Abstract
The consistent detection and quantification of protein post-translational modifications (PTMs) across sample cohorts is an essential prerequisite for the functional analysis of biological processes. Data-independent acquisition (DIA), a bottom-up mass spectrometry based proteomic strategy, exemplified by SWATH-MS, provides complete precursor and fragment ion information of a sample and thus, in principle, the information to identify peptidoforms, the modified variants of a peptide. However, due to the convoluted structure of DIA data sets the confident and systematic identification and quantification of peptidoforms has remained challenging. Here we present IPF (Inference of PeptidoForms), a fully automated algorithm that uses spectral libraries to query, validate and quantify peptidoforms in DIA data sets. The method was developed on data acquired by SWATH-MS and benchmarked using a synthetic phosphopeptide reference data set and phosphopeptide-enriched samples. The data indicate that IPF reduced false site-localization by more than 7-fold in comparison to previous approaches, while recovering 85.4% of the true signals. IPF was applied to detect and quantify peptidoforms carrying ten different types of PTMs in DIA data acquired from more than 200 samples of undepleted blood plasma of a human twin cohort. The data approportioned, for the first time, the contribution of heritable, environmental and longitudinal effects on the observed quantitative variability of specific modifications in blood plasma of a human population.
Introduction
Proteins catalyze and control essentially all biochemical functions of a living cell. Discovery mass spectrometry methods have identified products from the predicted protein coding regions (open reading frames, ORFs) for numerous species, including the human species, to apparent saturation1. Yet, the number of proteoforms expressed from a particular genome by far exceeds the number of protein coding ORFs because a multitude of processes contribute to increasing proteomic diversity. Among these, post-translational modifications (PTMs) generate an enormous, but as yet unknown expansion of the expressed proteoforms as each protein contains many amino acid residues that are potentially modified. For the human proteome it has been estimated that these processes expand the core products of the ~20,000 ORFs to around 1 million different proteoforms2.
The detection of specific proteoforms has frequently been attempted by antibody-based methods3. For this, affinity reagents need to be optimized for each targeted species4. In reality, such reagents have frequently been of varying sensitivity and specificity5. Alternatively, “top-down” proteomics which uses mass spectrometry to assess intact proteins can differentiate individual proteoforms2, but is currently of limited throughput6. Thus, for many applications, liquid chromatography-coupled tandem mass spectrometry of proteolyzed proteins (LC-MS/MS; “bottom-up” proteomics) has been the method of choice for the unbiased, high-throughput identification and quantification of differentially modified peptides7,8, even though the information about proteoform association of thus identified peptides is lost during the step of enzymatic digestion.
Several “bottom-up” MS technologies have been developed that differ in their performance profiles9. They include discovery proteomics employing data-dependent acquisition (DDA)10, targeted proteomics by selected or parallel reaction monitoring (SRM11 or PRM12) and data-independent acquisition (DIA)13. DIA methods, exemplified by SWATH-MS, systematically fragment all precursor ions in a user defined retention time vs. precursor ion mass to charge (m/z) window, thus overcoming the stochastic precursor ion selection of DDA14. The favorable properties of DIA implemented on high resolution, accurate mass instruments include highly consistent detection of analytes across sample cohorts and accurate quantification over a dynamic range of more than 4.5 orders of magnitude15, and have contributed to the recent popularization of DIA-based methods16.
In bottom-up proteomics, the characterization of proteoforms relies on peptide level evidence. In analogy to Smith & Kelleher et al. 2, we herein use the term “peptidoform” to describe such specifically modified or mutated peptides with the same backbone amino acid sequence. Irrespective of the data acquisition method used, several significant challenges remain for peptidoform identification and quantification. They are i) correct identification of the peptide backbone sequence, ii) correct identification of the types of modified amino acids and, iii) correct localization of the modification(s) within the backbone sequence. Modification types have been identified by two different approaches. In the first, modified amino acids are included in the database search step as (optional) modification to the peptide sequence. In the second, peptide sequences are first identified and subsequently the modification mass is inferred from the measured precursor and fragment ion mass shifts to the theoretical masses of the unmodified peptide sequences (open or “blind” modification search)17. To address the problem of modification site-localization, algorithms have been developed which assess site-localization confidence on independent, annotated peptide spectrum matches (PSMs) in DDA data18. At a smaller scale, targeted analysis strategies for peptidoform assessment were previously used to infer site-localizations and modification types in SRM19 or PRM20 data.
In principle, the above methods and strategies could also be applied to the analysis of DIA data, either by the spectrum-centric approaches21–24 or by peptide-centric scoring25 based targeted data extraction methods as in SWATH-MS14,26. Particularly, the high degree of consistency of DIA data is expected to provide additional benefits for site-localization and quantification across all runs of a study. However, the structure of DIA methods presents additional challenges for peptidoform identification or detection that arise from the large precursor isolation windows used for data acquisition14. Specifically, if peptidoform precursors differ by modifications for which the m/z increment is below the width of the precursor isolation windows used, they are isolated together in the same window. This can lead to peak picking conflicts in the retention time (RT) dimension or lead to fragment ion interferences when they are co-eluting. For this reason, several studies focusing on peptide modifications in complex samples relied on manual inspection of extracted diagnostic fragment ions to differentiate peptidoforms27–30 or spectrum-centric assessment of the modified peptides22,24,31. However, manual inspection is prone to biases and does not scale to dozens or hundreds of samples with tens of thousands of peptides queried per sample. Further, the spectrum-centric approaches often have to apply a second peptide-centric scoring step that is dependent on very specific peptide query parameters31. Therefore, there is a critical need for algorithms that can automatically and confidently assign peptidoforms to detected peak groups in DIA data sets29.
Here we present IPF (Inference of PeptidoForms), an algorithm and software tool supporting the SWATH-MS14 methodology of data-independent acquisition and targeted data analysis. It is configured as a novel component of the OpenSWATH26 workflow supporting the analysis of peptidoforms. IPF offers the following features: i) IPF can generate peptide query parameters from various sources, such as DDA32 or DIA data, including pseudo spectra22 and open modification search results24,31; ii) IPF supports a targeted, hypothesis-driven approach to assign peptidoforms to candidate peak groups; iii) IPF adopts a multi-tier scoring approach, propagating the confidence of detection and site-localization from precursors and transitions to peptidoform-level using a Bayesian hierarchical model, and iv) IPF integrates seamlessly into the existing workflows to support peptidoform-specific large-scale experiments.
We benchmarked IPF performance on a “ground truth” sample consisting of a set of synthetic phosphopeptides and assessed the applicability, scalability and consistency of detection on both, phosphopeptide-enriched and non-enriched samples. We further demonstrate the application of the algorithm to a longitudinal heritability study of peptidoforms of human blood plasma proteins. The previously acquired data set was derived from plasma samples collected at two time points from 36 pairs of monozygotic and 22 pairs of dizygotic twins33. The data allowed us to assess the heritability, environmental and longitudinal effects on the observed variability of 4532 peptidoforms, and to differentiate between inherited and environmentally induced quantitative changes in PTMs.
Results
IPF enables peptidoform characterization from SWATH-MS data sets
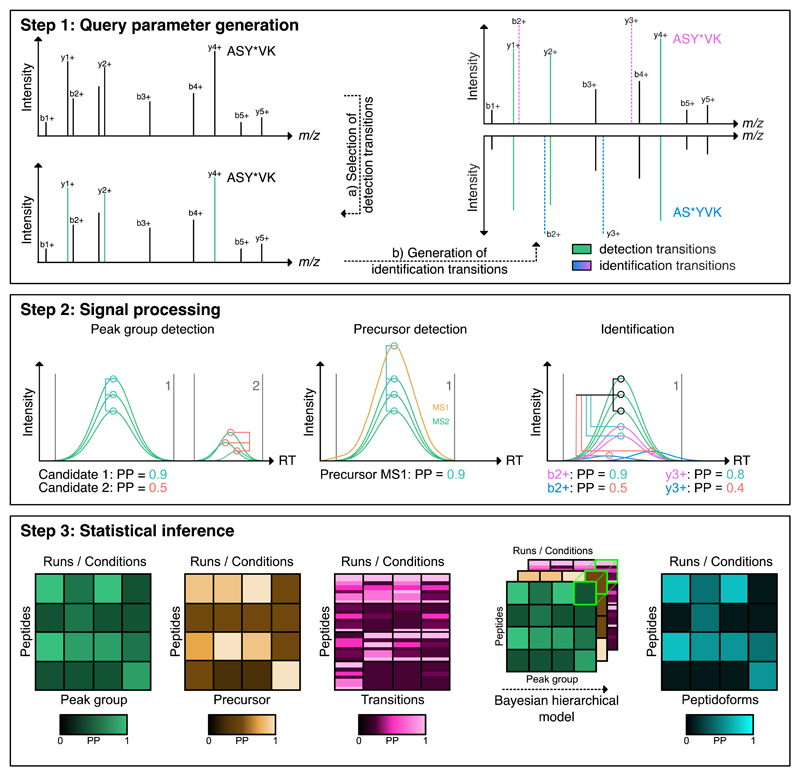
The IPF algorithm was developed for the detection of peptidoforms via peptide-centric scoring25 of DIA or SWATH-MS data and to support the consistent scoring of distinctive peptidoforms across sample cohorts. It extends the scoring systems commonly used for unmodified peptides to the peptidoform-level and includes the capability to validate multiple concurrent modification types and site-localizations on the same peptide. IPF consists of three main components (Fig. 1, Supplementary Notes I.A – I.C):
Figure 1. IPF analysis workflow overview.
Step 1: Query parameter generation: Based on a spectrum-centric workflow (DDA or DIA), peptide query parameters consisting of detection (most intense) and identification (site determining) transitions for all peptidoforms are generated. Step 2: Signal processing: Using a multi-tier scoring approach, the detection and identification transition-level and precursor ion chromatograms are extracted from the SWATH maps. The chromatograms on detection transition-level are used to find candidate peak groups against which the chromatograms of the precursor ion and identification transitions are scored. The multi-tier scoring estimates the posterior probability (PP) for the candidate peak groups using the detection transitions. In the step on precursor and transition-level, the PPs are estimated for the likelihood that individual precursor ions or identification transition-level chromatograms are originating from the peak group associated peptide. Step 3: Statistical inference: A Bayesian hierarchical model (BHM) integrates the precursor and transition PPs, according to residue specificity, to peptidoform PPs. In addition to the peptidoforms, the PP that the signal is a false positive is being updated.
Step 1: Query parameter generation
Peptide-centric scoring by targeted data extraction requires predefined peptide query parameters (also referred to as “Tier 3” assays34), consisting of specific transitions (precursor and product ion m/z), normalized fragment ion intensities and normalized retention time32. As first step, IPF uses spectral libraries or transition lists from prior spectrum-centric analyses of DDA or DIA data, processed by database and/or open modification searches as input to constitute sets of peptide query parameters. These empirically observed transitions are defined as “detection transitions”, because they can be used for the sensitive detection of peptides14. To increase specificity and to differentiate peak groups that could originate from closely related peptide species, IPF generates theoretical “identification transitions” using a defined model of modification residue specificity to probe the candidate peptidoform space, providing (weighted) evidence for or against particular peptidoforms (Supplementary Notes I.A, II.A, Fig. 1, Supplementary Fig. 1). The output of this step is a set of hybrid peptide query parameters for each peptidoform and precursor charge state, consisting of the different transition types, annotated with their specific scoring attributes.
Step 2: Signal processing
Targeted data extraction from the SWATH-MS data and peptide-centric scoring using the detection transitions is conducted as established for the standard OpenSWATH workflow26. In addition, chromatograms for identification transitions of precursor and fragment ions are extracted from MS1 and MS2 maps and scored individually against the chromatograms of the detection transitions within the boundaries of the detected peak groups (Fig. 1, Supplementary Fig. 1-2). The output of this step is the set of scores for candidate peak groups, their identification transition-level chromatograms and their precursor signals (Supplementary Notes I.B, II.B-C).
Step 3: Statistical inference and error-rate control
To infer the set of peptidoforms at a q-value or false discovery rate (FDR) threshold that is detected in the SWATH-MS data set, a statistical inference step is conducted by IPF. This is accomplished by processing the scored MS1-, MS2- and transition-level signals by a multi-level, semi-supervised learning algorithm, followed by peptidoform inference employing a Bayesian hierarchical model (Fig. 1, Supplementary Fig. 1). The confidence scores computed from candidate peak groups, precursor ions and individual transition-level chromatograms are propagated towards the identification of a peptidoform (Supplementary Notes I.C, II.D-F). The scored peak groups can then be used by TRIC35 to propagate the peptidoform-level detection confidence across multiple aligned runs to generate a more complete quantitative matrix (Supplementary Note II.G).
In summary, IPF extends the standard OpenSWATH workflow by providing confidence metrics for unmodified peptide queries as well as for other peptidoforms.
Benchmarking using a synthetic reference data set
To benchmark IPF, we performed SWATH measurements (termed synthetic phosphopeptide reference data set) on a collection of 579 heavy isotope labeled, unpurified synthetic phosphopeptides corresponding to Saccharomyces cerevisiae proteins involved in a range of cellular processes36 (Supplementary Table 1). The synthetic peptides were spiked in a 13-step dilution series (highest concentration: 0.002 µg/µl, see Methods) into a background consisting of an extract of the human cell line HEK-293 to enable assessment of the limits of detectability. The maximum dilution reached in the dilution series compared to the synthetic peptide mixture without background was 127-fold. Of the full set, 481 peptides were used as ground truth for benchmarking, because they were synthesized in only a single site-localized peptidoform. However, they contain multiple potentially modifiable residues (on average more than 3 modifiable residues, often near the actual modified residue).
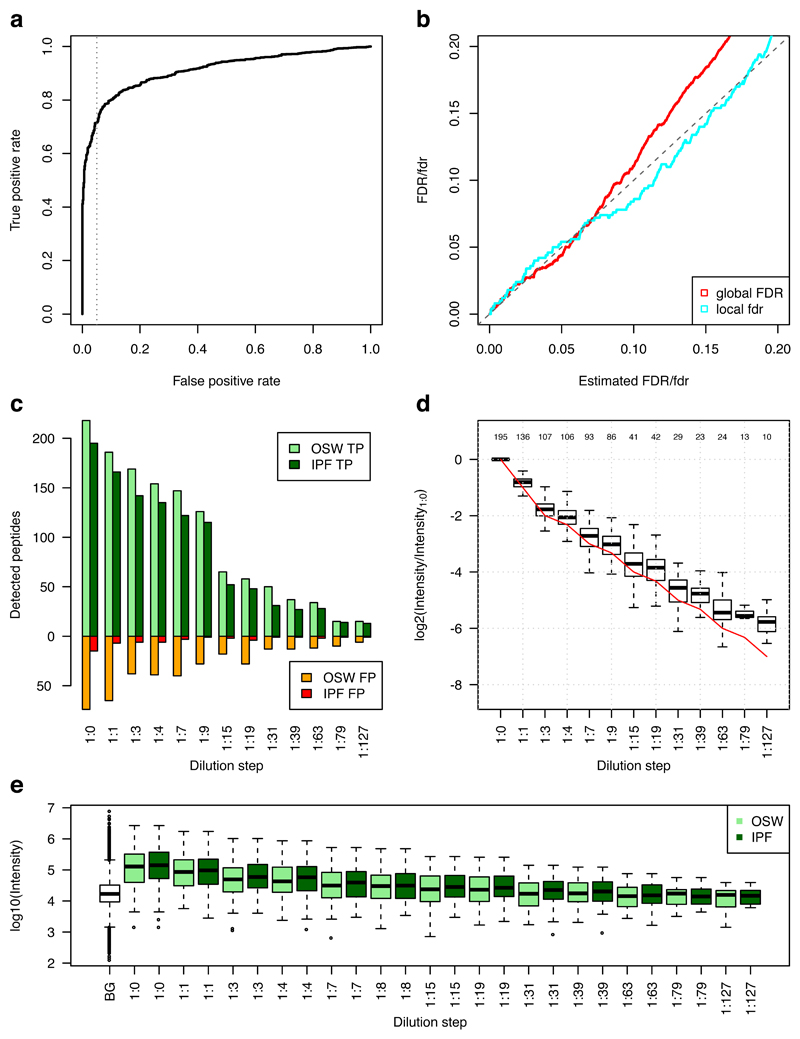
We first generated both a comprehensive DDA library of the phosphorylated ground truth peptides and a DIA library of the corresponding runs, including background proteome (Methods). Peptide query parameters for 297 ground truth peptides were derived from the DDA library, which IPF used to extract precursor and fragment ion chromatograms from the SWATH-MS data of the phosphopeptide dilution series in the HEK-293 human cell line background proteome. Figure 2a depicts the receiver operating characteristic (ROC) curve, where the cumulative positive (correct site-localizations) and negative detections over all 13 measurements were used as class labels. False identifications originated from erroneous phospho-site localization on correctly detected backbone sequences. At a false positive rate of 5%, a recall of 71.6% could be reached. The estimated global false discovery rate (global FDR) and local false discovery rate37,38 (local fdr) (Fig. 2b) compared to the ground truth indicate that the confidence propagation from transition- to peptidoform-level enabled accurate error control in the commonly used ranges of 1-5% fdr/FDR, with a small underestimation of the error in the higher ranges. Detectability at a global FDR of 5% over the whole dilution series shows a linear relationship of peptidoform detectability confidence to the abundance, with 195 correctly detected peptidoforms in the sample with the highest peptide concentration (Fig. 2c). In direct comparison to OpenSWATH, IPF produces a reduction of false site localizations by 87.0%, while recovering 85.4% of the true OpenSWATH signals at 5% estimated FDR. Quantification of correctly detected peptidoforms over the dilution series normalized to the undiluted sample indicates accurate quantification until the 1:15 dilution step, with a slight overestimation of the abundance in the more diluted samples (Fig. 2d). Confidence on the peptidoform-level in general requires slightly more intense signals than on the peptide sequence-level, indicating that the lowered sensitivity of IPF compared to OpenSWATH originates from ambiguous lower intensity signals (Fig. 2e). Supplementary Figure 3 depicts the equivalent results using a DIA library generated by DIA-Umpire22, where IPF achieved similar relative performance metrics compared to the DDA library, but due to the lower size of the input library, the absolute number of detected peptidoforms was smaller.
Figure 2. Benchmarking on the synthetic phosphopeptide reference data set.
Spiked-in synthetic yeast phosphopeptides36 were measured in a 13-step dilution series with a human cell line background. IPF was applied using a spectral library generated from DDA measurements of the synthetic peptides. a) The receiver operating characteristic (ROC) indicates high sensitivity at commonly used confidence thresholds with 71.6% recovery at 5% (grey dotted line) false positive rate. b) The estimated global false discovery rate (FDR) or q-values are plotted against the true FDR, computed using the ground truth. The dashed diagonal line indicates the optimum. The estimated local false discovery rate (fdr) or posterior error probability (PEP) is plotted against the actual fdr, approximated using the ground truth with a window size of 50037. IPF enables accurate estimation in the commonly used ranges of 1-5% fdr/FDR, with a small underestimation of the error in the higher ranges. c) The dilution series of synthetic spiked-in standard peptides against the constant human cell line digest and the detected true (green) and false (red) peptidoforms at 5% FDR are depicted. The light colors (OSW; OpenSWATH) represent the detectable peptide sequence-specific peptide query-level candidate peak group signals. The dark colors (IPF) represent the corrected, peptidoform-specific signals. A high gain in selectivity with a slight drop in sensitivity can be observed for IPF in comparison to OpenSWATH. d) The quantification of the peak groups (normalized against the 1:0 sample) is compared against the ground truth (red line). Until dilution step 1:15 the quantification is accurate, with a slight bias for overestimation at lower abundance dilution steps. The numbers above the boxplots indicate the number of peptides per dilution steps that are also present in the 1:0 step. e) The boxplots depict the intensities of correct peptidoforms and background (BG) peptides at 5% FDR. To achieve high confidence on peptidoform-level, IPF requires slightly higher signal intensities than OpenSWATH on peptide-level. Supplementary Figure 3 depicts the same plots for a library generated using DIA-Umpire.
We further used the results of the DIA-Umpire analysis to benchmark the site-localization component of IPF against established methods for spectrum-centric site-localization (Supplementary Note III.B, Supplementary Fig. 4). LuciPHOr39,40 is a recently developed algorithm for site localization. In addition to site-localization it also estimates the false localization rate (FLR). Using a version of IPF reduced to the second, site-localization layer of the Bayesian hierarchical model we analyzed the synthetic phosphopeptide reference data set to compare the correct and wrong site-localized peptides at estimated false localization rates with DIA-Umpire/LuciPHOr. IPF provided more sensitive results with a recovery of 66.7% of all true site-localized peptides at 5% false positive rate, compared to 55.3% as achieved by DIA-Umpire/LuciPHOr.
The above results demonstrate that IPF accurately determines peptidoforms with modification site-localization using spectral libraries generated both by DDA- and DIA-based methods. IPF reaches a favorable tradeoff between sensitivity and selectivity compared to the standard peptide-centric workflows, for example implemented in OpenSWATH.
Benchmarking using a data set generated from phosphopeptide-enriched samples
To assess the scalability of IPF for the analysis of thousands of peptides, we generated a data set of phosphopeptide-enriched samples of human U2OS cells. Cells were either treated with nocodazole or left untreated (control) and the resulting patterns were comparatively analyzed by IPF. Nocodazole arrests cells at the mitotic stage and thus has a substantial effect on signaling pathways involving phosphorylation41. We acquired ten biological replicates, processed in parallel, each for nocodazole-treated and control samples, both in DDA and DIA modes (see Methods). We then used the DDA data to generate a spectral library for quantitative analysis by IPF on the corresponding 20 DIA runs covering 4,298 phosphopeptides (Methods, Supplementary Notes IV.B)
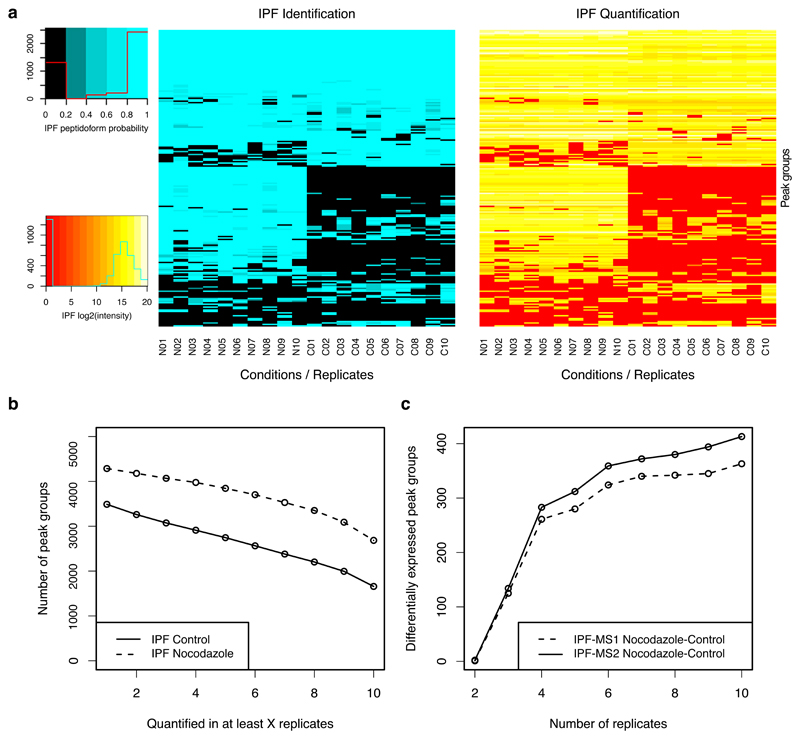
We next analyzed the quantitative data matrix produced by IPF across all ten replicates of each condition, considering only peak groups with at least one confident detection or quantification per biological condition (Fig. 3). For the nocodazole treated and the control samples, IPF achieved both consistent detection and quantification for 62.6% (nocodazole) and 47.5% (control) of all phosphopeptides (Fig. 3a-b). To investigate the effect that consistency of quantification has in dependency of the number of replicates, we conducted differential expression analysis using mapDIA42 on variable numbers of sampled replicates. Depending on the number of replicates, more than 400 differentially expressed peak groups were detected (Fig. 3c, significance thresholds: FDR < 0.01 & log2(FC) > 2). Considering only three replicates, IPF identified 134 significant peak groups. These results demonstrate that IPF can achieve consistent detection and quantification for enriched phosphopeptide data sets across multiple samples.
Figure 3. Benchmarking using a data set generated from phosphopeptide-enriched samples.
Enriched phosphopeptide samples of a human U2OS cell line treated with nocodazole and without treatment (control) were measured in both DDA and DIA modes in each 10 replicates. a) 200 peptides were randomly selected and the corresponding detected peak groups and peptide precursors are visualized in a heatmap (sorted by a hierarchical dendrogram for identification/detection by rows) for detection confidence (blue) and quantification (red-yellow; including alignment). IPF achieved a high level of completeness for both detection and quantification in individual experimental conditions (Nocodazole N01-N10: 62.6%; Controls C01-C10: 47.5%). b) The consistency of quantification for all intersecting peptides is depicted. c) Differential expression analysis was conducted using mapDIA (significance thresholds: FDR < 0.01 & log2(FC) > 2). For IPF on both MS2 peak group (IPF-MS2) and MS1 (IPF-MS1) precursor levels, the same peptide/precursor-level model and parameters were used.
Analysis of phosphopeptide-enriched samples is a frequent objective of discovery proteomics workflows. The performance of IPF should thus be assessed in comparison to the state-of-the-art; however, comparing algorithms based on different concepts and requiring different input data (DDA vs. DIA) is challenging (Supplementary Notes IV.A). Using the DDA and DIA data of the phosphopeptide-enriched samples described above and non-enriched samples of a previously published 14-3-3 scaffold protein interactome study43, we conducted assessments of a DDA-based workflow and IPF (Supplementary Notes IV.B-C, Supplementary Fig. 5-6). The results suggest that within the parameter space tested, IPF using DIA data substantially improves the consistency of detection and quantification of phosphopeptides compared to DDA-based workflows.
Assessment of variance components of peptidoform abundance in human blood plasma
To demonstrate the utility of IPF for biological research requiring larger cohort sizes, we applied it to a longitudinal twin study consisting of 116 individuals (58 twin pairs), who donated blood plasma twice within a time span of 2-7 years. The study was designed to assess the effects of heritability and environment on blood plasma protein abundance33. We revisited the previously acquired data set of this large sample cohort to investigate the biological variability of 10 selected post-translational modifications (oxidation, deamidation, carbamylation, formylation, acetylation, methylation, carboxylation, ubiquitination, nitrosylation, phosphorylation) that were previously associated with blood plasma proteins44–48. The samples further contained 73 spiked-in stable isotope labelled (SIS) peptides, corresponding to 37 plasma proteins33,49, with levels generally adjusted to the endogenous peptides in the human plasma proteome49. The two isotope labels for arginine and lysine and static carbamidomethylation modification were added to the list of included modification types.
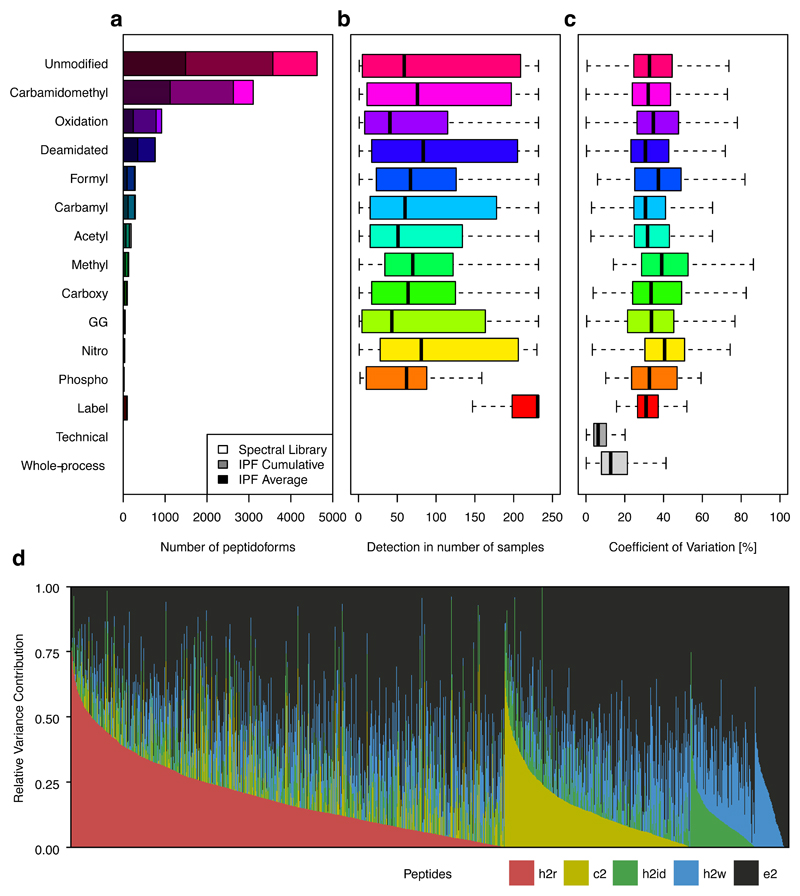
As a first step we generated a spectral library by searching DDA data from chromatographic fractions of a pooled sample. This library was used for the analysis of the 232 samples of the twin study (plus 10 technical and whole-process replicates) by IPF. Figure 4a depicts the spectral library and IPF cumulative and average detection statistics. In summary, of the 9,272 peptidoforms covered by the library, 7653 (82.5%) could be detected by IPF cumulatively across all samples with an average of 3153.7 (34.0%) per run. Of all the peptidoforms, 49.9% were unmodified, 48.5% carried PTMs of likely artefactual origin and 5.5% were modified due to likely biological causes (Methods, Supplementary Table 2). Importantly, peptidoforms can carry both artefactual and biological PTMs.
Figure 4. Assessment of variance components of peptidoform abundance in human blood plasma.
a) The overlaid histogram of unmodified and modified peptides grouped according to modification type contained in the spectral library, confidently detected in at least one sample (shaded grey) and the average number of detected peptidoforms per sample (shaded black). b) The detectability in number of samples of peptidoforms grouped by modification types indicates a median detectability in ~50-100 samples. c) The coefficient of variation (CV; computed only if the peptidoform was detected in at least 2 runs) of the quantile normalized peak group intensities across different modification types is depicted. Technical and whole-process variability are consistently lower than the intensities of unmodified and modified peptides across the cohort, indicating that biological variance constitutes a major component of the total variance. d) The cumulative relative variance contributions (RVC) of the heritable (h2r), common environment (c2), individual environment (h2id) and longitudinal (h2w) effects per peptidoform peak group grouped and ordered by effect type and contribution are depicted.
Different peptidoforms and modification types can be differentially expressed between samples and conditions based on sample-specific biochemical reactions, protein abundance and other effects. While in general, modified peptidoforms are expressed over a similar range than unmodified peptidoforms (Supplementary Note V.A, Supplementary Fig. 7-8), abundance differences can be of the on/off type or quantitatively different between individuals. Accordingly, Figure 4b depicts the distribution of the observed detectability (in number of samples) grouped according to modification type. The spiked-in SIS peptides could be detected consistently across almost all the twin samples. In contrast, the median detectability of endogenous unmodified and modified peptides was more variable across the data set, ranging from ~50 – 100 samples, suggesting significant variability of peptidoform-level abundance in undepleted blood plasma samples. We assessed the quantitative variation of the peptidoforms across the samples (Fig. 4c): Technical and whole-process replicates indicate a median coefficient of variation (CV) of <10% and <20%, respectively (Supplementary Fig. 9-10) 26. In contrast, peptides subject to biological variation, showed a median CV of 30 – 50%. Our data thus provides a complex snapshot of the peptidoforms of blood plasma proteins in a human cohort.
To understand the root causes, i.e. heritable, common and individual environmental and longitudinal effects of biological variation of endogenous human plasma peptidoforms, we utilized a linear mixed model approach to fit our data after imputation of background intensities (Methods). Queried peptidoforms were only considered if they were detected in at least 20 samples of the data set, resulting in 4532 peptidoforms targeted by 5829 queried peptides. Of these, 1755 peptidoforms (1954 peptide queries, e.g. different precursors and site-localizations contained within the library) contained at least one significant (q-value < 1%) component that could be ascribed to the factors generating biological variance, specifically heritability, familial environment, individual environment and the 2-7 year longitudinal visits (Fig. 4d, Supplementary Table 3, Supplementary Data 1). Among all peptide queries, the longitudinal component (variance between two longitudinal visits of the same individual33) cumulatively was found to be the major component contributing to the biological variability (h2w: 15.3%), followed by heritability (h2r: 12.0%), the individual environmental (h2id: 8.2%), and the common environmental effects (c2: 7.8%), with the unexplained effects accounting for 56.7% on average. These results are consistent with the original study on the protein level33, where the average component effects were of similar magnitude (h2w: 13.5%, h2r: 13.6%, h2id: 11.6%, c2: 10.8%, e2: 50.5%).
We further investigated the effects of the quantitative peptidoform variability on selected key blood plasma proteins in more detail. Human serum albumin (ALBU), the major protein constituent accounting for 55% of the plasma protein mass50, was represented by 440 peptidoforms (9.7%), whereas all the other plasma proteins were covered by 11.6 peptidoforms on average (Supplementary Table 4). In general, the individual peptidoforms were similarly affected by the different root causes of observed variability as compared to other proteins, suggesting that peptidoform diversity was mainly depending on the general ALBU protein abundance level in plasma (Supplementary Note V.C, Supplementary Fig. 11-12). Further, to demonstrate the heritable or familial environmental components of different peptidoforms that can be efficiently dissected using the data, we investigated allele variants of ApoE, a protein associated with the high-density lipoprotein (HDL) class. We confirmed that its main biological effects can be mainly attributed to the familial environmental components (Supplementary Note V.D, Supplementary Fig. 13).
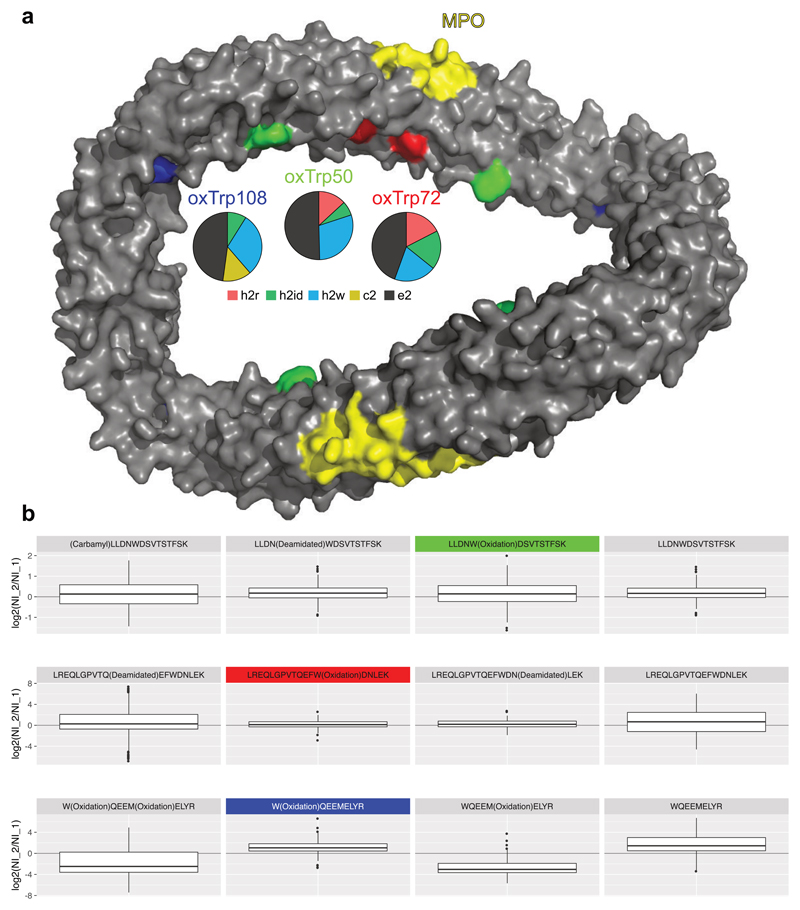
ApoA1 and other members of the high density lipoprotein (HDL) complex have previously been found to be affected by oxidative modifications introduced by myeloperoxidase (MPO)51,52. Oxidative modifications at specific sites, particularly oxTrp7252, were found to inhibit the cholesterol acceptor function of ApoA1, resulting in a dysfunctional protein that is associated with atherosclerosis and cardiovascular disease52. In this context, we investigated the heritable and environmental effects on oxidative modifications on ApoA1 within the twin cohort. In total, the variance components of 12 oxidized (4 tryptophan, 7 methionine, 1 tryptophan + methionine) peptidoforms could be decomposed in the data set, that were constituted of 3 tryptophan and 4 methionine sites from different regions of the protein structure53. We found oxTrp72 among the oxidized tryptophan peptidoforms, along with two other known sites52 in close proximity (Fig. 5a), oxTrp50 and oxTrp108, and found that their peptidoform abundance levels are longitudinally upregulated with a relative variance contribution of 19.8–29.7% (Fig. 5b, Supplementary Fig. 13). While also the abundances of other peptidoforms, including the unmodified ones, are longitudinally upregulated, the longitudinal regulation is different for peptidoforms having oxidized methionine residues. Two previously oxidized methionine residues, hypothetical biomarkers for atherosclerosis54, oxMet86 and oxMet112, were detected and quantified in our data as well. In contrast to the constant or upregulated oxidized tryptophan peptidoforms (Fig. 5b, Supplementary Fig. 14), their peptidoform abundance levels show an inverse longitudinal effect that could also be explained to be technical artifacts: The samples of the first visit were stored for a longer period of time and thus contained higher fractions of spontaneous methionine oxidations. These data thus support the hypothesis that oxTrp50 and oxTrp108, in contrast to the potential technical artefacts oxMet86 and oxMet112, are biochemically induced by MPO under a similar mechanism as oxTrp72 and might be useful as candidate biomarkers for proatherogenic processes52. Intriguingly, IPF successfully dissected the differential technical and biological variation of oxMet112 and oxTrp108, which was measured by two isobaric peptidoforms of the same peptide backbone sequence.
Figure 5. Oxidative tryptophan modifications of ApoA1.
a) The asymmetric units form an antiparallel four-helix bundle in an elliptical ring shape53. The myeloperoxidase (MPO) binding site is highlighted in yellow. On the opposite site, oxTrp72 (h2w (longitudinal): 19.8%, h2id (individual environment): 18.3%, h2r (heritable): 17.5%, c2 (common environment): 0%, e2 (unexplained): 44.4%), a potential biomarker for dysfunctional ApoA1 is highlighted in red. oxTrp50 (green; h2w: 29.7%, h2r: 13.2%, h2id: 6.8%, c2: 0%, e2: 50.3%) and oxTrp108 (blue; h2w: 29.7 %, c2: 13.5%, h2id: 8.9%, h2r: 0%, e2: 47.9%) are in close spatial proximity to oxTrp72. For all three modification sites, the longitudinal effect is the major component. b) The boxplots depict the peptidoform abundance fold changes between timepoints 2 (later) and 1 (log2(NI_2/NI_1); NI: quantile normalized intensity) for all individuals. oxTrp50 (green) and oxTrp72 (red) are stable in abundance over both time points with a slight increase in intensity, which is also present for the other related peptidoforms. The oxTrp108 (blue) peptidoforms show the largest increase in abundance when comparing the second to the first visit. However, the methionine oxidized peptidoforms (oxMet112) show a decrease, which might be induced by the longer time of sample storage and thus spontaneous methionine oxidation for the samples at the first visits. For all sites the unmodified peptidoforms also show a longitudinal increase, indicating a longitudinally increasing total protein abundance.
Discussion
Peptidoforms carrying biologically relevant post-translational modifications are often more difficult to measure consistently across many samples because their abundance is frequently more variable than the abundance of dominating other peptidoforms, e.g. the non-modified peptide. Targeted data extraction was demonstrated to improve the consistency of peptide detection and quantification, particularly for large-scale DIA analysis22,26. Although the DIA analysis of peptidoforms can provide excellent quantitative performance, the medium- to large- precursor-isolation window configurations can result in the co-isolation of different peptidoforms22,55, making their discrimination very challenging, particularly in cases where the peptidoforms differ by site-localization.
Peptide-centric scoring using peptide query parameters is very sensitive but is frequently not selective enough for targeting peptidoforms. IPF improves the selectivity by integrating evidence on different levels to a single peptidoform confidence for each peak group. It is thus conceptually related to spectrum-centric site-localization applicable to DIA data, e.g. DIA-Umpire22,56/LuciPHOr39,40 or SWATHProphetPTM31, but with a focus on consistent detection and quantification. This is useful for TRIC35, which transfers the detection confidence across different runs based on the retention times and detection confidence of the candidate peak groups. If in a certain fraction of measurements, the peptidoform detection is less confident because of bad or missing precursor signals or missing site-determining transition-level chromatograms, TRIC can recover some of these signals. The seamless integration of IPF with TRIC confidently extends the list of peptidoforms detected and quantified in individual measurements to a quantitative matrix over large sample cohorts.
The functions performed by IPF are important for large-scale, complex data sets, as demonstrated by the peptidoform-level analysis of the twin plasma data set. Despite of its promise for clinical applications, blood plasma presents one of the most analytically challenging human-derived proteomes, due to its different tissue proteome subsets and the large dynamic range with only 22 proteins accounting for ~99% of the total protein mass50,57. IPF enabled us to study the heritable and environmental components of peptidoforms carrying different types of PTMs. We found that most peptidoforms are expectedly co-regulated with the generating proteins; however, our approach enables further discrimination of the effects on peptidoform-level into biological and technical causes, as exemplified by ApoA1 oxidation.
In conclusion, we describe, benchmark and apply a new algorithm for targeted data analysis of DIA data sets that is geared towards the consistent detection and quantification of peptidoforms based on low- to high-confidence spectral libraries from supporting hypothesis generating workflows based on DDA and DIA. Our generic approach is scalable to hundreds of samples, optimizing for peptidoform-level selectivity while maintaining high sensitivity. The validation based on the synthetic phosphopeptide reference data set established the accurate error-rate control capabilities of IPF. The application to a data set generated from enriched phosphopeptide samples proved the general applicability and improvements of IPF for commonly employed experimental designs, such as phosphoproteomic profiling. Further, the application to the challenging twin blood plasma data set demonstrates the utility for practical applications in complex samples. We expect that the availability of the algorithm in the open source workflow OpenSWATH, the generic utility for all modification types and scalability will enable confident quantification of PTMs in large-scale studies using DIA data.
Online Methods
Synthetic phosphopeptide reference data set
Sample preparation
Crude synthetic peptides
To generate the synthetic phosphopeptide reference data, a set of 579 synthetic, unpurified, heavy-isotope labeled phosphopeptides (Thermo Scientific Biopolymers) was used. These phosphopeptides represent biologically relevant sequences from S. cerevisiae proteins, and include previously published markers of cellular processes36. The complete peptide set contains a mixture of singly and doubly phosphorylated sequences with in average more than 3 modifiable residues per peptide (serines, threonines or tyrosines, often near each other). The complete sequence list can be found in Supplementary Table 1. All peptides were mixed with equal volumes and the concentrations were estimated to be around 0.002 µg/µl based on the vendor estimates of the unpurified peptides. The resulting peptide mix was either analyzed directly in DDA mode for spectral library generation or spiked into a human cell line background proteome in a 13-step dilution series and analyzed in SWATH mode for the generation of the synthetic phosphopeptide reference data set (see below).
Human cell line background proteome
HEK-293 cell pellets were lysed on ice by using a lysis buffer containing 8 M urea (EuroBio), 40 mM Tris-base (Sigma-Aldrich), 10 mM DTT (AppliChem) and complete protease inhibitor cocktail (Roche). The resulted mixtures were sonicated in 4 °C for 5 mins using a VialTweeter device (Hielscher-Ultrasound Technology) with full power and centrifuged at 21,130 g, 4 °C for 1 h to remove the insoluble material. The supernatant protein mixtures were transferred and the protein amount was determined with a Bradford assay (Bio-Rad). Aliquots of 2 mg protein mixtures were reduced by 5 tris(carboxyethyl)phosphine (Sigma-Aldrich) and alkylated by 30 mM iodoacetamide (Sigma-Aldrich). Then 5 volumes of precooled precipitation solution containing 50% acetone, 50% ethanol, and 0.1% acetic acid were added to the protein mixture and kept at -20 °C overnight. The mixture was centrifuged at 20,400 g for 40 min. The pellets were further washed with 100% acetone and 70% ethanol with centrifugation at 20,400 g for 40 min. The samples were then resolved by 100mM NH4HCO3 and were digested with sequencing-grade porcine trypsin (Promega) at a protease/protein ratio of 1:50 overnight at 37 °C58. Digests were combined and purified with Sep-Pak C18 Vac Cartridge (Waters). Peptide amount was determined by using Nanodrop ND-1000 (Thermo Scientific). An aliquot of retention time calibration peptides from the iRT-Kit (Biognosys) was spiked into the total mixture of the sample at a ratio of 1:20 (v/v) to correct relative retention times between runs59.
Dilution series of synthetic peptides
The heavy-labeled synthetic phosphopeptide mix described above was spiked in 13 defined dilution steps into a HEK293 total cellular proteome background (final constant background concentration in all 13 dilution samples = 0.5 µg/µl). The 13 dilution steps of the heavy-labeled synthetic phosphopeptides were: 0, 1:1, 1:3, 1:4, 1:7, 1:9, 1:15, 1:19, 1:31, 1:39, 1:63, 1:79 and 1:127. This dilution range was judged to be appropriate because at the final lowest concentration step the phosphopeptide amounts loaded onto the column were approximately estimated to be in the range of tens of attomoles.
DDA mass spectrometry
The synthetic phosphopeptide mix (without added background added) was measured on a SCIEX 5600+ TripleTOF mass spectrometer operated in DDA mode in technical triplicates. The mass spectrometer was interfaced with an Eksigent NanoLC Ultra 2D Plus HPLC system as previously described14,43,60. Peptides were directly injected onto a 20-cm PicoFrit emitter (New Objective, self-packed to 20 cm with Magic C18 AQ 3-μm 200-Å material), and then separated using a 120-min gradient from 2–35% (buffer A 0.1% (v/v) formic acid, 2% (v/v) acetonitrile, buffer B 0.1% (v/v) formic acid, 90% (v/v) acetonitrile) at a flow rate of 300 nL/min. MS1 spectra were collected in the range 360–1,460 m/z for 500 ms. The 20 most intense precursors with charge state 2–5 which exceeded 250 counts per second were selected for fragmentation, and MS2 spectra were collected in the range 50–2,000 m/z for 150 ms. The precursor ions were dynamically excluded from reselection for 20 s. Acquired file names:
| Type | Filename |
|---|---|
| Replicate 1 | chludwig_K141203_001_IDA |
| Replicate 2 | chludwig_K141203_002_IDA |
| Replicate 3 | chludwig_K141203_003_IDA |
DIA mass spectrometry
The 13-step dilution series of the synthetic heavy phosphopeptide mix (spiked into a constant human background) was measured in SWATH-MS mode on the same LC-MS/MS systems used for DDA measurements in technical triplicates14,43,60. In SWATH-MS mode the SCIEX 5600+ TripleTOF instrument was specifically tuned to optimize the quadrupole settings for the selection of 64 variable wide precursor ion selection windows. The 64-variable window schema was optimized based on a normal human cell lysate sample, covering the precursor mass range of 400–1,200 m/z. The effective isolation windows can be considered as being 399.5~408.2, 407.2~415.8, 414.8~422.7, 421.7~429.7, 428.7~437.3, 436.3~444.8, 443.8~451.7, 450.7~458.7, 457.7~466.7, 465.7~473.4, 472.4~478.3, 477.3~485.4, 484.4~491.2, 490.2~497.7, 496.7~504.3, 503.3~511.2, 510.2~518.2, 517.2~525.3, 524.3~533.3, 532.3~540.3, 539.3~546.8, 545.8~554.5, 553.5~561.8, 560.8~568.3, 567.3~575.7, 574.7~582.3, 581.3~588.8, 587.8~595.8, 594.8~601.8, 600.8~608.9, 607.9~616.9, 615.9~624.8, 623.8~632.2, 631.2~640.8, 639.8~647.9, 646.9~654.8, 653.8~661.5, 660.5~670.3, 669.3~678.8, 677.8~687.8, 686.8~696.9, 695.9~706.9, 705.9~715.9, 714.9~726.2, 725.2~737.4, 736.4~746.6, 745.6~757.5, 756.5~767.9, 766.9~779.5, 778.5~792.9, 791.9~807, 806~820, 819~834.2, 833.2~849.4, 848.4~866, 865~884.4, 883.4~899.9, 898.9~919, 918~942.1, 941.1~971.6, 970.6~1006, 1005~1053, 1052~1110.6, 1109.6~1200.5 (including 1 m/z window overlapping). SWATH MS2 spectra were collected from 50 to 2,000 m/z. The collision energy (CE) was optimized for each window according to the calculation for a charge 2+ ion centered upon the window with a spread of 15 eV. An accumulation time (dwell time) of 50 ms was used for all fragment-ion scans in high-sensitivity mode and for each SWATH-MS cycle a survey scan in high-resolution mode was also acquired for 250 ms, resulting in a duty cycle of ~3.45 s. Per MS injection 2 µg of protein amount (i.e., 4µL of the final dilution mixture) was loaded onto the HPLC column. Acquired file names:
| Dilution (PSGS) | Filename |
|---|---|
| Dilution 1:0 | chludwig_K150309_013_SW |
| Dilution 1:1 | chludwig_K150309_012_SW |
| Dilution 1:3 | chludwig_K150309_010_SW |
| Dilution 1:4 | chludwig_K150309_011_SW |
| Dilution 1:7 | chludwig_K150309_008_SW |
| Dilution 1:9 | chludwig_K150309_009_SW |
| Dilution 1:15 | chludwig_K150309_006b_SW |
| Dilution 1:19 | chludwig_K150309_007b_SW |
| Dilution 1:31 | chludwig_K150309_004b_SW |
| Dilution 1:39 | chludwig_K150309_005b_SW |
| Dilution 1:63 | chludwig_K150309_002b_SW |
| Dilution 1:79 | chludwig_K150309_003b_SW |
| Dilution 1:127 | chludwig_K150309_001b_SW |
Spectral library and peptide query parameter generation
We generated a peptide sequence and precursor-specific (1% peptide sequence FDR) spectral library for the phosphorylated peptides. Peptide-centric scoring of DIA data benefits from using saturated, optimized spectral libraries that best cover the target peptides26. For this reason, we searched the 45 DDA runs of the original study36 and acquired three new replicate DDA runs of a pooled sample of the synthetic library without background proteome (see above). Then, consensus spectra for different site-localized peptidoforms were generated from the search output. To increase the coverage, the peptide identifications were not post-processed by a site-localization algorithm. This resulted in over-annotation of the peptides potentially contained in the sample because spectra originating from the same peptidoform could be assigned to different (incorrect) site-localizations. The library was used to derive sets of peptide query parameters for 554 peptidoforms (e.g. PEPT(Phospho)IDEK), mapping to 297 combinations of peptide sequences and numbers of phosphorylated residues (e.g. PEPTIDEK + 1 phosphorylation). In parallel, to assess the application of IPF on spectral libraries generated directly from DIA data, a second library was generated by DIA-Umpire22 using the 13-step dilution series DIA data, which includes a HEK-293 proteome background. The thus generated library contained peptide query parameters for 169 peptidoforms, mapping to 122 combinations of peptide sequences and numbers of phosphorylated residues (Methods, Supplementary Fig. 3).
DDA database search and spectral library generation
All original36 and new raw instrument data acquired in DDA mode were centroided and converted to mzXML using qtofpeakpicker (ProteoWizard61 3.0.10200) as described previously32. The phosphopeptide sequences were appended with a set of contaminant proteins, iRT peptide sequences and pseudo-reverse decoys. The files were searched using Comet62 (2015.02) using the default parameters for high mass accuracy instruments: peptide mass tolerance: 20 ppm (monoisotopic), isotope error enabled, fully tryptic digestion with max 5 missed cleavages, static C (Carbamidomethyl), variable M (Oxidation), variable K (Label:13C(6)15N(2)), variable R(Label:13C(6)15N(4)), variable STY (Phospho), max variable mods: 5. PeptideProphet38,63 (TPP64 5.0.0) with parameters -dDECOY_ -OAPdlIwt was run on all search results together and iProphet65 was used to combine all results. SpectraST66 (TPP 5.0.0) was used to generate a spectral library of all peptide identifications at iProphet FDR 1% with the following parameters: -cP0.8326 -c_IRR -c_IRTirtkit.txt -cICID-QTOF -c_RDYDECOY -cAC –cM. All peptides except the synthetic phosphopeptides were excluded from the library. OpenMS67 (OpenMS 2.1) was used for all following steps: ConvertTSVToTraML was used to convert the SpectraST MRM file to a TraML. OpenSwathAssayGenerator was applied on the TraML with following parameters: -swath_windows_file swath64.txt -allowed_fragment_charges 1,2,3,4 -enable_ms1_uis_scoring -max_num_alternative_localizations 2000-enable_identification_specific_losses -enable_identification_ms2_precursors. OpenSwathDecoyGenerator was applied to append decoys to the target peptide query parameters using the following parameters: -method shuffle -append -mz_threshold 0.1 -remove_unannotated. All OpenMS tools were executed using the modified chemistry parameters for phosphorylation (OpenMS.phospho.params) (ProteomeXchange repository).
DIA database search and spectral library generation
All raw instrument data acquired in DIA mode were centroided and converted using the SCIEX MS Data Converter (Beta Version 1.3) and msconvert (ProteoWizard61 3.0.7162) using the parameters as suggested22. The signal extraction module of DIA-Umpire22 (1.2, 2014.10) was applied to the 13-step dilution series SWATH-MS data set using recommended parameters. The ORF protein translation FASTA database for yeast was obtained from the Saccharomyces Genome Database68 (2015-02-24) and appended with the non-redundant reviewed human protein FASTA obtained from the UniProtKB/Swiss-Prot69 (2015-02-23) and the iRT peptide sequences and pseudo-reverse decoys. The files were searched using Comet62 (2015.02) using the parameters recommended for DIA-Umpire22: peptide mass tolerance: 40 ppm (monoisotopic), isotope error enabled, fully tryptic digestion with max 2 missed cleavages, static C (Carbamidomethyl), variable M (Oxidation), variable K (Label:13C(6)15N(2)), variable R(Label:13C(6)15N(4)), variable STY (Phospho), max variable mods: 5. PeptideProphet38,63 (TPP64 4.8.0) with parameters -dDECOY_ -OAPdlIwt was run independently per file and iProphet65 was used to combine all quality tier Q1 & Q3 (Q2 were excluded) results. SpectraST66 (TPP 4.8.0) was used to generate a spectral library of all peptide identifications at iProphet FDR 1% with the following parameters: -cP0.7850 -c_IRR -c_IRTirtkit.txt -cICID-QTOF -c_RDYDECOY -cAC –cM. OpenMS (https://github.com/grosenberger/OpenMS/tree/feature/ipf @7924fc3) was used for all following steps: ConvertTSVToTraML was used to convert the SpectraST MRM file to a TraML. OpenSwathAssayGenerator was applied on the TraML with following parameters: -swath_windows_file swath64.txt -allowed_fragment_charges 1,2,3,4 -enable_ms1_uis_scoring -max_num_alternative_localizations 20 -enable_identification_specific_losses -enable_identification_ms2_precursors. OpenSwathDecoyGenerator was applied to append decoys to the target peptide query parameters using the following parameters: -method shuffle -append -mz_threshold 0.1 -remove_unannotated. All OpenMS tools were executed using the modified chemistry parameters for phosphorylation (OpenMS.phospho.params) (ProteomeXchange repository).
OpenSWATH and PyProphet
OpenSwathWorkflow (OpenMS 2.1) was run with the following parameters -min_upper_edge_dist 1 -mz_extraction_window 0.05 -rt_extraction_window 600 -extra_rt_extraction_window 100 -min_rsq 0.95 -min_coverage 0.6 -use_ms1_traces -enable_uis_scoring -Scoring:uis_threshold_peak_area 0 -Scoring:uis_threshold_sn -1 -Scoring: stop_report_after_feature 5 -tr_irt hroest_DIA_iRT.TraML. The following subset of scores was used on MS2-level: xx_swath_prelim_score library_corr yseries_score xcorr_coelution_weighted massdev_score norm_rt_score library_rmsd bseries_score intensity_score xcorr_coelution log_sn_score isotope_overlap_score massdev_score_weighted xcorr_shape_weighted isotope_correlation_score xcorr_shape. All MS1 and UIS scores were used for PyProphet.
PyProphet (https://github.com/grosenberger/pyprophet/tree/feature/ipf @93ec307) was run on a concatenated file of all 13 runs containing only the unambiguous phosphopeptides with the following parameters:
--final_statistics.emp_p --qvality.enable --qvality.generalized
--ms1_scoring.enable --uis_scoring.enable
--semi_supervised_learner.num_iter=20 --xeval.num_iter=20
--ignore.invalid_score_columns.
PyProphet in site-localization mode only (https://github.com/grosenberger/pyprophet/tree/feature/ipf_flr @b92ccf9) was run on a concatenated file of all 13 runs with the following parameters:
--final_statistics.emp_p --qvality.enable --qvality.generalized
--ms1_scoring.enable --uis_scoring.enable
--semi_supervised_learner.num_iter=20 --xeval.num_iter=20
--ignore.invalid_score_columns --uis_scoring.disable_precursor_inference.
LuciPHOr
LuciPHOr39,40 2 (1.2014Oct10-iprophet) was modified to support iProphet65 posterior probabilities, required for DIA-Umpire pseudo spectra identifications. Static, variable and target residue modifiability was defined as for the database search and neutral losses for phosphorylation were allowed as suggested. The fragment ion tolerance was set to 0.025 Da and the algorithm was run in CID mode. All other parameters were set as suggested by the default settings.
Validation using the synthetic phosphopeptide reference data set
To validate the IPF results on the synthetic phosphopeptide reference data set, the synthesized reference peptide sequences were used as ground truth. To specifically assess the global and local false discovery rates, only those detected peak groups were used where the unmodified peptide sequence, modification types (except variable methionine oxidation) and modification numbers matched the synthesized peptides exactly. If multiple peptide queries resulted in peak groups for the same peptidoform (or equal site-localization), i.e. when multiple precursors could be detected, only the best scoring one would be used. All perfect matches to the ground truth were counted as true, all others as false positives.
Statistics and visualization
Visualization of Fig. 2 and Supplementary Fig. 3 was conducted using the boxplot function of the R package “graphics” with the default parameters: The box borders represent the 25th and 75th quantile, respectively. The bold bar represents the 50th quantile. The whiskers are defined as following: upper whisker: min(max(x), Q_3 + 1.5*IQR); lower whisker: max(min(x), Q_1-1.5*IQR), with Q_1 and Q_3 representing the 25th and 75th quantile, respectively, and IQR representing the inter-quantile range Q_3-Q_1.
Data set generated from phosphopeptide-enriched samples
Sample preparation
Phospho-enrichment of human cell line and sample preparation
U2OS cells were grown in DMEM media (Life Technologies) supplemented with 10% FCS (BioConcept), 1% penicillin-streptomycin (Life Technologies). Cells cultures (approximately 3-4 million cells per plate) were treated with nocodazole (Sigma-Aldrich) at a final concentration of 100ng/ml for 18 hours. Treated and untreated samples (ten replicates respectively) were collected, washed with PBS and frozen in liquid nitrogen. To include variation of the sample preparation, we directly processed all the cells from each plate for the protein digestion (see above) and phosphoproteomic analysis. Phosphopeptide enrichment was performed from approximately 1mg of total peptide mass according to a modified protocol70 using TiO2 resin (GL Sciences). In short, the dried peptides were dissolved in a loading buffer (80% ACN, 5% TFA, 1M glycolic acid) and vortexed in a shaker for 10 min at 25°C, 1400 rpm. Peptide mixtures were added to equilibrated enrichment resin preloaded in a 200ul tip with a C8 plug (3M Empore). The resin was then washed once with the loading buffer, once with 80% ACN, 0.1% TFA, and once with 50% ACN, 0.1% TFA. The peptides were eluted with a 0.6 M NH4OH solution (Sigma-Aldrich), followed by an elution from the C8 plug with 50% ACN, 0.1% FA. The pH of the eluates was adjusted to <3 and the phosphopeptide samples were desalted using C18 ultramicrospin columns (Nest). Samples were re-suspended as described above. Phosphopeptide mixture originating from ~10% of the starting cell materials was injected for shotgun and SWATH measurements.
DDA mass spectrometry
The samples were measured on a SCIEX 6600 TripleTOF mass spectrometer operated in DDA mode in ten replicates for each experimental condition. The samples were measured as described above with a modified LC gradient of 2-30% buffer during the same acquisition time. Acquired file names:
| Treatment | Replicate | Filename |
|---|---|---|
| Control | 1 | yanliu_I170114_001_PhosCyc1_shotgun |
| 2 | yanliu_I170114_005_PhosCyc2_shotgun | |
| 3 | yanliu_I170114_009_PhosCyc3_shotgun | |
| 4 | yanliu_I170114_013_PhosCyc4_shotgun | |
| 5 | yanliu_I170114_017_PhosCyc5_shotgun | |
| 6 | yanliu_I170114_021_PhosCyc6_shotgun | |
| 7 | yanliu_I170114_025_PhosCyc7_shotgun | |
| 8 | yanliu_I170114_029_PhosCyc8_shotgun | |
| 9 | yanliu_I170114_033_PhosCyc9_shotgun | |
| 10 | yanliu_I170114_037_PhosCyc10_shotgun | |
| Nocodazole | 1 | yanliu_I170114_002_PhosNoco1_shotgun |
| 2 | yanliu_I170114_007_PhosNoco2_shotgun | |
| 3 | yanliu_I170114_011_PhosNoco3_shotgun | |
| 4 | yanliu_I170114_015_PhosNoco4_shotgun | |
| 5 | yanliu_I170114_019_PhosNoco5_shotgun | |
| 6 | yanliu_I170114_023_PhosNoco6_shotgun | |
| 7 | yanliu_I170114_027_PhosNoco7_shotgun | |
| 8 | yanliu_I170114_031_PhosNoco8_shotgun | |
| 9 | yanliu_I170114_035_PhosNoco9_shotgun | |
| 0 | yanliu_I170114_039_PhosNoco10_shotgun |
DIA mass spectrometry
The samples were measured in SWATH-MS mode as described above on the same LC-MS/MS system used for DDA measurements in ten replicates for each experimental condition with the following difference: The MS1 acquisition time for each cycle was set to 200ms and the MS2 scan range was set to 300-2000 m/z. Acquired file names:
| Treatment | Replicate | Filename |
|---|---|---|
| Control | 1 | yanliu_I170114_003_PhosCyc1_SW |
| 2 | yanliu_I170114_006_PhosCyc2_SW | |
| 3 | yanliu_I170114_010_PhosCyc3_SW | |
| 4 | yanliu_I170114_014_PhosCyc4_SW | |
| 5 | yanliu_I170114_018_PhosCyc5_SW | |
| 6 | yanliu_I170114_022_PhosCyc6_SW | |
| 7 | yanliu_I170114_026_PhosCyc7_SW | |
| 8 | yanliu_I170114_041_PhosCyc8_SW_rep | |
| 9 | yanliu_I170114_034_PhosCyc9_SW | |
| 10 | yanliu_I170114_038_PhosCyc10_SW | |
| Nocodazole | 1 | yanliu_I170114_004_PhosNoco1_SW |
| 2 | yanliu_I170114_008_PhosNoco2_SW | |
| 3 | yanliu_I170114_012_PhosNoco3_SW | |
| 4 | yanliu_I170114_016_PhosNoco4_SW | |
| 5 | yanliu_I170114_020_PhosNoco5_SW | |
| 6 | yanliu_I170114_024_PhosNoco6_SW | |
| 7 | yanliu_I170114_028_PhosNoco7_SW | |
| 8 | yanliu_I170114_032_PhosNoco8_SW | |
| 9 | yanliu_I170114_036_PhosNoco9_SW | |
| 10 | yanliu_I170114_040_PhosNoco10_SW |
Spectral library and peptide query parameter generation
The MaxQuant workflow71 provides integrated site-localization of the phosphopeptides and MS1 precursor-level label-free quantification by alignment and matching between runs and can thus generate a quantitative matrix at the level of phosphopeptides for DDA data sets. In total, MaxQuant identified 10,051 unique peptide sequences carrying phosphorylated residues, filtered at 1% PSM FDR for the whole data set, in at least one of the ten replicates of each experimental condition. From these results, we generated a library covering a subset of 8,013 phosphorylated unique peptide sequences for the analysis of the corresponding 20 DIA runs using IPF, because not all PSMs fulfilled the criteria for peptide queries (e.g. at least six annotated fragment ions).
MaxQuant analysis and spectral library generation
All raw data was analyzed in a combined setting with MaxQuant (1.5.6.5) using primarily the default parameters71: The non-redundant reviewed human protein FASTA was obtained from the UniProtKB/Swiss-Prot69 (2016-12-19) and appended with iRT peptide sequences and searched with static C (Carbamidomethyl), variable M (Oxidation) and variable STY (Phospho) modifications. “Match-between-runs” and the MaxLFQ algorithm were enabled. All specific parameters are provided in the file mqpar.xml in the ProteomeXchange repository.
To derive peptide query parameters, we selected the best scoring spectrum per peptidoform as reported by Andromeda in the file “msms.txt”. RT calibration was conducted using the spiked-in iRT-kit per run. OpenSwathAssayGenerator and OpenSwathDecoyGenerator (OpenMS 2.1) were run as described above. For all other analyses, we used the reported confidence values and intensities from the file “Phospho (STY)Sites.txt”.
OpenSWATH, PyProphet and TRIC
OpenSwathWorkflow (OpenMS 2.1) and PyProphet (https://github.com/grosenberger/pyprophet/tree/feature/ipf @93ec307) were used as described above.
TRIC (msproteomicstools/master @d2b5e17) was run with the following parameters:
feature_alignment.py: --file_format openswath --fdr_cutoff 0.01 --max_fdr_quality 0.2 --mst:useRTCorrection True --mst:Stdev_multiplier 3.0 --method LocalMST --max_rt_diff 30 --alignment_score 0.0001 --frac_selected 0 --realign_method lowess_cython --disable_isotopic_grouping
Statistics and visualization
The site-localization and peptidoform FDR for MaxQuant and IPF were computed for phosphopeptides only as described in Supplementary Notes II.F.
Visualization of Fig. 3 and Supplementary Fig. 5 was conducted using the heatmap.2 function of the R package “gplots” with the default parameters, but sorted by generation of a dendrogram on the row-level only. The intersection of peptides containing phosphorylated residues between IPF and MaxQuant was used for all comparisons. For the quantification heatmaps, the dendrograms were applied from the corresponding heatmaps for identification/detection.
Fig. 3c and Supplementary Fig. 5c depict only peak groups / peptide precursors reported as differentially expressed by mapDIA (significance thresholds: FDR < 0.01 & log2(FC) > 2). mapDIA42 (3.0.1) was used with the following parameters:
LEVEL=1, EXPERIMENTAL_DESIGN=IndependentDesign, NORMALIZATION=tis, MIN_OBS=1, MIN_DE=.01, MAX_DE=.99.
Analysis of the 14-3-3β data set
Spectral library and peptide query parameter generation
The 18 DDA runs were supplemented with 2 phosphopeptide-enriched DDA runs to enable mapping of potentially unidentified MS1 features of the biologically-relevant samples with identified MS/MS spectra from the two enriched runs. Of all confidently identified peptides (PSM FDR: 1%), MaxQuant cumulatively identified 1,286 unique phosphorylated peptide sequences, of which we could generate a spectral library for 1,068 peptides that was used for the analysis of the corresponding 18 DIA runs by IPF.
MS data analysis
The analysis of the 14-3-3β data set43 was conducted identically as for the enriched phosphopeptide data set. Normalization was conducted using the aLFQ (https://github.com/grosenberger/aLFQ/tree/develop @94dfd2b) function normalizeBetweenRuns against the peptides of the 14-3-3β bait protein.
Statistics and visualization
The site-localization and peptidoform FDR for MaxQuant and IPF were computed for phosphopeptides only as described in Supplementary Notes II.F.
mapDIA42 (3.0.1) was used with the following parameters:
Phosphopeptide-level: Level=1, EXPERIMENTAL_DESIGN=ReplicateDesign, MIN_OBS=1, MIN_DE=.01, MAX_DE=.99
Protein-level: Level=2, EXPERIMENTAL_DESIGN=ReplicateDesign, SDF=2, MIN_CORREL=0.2, MIN_OBS=1, MIN_PEP_PER_PROT=1, MAX_PEP_PER_PROT=5, MIN_DE=.01, MAX_DE=.99
The 14-3-3β binding motifs computed as part of the original study43 were used to map motifs to phosphopeptides. Visualization of Supplementary Fig. 6b-c was conducted using the boxplot function of the R package “graphics” with the default parameters as described above.
Assessment of variance components of post-translational modifications in human blood plasma
Spectral library and peptide query parameter generation
We generated a spectral library by searching DDA data from strong anion exchange chromatography fractions of trypsinized, depleted plasma samples. The searches were carried out separately for each type of modification. This resulted in a cumulative library covering 467 non-redundant proteins, 6,928 peptide sequences and 9,272 peptidoforms at 0.2% iProphet65 peptide FDR, consistent with the proteome coverage of the original study33. Of all the peptidoforms, 49.9% were unmodified and 33.5% contained carbamidomethylated residues. The other modification types were respectively present at the level of 0 – 9.9% of the detected peptides (Fig. 4a). Based on residue specificity (see below), we conservatively grouped PTMs (preferring artefactual causes) to originate from artefactual or biological effects. 4,495 peptidoforms contain PTMs that are likely technical artefacts, whereas 511 are attributed with biological effects (Supplementary Table 2).
DDA database search and spectral library generation
All raw instrument data acquired in DDA mode were centroided and converted to mzXML using msconvert (ProteoWizard61 3.0.7162). The non-redundant reviewed human protein FASTA was obtained from the UniProtKB/Swiss-Prot69 (2015-02-23) and appended with iRT peptide sequences and pseudo-reverse decoys. In addition, a second FASTA was generated based on the first with additional sequence variants of the human ApoE protein (ProteomeXchange repository). The files were searched using Comet62 (2015.02) using the default parameters for high mass accuracy instruments: peptide mass tolerance: 20 ppm (monoisotopic), isotope error enabled, fully tryptic digestion with max 2 missed cleavages, static C (Carbamidomethyl), variable M (Oxidation), max variable mods: 5. Based on these parameters, for each additional variable modification type, a separate search was conducted: MW (Oxidation), c (Amidated), NQ (Deamidated), Kn (Carbamyl), K (Label:13C(6)15N(2)), R (Label:13C(6)15N(4)), KnST (Formyl), EK (Carboxy), Kn (Acetyl), Y (Nitro), KRDEc (Methyl), STY (Phospho), Y (Sulfo), K (GG), KR (Dimethyl), ApoE. PeptideProphet38,63 (TPP64 4.8.0) with parameters -dDECOY_ -OAPdlIwt was run independently per file and iProphet65 was used to combine all results. The heavy isotope labeled forms of K and R were included because heavy peptide standards with established SRM methods49 were spiked in the plasma digest for SWATH-MS quantification. Specifically, these heavy peptide standards included 73 spiked-in SIS peptides, corresponding to 37 plasma proteins33,49, with levels generally adjusted to the endogenous peptides in the human plasma proteome49. Furthermore, Amidation, sulfonation and dimethylation were excluded at later stages of the analysis, because they did either not generate enough suitable detection transitions (amidation), were either not identified with confidence (sulfonation; potential conflict with phosphorylation due to very similar mass and residue specificity), or resulted in only one detected peptide (dimethylation). SpectraST66 (TPP/SVN r7019, custom build with disabled hardcoded modifications) was used to generate a spectral library of all peptide identifications at iProphet FDR 0.2% with the following parameters: -cP0.9 -c_IRR -c_IRTirtkit.txt -cICID-QTOF -c_RDYDECOY -cAC –cM. OpenMS version (https://github.com/grosenberger/OpenMS/tree/feature/ipf @7924fc3) was used for all following steps: ConvertTSVToTraML was used to convert the SpectraST MRM file to a TraML. OpenSwathAssayGenerator was applied on the TraML with following parameters: -swath_windows_file swath32.txt -allowed_fragment_charges 1,2,3,4 -enable_ms1_uis_scoring -max_num_alternative_localizations 20 -enable_identification_specific_losses -enable_identification_ms2_precursors. OpenSwathDecoyGenerator was applied to append decoys to the target peptide query parameters using the following parameters: -method shuffle -append -mz_threshold 0.1 -remove_unannotated. All OpenMS tools were executed using the modified chemistry parameters for the extended modification set (OpenMS.extended.params) (ProteomeXchange repository).
OpenSWATH, PyProphet and TRIC
Peptide queries based on the spectral library were run using IPF against the SWATH-MS data set consisting of 232 samples (with additional replicates, see Methods). To improve the consistency of the quantitative matrix, the inter-run alignment algorithm TRIC35 was applied based on the peptidoform-level confidence estimates of IPF to transfer the confidence to aligned peak groups and to impute background noise values (Methods). Based on the detected peak groups for all runs, this increased the matrix consistency (all peak groups vs all runs) by 10.3%. Cumulatively, 82.5% of the peptidoforms present in the library could be detected in at least one sample and 50.3% were detected in at least 20 samples with a run-specific IPF peptide query FDR threshold at 1% (Supplementary Table 2). This recovery rate corresponds well with expectations in cases in which SWATH-MS data from undepleted and unfractionated plasma digest is queried with a sample-specific library generated from multiple fractions of the same sample 26.
All steps were applied to the complete data set consisting of the 232 samples and additionally the 6 technical and 4 whole process replicates and 4 repeat runs.
OpenSwathWorkflow (https://github.com/grosenberger/OpenMS/tree/feature/ipf @7924fc3) was run with the following parameters -min_upper_edge_dist 1 -mz_extraction_window 0.05 -rt_extraction_window 600 -extra_rt_extraction_window 100 -min_rsq 0.95 -min_coverage 0.6 -use_ms1_traces -enable_uis_scoring -Scoring:uis_threshold_peak_area 0 -Scoring:uis_threshold_sn -1 -Scoring: stop_report_after_feature 5 -tr_irt hroest_DIA_iRT.TraML. The following subset of scores was used on MS2-level: xx_swath_prelim_score library_corr yseries_score xcorr_coelution_weighted massdev_score norm_rt_score library_rmsd bseries_score intensity_score xcorr_coelution log_sn_score isotope_overlap_score massdev_score_weighted xcorr_shape_weighted isotope_correlation_score xcorr_shape. All MS1 and UIS scores were used for PyProphet.
PyProphet (https://github.com/grosenberger/pyprophet/tree/feature/ipf @93ec307) was run on all runs independently with the following parameters:
--final_statistics.emp_p --qvality.enable --qvality.generalized
--ms1_scoring.enable --uis_scoring.enable --uis_scoring.expand_peptidoforms
--xeval.num_iter=10 --ignore.invalid_score_columns.
TRIC (msproteomicstools/master@d2b5e17) was run with the following parameters:
feature_alignment.py: --file_format openswath --fdr_cutoff 0.01 --max_fdr_quality 0.2 --mst:useRTCorrection True --mst:Stdev_multiplier 3.0 --method LocalMST --max_rt_diff 30 --alignment_score 0.0001 --frac_selected 0 --realign_method lowess_cython --disable_isotopic_grouping
requantAlignedValues.py: --disable_isotopic_grouping --disable_isotopic_transfer --realign_runs lowess_cython --method singleShortestPath --do_single_run
Residue modifiability and differentiation between artefactual and biological PTMs
Differentiation between biological and artefactual causes is challenging for most modification types, because many can originate from both causes, depending on the residue and context. However, to provide an approximate assessment, we generated a manually refined list of UniMod residue modifiability:
Artefactual PTMs: C (Carbamidomethyl), Kn (Carbamyl), K (Carboxy), NQ (Deamidated), KST (Formyl), K (Label:13C(6)15N(2)), R (Label:13C(6)15N(4)), DE (Methyl), MW (Oxidation), c (Methyl)
Biological PTMs: Kn (Acetyl), E (Carboxy), KR (Dimethyl), K (GG), KR (Methyl), Y (Nitro), STY (Phospho), Y (Sulfo), n (Formyl)
Variance decomposition of peptidoform level
Quantitative variance decomposition on peptide query / peptidoform level was conducted as previously reported33 with normalization of the peptidoform intensities by a rank normal transformation per run. The input data was restricted to ensure that each peptidoform could be detected in at least 20 samples below the threshold (1% IPF peptide query peptidoform FDR). The heritability analysis was conducted using SOLAR72 (7.6.4). The significance of the components was computed by assuming a X2 distribution of and one degree of freedom. Correction for multiple testing was conducted using Storey’s q-value73.
Statistics and visualization
Visualization of Fig. 4, 5b and Supplementary Fig. 9, 10, 12 and 14 was conducted using the boxplot function of the R package “graphics” or “ggplot2” as described above. Fig. 5b and Supplementary Fig. 12 and 14 indicate outliers as dots. The coefficient of variation (CV) values visualized in Fig. 4b and Supplementary Fig. 9, 10 were computed using the cv function of the R package “raster”. Only CV values where peptidoforms were detected in at least 2 samples were considered, all others were omitted.
Supplementary Fig. 11 indicates the quantitative variability of each peptidoform as mean with the error bars indicating the standard deviation.
Supplementary Material
Acknowledgments
H.L.R was founded by the Swiss National Science Foundation (SNSF grant P2EZP3 162268). R.A. was supported by ERC Proteomics v3.0 (AdG-233226 Proteomics v.3.0 and AdG-670821 Proteomics 4D) and the Swiss National Science Foundation (SNSF) (31003A_166435).
We would like to thank L. Gillet and A. Leitner for insightful discussions on post-translational modification and SWATH-MS. U2OS cells were kindly provided by Y. Shiloh, Tel Aviv University, Israel. We are grateful to all twin registry participants recruited in this study. For the unit of Twins UK, this study was funded by the Wellcome Trust and EC’s Seventh Framework Programme (FP7/2007-2013) and also received support from the National Institute for Health Research-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. Further acknowledgments go to the Scientific IT Support team (SIS) of ETH Zurich for support and maintenance of the lab-internal computing infrastructure, the HPC team (Brutus) and the OpenMS and PyProphet developers for including IPF in the OpenMS and PyProphet frameworks. We thank the PRIDE team for proteomic data deposition.
Footnotes
Author Contributions
G.R. developed and implemented IPF and analyzed the synthetic phosphopeptide reference, enriched phosphopeptide, 14-3-3β and twin study data. Y.L. provided and analyzed the twin study data. H.L.R. developed and implemented the MS1 scoring and quantification in OpenSWATH. C.L. provided the synthetic phosphopeptide reference sample and acquired the data. A. Buil and G.R. conducted the heritability analysis of the twin study. A. Bensimon and Y.L. conducted the enriched phosphopeptide experiment and acquired the data. M.S. provided the synthetic phosphopeptide reference sample. B.C.C analyzed the 14-3-3β data. All authors provided critical input on the project. G.R., Y.L. and R.A. wrote the paper with feedback from all authors. L.M. supervised the development of IPF and conducted the protein-level PTM meta-analysis. R.A. designed and supervised the study.
Competing Financial Interests Statement
R.A. holds shares of Biognosys AG, which operates in the field covered by the article. The remaining authors declare no competing financial interest.
Source Code Availability
IPF is available as platform-independent open source software (http://www.openswath.org) under the Modified BSD License and implemented as part of OpenMS67 (http://www.openms.org) and PyProphet74 (https://github.com/PyProphet).
Data Availability and Accession Code Availability Statements
The synthetic phosphopeptide reference mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE75 partner repository (http://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD004573.
The enriched U2OS phosphopeptide mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE75 partner repository (http://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD006056.
The 14-3-3β phosphopeptide interactomics mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE75 partner repository (http://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD006057.
The twin study mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE75 partner repository (http://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD004574.
References
- 1.Deutsch EW, et al. State of the Human Proteome in 2014/2015 As Viewed through PeptideAtlas: Enhancing Accuracy and Coverage through the AtlasProphet. J Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00500. 150724142438005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith LM, Kelleher NL. Consortium for Top Down Proteomics. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhlen M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419–1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 4.Edwards AM, et al. Too many roads not taken. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- 5.Marx V. Finding the right antibody for the job. Nat Methods. 2013;10:703–707. doi: 10.1038/nmeth.2570. [DOI] [PubMed] [Google Scholar]
- 6.Chait BT. Mass Spectrometry: Bottom-Up or Top-Down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 7.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 8.Doll S, Burlingame AL. Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem Biol. 2015;10:63–71. doi: 10.1021/cb500904b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 10.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 11.Picotti P, Aebersold R. Selected reaction monitoring–based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 12.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics. Mol Cell Proteomics. 2012;1:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman JD, Goodlett DR, Masselon CD. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 14.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717–O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro P, et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat Biotechnol. 2016;34:1130–1136. doi: 10.1038/nbt.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerr A. DIA mass spectrometry. Nat Methods. 2015;12:35–35. [Google Scholar]
- 17.Na S, Paek E. Software eyes for protein post-translational modifications. Mass Spectrom Rev. 2015;34:133–147. doi: 10.1002/mas.21425. [DOI] [PubMed] [Google Scholar]
- 18.Chalkley RJ, Clauser KR. Modification Site Localization Scoring: Strategies and Performance. Mol Cell Proteomics. 2012;11:3–14. doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira AP, et al. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol. 2012;8:623. doi: 10.1038/msb.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abelin JG, et al. Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Mol Cell Proteomics. 2016;15:1622–1641. doi: 10.1074/mcp.M116.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 22.Tsou C-C, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, et al. Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files. Nat Methods. 2015;12:1105–1106. doi: 10.1038/nmeth.3593. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat Methods. 2015;12:1106–1108. doi: 10.1038/nmeth.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ting YS, et al. Peptide-Centric Proteome Analysis: An Alternative Strategy for the Analysis of Tandem Mass Spectrometry Data. Mol Cell Proteomics. 2015;14:2301–2307. doi: 10.1074/mcp.O114.047035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 27.Sidoli S, et al. SWATH Analysis for Characterization and Quantification of Histone Post-translational Modifications. Mol Cell Proteomics. 2015;14 doi: 10.1074/mcp.O114.046102. mcp.O114.046102–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krautkramer KA, Reiter L, Denu JM, Dowell JA. Quantification of SAHA-Dependent Changes in Histone Modifications Using Data-Independent Acquisition Mass Spectrometry. J Proteome Res. 2015;14 doi: 10.1021/acs.jproteome.5b00245. 150629103741000–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter CJ, Bereman MS. Data-independent-acquisition mass spectrometry for identification of targeted-peptide site-specific modifications. Anal Bioanal Chem. 2015;407:6627–6635. doi: 10.1007/s00216-015-8819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence RT, Searle BC, Llovet A, Villén J. Plug-and-play analysis of the human phosphoproteome by targeted high-resolution mass spectrometry. Nat Methods. 2016;13:431–434. doi: 10.1038/nmeth.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller A, et al. Opening a SWATH Window on Posttranslational Modifications: Automated Pursuit of Modified Peptides. Mol Cell Proteomics. 2016;15:1151–1163. doi: 10.1074/mcp.M115.054478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert OT, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786–786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carr SA, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Röst HL, et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat Methods. 2016;13:777–783. doi: 10.1038/nmeth.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soste M, et al. A sentinel protein assay for simultaneously quantifying cellular processes. Nat Methods. 2014;11:1045–1048. doi: 10.1038/nmeth.3101. [DOI] [PubMed] [Google Scholar]
- 37.Choi H, Ghosh D, Nesvizhskii AI. Statistical Validation of Peptide Identifications in Large-Scale Proteomics Using the Target-Decoy Database Search Strategy and Flexible Mixture Modeling. J Proteome Res. 2007;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 38.Choi H, Nesvizhskii AI. Semisupervised Model-Based Validation of Peptide Identifications in Mass Spectrometry-Based Proteomics. J Proteome Res. 2008;7:254–265. doi: 10.1021/pr070542g. [DOI] [PubMed] [Google Scholar]
- 39.Fermin D, Walmsley SJ, Gingras A-C, Choi H, Nesvizhskii AI. LuciPHOr: algorithm for phosphorylation site localization with false localization rate estimation using modified target-decoy approach. Mol Cell Proteomics. 2013;12:3409–3419. doi: 10.1074/mcp.M113.028928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fermin D, Avtonomov D, Choi H, Nesvizhskii AI. LuciPHOr2: site localization of generic post-translational modifications from tandem mass spectrometry data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagano K, et al. Phosphoproteomic analysis of distinct tumor cell lines in response to nocodazole treatment. Proteomics. 2009;9:2861–2874. doi: 10.1002/pmic.200800667. [DOI] [PubMed] [Google Scholar]
- 42.Teo G, et al. mapDIA: Preprocessing and statistical analysis of quantitative proteomics data from data independent acquisition mass spectrometry. J Proteomics. 2015;129:108–120. doi: 10.1016/j.jprot.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins BC, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 44.Zhu P, Bowden P, Zhang D, Marshall JG. Mass spectrometry of peptides and proteins from human blood. Mass Spectrom Rev. 2011;30:685–732. doi: 10.1002/mas.20291. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Zubarev RA. Mass spectrometric analysis of asparagine deamidation and aspartate isomerization in polypeptides. Electrophoresis. 2010;31:1764–1772. doi: 10.1002/elps.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zawadzka AM, et al. Variation and quantification among a target set of phosphopeptides in human plasma by multiple reaction monitoring and SWATH-MS2 data-independent acquisition. Electrophoresis. 2014;35:3487–3497. doi: 10.1002/elps.201400167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abello N, Kerstjens HAM, Postma DS, Bischoff R. Protein Tyrosine Nitration: Selectivity, Physicochemical and Biological Consequences, Denitration, and Proteomics Methods for the Identification of Tyrosine-Nitrated Proteins. J Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 48.Gu H, et al. Quantitative Profiling of Post-translational Modifications by Immunoaffinity Enrichment and LC-MS/MS in Cancer Serum without Immunodepletion. Mol Cell Proteomics. 2016;15:692–702. doi: 10.1074/mcp.O115.052266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hüttenhain R, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4:142ra94–142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y, et al. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J Clin Invest. 2013;123:3815–3828. doi: 10.1172/JCI67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, et al. An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat Med. 2014;20:193–203. doi: 10.1038/nm.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pankhurst G, et al. Characterization of specifically oxidized apolipoproteins in mildly oxidized high density lipoprotein. J Lipid Res. 2003;44:349–355. doi: 10.1194/jlr.M200256-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Egertson JD, et al. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsou C-C, Tsai CF, Teo G, Chen YJ, Nesvizhskii AI. Untargeted, spectral library-free analysis of data independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics. 2016 doi: 10.1002/pmic.201500526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T, et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim SC, et al. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res. 2006;5:3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 59.Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, et al. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 61.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 63.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 64.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shteynberg D, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam H, et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Röst HL, et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 68.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009–bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou H, et al. Robust phosphoproteome enrichment using monodisperse microsphere–based immobilized titanium (IV) ion affinity chromatography. Nat Protoc. 2013;8:461–480. doi: 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- 71.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 72.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teleman J, et al. DIANA-algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2014;31:555–562. doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- 75.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.