Abstract
Extended laboratory culture and antimicrobial susceptibility testing timelines hinder rapid species identification and susceptibility profiling of bacterial pathogens associated with bovine respiratory disease, the most prevalent cause of cattle mortality in the United States. Whole-genome sequencing offers a culture-independent alternative to current bacterial identification methods, but requires a library of bacterial reference genomes for comparison. To contribute new bacterial genome assemblies and evaluate genetic diversity and variation in antimicrobial resistance genotypes, whole-genome sequencing was performed on bovine respiratory disease–associated bacterial isolates (Histophilus somni, Mycoplasma bovis, Mannheimia haemolytica, and Pasteurella multocida) from dairy and beef cattle. One hundred genomically distinct assemblies were added to the NCBI database, doubling the available genomic sequences for these four species. Computer-based methods identified 11 predicted antimicrobial resistance genes in three species, with none being detected in M. bovis. While computer-based analysis can identify antibiotic resistance genes within whole-genome sequences (genotype), it may not predict the actual antimicrobial resistance observed in a living organism (phenotype). Antimicrobial susceptibility testing on 64 H. somni, M. haemolytica, and P. multocida isolates had an overall concordance rate between genotype and phenotypic resistance to the associated class of antimicrobials of 72.7% (P < 0.001), showing substantial discordance. Concordance rates varied greatly among different antimicrobial, antibiotic resistance gene, and bacterial species combinations. This suggests that antimicrobial susceptibility phenotypes are needed to complement genomically predicted antibiotic resistance gene genotypes to better understand how the presence of antibiotic resistance genes within a given bacterial species could potentially impact optimal bovine respiratory disease treatment and morbidity/mortality outcomes.
Keywords: Histophilus somni, Mycoplasma bovis, Mannheimia haemolytica, Pasteurella multocida
Historically, bacterial pathogens were identified by biochemical, antigen, or PCR-based methods, with results taking from days to weeks. Culture-independent diagnostic testing (CIDT), typically used to identify slow-growing agents or those that cannot be cultured in vitro (Janda and Abbot 2014), is gaining attention due to the availability of whole-genome sequence (WGS) data. WGS can reveal a pathogen’s identity and uncover other clinically useful information typically garnered through multiple diagnostic tests (Gilmour et al. 2013).
Employment of WGS for pathogen identification in Salmonella (Mohammed and Cormican 2015), Klebsiella pneumoniae (Marsh et al. 2015), Neisseria meningitidis (Mustapha et al. 2015), and Escherichia coli (Berenger et al. 2015) outbreaks demonstrated the effectiveness of this approach, but highlighted significant limitations due to the paucity of published pathogen genome sequences. It was recognized that broad sampling and establishing a global genomic database from isolates would be required to understand the genetic diversity and distribution of traits within a pathogen population (Mutreja et al. 2011; Koser et al. 2014).
CIDT could be useful in characterizing pathogens associated with bovine respiratory disease (BRD) complex, the most prevalent and costly cause of mortality for cattle in the United States (Chirase and Green 2000; USDA 2011a). Several bacterial pathogens are associated with BRD, with Histophilus somni, Mycoplasma bovis, Mannheimia haemolytica, and Pasteurella multocida being the most common (Griffin et al. 2010). Complete bacterial genomes are available for two strains of H. somni, five of M. bovis, 10 of M. haemolytica, and 11 of P. multocida (Table 1).
Table 1. Known reference sequences for the species sequenced in this study.
| Strain | GenBank Accession | Publication |
|---|---|---|
| 2336 | GCA_000019405.1 | Siddaramappa et al. (2011) |
| 129PT | GCA_000011785.1 | Challacombe et al. (2007) |
| Hubei1 | GCA_000219375.1 | Li et al. (2011) |
| HB0801 | GCA_000270525.1 | Qi et al. (2012) |
| CQ-W70 | GCA_000696015.1 | None |
| NM2012 | GCA_000183385.1 | Wise et al. (2010) |
| PG45 | GCA_001043135.1 | None |
| M42548 | GCA_000376645.1 | Eidam et al. (2013) |
| 89010807N_lktA | GCA_000963675.1 | Heaton et al. (2015) |
| D153 | GCA_000422145.1 | Hauglund et al. (2013) |
| D171 | GCA_000427275.1 | Hauglund et al. (2015a) |
| D174 | GCA_000422095.1 | Hauglund et al. (2015b) |
| 89010807N | GCA_000963635.1 | Heaton et al. (2015) |
| USDA-ARS-SAM-185 | GCA_000349785.1 | Harhay et al. (2013) |
| USDA-ARS-USMARC-183 | GCA_000349765.1 | Harhay et al. (2013) |
| USDA-ARS-USMARC-184 | GCA_000819525.1 | None |
| USMARC_2286 | GCA_000439735.1 | Koren et al. (2013) |
| 36950 | GCA_000234745.1 | Michael et al. (2011) |
| 3480 | GCA_000259545.1 | None |
| ATCC_43137 | GCA_000754275.1 | Davenport et al. (2014) |
| HB01 | GCA_001663095.1 | Peng et al. (2016) |
| HB03 | GCA_000512395.1 | None |
| HN06 | GCA_000255915.1 | Liu et al. (2012) |
| OH1907 | GCA_000973565.1 | None |
| OH4807 | GCA_000973525.1 | None |
| PMTB2.1 | GCA_001578435.1 | None |
| Pm-3 | GCA_001721885.1 | None |
| Pm70 | GCA_000006825.1 | May et al. (2001) |
It is important to consider antimicrobial resistance and the spread of antimicrobial resistance genes (ARG) in treatment decisions associated with common diseases such as BRD (Koser et al. 2014). Understanding the epidemiology of antimicrobial resistance in BRD pathogens in healthy cattle, as well as treated and untreated sick cattle, is crucial to understanding the role of resistance in BRD treatment failure. In addition, the increasing use of WGS in antimicrobial resistance surveillance necessitates a better understanding of the degree of concordance between phenotypic susceptibility and resistance genotype.
The primary objective of this study was to perform WGS on bacterial isolates collected from untreated BRD-affected beef and dairy cattle, and unaffected dairy cattle, to contribute additional bacterial genome sequences that could be used for CIDT in the future. Secondary study objectives were to perform bioinformatic analyses and susceptibility testing to analyze concordance between phenotypic susceptibility and resistance genotype. Identification of distinguishing genomic characteristics, such as ARG, of different strains of the major BRD pathogens may facilitate a better understanding of these pathogens and their host interactions with the goal of identifying improved treatment methods specific to the host species, ARG genotype, and pathogen combination.
Materials and Methods
Cattle sampling
Personnel at a northern California feedlot and a central California dairy calf nursery cared for the animals used in this study. In the calf nursery, preweaned Holstein dairy calves 22–60 d of age, who had never been treated for BRD nor treated with antimicrobials for any reason within the previous 10 d, were enrolled in a matched case-control study between July 2011 and January 2012, as described previously (Love et al. 2014; Neibergs et al. 2014). Upon morning observation, BRD suspect calves and matched adjacent control calves were scored using a clinical scoring system that assigns numerical values from 0 to 3 based on severity of symptoms for rectal temperature, cough, nasal discharge, ocular discharge, ear or head tilt or droop, and fecal consistency, with a score of ≥5 classified as a case (McGuirk 2008). Guarded deep nasopharyngeal swabs were collected from cases and control calves and placed into 1 ml of bacterial transport media (Brucella broth with 15% glycerol) for the detection of bacteria as described (Love et al. 2014). All samples were stored on wet ice in the dark until completion of sampling and delivery to the lab for aerobic bacteria and Mycoplasma culturing (Fulton and Confer 2012). Similar procedures were followed for beef cattle diagnosed as BRD cases on a California feedlot. Sampling was performed in compliance with approved Institutional Animal Care and Use protocols at the University of California, Davis (protocols 16431 and 17974).
Bacterial isolates
Aerobic cultures from dairy calves were set up on sheep blood agar and chocolate agar plates and incubated at 37° in 7% CO2 for 2–4 d. Swabs for Mycoplasma isolation were swished in Mycoplasma D broth and incubated aerobically at 37° for 2–3 d, then plated to Mycoplasma N plates and incubated at 37° in 7% CO2 for up to 7 d. Bacteria from dairy cattle were isolated by morphology and subjected to testing for species identification at the California Animal Health and Food Safety Laboratory System Tulare laboratory. Urease detection, triple sugar iron agar, oxidase, indole production, hemolysis on sheep blood agar, and Gram stain testing were performed on M. haemolytica, P. multocida, and H. somni isolates. β-Galactosidase testing was performed on P. multocida and M. haemolytica and trehalose fermentation testing was performed on all M. haemolytica samples. Catalase and motility were also performed on H. somni isolates. M. bovis samples were confirmed as Mycoplasma by colony morphology, Dienes stain, and digitonin susceptibility prior to identification.
Ninety-four isolates of the four target organisms were frozen at −80°. Isolates were preferentially selected from cases rather than controls, generally from samples with moderate to large numbers of the organism in pure or predominant culture, and selection was spread out over the 6 months of the study. Four nonhemolytic M. haemolytica isolates were identified and removed from further analysis. Mycoplasma spp. were picked based on rapid growth and classic appearance for M. bovis, with only one morphotype present to prevent mixed cultures. In total, 74 case samples and 20 control samples were submitted for sequencing: H. somni (n = 16; 13 cases, three controls), M. bovis (n = 20; 17 cases, three controls), M. haemolytica (n = 28; 20 cases, eight controls), and P. multocida (n = 30; 24 cases, six controls). Species-identified isolates from dairy cattle were streaked onto various media and cultured at 37° for 1–5 d, depending on the species (Table 2). A single colony was added to 5 ml of PPLO broth for M. bovis isolates and BHI broth for the other three species, and incubated at 37° for 1–5 d prior to DNA isolation.
Table 2. Growth conditions for each of the four species used during this study.
| Species | Plate Media | Days for Growth | Broth Media | Days for Growth | Shaker | Environment |
|---|---|---|---|---|---|---|
| Histophilus somni | Columbia agar | 1–2 | BHI | 1–2 | N/A | Anaerobic |
| Mycoplasma bovis | PPLO | 5 | PPLO | 5 | N/A | 5% CO2 |
| Mannheimia haemolytica | Sheep’s blood agar | 1–2 | BHI | 1–2 | 250 rpm | Facultative |
| Pasteurella multocida | Sheep’s blood agar | 1–2 | BHI | 1–2 | 250 rpm | Facultative |
BHI, brain–heart infusion; PPLO, pleuropneumonia-like organism.
One swab from each of the beef cattle sampled was plated on sheep blood agar, Columbia agar, and PPLO agar plates, and cultured at 37° for 1–5 d. Ten single colonies were isolated by morphology, replated, and cultured as described. A total of 10 samples were submitted from beef feedlot cattle: M. haemolytica (n = 2; two cases) and P. multocida (n = 8; eight cases).
DNA isolation
A 2.5 ml culture was centrifuged at 13,000 rpm for 5 min for H. somni, M. haemolytica, and P. multocida, and for 20 min for M. bovis. The supernatant was discarded and the pellet was resuspended in 200 μl of PBS. DNA was extracted using the QIAamp Cador Pathogen Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions.
PCR identification
PCR with species-specific primers was used to identify the four bacterial species from isolates obtained from beef cattle, and to confirm the biochemical (aerobic) and morphologic (Mycoplasma) identification of the isolates from the dairy cattle. Primers were designed in Primer3 (Koressaar and Remm 2007; Untergrasser et al. 2012) to targeted genes specific to each species (Table 3). PCR protocols for M. haemolytica (Alexander et al. 2008) and P. multocida (Ullah et al. 2009) were performed as described previously. Protocols for H. somni and M. bovis were developed during this study. PCR was performed on an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) with 1× GoTAQ Green Master Mix (Promega Biosciences LLC, San Luis Obispo, CA), 10 μM of each primer and 10 ng of DNA for 5 min at 95°, 35 cycles of 30 sec at 95°, 30 sec at 56°, and 45 sec at 72°, followed by 10 min at 72°. Products were visualized on a 1.5% agarose gel using a ChemiDoc-ItTS2 Imager (UVP, LLC, Upland, CA), purified using the QIAquick PCR Purification Kit (Qiagen), Sanger sequenced (www.davissequencing.com) and aligned to their respective reference genomes using Bowtie2–default v2.2.8 (Langmead and Salzberg 2012) to verify that the appropriate target regions were amplified.
Table 3. Primers and PCR amplification protocols used in this study.
| Species | Primer | Sequence (5′-3′) | Tm (°) |
|---|---|---|---|
| Histophilus somni | Lob1F | AAATGGAAACACCCAACCAA | 56.3 |
| Lob1R | CAAACGCTTGATACGCTCA | 58.4 | |
| Mycoplasma bovis | MB528F | CATGCGAAAACACTGCAACT | 58.4 |
| MB528R | CCACGCATTCAGAATTGAAA | 56.3 | |
| Mannheimia haemolytica [Alexander et al. 2008] | LktF | GCAGGAGGTGATTATTAAAGTGG | 61 |
| LktR | CAGCAGTTATTGTCATACCTGAAC | 61.2 | |
| Pasteurella multocida [Ulhah et al. 2009] | KMT1SPG | GCTGTAAACGAACTCGCCAC | 62.4 |
| KMT1T7 | ATCCGCTATTTACCCAGTGG | 60.4 | |
| Positive control | 16SF | GCTAACTCCGTGCCAGCAG | 64.5 |
| 16SR | CGTGGACTACCAGGGTATCTAATC | 65.6 |
Library construction and sequencing
The PCR-confirmed DNA samples were sonicated on a Diagenode Bioruptor Pico Ultrasonicator (Diagenode Inc., Denville, NJ) to shear the DNA into 250 bp fragments. Next-generation sequencing libraries were created using the Kapa Genomic DNA Library Preparation kit (Kapa Biosystems, Wilmington, MA). The NEBNext Multiplex Oligos for Illumina kit (New England BioLabs Inc., Ipswich, MA) was used for indexing. The samples were sequenced (whole-genome shotgun, paired-end, 100 bp) on an Illumina HiSeq2500 sequencer at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley (supported by NIH S10 Instrumentation grants S10RR029668 and S10RR027303). A total of 104 isolates were whole-genome sequenced, along with five technical replicates.
Read cleanup and de novo assembly
Raw reads were run through expHTS v0.0.Oct212015 (Street et al. 2015), which screens for contaminants using a mapped based approach, de-duplicates reads using Super-Deduper v2.0, performs quality trimming using Sickle v1.40, overlaps paired-end reads using Flash2 v2.2.00, and performs a quality analysis. Cleaned reads were then de novo assembled using the SPAdes assembler v3.5.0 (Bankevich et al. 2012) by running through the expHTS pipeline. Contigs were aligned to GenBank using BLAST (Altschul et al. 1990).
Mapping assembly and genetic diversity
Available reference sequences were obtained from NCBI for each of the four species of interest (Table 1). Sequence reads from each isolate were aligned to their respective reference sequences using Bowtie2–default v2.2.8. SAM files were converted to BAM files and sorted using SAMtools v1.3 (Li et al. 2009). Consensus sequences were called using ANGSD v0.915 (Korneliussen et al. 2014) using -doFasta 3, which uses the base with the highest effective depth, and a minimum MAPQ score = 20. Average coverage for each consensus sequence was determined using SAMtools Depth v1.3. Consensus sequences were aligned and genetic distance trees, which were bootstrapped 100 times, were generated using SeaView v4.6.1 (Gouy et al. 2010). M. haemolytica sample sequences mapped to reference sequence USA-ARS-USMARC-183 were genotyped using previously described methods (Clawson et al. 2016). Statistical significance of genotype and the probability of being a case or control was assessed using the Fisher’s exact test.
Antimicrobial resistance
Raw reads and de novo assembled contigs from each sample were aligned to the Resfinder database (v3-20-2017; Zankari et al. 2012) using Short Read Sequence Typing for Bacterial Pathogens (SRST2; Inouye et al. 2014) v0.2.0 and ABRicate v0.5-dev (https://github.com/tseemann/abricate), respectively, using default settings to identify resistance genes. For gene variants that confer resistance, de novo assembled contigs were aligned to the Comprehensive Antibiotic Resistance Database (CARD; Jia et al. 2017) variant model database using the BLAST-like alignment tool (BLAT; Kent 2002) v35, and visually inspected for resistance-conferring SNPs. Raw reads from each sample were aligned to the PlasmidFinder database (v3-23-2017; Carattolli et al. 2014) using the SRST2 v0.2.0 using default settings. The NCBI reference sequences were aligned to the Resfinder database using ABRicate.
Susceptibility testing
Microdilution susceptibility testing of isolates was performed at the Kansas State Veterinary Diagnostic Laboratory using the Sensititre bovine/porcine plate (Thermo Scientific; plate code BOPO6F, Supplemental Material, Table S1), which tests for susceptibility to 18 drugs across nine antimicrobial classes: (i) β-lactam [Ampicillin (AMP), Ceftiofur (CEFT), Penicillin (PEN)]; (ii) tetracycline [Chlortetracycline (CHLOR), Oxytetracycline (OXY)]; (iii) fluoroquinolone [Danofloxacin (DAN), Enrofloxacin (ENRO)]; (iv) phenicol [Florfenicol (FLOR)]; (v) aminoglycoside [Gentamicin (GEN), Neomycin (NEO), Spectinomycin (SPEC)]; (vi) sulfonamide [Sulfadimethoxine (SUL)]; (vii) trimethoprim/sulfonamide [Trimethoprim-Sulfamethoxazole (TMS)]; (viii) macrolides-lincosamides (MLS) [Clindamycin (CLIN), Tilmicosin (TIL), Tulathromycin (TUL) Tylosin tartrate (TYL)]; and (ix) pleuromutilans [Tiamulin (TIA)] (Table 4). Breakpoints were chosen based on cattle-specific Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI 2013) when possible. CLSI swine-specific breakpoints were used for AMP and TIA, canine-specific breakpoints for CLIN and canine- and equine-specific breakpoints for GEN, and human-specific breakpoints for SUL and TMS. The CLSI breakpoints were not available for NEO and TYL, and therefore human-specific breakpoints from the National Antimicrobial Resistance Monitoring System were used (Table 4; FDA 2013). Wherever possible, breakpoints for the actual pathogens tested in this study were utilized. Based on these breakpoints, isolates were classified as susceptible, intermediate, or resistant. For analysis purposes, susceptible and intermediate isolates were considered to be susceptible. Crude prevalence of phenotypic resistance for each drug tested was calculated by bacterial species and across all three species. In addition, prevalence of multidrug resistant (MDR) phenotypes (defined as resistance to more than two antimicrobial drug classes) was calculated and MDR patterns were assessed.
Table 4. Mapping of identified resistance genes to antimicrobial drugs included on the broth microdilution phenotypic susceptibility panel.
| Resistance Gene | Antibiotic (Abbreviation) | Antibiotic Class | CLSI Approved Breakpoint | Breakpoint (mg/ml) | ||
|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||||
| tetH | Chlortetracycline (CHLOR) | Tetracycline | Yes | ≤2 | 4 | ≥8 |
| Oxytetracycline (OXY) | Tetracycline | Yes | ≤2 | 4 | ≥8 | |
| aph3-Ia | Neomycin (NEO) | Aminoglycoside | Noa | ≤2 | 4 | ≥8 |
| strA | None tested | N/A | N/A | N/A | N/A | N/A |
| strB | None tested | N/A | N/A | N/A | N/A | N/A |
| ROB-1 | Ampicillin (AMP) | β-Lactam | Yesb | ≤0.5 | 1 | ≥2 |
| Ceftiofur (CEFT) | β-Lactam | Yes | ≤2 | 4 | ≥8 | |
| Penicillin (PEN) | β-Lactam | Yes | ≤0.25 | 0.5 | ≥1 | |
| sul2 | Sulfadimethoxine (SUL) | Sulfonamide | Yesc | ≤256 | — | ≥512 |
| Trimethoprim-sulfamethoxazole (TMS) | Sulfonamide | Yesd | <0.5 and 9.5 | 1–2 and 19–38 | 4 and 76 | |
| erm42/ermF | Clindamycin (CLIN) | MLS | Yese | ≤0.5 | 1 - 2 | ≥4 |
| Tilmicosin (TIL) | MLS | Yesf | ≤8 | 16 | ≥32 | |
| Tulathromycin (TUL) | MLS | Yes | ≤16 | 32 | ≥64 | |
| Tylosin tartrate (TYL) | MLS | Nog | ≤8 | 16 | ≥32 | |
| cat2 | Florfenicol (FLOR) | Phenicol | Yes | ≤2 | 4 | ≥8 |
| floR | Florfenicol (FLOR) | Phenicol | Yes | ≤2 | 4 | ≥8 |
| dfrA14 | Trimethoprim-sulfamethoxazole (TMS) | TMS | Yesd | <0.5 and 9.5 | 1–2 and 19–38 | 4 and 76 |
| None identified | Danofloxacin (DAN) | Fluoroquinolone | Yesc | ≤0.25 | — | — |
| None identified | Enrofloxacin (ENRO) | Fluoroquinolone | Yes | ≤0.25 | 0.5–1 | ≥2 |
| None identified | Gentamicin (GENT) | Aminoglycoside | Yesh | ≤2 | 4 | ≥8 |
| None identified | Spectinomycin (SPEC) | Aminoglycoside | Yes | ≤32 | 64 | ≥128 |
| None identified | Tiamulin (TIA) | Pleuromutilin | Yesi | ≤16 | — | ≥32 |
MLS, macrolide-lincosamide-streptogramin.
For Neomycin, we used NARMS 2011 breakpoints for aminoglyosides.
For Ampicillin, we used CLSI breakpoints for swine that were validated for the following bacteria: Actinobacillus pleuropneumoniae and P. multocida.
For Danofloxacin, we used CLSI breakpoints for cattle, but only validated for the following bacteria: M. haemolytica and P. multocida.
For Sulfadimethoxine and TMS, we used CLSI breakpoints for humans with systemic disease and validated in Enterobacteriaceae.
For Clindamycin, we used CLSI breakpoints for canines that were validated for the following bacteria: Staphylcoccus spp. and β-hemolytic Streptococci.
For Tilmicosin, we used CLSI breakpoints for cattle, but only validated for the following bacteria: M. haemolytica.
For Tylosin, we used the same breakpoints as in the following document: https://www.ars.usda.gov/SP2UserFiles/Place/60400520/NARMS/ABXEntero.pdf.
For Gentamicin, we used canine and equine breakpoints validated in Pseudomonas aeruginosa and Enterobacteriaceae and the equine breakpoint for Actinobacillus spp.
For Tiamulin, we used CLSI breakpoints for swine that were validated for the following bacteria: Actinobacillus pleuropneumoniae.
In order to compare phenotypic and genotypic susceptibility results, identified resistance genes were mapped to the antimicrobial drug classes to which they are known to confer resistance (Table 4). Proportion of isolates with concordant and discordant genotype–phenotype results were calculated for each antimicrobial drug and organism.
Statistical significance of discordance was assessed using the mid-P exact for binomial paired data with a two-tailed P-value (Edwards 1948), unless there were at least 20 discordant observations or the marginal counts were equal to zero, in which case McNemar’s test with continuity correction was used (Fagerland et al. 2013). If there was a lack of observations in any of the genotype–phenotype concordance–discordance combinations, 0.5 was added to the count in order to enable calculation of the test statistic. OpenEpi was used for all calculations (Dean et al. 2013). A two-tailed P-value was used and the α level was 0.05.
Data availability
Sequence reads are available in the NCBI Sequence Read Archive as Bioproject PRJNA306895 and SRA accession number SRP095705. De novo assemblies are available as NCBI whole genome shotgun sequences (accession numbers: LQCX01-LQGS01). GenBank accession numbers are SRR5133037–SRR5133136. Additional supporting data are provided in Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, File S1, Table S1, Table S2, Table S3, Table S4, and Table S5.
Results
Sequencing and assembly
Laboratory-confirmed isolates of H. somni (n = 16; 13 cases, three controls), M. bovis (n = 20; 17 cases, three controls), M. haemolytica (n = 28; 20 cases, eight controls), and P. multocida (n = 30; 24 cases, six controls) from dairy cattle; and M. haemolytica (n = 2; two cases) and P. multocida (n = 8; eight cases) from beef cattle were sequenced, and raw reads were processed and assembled (Table 5 and Table S2). Staphylococcus aureus contamination was identified in one of the P. multocida samples isolated from an affected dairy calf and from two of the beef cattle samples, as well as in one of the beef cattle M. haemolytica samples. These samples were excluded from further analysis. A total of 100 sequences—16 H. somni (all dairy), 20 M. bovis (all dairy), 29 M. haemolytica (28 dairy, one beef), and 35 P. multocida (29 dairy, six beef)—were selected for further analysis. Raw reads from these 100 samples were mapped to each of their respective reference sequences and subjected to further analysis.
Table 5. Average assembly statistics for each species sequenced in this study from de novo assembled contigs.
| Species | Contig | Coverage | Total Length | N50 | Min. Contig | Max. Contig |
|---|---|---|---|---|---|---|
| Histophilus somni | 77 | 62.46 | 2,136,280 | 89,438 | 1094 | 294,083 |
| Mycoplasma bovis | 74 | 72.41 | 943,236 | 27,508 | 1061 | 59,658 |
| Mannheimia haemolytica | 105 | 35.68 | 2,637,528 | 70,779 | 1068 | 282,147 |
| Pasteurella multocida | 33 | 29.33 | 2,317,940 | 171,866 | 1099 | 469,530 |
Total length is the total of all contig lengths; N50 is 50% of the mass of distribution of the contig lengths; Min. contig and Max. contig are the length of the shortest and longest contigs in each assembly.
Of the 29 M. haemolytica samples, 22 were classified as genotype 2 and the other seven were classified as genotype 1, as recently described by Clawson et al. (2016). Of the 22 samples classified as genotype 2, 19 were obtained from cases (86.4%; P < 0.01); while of the seven samples classified as genotype 1, five were obtained from controls (71.4%; P < 0.01). Samples classified as genotype 2 have largely been associated with those taken from BRD-affected animals, while samples classified as genotype 1 have been associated with nonclinical isolates of M. haemolytica. There have been some genotype 1 samples obtained from BRD-affected animals, as well as genotype 2 samples obtained from non-BRD–affected animals (Clawson et al. 2016).
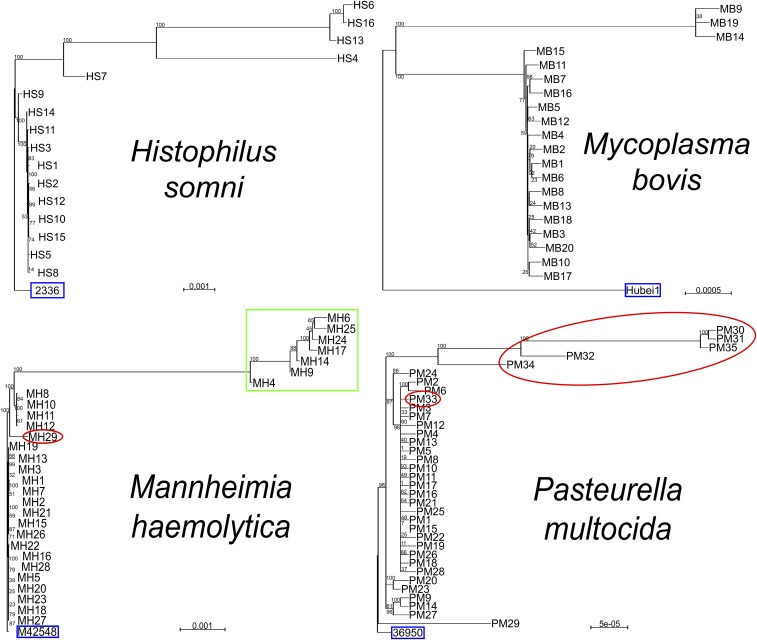
Genetic distance trees were generated to visualize the genetic distance between the genomic sequences and mapped assemblies. Overall, the sequence assemblies for the samples collected in this study increased the genetic diversity for available WGS for three out of the four species analyzed, H. somni, M. bovis, and P. multocida (Figure 1). Five of the H. somni samples contained sequences that were genomically distant from both available reference sequences, while the other 11 samples were genomically distant from reference sequence 129PT (Figure S1a). While the M. bovis samples showed little variation among themselves, all 20 samples were genomically distant from the five reference sequences (Figure S1b). The M. haemolytica sample population was the only species that revealed little genetic variation between the samples collected and the available reference sequences (Figure S1c). The seven samples classified as genotype 1 clustered together away from the other 22 samples (Figure S1c). The P. multocida sample population showed significant variation between the samples collected and six of the 11 available reference sequences (Figure S1d). For samples aligned to P. multocida reference sequences 3480, HN06, OH1905, OH4807, PMTB2.1, and Pm70, the study samples clustered together away from the reference sequence, showing a large distance between the sample group and the reference sequences. For the remaining five reference sequences, 36950, ATCC_43137, HB01, HB03, and Pm-3, there was little genetic diversity between the samples collected and the reference sequences.
Figure 1.
Genetic distance trees from mapped assemblies of the four bacterial species, Histophilus somni aligned to reference sequence 2336, Mycoplasma bovis aligned to Hubei1, Mannheimia haemolytica aligned to reference sequence M42548, and Pasteurella multocida aligned to reference sequence 36950. Red circle, beef cattle; blue box, reference sequence; green box, M. haemolytica genotype 1 (Clawson et al. 2016). Scale bar represents number of substitutions per site.
ARGs
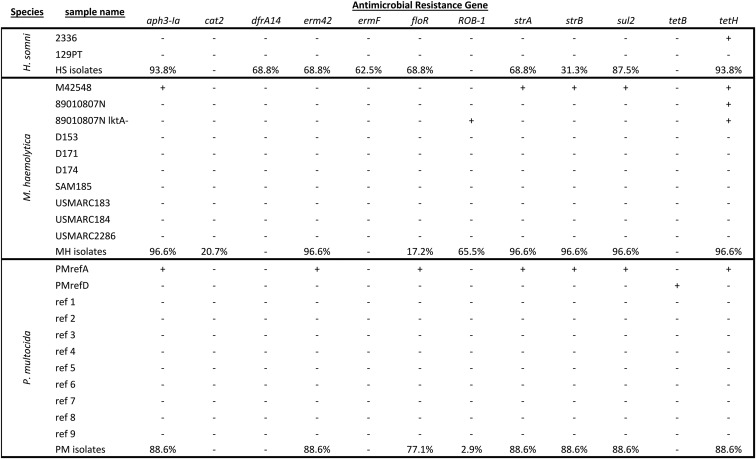
Raw read and de novo assembled contigs were aligned to the Resfinder database using SRST2 and ABRicate, respectively. Table 4 lists known ARG detected and the antimicrobial(s) to which the genes can confer resistance. Phenotypic antimicrobial susceptibility testing results were obtained for 64 isolates (13 H. somni, 26 M. haemolytica, and 25 P. multocida) for 18 antimicrobials from nine classes. The remaining three samples of H. somni, three samples of M. haemolytica and 10 samples of P. multocida failed to grow in the susceptibility testing positive control media and were excluded from analysis. Using in silico ARG prediction methods, 11 total ARG were detected, with two genes specific to H. somni isolates (ermF and dfrA14), two genes specific to M. haemolytica isolates (ROB-1 and cat2), and the other seven detected in H. somni, M. haemolytica, and P. multocida (Figure 2). No ARG were detected in any of the M. bovis samples, and therefore, these samples were not subjected to susceptibility testing. Raw reads were mapped to the PlasmidFinder database using SRST2 to determine whether ARG present were on plasmids or integrated into the genome. No plasmids containing known ARG sequences were detected using this method. Additionally, alignment of de novo assembled contigs to the CARD protein variant model database did not reveal any variants in genes known to confer resistance to antimicrobials.
Figure 2.
Prevalence of resistance genes among isolates of Histophilus somni (N = 16), Mannheimia haemolytica (N = 29), and Pasteurella multocida (N = 35), as well as available reference sequences. +, present; −, absent.
H. somni ARG:
In silico ARG identification was performed for the two H. somni reference sequences 2336 and 129PT (Figure 2). No ARG were predicted in reference sequence 129PT and only tetH was predicted in reference sequence 2336. Nine ARG were detected in the H. somni isolates (Figure S2). Two aminoglycoside resistance genes that confer resistance to the drugs KAN and NEO (aph3-Ia) and streptomycin (strA, strB) were detected. Two macrolide-lincosamide-streptogramin B (MLSb) resistance genes (erm42 and ermF) were also detected, as was a phenicol resistance gene (floR), a sulfonamide resistance gene (sul2), a trimethoprim resistance gene (dfrA14), and a tetracycline resistance gene (tetH) (Figure 2). There was a 79.2% concordance rate between genes identified using raw reads and de novo assembled contigs.
M. haemolytica ARG:
Different ARG were identified across M. haemolytica reference sequences. The genes aph3-Ia, strB, sul2, tetH and the β-lactamase gene, ROB-1, were detected in reference sequence M42548; ROB-1 was detected in reference sequence 89010807N_lktA; tetH was detected in reference sequence 89010807N_lktA and reference sequence 89010807N; no ARG were detected in any of the other available reference sequences (n = 7, Figure 2). Seven of the eight ARG detected in M. haemolytica isolates (aph3-Ia, erm42, floR, strA, strB, sul2, and tetH) were also found in H. somni and P. multocida (Figure 2). No ARG were detected in the only M. haemolytica sample sequenced from BRD-affected beef cattle (Figure S3). Two of the genes (erm42 and floR) were not detected in any reference sequence but have been previously reported (Clawson et al. 2016). There was an 89.3% concordance rate between genes identified using raw reads and de novo assembled contigs. There was complete linkage of ARG aph3-Ia, strA and strB in 28 out of 29 samples, showing homology to the conserved backbone of ICEMh1 and ICEPmu1 (Figure S3). The presence of the conserved ICE backbone has been associated with samples classified as genotype 2. Here, 21 out of 22 samples classified as genotype 2 contained regions homologous to the ICE backbone and all seven samples classified as genotype 1 also contained regions homologous to the ICE backbone. Additionally, the phenicol resistance gene cat2 was identified in six out of 29 samples. Six of the seven samples classified as genotype 1 contained the cat2 ARG, while the other 22 samples, classified as genotype 2, did not contain the cat2 ARG (Figure S3).
P. multocida ARG:
The available P. multocida reference sequences had similar ARG variation to that seen in the M. haemolytica reference sequences. All seven of the ARG (tetH, aph3-Ia, erm42, strA, strB, floR, and sul2) detected in the P. multocida samples were also detected in reference sequence 36950 (Michael et al. 2011). The gene tetB was detected in reference sequence HB01, but no ARG were detected in any of the other nine reference sequences (Figure 2). Seven of the 11 ARG were detected in the P. multocida samples originating from dairy cattle (Figure 2). Sequences from two samples obtained from beef cattle contained four and five of seven ARG, tetH, aph3-Ia, strA, strB and tetH, aph3-Ia, erm42, strA, strB, respectively, but there were no ARG detected in the other four samples obtained from beef cattle (Figure S4). One of the 35 samples contained the β-lactamase gene, ROB-1. There was a 76.5% concordance rate between genes identified using raw reads and de novo assembled contigs. Like M. haemolytica samples, there was complete linkage of ARG aph3-Ia, strA and strB in 31 out of 35 samples, showing homology to the conserved backbone of ICEMh1 and ICEPmu1.
Phenotypic susceptibility testing results
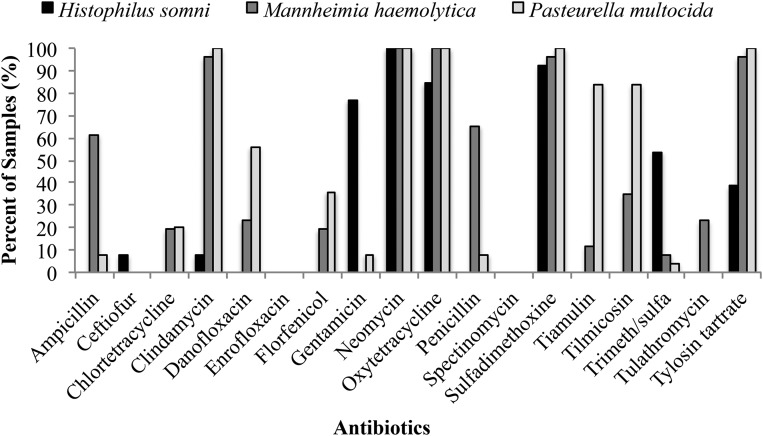
All 64 isolates displayed phenotypic resistance to NEO at a resistant breakpoint of ≥32 μg/ml (File S1 and Table 6). All 25 of the P. multocida isolates were also resistant to TYL, OXY, SUL, and CLIN, while all 26 of the M. haemolytica were resistant to OXY (Figure 3). None of the isolates displayed phenotypic resistance to SPEC or ENRO, and a single H. somni isolate was resistant to CEFT (Table 6 and Table S3). M. haemolytica was the only organism that displayed resistance to TUL (five out of 26 isolates). None of the isolates from any species were susceptible to all drugs tested; 61 were MDR (defined as exhibiting phenotypic resistance to more than two antimicrobial classes tested) and the remaining three were resistant to only two classes of antimicrobials (Figure S5).
Table 6. Concordance of phenotypic and susceptibility results for all three organisms (64 isolates).
| Pheno: S | Pheno: R | |||||||
|---|---|---|---|---|---|---|---|---|
| Class | Antibiotic | Geno:R | Geno:S | Geno:R | Geno:S | Discordant (%) | Concordant (%) | P-Value |
| β-Lactam | Ampicillin | 4 | 42 | 15 | 3 | 11 | 89 | 0.73 |
| β-Lactam | Ceftiofur | 19 | 44 | 0 | 1 | 31 | 69 | <0.001 |
| β-Lactam | Penicillin | 4 | 41 | 15 | 4 | 13 | 88 | 0.99 |
| Tetracycline | Chlortetracycline | 53 | 1 | 10 | 0 | 83 | 17 | <0.001 |
| Tetracycline | Oxytetracycline | 1 | 1 | 62 | 0 | 2 | 98 | 0.50 |
| Fluoroquinolone | Danofloxacin | 0 | 44 | 0 | 20 | 31 | 69 | <0.001 |
| Fluoroquinolone | Enrofloxacin | 0 | 64 | 0 | 0 | 0 | 100 | 0.32 |
| Phenicol | Florfenicol | 24 | 26 | 14 | 0 | 38 | 63 | <0.001 |
| Aminoglycoside | Gentamicin | 0 | 52 | 0 | 12 | 19 | 81 | 0.002 |
| Aminoglycoside | Neomycin | 0 | 0 | 63 | 1 | 2 | 98 | 1.0 |
| Aminoglycoside | Spectinomycin | 0 | 64 | 0 | 0 | 0 | 100 | 0.37 |
| Sulfonamide | Sulfadimethoxine | 1 | 1 | 60 | 2 | 5 | 95 | 0.63 |
| Trimeth/Sulf | TMS | 33 | 21 | 7 | 3 | 56 | 44 | <0.001 |
| Pleuromutilin | Tiamulin | 0 | 40 | 0 | 24 | 37 | 63 | <0.001 |
| MLS | Clindamycin | 7 | 6 | 31 | 20 | 42 | 58 | 0.02 |
| MLS | Tilmicosin | 13 | 21 | 25 | 5 | 28 | 72 | 0.06 |
| MLS | Tulathromycin | 33 | 26 | 7 | 3 | 52 | 48 | <0.001 |
| MLS | Tylosin tartrate | 5 | 4 | 33 | 22 | 42 | 58 | <0.001 |
Pheno, phenotype; S, susceptible; R, resistant; Geno, genotype; Trimeth/Sulf, trimethoprim/sulfonamide; TMS, trimethoprim-sulfamethoxazole; MLS, macrolide-lincosamide-streptogramin.
Figure 3.
Prevalence of phenotypic resistance to all drugs tested among isolates of Histophilus somni (N = 13), Mannheimia haemolytica (N = 26), and Pasteurella multocida (N = 25).
Genotype–phenotype concordance
Concordance rates for antimicrobial resistance genotype and phenotype varied widely for each antimicrobial drug (Table 6). Across all three organisms, ENRO and NEO exhibited 100% concordance, with all isolates displaying phenotypic susceptibility and none containing genes typically associated with resistance to these antimicrobials, i.e., adenylyltransferases AAD(3″) and AAD(9) for SPEC and parC, parE, gyrA, and gyrB for ENRO. As with NEO, SPEC also exhibited high genotype–phenotype concordance (98%), but in contrast with NEO, this concordance was driven by presence of aph3-Ia and corresponding phenotypic resistance. Concordance to OXY was also very high across all three organisms; of the 64 isolates tested, 62 displayed phenotypic resistance to OXY and contained the tetH resistance gene. All three organisms exhibited statistically significant phenotype–genotype discordance for CHLOR and TIL (P < 0.05; Table S3, Table S4, and Table S5). CHLOR discordance was driven by phenotypic susceptibility in the presence of tetH (opposite the results for OXY). For H. somni and P. multocida, TIL discordance resulted from phenotypic susceptibility in the presence of ermF and/or erm42. Interestingly, erm42 has previously been identified in MDR P. multocida isolates from cases of BRD, but has yet to be described in H. somni (Kadlec et al. 2011). Unlike TIL discordance in H. somni and P. multocida, discordance in M. haemolytica was due largely to phenotypic resistance without identification of a known macrolide resistance gene.
H. somni and M. haemolytica demonstrated significant discordance for CLIN, however this discordance was driven by different dynamics (Table S3 and Table S4). Within H. somni, discordance resulted from phenotypic susceptibility in the presence of both erm42 and ermF, whereas phenotypic resistance and a lack of any known macrolide resistance genes drove the discordance in M. haemolytica.
H. somni and P. multocida exhibited significant discordance for FLOR, which in both organisms resulted from phenotypic susceptibility despite presence of the floR resistance gene. In addition, TUL was significantly discordant in both H. somni and P. multocida (P < 0.05; Table S3, Table S4, and Table S5), in both cases due to presence of erm42 and/or ermF without corresponding phenotypic resistance.
Phenotype and genotype results for DAN and TMS were significantly discordant in both M. haemolytica and P. multocida (P < 0.05; Table S4 and Table S5). For DAN, this discordance was due to phenotypic resistance without identification of a known danofloxacin resistance gene. Conversely, TMS discordance was driven by phenotypic susceptibility despite presence of the sul2 gene.
In addition, H. somni exhibited significant discordance for GENT, with 10 out of 13 isolates displaying phenotypic resistance but none containing a resistance gene known to confer resistance to this drug. All 10 of these isolates contained aph3-Ia, but this gene is typically associated with resistance to the aminoglycosides kanamycin, NEO, and paromomycin within Enterobacteriaceae spp. (Livermore et al. 2001). The breakpoint used for GENT was developed for Pseudomonas aeruginosa and Enterobacteriaceae in canines and equids, and Actinobacillus spp. in equids (CLSI 2013), and it is unknown whether these breakpoints are appropriate for H. somni in cattle.
M. haemolytica isolates also displayed statistically significant discordance for both CEFT and TYL (P < 0.05; Table S4). In the case of TYL, 25 of the 26 isolates were phenotypically resistant, but only six of these 25 isolates contained erm42. CEFT discordance resulted from the fact that none of the isolates whose genomes contained ROB-1 experienced CEFT resistance due to the presence of this β-lactamase gene.
P. multocida isolates were also significantly discordant for the pleuromutilin TIA (P < 0.05; Table S5), with 21 out of 25 isolates displaying phenotypic resistance despite lack of an identified pleuromutilin resistance gene within the isolates’ genomes.
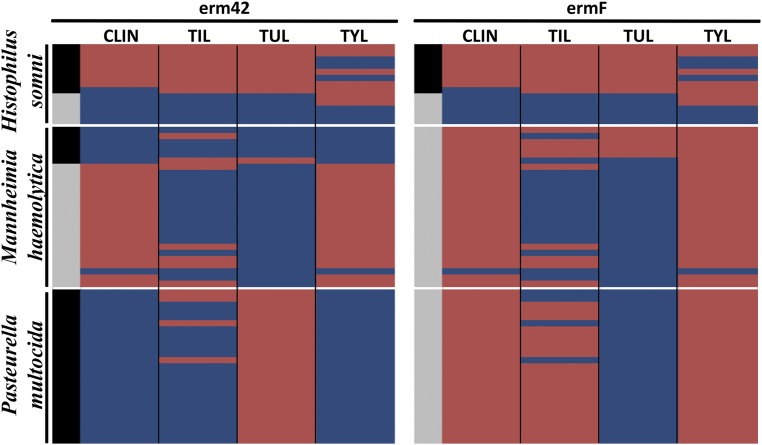
Macrolide resistance is of particular concern, as BRD is typically treated with either tetracyclines or macrolides. We identified two MLSb resistance genes within the 64 samples, erm42 and ermF. The genotype–phenotype concordance results differed greatly by the combination of gene, bacteria, and macrolide drug in question, as illustrated in Figure 4. When averaged across all three bacterial species, the genotype–phenotype discordance rates were statistically significant for all four macrolides tested (Table 6). However, discordance rates and significance varied by organism (Table S3, Table S4, and Table S5). Additionally, there was not a strong correlation between the number of ARG detected in each sample and the number of antimicrobials that each sample was resistant to (Figure S6).
Figure 4.
Heatmap showing concordance (blue) and discordance (red) between erm42 and ermF presence (black) or absence (gray) and phenotypic resistance to the four macrolide drugs included in the susceptibility testing panel (columns), by isolate (rows) within organism. CLIN, clindamycin; TIL, tilmicosin; TUL, tulathromycin; TYL, tylosin tartrate.
Discussion
Genome assembly and diversity
Additional WGS from BRD pathogens are needed to help understand genetic diversity and its role in pathogenesis, genotype–phenotype associations, and the relationship between genomic features and clinical disease. This study doubled the number of available genomic sequences for these four species. In addition to expanding current WGS databases, standardization of genome analysis methods for genomic epidemiological interpretation is important (Gilmour et al. 2013). As we have shown in this study, results obtained from de novo assemblies vs. consensus sequences constructed from mapping to reference genomes can be highly variable. This is demonstrated not only by the differences in genetic distances for each consensus sequence called when mapped to their respective reference sequences (Figure S1), but also by variability in ARG (Figure 2). However, there are many benefits to using a reference-based mapping approach, particularly when many samples need to be sequenced and analyzed. One such advantage is the computing power needed for consensus mapping is much lower as compared to de novo assembly and phylogenomic analyses using mapped assemblies yield similar results to those produced by de novo assemblies (Fitz-Gibbon et al. 2017). Genome assembly via mapping of reads to reference sequences enabled us to efficiently characterize variation within our study population, even if there is a slight bias toward the reference sequences (Figure S1).
Little variation was detected between samples mapped to each of the 10 M. haemolytica reference sequences. This is likely due to similarity in strains from cattle across the Midwest and California, as nine of the 10 reference sequences were obtained from cattle in the Midwest. Similarly, P. multocida samples clustered much more closely to reference sequences 3480, HN06, OH1905, OH4807, PMTB2.1, and Pm70, as each of these samples were obtained from cattle in California or the Midwest. In contrast, P. multocida samples in our study clustered more distantly from reference sequences 36950, ATCC_43137, HB01, HB03, and Pm-3, which were obtained from cattle in Germany or China. In this dataset, there was a clear distinction between P. multocida isolates collected from the beef and dairy cattle populations with the exception of one sample. However, it is important to note that only six of the 35 P. multocida samples analyzed were obtained from BRD-affected beef cattle. Further investigations with larger sample sizes from broader geographical ranges will be required to validate these observations.
Predicting antimicrobial resistance
The ability to predict ARG in silico would have a significant impact on our ability to diagnose and treat pathogenic diseases and monitor the spread of ARG (Koser et al. 2014). In a study of animals previously treated with antimicrobials, resistance was identified in 72% of M. haemolytica and 50% of P. multocida isolates, with 13 ARG present in H. somni (Klima et al. 2014). Other studies in healthy cattle suggest that the prevalence of antimicrobial resistance in BRD-associated bacteria may be relatively low (Noyes et al. 2015). M. haemolytica and P. multocida harbor integrative and conjugative elements that enable the transfer of gene blocks between the species (Klima et al. 2014) and confer resistance to antimicrobials (Michael et al. 2011; Eidam et al. 2015). M. bovis lacks a cell wall and is resistant to drugs that target the cell wall of Gram-negative bacteria; to date no ARG have been found in any strain of M. bovis (Maunsell et al. 2011).
In this study, we detected 11 antibiotic resistance genes within the sample population, including three aminoglycoside resistance genes, two macrolide-lincosamide-streptogramin resistance genes, two phenicol and one each of β-lactam, sulfonamide, fluoroquinolone, tetracycline, and trimethoprim resistance genes. Tetracyclines and macrolides are often used as first-line agents in the treatment of BRD. The prevalence of tetH and erm42 were relatively high in this population, and the presence of these ARG within BRD-associated bacterial isolates could render BRD treatment more challenging (Livermore et al. 2001). However, prevalence of resistance to antimicrobials typically used as second- or third-line treatments was relatively low, i.e., floR and cat2 for florfenicol. As a result, several alternative, commonly used antimicrobials for BRD treatment (USDA 2011b) could still be effective based on our findings.
Phenotypic antimicrobial resistance and the presence of ARG have both been demonstrated for H. somni, M. haemolytica, and P. multocida (Klima et al. 2014; DeDonder and Apley 2015). It is known that antimicrobials that target the cell wall, such as β-lactams, are not effective against M. bovis due to the lack of a cell wall. While we did not identify ARG in any of the M. bovis genomes in this study, phenotypic resistance within this organism has previously been demonstrated (Kawai et al. 2014; Kong et al. 2015). Each of the other three species has shown resistance to tetracyclines (Kehrenberg et al. 2001; D’Amours et al. 2011), which is the primary resistance gene found in H. somni samples in previous studies, but the majority of resistance genes previously found have been in P. multocida and M. haemolytica.
Previous studies have shown macrolide-lincosamide-streptogramin, kanamycin, NEO, and streptomycin resistance in H. somni samples (DeDonder and Apley 2015), but there were no ARG found in H. somni reference sequences other than tetH. However, recent studies have shown the presence of seven out of nine ARG found in H. somni samples during this study (Klima et al. 2014), showing the variation of ARG found in different strains. Recent studies showing the presence of known ARG, as well as phenotypic antimicrobial resistance, have primarily been done in diseased animals from feedlots around the Midwest, while the reference sequence samples were obtained from diseased animals in California. Our study shows a similarity in ARG to the recent study of samples obtained from feedlots in Texas, Nebraska, and Alberta, Canada (Klima et al. 2014). Importantly, eight of nine ARG found in H. somni and three of eight ARG found in M. haemolytica were not found in any of the previously available reference genome sequences.
It is important to note that there was slightly more ARG detected in each sample using raw reads compared to ARG detected using de novo assembled contigs. Using raw reads for gene detection has been shown to be superior to assembly-based approaches. This is primarily because assembly-based approaches are dependent on the quality of the sequencing, the assembly method used, and the quality of the assembly itself (Clausen et al. 2016).
The grouping of ARG within each sample is not surprising, as these genes would likely increase the survival rate of the bacteria, thus representing a substantial fitness benefit and eventually becoming fixed in the population (Griffin et al. 2010). The fact that three out of four species of bacteria associated with BRD share many of the same ARG is likewise unsurprising, as infection is highly associated with the presence of multiple pathogens (Griffin et al. 2010). Additionally, previous studies have shown there is a high incidence of horizontal gene transfer of ARG-containing plasmids between M. haemolytica and P. multocida (Klima et al. 2014), and the presence of the integrative and conjugative elements found in these two species have been associated with isolates recovered from BRD-affected cattle (Neibergs et al. 2014). Here, we show that 97% of M. haemolytica samples and 86% P. multocida samples obtained from BRD-affected cattle contain regions homologous to the ICE backbone. It was interesting that no ARG were detected in five of the seven samples collected from untreated beef cattle, other than the two P. multocida samples. While both M. haemolytica and P. multocida are known to exist commensally in the respiratory tract of animals not affected by BRD (Neibergs et al. 2014), the significance of these findings needs to be substantiated by a larger number of more diverse samples.
While in silico analysis can be used to identify ARG within WGS data, it does not reveal the functionality or effectiveness of the ARG in vivo. Therefore, antimicrobial susceptibility testing should be performed alongside in silico methods in order to understand the expression of these ARG, at least within laboratory testing conditions. In addition, extensive metadata should be collected for each sample, including the eventual clinical outcome for the animal from which the sample was taken. These types of data would help us to understand how knowledge of the presence of ARG within a given genome could potentially predict treatment and morbidity/mortality outcomes.
Genotype–phenotype concordance
The genotype–phenotype results from this study demonstrate the high degree of variability in concordance rates between different drugs, genes, and organisms. The ability of antimicrobial resistance genotype to predict phenotypic susceptibility has been under much discussion recently, particularly in the context of increased use of WGS as part of national food safety monitoring systems (Allard et al. 2016; Burrell et al. 2016). Some of the discordance rates presented in this study were higher than rates previously reported (Card et al. 2013; Agyekum et al. 2016). However, the available literature does not include results for the three organisms included in this study, which suggests that genotype–phenotype concordance rates may be highly species-specific. Alternatively, the breakpoints that we utilized for this study may not have been appropriate for H. somni, M. haemolytica, and P. multocida, especially since many of the breakpoints were not formulated for the relevant drug-bacterium-host combinations (Table 4). It has been shown that when changing the specific parameters of drug-bacterium-host, the CLSI-approved breakpoints become invalid and in cases were phenotypic resistance was seen, but no known ARG was detected, it becomes difficult to discern actual phenotypic resistance from inappropriate reporting (DeDonder and Apley 2015). However, even among breakpoints that were relevant, we found varying levels of genotype–phenotype concordance. Host- and organism-appropriate breakpoints were used for ENRO, TUL, FLOR, CHLOR, OXY, CEFT, and SPEC. While concordance rates were >98% for ENRO, OXY, and SPEC, there was significant discordance for CEFT, CHLOR, FLOR, and TUL (P < 0.05; Table 6). Therefore, issues concerning breakpoints do not fully account for the relatively high rates of genotype–phenotype discordance observed.
In many cases, the presence of a resistance gene correlated well with certain drugs within the relevant antimicrobial class, but not others. For instance, within M. haemolytica isolates the presence of the ROB-1 gene correlated well with phenotypic resistance to AMP and PEN, but not CEFT, to which there was no phenotypic resistance. Therefore, these results suggest that ROB-1 may not cause resistance to third-generation cephalosporins in M. haemolytica; a similar finding has been demonstrated with ROB-1 in Canadian Haemophilus influenzae human isolates (Karlowsky et al. 2000). Phenotypic NEO resistance was present across all 64 isolates, and 63 of the isolates also contained aph3-Ia, suggesting that this gene may be a very good predictor of neomycin resistance in H. somni, M. haemolytica, and P. multocida.
Resistance to TYL was nearly 100% within M. haemolytica and P. multocida isolates. However, presence of the erm42 gene was only predictive of resistance in P. multocida (Figure 4). Therefore, additional unrecognized mechanisms or genes are likely responsible for resistance in the other two species, although inappropriate breakpoints cannot be discounted. In contrast, the presence of erm42 predicted TUL resistance well within M. haemolytica, in that all five resistant isolates contained erm42 and none of the susceptible isolates did. However, the presence of erm42 in all 25 P. multocida isolates did not result in TUL resistance (Figure 4). These results highlight the importance of considering the specific antimicrobial drug, resistance gene, organism, and host when investigating the ability of genotype to predict phenotype (or vice versa).
Results of our study demonstrate the complexity of genome-wide dynamics in mediation of resistance, and emphasize the fact that simple presence/absence of individual genes does not always predict the behavior of a given organism under varying levels of antimicrobial drug exposures. More research is needed to develop reliable and accurate genomic predictors of antimicrobial resistance, and this research will likely need to be targeted toward specific drug-bacterium-host combinations. Until such genomic predictors are identified and refined, the scientific, medical, public health, and regulatory communities should be cognizant of the potential limitations of relying solely on genomic data to guide treatment and programmatic or regulatory decisions. Such a conservative approach is especially relevant to the veterinary community, where WGS of pathogens with high animal health importance has been limited.
The sequences produced as part of this work represent a first step toward increased understanding of genome-mediated antimicrobial resistance dynamics within these important cattle pathogens. However, the variability in our results highlights the continuing criticality of collecting pertinent host, production, and health outcome information (i.e., metadata) for each isolate. Without such information, the scientific community will be unable to evaluate the implications of positive identification of resistance genes within WGS data, and improve BRD morbidity/mortality outcomes by predicting the most effective treatment regime for BRD based on the specific gene-bacterium-host combination.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.1137/-/DC1.
Acknowledgments
Library preparation was performed at the University of California Davis DNA Technologies Core Facility. The authors would like to thank the University of California Davis Bioinformatics Core Facility for assistance with data analysis. The authors would also like to thank Paul Rossitto, James Moller, Bryan Welly, Ellen Lai, and Nadia Torok for assistance with sample collection, and Julie Cleek, Toni Ortiz, and Muhammad Ilyas for bacterial work-up and biochemical identification. Dairy cattle samples were collected as part of the US Department of Agriculture, Agriculture and Food Research Initiative competitive grant no. 2011-68004-30367. The sequencing and antimicrobial resistance phenotyping was funded by a grant from the Russell L. Rustici Rangeland and Cattle Research Endowment.
Footnotes
Communicating editor: M. Watson
Literature Cited
- Agyekum A., Fajardo-Lubian A., Ai X., Ginn A. N., Zong Z., et al. , 2016. Predictability of phenotype in relation to common betalactam resistance mechanisms in Escherichia coli and Klebsiella pneumoniae. J. Clin. Microbiol. 15: 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T. W., Cook S. R., Yanke L. J., 2008. A multiplex polymerase chain reaction assay for the identification of Mannheimia haemolytica, Mannheimia glucosida and Mannheimia ruminalis. Vet. Microbiol. 130: 165–175. [DOI] [PubMed] [Google Scholar]
- Allard M. W., Strain E., Melka D., Bunning K., Musser S. M., et al. , 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 54: 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenger B. M., Berry C., Peterson T., Fach P., Delannoy S., et al. , 2015. The utility of multiple molecular methods including whole genome sequencing as tools to differentiate Escherichia coli O157:H7 outbreaks. Euro Surveill. 20: 30073. [DOI] [PubMed] [Google Scholar]
- Burall L. S., Grim C. J., Mammel M. K., Datta A. R., 2016. Whole genome sequence analysis using JSpecies tool establishes clonal relationships between Listeria monocytogenes strains from epidemiologically unrelated Listeriosis outbreaks. PLoS One 11: e0150797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby L. M., Lund O., et al. , 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58: 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card R., Zhang J., Das P., Cook C., Woodford N., et al. , 2013. Evaluation of an expanded microarray for detecting antimicrobial resistance genes in a broad range of gram-negative bacterial pathogens. Antimicrob. Agents Chemother. 57: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe J. F., Duncan A. J., Brettin T. S., Bruce D., Chertkov O., et al. , 2007. Complete genome sequence of Haemophilus somnus (Histophilus somni) strain 129Pt and comparison to Haemophilus ducreyi 35000HP and Haemophilus influenzae Rd. J. Bacteriol. 189: 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirase N. K., Green L. W., 2000. Influence of oral natural interferon-a on performance and rectal temperature of newly received beef steers. Proc. Am. Soc. Anim. Sci. West. Sec. 51: 411–414. [Google Scholar]
- Clausen P. T., Zankari E., Aarestrup F. M., Lund O., 2016. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J. Antimicrob. Chemother. 71: 2484–2488. [DOI] [PubMed] [Google Scholar]
- Clawson M. L., Murray R. W., Sweeney M. T., Apley M. D., DeDonder K. D., et al. , 2016. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antimicrobial resistance genes. BMC Genomics 17: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI, 2013 Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, Ed. 4. Approved Standard VET01–A4. CLSI, Wayne, PA. [Google Scholar]
- D’Amours G. H., Ward T. I., Mulvey M. R., Read R. R., Morck D. W., et al. , 2011. Genetic diversity and tetracycline resistance genes of Histophilus somni. Vet. Microbiol. 150: 362–372. [DOI] [PubMed] [Google Scholar]
- Davenport K. W., Daligault H. E., Minogue T. D., Bishop-Lilly K. A., Bruce D. C., et al. , 2014. Complete genome sequence of type strain Pasteurella multocida subsp. multocida ATCC 43137. Genome Announc. 2: e01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, A. G., K. M. Sullivan, and M. M. Soe, 2013 OpenEpi: Open Source Epidemiologic Statistics for Public Health, 2013. Available at: www.OpenEpi.com. Accessed: February 26, 2017.
- DeDonder K. D., Apley M. D., 2015. A literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease. Anim. Health Res. Rev. 16: 125–134. [DOI] [PubMed] [Google Scholar]
- Edwards A. L., 1948. Note on the “correction for continuity” in testing the significance of the difference between correlated proportions. Psychometrika 13: 185–187. [DOI] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Brenner M. G., Kadlec K., Liesegang H., et al. , 2013. Complete genome sequence of Mannheimia haemolytica strain 42548 from a case of bovine respiratory disease. Genome Announc. 1: e00318-e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Leimbach A., Michael G. B., Kadlec K., et al. , 2015. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 70: 93–97. [DOI] [PubMed] [Google Scholar]
- Fagerland M. W., Lydersen S., Laake P., 2013. The McNemar test for binary matched-pairs data: mid-p and asymptotic are better than exact conditional. BMC Med. Res. Methodol. 13: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA , 2013. National Antimicrobial Resistance Monitoring System – Enteric Bacteria (NARMS): 2011 Executive Report. U.S. Department of Health and Human Services, Food and Drug Administration, Rockville, MD. [Google Scholar]
- Fitz-Gibbon S., Hipp A., Pham K., Manos P., Sork V. L., 2017. Phylogenomic inferences from reference-mapped and de novo assembled short- read sequence data using RADseq sequencing of California white oaks (Quercus subgenus Quercus). Genome 10.1139/gen-2016-0202 [DOI] [PubMed] [Google Scholar]
- Fulton R. W., Confer A. W., 2012. Laboratory test descriptions for bovine respiratory disease diagnosis and their strengths and weaknesses: gold standards for diagnosis, do they exist? Can. Vet. J. 53: 754–761. [PMC free article] [PubMed] [Google Scholar]
- Gilmour M. W., Graham M., Reimer A., Van Domselaar G., 2013. Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genomics 16: 25–30. [DOI] [PubMed] [Google Scholar]
- Griffin D., Chengappa M., Kuszak J., McVey D. S., 2010. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 26: 381–394. [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O., 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Harhay G. P., Koren S., Phillippy A. M., McVey D. S., Kuszak J., et al. , 2013. Complete closed genome sequences of Mannheimia haemolytica serotypes A1 and A6, isolated from cattle. Genome Announc. 1: e00188-e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauglund M. J., Tatum F. M., Bayles D. O., Maheswaran S. K., Briggs R. E., 2013. Genome sequences of Mannheimia haemolytica serotype A1 strains D153 and D193 from bovine pneumonia. Genome Announc. 1: e00848-e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauglund M. J., Tatum F. M., Bayles D. O., Maheswaran S. K., Briggs R. E., 2015a Genome sequences of Mannheimia haemolytica serotype A2 isolates D171 and D35, recovered from bovine pneumonia. Genome Announc. 3: e00093-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauglund M. J., Tatum F. M., Bayles D. O., Maheswaran S. K., Briggs R. E., 2015b Genome sequences of serotype A6 Mannheimia haemolytica isolates D174 and D38 recovered from bovine pneumonia. Genome Announc. 3: e00086-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton M. P., Harhay G. P., Smith T. P., Bono J. L., Chitko-McKown C. G., 2015. Complete closed genome sequences of a Mannheimia haemolytica serotype A1 leukotoxin deletion mutant and its wild-type parent strain. Genome Announc. 3: e00417-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Dashnow H., Raven L., Schultz M. B., Pope B. J., et al. , 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6: 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M., Abbot S. A., 2014. Culture-independent diagnostic testing: have we opened Pandora’s box for good? Diagn. Microbiol. Infect. Dis. 80: 171–176. [DOI] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., et al. , 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45: D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K., Brenner M. G., Sweeney M. T., Brzuszkiewicz E., Liesegang H., et al. , 2011. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob. Agents Chemother. 55: 2475–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Verma G., Zhanel G. G., Hoban D. J., 2000. Presence of ROB-1 betalactamase correlates with cefaclor resistance among recent isolates of Haemophilus influenzae. J. Antimicrob. Chemother. 45: 871–875. [DOI] [PubMed] [Google Scholar]
- Kawai K., Higuchi H., Iwano H., Iwakuma A., Onda K., et al. , 2014. Antimicrobial susceptibilities of Mycoplasma isolated from bovine mastitis in Japan. Anim. Sci. J. 85: 96–99. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Salmon S. A., Watts J. L., Schwarz S., 2001. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMHT1. J. Antimicrob. Chemother. 48: 631–640. [DOI] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C. L., Zaheer R., Cook S. R., Booker C. W., Hendrick S., et al. , 2014. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 52: 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L. C., Goa D., Jia B. Y., Wang Z., Gao Y. H., et al. , 2015. Antimicrobial susceptibility and molecular characterization of macrolide resistance of Mycoplasma bovis isolates from multiple provinces in China. J. Vet. Med. Sci. 78: 293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Harhay G. P., Smith T. P., Bono J. L., Harhay D. M., et al. , 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 14: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M., 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser C. U., Ellington M. J., Peacock S. J., 2014. Whole genome sequencing to control antimicrobial resistance. Trends Genet. 30: 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng H., Liu Y., Jiang Y., Xin J., et al. , 2011. The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS One 6: e20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Yang M., Xu Z., Zheng H., Liang W., et al. , 2012. Complete genome sequence of Pasteurella multocida HN06, a toxigenic strain of serogroup D. J. Bacteriol. 194: 3292–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Winstanley T. G., Shannon K. P., 2001. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 48: 87–102. [DOI] [PubMed] [Google Scholar]
- Love W. J., Lehenbauer T. W., Kass P. H., Van Eenennaam A. L., Aly S. S., et al. , 2014. Development of a novel clinical scoring system for on-farm diagnosis of bovine respiratory disease in pre-weaned dairy calves. PeerJ 2: e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. W., Krauland M. G., Nelson J. S., Schlackman J. L., Brooks A. M., et al. , 2015. Genomic epidemiology of an endoscope-associated outbreak of Klebsiella pneumoniae Carbapenemase (KPC)-producing K. pneumoniae. PLoS One 10: e0144310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunsell F. P., Woolums A. R., Francoz D., Rosenbusch R. F., Step D. L., et al. , 2011. Mycoplasma bovis infections in cattle. J. Vet. Intern. Med. 25: 772–783. [DOI] [PubMed] [Google Scholar]
- May B. J., Zhang Q., Li L. L., Paustian M. L., Whittam T. S., et al. , 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98: 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuirk S. M., 2008. Disease management of dairy calves and heifers. Disease management of dairy calves and heifers. Vet. Clin. North Am. Food Anim. Pract. 24: 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., et al. , 2011. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 67: 91–100. [DOI] [PubMed] [Google Scholar]
- Mohammed M., Cormican M., 2015. Whole genome sequencing provides possible explanations for the difference in phage susceptibility among two Salmonella Typhimurium phage types (DT8 and DT30) associated with a single foodborne outbreak. BMC Res. Notes 8: 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha M. M., Marsh J. W., Krauland M. G., Fernandez J. O., de Lemos A. P., et al. , 2015. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2: 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja A., Kim D. W., Thomson N. R., Connor T. R., Lee J. H., et al. , 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477: 462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neibergs H. L., Seabury C. M., Wojtowicz A. J., Wang Z., Scraggs E., et al. , 2014. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics 15: 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes N. R., Benedict K. M., Gow S. P., Booker C. W., Hannon S. J., et al. , 2015. Mannheimia haemolytica in feedlot cattle: prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J. Vet. Intern. Med. 29: 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Liang W., Liu W., Wu B., Tang B., et al. , 2016. Genomic characterization of Pasteurella multocida HB01, a serotype A bovine isolate from China. Gene 581: 85–93. [DOI] [PubMed] [Google Scholar]
- Qi J., Guo A., Cui P., Chen Y., Mustafa R., et al. , 2012. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS One 7: e38239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddaramappa S., Challacombe J. F., Duncan A. J., Gillaspy A. F., Carson M., et al. , 2011. Horizontal gene transfer in Histophilus somni and its role in the evolution of pathogenic strain 2336, as determined by comparative genomic analyses. BMC Genomics 12: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street D. A., Petersen K. R., Gerritsen A. T., Hunter S. S., Settles M. L., 2015. expHTS: analysis of high throughput sequence data in an experimental framework. Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, Atlanta, GA. pp. 523–524. [Google Scholar]
- Ullah W., Abubakar M., Arshed M. J., 2009. Differentiation of closely related vaccinal strains of Pasteurella multocida using polymerase chain reaction (PCR). VetScan. 4: 36. [Google Scholar]
- Untergrasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., et al. , 2012. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA, 2011a National Agricultural Statistics Service Agricultural Statistics Board (NASS SAB) Cattle Death Loss. Available at: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1625. Accessed: February 26, 2017.
- USDA, 2011b Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1,000 or More Head. USDA–APHIS–VS–CEAH–NAHMS. Fort Collins, CO #638.0913. [Google Scholar]
- Wise K. S., Calcutt M. J., Foecking M. F., Roske K., Madupu R., et al. , 2010. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect. Immun. 79: 982–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., et al. , 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67: 2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads are available in the NCBI Sequence Read Archive as Bioproject PRJNA306895 and SRA accession number SRP095705. De novo assemblies are available as NCBI whole genome shotgun sequences (accession numbers: LQCX01-LQGS01). GenBank accession numbers are SRR5133037–SRR5133136. Additional supporting data are provided in Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, File S1, Table S1, Table S2, Table S3, Table S4, and Table S5.