Significance
Viruses often carry genes acquired from their host. In the present work, we show that a virus of a marine alga carries a gene encoding a transporter protein that mediates nutrient uptake. We confirm that the viral transporter protein is expressed during infection and show that the protein functions to take up sources of nitrogen. This is important because acquisition of nutrients often determines the ecological success of phytoplankton populations. This work demonstrates how a virus can amend host–viral dynamics by modulating acquisition of nutrients from the environment.
Keywords: Phycodnaviridae, NCLDV, prasinophytes, Mamiellophyceae, lateral gene transfer
Abstract
Phytoplankton community structure is shaped by both bottom–up factors, such as nutrient availability, and top–down processes, such as predation. Here we show that marine viruses can blur these distinctions, being able to amend how host cells acquire nutrients from their environment while also predating and lysing their algal hosts. Viral genomes often encode genes derived from their host. These genes may allow the virus to manipulate host metabolism to improve viral fitness. We identify in the genome of a phytoplankton virus, which infects the small green alga Ostreococcus tauri, a host-derived ammonium transporter. This gene is transcribed during infection and when expressed in yeast mutants the viral protein is located to the plasma membrane and rescues growth when cultured with ammonium as the sole nitrogen source. We also show that viral infection alters the nature of nitrogen compound uptake of host cells, by both increasing substrate affinity and allowing the host to access diverse nitrogen sources. This is important because the availability of nitrogen often limits phytoplankton growth. Collectively, these data show that a virus can acquire genes encoding nutrient transporters from a host genome and that expression of the viral gene can alter the nutrient uptake behavior of host cells. These results have implications for understanding how viruses manipulate the physiology and ecology of phytoplankton, influence marine nutrient cycles, and act as vectors for horizontal gene transfer.
Phytoplankton underpin the biogeochemistry of the surface oceans and drive the marine carbon and nitrogen cycles (1). Nutrients fuel cyanobacteria and eukaryotic single-celled algae with the elements essential for organic matter biosynthesis, especially nitrogen (N) and phosphorus (P); hence nutrient availability exerts a bottom–up control on phytoplankton cell growth and oceanic productivity (2, 3). In particular, N and P are found in low concentrations over much of the open ocean, limiting phytoplankton growth rates. Nutrient limitation, including colimitation by several nutrients, results in competition among phytoplankton. In oligotrophic environments, and transiently nutrient-depleted environments, phytoplankton species have evolved a range of strategies to optimize nutrient acquisition (4). The genetic repertoire of N and P transporters shows evidence of gene duplication, differential loss, and horizontal gene transfer (HGT) in phytoplankton genomes as well as contrasting gene expression levels (5–9), suggesting that the evolution of these genes has been driven by adaptation to environmental limitation. Ammonium () and nitrate () are commonly available N sources for marine phytoplankton (10, 11) and, as such, the gene repertoires of cyanobacterial and eukaryotic phytoplankton are configured toward utilization of these two forms of inorganic N sources (12).
In addition to bottom–up nutrient limitation, phytoplankton community structures are influenced by top–down controls (13). For phytoplankton communities these include predation by grazers and viral infection (e.g., ref. 14). Marine viruses are the most abundant biological entities in the oceans; they are estimated to induce 1023 infections daily (15) and are thought to control phytoplankton abundance, biomass, and species composition through taxon-specific infections (16, 17). There is a growing appreciation of the role of viruses in oceanic nutrient cycles, based on lytic infections leading to the release of dissolved and particulate organic matter, which is then recycled by other microorganisms (15, 18). This virus-mediated process, referred to as the viral shunt, was suggested as a mechanism that maintains availability of organic matter in the euphotic zone by lysing cells before they sink (15, 18). However, the influence of viruses on oceanic ecosystems extends beyond top–down host mortality. Genomic and metagenomic analyses show that marine viruses—either phages or eukaryotic viruses—harbor host-derived genes encoding a diverse range of putative functions (19–21), including whole biochemical pathways (22). These “auxiliary metabolic genes” (AMGs) may allow the virus to manipulate the host via metabolic reprogramming during infection and have been experimentally shown to alter the central carbon metabolism and pigment biosynthesis of cyanobacteria (23, 24) and to “reprogram” lipid biosynthesis of eukaryotic algal cells (25–27). AMGs are thought to modulate host function to improve fitness of the virus and, in some cases, temporarily the host. For example, it was hypothesized that virally encoded putative phosphate transporters increase accumulation of P in host cells (28, 29), which may in turn increase virus fitness given that P-depleted phytoplankton cells limit viral proliferation (e.g., refs. 30 and 31).
Given the evidence of HGT for genes involved in both N and P metabolisms (7, 9, 32), the presence of host-derived phosphate transporters in phytoplankton viral genomes (29), and the importance of nutrient availability for phytoplankton and viral replication (33), we hypothesize that functional N transporters would also be found in genomes of phytoplankton viruses. Indeed, a recent analysis of marine viral metagenomes extended the catalog of functions encoded by AMGs and reported the presence of genes putatively encoding transporters in metagenomic assemblies which harbored phage genes (21). To confirm the existence of potential virus-mediated N-uptake processes, we searched available viral genomes for the presence of N transporters. Here we report the identification of a host-derived N transporter harbored by an algal virus, OtV6. This virus infects the green alga Ostreococcus tauri and we show that the viral transporter is transcribed during the infection cycle. Cloning and phenotype analysis in yeast demonstrate that the viral protein transports , methylammonium, and potentially a range of alternative N sources and that the viral transporter mediates a higher rate of methylammonium uptake at low environmental concentrations compared with the O. tauri homolog. Algal culture experiments show viral infection alters host nutrient uptake dynamics.
Results
OtV6 Genome Harbors a Putative Transporter.
To identify viral transporter proteins putatively involved in N uptake, all available viral amino acid sequences were screened using similarity searches based on hidden Markov models (HMM) encompassing the main N transporter protein families. These HMM searches discovered a single viral protein potentially involved in direct N uptake. This viral protein sequence [UniProtKB (34) identifier: H8ZJB2] generated a significant hit with the Amt/Mep/Rh superfamily HMM (SI Appendix, Fig. S1A). Members of this superfamily are integral membrane proteins involved in the electrogenic transport of ammonium ions, either the direct transport of or ammonia (NH3)/H+ cotransport (35). Several proteins from this superfamily have been shown to mediate the uptake of methylammonium, which can be used as a radiolabeled tracer (14CH3) to infer uptake rates (36). The viral transporter identified is encoded in the genome of OtV6, a virus belonging to the Phycodnaviridae family of nucleocytoplasmic large dsDNA viruses [NCLDV (37)]. We name this viral putative transporter vAmt (viral ammonium transporter).
Phycodnaviridae infect a broad range of eukaryotic algae (38). To date, 12 genome sequences of viruses infecting the prasinophyte alga Ostreococcus—a widely distributed marine Mamiellophyceae (39, 40) and the smallest known free-living eukaryotic cell—are available (41–46). These include viruses that infect one of three species: O. tauri, Ostreococcus lucimarinus, and Ostreococcus mediterraneus. OtV6 infects a distinct population of O. tauri [shown to be resistant to another virus, OtV5 (44)], an alga originally isolated from a coastal northwest Mediterranean lagoon (47).
OtV6 Is Evolutionarily Distinct from Other Ostreococcus Viruses.
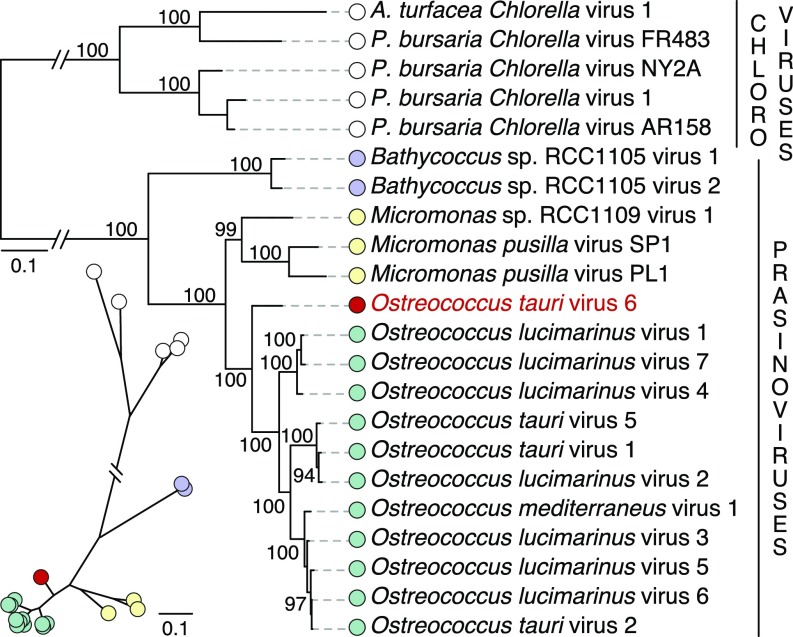
To determine the phylogenetic position of OtV6 among the Phycodnaviridae that infect green algae, we used the OtV6 genomic data (44) for a maximum-likelihood (ML) phylogenetic analysis. The ML tree reconstruction was based on a concatenated alignment of 22 conserved protein sequences (46) with a sampling of 7,668 sites. In the resulting ML phylogeny, OtV6 branched at the base of all other Ostreococcus viruses (Fig. 1); both the basal position of OtV6 and the clustering of all other Ostreococcus viruses in a single clade were strongly supported (100% bootstrap support). This intermediate phylogenetic position was also found in a ML phylogenetic tree of the viral DNA polymerase B (915 sites; SI Appendix, Fig. S2) a gene commonly used as a marker for NCLDV phylogenetic and diversity analyses (48–50).
Fig. 1.
OtV6 branches basal to all other available Ostreococcus spp. viral genomes. ML phylogenetic tree of green algal viruses is inferred from a concatenated sequence alignment of 22 core proteins shared among these viruses (7,668 sites) under the LG+G+F model. The unrooted version of this tree is presented below the midpoint-rooted tree. A red circle indicates OtV6 branch; other colored circles represent the taxonomy of the viral hosts: green (Ostreococcus), purple (Bathycoccus), yellow (Micromonas), and white (chloroviruses, viruses of Chlorella, as outgroup clade). Node support was calculated from 1,000 nonparametric bootstrap replicates; only bootstrap values >90% are shown. (Scale bar represents the number of estimated substitutions per site.) The branch connecting the prasinoviruses to the chloroviruses was truncated for display.
The phylogenies of both the viral core protein set and DNA polymerase B trees demonstrate that OtV6 is positioned almost equidistant between the sampled members of the Ostreococcus virus and Micromonas virus clades. By comparing the amino acid conservation levels of the 250 ORF sequences of OtV6 with those of the other 11 Ostreococcus viruses and 3 Micromonas viruses, we found that 20% of OtV6 ORFs were more similar to Micromonas virus homologs than to ones from Ostreococcus viruses (SI Appendix, Fig. S3). In addition, 13 OtV6 ORFs had homologs in the genomes of Micromonas viruses, which were absent from all other Ostreococcus viruses. Given current sampling of Phycodnaviridae genomes, these results suggest that OtV6 represents an intermediate prasinovirus lineage.
vAmt Is Virally Encoded and Expressed During Infection.
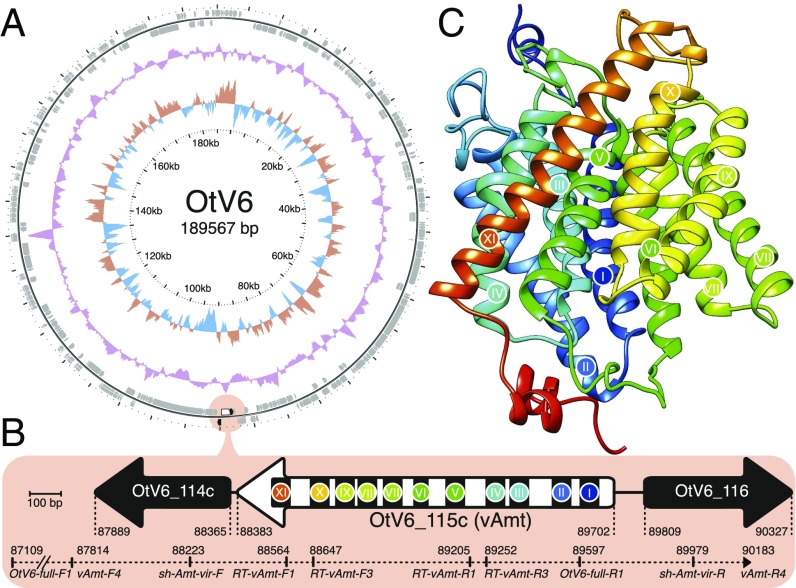
Sequence searches and ML phylogenetic tree reconstructions confirmed the viral provenance of both vAmt flanking genes (OtV6_114c and OtV6_116; SI Appendix, Fig. S4). To rule out contamination and genome assembly artifacts, we confirmed the presence of the vAmt-encoding gene (OtV6_115c) on the OtV6 genome by targeted PCR amplification (Fig. 2 A and B). Three sets of PCR primers were designed to amplify through the complete vAmt-encoding gene and its 5′ and 3′ flanking genes (Fig. 2B and SI Appendix, Table S1; GenBank identifier: KX254356). These results confirm that the transporter-encoding gene is linked to the viral genes OtV6_114c and OtV6_116 and therefore residing on the viral genome. In addition, no spliceosomal introns were identified in the vAmt-encoding gene, while all Ostreococcus spp. homologs have one intron with the exception of Ostreococcus sp. RCC809, which has no introns (SI Appendix, Fig. S1B).
Fig. 2.
vAmt genomic context and putative protein structure. (A) Circular depiction of the OtV6 genome. Oriented ORFs are mapped in gray on the outer ring. The light red circle indicates the vAmt locus along with neighboring ORFs. The colored plots drawn as intermediate rings display the biases in dinucleotide uses across the OtV6 genome: The red and blue plot shows the A+T skew, and the purple plot shows the G+C content. (B) vAmt locus and neighboring ORFs. A white arrow represents the vAmt ORF (OtV6_115c), and black arrows represent the 5′ and 3′ vAmt flanking ORFs (OtV6_114c and OtV6_116, respectively; SI Appendix, Fig. S4 displays the phylogenies of these flanking ORFs demonstrating viral provenance). The 11 predicted transmembrane domains along the vAmt ORF are indicated by black rectangles with colored circles and numbered with Roman numerals. ORF lengths were scaled according to the 100-bp scale bar. Genomic coordinates indicate the 5′ and 3′ boundaries of each depicted ORF. The PCR-amplified region is represented by a dashed line along with the corresponding amplicon genomic coordinates. (C) vAmt protein structure prediction. The predicted protein structure is composed of 11 helices corresponding to the transmembrane domains indicated by colored circles and Roman numerals, as in B (see SI Appendix, Fig. S6 for sequence and structural comparisons of both host and viral copies).
Next we sought to confirm that the vAmt-encoding gene is expressed during infection. We isolated RNA from infected and uninfected O. tauri cultures in parallel. Two different sets of reverse transcription (RT)-PCR primers directed against the vAmt-encoding transcript (RT-vAmt primer sets; Fig. 2B and SI Appendix, Table S1) amplified products in the infected cultures; no amplicons were found in the uninfected cultures (SI Appendix, Fig. S5). We sequenced both RT-PCR amplicons from the infected cultures and confirmed that they corresponded to the vAmt-encoding gene and that this viral gene is expressed during infection.
vAmt Is an Transporter.
The vAmt ORF is located on the reverse strand of the OtV6 genome (Fig. 2A) and encodes a putative protein of 439 aa. Sequence similarity searches of UniProtKB demonstrate the Amt1.1 sequence from the OtV6 host O. tauri (UniProtKB identifier: A0A096PA30) has the highest similarity with 75.8% amino acid similarity and 62.3% nucleotide identity (SI Appendix, Fig. S6A). In the curated SwissProt database (34), the best match to the viral protein was Arabidopsis thaliana Amt1.3 sequence (Q9SQH9; 65.9% amino acid similarity with vAmt), which was shown to mediate uptake in N-replete and N-deplete conditions (51).
The vAmt predicted protein secondary structure has 11 transmembrane domains, a structural feature shared by other transporter proteins including the O. tauri Amt1.1 (Fig. 2B and SI Appendix, Figs. S6A and S7). These transmembrane domains corresponded to 11 -helices, which have been shown for several transporters to cross the membrane making up a conserved hydrophobic pore and to contribute to the overall channel stability of the transporter, as revealed by crystal structures (52–54). In addition, the vAmt has a predicted extracellular N-terminal and cytosolic C-terminal topology, a topology also found for eukaryotic Amt proteins (35) (SI Appendix, Fig. S7; see SI Appendix, Fig. S1C for comparison of Amt homolog C termini). Furthermore, vAmt possesses another hallmark of transporters: two conserved histidine residues in the hydrophobic pore consistently found in helices V and X (SI Appendix, Fig. S6A) and shown to be essential for transport activity (55). In addition to the high level of sequence conservation between the vAmt and its O. tauri Amt1.1 homolog, protein structures inferred from viral (Fig. 2C) and host (SI Appendix, Fig. S6B) homologs showed a high level of structural similarity (SI Appendix, Fig. S6C).
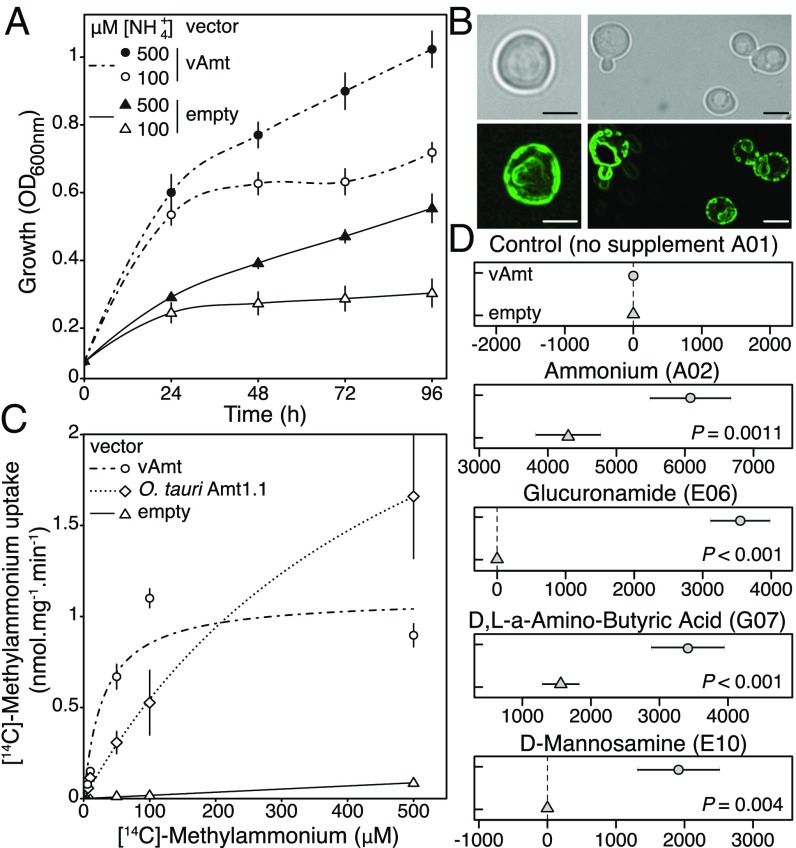
To confirm the vAmt is a functional transporter, we cloned the vAmt-encoding gene (pAG416 GPD vAmt) and transformed it into the yeast Saccharomyces cerevisiae mutant 31019b (56), which has the three known native yeast Mep transporters deleted (mep1mep2mep3). We compared growth of this mutant with parallel cultures of the same strain but containing only an empty vector (pAG416 GPD) and under culture conditions where 100 M or 500 M was available as the sole N source. Complementation of the yeast mutant with vAmt increased growth rate and culture density, consistent with vAmt encoding a functional transporter (Fig. 3A and SI Appendix, Fig. S8A). For comparison we showed that addition of pAG416 GPD vAmt did not facilitate growth of the YNVW1 S. cerevisiae mutant (57), which has its urea transporter dur3 gene deleted (dur3), on medium containing 100 M or 500 M urea as sole N source (SI Appendix, Fig. S8), indicating that vAmt does not transport urea in addition to . GFP tagging of either the vAmt C or N termini identifies a plasma membrane localization, with some additional patchy localization, in yeast (Fig. 3B). Similar patterns of localization have been reported for plant Amt transporters, when expressed and tagged in yeast (58).
Fig. 3.
vAmt is a functional transporter that takes up alternative substrates, is localized to the cell membrane, and mediates an uptake at higher rate for low substrate concentrations than Ostreococcus Amt1.1 homolog when expressed in yeast. (A) Culture optical density (OD600nm) of an uptake defective yeast mutant (strain 31019b) transformed either with an empty vector or with a vAmt-containing vector in two concentrations (100 M and 500 M). Error bars represent SEs based on culture triplicates. The lines represent local polynomial regression fits (see SI Appendix, Fig. S8 for the equivalent results of an additional experiment with O. tauri Amt1.1). Growth of the empty vector transformed mutant is facilitated by passive diffusion of across the yeast membrane and/or scavenging of intracellular N stocks. (B) vAmt GFP fusion proteins expressed in yeast, cloned in frame in N terminus (Left) or in C terminus (Right). (Scale bars, 3-m distances.) (C) [14C]-Methylammonium uptake rates in mutant strain 31019b complemented with vAmt or the O. tauri Amt1.1 homolog. Lines show Michaelis–Menten curves; error bars represent SEs based on culture triplicates. (D) Omnilog comparison of mean point estimates and their 95% confidence intervals for N sources showing significant increase in respiration (area under the curve) of vAmt-transformed 31019b yeast cultures compared with non–vAmt-transformed 31019b cultures; data were normalized by subtracting the negative control. See SI Appendix, Fig. S8 for complementation experiments confirming culture growth phenotypes in all of these results apart from D,L-a-amino butyric acid where complementation is not confirmed; see SI Appendix, Fig. S9 for OmniLog Phenotype Microarray respiration curves.
Comparison of radiolabeled methylammonium uptake rates in yeast mutants (strain 31019b) complemented with either the vAmt-encoding or the O. tauri Amt1.1-encoding genes demonstrated that the viral variant encodes a protein mediating the uptake of methylammonium with a higher rate at lower concentrations (e.g., 50–100 M) compared with its O. tauri homologous protein which showed an improved uptake rate at higher methylammonium concentrations (e.g., 500 M; Fig. 3C). Strikingly, these analyses demonstrate that both transporter proteins have distinct substrate affinities and kinetics; specifically, O. tauri Amt1.1 transporter has a of 520 M (250 SE; = 3.38 0.91 nmolmgmin−1) and the vAmt has a of 30 M (10; = 1.1 0.1 nmolmgmin−1). These distinct properties of host and viral transporters indicate that production of the vAmt protein during infection has the potential to alter the N-uptake dynamics of infected cells.
Using the OmniLog system (59), we investigated the range of N substrates that showed an increased respiration rate phenotype in yeast. Comparisons of the -uptake–deficient 31019b strain carrying pAG416 GPD vAmt with 31019b carrying only an empty vector (pAG416 GPD) were used to identify potential alternative substrates of the vAmt transporter that resulted in increased respiratory rates. In addition to , this phenotype assay identified a significant increase in respiratory rate on three alternative N sources: D,L-a-amino butyric acid, glucuronamide, and D-mannosamine (Fig. 3D and SI Appendix, Fig. S9). Two of these three alternative vAmt substrates were further confirmed by functional complementation experiments in yeast cultures (SI Appendix, Fig. S8). We note that this method likely underestimates the range of substrates transported by vAmt because alternative substrates may have no “metabolic” effect in yeast but could be used in other cellular backgrounds such as O. tauri.
Viral Infection Alters the Uptake Rate of Methylammonium by Ostreococcus Cells.
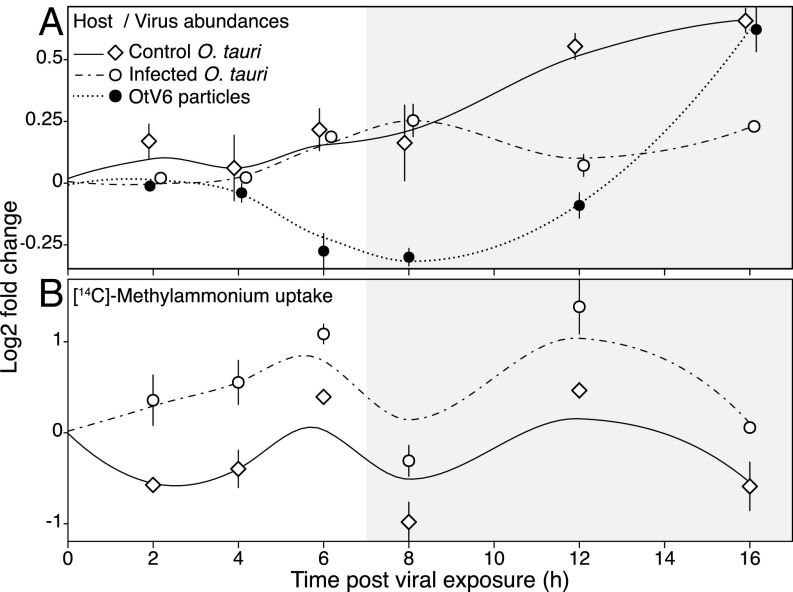
Next we carried out infection experiments of O. tauri cultures with OtV6, to determine whether infection and expression of the vAmt-encoding gene altered the uptake rates of the host alga (relative to noninfected, control cultures; n = 3). To this aim, we conducted an infection time-course experiment. For 16 h postinfection (hpi), we monitored O. tauri and virus-like particle (VLP) abundances [enumerated by flow cytometry (FCM)] and methylammonium uptake rates (Fig. 4 and SI Appendix, Tables S2 and S3).
Fig. 4.
Nitrogen source uptake during viral infection. Temporal dynamics of host cell and viral particle abundances (A) and [14C]-methylammonium uptake rates (B) in OtV6-infected and noninfected O. tauri cultures. Three O. tauri cultures were exposed to OtV6 (time point 0 hpi) and three other noninfected cultures were used as controls. For each panel, (log2)-fold changes are based on time point 0 hpi (i.e., time of viral inoculation for infected O. tauri cultures); plots summarize the fold change distribution of three cultures for a given time point during the experiment, and trend lines were estimated by local polynomial regressions (loess). Error bars represent SEs. Shaded overlays indicate incubator dark period (“night”). O. tauri cells and virus-like particles were enumerated using flow cytometry; uptake rates were estimated by tracing the isotope-labeled [14C]-methylammonium in cultures taken as subsamples throughout the time-course experiment. See SI Appendix, Tables S2 and S3 for two-sample t-test results.
Based on FCM estimates of cell and VLP abundances, the ratio OtV6 particle to O. tauri cell [as a proxy for multiplicity of infection (60)] at the start of the experiment was 0.42 ± 0.06 SE; algal growth was not significantly affected by viral infection for the first 12 hpi (Fig. 4A). At 12 hpi, temporal change in cell abundance significantly differed between control and infected algal cultures (two-sample t test, t4 = 7.03, P = 0.002; SI Appendix, Table S2), with a sharp decrease of 11.8% on average in abundance for the infected cultures (corresponding to a loss of 0.59 ×107 cells⋅ml−1 ± 0.12 SE); in contrast, the control O. tauri cultures were steadily growing, with an average gain of 33% between 8 hpi and 12 hpi (a gain of 1.44×107 cells⋅ml−1 ± 0.53 SE). This drop in O. tauri cell number in infected cultures indicates occurrences of cell burst and lysis caused by viral shedding. The OtV6 latent period is thus between 8 hpi and 12 hpi, a time frame comparable to that of other prasinoviruses of Ostreococcus and Micromonas spp. (31, 41). FCM enumeration of VLPs confirmed the reduction in VLPs before 12 hpi, consistent with virus adsorption followed by increase in VLPs after 12 hpi, which is in turn consistent with OtV6 particle release (Fig. 4A and SI Appendix, Table S3).
During the infection experiment, we used two methods to track flux in O. tauri cells. First, we determined [14C]-methylammonium uptake rates of standardized subsamples of O. tauri cells throughout the time-course experiments. This analysis demonstrated significant increases in methylammonium uptake rates for infected cells, consistent with viral infection altering the uptake phenotype of the phytoplankton host (Fig. 4B). We also monitored by fluorometry concentration in the cell-free culture media, which showed depletion at 8 hpi in concentration compared with 0 hpi in the infected cultures, although not significant (SI Appendix, Fig. S10 and Table S2).
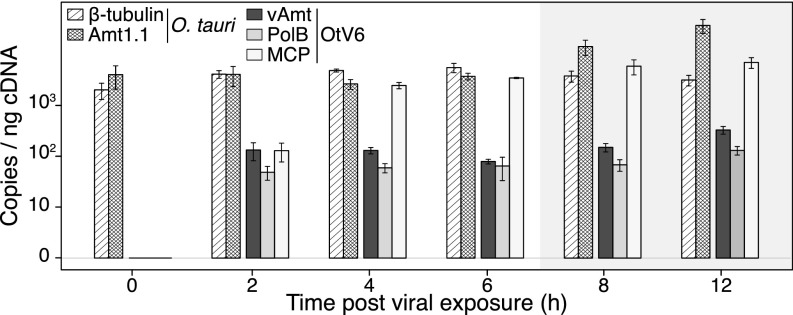
In addition, we used quantitative PCR (qPCR) to quantify the transcript levels of several O. tauri and OtV6 genes (SI Appendix, Table S4) during the latent period of infected cultures (i.e., 0–12 hpi). The qPCR shows that transcription of the vAmt-encoding gene is maintained relatively constant during infection and at a similar level to that of the viral DNA polymerase B gene, while transcription of the gene encoding the major capsid protein increases until 4 hpi and is then expressed at a relatively high level throughout 4–12 hpi (Fig. 5). Taken together, VLP and host cell dynamics, along with gene transcription, all indicate a substantial viral infection of O. tauri cells resulting in an altered methylammonium uptake phenotype.
Fig. 5.
OtV6-infected O. tauri gene expression. Transcript levels were RT-qPCR monitored in infected O. tauri cultures (n = 3; same infected cultures as presented in Fig. 4) and expressed as copies per nanogram of cDNA library (displayed on a log10 scale, y axis). Hatched bars show transcript copy numbers of two O. tauri genes (-tubulin and the transporter Amt1.1); solid color bars show copy numbers for three OtV6 genes (dark gray, viral transporter, vAmt; light gray, DNA polymerase B; white, major capsid protein). Error bars represent SEs. Shaded overlays indicate incubator dark period (night).
OtV6 Acquired vAmt from an Ostreococcus Host.
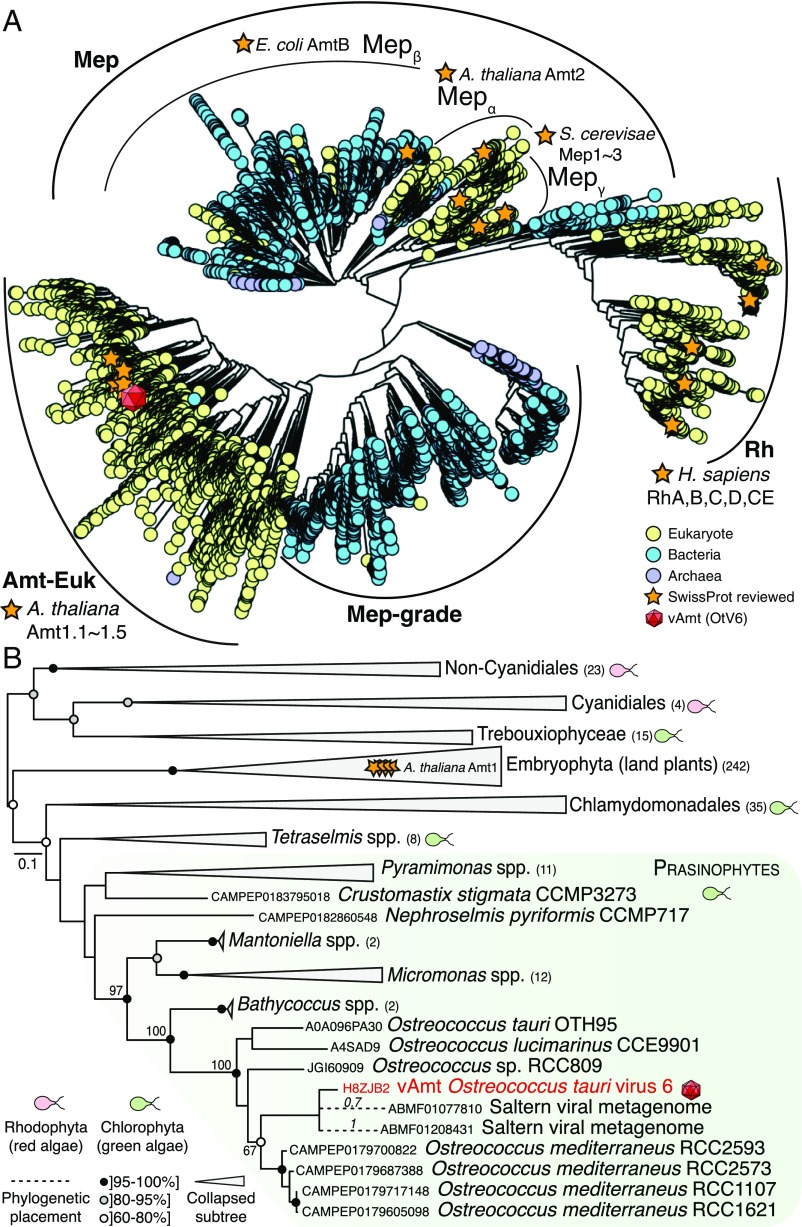
We sought to explore the ancestry of the vAmt-encoding gene by conducting phylogenetic tree reconstruction using a comprehensive sampling of protein sequences of the Amt/Mep/Rh superfamily (Fig. 6A). Proteins of this superfamily are found across all three domains of life and are involved in the transport (uptake and excretion) of ammonium ions through cell membranes (35). This superfamily of membrane proteins has a complex evolutionary history marked by gene duplications, losses, and HGT events, probably driven by environmental selection linked to N availability (7, 32). In a phylogenetic study, McDonald et al. updated the Amt/Mep/Rh superfamily phylogeny (32), showing a partitioning into distinct clades: the monophyletic groups “Amt-Euk” (originally named Amt1) and “Mep,” as well as two distantly related clades, “Rh” (Rhesus) and a cluster of archaeal and bacterial homologs grouped into “Mep-grade.” The Amt-Euk clade is composed of eukaryotic sequences, with a large representation of phytoplankton and plant species. For the host of OtV6, O. tauri, four Amt/Mep/Rh transporters were identified in its genome, two Amt-Euk (eukaryotic origin; Amt1.1 and -1.2) and two Mepα [probably acquired via HGT; Amt2.1 and -2.2 (7)].
Fig. 6.
vAmt phylogenetic ancestry shows it is derived by HGT from the host lineage. (A) Amt/Mep/Rh superfamily phylogenetic tree. This large-scale approximate ML tree was inferred under the WAG+G model. Putative homologs were recruited based on a similarity search using the Pfam HMM corresponding to the Amt/Mep/Rh superfamily (PF00909) against UniRef100 (i.e., nonredundant version of UniProtKB), MMETSP protist transcriptomes, and predicted proteomes from various protist genome projects; the final curated alignment was composed of ∼20,000 protein sequences encompassing 374 sites. Curved black lines indicate the phylogenetic positions of the main clades (Amt-Euk, Mep, Mep-grade, and Rh) as well as Mep subclades (, , and ). The red capsid graphics show the phylogenetic position of the vAmt within the superfamily tree, positioned within the Amt-Euk clade. For contextual reference, orange stars represent SwissProt reviewed protein entries; yellow, blue, and purple circles represent eukaryotic, bacterial, and archaeal proteins, respectively. See SI Appendix, Fig. S11 for additional information, including local support values, MMETSP sequence positions, and scale bar. (B) vAmt evolutionary relationships with Amt-Euk (Amt1) homologs. This ML phylogenetic tree was inferred under the LG+I+G+F model, based on a multiple alignment of 364 proteins totaling 429 sites. These vAmt homologs were recruited and selected based on the Amt/Rh/Mep superfamily phylogenetic tree reconstruction. Green and red cell schematics represent green and red algal lineages. vAmt is highlighted in red with capsid graphics and branched within the prasinophyte green algae, delimited by a green frame. Code numbers in front of species names represent sequence identifiers from the MMETSP transcriptomes (O. mediterraneus, Nephroselmis pyriformis, and Crustomastix stigmata), UniProtKB (O. lucimarinus and O. tauri), and the Ostreococcus sp. RCC809 genome project available at the DoE-Joint Genome Institute. Numbers in parentheses beside clade names are the number of sequences present in collapsed nodes. Branch node supports were computed from 1,000 nonparametric bootstrap replicates. Gray and black circles correspond to bootstrap values of ]80, 95%] and ]95, 100%], respectively. The dashed branches represent the phylogenetic placement of two short environmental sequences, with placement posterior probabilities indicated on their corresponding branches; both sequences originate from a saltern viral metagenome (NCBI BioProject: PRJNA28353), and their GenBank sequence identifiers are provided. See SI Appendix, Fig. S12 for additional data and uncollapsed branch information.
To determine which clade the vAmt groups within, a large-scale, approximate ML phylogenetic tree reconstruction was conducted based on an alignment of all available, nonredundant Amt/Mep/Rh protein sequences, including those from the marine microbial eukaryotic transcriptome sequencing project [MMETSP (61)] and which includes several Ostreococcus transcriptomes such as O. mediterraneus. The resulting Amt/Mep/Rh superfamily phylogeny (based on an alignment of 374 sites; Fig. 6A and SI Appendix, Fig. S11) demonstrates that the vAmt branched within the Amt-Euk clade. To determine the branching position of the vAmt sequence more accurately, a ML phylogenetic tree was reconstructed using a subset of Amt-Euk homologs (364 sequences, alignment composed of 429 sites; Fig. 6B and SI Appendix, Fig. S12). This phylogeny shows the vAmt branching with the Ostreococcus Amt1.1 family radiation, demonstrating that the OtV6 vAmt-encoding gene is derived from its host lineage, most likely via host-to-virus HGT. In addition to the recent identification of putative Amt transporters on phage contigs assembled from marine metagenomes (21), our phylogenetic analyses also demonstrated that two metagenomic sequences retrieved from the viral fraction of aquatic samples [Pacific solar salterns, San Diego (62)] branch with the OtV6 vAmt (Fig. 6B and SI Appendix, Fig. S12), suggesting a wider geographical distribution of viral transporters.
Discussion
Here we demonstrate that a virus of the marine phytoplankton O. tauri, representing a distinct lineage of Ostreococcus virus (Fig. 1), harbors a gene encoding a transporter protein that is expressed during infection (Fig. 5 and SI Appendix, Fig. S5) and functions to take up (Fig. 3A and SI Appendix, Fig. S8A) and a range of alternative fixed N sources (Fig. 3D and SI Appendix, Figs. S8 and S9). Phylogenetic analyses show that the viral transporter is the result of a HGT event and was acquired from the host Ostreococcus lineage (Fig. 6B).
Viral HGT and Functional Compatibility.
Many host-derived AMGs, including the vAmt-encoding gene reported here, are highly similar at the sequence level to their host homologs. This has two implications: First, it complicates the analysis of viral metagenomes, as the process of host-to-virus HGT is likely to “contaminate” viral metagenomes with sequences that appear as though they should belong to larger sample filtration fractions (i.e., the fraction containing host genomes). This can be ameliorated by the use of metagenome assembly methods that control for host-derived genes harbored by viral genomes (21). Second, it confirms that viruses are acquiring genes from their host lineage and therefore acting as vectors for gene transfers. HGT and viral integration of genes that function in how host cells interact with their environment demonstrate a selective scenario that can drive transfer of genes encoding functional traits. Indeed, bacterial genomic islands often contain transporter-encoding genes and have been shown to undergo gene gain at high relative rates [for instance, in the pico-cyanobacterium Prochlorococcus (63)]. Genes that encode transporter proteins are relevant because they encode the proteins responsible for nutrient uptake and are “functionally compatible.” We define a functionally compatible gene as a single gene that encodes a complete trait and also requires little protein–protein interaction network complexity to result in a function (64, 65). As such, these gene classes are easily lost, duplicated, or (re)acquired during the diversification of a lineage, leading to these genes having complex evolutionary histories often involving HGT events (7, 29, 32, 66).
The Role of vAmt During Infection.
Both O. tauri and O. lucimarinus are able to grow with as the sole N source (67). Here we demonstrate that the vAmt-encoding gene harbored by OtV6 is transcribed during viral infection (Fig. 5 and SI Appendix, Fig. S5), and (methylammonium) uptake in infected Ostreococcus cells is enhanced compared with that in uninfected cells (Fig. 4B). Interestingly, expression in a yeast mutant demonstrates that the viral vAmt transporter has a higher affinity (defined here as [14C]-methylammonium uptake rate) at low environmental substrate concentrations compared with the algal transporter homolog, which has a higher relative uptake rate at higher environmental concentrations. Competition for is fierce in many marine systems, given that it is often the preferred source of inorganic N for phytoplankton cells (10). Expression of the vAmt-encoding gene during O. tauri infection may increase host uptake rate and therefore may provide an advantage to infected host cells over noninfected algae in terms of N uptake, allowing an infected population to temporarily outcompete other phytoplankton species, including uninfected sister cells. As such, production of vAmt transporter proteins is likely to act to fulfill the extended N requirements of viral replication within the infected host cell to sustain the physiological burden of viral replication through the uptake of various organic N sources.
Using comparative culture screening, we show that the vAmt protein potentially mediates the uptake of alternative organic N sources in addition to . Amt transporter proteins are known to mediate the uptake of at least one alternative to , methylammonium (68). Most knowledge regarding substrate affinities of protein transporters derives from annotation resulting from sequence homology searches and few experimental studies; it is thus important to take into account, when assessing the significance of transporter proteins in an ecological context, that these proteins may display affinities to additional substrates not described via bioinformatic annotation (69).
Viral Manipulation of Host Elemental Composition.
Harboring genes coding for functional nutrient transporters implies that phytoplankton viruses manipulate the elemental composition of their host cells during infection. This viral stoichiometric alteration would confer a strong advantage to viruses able to interfere with the hosts’ nutrient uptake capabilities by encoding their own transporter repertoires. Availability of nutrients to phytoplankton hosts has been shown to be of critical importance for phytoplankton virus proliferation (33). Infection experiments on phytoplankton cultures depleted in P and N have shown that low concentrations of these nutrients can significantly hamper viral cycle dynamics in terms of latent period and burst size (i.e., the number of viral progeny per lysed cell) as well as decreasing the infectivity of viral progenies; such constraints on viral replication have been shown for various clades of Phycodnaviridae (e.g., refs. 30 and 70), including for a Micromonas virus, a close relative of OtV6 (Fig. 1).
Although most studies have focused on the effect of low P availability on phytoplankton virus replication, recent studies have shown the importance of N availability for the quality of viral replication (31, 70). In particular, the burst size of PBCV1, a virus of the green alga Chlorella, was shown to decrease with higher C:N ratio of the algal host cell (70). Furthermore, the effect of N depletion was shown to be more detrimental to viral proliferation than low P availability for a Phycodnaviridae–phytoplankton system (31). These results are somewhat anticipated because of the stoichiometric requirements of phytoplankton virus replication, which are exacerbated by the strong discrepancy in C:N:P ratios between the viral particle and the host phytoplankton cell (71). Viruses exhibit much lower C:P and C:N ratios than their hosts, as shown for PBCV1’s 17C:5N:1P composition (72), compared with the Redfield ratio of 106C:16N:1P, the elemental composition of an average phytoplankton (71).
In the case of the Phycodnaviridae EhV, a virus of the coccolithophore Emiliania, it was noted that viral genomes were being overproduced during the infection cycle, relative to the number of capsids being produced by the infected phytoplankton (73). Such a discrepancy between genome and capsid productions may be linked to a shortage in amino acids and/or lipids, a constituent of EhV lipid membrane, limiting viral production (73, 74). This is predictable for Phycodnaviridae given the N requirements of PBCV1 capsid, which is composed of ∼5,050 major capsid proteins (MCPs) (75). Based on the MCP protein sequence of PBCV1, the elemental composition of the complete PBCV1 capsid would be C2161H3301N577O657S9, that is, a C:N ratio of 4:1. Hence, the ability of OtV6 to express its gene encoding vAmt and enhance or maintain uptake would allow the host to fulfill the requirements in N imposed by the viral replication, in turn providing a strong fitness advantage to OtV6 relative to marine viruses without a N transporter gene repertoire. Further work will be required to understand the role of virally encoded nutrient transporters in the dynamics of viral replication, especially in contrasting nutrient concentrations.
Blurring Top–Down and Bottom–Up Controls.
Consistent with the idea that viral proteins act to amend host N metabolic function, a putative glutamine synthetase gene has also been identified in the genomes of NCLDVs such as Mimivirus and Mamavirus (76). These data suggest that viral reprogramming of host N metabolism may be a wider phenomenon (21, 77), as has been shown for photosynthetic function (78), sulfur oxidation (79), and lipid (25–27) and phosphate metabolism (80). Evidence that a viral lineage can acquire host genes to amend host nutrient uptake has implications for our understanding of phytoplankton ecology. Specifically, this phenomenon blurs the lines between bottom–up and top–down regulation of phytoplankton communities, because here a top–down viral agent has acquired the host genes which allow it to amend phytoplankton nutrient acquisition from the environment, which is normally considered a bottom–up process.
Materials and Methods
vAmt Identification, Sequence, and Structure Analyses.
To identify a viral transporter sequence putatively involved in uptake, all protein sequences available in UniProtKB (34) and the NCBI nonredundant Reference Sequence database (81) were searched using “hmmsearch,” part of the HMMer v3 software suite (82), with the Pfam (83) HMM corresponding to the transporter superfamily (Pfam release 3.0 identifier: PF00909). Hits were filtered using E-value (1e−10) and gathering cutoffs. This large-scale search identified only a single viral encoded putative transporter protein, OtV6 vAmt (HMMer search statistics with PF00909 model: score, 418.4; E value, 1). To avoid missing additional putative viral transporters—potentially due to inaccurate viral gene modeling—a HMM-based search was repeated on six-frame translations of all viral genomes available at the NCBI genomic sequence repository (ORF minimal size: 60-aa residues). In addition to the Pfam-based HMM searches, further searches using the TIGRfam (84) transporter HMM (TIGR00836; release 15) were conducted using the aforementioned protein sequence datasets. These additional searches did not recover any other viral sequences matching the transporter HMMs.
Transmembrane domains of the vAmt and O. tauri Amt1.1 protein sequences were identified using TMHMM v2 (85). Protein structures were predicted using I-TASSER v4.2 (86); the C scores of the vAmt and O. tauri Amt1.1 3D structure models were 1.36 and 0.72, respectively. O. tauri Amt1.1 and vAmt predicted structures were aligned using MatchMaker [UCSF Chimera (87)].
Phylogenetic Tree Reconstructions.
The Amt/Mep/Rh superfamily phylogeny was reconstructed using an approximate ML method as implemented in FastTree v2.1 (88) (compiled using the double-precision flag) and based on an alignment of 19,493 protein sequences sampled from UniProt100 [clustered at 100% sequence identity (34) to reduce redundancy]. HMM searches were also conducted against predicted protein sequences from the MMETSP transcriptomic data (61) and from protist genome projects available at the DoE-Joint Genome Institute. All identified putative Amt/Mep/Rh sequences were then aligned using “hmmalign” (82) and sequences shorter than 50-aa residues were discarded. Alignment sites composed of more than 50% of gaps were discarded. Misaligned and/or false positive sequences were detected using preliminary phylogenetic reconstructions with FastTree (default parameters). Sequences that resulted in very long branches were then removed, allowing a final tree calculation using the “accurate” mode (-slownni) with the WAG+G substitution matrix.
For the ML phylogeny, different strategies were applied for putative homolog sampling. For the viral DNA polymerase B and the vAmt flanking ORFs, homologous protein sequences were identified with BLASTP (89) similarity searches against UniProtKB. The homologous sequences were then aligned using the iterative refinement method E-INS-i as implemented in MAFFT v7.2 (90), edited with the trimAl v1.4 “strict” algorithm (91), and manually inspected and corrected. For the phylogeny based on the 22 core proteins shared among green algal viruses (46), the OtV6 predicted proteome was parsed using BLASTP and homologous sequences from other Ostreococcus spp. viruses were used as search queries. The resulting 22 OtV6 core proteins were then concatenated, aligned, and added to the original viral core protein alignment using MAFFT.
For the vAmt/Amt-Euk ML phylogenetic subtree, vAmt homologs were identified using the previous large-scale, superfamily phylogenetic reconstruction described above. Sequences branching with vAmt were identified; corresponding full-length sequences were retrieved and aligned with MAFFT E-INS-i and manually inspected. After homolog sequence selection and alignment, the same methodology was applied for all ML phylogenetic tree reconstructions. The most likely tree was identified from 100 ML reconstructions, using RAxML v8.2 (92), and under the substitution model best fitting the data, as identified by ProtTest v3 (93) using the Akaike information criterion; branch support values were based on 1,000 nonparametric bootstrap replicates.
Metagenome Screening and Phylogenetic Mapping.
To investigate the wider distribution of vAmt-like homologs, we screened metagenomes for similar sequences in the Global Ocean Sampling and Tara Oceans giant virus datasets (50) as well as all aquatic metagenomes available at iMicrobe (94). Putative vAmt homologs were aligned to alignment corresponding to the vAmt subtree (Fig. 6B and SI Appendix, Fig. S12) using MAFFT (without altering the original alignment) and then phylogenetically mapped onto the ML tree using pplacer v1.1 (95). Within this tree, two Amt-Euk sequences mapped to the vAmt branch.
vAmt Genomic Amplification.
To confirm the vAmt-encoding gene was harbored by OtV6, we designed specific primer pairs (listed in SI Appendix, Table S1) and the vAmt gene along with its 5′ and 3′ flanking regions were amplified, represented by an ∼2,500-bp sequence. For each PCR, a negative control (distilled H2O) was included. PCRs (25 L total volume) were conducted using GoTaq Green Master Mix (Promega) with 1 L of virus OtV6. Cycling reactions were as follows: 5 min at 95 °C; followed by 30 cycles of 30 s at 95 °C, 30 s at 54 °C, and 120 s at 72 °C; and with an additional 10-min extension at 72 °C. The PCRs were checked on a 1% agarose gel stained with GelRed (Biotium). The positive PCRs were then directly cloned using the StrataClone PCR cloning kit (Agilent Technologies) according to the manufacturer’s instructions. One clone per library was selected and double-strand sequenced using universal M13 primers. All amplicon sequencing was performed externally by Eurofins Genomics.
Ostreococcus Culture Infection and vAmt RT-PCR.
O. tauri cells from the RNP2 population [OtV5 resistant (44)] were grown in L1 medium exponentially under 12:12 light (32.3 molms−1; 1,700 lux). Infection experiments proceeded as follows: A total of 5 mL of O. tauri at exponential growth was infected using 250 L of purified OtV6; as a negative control, 5 mL of O. tauri culture was incubated with 250 L of L1 medium. To test for the expression of the vAmt-encoding gene during the infection cycle, the O. tauri cultures were sampled 12 h after OtV6 inoculation; 1 mL of the infected and uninfected cultures was sampled and centrifuged at 6,200 g during 10 min. After removal of the supernatant, the pellets were flash frozen and stored at −80 °C. RNA was extracted using the RNeasy Plus Universal Kits (Qiagen). An extra step to remove all genomic DNA was added after the RNA extraction protocol, using the RTS DNase kit (MO BIO Laboratories, Qiagen).
Using three sets of primers that amplify an overlapping region of the vAmt-encoding gene (OtV6-full-F1/R1, vAmtF4/R4, and sh-Amt-vir-F/R; Fig. 2B and SI Appendix, Table S1), we first checked for the presence of DNA contamination in the RNA samples, using PCR amplification. For every PCR, we included a negative control (distilled H2O) and a positive control (purified OtV6). PCRs were performed in 25 μL total volume, using GoTaq Green Master Mix with 1 L of RNA sample. Cycling reactions were as follows: 5 min at 95 °C; followed by 35 cycles of 30 s at 95 °C, 30 s at 50 °C, and 45 s at 72 °C; with an additional 10-min extension at 72 °C. The PCRs were checked on a 1% agarose gel stained with GelRed. The RT-PCRs were conducted using OneTaq One-Step RT-PCR (Qiagen). A total of 1 L of RNA was mixed in 50 L with 10 L of Buffer 5, 2 L of 10 mM dNTP, 1 L of each primer (0.2 mM final concentration), 2 L enzyme mix, and finally 0.25 L of RNaseOUT (10 units/L; Invitrogen, Life Technologies). Negative controls were made with 1 L of water instead of the RNA samples. Each PCR amplification was checked on a 1% agarose gel stained with GelRed. Positive RT-PCRs were cloned using a Strataclone PCR cloning kit; one clone per library was selected and double-strand sequenced using M13 primers.
Cloning and Functional Analysis of vAmt in Yeast.
The vAmt ORF was synthesized de novo by Genscript, codon optimized for expression in S. cerevisiae, and fused to a C-terminal GFP tag in vector p426 GPD. For complementation assays, the vAmt ORF was amplified with primers vAmt-attF/R (SI Appendix, Table S1), using Phusion polymerase (New England Biolabs) to remove the GFP-coding region; cloned into pDONR221 using Gateway recombination (Life Technologies); and mobilized into pAG416 GPD (low-copy constitutive expression vector). For fluorescence microscopy, the ORF was mobilized into pAG426 GPD EGFP (N-terminal EGFP vector) or the original p426 GPD EGFP (C-terminal) construct was used.
For yeast transformations, competent cells were prepared as described in Thomson et al. (96), mixed with 500 ng of plasmid DNA, and pulsed at 1.5 kV in an Eppendorf electroporator. S. cerevisiae strain 31019b was transformed with pAG416 GPD vAmt or pAG416 GPD empty vector, as described above. Transformed yeast 31019b cultures were grown to stationary phase at 30 °C and centrifuged at 3,200 g for 2 min, washed twice in water, and diluted to OD600nm 0.1 in 10 mL yeast nitrogen base (YNB) liquid medium [0.19% YNB without amino acids and without (NH4)2SO4; Formedium] containing a final concentration of 0.1 mM or 0.5 mM (NH4)2SO4, D-mannosamine, D-glucuronamide, or D,L-a-amino butyric acid. Cells were then incubated at 30 °C and OD600nm measurements were taken at 24-h intervals. This method was also used to assess growth of S. cerevisiae YNVW1 (dur3) and 23346c (wild type) on urea, with YNB supplemented with urea (0.1–2 mM) instead of (NH4)2SO4. R v3 and ggplot2 (97) were used for statistical analyses and plots.
Spinning-Disc Confocal Microscopy.
GFP constructs were transformed into S. cerevisiae strain BY4742 as described above, grown to midlog phase, and suspended in PBS. Spinning-disc confocal microscopy of EGFP-labeled cells was performed using an Olympus IX81 inverted microscope and a CSU-X1 Spinning-Disc unit (Yokogawa). A 60/1.35 oil or 100/1.40 oil objective was used with a 488-nm solid-state laser to excite the EGFP fluorophore. A Photometrics CoolSNAP HQ2 camera (Roper Scientific) was used for imaging with the VisiView software (Visitron Systems).
Transporter Protein Phenotyping.
To prepare cells for OmniLog Phenotype Microarray (PM) plates (Biolog), each yeast strain (S. cerevisiae 31019b transformed with either pAG416 GPD or pAG416 GPD vAmt) was grown on YNB complemented with potassium nitrate and uracil at 30 °C for 48–72 h. Colonies were suspended in Yeast Nutrient Supplement solution (Biolog) and adjusted to 62% turbidity. A total of 250 L of cell suspension was made up to a final volume of 12 mL of inoculating fluid, containing 1 IFY-0, 1 Dye Mix D, 50 mM D-glucose, 1 mM disodium pyrophosphate, and 2 mM sodium sulfate, and 100 L was inoculated into each well of a PM3 MicroPlate (N sources). Assays were run in triplicate using independently obtained transformants. OmniLog Phenotype Microarray outputs were analyzed by normalizing each individual plate against well A01 (negative control) to control for any background growth as a result of the inoculation solution and then analyzed using the R package opm (98). Data were aggregated using the “opm-fast” method, analyzed using the area under curve (AUC) parameter, and tested by t test to detect significant increase in respiration rates in the pAG416 GPD vAmt strain.
Yeast Methylammonium Uptake Assays.
S. cerevisiae strains were grown in 25 mL minimal proline medium (99) for 16 h at 30 °C with shaking. Cells were then diluted and grown until early log phase and harvested by centrifugation at 1,000 g for 3 min. Cells were washed once and suspended in 2 mL medium lacking proline. The dry weight of each sample was noted, before 90-L aliquots of cells were exposed to five concentrations (5–500 M) of methylamine [14C] hydrochloride for 1 min at 24 °C. Reactions were stopped by the addition of 1 mL 120 mM methylamine hydrochloride. Background adsorption was also calculated by exposing cells to 1 mL of 120 mM unlabeled methylamine hydrochloride before the addition of the radiolabeled substrate. Cells were collected by centrifugation at 14,000 g for 3 min, washed, and suspended in 500 L deionized water, and then radioactivity was determined by liquid scintillation counting in a liquid scintillation analyzer (LS 6500; Beckman Coulter) after addition of 2.5 mL Emulsifier-Safe scintillation mixture solution (Perkin-Elmer). The R package drc (100) was used to calculate Michaelis–Menten kinetics and curves.
Infection Time-Course Experiments.
Two 15-mL cultures of O. tauri were inoculated with OtV6 viruses and the infection was allowed to proceed until culture bleaching after 5 d. Cell debris were removed by centrifugation at 3,200g for 20 min followed by filtering through a 0.2-m syringe filter. Viruses were concentrated 10-fold using a 50-kDa Amicon Ultra-15 Centrifugal Filter (Merck Millipore) followed by centrifugation at 3,200g for 2.5 min. A total of 18 mL O. tauri cultures were cultured using 10 mL preculture and 8 mL media, 2 d before the experiment. To begin the infection time-course experiments, 750 L OtV6 was added and at each time point, 1 mL sample was pelleted and frozen in liquid nitrogen for RNA preservation and the supernatant was stored at −80 °C for fluorometric detection. Two 200-L samples were also taken for flow cytometry: (i) 2 L 25% glutaraldehyde was added and the sample incubated for 15 min before freezing in liquid nitrogen for O. tauri counts and (ii) 8 L 25% glutaraldehyde was added and the sample incubated for 30 min at 4 °C before freezing for viral counts. O. tauri and VLP abundances were monitored by a FACSCanto flow cytometer (BD Biosciences) according to their right-angle scatter and the fluorescence emission due either to the chlorophyll a pigment for O. tauri (101) or to SYBR Green I (Roche Diagnostics) staining for VLPs (102). A 900-L sample was also removed for assessing methylammonium uptake at each time point. Cells were harvested by centrifugation at 5,000g for 3 min and suspended in 450 L media. For each sample, the uptake rate for methylamine [14C] hydrochloride was determined as previously described. The orthophtaldialdehyde (OPA) method (103) was used for fluorometric detection in cell-free medium (supernatant, see above). A total of 20 L of the working reagent (OPA, borate buffer, and sodium sulfite) was added to 80 L of the culture sample (or standard); samples and standards were processed on a 96-well plate, which was incubated in the dark at 25 °C with shaking for 2–3 h. Fluorescence was then read at 355 nm excitation and 420 nm emission on an Infinite M200 plate reader (Tecan).
Quantitative PCR.
RNA was extracted using the RNeasy Plus Universal Mini kit (Qiagen) following the manufacturer’s instructions and incorporating a 5-min elution step using 30 L RNase-free water. Residual DNA was removed using the Turbo DNA-free kit (Ambion, Life Technologies) and 6 L RNA reverse transcribed using the Superscript III First Strand Synthesis SuperMix (Invitrogen) and oligo(dT)20 primers, following the manufacturer’s instructions. cDNA was quantified using a Qubit ssDNA Assay Kit (Life Technologies) and stored at −20 °C before performing qPCR. Plasmids for each gene of interest were generated by Phusion polymerase (New England Biolabs) PCR using cDNA templates, followed by A-tailing using Taq polymerase and cloning using a Strataclone PCR cloning kit (Agilent Technologies). Each plasmid was confirmed by sequencing (Eurofins Genomics) and serial dilutions (108 to 10 copies) were used to generate standard curves for each primer pair and probe. Efficiencies ranged from 96% to 102% (SI Appendix, Table S4). qPCRs were performed in a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). Each 25-L reaction contained 12.5 L TaqMan Gene Expression Master Mix (Thermo Fisher Scientific), 900 nM each primer, 250 nM hydrolysis probe, and 1 L cDNA per plasmid DNA and was performed in duplicate alongside no-template and minus-RT controls. Cycling conditions were as follows: UDG activation for 2 min at 50 °C and DNA polymerase activation for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. ROX was used as an internal reference dye for analysis of CT values, which were determined using StepOne Software v2.3 (Thermo Fisher Scientific), and standard curves were used for quantification of each gene.
Data Access.
Phylogenetic and experimental data are available at the Zenodo data repository: zenodo.org/record/61901. PCR-amplified sequence assembly of the vAmt-encoding locus was deposited in GenBank (KX254356).
Supplementary Material
Acknowledgments
We thank C. Salmeron and the cytometry platform of the Observatoire Océanologique de Banyuls-sur-Mer, as well as C. Lambert and the cytometry core facilities of the Laboratoire des Sciences de l’Environnement Marin (Institut Universitaire Européen de la Mer) for assistance in FCM counts. We are grateful to Dr. A-M. Marini (Université Libre de Bruxelles) and to Dr. G. P. Bienert (Leibniz Institute of Plant Genetics and Crop Plant Research) for yeast mutant strains. We thank Prof. K. Haynes, Dr. F. Maguire, Dr. F. Savory, Prof. N. Smirnoff and Dr. J. G. Wideman (University of Exeter) for helpful comments. A.M. and T.A.R. are funded by the Royal Society, through Newton and University Research fellowships, respectively. This work is supported in part by research grants from The Gordon and Betty Moore Foundation (GBMF5514), Leverhulme Trust (PLP-2014-147), and the University of Exeter. The University of Exeter OmniLog facility is supported by a Wellcome Trust Institutional Strategic Support Award WT105618MA. Phylogenetic reconstructions were computed on the Data Intensive Academic Grid (National Science Foundation, MRI-R2 Project DBI-0959894).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KX254356). Phylogenetic and experimental data are available at the Zenodo data repository,zenodo.org/record/61901.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708097114/-/DCSupplemental.
References
- 1.Falkowski P. Ocean science: The power of plankton. Nature. 2012;483:S17–S20. doi: 10.1038/483S17a. [DOI] [PubMed] [Google Scholar]
- 2.Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349–355. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 3.Moore C, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–710. [Google Scholar]
- 4.Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. The role of functional traits and trade-offs in structuring phytoplankton communities: Scaling from cellular to ecosystem level. Ecol Lett. 2007;10:1170–1181. doi: 10.1111/j.1461-0248.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 5.Hildebrand M. Cloning and functional characterization of ammonium transporters from the marine diatom Cylindrotheca fusiformis (Bacillariophyceae) J Phycol. 2005;41:105–113. [Google Scholar]
- 6.Allen AE, Vardi A, Bowler C. An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr Opin Plant Biol. 2006;9:264–273. doi: 10.1016/j.pbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 7.McDonald SM, Plant JN, Worden AZ. The mixed lineage nature of nitrogen transport and assimilation in marine eukaryotic phytoplankton: A case study of Micromonas. Mol Biol Evol. 2010;27:2268–2283. doi: 10.1093/molbev/msq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dagenais-Bellefeuille S, Morse D. Putting the N in dinoflagellates. Front Microbiol. 2013;4:369. doi: 10.3389/fmicb.2013.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terrado R, Monier A, Edgar R, Lovejoy C. Diversity of nitrogen assimilation pathways among microbial photosynthetic eukaryotes. J Phycol. 2015;51:490–506. doi: 10.1111/jpy.12292. [DOI] [PubMed] [Google Scholar]
- 10.Mulholland MR, Lomas MW. Nitrogen uptake and assimilation. In: Capone D, Bronk D, Mulholland M, Carpenter E, editors. Nitrogen in the Marine Environment. Academic; New York: 2008. pp. 303–384. [Google Scholar]
- 11.Glibert PM, et al. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol Oceanogr. 2016;61:165–197. [Google Scholar]
- 12.Gobler CJ, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci USA. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worm B, Lotze HK, Hillebrand H, Sommer U. Consumer versus resource control of species diversity and ecosystem functioning. Nature. 2002;417:848–851. doi: 10.1038/nature00830. [DOI] [PubMed] [Google Scholar]
- 14.Pasulka AL, Samo TJ, Landry MR. Grazer and viral impacts on microbial growth and mortality in the Southern California current ecosystem. J Plankton Res. 2015;37:320–336. [Google Scholar]
- 15.Suttle CA. Marine viruses—Major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 16.Brussaard CP. Viral control of phytoplankton populations—A review. J Eukaryot Microbiol. 2004;51:125–138. doi: 10.1111/j.1550-7408.2004.tb00537.x. [DOI] [PubMed] [Google Scholar]
- 17.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 18.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. BioScience. 1999;49:781–788. [Google Scholar]
- 19.Breitbart M. Marine viruses: Truth or dare. Annu Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 20.Hurwitz BL, Hallam SJ, Sullivan MB. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013;14:R123. doi: 10.1186/gb-2013-14-11-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roux S, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 22.Monier A, et al. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009;19:1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson LR, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dammeyer T, Bagby SC, Sullivan MB, Chisholm SW, Frankenberg-Dinkel N. Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr Biol. 2008;18:442–448. doi: 10.1016/j.cub.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 25.Rosenwasser S, et al. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell. 2014;26:2689–2707. doi: 10.1105/tpc.114.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malitsky S, et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol. 2016;210:88–96. doi: 10.1111/nph.13852. [DOI] [PubMed] [Google Scholar]
- 27.Ziv C, et al. Viral serine palmitoyltransferase induces metabolic switch in sphingolipid biosynthesis and is required for infection of a marine alga. Proc Natl Acad Sci USA. 2016;113:E1907–E1916. doi: 10.1073/pnas.1523168113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson WH, Carr NG, Mann NH. The effect of phosphate status on the kinetics of cyanophage infection in the oceanic cyanobacterium Synechococcus sp. wh78031. J Phycol. 1996;32:506–516. [Google Scholar]
- 29.Monier A, et al. Phosphate transporters in marine phytoplankton and their viruses: Cross-domain commonalities in viral-host gene exchanges. Environ Microbiol. 2012;14:162–176. doi: 10.1111/j.1462-2920.2011.02576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maat DS, Crawfurd KJ, Timmermans KR, Brussaard CP. Elevated CO2 and phosphate limitation favor Micromonas pusilla through stimulated growth and reduced viral impact. Appl Environ Microbiol. 2014;80:3119–3127. doi: 10.1128/AEM.03639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maat DS, Brussaard CP. Both phosphorus-and nitrogen limitation constrain viral proliferation in marine phytoplankton. Aquat Microb Ecol. 2016;77:87–97. [Google Scholar]
- 32.McDonald TR, Dietrich FS, Lutzoni F. Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: Toward a new functional and evolutionary classification. Mol Biol Evol. 2012;29:51–60. doi: 10.1093/molbev/msr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mojica KD, Brussaard CP. Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol Ecol. 2014;89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 34.UniProt Consortium Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade SL, Einsle O. The Amt/Mep/Rh family of ammonium transport proteins. Mol Membr Biol. 2007;24:357–365. doi: 10.1080/09687680701388423. [DOI] [PubMed] [Google Scholar]
- 36.von Wirén N, Gazzarrini S, Gojon A, Frommer WB. The molecular physiology of ammonium uptake and retrieval. Curr Opin Plant Biol. 2000;3:254–261. [PubMed] [Google Scholar]
- 37.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Short SM. The ecology of viruses that infect eukaryotic algae. Environ Microbiol. 2012;14:2253–2271. doi: 10.1111/j.1462-2920.2012.02706.x. [DOI] [PubMed] [Google Scholar]
- 39.Marin B, Melkonian M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear-and plastid-encoded rRNA operons. Protist. 2010;161:304–336. doi: 10.1016/j.protis.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Monier A, Worden AZ, Richards TA. Phylogenetic diversity and biogeography of the Mamiellophyceae lineage of eukaryotic phytoplankton across the oceans. Environ Microbiol Rep. 2016;8:461–469. doi: 10.1111/1758-2229.12390. [DOI] [PubMed] [Google Scholar]
- 41.Derelle E, et al. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One. 2008;3:e2250. doi: 10.1371/journal.pone.0002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weynberg KD, Allen MJ, Ashelford K, Scanlan DJ, Wilson WH. From small hosts come big viruses: The complete genome of a second Ostreococcus tauri virus, OtV-1. Environ Microbiol. 2009;11:2821–2839. doi: 10.1111/j.1462-2920.2009.01991.x. [DOI] [PubMed] [Google Scholar]
- 43.Weynberg KD, Allen MJ, Gilg IC, Scanlan DJ, Wilson WH. Genome sequence of Ostreococcus tauri virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean. J Virol. 2011;85:4520–4529. doi: 10.1128/JVI.02131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas R, et al. Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ Microbiol. 2011;13:1412–1420. doi: 10.1111/j.1462-2920.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- 45.Moreau H, et al. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J Virol. 2010;84:12555–12563. doi: 10.1128/JVI.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derelle E, et al. Diversity of viruses infecting the green microalga Ostreococcus lucimarinus. J Virol. 2015;89:5812–5821. doi: 10.1128/JVI.00246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derelle E, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monier A, et al. Marine mimivirus relatives are probably large algal viruses. Virol J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monier A, Claverie JM, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hingamp P, et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013;7:1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loqué D, et al. Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 2006;48:522–534. doi: 10.1111/j.1365-313X.2006.02887.x. [DOI] [PubMed] [Google Scholar]
- 52.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 53.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Javelle A, et al. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 56.Marini AM, Soussi-Boudekou S, Vissers S, André B. A family of ammonium transporters in Saccharomyces Cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu LH, Ludewig U, Frommer WB, von Wirén N. AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell. 2003;15:790–800. doi: 10.1105/tpc.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bu Y, et al. Identification and characterization of a PutAMT1; 1 gene from Puccinellia tenuiflora. PLoS One. 2013;8:e83111. doi: 10.1371/journal.pone.0083111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown CM, Bidle KD. Attenuation of virus production at high multiplicities of infection in Aureococcus anophagefferens. Virology. 2014;466:71–81. doi: 10.1016/j.virol.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 61.Keeling PJ, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 63.Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richards TA, Talbot NJ. Horizontal gene transfer in osmotrophs: Playing with public goods. Nat Rev Microbiol. 2013;11:720–727. doi: 10.1038/nrmicro3108. [DOI] [PubMed] [Google Scholar]
- 65.Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ignacio-Espinoza JC, Sullivan MB. Phylogenomics of T4 cyanophages: Lateral gene transfer in the ‘core’ and origins of host genes. Environ Microbiol. 2012;14:2113–2126. doi: 10.1111/j.1462-2920.2012.02704.x. [DOI] [PubMed] [Google Scholar]
- 67.Worden AZ, et al. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–179. [Google Scholar]
- 68.Stevenson R, Silver S. Methylammonium uptake by Escherichia coli: Evidence for a bacterial NH+ transport system. Biochem Biophys Res Commun. 1977;75:1133–1139. doi: 10.1016/0006-291x(77)91501-7. [DOI] [PubMed] [Google Scholar]
- 69.Johnson DA, et al. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 2008;4:e1000211. doi: 10.1371/journal.pgen.1000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng YS, Labavitch J, VanderGheynst JS. Organic and inorganic nitrogen impact Chlorella variabilis productivity and host quality for viral production and cell lysis. Appl Biochem Biotechnol. 2015;176:467–479. doi: 10.1007/s12010-015-1588-0. [DOI] [PubMed] [Google Scholar]
- 71.Jover LF, Effler TC, Buchan A, Wilhelm SW, Weitz JS. The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat Rev Microbiol. 2014;12:519–528. doi: 10.1038/nrmicro3289. [DOI] [PubMed] [Google Scholar]
- 72.Clasen JL, Elser JJ. The effect of host Chlorella NC64A carbon: Phosphorus ratio on the production of Paramecium bursaria Chlorella virus-1. Freshwat Biol. 2007;52:112–122. [Google Scholar]
- 73.Nissimov JI, Napier JA, Allen MJ, Kimmance SA. Intragenus competition between coccolithoviruses: An insight on how a select few can come to dominate many. Environ Microbiol. 2016;18:133–145. doi: 10.1111/1462-2920.12902. [DOI] [PubMed] [Google Scholar]
- 74.Maat DS, van Bleijswijk JD, Witte HJ, Brussaard CP. Virus production in phosphorus-limited Micromonas pusilla stimulated by a supply of naturally low concentrations of different phosphorus sources, far into the lytic cycle. FEMS Microbiol Ecol. 2016;92:fiw136. doi: 10.1093/femsec/fiw136. [DOI] [PubMed] [Google Scholar]
- 75.Wilson W, Van Etten JL, Allen M. The Phycodnaviridae: The story of how tiny giants rule the world. In: Van Etten JL, editor. Lesser Known Large dsDNA Viruses. Springer; Berlin: 2009. pp. 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colson P, et al. Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of mimivirus genes. Genome Biol Evol. 2011;3:737–742. doi: 10.1093/gbe/evr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sullivan MB, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. 2010;12:3035–3056. doi: 10.1111/j.1462-2920.2010.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharon I, et al. Photosystem I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anantharaman K, et al. Sulfur oxidation genes in diverse deep-sea viruses. Science. 2014;344:757–760. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 80.Zeng Q, Chisholm SW. Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol. 2012;22:124–128. doi: 10.1016/j.cub.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 81.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2013;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Price MN, Dehal PS, Arkin AP. FastTree 2 –approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hurwitz B. Plant and Animal Genome XXII Conference. Plant and Animal Genome; San Diego: 2014. iMicrobe: Advancing clinical and environmental microbial research using the iPlant cyberinfrastructure. [Google Scholar]
- 95.Matsen FA, Kodner RB, Armbrust EV. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thompson JR, Register E, Curotto J, Kurtz M, Kelly R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast. 1998;14:565–571. doi: 10.1002/(SICI)1097-0061(19980430)14:6<565::AID-YEA251>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 97.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 98.Vaas LA, et al. opm: An R package for analysing OmniLog® phenotype microarray data. Bioinformatics. 2013;29:1823–1824. doi: 10.1093/bioinformatics/btt291. [DOI] [PubMed] [Google Scholar]
- 99.Jacobs P, Jauniaux JC, Grenson M. A cis-dominant regulatory mutation linked to the argb-argc gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980;139:691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- 100.Ritz C, et al. Bioassay analysis using r. J Stat Software. 2005;12:1–22. [Google Scholar]
- 101.Trask B, Van den Engh G, Elgershuizen J. Analysis of phytoplankton by flow cytometry. Cytometry. 1982;2:258–264. doi: 10.1002/cyto.990020410. [DOI] [PubMed] [Google Scholar]
- 102.Brussaard CP. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.