ABSTRACT
Background: Researches on immunotherapy of glioma has been increasing exponentially in recent years. However, autoimmune-like side effects of current immune checkpoint blockade hindered the clinical application of immunotherapy in glioma. The discovery of the TIM-3, a tumor-specific immune checkpoint, has shed a new light on solution of this dilemma. We aimed at investigating the role of TIM-3 at transcriptome level and its relationship with clinical practice in glioma.
Methods: A cohort of 325 glioma patients with RNA-seq data from Chinese Glioma Genome Atlas (CGGA project) was analyzed, and the results were well validated in TCGA RNA-seq data of 699 gliomas. R language was used as the main tool for statistical analysis and graphical work.
Results: TIM-3 was enriched in glioblastoma (the most malignant glioma) and IDH-wildtype glioma. TIM-3 can act as a potential marker for mesenchymal molecular subtype according to TCGA transcriptional classification scheme in glioma. TIM-3 was closely related to immune functions in glioma, especially T cell mediated immune response to tumor cell and T cell mediated cytotoxicity directed against tumor cell target. Moreover, TIM-3 and PD-L1 played almost exactly the same inflammatory activation functions in glioma. Clinically, high expression of TIM-3 was an independent indicator of poor prognosis.
Conclusion: The expression of TIM-3 is closely related to the pathology and molecular pathology of glioma. Meanwhile, in glioma TIM-3 plays a specific role in T cell tumor immune response. Therefore, TIM-3 is a promising target for immunotherapeutic strategies, providing an alternative treatment when glioma gains resistance to antibodies of PD-1/PD-L1.
KEYWORDS: Glioma, immune response, inflammatory activation, prognosis, TIM-3
Introduction
Glioma represents the most common and malignant brain tumor in adults, characterized by a high recurrence rate and high fatality rate.1,2 Despite multimodal conventional therapy, including neurosurgical resection and radiotherapy with concomitant and adjuvant alkylating agent temozolomide chemotherapy, glioma remains a leading cause of death in human cancer. Within decades, amounts of researches on molecular markers and molecular targeted drugs produced very limited effect in prolonging life expectancies of glioma patients. The discovery of intracranial lymphatic system has brought a new theoretical basis and new hope for brain tumor immunotherapy.3
Driven by the success of immune checkpoint blockade in other cancers, researches on immunotherapy of glioma has increased exponentially in the past few years.4,5 However, most glioma have been proved to be refractory to current immunotherapies. This has raised our interest in finding novel immune checkpoints directly targeting tumor cells in glioma.
T cell immunoglobulin domain and mucin domain 3 (TIM3) is a membrane protein selectively expressed on interferon-gamma-secreting CD4+ T helper 1 (Th1) and CD8+ T cytotoxic (Tc1) cells.6 Emerging data suggest that Tim-3 takes a center stage in T cell exhaustion by triggering cell death: T cells fail to exert effective functions such as cytotoxicity and cytokine secretion in response to antigen and tumor cell stimulation.7,8 TIM-3, as a key immune checkpoint in tumor-induced immune suppression, exhibits several unique features that make it an intriguing candidate for the next wave of therapies in cancer.9
As a novel immune checkpoint, TIM-3 is gaining more and more attention in both solid and hematologic malignancies.10 However, we reviewed the current evidence and failed to find a single comprehensive report about TIM-3 in glioma. Taking advantage of Chinese Glioma Genome Atlas (CGGA) data set, we gathered RNA-seq data of 325 glioma samples to take an integrative investigation of TIM-3 in glioma. Moreover, our findings were well validated in another RNA-seq data set of 699 gliomas obtained from TCGA network (http://cancergenome.nih.gov). This is the first integrative study characterizing TIM-3 expression in whole grade glioma molecularly and clinically. And we believe that TIM-3 will be a hot ticket in glioma immunotherapy.
Patients and methods
Patients and samples
This study was approved by the Beijing Tiantan Hospital institutional review board (IRB), and wrote informed consent was obtained from each patient. Only samples with greater than 80% tumor cells were selected. Transcriptome sequencing data of glioma samples were from CGGA generating with Illumina Hiseq platform. Overall survival was estimated from the date of diagnosis to the date of either death or last follow-up. Methods to detect IDH mutation state has been described in our previous study.11 The Cancer Genome Atlas (TCGA) mRNA-seq database was downloaded from public databases (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). The primary GSC (glioma stem-like cells) gene expression array data set was downloaded from GSE67089.
Gene set variation analysis (GSVA) analysis
After Spearman correlation analysis, gene ontology (GO) analysis of the most correlated genes was constructed by Heatmap. The GO geneset was downloaded from AmiGO 2 Web portal (http://amigo.geneontology.org/amigo/landing). Inflammatory-related metagenes has been described in our previous study.5
Statistical analysis
The prognostic value of TIM-3 was estimated by Kaplan–Meier analysis and Cox proportional hazard model analysis using SPSS statistical software (version 19). Other statistical computations and figures drawing were performed with several packages (ggplot2, pheatmap, pROC and corrgram) in the statistical software environment R, version 3.3.2 (http://www.r-project.org). For all statistical methods, p < 0.05 was considered as significant difference.
Results
TIM-3 was enriched in glioblastoma and IDH-wildtype glioma
To characterize the expression pattern of TIM-3 in glioma, we examined the RNA-sequencing data of glioma from CGGA and TCGA database. Compared to WHO grade II and grade III glioma, glioblastoma (WHO IV) showed the highest TIM-3 expression in CGGA database (Fig. 1A). This result was well validated in TCGA RNA-seq data (Fig. 1C). These results suggested that high expression of TIM-3 was a sign of high malignancy of glioma, in consistence with other malignant tumors reported previously.12-14 It is acknowledged that IDH mutation plays a very important role in the development and progression of glioma.15,16 Consequently, we also explored the relationship between TIM-3 expression level and IDH mutation status and found that TIM-3 was highly enriched in IDH wildtype glioma in both CGGA and TCGA data set (Figs. 1B and D and Fig. S1). This finding indicated that TIM-3 check point related immune responses were more prevalent in IDH wildtype glioma. Moreover, we found that TIM-3 was specifically expressed in GSC cells (Fig. S2), which was also found in acute myeloid leukemia.17
Figure 1.
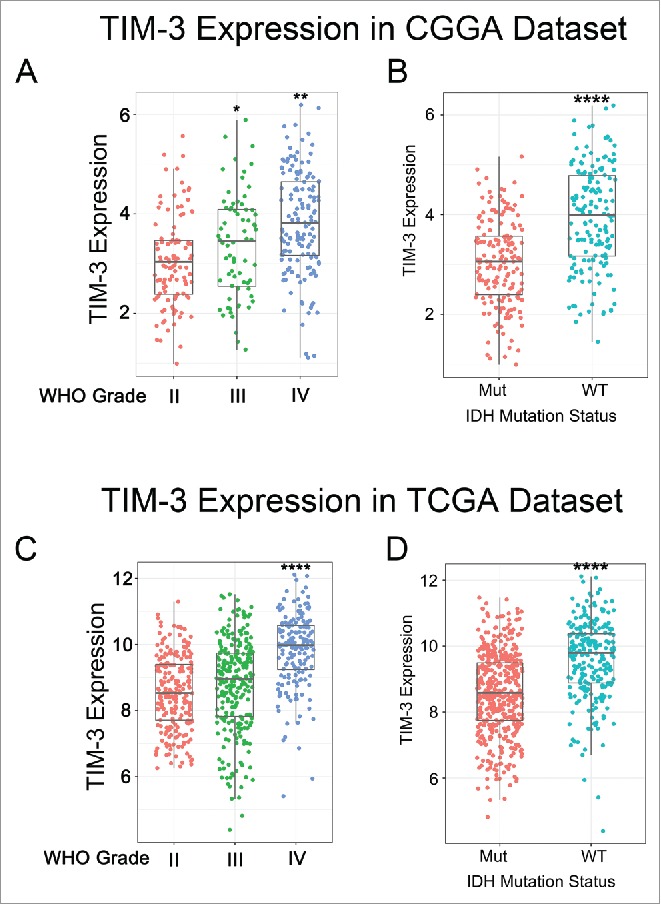
TIM-3 was significantly upregulated in glioblastoma and IDH-wildtype glioma. (A, C) TIM-3 was significantly increased in glioblastoma (WHO IV) in CGGA and TCGA data set. (B, D) TIM-3 was significantly increased in IDH-wildtype gliomas in CGGA and TCGA data set. *, ** and **** indicate p < 0.05, p < 0.01 and p < 0.0001, respectively.
TIM-3 was a potential marker for mesenchymal molecular subtype glioma
To find out the molecular expression pattern of TIM-3, we asked the distribution of TIM-3 in different molecular subtypes defined by TCGA network.18 When compared with other three subtypes respectively, TIM-3 was significantly upregulated in mesenchymal subtype in CGGA cohort as well as in TCGA cohort (Figs. 2A and C). To further validate this finding, ROC curves for TIM-3 expression and mesenchymal subtype of all grade glioma were performed. Surprisingly, area under the curve (AUC) were up to 90.5% and 88.9% in CGGA and TCGA data set, respectively (Figs. 2B and D). These results suggested that TIM-3 was highly specifically expressed in mesenchymal subtype glioma. Based on these findings, we inferred that TIM-3 may play important biologic functions in glioma. To validate our hypothesis, biologic function analyses need to be performed.
Figure 2.
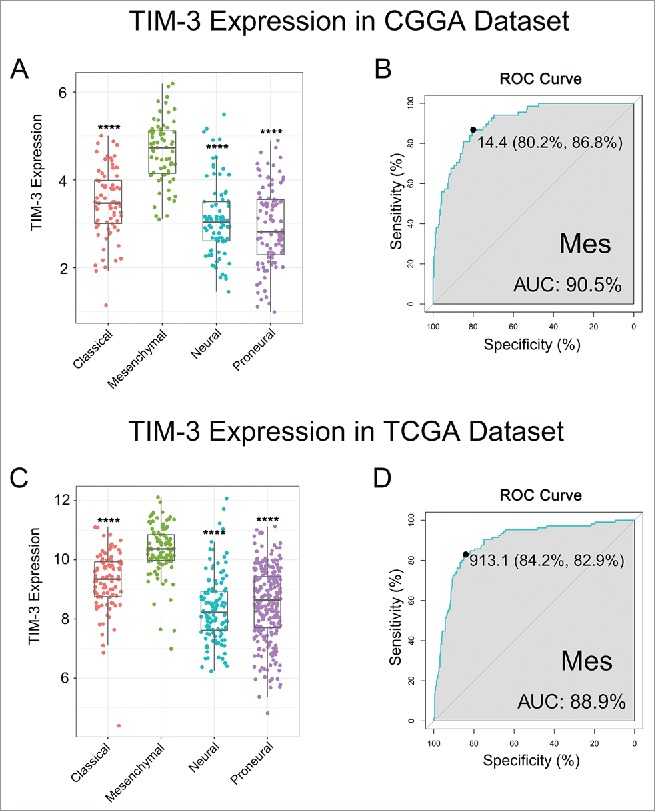
TIM-3 was highly enriched in mesenchymal molecular subtype glioma. (A, C) TIM-3 expression pattern in different molecular subtypes in CGGA and TCGA data set. (B, D) ROC curve analysis showed that TIM-3 had highly sensitivity and specificity to predict mesenchymal subtype in CGGA and TCGA database. **** indicates p < 0.0001.
TIM-3 was closely related to immune functions in glioma
To clarify the biologic role of TIM-3 in glioma, we performed GO analysis. First, we created a gene list that strongly correlated with TIM-3 by Pearson correlation analysis (Pearson |R| > 0.6). Finally, there were 390 genes in CGGA gene list and 495 genes in TCGA gene list. Then, we explored the biofunction of these genes respectively by GO analysis in DAVID Bioinformatics Resources 6.8. When the gene function was sorted by p value in increasing order, genes most relevant to TIM-3 were mostly involved in immune response and inflammatory response in both CGGA and TCGA database (Figs. 3A and B). To further explore the immune function of TIM-3 in specific immune functions, we then studied the role of the specific immune roles of TIM-3 in immune response and inflammatory activities.
Figure 3.

TIM-3 was closely related to immune functions in glioma. (A, B) Gene ontology analysis showed that TIM-3 was mostly involved in immune response and inflammatory response in CGGA and TCGA database. (C, D) Most immune response related genes were significantly positively correlated with TIM-3 expression, while there were still a small number of genes were significantly negatively correlated with Tim-3 expression.
TIM-3 related immune response
To further explore the role of TIM-3 in the immune response in glioma, we found out certain genesets related to the immune response from the AmiGO 2 Web portal. When removed insignificant genes (gene expression was 0 more than half patients), the remaining 1,540 genes were eligible for subsequent analysis. We selected the genes that were most relevant to TIM-3(Pearson |R| > 0.4) for the heatmap drawing. Among the 411 selected-genes, 397 genes were significantly positively correlated with TIM-3 expression and 14 genes were significantly negatively correlated with Tim-3 expression (Figs. 3C and D). A detailed list of these genes was shown in Table S1. Above analysis found that TIM-3 was positively correlated with most immune responses and negatively correlated with a small number of immune responses in glioma. A thorough study of this result may reveal the important role of TIM-3 in gliomas.
The relationship between TIM-3 and T cell immunity
TIM-3 promotes tumor progression in a variety of tumors by inhibiting T cell immune function.19,20 Whether it has the same functions in gliomas remain an enigma. To elucidate the relationship between TIM-3 and T cell immune in glioma, GSVA analysis was performed. As shown in Figs. 4A and B, TIM-3 was positively correlated with T-helper 1/2 type immune response, T-helper 1/2 cell cytokine production and natural killer cell mediated cytotoxicity directed against tumor cell target. Meanwhile, it was negatively correlated with T cell mediated immune response to tumor cell and T cell mediated cytotoxicity directed against tumor cell target. Importantly, this result can be verified mutually in CGGA and TCGA databases. This also confirmed that TIM-3 played an inhibitory role in T cell immune to tumor in glioma. The special immune function of TIM-3 revealed the mechanism of this gene in glioma.
Figure 4.
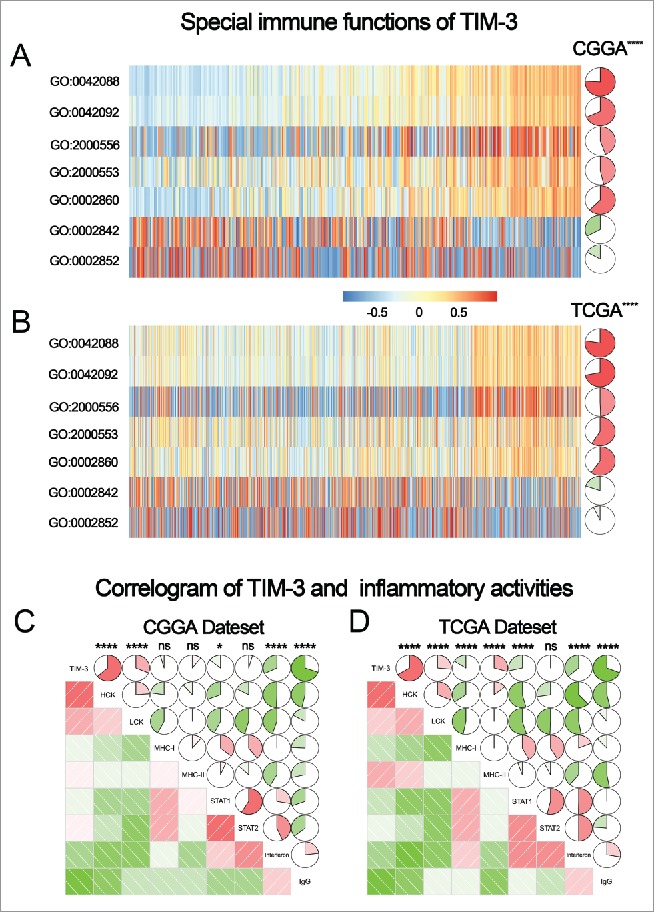
TIM-3-related T cell immunity and inflammatory activities in glioma. (A, B) The relationship between TIM-3 and T cell immunity in CGGA and TCGA data set. (C, D) The relationship between TIM-3 and inflammatory activities in glioma. The correlation between TIM-3 and other functions was analyzed by pearson correlation analysis. ns, * and **** indicate no statistical difference, p < 0.05 and p < 0.0001, respectively. GO:0042088: T-helper 1 type immune response, GO:0042092: T-helper 2 type immune response, GO:2000556: positive regulation of T-helper 1 cell cytokine production, GO:2000553: positive regulation of T-helper 2 cell cytokine production, GO:0002860: positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target, GO:0002842: positive regulation of T cell mediated immune response to tumor cell, GO:0002852: regulation of T cell mediated cytotoxicity directed against tumor cell target.
The relationship between TIM-3 and inflammatory activities
As revealed above, TIM-3 also played an important role in the inflammatory response in glioma. We used the method described previously to specifically analyze the role of TIM-3 in the glioma inflammatory response.5 To our surprise, TIM-3 showed exactly similar pattern to inflammation activities as PD-L1 we reported earlier5 (Figs. 4C and D). Both genes were positively associated with HCK, LCK and MHC-I, but were negatively associated with IgG. These results suggested that TIM-3 and PD-L1 play almost exactly the same inflammatory activation functions (they were macrophages and T cells signaling transduction suppressors, but not much involved in B linage related immune responses) in glioma. This finding was consistent with the above results and further confirmed the important immune function of TIM-3 in glioma.
TIM-3 predicted worse survival in glioma
As TIM-3 showed robust negative relationship with T cell tumor immune response, we additional analysis the prognostic value of TIM-3. We analyzed the prognosis of a total of 1,024 glioma patients by Kaplan–Meier method. Similar with most malignancies, overexpression of TIM-3 predicted significantly poor overall survival in both CGGA and TCGA databases (Figs. 5A and B). Furthermore, Cox regression analysis was preformed additionally, verifying the independence of the clinical prognostic significance of TIM-3 in glioma. In the above two databases, it was showed that TIM-3, Age at Diagnosis, WHO Grade, KPS Score and IDH Status were significantly associated with overall survival. On multivariate analysis, the expression of TIM-3 was also a significant factor after adjusting for the clinical factors mentioned above (Table 1 and Table S2). These findings indicated that TIM-3 predicted poor prognosis in glioma due to suppressive effect on T-cell-related immune response, especially T cell immune to tumor.
Figure 5.
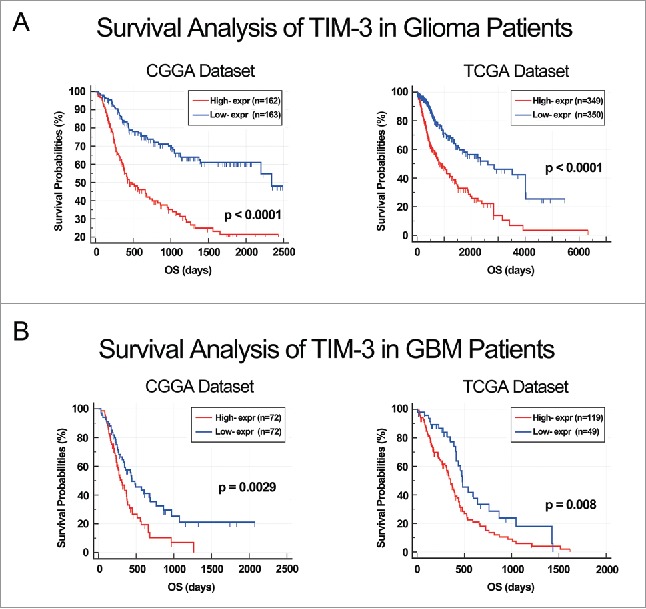
ALDH1A3 was a prognostic factor in glioma and GBM patients. (A) Kaplan–Meier survival analysis showed that high expression of TIM-3 conferred a worse prognosis in glioma patients. (B) Kaplan–Meier survival analysis showed that high expression of TIM-3 conferred a worse prognosis in GBM patients.
Table 1.
Univariate and multivariate analysis of clinical prognostic parameters in CGGA Dataset.
| Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| Variable | HR (95% CI) | p value | HR (95% CI) | p value |
| TIM-3 expression | 1.038 | <0.0001 | 1.023 | 0.005 |
| (1.027–1.050) | (1.007–1.040) | |||
| Age at diagnosis | 1.038 | <0.0001 | 0.991 | 0.452 |
| (1.023–1.054) | (0.969–1.014) | |||
| WHO grade | 3.477 | <0.0001 | 2.752 | <0.0001 |
| (2.716–4.452) | (1.961–3.862) | |||
| KPS score | 0.972 | <0.0001 | 0.972 | <0.0001 |
| (0.962–0.982) | (0.960–0.984) | |||
| IDH mutation | 0.228 | <0.0001 | 0.763 | 0.348 |
| (0.158–0.329) | (0.433–1.344) | |||
Discussion
Glioma is one of the highly fatal diseases that affects human health severely. Aside from temozolomide, for decades, no major breakthrough was achieved to improve the prognosis of glioma patients.2,21 Therefore, new therapeutic approaches are urgently needed. In recent years, as a novel therapeutic approach of glioma patients, immunotherapy has shown a promising prospect.22 At present, researches on the field of neural tumor immunotherapy are mainly focus on CTLA-4 and PD-1/PD-L1 blockade.23,24 However, since the autoimmune-like side effects of current immune checkpoint blockade, immunotherapy is difficult to be widely applied. This is the so-called “Cancer Immunology at the Crossroads.”10 Fortunately, at this critical moment, people discovered a novel immune checkpoint receptor-TIM-3. This novel immune checkpoint exhibits several unique advantages that make it an intriguing candidate for immunotherapy of glioma in the future.
Here, we analyzed the TIM-3 expression in the RNA-seq data of 1,024 glioma patients. As expected, TIM-3 expression was significantly upregulated in higher malignant pathological type gliomas. Moreover, we also found that high expression of TIM-3 was highly enriched in the phenotype of known malignant molecules, such as IDH wildtype state, mesenchymal phenotype and glioma stem cells. All these results indicated that TIM-3 expression was associated with more malignant biologic process as other solid and hematologic malignancies.25,26 It is most likely that these malignant biologic behaviors have contributed to tumor recurrence and therapy resistance. Revealing mechanism of TIM-3 in glioma may be the key to triumphing over this deadly disease.
Through an in-depth analysis of the biologic functions of TIM-3 in glioma, we found that TIM-3 played an important role in the glioma immune response, especially in tumor-induced immune suppression. This special effect of TIM-3 has also been observed in other tumors.7,8,20 This may be accused to the unique presence of TIM-3 Tregs in tumor tissue, which was a unique advantage over other immune checkpoints.27-29 Furthermore, another unique advantage of TIM3 was that intracellular tail of TIM-3 did not contain any immunoreceptor tyrosine-based inhibition motifs (ITIM) or immunoreceptor tyrosine-based switch motifs (ITSM).30 These advantages effectively avoided the major deficiencies of current immunotherapy and highlighted its value in glioma immunotherapy. As an immune inhibitory receptor, suppressing the inflammatory response and autoimmunity are the natural property of TIM-3.31,32 We found that TIM-3 played almost exactly the same role in inflammatory response as PD-L1 in glioma. Together, these data indicated that TIM-3 and PD-L1 played a synergistic role in tumor inflammatory response to promote the development of a severe dysfunctional phenotype in T cells in glioma as reported in other cancers.33,34 Thus, theoretically, antibodies derived to block the stimulation of TIM-3 should have the same effect as PD-L1 suppression. Multiple preclinical and clinical targeting TIM-3 have yielded consistent results with the theory in various tumors, particularly melanoma and non-small-cell lung cancer.35,36 A research published in the latest issue of Clin Cancer Res reported that TIM-3/PD-1 co-blockade was more effective than either TIM-3 or PD-1 blockade alone at restoring inflammation activation and improving survival time from glioma-bearing murine.37 This gratifying discovery provided a reliable support for our research. Furthermore, TIM-3 will also have a profound impact on studies of glioma stem cells if it can be a therapeutic target of glioma stem cells. This hypothesis has already been validated in acute myeloid leukemia stem cells.38 Whether it is the same case in glioma stem cells or not, still need further studies. Collectively, these data strongly support the potential of TIM-3 blockade for the immunotherapy of glioma.
The future of glioma treatment most likely resides in combinatorial approaches, with administration of conventional treatments (surgery, radiochemotherapy) and immunotherapy complement each other. Our works will greatly promote the immunotherapy of glioma into a new era.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ms. Yuling Yang and Kun Yao for tissue sample collection and clinical data retrieval.
Funding
This work was supported by grants from National Key Research and Development Plan (No. 2016YFC0902500), National Natural Science Foundation of China (No. 81672479, 81502495, 81502606), Beijing Youth Talent Project (2015000021223ZK28), Beijing Nova Program (No. xx2014B062) and Fok Ying Tung Education Foundation (No. 141032).
References
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin 2010; 60(3):166-93; PMID:20445000; https://doi.org/ 10.3322/caac.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X, Jiang C, Kang C, Li X, Chen L et al.. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 2016; 375(2):263-73; PMID:26966000; https://doi.org/ 10.1016/j.canlet.2016.01.024 [DOI] [PubMed] [Google Scholar]
- 3.Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee K et al.. Structural and functional features of central nervous system lymphatic vessels. Nature 2015; 523(7560):337-41; PMID:26030524; https://doi.org/ 10.1038/nature14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366(26):2443-54; PMID:22658127; https://doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Zhang C, Liu X, Wang Z, Sun L, Li G, Liang J, Hu H, Liu Y, Zhang W et al.. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology 2016; 5(11):e1196310; PMID:27999734; https://doi.org/ 10.1080/2162402X.2016.1196310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu C, Anderson AC, Kuchroo VK. TIM-3 and its regulatory role in immune responses. Curr Top Microbiol Immunol 2011; 350:1-15. PMID:20700701; https://doi.org/ 10.1007/82_2010_84 [DOI] [PubMed] [Google Scholar]
- 7.Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev 2017; 276(1):97-111; PMID:28258697; https://doi.org/ 10.1111/imr.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattei F, Schiavoni G. TIM-3 as a molecular switch for tumor escape from innate immunity. Front Immunol 2012; 3:418; PMID:23316202; https://doi.org/ 10.3389/fimmu.2012.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero D. Immunotherapy: PD-1 says goodbye, TIM-3 says hello. Nat Rev Clin Oncol 2016; 13(4):202-3; PMID:26977783; https://doi.org/ 10.1038/nrclinonc.2016.40 [DOI] [PubMed] [Google Scholar]
- 10.Anderson AC. Tim-3: An emerging target in the cancer immunotherapy landscape. Cancer Immunol Res 2014; 2(5):393-8; PMID:24795351; https://doi.org/ 10.1158/2326-6066.CIR-14-0039 [DOI] [PubMed] [Google Scholar]
- 11.Yan W, Zhang W, You G, Bao Z, Wang Y, Liu Y, Kang C, You Y, Wang L, Jiang T. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One 2012; 7(1):e30339; PMID:22291938; https://doi.org/ 10.1371/journal.pone.0030339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Huang Y, Tan L, Yu W, Chen D, Lu C, He J, Wu G, Liu X, Zhang Y. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol 2015; 29(2):635-41; PMID:26428847; https://doi.org/ 10.1016/j.intimp.2015.09.017 [DOI] [PubMed] [Google Scholar]
- 13.Zhou E, Huang Q, Wang J, Fang C, Yang L, Zhu M, Chen J, Chen L, Dong M. Up-regulation of Tim-3 is associated with poor prognosis of patients with colon cancer. Int J Clin Exp Pathol 2015; 8(7):8018-27; PMID:26339368 [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J, Jiang B, Zhao H, Huang Q. Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma 2014; 61(1):35-40; PMID:24195506; https://doi.org/ 10.4149/neo_2014_006 [DOI] [PubMed] [Google Scholar]
- 15.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ et al.. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360(8):765-73; PMID:19228619; https://doi.org/ 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HY, Tang K, Liang TY, Zhang WZ, Li JY, Wang W, Hu HM, Li MY, Wang HQ, He XZ et al.. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res 2016; 35:86; PMID:27245697; https://doi.org/ 10.1186/s13046-016-0362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, Majeti R. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci U S A 2011; 108(12):5009-14; PMID:21383193; https://doi.org/ 10.1073/pnas.1100551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP et al.. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17(1):98-110; PMID:20129251; https://doi.org/ 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabatos CA, Chakravarti S, Cha E, Schubart A, Sánchez-Fueyo A, Zheng XX, Coyle AJ, Strom TB, Freeman GJ, Kuchroo VK. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat Immunol 2003; 4(11):1102-10; PMID:14556006; https://doi.org/ 10.1038/ni988 [DOI] [PubMed] [Google Scholar]
- 20.Cai C, Xu YF, Wu ZJ, Dong Q, Li MY, Olson JC, Rabinowitz YM, Wang LH, Sun Y. Tim-3 expression represents dysfunctional tumor infiltrating T cells in renal cell carcinoma. World J Urol 2016; 34(4):561-7; PMID:26253654; https://doi.org/ 10.1007/s00345-015-1656-7 [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Su HK, Zhao HF, Chen ZP, To SS. Progress in the application of molecular biomarkers in gliomas. Biochem Biophys Res Commun 2015; 465(1):1-4; PMID:26253473; https://doi.org/ 10.1016/j.bbrc.2015.07.148 [DOI] [PubMed] [Google Scholar]
- 22.Han SJ, Zygourakis C, Lim M, Parsa AT. Immunotherapy for glioma: Promises and challenges. Neurosurg Clin N Am 2012; 23(3):357-70; PMID:22748649; https://doi.org/ 10.1016/j.nec.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 23.Belcaid Z, Phallen JA, Zeng J, See AP, Mathios D, Gottschalk C, Nicholas S, Kellett M, Ruzevick J, Jackson C et al.. Focal radiation therapy combined with 4-1BB activation and CTLA-4 blockade yields long-term survival and a protective antigen-specific memory response in a murine glioma model. PLoS One 2014; 9(7):e101764; PMID:25013914; https://doi.org/ 10.1371/journal.pone.0101764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Zhang Z, Liu Y, Xue Y, Parney I. Programmed death-ligand 1 (PD-L1) may play a role in malignant glioma infiltration. Med Hypotheses 2015; 85(2):127-9; PMID:25936667; https://doi.org/ 10.1016/j.mehy.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Yin N, Zhang Z, Zhang Y, Zhang G, Chen W. Upregulation of T-cell immunoglobulin and Mucin-Domain Containing-3 (Tim-3) in monocytes/macrophages associates with gastric cancer progression. Immunol Invest 2017; 46(2):134-148; PMID:27911104; https://doi.org/ 10.1080/08820139.2016.1229790 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, Munger ME, Veenstra RG, Weigel BJ, Hirashima M, Munn DH, Murphy WJ, Azuma M, Anderson AC, Kuchroo VK et al.. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood 2011; 117(17):4501-10; PMID:21385853; https://doi.org/ 10.1182/blood-2010-10-310425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakuishi K, Ngiow SF, Sullivan JM, Teng MW, Kuchroo VK, Smyth MJ, Anderson AC. TIM3(+)FOXP3(+) regulatory T cells are tissue-specific promoters of T-cell dysfunction in cancer. Oncoimmunology 2013; 2(4):e23849; PMID:23734331; https://doi.org/ 10.4161/onci.23849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012; 486(7404):549-53; PMID:22722857; https://doi.org/ 10.1038/nature11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C et al.. A special population of regulatory T cells potentiates muscle repair. Cell 2013; 155(6):1282-95; PMID:24315098; https://doi.org/ 10.1016/j.cell.2013.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, Angin M, Wakeham A, Greenfield EA, Sobel RA et al.. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3–mediated cell death and exhaustion. Nat Med 2012; 18(9):1394-400; PMID:22863785; https://doi.org/ 10.1038/nm.2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Ma CJ, Wang JM, Ji XJ, Wu XY, Moorman JP, Yao ZQ. Tim-3 regulates pro- and anti-inflammatory cytokine expression in human CD14+ monocytes. J Leukoc Biol 2012; 91(2):189-96; PMID:21844165; https://doi.org/ 10.1189/jlb.1010591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramello MC, Tosello BJ, Canale FP, Mena HA, Negrotto S, Gastman B, Gruppi A, Acosta Rodríguez EV, Montes CL. Tumor-induced senescent T cells promote the secretion of pro-inflammatory cytokines and angiogenic factors by human monocytes/macrophages through a mechanism that involves Tim-3 and CD40L. Cell Death Dis 2014; 5:e1507; PMID:25375372; https://doi.org/ 10.1038/cddis.2014.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Cao C, Piao H, Li Y, Tao Y, Zhang X, Zhang D, Sun C, Zhu R, Wang Y et al.. Tim-3 protects decidual stromal cells from toll-like receptor-mediated apoptosis and inflammatory reactions and promotes Th2 bias at the maternal-fetal interface. Sci Rep 2015; 5:9013; PMID:25757669; https://doi.org/ 10.1038/srep09013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Chen G, Li Y, Wang R, Wang L, Lin Z, Gao X, Feng J, Ma Y, Shen B et al.. Involvement of T cell Ig Mucin-3 (Tim-3) in the negative regulation of inflammatory bowel disease. Clin Immunol 2010; 134(2):169-77; PMID:19913460; https://doi.org/ 10.1016/j.clim.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 35.Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med 2010; 207(10):2175-86; PMID:20819923; https://doi.org/ 10.1084/jem.20100637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, Zhu Y, Li G, Huang H, Zhang G, Wang F, Sun J, Yang Q, Zhang X, Lu B. TIM-3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS One 2012; 7(2):e30676; PMID:22363469; https://doi.org/ 10.1371/journal.pone.0030676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JE, Patel MA, Mangraviti A, Kim ES, Theodros D, Velarde E, Liu A, Sankey EW, Tam A, Xu H et al.. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res 2017; 23(1):124-136; PMID:27358487; https://doi.org/ 10.1158/1078-0432.CCR-15-1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki Y et al.. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell 2010; 7(6):708-17; PMID:21112565; https://doi.org/ 10.1016/j.stem.2010.11.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


