Summary
Huntington’s disease (HD) is caused by an expanded CAG repeat in the Huntingtin (HTT) gene. The mechanism(s) by which mutant HTT (mHTT) causes disease is unclear. Nucleocytoplasmic transport, the trafficking of macromolecules between the nucleus and cytoplasm is tightly regulated by nuclear pore complexes (NPCs) made up of nucleoporins (NUPs). Previous studies offered clues that mHTT may disrupt nucleocytoplasmic transport and a mutation of a NUP can cause HD-like pathology. Therefore, we evaluated the NPC and nucleocytoplasmic transport in multiple models of HD including mouse and fly models, neurons transfected with mHTT, HD iPSC-derived neurons and human HD brain regions. These studies revealed severe mislocalization and aggregation of NUPs and defective nucleocytoplasmic transport. HD repeat-associated non-ATG (RAN) translation proteins also disrupted nucleocytoplasmic transport. Additionally, overexpression of NUPs and treatment with drugs that prevent aberrant NUP biology also mitigated this transport defect and neurotoxicity, providing future novel therapy targets.
Keywords: Huntington’s disease, C9ORF72, Neurodegeneration, Nucleocytoplasmic Transport, Nuclear Pore Complex, RAN Translation, Induced Pluripotent Stem Cell, O-GlcNAc, Thiamet-G, KPT-350
INTRODUCTION
Huntington’s disease (HD) is the most common inherited neurodegenerative disorder, caused by an expanded CAG repeat in the first exon of the Huntingtin (HTT) gene coding for a polyglutamine (polyQ) tract within the protein Huntingtin (HTT) (Finkbeiner, 2011). This autosomal dominant disease results in the selective degeneration of striatal medium spiny projection neurons and cortical pyramidal neurons as well as the formation of intracellular neuronal aggregates. How mutant HTT (mHTT) can induce this selective neuronal loss despite ubiquitous expression throughout the nervous system is unknown. Disease onset and severity are dependent on CAG repeat length with a longer expansion resulting in an earlier onset and greater severity of illness. The HTT gene in asymptomatic individuals contains less than 35 CAG repeats whereas pathologic expansions contain greater than 39. A juvenile variant exists in which expansion lengths in excess of ~60 or more can result in symptom onset earlier than 20 years of age (Vonsattel and DiFiglia). The underlying mechanisms by which mHTT causes neurodegeneration have not been fully elucidated. Studies have provided clues that nuclear pore complex (NPC) dysfunction may be a pathogenic contributor (Basel-Vanagaite et al., 2006; Hosp et al., 2015; Liu et al., 2014; Suhr et al., 2001; Woerner et al., 2016) although very little is known about the NPC and the role of nucleocytoplasmic transport defects in human HD or in models of the disease.
The NPC is one of the largest molecular complexes in eukaryotic cells and serves as the main transport conduit between the nucleus and the cytoplasm. NPCs span the entire nuclear envelope and consist of multiple copies of approximately 30 different protein subunits called nucleoporins (NUPs). NUPs differ in functions including protein import, protein export, RNA export and membrane anchoring and are organized into 5 unique anatomical regions of the NPC (cytoplasmic ring/filaments, central channel, nuclear ring/basket, transmembrane, and scaffold) (Raices and D'Angelo, 2012; Wente and Rout, 2010). The NPC has nucleocytoplasmic transport-independent functions such as regulating genome organization, gene expression, cell differentiation and development, and RNA processing and quality control (Raices and D'Angelo, 2012; Raices and D'Angelo, 2017). During nuclear import, cargo is released into the nucleus when the transport receptor interacts with Ran-GTP, a GTP-binding nuclear protein. During nuclear export, cargo is released into the cytoplasm upon GTP hydrolysis of Ran-GTP by RanGAP1, a GTPase-activating protein located on the cytoplasmic filaments of the NPC that is also required for nuclear import. Higher levels of Ran-GTP in the nucleus compared to the cytoplasm is essential for fueling active transport through the NPC and defining nucleocytoplasmic transport directionality (Floch et al., 2014). The proper maintenance of this nuclear/cytoplasmic (N/C) Ran gradient by RanGAP1 is critical, and its loss has been shown to cause cell death within minutes (Hetzer et al., 2002). Finally, molecules less than ~40 kDa can freely migrate through the NPC via passive transport.
Recent work from our group and others has shown that nucleocytoplasmic transport is disrupted by a hexanucleotide repeat expansion (HRE) in the C9orf72 gene, the most common mutation in Amyotrophic Lateral Sclerosis (ALS), Frontotemporal Dementia (FTD) and a common cause of HD phenocopies (Freibaum et al., 2015; Hensman Moss et al., 2014; Jovičić et al., 2015; Zhang et al., 2015). A direct interaction between the HRE-containing RNAs and RanGAP1 can cause a disruption of the Ran energy gradient required for active transport. In addition, nucleocytoplasmic transport function can be disrupted by Repeat Associated Non-ATG (RAN) Translation produced polydipeptides encoded by the HRE RNAs, which cause NUPs to aggregate (Zhang et al., 2016) or block the central channel of the nuclear pore complex (Shi et al., 2017). RAN translation occurs not only in diseases with noncoding region repeat expansions like C9orf72 HRE, but recent work has shown that RAN translation also occurs in diseases with coding region expansions including Huntington’s disease (Bañez-Coronel et al., 2015).
HD is pathologically characterized by intracellular inclusions in the striatum and cortex, which consist of aggregates of HTT, HD-RAN proteins and other proteins including nucleoporin 62 (NUP62) (Suhr et al., 2001). NUP62, located in the central channel of the NPC, plays a direct role in nuclear import of proteins containing a nuclear localization signal (NLS) as well as roles in transcription and chromatin organization (Capelson et al., 2010; Kalverda et al., 2010; Liang and Hetzer, 2011). It is also a member of the phenylalanine-glycine (FG) repeat containing NUPs, and the FG-rich domain makes NUP62 important for controlling nuclear pore permeability and selective trafficking of macromolecules to and from the nucleus. Many transport receptors like importin-β and NTF2 can traffic cargo containing an NLS or NES (nuclear export signal) through the nuclear pore complex via dynamic hydrophobic interactions with the FG-rich domain of FG-NUPs (Wente and Rout, 2010). Intriguingly, a mutation in NUP62 causes autosomal recessive infantile bilateral striatal necrosis (IBSN), a fatal disorder characterized by striatal and globus pallidus degeneration (Basel-Vanagaite et al., 2006). Also, mHTT exhibits preferential binding to RanGAP1 and Ribonucleic Acid Export 1 (RAE1), an mRNA export factor (Hosp et al., 2015). In addition, nuclear membrane distortions can be observed in various cell culture models, transgenic animal models and ultrastructural studies of HD brain (Brettschneider et al., 2015; Cornett et al., 2005). Perinuclear inclusions of mHTT disrupt the nuclear membrane and cause striatal cell death in mouse and cell models of HD (Liu et al., 2014). Finally, cytoplasmic protein aggregates can disrupt nucleocytoplasmic trafficking of protein and RNA (Woerner et al., 2016). Given these findings, we hypothesized that mHTT may directly disrupt nucleocytoplasmic transport at the NPC.
We screened several NUPs and NPC-associated proteins from all anatomical regions of the NPC in the R6/2 and zQ175 mouse models of HD and identified a subset that colocalize with mHtt intracellular aggregates. These NUPs also aggregate or mislocalize in HD and juvenile HD (JHD) post-mortem human tissue. Using HD iPSC-derived neurons, primary neurons transfected with full length mHTT, and a Drosophila model of HD, we show that several NUPs are severely mislocalized, that there are deficits in both passive and active nucleocytoplasmic transport, and demonstrate correction of these deficits and neurotoxicity using small molecules and overexpression constructs that target the nucleocytoplasmic transport pathway. Finally, we demonstrate that the recently discovered HD-RAN proteins can also directly disrupt nucleocytoplasmic transport. These data suggest that mHTT disrupts nucleocytoplasmic transport directly at the NPC and that targeting this pathway may provide a therapeutic route for HD.
RESULTS
Nucleoporins Aggregate and Colocalize with mHtt in the R6/2 Mouse Model of HD
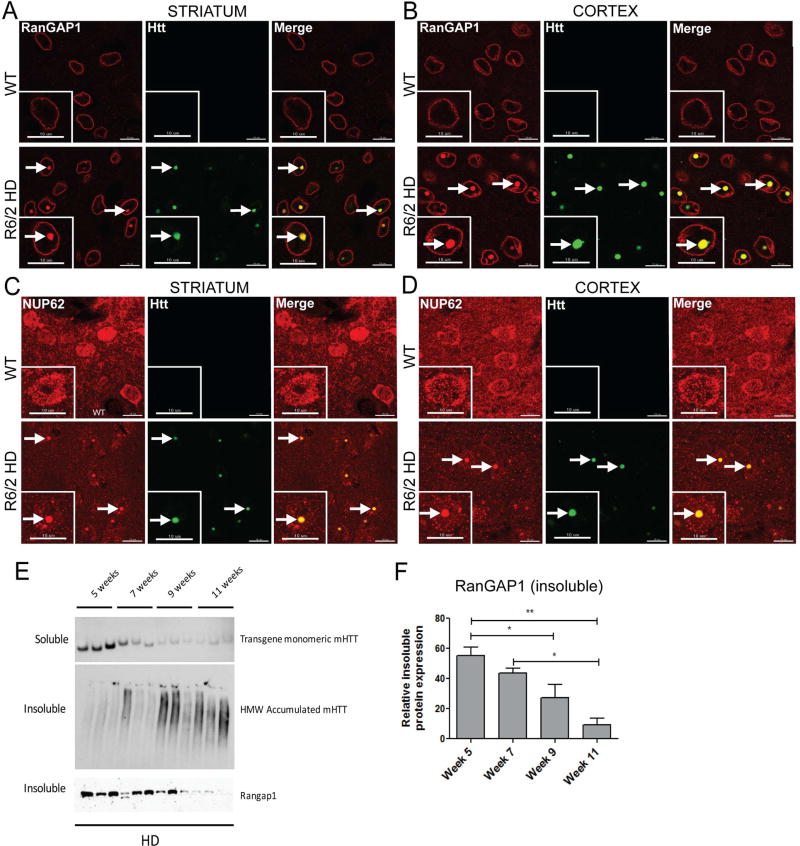
To analyze the integrity of the NPC in HD, we assessed the localization of the majority of NUPs and NPC-associated proteins (Table S1) in 10-week-old R6/2 mice using immunofluorescence. The R6/2 mouse model expresses exon 1 of human HTT with highly expanded CAG repeats (approximately 125–160 CAGs) and exhibits HD-like symptomology with motor symptom onset at 5 weeks, body weight decline onset at 10 weeks, and a lifespan ranging from 10–13 weeks (Pouladi et al., 2013). These animals also develop accumulation of aggregated mHtt. We frequently found that RanGAP1 and NUP62 form intranuclear inclusions that colocalize with EM48+ mHTT aggregates in neurons of the striatum and cortex of these mice (Figure 1A–D, Figure S1).
Figure 1. Nucleoporins Aggregate and Colocalize with mHtt in the R6/2 Mouse Model of HD.
(A,B) Coronal brain sections from 10 week old WT and TG R6/2 mice showing aggregates of RanGAP1 (red) that colocalize with EM48+ mHtt aggregates (green) in the striatum and cortex of TG R6/2 mice. n=5/group. (C,D) Coronal brain sections from WT and TG R6/2 mice showing aggregates of NUP62 (red) that colocalize with EM48+ mHtt aggregates (green) in the striatum and cortex of TG R6/2 mice. n=5/group. (E,F) Insoluble RanGAP1 levels decrease with disease progression in striatum of R6/2 mice. Insoluble RanGAP1 and soluble mHtt transgene levels are significantly reduced and insoluble HMW accumulated mHtt is increased in R6/2 mice from weeks 5 to 11. All data are expressed as western densitometry quantitation. Protein expression was validated for protein loading prior to antibody incubation using reversible protein stain and each samples’ corresponding soluble α-tubulin expression. Data represent mean ± SEM. *p < 0.05, **p < 0.01; One-way ANOVA followed by Bonferroni post-testing testing was applied. n = 3/time point. Scale bars, 10µm (A–D). See also Table S1, Figures S1 and S2.
We next examined insoluble and soluble mHtt and RanGAP1 levels in the striatum throughout disease progression in the R6/2 mice. We previously showed the presence of an insoluble HMW mHtt species along with other modified proteins in the detergentinsoluble fraction from R6/2 striatum (Ochaba et al., 2016), therefore detergent-soluble and detergent-insoluble proteins were evaluated. The detergent-soluble fraction contains mainly cytoplasmic proteins, monomeric forms of Htt (including the R6/2 mHTT fragment encoding human transgene), endogenous mouse full-length Htt, and soluble oligomeric species of Htt, which do not fully resolve on standard PAGE gels (O'Rourke et al., 2013; Sontag et al., 2012). In contrast, the detergent-insoluble fraction contains primarily nuclear proteins such as HMW mHtt species (likely multimers or potentially insoluble oligomers and fibrils), and insoluble accumulated forms of SUMO-and ubiquitin-modified proteins. As anticipated, levels of soluble monomeric transgene mHtt protein significantly decreased throughout disease progression (Figure 1E), with a corresponding age-dependent increase of an insoluble, HMW accumulated species of mHtt (Figure 1E). Notably, levels of insoluble RanGAP1 significantly decreased throughout the progression of the disease, with little RanGAP1 observed by 11 weeks of age in this fraction from R6/2 striatum (Figure 1E,F). This likely reflects a decrease in the levels of nuclear envelope RanGAP1.
If chronic expression of mHtt causes the mislocalization and nuclear envelope reduction of RanGAP1, then a molecular intervention that reduces the aberrant accumulation of HMW mHtt species in R6/2 mice might restore nuclear envelope RanGAP1 levels. Therefore, we evaluated striatal tissues from mice that had been treated with an miRNA directed against PIAS1, an E3-SUMO ligase that modulates SUMO-1 and SUMO-2/3 modification of HTT (O'Rourke et al., 2013; Ochaba et al., 2016). Reduction of PIAS1 through intrastriatal injection of a PIAS1 directed miRNA in vivo in R6/2 mice reduces formation of the HMW species of mHtt. Using the same fractionation procedure, the detergent insoluble fraction of non-transgenic (NT) and R6/2 striatum plus and minus PIAS1 knockdown (miPIAS1.3 treatment) at 10 weeks was analyzed for the presence of insoluble RanGAP1 (Figure S2A,B). Treatment significantly restored levels of nuclear RanGAP1 to normal (NT) levels in R6/2 striatum compared to miSAFE (control) treated mice. There were no significant differences either by genotype or treatment for soluble RanGAP1 levels (Figure S2A). We also assessed whether RanGAP1 was correspondingly reduced in mHtt aggregates using a filter retardation assay to examine levels of RanGAP1 which may be caught up in insoluble fibrils. We found that RanGAP1 levels were higher in aggregates resolved on filter blots in miSAFE treated R6/2 striatum than in miPIAS1.3 (Figure S2C), suggesting again the restoration of RanGAP1 dyshomeostasis upon PIAS1 knockdown and that RanGAP1 may be sequestered in aberrant protein inclusions and complexes in the disease state.
Nucleoporins Aggregate and Colocalize with mHtt in the zQ175 Mouse Model of HD
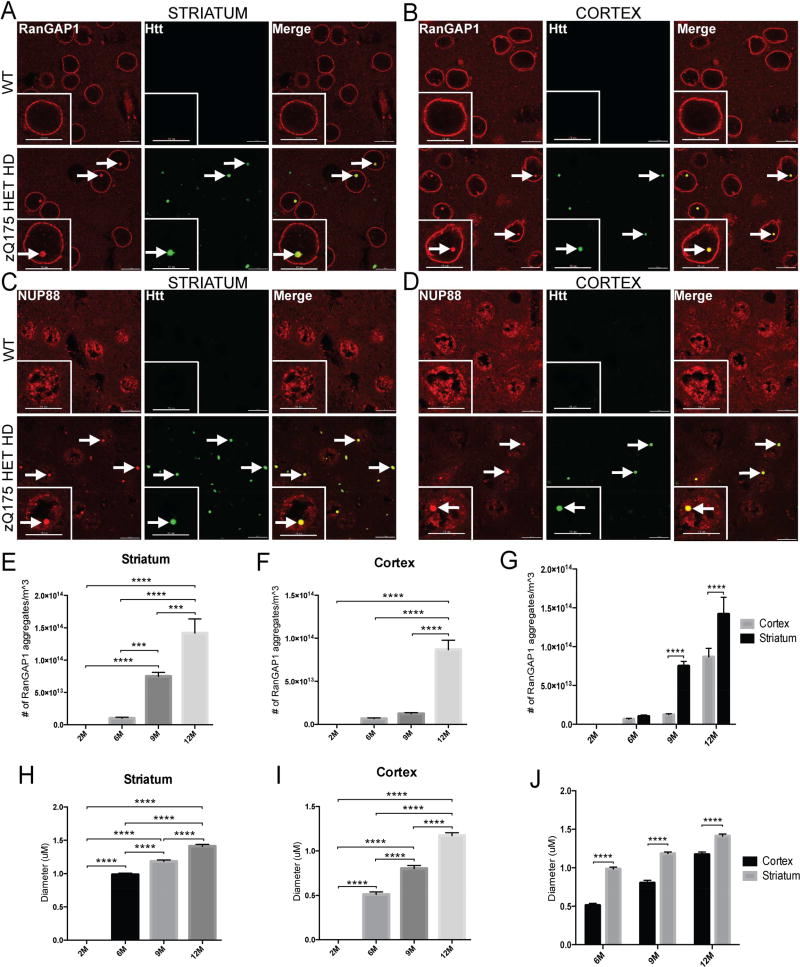
We next assessed the localization of many NUPs and NPC-associated proteins (Table S1) in the zQ175 mouse model of HD at 2 months, 6 months, 9 months and 12 months. This knock-in mouse model contains the human HTT exon 1 sequence with a ~193 CAG (193+/−7) repeat tract, replacing the mouse Htt exon1 within the native mouse Htt gene (Pouladi et al., 2013). Heterozygous zQ175 mice begin exhibiting motor symptoms after 3 months of age and demonstrate significant brain atrophy by 8 months. We found that RanGAP1 and nucleoporin 88 (NUP88) form intracellular inclusions that colocalize with EM48+ mHtt aggregates in the striatum and cortex of these mice (Figure 2A–D). On average, approximately 70% of all RanGAP1 aggregates colocalize with EM48+ mHtt aggregates at 12 months (Figure S3). Using the spot detection function in IMARIS software, these RanGAP1 aggregates increase in frequency with age in both the striatum and cortex but are significantly more prevalent in the striatum (Figure 2E–G). Similarly, quantification of the size of these inclusions indicates that the diameter increases with age in both brain regions but is significantly greater in the striatum (Figure 2H–J).
Figure 2. Nucleoporins Aggregate and Colocalize with mHtt in the zQ175 Mouse Model of HD.
(A,B) Coronal brain sections from 12 month old WT and HET zQ175 mice showing aggregates of RanGAP1 (red) that colocalize with EM48+ mHtt aggregates (green) in the striatum and cortex of HET zQ175 mice. (C,D) Coronal brain sections from 12 month old WT and HET zQ175 mice showing aggregates of NUP88 (red) that colocalize with EM48+ mHtt aggregates (green) in the striatum and cortex of HET zQ175 mice. (E,F,G) Quantification of number of RanGAP1 aggregates in the striatum and cortex of HET zQ175 mice at 2, 6, 9 and 12 months. (H,I,J) Quantification of diameter of RanGAP1 aggregates in the striatum and cortex of HET zQ175 mice at 2, 6, 9 and 12 months. Data (E–J) are presented as mean ± SEM. ***P<0.001 and ****P<0.0001 as analyzed by one-way ANOVA followed by Tukey’s post-hoc analysis. N = 3/age group. Scale bars, 10µm (A–D). See also Table S1, Figure S3.
Nucleoporin Pathology in Human HD and JHD Brain Tissue
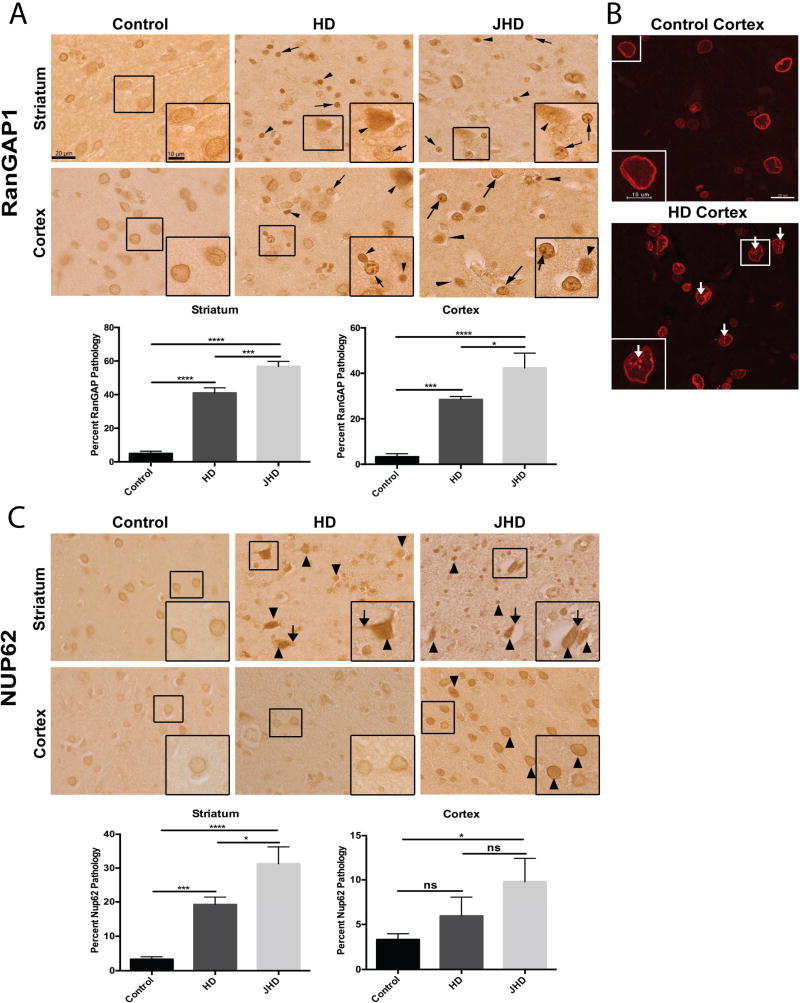
To determine whether RanGAP1 and NUP62 mislocalization and/or aggregation occurs in human disease, we assessed post-mortem human brain tissue from HD and JHD patients. Brain cells in HD and JHD frontal cortex and striatum commonly exhibit either a mislocalization (arrow heads) or aggregation (arrows) of RanGAP1 compared to smooth perinuclear staining observed in controls (Figure 3A,B and Table S2). Similar pathology was detected in HD and JHD cerebellum (Figure S4A). Quantitatively, the pathologic mislocalization or aggregation of RanGAP1 was more common in JHD than in adult-onset HD brain regions, which might be predicted given that greater mHTT CAG repeats are found in JHD. We then investigated whether NUP62 also exhibited abnormal localization and/or aggregation. Although we did not detect NUP62 aggregation, NUP62 was dramatically mislocalized to either the cytoplasm (arrows) or inside the nucleus (arrow heads) in HD and JHD striatum, but not in cortex (except slightly for JHD) (Figure 3C) or cerebellum (Figure S4B). This is consistent with the observations that human mutations in NUP62 cause selective striatal pathology in infantile bilateral striatal necrosis (IBSN) (Basel-Vanagaite et al., 2006). NUP62 cytoplasmic or intranuclear mislocalization was also more pronounced in JHD than in adult-onset HD brains regions (Figure 3C). Additionally, we saw no pathology in translocated promoter region (TPR) (not shown), a NUP located in the nuclear basket of the NPC and involved in nuclear export of N-terminal Htt (Cornett et al., 2005). This was expected given that polyQ expansion and aggregation of mHtt decreases its interaction with TPR (Cornett et al., 2005) and would presumably render TPR unaffected.
Figure 3. Nucleoporin Pathology in Human HD and JHD Brain Tissue.
(A) Immunohistochemical RanGAP1 staining in non-neurological disease control (n=10), HD (n=5) and JHD (n=5) striatum and frontal cortex showing aberrant nuclear aggregates (arrows) and intense nuclear mislocalization (arrowheads). Quantitation of percent of RanGAP1-positive cells with RanGAP1 pathology (nuclear aggregates or intense nuclear mislocalization) for each brain region shown on the right of the representative images. (B) Immunofluorescence RanGAP1 staining in non-neurological disease control and HD frontal cortex showing aberrant nuclear aggregates (arrows). (C) Immunohistochemical NUP62 staining in non-neurological disease control (n=10), HD (n=5), and JHD (n=5) striatum and frontal cortex showing intense cytoplasmic (arrows) and nuclear (arrowheads) mislocalization. Quantitation of percent of NUP62-positive cells with NUP62 pathology (intense nuclear and cytoplasmic mislocalization) for each brain region shown on the right of the representative images. Data are presented as mean ± SEM. *p < 0.05, ***P<0.001, and ****P<0.0001 as analyzed by one-way ANOVA followed by Tukey’s post-hoc analysis. Scale bars, 20µm (A–C); 10µm (A–C zoom inset). See also Table S2, Figure S4.
Nucleocytoplasmic Transport Defects in Human HD iPSC-Derived Neurons
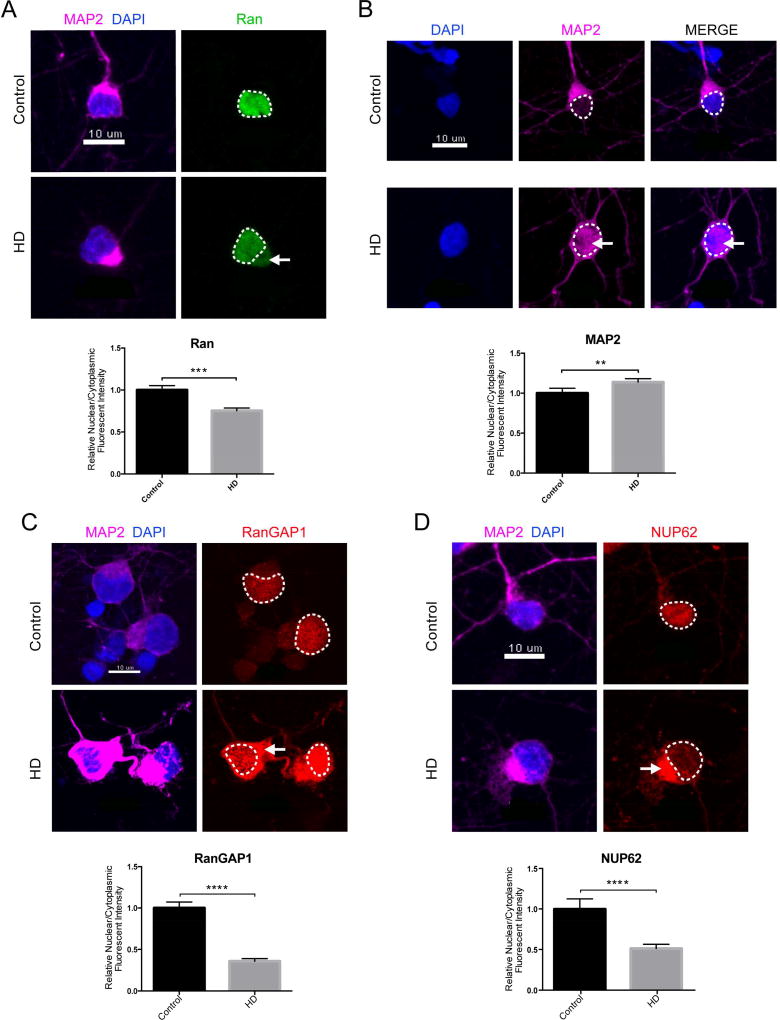
To validate our observations in human HD patient neurons (Table S3), we quantified nuclear and cytoplasmic (N/C) endogenous Ran in HD iPSC-derived neurons (iPSN) via immunofluorescence. High levels of Ran-GTP in the nucleus compared to the cytoplasm are required to fuel canonical active transport through the NPC. We observed a significant reduction in the N/C ratio of endogenous Ran in our HD iPSN lines, suggesting that active transport is deficient in HD (Figure 4A). We next assessed passive transport through the NPC by quantifying N/C MAP2 via immunofluorescence. MAP2 (~70 kDa) is a cytoplasmic protein that is normally excluded from the nucleus by the NPC (~40 kDa passive diffusion limit) (Izant and McIntosh, 1980). We found a significant increase in the N/C ratio of endogenous MAP2 indicating that the NPC may be leaky and compromised in HD (Figure 4B). We next quantified the N/C ratio of RanGAP1 and NUP62 via immunofluorescence and found significant decreases in both, providing additional evidence that these NUPs and NPC-associated proteins are severely mislocalized in HD (Figure 4C–D).
Figure 4. Nucleocytoplasmic Transport Defects in Human HD iPSC-Derived Neurons.
(A) iPSC-derived neurons (iPSNs) from control and HD patients showing mislocalization of Ran (green) to the cytoplasm in HD iPSNs. Quantification of N/C Ran gradient in neurons from 1 control (110 neurons) and 2 HD (144 neurons) iPSN lines when normalized to control shown below representative image. N/C Ran ratio is reduced in HD neurons. Bar indicates mean N/C Ran. (B) iPSNs from control and HD patients showing leakage of MAP2 (magenta) to the nucleus in HD iPSNs. Quantification of N/C MAP2 gradient in neurons from 1 control (110 neurons) and 2 HD (296 neurons) iPSN lines when normalized to control shown below representative image. Bar indicates mean N/C MAP2. (C) iPSNs from control and HD patients showing mislocalization of RanGAP1 (red) to the cytoplasm in HD iPSNs. Quantification of N/C RanGAP1 gradient in neurons from 1 control (30 neurons) and 2 HD (110 neurons) iPSN lines when normalized to control shown below representative image. Bar indicates mean N/C RanGAP1. (D) iPSNs from control and HD patients showing mislocalization of NUP62 (red) to the cytoplasm in HD iPSNs. Quantification of N/C NUP62 gradient in neurons from 1 control (100 neurons) and 2 HD (186 neurons) iPSN lines when normalized to control shown below representative image. Bar indicates mean N/C NUP62. Data are presented as mean ± SEM. Each independent experiment represents the average of 6 wells total per condition per line repeated over three separate differentiations. **P<0.01, ***P<0.001, ****P<0.0001 as analyzed by unpaired Students t test with Welch’s correction. Scale bars, 10µm (A–D). See also Table S3.
Nucleocytoplasmic Transport Defects in Primary Neurons Transfected with Full-Length mHTT
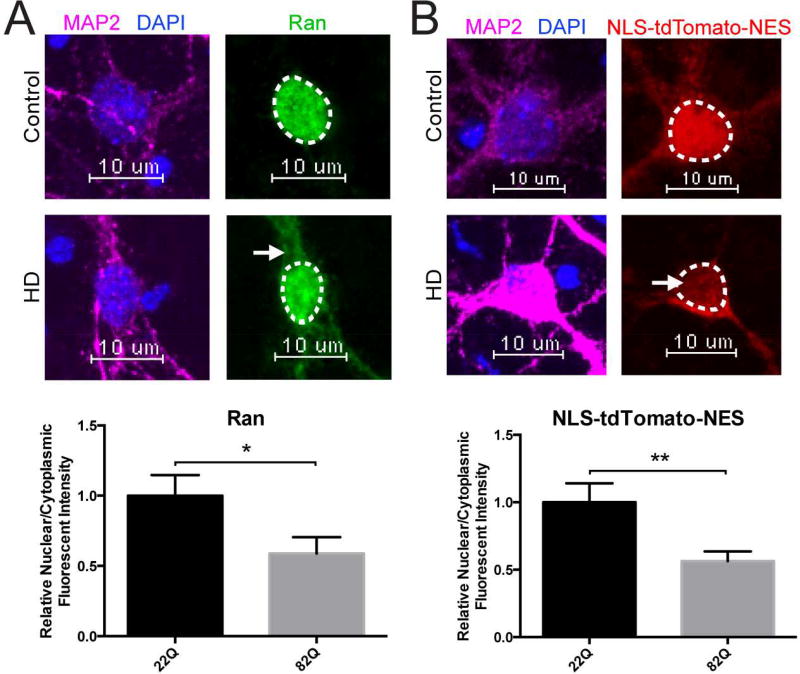
As an additional model system, we investigated the disruption of neuronal nucleocytoplasmic transport by employing rodent cortical neurons transfected at DIV5 with full-length HTT 22Q (control) or HTT 82Q (HD) and examined active nucleocytoplasmic transport via assessment of the Ran N/C gradient. We observed a marked mislocalization of Ran (green) to the cytoplasm in primary cortical neurons transfected with HTT 82Q, with an approximate 50% decrease in the Ran N/C gradient upon quantification compared to cortical neurons transfected with HTT 22Q (Figure 5A). To directly measure the nucleocytoplasmic transport of a protein reporter, we co-transfected primary cortical neurons at DIV5 with HTT 22Q or HTT 82Q and NLS-tdTomato-NES, a tdTomato protein fused with a classical Nuclear Localization Signal (NLS) and Nuclear Export Signal (NES), and measured the N/C ratio of NLS-tdTomato-NES. Again, we observed mislocalization of NLS-tdTomato-NES (red) to the cytoplasm in primary cortical neurons transfected with HTT 82Q and an approximate 50% loss of nuclear stain (Figure 5B). Collectively, these results may indicate that nuclear import is especially deficient in HD.
Figure 5. Nucleocytoplasmic Transport Defects in Primary Neurons Transfected with Full-Length mHTT.
(A) Primary cortical neurons transfected at DIV5 with HTT 22Q (control) or HTT 82Q showing mislocalization of Ran (green) to the cytoplasm in primary cortical neurons transfected with HTT 82Q. Quantification of N/C Ran gradient in neurons transfected with 22Q (25 neurons) and 82Q (23 neurons) when normalized to control shown below representative image. Bar indicates mean N/C Ran. Experiment represents the average of 3 wells. (B) Primary cortical neurons cotransfected at DIV5 with HTT 22Q (control) or HTT 82Q (HD) and NLS-tdTomato-NES showing mislocalization of NLS-tdTomato-NES (red) to the cytoplasm in primary cortical neurons transfected with HTT 82Q. Quantification of N/C NLS-tdTomato-NES gradient in neurons transfected with 22Q (48 neurons) and 82Q (54 neurons) when normalized to control shown below representative image. N/C NLS-tdTomato-NES is reduced in primary cortical neurons transfected with 82Q. Bar indicates mean N/C NLS-tdTomato-NES. Experiment represents average of 9 wells. Data (A–B) are presented as mean ± SEM. *p < 0.05 and **P<0.01 as analyzed by unpaired Students t test with Welch’s correction. Scale bars, 10µm (A–B).
HD-RAN Proteins Disrupt Nucleocytoplasmic Transport
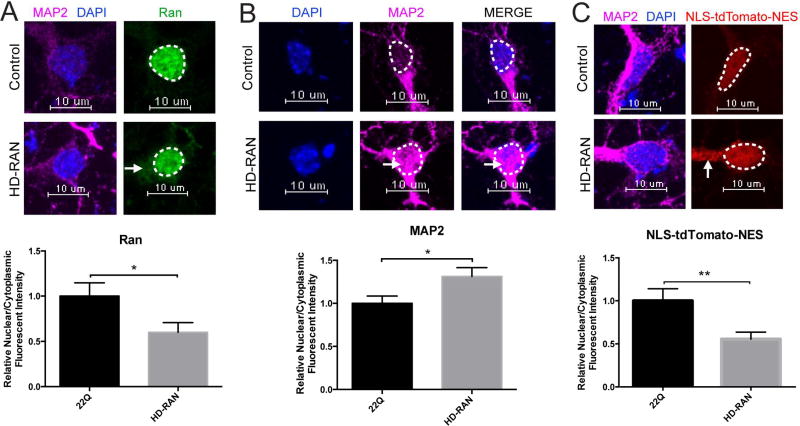
It was recently demonstrated that in addition to the HTT polyGln-expansion protein, four additional homopolymeric expansion proteins (polyAla, polySer, polyLeu, and polyCys) are generated in HD (Bañez-Coronel et al., 2015) from the nucleotide repeat. These sense and antisense HD repeat-associated non-ATG translation (HD-RAN) proteins are also toxic to neural cells. In order to initially assess whether HD-RAN proteins can alter nucleocytoplasmic transport, we transfected cortical primary neurons with a CAG HTT-exon1 minigene with a 6× STOP codon cassette (two stops in each frame) upstream of HTT exon1 (Bañez-Coronel et al., 2015) to generate these HD-RAN proteins (Figure S5) and quantified N/C Ran (active transport) and N/C MAP2 (passive transport) via immunofluorescence. We observed a significant reduction in the N/C ratio of endogenous Ran (Figure 6A) and a significant increase in the N/C ratio of endogenous MAP2 (Figure 6B) indicating that HD-RAN proteins can cause defects in both active and passive nucleocytoplasmic transport. Finally, we co-expressed the aforementioned HD-RAN construct with a tdTomato protein tagged with an NLS and NES and quantified N/C NLS-tdTomato-NES. We observed a significant decrease in the N/C ratio of NLS-tdTomato-NES (Figure 6C) indicating that HD-RAN proteins may also contribute to defects in nuclear import.
Figure 6. HD-RAN Proteins Disrupt Nucleocytoplasmic Transport.
(A) Primary cortical neurons transfected at DIV5 with HTT 22Q (control) or 6×Stop-(CAG)80 (HD-RAN) showing mislocalization of Ran (green) to the cytoplasm in primary cortical neurons transfected with 6×Stop-(CAG)80. Quantification of N/C Ran gradient in neurons transfected with 22Q (25 neurons) and 6×Stop-(CAG)80 (27 neurons) when normalized to control shown below representative image. Bar indicates mean N/C Ran. Experiment represents average of 2 wells. (B) Primary cortical neurons transfected at DIV5 with HTT 22Q (control) or 6×Stop-(CAG)80 (HD-RAN) showing leakage of MAP2 (magenta) to the nucleus in primary cortical neurons transfected with 6×Stop-(CAG)80. Quantification of N/C MAP2 gradient in neurons transfected with 22Q (50 neurons) and 6×Stop-(CAG)80 (54 neurons) when normalized to control shown below representative image. Bar indicates mean N/C MAP2. Experiment represents average of 6 wells. (C) Primary cortical neurons cotransfected at DIV5 with HTT 22Q (control) or 6×Stop-(CAG)80 (HD-RAN) and NLS-tdTomato-NES showing mislocalization of NLS-tdTomato- NES (red) to the cytoplasm in primary cortical neurons transfected with 6×Stop-(CAG)80. Quantification of N/C NLS-tdTomato-NES gradient in neurons transfected with 22Q (48 neurons) and 6×Stop-(CAG)80 (51 neurons) when normalized to control shown below representative image. Bar indicates mean N/C NLS-tdTomato-NES. Experiment represents average of 6 wells. Data (A–C) are presented as mean ± SEM. *p < 0.05 and **P<0.01 as analyzed by unpaired Student’s t test with Welch’s correction. Scale bars, 10µm (A–C). See also Figure S5.
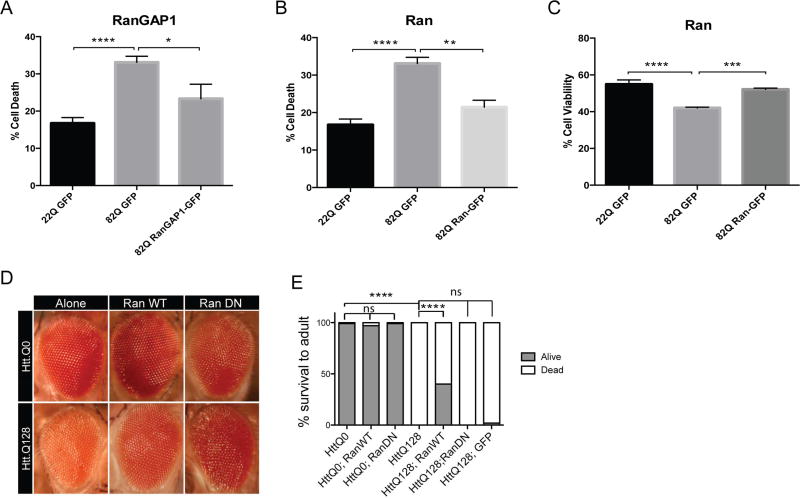
Overexpression of Ran and RanGAP1 are Neuroprotective in HD
To assess the extent to which the nucleocytoplasmic transport pathway may contribute to HD, we overexpressed a functional RanGAP1-GFP fusion protein in primary mouse cortical neurons that were co-transfected with either full-length HTT 23Q (control) or HTT 82Q (HD) and eGFP and performed a nuclear condensation assay to assess cell death (Figure 7A). We found that overexpression of RanGAP1-GFP significantly reduced cell death in primary cortical neurons that were transfected with HTT 82Q (Figure 7A). Overexpression of a functional Ran-GFP fusion protein also significantly reduced cell death (Figure 7B) and significantly increased cell viability as assessed with the alamarBlue Assay (Figure 7C). Finally, to test whether overexpression of Ran could modulate the mHTT phenotype in vivo, we tested the genetic interaction of Ran with Htt in an HD expanded repeat Drosophila model (Figure 7D). Using the GAL4/UAS system to express an N-terminal fragment of the human HTT protein in Drosophila tissues results in HTT aggregate formation and neurodegeneration (Lee et al., 2004). Expression of HTT 128Q but not HTT 0Q in the eye using GMR-GAL4, produced disruption in the external eye morphology including ommatidial disorganization and loss of pigment consistent with previous observations, a phenotype not observed upon expression of HTT 0Q. These phenotypes are rescued when we overexpress wildtype (WT) Ran and enhanced when we express a dominant negative (DN) form of Ran (Figure 7D). To confirm this genetic interaction in another neuronal subtype, we expressed HTT 128Q in motor neurons using the OK371-GAL4 driver causing complete pupal lethality (Figure 7E). However, when WT Ran without GFP or DN Ran, is coexpressed with Htt 128Q, there is a marked rescue of lethality with ~40% of flies surviving to adulthood.
Figure 7. Overexpression of Ran and RanGAP1 are Neuroprotective in HD.
(A,B) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant cell death compared to neurons transfected with HTT 23Q and eGFP, and cell death is significantly reduced when overexpressing RanGAP1-GFP or Ran-GFP. Experiment represents the average of 4 wells per condition. (C) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes a significant reduction in cell viability compared to neurons transfected with HTT 23Q and eGFP, and is rescued when overexpressing Ran-GFP. Experiment represents the average of 4 wells total per condition over the course of two separate experiments. Data (A–C) are presented as mean ± SEM. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 as analyzed by One-way ANOVA followed by Tukey’s post-hoc analysis. (D) Overexpression in the Drosophila eye of HTT.Q0 or HTT.Q128 using GMR-Gal4. Co-expressing wildtype (WT) Ran rescues this disorganization, whereas co-expression of a dominant negative (DN) Ran allele enhances this disorganization phenotype. (E) Overexpression of HTT.Q128 in motor neurons using OK371-Gal4 causes lethality at the pupal stage (Fisher’s exact test, p <.0001), whereas overexpression of HTT.Q0 has no phenotype alone or with Ran alleles. Co-expression of RanWT is sufficient to partially rescue the lethality caused by HTT.Q128 expression (Fisher’s exact test, p <.0001), whereas co-expression of RanDN or GFP alone was not sufficient to rescue.
Pharmacological Rescue of Nucleocytoplasmic Transport Defects and Neurotoxicity in HD
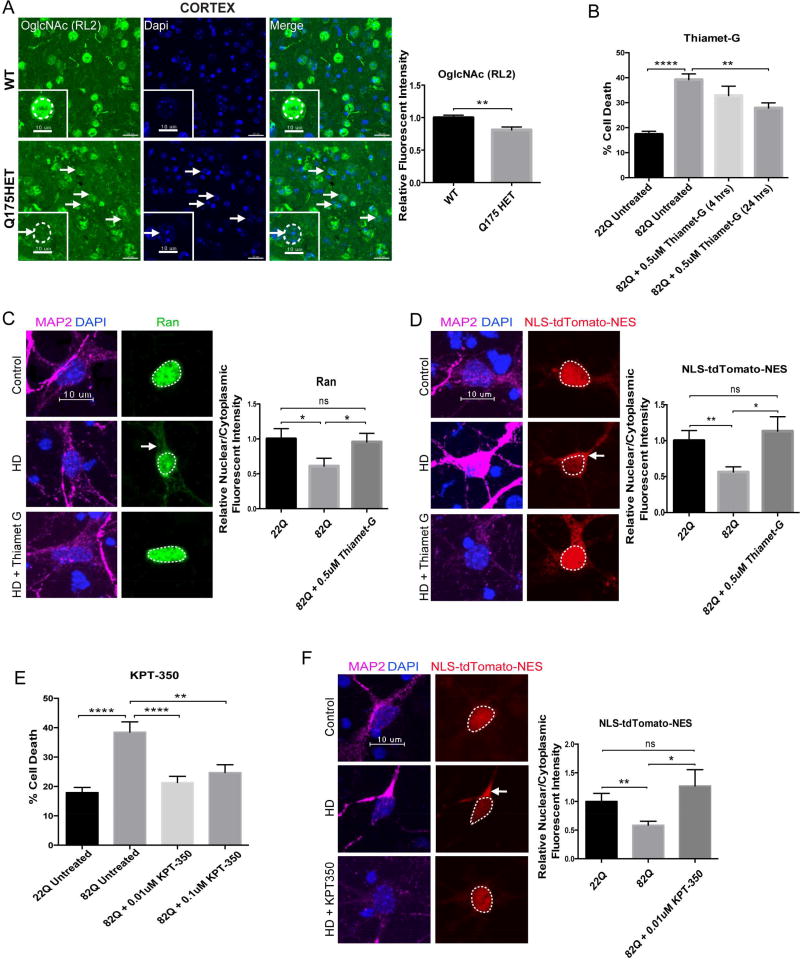
We next evaluated two additional pathways that might regulate function of NUPs and NPC-associated proteins and restore their proper homeostasis. First, O-GlcNAcylation is a post-translational modification in which β-N-acetylglucosamine (GlcNAc) is added to serine or threonine residues within intracellular proteins (Zachara and Hart, 2006). Interestingly, NUPs are the most heavily O-GlcNAc-modified proteins and are among the best substrates of O-GlcNAc transferase (OGT), the enzyme that catalyzes the addition of O-GlcNAc to proteins (Li and Kohler, 2014; Yang, 2016). Because O-GlcNAcylation is essential for NPC integrity and function as well as maintenance of the pore selectivity filter (Zhu et al., 2016), and 10 O-GlcNAc residues have been detected on NUP62, we quantified the nuclear levels of O-GlcNAc found specifically on NUPs using immunofluorescence in the cortex of 12 month old zQ175 animals (Figure 8A). Using the RL2 antibody that predominantly recognizes this post-translational modification on NUPs (Li and Kohler, 2014), we observed that O-GlcNAc is significantly decreased in cortical cells of Het zQ175 mice (Figure 8A). This suggests that NUPs like NUP62 which is mislocalized in HD cultured neurons (Figure 4D) and in rodent (Figure 1C,D) and human brain (Figure 3C), may be susceptible to mislocalization due to decreased levels and/or misregulation of O-GlcNAc. We therefore treated primary cortical neurons cotransfected with full length HTT 23Q (control) or HTT 82Q (HD) and eGFP with Thiamet-G (0.5uM). Thiamet-G is a potent selective inhibitor of O-GlcNAcase (OGA) that catalyzes the removal of O-GlcNAc, resulting in elevated levels of O-GlcNAc (Yuzwa et al., 2014). We observed that treatment with Thiamet-G significantly reduced cell death in primary cortical neurons transfected with Htt 82Q (Figure 8B) and increased cell viability (Figure S6A). Treatment with Thiamet-G also rescued nucleocytoplasmic trafficking defects with the nuclear restoration of both endogenous Ran (Figure 8C) and exogenous NLS-tdTomato-NES (Figure 8D).
Figure 8. Pharmacological Rescue of Nucleocytoplasmic Transport Defects and Neurotoxicity in HD.
(A) Coronal brain sections from WT and HET zQ175 mice showing decreased relative nuclear fluorescent intensity of O-GlcNAc (RL2) in the cortex of 12M HET zQ175 mice. Quantification of relative nuclear fluorescent intensity of O-GlcNAc (RL2) in cortical cells from 3 WT (150 neurons; 50 neurons each) and 3 zQ175 HET (150 neurons; 50 neurons each) when normalized to WT shown next to representative image. Bar indicates mean nuclear O-GlcNAc (RL2). Data is presented as mean ± SEM. **P<0.01 as analyzed by unpaired Student’s t test with Welch’s correction. (B) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant cell death compared to neurons transfected with HTT 23Q and eGFP, and cell death is significantly reduced when treating cells with 0.5uM Thiamet-G for 24 hours beginning 24 hours after transfection. Experiment represents the average of 4 wells. (C) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant mislocalization of Ran (green) to the cytoplasm, which is rescued back to control levels upon treatment with 0.5uM Thiamet-G for 4 hours beginning 44 hours after transfection. Quantification of N/C Ran gradient in neurons transfected with 22Q (25 neurons), 82Q (23 neurons), and 82Q + Thiamet G (30 neurons) when normalized to control shown next to representative image. Bar indicates mean N/C Ran. Experiment represents the average of 3 wells. (D) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant mislocalization of exogenous NLS-tdTomato-NES (red) to the cytoplasm, which is rescued back to control levels upon treatment with 0.5uM Thiamet-G for 4 hours beginning 44 hours after transfection. Quantification of N/C NLS-tdTomato-NES gradient in neurons transfected with 22Q (48 neurons), 82Q (54 neurons), and 82Q + Thiamet G (32 neurons) when normalized to control shown next to representative image. Bar indicates mean N/C NLS-tdTomato-NES. Experiment represents average of 9 wells. (E) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant cell death compared to neurons transfected with HTT 23Q and eGFP, and cell death is significantly reduced when treating cells with either 0.01uM or 0.1uM KPT-350 at the time of transfection for 48 hours. Experiment represents the average of 4 wells per condition. (F) Overexpression of HTT 82Q and eGFP in primary cortical neurons causes significant mislocalization of endogenous NLS-tdTomato-NES (red) to the cytoplasm, which is rescued back to control levels upon treatment with 0.01uM KPT-350 at the time of transfection for 48 hours. Quantification of N/C NLS-tdTomato-NES gradient in neurons transfected with 22Q (48 neurons), 82Q (54 neurons), and 82Q + KPT-350 (20 neurons) when normalized to control shown next to representative image. Bar indicates mean N/C NLS-tdTomato-NES. Experiment represents average of 3 wells. Data (B–F) are presented as mean ± SEM. *P<0.05, **P<0.01, and ****P<0.0001 as analyzed by one-way ANOVA followed by Tukey’s post-hoc analysis. Scale bars, 20µm (A); 10µm (A zoom inset, C, D, F). See also Figure S6.
Next, we assessed the therapeutic potential of KPT-350, a potent inhibitor of Exportin-1 (XPO1 or CRM1), a NPC-associated protein responsible for the nuclear export of proteins bearing a leucine-rich nuclear export signal (NES) and RNAs (Wente and Rout, 2010). KPT-350 was recently shown to be neuroprotective in models of inflammatory demyelination (Haines et al., 2015) and KPT-276, a closely related molecule, was shown to be neuroprotective in C9orf72-ALS models (Zhang et al., 2015). Treatment with KPT-350 (0.01 and 0.1uM) significantly reduced cell death in primary cortical neurons transfected with HTT 82Q (Figure 8E) and significantly increased cell viability (Figure S6B). Treatment with KPT-350 also rescued nucleocytoplasmic trafficking defects as seen with nuclear restoration of exogenous NLS-tdTomato-NES (Figure 8F). This suggests that inhibiting nuclear export may be neuroprotective and able to mitigate nucleocytoplasmic transport defects in HD by compensating for disrupted nuclear import.
DISCUSSION
Using mouse models, post-mortem human tissue, a drosophila model, human patient-derived neurons, and primary neurons transfected with full-length mHTT, we demonstrate that products of the CAG triplet repeat expansion within the HD gene cause disruption of the nuclear pore complex and nucleocytoplasmic transport. First, multiple proteins of the NPC, particularly those from the cytoplasmic ring/filaments and central channel regions, mislocalize and/or aggregate in the striatum and cortex of both an N-terminal mutant HTT transgenic and full-length mutant Htt knock-in mouse model of HD and human HD and JHD post-mortem tissue, in iPS-neurons (iPSNs) from HD human patients, and primary neurons transfected with full-length mHTT. These nucleoporin aggregates colocalize with mHtt and increase in frequency and diameter with disease progression. Second, the insoluble levels of one of these proteins, RanGAP1, which is essential for maintaining the energy gradient (Ran) that fuels active nucleocytoplasmic transport, decreases in the nuclear enriched fraction with disease progression. This can be rescued when knocking down levels of PIAS1, which was recently shown to decrease aberrant mHtt accumulation (Ochaba et al., 2016) and thus may free up proteins like NUPs that are caught up in insoluble protein aggregates and complexes. Third, iPSNs from human HD patients and primary neurons transfected with full-length mHTT show a disruption in the Ran gradient and increased permeability of the NPC indicating that both active and passive nucleocytoplasmic transport is disrupted in HD. Also, exogenous NLS-tdTomato-NES mislocalizes to the cytoplasm in primary neurons transfected with full-length mHTT suggesting that nuclear protein import may be especially deficient in HD. Additionally, recently discovered HD-RAN proteins can also contribute to deficits in nucleocytoplasmic transport. Finally, we can rescue cell death, increase cell viability, and restore nucleocytoplasmic transport function when overexpressing RanGAP1 or Ran in primary neurons transfected with full-length mHTT and in a Drosophila model of HD as well as when treating with Thiamet-G, an inhibitor that increases O-GlcNAc levels that highly regulate NPC homeostasis, or KPT-350, an inhibitor of canonical nuclear export.
There are prior clues to support our hypothesis that a disruption of nuclear events by mHTT is important for expanded CAG-repeat mediated pathophysiology. Previous studies showed that adding an exogenous NLS to polyglutamine expanded proteins such as small HTT amino-terminal fragments (Schilling et al., 2004) and full length HTT (Saudou et al., 1998) results in increased toxicity in cell and animal models, suggesting that nuclear localization of HTT polyglutamine expansions contributes to its toxicity. Additionally, adding an NES to full-length HTT, restricting it to the cytoplasm, reduces toxicity. However, the real target of nuclear disruption remained unknown. The comprehensive studies that we have amassed in human brain and multiple human cell and animal models, in vivo and in vitro, strongly suggests that mHTT compromises the gatekeeper of the nucleus, the NPC, and ultimately nucleocytoplasmic transport function. This pathophysiology may extend to other polyglutamine diseases given that nuclear shuttling and accumulation is common to the majority of polyglutamine disease proteins such as ataxin-2 in Spinocerebellar ataxia 2 (SCA2) (La Spada and Taylor, 2010). This may partially explain why the R6/2 mouse model exhibits such an extreme phenotype. This model uniquely expresses just the N-terminal fragment of HTT which contains no classic NLS or HEAT repeats but may be small enough to readily cross the NPC into the nucleus and potentially sequester significantly more NUPs compared to full-length Htt mouse models. In fact, the current study indicates that NUP aggregates are larger in diameter in the R6/2 mouse model compared to zQ175, alternatively, this could also be due to different expression levels of HTT. Further comparative studies in different animal models are needed.
Nucleocytoplasmic transport, the trafficking of protein and RNA between the nucleus and cytoplasm, is critical for the proper functioning of cells. This regulated and efficient communication of macromolecules is essential for signal transduction, neuronal plasticity, glial function and overall cellular survival (Dickmanns et al., 2015; Schachtrup et al., 2015). Altered nucleocytoplasmic distribution of proteins such as transcription factors is commonly seen across several neurodegenerative diseases and may be the result of defective nucleocytoplasmic transport. Examples include mislocalization of activating transcription factor 2 (ATF2), nuclear factor E2-related factor 2 (Nrf2), and importin-α1 in Alzheimer’s disease (AD) (Lee et al., 2006; Yamada et al., 1997), phosphorylated cyclic AMP response element-binding protein (pCREB) and phosphorylated extracellular signal-regulated kinase 1/2 (pERK1/2) in Parkinson’s disease (PD) (Chalovich et al., 2006; Ferrer et al., 2001), and TAR DNA binding protein 43 (TDP-43) in Frontotemporal Dementia (FTD) and Amyotrophic Lateral Sclerosis (ALS) (Neumann et al., 2006). Recent work from our group and others, using human, fly and yeast models revealed that an expanded hexanucleotide (GGGGCC) repeat (HRE) within a non-coding region of the C9orf72 gene causes dysfunction in nuclear import, which may explain the hallmark cytoplasmic accumulation of TDP-43 in ALS and FTD (Zhang et al., 2015). Given these prior studies in ALS, FTD and now HD, as well as a recent study demonstrating that certain cytoplasmic protein aggregates can interfere with nucleocytoplasmic trafficking of protein and RNA (Woerner et al., 2016), dysfunction in nucleocytoplasmic transport may be a shared common defect across multiple neurodegenerative disorders. Whether this pathway contributes to sporadic forms of these age-related degenerative diseases awaits further research.
Interestingly, altering NPC function has been shown to impact longevity (Lord et al., 2015). The NPC regulates not only nucleocytoplasmic transport but also genome organization, gene expression, cell differentiation, development and may be fundamental to brain aging. Some of the longest-lived proteins in the mammalian brain are components of the NPC and thus may represent a “weak link” in the aging proteome (Savas et al., 2012; Toyama et al., 2013). Because these NUPs persist for so long without being recycled, they have been shown to accumulate molecular damage over time, and this age-related deterioration results in an increase in nuclear permeability and the leaking of cytoplasmic proteins and toxins inside the nucleus (D'Angelo et al., 2009). In fact, the current study provides evidence of nuclear leakage of MAP2, a cytoplasmic protein, in several in vitro models of HD indicating a dysfunctional NPC. This could result in alterations in DNA and cellular aging. Given that aging is an independent risk factor for neurodegeneration, defects in the NPC may underlie neurodegeneration, and this injury may be mitigated by utilizing small molecules that act on this pathway such as Thiamet-G and KPT-350 as detailed in the current studies. In fact, Thiamet-G has already been shown to be neuroprotective in models of AD (Borghgraef et al., 2013; Yuzwa et al., 2014; Yuzwa et al., 2012) and KPT-350 and related SINE (Selective Inhibitor of Nuclear Export) compounds were recently shown to be neuroprotective in models of C9orf72-ALS (Zhang et al., 2015) and inflammatory demyelination (Haines et al., 2015). Therefore, these compounds may also prove therapeutic for other diseases that involve defects in NPC and nucleocytoplasmic transport function.
A question that often arises when studying neurodegeneration is why only some cells undergo injury when the mutant proteins or pathways are often ubiquitous? Very little is known about the neurobiology of the NPC in the CNS. Although the overall structure of the NPC is conserved across different cell types, studies suggest that cells may express unique combinations of NUPs to generate NPCs with specialized functions (Ori et al., 2013). There is evidence to suggest that individual NUPs may be responsible for the trafficking of unique macromolecules, thus adding a layer of diversity and specificity to the NPC (Raices and D'Angelo, 2012). This is best supported by the fact that mutations in various NUPs result in tissue-specific diseases. For instance, a missense mutation in NUP62 (Q391P) causes infantile bilateral striatal necrosis (IBSN), an autosomal recessive neurodegenerative disease that results in the selective degeneration of the corpus striatum in infants (Basel-Vanagaite et al., 2006). This suggests that NUP62 may play a critical role for the proper functioning of the corpus striatum. We now show that NUP62 mislocalizes and/or aggregates in several HD animal models, HD human tissue, and HD iPSC-derived neurons. In fact, NUP62 pathology was primarily found in HD human striatum, not cortex or cerebellum. Purified HTT fragment polyglutamine aggregates have been shown to sequester NUP62 (Suhr et al., 2001). The mechanism that underlies sequestration of NUP62 may mediate the selective degeneration of the striatum in HD. Future studies could address whether the composition of the NPC is different across the central nervous system at a regional and cellular level, yielding NPCs with distinct properties and specialized functions given brain cell type-specific constraints and context-dependent needs. If the composition of the NPC differs across CNS cell types and tissues, this may provide a plausible explanation for tissue-specific neurological diseases that arise from mutations in various NUPs like GLE1 in fetal motoneuron disease (Nousiainen et al.) and rare forms of ALS (Kaneb et al., 2015) and NUP62 in IBSN.
Our findings highlighting the contribution of O-GlcNAc and NPC proteins in HD provide a new avenue for investigation and therapeutics discovery. While we cannot rule out that our Thiamet-G treatment may have also modified other O-GlcNAc-modified proteins that indirectly altered nucleocytoplasmic transport to mitigate the injury observed, the current studies do show that altering nucleocytoplasmic transport function is one such candidate and not previously studied. Also, the O-GlcNAc antibody used in the current study, “RL2,” predominantly recognizes the O-GlcNAc modification on NPC proteins and these NUPs are the most heavily O-GlcNAc-modified proteins and are among the best substrates for O-GlcNAc transferase (OGT) (Li and Kohler, 2014; Yang, 2016). For example, down-regulation of O-GlcNAcylation selectively affects NUP62 and NUP88 but not TPR, in vitro, (Mizuguchi-Hata et al., 2013) which directly supports what we observe in our HD models. NUP88 homeostasis is also regulated through its interaction with O-GlcNAcylated NUP62, which suggests that the NPC defect in HD could be initiated with NUP62 mislocalization. O-GlcNAc also helps maintain NPC homeostasis by preventing the inappropriate aggregation and degradation of NUPs by decreasing ubiquitination or disturbing the ubiquitin-proteosomal degradation pathway (Zhu et al., 2016). Given the relationship between O-GlcNAc and NUP homeostasis and the fact that O-GlcNAc integrates metabolic information (Bond and Hanover, 2015; Hart, 2014), it is enticing to speculate whether distinct metabolic profiles across different regions of the brain (Magistretti and Allaman, 2015) may dictate NPC composition via O-GlcNAc signaling. Lastly, recent work showed that cytoplasmic aggregates such as tau can disrupt nucleocytoplasmic transport of protein and RNA (Woerner et al., 2016). Given we now show that Thiamet-G can restore nucleocytoplasmic transport function, it may also provide an additional explanation as to why Thiamet-G has already been shown to be neuroprotective against tauopathy (Borghgraef et al., 2013).
Overall, the current study provides compelling and comprehensive new data using multiple model systems and human brain, implicating aberrant NPC function, specifically its role in regulating nucleocytoplasmic transport, in the pathogenesis of HD and potentially other polyQ-expansion and neurodegenerative disorders. The mHTT polyQ protein is sufficient to induce nucleocytoplasmic trafficking deficits, however, future studies are required to further elucidate the contribution of newly discovered HD-RAN proteins in altering nucleocytoplasmic transport function. Although we demonstrate that the HD-RAN proteins can generally alter nucleocytoplasmic transport, it is still unknown as to which one of these specific protein(s) (polyAla, polySer, polyLeu, and polyCys) is/are mostly mediating this dysfunction. Interestingly, HD-RAN proteins appear to be more abundant in the cerebellum than the polyQ aggregates (Bañez-Coronel et al., 2015) and thus may be selectively responsible for the mislocalization or aggregation of RanGAP1 detected in HD but mostly JHD cerebellum in the current studies. This is consistent with recent reports demonstrating cerebellar pathology and atrophy in both adult and juvenile HD cases, which may explain some of the balance and gait abnormalities found in patients (Rub et al., 2013).
Lastly, future work should not only focus on further elucidating the role of nucleocytoplasmic transport in HD but also on other critical functions mediated by the NPC such as potential genome organization and gene expression alterations. Given the crucial role that the NPC plays in regulating the transport of macromolecules to and from the nucleus and local transcription, these new studies suggest that defects in the NPC may be playing a substantial role in HD pathogenesis and provide a new target for therapeutic interventions.
STAR★Methods
Contact for Reagent and Resource Sharing
Requests for additional information can be directed to Dr. Jeffrey D. Rothstein (jrothsteinjhmi.edu).
Experimental Model and Subject Details
Ethics Statement
All animal care, housing, and experiments were performed in compliance with guidelines established by the Research Animal Resources (RAR) at the Johns Hopkins University School of Medicine and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Research., 1996). Control and HD autopsy tissue was collected at Johns Hopkins with informed consent of patients or their relatives and approval of local institutional review boards.
HD Mouse Models
For all experiments, mice from both genders were used and group housed in standard cages under a 12 hr light/dark cycle with ad libitum access to water and food. Research Animal Resources (RAR) is Johns Hopkins’ centralized provider of veterinary medical care and research animal support. RAR ensures that mice are regularly checked for standard pathogens and health status. Health reports can be provided upon request. All mice had a standard specific-pathogen-free and immune status.
R6/2 mice
R6/2 transgenic mice and their NT littermates were obtained from either Jackson Laboratory (Bar Harbor, ME) or breeding colonies maintained at Johns Hopkins University School of Medicine (line 2810 ~150 ± 5 CAG repeats) or University of California, Irvine (line 6494 ~120 ± 5 CAG repeats). Offspring were genotyped using tail DNA. 10 week old R6/2 transgenic mice and their NT littermates were used for the immunofluorescence studies. 5, 7, 9, and 11 week old R6/2 transgenic mice and their NT littermates were used for the soluble/insoluble fractionation experiments and filter retardation assay.
zQ175 mice
zQ175 mice and wild-type littermate controls were acquired from Jackson Laboratory (Bar Harbor, ME). Genotyping and CAG repeat count were determined at Laragen Inc. (Culver City, CA, USA) by PCR of tail snips. The CAG repeat length was 193 ± 7 in heterozygous zQ175 mice used in the study. 2, 6, 9, and 12 month old zQ175 mice and wild-type littermate controls were used in this study. All mice were anesthetized and perfused transcardially with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. Brains were post-fixed overnight followed by immersion in 30% sucrose for 24 h. Coronal brain sections (40 µm) were cut on a cryostat and used for immunofluorescence. All animals have been described previously.
iPS Cell Culture
Human patient demographic information for control and HD iPSC lines is provided in Supplementary Table 3. Products were purchased from Thermo Fisher Scientific (Waltham, MA USA), unless otherwise specified. A control unaffected and apparently healthy non-disease GM03814 human fibroblast cell line was obtained from the Coriell Institute for Medical Research, under their consent and privacy guidelines as described. The parental fibroblast for the 109Q (Coriell, ND39258) repeat HD line was obtained from John’s Hopkins University under Dr. Russell Margolis’s IRB protocol #NA00018358 and the parental fibroblast line for the 53Q repeat HD line was obtained from UC Irvine IRB protocol #2008-6556. All procedures were performed in accordance with the institutional review board’s guidelines at the Cedars-Sinai Medical Center under the auspices of IRB-SCRO Protocols Pro00028429 (Transplantation of iPSC-derived human neural progenitors), Pro00021505, and Pro00032834. Upon iPSC generation at Cedars Sinai, they were renamed CS14CTR-28nXX, CS03iHD53nXX, and CS109iHD-109nXX to reflect, 1) last two digits of parental lines identifier, 2) non-disease or HD line, 3) CAG repeat number, and 4) XX is the clone number (PMID: 21474098). These iPSC lines are referred herein as 28Q, 53Q and 109Q, respectively. Fibroblasts were reprogrammed into non-integrating and virus-free iPSC lines with non-integrating episomal factors as described previously (PMID:25740845). iPSC colonies were maintained at 37°C, 95% air/5% CO2, in mTeSR™1 complete media (StemCell technologies, Vancouver, Canada) on hESC qualified Matrigel (Corning, Union City, CA, USA) coated, 6 well tissue culture dishes (Corning, USA. Cells were passaged every 3–4 days using Versene™.
Primary Neuron Culture
Primary mouse cortical and striatal neurons were prepared as described previously (Watkin et al 2015). Cortices and striatum of CD1 mice at embryonic day 17 were dissected out, digested with trypsin (0.05% Trypsin EDTA, GIBCO) for 10 minutes and mechanically dissociated. Neurons were resuspended in Neurobasal medium supplemented with B27 (Invitrogen) plated at 1×106 cell/cm2 on poly-D-lysine coated 24 well plates (Corning). Cells were kept in Neurobasal medium supplemented with B27 and 2mM GlutaMAX (Gibco, Rockville, MD, USA) in 5% CO2 at 37’C until the day of the experiment.
Drosophila Genetics
Flies were raised on standard food at 25°C unless otherwise noted. OK371-GAL4 and GMR-GAL4, and UAS-NLS-NESP12-GFP (referred to as UAS-GFP) were from Bloomington Drosophila Stock Center. UAS-Ran (Kumita et al.)-HA and UAS-Ran[T24N]-HA (dominant negative) were obtained from Kim McKim. UAS-Htt.Q128 and UAS-Htt.Q0 were obtained from Troy Littleton. To assess genetic interaction, OK371-GAL4; UAS-Ran* and GMR-GAL4; UAS-Ran* were crossed to either UAS-Htt.Q128 or UAS-Htt.Q0. For the eye phenotype, at least 20 flies per genotype were examined for ommatidial disorganization, loss of interommatidial bristles, photopigment loss, retinal collapse, and presence of necrotic spots. For pupal lethality assay, pupal cases were scored as eclosed (alive) or non-eclosed (dead) 15 days after the cross had been set.
Method Details
Immunofluorescence in Mouse Brain Tissue
40 µm-thick free-floating coronal sections were obtained from R6/2 mouse brains at 10 weeks of age and control and heterozygous zQ175 mouse brains at 2, 6, 9, and 12 months of age. Sections were incubated in blocking solution (1× PBS with 0.2% Triton X-100 and 10% normal goat serum (NGS)) for 1 hour. Next, sections were incubated with primary antibodies in blocking solution overnight at 4°C. Primary antibodies used are as follows: anti-RanGAP1 (1:100, Santa Cruz H-180 Cat# SC-25630), anti-NUP62 (1:50, Santa Cruz H-122 Cat#SC25523), anti-NUP88 (1:100, Abcam Cat#AB79785), and anti-O-GlcNAc (RL2) (1:1000, generously provided by Dr. Natasha Zachara and the Johns Hopkins O-GlcNAc Core). Summary of primary antibodies used to detect other NUPs and NPC-associated proteins are described in Table S1. After incubating overnight, sections were washed 3 times in 1× PBX (1× PBS with 0.1% Triton X-100) at room temperature (RT) for 30 minutes (10 minutes/wash). Sections were then incubated with goat secondary antibodies conjugated to Alexa Fluor 488 (1:1000), 555 (1:1000) and 633 (1:500) (Life Technologies) in blocking solution at RT for 1 hour. Sections were washed twice with 1× PBX for 20 minutes (10 minutes/wash), washed with 1× PBS and Hoescht dye (1:5000, Cell Signaling Technology Cat#4082S) for 10 minutes, and then transferred to a final 1× PBS wash. Sections were mounted using ProLong Gold Antifade Mountant with DAPI (Life Technologies).
Immunocytochemistry in Primary and iPS Neurons
Immunocytochemistry in neuronal cell lines has been described previously (Zhang et al., 2015). In brief, neurons were first washed with 1× PBS (1×), fixed with 4% PFA (diluted in 1× PBS) for 13 minutes, washed with 1× PBS (1×), permeabilized with 0.3% TritonX-100 (diluted in 1× PBS) for 10 minutes, washed with 1× PBS (2×), blocked with 1× PBS and 5% NGS for 1 hour, and incubated with primary antibodies in blocking solution overnight at 4°C. Primary antibodies used are as follows: anti-RanGAP1 (1:100, Santa Cruz H-180, Cat# SC-25630), anti-NUP62 (1:100, Santa Cruz H-122, Cat#SC25523), anti-NUP88 (1:200, Abcam, Cat#AB79785), anti-Ran (1:200, BD Biosciences, Cat#610341), anti-MAP2 (1:1000, Synaptic Systems, Cat#188004), and HD-RAN C-terminal antibody anti-polySer-Ct (1:500) (Bañez-Coronel et al., 2015). After incubating overnight, neurons were washed 3× with 1× PBS at room temperature (RT), incubated with goat secondary antibodies conjugated to Alexa Fluor 488 (1:1000), 555 (1:1000) and 633 (1:500) (Life Technologies) in blocking solution at RT for 1 hour. Neurons were then washed 3× with 1× PBS and mounted using ProLong Gold Antifade Mountant with DAPI (Life Technologies).
Immunohistochemistry in Human Brain Tissue
Immunohistochemistry (IHC) of the human autopsy tissue has been described previously (Zhang et al., 2015). To determine if nuclear pore pathology is present in HD we performed IHC to analyze the subcellular distribution of RanGAP1 as previously described (Zhang et al., 2015). Paraffin embedded sections of human brains from 5 adult HD, 5 juvenile HD (JHD) and 10 age/sex matched controls were subjected to immunohistochemical staining. Human patient demographic information for postmortem control and HD/JHD brain tissue is provided in Table S2. Paraffin sections (5um) from cerebellum, caudate and putamen (collectively labeled striatum), and medial frontal cortex were deparaffinized in xylene then rehydrated in a graded series of ethanol (100%, 100%, 95%, and 70%). Following rehydration, sections were steamed for 1 hour in HC-TekTM Epitope Retrieval Solution. Sections were then treated with 50/50 mixture of acetone/methanol to block endogenous peroxidase activity. Afterwards, sections were permeabilized with PBST (0.3% Triton X100) then blocked overnight (4C) in 10% normal goat serum PBST (0.2% Triton X100). To determine subcellular distribution of RanGAP1 and NUP62, sections were stained overnight at 4C with anti-RanGAP1 (1:100, Santa Cruz H-180 Cat# SC-25630) and anti-NUP62 (1:50, Santa Cruz H-122 Cat#SC25523). Sections were stained with goat anti-Rabbit-biotinylated (1:200, Vector Laboratories) at RT for 1 hr. Following secondary incubation sections were treated with Vectastain Elite ABC HRP Kit (Vector Laboratories, Cat#PK-6100) then developed with DAB (Vector Laboratories, Cat#SK-4100) for 2 min 30 sec. After DAB development, sections were mounted in ProLong Gold Antifade Mountant (Life Technologies). Once the mounting media cured, slides were sealed with nail polish and then imaged using brightfield on the Zeiss Apotome as described in the Histological Quantification Strategy (below).
Immunofluorescence in Human Brain Tissue
Human patient demographic information for postmortem control and HD/JHD brain tissue is provided in Table S2. Paraffin sections (5um) were deparaffinized in xylene (3×/5min each), then rehydrated in a graded series of ethanol (100%, 100%, 95%, and 70%/5min each), and washed in dH2O (3×/5min each). Following rehydration, sections were steamed for 1 hour in HC-TekTM Epitope Retrieval Solution. Sections were then washed with 1× PBS (3×/5min each) and treated with a 50/50 mixture of acetone/methanol to block endogenous peroxidase activity (10min). Afterwards, sections were washed with 1× PBS (2×/5min each), permeabilized with 0.4% Triton X100 in 1× PBS (8min), washed with 1× PBS (3×/5min) and blocked with Serum-free Protein Block (DAKO, X0909/2hrs). Next, sections were incubated with anti-RanGAP1 (1:100, Santa Cruz H-180 Cat# SC-25630) diluted in Antibody Diluent with Background Reducing Components (DAKO, S3022) overnight at 4°C. After incubating overnight, sections were washed in 1× PBS (3×/5min each) then incubated with goat secondary antibody conjugated to Alexa Fluor 555 (1:1000) (Life Technologies) in Antibody Diluent with Background Reducing Components at RT for 1 hour. Sections were then washed with 1× PBS (2×/5min each), 1× PBS and Hoescht dye (dilution 1:5000/5 min), and then transferred to a final 1× PBS wash (2×/5min each). Sections were mounted using ProLong Gold Antifade Mountant with DAPI (Life Technologies). Once the mounting media cured, slides were sealed with nail polish and subjected to confocal microscopy.
Histological Quantification Strategy
RanGAP1
To assess RanGAP1 pathology in HD/JHD human brain tissue, we randomly sampled 3–4 fields of view at 40× magnification from 3 different brain regions (cerebellum, striatum, frontal cortex) from 5 HD, 5 JHD, and 10 age/ sex-matched controls. Images were taken primarily in gray matter regions anatomically. To best assess pathology across the tissue, we quantified the distribution of RanGAP1 in all cells stained positive for RanGAP1. Grey matter neurons and glia that displayed positive RanGAP1 staining were included in the counts. The number of cells containing either mislocalized and/or aggregated RanGAP1 were included in the analysis. The percentage of cells containing abnormal subcellular localization or aggregation of RanGAP1 out of the total RanGAP1-positive cells were quantified for each section (N=~300 cells). To determine statistical significance, one-way ANOVA with Tukey’s post hoc analysis was performed and displayed as average percent.
(*p<0.05,**p<0.002,***p<0.0001).
NUP62
To quantify the extent of NUP62 pathology in HD/JHD human brain tissue we applied a similar method as stated above for RanGAP1. Cells in each brain region with mislocalization of the NUP62 protein from the nuclear envelope to either the cytoplasm or an intense nuclear staining were considered abnormal. Similarly, we performed a comprehensive sampling of multiple areas from each brain region noted in the RanGAP1 study. Percent of total cells displaying NUP62 were counted using ImageJ cell counter. The percentage of cells containing abnormal subcellular localization of NUP62 out of the total NUP62-positive cells were quantified for each section (N=~300 cells). To determine statistical significance, one-way ANOVA with Tukey’s post hoc analysis was performed and displayed as average percent.
(*p<0.05,**p<0.002,***p<0.0001).
Neuronal Differentiation
iPSC colonies were differentiated as described previously (PMID:26718628). Briefly, iPSCs were differentiated after reaching 60–70% confluency, cells were washed with PBS pH7.4 and medium was switched to SLI medium (Advanced DMEM/F12 (ADF) supplemented with 2 mM Glutamax™, 2% B27 without vitamin A (all Life Technologies), 10 µM SB431542 (Stem Cell Technologies), 1 µM LDN 193189 (Miltenyi Biotec) and 1.5 µM IWR1 (Tocris)) and cultured for 8 days with daily medium changes. At day 4 the cells were passaged 1:2 with accutase. At day 8, cells were passaged as above and transferred into LI medium (ADF supplemented with 2 mM Glutamax™, 2% B27 without vitamin A, 0.2 µM LDN 193189 and 1.5 µM IWR1. Cells were cultured in LI medium, with daily medium changes, until day 16, when they were dissociated from the dishes as above and frozen in Cryostor CS10 (StemCell Technologies) containing 10 µM Y27632 dihydrochloride until needed. Vials of day 16 cells were thawed in a 37 °C water bath, washed in ADF with 10 µM Y27632 dihydrochloride, centrifuged at 250 ×g for 3 minutes, and re-suspended in 1 ml SCM1 medium (ADF supplemented with 2 mM Glutamax™, 2% B27 supplement, 2 µM PD 0332991 (Tocris, USA), a CDK4/6 inhibitor, 10 µM DAPT (Tocris, USA), 10 ng/ml brain-derived neurotrophic factor (BDNF, Peprotech), 10 µM Forskolin, 3 µM CHIR 99021, 300 µM γ-amino butyric acid (GABA, all Tocris), supplemented with CaCl2 to final concentration of 1.8 mM) and 200 µM Ascorbic acid (both Sigma-Aldrich). A live/dead cell count was performed with trypan blue (Sigma-Aldrich) using a hemocytometer to allow accurate plating of 80,000 cells per well of a 4 well chamber slide in 80 µl of SCM1 medium on 100 µg/ml poly-D-lysine (Sigma-Aldrich)-/hESC-qualified Matrigel coated wells and allowed to settle for 60 minutes, prior to adding 500 µl SCM1 medium. A 50% medium change was performed every 2–3 days, then fully changed on day 23 to SCM2 medium (1:1 ADF:Neurobasal A supplemented with 2 mM Glutamax™, 2% B27 supplement, 2 µM PD 0332991, 10 ng/ml BDNF, 3 µM CHIR 99021, 1.8 mM CaCl2 and 200 µM Ascorbic acid). 1:1 medium changes continued every 2–3 days. The cells were then fixed on day 37 with 4% paraformaldehyde for 10 minutes at room temperature and shipped at 4°C in 1 × PBS pH 7.4.
Microscopy and Image Analysis
For all confocal imaging a Zeiss LSM 800 was used and quantifications were performed as previously described (Zhang et al., 2015). In brief, Z-stack images were taken at 1.0 uM intervals at identical settings within given experiments and individual planes were then projected into maximum intensity images for fluorescence intensity quantification using ImageJ. To determine quantity and diameter of aggregates, and percent colocalization using Z-stack images, IMARIS software spot detection in conjunction with the colocalization channel in 3D View was used in an unbiased manner.
Constructs
The HTT 22Q and 82Q constructs are full-length HTT expression plasmids described previously (Ratovitski et al., 2015). To generate HD-RAN proteins, a 6×Stop-(CAG)80 construct was used as previously described (Bañez-Coronel et al., 2015). In brief, this is a CAG and CAA HTT-exon1 minigenes with a 6× STOP codon cassette (two stops in each frame) upstream of HTT exon1 and C-terminal epitope tags in each of the three reading frames.
Cell Death and Viability Assays
Nuclear Condensation Assay (% cell death)
Neurons were cotransfected at DIV5 with HTT 23Q or HTT 82Q and eGFP (10:1 ratio) with lipofectamine 2000 per the manufacturer’s recommendations. After 48h of expression, cells were fixed with 4% paraformaldehyde for 30 minutes and nuclei were stained with Hoechst 33258 (bis-benzimide, Sigma-Aldrich). Treatments with KPT-350 were started at the time of transfection and neurons were exposed to two different doses (0.01uM and 0.1uM). Treatments with Thiamet-G (0.5uM) were started either 24 hours or 44 hours after transfection. Image acquisition was done using the Axiovision imaging software on an Axiovert 100 inverted microscope (Carl Zeiss). Analysis and quantification were performed using Volocity (Perkin-Elmer). Dapi intensity of GFP positive cells was measured. Cells were considered alive when the nuclear intensity was less than 200% of the intensity of control healthy cells. Results are shown as percentage of dead cells and each independent experiment represents the average of 4 wells per condition.
alamarBlue Assay (% cell viability)
The alamarBlue Cell Viability Reagent (Cat# DAL1025) was purchased from Thermo Fisher Scientific and the cell viability assay was performed as recommended by the manufacturer.
Soluble/Insoluble Fractionation Experiments
Whole mouse striatum tissue was processed for soluble/insoluble fractionation as previously described (Ochaba et al., 2016). Protein concentration was determined by BCA assay (Pierce, Rockford), and 30g of protein was reduced, loaded on 3–8% bis-acrylamide gels/4%–12% bis-tris mini gels (Life Technologies) for SDS-PAGE, transferred to nitrocellulose membrane, and nonspecific proteins were blocked with SuperBlock Blocking Buffer (Thermo Scientific). Primary antibodies used were: Anti-Huntingtin (Millipore Cat# MAB5492 RRID:AB_347723), Anti-RanGAP1 (Santa Cruz Cat# sc-25630) and anti-α-tubulin (Sigma-Aldrich Cat# T6074 RRID:AB_477582). Blots were developed using Pico/Dura Western Blotting Detection System (Pierce) and exposed to film for images. Protein quantification was performed using Scion Image analysis software. Band densities were normalized to α-tubulin.
Filter Retardation Assay
A total of 30 µg of detergent-soluble and detergent-insoluble protein in 200 µl of 2% SDS was boiled for 5 min and run through a dot blot apparatus under a vacuum onto a cellulose acetate membrane. Membrane was then washed 3× with 0.1% SDS and then blocked in 5% milk and subject to western blot analysis (previously described by (Sontag et al., 2012; Wanker et al., 1999).
Experimental Design
Multiple independent experiments were carried out using several sample replicates as detailed in the figure legends. For example, in Figure 4 involving the HD iPSC-derived neurons, each independent experiment represents the average of 6 wells total per condition per line repeated over three separate differentiations. Sample sizes were not chosen based on a pre-specified effect size. All quantifications and experiments were performed blinded. A strategy for randomization and/or stratification was not carried out.
Quantification and Statistical Analysis
Statistical analysis was performed using the Student’s t-test or One-Way Analysis of Variance with the Turkey’s or Dunnet’s posthoc test and the Prism 7 software (GraphPad Software, Inc).
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-RanGAP1 | Santa Cruz | Cat#SC25630; RRID: AB_2176978 |
| Rabbit anti-NUP62 | Santa Cruz | Cat#SC25523; RRID: AB_2157645 |
| Rabbit anti-NUP88 | Abcam | Cat#AB79785; RRID: AB_2042496 |
| Mouse anti-Huntingtin | Millipore | Cat#MAB5492; RRID: AB_347723 |
| Mouse anti-O-GlcNAc (RL2) | Johns Hopkins O-GlcNAc Core; Dr. Natasha Zachara CardioPEG CoreC4 | NHLBI P01 HL107153 |
| Mouse anti-Ran | BD Biosciences | Cat#610341; RRID: AB_397731 |
| Guinea Pig anti-MAP2 | Synaptic Systems | Cat#188004; RRID: AB_2138181 |
| Rabbit anti-polySer-Ct | Laboratory of Dr. Laura Ranum; (Bañez-Coronel et al., 2015) | N/A |
| Mouse anti-α-Tubulin | Sigma-Aldrich | Cat#T6074; RRID: AB_477582 |
| Alexa Fluor 555 Goat anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A21428; RRID: AB_2535849 |
| Alexa Fluor 488 Goat anti-Mouse IgG (H+L) | Thermo Fisher Scientific | Cat#A11001; RRID: AB_2534069 |
| Alexa Fluor 633 Goat anti-Guinea Pig IgG (H+L) | Thermo Fisher Scientific | Cat#21105; RRID: AB_2535757 |
| Biotinylated Goat anti-Rabbit IgG | Vector Laboratories | Cat#BA-1000; RRID: AB_2313606 |
| Vectastain Elite ABC HRP Kit | Vector Laboratories | Cat#PK-6100; RRID: AB_2336819 |
| DAB Substrate Kit | Vector Laboratories | Cat#SK-4100; RRID: AB_2336382 |
| Biological Samples | ||
| Non-neurological control, JHD, and HD brain tissue | JHMI Brain Resource Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Thiamet-G | Johns Hopkins O-GlcNAc Core; Dr. Natasha Zachara CardioPEG CoreC4 | NHLBI P01 HL107153 |
| KPT-350 | Karyopharm Therapeutics/Sharon Tamir | https://www.karyopharm.com/ |
| Critical Commercial Assays | ||
| alamarBlue Cell Viability Assay | Thermo Fisher Scientific | Cat#DAL1025 |
| Experimental Models: Cell Lines | ||
| CS14iCTR28 | Coriell Institute for Medical Research | Cat#GM03814 |
| CS03iHD53 (parental fibroblast for the 53Q repeat HD line was obtained from UC Irvine IRB protocol#2008-6556) | Coriell Institute for Medical Research | Cat#submitted, pending |
| CS09iHD109 (parental fibroblast for the 109Q repeat HD line was obtained from John’s Hopkins University IRB protocol#NA00018358) | Coriell Institute for Medical Research | Cat#ND39258 |
| Primary mouse cortical neurons | Laboratories of Drs. Christopher Ross and Jeffrey Rothstein | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | RRID: IMSR_JAX: 006494 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/1J | The Jackson Laboratory | RRID: IMSR_JAX: 002810 |
| Mouse: zQ175: B6J.129S1-Htttm1Mfc/190ChdiJ | The Jackson Laboratory | RRID: IMSR_JAX: 027410 |
| Drosophila: OK371-GAL4, GMR-GAL4, UAS-NLS-NES P12-GFP (referred to as UAS-GFP) | Bloomington Drosophila Stock Center | http://fly.bio.indiana.edu/ |
| UAS-Ran-HA and UAS-Ran[T24N]-HA (dominant negative) | Dr. Kim McKim | https://www.waksman.rutgers.edu/mckim/home |
| UAS-Htt.Q128 and UAS-Htt.Q0 | Dr. Troy Littleton | https://biology.mit.edu/people/j_troy_littleton |
| Recombinant DNA | ||
| Construct: 6xStop-(CAG)80 | Laboratory of Dr. Laura Ranum; (Bañez-Coronel et al., 2015) | N/A |
| Constructs: Full-length HTT 22Q and 82Q | Laboratory of Dr. Christopher Ross; (Ratovitski et al., 2015) | N/A |
| Software and Algorithms | ||
| Fiji Software | Fiji Contributors | http://fiji.sc/ RRID: SCR_002285 |
| Imaris | Bitplane | http://www.bitplane.com/ RRID: SCR_007370 |
| Volocity 3D Image Analysis Software | Perkin-Elmer | http://www.perkinelmer.com/pages/020/cellularimaging/products/volocity.xhtml RRID: SCR_002668 |
| Scion Image Analysis Software | Scion Corporation | http://www.scioncorp.com/pages/product_prices.htm#Software RRID: SCR_008673 |
| Axiovision Imaging Software | Carl Zeiss | http://www.zeiss.com/microscopy/en_de/products/microscope-software/axiovision-for-biology.html RRID:SCR_002677 |
| ZEN Digital Imaging for LSM 800 | Carl Zeiss | http://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html#introduction RRID: SCR_013672 |
| GraphPad Prism 7 | GraphPad Software | http://www.graphpad.com/ RRID: SCR_002798 |
| Other | ||
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668027 |
| SuperBlock Blocking Buffer | Thermo Fisher Scientific | Cat#37515 |
| Protein Block Serum-Free | Dako | Cat#X0909 |
| Antibody Diluent with Background Reducing Components | Dako | Cat#S3022 |
| HC-TekTM Epitope Retrieval Solution | IHC World | Cat#IW-1100 |
| Zeiss Axiovert 100 Inverted Microscope | Carl Zeiss | N/A |
| Zeiss LSM 800 Microscope | Carl Zeiss | N/A |
| Zeiss Apotome | Carl Zeiss | N/A |
Highlights.
HD human and mouse brain share profound nuclear pore complex pathology
HD iPS neurons mirror dysregulated HD human brain nuclear pore complex pathology
Aberrant nucleocytoplasmic transport is identified in various in vitro HD models
HD neurotoxicity can be mitigated with rescue of nucleocytoplasmic transport defects
Acknowledgments
Funded by grants from NIH (JDR, CR, LT, LR, SHS, TEL), National Science Foundation Graduate Research Fellowship Award (JCG), Thomas Shortman Training Fund Graduate Award (JCG), Axol Science Award (JCG), NIH Training in Neurotherapeutics Discovery and Development for Academic Scientists (JCG), MDI Biological Laboratory QFM Chroma Fellowship Award (JCG), National Science Foundation (JO), CIRM Training Grant (CG) and the Johns Hopkins Brain Science Institute (JDR). Drs. Ross, Troncoso, and Pletnikova provided human tissue and demographics. We wish to thank Michael J. Matunis for helpful discussions, Svetlana Videnskey for construct preparations and technical support, Natasha Zachara and the CardioPEG Core at Johns Hopkins (NHLBI PO1 HL107153) for providing Thiamet-G, Uzma Hussain for technical assistance and Sharon Tamir/Karyopharm Therapeutics for supplying KPT-350.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.C.G. designed, performed, and analyzed all the experiments including rodent, iPSC and human tissue studies and wrote the manuscript. J.G.D. optimized and performed histological analysis with assistance from J.G. and J.T.P. Additional experimental and data analytics support for human tissue and cell culture studies (J.C.T., O.P., L.P.W.R.); iPSC studies (C.G., L.M.T.); primary cultured rodent neurons (N.A., C.A.R.); fly studies (K.C.C., K.Z., H.W., T.E.L.); various HD mouse studies (I.A., Q.P., W.D., S.H.S.), and PIAS1 experiments (J.O., E.M., L.M.T.). J.D.R. oversaw project development, experimental design, data interpretation and manuscript writing. All authors contributed to revising and editing the manuscript.
None of the authors have competing financial interests.
References
- Bañez-Coronel M, Ayhan F, Tarabochia Alex D, Zu T, Perez Barbara A, Tusi Solaleh K, Pletnikova O, Borchelt David R, Ross Christopher A, Margolis Russell L, et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Muncher L, Straussberg R, Pasmanik-Chor M, Yahav M, Rainshtein L, Walsh CA, Magal N, Taub E, Drasinover V, et al. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann Neurol. 2006;60:214–222. doi: 10.1002/ana.20902. [DOI] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. 2015 doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghgraef P, Menuet C, Theunis C, Louis JV, Devijver H, Maurin H, Smet-Nocca C, Lippens G, Hilaire G, Gijsen H, et al. Increasing brain protein O-GlcNAc-ylation mitigates breathing defects and mortality of Tau.P301L mice. 2013 doi: 10.1371/journal.pone.0084442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Tredici KD, Lee VM-Y, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nature reviews Neuroscience. 2015;16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalovich EM, Zhu JH, Caltagarone J, Bowser R, Chu CT. Functional repression of cAMP response element in 6-hydroxydopamine-treated neuronal cells. J Biol Chem. 2006;281:17870–17881. doi: 10.1074/jbc.M602632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett J, Cao F, Wang C-E, Ross Ca, Bates GP, Li S-H, Li X-J. Polyglutamine expansion of huntingtin impairs its nuclear export. Nature genetics. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- D'Angelo Ma, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmanns A, Kehlenbach RH, Fahrenkrog B. Nuclear Pore Complexes and Nucleocytoplasmic Transport: From Structure to Function to Disease. International Review of Cell and Molecular Biology. 2015;320:171–233. doi: 10.1016/bs.ircmb.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Blanco R, Carmona M, Puig B, Barrachina M, Gomez C, Ambrosio S. Active, phosphorylation-dependent mitogen-activated protein kinase (MAPK/ERK), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), and p38 kinase expression in Parkinson's disease and Dementia with Lewy bodies. J Neural Transm (Vienna) 2001;108:1383–1396. doi: 10.1007/s007020100015. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. Huntington's Disease. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floch AG, Palancade B, Doye V. Fifty years of nuclear pores and nucleocytoplasmic transport studies: multiple tools revealing complex rules. Methods in cell biology. 2014;122:1–40. doi: 10.1016/B978-0-12-417160-2.00001-1. [DOI] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee K-H, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015 doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JD, Herbin O, Hera BD, Vidaurre OG, Moy GA, Sun Q, Yee H, Fung J, Albrecht S, Alexandropoulos K, et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. 2015 doi: 10.1038/nn.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW. Minireview series on the thirtieth anniversary of research on O-GlcNAcylation of nuclear and cytoplasmic proteins: Nutrient regulation of cellular metabolism and physiology by O-GlcNAcylation. The Journal of biological chemistry. 2014;289:34422–34423. doi: 10.1074/jbc.R114.609776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensman Moss DJ, Poulter M, Beck J, Hehir J, Polke JM, Campbell T, Adamson G, Mudanohwo E, McColgan P, Haworth A, et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology. 2014;82:292–299. doi: 10.1212/WNL.0000000000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol. 2002;4:E177–184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- Hosp F, Vossfeldt H, Heinig M, Vasiljevic D, Arumughan A, Wyler E, Landthaler M, Hubner N, Wanker Erich E, Lannfelt L, et al. Quantitative Interaction Proteomics of Neurodegenerative Disease Proteins. Cell Reports. 2015;11:1134–1146. doi: 10.1016/j.celrep.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izant JG, McIntosh JR. Microtubule-associated proteins: a monoclonal antibody to MAP2 binds to differentiated neurons. Proc Natl Acad Sci U S A. 1980;77:4741–4745. doi: 10.1073/pnas.77.8.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nature Neuroscience. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann aW, Belzil VV, Jao L-E, Leblond CS, Girard SL, Daoud H, Noreau a, Rochefort D, Hince P, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Human Molecular Genetics. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumita JR, Helmfors L, Williams J, Luheshi LM, Menzer L, Dumoulin M, Lomas DA, Crowther DC, Dobson CM, Brorsson AC. Disease-related amyloidogenic variants of human lysozyme trigger the unfolded protein response and disturb eye development in Drosophila melanogaster. FASEB J. 2012;26:192–202. doi: 10.1096/fj.11-185983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature reviews Genetics. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Ueda M, Miyamoto Y, Yoneda Y, Perry G, Smith MA, Zhu X. Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain Res. 2006;1124:1–4. doi: 10.1016/j.brainres.2006.09.084. [DOI] [PubMed] [Google Scholar]
- Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington's disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic (Copenhagen, Denmark) 2014;15:347–361. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol. 2011;23:65–70. doi: 10.1016/j.ceb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K-Y, Shyu Y-C, Barbaro Ba, Lin Y-T, Chern Y, Thompson LM, James Shen C-K, Marsh JL. Disruption of the nuclear membrane by perinuclear inclusions of mutant huntingtin causes cell-cycle re-entry and striatal cell death in mouse and cell models of Huntington's disease. Human Molecular Genetics. 2014;24:1602–1616. doi: 10.1093/hmg/ddu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CL, Timney BL, Rout MP, Wente SR. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. The Journal of cell biology. 2015;208:729–744. doi: 10.1083/jcb.201412024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- Mizuguchi-Hata C, Ogawa Y, Oka M, Yoneda Y. Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochimica et biophysica acta. 2013;1833:2682–2689. doi: 10.1016/j.bbamcr.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nousiainen HO, Kestila M Fau - Pakkasjarvi N, Pakkasjarvi N Fau - Honkala H, Honkala H Fau - Kuure S, Kuure S Fau - Tallila J, Tallila J Fau - Vuopala K, Vuopala K Fau - Ignatius J, Ignatius J Fau - Herva R, Herva R Fau - Peltonen L, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JG, Gareau JR, Ochaba J, Song W, Rasko T, Reverter D, Lee J, Monteys AM, Pallos J, Mee L, et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013;4:362–375. doi: 10.1016/j.celrep.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Monteys AM, Rourke JGO, Reidling JC, Steffan JS, Davidson BL, Thompson LM, Ochaba J, Monteys AM, Rourke JGO, et al. PIAS1 Regulates Mutant Huntingtin Accumulation and Huntington ’ s Disease-Associated Phenotypes Article PIAS1 Regulates Mutant Huntingtin Accumulation and Huntington ’ s Disease-Associated Phenotypes In Vivo. Neuron. 2016;90:507–520. doi: 10.1016/j.neuron.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Molecular systems biology. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouladi Ma, Morton aJ, Hayden MR. Choosing an animal model for the study of Huntington's disease. Nature reviews Neuroscience. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- Raices M, D'Angelo Ma. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nature reviews Molecular cell biology. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Raices M, D'Angelo MA. Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol. 2017;46:26–32. doi: 10.1016/j.ceb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA. PRMT5-mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD) Cell Cycle. 2015;14:1716–1729. doi: 10.1080/15384101.2015.1033595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Research., N.R.C.U.I.f.L.A. Guide for the Care and Use of Laboratory Animals. National Academies Press (US); 1996. [PubMed] [Google Scholar]
- Rub U, Hoche F, Brunt ER, Heinsen H, Seidel K, Del Turco D, Paulson HL, Bohl J, von Gall C, Vonsattel JP, et al. Degeneration of the cerebellum in Huntington's disease (HD): possible relevance for the clinical picture and potential gateway to pathological mechanisms of the disease process. Brain Pathol. 2013;23:165–177. doi: 10.1111/j.1750-3639.2012.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science (New York, NY) 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtrup C, Ryu JK, Mammadzada K, Khan AS, Carlton PM, Perez A, Christian F, Le Moan N, Vagena E, Baeza-Raja B, et al. Nuclear pore complex remodeling by p75NTR cleavage controls TGF-β signaling and astrocyte functions. Nature Neuroscience. 2015;18 doi: 10.1038/nn.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling G, Savonenko AV, Klevytska A, Morton JL, Tucker SM, Poirier M, Gale A, Chan N, Gonzales V, Slunt HH, et al. Nuclear-targeting of mutant huntingtin fragments produces Huntington's disease-like phenotypes in transgenic mice. Hum Mol Genet. 2004;13:1599–1610. doi: 10.1093/hmg/ddh175. [DOI] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, McKnight SL. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Lotz GP, Yang G, Sontag CJ, Cummings BJ, Glabe CG, Muchowski PJ, Thompson LM. Detection of Mutant Huntingtin Aggregation Conformers and Modulation of SDS-Soluble Fibrillar Oligomers by Small Molecules. J Huntingtons Dis. 2012;1:119–132. doi: 10.3233/JHD-2012-129004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr ST, Senut MC, Whitelegge JP, Faull KF, Cuizon DB, Gage FH. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. Journal of Cell Biology. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harbor perspectives in biology. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, Hipp MS. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yoshiyama Y, Kawaguchi N. Expression of activating transcription factor-2 (ATF-2), one of the cyclic AMP response element (CRE) binding proteins, in Alzheimer disease and non-neurological brain tissues. Brain Res. 1997;749:329–334. doi: 10.1016/S0006-8993(96)01356-X. [DOI] [PubMed] [Google Scholar]
- Yang ARaW. O-GlcNAc-ylation in the Nuclear Pore Complex. 2016 doi: 10.1007/s12195-016-0440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA, Shan X, Jones BA, Zhao G, Woodward ML, Li X, Zhu Y, McEachern EJ, Silverman MA, Watson NV, et al. Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic tau/APP mutant mice. Molecular Neurodegeneration. 2014;9:42. doi: 10.1186/1750-1326-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8 doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015 doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu TW, Madden Z, Yuzwa SA, Murray K, Cecioni S, Zachara N, Vocadlo DJ. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J Mol Cell Biol. 2016;8:2–16. doi: 10.1093/jmcb/mjv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.