ABSTRACT
Leishmania tropica, a unicellular eukaryotic parasite present in North and East Africa, the Middle East, and the Indian subcontinent, has been linked to large outbreaks of cutaneous leishmaniasis in displaced populations in Iraq, Jordan, and Syria. Here, we report the genome sequence of this pathogen and 7,863 identified protein-coding genes, and we show that the majority of clinical isolates possess high levels of allelic diversity, genetic admixture, heterozygosity, and extensive aneuploidy. By utilizing paired genome-wide high-throughput DNA sequencing (DNA-seq) with RNA-seq, we found that gene dosage, at the level of individual genes or chromosomal “somy” (a general term covering disomy, trisomy, tetrasomy, etc.), accounted for greater than 85% of total gene expression variation in genes with a 2-fold or greater change in expression. High gene copy number variation (CNV) among membrane-bound transporters, a class of proteins previously implicated in drug resistance, was found for the most highly differentially expressed genes. Our results suggest that gene dosage is an adaptive trait that confers phenotypic plasticity among natural Leishmania populations by rapid down- or upregulation of transporter proteins to limit the effects of environmental stresses, such as drug selection.
KEYWORDS: CNV, Leishmania, RNA-seq, gene dosage, gene expression
IMPORTANCE
Leishmania is a genus of unicellular eukaryotic parasites that is responsible for a spectrum of human diseases that range from cutaneous leishmaniasis (CL) and mucocutaneous leishmaniasis (MCL) to life-threatening visceral leishmaniasis (VL). Developmental and strain-specific gene expression is largely thought to be due to mRNA message stability or posttranscriptional regulatory networks for this species, whose genome is organized into polycistronic gene clusters in the absence of promoter-mediated regulation of transcription initiation of nuclear genes. Genetic hybridization has been demonstrated to yield dramatic structural genomic variation, but whether such changes in gene dosage impact gene expression has not been formally investigated. Here we show that the predominant mechanism determining transcript abundance differences (>85%) in Leishmania tropica is that of gene dosage at the level of individual genes or chromosomal somy.
INTRODUCTION
The leishmaniases are a group of vector-borne parasitic diseases caused by over 20 different species of flagellated protozoa belonging to the genus Leishmania; these parasites are transmitted to humans as extracellular promastigotes by phlebotomine sand flies and proliferate as obligate intracellular amastigotes in phagocytic cells of the human immune system. Human leishmaniases present clinically as cutaneous, mucocutaneous, or visceral disease. Approximately 200,000 to 400,000 new cases of visceral leishmaniasis (VL) and 700,000 to 1,300,000 new cases of cutaneous leishmaniasis (CL) occur each year, although epidemiological data are often unreliable or incomplete in many of the countries where Leishmania spp. are endemic (1).
Leishmania tropica is a species that has been recently linked to massive epidemics of CL in refugee camps in Syria and neighboring areas of the Middle East, including Iraq, Jordan, Israel, Palestine, and Afghanistan, due to ongoing armed conflict and civil unrest in the region (2–4). This species is highly prevalent in this geographic region and follows an anthroponotic, or human-to-human, transmission cycle, which sets this species apart from the zoonotic and coendemic species Leishmania major. L. tropica appears to harbor considerably higher genetic diversity than L. major, as determined by microsatellite marker analysis (5), but the role of hybridization in shaping its evolution remains controversial (6), due to the lack of high-resolution genomic studies in this species. Considerable variation in the response of Leishmania to treatment has been documented in affected patients, with cutaneous lesions due to L. tropica being generally less responsive to treatment and more prone to form satellite lesions than those due to L. major, but the genetic determinants for this variation are still largely unknown (7, 8).
Leishmania parasites are characterized by unique genetic regulatory mechanisms that include two important features: (i) RNA editing of the genes that encode kinetoplasts and (ii) the absence of promoter-mediated regulation at the level of transcription initiation of nuclear genes. Changes in steady-state transcript levels within the cell are primarily ascribed to differences in the maturation and stability of individual mRNAs, which are largely mediated by RNA-binding proteins (9). Reflecting this unusual mechanism of gene regulation, studies comparing gene expression between promastigote and amastigote stages have found only a small number of genes to be differentially expressed (10). No studies to date have systematically investigated the effects of structural variation on global transcriptional patterns, although evidence exists to suggest that aneuploidy and/or gene copy number variation (CNV) is correlated with increased expression of individual genes involved in drug resistance (11, 12). Leishmania parasites have the ability to tolerate extensive aneuploidy at the level of chromosomal “somy” (a general term covering disomy, trisomy, tetrasomy, etc.) (13, 14), but whether Leishmania spp. employ compensatory mechanisms to downregulate genes on supernumerary chromosomes (i.e., chromosomes with a somy larger than 2) has not been investigated.
More recent studies employing high-throughput RNA sequencing (RNA-seq) approaches during differentiation of the L. major promastigote vector stages have identified discrete gene expression signatures associated with life cycle progression to the infectious form (15). While there appear to be significant differences in gene expression levels during life cycle progression between different Leishmania species (16), no studies have addressed intraspecific transcriptional variations in Leishmania. We sought to systematically investigate whether the genomic plasticity observed among natural populations of Leishmania isolates results in global changes in gene expression, in the absence of conventional transcriptional control.
Here, we describe the first comprehensive, high-resolution study of intraspecific differences in genetic diversity and gene expression of the Old World species, L. tropica, and show that natural isolates of L. tropica possess elevated levels of admixture and heterozygosity. Our study sheds insight into the population genetic structure of this complex human pathogen, which appears to be punctuated with genetic marks of extensive hybridization, and we show that global gene expression differences can be explained by gene dosage and structural variation. Our study provides important insights into how differential gene expression can determine intraspecific phenotypic diversity in this important human pathogen and how this diversity in turn underpins variations in transmissibility, tissue tropism, clinical disease, and drug susceptibility.
RESULTS
An L. tropica reference genome for comparative intraspecific structural variation analysis.
For de novo sequencing, we obtained a DNA isolate from the Leishmania tropica LRC-L590 strain (WHO strain identifier MHOM/IL/1990/P283), a strain capable of differentiating into axenic amastigotes (17). We generated reads of 100 bp in length (via Illumina technology) of various insert sizes (200-bp fragments with overlapping reads of 3 and 8 kb), with a total sequence coverage of ~250×. In terms of assembly size and contiguity, measured as the N50 length, the L. tropica L590 genome (Leishmania_tropica_L590-2.0.2) assembled to a size of 31 Mb, with N50 contig and scaffold lengths of 32 and 303 kb, respectively. These results are similar to earlier assemblies of Leishmania genomes with a comparable degree of genic completeness (13). It should be noted that the Leishmania_tropica_L590-2.0.2 reference genome used in our studies is nearly identical but does not have the same sequence coordinates as the version available from the TriTrypDB database (version 2014/12/16).
Sequencing 14 geographically dispersed L. tropica field isolates.
Whole-genome sequencing of 14 natural isolates collected between 1974 and 2009 from across the geographic range of L. tropica (see Table S1 in the supplemental material) was undertaken to assess intraspecific population genetic diversity, heterozygosity, ploidy, and gene copy number variation. The average read depth coverage was 50-fold in disomic chromosome regions. Across the 14 isolates, a total of 268,518 biallelic single-nucleotide variants (SNPs) were mapped, representing an ~1% polymorphism rate among alleles circulating within the species, which is high relative to that in other Old World Leishmania species investigated so far (13, 18, 19). To put this into perspective, only 156,274 interspecific biallelic SNPs were found to separate L. infantum from L. donovani (20), indicating that intraspecific genetic diversity among circulating isolates of L. tropica is significant and greater than that found within the L. donovani complex.
Sample name, country of origin, WHO international identifier code, and ENA accession number of sequenced samples (ENA accession numbers for RNA-seq triplicates range from ERS763603 to ERS763656) Download TABLE S1, PDF file, 0.03 MB (34.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SNP heterozygosity is high in L. tropica isolates.
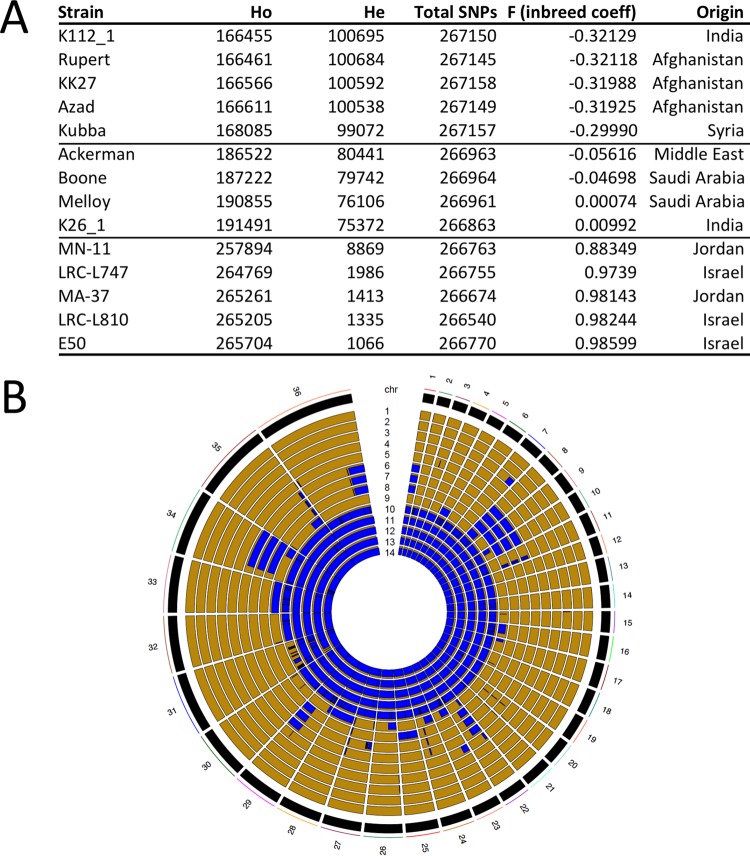
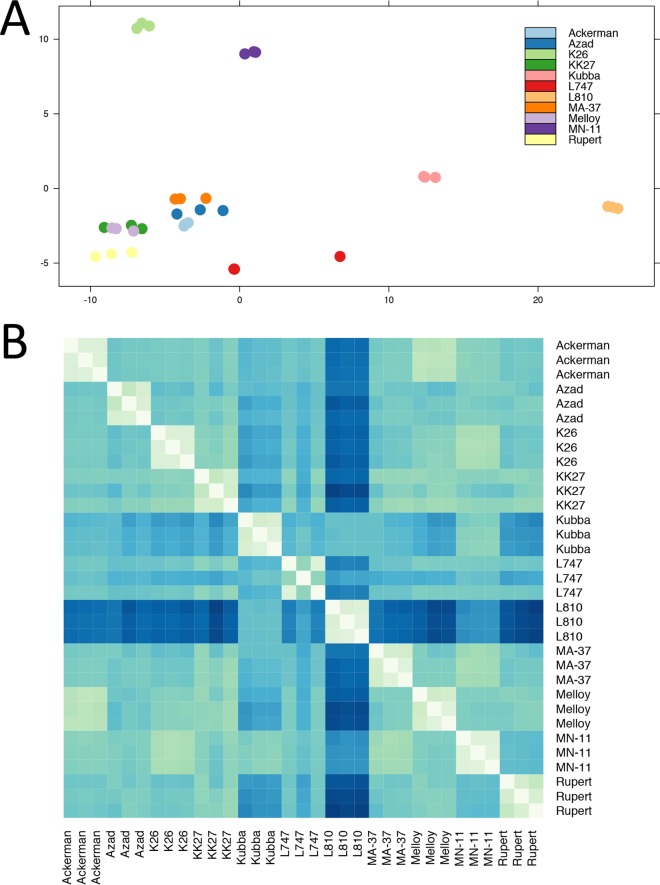
The degree of heterozygosity (He) and the allelic diversity were determined in order to investigate evolutionary pressures that could account for the high SNP diversity. Based on the number of heterozygous SNPs present, the 14 isolates were separated into three subpopulations. The first subpopulation of isolates (n = 5; KK27, Rupert, Kubba, Azad, and K112) possessed a remarkably high level of He compared to other Old World species (ranging from 99,072 to 100,695 heterozygous SNPs) (Fig. 1A), as reflected by its negative inbreeding coefficient (approximately −0.32). The heterozygous SNPs were distributed genome-wide, a pattern consistent with outcrossing by full-genome hybridization. The second subpopulation (n = 4; Ackerman, Melloy, Boone, and K26) had a reduced He (75,372 to 80,441 SNPs) that was not equally distributed throughout the genome and was restricted to either entire chromosomes or large blocks within individual chromosomes (Fig. 1A and B). In contrast, the third subpopulation (n = 5; E50, MA-37, MN-11, L747, and L810) possessed only low levels of He (1,066 to 8,869 SNPs) that were randomly distributed throughout the genome and were likely the result of stepwise accumulation of independent mutations in each homologous chromosome pair (Fig. 1A and B).
FIG 1 .
Heterozygosity and homozygosity among a total of 268,518 polymorphic sites identified by analyzing the genomes of 14 isolates of L. tropica. (A) Heterozygosity and homozygosity among the total 268,518 polymorphic positions in 14 isolates of L. tropica. The F statistic (inbreeding coefficient) and the number of heterozygous (He) and homozygous (Ho) positions were calculated using VCFtools. The isolates were sorted into low, intermediate, and high homozygosity groups, depending on the Ho. (B) LROH in 14 clinical isolates of L. tropica. The RCircos plot shows LROH for each of the 36 chromosomes, with homozygosity in blue and heterozygosity in gold. Note that no two isolates had the same exact pattern of LROH, despite many similarities among individual geographic groups (several isolates from Israel and Jordan were mostly homozygous, whereas the majority of the other isolates were largely heterozygous). The isolates represented in each panel are ordered in concentric rings, according to the F value (inbreeding coefficient) from panel A and numbered from 1 to 14, respectively: 1, K112_1 (India); 2, Rupert, Afghanistan; 3, KK27 (Afghanistan); 4, Azad, Afghanistan; 5, Kubba (Syria); 6, Ackerman (Israel); 7, Boone, Saudi Arabia; 8, Melloy (Saudi Arabia); 9, K26_1 (India); 10, MN-11 (Jordan); 11, L747 (Israel); 12, MA-37 (Jordan); 13, L810 (Israel); 14, E50 (Israel).
The excessive He could not be explained by ploidy or an excess of “private” SNPs, as the majority of heterozygous SNP positions were shared among strains and found on disomic chromosomes. Importantly, for the 4 isolates that were predominantly He but also possessed many long runs of homozygosity (LROH), each had significant differences from the others in terms of the size and distribution of its LROHs (Fig. 1B). Isolate K26, for example, possessed blocks or entire chromosomes of homozygous SNPs in 13/33 disomic chromosomes. A similar incidence of homozygous SNP blocks was observed in the Ackerman isolate, but this was for a different set of 11/32 disomic chromosomes (Fig. 1B). Overall, no two parasite isolates were identical in their pattern of LROH. Although the Ackerman, Melloy, and Boone isolates had very similar patterns of LROH, they could be distinguished from each other at chromosomes 25 and 27. Specifically, Boone was distinct from Ackerman and Melloy, which shared the same ancestry, in two large haploblocks on chromosome 27 (totaling over 267,600 nucleotides), whereas Melloy was distinct from Boone and Ackerman at chromosome 25 (Fig. 1B and data not shown).
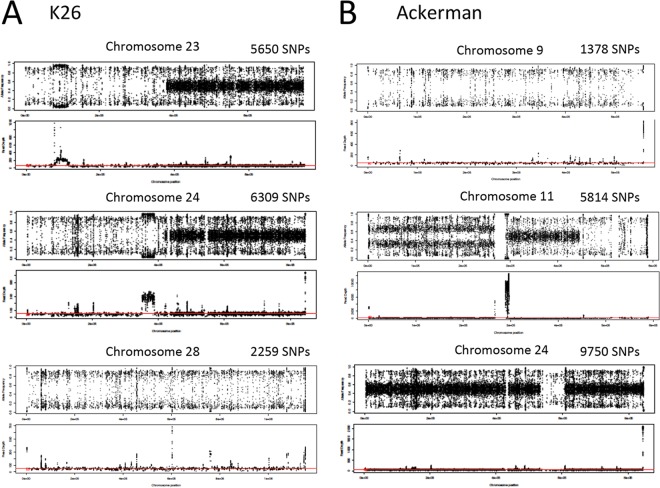
In support of genetic hybridization to explain the extant He, estimation of alternative allele frequencies across the 36 chromosomes established that, for the majority of isolates that showed substantial He (9/14), each had two different “sequence types” (from each other) or haplotypes that were also different from the reference L590 genome at all chromosomes bearing He (Fig. 2). For example, the allele frequency plots depicted for disomic chromosomes from isolate K26 showed that the second halves of chromosomes 23 and 24 were heterozygous and each parental allele was present in about half of the overall reads, such that the SNP variant positions were in a 1:1, or 0.5, ratio and showed a “hybrid” allele frequency line at 0.5 (Fig. 2A). On the other hand, the entire chromosome 28 and the first half of chromosomes 23 and 24 each possessed two haplotypes that were homozygous, or similar to each other, but shared a different ancestry from L590 (depicted by 2,259 SNP variant sites on chromosome 28); alleles at these loci are therefore at frequencies of either 1 or 0 (i.e., they show only a few data points in the y axis range between 0.1 and 0.9) (Fig. 2A; Fig. S3). The same was true for the Ackerman isolate. The entire chromosome 9 (which had 1,378 SNP variant sites relative to L590), the last third of chromosome 11, and the middle of chromosome 24 (Fig. 2B; Fig. S3) each possessed two haplotypes that were homozygous but shared a different ancestry from L590. On either side of the LROH in the middle of chromosome 24, the “hybrid” line at 0.5 identified two different haplotypes that were variant from L590. Of note, the first half of chromosome 11 is trisomic, and because it has only two haplotypes, the allele frequencies cluster at approximately 0.33 and 0.67, signifying that two of the three chromosomes have one allelic variant, and only one of the three chromosomes has the other variant. Integration of the genomic regions containing blocks or even entire chromosomes rich in heterozygous sites (which represent a mixed ancestry of two parental haplotypes) with that of homozygous sequence haploblocks that were introgressed into this background of He suggests that mechanisms promoting genetic admixture, such as meiotic recombination and gene conversion, have significantly impacted the population genetic histories for, at the very least, the 9 heterozygous natural isolates of L. tropica.
FIG 2 .
Heterozygous allele frequencies and read depths for selected chromosomes of two L. tropica isolates, K26 (India) and Ackerman (Israel). (A) K26 (chromosomes 23, 24, and 28); (B) Ackerman (chromosomes 9, 11, and 24). Variants called homozygous differences from the L590 reference scored either 1 or 0 and are depicted in the 0.1 or 0.9 y axis range. Variants called heterozygous show allele frequencies around 0.5 in the hybrid line for all disomic chromosomes. Variants called heterozygous on trisomic chromosomes show allele frequencies around 0.33 and 0.67 in the hybrid line. Allele frequency and the total number of biallelic SNPs are shown in the upper panel, and read depth across the chromosome is shown in the lower panel for each chromosome. Median read depth, marked by a red line, was approximately 50 reads for disomic chromosomes.
FST analysis identifies high allelic diversity and genetic admixture.
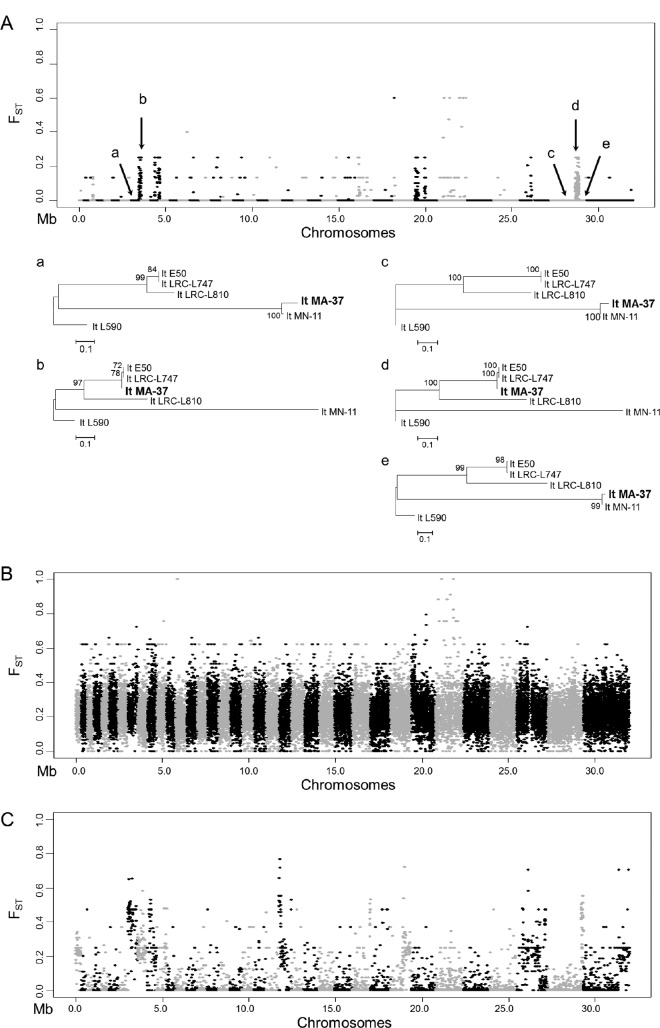
To determine the overall genetic divergence among the 5 geographically clustered isolates that possessed primarily homozygous differences (in which both haplotypes were similar to each other but different from the L590 reference genome), a genome-wide, pairwise FST analysis was carried out. FST is a statistical measure that identifies changes in allele frequencies across genetic distances within a population and can be used to demarcate regions undergoing significant genetic differentiation. High intraspecific allelic diversity was identified within the homozygous population (referred to here as Pop1), with at least 4 distinct ancestries identified across the 5 isolates (Fig. 3A). Punctuating across the 36 chromosomes were numerous admixture blocks of localized intraspecific genetic diversity in which FST values were high (i.e., >0.25; see the large haploblocks identified on chromosomes 8, 10, 14, 16, 18, 21, 25, 26, 27, 29, 30, 33, and 35). Separate phylogenetic trees generated in the “a” through “e” haploblocks on chromosomes 8 and 35 identified incongruence in the tree topologies, which was the result of admixture swapping among the 4 distinct haplotypes, and this established unequivocally that MA-37 underwent genetic hybridization within the homozygous population (Fig. 3A).
FIG 3 .
Genome-wide FST estimates and localized phylogenetic analysis indicated extensive genetic hybridization among isolates of L. tropica. (A) Pairwise FST estimates indicate localized high intrapopulation genetic diversity within the population of L. tropica clinical isolates that were predominantly homozygous (referred to as Pop1 in the main text). FST estimates were obtained with VCFtools, comparing isolates from Jordan (MA-37 and MN-11) to isolates from Israel (E50, LRC-L810, and LRC-L747). Regions surrounding peaks in FST values are highlighted in the plot as follows: a, LmjF.08 381000 to 439000; b, LmjF.08, 439000 to 550000; c, LmjF.35 1168000 to 1396000; d, LmjF.35 1396000 to 1624000; e, LmjF.35 1624000 to 1852000. These regions were chosen to generate phylogenetic trees to determine the allelic diversity present and its inheritance patterns across the homozygous strains. The y axis shows the pairwise FST values, whereas the x axis indicates the positions of the 36 chromosomes. Each dot represents the FST value in a 1-kb window. Separate phylogenetic trees using the SNP information from regions a to e reflect a swapping of ancestral haploblocks among the homozygous populations of L. tropica. Neighbor-joining phylogenetic trees were constructed using MEGA with 50% majority rules. (B) FST plot showing the genome-wide pairwise FST values identified when the homozygous population (n = 5; E50, LRC-L810, MA-37, LRC-L747, and MN-11) was compared against all other isolates that were heterozygous (n = 9; K112_1, Rupert, KK27, Azad, Kubba, Ackerman, K26_1, Boone, and Melloy). x and y axes are labeled as described above, and each dot represents the FST value in a 1-kb window. As expected, the FST values are higher for comparison of these two very divergent populations. (C) FST plot showing the genome-wide pairwise FST values identified within the heterozygous population comparing those that were heterozygous throughout (n = 5; K112_1, Rupert, KK27, Azad, and Kubba) with those that were heterozygous but possessed LROH (n = 4; Ackerman, K26_1, Boone, and Melloy). x and y axes are labeled as described above, and each dot represents the FST value in a 1-kb window. High localized FST values again suggest extensive haploblock swapping, as in panel A.
FST was next calculated for Pop1 against all 9 remaining isolates, which possessed high He levels. Not surprisingly, FST was high throughout (Fig. 3B), which showed that genetic mechanisms, such as genetic hybridization, likely favor the generation of these heterozygous genotypes, although preferential maintenance of these lines by mating incompatibility could not be ruled out.
When the FST was calculated between the heterozygous strains that possessed the highest He (Pop2; isolates KK27, Rupert, Kubba, Azad, and K112) with those that had LROH (Pop3; Ackerman, Melloy, Boone, and K26), distinct admixture blocks were identified which, in combination with rearrangement of maximum likelihood tree topologies across linked markers (data not shown), established that allelic swapping had occurred between the isolates in a manner similar to Pop1 and that many of the genotypes with high He were comprised of two distinct parental haplotypes that were shared, or recombined, across the isolates examined.
Somy differences among L. tropica isolates.
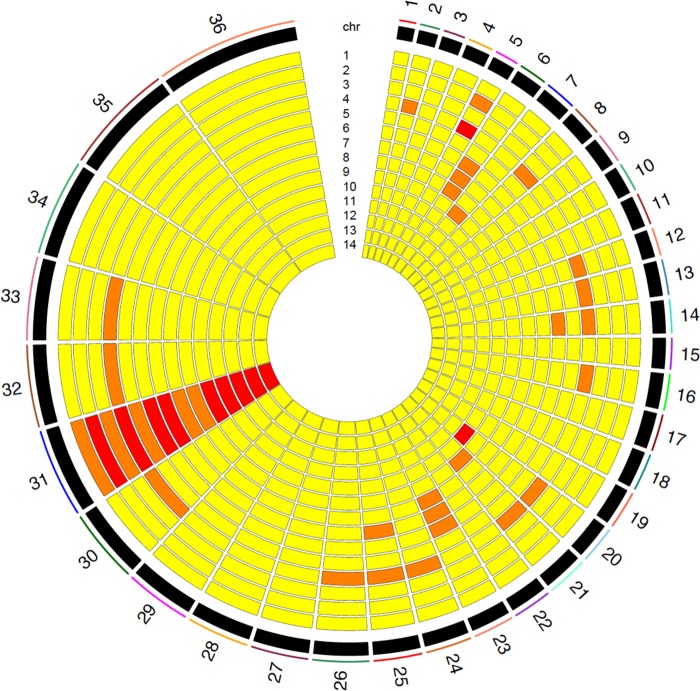
The somy of each chromosome was estimated using a read depth-based algorithm developed in-house specifically for short read sequence data generated from Leishmania DNA (see Materials and Methods). L. tropica, like L. major, has a 36-chromosome karyotype, with chromosome 1 being the shortest and chromosome 36 being the longest. Chromosome 31 was either trisomic, tetrasomic, or hexasomic in all isolates. Most chromosomes varied between the disomic and trisomic state (Fig. 4). The field isolate with the most variation in chromosome number (Azad, from Afghanistan) had 20 disomic chromosomes, 14 trisomic chromosomes, and 2 tetrasomic chromosomes. Seven field isolates were nearly diploid, with only one chromosome, chromosome 31, being present in either the tetrasomic (MA-37, E50, L747, and L810) or trisomic state (K112, KK27, and Kubba). To investigate whether gene dosage, either at the level of chromosomal somy or at the level of individual genes, impacts gene expression, we generated paired genome-wide DNA-seq with RNA-seq transcriptomic data for 11 of the L. tropica clinical isolates.
FIG 4 .
Extensive aneuploidy in 14 geographically diverse isolates of L. tropica. Estimated somy at each of the 36 chromosomes in the L. tropica genome, with increasing somy indicated from yellow (somy = 2), to orange (somy = 3), to red (somy = 4).
RNA-seq results for 11 L. tropica clinical isolates.
RNA purified from triplicate cultures of 11 L. tropica isolates was sequenced to a depth of 50 to 80×. Principal-component analysis (PCA) of the raw read counts normalized by library size for the 11 sets of triplicates showed tight clustering for each set of triplicates (Fig. 5A). However, all replicates for one of the isolates (L810, from Israel) clustered together, but separately from all other isolates along the first principal component, as well as additional principal components beyond the first two (data not shown).
FIG 5 .
Isolate L810 (Israel) showed an aberrant expression signature compared to all other isolates. (A) PCA plot of expression data, showing the first two principal components on the x and y axes for all strains except Boone (one of the three replicates showed some characteristics of an outlier; see the main text). Each set of triplicates is represented in a different color, depending on the isolate of origin. Note the close clustering of each set of triplicates, indicating accurate biological replication. The replicates for isolate L810 clustered closely together, but quite distantly from the rest of the isolates. (B) Heat map of Euclidean distances between variance-stabilized expression values for each pair of samples, with larger Euclidean distances represented by darker shades of blue. Expression signatures within each set of triplicates were very similar to each other (represented in light colors along the diagonal axis). Isolate L810 showed an aberrant expression signature, with higher distance values in all pairwise comparisons (average distance values were closer to 50, instead of 33, as with the rest of the isolates).
In order to confirm the divergent expression signature of the L810 isolate, Euclidean distances were calculated using variance-stabilized relative log expression (RLE)-normalized read counts. A heat map of these distances showed that L810 is highly divergent from the rest of the isolates in our set of samples (Fig. 5B; for L810, mean distance of 49.97 and standard deviation [SD] of 15.82; mean for all other isolates of 33.50 and SD of 11.42), which was consistent with the clustering seen in our PCA. Given the divergent pattern observed for L810, we performed pairwise likelihood ratio tests between L810 and the rest of the samples (separately), considered as two separate conditions. We identified 40 gene transcripts that were significantly differentially expressed (DE) in L810 and were either upregulated or downregulated by 2-fold or more compared to all other lines (Table S2; Fig. S2). The most upregulated transcript in L810 (>2 log-fold change) in comparison to all other samples was LmjF.17.0190, a receptor-type adenylate cyclase (see Table S3 for the complete list of differentially expressed genes). The majority of the downregulated transcripts in L810 belonged to unknown proteins. The two most downregulated transcripts in L810 (>6 log-fold change) were a hypothetical protein of unknown function and an acyltransferase-like protein (LmjF.35.0450 and LmjF.04.1050, respectively, in the L. major-based annotation).
The top 30 most highly expressed genes in the set of 11 isolates analyzed by RNA-seq. The top cluster of highly expressed genes contains β-tubulin (LmjF.21.1860), a well-described marker of the promastigote stage, and several genes encoding nucleoside and amino acid transporter proteins (LmjF.36.1940, LmjF.15.1240, LmJF.15.1230, LmjF.07.1160, LmjF.36.6300, LmjF.31.0350, and LmjF.06.1260). Most other highly expressed genes encode ribosomal subunit proteins. See Table S2 for a full list of the genes. Download FIG S1, PDF file, 0.02 MB (18.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in L180 compared to all other isolates. Twofold changes in expression are marked with horizontal blue lines. Genes in red are those that were attained statistical significance. The complete list of statistically significant, differentially expressed genes with more than a 2-fold change in expression is provided in Table S3. Download FIG S2, PDF file, 0.4 MB (396.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The top 30 most highly expressed genes in the 11 samples that were analyzed by RNA-seq (VSE stands for variance-stabilized expression and is a normalization procedure for expression values built into the R package DEseq) Download TABLE S2, PDF file, 0.03 MB (35.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in the L810 isolate compared to all other isolates that are upregulated or downregulated more than 2-fold (the log FC stands for the log fold change in expression level) Download TABLE S3, PDF file, 0.03 MB (35.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Somy and gene dosage (CNV) alter gene expression.
To investigate gene dosage effects, visual inspection of raw read counts from RNA-seq for each isolate confirmed that supernumerary chromosomes had higher absolute expression values than disomic chromosomes from the same sample, even prior to normalization by library size (see Materials and Methods). To confirm this observation, two pairwise comparisons were performed (K26 versus Ackerman and Azad versus KK27). Each of the four samples had similar genetic backgrounds but showed considerable variation in karyotype, with somy differing at 6 chromosomes between K26 and Ackerman and at 16 chromosomes between Azad and KK27. Using a false discovery rate (FDR) of <0.05 and a threshold of a 2-fold or greater change (i.e., a log fold change greater than 1), a total of 3,639 significant DE genes were found between K26 and Ackerman (46.28% of all genes), and 5,231 significant DE genes were found between Azad and KK27 (66.52% of all genes). We then proceeded to study these differences in gene expression in the context of observed copy number variation and somy differences between these lines.
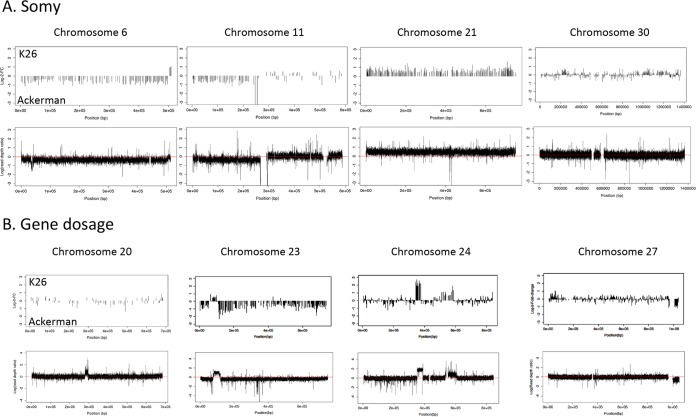
Superimposing expression information on known variations in somy revealed changes in the expression of genes on supernumerary chromosomes that consistently shifted in the direction of the chromosome with larger somy. This effect was dose dependent, as would be predicted when gene dosage affects the absolute level of transcripts present in each cell (Fig. 6A). Indeed, for chromosome 6 and the first half of chromosome 11, in which isolate Ackerman was trisomic compared to isolate K26, higher absolute expression values were detected in Ackerman. Conversely, for chromosome 21, in which K26 was trisomic compared to Ackerman, K26 had higher expression values chromosome-wide (Fig. 6A). However, when somy was equal, net expression differences were still identified, as can be seen by blocks of up- and downregulated gene transcripts in the pairwise comparisons between K26 and Ackerman on chromosome 30.
FIG 6 .
The pairwise comparison between K26 (India) and Ackerman (Israel) detected gene dosage effects due to copy number variation and somy differences. (A) Somy. The top graph represents the log fold changes in expression at genes along the chromosome, whereas the bottom graph represents the log scale ratios between the read depths in K26 (upper quadrant) and Ackerman (lower quadrant). The x axis represents base pair positions along the chromosome in both graphs. (B) Gene dosage effects due to copy number variation on the transcription of genes. Three large CNVs spanning 10 or more DE genes on chromosome 23, 24, and 27 are shown (CNVR159-178 on chromosome 27, CNVR114 on chromosome 24, and CNVR240 on chromosome 23) (Table S5). A smaller CNV on chromosome 24, upstream of the larger CNV, and one in the middle of chromosome 20 are also shown. Gene dosage effects in the two CNVs on chromosome 24 appear to behave in a dose-dependent fashion, with a 2-fold increase in copy number (consistent with a homozygous duplication) leading to a 2-fold increase in expression and a 1-fold increase in copy number (consistent with a heterozygous duplication), leading to a 1-fold increase in expression. The CNV on chromosome 20 does not appear to impact gene expression.
To investigate the degree to which gene copy number variation impacted gene expression, read depth information from whole-genome sequencing was scanned for evidence of CNVs. In general, relative log fold changes in expression in genes in these CNV regions mimicked sequencing read depth differences between the K26 and Ackerman lines in a dose-dependent fashion, in a manner similar to that observed for differences in chromosomal somy (Fig. 6B). For example, regions with higher relative read depths in K26 showed positive log fold changes in gene expression in K26 relative to Ackerman. The converse was also true: regions that had higher relative read depths in Ackerman showed negative log fold changes in gene expression (Fig. S3).
Chromosomal plots of read depth and log fold changes in gene expression in the K26 versus Ackerman strain, highlighting gene dosage effects due to local CNVs and somy differences. Positive values indicate higher values for K26 than for Ackerman, and negative values indicate the opposite. Axes are labeled as described for Fig. 6. Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A total of 419 CNVs were identified between K26 and Ackerman (median size, 2,838 bp, comprising 3.7% of the genome), overlapping 247 DE genes. A total of only 16 CNVs were identified between Azad and KK27 (median size, 1,274 bp, comprising 0.1% of the genome), overlapping 5 significant DE genes. Given the larger number of CNV calls made in the K26 versus Ackerman comparison, we focused on this pair of isolates to assess gene dosage effects in CNVs (Fig. 6B). Five large CNV regions on chromosomes 20, 23, 24, and 27 were found between these two lines. While the CNV regions on chromosomes 20, 23, and 24 had higher copy numbers in K26, and the CNV region on chromosome 27 had a higher copy number in Ackerman. Among these 5 CNVs, localized CNVs accounted for a net change in gene expression for chromosomes 23, 24, and 27, but this relationship was not absolute, as the CNV in chromosome 20 did not result in a net increase in localized gene expression for K26 (Fig. 6B), possibly due to the small number of genes on that chromosome.
Out of a total of 46 DE genes contained in the four largest CNVs with gene expression differences, 12 were predicted to have a transmembrane domain according to their annotation in L. major, suggesting enrichment of transmembrane proteins in large CNV regions (the probability of obtaining this number of genes carrying at least 1 transmembrane domain by chance was 0.0403, which was significant at the P < 0.05 threshold). Among the DE genes identified in these CNVs were several ABC transporters previously implicated in drug resistance, such as multidrug resistance protein A (MRPA; LmjF.23.0250) (Table S5).
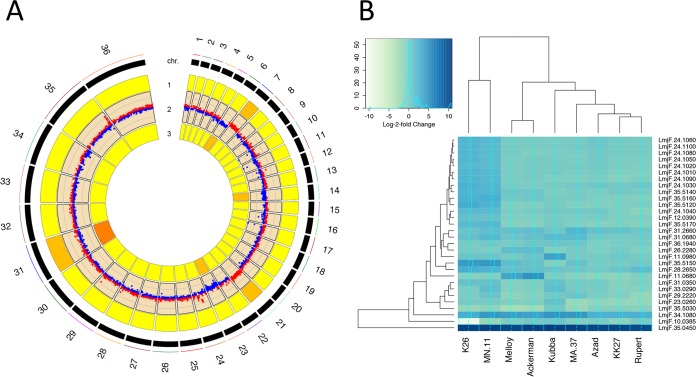
These global patterns in gene expression were next visualized genome-wide (Fig. 7A), and this confirmed the trends observed for individual chromosomes (Fig. 6). Specifically, in those chromosomes with somy differences, net expression differences were visualized as shifts in color between red (relatively higher expression in K26) and blue (relatively higher expression in Ackerman), toward the strain possessing greater somy (e.g., chromosomes 6, 7, 14, 21, and 23). For those with CNV differences, localized log fold changes in gene expression were observed as punctuated shifts in the direction of the strain with higher CNV, e.g., chromosomes 2, 11, 12, 16, 17, 22, 24, and 29 (Fig. 7A; Fig. S3). Overall, 167 genes were DE more than 2-fold, with approximately 20% of this differential expression due to CNVs and 43% due to somy. These findings corresponded to nearly 87% of highly differentially expressed coding sequences at the 2-fold threshold that was directly attributed to structural variation.
FIG 7 .
Global changes in transcription highlight significant gene dosage effects, with differential expression of membrane-bound transporter genes in a pairwise comparison between K26 (India) and Ackerman (Israel). (A) RCircos plot illustrating gene dosage effects on transcription genome-wide due to aneuploidy in a pairwise comparison between K26 and Ackerman. These isolates were selected due to the large number of chromosomes that differed for somy between each other. Tracks 1 and 3 represent somy for K26 and Ackerman, respectively, while the results shown in track 2 were obtained using edgeR to graphically represent differentially expressed genes, with upregulated genes in red (log fold change > 0) and downregulated genes in blue (log fold change < 0). Log fold changes in expression mirror differences in somy, indicating substantial gene dosage effects in L. tropica. (B) Heat map of the 30 most significantly differentially expressed genes, represented with their log fold changes in expression. A darker shade of blue indicates higher expression. Note the large cluster of DE genes on chromosome 24 that were upregulated in isolates K26 and MN11. This cluster is part of the CNV in isolate K26 (shown in Fig. 6; CNVR114). Other notable genes include LmjF.35.5150 and LmJF.10.0385, which appear to be amplified and deleted, respectively, in K26. These genes code for a biopterin transporter and a folate transporter that are known to act in concert in folate metabolism. A gene encoding a hypothetical MFS general transporter (LmjF.11.0680) was also upregulated in isolates Melloy and Ackerman.
Differential gene expression analysis across 11 isolates.
Our pairwise comparison between the K26 and Ackerman isolates suggested that gene dosage confers a degree of genome plasticity, for example, by allowing strains to alter expression of transporter genes to confer drug resistance. To confirm this, we first identified the most highly expressed genes found in the axenic procyclic promastigote stage (Fig. S1). In all isolates, the most highly expressed gene was for β-tubulin (LmjF.21.1860 in the L. major annotation), a well-known promastigote-specific marker present in multiple orthologous copies in the parasite genome (21). Also in a cluster of the most highly expressed genes were several transporter proteins, including an inosine/guanosine transporter (LmjF.36.1940) and two putative nucleoside transporter proteins (LmjF.15.1230 and LmjF.15.1240). At the top of the next cluster of highly expressed genes were several transporters, including two amino acid transporters (LmjF.07.1160 and LmjF.31.0350), a glucose transporter (LmJF.36.6300), and a pteridine transporter (LmjF.06.1260) (see Table S2 for the complete list of genes). Among other highly expressed genes were many ribosomal components involved in translation of mRNA, such as 40S and 60S and a nuclear RNA-binding domain protein (LmjF.32.0750).
In order to measure differential gene expression among the rest of the parasite lines, isolate L747 was chosen as the baseline to which all other samples were compared for calculation of log fold changes in expression (see Materials and Methods for our rationale). The top 30 genes with the smallest P values were selected (the FDR was approximately 0 for all selected genes) to generate a workable set of significant DE genes that varied among the remaining 10 parasite lines (Fig. 7B).
The most upregulated of the DE genes included a hypothetical protein with unknown function (LmjF.35.0450), which was found to be strongly downregulated in L810, and an amastin-like surface protein (LmjF.34.1080). The top 30 DE genes with the smallest P values included 9 genes in total with predicted transmembrane domains. Of the 1,546 genes containing 1 or more transmembrane domains in L. major, as annotated in TriTypDB, 1,344 were also present as homologues in the L. tropica annotation used for this study. The probability that at least 9 genes coding for membrane-associated protein products would be present by chance in a random sample of 30 genes in L. tropica is 0.02301 (P < 0.05 by Fisher exact test), confirming an enrichment of transmembrane proteins in our set of highly significant DE genes.
Comparing log fold changes in expression across samples for these 30 genes showed a cluster of 9 highly significant DE genes, arranged as an array on chromosome 24 (LmjF.24.1010 to LmjF.24.1100), which was upregulated in two isolates (K26 and MN-11). The protein products encoded by this cluster of differentially expressed genes on chromosome 24 appeared to have highly heterogeneous functions (see Table S4 for a complete list of genes and associated predicted protein products), but they included a hypothetical multipass transmembrane protein (LmjF.24.1090).
The top 30 most significant (lowest P value) differentially expressed genes across all isolates (a total of 9 genes were transmembrane, with significant enrichment compared to the total number of transmembrane genes in the genome [P < 0.05]) Download TABLE S4, PDF file, 0.03 MB (34.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in the 4 largest CNV regions identified in the Ackerman-K26 comparison based on CNV-seq (tThere were a total of 10 genes in CNVR114 [chromosome 24], 11 genes in CNVR181 [chromosome 11], 14 genes in CNVR159-178 [chromosome 27], and 11 in CNVR240 [chromosome 23]; the CNV region on chromosome 11 is so extensive that it overlaps approximately half of the chromosome [Fig. 6A]) Download TABLE S5, PDF file, 0.04 MB (40.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The same two samples that showed higher transcript levels in a region on chromosome 24 (MN-11 and K26) also had very dramatic upregulation in the transcript levels of a biopterin transporter protein, relative to other samples (BT1; LmjF.35.5150 in the L. major annotation), with an apparent downregulation of folate transporter protein transcripts (FT1; LmjF.10.0385 in the L. major annotation) that was especially evident in one of the two parasite lines, K26. Upon closer inspection of the sequence data, there was a clear deletion of FT1 and amplification of BT1 in K26 (Fig. S4). Other notable genes that were differentially expressed in a minority of lines were LmjF.11.0980 in Kubba, a hypothetical protein containing a HIT zinc finger domain, and LmjF.11.0680 in Melloy and Ackerman, a hypothetical transmembrane protein containing an MFS general substrate transporter domain.
Read depth plots of the chromosomal region surrounding the BT1 (LmjF.35.5150) and FT1 (LmjF.10.0385) genes in isolates K26 (in gold) and Ackerman (in blue). Red vertical bars mark the start and end of each gene. In K26, there is a clear homozygous deletion of FT1 (read depth of 0) and a possible heterozygous duplication of BT1 (read depth, ~75 to 90 across the region, suggesting that the gene is present in triplicate), while in the Ackerman line both genes are present in duplicate (read depth, ~50 to 60). It is important to note that both of these genes are present in multiple orthologous copies in most Leishmania species. Download FIG S4, PDF file, 0.2 MB (194.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
The present study describes the first comprehensive, high-resolution investigation of intraspecific genetic diversity and heterozygosity within the Old World Leishmania species L. tropica, a species that is responsible for large outbreaks of cutaneous leishmaniasis in the Middle East. We identified significant intraspecific heterogeneity and evidence of full-genome hybridization, which had previously been suggested only by low-resolution techniques. Moreover, it formally established that structural genomic variation, at the level of somy and copy number variation, is responsible for the majority of intraspecific gene expression differences within L. tropica isolates and presumably in other species within the genus. Significant variation was identified in the expression of membrane-bound transporter proteins, suggesting that gene dosage has phenotypic consequences and that such variation is functional and evolutionarily adaptive and may affect clinical disease and drug susceptibility for this organism in the Middle East region.
In most eukaryotes, the predominant mechanism regulating transcript abundance is at the level of promoter activity and transcription initiation. However, kinetoplastids possess genomes that are organized into polycistronic gene clusters that are constitutionally transcribed into mRNA and are thought to rely principally on posttranscriptional control of gene expression at the level of mRNA trans-splicing and polyadenylation (22, 23). In effect, regulation of message turnover is thought to cause the majority of gene expression differences, and this form of regulation is controlled by cap removal and shortening of the poly(A) tail by a variety of cellular decapping enzymes and deadenylases, followed by degradation of the RNA molecule by exonucleases (24, 25). Evidence for these exosome complex-mediated processes are abundant in kinetoplastids (26–30). Regulatory sequence elements in neighboring untranslated regions (UTRs) are likewise thought to determine the trans-splicing efficiency of protein-coding transcripts, as well as their half-lives within the cell, via interactions with RNA-binding proteins.
While message stability and posttranscriptional regulatory networks most certainly account for some degree of developmental and strain-specific gene expression, our results formally establish that the predominant mechanism determining transcript abundance differences between isolates of the same species is that of gene dosage. Pairwise comparisons of gene expression determined by whole-genome RNA-seq in four lines allowed direct observation of how differences in chromosome number and in subchromosomal amplifications and deletions, referred to as copy number variants, affect differential gene expression. Transcript levels analyzed from the four lines (K26, Ackerman, KK27, and Azad) specifically correlated with estimated chromosome number, with higher expression levels seen for genes on chromosomes with higher somy. No downregulation of transcript abundance of genes on supernumerary chromosomes was observed, and the majority (>85%) of genome-wide differences in gene expression were attributed to differences in gene dosage.
Focus on subchromosomal structural variations in the pairwise comparison between K26 and Ackerman, which had the highest number of CNVs, identified similar localized gene dosage effects within a chromosome. Importantly, transporter proteins were overrepresented in the four largest CNVs examined. Of note, a large gene cluster on chromosome 24 that was differentially expressed in K26 and MN-11 overlapped entirely with a large CNV (CNVR 114), implicating gene dosage as the underlying mechanism responsible for the differences in gene expression between these two lines. CNVs may thus represent an evolutionarily important adaptation to confer a degree of plasticity in the regulation and functional expression of parasite genes or gene clusters involved in, for example, drug resistance, as has been observed in antimony-resistant L. infantum (31) and in methotrexate-resistant L. donovani and L. tropica (32, 33). This is especially pronounced in some regions of the genome that appear to be more prone to deletion or amplification by virtue of being flanked with repetitive sequences that facilitate formation of both linear and circular amplicons via RAD51 recombinase-dependent and -independent mechanisms (12).
Differentially expressed genes in 10 isolates (i.e., genes that varied more across triplicates than within triplicates) were enriched in transmembrane proteins, with significant overrepresentation of transporter proteins. These surface proteins are known to play an important role in the uptake of essential nutrients from the external environment, as well as the import of drug compounds into the cell (34–36). The BT1 and FT1 transporters, for instance, are among the best-studied examples associated with in vitro resistance to the antifolate drug methotrexate (37), and they appeared to be deleted and duplicated in one of the clinical isolates examined (K26). These two transporter proteins are known to play a role in pterin/folate metabolism in Leishmania parasites, and concurrent downregulation of FT1 and upregulation of BT1 has been implicated in antifolate drug resistance.
Recently, mutations in aquaglyceroporin genes responsible for transport of trivalent antimonials into the cell have been implicated in antimonial drug resistance in India (38), and the LiMT/LiRos3 transport system has been implicated in experimental models of miltefosine resistance (39). Our study findings suggested that decreased susceptibility or complete resistance to antileishmanial compounds may similarly occur via structural variations in transporter genes. The redundancy observed in the function of many of these transporters and the relative ease with which their transcript levels can be regulated via amplification or deletion of the corresponding protein-coding gene suggest that the natural variation observed in our study may be an important preadaptation for survival in nutrient-poor environments or environments otherwise hostile to the parasite.
RNA-seq analysis of the L. tropica isolates also found one isolate (LRC-L810) with a very different expression signature from all other isolates. Interestingly, this sample was isolated from an infected sand fly in northern Israel, and subsequent follow-up studies found that this strain and other strains genetically similar to it were preferentially transmitted by a different vector species than genetically dissimilar L. tropica isolates originating from the same region (40). The most highly upregulated gene in this isolate compared to all other isolates was LmjF.17.0190, a receptor-type adenylate cyclase. Proteins that fall into this functional category have been linked in African trypanosomes to differentiation of the parasite from the epimastigote into trypomastigote form (41), inhibition of the host immune response (42), and parasite motility in the insect stages (43). Targeted gene knockout studies may shed additional light on the function of this gene for vector competency or specialization in L. tropica.
The capacity of Leishmania to support extensive aneuploidy and localized CNV (in the form of tandem arrays of duplicated genes) without any observable defect in fitness has been reported previously. Changes in somy and gene dosage appear to represent an evolutionary adaptation within Leishmania, one that allows isolates to dramatically alter their gene expression profiles for host or vector niche specialization, parasite growth, and survival. Numerous examples exist whereby clones under selection for targeted gene deletions (44), during selection for drug resistance (11), upon extended passage in vitro (45), or produced by experimental genetic hybridization (46–48) have all been shown to alter their ploidy. In yeast, polyploidy is known to have strong selective advantages, which include promoting phenotypic variations and genome plasticity to adapt to environmental stresses (49, 50). The work reported here establishes that the majority of differential gene expression among Leishmania clinical isolates is the result of differences in their gene dosage, at the level of ploidy and CNVs. However, the mechanisms generating differences in gene dosage and structural variation are less clear.
Our results highlight the specific need to quantify the exact nature and relative contribution of mitotic versus meiotic processes involved in generating aneuploidy, given the functional and phenotypic consequences related to regulation of mRNA expression by gene dosage. Possible mechanisms proposed to generate somy and CNV differences include nondisjunction during mitosis, which results in unequal segregation of homologous chromosomes during replication and division (13), genetic hybridization, including meiotic nondisjunction and gene conversion processes (46), and recombination-directed replication (51), a form of gene conversion that can be periodically induced to alter ploidy and gene expression, as has been proposed for Leishmania and demonstrated during Tetrahymena and Cryptococcus growth (52, 53). It remains unclear, however, whether the type of mosaicism found in Leishmania can be explained simply by chromosomal replication defects or whether genetic hybridization followed by genome erosion plays a significant role in the process.
Single-cell whole-genome sequencing and single-cell RNA-seq approaches may prove useful in dissecting whether unequal chromosome segregation during mitotic cell division is the most significant cause of mosaic aneuploidy within a single host-defined population of parasites. Regardless of the exact mechanism, such mosaicism in Leishmania may have evolved in order to maximize genomic variation within a single host, thereby allowing the parasite to rapidly evolve clones in situ that are capable of withstanding selective pressures, such as drug selection.
Meiotic reproduction in fungi is commonly associated with de novo generation of aneuploidy and is primarily responsible for conferring advantageous genotypic and phenotypic plasticity during fungal adaptation to novel environments (54). Analysis of natural recombinants and experimental generation of hybrid Leishmania lines likewise produces significant structural variation, including aneuploidy, and is known to enhance the transmission potential, fitness, and disease potential of these hybrids (46, 55). In the model yeast Saccharomyces cerevisiae, results similar to those presented here for L. tropica have been reported; aneuploidy is capable of causing gene expression and proteomic changes that confer advantageous phenotypic properties (50, 56).
The extent to which genetic hybridization in L. tropica impacts aneuploidy, gene expression, and phenotypic plasticity has not been examined here. In an effort to establish whether genetic hybridization impacts the natural population genetics of L. tropica, we employed an iterative, genome-wide analysis of the FST values and identified high He levels in a pattern that was consistent with outcrossing by full-genome hybridization, as observed in natural populations of other Leishmania species (57). We showed that many genotypes with high He are comprised of two distinct parental haplotypes that are shared, or recombined, across the isolates. Our data did not support mating incompatibility and the accumulation of independent mutations in each homologous chromosome pair as the mechanism for maintaining He, because the number of orphan alleles observed for each chromosome pair was limited. Allelic sharing between isolates was clear, with heterozygous genotypes being made up of two parental alleles that were present in homozygous form in other sampled isolates. We also found many localized areas that possessed high intraspecific genetic diversity compatible with allelic swapping among defined blocks of parental haplotypes within the population studied. Collectively, our analysis highlighted the potential for genetic admixture as a viable mechanism for generating aneuploidy and gene expression differences among natural isolates. Future studies will utilize experimental crosses and backcrosses between homozygous and/or heterozygous clones to determine the extent to which meiotic processes impact gene dosage and gene expression differences between hybrid progeny.
In addition to noting the importance of gene dosage effects and trans-regulators in determining transcript stability, it is crucial to also stress that transcript abundance in Leishmania generally correlates poorly with cellular protein levels (58). Additional downstream processes may be shaping the proteomic landscape at the level of protein translation from the initial cellular pool of mRNA transcripts. The mechanisms giving rise to aneuploidy and gene copy number variation in natural Leishmania populations remain poorly understood, and our study emphasizes the need to understand the processes shaping genetic diversity and aneuploidy and how they may contribute to genomic plasticity and adaptive evolution.
In conclusion, by using a combination of whole-genome DNA-seq coupled with RNA-seq, we produced high-quality data sets to map gene dosage and chromosomal somy as major factors that control steady-state mRNA transcript levels in isolates of L. tropica parasites. The availability of a high-quality draft genome allowed the assessment of natural variation and admixture as probable mechanisms for generating the types of structural variations and locus-specific gene dosage effects that underpin differential expression of genes associated with phenotypic plasticity and niche specialization in this important but neglected human pathogen.
MATERIALS AND METHODS
Reference genome sequencing.
The DNA for shotgun de novo sequencing was derived by utilizing CsCl/ethidium bromide density gradient centrifugation to minimize the amount of kinetoplast (mitochondrial) DNA. Sequencing was performed on Illumina HiSeq2000 instruments. All Illumina sequences were de novo assembled with the ALLPATHS program (59) using the default parameters. Pseudochromosome files were generated for scaffold assembly, which was done with ABACAS2, a currently unpublished successor to ABACAS (60), using alignments to the Leishmania major Friedlin reference at a minimum alignment length of 500 bp and at least 85% identity.
Sequencing and RNA-seq of field isolates.
All 14 low-passage-number samples (Table S1) were axenically expanded in vitro. Each isolate had previously been culture adapted and cryopreserved in dimethyl sulfoxide under liquid nitrogen storage conditions (−60°C). Each frozen stock was thawed and cultured for 1 to 3 days in complete M199 promastigote medium until the parasite density reached 1 × 106/ml. Each parasite culture was then split into three separate culture flasks for biological replication and serially passaged every 24 h for 3 days, following well-established procedures (61), to maintain log-phase growth and density and to synchronize the culture in the proliferative promastigote developmental stage. After 72 h, each set of replicates was pelleted and the RNA was extracted following the TRIzol protocol. RNA and DNA samples were used as the starting material to prepare Illumina libraries following the manufacturer’s specifications. For RNA, the TruSeq stranded mRNA prep kit was used, which relies on 3′ poly(A) tail pulldown to isolate RNA species of interest. Purified RNA species were then prepped into paired-end libraries with an average insert size of 250 bp and sequenced on the Illumina HiSeq 2500 platform for 75 cycles. DNA samples were sequenced for 100 cycles using paired-end libraries with an average insert size of 500 bp, with individual isolates being multiplexed over two lanes on the Illumina HiSeq 2500 platform to increase coverage.
Mapping and analysis of whole-genome sequence data.
Short-read genomic sequence data were mapped to a draft reference genome for L. tropica by using the SMALT program (https://sourceforge.net/projects/smalt/) with a sequence match threshold of 80% and using a 13-kmer seed. Minor allele frequency and depth of coverage were called with the GATK DepthOfCoverage tool (Broad Institute), and the resulting read depth information was manipulated using custom bash, perl, and R scripts to generate the desired plots. Short haplotypes were assembled using the physical phasing step-by-step procedures implemented in freebayes (available from https://github.com/ekg/freebayes) (62) to call, filter, and phase high-quality SNPs. These high-quality partially phased SNPs were then used to find long runs of homozygosity by using VCFtools and the LROH output statistic.
An in-house-developed expectation-maximization (EM) algorithm was used to estimate somy for each chromosome, starting from the expected haploid read depth. The EM algorithm to estimate somy uses a likelihood function which models the median read depth (RD) of each chromosome from a single sample as coming from a Poisson distribution. The mean of that Poisson distribution is defined as the product of the somy for that chromosome, as an integer, multiplied by the haploid RD, which is the same for all chromosomes in a particular sample. The unknown parameters are the haploid RD and the somy for each of the 36 chromosomes. The maximization step uses the current estimate of the somy vector to maximize the likelihood function for the haploid RD. In the expectation step, the maximum-likelihood estimate (MLE) of the haploid RD is then used to calculate the most likely value of the somy for each chromosome, based on its Poisson distribution. Allele frequency and read depth plots generated using custom R scripts were visually inspected and used to confirm the estimated somy for each chromosome. Circular chromosomal plots were generated using RCircos (63).
FST and F (inbreeding coefficient) values.
FST was calculated using VCFtools (64), which employs the FST estimate from Weir and Cockerham’s 1984 paper (65). The SNP file containing allele information for all 268,518 polymorphic positions in a .vcf format was imported into VCFtools and used to calculate F statistics on a 1-kb sliding window basis.
Phylogenetic tree analysis.
Clustal W/X (66) was used to align variant information from the .vcf file using default settings. Aligned variant sequence data in GCG/MSF format were imported into Molecular Evolutionary Genetic Analysis (MEGA) version 6 for neighbor-joining analyses (67) using both the distance and parsimony methods. One thousand bootstrap replicates were used, and consensus trees were drawn according to the bootstrap 50% majority rule, with a root to the L590 reference strain.
Mapping and analysis of RNA-seq data.
Despite the nearly complete absence of cis-splicing in Leishmania, short reads were mapped to the reference draft assembly for the L. tropica L590 strain by using Tophat (68), a splice-sensitive aligner based on the Bowtie algorithm. Since the RNA-seq paired-end library preparation protocol is strand sensitive, the option fr-firststrand was used during mapping to preserve sense/antisense directionality of sequence information. Given the large degree of gene conservation and synteny between homologous regions in Leishmania species (13), the draft assembly was scaffolded against the GeneDB release of the L. major Friedlin reference genome by using ABACAS v2.0 (60), with a minimum alignment length of 500 bp and at least 85% identity. (These parameters were empirically determined to maximize the total length of the L. tropica assembly that could be scaffolded.) Gene annotations were also transferred from the L. major reference genome by using RATT (69), as L. major is the most closely related Leishmania species with a well-annotated reference genome. This resulted in a total of 7,863 genes in L. tropica.
Reads overlapping feature annotations were counted with HTSeq 0.6.1, using the htseq-count function (70) and the “intersection nonempty” command option. Raw gene counts were imported into R statistical software and analyzed with the DEseq package (71). Custom scripts were used for the statistical analyses and to generate the figures. The Bayes empirical dispersion for each gene was calculated using RLE-normalized read counts, treating all samples as if they were replicates under the same condition. A variance-stabilizing transformation was then applied to the count data as implemented in DEseq. Euclidean distances were calculated on the variance-stabilized expression values for each pair of samples, and pairwise Euclidean distance values were plotted in a heat map to visualize differences in expression signatures between samples. Principal-component analysis was also performed on the variance-stabilized expression values. The top 30 genes across all samples with the highest mean RLE-normalized read counts were identified, and their variance-stabilized expression values for each sample were used to generate a heat map.
Differential gene expression.
Raw counts from HTseq analysis were normalized following the standard edgeR workflow. Raw read counts from HTseq were normalized by library size using the weighted trimmed mean of M-values (TMM) method implemented in edgeR (72). The isolate L747 was used as the intercept for calculation of fold change relative to this baseline expression level, given its similarity to most other samples by the Euclidean distance metric. The isolate L747 also had a nearly diploid karyotype and therefore provided the best baseline to measure deviations due to gene dosage effects.
A generalized linear model (GLM) for negative binomially distributed count data was built, with each set of triplicates modeled as a separate condition. Common, trended, and tagwise dispersions were calculated with the Cox-Reid estimator. Given the multifactorial model of the experiment, the negative binomial GLM was fitted with the tag-wise dispersion to allow for the possibility that dispersion might vary across genes. The likelihood ratio test was then used to compare each set of triplicates to the baseline and identify genes that were differentially expressed in any of the groups in a test analogous to a one-way analysis. P values for differentially expressed genes in any of the groups were calculated using the F distribution and then adjusted using the BH method.
Pairwise exact tests were also performed between two pairs of isolates (K26 versus Ackerman and KK27 versus Azad) to identify genes with differential expression in the context of observed variation in somy and CNVs. These two pairs of isolates were selected based on the fact that many of their chromosomes differed in somy between each other, allowing for a better assessment of gene dosage effects.
Copy number variation.
To estimate the effect of gene dosage on relative expression levels, copy number variants between each pair of isolates were identified and annotated using the CNV-seq pipeline (73). This pipeline identifies localized regions in which read depth normalized across the length of the chromosome differs significantly between two samples. Information on differentially expressed genes from pairwise exact tests between specific isolates was superimposed on these CNV regions by using custom R and bash scripts.
Accession number(s).
All sequence data have been deposited in the European Nucleotide Archive short read sequence repositories for the RNA-seq triplicates and assigned accession numbers ERS763603 to ERS763656.
ACKNOWLEDGMENTS
We acknowledge Deborah E. Dobson and Natalia S. Akopyants, who prepared and performed quality control for the DNA for the L. tropica reference strain L590 in the laboratory of S.M.B. at Washington University in St. Louis. We also thank Ehud Inbar, Phil Lawyer, and Audrey Romano in the lab of D.L.S. at NIAID for parasite culture assistance and stimulating initial discussion of results, as well as Adam Reid in the lab of M.B. at the Wellcome Trust Sanger Institute for help in interpreting the RNA-seq data.
This study was financially supported in part by the Intramural Research Program of the NIH and NIAID, by NIH grant AI29646 to S.M.B., and by NIH-NHGRI grant 5U54HG00307907 to Richard K. Wilson, Director of the Genome Institute at Washington University. M.E.G. is a scholar of the Canadian Institute for Advanced Research Integrated Microbial Biodiversity Program.
S.A.I., M.E.G., J.A.C., M.B., and D.L.S. developed the idea for the study. W.C.W., S.M.B., and J.A.C. generated the first version of the reference genome scaffolds. S.A.I. generated all data and performed the majority of the analyses. C.D. developed the EM algorithm for somy estimation from Leishmania short read sequence data, and A.K. performed the population analysis. M.J.S. oversaw sequencing of the samples by the production team at the Sanger Institute. S.A.I., M.E.G., and J.A.C. wrote the manuscript.
We acknowledge funding from the Wellcome Trust via their core support for the Wellcome Trust Sanger Institute (grant 206194).
Footnotes
Citation Iantorno SA, Durrant C, Khan A, Sanders MJ, Beverley SM, Warren WC, Berriman M, Sacks DL, Cotton JA, Grigg ME. 2017. Gene expression in Leishmania is regulated predominantly by gene dosage. mBio 8:e01393-17. https://doi.org/10.1128/mBio.01393-17.
REFERENCES
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M; WHO Leishmaniasis Control Team . 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alawieh A, Musharrafieh U, Jaber A, Berry A, Ghosn N, Bizri AR. 2014. Revisiting leishmaniasis in the time of war: the Syrian conflict and the Lebanese outbreak. Int J Infect Dis 29:115–119. doi: 10.1016/j.ijid.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 3.Du R. 2016. Old World cutaneous leishmaniasis and refugee crises in the Middle East and North Africa. PLoS Negl Trop Dis 10:e0004545. doi: 10.1371/journal.pntd.0004545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azmi K, Krayter L, Nasereddin A, Ereqat S, Schnur LF, Al-Jawabreh A, Abdeen Z, Schönian G. 2017. Increased prevalence of human cutaneous leishmaniasis in Israel and the Palestinian Authority caused by the recent emergence of a population of genetically similar strains of Leishmania tropica. Infect Genet Evol 50:102–109. doi: 10.1016/j.meegid.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Krayter L, Bumb RA, Azmi K, Wuttke J, Malik MD, Schnur LF, Salotra P, Schönian G. 2014. Multilocus microsatellite typing reveals a genetic relationship but, also, genetic differences between Indian strains of Leishmania tropica causing cutaneous leishmaniasis and those causing visceral leishmaniasis. Parasit Vectors 7:123. doi: 10.1186/1756-3305-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rougeron V, De Meeûs T, Bañuls AL. 2017. Reproduction in Leishmania: a focus on genetic exchange. Infect Genet Evol 50:128–132. doi: 10.1016/j.meegid.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, Ouellette M. 2006. Unresponsiveness to glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med 3:e162. doi: 10.1371/journal.pmed.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plourde M, Coelho A, Keynan Y, Larios OE, Ndao M, Ruest A, Roy G, Rubinstein E, Ouellette M. 2012. Genetic polymorphisms and drug susceptibility in four isolates of Leishmania tropica obtained from Canadian soldiers returning from Afghanistan. PLoS Negl Trop Dis 6:e1463. doi: 10.1371/journal.pntd.0001463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton CE. 2016. Gene expression in kinetoplastids. Curr Opin Microbiol 32:46–51. doi: 10.1016/j.mib.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Alcolea PJ, Alonso A, Gómez MJ, Postigo M, Molina R, Jiménez M, Larraga V. 2014. Stage-specific differential gene expression in Leishmania infantum: from the foregut of Phlebotomus perniciosus to the human phagocyte. BMC Genomics 15:849. doi: 10.1186/1471-2164-15-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ubeda JM, Légaré D, Raymond F, Ouameur AA, Boisvert S, Rigault P, Corbeil J, Tremblay MJ, Olivier M, Papadopoulou B, Ouellette M. 2008. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol 9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubeda JM, Raymond F, Mukherjee A, Plourde M, Gingras H, Roy G, Lapointe A, Leprohon P, Papadopoulou B, Corbeil J, Ouellette M. 2014. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol 12:e1001868. doi: 10.1371/journal.pbio.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, Harris D, Her Y, Herzyk P, Imamura H, Otto TD, Sanders M, Seeger K, Dujardin JC, Berriman M, Smith DF, Hertz-Fowler C, Mottram JC. 2011. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachaud L, Bourgeois N, Kuk N, Morelle C, Crobu L, Merlin G, Bastien P, Pagès M, Sterkers Y. 2014. Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect 16:61–66. doi: 10.1016/j.micinf.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Inbar E, Hughitt VK, Dillon LA, Ghosh K, El-Sayed NM, Sacks DL. 2017. The transcriptome of Leishmania major developmental stages in their natural sand fly vector. mBio 8:e00029-17. doi: 10.1128/mBio.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. 2008. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics 9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasereddin A, Schweynoch C, Schonian G, Jaffe CL. 2010. Characterization of Leishmania (Leishmania) tropica axenic amastigotes. Acta Trop 113:72–79. doi: 10.1016/j.actatropica.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Lukes J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KW, Jirků M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Downing T, Stark O, Vanaerschot M, Imamura H, Sanders M, Decuypere S, de Doncker S, Maes I, Rijal S, Sundar S, Dujardin JC, Berriman M, Schönian G. 2012. Genome-wide SNP and microsatellite variation illuminate population-level epidemiology in the Leishmania donovani species complex. Infect Genet Evol 12:149–159. doi: 10.1016/j.meegid.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schönian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong D, Wallach M, Keithly J, Melera PW, Chang KP. 1984. Differential expression of mRNAs for alpha- and beta-tubulin during differentiation of the parasitic protozoan Leishmania mexicana. Proc Natl Acad Sci U S A 81:5782–5786. doi: 10.1073/pnas.81.18.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton CE. 2002. Life without transcriptional control? From fly to man and back again. EMBO J 21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker R, Song H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 25.Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 26.Milone J, Wilusz J, Bellofatto V. 2002. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res 30:4040–4050. doi: 10.1093/nar/gkf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haile S, Estevez AM, Clayton C. 2003. A role for the exosome in the in vivo degradation of unstable mRNAs. RNA 9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estévez AM, Clayton C. 2006. Roles of a Trypanosoma brucei 5′ → 3 exoribonuclease homolog in mRNA degradation. RNA 12:2171–2186. doi: 10.1261/rna.291506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C. 2009. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res 37:5511–5528. doi: 10.1093/nar/gkp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. 2008. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leprohon P, Légaré D, Raymond F, Madore E, Hardiman G, Corbeil J, Ouellette M. 2009. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res 37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur K, Coons T, Emmett K, Ullman B. 1988. Methotrexate-resistant Leishmania donovani genetically deficient in the folate-methotrexate transporter. J Biol Chem 263:7020–7028. [PubMed] [Google Scholar]
- 33.Coderre JA, Beverley SM, Schimke RT, Santi DV. 1983. Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc Natl Acad Sci U S A 80:2132–2136. doi: 10.1073/pnas.80.8.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal G, Mandal S, Sharma M, Charret KS, Papadopoulou B, Bhattacharjee H, Mukhopadhyay R. 2015. Species-specific antimonial sensitivity in Leishmania is driven by post-transcriptional regulation of AQP1. PLOS Negl Trop Dis 9:e0003500. doi: 10.1371/journal.pntd.0003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leprohon P, Légaré D, Girard I, Papadopoulou B, Ouellette M. 2006. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot Cell 5:1713–1725. doi: 10.1128/EC.00152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. 2005. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol 57:1690–1699. doi: 10.1111/j.1365-2958.2005.04782.x. [DOI] [PubMed] [Google Scholar]
- 37.Cunningham ML, Beverley SM. 2001. Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol Biochem Parasitol 113:199–213. doi: 10.1016/S0166-6851(01)00213-4. [DOI] [PubMed] [Google Scholar]
- 38.Imamura H, Downing T, Van den Broeck F, Sanders MJ, Rijal S, Sundar S, Mannaert A, Vanaerschot M, Berg M, De Muylder G, Dumetz F, Cuypers B, Maes I, Domagalska M, Decuypere S, Rai K, Uranw S, Bhattarai NR, Khanal B, Prajapati VK, Sharma S, Stark O, Schönian G, De Koning HP, Settimo L, Vanhollebeke B, Roy S, Ostyn B, Boelaert M, Maes L, Berriman M, Dujardin JC, Cotton JA. 2016. Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. Elife 5:e12613. doi: 10.7554/eLife.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondelaers A, Sanchez-Cañete MP, Hendrickx S, Eberhardt E, Garcia-Hernandez R, Lachaud L, Cotton J, Sanders M, Cuypers B, Imamura H, Dujardin JC, Delputte P, Cos P, Caljon G, Gamarro F, Castanys S, Maes L. 2016. Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS One 11:e0154101. doi: 10.1371/journal.pone.0154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soares RP, Barron T, McCoy-Simandle K, Svobodova M, Warburg A, Turco SJ. 2004. Leishmania tropica: intraspecific polymorphisms in lipophosphoglycan correlate with transmission by different Phlebotomus species. Exp Parasitol 107:105–114. doi: 10.1016/j.exppara.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Fraidenraich D, Peña C, Isola EL, Lammel EM, Coso O, Añel AD, Pongor S, Baralle F, Torres HN, Flawia MM. 1993. Stimulation of Trypanosoma cruzi adenylyl cyclase by an alpha D-globin fragment from Triatoma hindgut: effect on differentiation of epimastigote to trypomastigote forms. Proc Natl Acad Sci U S A 90:10140–10144. doi: 10.1073/pnas.90.21.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmon D, Vanwalleghem G, Morias Y, Denoeud J, Krumbholz C, Lhommé F, Bachmaier S, Kador M, Gossmann J, Dias FB, De Muylder G, Uzureau P, Magez S, Moser M, De Baetselier P, Van Den Abbeele J, Beschin A, Boshart M, Pays E. 2012. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science 337:463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- 43.Lopez MA, Saada EA, Hill KL. 2015. Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Eukaryot Cell 14:104–112. doi: 10.1128/EC.00217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz AK, Titus R, Beverley SM. 1993. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Natl Acad Sci U S A 90:1599–1603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martínez-Calvillo S, Stuart K, Myler PJ. 2005. Ploidy changes associated with disruption of two adjacent genes on Leishmania major chromosome 1. Int J Parasitol 35:419–429. doi: 10.1016/j.ijpara.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL. 2009. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inbar E. 2013. The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet 9:e1003672. doi: 10.1371/journal.pgen.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romano A, Inbar E, Debrabant A, Charmoy M, Lawyer P, Ribeiro-Gomes F, Barhoumi M, Grigg M, Shaik J, Dobson D, Beverley SM, Sacks DL. 2014. Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc Natl Acad Sci U S A 111:16808–16813. doi: 10.1073/pnas.1415109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, Pellman D. 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marques CA, Dickens NJ, Paape D, Campbell SJ, McCulloch R. 2015. Genome-wide mapping reveals single-origin chromosome replication in Leishmania, a eukaryotic microbe. Genome Biol 16:230. doi: 10.1186/s13059-015-0788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee PH, Meng X, Kapler GM. 2015. Developmental regulation of the Tetrahymena thermophila origin recognition complex. PLoS Genet 11:e1004875. doi: 10.1371/journal.pgen.1004875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun S, Billmyre RB, Mieczkowski PA, Heitman J. 2014. Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet 10:e1004849. doi: 10.1371/journal.pgen.1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volf P, Benkova I, Myskova J, Sadlova J, Campino L, Ravel C. 2007. Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int J Parasitol 37:589–593. doi: 10.1016/j.ijpara.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 57.Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, Svobodova M, Volf P, Berriman M, Cotton JA, Smith DF. 2014. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10:e1004092. doi: 10.1371/journal.pgen.1004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lahav T, Sivam D, Volpin H, Ronen M, Tsigankov P, Green A, Holland N, Kuzyk M, Borchers C, Zilberstein D, Myler PJ. 2011. Multiple levels of gene regulation mediate differentiation of the intracellular pathogen Leishmania. FASEB J 25:515–525. doi: 10.1096/fj.10-157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A 108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler RJ, Gluenz E, Gull K. 2011. The cell cycle of Leishmania: morphogenetic events and their implications for parasite biology. Mol Microbiol 79:647–662. doi: 10.1111/j.1365-2958.2010.07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [qbio.GN] http://arXiv.org/abs/1207.3907.
- 63.Zhang H, Meltzer P, Davis S. 2013. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 66.Higgins DG, Thompson JD, Gibson TJ. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266:383–402. [DOI] [PubMed] [Google Scholar]
- 67.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: rapid annotation transfer tool. Nucleic Acids Res 39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders S, Pyl PT, Huber W. 2015. HTSeq-a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie C, Tammi MT. 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample name, country of origin, WHO international identifier code, and ENA accession number of sequenced samples (ENA accession numbers for RNA-seq triplicates range from ERS763603 to ERS763656) Download TABLE S1, PDF file, 0.03 MB (34.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The top 30 most highly expressed genes in the set of 11 isolates analyzed by RNA-seq. The top cluster of highly expressed genes contains β-tubulin (LmjF.21.1860), a well-described marker of the promastigote stage, and several genes encoding nucleoside and amino acid transporter proteins (LmjF.36.1940, LmjF.15.1240, LmJF.15.1230, LmjF.07.1160, LmjF.36.6300, LmjF.31.0350, and LmjF.06.1260). Most other highly expressed genes encode ribosomal subunit proteins. See Table S2 for a full list of the genes. Download FIG S1, PDF file, 0.02 MB (18.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in L180 compared to all other isolates. Twofold changes in expression are marked with horizontal blue lines. Genes in red are those that were attained statistical significance. The complete list of statistically significant, differentially expressed genes with more than a 2-fold change in expression is provided in Table S3. Download FIG S2, PDF file, 0.4 MB (396.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The top 30 most highly expressed genes in the 11 samples that were analyzed by RNA-seq (VSE stands for variance-stabilized expression and is a normalization procedure for expression values built into the R package DEseq) Download TABLE S2, PDF file, 0.03 MB (35.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in the L810 isolate compared to all other isolates that are upregulated or downregulated more than 2-fold (the log FC stands for the log fold change in expression level) Download TABLE S3, PDF file, 0.03 MB (35.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Chromosomal plots of read depth and log fold changes in gene expression in the K26 versus Ackerman strain, highlighting gene dosage effects due to local CNVs and somy differences. Positive values indicate higher values for K26 than for Ackerman, and negative values indicate the opposite. Axes are labeled as described for Fig. 6. Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The top 30 most significant (lowest P value) differentially expressed genes across all isolates (a total of 9 genes were transmembrane, with significant enrichment compared to the total number of transmembrane genes in the genome [P < 0.05]) Download TABLE S4, PDF file, 0.03 MB (34.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Differentially expressed genes in the 4 largest CNV regions identified in the Ackerman-K26 comparison based on CNV-seq (tThere were a total of 10 genes in CNVR114 [chromosome 24], 11 genes in CNVR181 [chromosome 11], 14 genes in CNVR159-178 [chromosome 27], and 11 in CNVR240 [chromosome 23]; the CNV region on chromosome 11 is so extensive that it overlaps approximately half of the chromosome [Fig. 6A]) Download TABLE S5, PDF file, 0.04 MB (40.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Read depth plots of the chromosomal region surrounding the BT1 (LmjF.35.5150) and FT1 (LmjF.10.0385) genes in isolates K26 (in gold) and Ackerman (in blue). Red vertical bars mark the start and end of each gene. In K26, there is a clear homozygous deletion of FT1 (read depth of 0) and a possible heterozygous duplication of BT1 (read depth, ~75 to 90 across the region, suggesting that the gene is present in triplicate), while in the Ackerman line both genes are present in duplicate (read depth, ~50 to 60). It is important to note that both of these genes are present in multiple orthologous copies in most Leishmania species. Download FIG S4, PDF file, 0.2 MB (194.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.