Abstract
Insertion mutations in EGFR and HER2 both occur at analogous positions in exon 20. Non-small cell lung cancer (NSCLC) patients with tumors harboring these mutations seldom achieve clinical responses to dacomitinib and afatinib, two covalent quinazoline-based inhibitors of EGFR or HER2, respectively. In this study, we investigated the effects of specific EGFR and HER2 exon 20 insertion mutations from NSCLC patients that had clinically achieved a partial response after dacomitinib treatment. We identified Gly770 as a common feature among the drug-sensitive mutations. Structural modeling suggested that this mutation may facilitate inhibitor binding to EGFR. Introduction of Gly770 into two dacomitinib-resistant EGFR exon 20 insertion mutants restored sensitivity to dacomitinib. Based on these findings we used afatinib to treat a NSCLC patient whose tumor harbored the HER2 V777_G778insGSP mutation and achieved a durable partial response. We further identified secondary mutations in EGFR (T790M or C797S) and HER2 (C805S) that mediated acquired drug resistance in drug-sensitive EGFR or HER2 exon 20 insertion models. Overall, our findings identified a subset of EGFR and HER2 exon 20 insertion mutations that are sensitive to existing covalent quinazoline-based EGFR/HER2 inhibitors, with implications for current clinical treatment and next-generation small molecule inhibitors.
Keywords: Epidermal growth factor receptor, human epidermal growth factor receptor 2, exon 20, drug resistance, tyrosine kinase inhibitor
Introduction
Genotype directed therapy is now the standard of care for subsets of patients with advanced non-small cell lung cancer (NSCLC) with tumors harboring oncogenic alterations. This is best exemplified in NSCLC patients with epidermal growth factor receptor (EGFR) mutant or anaplastic lymphoma kinase (ALK) rearranged lung cancers. In both instances, treatment with EGFR or ALK tyrosine kinase inhibitors (TKIs), respectively, is more effective and associated with a higher response rate (RR) and progression free survival (PFS) than platinum based chemotherapy (1–5).
The vast majority of clinical trials evaluating EGFR TKI therapy as initial treatment for advanced or recurrent EGFR mutant NSCLC have included only patients harboring the common drug sensitive EGFR exon 19 deletion and L858R mutations (2, 5, 6). Collectively, these two mutations account for 85% of all EGFR mutations (7). The remaining 15% of EGFR mutations are comprised of rarer point mutations in exon 18 (G719X) or exon 21 (L861Q) and the exon 20 insertion mutations (7). The exon 20 insertions comprise approximately 4 to 10% of all EGFR mutations and the majority occur after residue M766 of EGFR (8–11). Unlike other EGFR mutations, patients with EGFR exon 20 insertions rarely respond to gefitinib or erlotinib. A review of 84 patients with exon 20 insertions across different series treated with either gefitinib or erlotinib demonstrated a RR of only 11% with a PFS of 2.4 months (12). Similarly, treatment with afatinib in this patient population is also associated with a low RR and PFS (8.7% and 2.7 months, respectively) (13). Overall survival of patients with EGFR exon 20 insertion mutations is similar to that of patients without EGFR mutant NSCLC but inferior to that of patients with EGFR exon 19 deletion or L858R advanced NSCLC (9).
Notably, EGFR exon 20 insertion mutations occur in a structurally analogous position as exon 20 insertion mutations in HER2. Mutations in HER2 are oncogenic both in vitro and in vivo (14–16). Unlike EGFR exon 20 mutations, the spectrum of HER2 exon 20 mutations is more narrow, with the A775_G776insYVMA mutation accounting for most of the mutations seen in NSCLC (17–21). As with EGFR exon 20 mutations, there has been limited success in treating patients with HER2 exon 20 mutant NSCLC (22). Strategies to date have included the use of either single agent HER2 kinase inhibitors or a combination of a HER2 kinase inhibitor with agents targeting downstream signaling. A recent randomized phase II trial compared neratinib to the combination of neratinib and temsirolimus in patients with HER2 mutation positive NSCLC. While none of the patients treated with neratinib alone responded (RR: 0%), 3 of 14 (RR: 21%) patients treated with the combination of neratinib/temsirolimus had a PR (23). Collectively, for both EGFR and HER2 exon 20 insertion NSCLC patients, there remains a critical need to develop more effective therapies.
Despite the general lack of efficacy of EGFR or HER2 kinase inhibitors in EGFR or HER2 exon 20 mutant cancers, it is notable that a small but distinct group of patients have had substantial clinical benefits following treatment with EGFR and/or HER2 inhibitors. For example, patients harboring the rare exon 20 A763_Y764insFQEA insertion mutation remain sensitive to erlotinib (8). Further inquiry into the relationship between a specific mutation(s) and corresponding drug sensitivity may provide both biological insights into drug efficacy and identify subsets of patients who could benefit from a treatment strategy using existing drugs.
Dacomitinib is a covalent inhibitor of both EGFR and HER2. In patients harboring EGFR exon 19 deletion or L858R mutations, dacomitinib led to a RR of 76% and PFS of 18.2 months (24). The activity in patients with either EGFR or HER2 exon 20 insertions has also been evaluated. In the phase I study of dacomitinib, 6 patients with EGFR exon 20 insertions were treated and 1 of 6 patients had a sustained PR (25). In a phase II study, 3 of 26 patients with HER2 mutant NSCLC (12%) had a partial response (26). None of the three responders harbored the common A775_G776insYVMA HER2 mutation. This heterogeneity in clinical responses among patients with different EGFR or HER2 exon 20 insertion mutations treated with dacomitinib prompted us to study the different mutations in vitro; identify common features among the dacomitinib sensitive mutants; and determine whether the in vitro findings would be reflective of the differences observed clinically.
Materials and Methods
Patients
EGFR and HER2 exon 20 mutations were identified from patients with NSCLC at the Dana-Farber Cancer Institute as part of a routine genotyping effort. The methods of detection included Sanger sequencing or targeted next generation sequencing and have been previously described (27–29). All patients provided written informed consent, were conducted in accordance with the Declaration of Helsinki, and were approved by the Institutional Review Board at DFCI.
Expression constructs and cell culture
The full length wild type cDNAs of EGFR and HER2 were cloned into pDNR-Dual (BD Biosciences). All EGFR and HER2 mutations were introduced using site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Tech) with mutant-specific primers according to the manufacturer’s instructions. Generation of retroviruses, infection into Ba/F3 and NIH-3T3 cells, and selection of stably expressing cell populations was performed as previously described (30, 31). Ba/F3 cells were a generous gift from the lab of Dr. David Weinstock at DFCI (in 2014). Transformed Ba/F3 cells were maintained in RPMI1640 supplemented with 10% FBS, streptomycin and penicillin. Ba/F3 cells were not authenticated, since their STR profile has not been made publicly available. All cell lines used in the study tested negative for mycoplasma as determined by the Mycoplasma Plus PCR Primer Set (Agilent) in August 2016. All experiments were performed within 2 to 3 passages following thawing of the cell lines.
Cell proliferation and growth assay
Inhibition of growth was assessed by MTS assay according to previously established methods (31, 32). Ba/F3 cells or NSCLC cell lines were exposed to treatment for 72 hours. All experimental points were set up in six wells. The data were graphically displayed using GraphPad Prism version 5.0 for Windows (GraphPad Software Inc.).
Antibodies and western blotting
Cells grown under the previously specified conditions were lysed in a Lysis Buffer (Cell Signaling Technology). Western blot analyses were conducted after separation by SDS-PAGE electrophoresis and transfer to polyvinylidene difluoride-P immobilon membranes (Millipore). Immunoblotting was conducted according to the antibody manufacturer’s recommendations. Anti-phospho-Akt (Ser473), total AKT, phospho EGFR (Tyr1068), total EGFR, phospho Erk (Thr202/Tyr204), total Erk, phospho HER2 (Tyr1221/1222), total HER2, and PARP antibodies were obtained from Cell Signaling Technology. Anti-α-Tubulin antibody was purchased from Sigma-Aldrich.
Soft agar colony formation assay
NIH-3T3 cells expressing various HER2 mutations were suspended in growth medium containing 0.35% Noble agar (Sigma-Aldrich) with various concentration of each drug and plated on a bottom layer of 0.5% agar in 6-well plates. Plates were incubated for two weeks. Then, the cells were stained with 0.005% crystal violet, and plates were returned to the incubator for 3 hours. The number of viable colonies was quantified using ImageJ software.
Generation of drug resistant cells
N-ethyl-N-nitrosourea (ENU) mutagenesis was carried out as previously described (30, 31). Ba/F3 cells expressing InsGY mutation in EGFR and InsYVMA mutation in HER2 were exposed to 50 μg/mL of ENU (Sigma Aldrich) for 24 hours. Cells were then washed three times and expanded in growth media. After expansion, cells were cultured in 96-well plates in the presence of respective drugs (100nM and 1uM dacomitinib for EGFR, and 200 nM neratinib and 200 nM dacomitinib for HER2). Culture medium containing each drug was changed every few days and resistant wells were expanded. Individual drug resistant clones were isolated and confirmed to be drug resistant. DNA was extracted from resistant cells and sequencing of tyrosine kinase domain of each gene was performed.
Generation of patient-derived cell lines
DFCI58 was established from a murine xenograft model and grown in ACL-4 media (Invitrogen, Carlsbad, CA) with 10% FBS. Balb/c nude mice were used for subcutaneous transplantation of tumor cells from a pleural effusion. Harvested xenografts were finely minced with autoclaved scissors, filtered through a 100 μm Nylon Cell Strainer and seeded in complete ACL-4 media. The pleural effusion-derived cell line DFCI127 was directly developed initially in complete ACL-4 media and subsequently transferred and maintained in complete RPMI1640 media.
Results
Spectrum of EGFR and HER2 mutations in NSCLC patients
Between 2004 and 2015, 2789 patients were tested for EGFR mutations and between 2009 and 2015, 1901 patients were tested for HER2 mutations. EGFR mutations were detected in 678 patients (24%). Forty four EGFR mutations were insertion mutations in exon 20 (7% of EGFR mutant cases and 1.7% of all tested cases). Forty seven HER2 mutations were detected in exon 20 (2.5% of all tested cases).
Among the 44 EGFR exon 20 mutations, there were 19 different mutations (Table 1). The most common one was a duplication of codon 768 to 770 resulting in D770_N771insSVD. The second most common one was a duplication of codon 767 to 769 resulting in V769_D770insASV. These two insertions are very similar in their locations and together accounted for 36.3% (16 in 44 cases) of all EGFR exon 20 insertions. Eleven of 19 variants were identified in only 1 patient each. Some of these patients were enrolled in clinical trials of dacomitinib treatment, and one patient with D770delinsGY showed partial response (25).
Table 1.
Summary of EGFR and HER2 exon 20 insertion mutations.
| EGFR | Number (N=44) | Amino acid sequence |
|---|---|---|
| (codon number) | 765 770 775 | |
| (wild type sequence) | EAYVMASVDNPHVCRLL | |
|
| ||
| D770_N771insSVD | 10 | EAYVMASVDSVDNPHVCRLL |
| V769_D770insASV | 6 | EAYVMASVASVDNPHVCRLL |
| H773_V774insH | 4 | EAYVMASVDNPHHVCRLL |
| A763_Y764insFQEA | 3 | EAFQEAYVMASVDNPHVCRLL |
| D770delinsGY | 3 | EAYVMASVGYNPHVCRLL |
| V774_C775insHV | 3 | EAYVMASVDNPHVHVCRLL |
| H773_V774insNPH | 2 | EAYVMASVDNPHNPHVCRLL |
| H773_V774insPH | 2 | EAYVMASVDNPHPHVCRLL |
| Y764_V765insHH | 1 | EAYHHVMASVDNPHVCRLL |
| D770_N771insGL | 1 | EAYVMASVDGLNPHVCRLL |
| D770_N771insSVG | 1 | EAYVMASVDSVGNPHVCRLL |
| D770delinsVG | 1 | EAYVMASVVGNPHVCRLL |
| N771_P772insH | 1 | EAYVMASVDNHPHVCRLL |
| N771_P772insV | 1 | EAYVMASVDNVPHVCRLL |
| N771delinsGY | 1 | EAYVMASVDGYPHVCRLL |
| N771delinsTH | 1 | EAYVMASVDTHPHVCRLL |
| P772_H773insDNP | 1 | EAYVMASVDNPDNPHVCRLL |
| P772_H773insPNP | 1 | EAYVMASVDNPPNPHVCRLL |
| H773_V774insAH | 1 | EAYVMASVDNPHAHVCRLL |
| HER2 | Number (N=48) | Amino acid sequence |
|---|---|---|
| (codon number) | 775 780 785 | |
| (wild type sequence) | EAYVMAGVGSPYVSRLL | |
|
| ||
| A775_G776insYVMA | 33 | EAYVMAYVMAGVGSPYVSRLL |
| G776delinsVC | 5 | EAYVMAVCVGSPYVSRLL |
| P780_Y781insGSP | 5 | EAYVMAGVGSPGSPYVSRLL |
| M774delinsWLV | 1 | EAYVWLVAGVGSPYVSRLL |
| A775_G776insSVMA | 1 | EAYVMASVMAGVGSPYVSRLL |
| A775_G776insI | 1 | EAYVMAIGVGSPYVSRLL |
| G776delinsLC | 1 | EAYVMALCVGSPYVSRLL |
| G778_S779InsCPG | 1 | EAYVMAGVGCPGSPYVSRLL |
In contrast to EGFR, only 8 different variants of HER2 exon 20 mutations were identified (Table 1). The most common mutation resulted in a duplication of codon 772 to 775 (A775_G776insYVMA) and was found in 33 of 48 cases of all HER2 exon 20 insertions (68.8%). Five of the 8 variants were detected only once. Some HER2 exon 20 patients were also enrolled into clinical trials of dacomitinib, and two patients with a P780_Y781insGSP and one patient with a M774delinsWLV achieved a partial response (26).
EGFR exon 20 insertions and sensitivity to quinazoline based EGFR inhibitors
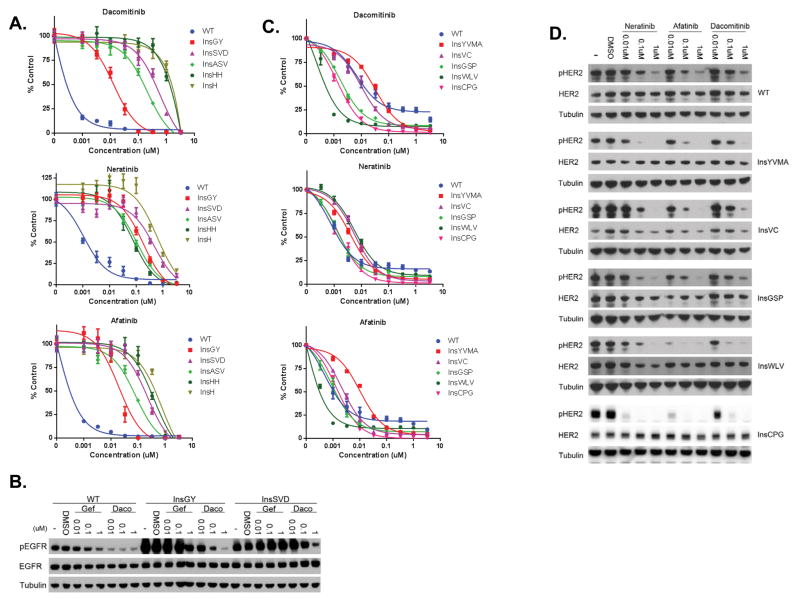
In order to study whether the various EGFR exon 20 insertion mutations impart differential drug sensitivities to quinazoline based EGFR inhibitors, we engineered and expressed the mutations in Ba/F3 cells. We generated Ba/F3 cells harboring 5 different EGFR exon 20 mutations including the three most common mutations (D770_N771insSVD (InsSVD), V769_D770insASV (InsASV), and H773_V774insH (InsH)), the dacomitinib-sensitive mutant (D770delinsGY (InsGY)), one rare mutant (Y764_V765insHH (InsHH)), and wild-type EGFR (EGFR WT). All exon 20 EGFR mutant Ba/F3 cells were able to grow without interleukin-3 (IL-3) with the exception of EGFR WT Ba/F3 cells which required 10 ng/ml of EGF to sustain growth (data not shown). All of the EGFR exon 20 insertion containing Ba/F3 cells were resistant to gefitinib while the EGFR WT cells remained sensitive (data not shown). Dacomitinib was a more potent inhibitor of WT EGFR Ba/F3 cells (Figure 1A and Figure S1). In addition, the InsGY Ba/F3 cells were significantly more sensitive to dacomitinib (IC50 17.5 nM) than the other four exon 20 insertion Ba/F3 cell lines (p = 0.013; mean IC50 in InsGY mutant (n=4) vs. mean IC50 in other exon 20 insertion mutants; t-test). Notably, this particular insertion was the one identified in a NSCLC patient who had clinically achieved a dramatic and sustained response to dacomitinib (25). The InsGY Ba/F3 cells were also slightly more sensitive to afatinib while all five exon 20 insertion mutations were relatively more resistant to neratinib. The differential sensitivity of dacomitinib in the InsGY cells was mirrored by its ability to inhibit pEGFR in these cells (Figure 1B).
Figure 1.
EGFR and ERBB2 exon 20 insertions and sensitivity to covalent quinazoline based EGFR inhibitors. A. Ba/F3 cells expressing wild type EGFR or different exon 20 insertion mutations. Cells were treated with different drugs at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. Ba/F3 cells expressing different EGFR exon 20 mutants were treated with gefitinib or dacomitinib at indicated concentrations for 6 hours. Cell extracts were immunoblotted to detect the indicated proteins. C. Ba/F3 cells expressing wild type HER2 or different exon 20 insertion mutations. Cells were treated with different drugs at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. D. Ba/F3 cells expressing different HER2 exon 20 mutants were treated with gefitinib or dacomitinib at indicated concentrations for 6 hours. Cell extracts were immunoblotted to detect the indicated proteins.
HER2 exon 20 insertions and sensitivity to quinazoline based EGFR inhibitors
We also generated and studied WT and 5 different HER2 exon 20 insertion Ba/F3 cell lines. The mutations included the most common HER2 mutations (A775_G776insYVMA (InsYVMA) and G776delinsVC (InsVC)); two mutations identified in patients who clinically responded to dacomitinib (P780_Y781insGSP (InsGSP) and M774delinsWLV (InsWLV)), and one rare mutation (G778_S779InsCPG (InsCPG)). All mutants, including WT HER2 were able to grow without IL-3 (data not shown). We evaluated the efficacy of neratinib, dacomitinib and afatinib in all six HER2 exon 20 insertion mutant Ba/F3 cell lines (Figure 1C). In general, the HER2 mutant Ba/F3 cells were more sensitive to all three drugs compared to the EGFR exon 20 mutant Ba/F3 cells (Figure 1A and Figure S1). The InsYVMA Ba/F3 cells were the most resistant with IC50 values higher than that seen for WT HER2 (Figure 1C). These cells were more sensitive to neratinib although not as sensitive as those the WT HER2 cells (Figure 1C). As previously noted, the dacomitinib sensitive Ba/F3 cell lines (InsGSP and InsWLV) contained mutations previously associated with clinical sensitivity to dacomitinib (26). Ba/F3 cells harboring the InsCPG were similarly sensitive to all three covalent inhibitors (Figure 1C). The IC50 values of the Ba/F3 cells harboring the dacomitinib sensitive mutations (InsGSP, InsWLV and InsCPG) were significantly lower than those containing the more resistant HER2 mutants or WT HER2 (p = 0.031; mean IC50 of sensitive vs. resistant mutants; t-test). Immunoblot analyses demonstrating inhibition of HER2 phosphorylation corresponded to the relative cellular sensitivities for each of the tested drugs (Figure 1D). We also performed colony formation assays in soft agar and assessed drug sensitivities of the different HER2 mutant 3T3 cells (Figures S2A and S2B). Similar to the Ba/F3 cells, neratinib was most effective at inhibiting the growth of the InsGSP and InsYVMA colonies while dacomitinib was most effective against the InsWLV cells (Figures S2A and S2B).
Generation and evaluation of patient derived EGFR exon 20 cell lines
Unlike the common EGFR activating mutations (L858R and Exon 19 deletions), there are no patient derived EGFR exon 20 mutant cell lines that contain the common EGFR exon 20 insertion mutations. Accordingly, we established two patient-derived cell lines (DFCI58 and DFCI127) harboring EGFR exon 20 insertion mutations (Figure S3A). DFCI58 was established from the adenocarcinoma of a 60-year-old male. He was diagnosed with stage IV disease and and an EGFR H773_V774insNPH exon 20 mutation was identified. Some of this effusion was collected prior to any systemic therapy, propagated in a murine xenograft, and used for establishment of the cell line. DFCI127 was established from the adenocarcinoma of a 42-year-old female. She was diagnosed with stage IV disease and had an EGFR P772_H773insPNP exon 20 mutation. She was treated with carboplatin/pemetrexed/bevacizumab, pemetrexed/bevacizumab, and docetaxel. Following progression on docetaxel, a thoracentesis was performed which was used to establish the cell line.
We evaluated the efficacy of different EGFR inhibitors in both the DFCI 58 and DFCI 127 cell lines (Figures S3B and S3C). Although higher concentrations of gefitinib had some efficacy in both cell lines, afatinib and dacomitinib were substantially more effective at inhibiting cell growth as well as phosphorylation of EGFR, AKT and ERK1/2. Immunoblot analysis revealed higher levels of cleaved PARP in response to dacomitinib treatment than to gefitinib treatment (Figure S3C). These data suggested that the apparent inhibition of cell growth induced by the dacomitinib treatment was mediated by enhanced apoptosis in both DFCI cell lines.
Structural insights into inhibitor sensitivity of exon20 insertion mutants
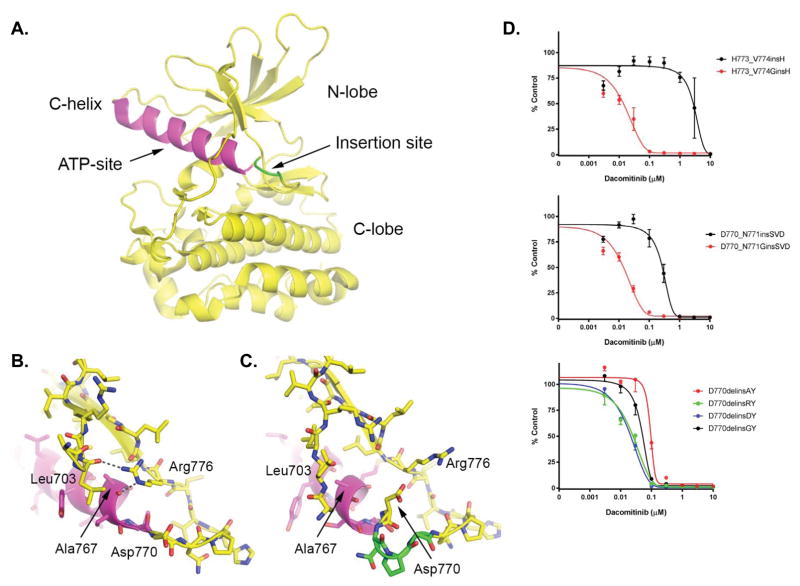
The structural basis for the inhibitor resistance of most exon 20 insertion mutants is poorly understood. Only one crystal structure is available for an exon 20 variant (the insNPG mutant) and it reveals an inhibitor binding site that is essentially indistinguishable from that of the inhibitor-sensitive L858R mutant (8). This insertion and most other insertions encoded within exon20 are not directly in contact with the ATP-site of the kinase (Figure 2A), suggesting that inhibitor resistance may arise, at least in part, from differences in conformational dynamics of the kinase.
Figure 2.
Structural context of exon20 insertion mutants and impact on dacomitinib sensitivity. A. Previously determined structure of the EGFR insNPG exon20 insertion mutant (PDB ID: 4LRM), highlighting the architecture of the kinase domains and the typical location of exon20 insertion mutants. The insNPG insertion is highlighted in green and the C-helix is shaded magenta. The ATP-site, which is also the site of binding of inhibitors, lies between the N- and C-lobes of the kinase as indicated. B. In the inactive conformation of the EGFR kinase, Arg 776 hydrogen bonds with the carbonyl of Ala767 at the end of the C-helix, and with Leu703 in the N-terminal portion of the kinase. We propose that this interaction is important for the stability of the inactive conformation of the kinase, as well for the transition between the inward (active) and outward (inactive) positions of the C-helix which may in turn control access of inhibitors to the adjacent ATP-site. (Panel drawn from PDB ID 1XKK.) C. Detailed view of the insertion site in the insNPG mutant (PDB ID 4LRM), colored as in panel A. The insertion reorients Asp770 such that it blocks access of Arg776 to the end of the C-helix. Other inhibitor-resistant mutants may have a similar effect on the position of Asp770, but at present crystal structures are available only for insNPG. In all of the inhibitor-sensitive insertion mutants studied here, a glycine residue is inserted in place of Asp770, potentially restoring the ability of Arg776 to rearrange in communication with the C-helix. D. Ba/F3 cells expressing different modified exon 20 insertion mutations. In the top two, Asp770 has ben replaced by a Gly. In the bottom, Gly770 has been replaced by either Asp, Ala or Arg. Cells were treated with dacomitinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls.
We next examined our dacomitinib-sensitive versus dacomitinib-resistant exon20 mutants, in light of available structural information, to gain insight into possible determinants of inhibitor sensitivity. In doing so, we noticed that all of the inhibitor-sensitive mutants contained a glycine in a position two residues beyond the end of the C-helix (Table 2 and Figure 2A). In wild type EGFR, this residue is Asp770. This residue lies in a region that is rearranged in the transition between the active and inactive conformations of the kinase. EGFR activity is regulated by repositioning of the C-helix, which rotates into the ATP-site in the active state while rotating out in the inactive state. These conformational transitions are also likely to be important for binding of either substrate or ATP-site inhibitors. In EGFR, Asp770 and the exon 20 insertions lie at the pivot-point of the C-helix. As part of the switch to the inactive conformation, the side chain of Arg776 rearranges to hydrogen bond with the carbonyl group of Ala767 at the end of the C-helix (Figure 2B). While this position is accessible in wild type EGFR, it is blocked by Asp770 in the insertion mutant (Figure 2C). The insertion residues reposition Asp770, thus sterically blocking Arg776 from accessing the end of the C-helix. Accordingly, we hypothesized that replacement of Asp770 with a glycine, as occurs in the inhibitor-sensitive mutants (Table 2), might restore access of Arg776 and thereby facilitate restoration of wild-type C-helix conformational changes and inhibitor binding.
Table 2.
Summary of drug responsive EGFR and HER2 exon 20 insertion mutations harboring a shared glycine.
| Mutation | Amino acid sequence |
|---|---|
| EGFR | |
| D770delinsGY |
|
| HER2 | |
| P780_Y781insGSP |
|
| M774delinsWLV |
|
| G778_S779InsCPG |
|
| Patient | |
| V777_G778insGSP |
|
To test this hypothesis, we mutated Asp770 to glycine in the context of the dacomitinib resistant Ba/F3 cell lines harboring the H773_V774 insH or D770_N771 insSVD mutants (Table S1). Both mutants led to transformation and IL-3 independent growth (data not shown). In addition, the proliferation rates were similar with or without the glycine substitution (data not shown). However, the glycine mutation markedly sensitized both variants to inhibition by dacomitinib (Figure 2D). In the D770_N771 insSVD mutation, the Asp770Gly mutation shifted the IC50 10-fold (p = 0.046; mean IC50 (n=3) insSVD vs. GinsSVD; t-test). The effect in the H773_V774 insH variant was more dramatic, with an IC50 of 5 nM (figure 2D) (p < 0.001; mean IC50 (n=3) insH vs. GinsH; t-test). We also carried out the converse experiment using the D770delinsGY mutant cell line by mutating the inserted glycine to alanine, arginine, or aspartic acid (Table S1). Notably, contrary to our hypothesis, these substitutions did not uniformly confer resistance; the alanine mutant was slightly more resistant while the arginine and aspartic acid mutants exhibited increased sensitivity (Figure 2D).
Treatment of HER2 mutant patient with afatinib
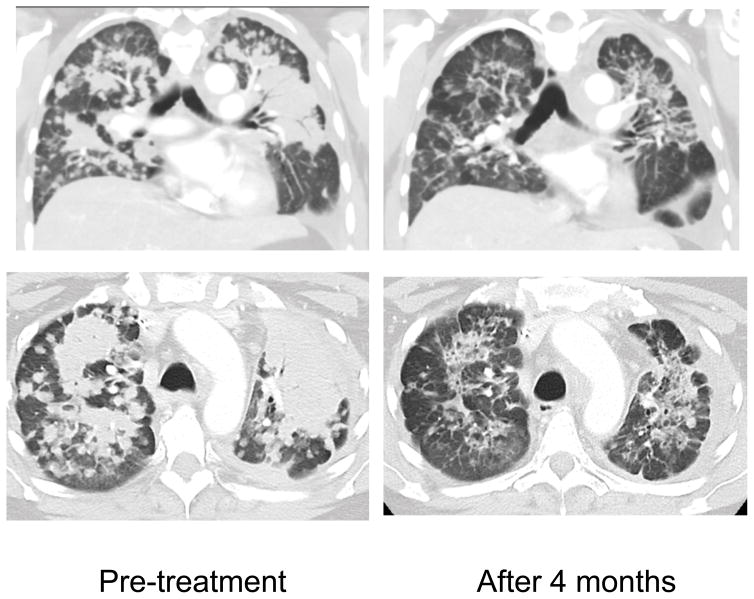
This patient was a 45 year old female never-smoker who initially presented with advanced lung adenocarcinoma metastatic to the bone. Routine targeted next generation sequencing of her tumor identified a HER2 V777_G778insGSP mutation (Table 2). Over the course of two years, her disease was controlled on first-line cisplatin/pemetrexed chemotherapy (7 months) and then second-line docetaxel (15 months). Because of her HER2 mutant NSCLC, she then initiated third-line vinorelbine/trastuzumab x2 cycles without any evidence of response in bilateral pulmonary disease, complicated by subsequent development of symptomatic brain metastases requiring whole brain radiation. Following radiation she was increasingly symptomatic with fatigue and ambulatory hypoxia requiring 2–4L of continuous O2 supplementation. The HER2 V777_G778insGSP mutation leads to the same changes in the HER2 protein as does the HER2 P780_Y781insGSP mutation (Table 2). Because of the potential sensitivity of this mutant to covalent EGFR/HER2 inhibitors, the patient was treated with 30 mg of afatinib given once daily. She experienced significant tumor shrinkage (Figure 3), accompanied by a decrease in the requirement for supplemental oxygen. The response was sustained for 7 months with eventual clinical progression due to leptomeningeal carcinomatosis.
Figure 3.
Treatment of a patient with ERBB2 V777_G778insGSP NSCLC with afatinib. Coronal and axial images obtained pre-treatment and following 4 months of afatinib treatment.
Identification of mutations that cause drug resistance in drug sensitive EGFR or HER2 exon 20 mutant cells
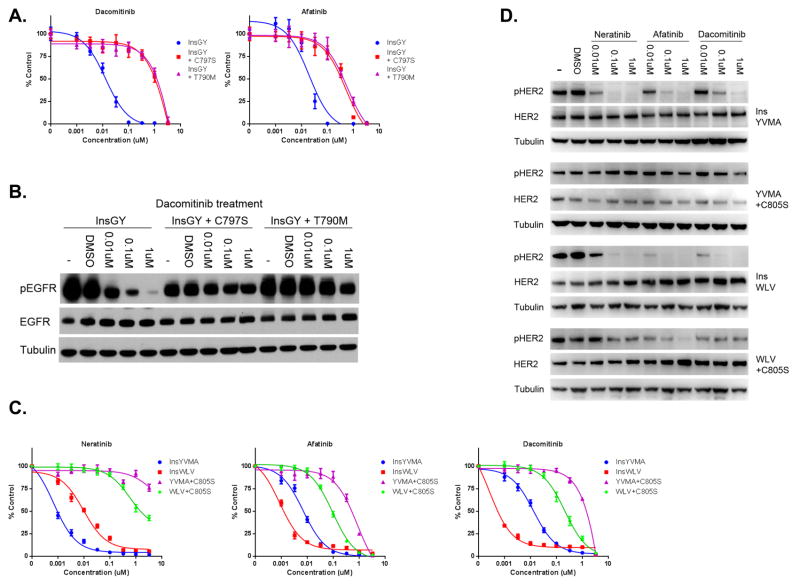
In order to identify how drug sensitive cancers with EGFR or HER2 exon 20 insertion mutations may develop acquired drug resistance, we performed an N-ethyl-N-nitrosourea (ENU) mutagenesis assay. This approach has previously led to identification of clinically relevant drug resistance mutations to EGFR TKIs (30, 31). We used the EGFR exon 20 InsGY cells and selected resistant clones in the presence of either 100 nM or 1 μM dacomitinib following ENU exposure. Seventeen (17) drug resistant clones were expanded and identified to be drug resistant; 12 from the 100 nM and 5 from the 1 μM treated cells. Sequencing of EGFR revealed a T790M mutation in 10/12 100 nM and 5/5 1 μM treated cells. The remaining 2 drug resistant clones from the 100 nM treated group did not contain any additional EGFR mutations (data not shown). We next generated Ba/F3 cells using EGFR InsGY that contained either T790M or C797S in cis. EGFR C797 is the site of covalent binding of neratinib, dacomitinib and afatinib. Mutations in C797 (to C797S) have recently been demonstrated to lead to resistance to mutant selective EGFR inhibitors both in vitro (WZ4002, rociletinib, osimertinib) and in lung cancer patients (31, 33). Both T790M and C797S resulted in drug resistance to dacomitinib and afatinib (Figure 4A). Immunoblot analysis revealed EGFR phosphorylation was not inhibited by dacomitinib in Ba/F3 cells harboring either the T790M or C797S mutations when compared to the parental InsGY cells (Figure 4B).
Figure 4.
Mutations at the site of drug binding cause resistance in drug sensitive EGFR or HER2 exon 20 insertion mutant models. A. EGFR InsGY Ba/F3 cells harboring either a concurrent T790M or a C797S are resistant to dacomitinib or afatinib. Cells were treated with dacomitinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. B. Ba/F3 cells expressing the respective constructs from A. were treated with different drugs at indicated concentrations for 6 hours. Cell extracts were immunoblotted to detect the indicated proteins. C. HER2 InsYVMA or InsWLV Ba/F3 cells harboring a concomitant C805S mutation are resistant to neratinib, afatinib and dacomitinib. Cells were treated with different drugs at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. D. Ba/F3 cells expressing the respective constructs from A. were treated with different drugs at indicated concentrations for 6 hours. Cell extracts were immunoblotted to detect the indicated proteins.
We analogously performed an ENU mutagenesis study using the HER2 InsYVMA Ba/F3 cells. Twelve (12) drug resistant clones were identified following selection in either 200 nM of neratinib (n=5) or 200 nM of dacomitinib (n=7). Sequencing of HER2 from the resistant cells revealed a secondary C805S mutation in all 12 clones that was not present in the drug sensitive cells. C805 is the EGFR C797 analogous cysteine residue (Figure S4). We further generated Ba/F3 cells co-expressing a HER2 activating mutation (InsYVMA or InsWLV) and C805S. Cell proliferation assay revealed these two cell lines were resistant to all three drugs compared to parental cells (Figure 4C). Immunoblot analysis revealed HER2 phosphorylation was not completely inhibited in the resistant cells (Figure 4D).
Discussion
Lung cancer patients harboring EGFR or HER2 exon 20 mutant tumors represent a unique subset of patients for whom there are currently no effective or approved targeted therapies. However, there is significant clinical and biochemical heterogeneity among EGFR or HER2 exon 20 mutations. Although the vast majority are associated with resistance to EGFR inhibitors, previous studies have identified a unique EGFR exon 20 mutation (A763_Y764insFQEA) that remained sensitive to erlotinib (8). In the present study, we identify four additional rare exon 20 mutations in EGFR (InsGY) and HER2 (InsGSP, InsWLV and InsCPG) that are uniquely more sensitive to the covalent EGFR/HER2 inhibitor dacomitinib than the more common EGFR or HER2 exon 20 mutations (Figure 1A and 1C). Three of these mutations were initially identified from the tumors of lung cancer patients who had clinically responded to dacomitinib while the fourth was identified as an in vitro mutation conferring drug sensitivity (Figure 1C). With the increased systematic use and availability of comprehensive tumor sequencing, even patients with rare EGFR mutations will continue to be identified, thus it will remain important to determine the correlation between specific mutations and their sensitivity to existing and available treatments.
In the phase II trial of dacomitinib, the mean trough concentration following dosing with 45mg of dacomitinib once daily was 56.7–74.7ng/mL (approximately 120–160nmol/L) (24). Of the EGFR exon 20 mutations tested, only the IC50 value of the InsGY mutation was below this concentration (17.5nM; Figures 1A and S1), suggesting that this glycine bearing mutation may have been the reason for the unique clinical sensitivity to dacomitinib in this patient. The findings for HER2 exon 20 mutations are similar albeit more subtle (Figure S1). The HER2 mutations identified from dacomitinib responding patients were also the most sensitive to dacomitinib in vitro (Figure 1C) but the differences in the IC50 values between these and mutations from patients with no response (such as HER2 InsYVMA) were not as pronounced as in those with the EGFR exon 20 mutations (Figure 1C). Collectively these findings raise the possibility that more potent and specific inhibitors of EGFR or HER2 exon 20 insertion mutations—with IC50 values closer to the Ba/F3 cell lines harboring dacomitinib sensitive mutations—may be clinically effective against a larger fraction of these cancers.
Remarkably, by studying the drug sensitive and resistant EGFR and HER2 exon 20 insertion mutations, we uncovered a common feature among the drug sensitive mutations: the presence of a glycine at position 770 (Table 2). Furthermore, by introducing a glycine into position 770 of two dacomitinib resistant EGFR exon 20 mutations, we were able to substantially enhance the efficacy of dacomitinib (Figure 2D). To date, no features among the different EGFR or HER2 exon 20 mutations have been identified that could predict clinical sensitivity to existing covalent EGFR or HER2 inhibitors. While our model linking rearrangements of Arg776 and confirmation of the C-helix to inhibitor sensitivity remains speculative, taken together, our findings show that inhibitor resistance is not an inherent feature of activating exon 20 insertions in this region. Indeed, glycine mutations can restore inhibitor sensitivity without abrogating transforming activity. These mutations could act by removing the steric hindrance imposed by Asp770, or by simply increasing the flexibility of the inserted loop. Detailed structural and biophysical studies will be required to directly ascertain the effects of sensitive versus resistant exon20 insertion mutants on the dynamics of the C-helix, the kinetics of inhibitor binding, and the coupling between the two. Based on our findings, we treated a patient with a HER2 mutant tumor harboring the glycine in position 770 (Table 2), who had previously failed a prior HER2 targeted therapy (trastuzumab). She ultimately had a clinically significant and durable response on afatinib treatment (Figure 3). Additional prospective clinical validation, specifically among EGFR and HER2 exon 20 mutant patients whose tumors share this glycine residue at position 770, will be necessary to further support our preclinical findings.
Our studies also reveal how the rare but drug sensitive EGFR or HER2 exon 20 mutant cancers may develop acquired drug resistance. In both cases described here, mutation in the covalent binding site of either EGFR (C797S) or HER2 (C805S) is sufficient to lead to drug resistance (Figure 4). For EGFR, the T790M secondary mutation also results in drug resistance; to a similar degree as C797S. However, the impact of the C805S mutation may be different than that recently described for the EGFR C797S mutation. Cells expressing an EGFR activating mutation and C797S in the absence of T790M are resistant to mutant selective EGFR inhibitors (WZ4002, osimertinib and rociletinib) but remain sensitive to afatinib (31). In the case of HER2, unlike EGFR, the efficacy of afatinib is more likely dependent on covalent binding than on non-covalent inhibition. As HER2 patients treated with afatinib, neratinib or dacomitinib develop acquired resistance, it will be interesting and clinically significant to determine whether C805S will also occur clinically as an acquired drug resistance mutation. In addition, these observations should inspire the development of alternative strategies to inhibit EGFR and HER2 even in the presence of these resistance mutations as such approaches may be clinically effective for a growing group of patients.
Furthermore, there continues to be a need to develop effective therapies for patients with the more common EGFR exon 20 mutation (InsSVD and InsASV, accounting for ~ 1/3 of all exon 20 mutations) and HER2 exon 20 mutations (InsYVMA, accounting for ~ 2/3 of all exon 20 mutations). Current approaches include using new EGFR/HER2 kinase inhibitors (AP32788; NCT02716116) or the use of Ado-Trastuzumab Emtansine (NCT02675829). Our studies highlight the heterogeneity among EGFR and HER2 exon 20 insertion mutations which may impact the efficacy of specific therapies. As additional therapies are developed for this subset of patients, it will be important to continue to correlate efficacy (or lack thereof) with specific genomic variants of EGFR and HER2.
Supplementary Material
Acknowledgments
Source of Funding: The National Cancer Institute of the NIH (R01CA135257 (P.A. Jänne and G.R. Oxnard) and R01CA114465 (P.A. Jänne)), the Cammarata Family Foundation Fund (P.A. Jänne), the Denise and Kevin Hanlon Family Fund for Lung Cancer Research (P.A. Jänne) and HER2 Research LLC (P.A. Jänne).
References
- 1.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 3.Solomon BJ, Mok T. First-line crizotinib in ALK-positive lung cancer. N Engl J Med. 2015;372:782. doi: 10.1056/NEJMc1415973. [DOI] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 8.Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–84. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–24. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naidoo J, Sima CS, Rodriguez K, Busby N, Nafa K, Ladanyi M, et al. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer. 2015;121:3212–20. doi: 10.1002/cncr.29493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–8. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 14.Shimamura T, Ji H, Minami Y, Thomas RK, Lowell AM, Shah K, et al. Non-small-cell lung cancer and Ba/F3 transformed cells harboring the ERBB2 G776insV_G/C mutation are sensitive to the dual-specific epidermal growth factor receptor and ERBB2 inhibitor HKI-272. Cancer Res. 2006;66:6487–91. doi: 10.1158/0008-5472.CAN-06-0971. [DOI] [PubMed] [Google Scholar]
- 15.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Perera SA, Li D, Shimamura T, Raso MG, Ji H, Chen L, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A. 2009;106:474–9. doi: 10.1073/pnas.0808930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–6. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 18.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 19.Buttitta F, Barassi F, Fresu G, Felicioni L, Chella A, Paolizzi D, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer. 2006;119:2586–91. doi: 10.1002/ijc.22143. [DOI] [PubMed] [Google Scholar]
- 20.Tomizawa K, Suda K, Onozato R, Kosaka T, Endoh H, Sekido Y, et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer. 2011;74:139–44. doi: 10.1016/j.lungcan.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910–8. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herter-Sprie GS, Greulich H, Wong KK. Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Frontiers in oncology. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besse B, Soria J, Yao B, Kris M, Chao B, Cortot A, et al. Neratinib (N) with or without temsirolimus (TEM) in patients (pts) with non-small cell lung cancer (NSCLC) carrying HER2 somatic mutations: An international randomized phase II study. Ann Oncol. 2014:25. [Google Scholar]
- 24.Janne PA, Ou SH, Kim DW, Oxnard GR, Martins R, Kris MG, et al. Dacomitinib as first-line treatment in patients with clinically or molecularly selected advanced non-small-cell lung cancer: a multicentre, open-label, phase 2 trial. Lancet Oncol. 2014;15:1433–41. doi: 10.1016/S1470-2045(14)70461-9. [DOI] [PubMed] [Google Scholar]
- 25.Janne PA, Boss DS, Camidge DR, Britten CD, Engelman JA, Garon EB, et al. Phase I dose-escalation study of the pan-HER inhibitor, PF299804, in patients with advanced malignant solid tumors. Clin Cancer Res. 2011;17:1131–9. doi: 10.1158/1078-0432.CCR-10-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kris MG, Camidge DR, Giaccone G, Hida T, Li BT, O’Connell J, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol. 2015;26:1421–7. doi: 10.1093/annonc/mdv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardarella S, Ortiz TM, Joshi VA, Butaney M, Jackman DM, Kwiatkowski DJ, et al. The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J Thorac Oncol. 2012;7:1767–74. doi: 10.1097/JTO.0b013e3182745bcb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol. 2016;34:721–30. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ercan D, Choi HG, Yun CH, Capelletti M, Xie T, Eck MJ, et al. EGFR mutations and resistance to Irreversible pyrimidine based EGFR inhibitors. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70:10038–43. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–2. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.