (0.098) | 0.01638 (0.00427) | −1.09647 | 0.216 |
| Hawaii | 119 | 20 | 0.868 (0.016) | 0.02124 (0.00088) | −0.09236 | 1.449 |
| Palmyra | 11 | 7 | 0.909 (0.066) | 0.01851 (0.00423) | −0.42215 | 0.526 |
| Overall | 353 | 63 | 0.935 (0.008) | 0.01966 (0.00079) | −1.25991 | −22.17** |
| IPBD | ||||||
| Japan | 3 | 2 | 0.667 (0.314) | 0.00346 (0.00163) | NA | 1.061 |
| SE China | 29 | 14 | 0.899 (0.036) | 0.01365 (0.00110) | 0.99207 | −2.389 |
| S China | 6 | 3 | 0.733 (0.155) | 0.01195 (0.00351) | 0.99488 | 2.76 |
| Indonesia | 2 | 2 | 1 (0.5) | 0.01039 (0.00519) | NA | 1.386 |
| Overall | 40 | 18 | 0.924 (0.022) | 0.01395 (0.00084) | 0.66414 | −4.005* |
The SD for haplotype and nucleotide diversity is given in parentheses. Significant results (p < 0.05) are given in bold
* p < 0.05; ** p < 0.01
Fig. 4.
Median-joining network tree for CBD and IPBD mtDNA control region haplotypes. Each circle represents a unique haplotype. The size of the circle indicates the number of individuals having the haplotype and the colour shade indicates the proportion of each population within the haplotype. The number of hatch marks at the lines indicates the number of mutational steps separating the haplotypes. Solid circles indicate missing intermediate haplotypes
For IPBD, we identified 18 haplotypes defined by 19 variable sites in a total of 40 sequences from the coastal waters around Japan, Southeast China, and South China (GenBank accession numbers MF806001-18). The overall h was 0.924 and π was 0.014; the h and π of IPBD populations were lower than CBD populations in general, but samples sizes per population were small (Table 3). TtHap_59 was an IPBD haplotype from a putative CBD specimen collected from Hawaiian waters (M34066), and the introduction of this alien haplotype to the CBD population was regarded as a result of introgression in the distant past (Martien et al. 2012).
Population structure and expansion history for CBD inferred from mtDNA data
Most pairwise F ST and ΦST comparisons were statistically significant (Table 4). Fisher’s exact tests based on haplotype frequencies suggested that the five putative populations were well differentiated, except for the comparison between Northeast and Southeast China (Table S9). The clear differentiation between the Hawaiian Islands and Palmyra Atoll populations has been reported in the original paper (Martien et al. 2012); here, our analysis further reveals that Hawaiian Islands and Palmyra Atoll populations were also differentiated from the WNP populations. Within the WNP, the Northeast China population was the least differentiated, although the statistical insignificance could be largely due to deficient sample size, which was an issue for our Northeast China population sample.
Table 4.
Pairwise mtDNA F ST and φST comparisons among the five putative CBD populations in the western-central North Pacific Ocean
| n | East Japan | SE China | NE China | Hawaii | Palmyra | |
|---|---|---|---|---|---|---|
| East Japan | 160 | 0.041** | 0.020 | 0.114** | 0.107** | |
| SE China | 49 | 0.080** | 0.018 | 0.104** | 0.076** | |
| NE China | 14 | 0.020 | 0.005 | 0.134** | 0.135** | |
| Hawaii | 119 | 0.160** | 0.059** | 0.092** | 0.109** | |
| Palmyra | 11 | 0.533** | 0.304** | 0.434** | 0.243** |
The pairwise F ST value is above the diagonal and the pairwise φST value is below the diagonal
* p < 0.05; ** p < 0.01
A negative Tajima’s D was estimated for all putative populations, although none of the values were significantly different from zero (Table 3). A negative Fu’s Fs was estimated for the East Japan and Southeast China populations, but again, none of the estimates were statistically significant. The only exception was when all samples were pooled together, the Fu’s Fs estimate was negative and significantly different from zero (Table 3). The mismatch distributions for each putative population appeared to be multimodal (Fig. S4), even though fit to the expansion model could only be rejected for the Hawaiian Islands and Northeast China populations (Table S10).
Interspecific comparisons
FCA analyses showed a clear genetic difference between CBD and IPBD (Fig. S5, S6). Using STRUCTURE, both ΔK and LnP(K) values supported K = 2 (Table S11). The ancestry assignment test showed three CBD individuals, from Japan, Taiwan, and Philippines, respectively, that might have had an IPBD grandparent, although the probability was only between 7.1 and 11.9% (Fig. S6; Table S12).
Discussion
CBD population structure in the WNP
Miyashita (1993) suggested the CBD in the WNP is mainly distributed in 30°–42°N and west of 160°E, with a density gap at 142°–145°E as a boundary separating the “Japanese coastal” population (west of 142°E) and “Japanese offshore” population (east of 145°E). That boundary is tentatively supported by a telemetry study showing that the CBD population targeted by the Japanese coastal drive fishery (Kishiro and Kasuya 1993) is unlikely to utilise the waters further than 200 nautical miles from land (Tanaka 1987). Since most of the samples were collected from the dolphins caught in the coastal drive fishery, our East Japan sample very likely represents this Japanese coastal population. Our Geneland analysis further suggests that the range of this Japanese coastal population could be extended further south to the eastern coast of Taiwan (22°–25°N, east of 121°E), and we, therefore, call this the “East Coast Cluster” to avoid confusion. The east coasts of Taiwan and Japan (between 22° and 42°N) are together embedded in a unique oceanic biogeographic province, of which the main characteristic is sharing the speedy, warm, relatively high saline Kuroshio current flowing northeastward from Luzon in the Philippines to the east coast of Japan year-round (Wyrtki 1975; Spalding et al. 2012; Fig. 1). Despite the uncertainty of the habitat preference for this East Coast Cluster CBD population, it is very likely that the strong, constant Kuroshio Current plays a crucial role in defining their habitat (Tanaka 1987). Similar structure of connectivity along the east coasts of Taiwan and Japan is also seen in short-finned pilot whales (Globicephala macrorhynchus) (Chen et al. 2014), Risso’s dolphins (Grampus griseus), and Fraser’s dolphins (Lagenodelphis hosei) (Chen 2016). However, we acknowledge that further fine-scale population sub-division within the Cluster is possible, as it seems to be a common pattern for CBD elsewhere in the world (e.g., Mirimin et al. 2011; Martien et al. 2012; Richards et al. 2013; Gaspari et al. 2015a), but our sample size for eastern Taiwan was too small (n = 4) to reveal such pattern, if it does exist. In contrast to some earlier studies (e.g., Wiszniewski et al. 2010), we found no evidence for sex-biased dispersal, though we cannot rule this out as the test has relatively low power (Goudet 2002).
On the other hand, Kasuya et al. (1997) found a subtle difference in several life history traits (e.g., the body length at sexual maturity, the age of sexual maturity, and the interval of breeding) between CBD caught in Taiji (eastern Japan) and Iki (southwestern Japan), suggesting that CBD populations between the east and west coasts of Japan could be differentiated (cited in Kasuya 2011). This hypothesis is tentatively supported by Hayano (2013), who studied a 520 bp mtDNA control region sequences in 42 CBD from the east and west coasts of Japan and found that seven of the ten samples collected from the west coast were grouped in a unique phylogenetic cluster with a bootstrap support value of 71%. Our FCA and Geneland results also support the differentiation of CBD populations between the west and east coasts of Japan (i.e., between the Sea of Japan and the Pacific coast of Japan), although the sample size from the population west of Japan is too small for robust inference. This pattern has been reported for other cetaceans found in the same region, e.g., in minke whales (Balaenoptera acutorostrata; Abe et al. 2000) and Dall’s porpoises (Phocoenoides dalli; Hayano et al. 2003).
Our results further reveal that there may be another coastal CBD population in the vicinity of the Taiwan Strait (western Taiwan), although its relationship with the CBD population from West Japan (in the Sea of Japan) is ambiguous, and so we grouped the samples together as a West Coast Cluster. Earlier studies for bottlenose dolphins in the coastal region of WNP mainly focused on the ecological, morphometric, and genetic differences between CBD and IPBD (Gao et al. 1995; Wang et al. 1999, 2000; Yang et al. 2005; Kurihara and Oda 2007), providing limited insight into the population differentiation within the CBD species. Our study highlights the need for further careful investigations into CBD in this region, including the distribution, habitat preferences, or behaviours of the dolphins, to shed more light on the evolutionary mechanisms driving the CBD populations in the Asian coastal waters to differentiate.
Our FCA and Geneland results suggest a fairly distinct population of CBD inhabiting Philippine waters, though the sample size is too small for strong inference. One of the samples could possibly be a hybrid with other delphinid species; however, we have insufficient data to be precise about which one. Dolar et al. (2006) reported that the CBD population in central Philippine waters (Sulu Sea and the Tañon Strait) was found exclusively in shallow and intermediate waters inside of the shelf break, and that this preference may limit the dispersal of the Philippine population. Although various lines of evidence indicate that possibility, more data are needed to resolve this question.
Population dynamics of the CBD in the WNP
The N e estimated for the West Coast Cluster and the East Coast Cluster are only about a quarter to a tenth of the N e estimated for the more pelagic central Pacific populations (Martien et al. 2012). This agrees with an earlier report that suggests that the N e for coastal CBD populations tends to be smaller than for pelagic populations (Louis et al. 2014a). Our calculation showed the N e N −1 estimates for both West and East Coast Clusters are similar in magnitude, ranging between 0.042 and 0.118. This range is consistent with estimates proposed by meta-analysis of N e N −1for wildlife populations (Frankham 1995).
Tajima’s D and Fu’s Fs estimates were statistically insignificant and the mismatch distributions appeared multimodal in both demographic and spatial models (though not always significantly different from the model for expansion). In general, there was no strong evidence for population expansion.
Possible mechanisms that shape the CBD population structure in the WNP
The migrate analysis suggests that long-term gene flow between the East and West Coast Clusters is limited to less than one migrant per generation. On the other hand, the GeneClass analysis identified three contemporary first-generation migrants, suggesting the presence of ongoing gene flow between the two populations. Low levels of gene flow have been reported between the coastal and pelagic populations in the eastern North Atlantic Ocean (Louis et al. 2014a), and among the regional populations around the Hawaiian Islands (Martien et al. 2012). It has been proposed that this could be promoted by assortative mating due to the constrained preference of natal habitat, specialised diet, and possibly culture familiarity (Hoelzel et al. 1998; Möller et al. 2007; Cantor and Whitehead 2013; Louis et al. 2014a, b). In our case, although the strong correspondence between the population structure and contrasting oceanographic features (i.e., shallow continental shelves vs. sharp continental slopes) implies population-specific resource preference, the structure may have been promoted by historic isolation (possibly during the glacial period). We propose that the Kuroshio current could play an important role in “regulating” the frequency of gene exchange.
During the glacial period, the influence of the Kuroshio current on the coasts of the eastern Asian continent was weakened as the flows to the East China Sea and South China Sea were limited (Ijiri et al. 2005; Jiang et al. 2006). The Tsushima Warm Current, a branch of Kuroshio Current carrying warm water into the Sea of Japan through the Tsushima Strait, was suspended during the Last Glacial Maximum (Itaki et al. 2004). Therefore, the oceanography of the region may not have promoted connectivity during the glacial period as much as today, and the lack of warmer water introduced by the Kuroshio Current from the south could have generated more contrasting physical conditions between the shallower western coastal and the deep eastern continental slope habitats. This habitat distinction may have also reinforced the reduction of connectivity between the two populations during that period. In contrast, today, the current itself and its branch currents constantly drive the surface waters in and out the shallow coastal region (Cho et al. 2009; Matsuno et al. 2009; Jan et al. 2010), and it has been observed that the movement of CBD can be influenced by the flow of Kuroshio Current (Tanaka 1987). Our data suggest recent migration between these regions, which may reflect this environmental facilitation (though the populations remain differentiated).
Sympatric relationship between IPBD and CBD
Our mtDNA data agree with the previous studies showing clear phylogenetic differentiation between IPBD and CBD (Wang et al. 1999; Kakuda et al. 2002; Natoli et al. 2004; Yang et al. 2005; Kita et al. 2013; Moura et al. 2013). Our microsatellite data also exhibit a distinct difference between IPBD and CBD. The microsatellite data also provide some indication of limited gene flow between these two species, indicating three individuals that might have hybrid ancestry. The two species are known to have interbred and produced reproductively viable female hybrids in a captive environment (Hale et al. 2000), and potential descendants of hybrids between the two species are found in the CBD populations in Hawaiian and Japanese waters (Martien et al. 2012; Hayano 2013). However, we cannot eliminate the possibility that the interbreeding was between CBD and other delphinid species that are closely related to IPBD but live sympatrically with the CBD, such as pantropical spotted dolphin (Stenella attenuata), striped dolphin (Stenella coeruleoalba), spinner dolphin (Stenella longirostris), common dolphin (Delphinus sp.), or Fraser’s dolphin (LeDuc et al. 1999; Kingston et al. 2009; Möller et al. 2008; McGowen 2011; Amaral et al. 2012). This has the potential to produce misleading results when the signal for hybridisation is weak, as in the case of our study. In fact, among four potential hybridisations between the Tursiops congeneric species (three from this study and one from Martien et al. 2012), one of the putative hybrid animals was similar in appearance to Fraser’s dolphin, and two (one form eastern Taiwan and the other from Hawaiian Islands) were sampled from a region where the occurrence of IPBD has never been documented (Yang et al. 1999; Chou 2007; Baird et al. 2013). Since there is evidence of polyphyly among the Tursiops–Stenella–Delphinius complex of species (LeDuc et al. 1999; Kingston et al. 2009; McGowen 2011; Amaral et al. 2012), the evidence for hybridisation, therefore, needs to be interpreted carefully.
Our genetic data for IPBD were obtained from three putative aggregation sites in Taiwanese and Japanese waters: Amakusa (southwestern Japan), Mikura Islands (southeastern Japan), and western Taiwan; and the FCA result showed distinct clustering for those samples (Fig. S5). The microsatellite data are consistent with the population structure proposed based on mtDNA data (Hayano 2013), but to verify the hypothesis of IPBD population structure in these waters, further examination using more samples from the same and further sites is necessary.
Conservation implications
Our results indicate that there are at least two populations of CBD distributed parapatrically in the coastal waters around Taiwan and Japan, corresponding to the distribution of shallow continental shelf or deep continental slope habitats. Further sampling may well reveal further structure in the broader region. Although our analyses detected some recent immigration, the long-term estimates show limited gene flow between the two populations. This potentially agrees with earlier analyses that show that habitat specialisation plays an important role in differentiating inshore and offshore populations (Hoelzel et al. 1998; Möller et al. 2007; Louis et al. 2014b; Gaspari et al. 2015b). The two CBD populations are likely affected by different anthropogenic threats. For instance, the small-scale dolphin drive fishery appears to target primarily the East Coast Cluster CBD (see Kasuya 2011). On the west coast, habitat loss and degradation, pollution, acoustic disturbances, and fisheries interactions have been identified as risks for coastal cetacean species (Perrin et al. 2005; Jefferson et al. 2009; Choi et al. 2013; Slooten et al. 2013). It is, therefore, justifiable to manage them as separate CBD populations, and further investigations are needed to evaluate their resistance to those threats. We address our aim to improve understanding of the pattern and origin of population structure in this region by identifying previously unrecognised boundaries and illustrating the historical and environmental contexts. This contributes both towards the more effective conservation of species in this genus, and towards a better understanding of processes associated with population differentiation in this region.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Kelly Robertson at the Southwest Fishery Science Center, National Oceanic and Atmospheric Administration (USA), Shinsuke Tanabe, and staffs at the es-BANK, Joint Usage/Research Center LaMer, at the Center for Marine Environmental Studies, Ehime University (Japan), Lien-Siang Chou at the National Taiwan University, staffs and volunteers at the Taiwan Cetacean Society (Taiwan), and Tadasu K. Yamada at the National Museum of Science (Japan) for their assistance on sample collection, administration, and shipping. IC would like to thank American Genetic Association and University College, University of Durham for sponsoring conference travel grants. The study was funded by the International Whaling Commission Small Cetaceans Research Fund (2012/13) and the Government Scholarship (2011) for IC provided by the Ministry of Education, Government of Taiwan.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest and the consent was obtained from all participants of the study.
Ethical standard
All specimens were collected according to the national legislation, acquired from reputable academic institutions, transported to, and examined in the Molecular Ecology Group at Durham University, with valid official permits issued by the authorities of Japan, Taiwan, United States, and United Kingdom.
Data accessibility
Sequence data are available at Genbank under accession numbers MF805937-MF806018, and microsatellite DNA genotypes are available in the supplementary file in Table S13.
Footnotes
Reviewed by Undisclosed experts.
Electronic supplementary material
The online version of this article (doi:10.1007/s00227-017-3232-8) contains supplementary material, which is available to authorized users.
References
- Abe H, Goto M, Pastene LA (2000) Population structure in the WNP minke whale inferred from microsatellite analysis. Paper SC/F2 K/J10 presented in the Workshop to Review the Japanese Whale Research Programme under Special Permit for North Pacific Minke Whales (JARPN), February 2000
- Allendorf FW, Luikart G. Conservation and the genetics of populations. London: Wiley-Blackwell; 2006. [Google Scholar]
- Amano M. A report on the public symposium part 1 “Recent Advances in Zoogeography” at the Annual Meeting of the Mammalogical Society of Japan (2006): population differentiation of small odontocetes around Japan. Mamm Sci. 2007;47:115–119. [Google Scholar]
- Amaral AR, Jackson JA, Möller LM, Beheregaray LB, Coelho MM. Species tree of a recent radiation: the subfamily Delphininae (Cetacea, Mammalia) Mol Phylogenet Evol. 2012;64:243–253. doi: 10.1016/j.ympev.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Baird RW, Webster DL, Aschettino JM, Schorr GS, McSweeney DJ. Odontocete cetaceans around the main Hawaiian Islands: habitat use and relative abundance from small-boat sighting surveys. Aquat Mamm. 2013;39:253–269. [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. PNAS. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France). http://kimura.univ-montp2.fr/genetix/. Accessed 26 June 2015
- Bilgmann K, Möller LM, Harcourt RG, Kemper CM, Beheregaray LB. The use of carcasses for the analysis of cetacean population genetic structure: a comparative study in two dolphin species. PLoS ONE. 2011;6:e20103. doi: 10.1371/journal.pone.0020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell RL Jr., Funahashi N (2013) Status of small local populations of Indi-Pacific bottlenose dolphins (Tursiops aduncus) in the Japanese Archipelago. Paper SC/65a/SM26 presented to the International Whaling Commission, Scientific Committee (SC65a meeting, Jeju Island, Republic of Korea, 2013)
- Cantor M, Whitehead H. The interplay between social networks and culture: theoretically and among whales and dolphins. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120340. doi: 10.1098/rstb.2012.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Robb K, Taylor AC, McKechnie SW. Population genetic structure of the Burrunan dolphin (Tursiops australis) in coastal waters of south-eastern Australia: conservation implications. Conserv Genet. 2015;16:195–207. [Google Scholar]
- Chen I (2016) Population genetics of Risso’s dolphins (Grampus griseus), Fraser’s dolphins (Lagenodelphis hosei) and bottlenose dolphins (Tursiops spp.) in the North Pacific Ocean. Ph.D. thesis, University of Durham
- Chen I, Yu H-Y, Yang W-C, Nishida S, Isobe T, Tanabe S, Watson A, Chou L-S. The “Southern form” of short-finned pilot whale (Globicephala macrorhynchus) in tropical west Pacific Ocean off Taiwan. Raffles Bull Zool. 2014;62:188–199. [Google Scholar]
- Cho YK, Seo GH, Choi BJ, Kim S, Kim YG, Youn YH, Dever EP. Connectivity among straits of the northwest Pacific marginal seas. J Geophys Res. 2009;114:C06018. [Google Scholar]
- Choi M, An YR, Park KJ, Lee IS, Hwang DW, Kim J, Moon HB. Accumulation of butyltin compounds in finless porpoises (Neophocaena asiaeorientalis) from Korean coast: tracking the effectiveness of TBT regulation over time. Marine Poll Bull. 2013;66:78–83. doi: 10.1016/j.marpolbul.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Chou L-S (2007) The population estimation and habitats of cetacean in coastal waters of Taiwan. Final Report to Fishery Agency, Council of Agriculture, Republic of China No. 96AS-15.1.1-FA-F3(2). Available from Agriculture Science Information Center, 3F, No. 14, Wunjhou St. Taipei City 10648 Taiwan
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolar MLL, Perrin WF, Taylor BL, Kooyman GL, Alava MNR. Abundance and distributional ecology of cetaceans in the central Philippines. J Cetacean Res Manag. 2006;8:93–111. [Google Scholar]
- Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol. 2004;13:853–864. doi: 10.1046/j.1365-294x.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Baird SJ, Piry S, Ray N, Tolley KA, Duke S, Birkun A, Ferreira M, Jauniaux T, Llavona Á, et al. Rise of oceanographic barriers in continuous populations of a cetacean: the genetic structure of harbour porpoises in Old World waters. BMC Biol. 2007;5:30. doi: 10.1186/1741-7007-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Fu Y-X. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao AL, Zhou KY, Wang YM. Geographical variation in morphology of bottlenosed dolphins (Tursiops sp.) in Chinese waters. Aquat Mamm. 1995;21:121–135. [Google Scholar]
- Gaspari S, Holcer D, Mackelworth P, Fortuna C, Frantzis A, Genov T, Vighi M, Natali C, Rako N, Banchi E, et al. Population genetic structure of common bottlenose dolphins (Tursiops truncatus) in the Adriatic Sea and contiguous regions: implications for international conservation. Aquat Conserv. 2015;25:212–222. [Google Scholar]
- Gaspari S, Scheinin A, Holcer D, Fortuna C, Natali C, Genov T, Frantzis A, Chelazzi G, Moura A. Drivers of population structure of the bottlenose dolphin (Tursiops truncatus) in the eastern Mediterranean Sea. Evol Biol. 2015;42:177–190. [Google Scholar]
- Goudet J (2002) Fstat 2.9.3.2. The website of the population genetics laboratory at University of Lausanne. http://www2.unil.ch/popgen/softwares/fstat.htm. Accessed 26 June 2015
- Goudet J, Perrin N, Waser P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol. 2002;11:1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- Guillot G, Estoup A, Mortier F, Cosson JF. A spatial model for landscape genetics. Genetics. 2005;170:1261–1280. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale PT, Barreto AS, Ross GJB. Comparative morphology and distribution of the aduncus and truncatus forms of bottlenose dolphin Tursiops in the Indian and Western Pacific Oceans. Aquat Mamm. 2000;26:101–110. [Google Scholar]
- Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- Hayano A. Genetic composition of genus Tursiops in Japanese waters inferred from mitochondrial DNA analysis. Kaiyo Monthly. 2013;45:341–347. [Google Scholar]
- Hayano A, Amano M, Miyazaki N. Phylogeography and population structure of the Dall’s porpoise, Phocoenoides dalli, in Japanese waters revealed by mitochondrial DNA. Genes Genet Syst. 2003;78:81–91. doi: 10.1266/ggs.78.81. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR. Molecular ecology. In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. 2. San Diego: Academic; 2009. pp. 736–741. [Google Scholar]
- Hoelzel AR, Hancock JM, Dover GA. Evolution of the cetacean mitochondrial D-loop region. Mol Biol Evol. 1991;8:475–493. doi: 10.1093/oxfordjournals.molbev.a040662. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR, Potter CW, Best PB. Genetic differentiation between parapatric ‘nearshore’ and ‘offshore’ populations of the bottlenose dolphin. Proc R Soc Lond B Biol Sci. 1998;265:1177–1183. doi: 10.1098/rspb.1998.0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel AR, Hey J, Dahlheim ME, Nicholson C, Burkanov V, Black N. Evolution of population structure in a highly social top predator, the killer whale. Mol Biol Evol. 2007;24:1407–1415. doi: 10.1093/molbev/msm063. [DOI] [PubMed] [Google Scholar]
- Hollatz C, Vilaça ST, Redondo RAF, Marmontel M, Baker CS, Santos FR. The Amazon River system as an ecological barrier driving genetic differentiation of the pink dolphin (Inia geoffrensis) Biol J Linnean Soc. 2011;102:812–827. [Google Scholar]
- Ijiri A, Wang L, Oba T, Kawahata H, Huang CY, Huang CY. Paleoenvironmental changes in the northern area of the East China Sea during the past 42,000 years. Palaeogeogr Palaeoclimatol Palaeoecol. 2005;219:239–261. [Google Scholar]
- Itaki T, Ikehara K, Motoyama I, Hasegawa S. Abrupt ventilation changes in the Japan Sea over the last 30 ky: evidence from deep-dwelling radiolarians. Palaeogeogr Palaeoclimatol Palaeoecol. 2004;208:263–278. [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jan S, Tseng Y-H, Dietrich DE. Sources of water in the Taiwan Strait. J Oceanogr. 2010;66:211–221. [Google Scholar]
- Jefferson TA, Hung SK, Würsig B. Protecting small cetaceans from coastal development: impact assessment and mitigation experience in Hong Kong. Mar Policy. 2009;33:305–311. [Google Scholar]
- Jiang H, Björck S, Ran L, Huang Y, Li J. Impact of the Kuroshio Current on the South China Sea based on a 115,000 year diatom record. J Quat Sci. 2006;21:377–385. [Google Scholar]
- Kakuda T, Tajima Y, Arai K, Kogi K, Hishii T, Yamada TK. On the resident “bottlenose dolphins” from Mikura waters. Mem Natl Sci Mus (Tokyo) 2002;38:255–272. [Google Scholar]
- Kasuya T. Japanese whaling and other cetacean fisheries. Environ Sci Pollut R. 2007;14:39–48. doi: 10.1065/espr2006.09.346. [DOI] [PubMed] [Google Scholar]
- Kasuya T. Conservation biology of small cetaceans around Japan. Tokyo: University of Tokyo Press; 2011. [Google Scholar]
- Kasuya T, Izumisawa Y, Komyo Y, Ishino Y, Maejima Y. Life history parameters of bottlenose dolphins off Japan. IBI Rep. 1997;7:71–107. [Google Scholar]
- Kemper C. Osteological variation and taxonomic affinities of bottlenose dolphins, Tursiops spp., from South Australia. Aust J Zool. 2004;52:29–48. [Google Scholar]
- Kingston SE, Rosel PE. Genetic differentiation among recently diverged delphinid taxa determined using AFLP markers. J Hered. 2004;95:1–10. doi: 10.1093/jhered/esh010. [DOI] [PubMed] [Google Scholar]
- Kingston SE, Adams LD, Rosel PE. Testing mitochondrial sequences and anonymous nuclear markers for phylogeny reconstruction in a rapidly radiating group: molecular systematics of the Delphininae (Cetacea: Odontoceti: Delphinidae) BMC Evol Biol. 2009;9:245. doi: 10.1186/1471-2148-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishiro T, Kasuya T. Review of Japanese dolphin drive fisheries and their status. Rep Int Whal Commn. 1993;43:439–452. [Google Scholar]
- Kita YF, Hosomichi K, Suzuki S, Inoko H, Shiina T, Watanabe M, Tanaka A, Horie T, Ohizumi H, Tanaka S, et al. Genetic and family structure in a group of 165 common bottlenose dolphins caught off the Japanese coast. Mar Mamm Sci. 2013;29:474–496. [Google Scholar]
- Kogi K, Hishii T, Imamura A, Iwatani T, Dudzinski KM. Demographic parameters of Indo-Pacific bottlenose dolphins (Tursiops aduncus) around Mikura Island, Japan. Mar Mamm Sci. 2004;20:510–526. [Google Scholar]
- Krützen M, Valsecchi E, Connor RC, Sherwin WB. Characterization of microsatellite loci in Tursiops aduncus. Mol Ecol Notes. 2001;1:170–172. [Google Scholar]
- Kurihara N, Oda S. Cranial variation in bottlenose dolphins Tursiops spp. from the Indian and western Pacific Oceans: additional evidence for two species. Acta Theriol. 2007;52:403–418. [Google Scholar]
- Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet. 2006;7:295–302. [Google Scholar]
- LeDuc RG, Perrin WF, Dizon AE. Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Mar Mamm Sci. 1999;15:619–648. [Google Scholar]
- Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Louis M, Viricel A, Lucas T, Peltier H, Alfonsi E, Berrow S, Brownlow A, Covelo P, Dabin W, Deaville R, et al. Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-East Atlantic. Mol Ecol. 2014;23:857–874. doi: 10.1111/mec.12653. [DOI] [PubMed] [Google Scholar]
- Louis M, Fontaine MC, Spitz J, Schlund E, Dabin W, Deaville R, Caurant F, Cherel Y, Guinet C, Simon-Bouhet B. Ecological opportunities and specializations shaped genetic divergence in a highly mobile marine top predator. Proc R Soc Lond B Biol Sci. 2014;281:20141558. doi: 10.1098/rspb.2014.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther-Thieleking JL, Archer FI, Lang AR, Weller DW. Genetic differentiation among coastal and offshore common bottlenose dolphins, Tursiops truncatus, in the eastern North Pacific Ocean. Mar Mamm Sci. 2015;31:1–20. [Google Scholar]
- Martien KK, Baird RW, Hedrick NM, Gorgone AM, Thieleking JL, McSweeney DJ, Robertson KM, Webster DL. Population structure of island-associated dolphins: evidence from mitochondrial and microsatellite markers for common bottlenose dolphins (Tursiops truncatus) around the main Hawaiian Islands. Mar Mamm Sci. 2012;28:E208–E232. [Google Scholar]
- Matsuno T, Lee J-S, Yanao S. The Kuroshio exchange with the South and East China Seas. Ocean Sci. 2009;5:303–312. [Google Scholar]
- McGowen MR. Toward the resolution of an explosive radiation—A multilocus phylogeny of oceanic dolphins (Delphinidae) Mol Phylogenet Evol. 2011;60:345–357. doi: 10.1016/j.ympev.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Mirimin L, Miller R, Dillane E, Berrow SD, Ingram S, Cross TF, Rogan E. Fine-scale population genetic structuring of bottlenose dolphins in Irish coastal waters. Anim Conserv. 2011;14:342–353. [Google Scholar]
- Miyashita T. Abundance of dolphin stocks in the western North Pacific taken by the Japanese drive fishery. Rep Int Whal Commn. 1993;43:417–437. [Google Scholar]
- Möller LM, Wiszniewski J, Allen SJ, Beheregaray LB. Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar Freshw Res. 2007;58:640–648. [Google Scholar]
- Möller LM, Bilgmann K, Charlton-Robb K, Beheregaray L. Multi-gene evidence for a new bottlenose dolphin species in southern Australia. Mol Phylogenet Evol. 2008;49:674–681. doi: 10.1016/j.ympev.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Morisaka T, Shinohara M, Nakahara F, Akamatsu T. Geographic variations in the whistles among three Indo-Pacific bottlenose dolphin Tursiops aduncus populations in Japan. Fish Sci. 2005;71:568–576. [Google Scholar]
- Moura AE, Nielsen SCA, Vilstrup JT, Moreno-Mayar JV, Gilbert MT, Gray HW, Natoli A, Möller L, Hoelzel AR. Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst Biol. 2013;62:865–877. doi: 10.1093/sysbio/syt051. [DOI] [PubMed] [Google Scholar]
- Nanbu Y, Hirose J, Kubo N, Kishiro T, Shinomiya A. Location and number of individuals of Indo-Pacific bottlenose dolphins (Tursiops aduncus) in Kagoshima Bay. Mem Fac Fish Kagoshima Univ. 2006;55:51–60. [Google Scholar]
- Natoli A, Peddemors VM, Hoelzel AR. Population structure and speciation in the genus Tursiops based on microsatellite and mitochondrial DNA analyses. J Evol Biol. 2004;17:363–375. doi: 10.1046/j.1420-9101.2003.00672.x. [DOI] [PubMed] [Google Scholar]
- Oremus M, Garrigue C, Tezanos-Pinto G, Baker CS. Phylogenetic identification and population differentiation of bottlenose dolphins (Tursiops spp.) in Melanesia, as revealed by mitochondrial DNA. Mar Mamm Sci. 2015;31:1035–1056. [Google Scholar]
- Oremus M, Leqata J, Baker CS. Resumption of traditional drive hunting of dolphins in the Solomon Islands in 2013. R Soc Open Sci. 2015;2:140524. doi: 10.1098/rsos.140524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Parsons KM, Durban JW, Claridge DE, Herzing DL, Balcomb KC, Noble LR. Population genetic structure of coastal bottlenose dolphins (Tursiops truncatus) in the northern Bahamas. Mar Mamm Sci. 2006;22:276–298. [Google Scholar]
- Perrin WF, Reeves RR, Dolar MLL, Jefferson TA, Marsh H, Wang JY, Estacion J, editors. Report of the second workshop on the biology and conservation of small cetaceans and dugongs of South-East Asia. Bonn: UNEP/CMS Secretariat; 2005. [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N, Currat M, Excoffier L. Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol. 2003;20:76–86. doi: 10.1093/molbev/msg009. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Rice DW. Marine mammals of the world. Systematics and distribution. Lawrence: Society for Marine Mammalogy; 1998. [Google Scholar]
- Richards VP, Greig TW, Fair PA, McCulloch SD, Politz C, Natoli A, Driscoll CA, Hoelzel AR, David V, Bossart GD, et al. Patterns of population structure for inshore bottlenose dolphins along the eastern United States. J Hered. 2013;104:765–778. doi: 10.1093/jhered/est070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robards MD, Reeves RR. The global extent and character of marine mammal consumption by humans: 1970–2009. Biol Conserv. 2011;144:2770–2786. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel PE, Hansen L, Hohn AA. Restricted dispersal in a continuously distributed marine species: common bottlenose dolphins Tursiops truncatus in coastal waters of the western North Atlantic. Mol Ecol. 2009;18:5030–5045. doi: 10.1111/j.1365-294X.2009.04413.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura I, Rocha-Olivares A, Flores-Ramírez S, Rojas-Brachoc L. Conservation implications of the genetic and ecological distinction of Tursiops truncatus ecotypes in the Gulf of California. Biol Conserv. 2006;133:336–346. [Google Scholar]
- Sellas AB, Wells RS, Rosel PE. Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv Genet. 2005;6:715–728. [Google Scholar]
- Shinohara M, Domingo-Roura X, Takenaka O. Microsatellites in the bottlenose dolphin Tursiops truncatus. Mol Ecol. 1997;6:695–696. doi: 10.1046/j.1365-294x.1997.00231.x. [DOI] [PubMed] [Google Scholar]
- Shirakihara M, Shirakihara K, Tomonaga J, Takatsuki M. A resident population of Indo-Pacific bottlenose dolphins (Tursiops aduncus) in Amakusa, western Kyushu, Japan. Mar Mamm Sci. 2002;18:30–41. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slooten E, Wang JY, Dungan SZ, Forney KA, Hung SK, Jefferson TA, Riehl KN, Rojas-Bracho L, Ross PS, Wee A, et al. Impacts of fisheries on the critically endangered humpback dolphin Sousa chinensis population in the eastern Taiwan Strait. Endanger Species Res. 2013;22:99–114. [Google Scholar]
- Spalding MD, Agostini VN, Rice J, Grant SM. Pelagic provinces of the world: a biogeographic classification of the world’s surface pelagic waters. Ocean Coast Manag. 2012;60:19–30. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. Satellite radio tracking of bottlenose dolphins Tursiops truncatus. Nippon Suisan Gakkaishi. 1987;53:1327–1338. [Google Scholar]
- Tezanos-Pinto G, Baker CS, Russell K, Martien K, Baird RW, Hutt A, Stone G, Mignucci-Giannoni AA, Caballero S, Endo T, et al. A worldwide perspective on the population structure and genetic diversity of bottlenose dolphins (Tursiops truncatus) in New Zealand. J Hered. 2009;100:11–24. doi: 10.1093/jhered/esn039. [DOI] [PubMed] [Google Scholar]
- Wang JY (2000) Cetaceans in the waters of Kenting National Park and adjacent regions of southern Taiwan. Kenting National Park Conservation Research Report 107. Kenting National Park, Pingtung, Taiwan
- Wang JY, Yang S-C. Indo-Pacific bottlenose dolphin Tursiops aduncus. In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. 2. San Diego: Academic; 2009. pp. 602–608. [Google Scholar]
- Wang JY, Chou L-S, White BN. Mitochondrial DNA analysis of sympatric morphotypes of bottlenose dolphins (genus: Tursiops) in Chinese waters. Mol Ecol. 1999;8:1603–1612. doi: 10.1046/j.1365-294x.1999.00741.x. [DOI] [PubMed] [Google Scholar]
- Wang JY, Chou L-S, White BN. Differences in the external morphology of two sympatric species of bottlenose dolphins (genus Tursiops) in the waters of China. J Mammal. 2000;81:1157–1165. [Google Scholar]
- Wells RS, Scott MD. Common bottlenose dolphin Tursiops truncatus. In: Perrin WF, Würsig B, Thewissen JGM, editors. Encyclopedia of marine mammals. 2. San Diego: Academic; 2009. pp. 249–255. [Google Scholar]
- Whitaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM. Likelihood-Based estimation of micro-satellite mutation rates. Genetics. 2003;164:781–787. doi: 10.1093/genetics/164.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewski J, Beheregaray LB, Allen SJ, Möller LM. Environmental and social influences on the genetic structure of bottlenose dolphins (Tursiops aduncus) in Southeastern Australia. Conserv Genet. 2010;11:1405–1419. [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wyrtki K. Fluctuations of the dynamic topography in the Pacific Ocean. J Phys Oceanogr. 1975;5:450–459. [Google Scholar]
- Yang S-C, Liao H-C, Pan C-L, Wang JY. A survey of cetaceans in the waters of central-eastern Taiwan. Asian Mar Biol. 1999;16:23–34. [Google Scholar]
- Yang G, Ji G, Ren W, Zhou K. Pattern of genetic variation of bottlenose dolphins in Chinese waters. Raffles Bull Zool. 2005;53:157–164. [Google Scholar]
- Young NM, Iudicello S (2007) Worldwide bycatch of cetaceans. NOAA Technical Memorandum NMFS-OPR-36. Office of International Affairs, NMFS, NOAA, Silver Spring, MD
- Zhou K, Qian W. Distribution of the dolphins of the genus tursiops (sic) in the China Seas. Aquat Mamm. 1985;11:16–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available at Genbank under accession numbers MF805937-MF806018, and microsatellite DNA genotypes are available in the supplementary file in Table S13.