The largest numbers of commensal bacteria reside within our intestinal tract, with an increasing density from mouth to anus. Recently, a new estimate for the total number of bacteria (3.8×1013) in the 70 kg ‘reference man’ was reported.1 For human cells, the same authors revised past estimates to 3.0×1013 cells, out of which approximately 90% belong to the haematopoietic lineage. Hence, the widely cited 10:1 ratio of bacteria versus human cells received an update, showing that the number of bacteria in the body is actually of the same order as the number of human cells, and that the cumulative bacterial mass is about 200 g. Still, this large number of bacteria highlights their importance in maintaining health and metabolism. Different parts of the intestinal tract have different functions, tissue structure varies accordingly and gradients exist for several physicochemical parameters such as nutrients, pH or oxygen levels.2 Consequently, microbiota composition varies along the gut, but also between luminal and mucosa-attached communities of the same intestinal segment, and even along the crypt-villus axis in the epithelium. Thus, host–microbiota interactions are likely regionally specific and the local crosstalk determines intestinal function and physiology. Probably each human individual carries its own ‘microbial fingerprint’ (especially when considering genomic variation within the bacterial species’ populations), which is why forensic scientists started to exploit the use of this non-human organ.3
Recent large-scale analyses of population-based cohorts with >1000 samples validated that body mass index, age at sampling and gender are important covariates that need to be included in microbiome association analyses.4 In sum, Falony et al identified 18 covariates, including stool consistency, dietary factors and blood traits, cumulatively explaining 7.7% of the total variation in the gut microbiota, leaving 92.3% of the interindividual microbial variation unexplained. In a recent host-microbiome genome-wide association study, 42 genetic loci were identified that explain another 10% of microbiome variability.5 Although these percentages need to be confirmed in other cohorts and for other ethnicities, both studies show that (A) a large portion of microbiome variability remains to be explained and (B) any microbiome study needs to account for covariates to prevent false-positive and false-negative results. This means for the field that the meta-data for a particular sample is at least as important for the data analysis as the actual microbiome data itself.
The main subtypes of IBD are Crohn’s disease (CD) and UC, affecting together more than 2.5 million European individuals. More than 200 genetic susceptibility loci have been identified for IBD, a contribution that significantly improved our disease understanding. In contrast to the static human genome, microbiome composition is more dynamic and can thus be more easily modified by, for example, probiotics, prebiotics, antibiotics or faecal microbiota transfer (FMT) from healthy donors to patients by enema or via the nasoduodenal route. The latter was demonstrated to be perhaps the best method to treat life-threatening recurrent Clostridium difficile infections,6–8 but is a controversial method for IBD and is therefore still regarded as an experimental therapy. Nevertheless, a recent multicentre placebo-controlled trial in Australia (the Faecal Microbiota Transplantation in Ulcerative Colitis (FOCUS) study) demonstrated efficacy of FMT for treating UC (p=0.021; enemas given at 5 days a week for overall 8 weeks).9 Based on this finding, it was suggested that future work should focus on defining the optimum treatment intensity and the role of donor–recipient matching based on microbial profiles. The growing body of studies using FMT and gnotobiotics started to move the field of microbiota research away from correlation to causality by proving a functional involvement of the microbiota for health and disease.10–12
Microbiome studies in IBD have been consistent in finding reduced diversity in patients compared with healthy controls (HC).13 14 A lack of microbial diversity seems to be a general theme in several diseases (obesity, diabetes, asthma, atopic dermatitis, and so on), suggesting that a species-rich ecosystem is more robust against environmental influences, as functionally related microbes exist in an intact ecosystem that can compensate for the function of another species that has disappeared. Consequently, diversity seems to be a good indicator of a ‘healthy gut.’15 A large study that analysed the microbiota in 447 treatment-naïve patients with new-onset CD (treatment effects on the microbiota can likely be excluded in this unique cohort) also identified different bacteria, which were more abundant in patients compared with controls and vice versa.16 Gevers et al also developed a microbial dysbiosis index that was correlated with the paediatric CD activity index. The dysbiotic signal in their study was stronger in the mucosa-attached microbial communities compared with faecal bacteria, suggesting that disease-relevant and more adherent bacteria are probably ‘diluted’ in the stool. The recently published longitudinal IBD microbiome study of Halfvarson and colleagues revealed that the microbiome of patients with IBD varies more over time compared with controls, corroborating that patients with IBD harbour a disturbed microbial ecosystem.17 The highest volatility of the microbiota, that is, dynamic deviation from a ‘healthy plane,’ was observed for patients with CD with ileal inflammation (lower volatility in colonic CD and UC). In an elegant visualisation study, Swidsinski et al demonstrated that the number of mucosa-associated bacteria increases with disease severity18 and together with reduced diversity, an increase in bacterial load seems to be a key feature of several diseases.19 Notably, diet can rapidly alter the gut microbiome with effects on disease susceptibility such as pathogenic infection or chronic inflammation.20–22 For example, removal of carbohydrates from the diet promotes the growth of mucus-utilising bacteria and alters microbial biogeography by attracting luminal bacteria to the mucus layer, which increases competition for beneficial mucus-resident bacteria such as Akkermansia muciniphila or Faecalibacterium prausnitzii, which in turn shifts epithelial responses, in particular goblet cell function.23 24 Oral but even intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD.25 Recent studies even suggest that changes in nutrition during the course of human history may have led to decreased microbiota diversity and the increased incidence of chronic inflammatory diseases and obesity.24 26 27 Another recent study has highlighted the potential effect of a disturbed host–microbiota crosstalk in IBD. This is evidenced by a striking uncoupling of host transcriptional patterns and microbiota signatures in IBD, with the affected pathways indicating a loss of cometabolic functions.28 It therefore seems imperative to correct for dietary variance in microbiota-related data and to explore dietary modulation of the metabolic properties of the gut ecosystem (eg, enrichment of dietary fibre) as a possible therapeutic approach for IBD. Figure 1 provides a schematic overview on known gut microbiome changes in IBD.
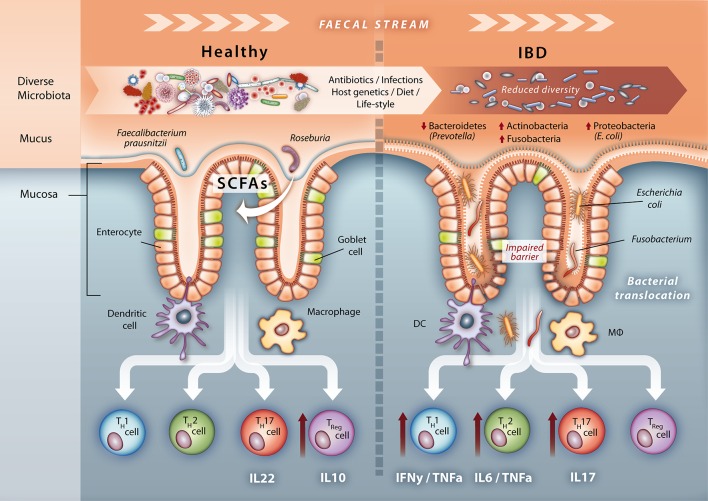
Figure 1.
Microbial signatures of a healthy gut and IBD. Under healthy homeostasis the microbiota is diverse. Goblet cells produce a thick colonic mucus layer, which creates a physical barrier against the microbiota, but also harbours a specific mucus-resident microbiota enriched in, for example, short-chain fatty acid producing bacteria Roseburia and Faecalibacterium prausnitzii. Immune sensory cells such as DC and MΦ sample microbial patterns and induce a T cell profile dominated by IL-10 producing Treg lymphocytes leading to homeostasis. However, the composition of the microbiota is less diverse in patients with IBD with fewer Bacteroidetes mainly attributed to loss of Prevotella species and expansion of Actinobacteria, Proteobacteria such as adherent invasive Escherichia coli and Fusobacteria. Mucosal function is also altered, for example, lipid metabolism, illustrating a cometabolism of the metaorganism. A reduction in Paneth and goblet cell number along with their impaired functions—secretion of antimicrobial substances and mucus—leads to decreased mucus thickness, reduced mucosal integrity and finally to an impaired barrier function. This increases bacterial translocation and stimulates activation of DCs and MΦs, which then induce an altered T cell profile with increased IFNγ/TNFα-producing Th1, IL-6/TNFα-producing Th2 and IL-17-producing Th17 lymphocytes resulting in a proinflammatory response and tissue damage, which in turn stabilises the dysbiotic microbiota and a chronic inflammatory tone. DC, dendritic cells; IFNγ, interferon gamma; IL, interleukin; MΦ, macrophages; TNFα, tumour necrosis factor alpha.
In the highlighted study, Pascal and colleagues29 report on an analysis of the faecal microbiome for a large IBD panel from four different countries (40 HC (unrelated HCs)/71 HR (healthy relatives from patients)/34 CD/74 UC from Spain, 54 CD from Belgium, 977 healthy twins from the UK and 158 patients with anorexia from Germany). In sum, they included about 1246 non-IBD and 162 IBD samples. The hypervariable region V4 of the 16S rRNA gene was targeted for microbiome profiling. Pascal et al replicate the findings of Gevers et al 16 that F. prausnitzii is reduced/absent in patients with CD and that patients have a higher abundance of Escherichia and Fusobacterium.
The key finding of their study is a ‘microbial signature’ that may be used to discriminate between CD and non-CD by eight prokaryotic groups: Faecalibacterium (genus from family Clostridiaceae), Peptostreptococcaceae (family from order Clostridiales), Anaerostipes (genus from family of Lachnospiraceae), Methanobrevibacter (genus from family Methanobacteriaceae), Christensenellaceae (family from order Clostridiales) and Collinsella (genus from family Coriobacteriaceae) being decreased in patients with CD; Fusobacterium (genus from family Fusobacteriaceae) and Escherichia (genus from family Enterobacteriaceae) having a higher relative abundance in patients with CD. The overall sensitivity of this diagnostic signature was 80%, with specificities of 89%–94% depending on the comparison. In comparison, assessing faecal calprotectin (a heterodimer of S100A8 (calgranulin A) and S100A9 (calgranulin B)), an established, non-invasive test that measures intestinal inflammation, has a sensitivity of 83% and specificity of 84% for distinguishing organic from functional intestinal diseases in symptomatic patients.30 However, diagnosis of IBD, and especially distinguishing between the different subtypes of IBD, still relies on a combination of diagnostic tests, including endoscopic and histological analyses, which showed high accuracy in a 12-month long follow-up, with only 1%–2% of patients with CD and UC being identified as false positives, but 9%–12% of patients with CD and UC being reclassified as another subform of IBD.31
While the study of Pascal et al employed a large sample panel in order to identify potential microbiomarkers of interest, a few words of caution are necessary. First of all, the findings still need to be validated by other centres using well-powered sample collections that include HC, CD, UC and IBD-related diseases, that is, differential diagnoses such as IBS. In a first attempt, Pascal et al applied their test to another cohort sequenced in another centre in France with another method (V3-V5 instead of V4). This led to a significantly lower sensitivity and specificity for the prediction of CD versus UC (60% and 68% respectively) and a lower sensitivity (60%) for the prediction of CD versus HC. While these results still suggest a non-random signal in the data, they also imply that larger and multicentre trials are clearly needed to produce a more robust and clinically useful diagnostic signature. It is noteworthy that Pascal et al included a very large UK control panel, making up almost half of their sample size, while they did not include a matched patient panel from the same geographical region. Anthropometric features as well as nutritional patterns can have a large impact on the gut’s microbial community and are a valuable resource in the investigation of connections between disease states and the microbiota. While these data will certainly lead to more complex models for the determination and distinguishing of dysbiotic states, they will most certainly lead to higher sensitivity and specificity of microbiome-based classifiers and should be considered for future approaches.
The regional specificity of the intestinal microbiota to different anatomical sites might partially explain why the authors failed for CD but were able to describe UC-specific microbiomarkers.2 16 Faeces may be a suitable surrogate for the colonic microbiota but not for that of the small intestine. Thus, the diagnosis of CD using microbiomarkers seems to require sampling of the local microbiota using biopsies.
To date, endoscopy is the gold standard to diagnose IBD and assess mucosal inflammation. Since endoscopy is an expensive and invasive procedure with the risk for complications, alternative scores, for example, the Harvey-Bradshaw index for CD32 or the simple clinical colitis activity index for UC,33 which include non-invasive measures such as general state of health, faecal properties and frequency or abdominal pain have been developed. These scores, however, do not correlate well with mucosal inflammation34 and therefore inflammatory markers such as C-reactive protein or faecal calprotectin are additionally used to evaluate disease activity. Still, levels of C-reactive protein are not specific for the intestine, but rather a general marker of systemic inflammation and faecal calprotectin correlates with UC but not CD and thus seems to be a marker of inflammation only for the colon.35 36 Thus, other non-invasive biomarkers are needed as surrogates for a healthy or inflamed gut ecosystem. Explorative metabolomics of faecal water showed already good differentiation of colonic versus ileal CD with a clear association of metabolites to the core metabolome or to specific microbiota,37 motivating thus also the use of high-resolution analytical technologies for the description of novel molecule biomarkers. The concept of determining non-invasive microbiomarkers in stool, which can aid gastroenterologists in the diagnosis of IBD and in distinguishing between different intestinal diseases, is a very attractive one and warrants future investigations.29 38 However, specific bacterial species that are clearly associated with disease activity are not yet identified. With IBD on the rise worldwide and a strong geographical influence on intestinal microbiota signatures,39 analysis of a core taxa signature (combination of bacterial taxa) in large transethnic cohorts or determining functional deficits of the microbial community seems to be required.16 29 40 More sophisticated sequencing-based approaches (see table 1) such as metagenomic/metatranscriptomic shotgun sequencing or metabolic modelling,41 which capture the functional capacity of the microbiota, overcome many limitations of 16S sequencing, yet should be complemented with phenotypic characterisation of individual bacteria using culture-based techniques, proteomics and metabolomics. With more complete microbiome reference data sets becoming available, and with larger international sample panels being collated—similar to the genetics community where several 10 000 DNAs and genome-wide genetic data sets were analysed in a joint effort (see www.ibdgenetics.org)—we are certain that microbial signatures of ‘caviar value’ may be derived in the future. Until then, we alert the scientists to the ‘caveats’ that come along with the field of biomarker research and call for more large-scale efforts by the community.
Table 1.
Overview of sequencing-based methods to characterise microbiota composition and function
| Full-length 16S rRNA gene sequencing | Targeted 16S rRNA gene amplicon sequencing | Metagenome sequencing | Metatranscriptomics | Single-cell analysis | |||
| Technique | Clade-specific amplification of 16S rRNA gene, vector-based cloning and sequencing | Long-read-based amplification and sequencing of large 16S rRNA gene fragments | PCR-based amplification of target followed by sequencing | Sequencing of entire DNA extracted from samples | Sequencing of entire RNA extracted from samples | Cultivation-based isolation of single bacterial clones | Emulsion or droplet-based single-cell amplification |
| Target | Entire 16S rRNA gene | Entire 16S rRNA gene | Variable regions, eg, V1-V2; V3-V4; V4; V6 |
All DNA molecules | All transcribed RNA molecules | Multiple complete genomes | Single bacterial cells |
| Potential bias | Gene copy-number bias | Amplification bias | Primer/region-specific amplification bias; gene copy-number bias | Transcriptionally more active bacteria are over-represented | Anaerobic and hard-to-culture bacteria are under-represented | More abundant taxa over-represented | |
| Sequencing technology | Sanger | PacBio or Oxford Nanopore | MiSeq | HiSeq/NextSeq | HiSeq/NextSeq | Any NGS technology | HiSeq/NextSeq |
| Advantages | Species-level resolution | High throughput; species-level resolution |
Low cost; high throughput |
Information on encoded functional repertoire; high-resolution taxonomic assignment; captures also viruses, fungi and archaea |
Information on actively expressed functional content | Reconstruction of multiple complete genomes from complex communities | Reconstruction of multiple complete genomes from complex communities |
| Disadvantages | No information on functional repertoire; low throughput; much hands-on time |
No information on functional repertoire; high error rates; higher cost per base than MiSeq |
No information on functional repertoire; low resolution on species level |
High cost; high computational burden; large amounts of unannotated data |
High cost; high computational burden; large amounts of unannotated data Removal of 16S rRNA required |
Tedious culturing and selection approaches (eg, media, oxygen); high cost |
High cost |
NGS, next generation sequencing.
Footnotes
Contributors: All authors edited the commentary and designed the figure.
Competing interests: None declared.
Patient consent: Parental/guardian consent obtained.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sommer F, Bäckhed F. Know your neighbor: Microbiota and host epithelial cells interact locally to control intestinal function and physiology. Bioessays 2016;38:455–64. 10.1002/bies.201500151 [DOI] [PubMed] [Google Scholar]
- 3. Franzosa EA, Huang K, Meadow JF, et al. . Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A 2015;112:E2930–E2938. 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falony G, Joossens M, Vieira-Silva S, et al. . Population-level analysis of gut microbiome variation. Science 2016;352:560–4. 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 5. Wang J, Thingholm LB, Skiecevičienė J, et al. . Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 2016;48:1396–406. 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamilton MJ, Weingarden AR, Unno T, et al. . High-throughput DNA sequence analysis reveals stable engraftment of gut Microbiota following transplantation of previously frozen fecal Bacteria. Gut Microbes 2013;4:125–35. 10.4161/gmic.23571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li SS, Zhu A, Benes V, et al. . Durable coexistence of donor and recipient strains after fecal Microbiota transplantation. Science 2016;352:586–9. 10.1126/science.aad8852 [DOI] [PubMed] [Google Scholar]
- 8. Manichanh C, Reeder J, Gibert P, et al. . Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 2010;20:1411–9. 10.1101/gr.107987.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paramsothy S, Kamm MA, Kaakoush NO, et al. . Multidonor intensive faecal Microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 2017;389:1218–28. 10.1016/S0140-6736(17)30182-4 [DOI] [PubMed] [Google Scholar]
- 10. Smirnov KS, Maier TV, Walker A, et al. . Challenges of metabolomics in human gut Microbiota research. Int J Med Microbiol 2016;306:266–79. 10.1016/j.ijmm.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 11. Zhao L. The gut Microbiota and obesity: from correlation to causality. Nat Rev Microbiol 2013;11:639–47. 10.1038/nrmicro3089 [DOI] [PubMed] [Google Scholar]
- 12. Saraswati S, Sitaraman R. Aging and the human gut microbiota-from correlation to causality. Front Microbiol 2014;5:764. 10.3389/fmicb.2014.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. . Reduced diversity of faecal Microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006;55:205–11. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ott SJ, Musfeldt M, Wenderoth DF, et al. . Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004;53:685–93. 10.1136/gut.2003.025403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sommer F, Anderson JM, Bharti R, et al. . The resilience of the intestinal Microbiota influences health and disease. Nat Rev Microbiol 2017 10.1038/nrmicro.2017.58 [DOI] [PubMed] [Google Scholar]
- 16. Gevers D, Kugathasan S, Denson LA, et al. . The treatment-naive microbiome in new-onset crohn’s disease. Cell Host Microbe 2014;15:382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Halfvarson J, Brislawn CJ, Lamendella R, et al. . Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. 10.1038/nmicrobiol.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swidsinski A, Ladhoff A, Pernthaler A, et al. . Mucosal flora in inflammatory bowel disease. Gastroenterology 2002;122:44–54. 10.1053/gast.2002.30294 [DOI] [PubMed] [Google Scholar]
- 19. Sommer F, Bäckhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol 2013;11:227–38. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 20. Desai MS, Seekatz AM, Koropatkin NM, et al. . A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016;167:e21:1339–53. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sonnenburg ED, Smits SA, Tikhonov M, et al. . Diet-induced extinctions in the gut Microbiota compound over generations. Nature 2016;529:212–5. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. David LA, Maurice CF, Carmody RN, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Earle KA, Billings G, Sigal M, et al. . Quantitative Imaging of Gut Microbiota spatial Organization. Cell Host Microbe 2015;18:478–88. 10.1016/j.chom.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab 2014;20:779–86. 10.1016/j.cmet.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee T, Clavel T, Smirnov K, et al. . Oral versus intravenous iron replacement therapy distinctly alters the gut Microbiota and metabolome in patients with IBD. Gut 2017;66:863–71. 10.1136/gutjnl-2015-309940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rampelli S, Schnorr SL, Consolandi C, et al. . Metagenome sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr Biol 2015;25:1682–93. 10.1016/j.cub.2015.04.055 [DOI] [PubMed] [Google Scholar]
- 27. Schnorr SL, Candela M, Rampelli S, et al. . Gut microbiome of the Hadza hunter-gatherers. Nat Commun 2014;5:3654. 10.1038/ncomms4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Häsler R, Sheibani-Tezerji R, Sinha A, et al. . Uncoupling of mucosal gene regulation, mRNA splicing and adherent microbiota signatures in inflammatory bowel disease. Gut 2016:gutjnl-2016-311651. 10.1136/gutjnl-2016-311651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pascal V, Pozuelo M, Borruel N, et al. . A microbial signature for crohn’s disease. Gut 2017;66:813–22. 10.1136/gutjnl-2016-313235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gisbert JP, McNicholl AG. Questions and answers on the role of faecal calprotectin as a biological marker in inflammatory bowel disease. Dig Liver Dis 2009;41:56–66. 10.1016/j.dld.2008.05.008 [DOI] [PubMed] [Google Scholar]
- 31. Moum B, Ekbom A, Vatn MH, et al. . Inflammatory bowel disease: re-evaluation of the diagnosis in a prospective population based study in south eastern Norway. Gut 1997;40:328–32. 10.1136/gut.40.3.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harvey RF, Bradshaw JM. A simple index of crohn’s-disease activity. Lancet 1980;1:514. [DOI] [PubMed] [Google Scholar]
- 33. Walmsley RS, Ayres RC, Pounder RE, et al. . A simple clinical colitis activity index. Gut 1998;43:29–32. 10.1136/gut.43.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Palmer KR. Complications of gastrointestinal endoscopy. Gut 2007;56:456–7. 10.1136/gut.2006.105577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut 2006;55:426–31. 10.1136/gut.2005.069476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. af Björkesten CG, Nieminen U, Turunen U, et al. . Surrogate markers and clinical indices, alone or combined, as indicators for endoscopic remission in anti-TNF-treated luminal Crohn’s disease. Scand J Gastroenterol 2012;47:528–37. 10.3109/00365521.2012.660542 [DOI] [PubMed] [Google Scholar]
- 37. Jansson J, Willing B, Lucio M, et al. . Metabolomics reveals metabolic biomarkers of crohn’s disease. PLoS One 2009;4:e6386 10.1371/journal.pone.0006386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tedjo DI, Smolinska A, Savelkoul PH, et al. . The fecal Microbiota as a biomarker for disease activity in Crohn’s disease. Sci Rep 2016;6:35216. 10.1038/srep35216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rehman A, Rausch P, Wang J, et al. . Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 2016;65:238–48. 10.1136/gutjnl-2014-308341 [DOI] [PubMed] [Google Scholar]
- 40. Papa E, Docktor M, Smillie C, et al. . Non-invasive mapping of the gastrointestinal Microbiota identifies children with inflammatory bowel disease. PLoS One 2012;7:e39242 10.1371/journal.pone.0039242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bauer E, Zimmermann J, Baldini F, Thiele I, et al. . BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol 2017;13:e1005544. 10.1371/journal.pcbi.1005544 [DOI] [PMC free article] [PubMed] [Google Scholar]