Abstract
It has long been appreciated that insulin action is closely tied to circadian rhythms. However, the mechanisms that dictate diurnal insulin sensitivity in metabolic tissues are not well understood. Retinol-binding protein 4 (RBP4) has been implicated as a driver of insulin resistance in rodents and humans, and it has become an attractive drug target in type II diabetes. RBP4 is synthesized primarily in the liver where it binds retinol and transports it to tissues throughout the body. The retinol–RBP4 complex (holo-RBP) can be recognized by a cell-surface receptor known as stimulated by retinoic acid 6 (STRA6), which transports retinol into cells. Coupled to retinol transport, holo-RBP can activate STRA6-driven Janus kinase (JAK) signaling and downstream induction of signal transducer and activator of transcription (STAT) target genes. STRA6 signaling in white adipose tissue has been shown to inhibit insulin receptor responses. Here, we examined diurnal rhythmicity of the RBP4/STRA6 signaling axis and investigated whether STRA6 is necessary for diurnal variations in insulin sensitivity. We show that adipose tissue STRA6 undergoes circadian patterning driven in part by the nuclear transcription factor REV-ERBα. Furthermore, STRA6 is necessary for diurnal rhythmicity of insulin action and JAK/STAT signaling in adipose tissue. These findings establish that holo-RBP and its receptor STRA6 are potent regulators of diurnal insulin responses and suggest that the holo-RBP/STRA6 signaling axis may represent a novel therapeutic target in type II diabetes.
Keywords: adipose tissue, circadian, insulin, Janus kinase (JAK), vitamin A, circadian, insulin
Introduction
Obesity and type II diabetes have become a global epidemic, affecting one-third of adults in the United States alone, and the numbers are on the rise (1–3). Insulin resistance is a major driver of type II diabetes as it results in diminished insulin-stimulated signaling cascades that regulate many metabolic processes, including the transport of glucose into skeletal muscle and adipose tissue and proper suppression of hepatic gluconeogenesis in the liver (4). Therefore, understanding the factors regulating insulin signaling under physiological and pathological circumstances is essential for discovering new treatments for type II diabetes.
It is well appreciated that disruption of circadian rhythms contributes to insulin resistance and the development of type II diabetes (5, 6). In mammals, the circadian clocks in the hypothalamic suprachiasmatic nucleus (SCN)2 and peripheral tissues coordinate insulin responsiveness and energy metabolism, and they synchronize endogenous physiological processes with the external environment. One of the main features of circadian rhythm is that they can be entrained by external cues termed Zeitgebers (ZT) such as light, temperature, or food. Circadian rhythms oscillate at a nearly 24-h period, and in mammals circadian oscillations of metabolic gene expression are primarily regulated by a network of transcriptional feedback loops (7). At the core of this mechanism are the heterodimeric transcription factors CLOCK and BMAL1, which activate the transcription of other clock genes, Period (Per) and Cryptochrome (Cry), via E-box elements in their promoters. PER and CRY dimerize in the cytosol and translocate to the nucleus, and as a negative feedback loop, they bind to CLOCK/BMAL1 that inhibits the ability of the complex to transactivate. Consequently, PER/CRY inhibit their own gene expression, thereby lifting repression of CLOCK/BMAL1, allowing for the positive arm of the loop to begin again (7). Additional nuclear transcription factors such as REV-ERBα (Nr1d1) act to fine-tune the core clock (8, 9). Therefore, it is through the temporal oscillation of circadian transcription factors that bind to the promoters of metabolic genes that regulate their circadian expression.
It is well established that insulin sensitivity follows a diurnal pattern (10, 11). Humans are more insulin-sensitive during the morning versus the evening (11). This phenomenon can partially explain why plasma glucose levels are maintained at a narrow range during distinct daily metabolic phases. During daytime feeding, tissues are more insulin-sensitive, and serum glucose is efficiently cleared and utilized. Alternatively, during the night-time fast, tissues are relatively insulin-resistant, which assists with glucose level maintenance (11, 12). Mice, however, are nocturnal mammals, and it is known that their hormonal rhythms, behavior, and insulin sensitivity are opposite relative to the light/dark cycle compared with humans (13, 14). It has been shown in humans that circadian disruption due to sleep disorders, shift work, or jet lag can promote obesity and insulin resistance, and it even increases the risk of breast and colon cancer (12, 15–18). Global and adipose-specific clock gene mutants display dysregulated glucose metabolism (19–21). However, it remains poorly understood what circadian factors ultimately control diurnal insulin sensitivity.
A gain of function mutation in retinol-binding protein 4 (RBP4) was discovered to increase the risk of type II diabetes by 80% in human homozygous carriers of the mutation (22). RBP4 is the serum carrier for vitamin A (retinol, ROH) that is produced and secreted primarily from the liver (23, 24). The retinol–RBP4 complex (holo-RBP) can be recognized by a cell-surface receptor known as stimulated by retinoic acid 6 (STRA6) (25). STRA6 is expressed in white adipose tissue (WAT) and skeletal muscle, yet it is not expressed in the liver (25–27). Upon binding of extracellular holo-RBP, STRA6 transports ROH from holo-RBP across the plasma membrane, and it also triggers a JAK2/STAT3 or STAT5 signaling cascade depending on cell type (28, 29). It has been reported that obese and insulin-resistant mice and humans show elevated serum levels of RBP4 (30, 31). Furthermore, RBP4 has been shown to promote insulin resistance in mice (30). The discovery that STRA6 activates JAK/STAT signaling provides a rationale for how RBP4 causes insulin resistance. Specifically, it was shown that STRA6 expression was essential for RBP-induced STAT target gene expression in WAT and skeletal muscle (28). STRA6-driven activation of JAK/STAT signaling activates transcription of the gene-encoding suppressor of cytokine signaling 3 (Socs3), a potent inhibitor of insulin receptor responses (28, 32, 33) Utilizing STRA6-null mice, it was demonstrated that STRA6 is required for RBP-induced insulin resistance (32).
Considering that RBP4 activation of STRA6 regulates insulin receptor responses in WAT, we hypothesized that STRA6 in white adipose tissue may regulate diurnal insulin responses under normal physiological conditions. Here, we directly tested whether STRA6 may display circadian patterns of expression and signaling in WAT, and we determined whether STRA6 is necessary for diurnal variations of insulin responses in WAT.
Results
Key players in vitamin A metabolism and signaling display circadian rhythmicity in white adipose tissue
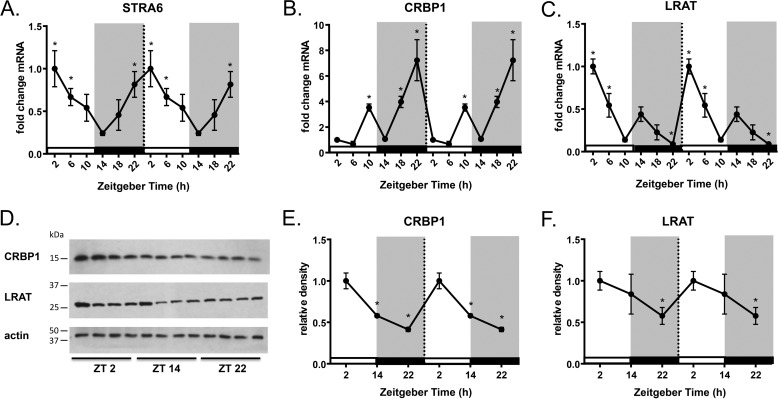
To investigate whether the holo-RBP/STRA6 signaling cascade is under circadian control in white adipose tissue, we collected tissue from ad libitum chow-fed mice every 4 h over a 24-h period. Stra6 mRNA expression displays circadian rhythmicity with a significant peak observed at ZT22-2 and a nadir observed at ZT14 with a 4-fold increase in expression compared with ZT2 (Fig. 1A). Increased Stra6 expression at ZT2 corresponds to the light/inactive phase when mice are known to be relatively insulin-resistant compared with the dark/active phase (34). Measurement of STRA6 protein expression is not currently feasible due to the lack of specific antibodies.
Figure 1.
Stra6, Crbp1, and Lrat display circadian patterns of expression in WAT. A–C, Stra6, Crbp1, and Lrat mRNA expression from epididymal WAT collected every 4 h over a 24-h period determined by quantitative real-time PCR. Data shown are double-plotted data. ZT0 refers to the hour mice are first exposed to light. D, Western blot analysis for CRBP1 and LRAT protein expression from epididymal WAT at indicated ZTs. E and F, quantification of Western blot analysis for CRBP1 and LRAT protein expression normalized by β-actin loading control. Data shown are double-plotted data. Data are represented as mean ± S.E. * indicates significance at least p < 0.05.
Both ROH uptake and cell signaling by STRA6 are critically linked to intracellular transport and metabolism of vitamin A. It has been demonstrated that both cellular retinol-binding protein 1 (CRBP1) and lecithin retinol acyltransferase (LRAT) are required for STRA6-dependent vitamin A uptake and signaling (35–37). CRBP1 (also referred to as RBP1) is a cytosolic ROH carrier protein that binds an intracellular portion of STRA6 and directly accepts ROH transferred from extracellular RBP through STRA6 (36). CRBP1 delivers ROH to the enzyme LRAT that catalyzes the conversion of ROH to retinyl esters for storage (38, 39). By storing ROH, LRAT creates an inward gradient of ROH across the cell. This allows for the regeneration of apo-CRBP1, which can re-bind to STRA6 and allow for continuing influx of ROH and STRA6 signaling (36). We hypothesized that Crbp1 and Lrat may also display circadian patterns of expression to further support STRA6 signaling. In WAT, Crbp1 mRNA expression has two significant peaks at ZT10 and ZT22, showing a 6-fold increase at ZT22 compared with ZT2 (Fig. 1B). CRBP1 protein expression is increased at ZT2 following the largest mRNA peak for CRBP1 at ZT22 (Fig. 1, D and E). Expression of the retinol-esterifying enzyme LRAT also shows circadian patterns of expression with a peak in both mRNA and protein at ZT2 compared with ZT14 (Fig. 1, C and F). The expression of CRBP1 and LRAT protein is synchronized with the peak in STRA6 mRNA at ZT2 expression during the light/inactive phase.
Serum RBP4 and transthyretin levels display circadian rhythmicity
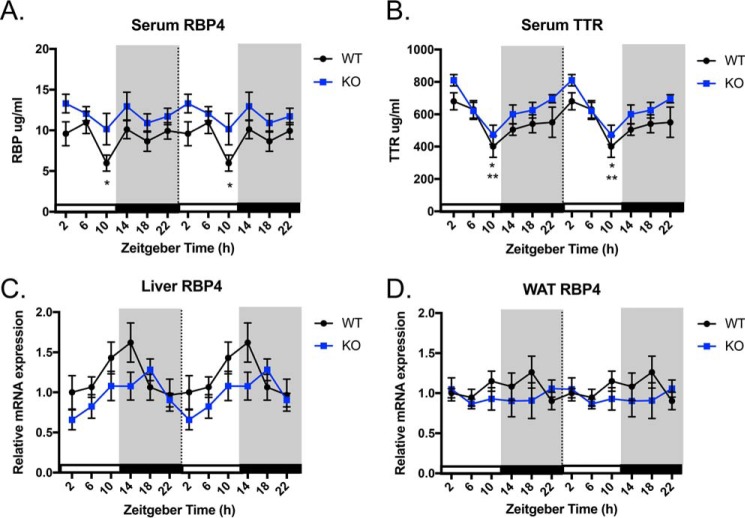
Because we observed circadian expression of Stra6 in WAT, we investigated whether RBP4, the ligand for STRA6, displayed circadian patterns in serum. In WT mice, serum RBP4 maintains constant levels across the day except for a significant decrease at ZT10 (Fig. 2A). STRA6-null mice do not display significant changes in serum RBP4 over a 24-h period. In serum, holo-RBP is found in complex with transthyretin (TTR) in a 1:1 molar ratio (40–42). The binding of holo-RBP to TTR prevents glomeruli filtration of the small RBP molecule by the kidney (43). RBP4 can only bind and exert its effects on insulin resistance through STRA6 when it is unbound or exceeds its serum carrier TTR (44). Therefore, we also investigated the level of circulating TTR. Serum TTR levels display a circadian pattern of expression with a peak at ZT2 and a significant nadir at ZT10 in both WT and STRA6-null mice (Fig. 2B).
Figure 2.
Serum RBP4 and TTR levels display circadian rhythmicity. A, serum RBP4 levels over a 24-h period in WT and STRA6-null mice. * indicates significant difference of at least p < 0.05 between WT at ZT10 and ZT14. B, serum TTR levels over a 24-h period in WT and STRA6-null mice. * indicates significant difference in WT between ZT2 and ZT10. ** indicates significant difference in KO between ZT2 and ZT10. C, liver Rbp4 mRNA expression over a 24-h period in WT and STRA6-null mice. D, WAT Rbp4 mRNA expression over a 24-h period in WT and STRA6-null mice. Data shown are double-plotted data. Data are represented as mean ± S.E.
Hepatocytes are the principal source of RBP4 that is secreted into the circulation (45). Because we observed serum RBP levels fluctuated in the serum of WT mice, we examined the pattern of Rbp4 expression in the liver. WT and STRA6-null mice do not display significant changes in liver Rbp4 mRNA expression over a 24-h period (Fig. 2C). Furthermore, Rbp4 mRNA expression does not display a circadian pattern in WAT for either WT or STRA6-null mice (Fig. 2D).
Circadian transcription factor rev-erbα binds the promoters of Stra6, Crbp1, and Lrat
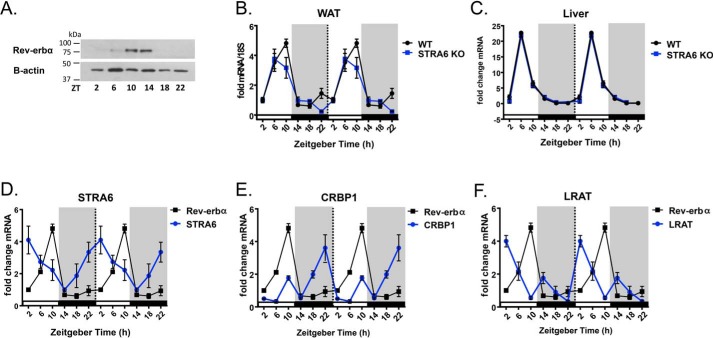
Because we observed circadian patterns of expression for STRA6, CRBP1, and LRAT in WAT, we investigated whether these genes were under transcriptional control by circadian clock proteins. Circadian patterns of gene expression are dependent on transcriptional regulation by several core clock proteins, including the nuclear transcription factor REV-ERBα (Nr1d1) (9, 46). REV-ERBα regulates the expression of the core circadian clock transcriptional machinery, but it also has been shown to directly regulate metabolic genes (9). REV-ERBα is an orphan nuclear receptor, and it lacks the C-terminal activation domain but binds the nuclear receptor co-repressor and therefore functions as a transcriptional repressor (47). REV-ERBα binds the response element termed the RAR-related orphan receptor α-response element (RORE) (46). ROREs are defined as nuclear receptor half-site motifs flanked 5′ by an (A/T)-rich sequence ((A/T)PuoGGTCA) (46). Bioinformatics analyses (AliBaba2.1) revealed the presence of putative ROREs in the promoters of mouse and human Stra6, Crbp1, and Lrat (Table 1). Similar to published literature (48), REV-ERBα protein expression in WAT displays significant expression at ZT10 and ZT14 (Fig. 3A). Likewise, WAT REV-ERBα mRNA expression is circadian with a significant peak flanking ZT10 (Fig. 3B). In the liver, REV-ERBα expression peaks at ZT6 and is low throughout the dark cycle (Fig. 3C). Importantly, the circadian nature of REV-ERBα is not altered in mice lacking STRA6 (Fig. 3, B and C). Examination of Rev-erbα mRNA expression in WAT compared with the Stra6, Crbp1, and Lrat mRNA expression revealed that the expression Rev-erbα is inverse to the expression of these genes (Fig. 3, D–F).
Table 1.
Putative RORE located on mouse and human STRA6, CRBP1, and LRAT
Promoter analysis was performed using Alibaba2.1. Locations of the putative RORE are denoted as distance from the start site of the gene base pair distal to the A of the ATG defined as −1.
| Gene | Species | Location | Putative sequence |
|---|---|---|---|
| STRA6 NM_001199040.1 | Human | −1651 | C(T/A)GGTCA |
| STRA6 NM_001199040.1 | Human | −3214 | A(A/A)GGTCA |
| STRA6 NM_001199040.1 | Human | −3746 | ATTT (G/T)GGTCA |
| Stra6 NM_009291.2 | Mouse | +241 | A(G/A)GGTCA |
| Stra6 NM_009291.2 | Mouse | −176 | TAA(C/A)GGTCA |
| Stra6 NM_009291.2 | Mouse | −339 | G(C/A)GGTCA |
| Stra6 NM_009291.2 | Mouse | −1543 | A(G/A)GGTCT |
| Stra6 NM_009291.2 | Mouse | −2244 | A(A/G)GGTCA |
| Stra6 NM_009291.2 | Mouse | −2688 | A(A/A)GGTGA |
| Stra6 NM_009291.2 | Mouse | −3708 | A(G/A)GGTCA |
| Stra6 NM_009291.2 | Mouse | −4040 | A(A/A)GGGCA |
| CRBP1 NC_000003.12 | Human | +241 | ATG(T/T)GGTCA |
| CRBP1 NC_000003.12 | Human | −64 | A(A/G)GGTCA |
| CRBP1 NC_000003.12 | Human | −480 | T(G/A)GGTCA |
| CRBP1 NC_000003.12 | Human | −854 | ATT/AGGTGA |
| Crbp1 NC_000075.6 | Mouse | −1162 | AG(T/A)GGTC(A/A)(A/A)GGTCA |
| Crbp1 NC_000075.6 | Mouse | −2142 | A(G/T)GGTCA |
| Crbp1 NC_000075.6 | Mouse | −2513 | (T/G)GGTTCA |
| Crbp1 NC_000075.6 | Mouse | −3519 | C(A/A)GGTCAACC |
| LRAT NC_000004.11 | Human | −609 | A(A/A)GGTCC |
| LRAT NC_000004.11 | Human | −728 | TTCGGCT |
| LRAT NC_000004.11 | Human | −914 | AGGTTC |
| Lrat NM_023624.4 | Mouse | −174 | GGGTCA |
| Lrat NM_023624.4 | Mouse | −405 | TTA(A/A)GGTCT |
| Lrat NM_023624.4 | Mouse | −1725 | A(A/A)GGTCA |
| Lrat NM_023624.4 | Mouse | −3218 | TG(C/T)GGTCA |
| Lrat NM_023624.4 | Mouse | −4060 | T(C/A)GGTCA |
Figure 3.
Stra6, Crbp1, and Lrat expression is inverse to Rev-erbα in WAT. A, Western blot analysis for REV-ERBα protein expression from epididymal WAT at indicated ZTs. B and C, Rev-erbα mRNA expression from epididymal WAT and liver from WT or STRA6-null mice collected every 4 h over a 24-h period determined by quantitative real-time PCR. D–F, mRNA expression of Rev-erbα superimposed on the mRNA expression pattern of Stra6, Crbp1, and Lrat from epididymal WAT collected every 4 h over a 24-h period determined by quantitative real-time PCR. Data shown are double-plotted data.
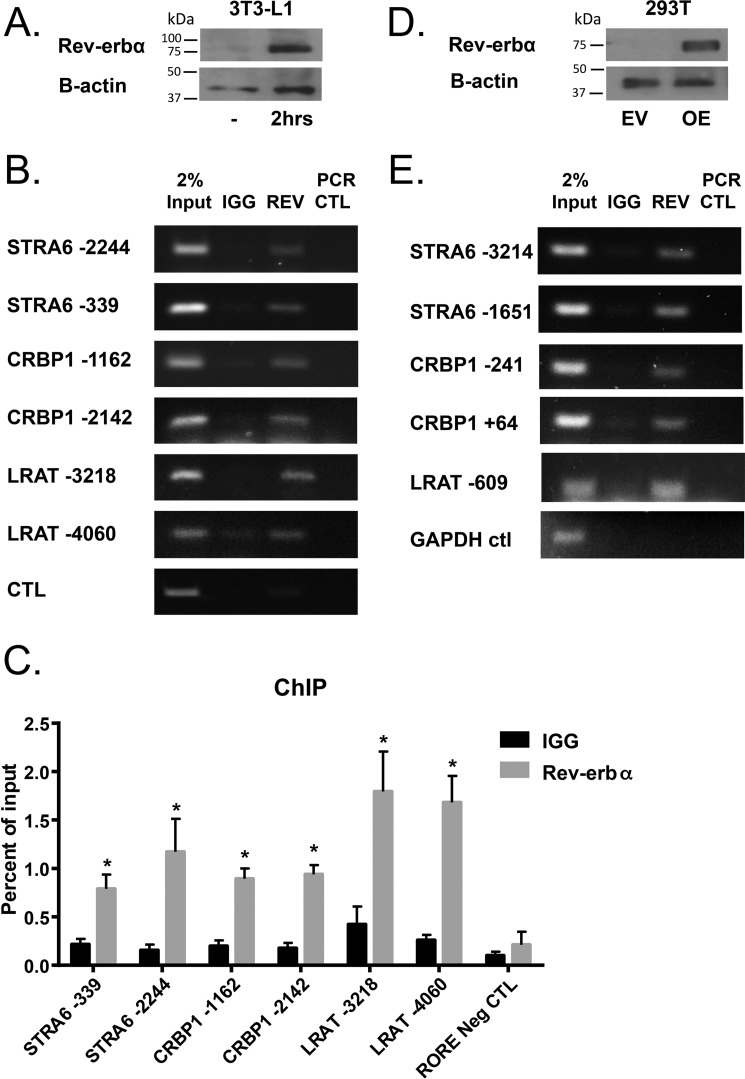
To determine whether REV-ERBα directly binds the promoters of Stra6, Crbp1, and Lrat, chromatin immunoprecipitation (ChIP) assays were carried out using the murine pre-adipocyte cell line NIH3T3-L1. Prior to the assay the cells were treated with adipocyte differentiation media (DMEM + 10% fetal bovine serum, 10 μg/ml insulin and 0.25 mmol/liter dexamethasone) for 2 h that served to increase the endogenous expression of REV-ERBα (Fig. 4A). We found that endogenous REV-ERBα binds to ROREs identified on Stra6, Crbp1, and Lrat (Fig. 4, B and C). REV-ERBα did not bind to a region −890 base pairs upstream from the start site on the STRA6 promoter that does not contain an RORE (Fig. 4, B and C). In agreement, we observed that REV-ERBα also binds to ROREs identified on human DNA, assessed by ChIP assays in the human 293T cell line overexpressing human REV-ERBα (Fig. 4, D and E).
Figure 4.
REV-ERBα binds to the promoters of Stra6, Crbp1, and Lrat. A, Western blot analysis for REV-ERBα protein expression in NIH3T3-L1 pre-adipocytes with or without treatment with differentiation media for 2 h. B, ChIP assays determined by semi-quantitative PCR in NIH3T3-L1 cells treated with differentiation media for 2 h to analyze the Rev-erbα recruitment to ROREs at sites denoted as distance from start site on the genes indicated. C, quantification of ChIP performed in triplicate. Data are represented as mean ± S.E. * indicates significance compared with IgG control of at least p < 0.05. D, Western blot analysis for REV-ERBα protein expression in 293T cells with or without overexpression of human REV-ERBα. E, ChIP assays determined by semi-quantitative PCR in 293T cells overexpressing REV-ERBα to analyze the REV-ERBα recruitment to ROREs at sites denoted as distance from start site on the genes indicated.
STRA6 regulates diurnal insulin responses in mice
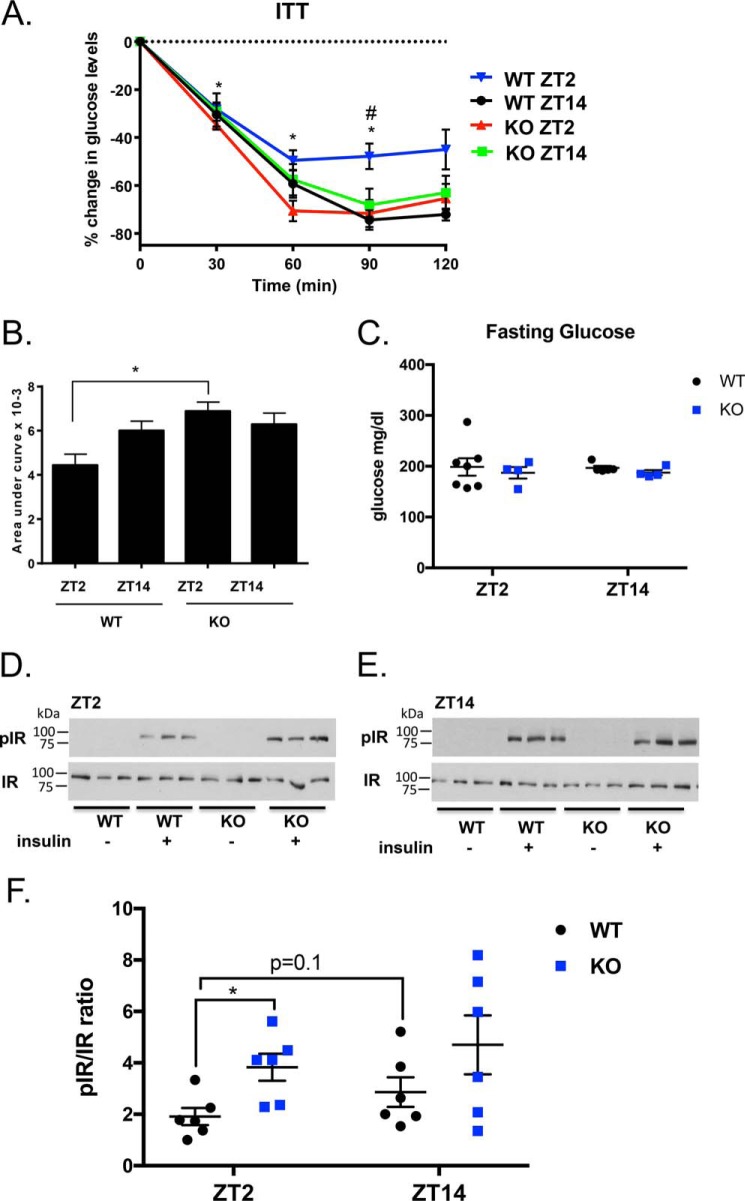
To determine whether STRA6 regulates whole-body diurnal patterns in insulin responsiveness, we performed insulin tolerance tests (ITT) (0.75 units/kg) in WT and STRA6-null male mice at ZT2 and ZT14 (Fig. 5, A–C). Serum glucose levels after a 4-h fast did not vary between genotypes at ZT2 or ZT14 (Fig. 5C). ITT tests show that WT mice display diurnal insulin sensitivity with increased insulin sensitivity at ZT14 (Fig. 5, A and B). These data agree with previously published reports that mice are more insulin-responsive during the dark phase relative to the light phase (34). In contrast, STRA6-null mice no longer display diurnal patterns in insulin sensitivity and remain insulin-sensitive to both ZT2 and ZT14 (Fig. 5, A and B).
Figure 5.
STRA6 regulates diurnal insulin responses in WAT. A, ITT (0.75 units/kg) in WT and STRA6-null male mice performed at ZT2 and ZT14. Data are represented as mean ± S.E. * denotes p < 0.02 comparing WT between time points ZT2 and ZT14. # denotes p < 0.05 comparing WT versus KO at the same time point. B, area above the curve data calculated from baseline from the ITT. C, serum glucose levels in male mice after a 4-h fast at ZT2 and ZT14 in WT and STRA6-null mice. D and E, Western blot analysis for pIR and IR from epididymal WAT collected from male mice before and after 5 min injection of insulin (1 μg/g) in WT and STRA6-null mice performed at ZT2 or ZT14. F, quantification of Western blots from D and E with n = 6 mice per group. Each blot was contained as an internal standard to accurately compare blot exposure and allow for quantification.
STRA6 signaling suppresses insulin receptor phosphorylation and promotes insulin resistance in mice (32). To determine whether the circadian expression of STRA6 regulates diurnal insulin receptor phosphorylation, male mice were fasted for 14 h, and tissue was collected before and after a 5-min bolus injection of insulin. WAT was collected, and Western blotting was performed to determine the ratio of phosphorylated insulin receptor compared with the total insulin receptor (pIR/IR). At ZT2, insulin treatment increases pIR/IR in WT and STRA6-null mice above basal pIR levels, which are nearly undetectable (Fig. 5D). In response to insulin at ZT2, STRA6-null mice have increased pIR/IR compared with WT mice (Fig. 5, D and F). In contrast, at ZT14 both WT and STRA6-null mice have similar responses to insulin (Fig. 5, E and F), corresponding to when STRA6 expression is low in WAT (Fig. 1A).
Activation of STRA6 by holo-RBP is diurnal
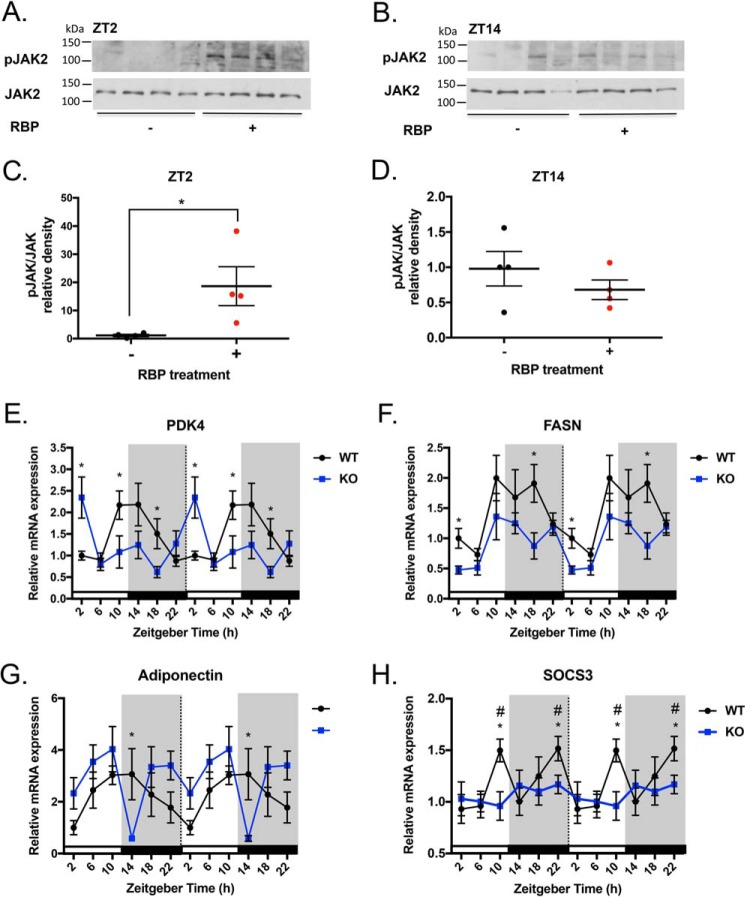
Upon the binding of extracellular holo-RBP, STRA6 propagates a JAK2/STAT5 signaling cascade (32, 35, 49). It is not known whether RBP4 activates STRA6 receptor signaling in a diurnal manner. To test this, male mice were injected with recombinant holo-RBP at either ZT0 (light/inactive phase) or ZT12 (dark/active phase). After 2 h, tissues were collected at the time points ZT2 and ZT14. We performed Western blotting to determine the ratio of phosphorylated JAK2 to total JAK2 expression (pJAK2/JAK2) in WAT at ZT2 and ZT14 (Fig. 6, A–D). Basal pJAK2/JAK2 in WAT is low (Fig. 6A). However, following holo-RBP treatment, pJAK2/JAK2 levels are increased on average 18-fold (Fig. 6, A and C). At ZT14 there are no differences in pJAK2/JAK2 before and after RBP injection (Fig. 6, B and D). These data indicate that the activation of STRA6-driven JAK2 signaling by holo-RBP is occurring at ZT2, when STRA6 expression is highest. In contrast, holo-RBP fails to stimulate JAK2 activation at ZT14 when STRA6 expression is lowest (Fig. 6, B and D).
Figure 6.
Effectors downstream of STRA6 signaling show diurnal variations. A and B, Western blot analysis for the ratio of pJAK/JAK protein levels from epididymal WAT collected from male mice injected with vehicle or recombinant holo-RBP (1 mg) mice after 2 h (n = 4). Tissue collection was performed at ZT2 and ZT14. C and D, quantification of Western blots for pJAK/JAK at ZT2 and ZT14. E–G, mRNA expression of Pdk4, Fasn, or adiponectin from epididymal WAT from WT or STRA6-null mice collected every 4 h over a 24-h period determined by quantitative real-time PCR. * denotes p < 0.05 comparing WT and STRA6 KO at the same time point. H, mRNA expression of Socs3 from epididymal WAT from WT or STRA6-null mice collected every 4 h over a 24-h period determined by quantitative real-time PCR. * denotes p < 0.02 comparing WT time points versus ZT2. # denotes p < 0.05 comparing WT versus STRA6 KO at same time point.
STRA6 regulates circadian expression of STAT5 target genes
STRA6 has been shown to specifically activate STAT5 in adipocytes (49). We investigated whether the expression of STRA6 altered the circadian expression of known STAT5 targets in WAT. Pyruvate dehydrogenase kinase 4 (Pdk4) is transcriptionally induced by STAT5 in mature adipocytes and acts to inhibit glycolysis (50). WT mice display a circadian pattern of Pdk4 expression with a peak at ZT10-14 and a valley at ZT22-6 (Fig. 6E). STRA6-null mice display an opposite pattern of expression, with a peak at ZT2 and significantly decreased levels compared with WT mice at ZT10 and ZT18 (Fig. 6E).
In vitro studies show that the lipogenic gene fatty-acid synthase (Fasn) is a direct target of STAT5 (51). In a mouse model with adipocyte-specific knockdown of STAT5, Fasn and other lipogenic genes were found to be decreased in WAT (52). We observed that Fasn expression displays a circadian pattern in both WT and STRA6-null mice, and the level of expression is significantly decreased in STRA6-null mice (Fig. 6F).
Adiponectin (Adipoq) is a direct target of STAT5, and studies show that STAT5 signaling represses adiponectin transcription (53). In WAT, adiponectin displays a circadian pattern of expression (Fig. 6G). In STRA6-null mice, the level of adiponectin in WAT is elevated compared with WT, except for a nadir at ZT14 (Fig. 6G).
In WAT, STRA6 signaling up-regulates the STAT target gene Socs3, a potent inhibitor of insulin receptor phosphorylation and its downstream responses (28, 33). We hypothesized that because STRA6 expression and activation are circadian, the expression of Socs3 in WAT would reflect STRA6 activity at the times when it is most highly expressed. In WT mice, Socs3 mRNA expression is increased at both ZT10 and ZT22 (Fig. 6H). The peak of Socs3 at ZT22 corresponds to the expression pattern of STRA6 when it is 4-fold higher at ZT22 versus ZT2 (Fig. 6H). In contrast to this, this rhythmic expression of Socs3 is completely abolished in STRA6-null mice (Fig. 6H).
STRA6-null mice have altered diurnal carbohydrate metabolism
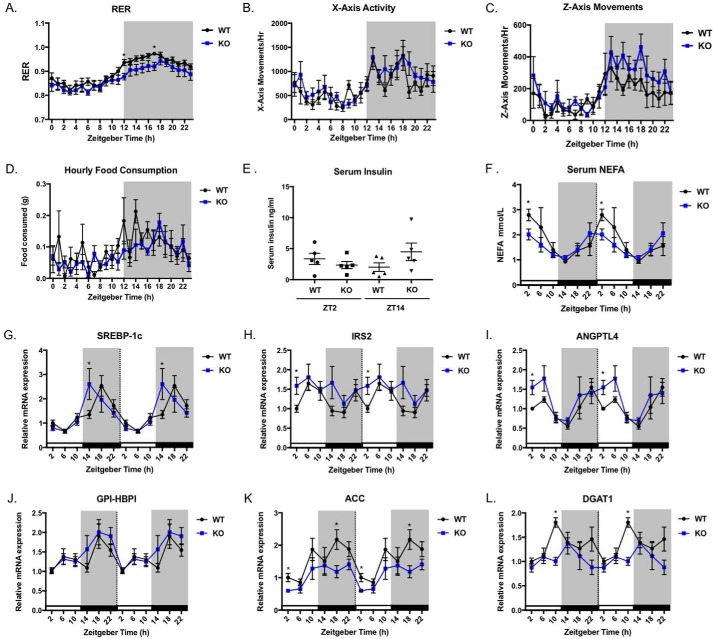
To investigate the metabolic phenotype of STRA6-null mice, we performed indirect calorimetry using male mice on a chow diet. We found the respiratory exchange ratio (RER) was significantly lower for STRA6-null mice during the dark/active phase compared with WT mice (Fig. 7A). A lower RER indicates that STRA6-null mice are using more fat as fuel compared with WT mice. This effect was not due to differences in body weight (data not shown) or the diurnal patterns of activity, feeding behavior, or insulin levels (Fig. 7, B–E). Because we observed increased fatty acid oxidation in STRA6-null mice, we investigated whether serum non-esterified fatty acids (NEFA) levels varied versus WT mice over a 24-h period. Although NEFA were circadian for both genotypes, levels were significantly lower in STRA6-null mice compared with WT at ZT2 (Fig. 7F).
Figure 7.
STRA6 regulates genes controlled by insulin action. A, respiratory exchange ratio for male mice on chow diet measured each hour over a 24-h period. * denotes p < 0.05 between WT and STRA6-null at the same time point. B, horizontal x axis movements in WT and STRA6-null mice measured each hour over a 24-h period. C, vertical z axis movements measured each hour over a 24-h period. D, hourly amounts of food consumed measured each hour over a 24-h period. E, serum insulin levels from ad libitum chow-fed WT and STRA6-null mice at ZT2 and ZT14. F, serum NEFA levels from ad libitum chow-fed WT and STRA6-null mice over a 24-h period. G–L, mRNA expression for the indicated genes in WT and STRA6-null mice over a 24-h period. * denotes p < 0.05 comparing WT versus STRA6 KO at same time point.
STRA6 regulates the expression of targets controlled by insulin action
Our data collectively show that STRA6-null mice are insulin-sensitive during the light/rest phase when WT mice are relatively insulin-resistant. We investigated whether STRA6 expression regulates known circadian targets in WAT that are controlled by insulin signaling. Sterol regulatory element-binding protein 1c (Srebp-1c), is stimulated by insulin signaling (54). In WAT, WT mice show a peak of Srebp-1c expression at ZT18 (Fig. 7G). STRA6-null mice show a peak for Srebp-1c 4 h in advance of WT, at ZT14 (Fig. 7G). Insulin receptor substrate 2 (Irs2) mediates downstream effects of insulin receptor phosphorylation and is also transcriptionally regulated by insulin (55, 56). We observe a circadian pattern of Irs2 expression in WT mice and a significant increase in Irs2 in STRA6-null mice at ZT2 (Fig. 7G).
Angiopoietin-like protein-4 (Angptl4) is highly expressed in adipose tissue, and it increases upon fasting and is decreased by insulin (57, 58). In WAT Angptl4 expression is circadian showing a valley at ZT14 and a peak at ZT22 (Fig. 7I). STRA6-null mice show increased Angptl4 expression at ZT2 compared with WT mice (Fig. 7I). Another gene regulated similarly to Angptl4 is glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (Gpi-hbp1) (57) This gene does display a circadian pattern of expression with increased levels during the dark phase (Fig. 7J). However, there were no differences in Gpi-hbp1 expression in WT and STRA6-null mice (Fig. 7J).
We observed that STRA6 expression regulates genes involved in fatty acid and triacylglycerol synthesis in WAT that are known to be circadian (59, 60). Acetyl-CoA carboxylase α (Acaca, also known as Acc) catalyzes the rate-limiting step for fatty acid synthesis (61). Acc mRNA displays a circadian pattern with a peak during the dark phase (Fig. 7K). STRA6-null mice display significantly decreased levels of Acc at ZT18 versus WT mice (Fig. 7K). Diacylglycerol O-acyltransferase 1 (Dgat1) catalyzes the last step in the formation of triglyceride (61). Dgat1 shows a circadian pattern in WT mice, peaking at ZT10 (Fig. 7L). However, in STRA6-null mice Dgat1 expression is decreased, and the circadian pattern of expression of Dgat1 is lost (Fig. 7L).
Discussion
It is well established that insulin sensitivity follows a diurnal pattern, which is crucial for maintaining blood glucose at steady-state concentrations to meet the needs of tissues before and after meals or during the overnight fast (12). This study identifies the holo-RBP receptor STRA6 as a regulator of diurnal insulin sensitivity and a major regulator of genes important for insulin action and insulin sensitivity. STRA6 is not expressed in the liver and therefore is regulating whole-body insulin sensitivity through extra-hepatic tissues (25–27). We show that Stra6 expression is circadian in WAT, peaking just previous to and during the light/rest phase when mice are known to be relatively insulin-resistant (34). Interestingly, proteins that are critical for STRA6 activity, CRBP1 and LRAT, also display circadian expression in WAT with increases during the light/rest phase, similarly to STRA6. The circadian expression of these genes along with STRA6 may serve to support signaling, although this will require further investigation. Therefore, with STRA6, CRBP1, and LRAT present early in the light phase, conditions are primed at this time of day for STRA6 activity.
Serum RBP4 levels are maintained across the day except for a decrease at ZT10. Levels of RBP4 do not exceed TTR at any time across the day, indicating that the liver may be secreting RBP4 and TTR in tandem. In addition, these data suggest that there is not a specific time of day when the levels of RBP4 exceed TTR, a circumstance that may trigger increased STRA6 activity. On the contrary, the levels of RBP4 are the same at ZT2 and ZT14 when we have performed insulin tolerance tests and observe the effects of STRA6 expression on diurnal insulin tolerance. This indicates that effects on insulin sensitivity at these times are not due to changes in RBP4 but are due to the circadian expression of its receptor STRA6.
Rbp4 mRNA expression patterns in the liver and WAT do not correlate with the decrease in serum RBP4 at ZT10. It is not known how the liver senses vitamin A status and regulates the secretion of holo-RBP (38). RBP secretion is thought to be tightly controlled, and we have observed that even a 12-h fast does not change RBP4 levels in the serum (data not shown).
Stra6 expression is highest during the light/rest phase when mice are relatively insulin-resistant. Insulin tolerance tests show that STRA6-null mice are more insulin-sensitive than WT mice at ZT2 during the light/rest phase. In contrast, WT and STRA6-null mice are similarly insulin-sensitive during the dark/active phase at ZT14. Therefore, the data demonstrate that it is primarily during the light/rest phase that expression of STRA6 is affecting insulin responses, corresponding to the pattern of STRA6 expression in WAT. Analogous to the insulin tolerance results, additional experiments show that in WAT of STRA6-null mice, insulin receptor phosphorylation levels are increased compared with WT at ZT2. However, WT and KO show similar IR phosphorylation at ZT14. These data demonstrate that STRA6 regulates the phosphorylation of the insulin receptor in a diurnal manner, affecting WT mice predominantly at ZT2, and not at ZT14 when STRA6 expression is low in WAT. Therefore, STRA6 plays a role regulating normal physiological diurnal insulin responses in the WAT, and it results in whole-body regulation of diurnal insulin sensitivity.
A key event in the STRA6 signaling cascade is the phosphorylation of JAK2, which binds to the cytoplasmic tail of STRA6 (36). Two hours after an injection of holo-RBP, JAK2 phosphorylation in WAT increased 18-fold during ZT2 when STRA6 expression is high, and not at all during ZT14 when STRA6 expression is low. Accordingly, STRA6 signaling in WAT in response to serum holo-RBP is diurnal and corresponds with light/rest phase when the effects of STRA6 on insulin sensitivity are observed.
We also examined the expression of a downstream effector of STRA6-mediated STAT5 signaling, Socs3. Our data show that WT mice display two peaks of Socs3 expression across the day in WAT. However, in STRA6-null mice Socs3 induction is completely abrogated. Although the Socs3 peak at ZT22 corresponds with STRA6 mRNA expression, it appears the circadian expression of Socs3 at both times is regulated by STRA6. Socs3 is an important negative regulator of insulin signaling and is a part of the inhibitory mechanisms employed by many cytokine receptors (33, 62, 63). It is possible that STRA6 regulates one of the peaks indirectly, due to cross-talk with another signaling receptor. One way that Socs3 inhibits insulin receptor responses is by binding to specific phosphotyrosine residues on IR and thereby preventing IRS1 or IRS2 phosphorylation and the downstream effects (62). Therefore, one mechanism by which STRA6 may regulate diurnal insulin receptor signaling is by up-regulating Socs3 during the light/rest phase. Other membrane receptors such as the leptin receptor up-regulate Socs3, and interestingly Socs3 also inhibits leptin signaling (64, 65). The adipocyte hormone resistin uses a similar mechanism via Socs3 in adipose tissue to inhibit insulin receptor phosphorylation (66). Moreover, modulating Socs3 expression in adipose tissue alone is effective for improving whole-body insulin sensitivity (63). Mice with knockdown of Socs3 expression specifically in adipocytes were protected from obesity-induced insulin resistance (63). Collectively, these data support the possibility that STRA6-driven up-regulation of Socs3 may be central to the regulation of diurnal insulin responses.
Because STRA6 signals via JAK2/STAT5, we determined whether STRA6 altered the expression of known STAT5 targets in mature adipocytes. The role of STAT5 in white adipose tissue has yet to be determined (52). In fact, relatively few STAT5 target genes have been identified in mature adipocytes to date (67). Pdk4 is induced by STAT5 in adipocytes and repressed by insulin signaling (50, 68). Therefore, Pdk4 mRNA levels are regulated by a balance of these two opposing mechanisms. WT and STRA6-null mice show a nearly opposite pattern of expression in WAT. During the day, WT mice display increasing Pdk4 beginning at ZT6, although this induction does not occur in STRA6-null mice. Pdk4 remains low in STRA6-null mice, until ZT2 when Pdk4 levels peak, and in an inverse pattern compared with WT mice. In addition to Pdk4, STRA6 regulates the basal gene expression of STAT5 target genes in adipocytes, Fasn, and adiponectin. Therefore, STRA6 expression regulates the circadian expression of known STAT5 target genes in WAT.
Studies employing the adipose-specific STAT5 knockdown mice display increased insulin sensitivity, decreased NEFA, and decreased expression of genes involved in fatty acid and triacylglycerol synthesis (52). STRA6-null mice display a similar phenotype, as they are more insulin-sensitive than WT mice at ZT2, have lower NEFA at ZT2, and have lower levels of Fasn, Acc, and Dgat expression. Therefore, the phenotype observed in the adipose-specific STAT5 knockdown mouse may be due in part to STRA6's ability to induce STAT5 signaling.
Our data show that STRA6 signaling in adipose tissue results in a large effect on whole-body insulin sensitivity. Adipocyte JAK/STAT signaling is well known for regulating systemic insulin responses and glucose metabolism (67). Although our experiments were carried out using a global STRA6 knock-out, it has been previously shown that even a partial knockdown of STRA6 in the adipocyte results in improved glucose tolerance (69). In addition, multiple groups have shown that the ability of STRA6 to induce insulin resistance in WAT is not due to changes in retinol uptake (32, 69). This indicates that the mechanism via which STRA6 mediates its effects on insulin resistance is not due to its ability to transport ROH, but rather via its ability to signal.
STRA6-null mice fed a chow diet have a more complex metabolic phenotype than previously considered. STRA6-null mice have altered RER and use more fat as fuel during the dark/active phase. The changes in RER are not due to changes in activity or feeding patterns. In addition, STRA6-null mice have a completely reorganized circadian expression of many genes involved in glucose and lipid metabolism in WAT. Not only does STRA6 regulate the circadian pattern of Pdk4, Fasn, Acc, Dgat, and Socs3 expression, it regulates the expression of genes induced by insulin. STRA6-null mice displayed a 4-h advanced peak for the insulin-inducible gene Srebp-1c and increased Irs2 expression at ZT2 compared with WT mice. Overall, the gene expression data support the idea that STRA6 mice are more insulin-sensitive. Now that we know that STRA6 has effects even in lean mice, future work investigating the circadian rhythmicity of circulating hormones and gene expression that regulate glucose tolerance, insulin action, lipolysis, and lipogenesis in a comprehensive study in STRA6-null mice could elucidate further the role of STRA6 in whole-body metabolism.
The daily rhythms of metabolic functions are governed by circadian clocks in the SCN and in other tissues such as liver, muscle, and adipose tissue (14, 48, 70). Studies employing the knock-out of clock genes such as Rev-erbα show metabolic perturbations (46, 71). It remains poorly understood what the mediators are downstream of clock proteins that ultimately control diurnal insulin sensitivity. We found that REV-ERBα expression is inverse to the expression of Stra6, Crbp1, and Lrat in WAT. Multiple canonical RORE were identified on the promoters of Stra6, Crbp1, and Lrat, and ChIP assays show that endogenous REV-ERBα binds to the promoters of these genes in a pre-adipocyte cell line. The fact that REV-ERBα regulates the circadian expression of a signaling protein like STRA6 provides one explanation of how circadian rhythms can have broad cellular effects on metabolism. However, additional studies are needed to determine whether STRA6 is necessary for REV-ERBα to regulate diurnal metabolic reprogramming in mice.
It has recently been reported that in mice, RBP4 displays a small increase in plasma at ZT4 (72). Our analysis of RBP4 in the serum revealed a different circadian pattern with a decrease at ZT10. Our data regarding the expression of Rbp4 mRNA in the liver and WAT are similar to the analysis presented in Ref. 72. The liver Rbp4 mRNA expression pattern does not correspond to either the serum peak of RBP4 at ZT4 described by Ma et al. (72) or the decrease in serum at ZT10 we observed, indicating RBP secretion into the serum by the liver is regulated by some other mechanism. Ma et al. (72) also showed that hepatic knockdown of RBP4 increased insulin sensitivity compared with WT mice at ZT4 but not at ZT16. Although this study shows a role for hepatic RBP4 in diurnal insulin sensitivity, there is no implication for how serum RBP4 exerts its effects. Plasma holo-RBP4 protein on its own has no intrinsic function except to carry ROH. Our data show that STRA6-null mice have increased insulin sensitivity compared with WT mice only at ZT2 when the protein is expressed, and not at ZT14 when expression is low. It is likely that loss or overexpression of RBP4 had no effect on insulin sensitivity during the dark/active phase in the previous study because STRA6 is not abundantly expressed at ZT14. The data from both studies highlight that the presence of STRA6 is essential for RBP's ability to induce insulin resistance.
The effects of circadian rhythmicity on glucose regulation have important clinical implications to understand the development and design of therapeutics for type II diabetes. Despite much progress in the understanding of insulin action, the molecular basis for the development of human insulin resistance or diurnal insulin responses remains unclear. Overall, our results identify the STRA6 signaling receptor as a novel regulator of diurnal insulin responses. Our data provide mechanistic insight into how STRA6 controls diurnal insulin sensitivity under normal daily physiological conditions. Treatments for insulin resistance have not been successful to date. There are reports showing agents that lower circulating RBP4 improve insulin sensitivity in obese mice, but as a whole RBP4 as a therapeutic agent is still unexplored (73, 74). The recently solved structure for zebrafish STRA6 may lead to the design of effective small molecular inhibitors in the future (75). Therefore, the RBP4/STRA6 axis may develop into an attractive pathway for early detection of or as a potential therapeutic target for pre-diabetes or type II diabetes.
Experimental procedures
Mouse experiments
Global STRA6-null mice were a gift from Norbert B. Ghyselink of the University of Strasbourg, Illkirch, France (76). The STRA6-null mouse expresses a truncated STRA6 protein, composed of the first 88 amino acid residues and is not functional as it lacks 10 of the 11 transmembrane domains (32). Mice were maintained on a 12-h light/dark cycle. Male and female mice 12–16 weeks of age were used for serum and tissue collection experiments for mRNA. Serum was collected by intracardiac puncture, and WAT was collected, flash-frozen, and stored at −80 °C until the time of analysis. Mice were housed, and experiments were performed in compliance with an IACUC protocol approved by the Case Western Reserve University and the Lerner College of Medicine, Cleveland Clinic.
Analysis of mRNA and of gene expression
RNA was isolated from WAT with RNeasy lipid tissue mini kit (Qiagen). cDNA was synthesized with a high capacity cDNA kit (Applied Biosystems). Quantitative real-time PCR was performed using TaqMan Gene Expression assay probes for Stra6 (Mm00486457_m1), Socs3 (Mm01249143), Crbp1 (Mm00441119_m1), Lrat (Mm00469972_m1), Nr1d1 (Hs00253876_m1, Mm00520708_m1), Dgat1 (Mm00515643_m1), and β-actin (Mm00607939_s1). The following were used: Rbp4 Fwd, 5′-GCA GGA GGA GCT GTG CCT AGA-3′, and Rev, 5′-GGA GGG CCT GCT TTG ACA GT-3′; Adipoq Fwd, 5′-CCC AAG GGA ACT TGT GCA GGT TGG ATG-3′, and Rev, 5′-GTT GGT ATC ATG GTA GAG AAG AAA GCC-3′; Pnpla3 Fwd, 5′-ATT CCC CTC TTC TCT GGC CTA-3′, and Rev, 5′-ATG TCA TGC TCA CCG TAG AAA GG-3′; Pdk4 Fwd, 5′-GGT ACG CCT GAA ACT CAG TCA-3′, and Rev, 5′-GGT CAG GGA GCA CAC CTT A-3′; Irs2 Fwd, 5′-TCC AGG CAC TGG AGC TTT-3′, and Rev, 5′-GGC TGG TAG CGC TTC ACT-3′; Srebp-1c Fwd, 5′-TCT CAC TCC CTC TGA TGC TAC-3′, and Rev, 5′- GCA ACC ACT GGG TCC AAT T-3′; Acaca Fwd, 5′-CCT GAG GAA CAG CAT CTC TAA C-3′, and Rev, 5′-GCC GAG TCA CCT TAA GTA CAT ATT-3; Fasn Fwd, 5′-GTC ACC ACA GCC TGG ACC GC-3′, and Rev, 5′-CTC GCC ATA GGT GCC GCC TG-3′. Gene expression was analyzed via the ddCt method (77).
Immunoblotting
Protein from WAT was extracted from homogenized tissue samples with Cell Lysis Buffer (catalog no. 9808, Cell Signaling) supplemented with Halt Protease and Phosphatase Inhibitors (Pierce). Total protein per sample was determined by the Bradford assay (Bio-Rad). 20–60 μg of protein was separated on 8–12% Tris-glycine acrylamide gels and transferred to nitrocellulose membranes. Antibodies against CRBP1 (ab154881, Abcam), LRAT(ab116784, Abcam), IR (Cell Signaling), pIR (Cell Signaling), REV-ERBα (13418S, Cell Signaling), JAK2 (3230S, Cell Signaling), and pJAK2 (3776S, Cell Signaling) were used at the dilution proposed by the manufacturer. An internal standard was used to compare individual blots when quantified. Western blottings were analyzed with ImageJ (National Institutes of Health) for quantification by densitometry.
Analysis of serum metabolites
Serum RBP4 was measured using a mouse RBP4 ELISA (R&D Systems). TTR was measured using a mouse pre-albumin ELISA (Innovative Research, Inc.) Serum insulin levels were determined by EZRMI mouse insulin ELISA (Millipore). Serum NEFA were measured with HR-Series NEFA-HR colorimetric kit (Wako).
Chromatin immunoprecipitation
ChIP assays were performed in human 293T cells (ATCC) or NIH3T3-L1 (ATCC) mouse pre-adipocytes treated for 2 h with a differentiation media containing (DMEM + 10% fetal bovine serum, 10 μg/ml insulin, and 0.25 mmol/liter dexamethasone). ChIP was performed using SimpleChIP® enzymatic chromatin immunoprecipitation kit (Cell Signaling) with antibodies for REV-ERBα (13418S, Cell Signaling) and IgG (sc-2025, Santa Cruz Biotechnology). PCR primers (Table 2) were designed for individual ROREs predicted by bioinformatics analysis (Alibaba2.1, Niels Grabe). Immunoprecipitations were performed in triplicate.
Table 2.
ChIP primer sequences
Primer sequences used for ChIP on ROREs are found on the promoters of STRA6, CRBP1, and LRAT. Locations of the RORE are denoted as distance from the start site of the gene base pair distal to the A of the ATG defined as −1.
| Gene | RORE | Species | 5′–3′ sequence |
|---|---|---|---|
| STRA6 | −1651 | Human | Fwd, GGCAAAACTTCCCCTAGGTC |
| Rev, TGTACCCCAGACTCCAACAC | |||
| STRA6 | −3214 | Human | Fwd, TTGCCTTGGAGGGGCTAAAG |
| Rev, TCTCAGGGGTTTGTGCCTTC | |||
| CRBP1 | +241 | Human | Fwd, CAACTGGCTCCAGTCACTCC |
| Rev, GCGCAGGTACTCCTCGAAAT | |||
| CRBP1 | −64 | Human | Fwd, TCCGGTCTCCTCTTCCTTTGT |
| Rev, GGATGTCGAAGGGTCAGGTTT | |||
| LRAT | −609 | Human | Fwd, CACCGAAGACTACCGCGAAG |
| Rev, CCGGTTGCACTACTGGCTTT | |||
| GAPDH | +3555 | Human | Fwd, TTGCCCTCAACGACCACTTT |
| Rev, TCAGGGCCCTTTTTCTGAGC | |||
| Stra6 | −339 | Mouse | Fwd, GCGGGCAGTTTGCACAAGAG |
| Rev, AGAGTGTTCTGCACCCCTGG | |||
| Stra6 | −2244 | Mouse | Fwd, GCTGAGTGTCTGTGACCCTT |
| Rev, ATCCAGTCTGGCAAAACCACT | |||
| Crbp1 | −1162 | Mouse | Fwd, CACAGGCTCCTAGGCTCTGA |
| Rev, CGTGGCTTCTGATCCTTGTT | |||
| Crbp1 | −2142 | Mouse | Fwd, AGGAGATGGCCAAAGTGG |
| Rev, ACAGAGGTTGCTCCAGGT | |||
| Lrat | −3218 | Mouse | Fwd, CTGAGTGTGGGCTTCCTTGA |
| Rev, GAAAAGTTGGACCCCAAGTCA | |||
| Lrat | −4060 | Mouse | Fwd, ATCTGAGCTCTGGAGCAGAC |
| Rev, CCTTTGACCTGAGTTTCTCATGC | |||
| Stra6 | −890 | Mouse | Fwd, AGCTCCTCTGGGAGGATAGG |
| Rev, AAGATGGATGGCTGCTGACC |
Recombinant holo-RBP injection
Recombinant RBP was made following the procedure described in Ref. 78. At 16 weeks old, male mice were injected with 1 mg of recombinant holo-RBP4 at ZT0 and ZT12. 2 hours post-injection, blood and WAT were collected, flash-frozen, and stored at −80 °C until the time of analysis.
Insulin injection
After a 14-h fast, 12–16-week-old male (n = 6) mice were anesthetized and infused with 0.75 units/kg insulin (Sigma) into the inferior vena cava. Tissues were collected before and after insulin injection.
Insulin tolerance testing
After a 4-h fast, 12–16-week-old male WT or STRA6-null were injected with an intraperitoneal injection of 0.75 units/kg insulin (Humalin R, Lilly) (n = 4–5). Whole-blood glucose was determined sequentially by sampling blood at 0, 30, 60, 90, and 120 min using a portable glucometer (Accu-Chek Performa II). ITT tests were performed at ZT2 and ZT14. Fasting glucose levels were determined from the sample at time 0.
Indirect calorimetry
WT and STRA6-null male mice 12 weeks of age were housed in individual metabolic chambers on a 12-h light/dark cycle. Respiratory gasses, locomotor activity, and food consumption were measured by the Comprehensive Lab Animal Monitoring System (CLAMS) (Columbus Instruments).
Statistical analysis
Data are expressed as means ± S.E. of the mean (S.E.). Statistical analyses were performed using two-tail Student's t test. Area under the curve was calculated for the ITT tests using Prism6, GraphPad. Differences were considered significant at p < 0.05.
Author contributions
N. N., C. M. G., and J. M. B. planned the project and designed experiments. C. M. G. and J. M. B. wrote the manuscript; C. M. G conducted mouse experiments, performed the molecular assays, and analyzed and interpreted data. C. M. G. and J. M. B. were involved in the editing of the final manuscript.
Acknowledgments
We thank Norbert B. Ghyselinck (Département de Génétique Fonctionnelle et Cancer, Institut de Génétique et de Biologie Moléculaire et Cellulaire, CNRS, INSERM, and Université de Strasbourg, Illkirch, France) for the generous gift of the STRA6-null mouse.
This work was supported in part by National Institutes of Health Grants R01 DK060684 (to N. N. and J. M. B.), R01 HL122283 (to J. M. B.), and P50 AA024333 (to J. M. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SCN
- suprachiasmatic nucleus
- LRAT
- lecithin-retinol acyltransferase
- N-CoR
- nuclear receptor corepressor
- Per
- period
- RORE
- RAR-related orphan receptor α-response element
- Rev-erbα
- nuclear receptor subfamily 1-group D-member 1
- ROH
- retinol
- WAT
- white adipose tissue
- ZT
- Zeitgeber
- NEFA
- non-esterified fatty acid
- TTR
- transthyretin
- Fwd
- forward
- Rev
- reverse
- RBP
- retinol-binding protein
- ITT
- insulin tolerance test
- IR
- insulin receptor
- RER
- respiratory exchange ratio
- ACC
- acetyl-CoA carboxylase
- DGAT
- diacylglycerol O-acyltransferase.
References
- 1. Ogden C. L., Carroll M. D., Kit B. K., and Flegal K. M. (2013) Prevalence of obesity among adults: United States, 2011–2012. NCHS Data Brief, 1–8 [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention (2014) National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, United States Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 3. Guo F., and Garvey W. T. (2016) Trends in cardiovascular health metrics in obese adults: National Health and Nutrition Examination Survey (NHANES), 1988–2014. J. Am. Heart Assoc. 5, e003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lebovitz H. E. (2001) Insulin resistance: definition and consequences. Exp. Clin. Endocrinol. Diabetes 109, S135–S148 [DOI] [PubMed] [Google Scholar]
- 5. Shi S. Q., Ansari T. S., McGuinness O. P., Wasserman D. H., and Johnson C. H. (2013) Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 23, 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maury E., Ramsey K. M., and Bass J. (2010) Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ. Res. 106, 447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Green C. B., Takahashi J. S., and Bass J. (2008) The meter of metabolism. Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guillaumond F., Dardente H., Giguère V., and Cermakian N. (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403 [DOI] [PubMed] [Google Scholar]
- 9. Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., and Lazar M. A. (2007) Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 10. Gibson T., and Jarrett R. J. (1972) Diurnal variation in insulin sensitivity. Lancet 2, 947–948 [DOI] [PubMed] [Google Scholar]
- 11. Saad A., Dalla Man C., Nandy D. K., Levine J. A., Bharucha A. E., Rizza R. A., Basu R., Carter R. E., Cobelli C., Kudva Y. C., and Basu A. (2012) Diurnal pattern to insulin secretion and insulin action in healthy individuals. Diabetes 61, 2691–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Cauter E., Polonsky K. S., and Scheen A. J. (1997) Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr. Rev. 18, 716–738 [DOI] [PubMed] [Google Scholar]
- 13. Hughey J. J., and Butte A. J. (2016) Differential phasing between circadian clocks in the brain and peripheral organs in humans. J. Biol. Rhythms 31, 588–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar Jha P., Challet E., and Kalsbeek A. (2015) Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol. Cell. Endocrinol. 418, 74–88 [DOI] [PubMed] [Google Scholar]
- 15. Nagaya T., Yoshida H., Takahashi H., and Kawai M. (2002) Markers of insulin resistance in day and shift workers aged 30–59 years. Int. Arch. Occup. Environ. Health 75, 562–568 [DOI] [PubMed] [Google Scholar]
- 16. Suwazono Y., Dochi M., Sakata K., Okubo Y., Oishi M., Tanaka K., Kobayashi E., Kido T., and Nogawa K. (2008) A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity 16, 1887–1893 [DOI] [PubMed] [Google Scholar]
- 17. Schernhammer E. S., Laden F., Speizer F. E., Willett W. C., Hunter D. J., Kawachi I., Fuchs C. S., and Colditz G. A. (2003) Night-shift work and risk of colorectal cancer in the nurses' health study. J. Natl. Cancer Inst. 95, 825–828 [DOI] [PubMed] [Google Scholar]
- 18. Schernhammer E. S., Laden F., Speizer F. E., Willett W. C., Hunter D. J., Kawachi I., and Colditz G. A. (2001) Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J. Natl. Cancer Inst. 93, 1563–1568 [DOI] [PubMed] [Google Scholar]
- 19. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., and Bass J. (2005) Obesity and metabolic syndrome in circadian clock mutant mice. Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., and Fitzgerald G. A. (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paschos G. K., Ibrahim S., Song W. L., Kunieda T., Grant G., Reyes T. M., Bradfield C. A., Vaughan C. H., Eiden M., Masoodi M., Griffin J. L., Wang F., Lawson J. A., and Fitzgerald G. A. (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Hoek M., Dehghan A., Zillikens M. C., Hofman A., Witteman J. C., and Sijbrands E. J. (2008) An RBP4 promoter polymorphism increases risk of type 2 diabetes. Diabetologia 51, 1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quadro L., Blaner W. S., Hamberger L., Novikoff P. M., Vogel S., Piantedosi R., Gottesman M. E., and Colantuoni V. (2004) The role of extrahepatic retinol-binding protein in the mobilization of retinoid stores. J. Lipid Res. 45, 1975–1982 [DOI] [PubMed] [Google Scholar]
- 24. Smith J. E., Muto Y., Milch P. O., and Goodman D. S. (1973) The effects of chylomicron vitamin A on the metabolism of retinol-binding protein in the rat. J. Biol. Chem. 248, 1544–1549 [PubMed] [Google Scholar]
- 25. Kawaguchi R., Yu J., Honda J., Hu J., Whitelegge J., Ping P., Wiita P., Bok D., and Sun H. (2007) A membrane receptor for retinol-binding protein mediates cellular uptake of vitamin A. Science 315, 820–825 [DOI] [PubMed] [Google Scholar]
- 26. Bouillet P., Sapin V., Chazaud C., Messaddeq N., Décimo D., Dollé P., and Chambon P. (1997) Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev 63, 173–186 [DOI] [PubMed] [Google Scholar]
- 27. Szeto W., Jiang W., Tice D. A., Rubinfeld B., Hollingshead P. G., Fong S. E., Dugger D. L., Pham T., Yansura D. G., Wong T. A., Grimaldi J. C., Corpuz R. T., Singh J. S., Frantz G. D., Devaux B., et al. (2001) Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 61, 4197–4205 [PubMed] [Google Scholar]
- 28. Berry D. C., Jin H., Majumdar A., and Noy N. (2011) Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc. Natl. Acad. Sci. U.S.A. 108, 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berry D. C., Levi L., and Noy N. (2014) Holo-retinol-binding protein and its receptor STRA6 drive oncogenic transformation. Cancer Res. 74, 6341–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Q., Graham T. E., Mody N., Preitner F., Peroni O. D., Zabolotny J. M., Kotani K., Quadro L., and Kahn B. B. (2005) Serum retinol-binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 436, 356–362 [DOI] [PubMed] [Google Scholar]
- 31. Graham T. E., Yang Q., Blüher M., Hammarstedt A., Ciaraldi T. P., Henry R. R., Wason C. J., Oberbach A., Jansson P. A., Smith U., and Kahn B. B. (2006) Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N. Engl. J. Med. 354, 2552–2563 [DOI] [PubMed] [Google Scholar]
- 32. Berry D. C., Jacobs H., Marwarha G., Gely-Pernot A., O'Byrne S. M., DeSantis D., Klopfenstein M., Feret B., Dennefeld C., Blaner W. S., Croniger C. M., Mark M., Noy N., and Ghyselinck N. B. (2013) The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J. Biol. Chem. 288, 24528–24539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi H., Tzameli I., Bjørbaek C., and Flier J. S. (2004) Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J. Biol. Chem. 279, 34733–34740 [DOI] [PubMed] [Google Scholar]
- 34. Prasai M. J., Mughal R. S., Wheatcroft S. B., Kearney M. T., Grant P. J., and Scott E. M. (2013) Diurnal variation in vascular and metabolic function in diet-induced obesity: divergence of insulin resistance and loss of clock rhythm. Diabetes 62, 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marwarha G., Berry D. C., Croniger C. M., and Noy N. (2014) The retinol esterifying enzyme LRAT supports cell signaling by retinol-binding protein and its receptor STRA6. FASEB J. 28, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berry D. C., O'Byrne S. M., Vreeland A. C., Blaner W. S., and Noy N. (2012) Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol. Cell. Biol. 32, 3164–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Amengual J., Golczak M., Palczewski K., and von Lintig J. (2012) Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 287, 24216–24227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Byrne S. M., and Blaner W. S. (2013) Retinol and retinyl esters: biochemistry and physiology. J. Lipid Res. 54, 1731–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Napoli J. L. (2016) Functions of intracellular retinoid binding-proteins. Subcell. Biochem. 81, 21–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Episkopou V., Maeda S., Nishiguchi S., Shimada K., Gaitanaris G. A., Gottesman M. E., and Robertson E. J. (1993) Disruption of the transthyretin gene results in mice with depressed levels of plasma retinol and thyroid hormone. Proc. Natl. Acad. Sci. U.S.A. 90, 2375–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bellovino D., Morimoto T., Tosetti F., and Gaetani S. (1996) Retinol binding protein and transthyretin are secreted as a complex formed in the endoplasmic reticulum in HepG2 human hepatocarcinoma cells. Exp. Cell Res. 222, 77–83 [DOI] [PubMed] [Google Scholar]
- 42. Pfeffer B. A., Becerra S. P., Borst D. E., and Wong P. (2004) Expression of transthyretin and retinol-binding protein mRNAs and secretion of transthyretin by cultured monkey retinal pigment epithelium. Mol. Vis. 10, 23–30 [PubMed] [Google Scholar]
- 43. van Bennekum A. M., Wei S., Gamble M. V., Vogel S., Piantedosi R., Gottesman M., Episkopou V., and Blaner W. S. (2001) Biochemical basis for depressed serum retinol levels in transthyretin-deficient mice. J. Biol. Chem. 276, 1107–1113 [DOI] [PubMed] [Google Scholar]
- 44. Berry D. C., Croniger C. M., Ghyselinck N. B., and Noy N. (2012) Transthyretin blocks retinol uptake and cell signaling by the holo-retinol-binding protein receptor STRA6. Mol. Cell. Biol. 32, 3851–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thompson S. J., Sargsyan A., Lee S. A., Yuen J. J., Cai J., Smalling R., Ghyselinck N., Mark M., Blaner W. S., and Graham T. E. (2017) Hepatocytes are the principal source of circulating RBP4 in mice. Diabetes 66, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., and Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harding H. P., and Lazar M. A. (1995) The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol. Cell. Biol. 15, 4791–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zvonic S., Ptitsyn A. A., Conrad S. A., Scott L. K., Floyd Z. E., Kilroy G., Wu X., Goh B. C., Mynatt R. L., and Gimble J. M. (2006) Characterization of peripheral circadian clocks in adipose tissues. Diabetes 55, 962–970 [DOI] [PubMed] [Google Scholar]
- 49. Berry D. C., and Noy N. (2012) Signaling by vitamin A and retinol-binding protein in regulation of insulin responses and lipid homeostasis. Biochim. Biophys. Acta 1821, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. White U. A., Coulter A. A., Miles T. K., and Stephens J. M. (2007) The STAT5A-mediated induction of pyruvate dehydrogenase kinase 4 expression by prolactin or growth hormone in adipocytes. Diabetes 56, 1623–1629 [DOI] [PubMed] [Google Scholar]
- 51. Hogan J. C., and Stephens J. M. (2005) The regulation of fatty acid synthase by STAT5A. Diabetes 54, 1968–1975 [DOI] [PubMed] [Google Scholar]
- 52. Kaltenecker D., Mueller K. M., Benedikt P., Feiler U., Themanns M., Schlederer M., Kenner L., Schweiger M., Haemmerle G., and Moriggl R. (2017) Adipocyte STAT5 deficiency promotes adiposity and impairs lipid mobilisation in mice. Diabetologia 60, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. White U. A., Maier J., Zhao P., Richard A. J., and Stephens J. M. (2016) The modulation of adiponectin by STAT5-activating hormones. Am. J. Physiol. Endocrinol. Metab. 310, E129–E136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boden G., Salehi S., Cheung P., Homko C., Song W., Loveland-Jones C., and Jayarajan S. (2013) Comparison of in vivo effects of insulin on SREBP-1c activation and INSIG-1/2 in rat liver and human and rat adipose tissue. Obesity 21, 1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Previs S. F., Withers D. J., Ren J. M., White M. F., and Shulman G. I. (2000) Contrasting effects of IRS-1 versus IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J. Biol. Chem. 275, 38990–38994 [DOI] [PubMed] [Google Scholar]
- 56. Hirashima Y., Tsuruzoe K., Kodama S., Igata M., Toyonaga T., Ueki K., Kahn C. R., and Araki E. (2003) Insulin down-regulates insulin receptor substrate-2 expression through the phosphatidylinositol 3-kinase/Akt pathway. J. Endocrinol. 179, 253–266 [DOI] [PubMed] [Google Scholar]
- 57. Kroupa O., Vorrsjö E., Stienstra R., Mattijssen F., Nilsson S. K., Sukonina V., Kersten S., Olivecrona G., and Olivecrona T. (2012) Linking nutritional regulation of Angptl4, Gpihbp1, and Lmf1 to lipoprotein lipase activity in rodent adipose tissue. BMC Physiol. 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kersten S., Mandard S., Tan N. S., Escher P., Metzger D., Chambon P., Gonzalez F. J., Desvergne B., and Wahli W. (2000) Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J. Biol. Chem. 275, 28488–28493 [DOI] [PubMed] [Google Scholar]
- 59. Shostak A., Meyer-Kovac J., and Oster H. (2013) Circadian regulation of lipid mobilization in white adipose tissues. Diabetes 62, 2195–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kohsaka A., Laposky A. D., Ramsey K. M., Estrada C., Joshu C., Kobayashi Y., Turek F. W., and Bass J. (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 [DOI] [PubMed] [Google Scholar]
- 61. Strable M. S., and Ntambi J. M. (2010) Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45, 199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ueki K., Kondo T., and Kahn C. R. (2004) Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol. 24, 5434–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Palanivel R., Fullerton M. D., Galic S., Honeyman J., Hewitt K. A., Jorgensen S. B., and Steinberg G. R. (2012) Reduced Socs3 expression in adipose tissue protects female mice against obesity-induced insulin resistance. Diabetologia 55, 3083–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bjorbak C., Lavery H. J., Bates S. H., Olson R. K., Davis S. M., Flier J. S., and Myers M. G. Jr. (2000) SOCS3 mediates feedback inhibition of the leptin receptor via Tyr985. J. Biol. Chem. 275, 40649–40657 [DOI] [PubMed] [Google Scholar]
- 65. Emilsson V., Arch J. R., de Groot R. P., Lister C. A., and Cawthorne M. A. (1999) Leptin treatment increases suppressors of cytokine signaling in central and peripheral tissues. FEBS Lett. 455, 170–174 [DOI] [PubMed] [Google Scholar]
- 66. Steppan C. M., Wang J., Whiteman E. L., Birnbaum M. J., and Lazar M. A. (2005) Activation of SOCS-3 by resistin. Mol. Cell. Biol. 25, 1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Richard A. J., and Stephens J. M. (2014) The role of JAK-STAT signaling in adipose tissue function. Biochim. Biophys. Acta 1842, 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Connaughton S., Chowdhury F., Attia R. R., Song S., Zhang Y., Elam M. B., Cook G. A., and Park E. A. (2010) Regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) gene expression by glucocorticoids and insulin. Mol. Cell. Endocrinol. 315, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zemany L., Kraus B. J., Norseen J., Saito T., Peroni O. D., Johnson R. L., and Kahn B. B. (2014) Downregulation of STRA6 in adipocytes and adipose stromovascular fraction in obesity and effects of adipocyte-specific STRA6 knockdown in vivo. Mol. Cell. Biol. 34, 1170–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Carrasco-Benso M. P., Rivero-Gutierrez B., Lopez-Minguez J., Anzola A., Diez-Noguera A., Madrid J. A., Lujan J. A., Martínez-Augustin O., Scheer F. A., and Garaulet M. (2016) Human adipose tissue expresses intrinsic circadian rhythm in insulin sensitivity. FASEB J. 30, 3117–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Delezie J., Dumont S., Dardente H., Oudart H., Gréchez-Cassiau A., Klosen P., Teboul M., Delaunay F., Pévet P., and Challet E. (2012) The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 26, 3321–3335 [DOI] [PubMed] [Google Scholar]
- 72. Ma X., Zhou Z., Chen Y., Wu Y., and Liu Y. (2016) RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 59, 354–362 [DOI] [PubMed] [Google Scholar]
- 73. Zemany L., Bhanot S., Peroni O. D., Murray S. F., Moraes-Vieira P. M., Castoldi A., Manchem P., Guo S., Monia B. P., and Kahn B. B. (2015) Transthyretin antisense oligonucleotides lower circulating RBP4 levels and improve insulin sensitivity in obese mice. Diabetes 64, 1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mody N., and Mcilroy G. D. (2014) The mechanisms of Fenretinide-mediated anti-cancer activity and prevention of obesity and type-2 diabetes. Biochem. Pharmacol. 91, 277–286 [DOI] [PubMed] [Google Scholar]
- 75. Chen Y., Clarke O. B., Kim J., Stowe S., Kim Y. K., Assur Z., Cavalier M., Godoy-Ruiz R., von Alpen D. C., Manzini C., Blaner W. S., Frank J., Quadro L., Weber D. J., Shapiro L., Hendrickson W. A., and Mancia F. (2016) Structure of the STRA6 receptor for retinol uptake. Science 353, aad8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ruiz A., Mark M., Jacobs H., Klopfenstein M., Hu J., Lloyd M., Habib S., Tosha C., Radu R. A., Ghyselinck N. B., Nusinowitz S., and Bok D. (2012) Retinoid content, visual responses, and ocular morphology are compromised in the retinas of mice lacking the retinol-binding protein receptor, STRA6. Invest. Ophthalmol. Vis. Sci. 53, 3027–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 78. Xie Y., Lashuel H. A., Miroy G. J., Dikler S., and Kelly J. W. (1998) Recombinant human retinol-binding protein refolding, native disulfide formation, and characterization. Protein Expr. Purif. 14, 31–37 [DOI] [PubMed] [Google Scholar]