Abstract
Manduca sexta stress response peptide-2 (SRP2) is predicted to be a 25-residue peptide (FGVKDGKCPSGRVRRLGICVPDDDY), which may function as an insect cytokine to regulate immune responses. Produced as an inactive precursor, endogenous proSRP2 is probably converted to active SRP2 by limited proteolysis in response to invading pathogens, along with prophenoloxidase and pro-Spätzle activation. In addition to immunity, SRP2 may control head morphogenesis or other developmental processes in the lepidopteran insect. We have examined the profiles of SRP2 gene expression in terms of immune induction capacity, tissue specificity, and developmental changes. To gain insights into its functions, we chemically synthesized SRP2, injected the peptide solution into naïve larvae, and detected significant up-regulation of several antimicrobial peptide genes. We determined the 3D molecular structure in solution of SRP2 by two-dimensional 1H-1H NMR spectroscopy. SRP2 has an ordered structure, which is composed of two short ß-strands at regions R12 - R15 and I18 - V20, one type-I’ ß-turn at region R15 - I18, and a half turn at region C8 - S10 in its well-defined core stabilized by a covalent disulfide bond between C8 and C19. The secondary and tertiary structures are further stabilized by hydrogen bonds. Possible relationships between the structure and function are also discussed.
Keywords: Cytokine, 2D nuclear magnetic resonance, epidermal growth factor, hemolymph protein, insect immunity, prophenoloxidase
1. INTRODUCTION
Analogous to innate immunity in vertebrates, insects rely on humoral and cellular responses to protect them from microbial infection [1]. In many moth species, proteolytic activation of plasmatocyte spreading peptides (PSPs, also known as GBPs for growth blocking peptides or PPs for paralytic peptides) from their precursors stimulates hemocyte encapsulation [2]. These active peptides have a conserved N-terminus of E-N-F and, therefore, are also called ENF factors. While ENF peptides are only found in lepidopterans, their evolution goes beyond moths and butterflies. Homologous peptides have been isolated from hemolymph of insects in six different orders [3]. GBP homologs also include: upstream ENF peptides (uENFs) [4] and stress response peptides (SRPs) [5]. The latter are identified in many insects including mosquitoes, moths, beetles and locusts. Since most of these peptides are activated during stress conditions such as wounding, infection, ligation, and heat/cold shock, SRP appears to be a proper general name for these phylogenetically related peptides. In addition to blocking growth, antimicrobial peptide induction [6], as well as hemocyte spreading and chemotaxis [7], SRPs play roles in brain morphogenesis [8] and thanatosis [9]. One of the five Drosophila SRPs induces diptericin and metchnikowin expression in an Imd-dependent manner [6a]. These peptides essentially fulfill the role of cytokines for these insects, partly based on their expression in the brain, midgut, fat body, and hemocytes. As with cytokines in vertebrates, many of these short peptides’ expression varies in different developmental stages of the organisms.
In contrast to the knowledge on SRPs’ diverse physiological functions, mechanisms for generating active SRPs from their inactive precursors are poorly understood. Evidence suggests that hemolymph proteases are responsible for the specific proteolysis. A system of serine proteases (SPs) exists in plasma of each one of the studied insects to mediate immune responses [10]. An SP inhibitor benzamidine blocks the processing of M. sexta proPSP in the larval hemolymph [11]. Prophenoloxidase (proPO) activation generates PO which catalyzes melanization. Since this defense response often accompanies hemocyte encapsulation and nodule formation, we postulate that the SP system activated upon recognition of pathogens is also involved in the SRP activation. With the genome sequence of M. sexta available, we identified 105 non-digestive SPs and most of them are predicted to have trypsin-like specificity [12]. To begin to study SRPs in this biochemical model insect, we performed a genome search and identified members of the SRP gene family, including PSP, uENF1, uENF2, and SRPs. While we do not yet know their exact physiological functions in M. sexta, the diverse functions observed in their homologs distributed in a wide range of taxonomic groups of insects have led us to undertake structural-functional studies of SRP2, which is one of the highly expressed SRPs in M. sexta.
The putative SRP2 consists of 25 amino acid residues (F-G-V-K-D-G-K-C-P-S-G-R-V-R-R-L-G-I-C-V-P-D-D-D-Y) with a single disulfide bond between Cys8-Cys19. Here we report the three-dimensional solution structure of SRP2 solved by two dimensional 1H-1H NMR spectroscopy. We found that the M. sexta SRP2 core region contains two short ß-strands stabilized by a type-I’ ß-turn and a disulfide bond. The overall secondary and tertiary structure is further stabilized by hydrogen bonds. We also discuss the expression profile of the SRP2 gene and its possible functions in immune signal transduction.
2. MATERIALS AND METHODS
2.1. Identification of SRP Genes in the Genome of M. sexta
The MCOT1.0 models [12] were matched with K/R-X1-15-C-X3-10-G-X1/2-C-X1-15, where X represents any amino acid residue other than C. This motif is modified from the GBP consensus sequence of C-X2-G-X4/6-G-X1/2-C-K/R [3a] by adding K/R in the front, since 68 of the 105 potential activating SPs are predicted to cleave after K/R residue [12]. The initial search results were manually examined to identify putative SRPs that are located in the carboxyl-terminus of secreted proteins shorter than 250 residues.
2.2. Insect Rearing and Injection, Total RNA Preparation, and cDNA Synthesis
M. sexta eggs, ordered from Carolina Biological Supply (Burlington, NC), were hatched at ambient temperature and larvae were reared on an artificial diet [13]. The 5th instar larvae (day 2) were separately injected with a mixture of bacteria (2×107 Escherichia coli cells and 20 μg Micrococcus luteus) (Sigma-Aldrich) and 20 μg curdlan (insoluble ß-1,3-glucan from Alcaligenes faecalis) (Sigma-Aldrich) suspended in H2O (30 μL per larva). Hemolymph samples were collected from cut prolegs of three larvae 24 h later and centrifuged at 5,000 g for 5 min to harvest hemocytes prior to dissection of fat body tissue. Total RNA was prepared from induced hemocytes and fat body using TRIZOL Reagent (Thermo Fisher Scientific). Control fat body and hemocyte total RNA samples were isolated from three 5th instar naïve larvae (day 3) by the same method. Other tissues (i.e., integument, muscle, midgut, trachea, Malpighian tubule, nerve tissue, and salivary gland) were dissected from 5–10 5th instar naïve larvae (day 3) for RNA preparation. To examine the developmental changes, fat body and hemocytes were prepared from larvae and pupae at different stages (4th instar to adult, 3–6 insects per stage) for total RNA isolation. These total RNA samples (1 μg each) were incubated with 1× iScript Reverse Transcription Supermix (Bio-Rad) in a 10 μL reaction at 42 °C for 30 min to synthesize cDNA. The reaction mixtures were heated at 95 °C for 3 min to denature the enzyme. To examine its possible role in the expression regulation of antimicrobial peptide (AMP) genes, 30 μL of phosphate-buffered saline (PBS, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) or SRP2 (0.1 μg/μL) in PBS was injected into each of six day 2, 5th instar larvae (3 control and 3 test). Fat body tissues were separately collected 6 h later for RNA isolation and cDNA synthesis.
2.3. Quantitative Polymerase Chain Reaction (qPCR)
The cDNA samples were incubated with 1× iTag Universal SYBR Green Supermix (Bio-Rad) along with specific primers (0.5 μM each) in triplicate. In each 10 μL reaction, each cDNA is equivalent to starting with 50 ng total RNA. The primers were: j037 (5’ CATGATC-CACTCCGGTGACC) and j038 (5’ CGGGAGCAT-GATTTTGACCTTAA) for rpS3; j1825 (5’ TATTCCTGATCGTGCTGGTC) and j1826 (5’ ACAATA-CATTCCGCCAGAGC) for diapausin-4; j1070 (5’ GCAGGCGACGACAAGAAC) and j1071 (5’ ATGCGTGTTGGTAAGAGTAGC) for attacin-1, 7, 8 and 10; j1072 (5’ CCGTGTTTTATTCTTCGTCTTC) and j1073 (5’ AATCCTTTGACCTGCACCC) for cecropin-6; j1827 (5’ GCTGTTGATCTGCGTGACAT) and j1828 (5’ TCCTCCTTTGAATCCACGTC) for defensin-2; j1074 (5’ GCAAGTCGGCAACAATGG) and j1075 (5’ ACCCTGTCCTGTCAGTTTG) for gloverin; j1829 (5’ CCTGTACTGCCCTCGATCAT) and j1830 (5’ TTGTATCCCGGGTGAGTAGC) for lebocin D; j1076 (5’ GTGTGCCTCGTGGAGAATG) and j1077 (5’ ATGCCTTGGTGATGTCGTC) for lysozyme-1; j1078 (TGCTTTCTTTAACCTTTGTCCTC) and j1079 (TATTC-TAACACAGCCTATAATGCG) for moricin-1. The PCR was carried out under the following thermal cycling conditions: denaturation at 95 °C for 3 min and 40 cycles of 95 °C for 10 seconds and 60 °C for 30 seconds. After the reactions were complete on a CFX Connect Real-Time PCR Detection System (Bio-Rad), melt curves of the products (106, 167, 135, 103, 151, 124, 200, 187 and 186 bp) were measured to ensure proper shape and TM values. Amplification efficiencies (E) were measured by amplifying a cDNA sample diluted to 10−1, 10−2, 10−3, 10−4 and 10−5 using the specific primer pairs under the same conditions, which were 93.8% for rpS3, 63.0% for diapausin-4, 105.3% for attacin-1, 7, 8 and 10, 90.0% for cecropin-6, 104.7% for defensin-2, 71.2% for gloverin, 88.1% for lebocin D, 121% for lysozyme-1, and 88.9% for moricin-1. Relative mRNA levels were calculated as: (1 + ErpS3)Ct, rpS3/(1 + Ex)Ct, x [14].
2.4. Chemical Synthesis of Putative M. sexta SRP2
The 25-residue SRP2 were prepared by stepwise solid-phase synthesis using 9-fluorenylmethoxycarbonyl (Fmoc) chemistry on an ABI model 431A automatic peptide synthesizer (Applied Biosystems; Foster City, CA). For peptide synthesis CLEAR amide resin (0.3 mmol g–1; Peptides International, Louisville, KY) and Nα-F-moc amino acids (Anas-pec Inc., San Jose, CA) were used. Following de-protection and cleavage, the linear peptide was cyclized by formation of a disulfide bond between C8 and C19. The peptide was purified to >95% purity by HPLC (System Gold HPLC; Beck-man Instruments, Inc., Fullerton, CA) with a Phenomenex reversed-phase C-18 column (Torrance, CA), and eluted using a linear gradient of 10–90% acetonitrile containing 0.1% trifluoroacetic acid at 1 mL min–1. HPLC-purified peptide was characterized by matrix-assisted-laser desorption time-of-flight (MALDI-TOF) mass spectrometric analysis using a Bruker Ultraflex II spectrometer (Bruker Daltronics, Billerica, MA). After characterization, peptide was lyophilized and stored as dry powder until use.
2.5. NMR Spectroscopy
1D and 2D 1H-1H NMR experiments were performed with the 11.75 Tesla Varian 500 MHz VNMRS system (Agilent Technologies Inc., Palo Alto, CA), operating at 499.84 MHz for 1H frequency. The NMR data were acquired at Biomolecular NMR Core Facility in the Department of Biochemistry and Biophysics at Kansas State University, using a 5 mm cryogenic triple resonance inverse detection pulse field gradient probe at 25 °C. The purified SRP2 was dissolved in 650 μL of 10% D2O and 90% triple distilled H2O to a final concentration of 2.2 mM for NMR analysis. A total of 512 increments of 4K data points were recorded for the 2D 1H-1H Double Quantum Filtered Correlation Spectroscopy (DQF-COSY) [15] experiments whereas 2D 1H-1H Total Correlation Spectroscopy (TOCSY) [16] and Nuclear Overhauser Effect Spectroscopy (NOESY) [17] experiments were performed using 2000 t2 and 256 t1 data points with spectral width of 12 ppm in each dimension, and 16 transient per increment. Spin-lock time of 80 ms at B1 field strength of 7 kHz was used for 2D 1H-1H TOCSY experiments using MLEV-17 pulse sequence [16], and mixing times of 100, 300 and 500 ms were used for 2D 1H-1H NOESY experiments. All data sets were collected in hyper-complex phase-sensitive mode. Before processing the t2 dimension of DQF-COSY data sets were zero filled to 8K and the t1 dimension of DQF-COSY data sets to 2K. For all other experiments, the linear prediction and zero filling to 4K data points in t2 dimension were used during NMR data processing. When necessary, spectral resolution was enhanced by Lorentzian-Gaussian apodization. Suppression of the solvent peak (HOD) was achieved using the WATERGATE pulse scheme during acquisition [18] and the residual water peak (4.78 ppm) was used as reference for chemical shift assignments. NMR data processing was done using VnmrJ3.2a (Agilent Technologies Inc., Palo Alto, CA) and analyzed using CCPNMR 2.4 [19] and Sparky, a graphical NMR Assignment and Integration program [20]. Side chain proton resonances were assigned by comparing cross peaks in the TOCSY spectra with those in the NOESY spectra acquired under similar experimental conditions. Amide proton resonances either appearing after 12 h or showing slow exchange in the 1D and 2D 1H-1H TOCSY spectra acquired in 100% D2O were considered for hydrogen bonding. For distance constraints determination, NOE cross peaks were classified as strong, medium, or weak intensity based on the observed number of contour lines.
2.6. Structure Calculations
NOESY spectra acquired with the different mixing times (100, 300 and 500 ms) were used to obtain inter-proton distance constraints. Initially, a mixing time of 300 ms was used for distance constraint calculations and the assigned peaks were also checked with the spectrum acquired with the 100 ms mixing time. In order to rule out the peaks due to spin diffusion, only NOE peaks present in the NOESY spectra acquired with 100 ms mixing time were considered. NOE-derived distance constraints were classified into three distance ranges (1.8–2.5, 1.8–3.5, and 1.8–5.0 Å) corresponding to strong, medium and weak NOE intensities. Upper bound distance constraint limits for methyl or non-stereo-specifically assigned methylene protons were corrected by adding 0.5 Å appropriately for center averaging [21]. Structure calculations were performed using CNS program version 1.3 [22] that uses a combination of simulated annealing in torsional space with refinement using molecular dynamics in a Cartesian space. Resulting in calculated structures with minimal restraint violation [23]. Force constants were scaled throughout the procedure using default parameters of CNS. The refinement protocol involved 100 ps restrained MD simulation at 300 K followed by energy minimization. A force constant of 50 kcal/mol/ 2 for the NOE derived distances constrains and 100 kcal/mol/ 2 for dihedral angles constraints, were imposed during molecular dynamics calculations. The force constant of NOE distances was increased to 100 kcal/mol/ 2 during final energy minimization. All NMR restraints, including NOE, dihedral angles and hydrogen bond derived distance constraints were used to calculate over 400 structures. The 20 lowest energy structures with no restraint violation were selected to obtain an ensemble of three dimensional NMR structures. The average structure was used to represent 3D solution NMR structure of M. sexta SRP2. The quality of calculated structure was analyzed using VMD [24] and PROCHECK-NMR programs [25]. Figures of the structures were generated using PyMOL software [26].
3. RESULTS
3.1. Identification and Features of Putative SRPs of M. sexta
Using a modified GBP consensus sequence [3a], we searched the MCOT1.0 assembly of 31,666 protein sequences, because this dataset consists of more gene models (18,089) than M. sexta Official Gene Set 2.0 does (15,451) [12]. After initial sequence matching, the hits were manually examined to ensure they are <250 residues long, with a signal peptide at the N-terminus, and a GBP-like sequence at the C-terminus. Eleven sequences satisfied the specifications (Fig. 1): uENF1, uENF2, and PSP precursors are encoded by three open reading frames located in a single transcript (data not shown). A shorter transcript also encodes the 130-residue PSP precursor as reported previously [11], indicating the existence of an alternative promoter. Eight other proteins are designated as precursors of SRP1 through SRP8, ranging from 72 to 144 residues long.
Figure 1. Sequence Alignment of a Family of Proteins with Their Carboxyl-Termini Similar to SRPs.

The last ten residues of the predicted pro-region and the putative SRPs at the C-terminal end are aligned. The predicted activation cleavage site is shown as a gap after the Arg/Lys (in bold) and the conserved recognition sites (R/K-X-Y-R/K) of intracellular processing proteases are marked red. The two invariable C residues (red, bold) may form a disulfide bond to stabilize the peptide structures. The two highly conserved G are shaded green. The D/E (blue, bold) residues after the 2nd C in most SRPs may provide negative charges to the C-terminal end (*). The three numbers following each sequence denote the sizes of active peptide, signal peptide, and entire pre-pro-peptide.
Following a signal peptide (15 to 23-residue), a pro-region (28–102 residues) and a carboxyl-terminal fragment (22–32 residues) were predicted in the mature proteins. While similarity was observed among some of these pro-segments, this seemed to just reflect the closer relationships of recently duplicated genes. Of the eleven pro-regions ending with R or K (Fig. 1), PSP, SRP4, SRP5, SRP7 and SRP8 contain the recognition sequence of intracellular processing enzymes (R/K-X-Y-R/K), where Y is often R/K[27]. This suggests that most of the five processed cytokine-like molecules are stored inside the cells and released in response to relevant physiological cues. In comparison to their pro-regions, the predicted C-terminal peptides are conserved in sequence and, perhaps, 2°/3° structure also. The eleven peptides all match the GBP consensus sequence of C-X2-G-X4/6-G-X1/2-C-R/K, except that the positively charged R/K was replaced by T/S/V/F in SRP1-5, 7 and 8 (Fig. 1). There are 5–10 residues before the first C. Of the 4–9 residues after the second C, 3 to 4 were negatively charged (i.e. D/E).
3.2. Immune Inducibility, Tissue Specificity, and Developmental Profiles of SRP2 Expression
To begin to study the functions of these peptides, we examined their mRNA levels in different tissues at various life stages represented by 52 RNA-Seq datasets [12] and in hemocytes and fat body from naïve and induced larvae represented by 4 other datasets [28]. Based on the results, we selected SRP2 to confirm their expression patterns. Quantitative PCR analysis demonstrated that SRP2 mRNA levels were significantly higher (p<0.05) in the induced fat body and hemocytes than their respective control samples (Fig. 2A). The mRNA levels in hemocytes were significantly higher than those in fat body. The higher expression in blood cells was confirmed when other tissues from day 3, 5th instar larvae were compared. The SRP2 transcripts in hemocytes were 2.5, 7.3, 11.5, 12.4 fold higher than those in trachea, nerve tissue, Malpighian tubules, and fat body (Fig. 2B), respectively. The mRNA levels in epithelial cells, midgut, muscles and salivary glands were negligible. We also determined the SRP2 expression in hemocytes and fat body at different life stages (Fig. 2C). In hemocytes, there was an up-regulation in the wandering stage before the larval-pupal molt and another mRNA peak occurred in the middle to late pupal stage before the pupal-adult molt. In fat body, the transcript levels were higher in day 0, 4th and 5th instar larvae than day 3 larvae of the same stages. The SRP2 mRNA level in fat body peaked in the end of 5th instar feeding phase and slightly decreased in the wandering stage. A similar expression pattern was observed in some AMP genes that are up-regulated in the wandering stage [29].
Figure 2. qRT-PCR Measurement of M. sexta SRP2 mRNA Levels. (A).
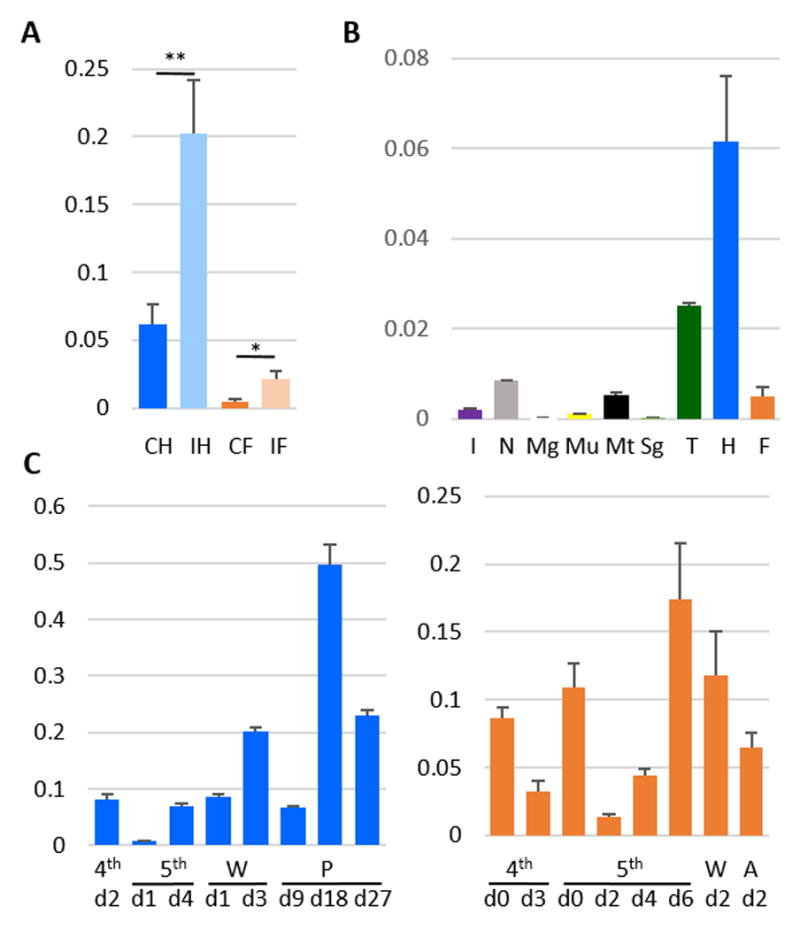
Immune inducibility. As described in Materials and methods, the total RNA samples of control and induced hemocytes (CH and IH) and fat body (CF and IF) were reverse transcribed to cDNA and analyzed by quantitative real-time PCR using the SRP2-specific primers. The relative mRNA levels normalized with rpS3 based on their Ct values in three biological replicates (3 larvae per sample) and plotted as a bar graph (mean ± SD, n = 3). The statistical significances were calculated using the Student’s t-test and significant increases are shown as * (p<0.05) and ** (p<0.01). (B) Tissue specificity. RNA samples of integument (I), nerve tissue (N), midgut (Mg), muscle (Mu), Malpighian tubule (Mt), salivary gland (Sg), trachea (T), hemocytes (H), and fat body (F) from day 3, fifth instar naïve larvae were prepared and analyzed by qPCR under the same conditions. (C) Developmental profiles. The mRNA levels in hemocytes (left) and fat body (right) at different life stages were determined using the same method. W, wandering; P, pupal; A, adult.
3.3. Induction of AMP Gene Expression by SRP2 Injection
To test whether SRP2 induces AMP synthesis, we chemically synthesized the 25-residue peptide and injected the peptide solution (3.0 μg) or PBS into day 2, 5th instar larvae. We analyzed the test and control RNA samples by qPCR and found five of the eight AMP genes were significantly induced (p<0.05) at 6 h after the SRP2 injection (Fig. 3). The cecropin-6, gloverin, lebocin D, lysozyme-1, and moricin-1 mRNA levels were 3.5, 3.1, 9.5, 4.2, and 14.0 fold higher than the control, respectively. While these increases were less dramatic than those caused by injection of a bacterial mixture [28] it implicates SRP2’s role in regulating the immune response in the tobacco hornworm. This is consistent with the result from Drosophila [6a] and justifies a structural study of this cytokine.
Figure 3. Induction of AMP Expression by Injection of PBS (grey) or SRP2 (red).
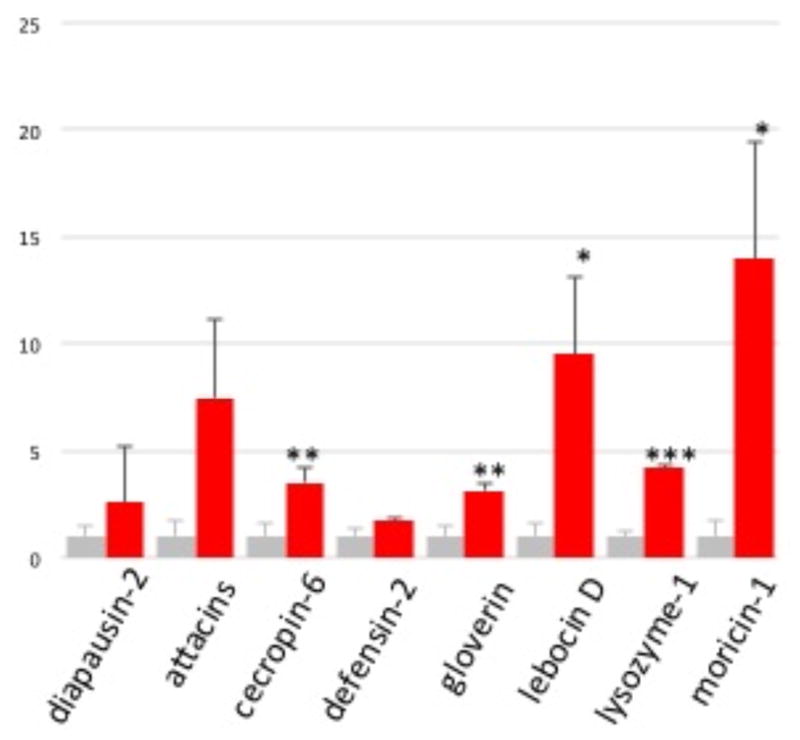
As described in Materials and methods, the test and control total RNA samples of fat body from the larvae injected with buffer on peptide were analyzed by qPCR using the primers specific for eight AMP genes. After normalization against rpS3, the average SRP2 mRNA levels in the control samples were adjusted to one, and relative levels in the SRP2 injected group were calculated accordingly. The statistical significances were calculated using the Student’s t-test and significant increases are shown as * (p<0.05), ** (p<0.01), or *** (p<0.001).
3.4. NMR Resonance Assignments and Secondary Structure
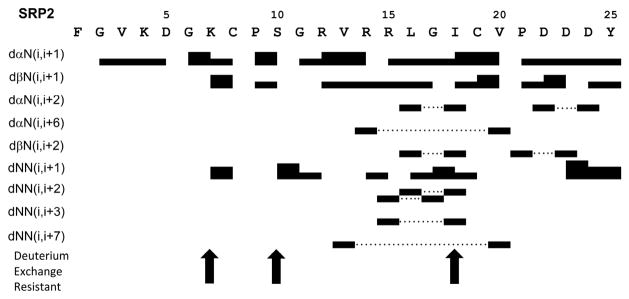
The amino acid spin system resonances and sequential proton chemical shift assignments for SRP2 were achieved through analyzing 2D 1H-1H TOCSY NMR spectra for intra residue proton-proton correlations and 2D 1H-1H NOESY spectra for inter residue proton-proton correlations as previously described [30]. The TOCSY spectra acquired for SRP2 at 25 °C gave good chemical-shift dispersion with limited spectral overlap, enabling assignments of the spin systems for all residues and suggesting the presence of a well-defined secondary and tertiary structure for this peptide. In these assignments, CaH (i) – CdH (i+1: P) (dad) or CaH (i) – CaH (i+1: P) (daa) NOEs instead of daN (i, i+1) were used for P9 and P21. Both prolines in SRP2 showed strong dad NOEs which were assigned to be trans configuration. The presence of any Pro in cis configuration would not be detected due to absence of daa NOE cross peaks in the 2D 1H-1H NOESY spectra. The 1H chemical shift assignments for M. sexta SRP2 have been deposited in BioMagResBank (http://www.bmrb.wisc.edu) under the accession number 26862. Fig. 4 shows the NH-CaH, NH-CßH, NH-C H, and NH-CdH cross peak assignments in the fingerprint region of the TOCSY spectrum used for residue identification. The 2D 1H-1H NOESY spectra allowed the identification of daN(i, i+1) proton connectivity for sequential assignments and the disambiguation of repeated amino acid residues. Further analysis of the NOESY spectra obtained with different mixing time revealed short, medium and long range connectivity including daN, dßN and dNN that provided structural information. An example of a NOESY spectrum showing dNN connectivity is provided in Supporting Information (Fig. S1). All the sequential and medium range distance NOE connectivity as well as deuterium exchange resistant data for residues are illustrated in Fig. 5. A number of non-sequential (weak daN((i, i+2) NOE between L16 and I18; weak dNN(i, i+3) NOE between R15 and I18) and long range (weak daN((i, i+6) NOE between R14 and V20; weak dNN(i, i+7) NOE between V13 and V20) NOEs characteristic of twisted antiparallel ß-sheets were observed for residues R12, V13, R14, R15, L16, I18, C19 and V20. This secondary structural observation was further supported by the down field shift of Ca proton resonances in chemical shift index ( CaH) values (Fig. S2) and larger (9.5 -15 Hz) coupling constant (3JNH-CaH) value for these residues. The random coil values for CaH protons were taken from the published data [31]. The SRP2 structure consists two short ß-strands at region R12-R15 (medium daN(i, i+1) NOEs from R12 to R14 and weak dNN(i, i+1) NOE between R14 and R15) and I18 – V20 (medium daN(i, i+1) NOEs from I18 to V20), a type-I’ ß-turn at region R15 – I18 (weak daN(i, i+1) NOE from L16 to G17 and medium dNN(i, i+1) NOE from G17 to I18) and a -turn at region C8 – S10 (carbonyl oxygen of i-residue C8 forming a hydrogen bond with slowly exchanged amide proton of i+2 residue S10, a characteristic of -turn). The short stretch of twisted anti-parallel ß-sheet was confirmed by observation of hydrogen bond between V13 - V20, and R15 - I18. Low preponderance of NOE cross peaks for N-terminal residues F1 – K7 and C-terminal residues D22 – Y25 suggested extended conformation for these regions.
Figure 4. Fingerprint Region (NH-CaH/Side chain protons) of 2D 1H-1H TOCSY Spectrum of SRP2 in H2O.
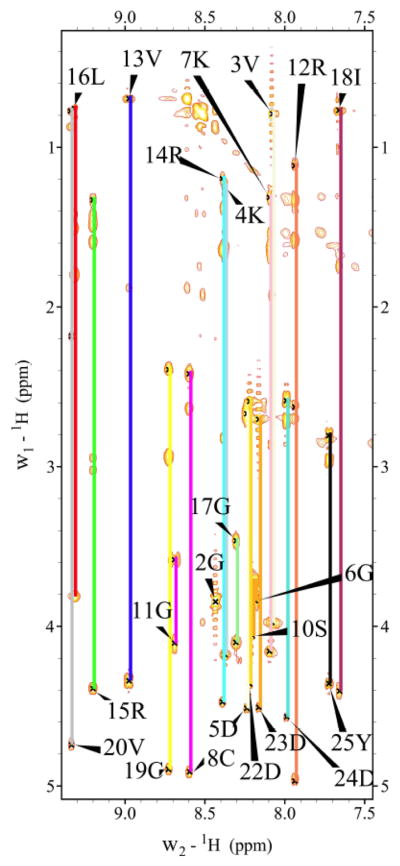
Individual amino acid spin systems connected with vertical lines and labeled with residue name and number.
Figure 5. Summary of the NOE connectivity for SRP2 in water.
The line thickness indicates peak intensity, dotted lines indicate nonsequential connections and arrows highlight residues that are resistant to deuterium exchange.
3.5. Tertiary Structure of M. sexta SRP2
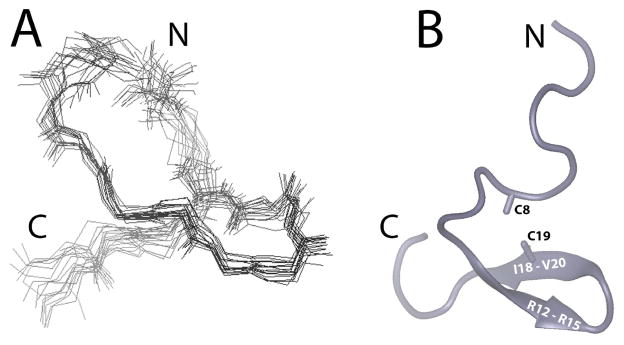
For tertiary structure calculations, a total of 224 constraints involving 106 intra-residue, 62 sequential, 17 medium range, 9 long range NOE and 8 hydrogen bond distance constraints as well as 20 dihedral angle constraints were used. Two additional distance constraints were included between S and Cß atoms of C8 and C19 to produce normal disulfide bond geometry (2.0 Å < dS-S < 2.5 Å, 2.2 Å < dCß-S < 3.5 Å). The structural statistics of SRP2 in water are summarized in Table 1. A simulated annealing molecular dynamics analysis was performed with these distance and dihedral constraints and found to a single family of conformers regardless of the starting structure. For these calculations, our starting structures were randomly chosen with extended conformation. From more than 400 conformers calculated, 20 structures with lowest energy were retained for further analysis. The torsion angle dynamics of all the 20 conformers were consistent with a pair of hydrogen bonds involving the backbone carbonyl of R15 and amide proton of I18. Evidence of two additional hydrogen bonds (I18 backbone car-bonyl to backbone amide proton of R15, and V20 backbone carbonyl to backbone amide proton of V13) were observed in a subset of the family of structures. The representative 20 superimposed structures (Fig. 6a) were in good agreement with experimental data with no distance violation larger than 0.3 Å and no dihedral angle violation larger than 5 degree. A Ramachandran plot produced by Procheck-NMR [25] showed that 72.1% of residues are in most favored stereo-chemically allowed regions and 23.2% are in additional allowed regions (Table 1). A superimposition of the 20 lowest energy structures showed a considerable degree of flexibility, however, alignments along residues 8–21, forming the core region for SRP2, of the 20 lowest energy structures showed backbone root mean square deviation value of 0.87 Å. The schematic ribbon diagram of the energy minimized average tertiary structure of M. sexta SRP2 is shown in Fig. 6b. Three hydrogen bonds that stabilized the tertiary structure of SRP2 were confirmed by the slowly exchanged amide protons from K7, S10, and I18.
Table 1.
Structural Statistics of the 20 Lowest Energy Structures for SRP2.
| NOE Constraints | Number |
|---|---|
| Total | 224 |
| Intra-residue | 106 |
| Sequential | 62 |
| Medium range (i–j = 5) | 17 |
| Long range (i–j = 5) | 9 |
| Disulfide bonds | 2 |
| Hydrogen bonds | 8 |
| Dihedral Constraints | 20 |
| Constraints/residue | 8.96 |
| Pairwise R.M.S.D. to Mean structure (residues 1–25) | |
| Backbone atoms (Å) | 2.45±0.48 |
| All nonhydrogen atoms (Å) | 3.13±0.47 |
| Pairwise R.M.S.D. to Mean structure (residues 8–21) | |
| Backbone atoms (Å) | 0.87±0.21 |
| All nonhydrogen atoms (Å) | 1.88±0.62 |
| Percentage of Residues in Regions of f- | Space |
| Allowed | 72.1% |
| Additionally Allowed | 23.2% |
| Generously Allowed | 3.8% |
| Disallowed | 0.9% |
Figure 6. Structural Representations of SRP2.
(A) Superimposition of the 20 lowest energy NMR structures of SRP2. (B) Ribbon diagram of the lowest energy structure of SRP2. The disulfide bond between C8 and C19 is indicated.
4. DISCUSSION
In this study, we have identified a family of eleven GBP-like genes in the genome of M. sexta, each coding for a protein with the pre-pro-structure. The signal peptide (i.e. pre-region) allows the pro-protein to be secreted into extracellular space or temporarily stored for later release through the secretory pathway, as these small proteins do not contain any transmembrane segment. In the former, the pro-proteins (precursors of uENF1, uENF2, PSP, SRP1-3, 6) are anticipated to be cleaved by extracellular SPs to become active. This mechanism is known for the proPSP activation [11]. In the latter, the pro-proteins (precursors of SRP4, SRP5, SRP7 and SRP8) may be further processed by intracellular convertases (e.g. furin) to become active peptides. They are probably released in response to particular physiological or pathological stimuli because, otherwise, we must assume that these four proSRPs are constitutively processed and released as not regulated active cytokines.
SRP2’s intensity and unique mRNA expression pattern (Fig. 2) [12, 28] makes it an excellent candidate for function exploration of this protein family. The qPCR results (Fig. 2) confirmed the data from the RNA-Seq analyses, suggesting that SRP2 takes part in antimicrobial defense of M. sexta. Fat body and hemocytes are major components of the immune system. After bacterial injection, the SRP2 transcript level increased 3.3-fold in hemocytes to 20.3% of rpS3’s, which is highly abundant in cells. In the fat body, the 4.4-fold increase brought SRP2 transcript to 2.2% of the rpS3’s level (Fig. 2A). As fat body has a much higher mass than hemocytes and acts as the major contributor of hemolymph proteome [32], it is unclear whether proSRP2 in plasma mainly comes from hemocytes or not. The proSRP2 made in trachea may also participate in immune responses, whereas its production in nerve tissue suggests a possible role as a neuropeptide upon activation (Fig. 2B). The up-regulation of SRP2 expression in the fat body in the end of 5th instar feeding phase (Fig. 2C) resembles that of the AMP genes [29].
Up-regulation of AMP gene expression after SRP2 injection implicates an immunological role (Fig. 3). Based on these observations, we postulate that the SP system in the hemolymph is triggered by bacterial infection to activate proSRP2 along with other precursors of immune factors (e.g. PO, Spätzle, and PSP). Remarkably, the predicted cleavage site (R*F) of proSRP2 is identical to that of proPO1 and proPO2, suggesting that proPO activating proteases (PAP1-3) can also activate proSRP1-3 (Fig. 1). In M. sexta, PO-mediated melanization occurs along with proHP6 and proHP8 activation [33]. Hemolymph protease-8 (HP8) generates active Spätzle-1 to induce AMP production via a putative Toll pathway [12, 33]. We propose SRP2, like Spätzle, may act as a cytokine to turn on another immune signaling pathways and induce AMP gene expression. In Drosophila, GBP (encoded by CG15917, one of the five SRP genes) induces an intracellular pathway via Imd to activate metchnik-owin and diptericin [6a]. Instead of the classical Imd pathway including Relish, the GBP binding signal may go through the JNK branch. It would be interesting to identify proSRP2-activating protease(s), the SRP2 receptor, and downstream signal transducers in M. sexta using a biochemical approach.
The analysis of the tertiary structure of SRP2 suggests that in aqueous solution this peptide possesses a well-defined structural core region with a short twisted anti-parallel ß-sheet that is stabilized by a type-I’ ß-turn and disulfide bond while the N- and C-termini are largely unstructured and apparently with substantial flexibility. Overall tertiary structure is stabilized by a combination of the hydrogen bonds within the ß-sheet structure and hydrophobic sidechain packing along with the covalent disulfide linkage. Within the core region of the molecule, one side of the structure is mainly composed of disordered charged residues, including R12, R14, and R15. The side chain of R12 showed NOE contacts to the side chains of C8, P9, and C19. The opposite side of the structure is composed of hydrophobic residues, including V13, L16, I18 and V20. These structural characteristics similar to M. sexta paralytic peptide structure [34] indicate that the SRP2 structural core adopts an epidermal growth factor (EGF)-like fold very similar to the carboxyl-terminal region of the vertebrate EGF though they have very low sequence similarities. Furthermore, this structural similarity may suggest that SRP2-receptor interactions are analogous to EGF and its receptor to trigger the biological activities of this stress response peptide.
5. CONCLUSION
There is a family of cytokine-like genes in the genome of M. sexta, which encode eleven small proteins with the pre-pro-structure. Extracellular serine proteases and intracellular processing enzymes are likely responsible for their proteolytic activation and/or release in response to physiological stimuli. Expression profiling has shown the tissues and developmental stages in which SRP2 gene is expressed. More importantly, the SRP2 gene transcription is up-regulated in hemocytes and fat body upon immune challenge and injection of the synthetic peptide significantly induced the expression of certain antimicrobial peptide genes. The latter suggest the existence of proSRP2 activating proteases, a SRP2 receptor, and an immune signal transduction pathway. The elucidation of the SRP2 solution structure represents the first step in our endeavor aimed at understanding its interactions with a putative receptor in the future.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM58634 and AI112662 (to HJ). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under project OKLO2450. Contribution number 17-028-J from the Kansas Agricultural Experiment Station.
ABBREVIATIONS
- AMP
antimicrobial peptide
- GBP
growth blocking peptide
- HP
hemolymph (serine) protease
- PO and proPO
phenoloxidase and its precursor
- PAP
proPO activating protease
- PP
paralytic peptide
- PSP
plasmatocyte spreading peptide
- SRP
stress response peptide
- uENF
upstream Glu-Asn-Phe factor
- SP and SPH
serine protease and its non-catalytic homolog
References
- 1.(a) Jiang H, Vilcinskas A, Kanost MR. Immunity in Lepidopteran Insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review of Immunology. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.Strand MR. The insect cellular immune response. Insect Sci. 2008;15(1):1–14. [Google Scholar]
- 3.(a) Matsumoto H, Tsuzuki S, Date-Ito A, Ohnishi A, Hayakawa Y. Characteristics common to a cytokine family spanning five orders of insects. Insect Biochem Molec. 2012;42(6):446–454. doi: 10.1016/j.ibmb.2012.03.001. [DOI] [PubMed] [Google Scholar]; (b) Duressa TF, Vanlaer R, Huybrechts R. Locust cellular defense against infections: Sites of pathogen clearance and hemocyte proliferation. Dev Comp Immunol. 2015;48(1):244–253. doi: 10.1016/j.dci.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Kanamori Y, Hayakawa Y, Matsumoto H, Yasukochi Y, Shimura S, Nakahara Y, Kiuchi M, Kamimura M. A Eukaryotic (Insect) Tricistronic mRNA Encodes Three Proteins Selected by Context-dependent Scanning. J Biol Chem. 2010;285(47):36933–36944. doi: 10.1074/jbc.M110.180398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi K, Matsumoto H, Ochiai M, Tsuzuki S, Hayakawa Y. Enhanced expression of stress-responsive cytokine-like gene retards insect larval growth. Insect Biochem Molec. 2012;42(3):183–192. doi: 10.1016/j.ibmb.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 6.(a) Tsuzuki S, Ochiai M, Matsumoto H, Kurata S, Ohnishi A, Hayakawa Y. Drosophila growth-blocking peptide-like factor mediates acute immune reactions during infectious and non-infectious stress. Sci Rep-Uk. 2012;2 doi: 10.1038/srep00210. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ishii K, Hamamoto H, Sekimizu K. Paralytic Peptide: An Insect Cytokine That Mediates Innate Immunity. Arch Insect Biochem. 2015;88(1):18–30. doi: 10.1002/arch.21215. [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa S, Oda Y, Kamiya M, Kamijima T, Aizawa T, Clark KD, Demura M, Kawano K, Strand MR, Hayakawa Y. A Novel Peptide Mediates Aggregation and Migration of Hemocytes from an Insect. Curr Biol. 2009;19(9):779–785. doi: 10.1016/j.cub.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 8.Tsuzuki S, Sekiguchi S, Kamimura M, Kiuchi M, Hayakawa Y. A cytokine secreted from the suboesophageal body is essential for morphogenesis of the insect head. Mech Develop. 2005;122(2):189–197. doi: 10.1016/j.mod.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Kiyotake H, Matsumoto H, Nakayama S, Sakai M, Miyatake T, Ryuda M, Hayakawa Y. Gain of long tonic immobility behavioral trait causes the red flour beetle to reduce anti-stress capacity. J Insect Physiol. 2014;60:92–97. doi: 10.1016/j.jinsphys.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 10.(a) Kanost MR, Jiang HB. Clip-domain serine proteases as immune factors in insect hemolymph. Curr Opin Insect Sci. 2015;11:47–55. doi: 10.1016/j.cois.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Park JW, Kim CH, Rui J, Park KH, Ryu KH, Chai JH, Hwang HO, Kurokawa K, Ha NC, Soderhall I, Soderhall K, Lee BL. Beetle Immunity. Adv Exp Med Biol. 2010;708:163–180. doi: 10.1007/978-1-4419-8059-5_9. [DOI] [PubMed] [Google Scholar]; (c) Veillard F, Troxler L, Reichhart JM. Drosophila melanogaster clip-domain serine proteases: Structure, function and regulation. Biochimie. 2016;122:255–269. doi: 10.1016/j.biochi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Jiang HB, Kanost MR. Biological activity of Manduca sexta paralytic and plasmatocyte spreading peptide and primary structure of its hemolymph precursor. Insect Biochem Molec. 1999;29(12):1075–1086. doi: 10.1016/s0965-1748(99)00086-7. [DOI] [PubMed] [Google Scholar]
- 12.Cao XL, He Y, Hu YX, Zhang XF, Wang Y, Zou Z, Chen YR, Blissard GW, Kanost MR, Jiang HB. equence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Molec. 2015;62:51–63. doi: 10.1016/j.ibmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Dunn PE, Drake DR. Fate of Bacteria Injected into Naive and Immunized Larvae of the Tobacco Hornworm Manduca-Sexta. J Invertebr Pathol. 1983;41(1):77–85. [Google Scholar]; (b) Yang F, Wang F, He Y, Jiang H. In search of a function of Manduca sexta hemolymph protease-1 in the innate immune system. Insect Biochem Molec. 2016;76:1–10. doi: 10.1016/j.ibmb.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieu I, Powers SJ. Real-Time Quantitative RT-PCR: Design, Calculations, and Statistics. Plant Cell. 2009;21(4):1031–1033. doi: 10.1105/tpc.109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wuthrich K. Improved Spectral Resolution in COSY H-1 NMR Spectra of Proteins Via Double Quantum Filtering. Biochemical and Biohpysical Research Communications. 1983;117(2):479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 16.Bax A, Davis DG. Mlev-17-Based Two-Dimensional Homonu-clear Magnetization Transfer Spectroscopy. J Magn Reson. 1985;65(2):355–360. [Google Scholar]
- 17.Kumar A, Ernst RR, Wuthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem Biophys Res Commun. 1980;95(1):1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 18.Piotto M, Saudek V, Sklenar V. Gradient-Tailored Excitation for Single-Quantum Nmr-Spectroscopy of Aqueous-Solutions. J Biomol Nmr. 1992;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 19.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas P, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 20.Goddard TK, DG . Sparky 3. 20 Vol. 15. University of California; San Fran-cisco: 2004. [Google Scholar]
- 21.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. pp. 93–113. [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Stein EG, Rice LM, Brunger AT. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J Magn Reson. 1997;124(1):154–64. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 24.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol Nmr. 1996;8(4):477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 26.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- 27.Veenstra JA. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch Insect Biochem Physiol. 2000;43(2):49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Zhang SG, Gunaratna RT, Zhang XF, Najar F, Wang Y, Roe B, Jiang HB. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem Molec. 2011;41(9):733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Cao X, Li K, Hu Y, Chen YR, Blissard G, Kanost MR, Jiang H. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem Mol Biol. 2015;62:23–37. doi: 10.1016/j.ibmb.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Bommineni YR, Dai H, Gong YX, Soulages JL, Fernando SC, Desilva U, Prakash O, Zhang G. Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. Febs J. 2007;274(2):418–28. doi: 10.1111/j.1742-4658.2006.05589.x. [DOI] [PubMed] [Google Scholar]; (b) Herrera AI, Al-Rawi A, Cook GA, Gao J, Iwamoto T, Prakash O, Tomich JM, Chen J. Structural characterization of two pore-forming peptides: consequences of introducing a C-terminal tryptophan. Proteins. 2010;78(10):2238–50. doi: 10.1002/prot.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xiao Y, Herrera AI, Bommineni YR, Soulages JL, Prakash O, Zhang G. The central kink region of fowlicidin-2, an alpha-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J Innate Immun. 2009;1(3):268–80. doi: 10.1159/000174822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–92. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Cao X, Zhang S, Rogers J, Hartson S, Jiang H. Changes in the Plasma Proteome of Manduca sexta Larvae in Relation to the Transcriptome Variations after an Immune Challenge: Evidence for High Molecular Weight Immune Complex Formation. Mol Cell Proteomics. 2016;15(4):1176–87. doi: 10.1074/mcp.M115.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An CJ, Kanost MR. Manduca sexta serpin-5 regulates prophenoloxidase activation and the Toll signaling pathway by inhibiting hemolymph proteinase HP6. Insect Biochem Molec. 2010;40(9):683–689. doi: 10.1016/j.ibmb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu XQ, Prakash O, Kanost MR. Structure of a paralytic peptide from an insect, Manduca sexta. J Pept Res. 1999;54(3):256–61. doi: 10.1034/j.1399-3011.1999.00136.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.