Abstract
Nanoscale materials are increasingly found in consumer goods, electronics, and pharmaceuticals. While these particles interact with the body in myriad ways, their beneficial and/or deleterious effects ultimately arise from interactions at the cellular and subcellular level. Nanoparticles (NPs) can modulate cell fate, induce or prevent mutations, initiate cell-cell communication, and modulate cell structure in a manner dictated largely by phenomena at the nano-bio interface. Recent advances in chemical synthesis have yielded new nanoscale materials with precisely defined biochemical features, and emerging analytical techniques have shed light on nuanced and context-dependent nano-bio interactions within cells. In this review, we provide an objective and comprehensive account of our current understanding of the cellular uptake of NPs and the underlying parameters controlling the nano-cellular interactions, along with the available analytical techniques to follow and track these processes.
Graphical Abstract
1 Introduction
The cell membrane (CM) protects intracellular components from the surrounding environment. More specifically, the CM maintains cell homeostasis, provides structural support, maintains ion concentration gradients, and controls the entry and exit of charged small molecules and nutrients.1–3 Almost all natural membranes, regardless of function, share a common general structure: a bilayer of amphiphilic lipids with hydrophilic heads and hydrophobic tails.2, 4 The amphiphilic properties of phospholipids make their bilayer assembly an efficient selective barrier, as “balanced” hydrophobicity/hydrophilicity is needed to permit a wide range of small biomolecules to enter the cell by passive diffusion. However, entry is regulated in some cases through other mechanisms (e.g., channel, receptor, or transporter).5
The development of nanoparticles (NPs) for a wide range of biomedical applications promises safer and more effective solutions to numerous medical issues.6 In this review, the word nanoparticles refers to an exogenous synthetic structure with nanoscale dimensions. For many NPs, their safe entry into cells is an important step in achieving high-yield prognostic and therapeutic efficacy. Moreover, the intracellular fate of NPs is critical to their success, considering that these carriers are intended to deliver specific molecules (i.e., genes, drugs, and contrast agents) to the cytosol, nucleus, or other specific intracellular sites. However, NPs’ efficient and controlled entry/trafficking into cells remains a major challenge. Besides their interactions with CMs, a more complete understanding of NPs’ cellular uptake and trafficking mechanisms is critical in designing efficient and safe nanomedicines by the careful tuning of the NP’s physicochemical properties to optimize cellular targeting, uptake, and trafficking.7–10
In this review, we will discuss the NPs’ journey inside the cell, with a focus on both extracellular and intracellular nano-bio interactions.
2 Cellular identity of nanoparticles and the effect of the microenvironment
Since NPs acquire different physicochemical properties in biological fluids such as blood and cell-culture media, we will first attempt to shed more light on this phenomenon. In biological fluids, the surface of NPs is dramatically modified by the adsorption of biomolecules including proteins, the so-called “protein corona”.11 Therefore, what cells “see” is corona-coated NPs rather than their pristine surfaces.12 More specifically, the composition of the protein layer (in terms of type, amount, and conformation of the proteins involved) is recognized as the biological identity of NPs. Three main factors affect the biological identity of NPs: 1) NP-related factors including the collective physicochemical properties of NPs such as size, polydispersity, shape, charge, surface chemistry, and surface hydrophobicity/hydrophilicity. 2) Biological factors including protein source (e.g., human serum, fetal bovine serum, rat serum) and protein abundance. 3) Experimental factors including adsorption temperature (incubating temperature and local temperature changes upon hyperthermic events), ionic strength, osmolarity, etc. We and others have thoroughly reviewed the effect of NPs’ physicochemical properties on their biological identity;13–16 in addition, we have also introduced and reviewed many biological and experimental factors.17–23 Therefore, the protein corona is not thoroughly discussed here, and readers are directed to the cited review papers for details. Before the NPs reach the exterior membranes of target cells, they must interact with the microenvironment around the target cells. Furthermore, that microenvironment, including fibrosis, extracellular matrix6, various microenvironmental factors, pH24, 25 and so on, can also change the properties of NPs and affect their interactions with the cell membrane and finally their intracellular fate. For the NPs designed for targeting tumor cells, the tumor microenvironment (TME) could have great influence on their cellular fate. Specifically, the highly aggressive replicative nature of tumor cells produces poor lymphatic drainage, extensive fibrosis, and a dense extracellular matrix, which will lead to elevated interstitial fluid pressures. The extravasation of NPs to distal regions in the tumor will be highly restricted by the high pressures in the TME, which can determine the population of tumor cells (e.g., the periphery or the core of tumors) that the NPs will interact with. The area of nano-cell interactions can also be affected by the interplay of various microenvironmental factors, such as bradykinin, vascular endothelial growth factor (VEGF), prostaglandins, and matrix metalloproteinases (MMPs). Moreover, characteristics of the microenvironment such as pH can affect the nano-cell interactions and the entry of NPs. As reported by Yuan et al.26, when they reach the TME with a low pH value of ~6.8, zwitterionic NPs can shed their anionic component leaving a positive charge on their surface, enhancing tumor cell entry. Another common example is that of oral-delivery NPs, such as insulin-loaded NPs for the treatment of diabetes. The microenvironment of the gastrointestinal (GI) tract, including extreme pH, enzymatic degradation, and poor permeability of the intestinal epithelium are critical in determining the NPs to be degraded or to cross the intestinal epithelium via transcytosis.27 Therefore, when we design NPs for different applications, the microenvironment of the target cells should be carefully considered, since it greatly influences the performance of NPs, determining where they go and what kind of cells they interact with.6, 24
3 Cell membrane interactions and nanoparticle entry into cells
When NPs reach the exterior membrane of a cell, they can interact with components of the plasma membrane or extracellular matrix and enter the cell, mainly through endocytosis. Endocytosis leads to the engulfment of NPs in membrane invaginations, followed by their budding and pinching off to form endocytic vesicles, which are then transported to specialized intracellular sorting/trafficking compartments. Depending on the cell type, as well as the proteins, lipids, and other molecules involved in the process, endocytosis can be classified into several types.28, 29 Five main different mechanisms of endocytosis will be discussed here: phagocytosis, clathrin-mediated endocytosis, caveolin-mediated endocytosis, clathrin/caveolae-independent endocytosis, and macropinocytosis. Some references may consider the last four mechanisms subtypes of the broadly defined process of pinocytosis. Compared to phagocytosis, which takes place mainly in professional phagocytes, pinocytotic mechanisms are more common and occur in many cell types.30
At the end of this section, a few other non-endocytic entry mechanisms are also briefly discussed.
3.1 Phagocytosis
Phagocytosis occurs primarily in professional phagocytes (e.g., macrophages, monocytes, neutrophils, and dendritic cells), which are responsible for host defense and the uptake of dead cells and cell debris. However, some other types of cells (e.g., fibroblasts, epithelial, and endothelial cells) also have phagocytic activity, but to a lower extent and are referred to as para-phagocytes.31
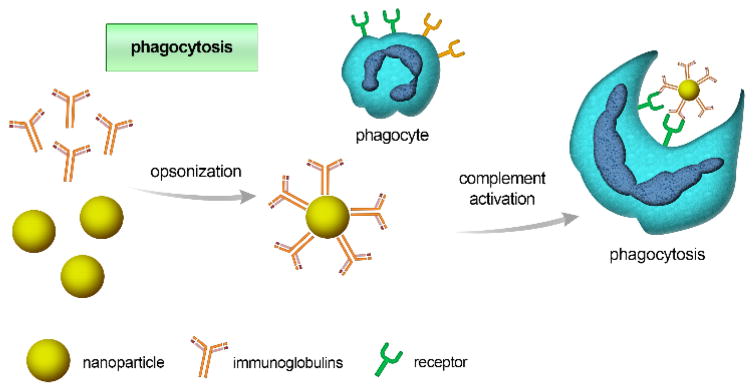
Phagocytosis of NPs is usually initiated by opsonization: opsonins such as immunoglobulins (i.e., antibodies), complement proteins, or other blood proteins (e.g., laminin and fibronectin) are adsorbed onto the NPs’ surface 31, 32 (see Fig. 1). Opsonized NPs are then recognized by, and attached to phagocytes via specific ligand-receptor interactions. This initializes a signaling cascade that can trigger actin assembly, the formation of cell surface extensions, and subsequent engulfing and internalization of particles, forming what is known as a “phagosome”.33 The aforementioned events take between 30 minutes to several hours, depending on cell type and the nature of the particle surface. Phagocyte receptors involved in this process include Fc receptors, complement receptors, and other receptors such as mannose/fructose receptors and scavenger receptors.
Fig. 1.
Schematic illustration of the opsonization process, initiated by the adsorption of immunoglobulins or other complement proteins (opsonins) to the nanoparticle’s surface. Opsonized particles are subsequently identified through receptors on phagocytic cells and internalized.
Uptake by the phagocytic route is governed by NP’s physiochemical characteristics including size, shape, and surface properties.33 The precise mechanism of phagocytosis, and subsequent events, also depend on the type of receptors involved; for example, Fc receptor-dependent phagocytosis results in the production of pro-inflammatory mediators, whereas complement receptor-dependent phagocytosis does not.34 Therefore, associated receptors affect not only NPs’ delivery, but also their toxicity (i.e. inflammatory response).
In general, larger particles experience more efficient uptake by phagocytes. For instance, when using radiolabelled albumin NPs in the size range of 200–1500 nm, larger particles clearly underwent higher phagocytic uptake when incubated with human mononuclear cells.35 This agrees with another study of polystyrene NPs ranging from 460–2100 nm in size, where maximal uptake by mouse peritoneal macrophages was reported in a size range of 1000–2000 nm.36, 37
In addition, shape was also found to have a profound effect on the cellular uptake of NPs.38, 39 For instance, a study by Arnida et al. compared the cellular uptake of PEGylated gold nanorods and nanospheres after incubation with murine macrophages for 6 hrs. Cells were washed, lysed, and analyzed for gold content. Gold nanorods accumulated to a lesser extent than nanospheres. These findings helped explain the in vivo part of the study, where, after injection in ovarian-tumor-bearing mice, gold nanorods achieved longer circulation, compared to nanospheres.38, 39
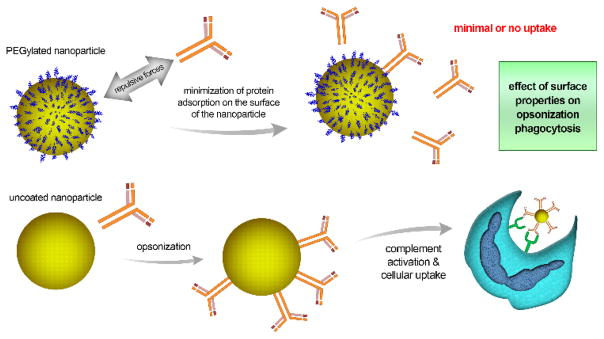
Another critical parameter controlling the uptake of NPs by phagocytes is their surface properties (Fig. 2), which first affect opsonization and then interactions with cellular membrane receptors that facilitate phagocytosis. Functionalization of NPs with sterically shielding polymers, such as hydrophilic PEG, can alter cellular uptake.40 PEGylated NPs can repel opsonization by preventing or minimizing protein adsorption to their surface. This can be explained by the conformation that PEG molecules adopt in solution: their extended form tends to create a repulsive barrier between NPs. Such a force can balance or overcome the attractive force for the intended opsonization. Interestingly, a minimum layer thickness is needed for such repulsion, which depends on the polymer’s molecular weight, conformation, and the density of chains adsorbed.41 PEGylation can increase the circulation half-life of NPs from a few minutes to several hours by avoiding uptake by the reticuloendothelial system (RES).42 An interesting example is the first FDA-approved anticancer liposome (Doxil®), in which PEGylation decreases uptake by phagocytes and thus increases the half-life of the liposomes loaded with doxorubicin, improving the overall pharmacokinetics of the nanocarrier.43 Conversely, NPs with charged or hydrophobic surfaces attract complement proteins and hence undergo greater uptake by phagocytes.33
Fig. 2.
Effect of surface properties on opsonization and subsequent internalization of nanoparticles into the cell. The Fig. compares PEG-coated nanoparticles to uncoated ones. The PEG shell repels complement proteins, minimizing protein adsorption and hence, cellular uptake. Accordingly, the uncoated nanoparticles undergo greater cellular uptake.
NPs highly susceptible to opsonization can preferentially accumulate in RES organs such as the liver and spleen. This accumulation can be exploited for the selective treatment of diseases affecting these organs such as hepatocarcinoma and liver infections.30 An interesting example is the loading of the anticancer agent doxorubicin into 200–300 nm polyalkylcyanoacrylate NPs. Upon delivery, these particles substantially accumulated in hepatic Kupffer cells, which then acted as a reservoir, slowly releasing doxorubicin as the particles degraded.44 When administered to mice, such NPs demonstrated lower systemic toxicity compared to the free form of the drug, as evident in longer animal survival rates and the absence of organ atrophy.45 Uptake of these particles was not achieved exclusively through the phagocytic route, suggesting other mechanisms might also be involved.30 This preparation has already passed phase I clinical trials 46 and is now in phase III clinical trials under the name Transdrug®.32 Another example is the encapsulation of the antifungal agent amphotericin B into nano-liposomes (AmBisome®) to treat leishmaniasis, an infection caused by a fungus that hides and survives in liver macrophages.47 Nevertheless, the rapid capture of NPs by hepatic Kupffer cells can be problematic upon delivery to non-macrophage populations; accordingly, prolonged circulation might be achieved by applying one or more macrophage-evading techniques, such as interference with protein adsorption and opsonization, managing splenic filtration, and various attempts at particle confinement within the vasculature.48
3.2 Clathrin-mediated endocytosis (CME)
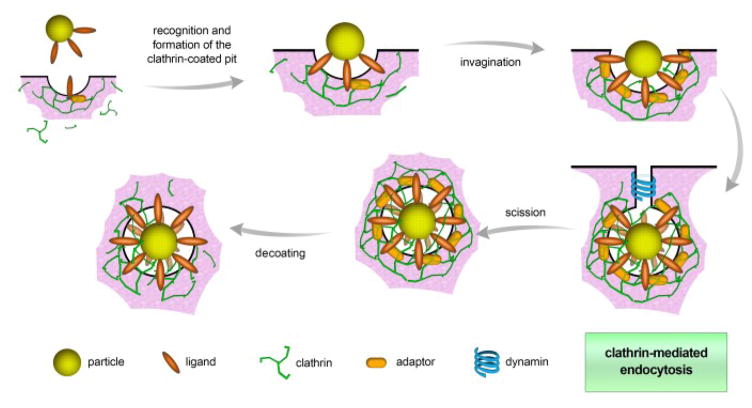
Clathrin-mediated endocytosis is the main mechanism by which cells obtain nutrients and plasma membrane components such as cholesterol via low-density lipoproteins, (LDLs) and iron via the transferrin carrier (Fig. 3). CME occurs either via receptor-specific uptake or by non-specific adsorptive uptake, which is also referred to as receptor-independent CME. In receptor-independent CME, uptake takes place without direct binding with membrane constituents; instead, non-specific hydrophobic or electrostatic interactions eventually initiate the uptake.49
Fig. 3.
Schematic of clathrin-mediated endocytosis. The process is initiated by ligand recognition and then the formation of clathrin-coated pits. With the aid of clathrin triskelions, a hexagonal lattice is formed, inducing the invagination of the plasma membrane. Afterwards, dynamin (a scission protein) releases the vesicle into the cytoplasm, where decoating takes place.
CME takes place at an area of the plasma membrane that is rich in clathrin; such domains cover about 0.5–2% of the cell surface.50 The clathrin assembly unit, known as the triskelion, has a three-legged structure consisting of three heavy and three light chains (Fig. 3).51 This unique protein and others (as detailed below) are responsible for spontaneous co-assembly into a complex architecture that generates and stabilizes membrane curvature and then the budding vesicle. Adaptor proteins are recognition sites for several different cargoes and sorting signals; they are employed in docking sites on the cytoplasmic face of the plasma membrane. Adaptor and accessory proteins are responsible for the coordination of clathrin nucleation at the sites of internalization in the membrane.52 Nucleation promotes the assembly of clathrin triskelions into curved lattices composed of pentagons and hexagons; this induces invagination of the membrane into clathrin-coated pits and consequently stabilizes the deformation points at the membrane.53 Accessory proteins (e.g., epsin, amphiphysin, SNX9) help generate and stabilize membrane curvature. Bin-amphiphysin-rvs BAR proteins can bind to dynamin, a membrane scission protein, and recruit it to the neck of the budding vesicle, releasing the vesicle into the cytoplasm.54 During CME, vesicles with diameter of 100–150 nm are formed, engulfing a volume of extracellular fluid proportional to the available internal volume of the formed vesicle. However, particles entering the cell by this route frequently end up in degradative lysosomes28, 55 and may not be suitable for coating NPs made of materials susceptible to degradation by lysosomal enzymes.
Several types of NPs were investigated for their uptake by the CME route. For instance, NPs made of D,L-polylactide (PLA) and poly(ethylene glycol co-lactide) (PEG-co-PLA) were found to be internalized via both the CME route and caveolae-mediated endocytosis (the caveolae-mediated route is discussed in more detail below). In addition, surface charge was observed to have a dominant effect over both the uptake mechanism and intracellular fate. For example, anionic particles demonstrated cellular uptake via both mechanisms in HeLa cervical cancer cells, while positively charged particles were strictly subject to CME.56 In the same context, the uptake mechanism of silica nanotubes (SNTs) was investigated using MDA-MB-231 cells via co-localization of clathrin (which served as a marker for CME) and Lysotracker (to track lysosomal uptake). Confocal microscopy indicated that positively charged particles achieved higher cellular interaction and uptake, compared to the unmodified bare SNTs. 57
3.3 Caveolae-dependent endocytosis
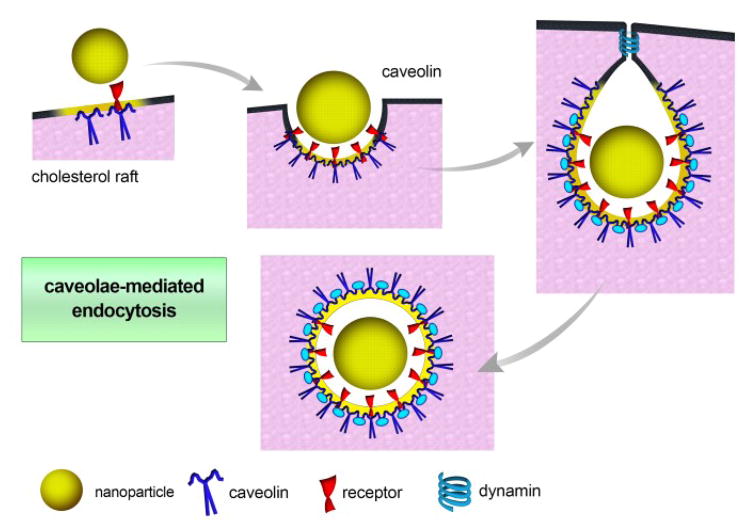
Caveolae-dependent endocytosis plays a critical role in many biological processes, such as cell signaling, transcytosis, and regulation of lipids, fatty acids, membrane proteins, and membrane tension. In addition, caveolae-mediated endocytosis is believed to be involved in a variety of diseases, including cancer, diabetes, and viral infections.58–65
Caveolae are flask-shaped membrane invaginations present in epithelial and non-epithelial cells, interspersed among regions of dense bodies anchoring the cytoskeleton. In the case of non-epithelial cells such as adipocytes and smooth muscle cells, caveolae constitute a substantial proportion of the cell membrane, increasing the surface area by up to 75% (Fig. 4).66, 67
Fig. 4.
Schematic of caveolae-mediated endocytosis. Caveolin proteins play the main role in curvature formation. As in CME, dynamin is the scission protein that allows for budding of the vesicle and release into the cell.
Caveolae are 50–80 nm in size and are lined with caveolin, a dimeric protein involved in the formation of their characteristic flask shape. Another caveolin protein known as caveolin 2 is involved in structural stabilization.58, 59, 61 Other proteins also play important roles in caveolae-mediated processes. For instance, cavin proteins aid membrane curvature, dynamin mediates scission and release of the budding vesicle, and vesicle-associated membrane protein (VAMP2) and synaptosome-associated protein (SNAP) are involved in subsequent vesicle fusion.68, 69 The commercially available nanotherapeutic Abraxane®, an albumin-bound form of paclitaxel, was found to be taken up by cancer cells by caveolae-mediated endocytosis. Albumin is of great added value to the preparation, as it binds to gp60, the albumin receptor present in the caveolae of endothelial cells, facilitating their transport to tumor interstitial spaces, where it exerts its action on cancer cells.30
Because particles entering the cell by such caveolin-dependent mechanisms can sometimes escape lysosomal degradation, this entry route is exploited by some pathogens such as viruses to escape degradation70. In terms of drug delivery, this pathway appears to be useful for the delivery of degradable materials such as genes and proteins.71 However, trafficking into acidic lysosomes could be the basis for engineering nanotherapeutics with acid-triggered release characteristics.72 For example, Sahay et al.73 investigated the caveolae-mediated cellular uptake of NPs loaded with the chemotherapeutic agent doxorubicin via pH-sensitive hydrazone bonds in an attempt to achieve intracellular drug release in the acidic environment of epithelial cancer cell lysosomes.73
3.4 Clathrin/caveolae independent endocytosis
Clathrin- and caveolae-independent endocytosis takes place in cells devoid of both clathrin and caveolae. These cells take up different cargos, such as cellular fluids, Interleukin-2, and growth hormones through other pathways, requiring a specific lipid composition (mostly cholesterol) without clathrin and caveolae.29, 74, 75
In addition, folic acid is also known to be internalized through this pathway.76 Bearing in mind the nature of cancer cell growth, including the increased demand for folic acid, folate bio-functionalization has frequently been addressed for targeting purposes. Folate-modified NPs are an excellent example of particles internalized by this route. Folate binds to its receptor, resulting in a non-destructive delivery of folate-functionalized NPs into the cytoplasm.77 Particles internalized by folate receptor-mediated endocytosis might also escape trafficking into lysosomes and often are retained in endocytic compartments or released directly into the cytoplasm.78
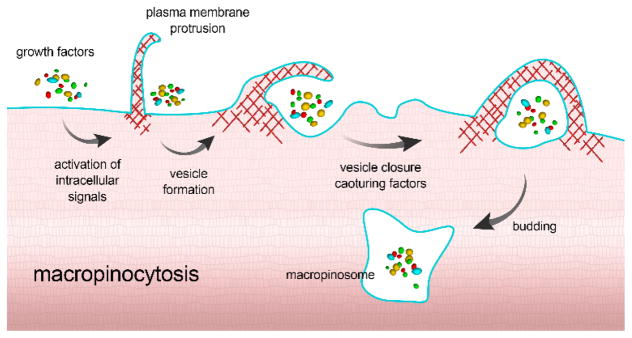
3.5 Macropinocytosis
Macropinocytosis is a unique pinocytosis process, since it does not involve the utilization of lipid rafts or pit-forming proteins. Here, large membrane extensions or ruffles are formed as a result of cytoskeleton rearrangement, which then fuse back onto the plasma membrane, creating a large vesicle (0.2–5 μm) that traps a large “gulp” of extracellular fluid (Fig. 5).79 In macropinocytosis, all particles and dissolved molecules in the extracellular fluid are taken into the endocytic vesicle, regardless of the presence of their specific receptors, making the process a form of nonspecific bulk fluid uptake.
Fig. 5.
Schematic illustration for the process of macropinocytosis. Upon recognition, intracellular signaling pathways are activated to promote the formation of large membrane extensions. These ruffles then fuse back to form a large vesicle entrapping the content of the extracellular fluid by engulfment with membrane extensions/processes.
Macropinocytosis is important in many physiological functions such as antigen presentation 80 and serves as a portal of entry for microbial pathogens, including many bacteria and viruses.79 Because this process involves the formation of large vesicles, it is considered important in the uptake of larger NPs that would not be possible via clathrin- or caveolae-dependent endocytosis.81
3.6 Other entry mechanisms
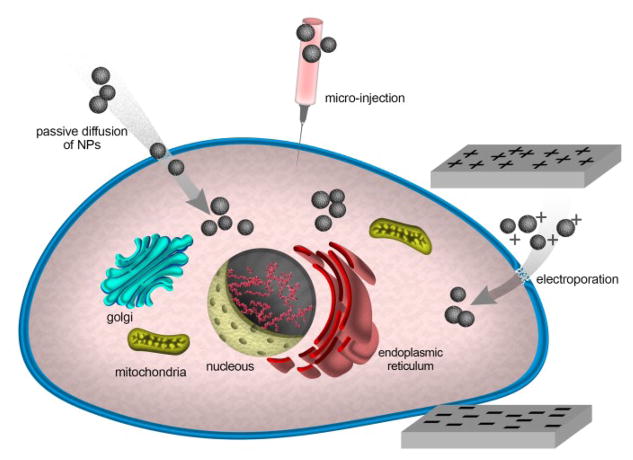
Although the different mechanisms of endocytosis discussed so far are widely recognized to be the major processes by which NPs enter cells, other entry mechanisms have also been reported, including passive diffusion, hole formation, direct microinjection, and electroporation (Fig. 6).8, 17, 82 Wang et al.83 reported that 4 nm D-penicillamine-coated quantum dots (DPA-QDs) were able to penetrate the plasma membrane of red blood cells (RBCs) through a non-endocytic passive penetration process. The authors showed that the interactions of the zwitterionic DPA-QDs with the lipid bilayer membrane led to its softening, rendering it more flexible and allowing the passive penetration of NPs without affecting the overall membrane integrity or causing any pore formation. Geiser et al.84 also showed that several nano-sized materials can cross cellular membranes by passive pathways such as diffusion and adhesive interactions, where thermal capillary waves and line tension play a significant role in controlling the entry of nanoparticles into cells. A range of cationic NPs made of both organic (peptides and polycationic polymers) and inorganic (Au-NH2, SiO2-NH2) materials have been shown to penetrate membrane-mimicking supported lipid bilayers (SLB) by disruption of lipid bilayers and nanoscale hole formation.85,86
Fig. 6.
Schematic illustration for some other non-endocytic entry mechanisms.
Artificially induced mechanisms have also been used to enable NP entry into cells. For instance, microinjection techniques have been used to directly inject silver NPs (20 nm) and metal oxide magnetite NPs (8 nm) into the cytoplasm of HeLa cells.87
Cytoplasmic microinjection has also been used to inject compacted DNA NPs into cancer cells.88 Although microinjection is a highly sophisticated technique and is not yet practical for the routine delivery of NPs into biological systems, it still represents a valuable tool in evaluating toxicity and other cellular effects of NPs; especially when the effects of extracellular biological factors (i.e., protein corona) are to be eliminated. Electroporation has also been used for physically promoting NP entry into cells. This technique is based on applying external high-voltage electrical impulses to cells, inducing the formation of transient membrane pores, through which NPs can pass. This technique has been used with different types of nanoparticles including oligonucleotide-conjugated gold nanoparticles and peptide-conjugated quantum dots.89,90
4 Physicochemical properties of nanoparticles affect their cellular uptake
It is now well understood that physicochemical properties of NPs (e.g., composition, size, shape, surface charge, surface functionalization, and surface hydrophobicity/hydrophilicity) play a key role in directing their interactions with cells. In this section, we focus on the effects of these properties on cell-NP interactions.
4.1 Effect of size and shape
The size of NPs plays an important role in cellular uptake.91 A key step in cellular uptake is the physical interaction between the NPs and the CM, which can lead to segregation and clustering of NPs on the cell surface and a subsequent CM response.92 Several theoretical models have been proposed to predict lower-threshold radius for NPs to be endocytosed. Chaudhuri et al.93 presented a two-step model, which includes cells and ligand-coated spherical NPs in thermodynamic equilibrium. The authors formulated an equation for the NP minimum radius (Rmin) required for full wrapping. (Rmin) depends on the energy released from ligand-receptor binding (adhesion strength) and the energy needed for bending the membrane (membrane rigidity). In addition, they incorporated possible electrostatic interactions between NPs and the CM before uptake to mimic real endocytosis conditions. Their results showed that NP clusters have higher rates of internalization than single NPs.
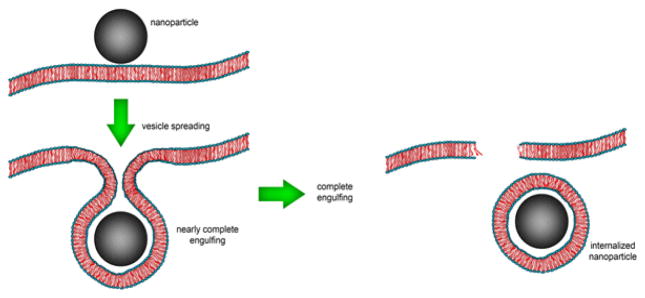
Furthermore, (Rmin) does not change with an increase in repulsive interaction values, whereas the maximum uptake is reduced, which can be explained by the balance of energetic terms. Smith et al.94 theoretically showed that a fully membrane-wrapped NP does not necessarily detach from the membrane. To complete the engulfing process and subsequent escape, the formation of phase-separated membrane domains (i.e., lipid raft) is needed. Le Bihan et al.95 demonstrated complete transmigration of silica-based NPs into large CM-mimicking unilamellar liposomes. Instead of a passive diffusion across the membrane, cryo-transmission electron microscopy (cryo-TEM) and cryoelectron tomography showed that NPs are internalized into the liposome through a three-step active process: i) vesicle spreading; ii) nearly complete engulfing; iii) completely internalized NPs (Fig. 7).
Fig. 7.
Schematic illustration of the internalization of NPs into the liposome through active process.
NP-membrane interactions can disturb the functionality and/or integrity of cellular membranes in a size-dependent manner. For example, De Planque et al.96 reported that membrane permeability and integrity depended largely on the size and surface chemistry of interacting NPs. In another study, Zhang et al.97 found a striking difference in morphological changes of giant unilamellar vesicles (GUV, a synthetic membrane-mimicking system) after interacting with silica NPs of different sizes. In the case of 18-nm silica NPs, the smooth and spherical GUVs transformed into crumpled “paper bags” with microscale holes, while the GUVs interacting with 182-nm silica NPs transformed into crumpled “pots” with a single micropore. Additionally, diffusion coefficient measurement revealed that small silica NPs considerably decreased lipid mobility (i.e., they have a freezing effect) on the membrane. This reduction in lipid mobility was time-dependent, suggesting that it does not proceed by a sharp first-order phase transition. On the other hand, larger silica NPs strongly increase lateral diffusion of GUVs, attributed to defects in the GUV membrane introduced by the wrapping mechanism.97
Quantitative and qualitative studies on the effect of NP size on endocytosis are needed to improve our understanding of NPs’ toxicity and to design effective NPs for diagnostic and therapeutic applications. Several experimental studies suggest that ~50 nm is the optimum size for NPs to achieve the highest cellular uptake in certain cells.98 However, theoretical models suggest a lower threshold, because most of these models are based on membrane deformation. For instance, Chithrani et al.98 qualitatively demonstrated that in the presence of serum proteins, the number of spherical Au NPs inside HeLa cells (introduced via receptor-mediated mechanisms) depends strongly on the NP size; 50-nm NPs entered cells more effectively than other NPs in the 14–100 nm range. The authors observed that the interaction between Herceptin-coated Au NPs (Her-Au NPs) and ErbB2 receptors of SK-BR-3 cells (i.e., receptor-mediated endocytosis) is greatly dependent on the size of the NPs; 40–50 nm Au NPs underwent much greater intracellular uptake;99 smaller NPs could not enhance the uptake process because of their inability to occupy multiple receptor binding sites and to firmly associate with receptors before being engulfed by the membrane (i.e., low binding avidity). Conversely, larger NPs, despite multivalent receptor binding, were too large for the membrane wrapping that is necessary for efficient endocytosis.
Shan et al.100 compared the uptake and unbinding forces of single Au NPs with diameters of 4, 12, and 17 nm and the HeLa cell. Their results indicated that both the uptake forces and unbinding forces increase with NP size, which can be traced back to the larger interaction area. NPs usually form heterogeneous aggregates, due to exposure to a high concentration of ions when they come into contact with biological media. 98, 101, 102 Aggregation of NPs can quantitatively affect cellular uptake based on the NPs’ physiochemical properties and the cell type. To properly mimic naturally forming aggregates in biological environments, Albanese et al.103 developed a method to prepare non-precipitating transferrin-coated aggregates of variously sized citrate-capped Au NPs. Cells were exposed to a constant number of monodisperse NPs (15 nm), and various states of aggregation (i.e., 26, 49, and 98 nm), and Au NP uptake was quantified. For HeLa and A549 cell lines, uptake of aggregated NPs was on average 25% lower than non-aggregated Au NPs. More importantly, the authors showed that aggregate uptake cannot be predicted based on the endocytosis of similar-sized single NPs, because i) aggregates are composed of small NP subunits, with higher surface curvature than larger spherical NPs, which reduces the density of targeting moieties on the aggregate’s surface; ii) the irregular shapes of aggregates can lead to aspect ratios larger than 1 (nonspherical shape); and iii) the asymmetrical structure of aggregates can also greatly influence binding avidity via multivalent receptor-to-ligand interactions. If aggregation of NPs increases their sedimentation rate, their uptake rate will be higher than that of single NPs.104 Other meaningful experimental studies also shed some light on the relationship between NP size and their intercellular distribution. For example, Williams et al.105 utilized a high content analysis (HCA) platform to locate size-tuned CdTe and CdSe/ZnS QDs in four different cells, demonstrating the QDs can enter the nucleus and finally nucleoli in all cell types examined, when they are small enough (2.1 nm). As the size of the QDs increased to 4.4 nm, penetration into cells was reduced. Nabiev et al.106 demonstrated that live human macrophages could rapidly uptake and use their active transport machineries to concentrate QDs in distinct cellular compartments specific to the size of QDs.
Another matter that generates significant uncertainty about the effects of NP size on endocytosis is how experimental data are generated. To eliminate this concern, a full-time dedicated study using a variety of complementary techniques is needed. Shapero et al.107 used flow cytometry (in addition to confocal and electron microscopy) to investigate cellular uptake, trafficking, and final localization of silica NPs of different sizes (50, 100, and 300 nm) inside A549 cells as a function of time. They showed that the uptake rate of silica NPs decreases with size; however, non-normalized flow cytometry can result in incorrect interpretation of kinetics profiles, due to the size-dependent fluorescence intensity of NPs.
In addition to size, the shape of NPs is another crucial factor in cellular uptake. Consistent with the previously mentioned report on the cellular uptake of rod-shaped and spherical gold NPs via murine macrophages38, several experimental studies have demonstrated that rod-shaped NPs undergo lower cellular uptake than spherical NPs.98, 108 For example, HeLa cells took up 74- and 14-nm spherical Au NPs at rates 5- and 3.75-fold higher than 74 × 14 nm rod-shaped Au NPs, respectively.98 Chithrani et al.108 presented two possible explanations: first, membrane wrapping for rod-shaped NPs takes longer than for spherical NPs; second, surfactant molecules adsorbed onto the longitudinal axis of nanorods impinge upon the ligand binding on the NP surface that facilitates cellular uptake. The aspect ratio, defined as the ratio of length to the width of nanorods, also has a significant effect on cellular uptake. In this case, cellular uptake (HeLa cell line) of rod-shaped Au NPs with a lower aspect ratio is greater than that of NPs with higher aspect ratio (i.e. 1.5 > 3.5 > 6). Qiu et al.109 reported that that Au rod-shaped NPs of shorter aspect ratio (ranging from 1 to 4, with sizes ~30 × 33 nm, 21 × 40 nm, 17 × 50 nm, and 14 × 51 nm, respectively) entered human breast adenocarcinoma cells (MCF-7) faster than longer NPs. The reason for this is that membrane wrapping time is longer for the larger rod-shaped NPs, as mentioned above, possibly due to formation of larger irregularly shaped aggregates. However, another study110 demonstrated that 150 × 450-nm rod-shaped (aspect ratio 3) cationic cross-linked PEG hydrogel NPs were internalized by HeLa cells 4-fold faster than 200 × 200 nm (aspect ratio 1) NPs. Moreover, the number of 100 × 300 nm NPs taken up was less than larger NPs with the same aspect ratio over a 4-h incubation period. The authors thus concluded that absolute size and/or volume also affects cellular uptake of rod-shaped NPs, due to the varying number of available receptor sites for binding. Dasgupta et al.111 used a simulation approach to show the role of shape and orientation in the cellular uptake of nanorods and nanocubes with different aspect ratios and edge curvature, respectively. For rod-like particles, they found that higher aspect ratio is not desirable for complete wrapping. NPs with small aspect ratios and flat tips enter tip-first in a “rocket” mode, whereas those with high aspect ratios and round tips enter via “submarine mode,” side-first with their long edge parallel to the membrane.111 Banerjee. et al. 112 recently investigated the cellular uptake of sphere-, rod-, and disc-shaped polystyrene NPs; rods and discs were fabricated by stretching 200-nm spheres in either one dimension (rods, hydrodynamic diameter = 394 nm) or two dimensions (discs, hydrodynamic diameter = 293 nm). While the volume of these NPs was kept constant, rods had twice as much surface area as spheres and about 1.5 times more surface area than discs. The results showed that rod and disc NPs were taken up by Caco-2 cells twice as quickly as spheres. Additionally, biotin conjugation increased the uptake of these NPs regardless of shape, but in a different manner; in fact, biotin-conjugation increased the uptake of rod NPs threefold and spheres/discs twofold. The authors linked the higher uptake of actively targeted rods compared to targeted discs and spheres to their larger surface-to-volume ratio, presenting more sites for biotin conjugation, thus increasing the probability of locating and interacting with cellular receptors for uptake.112
It has been shown that geometry can also dictate the uptake mechanism used by NPs, which determines intracellular fate. For example, Meng et al.113 synthesized mesoporous spherical (110 nm) and rod-shaped silica NPs with aspect ratios ranging from 1.5–1.7 (MSNP1) to 2.1–2.5 (MSNP2) to and 4–4.5 (MSNP3). Cytometry results for HeLa and A549 cells demonstrated remarkably higher cellular uptake of rod-shaped compared to spherical NPs; furthermore, among the rods, the intermediate aspect ratio (i.e. 2.1–2.5) was associated with the highest internalization. More importantly, the results qualitatively and quantitatively showed that greater internalization of MSNP2 is attributable to the activation of micropinocytosis, as evidenced by the enhanced activation of small GTP-binding proteins as well as the formation of actin cytoskeleton and filopodia.
Herd et al.114 investigated cellular uptake of three silica NP constructs: worm-like (232 × 1348 nm), cylindrical (214 × 428 nm), and spherical (178 nm). They found that the rate of uptake, especially in the early stages, depended on geometry. The authors linked the variations they found to the different internalization mechanisms undergone by NPs with different geometries. Furthermore, chemical inhibitor experiments suggested that clathrin-mediated endocytosis is the most favorable mechanism for spherical NPs, whereas their worm-like counterparts underwent micropinocytosis or phagocytosis. The authors’ speculation for this phenomenon is that the worms and cylinders are too large for clathrin-mediated endocytosis, and can interact only with the CM via either their 200 nm dimension or another dimension (400 and 1300 nm); i.e., within the size constraints of micropinocytosis and phagocytosis. Using a dissipative particle dynamics (DPD) simulation approach, Yang et al.115 reported that NPs with various shapes penetrate the lipid bilayer differently. Their results also suggested that NP penetration consists of a two-step rotation process, which, in addition to geometry, strongly depends on the initial orientation and the location of the NPs.
4.2 Effect of surface charge
Electrostatic interactions between charged NPs and the CM are of great biological importance.91 A number of theoretical studies on the interactions of NPs with membranes have highlighted the importance of NP surface charge. Li et al.116 employed molecular dynamics simulation to probe the effect of electrostatic attraction between cationic NPs and phospholipid membranes; the charged NPs showed a more favorable thermo-dynamical interaction than their uncharged counterparts. Moreover, the adhesion of positively charged NPs to the CM can promote membrane-wrapping phenomena. Computation of the average order parameter of lipid tails, which is a measure of the motional anisotropy of the particular bond investigated and yields its time-averaged orientation, also showed that the adhesion of anionic NPs exerts a stronger influence on the structure of membranes (i.e., formation of a high-density domain of lipid tails) compared to cationic NPs, which induce local disorders in the location of adhesion. In another molecular dynamics simulation report, the interactions of three kinds of Au NPs (cationic, hydrophobic, and anionic) with electronegative and electroneutral bilayers were probed.117 The results confirmed the dominant role of electrostatic interactions over hydrophobic interactions between NPs and the bilayer. More specifically, the positively charged NPs had a much stronger disruptive influence on the bilayers. Furthermore, both membrane penetration and disruption increase with increasing charge density, but in different manners; the degree of penetration increases remarkably at low charge densities up to an optimum value (cationic coverage of 50), while membrane disruption begins to increase quickly at higher charge densities.117 Nangia et al.118 showed that surface charge density can significantly affect the initial orientation of non-spherical NPs. For example, the computed results for rice-shaped NPs’ interaction with the lipid bilayer showed that NPs with a high positive surface charge become oriented parallel to the negatively charged lipid membrane, which maximizes adhesion and leads to substantial disruption of the bilayer. Additionally, the results emphasize the interplay of NP shape and surface charge density, which can be engineered to enhance the translocation rate by 60 orders of magnitude.118
A significant number of experimental studies have focused on interactions between NPs with various surface charges and lipid bilayer assemblies to gain a deeper understanding of the NP-membrane interaction. Leroueil et al.85 used AFM/SLB to study the degree of membrane disruption by a wide range of cationic NPs. Three general types of disruptions were detected: i) NPs that accumulate around the edges of pre-existing defects (e.g. membrane holes, membrane thinning, and/or membrane erosion), but do not induce new defects (e.g. PAMAM G3-NH2 dendrimers); ii) NPs that disrupt the bilayer mainly via diffusing to pre-existing defects and expanding them (e.g. amine-coated Au NPs (Au-NH2), the cell-penetrating peptide MSI-78, and PAMAM G5-NH2 dendrimers); iii) NPs that directly induce the formation of holes and defects in lipid bilayers, e.g. TAT sequence31 employed by HIV virus, PAMAM G7-NH2, polyethyleneimine (PEI), diethylaminoethyl-dextran (DEAE-DEX) and amine-coated silica NPs (silica-NH2). Li and Malmstadt119 showed that deformation and formation of pores (18–27 nm in size) of GUVs are elicited by relatively small cationic polystyrene (20 nm) NPs. Consistent with theoretical results, it has been shown that the strong electrostatic interaction between cationic NPs and the phosphate groups of the lipid maximize NP-membrane binding, increasing membrane surface tension and resulting in the formation of pores. Wang et al.120 showed that NPs (i.e., 20 nm polystyrene latex) with either negative or positive surface charges bind to lipid membranes and induce changes in the membrane phase state. More specifically, negatively charged NPs induced local gelation in fluid bilayers, while their positively charged counterparts induced gelled membranes to fluidize locally. This surface reconstruction of phospholipid membranes is attributed to NPs with either negative or positive surface charges that preferentially interact with N+/P− terminus of lipid membranes, respectively.
There are also several studies that have endeavored to more fully elucidate the role of surface charge on interactions between NPs and cellular membranes. For example, Cho et al.121 reported that the uptake rate of cationic Au NPs into SK-BR-3 cells is more than fivefold greater than their anionic counterparts. The authors also showed that half of cationic Au NPs diffuse into cells by generating holes or other disruptions in the CM (i.e., via non-endocytosis pathways), whereas anionic and neutral NPs are internalized into cells only via endocytosis pathways. Arizo et al.122 reported that positively charged Au NPs induce membrane depolarization for different cell types, i.e., ovarian cancer CP70 and A2780 cells, human bronchial epithelial cells (BECs), and human airway smooth muscle (ASM) cells. Membrane depolarization results in an increase in Ca2+ concentration inside the cell that can induce the modulation of intracellular pathways (e.g. inhibition of proliferation and reduction of viability in normal cells).
He et al.122 observed an increase in the uptake of chitosan NPs with higher surface charge (either positive or negative) by macrophages compared to neutral particles, suggesting the importance of electrostatic interaction in phagocytosis. However, in the case of non-phagocytic cells (human liver cell line L02 and human hepatoma cell line SMMC-7721), positively charged chitosan NPs were taken up by cells to a larger extent than were negatively charged NPs, which might be caused by the attractive/repulsive forces between the cationic/anionic NPs and the negatively charged CM. Hauck et al.123 studied the uptake of gold nanorods (18 × 40 nm) with a range of surface charges, from very positive (+ 37 mV) to very negative (−69 mV) into HeLa cells. Their results showed that the highest and lowest uptake were for nanorods with surface charges of + 37 mV and −69 mV, respectively, at all examined concentrations of gold in the media (i.e. 10, 20, 50, and 150 μM).
Hühn et al.124 explored how surface charge indirectly affects cell-NP interactions via the alteration of the protein corona around the NP. Positively charged NPs entered into different cell lines (i.e., 3T3 fibroblast cells and murine C17.2 neural progenitor cells/NPCs and human umbilical vein endothelial cells/HUVECs) more than negatively charged NPs, both with or without a protein corona. Furthermore, negatively and positively charged Au NPs (hydrodynamic radius ~7.9 and 5.1 nm, respectively) exposed to human serum albumin (HSA) have the same qualitative protein corona; i.e., the number of adsorbed HSA molecules per NP is not significantly affected by the difference in charge. The presence of proteins in the media decreases uptake of both the cationic and anionic NPs. However, differences in uptake depending on surface charge are still apparent. The authors’ speculation regarding this phenomenon is that the specific functional groups mediate the adsorption of different species of proteins with potentially varying orientation on the NP surface. In-depth assessments of NPs’ surface charge should be performed after their interactions with the cell media, since the protein corona forms at the surface of NPs and changes their surface properties.17–20, 125, 126 Many researchers have used serum-free media to avoid protein interactions with NPs.127 However, recent reports revealed that the protein corona can form even in serum-free media, mainly due to proteins secreted by cells.128
4.3 Effect of hydrophobicity
Hydrophobic and interfacial forces play important roles in the interaction between NPs and CM.91, 129 Several theoretical studies have demonstrated how the hydrophobicity of NPs affects their interactions with the lipid bilayer. Using a computer-based molecular dynamic simulation approach, Li et al.130 suggested that the hydrophobic NPs are thermodynamically stable around the middle of the hydrophobic core of the membrane. Moreover, the process of NP insertion leads to deformation and heterogeneity in the distribution of lipid molecules in the bilayer (i.e., hydrophobic mismatch), but does not cause membrane leakage. In contrast, the semi-hydrophilic NPs energetically prefer to be adsorbed on the surface of the bilayer rather than entering the core, which can induce membrane wrapping (i.e., endocytosis).
Fullerenes are well-known promising hydrophobic nanomaterials. Wong-Ekkabut et al.131 studied the thermodynamics and mechanisms of the permeation of fullerene aggregates through CMs using a computational approach. Their results suggest that fullerene clusters can easily penetrate and become embedded in a lipid membrane via passive transport, which is similar to the case with single fullerenes, but much more slowly. Interestingly, they did not find formation of stable clusters inside the bilayer or membrane disruption, even at high concentrations. However, fullerene permeation can influence cell function by altering the elastic properties of CMs. Alexeev et al.132 theoretically showed that the relative distribution of hydrophilic and hydrophobic moieties on Janus NPs’ surface influences the stability of pre-existing pores in a lipid bilayer. Specifically, Janus NPs have a hydrophobic portion that forms stable pores in amphiphilic membranes. Curtis et al.133 applied a simulation approach to visualize the molecular-level interaction between NPs and bilayer membranes. Hydrophilic NPs with diameters larger than 20 Å become wrapped, whereas NPs with a diameter of 10 Å become embedded in the bilayer surface and interact with the hydrophilic head groups of the lipid molecules. In the case of hydrophobic NPs with diameters 10–40 Å, these NPs do not undergo wrapping; instead they embed themselves within the inner hydrophobic core of the bilayers by directly penetrating the membrane.133
Understanding the interaction between NPs and a model CM can give insight into the potential effects of hydrophobic/hydrophilic NPs on living cells. Gopalakirshnan et al.135 reported that hydrophobic QDs (5 nm) were stably associated with and uniformly dispersed within/around the lipid bilayer of the vesicles, forming lipid/QD hybrids. The diffusion properties of the QDs within the membrane were also altered depending on the membrane state (i.e., arranged or disrupted lipid bilayer). These observations suggest that such hybrid vesicles could be promising nanocontainers for controlling the permeability of the membrane and be applied for delivery of small molecules into living cells.
Jing and Zhu136 reported that initiating pore formation (lipid-poor regions) on L-α-phosphatidylcholine (α-PC) SLBs with adsorbed hydrophobic polystyrene NPs occurs above a critical NP concentration that is independent of NP size. Furthermore, their results led to the conclusion that dragging lipid molecules from the SLB to adsorb and wrap on the NP surface is dominated by hydrophobic interactions, which can be enhanced by electrostatic interaction screening at increased ionic strength. Olubummo et al.137 reported that the location of QD NPs (2 nm) in mixed lipid/polymer membranes is dependent on hydrophobic, hydrophilic, or amphiphilic surface properties. Their findings indicate that hydrophobic QDs NPs can be selectively localized within the polymer domains of the mixed lipid/polymer membrane. In contrast, amphiphilic counterparts showed no specific localization in phase-separated lipid/polymer films.
Lee et al.138 reported the spontaneous incorporation of Au NPs functionalized with a mixed monolayer of hydrophilic and hydrophobic ligands (6 nm) into liposome walls (~2.5 nm thickness) to form NP-liposome complexes. In fact, hydrophobic and hydrophilic ligands redistribute themselves dynamically on the surface of Au NPs in response to interaction with the surfactant vesicle to initiate incorporation (i.e., hydrophobic ligands on the NP cluster and interact with the hydrophobic core of the bilayer, whereas hydrophilic parts remain in interaction with the aqueous solution).
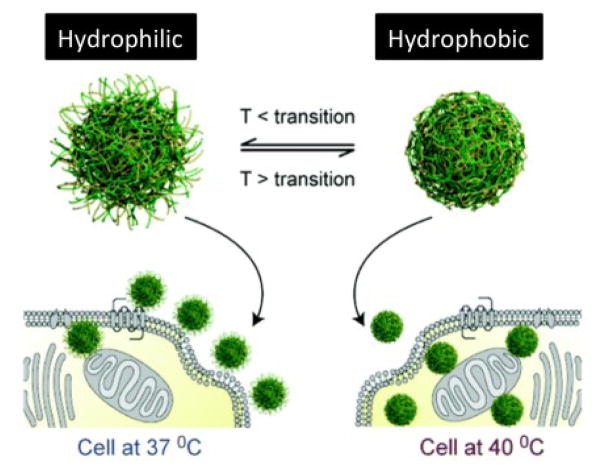
Furthermore, hydrophobicity can change the protein corona around the NP, which may indirectly modify cell-NP interaction. For example, Ge et al.139 showed that the adsorption amount and conformational changes of blood proteins (e.g., bovine fibrinogen, gamma globulin, bovine serum albumin, and transferrin) after interaction with single-wall carbon nanotubes (SWCNTs) are mainly governed by hydrophobic interactions between the surface and inner hydrophobic residues of proteins and the SWCNTs’ surface. However, those studies were performed using only a single protein; in real blood, there is a wide range of competing proteins. Therefore, more research is needed to probe the effects of NP hydrophobicity on the corona composition at the surface of NPs and their biological consequences. Moreover, the hydrophobicity of thermo-responsive NPs can be controlled by temperature (at 37 °C particles are hydrophilic but at 40 °C are hydrophobic). This approach was employed to increase the cellular uptake of thermo-responsive NPs by 20-fold at elevated temperature (40 °C), compared to the physiological temperature (Fig. 8).134, 140
Fig. 8.
NPs at temperatures below the thermal transition temperature of their outer shell exhibit hydrophilic chain-extended polymers (left-hand side) and enter cells less readily than the same NPs above their polymer thermal transition temperature (right-hand side). Reproduced with permission from Ref.134
4.4 Effect of surface functionality
Surface functionality is the principal parameter dominating NP interactions with cells and subsequent cell uptake. Since recent advances in manipulating NP surface to control interactions with cellular membrane have been reviewed by our team and others, the readers are referred to these publications for more details.24, 82, 141, 142 The current challenge in this field is to harness NPs and control their behavior by engineering their surfaces. A detailed understanding of the role of surface functionalization in the biological effect of NP is needed to facilitate efficient engineering of NPs for nanomedicine. One example that illustrates the complexity of this interaction is that even the coordination of ligands on the NP surface can significantly change their uptake pathways.143 To that end, changing the surface functionality of NPs can dictate their interactions with biological systems.144 For example, NPs can be equipped with a “homing device” that could guide the NPs to the intended target and specifically recognize the target site. The functionalized NPs could exploit the presence of overexpressed or unique receptors on the surface of cells. In this case, targeting small molecules, aptamers, peptides, proteins, and antibodies that can interact with these receptors is widely used to functionalize the NPs.145 The internalization of NPs can then occur via receptor-mediated endocytosis.146 Tao et al.147 reported a novel polydopamine-based surface functionalization method to develop aptamer-conjugated NPs for in vivo tumor targeting and finally achieved an enhanced therapeutic effect compared to the unfunctionalized NPs. The Mei group148 reported a folic acid (FA)-conjugated blended NP system that achieved active targeting of cervical cancer and enhanced the uptake efficiency of FA-NPs compared to unfunctionalized NPs. Alexis et al.149 reported the first example of a targeted NP-affibody bioconjugate for controlled drug delivery to HER-2-positive cancer cells.
A potential strategy for understanding the role of surface functionality in NP-cell interactions would be to systematically study NPs with the same size and shape, but differing surface functionalities. However, one of the major challenges in such investigations is the often low reproducibility of NP functionalization. In addition, the multi-parameter nature of such studies makes drawing definitive conclusions often rather difficult. NP cell-specific responses and the formation of non-specific protein coronas on NP surfaces also complicate research findings. Although a large number of studies have focused on studying the interactions of nanomaterials with cells and biological structures, there still is a long way to go before predicting NP–cell interactions becomes a reality.
5. Importance of cell type in the endocytosis and fate of nanoparticles
It has been suggested that various cell types may employ different endocytotic pathways to internalize the same NP.150, 151 Santos et al.151 utilized HeLa, 1321N1, and A549 cell lines to study the endocytotic pathways of carboxylated polystyrene NPs with 40 nm and 200 nm diameters. They found that 132N1 cells take up NPs mainly via clathrin-mediated endocytosis, while A549 cells use a caveolin-mediated pathway. It has been shown that caveolae-dependent endocytosis operates over one third of the cell membrane in some tissues.152 This mechanism is especially frequent in smooth muscle, endothelial cells, adipocytes, fibroblasts, and type I pneumocytes;152–155 while other cells such as neurons, leukocytes, and HepG2 cannot internalize substances by caveolae-dependent endocytosis because they lack caveolae1 protein.154, 156 Therefore, NPs taken up by these cells might experience a different fate. There is some evidence that the caveolae-dependent pathway can bypass lysosomes.150, 153, 155, 157 Most substances endocytosed by caveosomes are eventually delivered to the endoplasmic reticulum or the Golgi apparatus.158–163
Internalization of surrounding fluid occurs by large vacuoles through macropinocytosis.164 It happens in almost all cells, with a few exceptions such as brain microvessel endothelial cells.49, 164 Scientists do not yet agree on the final destiny of NPs captured in macropinosomes, but there is some evidence that it depends on cell type. For example, macropinosomes fuse with the lysosome in macrophages, whereas in A431 cells, they travel back to the surface and release their cargo to the extracellular space.164–166 Likewise, Falcone et al. showed that macropinosomes fuse with the cell membrane in human dendritic cells by exocytosis,167 while Kasahara et al.168, using a HeLa cell line overexpressing Src-tyrosine kinase, showed that high levels of c-Src kinase activity promote the formation of macropinosomes associated and merged with the lysosomal compartment.
In conclusion, it has been demonstrated that various types of cells employ different entry processes to internalize materials such as NPs, which are delivered to different cellular compartments. Table 1 offers a summary of studies of various cell types, which relate the intracellular fate of NPs to the endocytotic pathway employed during cellular uptake. Overall, the NPs’ intracellular fate is complicated, and the results obtained from studying a limited number of cell types cannot be generalized to all cells. Nevertheless, it is essential to take the effect of cell type on NP fate into account when NPs are to be used in vivo.
Table 1.
Intracellular fate of nanoparticles in various cell types.
| Cell type* | Origin | Nanoparticle | Purpose | Main endocytotic pathway | Fate/effect | Ref. |
|---|---|---|---|---|---|---|
| MDA435/LCC6/MDR1 | Human | Polymer-lipid hybrid nanoparticle (PLN) (290 nm) | To enhance cellular accumulation and retention of doxorubicin | Phagocytosis | Nuclear localization | 184 |
| MDA435/LCC6/WT | Human | Polymer-lipid hybrid nanoparticle (PLN) (290 nm) | To enhance cellular accumulation and retention of doxorubicin | Phagocytosis and pinocytosis | ---- | 184 |
| EMT6/AR1 | Mouse | Polymer-lipid hybrid nanoparticle (PLN) (290 nm) | To enhance cellular accumulation and retention of doxorubicin | Phagocytosis | Nuclear localization | 184 |
| EMT6/WT | Mouse | Polymer-lipid hybrid nanoparticle (PLN) (290 nm) | To enhance cellular accumulation and retention of doxorubicin | Phagocytosis and pinocytosis | ---- | 184 |
| MCF-7 | Human | Polymer-lipid hybrid nanoparticle (PLN) (200 nm) | To deliver doxorubicin to cancer cells and bypass multidrug resistance | ------- | Cytoplasm accumulation | 185 |
| MCF-7/ADR | Human | Polymer-lipid hybrid nanoparticle (PLN) (200 nm) | To deliver doxorubicin to cancer cells and bypass multidrug resistance | ------- | Nuclei accumulation | 185 |
| BOEC | Bovine | Polystyrene nanoparticles (Fluorescent) (44 nm) | Delivery or imaging | ATP-independent endocytosis | Cytoplasm and subsequent release to culture medium | 186 |
| HCF | Human | Polystyrene nanoparticles (Fluorescent) (44 nm) | Delivery or imaging | ATP-independent endocytosis | Cytoplasm and subsequent release to culture medium | 186 |
| HRCE | Human | Polystyrene nanoparticles (Fluorescent) (44 nm) | Delivery or imaging | Multiple mechanism of ATP-dependent and -independent internalization pathways | Cytosol and endocytotic vesicles | 187 |
| J774A.1 | Mouse | Polystyrene nanoparticles (40 nm) | Delivery of therapeutic agents | Clathrin-mediated endocytosis, phagocytosis, and macropinocytosis | ------- | 81 |
| A549 | Human | Polystyrene nanoparticles (40 nm) | Delivery of therapeutic agents | Clathrin-mediated and caveolin-mediated endocytosis | ------- | 81 |
| HepG2 | Human | Carboxylated polystyrene nanoparticles (20 nm) | Delivery of therapeutic agents | Not through early endosomes | Cytosol compartments and mitochondria | 188 |
| C3A | Human | Carboxylated polystyrene nanoparticles (20 nm) | Delivery of therapeutic agents | Not through early endosomes | Cytosol compartments and mitochondria | 188 |
| Primary IRHCs | Rat | Carboxylated polystyrene nanoparticles (20 nm) | Delivery of therapeutic agents | Not through early endosomes | Cytosol compartments but not mitochondria | 188 |
| RAW 264.7 | Mouse | NH2-labeled polystyrene (PS) nanoparticle (60 nm) | Delivery of therapeutic agents | LAMP-1_ endosome | Lysosome, mitochondrial damage, and cytotoxic (apoptosis) | 150 |
| BEAS-2B | Mouse | NH2-labeled polystyrene (PS) nanoparticle (60 nm) | Delivery of therapeutic agents | caveolin-mediated endocytosis | Caveolar endosome, cytosol, mitochondrial damage and cytotoxic (necrosis) | 150 |
| HMEC | Human | NH2-labeled polystyrene (PS) nanoparticle (60 nm) | Delivery of therapeutic agents | LAMP-1_endosome | Lysosome and cytotoxic at high concentration | 150 |
| HEPA-1 | Mouse | NH2-labeled polystyrene (PS) nanoparticle (60 nm) | Delivery of therapeutic agents | LAMP-1_endosome | Lysosome cytotoxic at high concentration | 150 |
| PC-12 | Rat | NH2-labeled polystyrene (PS) nanoparticle (60 nm) | Delivery of therapeutic agents | - | Endosome, Nontoxic | 150 |
| Hela | Human | Carboxylated polystyrene nanoparticles (40 nm and 200 nm) | Delivery or imaging | Clathrin-mediated endocytosis and macropinocytosis | ----- | 151 |
| 1321N1 | Human | Carboxylated polystyrene nanoparticles (40 nm and 200 nm) | Delivery or imaging | Clathrin-mediated endocytosis | ----- | 151 |
| A549 | Human | Carboxylated polystyrene nanoparticles (40 nm and 200 nm) | Delivery or imaging | Clathrin-mediated endocytosis | ----- | 151 |
| 293T | Human | Alginate–chitosan nanoparticle (157 nm) | DNA transfection | Clathrin-mediated endocytosis | Clathrin-dependent endosomes and lysosomes | 189 |
| COS-7 | Monkey | Alginate–chitosan nanoparticle (157 nm) | Successful DNA transfection | Clathrin-mediated endocytosis | Clathrin-dependent endosomes and lysosomes | 189 |
| CHO | Hamster | Alginate–chitosan nanoparticle (157 nm) | Failed DNA transfection | Caveolin-mediated endocytosis | Caveolin-mediated endosomes and not lysosomes | 189 |
| MCF-7 | Human | Carboxylated Quantum dots (QD655-COOH) (15–21 nm) | Live cell fluorescence imaging | Clathrin-mediated endocytosis | Localized in Lysosomes | 190 |
| MCF-10A | Human | Carboxylated Quantum dots (QD655-COOH) (15–21 nm) | Live cell fluorescence imaging | Clathrin-mediated endocytosis | Localized in lysosomes | 190 |
| Hela | Human | Carboxylated Quantum dots (QD655-COOH) (15–21 nm) | Intracellular imaging and therapeutic applications | Clathrin-mediated endocytosis | Slow release from endosomes to cytoplasm | 191 |
| H1299 | Human | Silica nanoparticles (50 nm) | Deliver therapeutic agents | Non-specific endocytosis | Lysosome | 192 |
| A549 | Human | Silica nanoparticles (50 nm, 100 nm and 300 nm) | Delivery or imaging | Not clathrin or caveolin mediated | Lysosomes | 107, 193 |
| THP-1 | Human | Silica nanoparticles (60 nm) | Deliver therapeutic agents | Clathrin-mediated and caveolin-mediated endocytosis | Vesicles and lysosome | 194 |
| A549 | Human | Silica nanoparticles (60 nm) | Deliver therapeutic agents | Clathrin-mediated endocytosis | Vesicles and lysosome | 194 |
| Hela | Human | Lipid nanoparticles (LNPs) (60 nm) | Deliver encaptured cargo | Macropinocytosis | Late endosomes, Lysosomes | 195 |
| NIH3T3 | Mouse | Lipid nanoparticles (LNPs) (60 nm) | Deliver cargo | Clathrin-mediated endocytosis | Lysosomes | 195 |
Information on the various cell lines: The P-glycoprotein-overexpressing human breast carcinoma cell line (MDA435/LCC6/MDR1), the human breast carcinoma cell line (wild type) (MDA435/LCC6/WT), the P-glycoprotein-overexpressing murine breast carcinoma cell line (EMT6/AR1), the murine breast carcinoma cell line (wild type) (EMT6/WT), human breast cancer cell line (Michigan Cancer Foundation 7) (MCF-7), the P-glycoprotein-overexpressing MCF-7/adriamycin cell line (MCF-7/ADR), bovine oviductal epithelial cells (BOEC), human colon fibroblasts (HCF), primary human renal cortical epithelial cells (HRCE), mouse macrophage cell line (J774A.1), human alveolar epithelial type II cell line (A549), human hepatocyte cell line (HepG2), human hepatocyte cell line (C3A), primary isolated rat hepatocyte couplets (Primary IRHCs), murine macrophage cell line (RAW 264.7), human bronchial epithelial cell line (BEAS-2B), the human microvascular endothelial cell (HMEC), mouse hepatocyte cell line (HEPA-1), a cell line derived from pheochromocytoma of the rat adrenal medulla (PC-12), human cervical cancer cell line (HeLa), human brain astrocytoma cell line (1321N1), human embryonic kidney cell line (293T), kidney cell line from African green monkey (COS-7), Chinese hamster ovary cell line (CHO), human non-tumorigenic epithelial cells (MCF-10A), human non-small cell lung carcinoma cell line (H1299), human monocytic cell line (THP-1), and mouse embryo fibroblast cell line (NIH3T3).
6 Intracellular trafficking
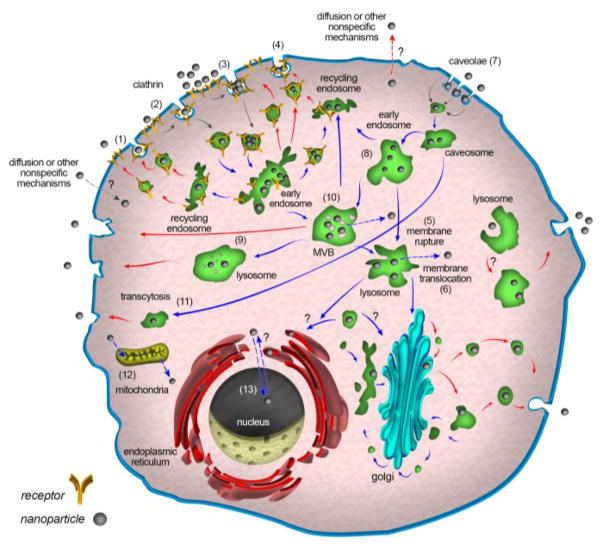
Intracellular trafficking of NPs has a critical role in the cellular fate of NPs and their therapeutic/imaging efficacy. The well-defined intracellular pathways of NPs are summarized in Fig. 9. For example, after the entry of NPs into cells via endocytic vesicles, their ultimate fate is usually determined by the intracellular sorting/trafficking mechanisms mediated mainly by a network of cellular endosomes in conjunction with the Golgi apparatus, endoplasmic reticulum (ER), and lysosomes90, 169 (see Fig. 9 for details). Endosomes are relatively large (up to 1 μm and even larger) intracellular membrane-bound compartments initially produced by the plasma membrane, and later they may fuse with Golgi-derived vesicles.170 Endosomes are usually found in the cytoplasm of most human cells and can be classified into three major types: early endosomes, recycling endosomes, and late endosomes (also called multi-vesicular bodies - MVB), which together constitute the major part of the intracellular endocytic pathway.171, 172
Fig. 9.
Schematic of endocytosis (gray arrows), intracellular trafficking (blue arrows), and cellular exocytosis (red arrows) of NPs. After cellular uptake, NPs are usually delivered to early endosomes, which are the main sorting stations in endocytosis; even vesicles related to non-receptor mediated entry mechanisms fuse with early endosomes. In the early endosome, some NPs are transported along with receptors to recycling endosomes and subsequently excreted; others that remain in early endosomes move slowly along microtubules toward the cell interior and fuse with late endosomes. Finally, late endosomes fuse with lysosomes, which are not necessarily the end of the pathway; some undergo exocytosis and release their undigested content by fusion with plasma membranes. On the pathway to multivesicular bodies (MVB) or even in lysosomes, a portion of NPs may escape from vesicular compartments to the cytoplasm; in addition, some NPs may begin by entering the cytoplasm via unspecific mechanisms. NPs in the cytoplasm or trapped in vesicles can enter the nucleus, mitochondria, endoplasmic reticulum (ER), and Golgi apparatus via unknown mechanisms. In fact, vesicles containing NPs can fuse with ER, Golgi, and other organelles. NPs that enter the ER or Golgi may leave the cell via vesicles related to the conventional secretion system. NPs that are localized in the cytoplasm can leave the cells via re-entering the vesicular system or directly via unspecific mechanisms. The “question marks” in the schematic denote unknown mechanisms.
After endocytic vesicles pinch off the plasma membrane, they become uncoated (if they were the coated type) and then fuse with early endosomes. Particles in endocytic vesicles become part of early endosomes, either freely in the lumen or inside smaller luminal vesicles. Early endosomes then act as a hub that guides further trafficking of the endocytic vesicles’ cargo into different cellular destinations.171, 173 Part of this cargo will be recycled to the plasma membrane in small vesicles, either directly or indirectly, by fusing with the recycling endosomes.174 The rest of the cargo remains in early endosomes, which then mature and differentiate into late endosomes. Late endosomes can either fuse with the plasma membrane (releasing their contents outside the cell in the form of exosomes) or they fuse with lysosomes, forming endolysosomes, whose contents become exposed to a variety of degradation enzymes such as lysosomal hydrolases.8,171
In some cases, the endocytosed NPs may escape the abovementioned endocytic pathway at any stage and become released into the cell cytoplasm; if such escape precedes fusion with the lysosomes, the particles can bypass lysosomal degradation and end up intact in the cytoplasm or in other intracellular compartments.175, 176 Another issue that should be considered when investigating the intracellular fate of NP’s is the process of autophagy. Autophagy is an intracellular degradation pathway that, distinct from the endocytic pathway, delivers certain cytoplasmic constituents to lysosomal degradation.177 This process poses a risk of recapturing the NPs after their initial escape to the cytoplasm and directing them again toward lysosomal degradation. On the other hand, since autophagy plays an essential role in maintaining cellular homeostasis, it is also important to consider the effect of NPs on this vital process, since several NP types have been suggested to either induce the process of autophagy or cause its dysfunction.178, 179
One of the major challenges in the cellular trafficking of NPs is the existence of several hidden factors, including “cell vision” and “protein corona”.126, 180 Cell vision refers to the mechanisms/behaviors that any particular cell can utilize in response to NPs. The cells can also secrete several proteins that may affect the biological identity of NPs. These two important issues can substantially change the intracellular trafficking of NPs and should be carefully considered in future reports. In addition, the type of employed protein source (e.g., fetal bovine serum) can significantly affect the intracellular trafficking of NPs.181, 182 Therefore, one can expect that a portion of the current data on in vivo intercellular trafficking of NPs is invalid, mainly because the biological identity of NPs is entirely different from that in in vitro studies.
Recent progress in the field of the biological identity of NPs revealed that even the type of disease may substantially change the biological identity of NPs and their biological fates, including their cellular uptake and toxicity.20–22, 183 Therefore, this “disease-specific” protein corona adds complications to the actual in vivo intracellular trafficking of NPs and needs more attention in future studies.
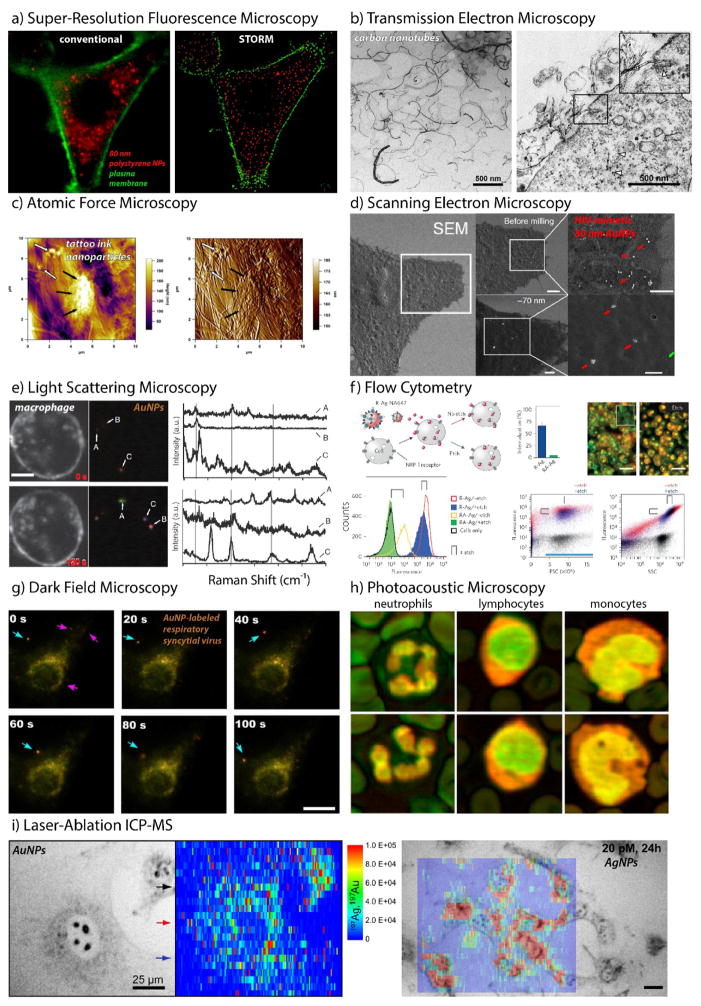
7 Probing Cellular Interactions of Nanoparticles
Our current understanding of cell-mediated NP trafficking is largely attributable to recent advances in chemical analysis and imaging. These methods provide not only information on the spatial distribution of particles within a given cell type, but also the corresponding bimolecular interactions and the phenotypic/genotypic consequences of these perturbations, as well as their time- and context-dependency. Imaging is perhaps the most salient of these techniques. Recent advances in super-resolution techniques, imaging cytometry, and high-content analytics have further expanded our appreciation of the complexity of NP trafficking and strengthened our capacity to control these cellular interactions and their specificity. Flow cytometric methods have also emerged as an indispensable tool in this regard, shedding light on the heterogeneity of cell-NPs interactions – even among monoclonal cell populations – and improving objectivity and statistical rigor through high-throughput single-cell analysis. As cell-based models give way to organoid and organ-on-a-chip technologies,196 new methods will need to be developed to address this increasing complexity. Highly automated phenotypic screening techniques are already commonplace among cell biologists, including methods for long-term, live cell imaging197 (e.g., IncuCyte® and CytoSMART™) and high-content image analysis198 (Cellomics™ and CellProfiler). High-throughput loss- and gain-of-function screens using RNA interference (RNAi),199 as well as CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa)200 will also doubtless lead to important new insights that will improve the diagnostic and therapeutic potential of nanoscale technologies. Although there are still very few reports regarding the latter methods, the following section summarizes some of the techniques currently employed to study NP cellular internalization and trafficking.
7.1 Super-Resolution Fluorescence Microscopy
Confocal laser scanning microscopy (CLSM) is distinguishable from the more common epifluorescence methods largely by its ability to exclude out-of-focus objects through point scanning.201 Due to its narrow depth of field, one can not only image cell association and trafficking with high z (axial) resolution, but also obtain serial optical sections with which to determine subcellular localization with improved accuracy. This technique has proven particularly useful when endosomal sequestration limits therapeutic potential – for example in the delivery of therapeutic nucleic acids such as plasmid DNA, small interfering RNA (siRNA), messenger RNA, and single guide RNA (sgRNA).202 CLSM can also shed light on particle-protein interactions, where co-localization can provide mechanistic information regarding intracellular trafficking routes and binding partners. Strano and coworkers,203 for example, used the intrinsic fluorescence from single-walled carbon nanotubes (SWNTs) and single-particle tracking software to monitor the cellular uptake and expulsion of SWNTS as a function of length by CLSM. They found that SWNT endocytosis was maximal near 50 nm diameter when scaled to account for diffusive interactions and the exocytosis rate of SWNTs slowed with increasing size (Table 2). Readers may note that fluorescence-, optical absorption/emission, and refractive index-based spectroscopic methods can also be used to quantify ensemble cellular uptake/efflux of nanomaterials or drugs (e.g. by HPLC,204 AAS:205 Table 2).
Table 2.
Summary of available reports on the exocytosis of NPs
| NPs | NPs’ Feature | Cell line* | Highlighted factor | Methods for exocytosis detection | Remark | Ref |
|---|---|---|---|---|---|---|
| Silica | 50 nm | H1299, NE083, NL20 | Effect of cell line | Confocal microscopy and TEM | The type of cell line is a dominant factor in the excretion profile of NPs. In the case of H1299 cells, the clusters of silica NPs in lysosomes are more easily exocytosed than single NPs in cytoplasm. | 192 |
| Silica | N/A | HUVECs, HeLa | Effect of cell line | Flow cytometry and confocal microscopy | Different retention and excretion abilities of normal and cancerous cell lines could lead to asymmetric cell-to-cell transfer of MSNs | 250 |
| Silica | 130 nm | A549, MDA-MB231, MCF-7, MDA-MB435, PANC-1, H9 | Effect of different abilities of lysosomal excretion/Effect of surface modification/Effect of extracellular calcium concentration | Flow cytometry and inductively coupled plasma-optical Emission spectroscopy (ICP-OES) | Lysosomal exocytosis plays a key role in the exocytosis process. Different surface modifications at the surface of NPs (phosphonate, folate, and PEI) change the exocytosis profile of NPs. The treatment of A549 cells with Ionomycin (an ionophore that transports calcium into cells) enhanced the exocytosis rate of the phosphonated NPs | 251 |
| Silica | 60, 180, 370, and 600 nm | HepG2 | Effect of NPs’ size | Flow cytometry | Smaller NPs are more easily cleared from HepG2 cells. Cellular exocytosis of these NPs was largely dependent on time. | 252 |
| Gold | 14, 30, 50, 74 and 100 nm (Rods/Spheres) | HeLa, SNB19, STO | Effect of cell line and Effect of size and shape of NPs | Inductively coupled plasma atomic emission spectroscopy (ICP-AES) and TEM | The proportion of rod-shaped NPs excreted from HeLa and SNB19 cells was much higher than spherical-shaped NPs. No observable difference in the amount of excreted rod-shaped and spherical-shaped NPs from STO cells. The exocytosis rate of 14 nm transferrin-coated Au NPs was two/five times faster than that of 74 nm and 100 nm NPs, respectively. (This trend was observed in three different cell lines) | 253 |
| Gold | Nanorods (length of 55 and width of 13 nm) | A549, 16HBE, MSC | Effect of cell line | Flow cytometry and inductively coupled plasma mass spectrometry (ICP-MS) | The cytotoxicity and exocytosis profile of NPs in cancer cells was different from that of normal and stem cells. | 254 |
| Gold | 35 nm and charge 18 mV | HUVECs | Effect of NPs surface modification | ICP-AES | Functionalization with non-targeting or targeting peptides yield different exocytosis profiles | 255 |
| Gold | 10 nm | HT-29 | Effect of extracellular calcium concentration | Absorbance plate reader | The concentration of excreted NPs from HT-29 cells was proportional to calcium concentration | 256 |
| Diamond | 100 nm | HeLa, 3T3-L1, and stromal cell lines | Effect of cell line | Flow cytometry | Exocytosis of the same NPs on various cells was significantly different. This observation could be important in the development of new NPSplatforms for cell-tracking and drug-delivery applications | 257 |
| SPIONs | 15 and 30 nm (trapped in porous silicon carriers) | J774 | Effect of NPs Size | SPECTRA max M2 plate reader | Smaller NPs were exocytose more easily from J774 cells compared to the larger NPs | 258 |
| Carbon nanotube | Rod (with length of 660, 430, 320 and 130 nm) | NIH-3T3 | Effect of nanotube length | Single-particle tracking (SPT) | Smaller NPs were exocytosed more easily than larger ones | 203 |
| PLGA NPs | 97 | VSMCs | Comparison between excretion rate of Lucifer yellow (molecular weight of ~450 Da) and PLGA (143,000 Da) | Confocal microscopy and HPLC | Smaller NPs or molecules were trapped in the rapid-recycling compartment, while the majority of larger molecules entered the slow-recycling compartment. Excretion of smaller NPs is thus much easier. The pre-incubation time of NPs before exocytosis did not show any considerable effect. Sodium azide and deoxyglucose reduced the exocytosis of PLGA NPs by 40% compared to the control group | 204 |
| Poly(methyl methacrylate)-block-poly(polyethylene glycol methyl ether methacrylate NPs | 20 and 25 nm | OVCAR-3 | Effect of crosslinking | Fluorescence spectroscopy | Crosslinked micelles were removed from cells more easily than non-crosslinked NPs | 259 |
| N-acetyl histidine-conjugated glycol chitosan NPs | 150–250 nm | HeLa | Effect of pre-incubation time | Confocal microscopy | The amount of excreted NPs was dependent on the pre-incubation time prior to removal of free NPs from the culture media | 260 |
| QDs | 22 to 25 nm (Cd/Se QDs) | HeLa and A549 Xenograft Tumor Models | Different binding stability between targeting moiety (transferrin) and the receptor | Fluorescence-activated cell sorting (FACS) and Fluorescence imaging | Binding between transferrin-QDs and receptor extended the intracellular NPS retention intervals | 261 |
| QDs | 65 nm (Si-QDs) | HUVECs | Effect of exposure time and NPs concentration | Confocal laser scanning microscopy (CLSM) | The exocytosis profile of Si-QDs depended on exposure time and exposed NPs concentration in cell culture | 262 |
| Silver | 6–20 nm | U251 | Effect of incubation time and NPs concentration | ICP-OES | Direct correlation between incubation time and NPS concentration with NPs excretion from cells | 263 |
| CuO | 20–40 nm | A549 | Effect of exposure time, NPs concentration, and intracellular distribution | Atomic absorption spectrometry (AAS) | The proportion of CuO NPs that were expelled from A549 was enhanced by increasing the exposure time and NPS concentration. A portion of NPs located in mitochondria and nucleus could not be excreted. | 205 |
| Maltodextrin | 60 nm | HBE | Effect of cholesterol depletion | Confocal microscopy | The exocytosis of these NPs stopped after application of filipin (an agent that extracts membranous cholesterol). | 264 |
Human lung carcinoma (H1299), human esophageal epithelial (NE083) and human bronchial epithelial (NL20) cells, human umbilical vein endothelial cells (HUVECs) and human fibroblast epithelial cells (HeLa), adenocarcinomic human alveolar basal epithelial cells (A549), breast cancer cell lines (MDA-MB231 and MCF-7), the melanoma cancer cell line (MDA-MB435), the pancreatic cancer cell line (PANC-1), and the human embryonic stem cell line (H9), hepatocellular cell line (HepG2), human glioblastoma (SNB 19) and mouse embryonic fibroblast (STO), human bronchial epithelial cells (16-HBE), mesenchymal stem cells(MSC), human colonic adenocarcinoma cell lines (HT-29), pre-adipocytes cells (3T3-L1), murine macrophages cell line (J774), mouse embryonic fibroblast cell line (NIH-3T3), human vascular smooth muscle cells (VSMCs), human ovarian cancer cell lines (OVCAR-3), human glioblastoma cells (U251)
While CLSM dramatically improved both lateral (xy) and axial (z) resolution, compared to epifluorescence methods, recently developed super-resolution techniques have further improved our ability to discriminate NPs trafficking with superior spatial and temporal resolution206 - in particular, stochastic super-resolution methods such as photo-activated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM). Both methods reconstruct the location of a fluorescent point source by discriminating emitters based on molecule-specific intensity fluctuations; the former based on photobleaching and the latter based on on/off state switching. Fig. 10a illustrates lateral resolution improvements obtained from STORM imaging of polystyrene NPs internalized within HeLa cervical carcinoma cells.207 Unlike confocal and conventional widefield methods, STORM can differentiate NPs of varying size (300 v 80 nm diameter) and functionality (color), providing previously unattainable information regarding aggregation state and location, as well as the capability to perform multiplex analyses in high-throughput assay formats.
Fig. 10. Probing Cellular Interactions of Nanoparticles.
a) Super-resolution fluorescence microscopy (STORM) of the trafficking of 80-nm polystyrene nanoparticles (red) in HeLa cervical carcinoma cells (plasma membrane, green) compared to conventional wide-field microscopic techniques. b) Transmission electron microscopy (TEM) images of carbon nanotubes (CNTs, left) interacting with lung alveolar cells (right) following intratracheal administration in C57BL/6 mice. c) Atomic force microscopy (AFM) imaging of tattoo ink nanoparticles in cryosectioned human skin. Large (black arrows) and small (white arrows) agglomerates, as well as the underlying collagen fibril network, are visible in AFM height (left) and amplitude (right) images. d) Focused ion beam scanning electron microscopy (FIB-SEM) of HIV-mimetic 80-nm Au NPs infecting cells expressing the glycosphingolipid receptor CD169. Cell surface-bound particles are diffusely spread, while intracellular nanoparticles appear sequestered through as-yet-undetermined mechanisms. e) Real-time, live-cell Raman scattering images of murine macrophages with internalized 50-nm gold nanoparticle probes. Surface-enhance Raman scattering (SERS) from individual nanoparticles is detected (left) and spectral features from nanoparticle region-of-interest (right) are reported. f) Flow cytometry of PPC-1 prostate cancer cells treated with fluorescent, peptide-targeted, and etchable silver nanoparticles. Both cell fluorescence and side-scattering increase with peptide targeting (R-) of silver nanoparticles, while both decrease in response to the etching of cell surface-bound particulates. g) Real-time, dark-field scattering microscopy of respiratory syncytial virus (RSV) trafficking and infection in larynx epidermal cells as imaged using gold nanoparticle surface labels. h) Photoacoustic (PA) microscopy of human leukocytes. Composite images (532 nm, green; 600 nm, red) illustrating chromatin, nuclear, and cytoplasmic morphology. Although not used here, nanoparticles commonly serve as strong PA contrast agents. i) Laser-ablation inductively coupled plasma mass spectrometry (ICP-MS) imaging (heatmap) of mouse fibroblasts (grayscale) incubated with Au (left) and Ag (right) nanoparticles. Reproduced with permission from (a) 207, (b) 209, (c) 243, (d) 214, (e) 217, (f) 221, (g) 225, (h) 228, and (i) 234. Copyright (a) 2016 American Chemical Society, (b) 2014 Købler et al., (c) 2015 Grant et al., (d) 2014 Nature Publishing Group, (e) 2013 Nature Publishing Group, (f) 2014 Nature Publishing Group, (g) 2014 Nature Publishing Group, (h) 2016 Strohm et al., and (i) 2012 American Chemical Society.
7.2 Transmission electron microscopy
Transmission electron microscopy (TEM) discriminates objects based on their interactions with a collimated electron beam.208 Due to the small de Broglie wavelength of electrons, TEM provides resolutions superior to photon-based methods, often in the order of a single atom. However, while powerful, it also suffers from many drawbacks: long sample prep times, low multiplexing capabilities, and low contrast among biological structures. Because TEM relies fundamentally on transmission, biological samples must also be sectioned (50–100 nm thick) with an ultramicrotome. Challenges notwithstanding, the method can provide a wealth of information regarding size/location, electron density, atomic periodicity, and elemental composition.206 Købler et al., for example, used TEM to investigate the possible biological interactions of carbon nanotubes (CNTs) ingested via inhalation (Fig. 10b).209 After intra-tracheally administering CNT solutions to C57BL/6 mice, the authors observed significant cell surface labeling and intracellular accumulation within alveolar cells and tissue-resident macrophages. Such insights are important to understanding the possible long-term consequences of CNT inhalation, where persistent inflammation has been shown to lead to cancer pathogenesis following exposure to other materials such as asbestos. Gold NPs have been routinely imaged by TEM, both as endocytosed probes coupled to various proteins and for immunogold labelling of cells and tissues. This method has been applied to studying leukocyte biology, and it has helped to demonstrate how activated leukocytes deliver specific cargos.210, 211
7.3 Atomic force microscopy
Atomic force microscopy (AFM) provides lateral resolution comparable to electron-based methods and uses a piezoelectric-driven scanning cantilever to measure interactions between a scanning probe tip (ca. 10 nm radius) and a sample, yielding topographical and mechanical information at high resolution.212 Unlike EM methods, which typically require vacuum conditions, AFM can be performed in the aqueous state, enabling maintenance of native hydration and biological architecture. While versatile, this technique often suffers from poor discrimination, and methods for labeling/multiplexing are not currently in widespread use. Fig. 10c demonstrates the unique structural information that AFM can provide. Here, tattoo ink NPs are directly imaged from cryosectioned human skin. Ink NPs and agglomerates were shown to be distributed throughout the dermis and surrounding collagen matrix, demonstrating not only the spatial distribution of these NPs but also the biomechanical properties of their surrounding environment.
7.4 Scanning electron microscopy
Scanning electron microscopy (SEM) maps the interactions of an electron beam with a sample based on the detection of secondary electrons emitted from its surface or backscattered electrons from its surface/subsurface.213 SEM provides lateral resolution on the order of 1.0 nm and can also provide elemental information useful in labeling experiments. Because transmission is not required as with TEM, SEM can image tissues of millimeter-scale thickness. By integrating focused ion beam (FIB) surface milling, biological tissues can also be imaged in serial sections, providing precise tomographic information. Yu et al., for example, used FIB-SEM to study the CD169 receptor-mediated uptake of HIV virus-like particles (VLPs, Fig. 10d).214 Because the cytoplasmic tail of CD169 lacks any known endocytic motifs, receptor-mediated HIV-1 uptake and subsequent trafficking were not fully clear. FIB-SEM imaging showed that cell surface-bound particles were diffusely spread, while intracellular NPs appeared sequestered in vesicles, suggesting the presence of a membrane-associated co-factor that promotes uptake/trafficking.
7.5 Light-scattering microscopy
Elastic and inelastic light scattering microscopy can provide considerable information regarding the local chemical environment within a biological sample.215, 216 The former can discriminate spectral absorption/scattering as well as orientation (e.g., collagen fibril orientation in polarized light microscopy), while the latter is particularly unique in offering information regarding solid-state and molecular vibrations. Palonpon et al. showed that molecular vibrations can be mapped in real time using live-cell Raman microscopy (Fig. 10e).217 Using 50-nm Au NPs as surface-enhanced Raman scattering218 (SERS) probes, individual NPs could be tracked in murine macrophages. The authors demonstrated the detection of exogenous (alkyne) DNA labels and were able to track the NPs’ trajectory and the surrounding chemical environment with confocal lateral and axial resolution.
7.6 Flow cytometry
Flow cytometry measures multicolor optical scattering and fluorescence (or impedance) from individual cells in suspension by focusing them through a fluidic channel in which cells are allowed to flow only in ‘single file’.219 Segmentation and electrostatic deflection of the cell-containing fluid column (i.e., flow-automated cell sorting, FACS), can also be performed in real time to physically sort cells based on their optical or electrical characteristics. Flow cytometry (analysis and sorting) can be performed with both live and fixed cells and routinely accommodates 10-color multiplexing (with spectral compensation) and measurement rates of thousands of cells per second. Statistically robust staining intensity values and distributions can be determined from heterogeneous cell populations in high throughput,220 while spatial distribution patterns and interaction partners can also be assessed using Förster resonance energy transfer (FRET) techniques. Ruoslahti and coworkers recently utilized flow cytometry to differentiate cell surface-association and internalization of ligand-targeted silver NPs using a cell membrane-impermeable chemical etchant to ‘de-stain’ surface-bound particles (Fig. 10f).221 The authors found that ~60% of peptide-targeted particles were internalized at 1 h in vitro, while comparable concentrations of untargeted NPs were only ~1% internalized. This technique was also capable of identifying tumor-specific intracellular NP delivery in vivo.
7.7 Dark-field microscopy
Dark-field illumination enables imaging using scattered rather than transmitted light.222 This method is particularly useful in imaging translucent objects whose refractive indices are similar to that of their surroundings (e.g., cells) and uses a beam stop, commonly a ‘spider stop’, to block the center of a conical light beam that impinges upon the sample at a low angle. Thus, only highly scattering objects are visible on a black background. In many cases, this technique can be more sensitive than fluorescence microscopy and is commonly used to track plasmonic NPs and probes thereof, in live cells.223, 224 Fig. 10g demonstrates the utility of this technique in monitoring respiratory syncytial virus (RSV) infection in larynx epidermal cells using virus surface-labeled Au NPs.225 The authors tracked RSV cell-entry kinetics and demonstrated that 13-nm Au NPs probes can serve as non-interfering, non-photobleaching dark-field imaging probes.
7.8 Photoacoustic microscopy
Photoacoustic (PA) imaging uses laser-induced thermoelastic expansion to map cells and tissues based on variations in subsequent ultrasound emission.226 The method typically employs non-ionizing laser pulses that convert light energy into sound following light absorption, heat generation, and local volume expansion of an object or contrast agent. The technique is most commonly used to macroscopically image tissues and vessels, but has more recently been employed to detect circulating tumor cells.227 Strohm recently demonstrated that PA imaging can also be used to image individual unstained cells at high resolution (Fig. 10h).228 The authors used two-color excitation to perform PA imaging on lymphocytes with micrometer-scale resolution. Because inorganic nanoscale particles are often efficient photothermal contrast agents229, 230 (e.g., CNTs, Au NP, etc), PA imaging is an emerging analytical method that efficiently differentiates probe-labeled cells and interrogates the trafficking of nanoscale materials across both microscopic- and macroscopic-length scales.
7.9 Surface-Enhanced Raman Scattering (SERS)
NPs of noble-metal such as gold, silver, and platinum can serve as excellent probes for Raman sensing (Fig. 10e). Raman tags using these NPs enhance signals by ~12 orders of magnitude, with higher stability of the tags (compared to fluorescent dyes) and high signal-to-noise ratio in biological settings.231 One research group investigated the use of Au NPs to probe the cellular ultra-structure and various sub-cellular environments in terms of spectral signature and signal strength.216
Another study used SERS tags to image the distribution of epidermal growth factor receptor (EGFR) by creating an intensity map. Malachite green signals were enhanced by spherical Au NPs 30–40 nm in size and functionalized with antibodies to EGFR; maps created were consistent with the expected distribution of Au-NPs and EGFR.232
7.10 Laser Ablation ICP-MS
Inductively coupled plasma–mass spectroscopy approaches are utilized to detect trace concentrations of metals as low as one part in a trillion. The sample is introduced by nebulization and converted to plasma at very high temperature. When combined with laser ablation, this approach can attain an even lower detection limit with quantitative and spatial distribution of silver NPs. LA-ICP-MS enabled the investigation of silver NP uptake and localization within a cell, as in Fig. 10f.233,234
7.11 Correlative microscopy
Correlative microscopy is the integration of two or more microscopy techniques to analyze the same sample (Fig. 11).235 The aim of correlative microscopy is to obtain more in-depth information about the sample using the advantages of individual techniques including light microscopy, electron microscopy, atomic force microscopy, and super-resolution microscopy. For example, super-resolution microscopy in a correlative microscopy can overcome the conventional limitation of the light microscopy, which is poor resolution below 300 nm. Therefore, the correlative microscopy may create exciting new opportunities in the field of nanomedicine, and more specifically in intracellular trafficking of NPs. More in-depth information on the correlative microscopy and its challenges and opportunities are available in our very recent review paper.235
Fig. 11.
Scheme showing the use of most important techniques in the correlative microscopy. The image is reproduced with permission from American Chemical Society. 235
7.12 X-ray adsorption near-edge spectroscopy (XANES)
This approach is employed to study the “chemical fate” of nanoparticles in biological fluids/media and also inside cells.236,237,238Chemical fate of nanoparticles is related herein to chemical dissolution, transformation, and speciation. For example, a recent study utilized XANES to show that silver nanoparticles (20 nm) were gradually transformed from nanoparticles into ionic silver (Ag+, Ag–O– and Ag–S– forms) inside cultured macrophages.239 This transformation mediated the observed toxicity of the nanoparticles. In another study, XANES was utilized to confirm the dissolution of 60 nm AgNPs into Ag+ inside primary murine macrophages, forming complexes with the thiolate group in the glutathione ligand.240 In a recent study, it was shown that XANES spectroscopy is capable of following the Zn speciation profiles of zinc oxide (ZnO) and copper oxide (CuO) nanoparticles. The authors showed that both types of nanoparticles are rapidly transformed/dissolved during a standard in vitro toxicological assay to soluble forms that are sequestered intracellularly.241 Rapid chemical transformation and degradation in an acidic lysosomal environment of Cu3BiS3 nanodots was characterized using XANES, confirming their excellent metabolism and rapid clearance, which is a prerequisite for clinical applications.242 Trends relevant to the use of nanomaterials in cellular trafficking research, organized by country and research funding source over the past decade, are provided in Fig. 12.
Fig. 12. Trends in nanomaterial cellular trafficking research over the past decade.
Web of Science query terms: (ts=(nanoparticle* OR nanomaterial*) AND ts=(uptake* OR endocyt* OR traffic* OR entry* OR corona*)) AND DOCUMENT TYPES: (Article)
8 Exocytosis of NPs
Exocytosis is involved in vital biological processes such as organizing membrane proteins (e.g., transporters, ion channels, and receptors) and lipids, excretion of essential molecules,244 and repairing the CM.245 The important pathways by which NPs are excreted from cells are fully described in our previous review paper.246 Though the NP’s journey inside the cell ends in exocytosis, the story is not finished, as the composition of the protein corona can undergo significant changes as a result,247 and changes in the protein corona may alter targeting outcomes. After exocytosis, NPs have been shown to acquire the ability to cross critical in vivo barriers, such as the blood-brain barrier, and cause unexpected cytotoxicity.248 Exosomes carrying inhaled nanoparticles out of alveolar cells and disseminating them into the systemic circulation have also been shown to induce systemic immune responses and subsequent inflammation; this further emphasizes the role of nanoparticles’ exocytosis in their potential toxicity in the human body.249
Improving our understanding of the exocytosis mechanisms involved in the excretion of NPs and the factors affecting them could help reduce toxicity. NP features such as size, shape, and surface modifications influence exocytosis along with cell type, NP concentration, incubation time, and organelle distribution. The effect of the above-mentioned factors and detailed associated information is summarized in Table 2. More details of the exocytosis of NPs can be found in our previous review paper.246
9 Artifacts and Crucial “ignored” parameters in Cellular Uptake Evaluation
Artifacts in experiments or result interpretation can yield incorrect and sometimes misleading conclusions.23, 126 For instance, in terms of nano-toxicological studies, few publications have tackled the toxicity of the NPs themselves versus the supernatant solution. For a preparation of gold nanorods, it was demonstrated that toxicity originated from agents in the supernatant solution (namely cetyltrimethylammonium bromide) CTAB rather than the NPs themselves.23
Artifacts are also expected in cellular uptake studies. In this regard, the stability of nanomaterials upon introduction to biological media in uptake studies or toxicological evaluation is a critical parameter in the early stages of experimental design. For example, NPs that are stable in their original solution may aggregate extensively upon exposure to biological medium, due to the presence of aggregation-inducing molecules/species (such as salt), which may significantly alter the uptake extent, rate, and mechanism in addition to the observed toxicity of NPs.23
Moreover, in the case of metallic NPs, inductively coupled plasma mass spectroscopy is a very popular technique for quantification of NP accumulation or uptake (with sensitivity down to the parts-per billion level) and was successfully used for the quantification of metal NPs per number of cells. Nevertheless, this technique cannot differentiate between NPs actually internalized and those attached to the surface. Xia et al.121 proposed a technique to distinguish between these two groups of NPs, relying on a selective etchant based on I2/KI to selectively dissolve the Au NPs on the surface of cells. Combined with ICP analysis, this technique has the potential to improve our understanding of the cellular uptake of Au NPs.
Other researchers investigated cellular uptake at two different temperatures: 4 °C and 37 °C. At lower temperatures, energy-dependent uptake and passive diffusion are blocked due to membrane rigidity.265 However, such comparisons can be used to draw inferences regarding the amount of particles adsorbed (at low temperature) on cells versus those internalized at physiological temperature.266
Another interesting study differentiated between cellular uptake resulting from the diffusion of particles and their sedimentation. In a typical in vitro study, cells are immobilized on the bottom of culture plates, and nanoparticles are dosed into the growth medium. The injected nanoparticles are assumed to diffuse by Brownian movement and interact with cells before cellular uptake/toxicity evaluation. However, aggregation of nanoparticles may be underestimated, and thus Brownian movement could be masked by the sedimentation effect, with the larger and heavier particles sedimenting first, possibly leading to the variability of the dose available on the cell surface. Thus, differential uptake may be related to variation in colloidal stability in biological media rather than differences in size, shape, or surface charge. With this in mind, careful consideration of NPs’ colloidal stability in biological fluids used in uptake/toxicity studies and their tendency regarding sedimentation is essential. 104
10 Conclusions and future perspectives
Although our knowledge regarding the cellular association and trafficking of nanoscale materials has advanced tremendously in the past several years, much progress remains to be realized in our ability to both understand and exploit context-dependent phenomena in various disease states and their pathogenesis. Additional work in the field will likely expand to both smaller and larger scales, at both sub-cellular and tissue levels. Recent advances in organ-on-a-chip technologies are expected to give rise to an improved understanding of macroscopic nano-bio interactions – for example, re-examination of the EPR effect – and provide insights into tissue-level phenomena such as transcytosis, exosomal transport, and synaptic cell signaling modulation. New methods of directing nano-bio interactions at subcellular compartments – for example, mitochondrial or epigenetic targeting – will likely expand our ability to control cell fate, while emerging methods of sub-cellular analysis such as super-resolution microscopy, cryo-electron microscopy, high content analytics, and imaging cytometry will provide new insights with which disease can be modulated or reverted using nanoscale technologies. As advances in chemical synthesis continue to yield new structures with precisely defined biochemical features, and as emerging analytical techniques continue to shed new light on nuanced and context-dependent nano-bio interactions, our ability to exploit and treat human diseases using synthetic nanomaterials will continue to grow as rapidly as the field has expanded over the past several years.
Supplementary Material
Acknowledgments
This work was supported by the grants US National Institutes of Health (NIH) CA151884 (O.C.F.) and EB015419 (O.C.F.).
Footnotes
Competing interests statement:
O.C.F. declares financial interests in Selecta Biosciences, Tarveda Therapeutics and Placon Therapeutics.
Notes and references
- 1.Shi Y, Massagué J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 2.McMahon HT, Gallop JL. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Gluck S, Hartwig J. J Cell Biol. 1987;105:1637–1648. doi: 10.1083/jcb.105.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edidin M. Nat Rev Mol Cell Biol. 2003;4:414–418. doi: 10.1038/nrm1102. [DOI] [PubMed] [Google Scholar]
- 5.Sabolić I, Katsura T, Verbavatz J-M, Brown D. J Membr Biol. 1995;143:165–175. doi: 10.1007/BF00233445. [DOI] [PubMed] [Google Scholar]
- 6.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Nat Rev Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 8.Chou LY, Ming K, Chan WC. Chem Soc Rev. 2011;40:233–245. doi: 10.1039/c0cs00003e. [DOI] [PubMed] [Google Scholar]
- 9.Pridgen EM, Alexis F, Farokhzad OC. Expert Opinion on Drug Delivery. 2015;12:1459–1473. doi: 10.1517/17425247.2015.1018175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC. J Control Release. 2014;190:485–499. doi: 10.1016/j.jconrel.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walkey CD, Chan WCW. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 12.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J Am Chem Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 13.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X-Q, Xu X, Bertrand N, Pridgen E, Swami A, Farokhzad OC. Adv Drug Deliv Rev. 2012;64:1363–1384. doi: 10.1016/j.addr.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmoudi M, Sant S, Wang B, Laurent S, Sen T. Adv Drug Deliv Rev. 2011;63:24–46. doi: 10.1016/j.addr.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Sharifi S, Behzadi S, Laurent S, Forrest ML, Stroeve P, Mahmoudi M. Chem Soc Rev. 2012;41:2323–2343. doi: 10.1039/c1cs15188f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahmoudi M, Tachibana A, Goldstone AB, Woo YJ, Chakraborty P, Lee KR, Foote CS, Piecewicz S, Barrozo JC, Wakeel A. Sci Rep. 2016;6:26960. doi: 10.1038/srep26960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi M, Lohse SE, Murphy CJ, Fathizadeh A, Montazeri A, Suslick KS. Nano Lett. 2014;14:6–12. doi: 10.1021/nl403419e. [DOI] [PubMed] [Google Scholar]
- 19.Mahmoudi M, Abdelmonem AM, Behzadi S, Clement JH, Dutz S, Ejtehadi MR, Hartmann R, Kantner K, Linne U, Maffre P, Metzler S, Moghadam MK, Pfeiffer C, Rezaei M, Ruiz-Lozano P, Serpooshan V, Shokrgozar MA, Nienhaus GU, Parak WJ. ACS Nano. 2013;7:6555–6562. doi: 10.1021/nn305337c. [DOI] [PubMed] [Google Scholar]
- 20.Behzadi S, Serpooshan V, Sakhtianchi R, Müller B, Landfester K, Crespy D, Mahmoudi M. Colloids Surf B Biointerfaces. 2014;123:143–149. doi: 10.1016/j.colsurfb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Hajipour MJ, Raheb J, Akhavan O, Arjmand S, Mashinchian O, Rahman M, Abdolahad M, Serpooshan V, Laurent S, Mahmoudi M. Nanoscale. 2015;7:8978–8994. doi: 10.1039/c5nr00520e. [DOI] [PubMed] [Google Scholar]
- 22.Hajipour MJ, Laurent S, Aghaie A, Rezaee F, Mahmoudi M. Biomater Sci. 2014;2:1210–1221. doi: 10.1039/c4bm00131a. [DOI] [PubMed] [Google Scholar]
- 23.Alkilany AM, Mahmoud NN, Hashemi F, Hajipour MJ, Farvadi F, Mahmoudi M. Chem Res Toxicol. 2016 doi: 10.1021/acs.chemrestox.6b00108. [DOI] [PubMed] [Google Scholar]
- 24.Blanco E, Shen H, Ferrari M. Nat Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y-Y, Mao C-Q, Du X-J, Du J-Z, Wang F, Wang J. Adv Mater. 2012;24:5476–5480. doi: 10.1002/adma.201202296. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Wu J, Shan W, Tao W, Zhao L, Lim J-M, D’Ortenzio M, Karnik R, Huang Y, Shi J, Farokhzad OC. Angew Chem, Int Ed. 2016;55:3309–3312. doi: 10.1002/anie.201509183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doherty GJ, McMahon HT. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 29.Kumari S, Mg S, Mayor S. Cell Res. 2010;20:256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahay G, Alakhova DY, Kabanov AV. J Control Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson JA. Nat Rev Mol Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aderem A, Underhill DM. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 33.Hillaireau H, Couvreur P. Cell Mol Life Sci. 2009;66:2873–2896. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill DM, Ozinsky A. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer V, von Briesen H, Andreesen R, Steffan A-M, Royer C, Tröster S, Kreuter J, Rübsamen-Waigmann H. Pharm Res. 1992;9:541–546. doi: 10.1023/a:1015852732512. [DOI] [PubMed] [Google Scholar]
- 36.Tabata Y, Ikada Y. Biomaterials. 1988;9:356–362. doi: 10.1016/0142-9612(88)90033-6. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Ibricevic A, Cohen JA, Cohen JL, Gunsten SP, Fréchet JM, Walter MJ, Welch MJ, Brody SL. Mol Pharm. 2009;6:1891–1902. doi: 10.1021/mp900215p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Champion JA, Mitragotri S. Pharm Res. 2009;26:244–249. doi: 10.1007/s11095-008-9626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 40.Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owens DE, Peppas NA. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Couvreur P, Vauthier C. Pharm Res. 2006;23:1417–1450. doi: 10.1007/s11095-006-0284-8. [DOI] [PubMed] [Google Scholar]
- 43.Barenholz YC. J Control Release. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 44.de Verdià A. Br J Cancer. 1997;76:198. doi: 10.1038/bjc.1997.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couvreur P, Kante B, Grislain L, Roland M, Speiser P. J Pharm Sci. 1982;71:790–792. doi: 10.1002/jps.2600710717. [DOI] [PubMed] [Google Scholar]
- 46.Kattan J, Droz J-P, Couvreur P, Marino J-P, Boutan-Laroze A, Rougier P, Brault P, Vranckx H, Grognet J-M, Morge X. Invest New Drugs. 1992;10:191–199. doi: 10.1007/BF00877245. [DOI] [PubMed] [Google Scholar]
- 47.Torrado J, Espada R, Ballesteros M, Torrado-Santiago S. J Pharm Sci. 2008;97:2405–2425. doi: 10.1002/jps.21179. [DOI] [PubMed] [Google Scholar]
- 48.Moghimi SM, Hunter AC, Murray JC. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 49.Conner SD, Schmid SL. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 50.Brown CM, Petersen NO. Biochem Cell Biol. 1999;77:439–448. [PubMed] [Google Scholar]
- 51.Ungewickell E, Branton D. Nature. 1981;289:420–422. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- 52.Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirst J, Robinson MS. Biochim Biophys Acta (BBA) - Molecular Cell Research. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 54.Praefcke GJK, McMahon HT. Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 55.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 56.Harush-Frenkel O, Debotton N, Benita S, Altschuler Y. Biochem Biophys Res Commun. 2007;353:26–32. doi: 10.1016/j.bbrc.2006.11.135. [DOI] [PubMed] [Google Scholar]
- 57.Nan A, Bai X, Son SJ, Lee SB, Ghandehari H. Nano Lett. 2008;8:2150–2154. doi: 10.1021/nl0802741. [DOI] [PubMed] [Google Scholar]
- 58.Pelkmans L, Helenius A. Traffic. 2002;3:311–320. doi: 10.1034/j.1600-0854.2002.30501.x. [DOI] [PubMed] [Google Scholar]
- 59.Rothberg KG, Heuser JE, Donzell WC, Ying Y-S, Glenney JR, Anderson RGW. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 60.Anderson RGW. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 61.Kurzchalia TV, Partan RG. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 62.Raucher D, Sheetz MP. Biophys J. 1999;77:1992–2002. doi: 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sens P, Turner MS. Phys Rev E. 2006;73:031918. doi: 10.1103/PhysRevE.73.031918. [DOI] [PubMed] [Google Scholar]
- 64.Anderson HA, Chen Y, Norkin LC. Mol Biol Cell. 1996;7:1825–1834. doi: 10.1091/mbc.7.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mora R, Bonilha VL, Marmorstein A, Scherer PE, Brown D, Lisanti MP, Rodriguez-Boulan E. J Biol Chem. 1999;274:25708–25717. doi: 10.1074/jbc.274.36.25708. [DOI] [PubMed] [Google Scholar]
- 66.Gabella G, Blundell D. Cell Tissue Res. 1978;190:255–271. doi: 10.1007/BF00218174. [DOI] [PubMed] [Google Scholar]
- 67.Taggart MJ. Physiology. 2001;16:61–65. doi: 10.1152/physiologyonline.2001.16.2.61. [DOI] [PubMed] [Google Scholar]
- 68.Nabi IR. Nat Cell Biol. 2009;11:789–791. doi: 10.1038/ncb0709-789. [DOI] [PubMed] [Google Scholar]
- 69.Schnitzer JE, Liu J, Oh P. J Biol Chem. 1995;270:14399–14404. doi: 10.1074/jbc.270.24.14399. [DOI] [PubMed] [Google Scholar]
- 70.Carver LA, Schnitzer JE. Nat Rev Cancer. 2003;3:571–581. doi: 10.1038/nrc1146. [DOI] [PubMed] [Google Scholar]
- 71.Rejman J, Conese M, Hoekstra D. J Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- 72.Karimi M, Ghasemi A, Zangabad PS, Rahighi R, Basri SMM, Mirshekari H, Amiri M, Pishabad ZS, Aslani A, Bozorgomid M. Chem Soc Rev. 2016;45:1457–1501. doi: 10.1039/c5cs00798d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sahay G, Kim JO, Kabanov AV, Bronich TK. Biomaterials. 2010;31:923–933. doi: 10.1016/j.biomaterials.2009.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirkham M, Fujita A, Chadda R, Nixon SJ, Kurzchalia TV, Sharma DK, Pagano RE, Hancock JF, Mayor S, Parton RG. J Cell Biol. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damm E-M, Pelkmans L, Kartenbeck J, Mezzacasa A, Kurzchalia T, Helenius A. J Cell Biol. 2005;168:477–488. doi: 10.1083/jcb.200407113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doherty GJ, McMahon HT. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 77.Lu Y, Low PS. Adv Drug Deliv Rev. 2012;64:342–352. [Google Scholar]
- 78.Kelemen LE. Int J Cancer. 2006;119:243–250. doi: 10.1002/ijc.21712. [DOI] [PubMed] [Google Scholar]
- 79.Lim JP, Gleeson PA. Immunol Cell Biol. 2011;89:836–843. doi: 10.1038/icb.2011.20. [DOI] [PubMed] [Google Scholar]
- 80.Norbury CC, Hewlett LJ, Prescott AR, Shastri N, Watts C. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 81.Kuhn DA, Vanhecke D, Michen B, Blank F, Gehr P, Petri-Fink A, Rothen-Rutishauser B. Beilstein J Nanotechnol. 2014;5:1625–1636. doi: 10.3762/bjnano.5.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 83.Wang T, Bai J, Jiang X, Nienhaus GU. ACS Nano. 2012;6:1251–1259. doi: 10.1021/nn203892h. [DOI] [PubMed] [Google Scholar]
- 84.Geiser M, Rothen-Rutishauser B, Kapp N, Schürch S, Kreyling W, Schulz H, Semmler M, Hof VI, Heyder J, Gehr P. Environ Health Perspect. 2005:1555–1560. doi: 10.1289/ehp.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Banaszak Holl MM. Nano Lett. 2008;8:420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- 86.Chen J, Hessler JA, Putchakayala K, Panama BK, Khan DP, Hong S, Mullen DG, DiMaggio SC, Som A, Tew GN, Lopatin AN, Baker JR, Holl MMB, Orr BG. J Phys Chem B. 2009;113:11179–11185. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Candeloro P, Tirinato L, Malara N, Fregola A, Casals E, Puntes V, Perozziello G, Gentile F, Coluccio ML, Das G, Liberale C, De Angelis F, Di Fabrizio E. Analyst. 2011;136:4402–4408. doi: 10.1039/c1an15313g. [DOI] [PubMed] [Google Scholar]
- 88.Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, Payne JM, Miller TJ, Brunovskis P, Fink TL. J Biol Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- 89.Jen C-P, Chen Y-H, Fan C-S, Yeh C-S, Lin Y-C, Shieh D-B, Wu C-L, Chen D-H, Chou C-H. Langmuir. 2004;20:1369–1374. doi: 10.1021/la036154k. [DOI] [PubMed] [Google Scholar]
- 90.Zhao F, Zhao Y, Liu Y, Chang X, Chen C, Zhao Y. Small. 2011;7:1322–1337. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- 91.Zhu M, Nie G, Meng H, Xia T, Nel A, Zhao Y. Acc Chem Res. 2013;46:622–631. doi: 10.1021/ar300031y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johannes L, Mayor S. Cell. 2010;142:507–510. doi: 10.1016/j.cell.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Chaudhuri A, Battaglia G, Golestanian R. Phys Biol. 2011;8:046002. doi: 10.1088/1478-3975/8/4/046002. [DOI] [PubMed] [Google Scholar]
- 94.Smith KA, Jasnow D, Balazs AC. J Chem Phys. 2007;127:084703. doi: 10.1063/1.2766953. [DOI] [PubMed] [Google Scholar]
- 95.Le Bihan O, Bonnafous P, Marak L, Bickel T, Trépout S, Mornet S, De Haas F, Talbot H, Taveau J-C, Lambert O. J Struct Biol. 2009;168:419–425. doi: 10.1016/j.jsb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 96.de Planque MR, Aghdaei S, Roose T, Morgan H. ACS Nano. 2011;5:3599–3606. doi: 10.1021/nn103320j. [DOI] [PubMed] [Google Scholar]
- 97.Zhang S, Nelson A, Beales PA. Langmuir. 2012;28:12831–12837. doi: 10.1021/la301771b. [DOI] [PubMed] [Google Scholar]
- 98.Chithrani BD, Ghazani AA, Chan WC. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 99.Jiang W, Kim BY, Rutka JT, Chan WC. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 100.Shan Y, Ma S, Nie L, Shang X, Hao X, Tang Z, Wang H. Chem Commun. 2011;47:8091–8093. doi: 10.1039/c1cc11453k. [DOI] [PubMed] [Google Scholar]
- 101.Rausch K, Reuter A, Fischer K, Schmidt M. Biomacromolecules. 2010;11:2836–2839. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- 102.Maiorano G, Sabella S, Sorce B, Brunetti V, Malvindi MA, Cingolani R, Pompa PP. ACS Nano. 2010;4:7481–7491. doi: 10.1021/nn101557e. [DOI] [PubMed] [Google Scholar]
- 103.Albanese A, Chan WC. ACS Nano. 2011;5:5478–5489. doi: 10.1021/nn2007496. [DOI] [PubMed] [Google Scholar]
- 104.Cho EC, Zhang Q, Xia Y. Nat Nanotechnol. 2011;6:385–391. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Williams Y, Sukhanova A, Nowostawska M, Davies AM, Mitchell S, Oleinikov V, Gun’ko Y, Nabiev I, Kelleher D, Volkov Y. Small. 2009;5:2581–2588. doi: 10.1002/smll.200900744. [DOI] [PubMed] [Google Scholar]
- 106.Nabiev I, Mitchell S, Davies A, Williams Y, Kelleher D, Moore R, Gun’ko YK, Byrne S, Rakovich YP, Donegan JF, Sukhanova A, Conroy J, Cottell D, Gaponik N, Rogach A, Volkov Y. Nano Lett. 2007;7:3452–3461. doi: 10.1021/nl0719832. [DOI] [PubMed] [Google Scholar]
- 107.Shapero K, Fenaroli F, Lynch I, Cottell DC, Salvati A, Dawson KA. Mol Biosyst. 2011;7:371–378. doi: 10.1039/c0mb00109k. [DOI] [PubMed] [Google Scholar]
- 108.Chithrani BD, Chan WC. Nano Lett. 2007;7:1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 109.Qiu Y, Liu Y, Wang L, Xu L, Bai R, Ji Y, Wu X, Zhao Y, Li Y, Chen C. Biomaterials. 2010;31:7606–7619. doi: 10.1016/j.biomaterials.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 110.Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc Natl Acad Sci U S A. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dasgupta S, Auth T, Gompper G. Nano Lett. 2014;14:687–693. doi: 10.1021/nl403949h. [DOI] [PubMed] [Google Scholar]
- 112.Banerjee A, Qi J, Gogoi R, Wong J, Mitragotri S. J Control Release. 2016;238:176–185. doi: 10.1016/j.jconrel.2016.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, Zhang H, Wang X, Lin S, Huang C. ACS Nano. 2011;5:4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herd H, Daum N, Jones AT, Huwer H, Ghandehari H, Lehr C-M. ACS Nano. 2013;7:1961–1973. doi: 10.1021/nn304439f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang K, Ma Y-Q. Nat Nanotechnol. 2010;5:579–583. doi: 10.1038/nnano.2010.141. [DOI] [PubMed] [Google Scholar]
- 116.Li Y, Gu N. J Phys Chem B. 2010;114:2749–2754. doi: 10.1021/jp904550b. [DOI] [PubMed] [Google Scholar]
- 117.Lin J, Zhang H, Chen Z, Zheng Y. ACS Nano. 2010;4:5421–5429. doi: 10.1021/nn1010792. [DOI] [PubMed] [Google Scholar]
- 118.Nangia S, Sureshkumar R. Langmuir. 2012;28:17666–17671. doi: 10.1021/la303449d. [DOI] [PubMed] [Google Scholar]
- 119.Li S, Malmstadt N. Soft Matter. 2013;9:4969–4976. [Google Scholar]
- 120.Wang B, Zhang L, Bae SC, Granick S. Proc Natl Acad Sci USA. 2008;105:18171–18175. doi: 10.1073/pnas.0807296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cho EC, Xie J, Wurm PA, Xia Y. Nano Lett. 2009;9:1080–1084. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- 122.Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, Rotello VM, Prakash Y, Mukherjee P. Nano Lett. 2010;10:2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hauck TS, Ghazani AA, Chan WC. Small. 2008;4:153–159. doi: 10.1002/smll.200700217. [DOI] [PubMed] [Google Scholar]
- 124.Hühn D, Kantner K, Geidel C, Brandholt S, De Cock I, Soenen SJH, Rivera_Gil P, Montenegro J-M, Braeckmans K, Müllen K, Nienhaus GU, Klapper M, Parak WJ. ACS Nano. 2013;7:3253–3263. doi: 10.1021/nn3059295. [DOI] [PubMed] [Google Scholar]
- 125.Zanganeh S, Spitler R, Erfanzadeh M, Alkilany AM, Mahmoudi M. Int J Biochem Cell Biol. 2016;75:143–147. doi: 10.1016/j.biocel.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Azhdarzadeh M, Saei AA, Sharifi S, Hajipour MJ, Alkilany AM, Sharifzadeh M, Ramazani F, Laurent S, Mashaghi A, Mahmoudi M. Nanomedicine. 2015;10:2931–2952. doi: 10.2217/nnm.15.130. [DOI] [PubMed] [Google Scholar]
- 127.Mirshafiee V, Kim R, Mahmoudi M, Kraft ML. Int J Biochem Cell Biol. 2015 doi: 10.1016/j.biocel.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 128.Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Acs Nano. 2014;8:5515–5526. doi: 10.1021/nn4061012. [DOI] [PubMed] [Google Scholar]
- 129.Rozenberg B, Tenne R. Prog Polym Sci. 2008;33:40–112. [Google Scholar]
- 130.Li Y, Chen X, Gu N. J Phys Chem B. 2008;112:16647–16653. doi: 10.1021/jp8051906. [DOI] [PubMed] [Google Scholar]
- 131.Wong-Ekkabut J, Baoukina S, Triampo W, Tang I-M, Tieleman DP, Monticelli L. Nat Nanotechnol. 2008;3:363–368. doi: 10.1038/nnano.2008.130. [DOI] [PubMed] [Google Scholar]
- 132.Alexeev A, Uspal WE, Balazs AC. ACS Nano. 2008;2:1117–1122. doi: 10.1021/nn8000998. [DOI] [PubMed] [Google Scholar]
- 133.Curtis EM, Bahrami AH, Weikl TR, Hall CK. Nanoscale. 2015;7:14505–14514. doi: 10.1039/c5nr02255j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abulateefeh SR, Spain SG, Thurecht KJ, Aylott JW, Chan WC, Garnett MC, Alexander C. Biomater Sci. 2013;1:434–442. doi: 10.1039/c2bm00184e. [DOI] [PubMed] [Google Scholar]
- 135.Gopalakrishnan G, Danelon C, Izewska P, Prummer M, Bolinger PY, Geissbühler I, Demurtas D, Dubochet J, Vogel H. Angew Chem Int Ed. 2006;45:5478–5483. doi: 10.1002/anie.200600545. [DOI] [PubMed] [Google Scholar]
- 136.Jing B, Zhu Y. J Am Chem Soc. 2011;133:10983–10989. doi: 10.1021/ja2040305. [DOI] [PubMed] [Google Scholar]
- 137.Olubummo A, Schulz M, Lechner B-D, Scholtysek P, Bacia K, Blume A, Kressler J, Binder WH. ACS Nano. 2012;6:8713–8727. doi: 10.1021/nn3023602. [DOI] [PubMed] [Google Scholar]
- 138.Lee H-Y, Shin SHR, Abezgauz LL, Lewis SA, Chirsan AM, Danino DD, Bishop KJM. J Am Chem Soc. 2013;135:5950–5953. doi: 10.1021/ja400225n. [DOI] [PubMed] [Google Scholar]
- 139.Ge C, Du J, Zhao L, Wang L, Liu Y, Li D, Yang Y, Zhou R, Zhao Y, Chai Z. Proc Natl Acad Sci USA. 2011;108:16968–16973. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Na K, Lee KH, Lee DH, Bae YH. Eur J Pharm Sci. 2006;27:115–122. doi: 10.1016/j.ejps.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 141.Mahmoudi M, Meng J, Xue X, Liang XJ, Rahman M, Pfeiffer C, Hartmann R, Gil PR, Pelaz B, Parak WJ, del Pino P, Carregal-Romero S, Kanaras AG, Tamil Selvan S. Biotechnol Adv. 2014;32:679–692. doi: 10.1016/j.biotechadv.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 142.Karakoti AS, Shukla R, Shanker R, Singh S. Adv Colloid Interface Sci. 2015;215:28–45. doi: 10.1016/j.cis.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 143.Yeh Y-C, Saha K, Yan B, Miranda OR, Yu X, Rotello VM. Nanoscale. 2013;5:12140–12143. doi: 10.1039/c3nr04037b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chompoosor A, Saha K, Ghosh PS, Macarthy DJ, Miranda OR, Zhu Z-J, Arcaro KF, Rotello VM. Small. 2010;6:2246–2249. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alexis F, Rhee J-W, Richie JP, Radovic-Moreno AF, Langer R, Farokhzad OC. Urol Oncol. 2008;26:74–85. doi: 10.1016/j.urolonc.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 146.Barreto JA, O’Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Adv Mater. 2011;23:H18–H40. doi: 10.1002/adma.201100140. [DOI] [PubMed] [Google Scholar]
- 147.Tao W, Zeng X, Wu J, Zhu X, Yu X, Zhang X, Zhang J, Liu G, Mei L. Theranostics. 2016;6:470. doi: 10.7150/thno.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tao W, Zhang J, Zeng X, Liu D, Liu G, Zhu X, Liu Y, Yu Q, Huang L, Mei L. Adv Healthc Mater. 2015;4:1203–1214. doi: 10.1002/adhm.201400751. [DOI] [PubMed] [Google Scholar]
- 149.Alexis F, Basto P, Levy-Nissenbaum E, Radovic-Moreno AF, Zhang L, Pridgen E, Wang AZ, Marein SL, Westerhof K, Molnar LK, Farokhzad OC. ChemMedChem. 2008;3:1839–1843. doi: 10.1002/cmdc.200800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xia T, Kovochich M, Liong M, Zink JI, Nel AE. ACS Nano. 2008;2:85–96. doi: 10.1021/nn700256c. [DOI] [PubMed] [Google Scholar]
- 151.dos Santos T, Varela J, Lynch I, Salvati A, Dawson KA. PLoS One. 2011;6:e24438. doi: 10.1371/journal.pone.0024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Parton RG, del Pozo MA. Nat Rev Mol Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 153.Parton RG, Simons K. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 154.Fujimoto T, Kogo H, Nomura R, Une T. J Cell Sci. 2000;113(Pt 19):3509–3517. doi: 10.1242/jcs.113.19.3509. [DOI] [PubMed] [Google Scholar]
- 155.Sottile J, Chandler J. Mol Biol Cell. 2005;16:757–768. doi: 10.1091/mbc.E04-08-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 157.Seto ES, Bellen HJ, Lloyd TE. Genes Dev. 2002;16:1314–1336. doi: 10.1101/gad.989602. [DOI] [PubMed] [Google Scholar]
- 158.Medina-Kauwe LK. Adv Drug Deliv Rev. 2007;59:798–809. doi: 10.1016/j.addr.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yousuf MA, Zhou X, Mukherjee S, Chintakuntlawar AV, Lee JY, Ramke M, Chodosh J, Rajaiya J. PLoS One. 2013;8:e77462. doi: 10.1371/journal.pone.0077462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nomura R. Uirusu. 2005;55:19–26. doi: 10.2222/jsv.55.19. [DOI] [PubMed] [Google Scholar]
- 161.Badizadegan K, Wolf AA, Rodighiero C, Jobling M, Hirst TR, Holmes RK, Lencer WI. Int J Med Microbiol. 2000;290:403–408. doi: 10.1016/S1438-4221(00)80052-1. [DOI] [PubMed] [Google Scholar]
- 162.Lencer WI, Hirst TR, Holmes RK. Biochim Biophys Acta. 1999;1450:177–190. doi: 10.1016/s0167-4889(99)00070-1. [DOI] [PubMed] [Google Scholar]
- 163.Pfeffer SR. Nat Cell Biol. 2001;3:E108–110. doi: 10.1038/35074625. [DOI] [PubMed] [Google Scholar]
- 164.Mercer J, Helenius A. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 165.Hewlett LJ, Prescott AR, Watts C. J Cell Biol. 1994;124:689–703. doi: 10.1083/jcb.124.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Swanson JA, Watts C. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 167.Falcone S, Cocucci E, Podini P, Kirchhausen T, Clementi E, Meldolesi J. J Cell Sci. 2006;119:4758–4769. doi: 10.1242/jcs.03238. [DOI] [PubMed] [Google Scholar]
- 168.Kasahara K, Nakayama Y, Sato I, Ikeda K, Hoshino M, Endo T, Yamaguchi N. J Cell Physiol. 2007;211:220–232. doi: 10.1002/jcp.20931. [DOI] [PubMed] [Google Scholar]
- 169.Cartiera MS, Johnson KM, Rajendran V, Caplan MJ, Saltzman WM. Biomaterials. 2009;30:2790–2798. doi: 10.1016/j.biomaterials.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huotari J, Helenius A. The EMBO journal. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jovic M, Sharma M, Rahajeng J, Caplan S. Histol Histopathol. 2010;25:99. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rauch J, Kolch W, Laurent S, Mahmoudi M. Chem Rev. 2013;113:3391–3406. doi: 10.1021/cr3002627. [DOI] [PubMed] [Google Scholar]
- 173.Grant BD, Donaldson JG. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Park M, Salgado JM, Ostroff L, Helton TD, Robinson CG, Harris KM, Ehlers MD. Neuron. 2006;52:817–830. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dominska M, Dykxhoorn DM. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 176.Martens TF, Remaut K, Demeester J, De Smedt SC, Braeckmans K. Nano Today. 2014;9:344–364. [Google Scholar]
- 177.Mizushima N. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 178.Ma X, Wu Y, Jin S, Tian Y, Zhang X, Zhao Y, Yu L, Liang X-J. ACS Nano. 2011;5:8629–8639. doi: 10.1021/nn202155y. [DOI] [PubMed] [Google Scholar]
- 179.Wang J, Yu Y, Lu K, Yang M, Li Y, Zhou X, Sun Z. Int J Nanomedicine. 2017;12:809. doi: 10.2147/IJN.S123596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mahmoudi M, Saeedi-Eslami SN, Shokrgozar MA, Azadmanesh K, Hassanlou M, Kalhor HR, Burtea C, Rothen-Rutishauser B, Laurent S, Sheibani S, Vali H. Nanoscale. 2012;4:5461–5468. doi: 10.1039/c2nr31185b. [DOI] [PubMed] [Google Scholar]
- 181.Laurent S, Burtea C, Thirifays C, Rezaee F, Mahmoudi M. J Colloid Interface Sci. 2013;392:431–445. doi: 10.1016/j.jcis.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 182.Schottler S, Klein K, Landfester K, Mailander V. Nanoscale. 2016;8:5526–5536. doi: 10.1039/c5nr08196c. [DOI] [PubMed] [Google Scholar]
- 183.Colapicchioni V, Tilio M, Digiacomo L, Gambini V, Palchetti S, Marchini C, Pozzi D, Occhipinti S, Amici A, Caracciolo G. Int J Biochem Cell Biol. 2016;75:180–187. doi: 10.1016/j.biocel.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 184.Wong HL, Bendayan R, Rauth AM, Xue HY, Babakhanian K, Wu XY. J Pharmacol Exp Ther. 2006;317:1372–1381. doi: 10.1124/jpet.106.101154. [DOI] [PubMed] [Google Scholar]
- 185.Li B, Xu H, Li Z, Yao M, Xie M, Shen H, Shen S, Wang X, Jin Y. Int J Nanomedicine. 2012;7:187–197. doi: 10.2147/IJN.S27864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fiorentino I, Gualtieri R, Barbato V, Mollo V, Braun S, Angrisani A, Turano M, Furia M, Netti PA, Guarnieri D, Fusco S, Talevi R. Exp Cell Res. 2015;330:240–247. doi: 10.1016/j.yexcr.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 187.Monti DM, Guarnieri D, Napolitano G, Piccoli R, Netti P, Fusco S, Arciello A. J Biotechnol. 2015;193:3–10. doi: 10.1016/j.jbiotec.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 188.Johnston HJ, Semmler-Behnke M, Brown DM, Kreyling W, Tran L, Stone V. Toxicol Appl Pharmacol. 2010;242:66–78. doi: 10.1016/j.taap.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 189.Douglas KL, Piccirillo CA, Tabrizian M. Eur J Pharm Biopharm. 2008;68:676–687. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 190.Xiao Y, Forry SP, Gao X, Holbrook RD, Telford WG, Tona A. J Nanobiotechnology. 2010;8:13. doi: 10.1186/1477-3155-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Duan H, Nie S. J Am Chem Soc. 2007;129:3333–3338. doi: 10.1021/ja068158s. [DOI] [PubMed] [Google Scholar]
- 192.Chu Z, Huang Y, Tao Q, Li Q. Nanoscale. 2011;3:3291–3299. doi: 10.1039/c1nr10499c. [DOI] [PubMed] [Google Scholar]
- 193.Lesniak A, Fenaroli F, Monopoli MP, Aberg C, Dawson KA, Salvati A. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 194.Hsiao I-Lun, Gramatke Annika Mareike, Joksimovic Rastko, Sokolowski Marek, Gradzielski M, Haase A. J Nanomed Nanotechnol. 2014:5. [Google Scholar]
- 195.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 196.Bhatia SN, Ingber DE. Nat Biotech. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 197.Hafner M, Niepel M, Chung M, Sorger PK. Nat Meth. 2016;13:521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Panarella A, Bexiga MG, Galea G, O’Neill ED, Salvati A, Dawson KA, Simpson JC. Sci Rep. 2016;6:28865. doi: 10.1038/srep28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dominguez AA, Lim WA, Qi LS. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lichtman JW, Conchello J-A. Nat Meth. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 202.Gilleron J, Paramasivam P, Zeigerer A, Querbes W, Marsico G, Andree C, Seifert S, Amaya P, Stöter M, Koteliansky V, Waldmann H, Fitzgerald K, Kalaidzidis Y, Akinc A, Maier MA, Manoharan M, Bickle M, Zerial M. Nucleic Acids Res. 2015;43:7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jin H, Heller DA, Sharma R, Strano MS. ACS Nano. 2009;3:149–158. doi: 10.1021/nn800532m. [DOI] [PubMed] [Google Scholar]
- 204.Panyam J, Labhasetwar V. Pharm Res. 2003;20:212–220. doi: 10.1023/a:1022219003551. [DOI] [PubMed] [Google Scholar]
- 205.Wang Z, Li N, Zhao J, White JC, Qu P, Xing B. Chem Res Toxicol. 2012;25:1512–1521. doi: 10.1021/tx3002093. [DOI] [PubMed] [Google Scholar]
- 206.Dreaden EC, Kong YW, Morton SW, Correa S, Choi KY, Shopsowitz KE, Renggli K, Drapkin R, Yaffe MB, Hammond PT. Clin Cancer Res. 2015;21:4410–4419. doi: 10.1158/1078-0432.CCR-15-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.van der Zwaag D, Vanparijs N, Wijnands S, De Rycke R, De Geest BG, Albertazzi L. ACS Appl Mater Interfaces. 2016;8:6391–6399. doi: 10.1021/acsami.6b00811. [DOI] [PubMed] [Google Scholar]
- 208.Kourkoutis LF, Plitzko JM, Baumeister W. Ann Rev Mater Res. 2012;42:33–58. [Google Scholar]
- 209.Købler C, Saber AT, Jacobsen NR, Wallin H, Vogel U, Qvortrup K, Mølhave K. Anal Bioanal Chem. 2014;406:3863–3873. doi: 10.1007/s00216-013-7566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Melo RC, Morgan E, Monahan-Earley R, Dvorak AM, Weller PF. Nat Protoc. 2014;9:2382–2394. doi: 10.1038/nprot.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wilson SM, Bacic A. Nat Protocols. 2012;7:1716–1727. doi: 10.1038/nprot.2012.096. [DOI] [PubMed] [Google Scholar]
- 212.Muller DJ, Dufrene YF. Nat Nano. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 213.Narayan K, Subramaniam S. Nat Meth. 2015;12:1021–1031. doi: 10.1038/nmeth.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yu X, Feizpour A, Ramirez N-GP, Wu L, Akiyama H, Xu F, Gummuluru S, Reinhard BM. Nat Commun. 2014;5:4136. doi: 10.1038/ncomms5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Boustany NN, Boppart SA, Backman V. Ann Rev Biomed Eng. 2010;12:285–314. doi: 10.1146/annurev-bioeng-061008-124811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kneipp J, Kneipp H, McLaughlin M, Brown D, Kneipp K. Nano Lett. 2006;6:2225–2231. doi: 10.1021/nl061517x. [DOI] [PubMed] [Google Scholar]
- 217.Palonpon AF, Ando J, Yamakoshi H, Dodo K, Sodeoka M, Kawata S, Fujita K. Nat Protocols. 2013;8:677–692. doi: 10.1038/nprot.2013.030. [DOI] [PubMed] [Google Scholar]
- 218.Dreaden EC, Near RD, Abdallah T, Talaat MH, El-Sayed MA. Appl Phys Lett. 2011;98:183115–183113. [Google Scholar]
- 219.Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Nat Immunol. 2014;15:128–135. doi: 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dreaden EC, Morton SW, Shopsowitz KE, Choi J-H, Deng ZJ, Cho N-J, Hammond PT. ACS Nano. 2014;8:8374–8382. doi: 10.1021/nn502861t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Braun GB, Friman T, Pang H-B, Pallaoro A, de Mendoza TH, Willmore A-MA, Kotamraju VR, Mann AP, She Z-G, Sugahara KN, Reich NO, Teesalu T, Ruoslahti E. Nat Mater. 2014;13:904–911. doi: 10.1038/nmat3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sönnichsen C, Alivisatos AP. Nano Lett. 2005;5:301–304. doi: 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- 223.Dreaden EC, Gryder BE, Austin LA, Tene Defo BA, Hayden SC, Pi M, Quarles LD, Oyelere AK, El-Sayed MA. Bioconjugate Chem. 2012;23:1507–1512. doi: 10.1021/bc300158k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dreaden EC, Mwakwari SC, Austin LA, Kieffer MJ, Oyelere AK, El-Sayed MA. Small. 2012;8:2819–2822. doi: 10.1002/smll.201200333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wan X-Y, Zheng L-L, Gao P-F, Yang X-X, Li C-M, Li YF, Huang CZ. Sci Rep. 2014;4:4529. doi: 10.1038/srep04529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zackrisson S, van de Ven SMWY, Gambhir SS. Cancer Res. 2014;74:979–1004. doi: 10.1158/0008-5472.CAN-13-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Galanzha EI, Shashkov EV, Kelly T, Kim J-W, Yang L, Zharov VP. Nat Nanotechnol. 2009;4:855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Strohm EM, Moore MJ, Kolios MC. Photoacoustics. 2016;4:36–42. doi: 10.1016/j.pacs.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Dreaden EC, Neretina S, Qian W, El-Sayed MA, Hughes RA, Preston JS, Mascher P. J Phys Chem C. 2011;115:5578–5583. [Google Scholar]
- 230.Weber J, Beard PC, Bohndiek SE. Nat Meth. 2016;13:639–650. doi: 10.1038/nmeth.3929. [DOI] [PubMed] [Google Scholar]
- 231.Raghavan V, Fan HM, Dockery P, Wheatley A, Keogh I, Olivo M. J Nanomed Nanotechnol. 2015:1. [Google Scholar]
- 232.Lucas L, Chen X, Smith A, Korbelik M, Zeng H, Lee P, Hewitt K. 2009 [Google Scholar]
- 233.Yu L, Andriola A. Talanta. 2010;82:869–875. doi: 10.1016/j.talanta.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 234.Drescher D, Giesen C, Traub H, Panne U, Kneipp J, Jakubowski N. Anal Chem. 2012;84:9684–9688. doi: 10.1021/ac302639c. [DOI] [PubMed] [Google Scholar]
- 235.Hauser M, Wojcik M, Kim D, Mahmoudi M, Li W, Xu K. Chem Rev. 2017 doi: 10.1021/acs.chemrev.6b00604. [DOI] [PubMed] [Google Scholar]
- 236.Chen C, Li Y-F, Qu Y, Chai Z, Zhao Y. Chem Soc Rev. 2013;42:8266–8303. doi: 10.1039/c3cs60111k. [DOI] [PubMed] [Google Scholar]
- 237.Wang B, Feng W, Zhao Y, Chai Z. Metallomics. 2013;5:793–803. doi: 10.1039/c3mt00093a. [DOI] [PubMed] [Google Scholar]
- 238.Wang B, Feng W, Chai Z, Zhao Y. Science China Chemistry. 2015;58:768–779. [Google Scholar]
- 239.Wang L, Zhang T, Li P, Huang W, Tang J, Wang P, Liu J, Yuan Q, Bai R, Li B, Zhang K, Zhao Y, Chen C. ACS Nano. 2015;9:6532–6547. doi: 10.1021/acsnano.5b02483. [DOI] [PubMed] [Google Scholar]
- 240.Veronesi G, Aude-Garcia C, Kieffer I, Gallon T, Delangle P, Herlin-Boime N, Rabilloud T, Carriere M. Nanoscale. 2015;7:7323–7330. doi: 10.1039/c5nr00353a. [DOI] [PubMed] [Google Scholar]
- 241.Ivask A, Scheckel KG, Kapruwan P, Stone V, Yin H, Voelcker NH, Lombi E. Nanotoxicology. 2017;11:150–156. doi: 10.1080/17435390.2017.1282049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu J, Wang P, Zhang X, Wang L, Wang D, Gu Z, Tang J, Guo M, Cao M, Zhou H, Liu Y, Chen C. ACS Nano. 2016;10:4587–4598. doi: 10.1021/acsnano.6b00745. [DOI] [PubMed] [Google Scholar]
- 243.Grant CA, Twigg PC, Baker R, Tobin DJ. Beilstein J Nanotechnol. 2015;6:1183–1191. doi: 10.3762/bjnano.6.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lodish HF. In: Molecular cell biology. 6. Lodish Harvey, et al., editors. W. H. Freeman; Basingstoke: 2008. [Google Scholar]
- 245.McNeil PL, Terasaki M. Nat Cell Biol. 2001;3:E124–E129. doi: 10.1038/35074652. [DOI] [PubMed] [Google Scholar]
- 246.Sakhtianchi R, Minchin RF, Lee K-B, Alkilany AM, Serpooshan V, Mahmoudi M. Adv Colloid Interface Sci. 2013;201–202:18–29. doi: 10.1016/j.cis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 247.Lundqvist M, Stigler J, Cedervall T, Berggård T, Flanagan MB, Lynch I, Elia G, Dawson K. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 248.Krol S, Macrez R, Docagne F, Defer G, Laurent S, Rahman M, Hajipour MJ, Kehoe PG, Mahmoudi M. Chem Rev. 2012;113:1877–1903. doi: 10.1021/cr200472g. [DOI] [PubMed] [Google Scholar]
- 249.Zhu M, Li Y, Shi J, Feng W, Nie G, Zhao Y. Small. 2012;8:404–412. doi: 10.1002/smll.201101708. [DOI] [PubMed] [Google Scholar]
- 250.Slowing II, Vivero-Escoto JL, Zhao Y, Kandel K, Peeraphatdit C, Trewyn BG, Lin VSY. Small. 2011;7:1526–1532. doi: 10.1002/smll.201002077. [DOI] [PubMed] [Google Scholar]
- 251.Yanes RE, Tarn D, Hwang AA, Ferris DP, Sherman SP, Thomas CR, Lu J, Pyle AD, Zink JI, Tamanoi F. Small. 2013;9:697–704. doi: 10.1002/smll.201201811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hu Ling, Mao Zhengwei, Zhang Yuying, Gao C. J Nanosci Lett. 2011;1:1–16. [Google Scholar]
- 253.Chithrani BD, Chan WCW. Nano Lett. 2007;7:1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- 254.Wang L, Liu Y, Li W, Jiang X, Ji Y, Wu X, Xu L, Qiu Y, Zhao K, Wei T, Li Y, Zhao Y, Chen C. Nano Lett. 2010;11:772–780. doi: 10.1021/nl103992v. [DOI] [PubMed] [Google Scholar]
- 255.Bartczak D, Sanchez-Elsner T, Louafi F, Millar TM, Kanaras AG. Small. 2011;7:388–394. doi: 10.1002/smll.201001816. [DOI] [PubMed] [Google Scholar]
- 256.Chen R, Huang G, Ke PC. Applied Physics Letters. 2010;97:093706. [Google Scholar]
- 257.Fang C-Y, Vaijayanthimala V, Cheng C-A, Yeh S-H, Chang C-F, Li C-L, Chang H-C. Small. 2011;7:3363–3370. doi: 10.1002/smll.201101233. [DOI] [PubMed] [Google Scholar]
- 258.Serda RE, Mack A, van de Ven AL, Ferrati S, Dunner K, Godin B, Chiappini C, Landry M, Brousseau L, Liu X, Bean AJ, Ferrari M. Small. 2010;6:2691–2700. doi: 10.1002/smll.201000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kim Y, Pourgholami MH, Morris DL, Lu H, Stenzel MH. Biomater Sci. 2013 doi: 10.1039/c2bm00096b. [DOI] [PubMed] [Google Scholar]
- 260.Park JS, Han TH, Lee KY, Han SS, Hwang JJ, Moon DH, Kim SY, Cho YW. J Control Release. 2006;115:37–45. doi: 10.1016/j.jconrel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 261.Wu L-C, Chu L-W, Lo L-W, Liao Y-C, Wang Y-C, Yang C-S. ACS Nano. 2013;7:365–375. doi: 10.1021/nn3043397. [DOI] [PubMed] [Google Scholar]
- 262.Ohta S, Inasawa S, Yamaguchi Y. Biomaterials. 2012;33:4639–4645. doi: 10.1016/j.biomaterials.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 263.AshaRani PV, Hande MP, Valiyaveettil S. BMC Cell Biol. 2009;10:65. doi: 10.1186/1471-2121-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Dombu CY, Kroubi M, Zibouche R, Matran R, Betbeder D. Nanotechnology. 2010:21. doi: 10.1088/0957-4484/21/35/355102. [DOI] [PubMed] [Google Scholar]
- 265.Summers HD, Rees P, Holton MD, Brown MR, Chappell SC, Smith PJ, Errington RJ. Nat Nanotechnol. 2011;6:170–174. doi: 10.1038/nnano.2010.277. [DOI] [PubMed] [Google Scholar]
- 266.Vranic S, Boggetto N, Contremoulins V, Mornet S, Reinhardt N, Marano F, Baeza-Squiban A, Boland S. Part Fibre Toxicol. 2013;10:1. doi: 10.1186/1743-8977-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.