Abstract
Optogenetics relies on the expression of specific microbial rhodopsins in the neuronal plasma membrane. Most notably, this includes channelrhodopsins, which when heterologously expressed in neurons function as light-gated cation channels. Recently, a new class of microbial rhodopsins, termed anion channel rhodopsins (ACRs), has been discovered. These proteins function as efficient light-activated channels strictly selective for anions. They exclude the flow of protons and other cations and cause hyperpolarization of the membrane potential in neurons by allowing the inward flow of chloride ions. In this study, confocal near-infrared resonance Raman spectroscopy (RRS) along with hydrogen/deuterium exchange, retinal analogue substitution, and site-directed mutagenesis were used to study the retinal structure as well as its interactions with the protein in the unphotolyzed state of an ACR from Guillardia theta (GtACR1). These measurements reveal that (i) the retinal chromophore exists as an all-trans configuration with a protonated Schiff base (PSB) very similar to that of bacteriorhodopsin (BR), (ii) the chromophore RRS spectrum is insensitive to changes in pH from 3 to 11, whereas above this pH the Schiff base (SB) is deprotonated, (iii) when Ser97, the homologue to Asp85 in BR, is replaced with a Glu, it remains in a neutral form (i.e., as a carboxylic acid) but is deprotonated at higher pH to form a blue-shifted species, (iv) Asp234, the homologue of the protonated retinylidene SB counterion Asp212 in BR, does not serve as the primary counteranion for the protonated SB, and (v) substitution of Glu68 with an Gln increases the pH at which SB deprotonation is observed. These results suggest that Glu68 and Asp234 located near the SB exist in a neutral state in unphotolyzed GtACR1 and indicate that other unidentified negative charges stabilize the protonated state of the GtACR1 SB.
TOC image
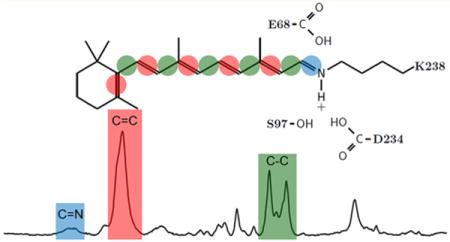
Channelrhodopsins (ChRs) are currently used extensively in neuroscience research1–7 and are potentially enabling tools for a growing list of biomedical applications.8–11 ChRs were first identified in 2002 as membrane-depolarizing phototaxis receptors in chlorophyte algae12,13 and found to function as light-gated cation channels when heterologously expressed in animal cells.14,15 In 2005, ChR genes were expressed in neurons and shown to enable their spatiotemporal photoactivation.16 Since then, thousands of publications have appeared reporting the use of ChRs in optogenetic applications.1,2
The most widely used ChRs are channelrhodopsin-2 from Chlamydomonas reinhardtii (CrChR2) and its derivatives. When CrChR2 is incorporated into neuronal membranes, absorption of blue light causes an inward current of protons and Na+. The subsequent depolarization of the membrane triggers an action potential when the light level and level of CrChR2 expression are sufficiently high.
Photosuppression (silencing) of nerve activity is also critical for various applications such as elucidating neural circuit function. Thus far, silencing has been achieved using outwardly directed microbial rhodopsin proton pumps such as archaerhodopsin-3 (AR3) or inwardly directed halorhodopsin (HR) anion pumps.17 These proteins hyperpolarize the membrane upon illumination due to active transport of positive charge out of the cell (AR3) or anions into the cell (HR). However, the current produced by these pumps is limited by translocation of only a single ion per photon absorbed, limiting greatly the efficiency of hyperpolarization.
Recently, a new class of natural ChRs, termed anion channel rhodopsins (ACRs), has been discovered.18 Unlike AR3 and HR, ACRs function as light-gated anion channels. They exclude the flow of protons and other cations and cause hyperpolarization of the membrane by allowing the inward flow of chloride into the neuron. ACRs exhibit faster kinetics and less than 1/1000th of the light intensity required for neuronal silencing by the most efficient currently available optogenetic proteins.18
A major advance at the molecular level was the elucidation of the three-dimensional (3D) structure at 2.3 Å resolution of a ChR chimera that combines elements of the first five transmembrane helices in CrChR1 and last two from CrChR2 (termed the C1C2 chimera).19 However, thus far, this is the only atomic-resolution ChR structure available. Furthermore, recent FTIR difference studies indicate that substantial differences in the structural changes, gating, and desensitization of the C1C2 chimera and CrChR2 exist.20
A key goal of this study is to compare and contrast aspects of the molecular mechanism of ACRs with known channelrhodopsins as well as with microbial rhodopsins in general. For this purpose, we have used confocal near-infrared resonance Raman spectroscopy (RRS) to characterize an ACR from Guillardia theta (GtACR1). GtACR1 was heterologously expressed and extracted from Pichia pastoris cells21 and reconstituted into model bilayer membranes. These measurements reveal information about the structure and interactions of the retinal chromophore of an ACR, including several unusual features compared to those of other microbial rhodopsins.
MATERIALS AND METHODS
Expression, Purification, and Reconstitution of ChRs
The 7TM domain of GtACR1 was expressed from P. pastoris18 and reconstituted using a procedure similar to that described in ref 22 for CaChR1. Cells were grown in BMGY (Buffered Minimal Glycerol Yeast) medium. Expression was induced by the addition of 0.5% methanol every 24 h in the presence of 5 μM all-trans retinal. Cells were grown for 2 days, harvested by low-speed centrifugation, and disrupted by a bead beater (BIOSPEC Products, model 1107900-101). Membrane fragments were collected by centrifugation for 40 min at 38000 rpm. The protein was partially purified on a Ni-NTA agarose column (Qiagen, Hilden, Germany) after solubilization by incubation overnight in 3% N-octyl D-glucopyranoside (OG). For membrane reconstitution, the protein was eluted in 20 mM HEPES (pH 7.4), 100 mM NaCl, 1% OG, and 300 mM imidazole, mixed with Escherichia coli polar lipids (ECPL) (Avanti Polar Lipids, Alabaster, AL) at a concentration of 5 μg/mL in 10% OG in a 1:10 ratio (GtACR1:ECPL), and incubated at 4 °C for 30 min. The solution was transferred into a dialysis membrane cassette (Thermo-Fisher, Slide-A-Lyser, 10K G2 type) with a 10 kDa cutoff and dialyzed against 50 mM K2HPO4 and 300 mM NaCl (pH 7.0) for 24 h. After a buffer change, the sample was dialyzed for an additional 4 h. The resulting GtACR1 reconstituted into membrane was then spun down at 15000g for 2 min. The pellet was washed twice with 5 mM phosphate buffer (pH 7.0) and washed twice with 5 mM K2HPO4 and 100 mM NaCl (pH 7.3). Similar procedures were used to express and reconstitute CaChR1 as previously described.22
Chromophore Substitution with A2 Retinal
A2 retinal (3,4-dehydroretinal) was purchased from Toronto Research Chemicals (catalog no. D230075, CAS Registry No. 472-87-7). GtACR1 containing a substituted A2 retinal instead of the native A1 was produced as described above except that a 0.5% methanol solution containing 10 μM A2 retinal instead of 5 μM A1 was added.
Near-IR Resonance Raman Spectroscopy
The reconstituted ChRs prepared as described above were used for resonance Raman studies. GtACR1 samples were measured by confocal near-IR resonance Raman spectroscopy using methods similar to those previously reported for AR3 and BR,23 and CaChR1 and CrChR2.22 Approximately 30 μg of the reconstituted GtACR1 was spun in a SCILOGEX D3024 centrifuge at 15000 rpm for 5 min, and the resulting pellet was resuspended in wash buffer (300 mM NaCl and 20 mM Tris/HEPES) adjusted to different pH values using HCl/NaOH titration. The solution was then repelleted and washed at least two additional times to form a final pellet. The final pellet was resuspended in a small amount of the wash buffer (<5 μL) and transferred using a 10 μL syringe (Hamilton Co., Reno, NV) to a 0.5 mm inner diameter square borosilicate glass capillary (Fiber Optic Center, New Bedford, MA) with one end sealed. The capillary was spun at a lower speed (10000 rpm for 1 min), and then the open side was sealed with Critoseal (Leica Microsystems, Buffalo Grove, IL). H/D exchange was achieved by suspending pelleted samples in D2O solutions prepared with 300 mM NaCl and 20 mM HEPES at its pH value (∼6.3).
RRS spectra were obtained at room temperature on a Renishaw inVia confocal Raman microscope equipped with a 20× objective with a numerical aperture (NA) of 0.4 using 785 nm laser excitation. Unless otherwise noted, the experiments were conducted with a power of 300 mW (∼25% throughput at the sample) at an effective pixel resolution of ∼1.2 cm−1. The system calibrates frequency accuracy using the 520.9 cm−1 band from an internal silicon chip. Data acquisition consisted of series of measurement cycles with each cycle consisting of a 30 s data acquisition period followed by a 20 s wait time in the dark. Depending on the signal-to-noise ratio, this cycle was repeated 30–1000 times. The spectra of the data acquisition period were then averaged. The empty capillary spectrum was subtracted from the averaged spectra to remove the fluorescence background. A multipoint linear baseline correction was performed to obtain the final reported spectra.
UV–Vis Spectroscopy
Approximately 50 μg of reconstituted GtACR1 was washed in 300 mM NaCl/20 mM HEPES buffer (pH 7). To reduce the scattering, the solution was sonicated in three cycles of 30 s in an ice bath with a 30 s rest between cycles. The solution was placed in a quartz cuvette (Thorlabs, Inc., Newton, NJ), and a Cary 6000 spectrometer equipped with a diffuse scattering apparatus (Agilent Technologies, Inc., Santa Clara, CA) was used to perform the UV–vis absorbance measurement using a 0.1 s per step size of 1 nm (total scan time of 1 min). Light-adapted sample measurements were performed immediately after illumination for >5 min with a 530 nm LED (Thorlabs, Inc.) operating at approximately 10 mW/cm2 at the sample. Dark-adapted sample measurements were performed after the sample had been kept in the dark for >30 min. To obtain the final reported spectra, baseline correction was performed by curve fitting to a combination of Rayleigh and Tyndall scattering curves to remove the scattering curve.
Spectral Analysis
Spectral subtractions, baseline corrections, and peak fitting were all performed using GRAMS/AI version 7.02 (ThermoFisher Scientific, Inc.), except for baseline correction for UV–vis spectra, which was performed using MATLAB (MathWorks, Natick, MA). The GRAMS/AI version 7.02 spectral analysis software package, which incorporates iterative χ2 minimization, was used to fit the subcomponent bands in the ethylenic and SB region of the RRS spectrum and in the visible absorption spectrum. RRS spectra were fitted from 1480 to 1600 cm−1 in the ethylenic region and from 1590 to 1700 cm−1 in the SB region with a linear baseline. For the ethylenic region, the curve fitting program found three Voigtian peaks, which resulted in an R2 value better than 0.99. The same routine was used for curve fitting the UV–vis spectrum in the range of 350–700 nm resulting in three Voigtian peaks with an R2 value better than 0.99.
RESULTS
UV–Visible Absorption
The UV–visible absorption of the dark-adapted spectrum of GtACR1 reconstituted into EPCL lipids is very similar to the spectrum of GtACR1 in 0.05% DDM detergent micelles21 and also the action spectrum previously reported.18 The unfitted λmax of the visible absorption spectrum is at 515 nm. Curve fitting reveals the major component of the absorption is at 520 nm, with smaller components at 467 and 419 nm (Figure S1). This compares closely to the visible absorption of CaChR1 that has a λmax at 519 nm with fitted components at 524, 474, and 415 nm.22 Light adaptation of GtACR1 for 10 min followed immediately by measurement of the UV–visible spectrum did not alter the absorption compared to the dark-adapted pigment except for a small drop in overall absorbance (Figure S1).
Similarity of the GtACR1 with the BR and CaChR1 Raman Spectra
The near-IR RRS spectrum of GtACR1 is very similar to CaChR1 and BR spectra (Figure 1), especially in key regions, including the ethylenic and fingerprint regions (see ref 24 for RRS spectra of BR along with complete band assignments). In an earlier near-IR RRS study, it was concluded that CaChR1 has an all-trans retinal configuration on the basis of its similarity to BR.22 We conclude GtACR1 also has an all-trans retinal configuration. In contrast, the spectrum of CrChR2 is significantly different, consistent with the existence of a mixed chromophore composition of all-trans and 13-cis isomers in this ChR.25
Figure 1.
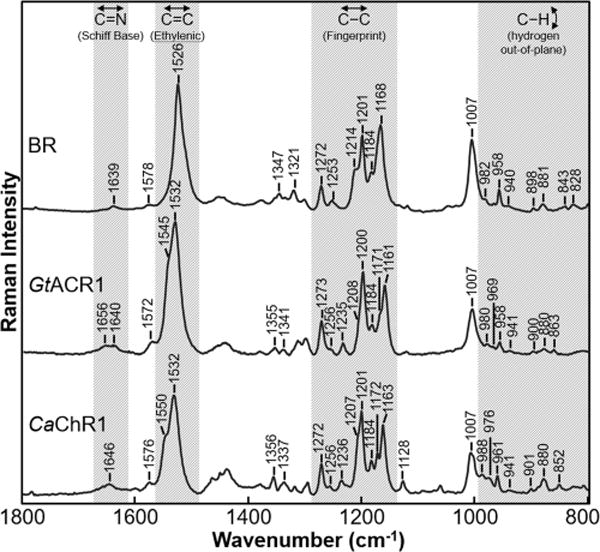
RRS spectra of the BR (in the purple membrane), GtACR1, and CaChR1 (reconstituted into E. coli polar lipids). Data were recorded at room temperature using a 785 nm probe laser with a power of 100 mW (40 mW measured at the sample) for BR and CaChR1 and with a power of 300 mW (70 mW measured at the sample) for GtACR1. In this figure and Figures 2–5 and Supplementary Figures 6 and 7, except where noted, spectra were not smoothed and were scaled using the intensity of the peaks in the fingerprint region and a background spectrum of the borosilicate capillary and buffer was subtracted from the sample. Additional details are given in Materials and Methods.
Fingerprint Region
The fingerprint region of retinal reflects mainly the vibration from the various mixed C–C stretching modes of the chromophore.24,26,27 A comparison of this region for BR, GtACR1, and CaChR1 (Figure 1) reveals bands with similar frequencies and relative intensities near 1161–1168, 1171, 1184, 1200, 1207–1214, 1253, and 1272 cm−1. As established by retinal isotope labeling combined with the Wilson-FG normal-mode analysis in the case of BR,24 these bands are highly characteristic of the C–C stretch modes of an all-trans configuration of a protonated SB (PSB) retinylidene chromophore. The most similar bands in the fingerprint region of these three spectra are the 1200, 1253, and 1273 cm−1 bands that are assigned in the case of BR to the C14–C15, C12–C13, and 11H in-plane bend plus lysine vibrations, respectively.24 This result indicates that the retinal structure in this region of the chromophore is very similar for all three proteins.
However, there are several small differences in the fingerprint region among BR, GtACR1, and CaChR1. For example, the small band near 1171 cm−1 in GtACR1 and CaChR1 does not appear in BR. This is most likely due to a frequency upshift of the band near 1161 cm−1 (1163 cm−1 in CaChR1) to 1168 cm−1 in BR. In the case of BR, the 1168 cm−1 band is assigned to a localized C10–C11 stretching mode.24 An increase in the bond strength (and hence stretching frequency) is expected to occur for BR because of its red-shifted λmax, which causes an increased level of delocalization of electron density in the polyene chain. In support of this explanation, AR3 with a λmax almost identical to that of BR, also displays a C10–C11 stretching mode near 1168 cm−1.24,28 In contrast, the more blue-shifted green proteorhodopsin (GPR; λmax ∼ 525 nm) and blue proteorhodopsin (BPR; λmax ∼ 480 nm) exhibit a band near 1162 cm−1 with a small shoulder near 1171 cm−1.29 Additional unexplained differences in the fingerprint region of GtACR1 and CaChR1 compared to BR and AR323 are the smaller 1207 cm−1 shoulder (relative to the more distinct shoulder at 1214 cm−1) and a new band appearing at 1235 cm−1.
Ethylenic C=C Stretch Region
Bands arising from the C=C stretching modes of the retinal chromophore appear in the 1500–1600 cm−1 region. In the case of GtACR1 and CaChR1, an intense band appears at 1532 cm−1 compared to the 1526 cm−1 band appearing in BR (Figure 1). The similarity of the ethylenic frequency between GtACR1 and CaChR1 and the difference with BR are expected because of the approximate inverse relationship between λmax and νC=C found for most microbial and animal rhodopsins.30–32
Curve fitting of this region reveals that in addition to the major component at 1532 cm−1, GtACR1 has a smaller component near 1545 cm−1, and as previously reported, CaChR1 has a second component at 1549 cm−1 (Figure S2).22 One possible explanation for two components is that two different forms of the protein exist in equilibrium in the unphotolyzed state. This would also be supported by the observation of multiple components in the visible absorption of GtACR1 and CaChR1. However, as discussed previously for the case of CaChR1,22 the second component in the visible absorption could also arise from vibronic coupling as commonly seen for other microbial rhodopsins such as NpSRII.33,34 Similarly, in the case of the RRS spectrum, the second component may arise from a second major ethylenic mode such as that seen in the L intermediate of BR.35,36 In general, normal-mode analysis of PSB all-trans retinals reveals multiple combination C=C stretching modes.24 Future experiments, including a study of the wavelength dependence of the GtACR1 photocycle using FTIR difference spectroscopy and the temperature dependence of the RRS spectrum, are planned to distinguish among these different possibilities.
Schiff Base C=N Stretch Region
The C=N SB stretching mode (both protonated and deprotonated) falls in the 1600–1700 cm−1 region. Because this region can reflect other vibrational modes, including nonresonance contributions from the protein amide I band,37 a retinal PSB C=N mode is assigned by using hydrogen/deuterium (H/D) exchange, which is expected to cause a frequency downshift of this band38 (but not an unprotonated retinal SB). Furthermore, the magnitude of the downshift provides information about the hydrogen bonding strength of the PSB.39,40
As seen in Figure 2, H/D exchange of GtACR1 causes the disappearance of a small band at 1640 cm−1 and the appearance of a new band at 1622 cm−1. The frequency and magnitude of the H/D-induced downshift are very similar to those of the PSB C=N mode assigned to BR (1639 cm−1 downshifts to 1622 cm−1)22 and also AR3.23 Thus, we conclude that the SB of GtACR1 is protonated and has a hydrogen bonding strength similar to those of BR and AR3. In contrast, CaChR1 has a higher SB C=N stretch frequency (1646 cm−1) and a larger H/D-induced downshift (26 cm−1) (Figure 2), indicative of stronger hydrogen bonding near the C=N bond. CrChR2 also displays a higher frequency for the C=N stretch (1659 cm−1) and a larger H/D-induced downshift (28 cm−1).25 It is also interesting to note that HRs from Halobacterium salinarum (HsHR) and from Natronobacterium pharaonis (NpHR) both exhibit C=N frequencies near 1632 cm−1 with much smaller H/D-induced downshifts of approximately 10–12 cm−1.41,42
Figure 2.
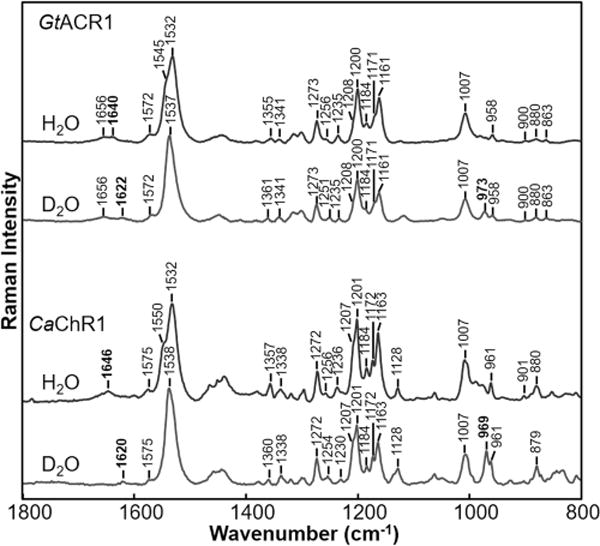
Comparison of resonance Raman spectra of GtACR1 and CaChR1 recorded in H2O and D2O. Data were recorded at room temperature using a 785 nm probe laser with a 100 mW power (40 mW measured at the sample) for CaChR1 and with a 300 mW power (70 mW measured at the sample) for GtACR1.
The second band observed in this region of the GtACR1 RRS spectrum at 1656 cm−1 most likely arises from the Amide I mode, which is normally the strongest band in the Raman spectrum of proteins that are not resonance-enhanced.43 One example is the Raman spectrum of opsin measured in a photoreceptor membrane where the retinylidene chromophore has been removed.37 Near-IR Raman spectroscopy of microbial rhodopsins with 785 nm excitation causes nonresonant contributions to appear such as from the strong amide I protein mode due to weaker resonance enhancement of retinal vibrations. In support of this argument, the putative amide I band does not appear when 532 nm excitation is used to measure the RRS spectrum (Figure S3).
Note that in this case photoproducts are likely to be produced by the 532 nm excitation. This is supported by the increase in intensity near 1534 cm−1 indicative of the formation of a blue-shifted photoproduct(s). In addition, this photo-product is likely to have a 13-cis-retinal configuration because of the increase in intensity of a band at 1184 cm−1 characteristic of a 13-cis-retinal isomer.28 In contrast, photoproducts are unlikely to be produced using 785 nm excitation even at high powers as indicated by the similarity of the RRS spectra recorded at 300 mW and at 1/1000th of this power (300 μW) (Figure S3).
NH In-Plane Bending Region
The NH in-plane bending (rocking) is also assigned to part of a band near 1350 cm−1 in BR. Note that this band also contains contributions from the C15H in-plane bending motion.24 The NH portion of the mode downshifts to 976 cm−1 upon H/D exchange.24 As seen in Figure 2, bands are observed in GtACR1 and CaChR1 in D2O at 973 and 969 cm−1 respectively. Along with the similar frequency and H/D-induced downshift of the C=N stretching mode, the similarity of the frequency of the ND in-plane bending mode indicates that the hydrogen bonding interactions near the PSB of these three proteins are similar.
HOOP Region
The region from 800 to 1000 cm−1 consists mainly of bands due to the retinal hydrogen-out-of-plane (HOOP) modes [sometimes termed wags (w)]. The intensity and frequency of these modes are highly sensitive to torsions around the single and double bonds that produce nonplanar polyene configurations.44–46 For polyenes with completely planar configurations, the HOOP modes are expected to have only weak RRS spectral intensity.44,47
Several GtACR1 bands are observed in this region at 863, 880, 900, 941, 958, 969, and 980 cm−1, which match well the frequency and intensity of BR and CaChR1 HOOP modes but not those of CrChR2.22 On the basis of earlier assignments of HOOP modes in BR,24 these bands provide information about the specific configuration and interaction of specific regions of the retinal, including 14w (880 cm−1), 10w (900 cm−1), NHw (940 cm−1), and 11w+12w (958), which must be very similar in the unphotolyzed states of all three proteins. One exception is the unassigned 863 cm−1 band that appears to be unique for GtACR1.
Effects of A2 Retinal Substitution
To further assign retinal vibrational bands in the near-IR RRS spectra, A2 retinal (3,4-dehydroretinal) was substituted for the native A1 retinal chromophore of GtACR1. As shown in Figure 3, this substitution causes a significant downshift in the major ethylenic band from 1532 to 1513 cm−1 along with the appearance of a second distinct band appearing at 1538 cm−1. Curve fitting reveals that as expected the major component of the visible absorption red-shifts to 552 nm along with several subcomponents contributing at 511, 481, and 432 nm (Figure S4). Similar red-shifts are observed in many (but not all) microbial rhodopsins upon substituting A2 for A1 retinal, including CaChR1 and CrChR2.48
Figure 3.
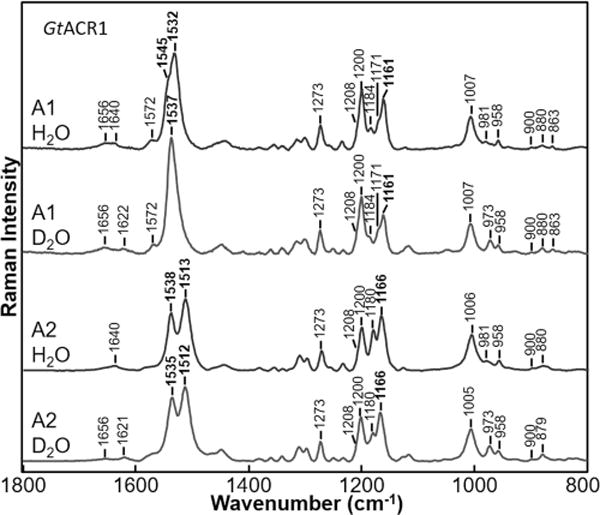
Comparison of resonance Raman spectra of GtACR1 with A1 and A2 retinals recorded in H2O and D2O. Data were recorded at room temperature using a 785 nm probe laser with a 100 mW power (40 mW measured at the sample) for BR and CaChR1 and with a 300 mW power (70 mW measured at the sample) for GtACR1.
Upon H/D exchange, the major component of the ethylenic mode at 1513 cm−1 downshifts slightly (∼1 cm−1), whereas the higher-frequency component at 1539 cm−1 downshifts 4 cm−1. This greater sensitivity of the high-frequency ethylenic mode may be explained by an increased level of coupling of this mode to the C=N stretch, which downshifts ∼18 cm−1 upon H/D exchange. A similar effect may also be occurring for the high-frequency ethylenic component of GtACR1 with A1 retinal at 1545 cm−1, thus explaining the apparent upshift of the overall ethylenic band to 1537 cm−1. In this case, the higher-frequency component downshifts, causing an apparent upshift of the entire ethylenic peak.
Interestingly, there is little effect of the A2 retinal substitution on the SB C=N stretch frequency for either H2O or D2O. This indicates that the increased level of electron delocalization in the polyene chain caused by the A2 substitution does not extend to the C=N bond. On the other hand, as discussed above, the frequency of the C10–C11 stretching mode at 1161 cm−1 24 upshifts to 1166 cm−1, obscuring the 1171 cm−1 band due to increased electron density in the single bond.
Effects of pH
The RRS spectrum of GtACR1 is insensitive to pH changes over the entire range of 3–11 (Figure 4). For example, only a small increase was detected in the frequency of the major component of the ethylenic band of less than ∼1 cm−1. Other regions of the spectrum discussed above, including those assigned to the C=N stretch, NH bend, fingerprint, and HOOP modes, also remain essentially unaltered. At pH 12, a gradual increase in intensity of a band at 1578 cm−1 is observed along with the almost complete disappearance of the 1532 cm−1 band at pH 12.5. In analogy with the high frequency of the BR M intermediate with an ethylenic C=C stretch frequency of 1567 cm−1, the 1578 cm−1 band is likely to correspond to the ethylenic stretch of an M-like blue-shifted species formed due to deprotonation of the SB. In detergent micelles, visible absorption measurements reveal a transition with a pK of ∼9 that involved the formation of an M-like product.49 The higher pH for this transition observed by RRS for reconstituted membranes is likely due to the combination of two factors: (i) a shift in pKa for this transition to a higher pH due to reconstitution of GtACR1 in ECPL lipid membranes compared to detergent micelles and (ii) the resonance enhancement from the 785 nm excitation that is expected to be weak for a blue-shifted deprotonated SB retinal species causing much smaller amounts of the low-pH form to be detected in the RRS spectrum even at high pH. Note that other vibrational modes, such as from the C=N stretch of the deprotonated SB, are also not expected to be detected due to the weak RRS enhancement.
Figure 4.
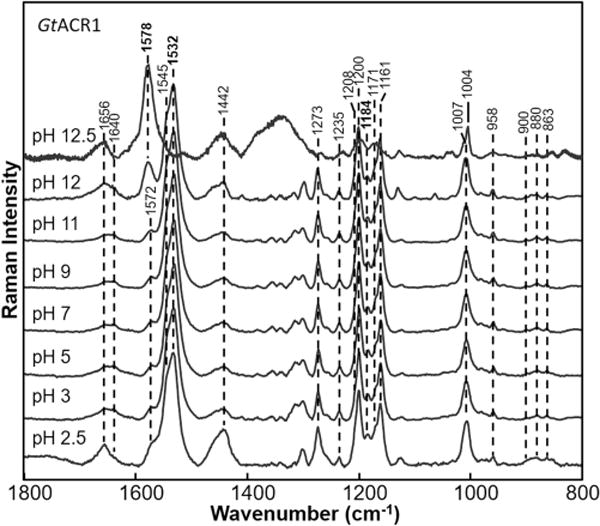
Resonance Raman spectra of GtACR1 recorded at various pH values ranging from 2.5 to 12.5. Data were recorded at room temperature using a 785 nm probe laser with a 300 mW power (70 mW measured at the sample). Spectra were not smoothed and were scaled using the intensity of the peaks in the fingerprint region (except for pH 12.5, which was scaled using the protein band near 1004 cm−1).
Changes are also detected below pH 3, including an increase for bands most likely originating from nonresonance contributions such as bands at 1656 cm−1 and near 1450 cm−1 that likely arise from the protein. The increased intensity of the 1184 cm−1 band characteristic of a 13-cis-retinal isomer28 also occurs, indicating that the retinal is no longer in a pure all-trans form.
Effects of Substitution of Glu for Ser97
Ser97 is in the position corresponding to Asp85, the primary SB counterion and proton acceptor in BR, and Glu169 in CaChR1. As in HsHR and NpHR, where Ser is found at this position, Ser97 is not expected to exist in an ionized form and thus cannot act as a counterion to the SB. The S97E mutation at pH 7 causes a 4 cm−1 downshift in the ethylenic υC=C (from 1532 to 1528 cm−1) (Figure 5). On the basis of the inverse correlation between λmax and υC=C, a red-shift in the visible λmax is expected, which was verified by visible absorption spectroscopy (Figure S5). Because introduction of a negative charge near the SB is expected to cause an increase in υC=C and a blue-shift in λmax, the result suggests that the Glu97 carboxylic acid side chain remains in a neutral state at least at pH 7. There is also almost no change in the fingerprint region, in-plane NH bending, and HOOP modes for S97E, indicating the chromophore remains in an all-trans configuration similar to that of wild-type GtACR1.
Figure 5.
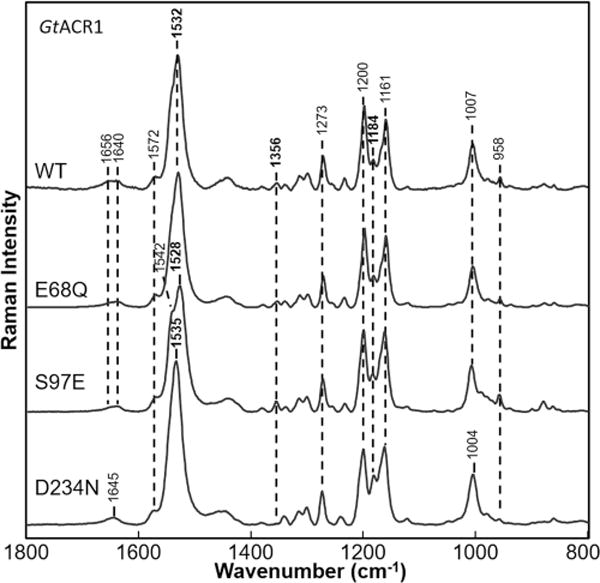
Comparison of resonance Raman spectra of GtACR1 WT and its mutants E68Q, S97E, and D234N. Data were recorded at room temperature using a 785 nm probe laser with a 300 mW power (70 mW measured at the sample).
However, at higher pH (Figure S6), a transition to a blue-shifted species is observed with an ethylenic υC=C at 1545 cm−1. This transition agrees with a large 35 nm blue-shift in the visible absorption maximum observed for this mutant with a pKa of ∼849 and most likely corresponds to deprotonation of Glu97. At pH >10.5, we also observe the appearance of a band at 1579 cm−1, which is similar to the case of WT, can be attributed to deprotonation of the SB and formation of an M-like intermediate.
Effects of Substitution of Asn for Asp234
Asp234 is the homologue of the SB counterion Asp212 in BR. Unlike Asp234, where the side chain could exist as a neutral carboxylic acid or negatively charged carboxylate, Asn remains neutral. Note, however, that it can still participate in hydrogen bonding with the SB as indicated for example for the D85N mutant in BR.50 In contrast to S97E, the D234N mutation causes a 3 cm−1 upshift in the ethylenic υC=C to 1535 cm−1 and a blue-shift of the visible λmax (Figure S5). As discussed above, the upshift of υC=C is inconsistent with a simple point charge model in which Asp234 serves as the primary counterion for the PSB. In this case, neutralization of Asp234 would be expected to cause a downshifted υC=C and red-shifted λmax. A change in peak intensity in the fingerprint region is also found possibly due to an increase in the intensity of the 1184 cm−1 band indicative of an increased level of the 13-cis isomer. The band at 1356 cm−1 assigned to the in-plane NH bending mode also disappears, indicative of a change in the hydrogen bonding of the SB. The SB υC=N appears upshifted to 1645 cm−1 compared to WT.
Effects of Substitution of Gln for Glu68
Glu68 is in the corresponding position of Glu90 in CrChR2. On the basis of the 3D structure of the C1C2 chimera, this residue is likely to be located sufficiently close to interact with the GtACR1 PSB,19 and its mutation has significant effects on the channel gating mechanism of GtACR1.21 However, the E68Q mutant measured at pH 7 produced no detectable change in the RRS spectrum relative to that of WT. Note that although visible absorption measurements in detergent micelles reveal a 3 nm red-shift,49 the corresponding shift in the ethylenic υC=C is expected to be too small to detect. This observation indicates that Glu68 most likely exists in a neutral form at neutral pH because otherwise a larger change would have occurred. A similar conclusion was also reached for the homologous residue E90 in CrChR2 based on UV–visible and FTIR measurements of WT CrChR2 and the E90Q mutant.22,51,52 Interestingly, the substitution increases the pH at which SB deprotonation is observed compared to that of WT. Like WT, E68Q is very stable up to pH 11 (Figure S7). However, at pH 12, unlike WT, no increase was found at 1578 cm−1, indicating that the SB has yet not undergone appreciable deprotonation. Instead, a small band appears at 1555 cm−1, which might correspond to a much smaller blue-shift. At pH 13, both WT and E68 appear to undergo complete bleaching because no retinal bands are detected in the RRS spectra (data not shown). These results suggest interaction of E68 with the PSB in agreement with visible absorption effects of the E68Q mutation.49
DISCUSSION AND CONCLUSIONS
More than 50 ChRs have thus far been discovered from chlorophyte algae.53 They constitute a phylogenetically diverse group that displays a range of spectroscopic, photocycle, cation selectivity, and electrical properties.54 Because of their function as light-activated cation channels, chlorophyte ChRs provide important tools for activating excitable cells such as neurons. The recent discovery of a new phylogenetically distinct class of type 1 rhodopsins from cryptophyte algae that function as anion channel rhodopsins (ACRs) considerably expands the repertoire of microbial rhodopsin function. In addition to providing an attractive system for studying passive chloride transport through membranes, ACRs hold great promise for effective optogenetic inhibition of neurons and other excitable cells.18
Our goal in this study is to investigate the retinal chromophore structure and retinal–protein interactions of ACRs. GtACR1 from G. theta is thus far the best characterized ACR.18,21,49 Some of the key residues in chlorophyte ChRs are conserved.18,21 However, Asp85, the helix 3 residue in BR that serves as the primary SB counterion and proton acceptor, is replaced with the neutral serine (Ser97) (Figure 6) located near the SB. This residue is also neutral (alanine) in the third ACR reported, PsuACR1.55 In contrast, nearly all chlorophyte ChRs thus far examined such as CrChR1, CrChR2, and CaChR1 have a glutamic acid residue at that position54,56 (one exception is Dunaliella ChR53).
Figure 6.
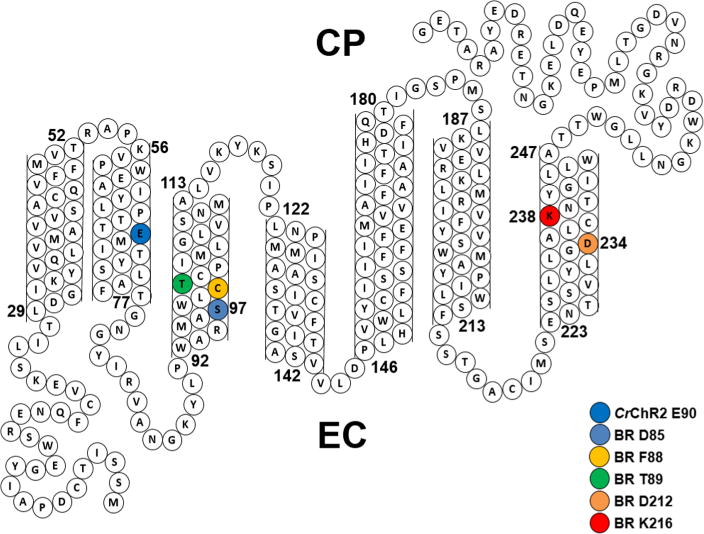
Sequence of GtACR1 and its predicted folding pattern. Highlighted residues include Ser97 and Asp234, which are homologues of Asp85 and Asp212, respectively, in BR that comprise the complex counterion to the SB.
HRs also have a neutral residue, usually threonine, at the BR Asp85 position.57 In this case, the negative charge provided by Asp85 in BR is replaced with a Cl− located near the SB.41,42,58–60 The acid purple form of BR produced at pH <2, where both Asp85 and Asp212 are neutral,61 also has a monovalent anion located close to the SB. It is possible that Asp234, the homologue for BR Asp212, serves as the primary SB counterion. Indeed, it was recently established for CaChR1 at pH 7 that Asp299 (the homologue of BR Asp212) serves as the primary SB counterion (and SB proton acceptor),62,63 while Glu169, the Asp85 homologue, is protonated (e.g., neutral).22,63,64 An alternate possibility is that another residue such as Glu68, a Cl− ion similar to HR, or a combination of these function as the primary counterion.
To investigate these different possibilities, we used near-IR confocal resonance Raman microscopy. The production of photointermediates during RRS measurements was minimized by the near-IR excitation wavelength (785 nm), which is significantly red-shifted relative to the λmax of GtACR1 at 515 nm (Figure S1). Importantly, preresonance with the retinal is still obtained, which significantly enhances retinal chromophore vibrations relative to other nonresonance vibrations of the protein and lipids. This effect has previously been demonstrated with other microbial rhodopsins such as proteorhodopsin, archaerhodopsin, and ChRs using excitation wavelengths red-shifted as far as 1060 nm.29,65,66 Confocal microscopy also allows small volumes of the sample (several microliters) to be measured in a stationary capillary, thus avoiding the use of larger volume flow and spinning cells.
Several conclusions can be drawn from the current results.
(1) GtACR1 Has an all-trans Chromophore with a Configuration Similar to That of BR and CaChR1
Although many microbial rhodopsins such as BR have exclusively an all-trans chromophore configuration, some microbial rhodopsins in their unphotolyzed state exhibit a mixture of different retinal isomers. For example, a recently discovered microbial rhodopsin termed middle rhodopsin (MR) can bind 11-cis retinal as does vertebrate rhodopsin.67 In the case of ChRs, RRS measurements of CrChR2 reveal a mixture of all-trans and 13-cis retinal in the unphotolyzed pigment.22,51,52 In contrast, CaChR1 was found to contain almost exclusively an all-trans retinal chromophore on the basis of near-IR RRS.22
We can conclude from the current RRS measurements that the unphotolyzed state of GtACR1 contains almost exclusively an all-trans-retinal chromophore. Especially striking is the agreement among the GtACR1, BR, and CaChR1 fingerprint, in-plane NH bend, and HOOP-mode regions. As discussed above, small differences in the fingerprint region of GtACR1 and BR are most likely due to their different visible λmax values at 515 and 570 nm, respectively.
(2) The GtACR1 SB Is Protonated and Has a Hydrogen Bonding Strength Similar to That of BR
RRS-based H/D exchange measurements conclusively show that the SB of GtACR1 is protonated in its unphotolyzed state over a wide pH range (see point 3 below). Furthermore, on the basis of the magnitude of the H/D-induced υC=N downshift, the hydrogen bonding strength is weaker than those of ChRs previously characterized using this method.22 For example, CaChR1 and CrChR2 have 26 and 28 cm−1 H/D-induced downshifts, respectively,22,68 compared to a value of 18 cm−1 for GtACR1. A weaker SB hydrogen bonding strength might be attributed to the presence of a neutral Ser97 residue instead of a negatively charged Asp or Glu residue located close to the SB. However, BR, which has a negatively charged Asp85, still exhibits approximately the same H/D downshift. Furthermore, RRS and UV–visible pH titration studies indicate that in CaChR1, Glu169, the Asp85 homologue, is protonated (e.g., neutral) in the unphotolyzed state of the protein.22,63,64 Thus, the overall cause of differences in measured SB hydrogen bonding strength between GtACR1 and other microbial rhodopsins is likely to be more complex and as described below may involve a number of factors, including the interaction of one or more water molecules and possibly a Cl− anion in the active center.
(3) The Protonation State of the SB and Nearby Residues Is Insensitive to pH Changes over a Wide pH Range
The RRS spectrum of GtACR1 over the pH range of 3–11 is largely unchanged. For example, the ethylenic υC=C shifts less than 1 cm−1. Furthermore, no changes are detected in the SB υC=N, fingerprint, in-plane NH bend, and HOOP-mode regions. This lack of changes indicates that the retinal chromophore remains in an all-trans protonated SB form and that nearby pH titratable groups such as Asp234 do not undergo changes in their protonation state (see below).
Part of this insensitivity to pH can be attributed to the presence of a neutral Ser97 at the Asp85 primary counterion position. Unlike Asp and Glu residues in BR and many ChRs, the neutral Ser OH side chain group is not expected to ionize over this pH range. In contrast, Asp234, which is also located near the SB, could undergo a change in protonation in this pH range. However, as discussed below, evidence indicates this residue exists in a neutral state at least at pH 7.
At pH 12, the intensity of the band at 1578 cm−1 increases, and eventually, the chromophore appears to completely bleach as indicated by an absence of strong RRS retinal vibrations (data not shown). The appearance of the 1578 cm−1 band is most likely due to deprotonation of the SB, which leads to formation of a blue-shifted GtACR1 species that absorbs near 380 nm typical of an unprotonated retinal SB. For example, at high pH, the BR mutant D85N (alkaline form) causes a visible absorption shift from 590 nm to near 400 nm accompanied by an upshift of the υC=C from 1515 to 1567 cm−1.50,69 As noted above, the actual formation of the deprotonated SB species may occur at a pH lower than that indicated by the appearance of the 1578 cm−1 band, because the resonance enhancement from the 785 nm excitation is expected to be weak for a blue-shifted deprotonated SB retinal species. This effect along with the expected increase in pKa for alkaline titrations in membranes versus detergent micelles may explain the lower pKa (∼9) observed for this transition based on visible absorption spectroscopy.49
At pH 2.5, changes were observed mainly in bands at 1656 cm−1 and near 1442 cm−1, which may be due to protein denaturation and a subsequent increase in nonresonance contributions such as the amide I mode. However, major bands assigned to the PSB retinal are still present. In contrast, protonation of the primary counterion Asp85 in BR occurs below pH 3, causing a red-shift in λmax from 570 to ∼600 nm and a downshift in νC=C from 1527 to 1518 cm−1.70 It is thus likely that no change in the protonation state occurs at least down to pH 2.5 for residues located close to the SB in GtACR1, including Asp234.
(4) Glu68, Ser97, and Asp234 Do Not Serve as the Primary Counterions for the SB at pH 7
RRS measurements indicate that unlike in BR, the homologues of Asp85 and Asp212 in GtACR1, Ser97 and Asp234, respectively, are unlikely to serve as primary counterions for the PSB. First, the high pKa of the Ser hydroxyl group rules out Ser97 ionization. Even when Ser97 is replaced with Glu97, our results indicate that it remains in a neutral form (i.e., as a carboxylic acid) at neutral pH and deprotonates at higher pH. Second, the magnitude and direction of υC=C and visible λmax shifts for D234N indicate that Asp234 does not strongly interact with the PSB in an ionized form. In fact, replacement of an ionized Asp234 with a neutral Asn234 is expected on the basis of a simple point charge model42,71 to cause a decrease in υC=C and an increase in visible λmax, while the opposite response is observed for D234N (Figure 6 and Figure S5).
An alternative candidate for the primary counterion for the SB is Glu68. On the basis of homology modeling with the C1C2 chimera, this residue is expected to be in a position that would allow interaction with the SB.19 However, the effects of the E68Q substitution on the RRS spectrum are not consistent with this residue interacting with the SB in an ionized state at neutral pH. Instead, the substitution appears to increase the pH for SB deprotonation compared to that of WT. One possible explanation is that because Glu68 already exists in a neutral form, substitution of a neutral Gln does not alter significantly the environment of the chromophore near the PSB at neutral pH. On the other hand, at higher pH, deprotonation of Glu68 may precede SB deprotonation. If this were true, then substitution with a nonionizable Gln residue might raise the pH required for SB deprotonation as observed.
In view of these results, an obvious possibility is that an anion such as chloride acts a Schiff base counterion similar to halorhodopsins. However, recent experiments reveal that the visible absorption and photocycle kinetics of GtACR1 are insensitive to deionization and replacement of Cl− with SO42−.49 In this regard, FTIR difference spectroscopy, which can be sensitive to alterations in anion–protein interaction(s),41 may be used to further investigate the effects on GtACR1 structure and photocycle conformational changes.
It is also important to note that the protonation of an Asp or Glu residue in the unphotolyzed state of GtACR1 does not necessarily exclude it from functioning as an acceptor group for the SB proton during the active photocycle. In fact, in a previous study of CaChR1, RRS and FTIR evidence supported a model in which Glu169 (the BR Asp85 homologue) is protonated in the unphotolyzed “ground” state, is deprotonated during the K photointermediate, and accepts a proton from the SB during formation of the M intermediate.63 A similar “two-step” process may apply to Glu68, which appears to be a likely candidate for the SB proton acceptor.49 For example, Gln substitution results in a 4–5-fold decrease in the degree of accumulation of the M intermediate and corresponding disappearance of fast channel closing at pH 7.4. In addition, this substitution eliminates a pH-dependent shift in the maximal absorption wavelength of the unphotolyzed WT.49 FTIR difference measurements currently underway are designed to further test this model.
Supplementary Material
Acknowledgments
We thank Sergey Mamaev for his advice and assistance with growth, purification, and expression of GtACR1 and Roberto Fernandez De Cordoba for constructing the snake diagram of GtACR1.
Funding
This work was supported by National Science Foundation Grant CBET-1264434 to K.J.R. and National Institutes of Health Grant R01GM027750, the Hermann Eye Fund, and Endowed Chair AU-0009 from the Robert A. Welch Foundation to J.L.S.
ABBREVIATIONS
- ACR
anion channelrhodopsin
- ChRs
channelrhodopsins
- RRS
resonance Raman spectroscopy
- FTIR
Fourier transform infrared
- GtACR1
ACR from G. theta
- CaChR1
channelrhodopsin-1 from Chlamydomonas augustae
- CrChR2
channelrhodopsin-2 from C. reinhardtii
- BR
bacteriorhodopsin
- HR
halorhodopsin
- H/D
hydrogen/deuterium
- SB
protonated Schiff base
- C1C2
chimera of CrChR1 and CrChR2
- PSB
protonated Schiff base
- HsHR
HR from H. salinarum
- NpHR
HR from N. pharaonis
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00104.
Examples of peak fitting for both UV-vis data and RRS data (Figures S1, S2, S4, and S5) and a comparison of RRS spectra taken with high and low laser powers and with 532 nm excitation (Figure S2) (PDF)
References
- 1.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyden ES. Optogenetics and the future of neuroscience. Nat Neurosci. 2015;18:1200–1201. doi: 10.1038/nn.4094. [DOI] [PubMed] [Google Scholar]
- 3.LaLumiere RT. A new technique for controlling the brain: optogenetics and its potential for use in research and the clinic. Brain Stimul. 2011;4:1–6. doi: 10.1016/j.brs.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Schoenenberger P, Scharer YP, Oertner TG. Channelrhodopsin as a tool to investigate synaptic transmission and plasticity. Exp Physiol. 2011;96:34–39. doi: 10.1113/expphysiol.2009.051219. [DOI] [PubMed] [Google Scholar]
- 5.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O’Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, Deisseroth K. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2011;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Han X. In vivo application of optogenetics for neural circuit analysis. ACS Chem Neurosci. 2012;3:577–584. doi: 10.1021/cn300065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doroudchi MM, Greenberg KP, Liu J, Silka KA, Boyden ES, Lockridge JA, Arman AC, Janani R, Boye SE, Boye SL, Gordon GM, Matteo BC, Sampath AP, Hauswirth WW, Horsager A. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol Ther. 2011;19:1220–1229. doi: 10.1038/mt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 11.Schoonheim PJ, Arrenberg AB, Del Bene F, Baier H. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J Neurosci. 2010;30:7111–7120. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridge KD. Algal rhodopsins: phototaxis receptors found at last. Curr Biol. 2002;12:R588–590. doi: 10.1016/s0960-9822(02)01099-0. [DOI] [PubMed] [Google Scholar]
- 14.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 15.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 17.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, Hegemann P, Maturana AD, Ishitani R, Deisseroth K, Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaguma A, Tsukamoto H, Kato HE, Kimura T, Ishizuka T, Oishi S, Yawo H, Nureki O, Furutani Y. Chimeras of channelrhodopsin-1 and –2 from Chlamydomonas reinhardtii exhibit distinctive light-induced structural changes from channelrhodopsin-2. J Biol Chem. 2015;290:11623–11634. doi: 10.1074/jbc.M115.642256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Gating mechanisms of a natural anion channelrhodopsin. Proc Natl Acad Sci U S A. 2015;112:14236–14241. doi: 10.1073/pnas.1513602112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogren JI, Mamaev S, Russano D, Li H, Spudich JL, Rothschild KJ. Retinal chromophore structure and Schiff base interactions in red-shifted channelrhodopsin-1 from Chlamydomonas augustae. Biochemistry. 2014;53:3961–3970. doi: 10.1021/bi500445c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saint Clair EC, Ogren JI, Mamaev S, Russano D, Kralj JM, Rothschild KJ. Near-IR resonance Raman spectroscopy of archaerhodopsin 3: effects of transmembrane potential. J Phys Chem B. 2012;116:14592–14601. doi: 10.1021/jp309996a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SO, Braiman MS, Myers AB, Pardoen JA, Courtin JML, Winkel C, Lugtenburg J, Mathies RA. Vibrational analysis of the all-trans-retinal chromophore in light-adapted bacteriorhodopsin. J Am Chem Soc. 1987;109:3108–3125. [Google Scholar]
- 25.Nack M, Radu I, Bamann C, Bamberg E, Heberle J. The retinal structure of channelrhodopsin-2 assessed by resonance Raman spectroscopy. FEBS Lett. 2009;583:3676–3680. doi: 10.1016/j.febslet.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 26.Smith SO, Hornung I, Van der Steen R, Pardoen JA, Braiman MS, Lugtenburg J, Mathies RA. Are C14-C15 single bond isomerizations of the retinal chromophore involved in the proton-pumping mechanism of bacteriorhodopsin? Proc Natl Acad Sci U S A. 1986;83:967–971. doi: 10.1073/pnas.83.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SO, Lugtenburg J, Mathies RA. Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J Membr Biol. 1985;85:95–109. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- 28.Smith SO, Pardoen JA, Lugtenburg J, Mathies RA. Vibrational analysis of the 13-cis-retinal chromophore in dark-adapted bacteriorhodopsin. J Phys Chem. 1987;91:804–819. [Google Scholar]
- 29.Kralj JM, Spudich EN, Spudich JL, Rothschild KJ. Raman spectroscopy reveals direct chromophore interactions in the Leu/Gln105 spectral tuning switch of proteorhodopsins. J Phys Chem B. 2008;112:11770–11776. doi: 10.1021/jp802629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergo V, Amsden JJ, Spudich EN, Spudich JL, Rothschild KJ. Structural changes in the photoactive site of proteorhodopsin during the primary photoreaction. Biochemistry. 2004;43:9075–9083. doi: 10.1021/bi0361968. [DOI] [PubMed] [Google Scholar]
- 31.Bergo V, Spudich EN, Spudich JL, Rothschild KJ. A Fourier transform infrared study of Neurospora rhodopsin: similarities with archaeal rhodopsins. Photochem Photobiol. 2002;76:341–349. doi: 10.1562/0031-8655(2002)076<0341:aftiso>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Bergo V, Spudich EN, Spudich JL, Rothschild KJ. Conformational Changes Detected in a Sensory Rhodopsin II-Transducer Complex. J Biol Chem. 2003;278:36556–36562. doi: 10.1074/jbc.M303719200. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi T, Yan B, Mazur P, Derguini F, Nakanishi K, Spudich JL. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II) Biochemistry. 1990;29:8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- 34.Chizhov I, Schmies G, Seidel R, Sydor JR, Luttenberg B, Engelhard M. The photophobic receptor from Natronobacterium pharaonis: temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys J. 1998;75:999–1009. doi: 10.1016/S0006-3495(98)77588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Argade PV, Rothschild KJ. Quantitative Analysis of Resonance Raman Spectra of Purple Membrane from Halobacterium halobium: L550 intermediate. Biochemistry. 1983;22:3460–3466. [Google Scholar]
- 36.Fodor SP, Pollard WT, Gebhard R, van den Berg EM, Lugtenburg J, Mathies RA. Bacteriorhodopsin’s L550 intermediate contains a C14-C15 s-trans-retinal chromophore. Proc Natl Acad Sci U S A. 1988;85:2156–2160. doi: 10.1073/pnas.85.7.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothschild KJ, Andrew J, DeGrip WJ, Stanley HE. Opsin Structure Probed by Raman Spectroscopy of Photorecptor Membranes. Science. 1976;191:1176–1178. doi: 10.1126/science.1257742. [DOI] [PubMed] [Google Scholar]
- 38.Smith SO, Myers AB, Mathies RA, Pardoen JA, Winkel C, van den Berg EM, Lugtenburg J. Vibrational analysis of the all-trans retinal protonated Schiff base. Biophys J. 1985;47:653–664. doi: 10.1016/S0006-3495(85)83961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baasov T, Friedman N, Sheves M. Factors affecting the C:N stretching in protonated retinal Schiff base: a model study for bacteriorhodopsin and visual pigments. Biochemistry. 1987;26:3210–3217. doi: 10.1021/bi00385a041. [DOI] [PubMed] [Google Scholar]
- 40.Lewis A, Spoonhower J, Bogomolni RA, Lozier RH, Stoeckenius W. Tunable laser resonance Raman spectroscopy of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1974;71:4462–4466. doi: 10.1073/pnas.71.11.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter TJ, Braiman MS. Anion-Protein Interactions during Halorhodopsin Pumping: Halide Binding at the Protonated Schiff Base. Biochemistry. 1994;33:1724–1733. doi: 10.1021/bi00173a015. [DOI] [PubMed] [Google Scholar]
- 42.Gerscher S, Mylrajan M, Hildebrandt P, Baron MH, Muller R, Engelhard M. Chromophore-anion interactions in halorhodopsin from Natronobacterium pharaonis probed by time-resolved resonance Raman spectroscopy. Biochemistry. 1997;36:11012–11020. doi: 10.1021/bi970722b. [DOI] [PubMed] [Google Scholar]
- 43.Parker FS. Applications of infrared, Raman and resonance Raman spectroscopy in biochemistry. Plenum Press; New York: 1983. [Google Scholar]
- 44.Eyring G, Curry B, Mathies R, Fransen R, Palings I, Lugtenburg J. Interpretation of the resonance Raman spectrum of bathorhodopsin based on visual pigment analogues. Biochemistry. 1980;19:2410–2418. doi: 10.1021/bi00552a020. [DOI] [PubMed] [Google Scholar]
- 45.Braiman M, Mathies R. Resonance Raman spectra of bacteriorhodopsin’s primary photoproduct: evidence for a distorted 13-cis retinal chromophore. Proc Natl Acad Sci U S A. 1982;79:403–407. doi: 10.1073/pnas.79.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothschild KJ, Marrero H, Braiman M, Mathies R. Primary photochemistry of bacteriorhodopsin: comparison of Fourier transform infrared difference spectra with resonance Raman spectra. Photochem Photobiol. 1984;40:675–679. doi: 10.1111/j.1751-1097.1984.tb05359.x. [DOI] [PubMed] [Google Scholar]
- 47.Aton B, Doukas AG, Callender RH, Becher B, Ebrey TG. Resonance Raman studies of the purple membrane. Biochemistry. 1977;16:2995–2999. doi: 10.1021/bi00632a029. [DOI] [PubMed] [Google Scholar]
- 48.Sineshchekov OA, Govorunova EG, Wang J, Spudich JL. Enhancement of the long-wavelength sensitivity of optogenetic microbial rhodopsins by 3,4-dehydroretinal. Biochemistry. 2012;51:4499–4506. doi: 10.1021/bi2018859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sineshchekov OA, Li H, Govorunova EG, Spudich JL. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc Natl Acad Sci U S A. 2016;113:E1993–E2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rath P, Marti T, Sonar S, Khorana HG, Rothschild KJ. Hydrogen Bonding Interactions With the Schiff Base of Bacteriorhodopsin: Resonance Raman Spectroscopy of the Mutants D85N and D85A. J Biol Chem. 1993;268:17742–17749. [PubMed] [Google Scholar]
- 51.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorenz-Fonfria VA, Resler T, Krause N, Nack M, Gossing M, Fischer von Mollard G, Bamann C, Bamberg E, Schlesinger R, Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci U S A. 2013;110:E1273–1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, Magnuson J, Hegemann P, Deisseroth K. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou SY, Govorunova EG, Ntefidou M, Lane CE, Spudich EN, Sineshchekov OA, Spudich JL. Diversity of Chlamydomonas channelrhodopsins. Photochem Photo-biol. 2012;88:119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Govorunova EG, Sineshchekov OA, Spudich JL. Proteomonas sulcata ACR1: A Fast Anion Channelrhodopsin. Photochem Photobiol. 2016;92:257–263. doi: 10.1111/php.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou SY. M.S. Thesis. The University of Texas; Houston: 2012. Homology Cloning, Heterologous Expression and Characterization of a New Channelrhodopsin. [Google Scholar]
- 57.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 58.Pande C, Lanyi JK, Callender RH. Effects of various anions on the Raman spectrum of halorhodopsin. Biophys J. 1989;55:425–431. doi: 10.1016/S0006-3495(89)82836-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter TJ, Braiman MS. FI-IR difference spectroscopy of halorhodopsin in the presence of different anions. In: Rigaud JL, editor. Structure and Functions of Retinal Proteins. 1992. pp. 233–236. [Google Scholar]
- 60.Song Y, Gunner MR. Halorhodopsin pumps Cl-and bacteriorhodopsin pumps protons by a common mechanism that uses conserved electrostatic interactions. Proc Natl Acad Sci U S A. 2014;111:16377–16382. doi: 10.1073/pnas.1411119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelemen L, Galajda P, Szaraz S, Ormos P. Chloride ion binding to bacteriorhodopsin at low pH: an infrared spectroscopic study. Biophys J. 1999;76:1951–1958. doi: 10.1016/S0006-3495(99)77354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Govorunova EG, Sineshchekov OA, Spudich JL. Role of a helix B lysine residue in the photoactive site in channelrhodopsins. Biophys J. 2014;106:1607–1617. doi: 10.1016/j.bpj.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogren JI, Yi A, Mamaev S, Li H, Spudich JL, Rothschild KJ. Proton Transfers in a Channelrhodopsin-1 Studied by Fourier Transform Infrared (FTIR) Difference Spectroscopy and Site-directed Mutagenesis. J Biol Chem. 2015;290:12719–12730. doi: 10.1074/jbc.M114.634840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogren JI, Yi A, Mamaev S, Li H, Lugtenburg J, DeGrip WJ, Spudich JL, Rothschild KJ. Comparison of the Structural Changes Occurring during the Primary Phototransition of Two Different Channelrhodopsins from Chlamydomonas Algae. Biochemistry. 2015;54:377–388. doi: 10.1021/bi501243y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rath P, Krebs MP, He Y, Khorana HG, Rothschild KJ. Fourier transform Raman spectroscopy of the bacteriorhodopsin mutant Tyr-185– > Phe: formation of a stable O-like species during light adaptation and detection of its transient N-like photoproduct. Biochemistry. 1993;32:2272–2281. doi: 10.1021/bi00060a020. [DOI] [PubMed] [Google Scholar]
- 66.Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 67.Inoue K, Reissig L, Sakai M, Kobayashi S, Homma M, Fujii M, Kandori H, Sudo Y. Absorption spectra and photochemical reactions in a unique photoactive protein, middle rhodopsin MR. J Phys Chem B. 2012;116:5888–5899. doi: 10.1021/jp302357m. [DOI] [PubMed] [Google Scholar]
- 68.Gellini C, Luttenberg B, Sydor J, Engelhard M, Hildebrandt P. Resonance Raman spectroscopy of sensory rhodopsin II from Natronobacterium pharaonis. FEBS Lett. 2000;472:263–266. doi: 10.1016/s0014-5793(00)01472-1. [DOI] [PubMed] [Google Scholar]
- 69.Nilsson A, Rath P, Olejnik J, Coleman M, Rothschild KJ. Protein conformational changes during the bacteriorhodopsin photocycle. A Fourier transform infrared/resonance Raman study of the alkaline form of the mutant Asp-85– > Asn. J Biol Chem. 1995;270:29746–29751. doi: 10.1074/jbc.270.50.29746. [DOI] [PubMed] [Google Scholar]
- 70.Smith SO, Mathies RA. Resonance Raman spectra of the acidified and deionized forms of bacteriorhodopsin. Biophys J. 1985;47:251–254. doi: 10.1016/s0006-3495(85)83899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kouyama T, Kanada S, Takeguchi Y, Narusawa A, Murakami M, Ihara K. Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis. J Mol Biol. 2010;396:564–579. doi: 10.1016/j.jmb.2009.11.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


